Abstract
Critical blind assessment of structure prediction techniques is crucial for the scientific community to establish the state of the art, identify bottlenecks, and guide future developments. In Critical Assessment of Techniques in Structure Prediction (CASP), human experts assess the performance of participating methods in relation to the difficulty of the prediction task in a biennial experiment on approximately 100 targets. Yet, the development of automated computational modeling methods requires more frequent evaluation cycles and larger sets of data. The “Continuous Automated Model EvaluatiOn (CAMEO)” platform complements CASP by conducting fully automated blind prediction evaluations based on the weekly pre-release of sequences of those structures, which are going to be published in the next release of the Protein Data Bank (PDB). Each week, CAMEO publishes benchmarking results for predictions corresponding to a set of about 20 targets collected during a 4-day prediction window. CAMEO benchmarking data are generated consistently for all methods at the same point in time, enabling developers to cross-validate their method’s performance, and referring to their results in publications. Many successful participants of CASP have used CAMEO—either by directly benchmarking their methods within the system or by comparing their own performance to CAMEO reference data. CAMEO offers a variety of scores reflecting different aspects of structure modeling, for example, binding site accuracy, homo-oligomer interface quality, or accuracy of local model confidence estimates. By introducing the “bestSingleTemplate” method based on structure superpositions as a reference for the accuracy of 3D modeling predictions, CAMEO facilitates objective comparison of techniques and fosters the development of advanced methods.
Keywords: benchmarking, CAMEO, CASP, continuous evaluation, homo-oligomer interface accuracy, ligand binding-site accuracy, model quality assessment, model confidence, oligomeric assessment, protein structure modeling, protein structure prediction
1 |. INTRODUCTION
Routine application of three-dimensional (3D) protein structure models in life science research requires fully automated, robust, reliable, and accurate modeling pipelines.1 However, the performance of prediction tools reported in the literature is often based on different background information, inconsistent benchmarking data sets, and distinct evaluation metrics, impeding quantitative comparison between methods. In the field of protein structure modeling, this well-known problem is successfully addressed by regular independent blind assessments in the form of the community experiment “Critical Assessment of Techniques in Structure Prediction” (CASP).2,3 CASP is organized every 2 years, with experts assessing methods based on approximately 100 prediction targets, and culminates in a meeting, where researchers compare the performance of the various approaches and discuss the latest developments. Yet, the development of automated server methods requires more frequent benchmarking on larger data sets in between CASP seasons to allow testing different hypotheses and to enable faster development cycles. The “Continuous Automated Model EvaluatiOn (CAMEO)”4 platform offers this functionality. The fully automated platform relies on the pre-release of sequences5 of structures, which are going to be published by the Protein Data Bank (PDB) 4 days later.6 These structures serve as references for the evaluations within CAMEO. The evaluation is performed across all participating servers at the same point in time, that is, all methods have access to the same background information such as template information or protein sequences in UniProt.7 Benchmarking results (predictions, reference structures and evaluation scores) are publicly available to document a method’s historic performance (eg, for reference in publications) and can be used as training data for further method development.
CAMEO significantly facilitates the development of modern protein structure prediction approaches: methods are transparently assessed by a variety of scores established by the community, representing different aspects of structure prediction. Superposition-free measures8–10 are crucial for fully automated assessment and are employed for aggregated scores as previously described. Although results for servers registered as “public servers” are visible for anyone, it is helpful for developers to be able to register new methods as anonymous “development servers,” visible only to other developers with a CAMEO account. This enables testing new hypotheses and “real-time” benchmarking to other cutting-edge methods without revealing the details of the method. For historic comparison, we encourage groups to keep the last and second last public server versions available to CAMEO and register major releases as a new server. CAMEO is an open platform inviting the wider community to propose scoring approaches, evaluation schemes, and new categories. In this article, we would like to focus on recent developments and results for the protein structure prediction (CAMEO 3D) and the local quality estimation (CAMEO QE) categories.
2 |. MATERIALS AND METHODS
2.1 |. Target set—3D
The CAMEO target set has been selected as previously described,4 where CAMEO deselects targets exhibiting high coverage in at least one template (“too easy”) and omits protein sequences that are shorter than 30 residues. CAMEO then submits the first 20 sequences every week. The CAMEO website allows accessing data in time frames “1 week,” “1 month,” “3 months,” “6 months,” and “1 year.” For this article, the target set here is composed of 248 targets spanning a 3-month period from 1 May 2018 to 28 July 2018. Due to its continuous cycle, CAMEO refers to the target structures as first published by the PDB in contrast to the PDB, which updates and sometimes even supersedes experimental structures, for example, 2018–05-12_00000055_1 (PDB id 5xpq, chain D), 2018–05-20_00000063_1 (PDB id 5xnu, chain B), and 2018–05-20_00000004_1 (PDB id 5ndl, chain B). We have excluded three targets from analyses in this article: 2018–06-16_00000009_1(PDB id 5o7b, chain A) due to its non-contiguous structure coverage, targets 2018–05-12_00000031_2 (PDB id 5nl1, chain L), and 2018–06-23_00000056_2 (PDBid 5wjc, chain B) turned out to be too short as large segments have not been crystallized.
Binding site accuracies have been determined for 94 targets. Oligomer-interface scores are available for 96 targets, where at least one biounit was assigned to be a homo-oligomer by the PDB deposition authors. In the case of targets with several functional forms (“biological assemblies,” biounits), the biounit yielding the highest score for the participant has been chosen (for a complete list of targets, refer to Table S1).
2.2 |. Score details—3D
CAMEO currently offers 18 different metrics to assess various aspects of protein structure modeling. Server response time is measured as the time between the CAMEO submission and the arrival of the response e-mail. We note that some servers may have priority queues for CAMEO to ensure modeling results are delivered within the 4-day window. Although this may be aimed at reflecting actual modeling time, it may not correspond to real-world user experience. Some metrics are available at the single chain level only, for example, GDT_HA,11 GDC11, MaxSub,12 TM-score,13 lDDT/lDDT Cα,9 RMSD, and model confidence. [Correction added on 11 Nov 2019, after first online publication: In the preceding sentence, GDC citation updated.] Other metrics measure the accuracy of homo-oligomer interfaces and the respective complexes like QS-score14 based measures, the MM-align15 based TM score, RMSD, and the lDDT-oligo score.
For the analysis of the homo-oligomer predictions, only those predictions have been included in the scoring that matched the number of chains in the reference biounit, that is, with the correct stoichiometry.
2.3 |. Comparison to reference baseline predictions—3D
The “NaiveBLAST” method employs a BLAST run against all available templates of the PDB at the time of submission for each target. If at least one template has been detected, it selects the first hit for a standard MODELLER run generating a protein structure prediction.
For the “bestSingleTemplate” method, templates are discovered by structural superposition of the target reference structures with all PDB structures using TM-align.16 The top 20 of the obtained structural alignments serve as input for the subsequent template-based modeling. Modeling is performed with SWISS-MODEL’s14 modeling engine ProMod317 (see Data S1 for details). [Correction added on 11 Nov 2019, after first online publication: In the preceding sentence, SWISS-MODEL citation updated.] Termini beyond the region covered by the template structure are modeled by a low-complexity Monte Carlo sampling approach. The final models are ranked by lDDT,9 and the top scoring model is selected for that particular target.
2.4 |. Target set—QE
The target set for model quality estimation refers to 3D protein structure predictions (rather than sequences) for the time period from 1 May 2018 to 28 July 2018. The QE target set was generated by collecting all models of the public servers in the CAMEO 3D category 32 hours after the submission of the 3D targets, amounting to 2798 QE targets (for details, please see Table S2). The targets that were excluded in the 3D category have also been disregarded for this analysis. The lDDT score ([0,100]) was used to classify residues as being of good (lDDT >= 60) or bad (lDDT < 60) quality. In an analysis of QE target contribution by public 3D modeling server, most servers produce models at all quality levels (Figure S3).
2.5 |. Score details—QE
To assess the quality of a given protein structure model, numerous scores have been employed such as accuracy of self-estimates (ASE)18 or cumulative rankings based on various scores such as GDT-TS,11 lDDT,9 CAD-score,8 or Spheregrinder.10 Here, we rank the method’s performance based on the partial receiver-operator characteristic (ROC) area under the curves (AUCs) for the false positive rate from 0.0 to 0.2 and the partial precision-recall (PR) AUCs for a recall between 0.8 and 1.0.
2.6 |. Comparison to reference baseline predictions—QE
The “BaselinePotential” server implements a classical distance-based statistical potential.9 Statistics have been extracted for pairwise distances between all chemically distinguishable heavy atoms in the 20 naturally occurring amino acids and histograms have been computed, neglecting all interactions from residues with a sequence separation of less than four residues. The resulting potential functions are applied on all pairwise interactions and per-residue scores are estimated by averaging and then smoothing all outcomes of interactions a residue is involved in2. The baseline server “naivePSIBLAST” assumes that conserved regions of a protein model are of higher quality than divergent regions. It searches the most recent version of the NCBI NR database with PSI-BLAST and estimates the sequence conservation from the position-specific scoring matrix.4
3 |. RESULTS AND DISCUSSION
3.1 |. 3D protein structure modeling
3.1.1 |. Target difficulty
The target set reported here encompassed 248 protein sequences, collected over the course of 3 months (1 May 2018 to 28 July 2018), reflecting the requirement of short evaluation cycles for continuously developed methods. To classify these targets, CAMEO applied a “post-diction” lDDT average across all received 3D structure predictions,4 where “Hard” targets are those with an average lDDT below 50, “Medium” targets are within an lDDT range from equal to 50 and up to 75, and “Easy” targets are equal to or above 75. Following this definition, 44 “Easy,” 138 “Medium,” and 69 “Hard” targets have been evaluated. The actual oligomeric state of a protein is unknown at the time of the CAMEO target selection and submission. CAMEO expects the prediction methods to infer this context independently and model the protein in the correct oligomeric state.
3.1.2 |. Improvement over reference methods
Among the various approaches to predict protein structures, utilizing previously determined structures of homologous proteins as templates, homology (comparative) modeling has so far produced protein structure models of the highest quality. Yet, recent advances in this round of CASP13 allow a glimpse into major improvements on template-free approaches.2,3 Here, we analyze the participants’ model with respect to two baseline reference methods. Both are aimed at aiding the comparison of each model to well-known approaches, thereby assisting the server algorithm developers to locate potential areas for further development. The first one represents a BLAST-based predictor (“NaiveBLAST”).4 Most of the methods produce higher quality models compared with NaiveBLAST, and the top five methods show a median improvement of more than 4.5 lDDT points (Figure 1). Reasons for improving the model may lie in a better template selection based on more sensitive profile-profile comparisons, better loop modeling, and multitemplate modeling in general. The second reference method is the structure-based “bestSingleTemplate” method, representing an upper limit for single template models. Here, only the top three methods improved more than 10% of the targets, with Robetta clearly in the lead (Figure 2). As opposed to the NaiveBLAST baseline, many targets have been predicted equal or worse (Figures S1 and S2). Even for the best methods, the limited improvement over the bestSingleTemplate approach clearly exhibits room for further development.
FIGURE 1.
Compared with the NaiveBLAST server in units lDDT, the medians are depicted by the horizontal bar in the boxes. The sort order is by the decreasing median. The number of targets used in the comparison by server is indicated at the top of each column, with a maximum of 228 out of a total of 248 targets returned by NaiveBLAST. The data set covers the time from 1 May 2018 to 28 July 2018
FIGURE 2.
Compared with the “bestSingleTemplate” method in units lDDT, the medians are depicted by the horizontal bar in the boxes. The sort order is by the decreasing median. The number of targets returned by each server is indicated, and the total number of targets is 248. The data set covers the time from 1 May 2018 to 28 July 2018
3.1.3 |. General performance analysis
CAMEO evaluates the structure predictions applying different scores for assessing different aspects of modeling, such as accuracy of a single protein chain, the homo-oligomeric interface, or the binding site (Table 1). Here, we categorized the data into the target domains “hard,” “medium,” and “easy.” The three best methods “Robetta,”19 “Raptor-X,”20 and “IntFOLD5-TS21 returned all hard targets (in total 62) with a very similar performance, both in average lDDT and SD, of 47.29±12.96, 45.25±13.57, and 44.79±12.82 (CAD-scores8: 0.55 ±0.09, 0.53±0.10, and 0.52±0.09), respectively. Although the performance is very similar on this specific target set, the response times vary greatly with “Raptor-X” clearly in the lead (7.8 hours). For the medium difficulty targets (in total 139), the situation is almost identical, albeit at a much higher average accuracy level with average lDDT values of 74.03±7.15, 71.50±7.28, and 72.53±6.99 (CAD-scores: 0.71 ±0.05, 0.69±0.05, and 0.69±0.06), respectively. Here, “SWISS-MODEL” (69.09±10.33 lDDT, 0.69 CAD-score, 99% of medium targets), “IntFOLD3-TS”22 (69.21±7.40 lDDT, 0.69 CAD-score, 98% of medium targets), and “HHPredB”23 (68.31±9.60 lDDT, 0.68 CAD-score, 100% medium targets) are close with regard to performance. “SWISS-MODEL” is sticking out with a very fast average response time of 12 minutes, closely followed by “HHPredB” (42 minutes), “PRIMO”24 (54 minutes), and “SPARKS-X”25 (114 minutes). For the easy targets (in total 47), 12 out of 16 servers showed lDDT scores in the range of 80 to 84, with “Robetta” (lDDT 84.55±4.04) and “SWISS-MODEL” (lDDT 83.88±5.38) in a narrow lead. Within a mere 4 units lDDT “Raptor-X,” “IntFOLD5-TS,” “HHPredB,” “PRIMO,” “M4T-SMOTIF-TF,”26 and “SPARKS-X” are close in performance.
TABLE 1.
Server comparison for the target domains ‘hard” (62), “medium” (139), and “easy” (47)
| Server name | Response time (hh:mm)—all | Returned fraction—hard | Mean lDDT—hard | Mean CAD-score—hard | ReturnedFraction—medium | Mean lDDT—medium | Mean CAD-score—medium | Returned fraction—easy | Mean lDDT—easy | Mean CAD-score—easy | Mean model confidence | Mean lDDT-BS—all | QS-score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Robetta | 32:12 | 0.98 | 47.29 (12.96) | 0.55 (0.09) | 1.00 | 74.03 (7.15) | 0.71 (0.05) | 1.00 | 84.55 (4.04) | 0.79 (0.03) | 0.81 (0.14) | 66.87 (20.05) | 0.26 (0.68) |
| RaptorX | 07:48 | 1.00 | 45.25 (13.57) | 0.53 (0.10) | 1.00 | 71.50 (7.28) | 0.69 (0.05) | 1.00 | 82.60 (3.30) | 0.77 (0.03) | 0.65 (0.08) | 66.60 (21.86) | - |
| IntFOLD5-TS | 30:30 | 1.00 | 44.79 (12.82) | 0.52 (0.09) | 1.00 | 72.53 (6.99) | 0.69 (0.06) | 1.00 | 83.73 (3.93) | 0.78 (0.03) | 0.83 (0.11) | 70.75 (22.79) | - |
| IntFOLD3-TSa | 31:06 | 1.00 | 40.80 (14.54) | 0.49 (0.10) | 0.98 | 69.21 (7.40) | 0.66 (0.06) | 1.00 | 82.74 (4.24) | 0.77 (0.04) | 0.85 (0.11) | 69.21 (22.62) | - |
| HHPredBa | 00:42 | 1.00 | 39.09 (12.89) | 0.49 (0.09) | 1.00 | 68.31 (9.60) | 0.68 (0.07) | 1.00 | 82.57 (3.87) | 0.78 (0.03) | 0.76 (0.12) | 66.52 (22.29) | - |
| SPARKS-X | 01:54 | 1.00 | 36.83 (11.01) | 0.48 (0.08) | 0.96 | 62.96 (9.59) | 0.62 (0.07) | 1.00 | 80.14 (4.30) | 0.75 (0.04) | 0.53 (0.07) | 62.40 (22.74) | - |
| IntFOLD4-TSa | 47:42 | 0.79 | 34.06 (13.75) | 0.40 (0.10) | 0.70 | 50.01 (7.35) | 0.48 (0.06) | 0.77 | 64.02 (3.93) | 0.60 (0.04) | 0.82 (0.13) | 50.29 (20.61) | - |
| SWISS-MODEL | 00:12 | 0.98 | 30.45 (18.39) | 0.35 (0.16) | 0.99 | 69.09 (10.33) | 0.67 (0.08) | 1.00 | 83.88 (5.38) | 0.79 (0.05) | 0.87 (0.09) | 67.55 (26.27) | 0.35 (0.71) |
| PRIMO_HHS_CL | 01:18 | 0.98 | 26.35 (14.60) | 0.34 (0.14) | 0.91 | 54.49 (15.95) | 0.55 (0.11) | 0.94 | 74.79 (6.50) | 0.70 (0.05) | 0.66 (0.09) | 58.09 (26.42) | - |
| PRIMO_HHS_3D | 01:30 | 0.95 | 24.91 (14.41) | 0.32 (0.13) | 0.91 | 55.05 (13.79) | 0.55 (0.10) | 0.94 | 74.25 (7.23) | 0.69 (0.06) | 0.67 (0.09) | 60.20 (24.45) | - |
| Phyre2a | 01:06 | 0.97 | 21.56 (15.95) | 0.30 (0.17) | 0.97 | 56.47 (12.56) | 0.62 (0.09) | 0.96 | 73.37 (8.75) | 0.73 (0.06) | 0.56 (0.10) | 58.36 (26.71) | - |
| PRIMO_BST_CL | 00:48 | 0.89 | 20.97 (13.02) | 0.28 (0.12) | 0.96 | 61.48 (13.52) | 0.60 (0.10) | 1.00 | 82.15 (5.46) | 0.76 (0.04) | 0.65 (0.09) | 59.03 (26.64) | - |
| PRIMO | 00:54 | 0.89 | 20.97 (13.02) | 0.28 (0.12) | 0.96 | 61.48 (13.52) | 0.60 (0.10) | 1.00 | 82.15 (5.46) | 0.76 (0.04) | 0.65 (0.09) | 59.03 (26.64) | - |
| M4T-SMOTIF-TF | 19:24 | 0.71 | 19.98 (16.10) | 0.25 (0.15) | 0.97 | 63.14 (17.29) | 0.61 (0.13) | 1.00 | 83.27 (3.34) | 0.77 (0.03) | 0.70 (0.10) | 63.42 (25.66) | - |
| PRIMO_BST_3D | 00:54 | 0.87 | 17.65 (13.21) | 0.24 (0.13) | 0.96 | 59.17 (14.88) | 0.58 (0.11) | 1.00 | 80.83 (7.13) | 0.75 (0.05) | 0.67 (0.09) | 57.74 (26.44) | - |
| NaiveBLASTa | 01:12 | 0.76 | 16.75 (18.03) | 0.20 (0.17) | 0.96 | 61.52 (14.76) | 0.59 (0.12) | 1.00 | 80.60 (6.18) | 0.75 (0.05) | 0.66 (0.10) | 58.93 (25.74) | - |
Notes: The overall sort order is given by the lDDT performance of the hard targets. Values in parenthesis are the SDs for the respective scores, except for QS-score,14 where the average across the modeled targets with correct assemblies is given for comparison. The data set covers the time from 1 May 2018 to 28 July 2018.
Method is not reflecting the current development and is kept in CAMEO for historic comparison.
A very important way to communicate a model’s quality to the end user is by assigning correct per-residue confidence estimates.27 “SWISS-MODEL,” “IntFOLD3-TS,” and “Robetta” are providing good to very good error estimates with “SWISS-MODEL” and “IntFOLD3-TS” peaking at 0.87 and 0.85 as analyzed by averaging per-model ROC AUCs.
CAMEO analyses the accuracy of the binding-site residues (“lDDT-BS”4), where on the current data set across 94 targets and 83 unique ligands (see Data S1), the top modeling groups are also producing good quality binding sites with lDDT-BS scores close to 70. The high SD is most likely owing to local inaccuracies and the fact that missing residues have a very pronounced effect on the rather small number of residues involved in forming the binding site.
For almost 40% of this data set, the biologically active form of a protein is a homo-oligomer, yet only two servers “Robetta” and “SWISS-MODEL” are routinely predicting homo-oligomers. “Robetta” returned 37 out of 96 assemblies correctly, while “SWISS-MODEL” returned 47. For 11 and 16 assemblies, respectively, the servers predicted wrong stoichiometries—these models have not been included in the analysis. Both servers failed to produce homo-oligomer models for 29 out of 96 (30%) of the targets.
3.2 |. Quality estimation
3.2.1 |. Analyses
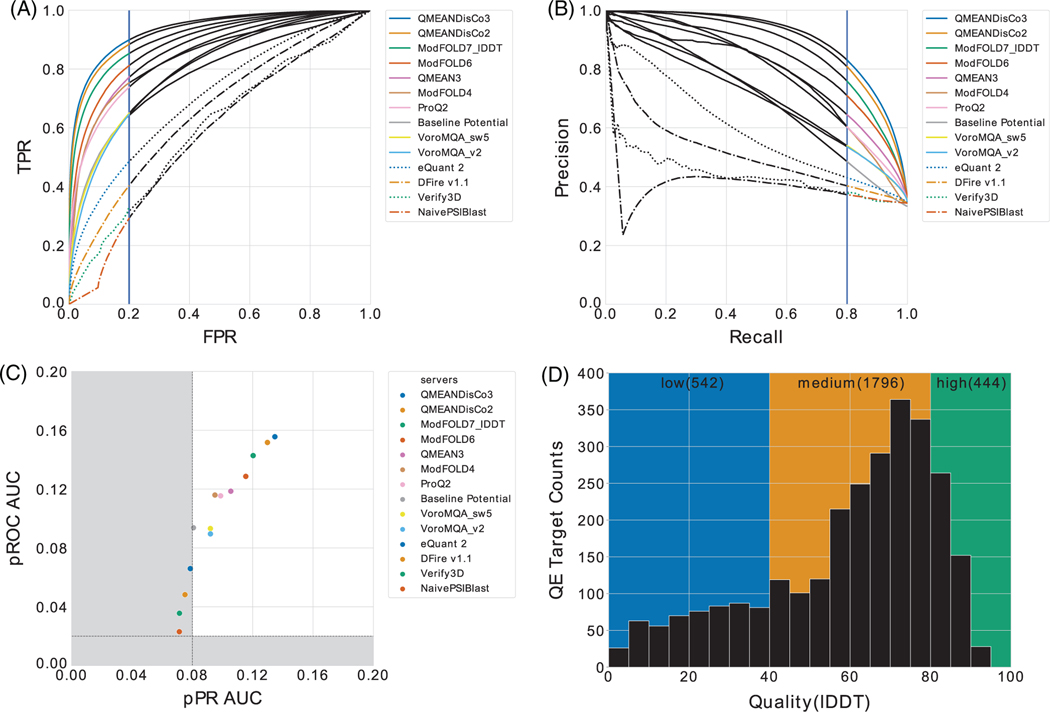
Closely related to computing reliable and biologically relevant protein structure models is robust model quality estimation—justifying its own category both at CASP and in CAMEO evaluations since many years. The CAMEO quality estimation (CAMEO QE) category focuses on per-residue evaluations, where the task is to distinguish residues of bad quality from high quality in protein structure models. Among many others, the ROCs and its AUC have a long history and have, thus, been implemented within CAMEO QE. PR curves are also featured to investigate potential bias in the data sets and to better capture performance in the case of a very low number of bad residues observed in high-quality structures. Here, in ROC space, a large change in the false positives has a small effect, while it is clearly captured in the PR space performance.28 Consequently CAMEO QE considers both analysis domains and offers a performance analysis based on partial ROC and partial PR. For the partial ROC analysis, we calculate the AUC only in the FPR range of 0.0 to 0.2 (Figure 3A) and for the partial PR analysis in a recall (TPR) range from 0.8 to 1.0 (Figure 3B). We condense the information in a scatterplot with the AUCs obtained for the partial ROC plotted against the AUCs’ partial PR analyses. Within the top performing regime of this performance domain “QMEANDisCo3”,27 “QMEANDisCo2,”29 “ModFOLD6,”30 and “ModFOLD7-lDDT”22 are clearly in the lead (Figure 3C), followed by “QMEAN3,” “ModFOLD4,”31 and “VoroMQA.”32 All these methods clearly outperform the “Baseline Potential” based on a full-atomic statistical potential of mean force as has been reported earlier4 (Table 2). In CAMEO, the current threshold of lDDT has been selected based on earlier unpublished investigations and discussions with the CAMEO participants. An analysis to study the stability of rankings dependent on different thresholds for lDDT and CAD score is provided as Data S1 (see Figures S4–S9). Methods that are similar in performance show minor variances in ranks across the thresholds. In PR space, the “Baseline Potential” showed the highest variation, likely due to its simplicity. For the lower two thresholds in ROC and partial ROC vs partial PR space, both lDDT and CAD score show similar behavior, while higher thresholds are reflecting the well-known issues of binary classifications. The findings confirm that for the question at hand, pROC AUC vs pPR AUC is the most stable ranking methods for both lDDT and CAD score. When considering the historic development of quality estimation methods, it is obvious that a tremendous performance gap exists between the original approaches, for example, PROSA33,34 or DFIRE35 and current methods (Figure 4). All analyses have been performed on a data set that is biased toward high-quality models (Figure 3D), aiming at reflecting a real-case modeling scenario.
FIGURE 3.
A, Partial precision-recall AUC, blue vertical line depicts the threshold of 0.2 FPR; B, partial ROC AUC, the blue vertical line indicates the threshold of 80% recall; C, pROC AUC vs the pPR AUC domain, applying an lDDT threshold of 60. The dashed lines represent the AUCs for the random predictor in the ROC domain and for the expected precision at 100% recall for the PR domain. Areas in grey are below these thresholds and would be considered performing worse than random. D, model quality distribution of the QE target set in units lDDT. The data set covers the time from 1 May 2018 to 28 July. AUC, area under the curves; ROC, receiver-operator characteristic
TABLE 2.
Performance ranking for participants of CAMEO QE by partial ROC AUC, where the maximum value is 0.2, as is for the partial PR AUC
| Server name | Returned fraction | Response time (hh:mm) | pROC AUC | pPR AUC | pROC AUC—all models | pPR AUC—all models |
|---|---|---|---|---|---|---|
| QMEANDisCo3 | 0.99 | 00:06 | 0.156 | 0.135 | 0.154 | 0.134 |
| QMEANDisCo2a | 0.99 | 00:42 | 0.152 | 0.130 | 0.150 | 0.129 |
| ModFOLD7_lDDT | 0.99 | 28:06 | 0.143 | 0.120 | 0.142 | 0.119 |
| ModFOLD6a | 0.71 | 72:48 | 0.129 | 0.115 | 0.092 | 0.082 |
| QMEAN3 | 0.99 | 00:48 | 0.119 | 0.105 | 0.118 | 0.104 |
| ModFOLD4a | 0.99 | 92:06 | 0.116 | 0.095 | 0.115 | 0.094 |
| ProQ2a | 0.99 | 00:30 | 0.115 | 0.099 | 0.114 | 0.098 |
| Baseline potential | 0.84 | 05:42 | 0.094 | 0.081 | 0.079 | 0.068 |
| VoroMQA_sw5 | 0.92 | 00:24 | 0.093 | 0.092 | 0.086 | 0.085 |
| VoroMQA_v2 | 0.99 | 00:18 | 0.090 | 0.092 | 0.089 | 0.091 |
| eQuant 2 | 0.99 | 00:30 | 0.066 | 0.079 | 0.065 | 0.078 |
| DFire v1.1a | 0.92 | 02:18 | 0.048 | 0.075 | 0.044 | 0.069 |
| Verify3Da | 0.92 | 03:24 | 0.036 | 0.071 | 0.033 | 0.065 |
| NaivePSIBlast | 0.92 | 04:36 | 0.023 | 0.071 | 0.021 | 0.065 |
Notes: Columns showing results for “all models” are including scores of 0 for missing predictions. The data set covers the time from 1 May 2018 to 28 July 2018.
Abbreviations: AUC, area under the curves; ROC, receiver-operator characteristic.
Method is not reflecting the current development and is kept in CAMEO for historic comparison
FIGURE 4.
Historic development of quality estimation tools. The improvements are impressive spanning early developments and recent approaches over the last 22 years, from well-known tools such as PROSA,33,34 Verify3D,36 DFIRE35 to the latest contestants such as ProQ3,37 ModFOLD7_lDDT22 and QMEANDisCo3.27 The years are assigned roughly to the best server of a particular year. The black empty circle illustrates estimated performance of QMEAN (Version 7.11)38 based on earlier CAMEO data. The blue star depicts the estimated performance of ProQ3 based on three months (17 May 2019 to 10 August 2019) of CAMEO data
4 |. CONCLUSION AND OUTLOOK
CAMEO continuously evaluates automated methods that either already run as a public productive server or are considered private development pipelines every week throughout the entire year. This distinguishes CAMEO from CASP and complements it at the same time, as many methods in CASP are not available as public services. CAMEO collected a large number of targets in the 3D (248) and QE (2798) category in just 3 months. This underlines the importance of a continuous evaluation, where server algorithm developers benefit from immediate feedback on their latest developments. These methods are often registered as anonymous servers, thereby hiding the identity until the performance improvements have been concluded. Apart from these anonymous server registrations, we encourage the independent re-registration of public servers following major release versions to retrospectively discuss improvements of algorithms more transparently. CAMEO is an open platform and features many scores from single chain accuracies to homo-oligomer interfaces reflecting different aspects of modeling.
CAMEO 3D introduced the “NaiveBLAST” baseline early on to estimate a lower bound of quality. Unsurprisingly, based on its simple approach, all actively developed servers show major improvements for more than 50% of the targets. Here, “Robetta” and “IntFOLD5-TS” are in the lead, closely followed by “SWISS-MODEL,” “RaptorX,” and “IntFOLD3-TS”. Besides “NaiveBLAST,” we have now introduced the “bestSingleTemplate” (bST) method, which yields an upper baseline for single template models. Consequently, the fraction of models scoring higher than bST is considerably lower than the “NaiveBLAST” method, where only three servers markedly improve their models compared with bST. The correct oligomeric state is crucial for biological relevance, yet with “SWISS-MODEL” and “Robetta” only two of the registered servers in CAMEO are currently modeling homo-oligomers. Here, “SWISS-MODEL” is in the lead at the moment, modeling more of the 96 targets with the correct assembly and with a higher QS-score. With a maximum of 31% correctly modeled homo-oligomer assemblies and no models for 29% of the targets from either server, this aspect clearly shows room for improvement.
For CAMEO QE, the emphasis lies on per-residue quality estimation. Global scores are not currently evaluated and the task of the predictors in this category is to find bad quality residues (local lDDT <60) in a data set of models ranging from low quality to high quality. Although ROC analyses are well suited for this kind of comparison, CAMEO also includes PR analyses to capture a large change in the false positives, which has a small effect in ROC space, while it is clearly captured in the PR space. We have introduced a combined partial ROC AUC and partial PR AUC analysis, where most servers exhibit a comfortable lead over the “Baseline Potential” predictor. In the respective time frame, “QMEANDisCo3” and “ModFOLD7-lDDT” clearly have the lead, with the Voronoi tessellation-based method “VoroMQA-v2” located in the midfield.
Recent advances have again pushed the boundaries as compared to the special issue 2 years ago,4 which marks the summit of continuous improvements over the last 22 years (Figure 4).
The continued development of CAMEO will focus on the selection and validation of target reference structures with the aim to maximize target diversity and target quality at the same time. Sets of previously established validation criteria will be employed for this task, which are based on the reports of the PDB validation pipeline.39 We envision to include ligand poses as well as assess heteromers in the near future stimulating the development of structure prediction methods even further.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank for the invaluable feedback that we obtained from observers and participants alike, in particular the Schwede group members for testing new CAMEO releases, open discussions, and critical feedback. We would like to thank the community for their support, their new scores, and prediction methods. We are grateful to the sciCORE team for providing support and computational resources. We would like to thank the RCSB PDB for publishing the pre-release data openly and their invaluable input on experimental structure matters. We are grateful for funding: from the SIB Swiss Institute of Bioinformatics toward the development of CAMEO and OpenStructure and the use of sciCORE computing infrastructure; from NIH and National Institute of General Medical Sciences (U01 GM093324–01) partially to CAMEO; and from ELIXIR EXCELERATE to CAMEO. Funding for open access charge was provided by the SIB Swiss Institute of Bioinformatics.
Funding information
ELIXIR Excelerate (H2020), Grant/Award Number: 676559; National Institute of General Medical Sciences, Grant/Award Number: U01 GM093324–01; Swiss Institute of Bioinformatics
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Schwede T, Sali A, Honig B, et al. Outcome of a workshop on applications of protein models in biomedical research. Structure. 2009;17: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croll TI, Sammito MD, Kryshtafovych A, Read RJ. Evaluation of template-based modeling in CASP13. Proteins. 2019;87(12):1113–1127. 10.1002/prot.25800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abriata LA, Tamò GE, Dal Peraro M. A further leap of improvement in tertiary structure prediction in CASP13 prompts new routes for future assessments. Proteins. 2019;87(12):1100–1112. 10.1002/prot.25787 [DOI] [PubMed] [Google Scholar]
- 4.Haas J, Barbato A, Behringer D, et al. Continuous Automated Model EvaluatiOn (CAMEO) complementing the critical assessment of structure prediction in CASP12. Proteins. 2018;86(Suppl 1):387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.wwPDB consortium. Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.UniProt Consortium T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olechnovič K, Venclovas C. The CAD-score web server: contact area-based comparison of structures and interfaces of proteins, nucleic acids and their complexes. Nucleic Acids Res. 2014;42:W259–W263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariani V, Biasini M, Barbato A, Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antczak PLM, Ratajczak T, Lukasiak P, Blazewicz J (2015) SphereGrinder—reference structure-based tool for quality assessment of protein structural models. IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 10.1109/bibm.2015.7359765 [DOI] [Google Scholar]
- 11.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siew N, Elofsson A, Rychlewski L, Fischer D. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16:776–785. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. [DOI] [PubMed] [Google Scholar]
- 14.Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep 2017;7:10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee S, Zhang Y. MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 2009;37:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studer G, Tauriello G, Bienert S, et al. Modeling of protein tertiary and quaternary structures based on evolutionary information. Methods Mol. Biol 2019;1851:301–316. [DOI] [PubMed] [Google Scholar]
- 18.Kryshtafovych A, Monastyrskyy B, Fidelis K, Schwede T, Tramontano A. Assessment of model accuracy estimations in CASP12. Proteins. 2018;86 Suppl 1:345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H, Kim DE, Ovchinnikov S, Baker D, DiMaio F. Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins. 2018;86(Suppl 1):283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Wang S, Bu D, Xu J. Protein threading using residue covariation and deep learning. Bioinformatics. 2018;34:i263–i273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGuffin LJ, Adiyaman R, Maghrabi AHA, et al. IntFOLD: an integrated web resource for high performance protein structure and function prediction. Nucleic Acids Res. 2019; 47(W1):W408–W413. 10.1093/nar/gkz322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGuffin LJ, Atkins JD, Salehe BR, Shuid AN, Roche DB. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res. 2015;43(W1):W169–W173. Available from:. 10.1093/nar/gkv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatherley R, Brown DK, Glenister M, Bishop ÖT. PRIMO: An Interactive Homology Modeling Pipeline. PLOS ONE. 2016;11:e0166698. 10.1371/journal.pone.0166698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Faraggi E, Zhao H, Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of query and corresponding native properties of templates. Bioinformatics. 2011;27:2076–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rykunov D, Steinberger E, Madrid-Aliste CJ, Fiser A. Improved scoring function for comparative modeling using the M4T method. J. Struct. Funct. Genomics 2009;10:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng J, Choe M-H, Elofsson A, et al. Estimation of model accuracy in CASP13. Proteins. 2019;87(12):1361–1377. 10.1002/prot.25767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis J, Goadrich M. (2006) The relationship between precision-recall and ROC curves. Proceedings of the 23rd International Conference on Machine Learning—ICML ‘06. 10.1145/1143844.1143874 [DOI] [Google Scholar]
- 29.Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maghrabi AHA, McGuffin LJ. ModFOLD6: an accurate web server for the global and local quality estimation of 3D protein models. Nucleic Acids Res. 2017;45:W416–W421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGuffin LJ, Buenavista MT, Roche DB. The ModFOLD4 server for the quality assessment of 3D protein models. Nucleic Acids Res. 2013; 41:W368–W372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olechnovič K, Venclovas Č. VoroMQA: Assessment of protein structure quality using interatomic contact areas. Proteins. 2017;85:1131–1145. [DOI] [PubMed] [Google Scholar]
- 33.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. [DOI] [PubMed] [Google Scholar]
- 34.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C. Accurate and efficient loop selections by the DFIRE-based all-atom statistical potential. Protein Sci. 2004;13:391–399. 10.1110/ps.03411904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenberg D, Lüthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277: 396–404. [DOI] [PubMed] [Google Scholar]
- 37.Uziela K, Menéndez Hurtado D, Shu N, Wallner B, Elofsson A. ProQ3D: improved model quality assessments using deep learning. Bioinformatics. 2017;33:1578–1580. [DOI] [PubMed] [Google Scholar]
- 38.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gore S, Velankar S, Kleywegt GJ. Implementing an X-ray validation pipeline for the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr 2012;68:478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.