Abstract
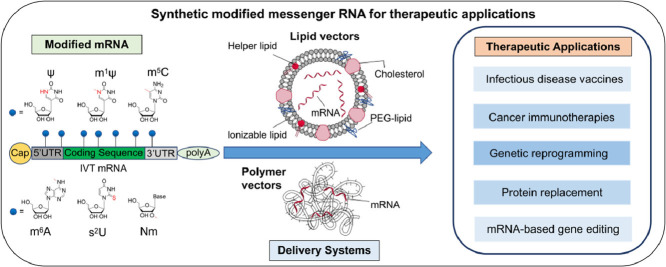
Synthetic modified messenger RNA (mRNA) has manifested great potentials for therapeutic applications such as vaccines and gene therapies, with the recent mRNA vaccines for global pandemic COVID-19 (corona virus disease 2019) attracting the tremendous attention. The chemical modifications and delivery vehicles of synthetic mRNAs are the two key factors for their in vivo therapeutic applications. Chemical modifications like nucleoside methylation endow the synthetic mRNAs with high stability and reduced stimulation of innate immunity. The development of scalable production of synthetic mRNA and efficient mRNA formulation and delivery strategies in recent years have remarkably advanced the field. It is worth noticing that we had limited knowledge on the roles of mRNA modifications in the past. However, the last decade has witnessed not only new discoveries of several naturally occurring mRNA modifications but also substantial advances in understanding their roles on regulating gene expression. It is highly necessary to reconsider the therapeutic system made by synthetic modified mRNAs and delivery vectors. In this review, we will mainly discuss the roles of various chemical modifications on synthetic mRNAs, briefly summarize the progresses of mRNA delivery strategies, and highlight some latest mRNA therapeutics applications including infectious disease vaccines, cancer immunotherapy, mRNA-based genetic reprogramming and protein replacement, mRNA-based gene editing.
Statement of significance
The development of synthetic mRNA drug holds great promise but lies behind small molecule and protein drugs largely due to the challenging issues regarding its stability, immunogenicity and potency. In the last 15 years, these issues have beensubstantially addressed by synthesizing chemically modified mRNA and developing powerful delivery systems; the mRNA therapeutics has entered an exciting new era begun with the approved mRNA vaccines for the COVID-19 infection disease. Here, we provide recent progresses in understanding the biological roles of various RNA chemical modifications, in developing mRNA delivery systems, and in advancing the emerging mRNA-based therapeutic applications, with the purpose to inspire the community to spawn new ideas for curing diseases.
Keywords: Synthetic modified mRNA, Therapeutic mRNA, RNA base modifications, mRNA vaccines, mRNA delivery
Graphical Abstract
1. Introduction
Messenger RNA (mRNA) lies in the middle layer of central dogma serving as the carrier in the genetic information flow from DNA to protein. Synthetic mRNA for therapeutic applications has attracted considerable interest from both academia and industry since it was first demonstrated to be transferred to and expressed in the mouse muscle in vivo in 1990 [1]. The real applications have been expanded to infectious disease vaccines, cancer immunotherapy, genetic engineering, protein replacement therapy, and mRNA-based gene editing [[2], [3], [4], [5], [6], [7], [8], [9]]. The field of mRNA therapeutics is gaining the momentum in recent years because the challenging issues like RNA instability, high innate immunogenicity, and targeted delivery have been substantially addressed by developing chemically modified mRNAs and sophisticated delivery strategies.
Synthetic mRNA can be cost-effectively produced in a large scale by the in vitro transcription technique, and be delivered for fast and transient expression in cytoplasm without the risk of integration into genome, however, the unmodified synthetic mRNA is not stable and stimulates the activation of cellular innate immunity. The synthetic unmodified mRNA largely suffers from the degradation issue due to the high activity of environmental RNases. Upon entering cells, the unmodified RNA molecules can induce the production of type I interferons, interleukin-6, and other proinflammatory cytokines through multiple signaling receptors such as endosomal Toll-like receptor (TLR) 3, 7, or 8, and cytoplasmic protein kinase R (PKR), 2′-5′-oligoadenylate synthetase (OAS), retinoic-acid inducible gene I (RIG-I) or melanoma differentiation-associated 5 (MDA5) [[10], [11], [12]].
In order to improve the stability and reduce the immunogenicity of synthetic mRNA, chemical modifications on the ribose, the RNA termini, and nucleobases have been proposed and examined, and a substantial success has been made in the last 15 years [[13], [14], [15], [16]]. It should be pointed out that we had limited knowledge on the roles of these mRNA modifications in the past, in spite of the observed reduced immunogenicity effect, because some modifications applied did not naturally occur in cellular mRNA or were not known to exist in cellular mRNA at the time of their uses. In the last decade, the rapid advances of emerging RNA epitranscriptome field have witnessed a discovery of several new chemical modifications on cellular mRNA and a substantial understanding on their roles in regulating gene expression [15]. Some of the synthetic mRNA modifications such as N 6-methyladenosine (m6A), pseudouridine (Ψ), 5-methylcytidine (m5C), and 2′-O-methylated nucleoside (Nm) in synthetic mRNA are also naturally occurring in cellular mRNA, therefore it is highly necessary to reconsider the therapeutic system made by synthetic modified mRNA. The cellular mRNA m6A modification found in 1970′s is the most extensively studied, and has been found to play an essential role in mRNA splicing, export, stability, and translation efficiency, significantly influencing the organismal physiology and pathology processes [[17], [18], [19], [20], [21]]. The cellular mRNA modifications including m5C, Ψ and Nm have also been actively investigated in recent years, and have revealed significant biological roles [[22], [23], [24], [25]]. Of note, m5C, Ψ and N 1-methylpseudouridine (m1Ψ) are the widely used modified nucleobases in therapeutic mRNA since they exhibited reduced immunogenicity and enhanced translation efficiency.
The delivery of macromolecular mRNA is another key issue for therapeutic applications. The RNA molecules are negatively charged polynucleotides. Various carriers for the in vivo delivery of mRNA molecules have been developed, including cationic lipid nanoparticles, cationic polymers and others [[26], [27], [28], [29], [30], [31], [32]]. The formulations of modified mRNA with specific vector have been thoroughly optimized to achieve a proper balance between the circulation time and penetration of the cell membrane barrier.
In this review, we will mainly discuss the roles of various chemical modifications on synthetic mRNAs, briefly summarize the progresses of mRNA delivery strategies, and highlight some latest therapeutic applications including infectious disease vaccines, cancer immunotherapies, genetic reprogramming, protein replacement, and mRNA-based gene editing. At last, the concerns and perspectives of mRNA therapeutics are discussed.
2. Chemical modifications of synthetic mRNA
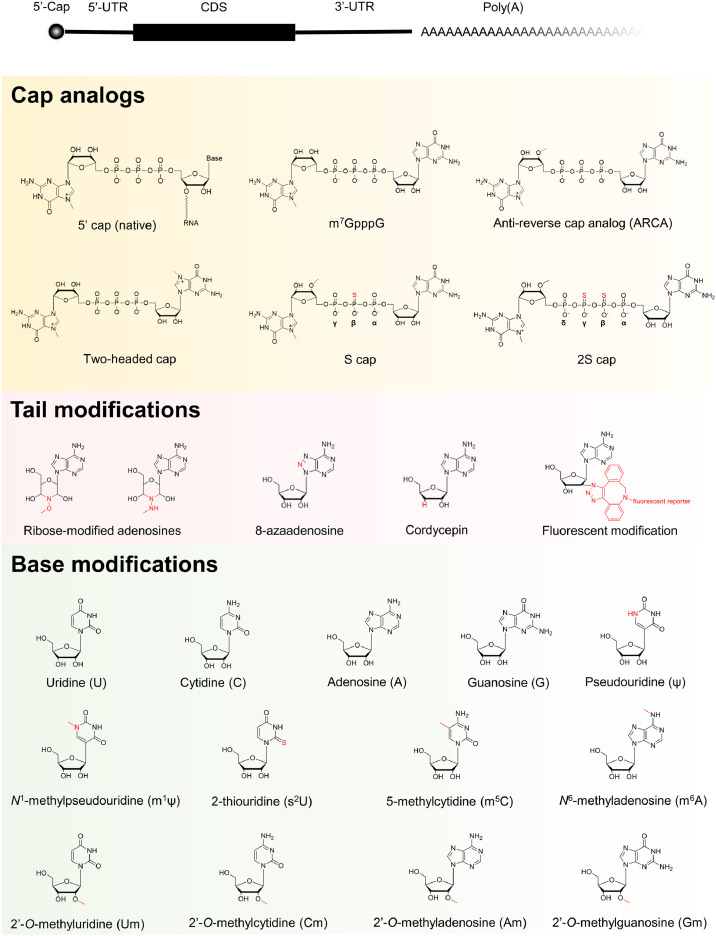
The cellular mRNA consists of elements including 5′-cap, 5′-untranslated region (5′-UTR), coding sequence region (CDS), 3′-UTR, and polyadenylated tail (poly(A)) (Fig. 1 ). The development of in vitro transcription technique has enabled a large scale production of synthetic mRNA, however, the unmodified synthetic mRNA suffers from poor stability and translation efficiency issues. The half-life and translation efficiency of in vitro transcribed mRNA (IVT mRNA) significantly affects the pharmacokinetic and pharmacodynamics properties of mRNA-based therapeutics. To enhance intracellular stability and translational efficiency of mRNA, variety chemical modifications compatible with the IVT system have been systematically examined, including modified termini, bases and ribose [2,[13], [14], [15], [16]] (Fig. 1).
Fig. 1.
An overview of essential structural elements and chemical modifications of the in vitro transcribed mRNA. UTR, untranslated region; CDS, coding sequence region.
2.1. Modifications of mRNA cap and tail
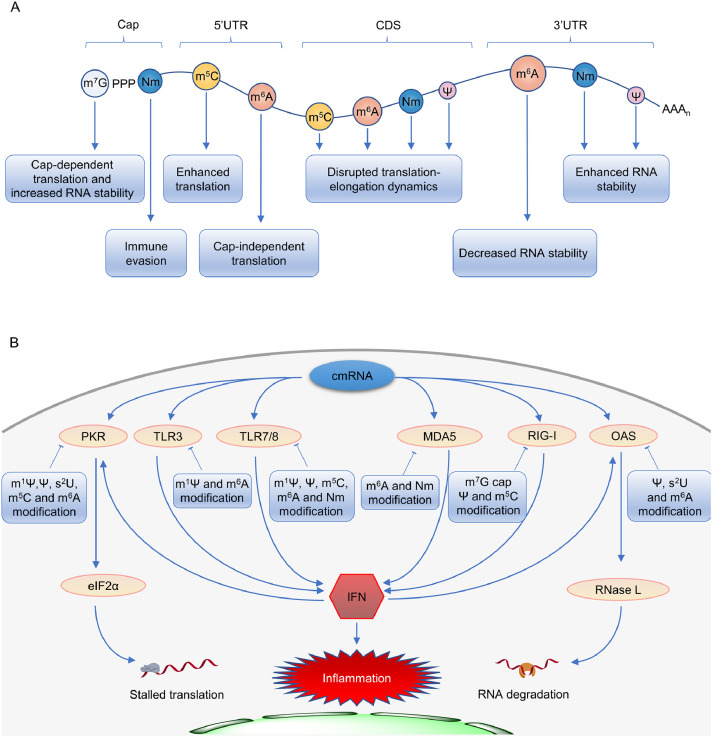
The 5′-cap and poly(A) tail are the hallmarks of eukaryotic mRNA. The native form of 5′-cap in cellular mRNA is featured with 7-methylguanosine (m7G) triphosphate group, which is formed in co-transcriptional process and is able to protect mRNA from 5′end degradation by cellular exonucleases [33]. During mRNA lifecycle inside the cell, there are intrinsic m7G-cap-dependent mRNA translation initiation mediated by the translation initiation factor 4E (eIF4E), and m7G-cap-associated mRNA decay mediated by the decapping complex DCP1/DCP2 [[33], [34], [35]] (Fig. 2 A).
Fig. 2.
Schematic illustrations of modified nucleosides and their functions on cellular and synthetic mRNAs. (A) The naturally occurring modified nucleosides on cellular mRNA and their biological functions. (B) The cellular innate immune responses to the chemically modified mRNA delivered to cells in a simplified view. m7G, 7-methylguanosine; Nm, 2′-O-methylated nucleoside; cmRNA, chemically modified RNA; PKR, protein kinase R; TLR, Toll-like receptor; MDA5, melanoma differentiation-associated 5; RIG-I, retinoic-acid inducible gene I; OAS, 2′-5′-oligoadenylate synthetase; IFN, interferon.
The cap mimics in synthetic mRNA were designed and made in order to enhance its translation efficiency and stability, as well as to reduce intracellular innate immunogenicity by impeding stimulation of RIG-I [36,37] (Fig. 2 B). The m7G can be capped to the 5′-end of IVT mRNA by Vaccinia virus capping enzymes, which are commercially available and widely used for post-transcriptional in vitro capping [38,39]. Alternatively, chemically synthesized dinucleotide m7GpppG or its analogs (Fig. 1) can serve as the first monomer to be directly incorporated into the RNA 5′-end during the in vitro transcription. The G marked in bold in m7GpppG or its extended form m7GpppNG is generally required. It should be noted that the cap dinucleotide (m7GpppG) is able to initiate the growth in two directions from 3′-OH groups on both m7G and G nucleoside moieties. To eliminate the unwanted growth from the 3′-OH in m7G, the anti-reverse cap analog (ARCA) [40] was designed with 3′-OH methylated to form dead end 3′-OMe. In addition, the symmetric two-m7G-headed cap [41] has been developed, and the outcome of either way of growth is the same. Furthermore, the reported 2′,4′-locked nucleic acid-modified [42] and 3′-O-benzyl-modified [43] caps provide the synthetic mRNA with resistance to decapping by the mRNA-decapping enzyme, leading to a longer half-life and more translation.
Apart from the modifications in guanosine, chemical manipulations of phosphate bridge have also yielded improved properties. The substitution of oxygen with sulfur at the β position of the triphosphate bridge in m7GpppG results in high affinity of the cap to eIF4E and low susceptibility to the decapping complex DCP1/DCP2 [[44], [45], [46]]. In addition, tetraphosphate bridge has been found to exhibit a higher affinity to eIF4E compared with the triphosphate bridge [47]. The 2S cap analog shown in Fig. 1 combines tetraphosphate bridge, disulphur-to-oxygen substitution, and 3′-OMe in ARCA, and shows enhanced translation in human immature dendritic cells and has superior performance to the previously published phosphate-modified cap analogs (m2 7,2′ O-GppspG) used in clinical trials [48]. In recent years, a naturally occurring Cap1 structure (e.g., m7GpppAmG) has been synthesized by a co-transcriptional capping method [49]. The reported efficiency of co-transcriptional capping is around 80−90%, and the uncapped mRNA can be removed by exonuclease Xrn1 [50,51].
The 3′-end of cellular mRNA in eukaryotes is decorated with poly(A) which plays a key role in regulating mRNA stability and translation efficiency [33]. The poly(A)-tailed RNA can be produced by directly transcribing from DNA template or be post-transcriptionally added to 3′-end of existing RNA by using a recombinant poly(A) polymerase [2]. The latter method is able to introduce chemically modified adenosines or mimics to the poly(A) tail, for instance, ribose-modified adenosines, cordycepin, 8-azaadenosine, and fluorophore-modified adenosine (Fig. 1). Most of these tail modifications stay at the last 3′-end of synthetic mRNA and can significantly reduce the possibility of mRNA degradation by 3′-to-5′ exonucleases. As a result, synthetic mRNA is endowed with a lengthened half-life and an increased translation efficiency in vitro and in vivo [[52], [53], [54]]. The fluorophore-labeled mRNA can be visualized during its metabolic lifecycle [54]. Of note, there are also other strategies to improve the translation and stability of IVT mRNA inside cells such as introduction of stabilizing elements [55,56], secondary structure [57], high GC content [58], and optimized codon [59,60], but these contents are not the focus of this review.
2.2. Modifications of pyrimidine bases
Cytosine (C), uracil (U), adenine (A) and guanine (G) are four canonical bases of mRNA. The AU-rich mRNAs generally exhibit a lower protein production yield than the GC-rich ones [58], and the U-rich RNA patches are known potent activators of the innate immune system through the TLRs [61]. It has been reported that 0.2−0.4% chemically modified bases (e.g., m5C, Ψ, m6A) [60] in synthetic RNA are sufficient to impede activation of TLR signaling and resistance to cleavage by RNase L [13,63]. The replacement of natural U/C with Ψ/m5C (Fig. 1) represented the best practice in the past, because they could reduce the immunogenicity of mRNA as well as increase its translation efficiency both in vitro and in vivo [62,[64], [65], [66], [67]]. Ψ impedes stimulation of TLR7/8, PKR, OAS and RIG-I-mediated signaling pathways [13,63,68] (Fig. 2 B). It should be emphasized that these cellular receptors sense not only the presence or absence of certain modifications but also the RNA strand feature (single-stranded or double-stranded RNA), and base modification location and combination [68]. The Ψ modification is the first RNA modification discovered in 1951 [[69], [70], [71]], and is the most abundant RNA modification with an estimated Ψ/U ratio of 7–9%. Compared to normal U, Ψ exhibits a stronger hydrogen bonding effect in Watson-Crick base pairing. Although Ψ has been known for decades to exist in tRNA, rRNA and snRNAs [72], its presence in cellular mRNA was recently identified [69]. Several pseudouridine synthases have been shown to modify human mRNA, including PUS1, PUS7, TRUB1, TRUB2 and RPUSD2 [23]. The Ψ-modified RNA could reduce its affinity to cytoplasmic RNA-binding protein PUM2 and nuclear splicing factor MBNL1 in vitro, but it is still unclear in vivo. The majority of Ψ’s in native mRNA response to environmental stimuli, while the biological functions of Ψ are not yet fully understood (Fig. 2 A). Some in vitro results suggest that Ψ may plays a key role in regulating translation. The presence of Ψ has been reported to alter the dynamics of the translating ribosomes possibly due to the stabilized mRNA secondary structure contributed by the enhanced H-bond of Ψ, which is equivalent to the GC content increase [57,69]. It should be pointed out that different systems give rise to different outcomes: in the rabbit reticulocyte system, the replacement of U with Ψ in the synthetic mRNA promotes translation, while in the wheat germ and the Escherichia coli systems, Ψ slightly repressed and blocked translation, respectively [73,74]. Another research shows that Ψ within the CDS of mRNA was not sufficient to reduce or enhance the translation efficiency in vivo [75].Together, the biological new findings about Ψ will definitely prompt us to reconsider the roles of Ψ modification in synthetic mRNA for therapeutic applications, and will also guide us to make new constructs with better performances. m5C has also been reported to naturally occur in mRNA [24]. NSUN2 and NUSN6 were found to exhibit m5C methyltransferase activity for mRNA. The nuclear ALYREF and DNA/RNA-binding YBX1 are capable of binding m5C to facilitate mRNA nuclear-cytoplasmic shuttling and improve mRNA stability, respectively. Another functional role of m5C in native mRNA refers to its influence on mRNA translation. A high m5C level in the CDS strongly impairs mRNA translation efficiency, while increased methylation in the 3′-UTR leads to improved translation [74,[76], [77], [78], [79]]. However, an opposite result was reported that the existence of numerous m5C's in CDS does not significantly affect the translation process [75] (Fig. 2 A). These progresses in m5C biology could help understand why synthetic m5C-modified mRNA has superior performance for therapeutic applications. The possible explanation is that m5C plays roles in avoiding the stimulation of cellular immune system through preventing the stimulation of TLR7/8, PKR and RIG-1 (Fig. 2 B), and stabilizing the synthetic mRNA from degradation, leading to a longer half-life [13,24,67,68].
Recently, m1Ψ modification (Fig. 1) has been designed and incorporated into synthetic mRNA, which outperform the current state-of-the-art Ψ-modified mRNA by offering reduced cellular innate immunogenicity (Fig. 2 B), improved cellular viability, and increased gene expression [65,80,81]. The high protein expression closely correlates with the increased secondary structure in m1Ψ-containing mRNA [57] and the attenuated eIF2α phosphorylation concurrent with diminished PKR activation which inherently inhibits the protein translation pathway [67,81]. Other uridine analogs like 5-methyluridine (m5U), 5-methyoxy-uridine (mo5U), and 2-thiouridine (s2U) (Fig. 1) could also reduce the mRNA immunogenicity, but find limited applications due to their lower translation performance than Ψ [37,49,66,73,82].
2.3. Modifications of purine bases
The m7G modification is well known to exist at mRNA 5′-cap, while it has been recently discovered to exist in the internal region of cellular mRNA and has exhibited a regulatory role to enhance mRNA translation [15,83]. So far, there has not been any trial for incorporation of m7G in synthetic mRNA for therapeutic applications.
Discovered in 1974, m6A is naturally occurring and the most prevalent internal modification in cellular mRNA [84]. In the last decade, the mRNA m6A field has experienced a revival and attracted a surge of interest because m6A has revealed significant biological functions in regulation of gene expression at a post-transcriptional level [[17], [18], [19], [20], [21]]. The m6A methylation is catalyzed by a nuclear methyltransferase complex (“writers”) consisting of METTL3, METTL14 and other auxiliary components WTAP/VIRMA/ZC3H13/HAKAI/RBM15. m6A is a highly dynamic and reversible mark, and can be removed by FTO and ALKBH5 (“erasers”) [85]. The m6A functions are mediated by “readers” proteins, for instance, YTH domain-containing proteins YTHDF1–3 and YTHDC1–2, and IGF2BPs [86,87]. The cellular m6A modification almost influences every step of mRNA in its lifecycle including pre-mRNA processing, export, decay, and translation. It is worth noticing that the location of m6A modification across mRNA determines its roles to a large extent, for examples, 5′-UTR m6A is able to initiate the cap-independent mRNA translation, CDS m6A impairs translation, and 3′-UTR m6A largely modulates mRNA stability [[87], [88], [89]] (Fig. 2 A). The translational inhibition was strongly dependent on the position of the m6A modification within a codon, and the strongest impact occurred when it is located at the first codon position [75]. This is possibly because the m6A in the first codon position can induce local perturbations within the decoding site during translation initiation, leading to a destabilization of the codon-anticodon helix [88].These regulatory effects are generally mediated by the effector reader proteins in a context-dependent manner; different cell lines and stress conditions may lead to different biological consequences.
Synthetic IVT mRNAs with a partial (e.g., 5%) or full replacement of A with m6A have revealed their ability to escape from recognition by innate immune system, however, their translatability is either not improved or completely abolished [73,90,91]. It is understandable that m6A modification reduces the binding of TLR3/7/8, PKR, MDA5, OAS and RIG-I to the synthetic mRNA, thus avoiding the stimulation of innate immune system [13,63,67,68,92] (Fig. 2 B). In terms of low translation efficiency, the possible reason is that m6A location in the synthetic IVT mRNA is randomized and its distribution pattern at CDS might disrupt the tRNA selection and translation-elongation dynamics [88]. Further, high density of m6A modification in CDS and 3′-UTR facilitates the YTHDF2-mediated mRNA decay [86,87]. These factors are likely dominating in the production of protein, although 5′-UTR m6A has a role for cap-independent translation initiation [89].
2.4. Modifications of ribose
The methylation at 2′-OH of ribose (2′-OMe) represents one of the most common RNA modifications found in a variety of cellular RNAs, and naturally occurs in all four canonical nucleosides (Nm: Am/Gm/Cm/Um) and other non-canonical nucleosides [25]. Recently, the Nm sites in mammalian cellular mRNA have been mapped transcriptome-wide at base resolution, however, its effector proteins and biological functions remain elusive [25,93]. From the structural point of view, 2′-OMe in Nm does not affect Watson-Crick base-pairing, while it increases the RNA resistance to degradation due to the abolishment of the hydrogen bonding and nucleophilic characters of 2′-OH [25] (Fig. 2 A). In addition, the absence of the proton at 2′-OH negatively affects TLR7/8 and MDA5 binding (Fig. 2 B), facilitating the escape of 2′-OMe-modified RNA from recognition by the innate immune system [61,94]. However, synthetic mRNA bearing 2′-OMe modification reveals a compromised translation efficiency when the modification occurs at the second nucleotide in the code [74,75], most likely due to the steric clash of the methyl group with the nucleotide A1824 in the eukaryotic18S rRNA [74,75,95]. Besides 2′-OMe, 2′-O-fluoro modification at ribose in synthetic mRNA has been shown to limit the inflammatory responses [96]. Interestingly, 2′-O-fluoro modified RNA was found to bind RIG-I with high affinity and to efficiently activate RIG-I receptor in vitro, while this signaling effect was not observed in vivo [68]. The ribose modifications await further investigations in the future.
3. Synthetic mRNA delivery systems
Efficient and safe delivery of synthetic mRNA represents one key challenge in the development of mRNA-based therapeutics. The synthetic RNA is a negatively charged macromolecular polynucleotide; its naked form is subjected to degradation by serum nucleases and is unable to readily pass through the cell membrane which contains the negatively charged lipid bilayer structure. Therefore, a vector or carrier (e.g., lipid and polymer) is generally needed to deliver the synthetic mRNA to the target cells, and the formulation of mRNA/vector into specific shape of nanoparticles in the presence of certain auxiliary components represents the major delivery strategy. An ideal delivery system should take the following issues into account: (i) biological safety, (ii) circulation time mainly influenced by renal clearance through glomerular filtration (8 nm cutoff) and mononuclear phagocyte system (MPS) clearance, and (iii) the performance of targeted cellular uptake and endosomal release [26].
Although viral vectors have been used to deliver DNA, they have limitations for mRNA delivery mainly due to their small packing size, immunogenicity, cytotoxicity, and complex production process [10,27,29]. In contrast, non-viral vector systems are widely used because they are safe and their productions are simple, economical and reproducible. Although protamine was used earlier to stabilize and deliver RNA, lipids and polymers have found the most applications. There are already a few excellent reviews summarizing various carrier materials [[26], [27], [28], [29], [30], [31], [32]]; we here only briefly discuss the two major classes of carriers, viz. lipids and polymers.
3.1. Lipid and lipidoid vectors
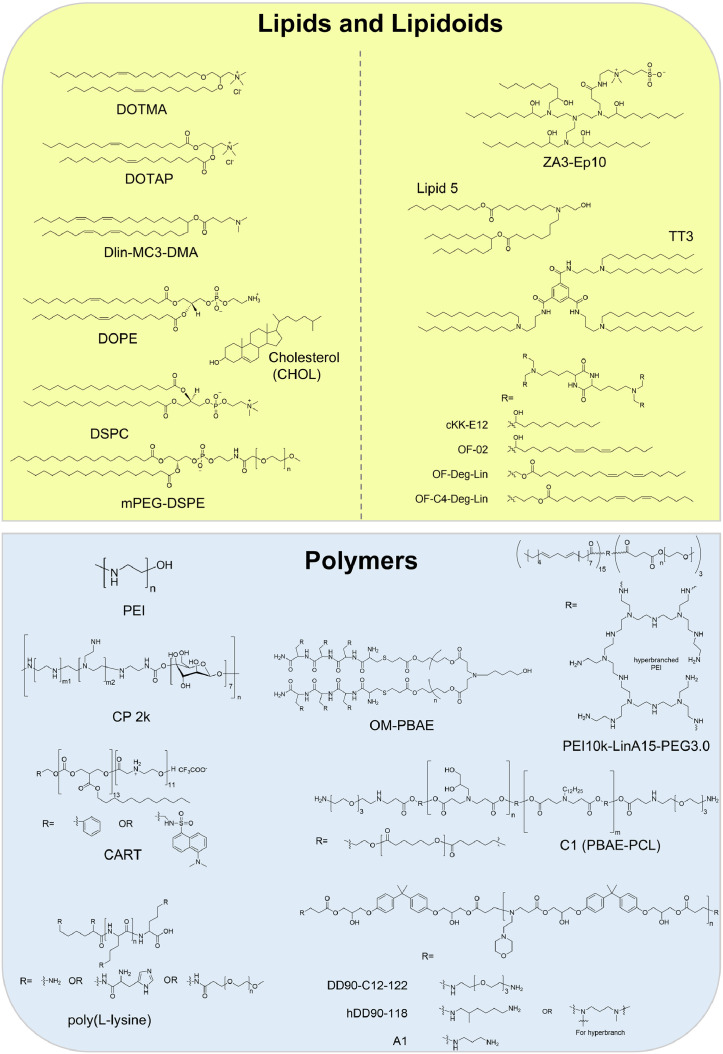
The lipid can be cationic, neutral, and zwitterionic, and it is the major class of vector material for mRNA delivery. The lipid molecules can be formulated with synthetic mRNAs into various nanostructures such as lipid nanoparticle (LNP) and liposome. The LNP has become one of the most commonly used mRNA delivery strategies due to its large payload, stability, and biocompatibility [97,98]. The molecular structure of a lipid is an important determinant of its cellular transfection efficiency and toxicity. The transfection efficiency of lipid is linked to its ability to complex with and release mRNA, and the toxicity is mainly dictated by the structure of its headgroup. The cationic lipid is able to mediate the electrostatic interaction between LNP and the cellular plasma or endosomal membrane, and facilitates cellular internalization and endosomal release of synthetic RNA [27,99]. The synthetic N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTMA) (Fig. 3 ) is the first cationic lipid used to deliver mRNA, which is applicable to human, rat, mouse, xenopus and drosophila cells in vitro [100]. Compared to DOTMA, its derivative named 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) (Fig. 3) exhibits higher cost-effectiveness and efficiency [29,101]. When a lipid has an ionizable headgroup instead of a permanently charged one, the toxicity in vivo could be reduced [16,102]. For instance, Dlin-MC3-DMA (Fig. 3), a component of Patisiran drug, has been approved by the FDA for small RNA interference therapy [103], and has also been used as a vehicle for transferring synthetic mRNA [104,105].
Fig. 3.
Structures of representative vectors for synthetic mRNA delivery.
Cationic lipids can be cytotoxic when used alone for mRNA formulation because they carry a high density of positive charge; the introduction of helper lipids into the formulation recipe could help address the issue. The helper lipids, for instance, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), phosphatidylcholine (PC), PEGylating lipid, and cholesterol (CHOL) (Fig. 3), are typically included as LNP components to enhance the particle stability and blood compatibility, and hence the RNA delivery efficiency [97,99]. DOPE and its derivatives [100,101,[104], [105], [106]] have a relatively small headgroup of phosphoethanolamine and two bulky and unsaturated oleoyl chains, and are featured with a cone-like shape. This lipid geometry can stabilize the non-bilayer hexagonal phase, which is found in the transition structures during membrane fusion and/or disruption. CHOL can stabilize lipid bilayers in the presence of serum proteins by filling in the gaps between phospholipids [[107], [108], [109]], and promote membrane fusion [110]. The modified PC's like distearoylphosphatidylcholine (DSPC) (Fig. 3) and hydrogenated soybean PC (HSPC), favor the formation of bilayer phase with high melting temperatures due to their cylindrical geometry. Therefore, they have been widely used to construct highly stable liposomes and LNPs with optimal in vivo serum stability [111]. The PEGylating lipid of mPEG-DSPE (methoxypolyethyleneglycol-distearoylphosphatidylethanolamine) (Fig. 3) can increase the LNP stability and resistance to serum protein opsonization and MPS clearance in vivo, leading to a longer circulation time for LNPs [98].
Hitherto, the combination of ionizable/cationic lipid and helper lipid to encapsulate synthetic mRNAs have represented the most popular choice to make LNP for both cellular and in vivo applications. The classic LNP contains four components: a cationic lipid, the modified PC or PE, CHOL and PEGylating lipid. For example, ionizable cationic lipid Dlin-MC3-DMA, PC, CHOL, and PEGylating lipid with a ratio of 50/10/38.5/1.5 (mol/mol) were utilized to encapsulate mRNA at an RNA to total lipid ratio of around 0.05 (wt/wt) [104,105,112,113]. There is a vast room to explore in order to find the best combinations.
In order to open up more possibilities for lipid-based vectors, lipidoids molecules have been developed for the delivery purpose [114]. Compared with conventional ionizable/cationic lipids, lipidoids introduce more artificial chemical constructs, e.g., structurally different backbone amine with reversible protonation, inverted ester linkage made by aliphatic alcohol and polyacid, and multiple lipid tails. For instance, cKK-E12 has a diketopiperazine core and has been shown to enhance the serum stability [[115], [116], [117], [118]] (Fig. 3). Several cKK-E12 derived lipidoids, including OF-02, OF-DegLin and OF-C4-Deg-Lin, have been proved to efficiently deliver mRNA for protein expression through systemic administration [[119], [120], [121]]. Recently, some other lipidoids have been reported for mRNA delivery, such as ZA3-Ep10, lipid 5, TT3, C12–200, 5A2-SC8, TNT derivatives, LP01, C14–113, MPA-A/MPA-B, 306Oi10, ssPalm/ssPalmO-Paz4-C2 and ATX-100 [2].
3.2. Polymer vectors
Polymer vectors have shown a great potential as another category of mRNA carriers, although not as clinically advanced as the lipid-based mRNA delivery system, (Fig. 3). Polymeric carriers exhibit significant nucleic acid delivery capabilities and higher stability, and can be used alone or together with helper lipids [122]. Polyethylenimine (PEI) is one of the major polymers used for nucleic acid delivery, which contains a large number of amine units [123]. The PEI can be complexed with mRNA through multiple amino group/phosphate group interaction, and its remaining positive charges after complexation facilitate the interaction of PEI/mRNA complex with the cell membrane and the entry into the target cell. Furthermore, amino groups are responsible for the “proton sponge effect” which can change the osmolarity of acidic endosome leading to the release of loaded mRNA. Due to these unique properties, PEI has been widely used for nucleic acids delivery both in vitro and in vivo [123]. However, the high density of positive charges and the non-degradable property induce the in vivo toxicity, which is attributed to the interactions between PEI and intracellular proteins, destabilization of lipid cellular membranes, and activation of the immune system [[124], [125], [126], [127]]. Chemical modifications of PEI with PEG, fatty acids or amino acids, and chemical cross-linking of low molecular weight PEI with biodegradable motifs, are two possible solutions to circumvent this issue [128]. Structurally modified PEI's such as cyclodextrin-polyethylenimine 2k conjugate (CP-2k) [129] and PEI10k-LinA15-PEG3.0 [130] have been developed for mRNA delivery in different tissues. However, the strong binding between PEI and mRNA makes the release of RNA from their complex difficult, leading to a low transfection efficiency. [131].
Poly(β-amino ester)s (PBAEs) exhibit lower toxicity, biodegradability and pH-responsive property; they potentially possess the same advantage to PEI in facilitating endosomal release of mRNA [132]. Some PBAEs like OM-PBAE [133], C1 (PBAE-PCL) [134], DD90-C12–122 [135], hDD90–118 [136] and A1 [137] have been used for mRNA delivery. Another type of biodegradable polyester, named charge-altering releasable transporter (CART), has also been developed as an mRNA carrier. Poly(amino acid)s are another type of synthetic polymers to deliver mRNA [[138], [139], [140], [141]], with poly(L-lysine), polyaspartamide [142,143] and polyglutamide [144] being the typical examples.
4. Therapeutic applications
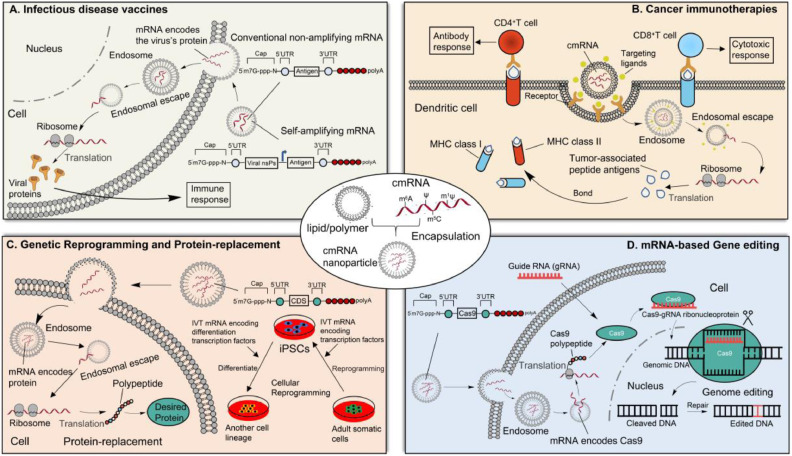
Benefited from the progresses in the knowledge of mRNA modifications and the delivery technologies, the therapeutic applications of synthetic modified mRNAs have developed rapidly in recent years. A broad range of the synthetic modified mRNA-based therapeutic applications are currently being explored, which can be divided into following major areas: infectious disease vaccines, cancer immunotherapy, mRNA-based gene editing, mRNA-based genetic reprogramming and protein replacement (Table 1 ). The mechanisms of the synthetic modified mRNA-based therapeutic applications are shown in Fig. 4 , in which modified mRNAs perform various functions in the different target cells.
Table 1.
Examples of synthetic modified mRNAs for therapeutic applications. LNP, lipid nanoparticle; CNE, cationic nanoemulsion; DENV, dengue virus; prM-E, premembrane and envelope glycoproteins; CAR, chimeric antigen receptors; IL-22BP, interleukin-22 binding protein; VSVMP, vesicular stomatitis virus matrix protein; MUC1, tumor-associated antigens mucin 1; CEA, carcinoembryonic; HER2, human epidermal growth factor receptor 2; MAA, melanoma-associated antigen; VEGF-A, vascular endothelial growth factor-A; CCR5, human C-C chemokine receptor 5; TALENs, transcription activator-like effector nucleases; PKLR, pyruvate kinase L/R gene; Cpf1, CRISPR from Prevotella and Francisella 1.
| Modification type | Delivery system/Method | mRNA encoding | Application | Year | Refs | ||
|---|---|---|---|---|---|---|---|
| m1Ψ | LNP | Spike protein | Infectious disease vaccines | COVID-19 | mRNA-1273 | 2020 | [[157], [160]] |
| m1Ψ | LNP | Spike protein | BNT162 | 2020 | [[159], [161]] | ||
| Unknown | LNP | Spike protein | ARCoV | 2020 | [162] | ||
| m1Ψ | LNP | Haemagglutinin | Influenza | 2019 | [[151], [165]] | ||
| m1Ψ | LNP | Structural proteins | DENV | 2020 | [167] | ||
| m1Ψ | LNP | Glycoproteins C, D, and E | HSV-2 | 2019 | [169,170] | ||
| Unknown | CNE | Glycoprotein G | Rabies | 2020 | [171] | ||
| m1Ψ | LNP | prM-E protein | ZIKV | 2017 | [113,168] | ||
| m1Ψ | LNP | Envelope | HIV-1 | 2019 | [172,173] | ||
| Unknown | T cells transfected ex vivo | CAR-HER2 | Cancer immunotherapy | Glioblastoma | 2013 | [179] | |
| Unknown | T cells transfected ex vivo | CAR-CD19 | Leukemia | 2011 | [180] | ||
| Unknown | DCs transfected ex vivo | IL-22BP; VSVMP | Colon cancer | 2018 | [181,182] | ||
| Unknown | LNP | MUC1, CEA, HER2 etc. | Renal cell cancer | 2011 | [183] | ||
| Unknown | DCs transfected ex vivo | MAA | Melanoma | 2013 | [177] | ||
| Ψ; m5C | Transfection ex vivo | Yamanaka factors | Genetic reprogramming and protein-replacement | Generating iPSCs | 2010 | [64] | |
| Unknown | Transfection ex vivo | VEGF-A | Differentiation from iPSCs | 2016 | [189] | ||
| Unknown | DCs transfected ex vivo | IL-4 | Autoimmune diabetes | 2010 | [190] | ||
| Unknown | Transfected ex vivo | IL-10 | Autoimmune myocarditis | 2012 | [191] | ||
| s2U; m5C | LNP | Surfactant protein B | Congenital lung disease | 2011 | [66] | ||
| Unknown | Transfection ex vivo | VEGF-A | Heart failure | 2020 | [29] | ||
| Unknown | Transfection ex vivo | Cas9 nuclease | mRNA-based gene editing | Editing CAR T cells | 2017 | [194] | |
| Ψ | Transfection ex vivo | Cas9 nuclease | Editing CAR T cells | 2018 | [195] | ||
| Unknown | Transfection ex vivo | Zinc-finger nucleases | Engineering animal models | 2009 | [201] | ||
| Unknown | Transfection ex vivo | Zinc-finger nucleases | Editing HIV-1 CCR5 gene | 2017 | [29] | ||
| Unknown | Transfection ex vivo | TALEN | Editing PKLR gene | 2019 | [203] | ||
| Unknown | Transfection ex vivo | Cpf1 | Engineering animal models | 2016 | [204] | ||
Fig. 4.
Working mechanisms of synthetic modified mRNA-based therapeutic applications. MHC, major histocompatibility complex; iPSCs, induced pluripotent stem cells.
4.1. Infectious disease vaccines
An important application of synthetic modified mRNAs is the vaccination for infectious diseases [145]. Over the past several years, numerous preclinical studies of mRNA vaccines have been executed, and mRNA vaccines become a promising therapeutic tool. There are currently two different types of mRNA vaccines: non-amplifying mRNA and self-amplifying mRNA, with the former conventional one finding more applications (Fig. 4 A). In the self-amplifying mRNA vaccines, the alphavirus nonstructural proteins (nsPs) can form the RNA-dependent RNA polymerase (RDRP) complex, which is responsible for the replication of mRNA [146]. Of note, the self-amplifying mRNA vaccines have shown enhanced antigen expression at lower doses compared to non-amplifying one, possibly leading to improved immunization [147]. However, the clinical data is limited to date and more studies are needed.
Compared to the conventional vaccines (virus vaccines, viral-vector vaccines, protein-based vaccines), mRNA vaccines exhibit several important advantages [148]. They are rapidly and scalably manufactured in a cell-free manner and better able to evade the adaptive immune response; they do not need to enter the nucleus or integrate themselves into the genome, which means higher safety profiles [101,145,[149], [150], [151]]. Many of mRNA vaccines are experiencing in the stages of preclinical and/or clinical trials (Table 1). More systematic and through studies are highly needed to evaluate the efficiencies and side effects of these mRNA vaccines. Collectively, mRNA vaccines have big potentials to solve many of the challenges in vaccine development for infectious diseases [97].
The global pandemic of the COVID-19 has caused tremendous public health crisis [152]; developing a COVID-19 vaccine that is safe, effective, and rapidly deployable is urgently needed. Recently, the innovation and development of technologies in mRNA sequences engineering, mRNA production, and mRNA vaccine delivery materials make the COVID-19 mRNA vaccines possible [153]. By April 2021, more than 14 mRNA vaccines are being developed against COVID-19 by researchers across the world [149,154,155]. The Phase III clinical trials of mRNA vaccines BNT162 (Pfizer/BioNTech Inc.) and mRNA-1273 (Moderna Inc.) [[156], [157], [158], [159], [160], [161]] have been finished (Table 1); the vaccine products have been approved for commercial use. However, both of them need strict cryopreservation and cold chain transportation. Along this line, a thermostable COVID-19 mRNA vaccine named ARCoV (Suzhou Abogen Biosciences Co.) is being developed in phase III clinical trial [162]. Most of the CDS regions of COVID-19 mRNA vaccines encode the coronavirus's spike protein, and most of the delivery systems are LNPs [7,31,148,154,163]. Modified nucleosides are very necessary for mRNA vaccines, which could enhance its stability and reduce adverse immunogenicity [97,148,153]. For example, m1Ψ modifications are incorporated into the mRNAs of BNT162 and mRNA-1273 vaccines [159,160], which can dampen innate immune sensing and increase mRNA translation in vivo [65,73,164]. The results of clinical phase III studies showed that both BNT162 and mRNA-1273 have high efficiency (>90%), revealing that the COVID-19 mRNA vaccines hold great promise.
Besides the COVID-19 mRNA vaccines, many other mRNA vaccines have shown protective immunization in previous preclinical studies against multiple infectious diseases including influenza [151,165,166], dengue virus (DENV) [167,168], herpes simplex virus type 2 (HSV-2) [169,170], Rabies [171], Zika virus (ZIKV) [113,168] and HIV-1 [172,173] (Table 1). Feldman et al. confirmed the safety and immunogenicity of the first mRNA vaccines against pandemic avian H10N8 and H7N9 influenza viruses, which consisted of chemically m1Ψ-modified mRNAs encoding the full-length hemagglutinin [151]. Recently, another m1Ψ-modified H1N1 mRNA vaccine was made, which could provide broad protection in mice [166]. In the HSV-2 mRNA vaccine, the m1Ψ-modified mRNA encodes glycoprotein C, D, and E [169,[169], [170]], while in a rabies vaccine, the glycoprotein G is chosen as antigen and its delivery system is a cationic nanoemulsion (CNE) [171]. Based on the LNP encapsulation strategy, an m1Ψ-modified anti-ZIKV mRNA vaccine encoding the premembrane and envelope (prM-E) glycoproteins was tested for its immunogenicity and protection in mice, and the vaccine elicited rapid and durable protective immunity [113,168]. Recently, an m1Ψ-modified HIV-1 mRNA vaccine encoding the HIV-1 envelope protein was investigated in mice, and an antigen-specific IgG response was observed [172,173]. There are many other ongoing clinical and preclinical trials for mRNA vaccines at present, most of which harness chemically modified mRNA.
4.2. Cancer immunotherapy
Synthetic modified mRNA-based delivery technologies have been widely utilized among both preclinical and clinical studies in the cancer immunotherapy field. The mRNA-based cancer immunotherapy can be divided into two major classes. The first class employs the direct injection of mRNA formulated with delivery materials into target cells like antigen presenting cells (APCs), especially dendritic cells (DCs) [174]. This is similar to that of mRNA vaccine at the cellular level, except that the mRNA encodes a tumor-associated peptide antigen (Fig. 4 B). Following the transfection of DCs with the IVT mRNA encoding an anti-tumor antigen and the subsequent translation, major histocompatibility complex (MHC) class I and II molecules can bind these antigenic epitopes and present them to CD8+ and CD4+ T cells, respectively, leading to the antigen-specific antibody response and cellular immunity, respectively [175,176] (Fig. 4 B). For example, in a phase IB clinical trial, the immunotherapy with DCs which were electroporated with a synthetic mRNA encoding a melanoma-associated antigen (MAA) was found to be safe and effectively stimulating CD8+ and CD4+ T cell responses in melanoma patients [177].
The second class of mRNA-based cancer immunotherapy involves the engineering of chimeric antigen receptors (CAR) T cells [178]. Specifically, synthetic mRNAs encoding CAR are transfected into T cells, and then CAR protein fragments are expressed in T cells. These modified T cells can identify specific tumor cells and kill them. So far, the mRNA-based cancer immunotherapy has been tried to cure many cancers, such as glioblastoma [179], leukemia [180], colon cancer [181,182], renal cell cancer [183], and melanoma [177]. Typical examples of mRNA-based cancer immunotherapy are shown in Table 1. For instance, T Cells electroporated with synthetic mRNA encoding anti-CD19 CAR (CAR-CD19) were used to cure CD19-marked leukemia cell in mice [180]. Various mRNA-based cancer immunotherapies have been evaluated in animal models, and most of them show efficacy and safety.
4.3. Genetic reprogramming and protein replacement
In 2006, Yamanaka et al. found that human somatic cell could be reprogrammed into induced pluripotent stem cells (iPSC) via the forced expression of four transcription factors OCT3/4, SOX2, MYC and KLF4 (now often referred to as Yamanaka factors) [184]. Since the discovery of iPSC, genetic reprogramming has been a topic of interest to regenerative medicine, and previous studies also confirmed that cell fate and functions could be redirected using synthetic modified mRNAs [[184], [185], [186]]. As shown in Fig. 4 C, synthetic modified mRNA encoding Yamanaka factors can be used for reprogramming of adult somatic cells to iPSC [64]. A striking reduction in immunogenicity was observed when the chemically modified Ψ and m5C proportionally replaced the canonical U and C in synthetic mRNAs, respectively [4,64]. In addition, the mRNA-encoded differentiation factors can also promote differentiation of iPSC to a desired cell type [187,188]. For example, in the treatment of myocardial infarction, intramyocardial injection of human vascular endothelial factor A (VEGF-A)-encoding mRNA in mice resulted in the differentiation of heart progenitor cells into endothelial cells, and improved heart function and long-term survival [189].
The protein-replacement therapy is the most straightforward and obvious application for the synthetic modified mRNAs, which can induce expression of desired proteins to replace proteins that are insufficient or aberrant in some diseases (Fig. 4 C). In protein-replacement therapy field, mRNAs are more effective than protein drugs, because a single mRNA molecule can be translated into many copies of proteins [7]. Several diseases (e.g., autoimmune diabetes and autoimmune myocarditis) are being studied with transfected synthetic mRNAs, but most of the studies are at the preclinical stage [190,191] (Table 1). Recently, Moderna and AstraZeneca carried out a phase II clinical trial, in which a naked IVT mRNA encoding VEGF-A is administered into heart failure patients through epicardial injection, and this study demonstrated the positive potential of IVT mRNA-encoded protein replacement therapy [29]. In comparison with the unmodified mRNA, the nucleoside-modified mRNA reveals high performances in the protein-replacement therapy due to their reduced immunogenicity and improved translation efficiency [62]. For example, Kormann et al. performed a protein-replacement therapy through intramuscular injection of modified mRNA into a mouse model having a lethal congenital lung disease caused by the absence of surfactant protein B (SPB). The results showed that the replacement of 25% of U and C with s2U and m5C, respectively, in mRNA synergistically decreased the immunogenicity, and the modified mRNA protected mouse from respiratory failure [66].
4.4. mRNA-based gene editing
In the recent years, clustered regularly spaced short palindromic repeat (CRISPR)-associated protein 9 (Cas9) system has become a powerful tool for site-specific editing of cellular genomes, which hold great promise to permanently cure gene-based diseases [192]. However, there is a concerned risk of off-target mutagenesis. One of the possible reasons is that the prolonged expression of nuclease enzyme Cas9 produced from DNA-based vectors leads to unwanted genome editing [193]. The synthetic mRNA-based gene editing technology is an attractive alternative, because the transient expression of mRNA can minimize the off-target effect (Fig. 4 D). Recently, the universal CAR T cells for cancer immunotherapy (described above) were developed via Cas9 mRNA-mediated gene editing [194,195] (Table 1). The conventional generation of CAR T cells requires a patient's own cells, which is expensive, time-consuming, and sometimes not feasible [196]. Remarkably, through elimination of some genes related to graft-versus-host disease and immunogenicity, the Cas9 editing technology can be used to generate universal CAR T cells with potentials for allogeneic cell therapy [197]. For example, Ren et al. used synthetic Cas9 mRNA to generate universal CAR T cells which were resistant to PD1 inhibition in Nalm6 tumor model (acute lymphoblastic leukemia) [194]. In another study, universal CAR T cells with a potent anti-leukemic effect were generated by Ψ-modified Cas9 mRNA [195].
Apart from Cas9, other nucleases produced from synthetic mRNA were also applied in gene editing, such as zinc finger nucleases (ZFNs) [198], transcription activator-like effector nucleases (TALENs) [199], and Cpf1 (CRISPR from Prevotella and Francisella 1) [200]. For example, in a rat model, the microinjection of ZFN-encoding mRNA in embryo was used to generate heritable knockout mutations at specific loci [201]. In addition, Sangamo Therapeutics is using a ZFN-encoding mRNA product to target the human C-C chemokine receptor 5 (CCR5, a co-receptor essential for HIV-1) gene for the treatment of ex vivo HIV-1 [29,202]. Recently, the TALEN mRNA system was used to correct the pyruvate kinase L/R gene (PKLR) in hematopoietic stem cells for the treatment of pyruvate kinase deficiency (PKD, a disease caused by mutations in the PKLR gene) [203]. Like Cas9, Cpf1 also revealed a gene-editing potential in animal model. The Cpf1 mRNA and its cognate CRISPR-derived RNA were microinjected into the mouse embryos, and a successful gene-edited mouse model was generated [204]. To sum, the application of synthetic modified mRNAs in gene editing is just beginning.
5. Conclusions and perspectives
We have the knowledge that synthetic mRNA undergoes transient protein expression after delivered to cytoplasm and can be completely degraded via physiological metabolic pathways, which can avoid the risk of genomic integration. This transient feature meets the need for many applications which require protein expression for only limited periods of time, such as gene editing, cell reprogramming, and some immunotherapies. However, the synthetic mRNA-based therapeutics also suffer from some drawbacks such as instability, immunogenicity and inefficient delivery, which have long hindered the clinical progression. Nowadays, these issues have been substantially addressed mainly due to our increased knowledge on understanding mRNA modifications and structures, and the advances of nucleic acid delivery technologies.
In the last decade, the field of naturally occurring chemical modifications on cellular mRNA, also known as RNA epigenetics or RNA epitranscriptomics, has been under the spotlight. Among these natural mRNA modifications, m6A is the most abundant and the most extensively studied regarding its functional roles in mRNA fate determination at a posttranscriptional level. The m5C and Ψ modifications were discovered to naturally exist in cellular mRNA in recent years, and are being actively investigated for its functional roles in control of gene expression. The cell itself may have modification code deposited on specific mRNA to regulate its metabolism and fate.
These progresses in understanding the roles of natural mRNA modifications inspire us to rethink about the therapeutic applications using synthetic modified mRNAs. It should be pointed out that a certain base modification tends to be site-specific in natural cellular mRNA at a low modified/unmodified ratio (less than 1%), while it is randomly distributed in synthetic mRNA at a higher modified/unmodified ratio (25−100%). It can be seen that the modification distribution and extent exert a big impact on the RNA functions. The m6A modification in cytoplasmic mRNA, which is enriched near stop codon and at 3′-UTR at 0.3−0.5%, plays pivotal roles in regulating mRNA decay and translation in a context-dependent manner, while the synthetic mRNAs with partial or full substitution of A with m6A are transfected into cell and reveal a very poor translation efficiency, possibly due to the activation of the cellular m6A-mediated decay pathway. The high translation efficiency of m5C- and Ψ-modified synthetic mRNAs is mainly attributed to their excellent stability by avoiding the activation of the RNA-binding protein-mediated degradation pathways. These results remind us that there is a vast room to explore in this field if we can synthesize site-specifically modified mRNA in a tailor-made way. However, it is a lack of such method to prepare a long mRNA molecule in a scalable fashion. Further, our understanding on the biology of natural mRNA modifications needs to be enhanced because there is a lot more knowledge unknown. Recently, there are an increased number of naturally occurring cellular mRNA modifications discovered within CDS, such as m7G [83], N 1-methyladenosine (m1A) [205,206], N 4-acetylcytidine (ac4C) [207], and 5-hydroxymethylcytosine (hm5C) [208]. The toolbox of RNA modifications is expanding. Although the synthetic mRNA-based therapeutics are rapidly developing, a major knowledge gap remains, that is the design rational for modified RNAs, which requires the full understanding of the biological mechanisms of various chemical modification formation on natural cellular mRNA.
With regard to the delivery vectors, new lipid and polymer materials with high biocompatibility, high load efficiency, and low immunogenicity are critically needed. Of note, we should consider more about the mRNA/vector formulation strategies. Although the LNP formulation strategy represents the best choice in current stage to deliver synthetic mRNA, there is still much room for improvement. How to balance the requirements in circulation time of LNP, tissue-specific targeting, and efficient cellular uptake and endosomal release of cargo is still a complex pharmacology issue. More auxiliary elements such as specific cell-targeting and environment-responsive ligands probably need to be incorporated on the surface of LNP.
Finally, the field of synthetic modified mRNA for therapeutic applications is gaining the momentum because of its great potentials. We have witnessed a remarkable progress in miscellaneous applications including infectious diseases vaccines, cancer immunotherapy, genetic reprogramming and protein replacement, and mRNA-based gene editing. The striking achievements are the two approved COVID-19 mRNA vaccines in which the mRNAs are m1Ψ-modified. Other therapeutic products are still in the preclinical or clinical stages and need a long-term validation of efficacy and safety. It is clear that some issues exist in many aspects of the real applications such as the design of modified mRNA constructs, scalable production and purification, the delivery system, immunogenicity, dosage, cell targeting selectivity, and cell transfection and cargo release efficiency. Although the challenges are faced, we see a bright vista.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank National Natural Science Foundation of China (21977087, 91853110, 22022702), National Key Research and Development Program of China (2017YFA0506800), the Fundamental Research Funds for the Central Universities, and Hundred Talents Program of Zhejiang University.
References
- 1.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. 4949 Pt 1. [DOI] [PubMed] [Google Scholar]
- 2.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., Xiao H., Liang X.J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40 doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 3.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28 [Google Scholar]
- 4.Warren L., Lin C. mRNA-Based Genetic Reprogramming. Mol. Ther. 2019;27(4):729–734. doi: 10.1016/j.ymthe.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27(4):757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon H., Kim M., Seo Y., Moon Y.S., Lee H.J., Lee K., Lee H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials. 2018;156:172–193. doi: 10.1016/j.biomaterials.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 7.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2(10):17. [Google Scholar]
- 8.Sergeeva O.V., Koteliansky V.E., Zatsepin T.S. mRNA-based therapeutics - advances and perspectives. Biochemistry (Mosc) 2016;81(7):709–722. doi: 10.1134/S0006297916070075. [DOI] [PubMed] [Google Scholar]
- 9.Vallazza B., Petri S., Poleganov M.A., Eberle F., Kuhn A.N., Sahin U. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond. Wiley Interdiscip. Rev. RNA. 2015;6(5):471–499. doi: 10.1002/wrna.1288. [DOI] [PubMed] [Google Scholar]
- 10.Bessis N., GarciaCozar F.J., Boissier M.C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11:S10–S17. doi: 10.1038/sj.gt.3302364. Suppl 1. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald K.A., Kagan J.C. Toll-like Receptors and the Control of Immunity. Cell. 2020;180(6):1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J., Chen Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 13.Freund I., Eigenbrod T., Helm M., Dalpke A.H. RNA modifications modulate activation of innate toll-like receptors. Genes (Basel) 2019;10(2) doi: 10.3390/genes10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badieyan Z.S., Evans T. Concise review: application of chemically modified mRNA in cell fate conversion and tissue engineering. Stem Cells Transl. Med. 2019;8(8):833–843. doi: 10.1002/sctm.18-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCown P.J., Ruszkowska A., Kunkler C.N., Breger K., Hulewicz J.P., Wang M.C., Springer N.A., Brown J.A. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA. 2020;11(5):e1595. doi: 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchida S., Perche F., Pichon C., Cabral H. Nanomedicine-Based Approaches for mRNA Delivery. Mol. Pharm. 2020;17(10):3654–3684. doi: 10.1021/acs.molpharmaceut.0c00618. [DOI] [PubMed] [Google Scholar]
- 17.Frye M., Harada B.T., Behm M., He C. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B.S., Nachtergaele S., Roundtree I.A., He C. Our views of dynamic N-6-methyladenosine RNA methylation. RNA. 2018;24(3):268–272. doi: 10.1261/rna.064295.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X.M., Zhou J. Multifaceted regulation of translation by the epitranscriptomic modification N(6)-methyladenosine. Crit. Rev. Biochem. Mol. Biol. 2021:1–21. doi: 10.1080/10409238.2020.1869174. [DOI] [PubMed] [Google Scholar]
- 21.Shulman Z., Stern-Ginossar N. The RNA modification N(6)-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 2020;21(5):501–512. doi: 10.1038/s41590-020-0650-4. [DOI] [PubMed] [Google Scholar]
- 22.Stockert J.A., Weil R., Yadav K.K., Kyprianou N., Tewari A.K. Pseudouridine as a novel biomarker in prostate cancer. Urol. Oncol. 2021;39(1):63–71. doi: 10.1016/j.urolonc.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borchardt E.K., Martinez N.M., Gilbert W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020;54:309–336. doi: 10.1146/annurev-genet-112618-043830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue C., Zhao Y., Li L. Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. Biomark. Res. 2020;8(1) doi: 10.1186/s40364-020-00225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayadi L., Galvanin A., Pichot F., Marchand V., Motorin Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim Biophys. Acta Gene Regul. Mech. 2019;1862(3):253–269. doi: 10.1016/j.bbagrm.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Dammes N., Peer D. Paving the Road for RNA Therapeutics. Trends Pharmacol. Sci. 2020;41(10):755–775. doi: 10.1016/j.tips.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and Challenges in the Delivery of mRNA-based Vaccines. Pharmaceutics. 2020;12(2) doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong Z., Mc Cafferty S., Combes F., Huysmans H., De Temmerman J., Gitsels A., Vanrompay D., Portela Catani J., Sanders N.N. mRNA therapeutics deliver a hopeful message. Nano Today. 2018;23:16–39. [Google Scholar]
- 29.Gomez-Aguado I., Rodriguez-Castejon J., Vicente-Pascual M., Rodriguez-Gascon A., Solinis M.A., Del Pozo-Rodriguez A. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials (Basel) 2020;10(2) doi: 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B., Zhang X., Dong Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019;11(2):e1530. doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019;27(4):710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni R., Feng R., Chau Y. Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers. Life (Basel) 2019;9(3) doi: 10.3390/life9030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labno A., Tomecki R., Dziembowski A. Cytoplasmic RNA decay pathways - Enzymes and mechanisms. Biochim. Biophys. Acta. 2016;1863(12):3125–3147. doi: 10.1016/j.bbamcr.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 34.Shirokikh N.E., Preiss T. Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. Wiley Interdiscip Rev RNA. 2018;9(4):e1473. doi: 10.1002/wrna.1473. [DOI] [PubMed] [Google Scholar]
- 35.Gagliardi D., Dziembowski A. 5′ and 3′ modifications controlling RNA degradation: from safeguards to executioners. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373(1762) doi: 10.1098/rstb.2018.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muttach F., Muthmann N., Rentmeister A. Synthetic mRNA capping. Beilstein J. Org. Chem. 2017;13:2819–2832. doi: 10.3762/bjoc.13.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., Endres S., Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 38.Paterson B.M., Rosenberg M. Efficient translation of prokaryotic mRNAs in a eukaryotic cell-free system requires addition of a cap structure. Nature. 1979;279(5715):692–696. doi: 10.1038/279692a0. [DOI] [PubMed] [Google Scholar]
- 39.Fuchs A.L., Neu A., Sprangers R. A general method for rapid and cost-efficient large-scale production of 5′ capped RNA. RNA. 2016;22(9):1454–1466. doi: 10.1261/rna.056614.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel "anti-reverse" cap analogs with superior translational properties. RNA. 2003;9(9):1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grudzien E., Stepinski J., Jankowska-Anyszka M., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel cap analogs for in vitro synthesis of mRNAs with high translational efficiency. RNA. 2004;10(9):1479–1487. doi: 10.1261/rna.7380904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kore A.R., Shanmugasundaram M., Charles I., Vlassov A.V., Barta T.J. Locked nucleic acid (LNA)-modified dinucleotide mRNA cap analogue: synthesis, enzymatic incorporation, and utilization. J. Am. Chem. Soc. 2009;131(18):6364–6365. doi: 10.1021/ja901655p. [DOI] [PubMed] [Google Scholar]
- 43.Shanmugasundaram M., Charles I., Kore A.R. Design, synthesis and biological evaluation of dinucleotide mRNA cap analog containing propargyl moiety. Bioorg. Med. Chem. 2016;24(6):1204–1208. doi: 10.1016/j.bmc.2016.01.048. [DOI] [PubMed] [Google Scholar]
- 44.Grudzien-Nogalska E., Jemielity J., Kowalska J., Darzynkiewicz E., Rhoads R.E. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA. 2007;13(10):1745–1755. doi: 10.1261/rna.701307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kowalska J., Lewdorowicz M., Zuberek J., Grudzien-Nogalska E., Bojarska E., Stepinski J., Rhoads R.E., Darzynkiewicz E., Davis R.E., Jemielity J. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA. 2008;14(6):1119–1131. doi: 10.1261/rna.990208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Tureci O., Sahin U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17(8):961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 47.Strenkowska M., Kowalska J., Lukaszewicz M., Zuberek J., Su W., Rhoads R.E., Darzynkiewicz E., Jemielity J. Towards mRNA with superior translational activity: synthesis and properties of ARCA tetraphosphates with single phosphorothioate modifications. New J. Chem. = Nouveau journal de chimie. 2010;34(5):993–1007. doi: 10.1039/b9nj00644c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strenkowska M., Grzela R., Majewski M., Wnek K., Kowalska J., Lukaszewicz M., Zuberek J., Darzynkiewicz E., Kuhn A.N., Sahin U., Jemielity J. Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential. Nucleic. Acids. Res. 2016;44(20):9578–9590. doi: 10.1093/nar/gkw896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaidyanathan S., Azizian K.T., Haque A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walczak S., Nowicka A., Kubacka D., Fac K., Wanat P., Mroczek S., Kowalska J., Jemielity J. A novel route for preparing 5 ' cap mimics and capped RNAs: phosphate-modified cap analogues obtained via click chemistry. Chem. Sci. 2017;8(1):260–267. doi: 10.1039/c6sc02437h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagarajan V.K., Jones C.I., Newbury S.F., Green P.J. XRN 5 ' ->3 ' exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta-Gene Regul. Mech. 2013;1829(6-7):590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabinovich P.M., Komarovskaya M.E., Ye Z.J., Imai C., Campana D., Bahceci E., Weissman S.M. Synthetic messenger RNA as a tool for gene therapy. Hum. Gene Ther. 2006;17(10):1027–1035. doi: 10.1089/hum.2006.17.1027. [DOI] [PubMed] [Google Scholar]
- 53.Gampe C., White A.C.S., Siva S., Zecri F., Diener J. 3′-Modification stabilizes mRNA and increases translation in cells. Bioorg. Med. Chem. Lett. 2018;28(14):2451–2453. doi: 10.1016/j.bmcl.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Anhauser L., Huwel S., Zobel T., Rentmeister A. Multiple covalent fluorescence labeling of eukaryotic mRNA at the poly(A) tail enhances translation and can be performed in living cells. Nucleic. Acids. Res. 2019;47(7):e42. doi: 10.1093/nar/gkz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makeyev A.V., Liebhaber S.A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8(3):265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adibzadeh S., Fardaei M., Takhshid M., Miri M., Rafiei Dehbidi G., Farhadi A., Ranjbaran R., Alavi P., Nikouyan N., Seyyedi N., Naderi S., Eskandari A., Behzad Behbahani A. Enhancing Stability of Destabilized Green Fluorescent Protein Using Chimeric mRNA Containing Human Beta-Globin 5′ and 3′ Untranslated Regions. Avicenna J. Med. Biotechnol. 2019;11:112–117. [PMC free article] [PubMed] [Google Scholar]
- 57.Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., McFadyen I.J. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. U. S. A. 2019;116(48):24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Courel M., Clement Y., Bossevain C., Foretek D., Vidal Cruchez O., Yi Z., Benard M., Benassy M.N., Kress M., Vindry C., Ernoult-Lange M., Antoniewski C., Morillon A., Brest P., Hubstenberger A., Roest Crollius H., Standart N., Weil D. GC content shapes mRNA storage and decay in human cells. Elife. 2019;8 doi: 10.7554/eLife.49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mauro V.P. Codon Optimization in the Production of Recombinant Biotherapeutics: Potential Risks and Considerations. BioDrugs. 2018;32(1):69–81. doi: 10.1007/s40259-018-0261-x. [DOI] [PubMed] [Google Scholar]
- 60.Plotkin J.B., Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12(1):32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kariko K., Buckstein M., Ni H.P., Weissman D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 63.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Kariko K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic. Acids. Res. 2011;39(21):9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., Daley G.Q., Brack A.S., Collins J.J., Cowan C., Schlaeger T.M., Rossi D.J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parr C.J.C., Wada S., Kotake K., Kameda S., Matsuura S., Sakashita S., Park S., Sugiyama H., Kuang Y., Saito H. N-1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020;48(6):9. doi: 10.1093/nar/gkaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29(2) doi: 10.1038/nbt.1733. 154-U96. [DOI] [PubMed] [Google Scholar]
- 67.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic. Acids. Res. 2010;38(17):5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Durbin A.F., Wang C., Marcotrigiano J., Gehrke L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio. 2016;7(5) doi: 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X., Ma S., Yi C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 2016;33:108–116. doi: 10.1016/j.cbpa.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., Leon-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., Fink G., Regev A. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ge J., Yu Y.-T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013;38(4):210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoernes T.P., Clementi N., Faserl K., Glasner H., Breuker K., Lindner H., Huttenhofer A., Erlacher M.D. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic. Acids. Res. 2016;44(2):852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoernes T.P., Heimdorfer D., Kostner D., Faserl K., Nussbaumer F., Plangger R., Kreutz C., Lindner H., Erlacher M.D. Eukaryotic Translation Elongation is Modulated by Single Natural Nucleotide Derivatives in the Coding Sequences of mRNAs. Genes (Basel) 2019;10(2) doi: 10.3390/genes10020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schumann U., Zhang H.-N., Sibbritt T., Pan A., Horvath A., Gross S., Clark S.J., Yang L., Preiss T. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020;18(1):40. doi: 10.1186/s12915-020-00769-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui X.A., Liang Z., Shen L.S., Zhang Q., Bao S.J., Geng Y.K., Zhang B., Leo V., Vardy L.A., Lu T.G., Gu X.F., Yu H. 5-Methylcytosine RNA Methylation in &ITArabidopsis Thaliana&IT. Mol. Plant. 2017;10(11):1387–1399. doi: 10.1016/j.molp.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 78.Huang T., Chen W.Y., Liu J.H., Gu N.N., Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 2019;26(5):380. doi: 10.1038/s41594-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 79.Li Q., Li X., Tang H., Jiang B., Dou Y.L., Gorospe M., Wang W.G. NSUN2-Mediated m5C Methylation and METTL3/METTL14-Mediated m6A Methylation Cooperatively Enhance p21 Translation. J. Cell. Biochem. 2017;118(9):2587–2598. doi: 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 81.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic. Acids. Res. 2017;45(10):6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li B., Luo X., Dong Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016;27(3):849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 83.Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X., He C. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell. 2019;74(6):1304–1316. doi: 10.1016/j.molcel.2019.03.036. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rottman F., Shatkin A.J., Perry R.P. Sequences containing methylated nucleotides at 5′ termini of messenger-RNAs - possible implications for processing. Cell. 1974;3(3):197–199. doi: 10.1016/0092-8674(74)90131-7. [DOI] [PubMed] [Google Scholar]
- 85.Iyer L.M., Zhang D., Aravind L. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays. 2016;38(1):27–40. doi: 10.1002/bies.201500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berlivet S., Scutenaire J., Deragon J.M., Bousquet-Antonelli C. Readers of the m(6)A epitranscriptomic code. Biochim Biophys. Acta Gene Regul. Mech. 2019;1862(3):329–342. doi: 10.1016/j.bbagrm.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 87.Patil D.P., Pickering B.F., Jaffrey S.R. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 2018;28(2):113–127. doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi J., Ieong K.-W., Demirci H., Chen J., Petrov A., Prabhakar A., O'Leary S.E., Dominissini D., Rechavi G., Soltis S.M., Ehrenberg M., Puglisi J.D. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016;23(2):110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu M., Zhang Z., Xue M., Zhao B.S., Harder O., Li A., Liang X., Gao T.Z., Xu Y., Zhou J., Feng Z., Niewiesk S., Peeples M.E., He C., Li J. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020;5(4):584–598. doi: 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B., Hur S., Chang H.Y. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell. 2019;76(1):96–109. doi: 10.1016/j.molcel.2019.07.016. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Qiu W., Zhang Q., Zhang R., Lu Y., Wang X., Tian H., Yang Y., Gu Z., Gao Y., Yang X., Cui G., Sun B., Peng Y., Deng H., Peng H., Yang A., Yang Y.G., Yang P. N(6)-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 2021;12(1):1582. doi: 10.1038/s41467-021-21904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dai Q., Moshitch-Moshkovitz S., Han D., Kol N., Amariglio N., Rechavi G., Dominissini D., He C. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods. 2017;14(7):695–698. doi: 10.1038/nmeth.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., Siddell S.G., Ludewig B., Thiel V. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Choi J., Indrisiunaite G., DeMirci H., Ieong K.W., Wang J., Petrov A., Prabhakar A., Rechavi G., Dominissini D., He C., Ehrenberg M., Puglisi J.D. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 2018;25(3):208–216. doi: 10.1038/s41594-018-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee Y., Urban J.H., Xu L., Sullenger B.A., Lee J. 2′Fluoro Modification Differentially Modulates the Ability of RNAs to Activate Pattern Recognition Receptors. Nucleic Acid Ther. 2016;26(3):173–182. doi: 10.1089/nat.2015.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nature Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jokerst J.V., Lobovkina T., Zare R.N., Gambhir S.S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (London, England) 2011;6(4):715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug. Deliv. Rev. 2016;99(Pt A):129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 100.Malone R.W., Felgner P.L., Verma I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. U. S. A. 1989;86(16):6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Husemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Tureci O., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 102.Rietwyk S., Peer D. Next-Generation Lipids in RNA Interference Therapeutics. ACS Nano. 2017;11(8):7572–7586. doi: 10.1021/acsnano.7b04734. [DOI] [PubMed] [Google Scholar]
- 103.Zhang X., Goel V., Robbie G.J. Pharmacokinetics of Patisiran, the First Approved RNA Interference Therapy in Patients With Hereditary Transthyretin-Mediated Amyloidosis. J. Clin. Pharmacol. 2019;60(5):573–585. doi: 10.1002/jcph.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jain R., Frederick J.P., Huang E.Y., Burke K.E., Mauger D.M., Andrianova E.A., Farlow S.J., Siddiqui S., Pimentel J., Cheung-Ong K., McKinney K.M., Kohrer C., Moore M.J., Chakraborty T. MicroRNAs Enable mRNA Therapeutics to Selectively Program Cancer Cells to Self-Destruct. Nucleic Acid Ther. 2018;28(5):285–296. doi: 10.1089/nat.2018.0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Szekely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2018;115(15) doi: 10.1073/pnas.1720542115. E3351-E3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., O'Hagan D.T., Singh M., Mason P.W., Valiante N.M., Dormitzer P.R., Barnett S.W., Rappuoli R., Ulmer J.B., Mandl C.W. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U. S. A. 2012;109(36):14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sakurai F., Nishioka T., Yamashita F., Takakura Y., Hashida M. Effects of erythrocytes and serum proteins on lung accumulation of lipoplexes containing cholesterol or DOPE as a helper lipid in the single-pass rat lung perfusion system. Eur. J. Pharm. Biopharm. 2001;52(2):165–172. doi: 10.1016/s0939-6411(01)00165-5. [DOI] [PubMed] [Google Scholar]
- 108.Tenchov B.G., MacDonald R.C., Siegel D.P. Cubic phases in phosphatidylcholine-cholesterol mixtures: cholesterol as membrane "fusogen". Biophys. J. 2006;91(7):2508–2516. doi: 10.1529/biophysj.106.083766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Semple S.C., Chonn A., Cullis P.R. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry. 1996;35(8):2521–2525. doi: 10.1021/bi950414i. [DOI] [PubMed] [Google Scholar]
- 110.Dabkowska A.P., Barlow D.J., Hughes A.V., Campbell R.A., Quinn P.J., Lawrence M.J. The effect of neutral helper lipids on the structure of cationic lipid monolayers. J. R. Soc. Interface. 2012;9(68):548–561. doi: 10.1098/rsif.2011.0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thewalt J.L., Bloom M. Phosphatidylcholine: cholesterol phase diagrams. Biophys. J. 1992;63(4):1176–1181. doi: 10.1016/S0006-3495(92)81681-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sedic M., Senn J.J., Lynn A., Laska M., Smith M., Platz S.J., Bolen J., Hoge S., Bulychev A., Jacquinet E., Bartlett V., Smith P.F. Safety Evaluation of Lipid Nanoparticle-Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet. Pathol. 2018;55(2):341–354. doi: 10.1177/0300985817738095. [DOI] [PubMed] [Google Scholar]
- 113.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., Wagner W., Granados A., Greenhouse J., Walker M., Willis E., Yu J.S., McGee C.E., Sempowski G.D., Mui B.L., Tam Y.K., Huang Y.J., Vanlandingham D., Holmes V.M., Balachandran H., Sahu S., Lifton M., Higgs S., Hensley S.E., Madden T.D., Hope M.J., Kariko K., Santra S., Graham B.S., Lewis M.G., Pierson T.C., Haynes B.F., Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Akinc A., Zumbuehl A., Goldberg M., Leshchiner E.S., Busini V., Hossain N., Bacallado S.A., Nguyen D.N., Fuller J., Alvarez R., Borodovsky A., Borland T., Constien R., de Fougerolles A., Dorkin J.R., Narayanannair Jayaprakash K., Jayaraman M., John M., Koteliansky V., Manoharan M., Nechev L., Qin J., Racie T., Raitcheva D., Rajeev K.G., Sah D.W.Y., Soutschek J., Toudjarska I., Vornlocher H.-P., Zimmermann T.S., Langer R., Anderson D.G. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rybakova Y., Kowalski P.S., Huang Y., Gonzalez J.T., Heartlein M.W., DeRosa F., Delcassian D., Anderson D.G. mRNA Delivery for Therapeutic Anti-HER2 Antibody Expression In Vivo. Mol. Ther. 2019;27(8):1415–1423. doi: 10.1016/j.ymthe.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yin H., Song C.Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K., Mou H., Oberholzer A., Ding J., Kwan S.Y., Bogorad R.L., Zatsepin T., Koteliansky V., Wolfe S.A., Xue W., Langer R., Anderson D.G. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35(12):1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid Nanoparticle Assisted mRNA Delivery for Potent Cancer Immunotherapy. Nano Lett. 2017;17(3):1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., Shi Y., Anderson D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11(1):2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fenton O.S., Kauffman K.J., McClellan R.L., Appel E.A., Dorkin J.R., Tibbitt M.W., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent In Vivo mRNA Delivery. Adv. Mater. 2016;28(15):2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F., Yang J.H., Oberli M.A., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv. Mater. 2017;29(33) doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 121.Fenton O.S., Kauffman K.J., McClellan R.L., Kaczmarek J.C., Zeng M.D., Andresen J.L., Rhym L.H., Heartlein M.W., DeRosa F., Anderson D.G. Customizable Lipid Nanoparticle Materials for the Delivery of siRNAs and mRNAs. Angew. Chem. Int. Ed. Engl. 2018;57(41):13582–13586. doi: 10.1002/anie.201809056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Faghfuri E., Pourfarzi F., Faghfouri A.H., Abdoli Shadbad M., Hajiasgharzadeh K., Baradaran B. Recent developments of RNA-based vaccines in cancer immunotherapy. Expert Opin. Biol. Ther. 2021;21(2):201–218. doi: 10.1080/14712598.2020.1815704. [DOI] [PubMed] [Google Scholar]
- 123.Demoulins T., Milona P., Englezou P.C., Ebensen T., Schulze K., Suter R., Pichon C., Midoux P., Guzman C.A., Ruggli N., McCullough K.C. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomedicine. 2016;12(3):711–722. doi: 10.1016/j.nano.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 124.Moghimi S.M., Symonds P., Murray J.C., Hunter A.C., Debska G., Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol. Ther. 2005;11(6):990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 125.Forcato D.O., Fili A.E., Alustiza F.E., Lazaro Martinez J.M., Bongiovanni Abel S., Olmos Nicotra M.F., Alessio A.P., Rodriguez N., Barbero C., Bosch P. Transfection of bovine fetal fibroblast with polyethylenimine (PEI) nanoparticles: effect of particle size and presence of fetal bovine serum on transgene delivery and cytotoxicity. Cytotechnology. 2017;69(4):655–665. doi: 10.1007/s10616-017-0075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Monnery B.D., Wright M., Cavill R., Hoogenboom R., Shaunak S., Steinke J.H.G., Thanou M. Cytotoxicity of polycations: Relationship of molecular weight and the hydrolytic theory of the mechanism of toxicity. Int. J. Pharm. 2017;521(1-2):249–258. doi: 10.1016/j.ijpharm.2017.02.048. [DOI] [PubMed] [Google Scholar]
- 127.Karimov M., Appelhans D., Ewe A., Aigner A. The combined disulfide cross-linking and tyrosine-modification of very low molecular weight linear PEI synergistically enhances transfection efficacies and improves biocompatibility. Eur. J. Pharm. Biopharm. 2021;161:56–65. doi: 10.1016/j.ejpb.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 128.Stegantseva M.V., Shinkevich V.A., Tumar E.M., Meleshko A.N. Multi-antigen DNA vaccine delivered by polyethylenimine and Salmonella enterica in neuroblastoma mouse model. Cancer Immunol. Immunother. 2020;69(12):2613–2622. doi: 10.1007/s00262-020-02652-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li M., Zhao M., Fu Y., Li Y., Gong T., Zhang Z., Sun X. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control. Release. 2016;228:9–19. doi: 10.1016/j.jconrel.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 130.Dunn A.W., Kalinichenko V.V., Shi D. Highly Efficient In Vivo Targeting of the Pulmonary Endothelium Using Novel Modifications of Polyethylenimine: An Importance of Charge. Adv. Healthc. Mater. 2018;7(23) doi: 10.1002/adhm.201800876. [DOI] [PubMed] [Google Scholar]
- 131.Zhang H., De Smedt S.C., Remaut K. Fluorescence Correlation Spectroscopy to find the critical balance between extracellular association and intracellular dissociation of mRNA complexes. Acta Biomater. 2018;75:358–370. doi: 10.1016/j.actbio.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 132.Lynn D.M., Langer R. Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. J. Am. Chem. Soc. 2000;122(44):10761–10768. [Google Scholar]
- 133.Fornaguera C., Guerra-Rebollo M., Angel Lazaro M., Castells-Sala C., Meca-Cortes O., Ramos-Perez V., Cascante A., Rubio N., Blanco J., Borros S. mRNA Delivery System for Targeting Antigen-Presenting Cells In Vivo. Adv. Healthc. Mater. 2018;7(17) doi: 10.1002/adhm.201800335. [DOI] [PubMed] [Google Scholar]
- 134.Capasso Palmiero U., Kaczmarek J.C., Fenton O.S., Anderson D.G. Poly(beta-amino ester)-co-poly(caprolactone) Terpolymers as Nonviral Vectors for mRNA Delivery In Vitro and In Vivo. Adv. Healthc. Mater. 2018;7(14) doi: 10.1002/adhm.201800249. [DOI] [PubMed] [Google Scholar]
- 135.Kaczmarek J.C., Patel A.K., Kauffman K.J., Fenton O.S., Webber M.J., Heartlein M.W., DeRosa F., Anderson D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. Int. Ed. Engl. 2016;55(44):13808–13812. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Patel A.K., Kaczmarek J.C., Bose S., Kauffman K.J., Mir F., Heartlein M.W., DeRosa F., Langer R., Anderson D.G. Inhaled Nanoformulated mRNA Polyplexes for Protein Production in Lung Epithelium. Adv. Mater. 2019;31(8) doi: 10.1002/adma.201805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kaczmarek J.C., Kauffman K.J., Fenton O.S., Sadtler K., Patel A.K., Heartlein M.W., DeRosa F., Anderson D.G. Optimization of a Degradable Polymer-Lipid Nanoparticle for Potent Systemic Delivery of mRNA to the Lung Endothelium and Immune Cells. Nano Lett. 2018;18(10):6449–6454. doi: 10.1021/acs.nanolett.8b02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Uchida S., Kinoh H., Ishii T., Matsui A., Tockary T.A., Takeda K.M., Uchida H., Osada K., Itaka K., Kataoka K. Systemic delivery of messenger RNA for the treatment of pancreatic cancer using polyplex nanomicelles with a cholesterol moiety. Biomaterials. 2016;82:221–228. doi: 10.1016/j.biomaterials.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 139.Chen Q., Qi R., Chen X., Yang X., Wu S., Xiao H., Dong W. A Targeted and Stable Polymeric Nanoformulation Enhances Systemic Delivery of mRNA to Tumors. Mol. Ther. 2017;25(1):92–101. doi: 10.1016/j.ymthe.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Van der Jeught K., De Koker S., Bialkowski L., Heirman C., Joe P.T., Perche F., Maenhout S., Bevers S., Broos K., Deswarte K., Malard V., Hammad H., Baril P., Benvegnu T., Jaffres P.A., Kooijmans S.A.A., Schiffelers R., Lienenklaus S., Midoux P., Pichon C., Breckpot K., Thielemans K. Dendritic Cell Targeting mRNA Lipopolyplexes Combine Strong Antitumor T-Cell Immunity with Improved Inflammatory Safety. Acs Nano. 2018;12(10):9815–9829. doi: 10.1021/acsnano.8b00966. [DOI] [PubMed] [Google Scholar]
- 141.Moignic A.Le, Malard V., Benvegnu T., Lemiegre L., Berchel M., Jaffres P.A., Baillou C., Delost M., Macedo R., Rochefort J., Lescaille G., Pichon C., Lemoine F.M., Midoux P., Mateo V. Preclinical evaluation of mRNA trimannosylated lipopolyplexes as therapeutic cancer vaccines targeting dendritic cells. J. Control. Release. 2018;278:110–121. doi: 10.1016/j.jconrel.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 142.Li J., Wang W., He Y., Li Y., Yan E.Z., Zhang K., Irvine D.J., Hammond P.T. Structurally Programmed Assembly of Translation Initiation Nanoplex for Superior mRNA Delivery. ACS Nano. 2017;11(3):2531–2544. doi: 10.1021/acsnano.6b08447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li J., He Y., Wang W., Wu C., Hong C., Hammond P.T. Polyamine-Mediated Stoichiometric Assembly of Ribonucleoproteins for Enhanced mRNA Delivery. Angew. Chem. Int. Ed. Engl. 2017;56(44):13709–13712. doi: 10.1002/anie.201707466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schumann C., Nguyen D.X., Norgard M., Bortnyak Y., Korzun T., Chan S., Lorenz A.S., Moses A.S., Albarqi H.A., Wong L., Michaelis K., Zhu X., Alani A.W.G., Taratula O.R., Krasnow S., Marks D.L., Taratula O. Increasing lean muscle mass in mice via nanoparticle-mediated hepatic delivery of follistatin mRNA. Theranostics. 2018;8(19):5276–5288. doi: 10.7150/thno.27847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sahin U., Kariko K., Tureci O. mRNA-based therapeutics - developing a new class of drugs. Nat. Rev. Drug Discovery. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 146.L. Kaariainen, T. Ahola, Functions of alphavirus nonstructural proteins in RNA replication, in: K. Moldave (Ed.), Progress in Nucleic Acid Research and Molecular Biology, Vol 71, Elsevier Academic Press Inc, San Diego, 2002, pp. 187–222. [DOI] [PMC free article] [PubMed]
- 147.Bloom K., van den Berg F., Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020 doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ahammad I., Lira S.S. Designing a novel mRNA vaccine against SARS-CoV-2: An immunoinformatics approach. Int. J. Biol. Macromol. 2020;162:820–837. doi: 10.1016/j.ijbiomac.2020.06.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580(7805):576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 150.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., Fotin-Mleczek M., Hoerr I., Clemens R., von Sonnenburg F. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390(10101):1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 151.Feldman R.A., Fuhr R., Smolenov I., Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson O., Pujar H.S., Laska M.E., Thompson J., Zaks T., Ciaramella G. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37(25):3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 152.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;580(7803):E7. doi: 10.1038/s41586-020-2202-3. E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pardi N., Hogan M.J., Weissman D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020;65:14–20. doi: 10.1016/j.coi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 154.Khan I., Ahmed Z., Sarwar A., Jamil A., Anwer F. The Potential Vaccine Component for COVID-19: A Comprehensive Review of Global Vaccine Development Efforts. Cureus. 2020;12(6):8. doi: 10.7759/cureus.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yi C., Yi Y., Li J. mRNA Vaccines: Possible Tools to Combat SARS-CoV-2. Virologica Sinica. 2020;35(3):259–262. doi: 10.1007/s12250-020-00243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martin C., Lowery D. mRNA vaccines: intellectual property landscape. Nat. Rev. Drug Discov. 2020;19(9) doi: 10.1038/d41573-020-00119-8. 578–578. [DOI] [PubMed] [Google Scholar]
- 157.K.S. Corbett, D. Edwards, S.R. Leist, O.M. Abiona, S. Boyoglu-Barnum, R.A. Gillespie, S. Himansu, A. Schafer, C.T. Ziwawo, A.T. DiPiazza, K.H. Dinnon, S.M. Elbashir, C.A. Shaw, A. Woods, E.J. Fritch, D.R. Martinez, K.W. Bock, M. Minai, B.M. Nagata, G.B. Hutchinson, K. Bahl, D. Garcia-Dominguez, L. Ma, I. Renzi, W.-P. Kong, S.D. Schmidt, L. Wang, Y. Zhang, L.J. Stevens, E. Phung, L.A. Chang, R.J. Loomis, N.E. Altaras, E. Narayanan, M. Metkar, V. Presnyak, C. Liu, M.K. Louder, W. Shi, K. Leung, E.S. Yang, A. West, K.L. Gully, N. Wang, D. Wrapp, N.A. Doria-Rose, G. Stewart-Jones, H. Bennett, M.C. Nason, T.J. Ruckwardt, J.S. McLellan, M.R. Denison, J.D. Chappell, I.N. Moore, K.M. Morabito, J.R. Mascola, R.S. Baric, A. Carfi, B.S. Graham, SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness, bioRxiv: the preprint server for biology (2020).
- 158.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O'Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.-P.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Cook A., Dodson A., Steingrebe K., Shi W. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Tuereci O., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Sahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine. Nature. 2020 doi: 10.1038/s41586-020-2639-4. BNT162b1 in adults. [DOI] [PubMed] [Google Scholar]
- 160.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schafer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A. E.J. Fritch, D.R. Martinez, K.W. Bock, M. Minai, B.M. Nagata, G.B. Hutchinson, K. Wu, C. Henry, K. Bahl, D. Garcia-Dominguez, L. Ma, I. Renzi, W.P. Kong, S.D. Schmidt, L. Wang, Y. Zhang, E. Phung, L.A. Chang, R.J. Loomis, N.E. Altaras, E. Narayanan, M. Metkar, V. Presnyak, C. Liu, M.K. Louder, W. Shi, K. Leung, E.S. Yang, A. West, K.L. Gully, L.J. Stevens, N. Wang, D. Wrapp, N.A. Doria-Rose, G. Stewart-Jones, H. Bennett, G.S. Alvarado, M.C. Nason, T.J. Ruckwardt, J.S. McLellan, M.R. Denison, J.D. Chappell, I.N. Moore, K.M. Morabito, J.R. Mascola, R.S. Baric, A. Carfi, B.S. Graham, SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Tureci O., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Sahin U., Gruber W.C. The New England journal of medicine; 2020. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., Guo Y., Sun S.H., Fan H., Zu S.L., Chen Q., He Q., Cao T.S., Huang X.Y., Qiu H.Y., Nie J.H., Jiang Y.H., Yan H.Y., Ye Q., Zhong X., Xue X.L., Zha Z.Y., Zhou D.S., Yang X., Wang Y.C., Ying B., Qin C.F. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182(5):1271. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Khan K., Dimtri F., Vargas C., Surani S. COVID-19: A Review of Emerging Preventative Vaccines and Treatment Strategies. Cureus. 2020;12(5):6. doi: 10.7759/cureus.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Laczko D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castano D., Amanat F., Muramatsu H., Oguin T.H., 3rd A.Ojha, Zhang L., Mu Z., Parks R., Manzoni T.B., Roper B., Strohmeier S., Tombacz I., Arwood L., Nachbagauer R., Kariko K., Greenhouse J., Pessaint L., Porto M., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Lin P.J.C., Tam Y.K., Sempowski G.D., Farzan M., Choe H., Saunders K.O., Haynes B.F., Andersen H., Eisenlohr L.C., Weissman D., Krammer F., Bates P., Allman D., Locci M., Pardi N. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53(4):724–732. doi: 10.1016/j.immuni.2020.07.019. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30(12):1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 166.Freyn A.W., da Silva J.R., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., Ferreira L.C.D., Weissman D., Krammer F., Coughlan L., Palese P., Pardi N., Nachbagauer R. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020;28(7):1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang M., Sun J., Li M., Jin X. Modified mRNA-LNP Vaccines Confer Protection against Experimental DENV-2 Infection in Mice. Mol. Therapy-Methods Clin. Dev. 2020;18:702–712. doi: 10.1016/j.omtm.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168(6):1114. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Awasthi S., Hook L.M., Pardi N., Wang F.S., Myles A., Cancro M.P., Cohen G.H., Weissman D., Friedman H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019;4(39):15. doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Egan K.P., Hook L.M., Naughton A., Pardi N., Awasthi S., Cohen G.H., Weissman D., Friedman H.M. An HSV-2 nucleoside-modified mRNA genital herpes vaccine containing glycoproteins gC, gD, and gE protects mice against HSV-1 genital lesions and latent infection. PLoS Pathog. 2020;16(7) doi: 10.1371/journal.ppat.1008795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stokes A., Pion J., Binazon O., Laffont B., Bigras M., Dubois G., Blouin K., Young J.K., Ringenberg M.A., Ben Abdeljelil N., Haruna J., Rodriguez L.-A. Nonclinical safety assessment of repeated administration and biodistribution of a novel rabies self-amplifying mRNA vaccine in rats. Regul. Toxicol. Pharmacol. 2020:113. doi: 10.1016/j.yrtph.2020.104648. [DOI] [PubMed] [Google Scholar]
- 172.Blakney A.K., McKay P.F., Yus B.I., Aldon Y., Shattock R.J. Inside out: optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019;26(9):363–372. doi: 10.1038/s41434-019-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Saunders K.O., Pardi N., Parks R., Santra S., Mu Z., Sutherland L., Scearce R., Barr M., Eaton A., Hernandez G., Goodman D., Hogan M.J., Tombacz I., Gordon D.N., Rountree R.W., Wang Y., Lewis M.G., Pierson T.C., Barbosa C., Tam Y., Shen X., Ferrari G., Tomaras G.D., Montefiori D.C., Weissman D., Haynes B.F. 2020. Lipid Nanoparticle Encapsulated Nucleoside-Modified mRNA Vaccines Elicit Polyfunctional HIV-1 Antibodies Comparable to Proteins in Nonhuman primates, bioRxiv: the Preprint Server For Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Trumpfheller C., Longhi M.P., Caskey M., Idoyaga J., Bozzacco L., Keler T., Schlesinger S.J., Steinman R.M. Dendritic cell-targeted protein vaccines: a novel approach to induce T-cell immunity. J. Intern. Med. 2012;271(2):183–192. doi: 10.1111/j.1365-2796.2011.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kreiter S., Konrad T., Sester M., Huber C., Tuereci O., Sahin U. Simultaneous ex vivo quantification of antigen-specific CD4 and CD8(+) T cell responses using in vitro transcribed RNA. Cancer Immunol. Immunother. 2007;56(10):1577–1587. doi: 10.1007/s00262-007-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kreiter S., Selmi A., Diken M., Sebastian M., Osterloh P., Schild H., Huber C., Tuereci O., Sahin U. Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J. Immunol. 2008;180(1):309–318. doi: 10.4049/jimmunol.180.1.309. [DOI] [PubMed] [Google Scholar]
- 177.Wilgenhof S., Van Nuffel A.M.T., Benteyn D., Corthals J., Aerts C., Heirman C., Van Riet I., Bonehill A., Thielemans K., Neyns B. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann. Oncol. 2013;24(10):2686–2693. doi: 10.1093/annonc/mdt245. [DOI] [PubMed] [Google Scholar]
- 178.Kalos M., June C.H. Adoptive T Cell Transfer for Cancer Immunotherapy in the Era of Synthetic Biology. Immunity. 2013;39(1):49–60. doi: 10.1016/j.immuni.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hegde M., Corder A., Chow K.K.H., Mukherjee M., Ashoori A., Kew Y., Zhang Y.J., Baskin D.S., Merchant F.A., Brawley V.S., Byrd T.T., Krebs S., Wu M.F., Liu H., Heslop H.E., Gottachalk S., Yvon E., Ahmed N. Combinational Targeting Offsets Antigen Escape and Enhances Effector Functions of Adoptively Transferred T Cells in Glioblastoma. Mol. Ther. 2013;21(11):2087–2101. doi: 10.1038/mt.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Barrett D.M., Zhao Y.B., Liu X.J., Jiang S.G., Carpenito C., Kalos M., Carroll R.G., June C.H., Grupp S.A. Treatment of Advanced Leukemia in Mice with mRNA Engineered T Cells. Hum. Gene Ther. 2011;22(12):1575–1586. doi: 10.1089/hum.2011.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang R., Men K., Zhang X.Y., Huang R., Tian Y.M., Zhou B.L., Yu C.H., Wang Y.T., Ji X., Hu Q.Y., Yang L. Delivery of a Modified mRNA Encoding IL-22 Binding Protein (IL-22BP) for Colon Cancer Gene Therapy. J. Biomed. Nanotechnol. 2018;14(7):1239–1251. doi: 10.1166/jbn.2018.2577. [DOI] [PubMed] [Google Scholar]
- 182.Men K., Zhang R., Zhang X.Y., Huang R., Zhu G.N., Tong R.S., Yang L., Wei Y.Q., Duan X.M. Delivery of modified mRNA encoding vesicular stomatitis virus matrix protein for colon cancer gene therapy. RSC Adv. 2018;8(22):12104–12115. doi: 10.1039/c7ra13656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rittig S.M., Haentschel M., Weimer K.J., Heine A., Muller M.R., Brugger W., Horger M.S., Maksimovic O., Stenzl A., Hoerr I., Rammensee H.-G., Holderried T.A.W., Kanz L., Pascolo S., Brossart P. Intradermal Vaccinations With RNA Coding for TAA Generate CD8(+) and CD4(+) Immune Responses and Induce Clinical Benefit in Vaccinated Patients. Mol. Ther. 2011;19(5):990–999. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 185.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 186.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H.G., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175) doi: 10.1038/nature06534. 141-U1. [DOI] [PubMed] [Google Scholar]
- 187.Preskey D., Allison T.F., Jones M., Mamchaoui K., Unger C. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem. Biophys. Res. Commun. 2016;473(3):743–751. doi: 10.1016/j.bbrc.2015.09.102. [DOI] [PubMed] [Google Scholar]
- 188.Ni Y., Zhao Y., Wong A., Wang J. Induction of Skeletal Myocytes, Myoblast and Satellite Cells Differentiation from iPSCs with mRNA. Mol. Biol. Cell. 2015;26 [Google Scholar]
- 189.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Spater D., Xu H.S., Tabebordbar M., Gorbatov R., Sena B., Nahrendorf M., Briscoe D.M., Li R.A., Wagers A.J., Rossi D.J., Pu W.T., Chien K.R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31(10):898. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Creusot R.J., Chang P., Healey D.G., Tcherepanova I.Y., Nicolette C.A., Fathman C.G. A Short Pulse of IL-4 Delivered by DCs Electroporated With Modified mRNA Can Both Prevent and Treat Autoimmune Diabetes in NOD Mice. Mol. Ther. 2010;18(12):2112–2120. doi: 10.1038/mt.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zimmermann O., Homann J.M., Bangert A., Mueller A.-M., Hristov G., Goeser S., Wiehe J.M., Zittrich S., Rottbauer W., Torzewski J., Pfitzer G., Katus H.A., Kaya Z. Successful Use of mRNA-Nucleofection for Overexpression of Interleukin-10 in Murine Monocytes/Macrophages for Anti-inflammatory Therapy in a Murine Model of Autoimmune Myocarditis. J. Am. Heart Assoc. 2012;1(6) doi: 10.1161/JAHA.112.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mali P., Yang L.H., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fu Y.F., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31(9):822. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ren J.T., Liu X.J., Fang C.Y., Jiang S.G., June C.H., Zhao Y.B. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin. Cancer Res. 2017;23(9):2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Georgiadis C., Preece R., Nickolay L., Etuk A., Petrova A., Ladon D., Danyi A., Humphryes-Kirilov N., Ajetunmobi A., Kim D., Kim J.S., Qasim W. Long Terminal Repeat CRISPR-CAR-Coupled "Universal" T Cells Mediate Potent Anti-leukemic Effects. Mol. Ther. 2018;26(5):1215–1227. doi: 10.1016/j.ymthe.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu X.J., Zhang Y.P., Cheng C., Cheng A.W., Zhang X.Y., Li N., Xia C.Q., Wei X.F., Liu X., Wang H.Y. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27(1):154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen Q., Zhang Y., Yin H. Recent advances in chemical modifications of guide RNA, mRNA and donor template for CRISPR-mediated genome editing. Adv. Drug. Deliv. Rev. 2021;168:246–258. doi: 10.1016/j.addr.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 198.Miller J.C., Holmes M.C., Wang J.B., Guschin D.Y., Lee Y.L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A., Gregory P.D., Pabo C.O., Rebar E.J. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 199.Hockemeyer D., Wang H.Y., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C., Zeitler B., Cherone J.M., Meng X.D., Hinkley S.J., Rebar E.J., Gregory P.D., Urnov F.D., Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., Koonin E.V., Zhang F. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Geurts A.M., Cost G.J., Freyvert Y., Zeitler B., Miller J.C., Choi V.M., Jenkins S.S., Wood A., Cui X.X., Meng X.D., Vincent A., Lam S., Michalkiewicz M., Schilling R., Foeckler J., Kalloway S., Weiler H., Menoret S., Anegon I., Davis G.D., Zhang L., Rebar E.J., Gregory P.D., Urnov F.D., Jacob H.J., Buelow R. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science. 2009;325(5939) doi: 10.1126/science.1172447. 433–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Badia R., Pauls E., Riveira-Munoz E., Clotet B., Este J.A., Ballana E. Zinc Finger Endonuclease Targeting PSIP1 Inhibits HIV-1 Integration. Antimicrob. Agents Chemother. 2014;58(8):4318–4327. doi: 10.1128/AAC.02690-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Quintana-Bustamante O., Fananas-Baquero S., Orman I., Torres R., Duchateau P., Poirot L., Gouble A., Bueren J.A., Segovia J.C. Gene editing of PKLR gene in human hematopoietic progenitors through 5 ' and 3 ' UTR modified TALEN mRNA. PLoS One. 2019;14(10) doi: 10.1371/journal.pone.0223775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kim Y., Cheong S.A., Lee J.G., Lee S.W., Lee M.S., Baek I.J., Sung Y.H. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016;34(8):808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- 205.Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C., Zheng G., Pan T., Solomon O., Eyal E., Hershkovitz V., Han D., Dore L.C., Amariglio N., Rechavi G., He C. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016;12(5):311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 207.Arango D., Sturgill D., Alhusaini N., Dillman A.A., Sweet T.J., Hanson G., Hosogane M., Sinclair W.R., Nanan K.K., Mandler M.D., Fox S.D., Zengeya T.T., Andresson T., Meier J.L., Coller J., Oberdoerffer S. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018;175(7):1872–1886. doi: 10.1016/j.cell.2018.10.030. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Delatte B., Wang F., Ngoc L.V., Collignon E., Bonvin E., Deplus R., Calonne E., Hassabi B., Putmans P., Awe S., Wetzel C., Kreher J., Soin R., Creppe C., Limbach P.A., Gueydan C., Kruys V., Brehm A., Minakhina S., Defrance M., Steward R., Fuks F. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351(6270):282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]