Summary
Kernel number per spike determined by the spike or inflorescence development is one important agricultural trait for wheat yield that is critical for global food security. While a few important genes for wheat spike development were identified, the genetic regulatory mechanism underlying supernumerary spikelets (SSs) is still unclear. Here, we cloned the wheat FRIZZY PANICLE (WFZP) gene from one local wheat cultivar. WFZP is specifically expressed at the sites where the spikelet meristem and floral meristem are initiated, which differs from the expression patterns of its homologs FZP/BD1 in rice and maize, indicative of its functional divergence during species differentiation. Moreover, WFZP directly activates VERNALIZATION1 (VRN1) and wheat HOMEOBOX4 (TaHOX4) to regulate the initiation and development of spikelet. The haplotypes analysis showed that the favourable alleles of WFZP associated with spikelet number per spike (SNS) were preferentially selected during breeding. Our findings provide insights into the molecular and genetic mechanisms underlying wheat spike development and characterize the WFZP as elite resource for wheat molecular breeding with enhanced crop yield.
Keywords: supernumerary spikelets, WFZP, VRN1, TaHOX4, spike architecture, bread wheat
Introduction
Bread wheat (Triticum aestivum; AABBDD) provides up to 20% of the calories to feed the human populations worldwide, and therefore, its yield is greatly important for global food security. Among the three yield components, kernel number per spike is directly determined by the spike or inflorescence development. In most of wheat cultivars, the spike bears one sessile spikelet per rachis node. At the double‐ridge stage of early wheat spike development, the inflorescence meristem (IM) generates spikelet meristems (SMs) that subsequently produce floral meristems (FMs) to form unbranched spike architecture (Dobrovolskaya et al., 2015). The IM will be terminated by a terminal SM shortly after a certain number of SMs are produced, when the SNS will be determined at the early floret differentiation stage (Li et al., 2019a). The timing of this conversion depends on the IM activity, and prolonged IM activity will result in increased spikelet number. Except for the normal spike architecture, some cultivars develop supernumerary spikelets (SSs) phenotype characterized by the spike bearing more spikelets per rachis node. The formation of SSs is usually due to the alteration of SM identity or impaired transition from SM to FM (Dobrovolskaya et al., 2015). Both prolonging IM maintenance and creation of SS spike architecture in common wheat cultivars are selectively strategy to increase kernel number per spike.
Up to now, several genes controlling IM maintenance have been identified in grass. In rice, ABERRANT PANICLE ORGANIZATION1 (APO1), APO2 and TAWAWA1 (TAW1) regulate the IM or primary branch meristems (pBMs) maintenance through repressing the precocious conversion of IM or pBM to secondary branch meristems (sBM) or SM (Ikeda et al., 2007; Ikeda‐Kawakatsu et al., 2012; Yoshida et al., 2013). In wheat, two MADS family genes VRN1 and FRUITFULL2 (FUL2) are found to influence IM maintenance (Li et al., 2019a). Mutations in any of both genes will delay the transition from IM to terminal SM and then increase SNS (Li et al., 2019a). Despite these genes were identified, the direct genetic interactions and molecular mechanisms underlying these genes are still unclear.
Previous studies revealed that SM maintenance and the transition from SM to FM are regulated by FZP in rice and its homologous gene BRANCHED SILKLESS1 (BD1) in maize (Chuck et al., 2002; Komatsu et al., 2003). FZP encodes an AP2/ERF transcription factor with transcriptional activator activity (Komatsu et al., 2003). FZP is specifically expressed at the axils of rudimentary glumes primordial to control floral fate. In null mutant fzp, spikelet is replaced by the branch‐like structure which bears fewer fertile spikelets, while in the FZP knockdown plants, most of spikelets are substituted by secondary branches, indicating that FZP inhibits the formation of axillary meristem (AxM) or ectopic SM (eSM) in a dose‐depend manner during SM‐to‐FM transition in rice (Bai et al., 2017; Komatsu et al., 2003). In bread wheat, mutations in WFZP result in SS phenotype showing multirow spike developed at a rachis node (Dobrovolskaya et al., 2015). Besides WFZP, wheat spikelet development is also regulated by VRN1, FUL2 and FUL3. In vrn1ful2 double mutant or vrn1ful2ful3 triple mutant, the SMs are replaced by other kind of meristems, demonstrating that these genes play critical roles in maintaining the SM identity (Li et al., 2019a).
To date, the expression regulation and interaction proteins of FZP have been well studied (Bai et al., 2017; Huang et al., 2018). However, the molecular and genetic mechanisms underlying the functions of FZP and its orthologs in inflorescence development remain unclear. In this study, we cloned TOO MANY SPIKELETS (TMS) gene, which is identical to WFZP, from one local wheat cultivar. WFZP determines the SNS and the FM fate, which is mediated partially by VRN1 and TaHOX4. In addition, haplotype analysis and geographic distribution of WFZP were also investigated, in which a G‐to‐A substitution at WFZP‐A promoter has potential to increase the SNS and the crop yield during breeding.
Results
YM44 produces supernumerary spikelets due to generation of secondary spikelets from primary spikelet meristem
To isolate the native gene sources that function in the wheat spike development, we screened hundreds of wheat cultivars grown in North China according to the spike architecture variations. One local cultivar, YM44, is arrested for its SSs phenotype (Figure 1a–c). Normally, common wheat is unbranched and thus the rachis node number is the same as the SNS. We found that YM44 spike generates more rachis nodes (26.2 ± 2.2) than Kenong9204 (KN9204) (20.4 ± 1.2), a typical winter wheat (Cui et al., 2014), indicative of prolonged IM activity of YM44 to produce SM (Figure 1b,d). Remarkably, branch‐like structure is produced from middle of main axis of YM44, on which several spikelets are initiated (Figure 1b,c), indicating that the SM and FM identity of YM44 may be impaired. Due to the SSs and prolonged IM activity, YM44 has higher spikelet and kernel number per spike compared with KN9204 (Figure 1d). Microanatomy observation by scanning electron microscopy showed that, at glume differentiation stage, both KN9204 and YM44 could produce normal glume primordia (Figure 1e,f), showing that the primary SM identity of YM44 may not be impaired. At floret differentiation stage, instead of FM, secondary SM was produced from YM44 primary SM to generate additional spikelet (Figure 1g–j), indicating that the FM fate is changed or the AxM activity is de‐repressed in YM44.
Figure 1.
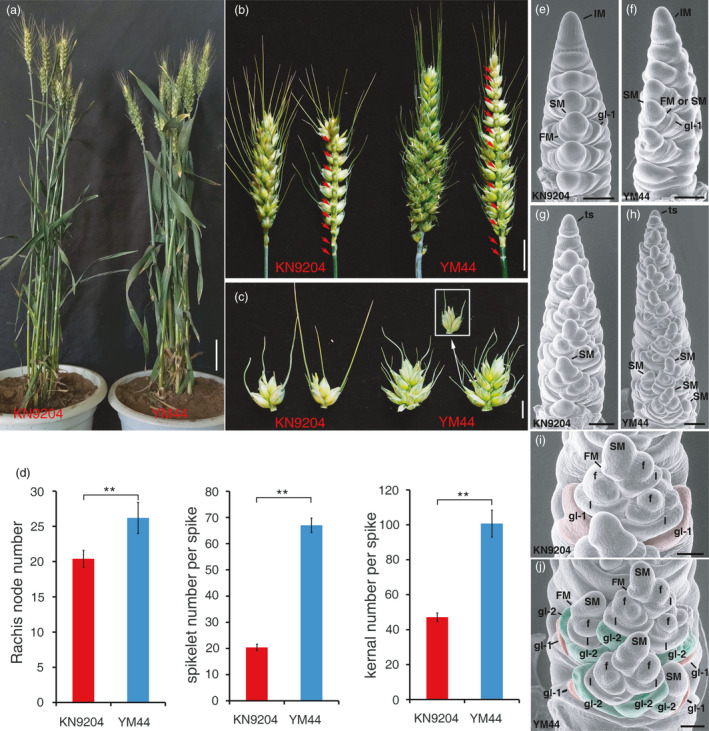
Phenotypic characterization of KN9204 and YM44. (a) Pictures of representative plants of KN9204 and YM44 before maturing. (b) Spikes of KN9204 and YM44 with red arrow showing the rachis node. (c) The spikelet of KN9204 and a supernumerary spikelet (SS) attached in one rachis node of YM44. The inset shows an intact spikelet initiated from primary SM. (d) Statistics comparison of rachis node number, spikelet number per spike and kernal number per spike between KN9204 and YM44. The error bars denote ± SE, **P < 0.01. (e–j) The electron scanning images of young spikes of KN9204 (e, g and i) and YM44 (f, h and j) at early floret differentiation stage (e and f) and later floret differentiation stage (g–j). IM, inflorescence meristem; SM, spikelet meristem; FM, floret meristem; f, floret with floret organ differed; gl‐1, glume initiated from primary SM (Red coloured in (i and j); gl‐2, glume initiated from secondary SM (Green coloured in (j); l, lemma. Bars = 5cm in A, 2 cm in B, 1 cm in C, 200 μm in e–h, 100 μm in I and j.
Cloning the gene responsible for supernumerary spikelets
To investigate the genetic reason underlying the SSs phenotype, we created a segregation population by crossing YM44 with KN9204. All the individuals in F1 population have normal spike, demonstrating that the SSs phenotype of YM44 is controlled by recessive loci. In the F2 population, 47 out of 884 individuals show SSs phenotype. The segregation ratio is in agreement with the expected segregation ratio of two recessive Mendelian factor (χ2 = 1.31, P = 0.25). Thus, we named the two loci as TOO MANY SPIKELETS‐1/2 (TMS‐1/2). We performed the linkage analysis using a set of SSR markers evenly distributed in wheat genome (https://wheat.pw.usda.gov/GG3/) and found that the SS phenotype was linked with the marker Xwmc522 and cfd56 which were located on chromosome 2A and 2D, respectively (Figure 2a). Then, the expression level of all genes near these two markers was examined using the RNA‐seq data from young spike at glume differentiation stage of KN9204 and YM44 (Li et al., 2018b and this work). Among these genes, WFZP‐D (TraesCS2D02G118200) was nearly no expression in YM44 that was confirmed by RT‐PCR (Figure 2b,c). We then sequenced the genomic regions of WFZP‐D and its homeologous gene WFZP‐A (TraesCS2A02G116900) that is near the marker Xwmc522. While there is no any sequence difference at WFZP‐D locus in YM44 and Chinese Spring reference gene, a 14 bp deletion just adjacent to the start codon of WFZP‐A was detected resulting in a frame‐shift (Figure 2d). Next, an Indel marker of WFZP‐A and a CAPS marker of WFZP‐D were developed based on the sequence difference between KN9204 and YM44, and the linkage of WFZP‐A, WFZP‐D and the SSs phenotype were further confirmed using these two markers (Figure S1). Thus, the TMS‐1 and TMS‐2 are identical to the reported WFZP‐A and WFZP‐D, respectively (Dobrovolskaya et al., 2015).
Figure 2.
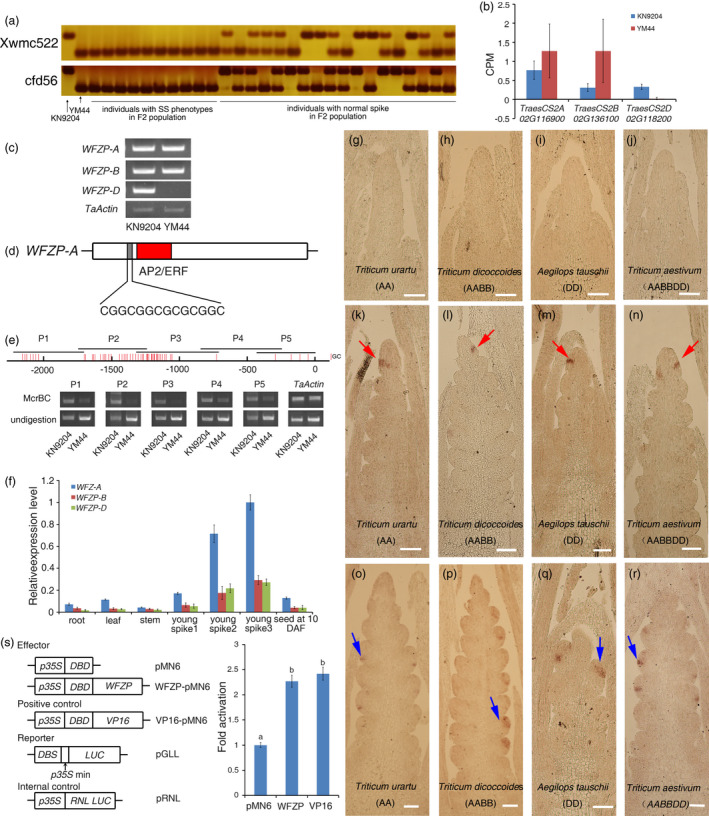
Cloning and characterization of TMS/WFZP. (a) Linkage analysis of SSR markers Xwmc522 and cfd56 and YM44 SS phenotype. (b, c) The expression level of WFZP‐A, B and D in the young spikes of KN9204 and YM44 examined by RNA‐seq (b) and Semi‐quantitative RT‐PCR (c). (d) The schematic diagram of the sequence of WFZP‐A in YM44. The grey region shows a 14bp‐deletion. The AP2/ERF domain is marked by red colour. (e) McrBC analysis of WFZP‐D promoter in KN9204 and YM44. A genomic region at TaActin locus was used as control. (f) Tissue‐specific expression analysis of WFZP‐A, B and D. The error bars denote ± SE. (g–r) In situ hybridization to examine the expression pattern of WFZP in Triticum urartu (g, k and o), Triticum dicoccoides (h, l and p), Aegilops tauschii (i, m and q) and bread wheat (j, n and r). Red arrows show signals in the top region of IM where the SM will be initiated and blue arrows show signals inside SM where the FM will be initiated. (S) Transactivation analysis of WFZP. VP16 was used as the positive control. The error bars denote ± SE. Different letters mean significant difference at P < 0.01. Bars = 200 μm in g–r.
To figure out why the WFZP‐D is silenced in YM44, we examined the DNA methylation status of WFZP‐D promoter region in KN9204 and YM44 using McrBC‐PCR method. The 2.2kb promoter region of WFZP‐D was divided into 5 overlapped fragments, and PCR results showed that genomic DNA in P2 and P3 regions containing a high proportion of GC were high methylated in YM44, but not in KN9204 (Figure 2e). The McrBC analysis in the descendants from the cross of KN9204 and YM44 implied that the DNA methylation at WFZP‐D promoter of YM44 could be stably transferred to next generation, and lead to the gene silencing (Figure S2). To further detect whether there is sequence variation that can influence the DNA methylation status, we sequenced 6.5 kb promoter region of WFZP‐D in YM44. The sequence was same as that of Chinese Spring, indicating that the sequence variation, if exists, maybe located at the flanking of WFZP‐D locus, which is worthy for further investigation.
WFZP has distinct expression patterns during spike development
To investigate the spatio‐temporal expression patterns of WFZP in wheat, we examined the stage‐specific expression level of WFZP by RT‐qPCR. All of WFZP‐A, B and D were highly expressed in young spikes at double‐ridge stage and floral differentiation stage, in line with its roles in spike development. Moreover, WFZP‐A had higher expression level compared with the other two copies at these developmental stages (Figure 2f). Next, the tissue‐specific expression patterns of WFZP in wheat young spike were detected by in situ hybridization. At the single ridge stage, no WFZP expression was detected at shoot apical meristem (SAM) (Figure 2j). Subsequently, at the later of double‐ridge stage before the IM is converted into terminal spikelet, a clear and distinct signal was detected at the apical region of IM where the SM will be initiated (Figure 2n), which is similar to the expression patterns of APO1, APO2 or TAW1 in IM or pBM of rice at the sBM and SM initiation stage (Ikeda et al., 2007; Ikeda‐Kawakatsu et al., 2012; Yoshida et al., 2013). However, this expression pattern was not detected for its orthologs, FZP and BD1, in rice and maize, respectively, indicative of the functional divergence of WFZP among these species. At the glume primordium differentiation stage when FM is generated, WFZP was expressed in the inner region of SM where the FM will be initiated, similar to the expression patterns of FZP/BD1 in rice or maize (Figure 2r; Chuck et al., 2002; Huang et al., 2018; Komatsu et al., 2003). To examine the WFZP expression pattern during evolution, we performed in situ hybridization using the ancestor species of bread wheat, including Triticum urartu (AA), Triticum dicoccoides (AABB) and Aegilops tauschii (DD). The distinct expression patterns at the apical region of IM and the inner region of SM could be detected in all of these species, indicating that the functional divergence of WFZP occurred before the Triticum formation, rather than during the polyploidization process (Figure 2g–r).
WFZP encodes a transcriptional activator
Previous studies showed that WFZP encodes a transcription factor belonging to the AP2/ERF family, and its homolog in rice is a transcriptional activator (Dobrovolskaya et al., 2015; Komatsu et al., 2003). We then used the dual‐luciferase reporter array system to examine the transcription activity of WFZP. Results showed that, just as FZP in rice, WFZP also functions as a transcriptional activator in wheat (Figure 2s).
WFZP controls plant height, spikelet number and grain weight in wheat
To investigate the roles of WFZP in wheat development, we introduced homeologous wfzp‐a and/or wfzp‐d into Kenong199 (KN199) by backcrossing KN199 with YM44 for 6 generations to create near‐isogenic lines (NIL). Meanwhile, the construct of UBI::WFZP‐3FLAG was created and transformed into KN199 (Figure S3) and Bdfzp mutant, in which a single amino acid mutation in conserved AP2/ERF domain of BdFZP also resulted in SS phenotype (Figure S4a,c). Consistent with the high homology of protein sequence of WFZP and BdFZP, the transgene could fully rescue the SSs phenotype of Bdfzp, demonstrating that WFZP is functional and it has conserved function between wheat and Brachypodium in the regulation of spike development (Figure S4a–c). Compared to KN199, KN199 wfzp‐a/d showed slightly decreased plant height, while the WFZP OE lines had dramatically reduced plant height (Figure 3a and Figure S5a). The possible reason may be the ectopic expression of WFZP under strong UBIQUITIN promoter.
Figure 3.
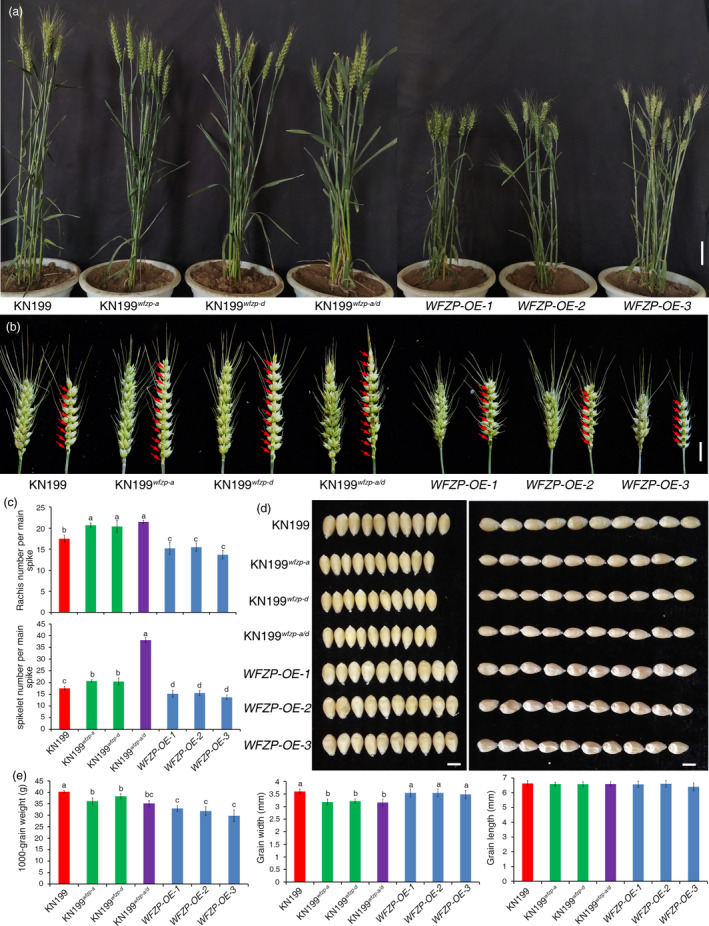
Phenotypic characterization of KN199, KN199 wfzp‐a , KN199 wfzp‐d , KN199 wfzp‐a/d and WFZP OE lines. (a) Representative plants of KN199, wfzp mutant and WFZP OE lines. (b) Spikes of KN199, wfzp mutant and WFZP OE lines. Red arrows show the rachis node. (c) Statistics comparison of rachis node number per main spike and spikelet number per main spike between KN199, wfzp mutant and WFZP OE lines. The error bars denote ± SE. Different letters mean significant difference at P < 0.01. (d) The comparison of grain width and grain length between KN199, wfzp mutant and WFZP OE lines. (e) Statistics comparison of 1000‐grain weight, grain width and grain length between KN199, wfzp mutant and WFZP OE lines. The error bars denote ± SE. Different letters mean significant difference at P < 0.01. Bars = 5cm in a, 2 cm in b and 0.5 cm in d.
Given the high expression level and the distinct expression patterns of WFZP in spike, we focused on the functions of WFZP in spike development. Both KN199 wfzp‐a and KN199 wfzp‐d as well as KN199 wfzp‐a/d plants produce more rachis nodes resulting in longer spike length than that of KN199 (Figure 3b,c and S5b), indicating that WFZP‐A and WFZP‐D repress SM initiation in a dose‐depend manner in line with its distinct expression pattern at the apical of IM (Figure 2n). Correspondingly, WFZP OE plants generate short spike length with less rachis nodes (Figure 3b and S5b). These results demonstrated that WFZP could influence IM activity, which differed from FZP/BD1, but a bit similar to APO1, APO2 and TAW1 (Ikeda et al., 2007; Ikeda‐Kawakatsu et al., 2012; Yoshida et al., 2013). While KN199 wfzp‐a and KN199 wfzp‐d have normally unbranched spike architecture with elevated SNS due to the increased rachis nodes, all KN199 wfzp‐a/d plants had SSs phenotype and dramatically increased SNS (Figure 3b,c), indicating that WFZP‐A and WFZP‐D additively control the identity of FM. Meanwhile, these findings also confirmed that the mutations at WFZP‐A and D loci are responsible for the SSs phenotype of YM44.
Previous studies showed that the SNS is negatively related to 100‐grain weight (TGW) (Ma et al., 2019). While overexpressing WFZP had no obvious effect on grain size, TGW of WFZP OE plants is significantly decreased due to deficient grain filling (Figure 3d,e). In the WFZP single or double mutant, TGW was also reduced due to the reduced grain width rather than grain length (Figure 3d,e), which differs from the role of FZP in rice to influence grain length (Bai et al., 2017). Cytological examination demonstrated that the cell number of the outer integument was decreased in these mutants (Figure S6a–c). Previous studies revealed that several genes such as GS3, GS5, GW2, GW5 and GW8 control grain size by regulating cell proliferation (Ren et al., 2018). We further examined the expressions of these genes in WFZP mutants and OE plants. RT‐qPCR results showed that the expressions of TaGW5 and TaGW8 (the negative and positive regulator for seed size, respectively) (Liu et al., 2017; Wang et al., 2012), were activated or repressed in the mutants, but nearly no change in the OE lines (Figure S6d,e). These results implied that mutation of WFZP may reduce the cell number of outer integument, leading to reduced grain width and grain weight by activating TaGW5 or repressing TaGW8 directly or indirectly.
WFZP regulates multiple biological processes during spike development
To investigate the molecular mechanism of WFZP in regulation of spike development, we collected the young spikes at glume differentiation stage of KN199, KN199 wfzp‐a/d and WFZP OE plants to perform RNA‐seq analysis with three biological replicates. The principle component analysis (PCA) results showed that the sequencing data were highly reproducible (Fig. S7A). Totally, compared with KN199, 7159 and 5917 differentially expressed genes (DEGs) were identified up‐ and down‐regulated in KN199 wfzp‐a/d , respectively, while there were 5452 and 5407 DEGs in WFZP OE lines (Figure S7b,c and Dataset S1). Next, we checked the expression levels of WFZP‐A, B and D in RNA‐seq and validated them by RT‐qPCR, and the higher expression level of WFZP‐A compared with WFZP‐B and D could also be found in the RNA‐seq, similar to that in KN9204 (Figure 2b, and Figure S8a–d).
We then analysed the DEGs between KN199 wfzp‐a/d and KN199. Gene ontology (GO) analysis results showed that many development‐related terms, hormone‐related terms and transcription‐related terms were enriched in the down‐regulated genes in KN199 wfzp‐a/d (Figure 4a). On the other hand, development‐related process was rarely enriched in the up‐regulated genes (Figure 4b). The 791 (cluster1) and 1395 (cluster2) genes with opposite expression trend in KN199 wfzp‐a/d and WFZP OE were also analysed (Figure S7b,c, Figure S9a,c). Similarly, many flower‐related terms were enriched in the cluster1, while no flower development‐related terms were enriched in cluster2 (Figure S9b,d). These results implied that the regulation of spike development by WFZP might depend on its activation activity, consistent with its transcriptional activity.
Figure 4.
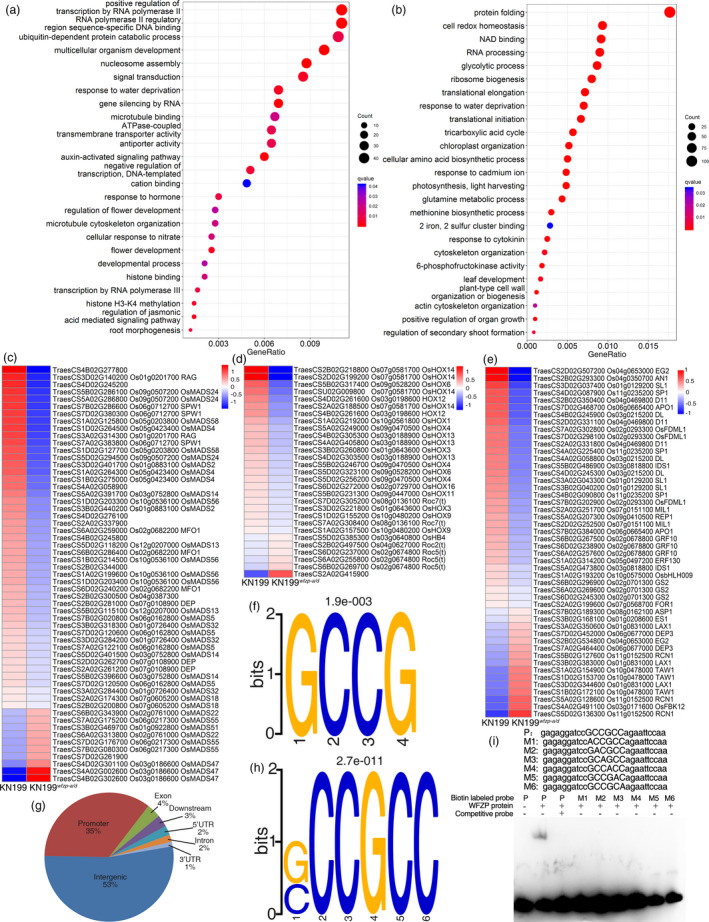
RNA‐seq and ChIP‐seq analysis of genes regulated by WFZP. (a) GO enrichment analysis of down‐regulated genes in KN199 wfzp‐a/d compared with KN199. (b) GO enrichment analysis of up‐regulated genes in KN199 wfzp‐a/d compared with KN199. (c–e) The MADS family TFs (c), HD‐ZIP family TFs (d) and other inflorescence development‐related genes (e) in the DEGs between KN199 wfzp‐a/d and KN199. (f) The binding motif identified by SAAB. (g) Classification of WFZP binding sites identified by ChIP‐seq in the wheat genome. (h) The binding motif identified by ChIP‐seq. (I) EMSA confirmation of WFZP binding to GCC‐box.
Since several transcription‐related processes were enriched in the down‐regulated genes in KN199 wfzp‐a/d , we checked all the transcription factors (TFs) in the DEGs. There were 421 and 291 TFs in the down‐ and up‐regulated DEGs, respectively (Dataset S2). Many MADS and HD‐ZIP family TFs were enriched in the down‐regulated DEGs, but only few were found in the up‐regulated DEGs (Figure S10a,b). Homology comparison showed that most of the homologous genes of down‐regulated MADS and HD‐ZIP TFs in rice could modulate panicle or flower development (Figure 4c,d and Dataset S3) (Agalou et al., 2008; Bhattacharjee et al., 2017; Dai et al., 2008; Shao et al., 2018). Besides, the homologous genes of other panicle development‐related genes, such as TAW1, RICE CENTRORADIALIS1 (RCN1), LAX PANICLE1 (LAX1), APO1, INDETERMINATE SPIKELET1 (IDS1) and ABERRANT SPIKELET AND PANICLE1 (ASP1) were also found in the DEGs (Figure 4e and Dataset S4). Many of these genes were grouped in cluster1 and cluster2, implying that they might be genetically regulated by WFZP and participated the regulation of spike development in wheat (Figure S11 and Dataset S5).
To identify the binding motif of WFZP, we purified GST‐WFZP protein and performed the selection and amplification binding (SAAB) assay in vitro. Results showed that GCCG was the binding element of WFZP (Figure 4f). To further identify the putative WFZP binding sites in planta, we carried out a chromatin immunoprecipitation sequencing (ChIP‐seq) analysis with anti‐FLAG antibody using WFZP OE plant. Totally, 168 peaks were detected in both biological replicates, with nearly half of them (79) located in the genic region (promoter, 5’UTR, exon, intron, 3’UTR and downstream) of 75 genes (Figure 4g and Dataset S6). Binding motif analysis showed that SCCGCC, which included GCC‐box bound by AP2/ERF family TFs (Chakravarthy et al., 2003), was the binding motif of WFZP in vivo (Figure 4H). Next, the electrophoretic mobility shift assay (EMSA) was performed using the GCCGCC sequence as probe to validate the binding of WFZP on GCC‐box. An obvious binding shift was observed when the GCCGCC sequence as probe was used, but no binding was found after adding competitive probe or using probes with any one nucleotide mutated to A in the GCCGCC sequence, indicating that the SCCGCC element is critical for the WFZP binding (Figure 4I).
VRN1‐A and TaHOX4‐A are the targets of WFZP in regulating spike development
To investigate the genetic network of WFZP in regulating spike development, we integrated RNA‐seq and ChIP‐seq data to identify the targets of WFZP. Four putative targets of WFZP were identified since their promoter or 5’UTR were bound by WFZP (Dataset S6), including two genes belonging to MADS and HD‐ZIP family TFs. They were TraesCS5A02G391700, the well‐known vernalization gene VRN1‐A, which modulates the vernalization process, IM activity and SM identity maintenance in wheat (Li et al., 2019a; Yan et al., 2003), and TraesCS5A02G249000, homolog of rice OsHOX4 gene that modulates panicle size (Agalou et al., 2008; Dai et al., 2008). There were four GCC‐box in the promoter or 5’UTR region of VRN1‐A, and one GCC‐box in the first exon of TaHOX4‐A, which were overlapped with the WFZP binding peaks (Figure 5a). Then, the binding of WFZP on the specific regions of VRN1‐A and TaHOX4‐A genes were validated by ChIP‐qPCR, and the activation of the two genes by WFZP was confirmed by RT‐qPCR, which were consistent with the RNA‐seq and ChIP‐seq analysis results (Figure 5b–d). Therefore, WFZP directly bound to VRN1‐A and TaHOX4‐A to regulate their expression.
Figure 5.
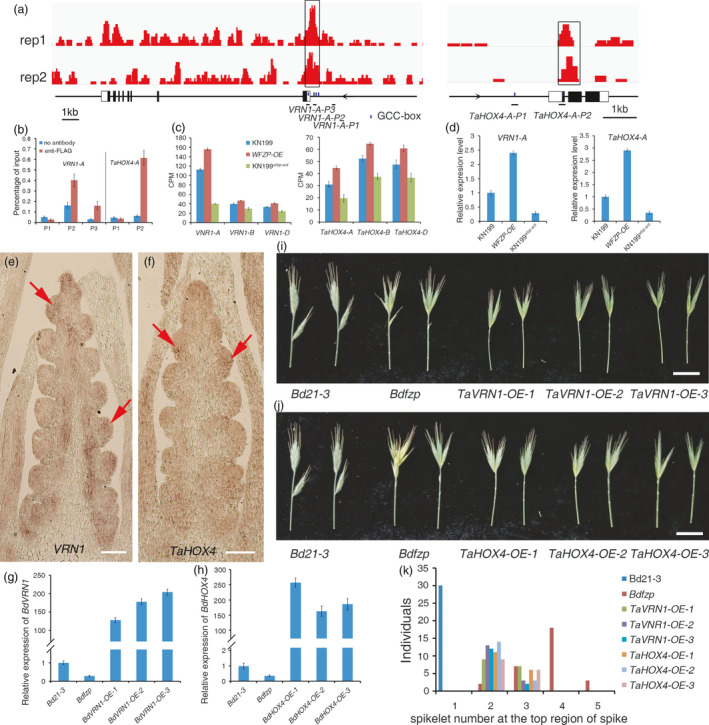
VRN1‐A and TaHOX4‐A mediated the function of WFZP in spike development. (a) the WFZP‐3FLAG ChIP‐seq peaks (two biological replicates) at VRN1‐A and TaHOX4‐A revealed in IGV; peaks, gene structures and regions for validation by ChIP‐qPCR were displayed from top to bottom rows, respectively. (b) ChIP‐qPCR validation of WFZP binding to VRN1‐A and TaHOX4. The error bars denote ± SE. (c, d) The expression level of VRN1, TaHOX4 in KN199, KN199 wfzp‐a/d and WFZP OE lines examined by RNA‐seq (c) and RT‐qPCR (d). The error bars denote ± SE. (e, f) In situ hybridization to examine the expression pattern of VRN1 (e) and TaHOX4 (f) in wheat. Red arrows show the strong signals in the SM where the FM will be initiated. (g) The expression level of BdVRN1 in Bd21‐3, Bdfzp and BdVRN1 OE lines under Bdfzp background detected by RT‐qPCR. The error bars denote ± SE. (h) The expression level of BdHOX4 in Bd21‐3, Bdfzp, BdHOX4 OE lines under Bdfzp background examined by RT‐qPCR. The error bars denote ± SE. (i) The spike of Bd21‐3, Bdfzp, TaVRN1 OE lines under Bdfzp background. (j) The spike of Bd21‐3, Bdfzp, TaHOX4 OE lines under Bdfzp background. (k) Distribution of number of spikelet in the terminal region of spike of Bd21‐3, Bdfzp, TaVRN1 OE and TaHOX4 OE lines under Bdfzp background. Bars = 200μm in e and f, 1.5cm in i and j.
We then performed the in situ hybridization to examine the expression patterns of both genes in young spikes. Results showed that, although the two genes had border expression regions in wheat spikes, strong and specific signals could be detected at the sites where the FMs are initiated, which overlapped with the expression regions of WFZP (Figures 2o–r and 5e,f). These results indicated that VRN1‐A and TaHOX4‐A may mediate the functions of WFZP in regulating spike development.
Given WFZP and BdFZP have conserved function and WFZP can rescue the SS phenotype of Bdfzp, we used the Bdfzp mutant to validate the genetic relationship of WFZP, VRN1‐A and TaHOX4. The high homology of protein sequence and reduced expression of BdVRN1 and BdHOX4 in Bdfzp mutant implied that both genes may be also activated by BdFZP, and they may have conserved function in wheat and Brachypodium (Figure 5g,h, Figure S12a,b). Next, we transformed UBI::TaVRN1‐A and UBI::TaHOX4‐A, respectively, into Bdfzp to overexpress each of the two genes, which was validated by RT‐qPCR (Figure 5g,h). Although the SS phenotype of Bdfzp was not fully rescued by each transgene, the spikelet number at the top region of spike was significantly reduced in the overexpression lines, indicating that reduced expression of BdVRN1 and BdHOX4 due to the Bdfzp mutation is responsible for the SS phenotype of Bdfzp (Figure 5i–k). These results demonstrated that TaVRN1‐A and TaHOX4‐A mediate the functions of WFZP in regulating the spike development in wheat.
Screening for favourable alleles of WFZP
To detect sequence variation and screen for favourable alleles of WFZP, we firstly sequenced the promoter and coding regions of WFZP‐A, B and D in 30 wheat accessions (Li et al., 2019b). Totally, two, four and seven haplotypes were found for WFZP‐A, B and D, respectively (Figure 6a, Table S1 and S2).
Figure 6.
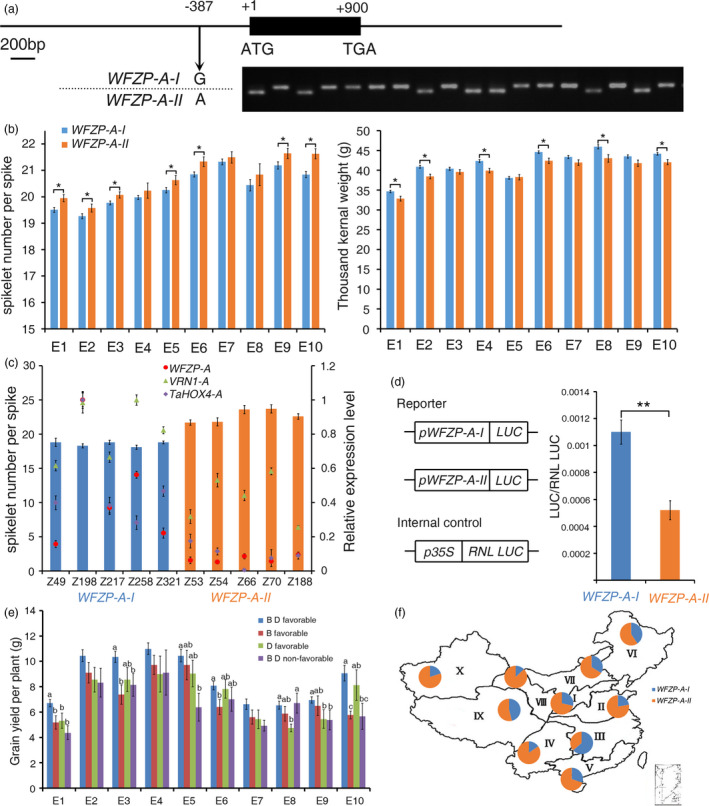
A SNP in the promoter region of WFZP‐A influence the expression level of WFZP‐A, spikelet number per spike and 1000‐grain weight. (a) dCAPs marker developed for genotyping the SNP in the −387 loci upstream of WFZP‐A start condon. (b) spikelet number per spike and 1000‐grain weight of cultivars with different haplotypes of WFZP‐A in ten growing environments. The error bars denote ± SE. *P < 0.05. (c) spikelet number per spike of cultivars with different haplotypes of WFZP‐A and their expression level of WFZP‐A, VRN1‐A and TaHOX4. Cultivar names could be seen in supplemental Dataset S7. The error bars denote ± SE. (d) Transient assay of WFZP‐A promoter activity. Schematic diagram in the left part showing the vectors used in this array. The error bars denote ± SE. **P < 0.01. (e) The grain yield per plant of cultivars with WFZP‐A‐II and different haplotype combinations of WFZP‐B and D. The error bars denote ± SE. Different letters mean significant difference at P < 0.01. (f) The distribution of WFZP‐A haplotypes in MCC in ten Chinese wheat ecological production zones.
Next, an association analysis between the haplotypes of WFZP‐A, B and D and several agronomic traits including SNS and TGW in a natural population with 323 accessions (Dataset S7) was performed (Li et al., 2019b). In ten environments, the associations of WFZP‐A with SNS and TGW were detected in eight and six environments, respectively; the associations of WFZP‐B and WFZP‐D with TGW were detected in nine and six environments, respectively; no significant association of WFZP‐B or WFZP‐D with SNS was detected (Table S3). These results showed that the natural sequence variations of WFZP‐A, B and D contribute to TGW, while only the natural sequence variation of WFZP‐A contributes to SNS in the testing population.
We next screened the favourable alleles of WFZP‐A, B and D for TGW and SNS. For WFZP‐B, WFZP‐B‐I and WFZP‐B‐III were favourable alleles for TGW, since cultivars with any of the two haplotypes have a higher TGW (Fig. S13A). For WFZP‐D, the favourable alleles for TGW were WFZP‐D‐III, WFZP‐D‐V and WFZP‐D‐VI (Figure S13b). In addition, WFZP‐A‐II was favourable alleles for SNS, while WFZP‐A‐I was favourable alleles for TGW, in agree with the negative correlation between TGW and SNS (Figure 6b). The variation between the two haplotypes resulted from one SNP at the promoter region (Figure 6a), which may influence the transcription level of WFZP‐A. As thus, we chose 10 cultivars with different haplotypes of WFZP‐A to detect its expression level. Results showed that cultivars harbouring WFZP‐A‐II haplotype had lower expression level of WFZP‐A, but higher SNS (Figure 6c), which was consistent with previous result that mutant at WFZP‐A loci could increase SNS. Correspondingly, the expression levels of VRN1‐A and TaHOX4‐A were lower in the WFZP‐A‐II cultivars than in the WFZP‐A‐I cultivars (Figure 6c), supporting our previous results that the two genes are target genes of WFZP. A dual‐luciferase reporter array system was used to further detect the contribution of this SNP in gene expression regulation. A higher ratio of LUC/RNL LUC was detected when the promoter of WFZP‐A‐I haplotype was used to drive LUC (Figure 6d), validating that the G/A SNP is important for the WFZP‐A expression regulation. Next, we analysed whether combination of WFZP‐A‐II (favourable haplotype for SNS) and WFZP‐B and D favourable haplotypes for TGW can elevate grain yield per plant. Results showed that cultivars with both WFZP‐A‐II and WFZP‐B and/or WFZP‐D favourable haplotypes usually had higher grain yield per plant compared with cultivars with WFZP‐A‐II haplotype and both WFZP‐B and D non‐favourable haplotypes for TGW (Figure 6e), demonstrating that the WFZP‐A favourable haplotype for SNS and WFZP‐B and D favourable haplotype for TGW could be used together to improve the grain yield of wheat.
The Chinese wheat production area is divided into ten major agro‐ecological production zones based on ecological conditions, cultivar type and growing season (Zhang et al., 2015). To determine which haplotype of WFZP‐A was selected during breeding in China, the geographic distributions of WFZP‐A haplotypes were evaluated using Chinese wheat mini‐core collection (MCC) from all ecological zones (Dataset S8). WFZP‐A‐II was predominantly occurred in most of Chinese wheat production zones (I, II, IV, V, VII, VIII, X), indicating that SNS is the favourable trait to ensure a convenient yield compared to TGW in these zones. In the Zone VI and Zone IX, which are Northeastern Spring Wheat Zone and Qinghai‐Tibetan Plateau Spring‐Winter Wheat Zone, respectively, both WFZP‐A haplotypes had similar frequency, while the WFZP‐A‐I mainly occurred in Middle and Lower Yangtze Valleys Autumn‐Sown Spring Wheat Zone (Zone III), indicating that TGW is important than SNS for the crop yield in this zone (Figure 6f).
Discussion
WFZP has conserved and diverged function in wheat
Previous researches revealed that FZP and its orthologs in maize, wheat and Brachypodium have conserved function in regulating the transition from SM to FM (Chuck et al., 2002; Derbyshire and Byrne, 2013; Dobrovolskaya et al., 2015; Komatsu et al., 2003). In our study, the SSs phenotype, distinct expression pattern of WFZP in the inner part of SM and complementary assay of Bdfzp mutant further validated this conserved function of WFZP as its homologs, It was reported that SM identity maintenance and transition from SM to FM depend on the expression dose of FZP in rice (Bai et al., 2017; Komatsu et al., 2003). Meanwhile, in previous study, WFZP‐D alone could regulate the SS phenotype, due to the higher expression level compared with its homologs in chromosome 2A and 2B (Dobrovolskaya et al., 2015). However, we found that the WFZP‐A and B had a higher and relative equal expression level compared with WFZP‐D in KN9204 and KN199, indicating that different cultivar shares diverse WFZP expression level. As a result, the WFZP‐A and D together regulated the SS phenotype (Figure 3a). Therefore, we speculated that except for the SS phenotype, there would be more abnormally developed florets in wfzp‐a/b/d triple mutant compared with wild‐type plants which is waiting for the further investigation.
Surprisingly, unlike the rice and maize inflorescence, wheat inflorescence expresses WFZP at the site where the SM is initiated at late of double‐ridge stage before IM termination (Figure 2k–n), a similar pattern to that of APO1, APO2, and TAW1 in IM or pBM in rice when sBM or SM are initiated, implying that WFZP may have the function of regulation AxMs formation in IM (Ikeda et al., 2007; Ikeda‐Kawakatsu et al., 2012; Yoshida et al., 2013). Consistently, impaired WFZP expression resulted in increased rachis node and overexpression of WFZP led to decreased spikelet number (Figure 3b). Since WFZP is not expressed in the whole IM like the earlier expression pattern of APO1, APO2 or TAW1 in rice, nor in the whole apical region of IM like that of VRN1 in wheat, we speculated that WFZP may not regulate the IM activity directly, but represses SM initiation or SM activity at distinct developmental stage in IM, and then influences IM activity indirectly in wheat. A possible work mode of WFZP in regulating wheat inflorescence architecture is that WFZP is induced expression at late of double‐ridge stage and early glume primordium differentiation stage to repress the SM initiation in IM and SM and subsequently promote FM initiation (Figure 7a). Therefore, which genes and how the genes regulate WFZP expression in wheat need to be further investigated and will contribute to wheat breeding.
Figure 7.
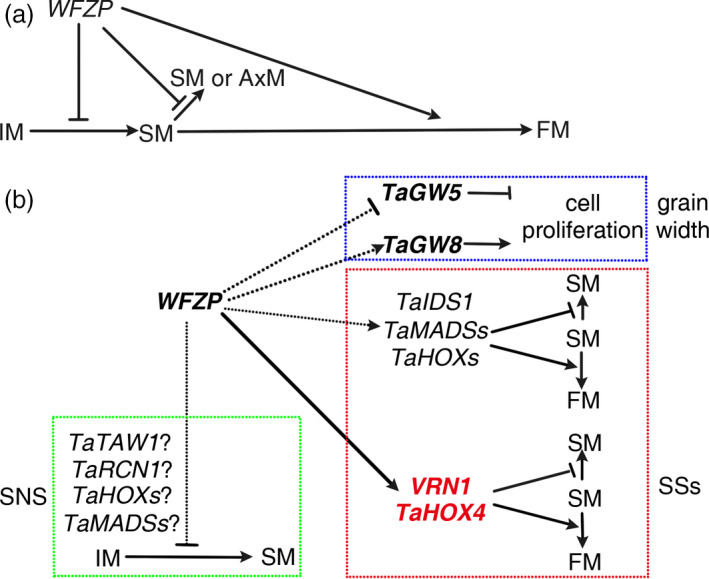
The working model of WFZP in wheat. (a) the model representing the function of WFZP in wheat. (b) the genetic network of WFZP in wheat.
The molecular mechanism of WFZP in the regulation of spike development and grain size in wheat
Up to now, the molecular mechanism and genetic network of WFZP and its ortholog were rarely studied. Here, we combined the RNA‐seq and ChIP‐seq data analysis to seek for the direct target of WFZP, and two genes, VRN1‐A and TaHOX4‐A were identified. Just as WFZP, VRN1 could also maintain the SM identity and promote the FM transition in wheat (Li et al., 2019a). TaHOX4‐A is homologous gene of rice HOX4 with higher expressed in FM and regulates spike length and panicle size (Dai et al., 2008). Both genes were expressed overlapping with WFZP, and were bound and activated by WFZP (Fig. 5A‐5F). Overexpression of both genes in Brachpodium Bdfzp mutant could reduce the spikelet number at the topic region (Figure 5i–k), implying that the IM activity to produce SM in Bdfzp were repressed by the transgene. Given the high protein conservation of WFZP and BdFZP and the similar SS phenotype of wfzp and Bdfzp, we reasoned that VRN1‐A and TaHOX4‐A function as the targets of WFZP in the modulation of SM development, mainly SM identity maintenance. Since the phenotype of Bdfzp could not be fully restored, the SM identity maintenance by WFZP may need participation of other genes, such as other MADS or HD‐ZIP family TFs or genes homologous to rice IDS1.
Besides the spike development, WFZP plays important role in control wheat grain size. Unlike FZP that regulates rice grain length, WFZP controls grain weight through regulating grain width (Figure 3d,e). While GW5 was de‐repressed expression, GW8 was increased expression in the wfzp‐a/d mutant (Fig. S6d,e). Given that WFZP is a transcriptional activator, how it functions in control grain size is waiting further investigation (Figure 7b).
Based on our RNA‐seq and ChIP‐seq result and genetic analysis, combined with homology comparison, a working model of molecular mechanism of WFZP is conceived as following. WFZP directly promotes the expression of VRN1‐A and TaHOX4‐A to inhibit secondary SM (or AxM) generation and promote SM‐to‐FM transition. Besides, WFZP may repress primary SM initiation from IM, and modulate SNS through regulating the expression TaTAW1, TaRCN1, TaHOXs and TaMADSs genes (Figure 7b).
WFZP was a valuable locus for wheat breeding
To date, improving the grain yield of wheat is still a challenging work in wheat breeding, and screening for valuable genes and favourable alleles is the important way to achieve this goal. Here we illustrated that mutations in WFZP‐A and D result in SSs phenotype and prolonged IM activity, both of which are contributive to modify SNS trait in wheat (Figure 3b,c). Besides the rare mutation, favourable alleles of WFZP such as the WFZP‐A‐II for a given agronomic trait could also be found in the natural resources (Figure 6a–f), and the favourable alleles of homeologous WFZP could be used together to improve the grain yield potential of wheat (Figure 6a–f).
Experimental procedures
Plant materials and growth conditions
The wheat materials used to test agronomic traits were planted in the field of Shijiazhuang in 2018‐2019. Each of the materials was planted in 3 blocks, and all the blocks have identical environments and growth conditions. The wheat materials for in situ hybridization and RNA‐seq analysis and all the Brachypodium materials were grown in greenhouse under long‐day conditions (16 h light/8 h dark) at 22°C after fully vernalization.
The natural population was planted at Shunyi and Changping in 2015 and 2016, respectively. The growing environments included drought stress (DS), well‐watered (WW) and heat stress (HS) as described in previous research (Zhang et al., 2015). The E1 to E10 indicated the individual environment at Shunyi in 2015 under DS + HS, DS, WW + HS and WW, Shunyi in 2016 under DS + HS, DS, WW + HS and WW, at Changping in 2016 under WW and DS.
Statistics analysis
For the agronomic traits comparison, 10 individuals of every material in each block were investigated. For quantitative RT‐PCR and dual‐luciferase reporter assay, three biological replicates with three technical repetition were examined. All these data were compared using student’s t test (for comparing two groups of data) or one‐way ANOVA analysis (for comparing more than two groups of data), and the mean ± SE was presented in our results. For the natural population, the agronomic traits were investigated as previously described (Li et al., 2019b; Zhang et al., 2015). The population structure has been analysed in the previous study (Li et al., 2019b). After genotyping WFZP‐A, B and D using primers listed in Table S4, a general linear model (GLM) which accounted for population structure (Q) in TASSEL V2.1 was used to perform association analysis.
Constructs
For the overexpression constructs, the CDS of WFZP‐D, VRN1‐A and TaHOX4 were ligated into modified pTCK303 vector using In‐Fusion method. For the transactivation analysis, the CDS of WFZP‐D were ligated into pMN6 vector using In‐Fusion method. For the transient assay, different promoter version of WFZP‐A were ligated into ligated into ENTR1A‐T vector using TA cloning method, then the CDS of firefly luciferase (LUC) were cloned in the downstream of WFZP‐A promoter at KpnI and NotI loci, and finally pWFZP‐A::LUC were cloned into pEarleyGate301 (PEG301) vector using gateway LR recombination method.
Generation of gene overexpressing wheat or Brachypodoium distachyon lines
The resulting construct of WFZP‐D‐OE was transformed into immature embryos of KN199 by particle bombardment. The constructs of VRN1‐A‐OE and TaHOX4‐A‐OE were transformed into the Brachypodium through Agrobacterium‐mediated transformation method as previously described (Alves et al., 2009).
Cytological observation
Young spikes of KN9204 and YM44 were dissected under the stereomicroscope and captured using TM3030 (Hitachi) according to the manufacturer’s instruction. The cell length and cell number of the outer integument were measured as previous described (Ma et al., 2015).
McrBC enzyme array
The McrBC enzyme array was performed as described (Wu et al., 2010). 500ng total genomic DNA of KN9204, YM44 and F2 descents was digested with McrBC (NEB). Equal amount of McrBC digested or undigested DNA was used for PCR amplification.
In situ hybridization assay
In situ hybridization was carried out as previously described (Liu et al., 2011). The CDS region of WFZP‐D, VRN1‐A, TaHOX4‐A were amplified using primers added SP6 or T7 promoter sequence for sense and antisense probes, respectively (listed in Table S4). After purifying the PCR products amplified by these primers, sense or antisense probes was synthesized by in vitro transcription using SP6 or T7 RNA polymerase.
RNA‐seq
The RNA‐seq of young spikes of YM44 was performed in parallel with our previous study (Li et al., 2018b). For the RNA‐seq of young spikes of KN199, WFZP OE line and KN199 wfzp‐a/d , plants were planted in parallel in greenhouse, and young spikes at glume differentiation stage were collected. Each sample contained 3 biological replicates. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). Libraries were generated using the standard protocol, and paired‐end sequencing libraries were sequenced on an Illumina NovaSeq 6000 sequencer.
ChIP‐seq
ChIP was performed as described previously (Liu et al., 2011) with some modifications. Young spikes of WFZP OE lines were harvested and ground into fine powder with liquid nitrogen. Plant chromatin was extracted as described previously (Guo et al., 2018). After the supernatant from sonicated chromatin was diluted with ChIP dilution buffer, precleaning was performed by incubating diluted chromatin with protein‐Aagarose beads/salmon sperm DNA. After 1h incubation, supernatant was incubated with anti‐FLAG M2 Affinity Gel overnight with rotation. After several washes, bound chromatin was eluted and then was reverse cross‐linked overnight. DNA was recovered by phenol‐chloroform extraction and precipitated by ethanol. DNA was finally dissolved in 10mM Tris buffer (pH = 8.0) for ChIP‐seq.
Analysis of RNA‐seq and ChIP‐seq data
Clean reads of RNA‐seq were aligned to wheat reference genome (IWGSC RefSeq v1.0) using Tophat2 (version 2.1.1). The htseq‐count script in HTSeq was used to count the number of reads uniquely mapped to each annotated gene. Then, the CPM was calculated and DEGs were identified using edgeR. Genes with a FDR < 0.05 were considered as DEGs. The transcription factor family analysis was performed as described previously (Li et al., 2018b). The homologous genes in rice of DEGs were gained from Ensembl Biomart.
Clean reads of ChIP‐seq were aligned to wheat reference genome (IWGSC RefSeq v1.0) using bowtie2 (version 2.3.4.1). The uniquely aligned reads were used to find peaks using MACS14 with default parameters except that duplicates were allowed just as described previously (Li et al., 2018a). The overlapped peaks were reserved for further analysis. The 1000 bp sequence (500 bp upstream and 500 bp downstream) around the peak summit were used for motif identification by MEME (Bailey et al., 2006) as described previously (Li et al., 2016). The overlapped peaks were merged by samtools for annotation. For the peak annotation, 3 kb region upstream the transcription start site was defined as promoter region, and 3 kb region downstream the transcription terminal site was defined as downstream region. The IGV was used to visualize ChIP‐seq read depths.
SAAB assay and EMSA
The SAAB assay was performed as described previously (Smith et al., 2002), and the internal 20bp oligonucleotide gained by SAAB array was used to analyse the binding motif by MEME. The EMSA assay was performed as described previously (Zhang et al., 2018). Biotin‐labelled probes mixed without or with competitive probes were incubated with purified GST‐WFZP‐D proteins in 1 × binding buffer at room temperature for 20 min. Biotin‐labelled probes were detected using a Light Shift Chemiluminescent EMSAkit (Thermo Scientific, Waltham, MA, USA).
Transactivation analysis
The transcriptional activity analysis was performed as described previously (Xie et al., 2014). WFZP‐pMN6 or pMN6 vector together with pGLL and pRNL vector were transformed into wheat protoplasts. VP16 served as a positive control. LUC and RNL LUC activities were measured using TransDetect Double‐Luciferase Reporter Assay Kit (TransGen Biotech: Beijing, China) on a Packard TopCount luminometer.
Transient assay of WFZP‐A promoter
The transient assay of WFZP‐A promoter was detected using dual‐Luciferase system as described previously (Zhao et al., 2018). Two kinds of pWFZP‐A::LUC constructs vector were transformed into tobaccos together with 35S::RNL LUC. LUC and RNL LUC activities were measured using TransDetect Double‐Luciferase Reporter Assay Kit on a Multiscan Spectrum.
Accession numbers
The RNA‐seq data of KN9204 have been submitted in the Gene Expression Omnibus (GEO) database under accession number GSE83287 in our previous paper (Li et al., 2018b). The RNA‐seq data of YM44, RNA‐seq data of KN199, WFZP OE and KN199 wfzp‐a/d and ChIP‐seq data have been deposited in the Sequence Read Archive (SRA) database under accession numbers PRJNA640732, PRJNA635231 and PRJNA635237, respectively.
Conflict of interest
The authors declare no competing interests.
Author contributions
X.L., A.Z. and X.F. conceived the project. X.L. and R.J. designed the experiments. Y.L., M.Z., L.L., L.G., X.G., D.Z. and A.B. performed the experiments. Y.L. analysed the RNA‐seq and ChIP‐seq data. X.L., B.D., H.X., S.C. and J.L. analysed the experimental data. X.L. and Y.L. wrote the article with contributions of all the authors.
Supporting information
Figure S1 Linkage analysis of WFZP‐A, WFZP‐D and YM44 SSs phenotype.
Figure S2 McrBC analysis of the promoter of WFZP‐D in the P3 region in the individuals of F2 crossed by KN9204 and YM44.
Figure S3 Western blotting of WFZP OE lines expressing the protein of WFZP fused with 3 FLAGs.
Figure S4 Functional verification of WFZP.
Figure S5 Statistics comparison of plant height (a) and spike length (b) between KN199, KN199 wfzp‐a , KN199 wfzp‐d , KN199 wfzp‐a/d and WFZP OE lines.
Figure S6 Effects of WFZP on seed coat cell proliferation.
Figure S7 The overall view of RNA‐seq data.
Figure S8 The expression level of WFZP‐A, B and D in KN199, KN199 wfzp‐a/d and WFZP OE lines detected by RNA‐seq (a) and qPCR (b–d).
Figure S9 GO enrichment analysis of genes in the overlapped DEGs of KN199 wfzp‐a/d up‐regulated/down‐regulated genes and WFZP‐OE down‐regulated/up‐regulated genes.
Figure S10 Transcription factors in the KN199 wfzp‐a/d _vs_KN199 down‐regulated genes (a) and KN199 wfzp‐a/d _vs_KN199 up‐regulated genes (b).
Figure S11 Heatmap of MADS family TFs, HD‐ZIP family TFs and other development‐related genes in the overlapped DEGs of KN199 wfzp‐a/d up‐regulated/down‐regulated genes and WFZP‐OE down‐regulated/up‐regulated genes.
Figure S12 The protein alignment of TaVRN1‐A and BdVRN1 (a) and TaHOX4‐A and BdHOX4 (b).
Figure S13 1000‐grain weight of cultivars with different haplotypes of WFZP‐B (a) and WFZP‐D (b) in ten growing environments. The error bars denote ± SE.
Table S1 The polymorphism of WFZP‐B.
Table S2 The polymorphism of WFZP‐D.
Table S3 Association analysis of WFZP with SNS and TGW.
Table S4 Primers used in this study.
Dataset S1 DEGs between WFZP‐OE, KN199 wfzp‐a/d and KN199.
Dataset S2 Transcription factors (TFs) in the DEGs between KN199 wfzp‐a/d and KN199.
Dataset S3 The expression level and homolog genes of MADS and HD‐ZIP family TFs in the DEGs between KN199 wfzp‐a/d and KN199.
Dataset S4 The expression level and homolog genes of other development‐related genes in the DEGs between KN199 wfzp‐a/d and KN199.
Dataset S5 The expression level and homolog genes of MADS and HD‐ZIP TFs and other development‐related genes in the DEGs between WFZP‐OE, KN199 wfzp‐a/d and KN199.
Dataset S6 The annotation of overlapped merged peaks.
Dataset S7 The haplotype of WFZP‐A, B and D in the natural population used for association analysis.
Dataset S8 The haplotype of WFZP‐A in MCC.
Acknowledgements
We thank Versailles INRA, France, for providing Bdfzp seeds. This work was supported by National Key Research and Development Program of China (2016YFD0100401 to X.L.), Research Fund for Talent Introduction of Hebei Province (2020HBQZYC004 to X.L.), National Basic Research Program of China (2014CB138100 to X.L.) and National Natural Science Foundation of China (31701423 to Y. L., 31871634 to M.Z. and 31970824 to X. L.).
Li, Y. , Li, L. , Zhao, M. , Guo, L. , Guo, X. , Zhao, D. , Batool, A. , Dong, B. , Xu, H. , Cui, S. , Zhang, A. , Fu, X. , Li, J. , Jing, R. and Liu, X. (2021) Wheat FRIZZY PANICLE activates VERNALIZATION1‐A and HOMEOBOX4‐A to regulate spike development in wheat. Plant Biotechnol. J, 10.1111/pbi.13535
Contributor Information
Ruilian Jing, Email: jingruilian@caas.cn.
Xigang Liu, Email: xgliu@sjziam.ac.cn.
References
- Agalou, A. , Purwantomo, S. , Oevernaes, E. , Johannesson, H. , Zhu, X. , Estiati, A. , de Kam, R.J. et al. (2008) A genome‐wide survey of HD‐Zip genes in rice and analysis of drought‐responsive family members. Plant. Mol. Biol. 66, 87–103. [DOI] [PubMed] [Google Scholar]
- Alves, S.C. , Worland, B. , Thole, V. , Snape, J.W. , Bevan, M.W. and Vain, P. (2009) A protocol for Agrobacterium‐mediated transformation of Brachypodium distachyon community standard line Bd21. Nat. Protoc. 4, 638–649. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Huang, Y. , Hu, Y. , Liu, H. , Zhang, B. , Smaczniak, C. , Hu, G. et al. (2017) Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants, 3, 885–893. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L. , Williams, N. , Misleh, C. and Li, W.W. (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee, A. , Sharma, R. and Jain, M. (2017) Over‐expression of OsHOX24 confers enhanced susceptibility to abiotic stresses in transgenic rice via modulating stress‐responsive gene expression. Front. Plant Sci. 8, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy, S. , Tuori, R.P. , D'Ascenzo, M.D. , Fobert, P.R. , Despres, C. and Martin, G.B. (2003) The tomato transcription factor Pti4 regulates defense‐related gene expression via GCC box and non‐GCC box cis elements. Plant Cell, 15, 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G. , Muszynski, M. , Kellogg, E. , Hake, S. and Schmidt, R.J. (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science, 298, 1238–1241. [DOI] [PubMed] [Google Scholar]
- Cui, F. , Fan, X. , Zhao, C. , Zhang, W. , Chen, M. , Ji, J. and Li, J. (2014) A novel genetic map of wheat: utility for mapping QTL for yield under different nitrogen treatments. BMC Genet. 15, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, M. , Hu, Y. , Ma, Q. , Zhao, Y. and Zhou, D.‐X. (2008) Functional analysis of rice HOMEOBOX4 (Oshox4) gene reveals a negative function in gibberellin responses. Plant Mol. Biol. 66, 289–301. [DOI] [PubMed] [Google Scholar]
- Derbyshire, P. and Byrne, M.E. (2013) MORE SPIKELETS1 is required for spikelet fate in the inflorescence of Brachypodium . Plant Physiol. 161, 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya, O. , Pont, C. , Sibout, R. , Martinek, P. , Badaeva, E. , Murat, F. , Chosson, A. et al. (2015) FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol. 167, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L. , Cao, X. , Liu, Y. , Li, J. , Li, Y. , Li, D. , Zhang, K. et al. (2018) A chromatin loop represses WUSCHEL expression in Arabidopsis . Plant J. 94, 1083–1097. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Zhao, S. , Fu, Y. , Sun, H. , Ma, X. , Tan, L. , Liu, F. et al. (2018) Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J. 96, 716–733. [DOI] [PubMed] [Google Scholar]
- Ikeda, K. , Ito, M. , NagasawaO, N. , Kyozuka, J. and Nagato, Y. (2007) Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F‐box protein, regulates meristem fate. Plant J. 51, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Ikeda‐Kawakatsu, K. , Maekawa, M. , Izawa, T. , Itoh, J.I. and Nagato, Y. (2012) ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. [DOI] [PubMed] [Google Scholar]
- Komatsu, M. , Chujo, A. , Nagato, Y. , Shimamoto, K. and Kyozuka, J. (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. [DOI] [PubMed] [Google Scholar]
- Li, D. , Fu, X. , Guo, L. , Huang, Z. , Li, Y. , Liu, Y. , He, Z. et al. (2016) FAR‐RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA, 113, 9375–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Fu, X. , Zhao, M. , Zhang, W. , Li, B. , An, D. , Li, J. et al. (2018b) A genome‐wide view of transcriptome dynamics during early spike development in bread wheat. Sci. Rep. 8, 15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Lin, H. , Chen, A. , Lau, M. , Jernstedt, J. and Dubcovsky, J. (2019a) Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinacy. Development 146, dev175398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Peng, Z. , Mao, X. , Wang, J. , Chang, X. , Reynolds, M. and Jing, R. (2019b) Genome‐wide association study reveals genomic regions controlling root and shoot traits at late growth stages in wheat. Ann. Bot‐London, 124, 993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Yue, Y. , Chen, H. , Qi, W. and Song, R. (2018a) The ZmbZIP22 transcription factor regulates 27‐kD gamma‐Zein gene transcription during maize endosperm development. Plant Cell, 30, 2402–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Chen, J. , Zheng, X. , Wu, F. , Lin, Q. , Heng, Y. , Tian, P. et al. (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants, 3, 17043. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Kim, Y.J. , Müller, R. , Yumul, R.E. , Liu, C. , Pan, Y. , Cao, X. et al. (2011) AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell, 23, 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , Ding, P. , Liu, J. , Li, T. , Zou, Y. , Habib, A. , Mu, Y. et al. (2019) Identification and validation of a major and stably expressed QTL for spikelet number per spike in bread wheat. Theor. Appl. Genet. 132, 3155–3167. [DOI] [PubMed] [Google Scholar]
- Ma, M. , Wang, Q. , Li, Z.J. , Cheng, H.H. , Li, Z.J. , Liu, X.L. , Song, W.N. et al. (2015) Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. Plant J. 83, 312–325. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Hu, J. , Xu, Q. , Cui, Y. , Zhang, Y. , Zhou, T. , Rao, Y. et al. (2018) FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 69, 4853–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, J. , Haider, I. , Xiong, L. , Zhu, X. , Hussain, R.M.F. , Overnas, E. , Meijer, A.H. et al. (2018) Functional analysis of the HD‐Zip transcription factor genes Oshox12 and Oshox14 in rice. PLoS One 13, e0199248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S. , Boschke, I. and Hake, S. (2002) Selective interaction of plant homeodomain proteins mediates high DNA‐binding affinity. Proc. Natl. Acad. Sci. USA, 99, 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Wu, K. , Yuan, Q. , Liu, X. , Liu, Z. , Lin, X. , Zeng, R. et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. [DOI] [PubMed] [Google Scholar]
- Wu, L. , Zhou, H. , Zhang, Q. , Zhang, J. , Ni, F. , Liu, C. and Qi, Y. (2010) DNA methylation mediated by a MicroRNA pathway. Mol. Cell, 38, 465–475. [DOI] [PubMed] [Google Scholar]
- Xie, Q. , Wang, P. , Liu, X. , Yuan, L. , Wang, L. , Zhang, C. , Li, Y. et al. (2014) LNK1 and LNK2 Are transcriptional coactivators in the arabidopsis circadian oscillator. Plant Cell, 26, 2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L. , Loukoianov, A. , Tranquilli, G. , Helguera, M. , Fahima, T. and Dubcovsky, J. (2003) Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA, 100, 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, A. , Sasao, M. , Yasuno, N. , Takagi, K. , Daimon, Y. , Chen, R. , Yamazaki, R. et al. (2013) TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. 110, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Liu, X. , Xu, W. , Chang, J. , Li, A. , Mao, X. , Zhang, X. and et al. (2015) Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Sci. Rep. 5, 12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Wang, R. , Zi, H. , Li, Y. , Cao, X. , Li, D. , Guo, L. et al. (2018) AUXIN RESPONSE FACTOR3 Regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell, 30, 324–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Li, M. , Xu, C. , Yang, X. , Li, D. , Zhao, X. , Wang, K. et al. (2018) Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J 96, 147–162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Linkage analysis of WFZP‐A, WFZP‐D and YM44 SSs phenotype.
Figure S2 McrBC analysis of the promoter of WFZP‐D in the P3 region in the individuals of F2 crossed by KN9204 and YM44.
Figure S3 Western blotting of WFZP OE lines expressing the protein of WFZP fused with 3 FLAGs.
Figure S4 Functional verification of WFZP.
Figure S5 Statistics comparison of plant height (a) and spike length (b) between KN199, KN199 wfzp‐a , KN199 wfzp‐d , KN199 wfzp‐a/d and WFZP OE lines.
Figure S6 Effects of WFZP on seed coat cell proliferation.
Figure S7 The overall view of RNA‐seq data.
Figure S8 The expression level of WFZP‐A, B and D in KN199, KN199 wfzp‐a/d and WFZP OE lines detected by RNA‐seq (a) and qPCR (b–d).
Figure S9 GO enrichment analysis of genes in the overlapped DEGs of KN199 wfzp‐a/d up‐regulated/down‐regulated genes and WFZP‐OE down‐regulated/up‐regulated genes.
Figure S10 Transcription factors in the KN199 wfzp‐a/d _vs_KN199 down‐regulated genes (a) and KN199 wfzp‐a/d _vs_KN199 up‐regulated genes (b).
Figure S11 Heatmap of MADS family TFs, HD‐ZIP family TFs and other development‐related genes in the overlapped DEGs of KN199 wfzp‐a/d up‐regulated/down‐regulated genes and WFZP‐OE down‐regulated/up‐regulated genes.
Figure S12 The protein alignment of TaVRN1‐A and BdVRN1 (a) and TaHOX4‐A and BdHOX4 (b).
Figure S13 1000‐grain weight of cultivars with different haplotypes of WFZP‐B (a) and WFZP‐D (b) in ten growing environments. The error bars denote ± SE.
Table S1 The polymorphism of WFZP‐B.
Table S2 The polymorphism of WFZP‐D.
Table S3 Association analysis of WFZP with SNS and TGW.
Table S4 Primers used in this study.
Dataset S1 DEGs between WFZP‐OE, KN199 wfzp‐a/d and KN199.
Dataset S2 Transcription factors (TFs) in the DEGs between KN199 wfzp‐a/d and KN199.
Dataset S3 The expression level and homolog genes of MADS and HD‐ZIP family TFs in the DEGs between KN199 wfzp‐a/d and KN199.
Dataset S4 The expression level and homolog genes of other development‐related genes in the DEGs between KN199 wfzp‐a/d and KN199.
Dataset S5 The expression level and homolog genes of MADS and HD‐ZIP TFs and other development‐related genes in the DEGs between WFZP‐OE, KN199 wfzp‐a/d and KN199.
Dataset S6 The annotation of overlapped merged peaks.
Dataset S7 The haplotype of WFZP‐A, B and D in the natural population used for association analysis.
Dataset S8 The haplotype of WFZP‐A in MCC.


