Significance
Opioid-induced respiratory depression (OIRD) is the direct cause of death from opioid overdose, which accounts for the current global opioid crisis. Here, we report that neurons expressing the μ-opioid receptors in the lateral parabrachial nucleus of the pontine respiratory group are necessary and sufficient for the pathogenesis of OIRD. Activating these neurons through endogenous or artificial G protein–coupled receptor signaling pathways rescues OIRD in intact mice, suggesting its therapeutic utility in OIRD patients.
Keywords: parabrachial nucleus, μ-opioid receptor, OIRD
Abstract
Opioid-induced respiratory depression (OIRD) causes death following an opioid overdose, yet the neurobiological mechanisms of this process are not well understood. Here, we show that neurons within the lateral parabrachial nucleus that express the µ-opioid receptor (PBLOprm1 neurons) are involved in OIRD pathogenesis. PBLOprm1 neuronal activity is tightly correlated with respiratory rate, and this correlation is abolished following morphine injection. Chemogenetic inactivation of PBLOprm1 neurons mimics OIRD in mice, whereas their chemogenetic activation following morphine injection rescues respiratory rhythms to baseline levels. We identified several excitatory G protein–coupled receptors expressed by PBLOprm1 neurons and show that agonists for these receptors restore breathing rates in mice experiencing OIRD. Thus, PBLOprm1 neurons are critical for OIRD pathogenesis, providing a promising therapeutic target for treating OIRD in patients.
The misuse of prescription and recreational opioids has taken nearly 4.5 million lives between 1999 and 2018 in the United States alone (1). The direct cause of death from opioid overdose is opioid-induced respiratory depression (OIRD) (2, 3). Currently, the only available antidote capable of reversing OIRD is naloxone, a nonselective opioid receptor antagonist. However, naloxone is associated with several disadvantages, namely the reappearance of OIRD because of its short half-life, the inability to reverse high-affinity opioid drugs (e.g., carfentanil and buprenorphine) because of its low binding affinity, and the potential to induce a catecholamine surge at high doses, which can cause cardiopulmonary arrest (4–6). Alternative nonopioid interventions are also promising candidates for rescuing OIRD (5). These include potassium channel blockers that regulate the excitability of carotid body cells (e.g., GAL-021) (7), ampakines that activate the amino-3-hydroxy-5-methyl-D-aspartate receptors in the respiratory centers (e.g., CX717) (8), and serotonin receptor agonists that activate the respiratory network (9, 10). Among them, ampakines made the most progress in clinical trials (search of “opioids” and “respiratory depression” via clinicaltrials.gov in January 2021) (11). Although some of these alternatives are promising for OIRD rescue, the safety of these interventions has not been well documented. Therefore, novel treatment strategies are needed, and more effective countermeasures can only be developed with a mechanistic understanding of OIRD pathogenesis.
Neural substrates that contribute to OIRD pathogenesis should satisfy two criteria. They should localize to the breathing control network and express the µ-opioid receptor (MOR, encoded by the Oprm1 gene), which has been demonstrated as the primary mediator of both the analgesic and respiratory effects of opioids (12, 13). Two candidate structures satisfy these criteria, namely the pre-Bötzinger complex (preBötC) and the parabrachial complex (14–22). The former lies in the ventrolateral medulla and is the primary generator of the respiratory rhythm (23–25). The parabrachial complex, including the lateral parabrachial, medial parabrachial, and Kölliker-Fuse (KF) nuclei, is located in the dorsolateral pons and modulates breathing (26–28) in response to homeostatic disturbances such as CO2/O2 imbalance and noxious stimuli (29–33). Recent studies have suggested the involvement of these two brain regions on OIRD pathogenesis by using either pharmacological or genetic approaches. Direct infusion of opioid agonists or antagonists into these brain areas induces or attenuates OIRD (14, 17, 34, 35). Region-specific genetic deletion of the Oprm1 genes by viral delivery of Cre recombinase into these areas of the Oprm1-floxed mice also attenuates OIRD (18, 19). However, the pharmacological approach lacks considerable specificity primarily because of the local diffusion of drugs and the difficulty of reproducible targeting, whereas the genetic deletion approach is often incomplete and prevents the possibility to directly control the neuronal activity in a spatially and temporally precise manner. As a result, discrepancies arise over the critical site of action responsible for OIRD (18, 19, 21, 22). To expand the scope of current research, our paper utilizes contemporary cell type–specific approaches to characterize the role of a defined neuronal population in morphine-induced respiratory depression. This will allow us to better understand its cellular and physiological mechanisms and make inroads in developing therapeutic strategies.
Here, we report that a genetically defined population of neurons that encodes the Oprm1 gene in the lateral parabrachial nucleus (PBLOprm1 neurons) is an important regulator of respiratory rhythm. Inhibition of these neurons by opioids leads to respiratory depression. Furthermore, we show that both chemogenetic activation of these neurons and pharmacological activation of endogenous G protein–coupled receptors (GPCRs) expressed specifically in these neurons completely rescues OIRD in mice.
Results
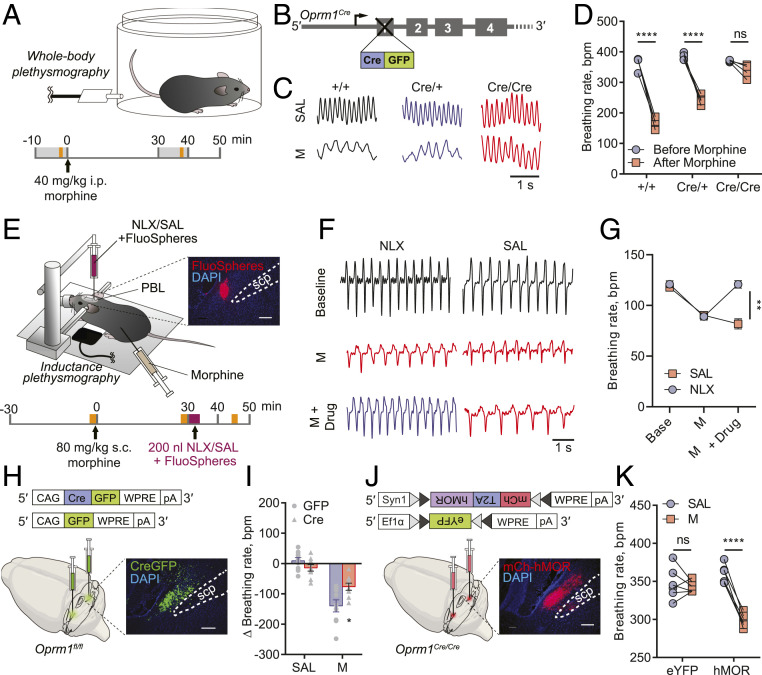
To examine the contributions of global MOR signaling to OIRD, we generated an Oprm1 reporter mouse line in which the endogenous Oprm1 gene was knocked out by inserting a Cre:green fluorescent protein (GFP) cassette upstream of the start codon (Oprm1Cre) (Fig. 1B and SI Appendix, Fig. S1A for validation) and measured the effects of systemic morphine injection on breathing rhythms via whole-body plethysmography (WBP) (Fig. 1A). After receiving a morphine injection (40 mg/kg, intraperitoneal [i.p.]), wild-type mice exhibited a significantly lower respiratory rate (363.1 ± 11.1 bpm to 168.1 ± 8.0 bpm, data are represented as mean ± SEM throughout the paper). In contrast, mice lacking one or both functional copies of the Oprm1 gene (Oprm1Cre/+ and Oprm1Cre/Cre) exhibited more moderate reductions in respiratory rate (384.2 ± 5.7 bpm to 242.2 ± 8.0 bpm and 370.6 ± 1.6 bpm to 337.9 ± 11 bpm, respectively) (Fig. 1 C and D). No significant differences in baseline respiratory rate were observed among genotypes, indicating that the knockout of Oprm1 gene does not influence the baseline breathing rate (Fig. 1D and SI Appendix, Fig. S1D). It should be noted that the differences in the morphine-induced respiratory responses across genotypes may be partially influenced by morphine pharmacokinetics, if opioid receptor deficiency, in turn, affects opioid metabolizers and transporters. Oprm1, therefore, plays a vital role in OIRD.
Fig. 1.
MOR signaling in the PBL mediates morphine-induced respiratory depression. (A) Schematics of breathing monitoring in awake mice with WBP before and after systemic morphine injection (refer to C, D, I, and K). The gray shade indicates the 10 min WBP recording, and the yellow shade indicates the 1 min episode used for data analysis. (B–D) Oprm1+/+, Oprm1Cre/+, and Oprm1Cre/Cre mice (B) displayed different breathing patterns (C) and rate changes (D) in response to systemic morphine injection. (E) Schematics of breathing monitoring in anesthetized mice with inductance plethysmography during systemic morphine injection and PBL-specific blockade of MOR signaling. The yellow shade indicates the 2 min plethysmography recording before morphine, after morphine, and after naloxone/saline injections; the purple shade indicates the naloxone/saline injection which lasts ∼4 min. Onset, histology of the coinjected FluoSpheres in the PBL. Scp, superior cerebellar peduncle. (F) Representative plethysmographs from naloxone (NLX) and saline (SAL) injected groups before morphine, after morphine, and after drug injections into the PBL. (G) Injecting naloxone (n = 5) but not saline (n = 5) into the PBL rescued morphine-induced respiratory depression. (H) Schematics of PBL-specific knockout of the Oprm1 gene by stereotaxically injecting AAV-Cre-GFP into the PBL of the Oprm1fl/fl mice. (I) PBL-specific Oprm1 knockout (n = 9) significantly attenuated OIRD compared to the GFP-injected control group (n = 9) after systemic morphine injection. (J) Schematics of PBL-specific rescue of the Oprm1 gene in Oprm1-null background by stereotaxically injecting AAV-FLEX-mCh-T2A-hMOR into the PBL of the Oprm1Cre/Cre mice. (K) Mice with PBL-specific rescue of Oprm1 (n = 6) displayed OIRD phenotype, whereas eYFP-injected control group (n = 6) remained insensitive after systemic morphine injection. Morphine doses were determined according to different genetic background or experimental conditions: 10 mg/kg (I); 40 mg/kg (C, D, and K); 80 mg/kg (F and G). Data are presented as mean ± SEM. Statistical tests: two-way ANOVA with Bonferroni’s multiple comparison post hoc test (D, G, I, and K). ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (Scale bar, 1 s for C and F or 200 μm for E, H, J.)
We then used pharmacological and genetic tools to manipulate opioid signaling specifically in the PBL. First, wild-type mice received a systemic injection of morphine followed by an infusion of naloxone directly into the PBL (Fig. 1E). To facilitate stereotaxic naloxone delivery, we lightly anesthetized mice with 1 to 1.5% isoflurane and monitored their breathing using a piezoelectric sensor that detects chest movements (36). After systemic injection of morphine (80 mg/kg, subcutaneous [s.c.], a high concentration was used in anesthetized animals), the breathing rate dramatically decreased from 121.0 ± 3.4 bpm to 89.1 ± 3.5 bpm, and subsequent stereotaxic injection of naloxone into the PBL recovered the breathing rate back to premorphine baseline (120.8 ± 3.7 bpm) (Fig. 1 F and G). We then explored whether the expression of the Oprm1 gene in the PBL is necessary and sufficient for OIRD. To assess necessity, we conditionally knocked out the Oprm1 gene using stereotaxic delivery of a recombinant adeno-associated virus (AAV) expressing Cre recombinase into the PBL of the Oprm1fl/fl mice, in which the Oprm1 gene is flanked by loxP sites (Fig. 1H). Compared to mice receiving the GFP-expressing control virus, PBL-specific knockout of the Oprm1 gene attenuated OIRD following systemic injection of morphine (10 mg/kg, i.p.), as measured by WBP (breathing rate changes in GFP group: −136.6 ± 21.1 bpm; Cre group: −76.4 ± 11.6 bpm) (Fig. 1I and SI Appendix, Fig. S2 A–C). To test sufficiency, we reintroduced the wild-type Oprm1 gene into the PBLOprm1 neurons of Cre-expressing Oprm1-null mice. This was achieved by injecting an AAV expressing Cre-dependent human MOR (AAVDJ-Syn1-FLEX-mCh-T2A-FLAG-hMOR-WPRE) (Fig. 1J) into the PBL of Oprm1Cre/Cre mice. Selective expression of hMOR in the PBL restored the OIRD phenotype (hMOR group, postsaline: 359.0 ± 5.1 bpm; postmorphine: 299.0 ± 3.6 bpm, 40 mg/kg, i.p.) compared to the control group (Fig. 1K and SI Appendix, Fig. S2D). Note that the breathing rate measured by the piezoelectric sensor under anesthesia (Fig. 1G) is ∼threefold lower than that measured in a plethysmography chamber (Fig. 1K). Together, these data strongly support the conclusion that the Oprm1 gene expression in the PBL mediates OIRD in mice.
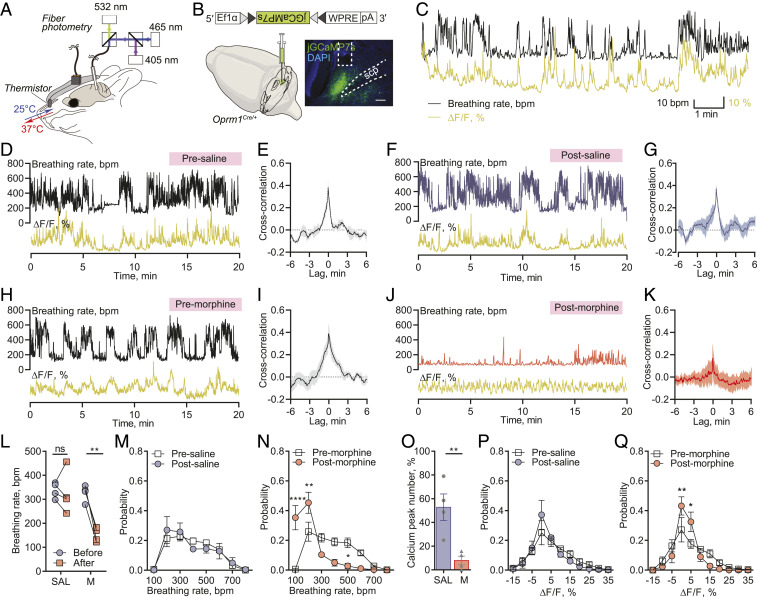
We then focused on PBLOprm1 neurons and explored their relationship with OIRD. PBLOprm1 neurons are mostly glutamatergic and express higher levels of the Oprm1 gene than the adjacent KF nucleus, which provides postinspiratory drive (18, 28, 36, 37) (SI Appendix, Fig. S1 B and C). We devised an experimental platform to simultaneously measure PBLOprm1 neuronal activity and respiration in awake, freely moving mice using fiber photometry calcium-mediated fluorescence measurements with a thermistor implanted into the nasal cavity (Fig. 2A). The thermistor-based respiration monitoring device detects the temperature change between inhaled and exhaled air and converts breath-to-breath fluctuations in resistance into voltage signals (38). We stereotaxically injected an AAV expressing Cre-dependent jGCaMP7s (AAV-DIO-jGCaMP7s) into the PBL, and then implanted an optic fiber into the PBL of Oprm1Cre/+ mice and a respiration sensor in the nasal cavity (Fig. 2B). PBLOprm1 activity and breathing rate exhibited a tight positive correlation (Fig. 2 C, D, and H). Moreover, morphine administration substantially decreased the breathing rate (326.5 ± 17.0 bpm to 151.8 ± 14.7 bpm, 40 mg/kg, i.p.) (Fig. 2 H, J, and L, and N) despite the morphine-induced increase in locomotor activity (SI Appendix, Fig. S3), which one would expect to elevate the breathing rate. Besides, morphine decreased calcium activity (Fig. 2 H, J, and O, and Q) as well as eliminated any spontaneous fluctuations in both breathing and calcium signals (Fig. 2 N and Q). The opposite effects on locomotion and calcium activity observed after morphine administration demonstrated that the breathing-associated fluctuation of PBLOprm1 activity is not affected by the movement, which is further demonstrated in our concurrent independent study (39). In contrast, saline injection largely preserved the original patterns of the signals (Fig. 2 D, F, L, M, O, and P). Using convergent cross mapping (CCM) (40), a statistical algorithm that makes predictions from independent time-series data, we predicted changes in respiratory rate using the calcium signal as input (76.2 ± 8.1%, premorphine predictability) (SI Appendix, Fig. S4). Interestingly, systemic morphine administration not only reduced the correlation coefficient (Fig. 2 I and K) but also completely abolished the predictive relationship between respiratory rate and the calcium signal (12.5 ± 20.2%, postmorphine predictability) (SI Appendix, Fig. S4 C and D). Conversely, the saline injection did not alter the correlation between these signals (Fig. 2 E and G and SI Appendix, Fig. S4 A and B). These results indicate that PBLOprm1 neurons are critically involved in modulating respiratory rate and that morphine suppresses both the PBLOprm1 neuronal activity and its coupling with the breathing rate.
Fig. 2.
Morphine disrupts the tight correlation between PBLOprm1 neuronal activity and respiratory rate. (A) Schematics of simultaneous recording of respiration and PBLOprm1 calcium activity with a thermistor sensor (implanted in the nasal cavity) and fiber photometry. (B) The genetically encoded calcium indicator, jGCaMP7s, was specifically expressed in the PBLOprm1 neurons by stereotaxic injection of AAV-DIO-jGCaMP7s into the PBL of the Oprm1Cre/+ mice. (C) Time-matched traces of PBLOprm1 activity and breathing rate as calculated from the thermistor sensor. (D, F, H, and J) Simultaneously recorded PBLOprm1 calcium activity and respiratory rate before and after systemic saline or morphine (40 mg/kg) injection. (E, G, I, and K) Cross-correlogram of calcium signal and respiratory rate in the presaline, postsaline, premorphine, and postmorphine groups (n = 4). (L) Morphine injection significantly depressed respiratory rate (n = 4), whereas saline injection (n = 4) did not significantly affect the respiratory rate. (M and N) Normalized distribution of breathing rate before and after saline (M) and morphine (N) injections (n = 4). (O) The number of calcium peaks was significantly decreased after morphine injection (n = 4) compared to saline injection (n = 4). (P and Q) Normalized distribution of calcium signals before and after saline (P) and morphine (Q) injections (n = 4). Data are presented as mean ± SEM. Statistical tests: unpaired t test (O) or two-way ANOVA with Bonferroni’s multiple comparison post hoc test (L, M, N, P, and Q). ns, not significant; *P < 0.05, **P < 0.01, ****P < 0.0001. (Scale bar, 200 μm.)
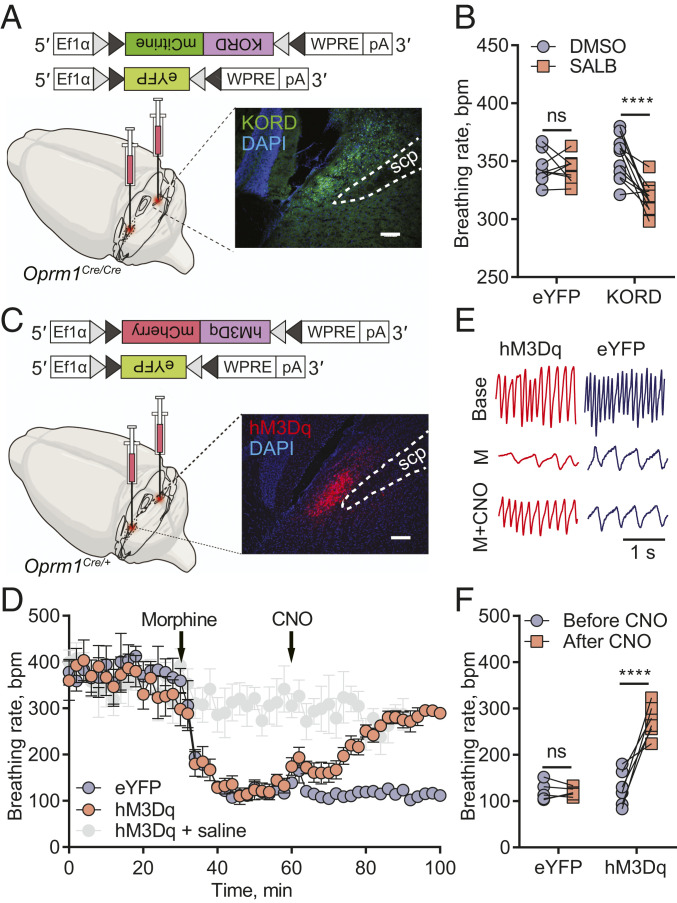
To further investigate PBLOprm1 neurons’ role in breathing regulation, we specifically manipulated their activity using chemogenetic tools, namely designer receptor exclusively activated by designer drugs (DREADD) (41). If PBLOprm1 neurons directly regulate breathing, inhibiting their activity via a Gi/o-coupled DREADD should recapitulate the OIRD phenotype. Furthermore, this should also happen in Oprm1 knockout mice, which failed to display OIRD when given systemic morphine (Fig. 1 C and D). We expressed a κ-opioid receptor-derived DREADD (KORD) (42) Cre-dependently and bilaterally in PBLOprm1 neurons of Oprm1Cre/Cre mice (Fig. 3A), to phenocopy the inhibition of MOR by morphine, because KOR and MOR have high structural similarity (43) and engage with conserved intracellular signaling pathways (44). Systemic injection of the synthetic ligand of KORD, salvinorin B (SALB, 7.5 mg/kg, i.p.), decreased the respiratory rate (352.8 ± 18.4 bpm to 317.8 ± 3.6 bpm) compared to the dimethyl sulfoxide (DMSO)-injected control group (344.8 ± 13.8 bpm to 342.4 ± 4.1 bpm) (Fig. 3B and SI Appendix, Fig. S5 A and B). We did not observe significant breathing amplitude changes upon SALB injection in either control or KORD-expressing animals (SI Appendix, Fig. S5 B and C).
Fig. 3.
Inhibition of PBLOprm1 neurons mimics OIRD, and chemogenetic activation of PBLOprm1 neurons rescues OIRD. (A) Schematics of mimicking OIRD through PBLOprm1 inhibition using an inhibitory KORD. Here, AAV-DIO-KORD-mCitrine is stereotaxically injected into the bilateral PBL of Oprm1Cre/Cre mice to express in the PBLOprm1 neurons. (B) SALB (7.5 mg/kg) injection significantly decreased respiratory rate compared to the DMSO control in the KORD-expressing mice (n = 11). In contrast, no changes were observed in eYFP-expressing mice (n = 8). (C) Schematics of rescuing OIRD through PBLOprm1 activation by an excitatory DREADD, hM3Dq. Here, AAV-DIO-hM3Dq-mCherry was expressed in the bilateral PBL of Oprm1Cre/+ mice. (D) Activation of PBLOprm1 neurons by injecting CNO (5 mg/kg) rescued the morphine-induced respiratory depression (40 mg/kg) to control levels in the hM3Dq-expressing group (n = 6) but not the eYFP-expressing group (n = 5). (E and F) Representative plethysmograph and quantitative analysis depicting OIRD rescued by CNO injection in the hM3Dq-expressing group (n = 7) but not in the eYFP-expressing group (n = 5). Statistical tests: two-way ANOVA with Bonferroni’s multiple comparison post hoc test. ns, not significant; ****P < 0.0001. (Scale bar, 200 μm.)
We next asked whether artificial activation of PBLOprm1 neurons could prevent OIRD in mice. We expressed hM3Dq, a metabotropic acetylcholine receptor-derived excitatory DREADD, in PBLOprm1 neurons via bilateral injection of an AAV encoding Cre-dependent hM3Dq into Oprm1Cre/+ mice (Fig. 3C). Additionally, we implanted a microthermistor sensor into the nasal cavity to monitor respiratory rhythm in awake, freely moving mice over the course of OIRD (SI Appendix, Fig. S5D). Upon systemic morphine injection (40 mg/kg, i.p.), the respiratory rate decreased significantly within 10 min (from 332.0 ± 25.6 bpm to 124.6 ± 9.1 bpm) (Fig. 3 D and E). We then systemically injected the hM3Dq agonist, clozapine-N-oxide (CNO, 5 mg/kg, i.p.), into the same animal to activate PBLOprm1 neurons. CNO injection increased the breathing rate in hM3Dq-expressing animals (268.0 ± 13.5 bpm) to a level comparable to controls that did not receive morphine (275.7 ± 24.9 bpm). Nevertheless, CNO did not rescue breathing rates in eYFP-expressing control animals (120.0 ± 4.3 bpm) (Fig. 3 D–F and SI Appendix, Fig. S5E). A complete rescue was also observed in minute volume (SI Appendix, Fig. S5 F and G). These data show that activation of PBLOprm1 neurons is sufficient to restore normal breathing rhythms in mice displaying OIRD.
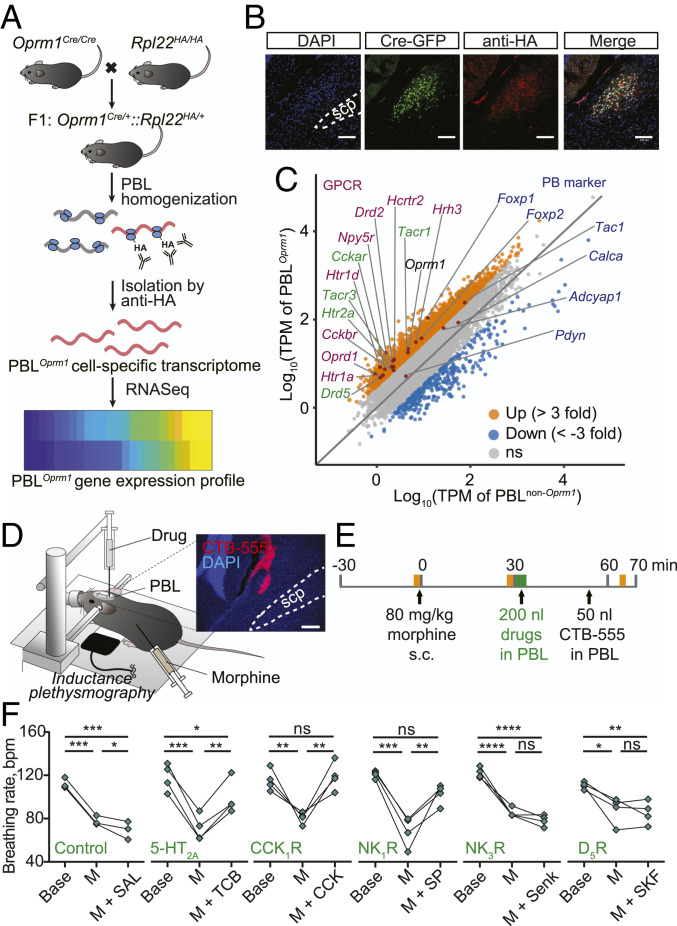
Having determined that chemogenetic activation of PBLOprm1 neurons could effectively prevent OIRD, we sought to identify endogenous signaling pathways that could be used to activate PBLOprm1 neurons. To identify the active transcriptome enriched in PBLOprm1 neurons, we sequenced ribosome-associated messenger RNAs (mRNAs) from the PBL of Oprm1Cre:RiboTag mice, which express hemagglutinin (HA)-tagged ribosomal protein Rpl22 in Oprm1+ neurons (Fig. 4 A and B) (45). We identified 69 mRNAs enriched over threefold in PBLOprm1 neurons than non-Oprm1 neurons in the PBL (Fig. 4C). Oprm1, as well as other major parabrachial markers (Tac1, Foxp2, and Adcyap1), were enriched, but Calca and Pdyn, markers for the external lateral and dorsal lateral parabrachial nucleus, were not (46, 47). Notably, the PBLOprm1 active transcriptome revealed several GPCRs expressed at high levels in these neurons, which we subsequently investigated as potential pharmacological targets to rescue OIRD.
Fig. 4.
Rescue of OIRD through PBLOprm1 stimulation via receptors identified by RiboTag RNA sequencing. (A) Schematics of obtaining PBLOprm1 active transcriptome. Oprm1Cre/Cre mice were crossed with RiboTag mice, which express hemagglutinin-tagged ribosomal protein L22 (Rpl22HA) in a Cre-dependent manner. After collecting the PBL, active mRNAs bound to the Rpl22HA were isolated with anti-HA antibody-mediated ribosome pulldown and sequenced. (B) Immunohistochemistry of the PBL region of the Oprm1Cre:Rpl22HA mice with anti-GFP and anti-HA antibodies showed that Cre:GFP and Rpl22HA were coexpressed in PBLOprm1 neurons. (C) A total of 14 GPCR mRNAs (magenta and green) were enriched more than threefold in PBLOprm1 neurons compared with non-Oprm1 neurons in the PBL. Conventional parabrachial markers (dark blue) such as Foxp2, Tac1, and Adcyap1 genes were enriched in PBLOprm1 neurons but not Pdyn and Calca. (D and E) Schematics of rescuing OIRD through artificial stimulation of receptors expressed on PBLOprm1 neurons by PBL-specific injection of the agonists. Yellow shade indicates the 2 min plethysmography recording before morphine (80 mg/kg), after morphine, and after drug/cholera toxin subunit B (CTB) injections; the green shade indicates the drug injection, which lasts ∼400 s. (Inset) Histology of CTB-555 for marking the injection site. (F) Breathing rate decreased upon systemic morphine injection and rescued by PBL-specific injection of the agonists of 5-HT2A, CCK1R, and NK1R. For the list of pharmacological agents, see SI Appendix, Table S1. Statistical tests: RM one-way ANOVA with Tukey’s multiple comparison post hoc test, ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (Scale bar, 100 μm for B or 200 μm for D.)
We selected five excitatory GPCRs for pharmacological manipulation and confirmed that they were expressed in PBLOprm1 neurons by RNA in situ hybridization (SI Appendix, Fig. S6 A and B). These GPCRs were 5-hydroxytryptamine receptor 2A (Htr2a), cholecystokinin A receptor (Cckar), tachykinin receptor 1 (Tacr1), tachykinin receptor 3 (Tacr3), and dopamine receptor D5 (Drd5). We systemically injected their agonists (TCB-2, CCK8S, Substance P, Senktide, and SKF83959, respectively) into the PBL of anesthetized mice after inducing OIRD with morphine (80 mg/kg, s.c.; breathing rate before morphine: 115.4 ± 1.6 bpm, after morphine: 77.9 ± 2.3 bpm; Fig. 4 D and E and SI Appendix, Table S1). Three of the five agonists tested (TCB-2, CCK8S, and Substance P) increased the respiratory rate (98.1 ± 7.9, 117.1 ± 6.4, 100.8 ± 4.6 bpm; Fig. 4F and SI Appendix, Fig. S6 C–E). Notably, these same drugs have been shown to modulate respiratory rhythm in different contexts by activating other respiratory centers, and some have been associated with an anti-morphine effect (48–50). Our findings suggest that activating PBLOprm1 neurons through endogenous signaling pathways may effectively treat OIRD in patients.
Discussion
We have identified a population of neurons in the pontine respiratory group, the PBLOprm1 neurons, as critical mediators of morphine-induced respiratory depression and candidates for its rescue in intact mice. Genetic deletion of Oprm1 from the PBL attenuated OIRD, while reintroducing human MOR into the PBLOprm1 neurons of Oprm1-null mice restored OIRD. By combining cell type–specific tools with breathing monitoring in awake behaving mice, we have demonstrated that PBLOprm1 neurons are regulators of the breathing rhythm and pattern. Systemic morphine administration dramatically abolishes the activity of these neurons and weakens their control on breathing. Furthermore, chemogenetic inhibition of PBLOprm1 neurons mimics OIRD, and prolonged optogenetic inhibition of PBLOprm1 neurons induces apnea [observations from our group (39)], which precedes cardiorespiratory collapse prior to overdose-induced death. By contrast, artificial activation of PBLOprm1 neurons, either chemogenetically or pharmacologically, successfully restores breathing in mice experiencing OIRD (SI Appendix, Fig. S7).
Investigating OIRD with Cell Type–Specific Approaches.
Recent studies have demonstrated the involvement of both pontine and medullary respiratory groups in OIRD (18, 19) without reaching a consensus on the key players. We think this is mostly due to the lack of specificity from the pharmacological and genetic deletion approaches and the intricacies of physiological preparations, both of which hampers reproducibility. In comparison, we exploited the Oprm1Cre mouse line, which provides access to a restricted neuronal population, and obtained multifaceted evidence supporting the role of the parabrachial nucleus in OIRD. In our view, OIRD is a synergistic outcome from impairment of the interconnected pontomedullary breathing network that widely expresses MOR both pre- and postsynaptically (15, 16, 35, 51, 52). For example, parabrachial neurons send tonic excitatory inputs to the preBötC (39, 47, 53, 54), and both of these areas express MOR (23, 55). Besides, the preBӧtC, as the breathing pattern generator, has been known to contribute to the OIRD phenotype, yet the level of its involvement varies across species and overdose models. Deletion of the Oprm1 gene in the preBӧtC by injecting AAV that encodes Cre recombinase into the preBӧtC of the Oprm1fl/fl mice attenuated the respiratory rate depression induced by i.p. morphine injection (18, 19). Interestingly, the effect largely depends on the dosage of opioids (18). Naloxone infusion into the preBӧtC prevented the respiratory rate depression induced by intravenous fentanyl/remifentanil injection in rats (20) but not in dogs (56) or rabbits (57). It would therefore be valuable to test the contribution of Oprm1-expressing neurons within the preBӧtC in regulating breathing rhythm with multiple OIRD models.
One challenge to elucidating the OIRD mechanism is how to access a group of neurons while simultaneously recording breathing over the course of OIRD. Traditional respiratory monitoring approaches in rodents, such as in situ or in vivo anesthetized preparations and plethysmography chambers, are more challenging to achieve this goal. Therefore, we utilized the implantable temperature sensor to monitor breathing in awake, unrestrained, behaving mice with unprecedented freedom to access the brain because of its open configuration. With this approach, we implemented chemogenetics and in vivo neural activity monitoring simultaneously with breathing rhythm recording. Similar techniques can be applied to the Oprm1-expressing neurons in the preBötC, KF, the postinspiratory complex (58), and other respiratory centers to resolve the long-standing debate on their contributions in OIRD (21, 22).
PBLOprm1 Neurons’ Role in Breathing Regulation and OIRD.
The parabrachial complex has long been reported to modulate respiration and sometimes exert opposing effects, depending on the location of artificial manipulation (26–28). Stimulation of the PBL results in tachypnea, whereas stimulation of the KF elicits prolonged postinspiration and bradypnea (18, 36, 37). Consistent with these previous reports, our monitoring and manipulation data suggest that PBLOprm1 neurons have substantial effect on the breathing rate (28). In addition to breathing, the PBL also regulates various homeostatic functions (47) and incorporates the alarm center, which senses deviations from the homeostasis and relays aversive signals to higher-order brain structures (46, 59). At the same time, by sending tonic excitatory outputs to medulla respiratory centers (39, 47, 53), the parabrachial neurons can increase breathing rhythm under conditions that immediately require more oxygen because of metabolic needs, such as hypoxia (32) and hypercapnia (31), in particular, hypercapnic arousal during sleep (29, 33, 60), or nonmetabolic behavioral demands, such as escaping from a threat (30). As a representative glutamatergic population in the PBL (SI Appendix, Fig. S1), Oprm1-positive neurons are likely to participate in similar physiological processes. Our data demonstrated that an overdose of morphine shuts down PBLOprm1 neurons' activity, preventing them from responding to conditions that decrease respiratory rhythm. These conditions may include hypoxia, hypercapnia, anesthesia, or sedative drug treatment. Indeed, the incidence of OIRD is dramatically increased when opioids are used together with sedatives and anesthetics (61), and the failure to increase respiratory behavior in response to hypercapnic gas is one characteristic of OIRD (3). On the contrary, a mild inhibition of PBLOprm1 neurons may explain many of the behavioral effects of lower doses of endogenous or exogenous opioids, such as slowed breathing, calmness, and reward (39). The current study used morphine as a starting point to induce OIRD, and further investigation is required to determine the involvement of PBLOprm1 neurons under the challenge of other types of sedative agents.
A Proof of Principle of Rescuing OIRD by Activating a Pontine Population.
PBLOprm1 neurons are among the key node most vulnerable to opioid action, and their activation is sufficient to restore the breathing rate and tidal volume following morphine administration. This activation can be achieved by exploiting both artificial and, more importantly, endogenous signaling pathways. Besides, manipulating PBLOprm1 neurons located in the dorsolateral pons is associated with lower risk than the respiratory rhythm generators within the deep medulla. We have provided a proof of concept of rescuing OIRD by activating endogenous Gq/s-coupled GPCRs expressed in PBLOprm1 neurons (SI Appendix, Fig. S7). In principle, cell type–specific manipulation can also be achieved through the collective activation of multiple GPCRs.
Our transcriptomic profiling data suggested that other neuromodulators, such as serotonin, cholecystokinin, and tachykinin, may also be involved in respiratory regulation with potential interplay with the opioid system (48–50). The same receptor that we identified, 5-HT2A receptor, has recently been confirmed to be expressed in the PBL and mediate hypercapnia-induced arousal by integrating serotonergic inputs from the dorsal raphe (60). It would be interesting to identify other neuromodulatory circuits that may be involved in the regulation of OIRD. Similar transcriptomic profiling strategies can also be applied to the broader breathing control network to exploit the therapeutic potential of GPCRs in reversing OIRD.
Materials and Methods
Experimental Animals.
The Oprm1Cre:GFP mouse line was generated from the laboratory of Richard Palmiter (see below). C57BL/6J (stock no. 000664), Oprm1fl/fl (stock no. 030074), RiboTag Rpl22HA/HA (stock no. 011029), and Ai14 Gt(ROSA)26Sortm14(CAG-tdTomato)Hze (stock no. 007914) mouse lines were obtained from The Jackson Laboratory. Homozygous RiboTag and Ai14 mice were crossed with homozygous Oprm1Cre mice for RiboTag and anatomy experiments, respectively. Both male and female mice were used in all studies. Animals were randomized to experimental groups, and no sex differences were noted. Mice were maintained on a 12 h light/dark cycle and provided with food and water ad libitum.
Generation of Oprm1Cre Mice.
A cassette encoding Cre:GFP was inserted just 5′ of the initiation codon in the first coding exon of the Oprm1 gene. The 5′ arm (∼3.1 kb with PacI and SalI sites at 5′ and 3′ ends, respectively) and 3′ arm (∼4.5 kb with XhoI and NotI sites at 5′ and 3′ ends, respectively) of the Oprm1 gene were amplified from a C57BL/6 bacterial artificial chromosome clone by PCR using Q5 Polymerase (New England Biolabs) and cloned into polylinkers of a targeting construct that contained mnCre:GFP, a FRT-flanked Sv40Neo gene for positive selection, and herpes simplex virus type 1 thymidine kinase (HSV-TK) and phosphoglycerate kinase 1 promoter-diphtheria toxin A chain (Pgk-DTA) genes for negative selection. The mnCre:GFP cassette has a Myc-tag and nuclear localization signals at the N terminus of Cre recombinase, which is fused to green fluorescent protein followed by a SV40 polyadenylation sequence. The construct was electroporated into G4 ES cells (C57BL/6 × 129 Sv hybrid), and correct targeting was determined by a Southern blot of DNA digested with KpnI using a 32P-labeled probe downstream of the 3′ arm of the targeting construct. Out of 84 clones analyzed, 12 were correctly targeted. One clone that was injected into blastocysts resulted in good chimeras that transmitted the targeted allele through the germline. Progeny were bred with Gt(Rosa)26Sor-FLP recombinase mice to remove the SV-Neo gene. Mice were then continuously backcrossed to C57BL/6 mice. Routine genotyping is performed with three primers: 5′ CCT TCC ACT CAG AGA GTG GCG (Oprm1 forward), 5′ CCT TCC ACT CAG AGA GTG GCG (Oprm1 reverse), and 5′ GGC AAA TTT TGG TGT ACG GTC AG (Cre reverse). The wild-type allele gives a band of ∼500 bp, while the targeted allele provides a band with ∼400 bp after 34 cycles with 20 s annealing at 60 °C.
Respiratory Measurements.
Inductance plethysmography.
Inductance plethysmography was performed by placing a piezoelectric film beneath the chest of an anesthetized animal, which converts the chest movement into a voltage signal. The PowerLab system with LabChart 8 software (ADInstruments, Inc.) was used for data acquisition, inspiratory and expiratory peak detection, and rate and amplitude calculation. Data were sampled at 100 or 400 Hz, low-pass filtered at 10 Hz, and smoothed with a 100 ms moving window. Automatic peak detection was validated with manual peak detection. Since the location of the sensor is subject to the slight movements of the animal’s body, the raw voltage value of the respiratory peak is less representative of the breathing amplitude.
WBP.
A custom-built WBP chamber was utilized for measuring respiratory changes. The PowerLab system with LabChart 8 software was used for data acquisition, inspiratory and expiratory peak detection, and rate and amplitude calculation. Data were sampled at 1 kHz, band-pass filtered at 1 to 10 Hz, and smoothed with a 100 ms moving window. Automatic peak detection was validated with manual peak detection.
Micro thermistor-based plethysmography.
A custom-built micro thermistor was implanted into the mouse nasal cavity to detect changes in temperature between inspiratory and expiratory airflow (18). The sensor was assembled using a negative temperature coefficient thermistor (TE Connectivity Ltd.), an interconnector (Mill-Max Manufacturing Corporation), and a voltage divider (Phidgets, Inc.). PowerLab was used for data acquisition, inspiratory and expiratory peak detection, and rate and amplitude calculation. Data were sampled at 1 kHz, filtered with a 0.4 to 25 Hz band-pass filter, and smoothed with a 50 ms moving window. Automatic peak detection was validated with manual peak detection.
Minute volume is approximated by first calculating the integral of the voltage channel for each breath, summing across 1 min, then normalizing against the average value of the first 30 min. Values are shown after normalization as the raw values vary considerably across animals. Values during the episodes when the sensor accidentally fell off because of the animal body rotation were excluded.
Stereotaxic Surgery.
Mice were anesthetized with isoflurane (5% induction, 1.5 to 2% maintenance with a nose cone; Dräger Vapor 2000, Draegar, Inc.) and placed onto a water recirculating heating pad throughout the surgery. Mice were placed on a stereotaxic frame (David Kopf Instruments), the skull was exposed, and the cranium was drilled with a micro motor handpiece drill (Foredom; one or two holes were drilled for viral injection(s) or pharmacological delivery, two holes for screws with implantation, one or two holes for optic fibers, and one hole for a micro thermistor). The virus or drug was injected unilaterally (right side) or bilaterally into the PBL (anteroposterior [AP], −1 mm from lambda; mediolateral [ML], ±1.5 mm; dorsoventral [DV], −3.5 mm, re-zero at the midline with the same AP). Injection of the virus or drug was administered with a glass pipette (tips broken for an inner diameter of 20 µm) connected to the Nanoject III Programmable Nanoliter Injector (Drummond Scientific) at a rate of 60 nL/min. Naloxone was injected at a rate of 100 nL/min with a syringe (65458-01, Hamilton) connected to an ultra-micropump (UMP-3, World Precision Instruments). A glass pipette or syringe needle was retracted from the brain slowly after 5 to 10 min. For implantation, optic fibers were implanted above the injection site with the DV noted below, and the micro thermistor head was carefully lowered into the hole above the nasal cavity (AP +3.5 from the nasal fissure, ML 0.3). The implants were covered with superglue and dental cement for stabilization. Behavioral experiments were performed 3 wk after virus injection and 1 wk after the microthermistor implantation unless otherwise noted.
For the PBL-specific conditional knockout of the Oprm1 gene, Oprm1fl/fl mice were bilaterally injected with 200 nL of AAVDJ-CAG-Cre-GFP (1.25E+12 GC/mL) or control AAVDJ-CAG-GFP (2.10E+12 GC/mL) (Salk Institute Viral Vector Core) into the PBL.
For PBL-specific rescue of the Oprm1 gene, Oprm1Cre/Cre mice were bilaterally injected with 200 nL of AAVDJ-Syn1-FLEX-mCh-T2A-FLAG-hMOR-WPRE (Addgene plasmid 166970) or control AAV1-hSyn-DIO-eYFP (2.12E+12 GC/mL) into the PBL.
For fiber photometry, Oprm1Cre/+ mice were unilaterally injected with 200 nL of AAV1-hSyn-DIO-jGCaMP7s (Addgene viral prep 104491-AAV1) (3.75 E+13 GC/mL) or control AAV1-hSyn-DIO-eYFP (2.12E+12 GC/mL) into the PBL, and a stainless-steel mono fiberoptic cannula (400 μm diameter, 0.37 numerical aperture [NA], Doric Lenses) was implanted 0.05 mm above the injection site.
For chemogenetics, 200 nL of AAV1-hSyn-DIO-hM3Gq-mCherry (6.56E+11 GC/mL) (Addgene plasmid 44361), AAV1-hSyn-DIO-KORD-mCitrine (Addgene plasmid 65471), or control AAV1-hSyn-DIO-eYFP (2.12E+12 GC/mL) was bilaterally injected into the PBL of Oprm1Cre/+ (for hM3Dq) or Oprm1Cre/Cre (for KORD) mice.
Pharmacology.
Morphine sulfate (Spectrum Chemical) was dissolved in 0.9% saline to make a 4 mg/mL working concentration. The final concentration of morphine was 10 mg/kg in PBL-specific Oprm1 knockout tests, 80 mg/kg in PBL-specific rescue of OIRD, and 40 mg/kg in all other experiments (SI Appendix, Table S1). The loss of the Oprm1 gene decreases morphine sensitivity (Fig. 1C, P < 0.0001 between each genotype pairs after morphine injection, two-way ANOVA with Bonferroni’s post hoc comparison), so we used a higher dose (40 mg/kg) in the Oprm1Cre background and a lower dose (10 mg/kg) in the wild-type Oprm1 background (Oprm1fl/fl). Anesthesia reduces the breathing rate and morphine responses, and therefore, a higher dose of 80 mg/kg was used in anesthetized mice.
For the characterization of morphine-induced respiratory depression with the WBP, animals were introduced into the WBP chamber for three 20 min habituation sessions before testing. During testing, mice were introduced into the WBP chamber 0 to 10 min before morphine injection and 30 to 40 min after morphine injection. For each recording session, after 5 to 8 min of chamber introduction, a stable pattern, defined as the baseline fluctuation less than 20 bpm within any minute, was reached and maintained until the end of the recording. The average value of a stabilized 1 min segment was analyzed. In each experiment, control and experimental groups were analyzed with the same criteria described above.
For PBL-specific rescue of OIRD with naloxone, wild-type mice were kept under isoflurane anesthesia until the breathing rate was stable (fluctuation < 10 bpm) for 10 min. Then, 80 mg/kg morphine (subcutaneous, s.c.) was systemically administered. After 30 min, 200 nL of 0.4 mg/mL naloxone (Somerset Therapeutics) and FluoSpheres (540/560, 10% vol/vol, Thermo Fisher) mixture or a control 0.9% saline-FluoSpheres mixture was stereotaxically injected bilaterally into the PBL at a rate of 100 nL/min. Breathing was analyzed from three 2 min episodes: immediately before morphine injection, 30 min after morphine injection, and 10 min after naloxone or saline injection.
For hM3Dq-dependent activation of PBLOprm1 neurons, Oprm1Cre/+ mice were attached with the patch cord to their respiration sensor and placed in an empty standard mouse cage. After 30 min, 40 mg/kg morphine was intraperitoneally injected into the test animal. After another 30 min, 5 mg/kg CNO was intraperitoneally injected into the test animal. For quantification of the breathing rate and minute volume, two time periods (0 to 10 min before and 20 to 30 min after CNO injection) were analyzed.
For endogenous receptor activation of PBLOprm1 neurons, wild-type mice were kept under 1 to 1.5% isoflurane anesthesia until breathing rate is stable (fluctuation < 10 bpm) for 10 min. Then, 80 mg/kg morphine (s.c.) was systemically administered. After 30 min, 200 nL of the pharmacological agent (SI Appendix, Table S1) was stereotaxically injected into the PBL bilaterally. 50 nL of Cholera Toxin Subunit B-555 was subsequently injected with the same coordinates to mark the injection site. Breathing was analyzed from three 2 min episodes: immediately before morphine injection, 30 min after morphine injection, and 30 min after drug injection.
Fiber Photometry.
Fiber photometry (1-site Fiber Photometry System, 405 and 465 nm, Doric Lenses, Inc.) with Doric Neuroscience Studio software was used to record PBLOprm1 neural activities. GCaMP isosbestic fluorescence (405-nm excitation) and calcium-dependent fluorescence (465 nm excitation) were recorded at a sampling rate of 12 kHz, and data were analyzed with the Doric Neuroscience Studio software. F0 was calculated by a least mean squares fitting of the 405 nm channel relative to the 465 nm channel, and ΔF/F was calculated as (F465-F405_fitted)/F405_fitted. Data were further analyzed with custom MATLAB scripts. Animals with fluctuating isosbestic signals indicating insecure implants were not used for the experiment.
For concurrent measurements of PBLOprm1 neural activity and respiration, animals were recorded in their home cage for 30 min before and 30 min after i.p. morphine injection at 40 mg/kg. Cross-correlation analysis between the calcium signals and respiration data were performed with the z-scored data, then with the MATLAB “xcorr” function using the normalized option such that autocorrelations at zero lag equal 1. Calcium peaks were detected with the MATLAB “findpeaks” function.
CCM.
State-space reconstruction models were generated using the framework of CCM (40), a nonlinear time-series embedding method (62) based on the Takens theorem and its generalized form (63) that builds low-dimensional manifolds from time series and makes predictions across variables. Analysis and predictions were calculated using the R package rEDM 0.7.2 (https://cran.r-project.org/web/packages/rEDM/) for evaluation and rEDM 0.7.4 (https://ha0ye.github.io/rEDM/) for model predictions in the RStudio environment. This program was run on a dual Intel Xeon Gold 6148 server with 384GB RAM or an Intel Core i9 2.4 GHz MacBook Pro with 32 GB RAM. Key parameters were determined individually by lagged coordinate embedding using the simplex function implementation in rEDM to optimize predictive skill as measured by the predicted over observed rho. Parameters include the delay tau, which gives the characteristic timescale of the time series, and the embedding dimensionality, which estimates the number of variables driving the system and approximates the real number of variables as given by the Whitney embedding theorem (64) as minimally equal to the real number n of variables, but no more than two times n + 1 (). The choice of tau was also informed by minimizing mutual information (65). This approximately corresponds to an autocorrelation of ∼0.3, which was applied if it maximized predictive skill across datasets. To prevent data contamination, an exclusion radius was applied that was larger than the respiration rate smoothing window of five timesteps. Whenever the data allowed, an exclusion radius of E*tau was applied, unless the data were insufficient to apply this upper bound. In this case, the exclusion radius would be made just larger than tau.
Chemogenetics.
For KORD-mediated inhibition, SALB (Cayman Chemical) was diluted in 100% DMSO to make a 15 mg/mL stock solution, and then further diluted in 0.9% saline (10% vol/vol) to make a 1.5 mg/mL working suspension. The control used consisted of DMSO diluted in 0.9% saline to make 10% vol/vol. Mice were injected with 5 µL/g of body weight. The final concentration was 7.5 mg/kg. Breathing recording was performed with the WBP 0 to 10 min before and 0 to 10 min after the DMSO/SALB injection as described in Pharmacology. Animals that did not show the viral expression on one side were excluded from the analysis.
For hM3Dq-mediated excitation, CNO) (Cayman Chemical) was diluted in 0.9% saline to make a 1 mg/mL working solution. No precipitate was observed in the solution, indicating that the specific polymorph of the drug exhibits sufficient water solubility. Mice were injected with 5 µL/g of body weight, and the final concentration was 5 mg/kg. Breathing recording and quantification were performed as described in Pharmacology.
RiboTag Profiling and Library Generation.
Isolation of polysome-associated mRNA using RiboTag was performed as previously described with minor modification (45, 66). Briefly, 250 µm thick slices containing the PBL were obtained using a VT1200S Vibratome (Leica). The region of interest was further dissected with surgical scissors. Tissues were transferred into 1.5 mL microcentrifuge tubes containing homogenization buffer (50 mM Tris, pH 7.5, 100 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM dithiothreitol (DTT), 200 U/mL RNasin, 1 mg/mL heparin, 100 μg/mL cycloheximide, and 1× protease inhibitor mixture) and mechanically dissociated and lysed using pellet pestles (7495211500-DS, DWK Life Sciences LLC). Lysates were centrifugated for 10 min at 12,000 rpm at 4 °C. Post-mitochondrial supernatants were transferred to fresh 1.5 mL microcentrifuge tubes. For immunoprecipitation, 4 μL of anti-hemagglutinin 1.1 antibody (BioLegend) was added to the lysate-containing tube and incubated for 4 h at 4 °C on a microtube rotator. After incubation, magnetic protein A/G beads (category no. 88803; Thermo Fisher Scientific) were added to the lysate with antibody prior to incubation on a microtube rotator at 4 °C overnight. The following day, the microcentrifuge tubes were placed into the magnetic stand on ice. The supernatant was removed from sample tubes and used for non-Oprm1 control subsequently. The magnetic beads were washed with high-salt buffer (50 mM Tris, pH 7.5, 300 mM KCl, 12 mM MgCl2, 1% Nonidet P-40, 1 mM DTT, and 100 μg/mL cycloheximide) to remove the nonspecific binding from immunoprecipitation. After washing, 350 μL of RLT plus buffer with β-mercaptoethanol from the RNeasy Mini Kit (Qiagen) was added. The extraction of total RNA was performed with the RNeasy Mini Kit. All RNA samples were quantified with the Qubit RNA Assay Kit (Invitrogen) and analyzed with the RNA 6000 Pico Kit (Agilent).
Isolated RNA was prepared using the Trio RNA-Seq (category no. 0507–08; NuGEN). Briefly, complementary DNA (cDNA) was synthesized from the total RNA using reverse transcriptase with oligo(dT) and resynthesized to produce double-stranded cDNA. After amplification of double-stranded cDNA, cDNA was purified with AMPure XP Beads (category no. A63881; Beckman Coulter), fragmented to the library, and classified using a barcoded adaptor. All libraries were quantified by qPCR and analyzed with the RNA 6000 Pico Kit.
RiboTag Profiling Analysis.
RNA library quality was confirmed using the 2100 Bioanalyzer (Agilent). Barcoded samples were pooled and sequenced on the NextSeq 500 (Illumina) with the 75 bp read length single-end format. Image analysis and base calling were conducted using the Illumina CASAVA-1.8.2 software. Sequencing read quality was evaluated with the FastQC package (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). FastQ reads were aligned to the reference genome (GRCm38.p6) using the STAR tool (version 2.7.2) (67). The quantification package RSEM (version 1.2.28) (68) was utilized to calculate gene expression from BAM files. In doing so, estimated count and TPM (transcripts per million) were computed. Fold changes were calculated from TPM values (estimated counts > 20) between HA-tag and controls. To visualize fold changes, we utilized the ggplot2 package from R. UP (>threefold change) and DOWN (<negative threefold change) were highlighted with orange and blue colors, respectively. Key genes for the parabrachial marker and GPCRs were further annotated.
Histology.
Mice were killed with CO2 at a flow rate of 1.2 L per minute (LPM), perfused intracardially with ice-cold phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde (PFA) in phosphate buffer (PB). The brain was extracted, postfixed in 4% PFA overnight, and dehydrated in 30% sucrose in PBS until sliced. Frozen brains were cut into 30- to 50-µm coronal slices using a CM 1950 cryostat (Leica) and stored in PBS before mounting or immunohistochemistry. The slices were mounted on Superfrost Microscope Slides (Fisher Scientific) with DAPI Fluoromount-G mounting media (Southern Biotech) for imaging.
Immunohistochemistry.
To validate the coexpression of Cre:GFP and Rpl22:HA in PBLOprm1 neurons, immunohistochemistry for HA and GFP was performed. To validate the conditional knockout of the Oprm1 gene in the PBL, we performed immunohistochemistry for MOR.
Sections were washed with PBST (PBS with 3% Tween-20, Fisher Bioreagents). After blocking with 3% normal donkey serum (NDS, Jackson ImmunoResearch, Inc.) for 1 h at room temperature and rinsing with PBST, the slices were incubated with the corresponding antibodies: mouse monoclonal anti-HA1.1 (1:1,000, BioLegend), chicken anti-GFP (1:1,000, Aves Labs, Inc.), and rabbit anti-MOR (1:1,000, ImmunoStar) antibodies at 4 °C overnight (anti-HA1.1 and anti-GFP) or for 24 h (anti-MOR). The next day, brain tissues were rinsed with PBST and incubated in Alexa Fluor 647-conjugated Donkey Anti-Mouse IgG, Alexa Fluor 488-conjugated Donkey Anti-Chicken IgY, and Alexa Fluor 647-conjugated Donkey Anti-Rabbit IgG (1:1,000, Jackson ImmunoResearch, Inc.) in 3% NDS for 90 min at room temperature. After washing with PBS, brain slices were mounted on Superfrost Microscope Slides with DAPI Fluoromount-G mounting media for imaging.
RNA In Situ Hybridization.
RNA in situ hybridization was performed using the RNAscope Fluorescent Multiplex Assay using the probes and kits purchased from Advanced Cell Diagnostics (ACD). Brains were collected from wild-type mice and immediately frozen with 2-methylbutane chilled with dry ice. Frozen brains were cut into 20 µm coronal slices with a CM 1950 cryostat and directly mounted onto the Superfrost Plus Microscope Slides (Fisher Scientific). Sample preparation, pretreatment, and signal detection were performed according to the ACD protocols. Probes used are listed below: Oprm1 (ACD #315841), Htr2a (#401291), Cckar (#313751), Drd5 (#494411), Tacr3 (#481671), Tacr1 (#428781), Cre (#402551), Slc17a6 (#319171), and Slc32a1 (#319191).
Two to three representative images from the PBL (AP = −5.0 to −5.2) were selected from n = 3 mice, and PBL cells within a field of view of 300 µm × 300 µm (for Oprm1 colocalization with Cre, Htr2a, Cckar, Tacr3, Tacr1, and Drd5) or 600 µm × 600 µm (for Oprm1 colocalization with Slc17a6 and Slc32a1) were used for quantification. Quantification of the colocalization is done manually with ImageJ software according to the ACDBio technical note. DAPI-stained nuclei were first identified, and then the cell contour was defined with a 2 µm radius surrounding the DAPI signals. Cells containing at least five puncta inside the imaginary boundary were labeled as positive.
Imaging.
Images for verification of injection and implantation site and anti-MOR staining for conditional knockout validation were taken with a BZ-X710 all-in-one fluorescence microscope with BZ-X viewer software under a 10×, 0.45 NA objective (Keyence). Images for anti-HA and anti-GFP immunostaining, RNAscope, and Oprm1Cre:Ai14 expression characterization were acquired with an FV3000 Confocal Laser Scanning Microscope with FV31S-SW software under a 20×, 0.75 NA or 40×, 0.95 NA UPLSAPO objective (Olympus). For comparison, images were processed with the same gain, offset, and exposure time.
Locomotor Activity Test.
To measure the locomotor activity after morphine injection, Oprm1Cre/+ mice were injected with 40 mg/kg morphine or saline intraperitoneally and placed back into the home cage. After 30 to 40 min, animals were introduced to the center of an open field arena (50 cm × 50 cm) and allowed to explore for 10 min. The velocity of the animal was analyzed using the video tracking software (EthoVision XT 12, Noldus Information Technology, Inc.).
Statistical Analysis.
All data are shown as mean ± SEM and analyzed using either a Student’s t test, one-way ANOVA with Tukey’s post hoc comparison, or two-way ANOVA with Bonferroni’s post hoc comparison. All statistical analyses were performed with Prism 6 (GraphPad Software, Inc.).
For a detailed list of resources, please refer to SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank the Han laboratory members for the critical discussion of the paper and D. O’Keefe and M. Shields for critical input on the manuscript. S.H. is supported by 1R01MH116203 from the National Institute of Mental Health, the Bridge to Independence award from the Simons Foundation Autism Research Initiative (SFARI #388708), and the Brain Research Foundation Fay/Frank Seed Grant. K.-F.L. is supported by MH114831, OD023076, AG062232, NS115183, and AG064049. M.R.B. is supported by R00DA034648, the Klingenstein-Simons Fellowship Award in Neuroscience, and the Rita Allen Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022134118/-/DCSupplemental.
Data Availability
The transcriptome datasets in the current study are deposited in a public repository (National Center for Biotechnology Information, PRJNA693293).
References
- 1.Hedegaard H., Miniño A. M., Warner M., Drug overdose deaths in the United States, 1999-2018. NCHS Data Brief 356, 1–8 (2020). [PubMed] [Google Scholar]
- 2.Pattinson K. T., Opioids and the control of respiration. Br. J. Anaesth. 100, 747–758 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Dahan A., Aarts L., Smith T. W., Incidence, reversal, and prevention of opioid-induced respiratory depression. Anesthesiology 112, 226–238 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Skolnick P., On the front lines of the opioid epidemic: Rescue by naloxone. Eur. J. Pharmacol. 835, 147–153 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Dahan A., et al., Averting opioid-induced respiratory depression without affecting analgesia. Anesthesiology 128, 1027–1037 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Boyer E. W., Management of opioid analgesic overdose. N. Engl. J. Med. 367, 146–155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roozekrans M., et al., Two studies on reversal of opioid-induced respiratory depression by BK-channel blocker GAL021 in human volunteers. Anesthesiology 121, 459–468 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Oertel B. G., et al., Selective antagonism of opioid-induced ventilatory depression by an ampakine molecule in humans without loss of opioid analgesia. Clin. Pharmacol. Ther. 87, 204–211 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Manzke T., et al., 5-HT4(a) receptors avert opioid-induced breathing depression without loss of analgesia. Science 301, 226–229 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Oertel B. G., et al., The partial 5-hydroxytryptamine1A receptor agonist buspirone does not antagonize morphine-induced respiratory depression in humans. Clin. Pharmacol. Ther. 81, 59–68 (2007). [DOI] [PubMed] [Google Scholar]
- 11.van der Schier R., Roozekrans M., van Velzen M., Dahan A., Niesters M., Opioid-induced respiratory depression: Reversal by non-opioid drugs. F1000Prime Rep. 6, 79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthes H. W., et al., Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature 383, 819–823 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Dahan A., et al., Anesthetic potency and influence of morphine and sevoflurane on respiration in μ-opioid receptor knockout mice. Anesthesiology 94, 824–832 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Miller J. R., et al., A subregion of the parabrachial nucleus partially mediates respiratory rate depression from intravenous remifentanil in young and adult rabbits. Anesthesiology 127, 502–514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cinelli E., Bongianni F., Pantaleo T., Mutolo D., Activation of μ-opioid receptors differentially affects the preBötzinger Complex and neighbouring regions of the respiratory network in the adult rabbit. Respir. Physiol. Neurobiol. 280, 103482 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Wei A. D., Ramirez J.-M., Presynaptic mechanisms and KCNQ potassium channels modulate opioid depression of respiratory drive. Front. Physiol. 10, 1407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prkic I., et al., Pontine μ-opioid receptors mediate bradypnea caused by intravenous remifentanil infusions at clinically relevant concentrations in dogs. J. Neurophysiol. 108, 2430–2441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varga A. G., Reid B. T., Kieffer B. L., Levitt E. S., Differential impact of two critical respiratory centres in opioid-induced respiratory depression in awake mice. J. Physiol. 598, 189–205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmutsky I., Wei X. P., Kish E., Yackle K., Opioids depress breathing through two small brainstem sites. eLife 9, e52694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montandon G., et al., PreBotzinger complex neurokinin-1 receptor-expressing neurons mediate opioid-induced respiratory depression. J. Neurosci. 31, 1292–1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montandon G., Horner R., CrossTalk proposal: The preBotzinger complex is essential for the respiratory depression following systemic administration of opioid analgesics. J. Physiol. 592, 1159–1162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lalley P. M., Pilowsky P. M., Forster H. V., Zuperku E. J., CrossTalk opposing view: The pre-Botzinger complex is not essential for respiratory depression following systemic administration of opioid analgesics. J. Physiol. 592, 1163–1166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Negro C. A., Funk G. D., Feldman J. L., Breathing matters. Nat. Rev. Neurosci. 19, 351–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J. C., Ellenberger H. H., Ballanyi K., Richter D. W., Feldman J. L., Pre-Bötzinger complex: A brainstem region that may generate respiratory rhythm in mammals. Science 254, 726–729 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Y., et al., Defining preBötzinger complex rhythm- and pattern-generating neural microcircuits in vivo. Neuron 91, 602–614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamberlin N. L., Saper C. B., Topographic organization of respiratory responses to glutamate microstimulation of the parabrachial nucleus in the rat. J. Neurosci. 14, 6500–6510 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutschmann M., Dick T. E., Pontine mechanisms of respiratory control. Compr. Physiol. 2, 2443–2469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarrete-Opazo A. A., et al., Endogenous glutamatergic inputs to the Parabrachial Nucleus/Kölliker-Fuse Complex determine respiratory rate. Respir. Physiol. Neurobiol. 277, 103401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur S., et al., A genetically defined circuit for arousal from sleep during hypercapnia. Neuron 96, 1153–1167.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M., Alheid G. F., Calandriello T., McCrimmon D. R., Parabrachial-lateral pontine neurons link nociception and breathing. Respir. Physiol. Neurobiol. 143, 215–233 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Song G., Poon C. S., Lateral parabrachial nucleus mediates shortening of expiration and increase of inspiratory drive during hypercapnia. Respir. Physiol. Neurobiol. 165, 9–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song G., Poon C. S., Lateral parabrachial nucleus mediates shortening of expiration during hypoxia. Respir. Physiol. Neurobiol. 165, 1–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaur S., et al., Glutamatergic signaling from the parabrachial nucleus plays a critical role in hypercapnic arousal. J. Neurosci. 33, 7627–7640 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders S. E., Levitt E. S., Kölliker-Fuse/Parabrachial complex mu opioid receptors contribute to fentanyl-induced apnea and respiratory rate depression. Respir. Physiol. Neurobiol. 275, 103388 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lalley P. M., Opiate slowing of feline respiratory rhythm and effects on putative medullary phase-regulating neurons. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1387–R1396 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Levitt E. S., Abdala A. P., Paton J. F., Bissonnette J. M., Williams J. T., μ opioid receptor activation hyperpolarizes respiratory-controlling Kölliker-Fuse neurons and suppresses post-inspiratory drive. J. Physiol. 593, 4453–4469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutschmann M., Herbert H., The Kölliker-Fuse nucleus gates the postinspiratory phase of the respiratory cycle to control inspiratory off-switch and upper airway resistance in rat. Eur. J. Neurosci. 24, 1071–1084 (2006). [DOI] [PubMed] [Google Scholar]
- 38.McAfee S. S., et al., Minimally invasive highly precise monitoring of respiratory rhythm in the mouse using an epithelial temperature probe. J. Neurosci. Methods 263, 89–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S., et al., Identification of a novel breathing circuit that controls pain and anxiety. bioRxiv [Preprint] (2020). 10.1101/2020.01.09.900738 (Accessed 10 January 2021). [DOI]
- 40.Sugihara G., et al., Detecting causality in complex ecosystems. Science 338, 496–500 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Roth B. L., DREADDs for neuroscientists. Neuron 89, 683–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vardy E., et al., A new DREADD facilitates the multiplexed chemogenetic interrogation of behavior. Neuron 86, 936–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Che T., et al., Structure of the nanobody-stabilized active state of the kappa opioid receptor. Cell 172, 55–67.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Hasani R., Bruchas M. R., Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology 115, 1363–1381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanz E., Bean J. C., Carey D. P., Quintana A., McKnight G. S., RiboTag: Ribosomal tagging strategy to analyze cell-type-specific mRNA expression in vivo. Curr. Protoc. Neurosci. 88, e77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmiter R. D., The parabrachial nucleus: CGRP neurons function as a general alarm. Trends Neurosci. 41, 280–293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang D., Grady F. S., Peltekian L., Geerling J. C., Efferent projections of Vglut2, Foxp2, and Pdyn parabrachial neurons in mice. J. Comp. Neurol. 529, 657–693 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niebert M., et al., Expression and function of serotonin 2A and 2B receptors in the mammalian respiratory network. PLoS One 6, e21395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellenberger H. H., Smith F. M., Sulfated cholecystokinin octapeptide in the rat: Pontomedullary distribution and modulation of the respiratory pattern. Can. J. Physiol. Pharmacol. 77, 490–504 (1999). [PubMed] [Google Scholar]
- 50.Hedner J., Hedner T., Wessberg P., Jonason J., Interaction of substance P with the respiratory control system in the rat. J. Pharmacol. Exp. Ther. 228, 196–201 (1984). [PubMed] [Google Scholar]
- 51.Arvidsson U., et al., Distribution and targeting of a mu-opioid receptor (MOR1) in brain and spinal cord. J. Neurosci. 15, 3328–3341 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurlé M. A., Mediavilla A., Flórez J., Differential respiratory patterns induced by opioids applied to the ventral medullary and dorsal pontine surfaces of cats. Neuropharmacology 24, 597–606 (1985). [DOI] [PubMed] [Google Scholar]
- 53.Yokota S., Kaur S., VanderHorst V. G., Saper C. B., Chamberlin N. L., Respiratory-related outputs of glutamatergic, hypercapnia-responsive parabrachial neurons in mice. J. Comp. Neurol. 523, 907–920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulwiler C. E., Saper C. B., Subnuclear organization of the efferent connections of the parabrachial nucleus in the rat. Brain Res. 319, 229–259 (1984). [DOI] [PubMed] [Google Scholar]
- 55.Erbs E., et al., A mu-delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct. Funct. 220, 677–702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mustapic S., et al., Clinically relevant infusion rates of μ-opioid agonist remifentanil cause bradypnea in decerebrate dogs but not via direct effects in the pre-Bötzinger complex region. J. Neurophysiol. 103, 409–418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stucke A. G., et al., Opioid-induced respiratory depression is only partially mediated by the preBötzinger complex in young and adult rabbits in vivo. Anesthesiology 122, 1288–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson T. M., et al., A novel excitatory network for the control of breathing. Nature 536, 76–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tokita K., Inoue T., Boughter J. D. Jr, Afferent connections of the parabrachial nucleus in C57BL/6J mice. Neuroscience 161, 475–488 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaur S., et al., Role of serotonergic dorsal raphe neurons in hypercapnia-induced arousals. Nat. Commun. 11, 2769 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta K., et al., Risk factors for opioid-induced respiratory depression and failure to rescue: A review. Curr. Opin. Anaesthesiol. 31, 110–119 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Sugihara G., May R. M., Nonlinear forecasting as a way of distinguishing chaos from measurement error in time series. Nature 344, 734–741 (1990). [DOI] [PubMed] [Google Scholar]
- 63.Deyle E. R., Sugihara G., Generalized theorems for nonlinear state space reconstruction. PLoS One 6, e18295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitney H., The imbedding of manifolds in families of analytic manifolds. Ann. Math. 37, 865–878 (1936). [Google Scholar]
- 65.Fraser A. M., Swinney H. L., Independent coordinates for strange attractors from mutual information. Phys. Rev. A Gen. Phys. 33, 1134–1140 (1986). [DOI] [PubMed] [Google Scholar]
- 66.Sanz E., et al., Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U.S.A. 106, 13939–13944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome datasets in the current study are deposited in a public repository (National Center for Biotechnology Information, PRJNA693293).