Abstract
Background
The spread of a novel severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) has affected both the public health and the global economy. The current study was aimed at analysing the genetic sequence of this highly contagious corona virus from an evolutionary perspective, comparing the genetic variation features of different geographic strains, and identifying the key miRNAs as well as their gene targets from the transcriptome data of infected lung tissues.
Methods
A multilevel robust computational analysis was undertaken for viral genetic sequence alignment, phylogram construction, genome-wide transcriptome data interpretation of virus-infected lung tissues, miRNA mapping, and functional biology networking.
Results
Our findings show both genetic similarities as well as notable differences in the S protein length among SARS-CoV-1, SARS-CoV-2 and MERS viruses. All SARS-CoV-2 strains showed a high genetic similarity with the parent Wuhan strain, but Saudi Arabian, South African, USA, Russia and New Zealand strains carry 3 additional genetic variations like P333L (RNA -dependant RNA polymerase), D614G (spike), and P4715L (ORF1ab). The infected lung tissues demonstrated the upregulation of 282 (56.51%) antiviral defensive response pathway genes and downregulation of 217 (43.48%) genes involved in autophagy and lung repair pathways. By miRNA mapping, 4 key miRNAs (hsa-miR-342-5p, hsa-miR-432-5p, hsa-miR-98-5p and hsa-miR-17-5p), targeting multiple host genes (MYC, IL6, ICAM1 and VEGFA) as well as SARS-CoV2 gene (ORF1ab) were identified.
Conclusion
Systems biology methods offer a new perspective in understanding the molecular basis for the faster spread of SARS-CoV-2 infection. The antiviral miRNAs identified in this study may aid in the ongoing search for novel personalized therapeutic avenues for COVID patients.
Keywords: COVID19, Genomic alignment, mRNA, miRNA, Network
1. Introduction
The spread of a novel coronavirus named severe acute respiratory syndrome (SARS-CoV-2) has devastated the public health and global economy since early 2020 [1]. Taxonomic studies have confirmed that SARS-CoV-2 has evolved from SARS-CoV-1 as a different strain. Phylogenetic analysis has suggested that SARS-CoV-2 is different from SARS-CoV-1 (similarity is 79%) and MERS-CoV (similarity is 50%), but shares the highest similarity with bat SARS-CoV1 (similarity is 96%). Although, all three belong to β-coronavirus genus, SARS-CoV2 has caused more global mortalities compared to SARS-CoV-1 or MERS [2]. Therefore, we compared the genetic sequences of these 3 viruses to better understand their molecular differences determining the viral transmission rate and pathogenicity.
SARS-CoV-2 virus has a single stranded RNA genome, whose size ranges from 29.8 kb to 29.9 kb [3,4]. The genomic repertoire of SARS-CoV-2 is as follows; 5′-UTR, replication complex genes (ORF1a and ORF1b) encoding 16 non-structural proteins, structural protein encoding genes like spike (S), envelope (E), membrane (M), and nucleocapsid (N), few accessory protein encoding ORFs (ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF9), 3′-UTRs and a Poly Adenine Tail [5]. Structural proteins facilitate the viral entry to host cells, its assembly and maturation, whereas non-structural proteins are mostly involved in genetic replication and transcription. The accessory proteins interfere with the host defence mechanisms [6]. The receptor binding domain (RBD) of the spike protein subunit S1, which directly interacts with host cellular receptor (ACE2) is conserved across different globally prevalent corona viruses, but recent reports indicate that genetic variations occurring in this sequence determines the host range and infectivity [7,8]. Hence, we carried out phylogenetic analysis of the RBD sequences across different geographic SARS-CoV-2 strains to characterize their genetic features.
The lung is the primary human organ infected by SARS-CoV-2 and most of the global fatalities have resulted mainly from pulmonary complications. Several genes involved in lung physiology and function show aberrant expression in viral infections [9]. However, the SARS-CoV-2 mediated transcriptomic changes and subsequent impact on inflammatory pathways are under intense investigation. Lung epithelial cells besides acting as physical barrier, also modulates lung leukocyte response and release microbicidal molecules including small peptides, reactive oxygen species (ROS) and microRNAs (miRNAs) [10]. The miRNAs are a group of small non-coding RNA molecules (18–22 nucleotides), which regulates the expression of one third of human genes including those involved in innate and adaptive immunity responses against viral infections [[11], [12], [13]]. The miRNAs specific to highly expressed immune response genes in the lung are likely to protect against the viral infection, conversely, their low expression confers susceptibility to infection. Hence, investigating the role of miRNAs involved in the host-SARS-CoV-2 interface is expected to provide valuable insights in identifying potential molecular therapeutic targets for controlling SARS-CoV-2 pathogenesis.
Given the growing evidence of the crucial role of endogenous miRNAs in determining infection susceptibility and mounting antiviral defensive mechanisms, this study used multiple bioinformatic resources to predict the miRNAs against differentially expressed genes (DEGs) in human lung epithelial cells infected with SARS-CoV-2. Additionally, this study has tried to identify miRNAs with predicted binding sites in the SARS-CoV-2 genome and compared them lung DEGs for interpreting their role in mitigating susceptibility to viral infections. The autophagy pathway is an essential component of host anti-viral defence mechanisms. However, some viruses, can exploit the autophagy machinery to support replication [14]. In this study, we also tried to explore the expression status of autophagy-related genes (ATG) in human lung tissues infected with SARS-CoV-2 to understand how the virus mitigates its host autophagy mechanism and establish infection [15]. We used multilevel computational and bioinformatic methods to analyse the genomic and transcriptomic interaction data of SARS-CoV-2 infected lung tissues.
2. Materials and methods
2.1. The retrieval of SARS-CoV2 genome sequence datasets
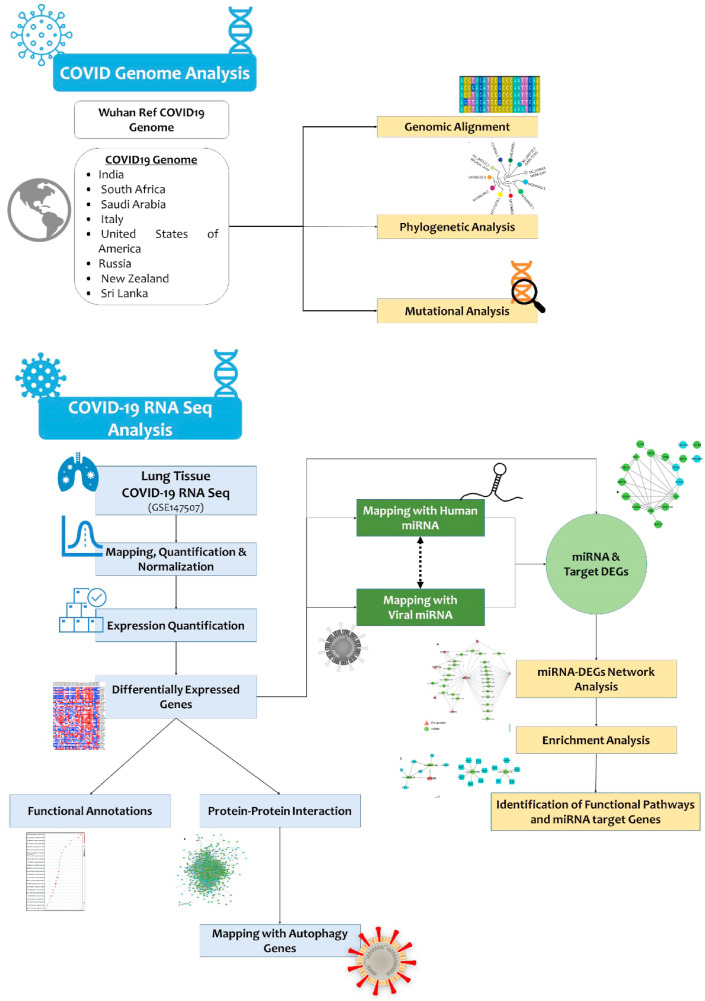
The summary of methods implemented in this study are shown in Fig. 1 . The genome sequences of different SARS-CoV-2 strains reported from different countries like Wuhan, China (Accession No.: NC_045512.2), Saudi Arabia (Accession No. MT630428.1), Italy (Accession No. MT066156), USA (Accession No.: MT339041), India (Accession No.: MT396248.2), New Zealand (Accession No.: MT706050.1), South Africa (Accession No.: MT324062), Russia (Accession No.:MT890462.1), Sri Lanka (Accession No.:MT371050.1), in addition to SARS-COV1 (Accession No.: NC_004718.3) and MERS-CoV (Accession No.: NC_019843) were collected from the NCBI database (https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs).
Fig. 1.
Multilevel systems biology workflow showing SARS-CoV-2 genome analysis, genetic variations analysis and RNA seq analysis of lung tissues led identification of key miRNAs and their target genes.
2.2. Evolutionary analysis of SARS-CoV-2 viruses and its subtyping
The Genome Detective Coronavirus Typing Tool (version 1.1.3) was used to identify different types of SARS-CoV-2 strains. This tool uses Basic local alignment (BLAST) method to detect genetic variations in each queried viral genome. Open source, Multiple Sequence Alignment Fast Fourier Transform Program (MAFFT) [16] was used to align and characterize the genomic sequences of different novel corona viruses. This tool's intake is up to 2000 sequences. The molecular evolution of geographical viral strains from parent Wuhan SARS-CoV-2 strain was explored through phylogenetic analysis using Phylogeny.fr tool [17].
2.3. Identification of differentially expressed genes in human lung tissues infected with SARS CoV-2
To understand the expression pattern of host genes upon SARS-CoV-2 infection, we analysed the RNA sequencing dataset (accession number is GSE147507 - Illumina NextSeq 500 Platform) [18,19] generated from the SARS-CoV-2 infected lung tissues [https://www.ncbi.nlm.nih.gov/geo/]. The full details of experimental protocol including the samples, RNA extraction and sequence analysis are provided in the original publication [20]. In brief, the study investigators have used the transformed lung alveolar (A549) cells and primary human lung epithelium cells (NHBE), which was then mock treated with SARS-CoV-2. RNASeq data normalization to log2 counts per million was performed with EdgeR package [21]. The genes showing an expression difference of with a false discovery rate of <0.05, in addition to Benjamini and Hochberg with a p value of <0.05 were considered to be differentially expressed transcripts. These DEGs were further explored in DAVID, an open source data mining tool, which systemically combines functionally descriptive data from Reactome, InterPRO, KEGG and Pfam databases [22]. This tool aids in gene discovery through biochemical pathway mapping, functional classification, and biological annotations [23].
2.4. Construction of a protein-protein interaction (PPI) network of lung DEGs
Protein interaction networks provide deeper insights into physical contacts holding the protein partners and determines the molecular relationship between host and pathogen. In this study, we used lung DEGs to construct protein-protein network using the STRING v.10.5 database [24]. We used Network Analyser, a plugin of Cytoscape v3.7.2, to identify the hub genes, which shows highest degree of connectivity in PPI network modules [25].
2.5. Identification of autophagy genes from SARS lung DEGs
We retrieved the autophagy-related genes (ATGs) from Human Autophagy Modulatory Database (HAMDB), which hosts the data of 841 chemical molecules, 797 proteins, 132 miRNAs involved in the autophagy pathway. The expression status of autophagy genes (ATGs) in lung transcriptome data was mapped by automated comparison of ATGs against DEG list.
Identification of miRNAs targeting differentially expressed genes.
The miRNA hits against lung DEGs were predicted using MicroRNA Enrichment Turned Network (MIENTURNET; http://userver.bio.uniroma1.it/apps/mienturnet/) [26]. With the input list of genes and mature miRNAs, this open source webtool provides the statistical analysis for over representation of miRNA-target gene interactions by fetching the computationally predicted and experimentally validated data from miRTarBase, miRDB and TargetScan databases. We have also created target gene-miRNA interaction co-expression network by using Cytoscape 3.6.1 [27]. Furthermore, the functional enrichment analysis of target genes and selected miRNAs using KEGG, REACTOME, WiKi Pathways and Disease Ontology (human) was performed. The statistically most enriched GO terms were visualized in ggplot2 [28].
2.6. The miRNA prediction hits for COVID-19 proteins
Initially, the nucleotide and amino acid sequences of different SARS-CoV2 strains sourced from NCBI were queried in the Virus Pathogen Resource (ViPR) (https://www.viprbrc.org/) database. The default search options like “human host” and “SARS-CoV2” were used to map the viral coding genes [29]. ViPR database provides comprehensive viral pathogen data (genetic and protein sequences), along with experimentally determined immune epitope data and anti-viral host responses. In the next steps, ViPR generated SARS-CoV2 genomic data were used to predict miRNA hits (at 65% association cut-off value) against different viral proteins using MirBase [30]. Thereafter, these anti-viral miRNAs were further used to explore target human gene hits. MirBase enables pre-miRNA and mature miRNA prediction workflows due to its default Konstanz Information Miner (KNIME) platform [31]. Furthermore, to better understand host-pathogen interactions, host microRNA and viral gene association network was generated using Cytoscape.
3. Results
3.1. Genomic comparison of SARS-CoV2 with SARS CoV1 and MERS
The nucleotide sequences of the 3 viral genomes revealed the higher identity of SARS-CoV-2 with the genome of SARS-CoV-1 than MERS-CoV. The SARS-CoV-2 shares 80% nucleotide sequence similarity with SARS-CoV-1 genome, and nearly 84% of coding regions are identical in both strains. The sequence length of 10 coding genes (S, E, M, N, 3a, p6, 7a, 7b, 9b and ORF14) of SARS-CoV-1 and CoV-2 were almost identical with only a few minor insertions or deletions. The 39 amino acid long ORF8a peptide was observed only in SARS-CoV-1, where it was found absent in SARS-CoV-2. The length of ORF8b protein in CoV-2 is 121 aa, which is 37 amino acids longer than the corresponding amino acid sequence of CoV1 (84 aa). The ORF3b protein is 132 amino acids long in CoV-2 (132 aa) compared to CoV-1 (154 aa). The genomic alignment of SARS-CoV-2 with MERS-CoV showed that both share 50.2% sequence and 69% coding region similarities. The major difference in sequence identity between CoV-2 and MERS-CoV, is due to the five unique accessory genes 3, 4a, 4b, 5, and 8b found in MERS-CoV. The striking differences in MERS-CoV and CoV-2 genomes, is in the length of S protein, which is 1353 aa in MERS-CoV 1273 aa in SARS-CoV-2. The comparison of CoV-1 and CoV-2 genomes has revealed the evidence of 1349 variations in different genes, including nsp 1–16 (888), spike protein (273), ORF14 (12), E (3), M (19), ORF7a (17), ORF7b (8), ORF8b (49), ORF9b (26), N (35) and 6 (19). SARS-CoV-2 vs MERS-CoV comparison showed 268 genetic variations in different genes like nsp 1–16 (125), spike protein (41), M (5), ORF7a (17), ORF7b (8), ORF8b (22), ORF9b (26), N (5) and 6 (19).
3.2. Genomic comparison of SARS CoV2 geographic strains
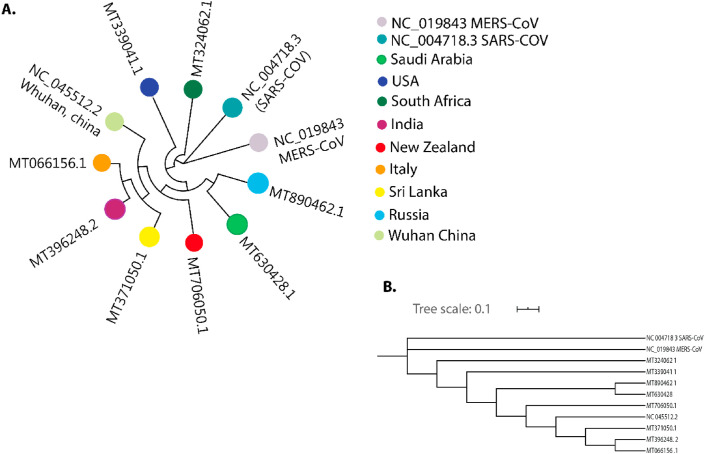
The nucleotide sequences of different geographic SARS-CoV-2 viral strains showed 99% sequence identity to Wuhan SARS-CoV-2 genome (Fig. 2 A). The phylogenetic analysis revealed that all nine SARS-CoV-2 strains are clustered together in a single clade compared to SARS-CoV-1 (Fig. 2B). The Saudi Arabian and Russian strains originated from single clade and presented 99.5% similarities among them. The genetic variation analysis of those SARS-CoV-2 viruses has revealed different types of genetic variations in 9 genes (ORF1ab, ORF1a, ORF3ab, nsp2, RNA dependant RNA polymerase, helicase, spike, envelope protein, and Nucleocapsid proteins; Table 1 ). No genetic variations in other 18 genes like nsp4, 3C-like proteinase, nsp6, nsp7, nsp8, nsp9, nsp10, 3′-to-5′ exonuclease, endoRNAse, 2′-O-ribose methyltransferase, leader protein, nsp11, M, ORF6, ORF7a, ORF7b, ORF8 and ORF10 were observed in 9 SARS-CoV2 strains analysed (Table S1). Interestingly, the viral strains from Saudi Arabia, South Africa, USA, Russia and New Zealand have 3 genetic variations: P333L (RNA -dependant RNA polymerase), D614G (spike), and P4715L (ORF1ab). We have also noticed few other shared variations like R203K and G204R in nucleocapsid protein among the viral strains from Saudi Arabia and Russia, and the S166A variation in the viral strains from Saudi and India. The G251V variation was shared by both Italian and Sri Lankan strains (Table S1, Table S2).
Fig. 2.
Phylogenetic analysis of SARS-CoV2 genomes from different geographical locations: (A) Genome Detective Coronavirus Subtyping Analysis (B) NJ bootstrap tree for reference SARS-CoV2 genomes.
Table 1.
Genetic variations of SARS-CoV-2 proteins.
| Protein | SARS-CoV2/Saudi Arabia | SARS-CoV2/USA SARS-CoV2 | SARS-CoV2/India SARS-CoV2 | SARS-CoV2/Italy SARS-CoV2 | SARS-CoV2/South Africa SARS-CoV2 | SARS-CoV2/Russia SARS-CoV2 | SARS-CoV2/Sri Lanka SARS-CoV2 | SARS-CoV2/New Zeland SARS-CoV2 |
|---|---|---|---|---|---|---|---|---|
| Orf1ab | P4715L (14408C>T) | T265I (1059C>T), P4715L (14408C>T) | A4489V (13730C>T), S5490A (16732T>G) | Nil | P4715L (14408C>T) | P4715L (14408C>T) | Nil | P4715L (14408C>T) |
| RNA-dependent RNA Polymerase | P323L (14408C>T) | P323L (14408C>T) | A97V (13730C>T) | A97V (13730C>T) | P323L (14408C>T) | P323L (14408C>T) | Nil | P323L (14408C>T) |
| Helicase | S166A (16732T>G) | Nil | S166A (16732T>G) | – | – | – | – | |
| S | D614G (23403A>G) | D614G (23403A>G) | – | – | D614G (23403A>G) | D614G (23403A>G) | – | D614G (23403A>G) |
| E | – | V75F (26467G>T) | – | – | – | – | – | |
| N | R203K (28881G>A 28882G>A), G204R (28883G>C) | – | P13L (28311C>T) | – | – | R203K (28881G>A 28882G>A), G204R (28883G>C) | – | |
| Orf3ab | – | Q57H (25563G>T) | – | G251V (26144G>T) | – | – | G251V (26144G>T) | |
| Nsp2 | – | T85I (1059C>T) | – | – | – | – | – | |
| Orf1a | – | T265I (1059C>T) | – | – | – | – | – |
3.3. Identification of differentially expressed genes (DEGs) in SARS-CoV-2 infected lungs and their functional enrichment analysis
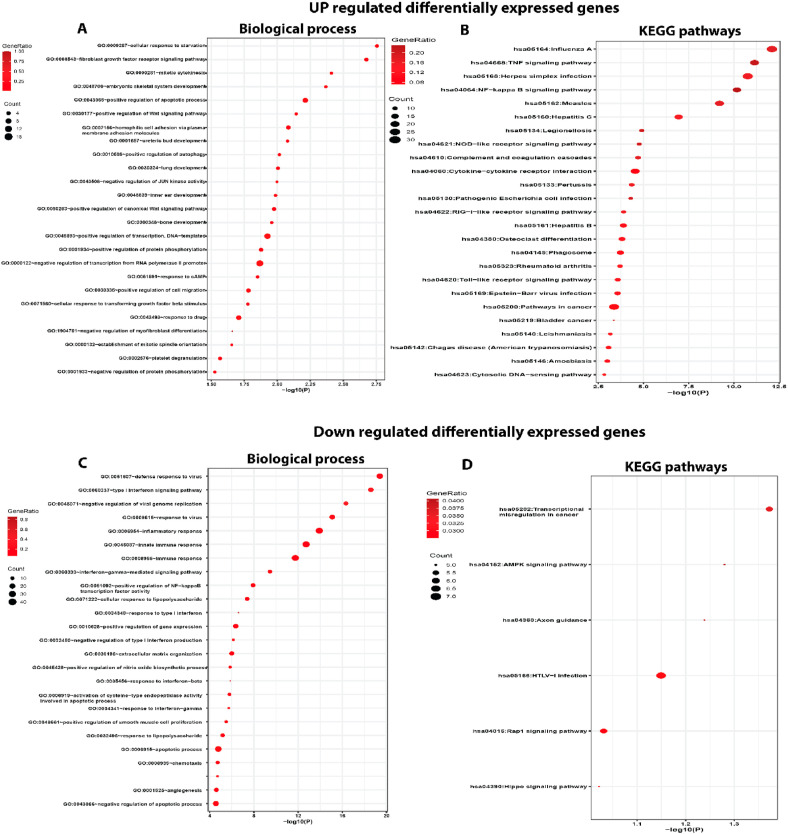
SARS-CoV-2 infected lung epithelial cells showed 499 differentially expressed genes (DEGs), of which 282 (56.51%) were up-regulated and 217 (43.48%) down-regulated with adjusted p < 0.05. These DEGs were further explored by GO enrichment and KEGG pathway analyses to understand their biological relevance in SARS-CoV2 infection (Table S3). The GO enrichment results of lung tissue DEGs are shown in Fig. 3 , where up-regulated DEGs from BP category reveal the dysregulation of genes involved in defensive response to viruses, regulation of viral genome replication, immune response, inflammatory response, interferon-gamma-mediated signalling pathway and cellular response to lipopolysaccharide with adjusted p-value <4.06 E−08 (Fig. 3A). The pathway analysis of the up-regulated DEGs show their involvement in influenza, TNF signalling pathway, NF-kappa beta signalling pathway, hepatitis C and cytokine-cytokine receptor interaction (p-value <2.01 E−05). The pathways in cancer were shown to be significantly down-regulated in response to SARS-CoV-2 infection (Fig. 3B). The enrichment of down-regulated DEGs under BP category reveals the involvement of genes related to cellular response to starvation, fibroblast growth factor receptor signalling pathway, mitotic cytokinesis, positive regulation of Wnt signalling pathway, positive regulation of autophagy (p-value <0.01) and lung development (Fig. 3C). The down-regulated DEGs in lung were enriched in KEGG pathways including transcriptional mis-regulation in cancer, AMPK signalling and Hippo signalling (p-value <0.05) (Fig. 3D).
Fig. 3.
Functional and pathway enrichment analysis DEGs in SARS-CoV-2 infected lung tissue (A) The biological process enrichment of up-regulated DEGs (B) Enriched KEGG pathway of the upregulated DEGs (C) The biological process enrichment analysis of down-regulated DEGs (D) Enriched KEGG pathways of the down-regulated DEGs.
3.4. Construction of protein interaction network of DEGs
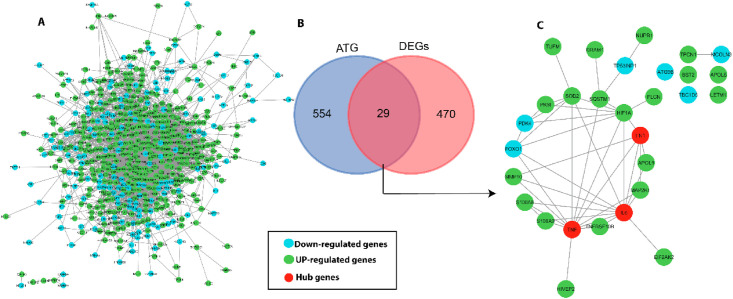
Using the STRING v.10.5 database, PPI networks of 499 DEGs were built and analysed. The protein network consists of 460 nodes and 2539 edges with an average local clustering co-efficient value of 0.452 (PPI enrichment p < 1.00e−16; Fig. 4 A). PPI network analysis identified 10 hub genes (highly connected genes in the network) like IL6, TNF, VEGFA, CXCL8, IL1B, STAT1, FN1, MYC, ICAM1 and IRF7, based on their centrality scores (>56 score) (Table 2 ).
Fig. 4.
(A) Protein-protein interaction (PPI) network of DEGs. The green circles represent up-regulated genes and cyan color circles for down-regulated genes (B) Venn diagram of 29 common genes between total DEGs and ATGs. (C) PPI regulatory network of 29 DEGs and autophagy related genes (ATG), red color circle represents hub genes.
Table 2.
Topological properties of top 10 hub genes.
| S. No. | Hub gene | Degree | Betweenness Centrality | Closeness Centrality | Clustering Coefficient | Autophagy gene |
|---|---|---|---|---|---|---|
| 1. | IL6 | 124 | 0.107528 | 0.461998 | 0.150407 | Yes |
| 2. | TNF | 121 | 0.084793 | 0.458475 | 0.157576 | Yes |
| 3. | VEGFA | 101 | 0.085286 | 0.44637 | 0.155842 | – |
| 4. | CXCL8 | 90 | 0.041332 | 0.435237 | 0.216479 | – |
| 5. | IL1B | 84 | 0.040898 | 0.43039 | 0.207975 | – |
| 6. | STAT1 | 81 | 0.046246 | 0.423649 | 0.250617 | – |
| 7. | FN1 | 80 | 0.077877 | 0.428345 | 0.179114 | Yes |
| 8. | MYC | 77 | 0.077608 | 0.43629 | 0.176692 | – |
| 9. | ICAM1 | 56 | 0.015464 | 0.405547 | 0.32987 | – |
| 10. | IRF7 | 56 | 0.014293 | 0.385602 | 0.335065 | – |
3.5. Mapping of host autophagy-related genes (ATG) in DEGs from SARS-CoV2 infected lung tissues
The retrieved ATGs (in reference to HAMdb) against 499 lung DEGs, revealed that 29 genes (4.97%; SOD2 SQSTM1 APOL6 TBC1D5 TPCN1 LETM1 NUPR1 FN1 TNF DRAM1 APOL1 TUFM S100A8 FOXO1 PDK4 ATG9B IL6 EIF2AK2 FLCN BST2 HIVEP2 TP53INP1 MCOLN3 TNFRSF10B HIF1A PKM MAP2K1 MMP10 and S100A9) are shared and have functional role in the autophagy pathway (Fig. 4B). The regulatory network of these 29 ATGs is shown in Fig. 4C. We further mapped the expression status of ATGs in lung DEGs. Of these 10 hub genes (identified from 499 lung DEGs), 3 hubs, namely, IL6, TNF and FN1 are up-regulated in SARS-CoV2 infected lungs and they are known to directly target ORF3a and ORF8 genes of SARS-CoV-2 genome (Table 2).
3.6. Identification of miRNAs targeting lung DEGs and SARS CoV-2 genome
3.6.1. Prediction of miRNAs of DEGs and their major dysregulated pathways in COVID infected lung tissues
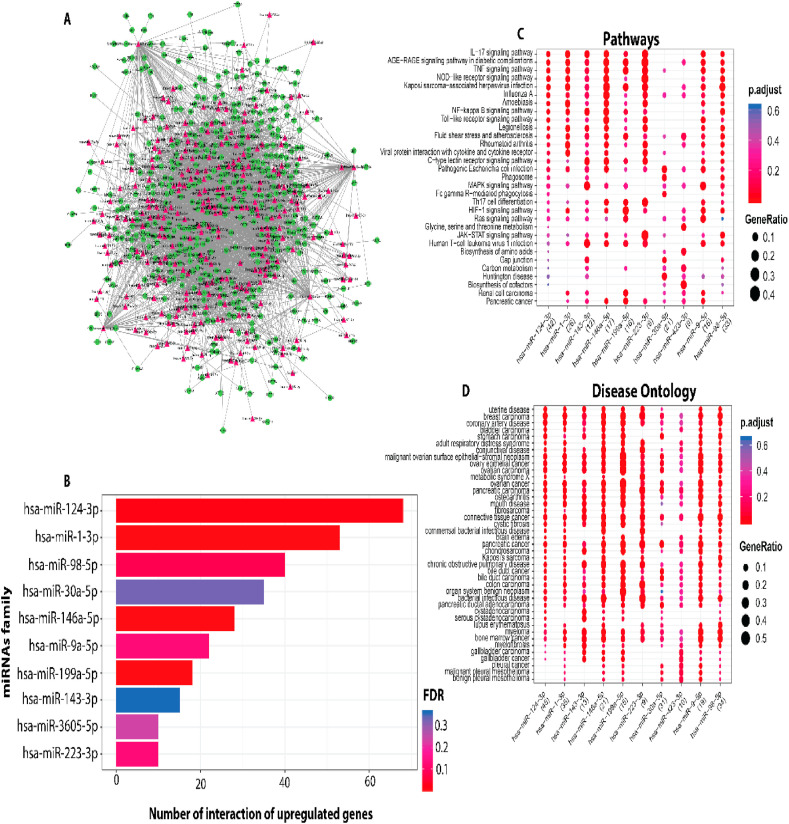
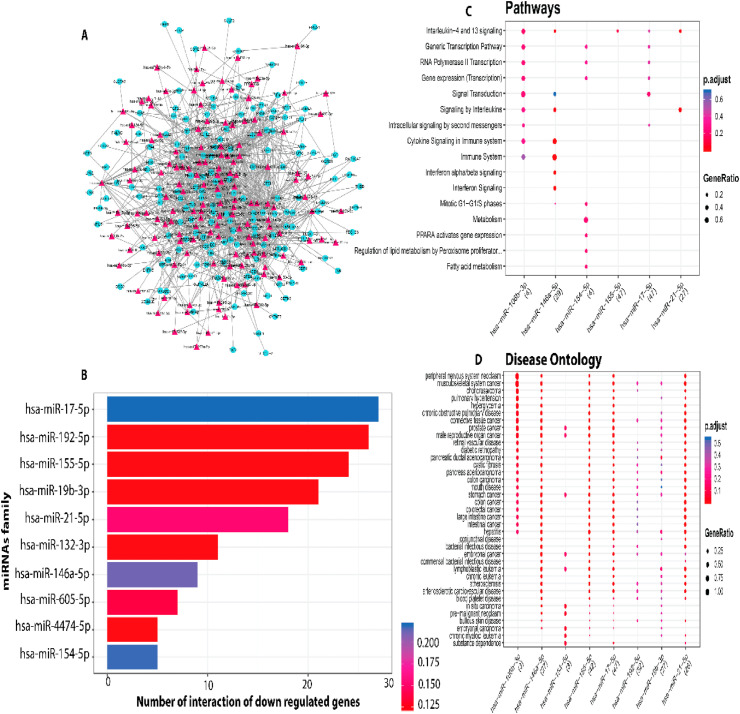
MIENTURNET (MicroRNA ENrichment TURned NETwork) webtool has predicted 494 miRNA hits against 38.67% of (193/499) lung DEGs, of which 110 (60.10%) were down-regulated and 83 (45.35%) were up-regulated (Table S5). The miRNA-mRNA network for up and down regulation in DEGs is shown in Fig. 5 A and Fig. 6 A, respectively (Table S6). The functional network of miRNA-up regulated DEGs hits revealed top 10 miRNAs (hsa-miR-124-3p, hsa-miR-1-3p, hsa-miR-98-5p, hsa-miR-30a-5p, hsa-miR-146a-5p, hsa-miR-9a-5p, hsa-miR-199a-5p, hsa-miR-143-3p, hsa-miR-3605-5p and hsa-miR-223-3p) based on their association scores (p < 0.05) (Fig. 5B, Table 3 ). Major dysregulated pathways targeted by these 10 miRNAs include signalling of IL−17, TNF, NOD−like receptor, NF−kappa Band MAPK genes and Fc gamma R−mediated phagocytosis (Fig. 5C–D). The regulatory network between down-regulated DEGs and miRNAs has revealed that the top miRNAs (hsa-miR-17-5p, hsa-miR-192-5p, hsa-miR-155-5p, hsa-miR-19b-3p, hsa-miR-21-5p, hsa-miR-132-3p, hsa-miR-146a-5p, hsa-miR-605-5p, hsa-miR-4474-5p, hsa-miR-154-5p) regulates Interleukin−4 and 13, cytokine signalling in immune system, and mitotic G1−G1/S phases, while PPARA activates gene expression pathways in the SARS-CoV2 infected lung tissues (Fig. 6B–D).
Fig. 5.
(A) The miRNA-mRNA network analysis of up-regulated DEGs involved in host biological processes in response to SARS-CoV-2 infection. Red diamonds represent miRNAs and green circles represent mRNA. (B) The miRNAs vs number of up-regulated DEGs (C) Enriched biological process of miRNAs. (D) biological pathway of the miRNAs.
Fig. 6.
(A) The miRNA-mRNA network analysis of downregulated DEGs involved in host biological processes in response to SARS-CoV-2 infection. Red diamonds represent miRNAs and cyan circles represents host mRNA. (B) The miRNAs vs number of downregulated DEGs (C) TEnriched biological process of miRNAs targeted to down-regulated DEGs. (D) Biological pathways of the miRNAs.
Table 3.
Predicted miRNAs targeted by hub gene. Bold miRNAs are top signification miRNA of upregulated and downregulated hub gene.
| S. No. | Hub gene | Predicted top 10 miRNA |
|---|---|---|
| 1. | IL6 | hsa-miR-202-3p, hsa-let-7c-3p, hsa-let-7b-5p, hsa-miR-34a-5p, hsa-let-7a-5p and hsa-miR-98-5p |
| 2. | TNF | hsa-miR-17-5p, hsa-miR-34a-5p and hsa-miR-203a-3p |
| 3. | VEGFA | hsa-miR-17-5p hsa-miR-330-3p hsa-miR-200b-3p hsa-miR-20b-5p hsa-miR-20a-5p hsa-miR-106a-5p hsa-miR-141-5p hsa-miR-374b-5p hsa-miR-125a-3p hsa-miR-126-5p hsa-miR-378a-3p hsa-miR-34a-5p hsa-miR-335-5p hsa-miR-203a-3p hsa-miR-369-3p |
| 4. | CXCL8 | hsa-miR-202-3p hsa-let-7b-5p hsa-let-7f-5p hsa-miR-335-5p hsa-let-7d-5p hsa-let-7a-5p hsa-let-7c-5p hsa-miR-203a-3p hsa-miR-98-5p hsa-let-7g-5p hsa-miR-106a-5p |
| 5. | IL1B | hsa-miR-106a-5p |
| 6. | STAT1 | hsa-miR-34a-5p hsa-miR-203a-3p |
| 7. | FN1 | hsa-miR-200b-3p hsa-let-7g-5p |
| 8. | MYC | sa-miR-34b-5p hsa-miR-17-5p hsa-miR-196a-5p hsa-let-7b-5p hsa-miR-148a-3p hsa-miR-129-2-3p hsa-miR-98-5p hsa-let-7g-5p hsa-miR-20a-5p hsa-miR-599 hsa-miR-449c-5p hsa-miR-125a-3p hsa-miR-126-5p hsa-miR-378a-3p hsa-let-7f-5p hsa-miR-34a-5p hsa-miR-335-5p hsa-let-7d-5p hsa-miR-320b hsa-let-7a-5p hsa-let-7c-5p hsa-miR-33b-5p |
| 9. | ICAM1 | hsa-miR-17-5p hsa-miR-335-5p hsa-miR-17-3p hsa-miR-98-5p hsa-miR-141-5p |
3.7. Prediction of miRNAs against SARS-CoV2 genes
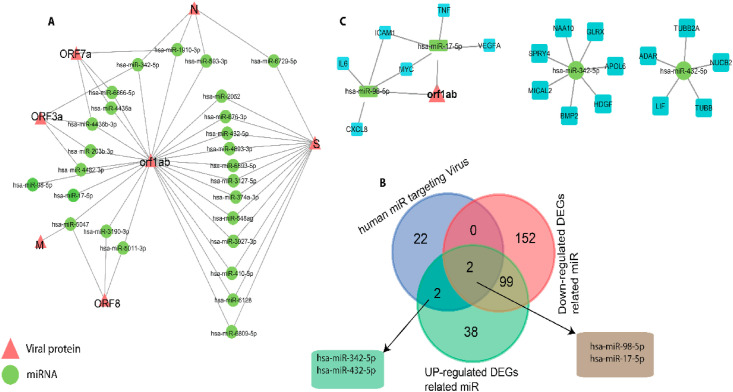
MirBase has predicted 24 potential human miRNAs (based on miRbase Score >68) targeting ORF1ab, S, ORF3a, M, ORF7a, ORF8, N, E and ORF10 genes of the SARS CoV2 virus (Fig. 7 A). The regulatory network analysis showed that miRNAs namely hsa-miR-2052, hsa-miR-676-3p, hsa-miR-432-5p, hsa-miR4693-3p, hsa-miR-6893-3p, hsa-miR-3127-5p, hsa-miR-374a-3p, hsa-miR-548a-3p, hsa-miR-3927a-3p, hsa-miR-410-5p, hsa-miR-6128, hsa-miR-6809 interacts with both ORF1ab and spike proteins of the virus (Fig. 7A). The data shown in the Table S7 highlights the miRNA hits against several SARS-CoV2 viral genes primarily responsible for entry, infection, viral biogenesis, entry and replication.
Fig. 7.
(A) Host-pathogen interaction of SARS-CoV-2 and host miRNA. Triangular red nodes represent SARS-COV-2 proteins, while circles (green) represent host miRNAs. (B) Venn diagram shows the 2 common miRNAs among up and down regulated DEGs related miRNAs and human miRNA targeting virus. (C) Regulatory network of hsa-miR-98-5p, hsa-miR-17-5p and hub genes (IL6, CXCL8, MYC, ICAM1, TNF, VEGFA) and orf1ab. The hsa-miRNA-342-5p and hsa-miRNA-432-5p are the common miRNAs targeting UP-regulated DEGs related miRNAs and human miRNA targeting virus.
3.8. Mapping shared miRNA targets from SARS-CoV-2 genome and lung DEGs
Of the 24 miRNAs targeting SARS-CoV-2 genes, 4 miRNAs (hsa-miR-342-5p, hsa-miR-432-5p, hsa-miR-98-5p and hsa-miR-17-5p) were predicted to target lung DEGs (Fig. 7A and B). Of those miRNAs, the hsa-miR-342-5p targets 7, hsa-miR-432-5p targets 5 DEGs (Fig. 7B), hsa-miR-98-5p targets 58 DEGs and hsa-miR-17-5p targets 69 DEGs of the SARS-CoV2 infected lung tissues (Table S6). Furthermore, the network analysis of 4 miRNAs and 138 DEGs has revealed that hsa-miR-342-5p, regulates 7 DEGs (GLARX, NAA10, SPRY4, MICAL2, BMP2, HDGF, APOL6), hsa-miR-432-5p, regulates 5 DEGs (TUBB2A, ADAR, LIF, TUBB, NUCB2), hsa-miR-98-5p regulates 4 DEGs (IL6, CXCL8, MYC and ICAM1) and 1 viral gene (orf1ab), and hsa-miR-17-5p regulate 7,5,4 and 4 DEGs respectively. Two of those miRNAs, hsa-miR-98-5p and hsa-miR-17-5p also regulate one viral protein ORF1ab (Fig. 7C).
4. Discussion
Our bioinformatic analysis has confirmed the shared genetic similarities among SARS-CoV1, SARS-CoV-2 and MERS-CoV [32]. All three use Spike structural protein to bind to host cell receptors and cellular protease for its activation. The S protein consists of 2 subunits (S1 and S2), of which S1 subunit (RBD) takes part in receptor binding and S2 subunit primarily facilitates viral fusion to cell membrane and genome entry to host cells [33]. Comparison of S gene sequences in SARS-CoV-2 SARS-CoV1, and MERS-CoV has confirmed its variable length among them [34]. Both SARS-CoV1 and SARS-CoV2 binds to ACE2 receptor, but they show different binding affinities [35]. The sequence similarity of spike protein, especially RBD of S2 subunit between both viruses is 73%–76%. MERS virus uses Dipeptidyl-peptidase 4 (DPP4) glycoprotein as the receptor [36]. Among the two receptors ACE2 show wider tissue distribution compared to DPP4 [37]. The higher transmissibility rate of SARS-CoV2 is attributed to furin-like cleavage sites, which facilitate the S protein priming and its high reproductive number (>3–5), which is relatively higher than SARS-CoV1 (1.6–1.9) and MERS (<1). Additional known factors underlying the rapid spread of CoV2 infection include the accumulation of single nucleotide variations, insertions, and deletions in S1 and S2 domains of the spike protein, which enhances the binding with ACE2, and allows effective invasion compared to both SARS-CoV1 and MERS.
The rapid global transmission of SARS-CoV-2 highlights the role of genetic variation or recombination driven viral evolution and adaptation in host systems. Therefore, we analysed genomic sequence of SARS-CoV2 strains from different countries to understand the viral origin and its evolution during the pandemic. In this study, we observed that SNP occurrence is not random, but it is more common in the genes, which are critical for virus [38]. In Saudi Arabian SARS-CoV2 strain (EPI_ISL_678216), 16 variations were observed as compared to the wuhan-hu1 SARS-CoV2 strain. These 16 variations include 6 synonymous, 7 missense, 2 downstream and 1 upstream variant. Majority of these variants were seen in ORF1ab that encodes 16 non-structural proteins, which contributes to the transcription of viral genes, replication, proteolytic processing and suppression of host gene expression connected to immune response. A recent large scale study on 3067 SARS-CoV-2 genomes collected from various geographical regions has identified 782 variations, of which 65.98% were non-synonymous, 28.39% were synonymous, and 5.63% were distributed in intergenic regions compared to the Wuhan-Hu-1/2019 reference sequence [39].
In this study, variant D614G (spike), localized to highly conserved B-cell epitope is the most common clade identified in viral strains from Saudi Arabia, South Africa, USA, Russia and New Zealand. The amino acids in this epitope are highly conserved and are likely to interfere with the effectivity of vaccines [40]. Almost all viral strains with D614G, have also accumulated other variants like P333L (RNA-dependent RNA polymerase) and P4715L (ORF1ab) in proteins responsible for SARS-CoV-2 replication. It is noteworthy that spike protein, RNA-dependent RNA polymerase and ORF1ab proteins are therapeutic targets for antiviral drugs like Remdesivir [41] and Arbidol [42]. Hence, accumulations of nucleotide variations in these genes could render SARS-CoV2 variants resistant to treatment. The fast-evolving viral genetic architecture necessitates more caution while developing new antiviral drugs or vaccines against the SARS-CoV-2 strains. We also speculate from this study that the existence of a country-specific genetic variation continuum may also be able to explain the current situation in these countries, such as disease incidence, epidemic control, degree and timing of exposure to a symptomatic carrier, etc. The probability of genetic variation occurrence is more at transcriptional level than at translational level, as evident from the existence of more codon variations than the actual amino acid sequence changes in the candidate protein.
The genome wide gene expression data is a proven resource to identify biological pathways underlying human diseases [43,44], including viral infections [45]. Few studies have reported gene expression changes in SARS‐CoV‐2 infected clinical samples using in vivo, in vitro or ex vivo approaches [46,47]. However, these studies have not been able to fully interpret the role of dysregulated genes at molecular and network levels. In this study, we used a series of comprehensive computational investigations to analyse the lung tissue response to SARS-CoV2 infection. Our analysis revealed the enrichment of TNF and NF-kappa beta signalling pathways in up-regulated genes, whose biological processes are connected to host defensive response, regulation of viral genome viral replication, immune response, inflammatory response and cellular response to lipopolysaccharide. These results support the previous findings, where corona virus infection is seen to activate innate immune response in the body by triggering the production of anti-viral interferon-gamma cytokine molecules [48,49]. Macrophage produced TNF is known to damage pulmonary vascular endothelial cells and cause pulmonary edema [50]. Inhibition of TNF-α is shown to reduce the lung damage in viral infections. One of the enriched pathways involves NF-kappa B signaling which has a critical role in innate and adaptive immunity [51].
We have also noted the enrichment of AMPK and Wnt signalling pathways in down-regulated genes, regulating cellular responses to starvation, mitotic cytokinesis, positive regulation of autophagy and lung development. Studies in transgenic mice models show the loss of alveolar tissue repair capacity in the lungs due to blockage of FGFR signalling caused by the viral infection [52]. Wnt signalling is essential for the repair and regeneration of human lung epithelial cells when infected with viruses [53]. Hence, it implies that SARS-CoV2 infection may dysregulate cell signalling and impairs the epithelial cell regeneration and repair of lung tissues. Corona viruses are known to modulate autophagy to enable their replication in host cells [54]. In this study, we have identified 29 autophagy genes from the total 499 lung DEGs involved in SARS-CoV2 infection. Protein interaction network analysis of DEGs-ATGs has confirmed the up-regulation of 3 autophagy genes (IL6, TNF and FN1), which are known to directly target ORF3a and ORF8 genes of SARS-CoV-2 genome. The SARS-CoV-2 spike protein is demonstrated to promote IL6 trans-signaling and initiate hyperinflammatory response by activation of AT1 axis in epithelial cells [55].
The miRNAs are known to regulate the expression of host genetic factors required for viral pathogenesis [56]. In this study, we observed that 193 (38.67%) of the 499 lung DEGs are targeted by a range of host miRNAs. Of these miRNAs, 83 (45.35%) can directly target IL-17, TNF, NOD-like receptor, NF-kB, MAPK and IL-4 and -13 signaling pathways, which are required for viral replication and disease pathogenesis. For example, TNF-α levels are known to be elevated throughout the SARS-CoV2 infection. In COVID19 patients, TNF-α induces the production of HA-synthase-2 in lung alveolar epithelium, which further increases fluid influx in lung alveoli and cause deoxygenation [57]. Similarly, IL-17 with IL-16 promotes the viral survival by countering the T cell function and cellular apoptosis [58].
We also found 110 miRNAs targeting down-regulated genes connected to pathways such as Interleukin−4 and 13 signaling and cytokine signaling in Immune system and Mitotic G1−G1/S phases involved in suppressing viral entry and controlling adverse inflammatory reactions in the lungs. Recent studies have suggested the involvement of host miRNAs in targeting SARS-CoV2 genes involved in immune signaling pathways [59,60]. However, there is dearth of information focusing on the identification of shared miRNAs acting on both human and viral genes. Therefore, in the present study, we performed the functional enrichment analysis of host and viral genes against the predicted miRNAs. We found that 24 predicted miRNAs target several genes (ORF1ab, S, ORF3a, M, ORF7a, ORF8, N, E and ORF10) that are primarily responsible for entry, infection, viral biogenesis, entry and replication of the SARS CoV2 virus.
Interestingly, 4 out of 24 anti-SARS-Cov2 miRNAs (hsa-miR-342-5p, hsa-miR-432-5p, hsa-miR-98-5p and hsa-miR-17-5p) are predicted to target many dysregulated genes in lung including MYC, IL6, ICAM1 and VEGFA. Its noteworthy, that MYC controls adaptive immunity by generating CD8+ T cells upon viral infection [61]. By targeting the 2C-coding region of the viral genome, miR-342-5p inhibits the biosynthesis of Coxsackievirus B3 (CVB3), an Enterovirus (EV71) [62]. Since there is strong evidence that host cells can use cellular miRNAs to combat viral infection, host miRNAs may be able to suppress SARS-Cov-2 replication. However, the role of microRNAs in SARS-Cov-2 infection and replication is unknown. The hsa-miR-17-5p was identified as another significant miRNA in our research. TNF expression in leukaemia cells is inhibited by hsa-miR-17, which binds to TNF's 3′-UTR [63,64]. During the COVID-19 infection, hsa-miR-17-3p was found in human alveolar basal epithelial cells (HBEpC) [64]. hsa-miR-98-5p was discovered as a promising candidate, and it was mechanistically validated as a regulator of TMPRSS2 transcription in two human endothelial cell types derived from the lung and the umbilical vein [63,64]. IL6 stimulates inflammatory reactions in mucosa during COVID infection [65]. ICAM-1, a major cell adhesion molecule modulates the viral replication by inducing the NF-kB protein expression [66]. VEGF, a proangiogenic factor is known for its involvement in vascular leakiness and inflammation in COVID infected lung tissues [67]. Our findings confirm the recent miRNA studies in COVID patients [[68], [69], [70]]. The effective targetability of both host and viral genes miRNAs presents them as novel avenues to control the COVID pandemic.
This study confirms the genetic lineage between SARS CoV2 with SARS CoV1 and MERS viruses. The genetic variation analysis of different SARS-CoV2 strains proves that dynamic molecular divergences taking place in orf1ab, orf1a, Spike surface glycoprotein, Envelope protein, Nucleocapsid protein and ORF8 genes, which offer advantage to sustain the environmental pressures when the strain enters a new geographic location. In this study, we confirm that infected lung tissues show upregulation of 282 (56.51%) antiviral defensive response pathway genes and downregulation of 217 (43.48%) genes involved in autophagy and lung repair pathways. Future studies are required to study the impact of SARS-CoV2 virus genetic variations on lung DEG dysregulations to establish viral genotype and COVID patient phenotype connections. The key miRNAs (hsa-miR-342-5p, hsa-miR-432-5p, hsa-miR-98-5p and hsa-miR-17-5p) targeting multiple host genes as well as SARS-CoV2 genes presents us a narrow window of novel therapeutic opportunity to target genes in COVID pathophysiology. However, in vitro, ex vivo and in vivo studies are required to validate the actual role of anti-SARS microRNAs in COVID patients.
Author contributions
Conceptualization: BB, NA, MZM, NS; Data curation: BB, MZM and NS; Formal Analysis: BB, FA, MZM; Funding acquisition: BB; Methodology: BB, NA, MZM, AA, ZA, RE and NS; Software: BB and MZM; Supervision: BB, NS and RE Validation: BB, NA and NS; Visualization: BB and MZM; Writing original draft and editing: BB, NA, MZM, AA, FA, ZA, RE and NS.
Declaration of competing interest
Authors declares that they have no conflict of interest.
Acknowledgements
The Project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. GCV19-40-1441. The authors, therefore, acknowledge with thanks DSR for technical and financial support. The co-Author Abdulhakeem S. Alamri would like to acknowledge TRUSP (2020/288).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104570.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rastogi M., Pandey N., Shukla A., Singh S.K. SARS coronavirus 2: from genome to infectome. Respir. Res. 2020;21:318. doi: 10.1186/s12931-020-01581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Q., Sadykov M., Mfarrej S., Hala S., Naeem R., Nugmanova R., Al-Omari A., Salih S., Mutair A.A., Carr M.J., Hall W.W., Arold S.T., Pain A. A genetic barcode of SARS-CoV-2 for monitoring global distribution of different clades during the COVID-19 pandemic. Int. J. Infect. Dis. 2020;100:216–223. doi: 10.1016/j.ijid.2020.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B., Tian E.K., He B., Tian L., Han R., Wang S., Xiang Q., Zhang S., El Arnaout T., Cheng W. Overview of lethal human coronaviruses. Signal Transduct. Target Ther. 2020;5:89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans S.E., Tseng C.K., Scott B.L., Höök A.M., Dickey B.F. Inducible epithelial resistance against coronavirus pneumonia in mice. Am. J. Respir. Cell Mol. Biol. 2020;63:540–541. doi: 10.1165/rcmb.2020-0247LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Zou X. An overview of RNA virus-encoded microRNAs. ExRNA. 2019;1:37. [Google Scholar]
- 12.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9 doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundhoff A., Sullivan C.S. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadovsky Y., Mouillet J.F., Ouyang Y., Bayer A., Coyne C.B. The function of TrophomiRs and other MicroRNAs in the human placenta. Cold Spring Harb. Perspect. Med. 2015;5 doi: 10.1101/cshperspect.a023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daamen A.R., Bachali P., Owen K.A., Kingsmore K.M., Hubbard E.L., Labonte A.C., Robl R., Shrotri S., Grammer A.C., Lipsky P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021;11:7052. doi: 10.1038/s41598-021-86002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 23.Wang N.N., Dong J., Zhang L., Ouyang D., Cheng Y., Chen A.F., Lu A.P., Cao D.S. HAMdb: a database of human autophagy modulators with specific pathway and disease information. J. Cheminf. 2018;10:34. doi: 10.1186/s13321-018-0289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–d368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assenov Y., Ramírez F., Schelhorn S.E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 26.Licursi V., Conte F., Fiscon G., Paci P. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinf. 2019;20:545. doi: 10.1186/s12859-019-3105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito K., Murphy D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst. Pharmacol. 2013;2:e79. doi: 10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C.N., Dietrich J., Klem E.B., Scheuermann R.H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–d162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazanetz M.P., Marmon R.J., Reisser C.B., Morao I. Drug discovery applications for KNIME: an open source data mining platform. Curr. Top. Med. Chem. 2012;12:1965–1979. doi: 10.2174/156802612804910331. [DOI] [PubMed] [Google Scholar]
- 32.Hu T., Li J., Zhou H., Li C., Holmes E.C., Shi W. Bioinformatics resources for SARS-CoV-2 discovery and surveillance. Briefings Bioinf. 2021;22:631–641. doi: 10.1093/bib/bbaa386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y.X., Sheng D.Q., Cheng L., Song X.Y. Current landscape of epigenetics in lung cancer: focus on the mechanism and application. J. Oncol. 2019;2019:8107318. doi: 10.1155/2019/8107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar D T., Shaikh N., Kumar S U., Doss C G.P., Zayed H. Structure-based virtual screening to identify novel potential compound as an alternative to Remdesivir to overcome the RdRp protein mutations in SARS-CoV-2. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.645216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Team V.-C.C.-R., Hoffmann M., Pohlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015 e1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkatakrishnan A.J., Puranik A., Anand A., Zemmour D., Yao X., Wu X., Chilaka R., Murakowski D.K., Standish K., Raghunathan B., Wagner T., Garcia-Rivera E., Solomon H., Garg A., Barve R., Anyanwu-Ofili A., Khan N., Soundararajan V. Knowledge synthesis of 100 million biomedical documents augments the deep expression profiling of coronavirus receptors. Elife. 2020;9 doi: 10.7554/eLife.58040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S.U., Priya N.M., Nithya S.R., Kannan P., Jain N., Kumar D.T., Magesh R., Younes S., Zayed H., Doss C.G.P. A review of novel coronavirus disease (COVID-19): based on genomic structure, phylogeny, current shreds of evidence, candidate vaccines, and drug repurposing. 3 Biotech. 2021;11:198. doi: 10.1007/s13205-021-02749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laamarti M., Alouane T., Kartti S., Chemao-Elfihri M.W., Hakmi M., Essabbar A., Laamarti M., Hlali H., Bendani H., Boumajdi N., Benhrif O., Allam L., El Hafidi N., El Jaoudi R., Allali I., Marchoudi N., Fekkak J., Benrahma H., Nejjari C., Amzazi S., Belyamani L., Ibrahimi A. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. PloS One. 2020;15 doi: 10.1371/journal.pone.0240345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 2020;9:324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020;56:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banaganapalli B., Mansour H., Mohammed A., Alharthi A.M., Aljuaid N.M., Nasser K.K., Ahmad A., Saadah O.I., Al-Aama J.Y., Elango R., Shaik N.A. Exploring celiac disease candidate pathways by global gene expression profiling and gene network cluster analysis. Sci. Rep. 2020;10:16290. doi: 10.1038/s41598-020-73288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahly N.N., Banaganapalli B., Sahly A.N., Aligiraigri A.H., Nasser K.K., Shinawi T., Mohammed A., Alamri A.S., Bondagji N., Elango R., Shaik N.A. Molecular differential analysis of uterine leiomyomas and leiomyosarcomas through weighted gene network and pathway tracing approaches. Syst. Biol. Reprod. Med. 2021:1–12. doi: 10.1080/19396368.2021.1876179. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Z., Huang S., Zhang Y., Sun C., Tang Y., Zhao Q., Zhou Q., Ju W., He X. Bioinformatics analysis on multiple Gene Expression Omnibus datasets of the hepatitis B virus infection and its response to the interferon-alpha therapy. BMC Infect. Dis. 2020;20:84. doi: 10.1186/s12879-019-4720-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kindler E., Thiel V., Weber F. Interaction of SARS and MERS coronaviruses with the antiviral interferon response. Adv. Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie T.A., Han M.Y., Su X.R., Li H.H., Chen J.C., Guo X.G. Identification of Hub genes associated with infection of three lung cell lines by SARS-CoV-2 with integrated bioinformatics analysis. J. Cell Mol. Med. 2020;24:12225–12230. doi: 10.1111/jcmm.15862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukhopadhyay S., Hoidal J.R., Mukherjee T.K. Role of TNFα in pulmonary pathophysiology. Respir. Res. 2006;7:125. doi: 10.1186/1465-9921-7-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hariharan A., Hakeem A.R., Radhakrishnan S., Reddy M.S., Rela M. The role and therapeutic potential of NF-kappa-B pathway in severe COVID-19 patients. Inflammopharmacology. 2021;29:91–100. doi: 10.1007/s10787-020-00773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chugh R.M., Bhanja P., Norris A., Saha S. Experimental models to study COVID-19 effect in stem cells. Cells. 2021:10. doi: 10.3390/cells10010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hancock A.S., Stairiker C.J., Boesteanu A.C., Monzón-Casanova E., Lukasiak S., Mueller Y.M., Stubbs A.P., García-Sastre A., Turner M., Katsikis P.D. Transcriptome analysis of infected and bystander type 2 alveolar epithelial cells during influenza A virus infection reveals in vivo Wnt pathway downregulation. J. Virol. 2018;92 doi: 10.1128/JVI.01325-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Randhawa P.K., Scanlon K., Rappaport J., Gupta M.K. Modulation of autophagy by SARS-CoV-2: a potential threat for cardiovascular system. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.611275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patra T., Meyer K., Geerling L., Isbell T.S., Hoft D.F., Brien J., Pinto A.K., Ray R.B., Ray R. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaufmann S.H.E., Dorhoi A., Hotchkiss R.S., Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018;17:35–56. doi: 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fara A., Mitrev Z., Rosalia R.A., Assas B.M. Cytokine storm and COVID-19: a chronicle of pro-inflammatory cytokines. Open Biol. 2020;10:200160. doi: 10.1098/rsob.200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou W., Jin Y.-H., Kang H.S., Kim B.S. Interleukin-6 (IL-6) and IL-17 synergistically promote viral persistence by inhibiting cellular apoptosis and cytotoxic T cell function. J. Virol. 2014;88:8479–8489. doi: 10.1128/JVI.00724-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan M.A.-A.-K., Sany M.R.U., Islam M.S., Islam A.B.M.M.K. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020:11. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan M.A., Sany M.R.U., Islam M.S., Islam A. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020;11:765. doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haque M., Song J., Fino K., Wang Y., Sandhu P., Song X., Norbury C., Ni B., Fang D., Salek-Ardakani S., Song J. C-Myc regulation by costimulatory signals modulates the generation of CD8+ memory T cells during viral infection. Open Biol. 2016;6:150208. doi: 10.1098/rsob.150208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng Z., Ke X., Wang M., He S., Li Q., Zheng C., Zhang Z., Liu Y., Wang H. Human microRNA hsa-miR-296-5p suppresses enterovirus 71 replication by targeting the viral genome. J. Virol. 2013;87:5645–5656. doi: 10.1128/JVI.02655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang S., Amahong K., Sun X., Lian X., Liu J., Sun H., Lou Y., Zhu F., Qiu Y. The miRNA: a small but powerful RNA for COVID-19. Briefings Bioinf. 2021;22:1137–1149. doi: 10.1093/bib/bbab062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merino G.A., Raad J., Bugnon L.A., Yones C., Kamenetzky L., Claus J., Ariel F., Milone D.H., Stegmayer G. Novel SARS-CoV-2 encoded small RNAs in the passage to humans. Bioinformatics. 2021;36:5571–5581. doi: 10.1093/bioinformatics/btaa1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gubernatorova E.O., Gorshkova E.A., Polinova A.I., Drutskaya M.S. IL-6: relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020;53:13–24. doi: 10.1016/j.cytogfr.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Othumpangat S., Noti J.D., McMillen C.M., Beezhold D.H. ICAM-1 regulates the survival of influenza virus in lung epithelial cells during the early stages of infection. Virology. 2016;487:85–94. doi: 10.1016/j.virol.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pang J., Xu F., Aondio G., Li Y., Fumagalli A., Lu M., Valmadre G., Wei J., Bian Y., Canesi M., Damiani G., Zhang Y., Yu D., Chen J., Ji X., Sui W., Wang B., Wu S., Kovacs A., Revera M., Wang H., Jing X., Zhang Y., Chen Y., Cao Y. Efficacy and tolerability of bevacizumab in patients with severe Covid-19. Nat. Commun. 2021;12:814. doi: 10.1038/s41467-021-21085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chow J.T., Salmena L. Prediction and analysis of SARS-CoV-2-targeting MicroRNA in human lung epithelium. Genes. 2020:11. doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Centa A., Fonseca A.S., Ferreira S., Azevedo M.L.V., Vaz de Paula C.B., Nagashima S., Machado-Souza C., Miggiolaro A., Baena C.P., de Noronha L., Cavalli L.R. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;320:L405–L412. doi: 10.1152/ajplung.00457.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saini S., Saini A., Thakur C.J., Kumar V., Gupta R.D., Sharma J.K. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Mol. Biol. Res. Commun. 2020;9:83–91. doi: 10.22099/mbrc.2020.36507.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.