Significance
The preparation of membrane-protein specimens for structural characterization commonly involves a detergent-solubilization step that is sometimes followed by the reincorporation of the protein into laboratory-made lipid nanodiscs. Whether the native conformation and arrangement of tightly bound lipids are retained despite solubilization or restored upon nanodisc formation cannot be predicted. To determine the structure of the bacterial pentameric ligand-gated ion channel ELIC in its native environment, we overexpressed it in Escherichia coli, fragmented the inner membranes into disc-shaped structures in the absence of detergents, purified discs containing single copies of the protein, and imaged the resulting particles using cryogenic electron microscopy. The obtained three-dimensional reconstruction of the unliganded channel (2.5-Å resolution) revealed previously unseen details of the lipid–protein interface.
Keywords: Cys-loop receptors, nicotinic receptors, cardiolipin, cryo-EM, styrene–maleic acid nanodiscs
Abstract
Although it has long been proposed that membrane proteins may contain tightly bound lipids, their identity, the structure of their binding sites, and their functional and structural relevance have remained elusive. To some extent, this is because tightly bound lipids are often located at the periphery of proteins, where the quality of density maps is usually poorer, and because they may be outcompeted by detergent molecules used during standard purification procedures. As a step toward characterizing natively bound lipids in the superfamily of pentameric ligand-gated ion channels (pLGICs), we applied single-particle cryogenic electron microscopy to fragments of native membrane obtained in the complete absence of detergent-solubilization steps. Because of the heterogeneous lipid composition of membranes in the secretory pathway of eukaryotic cells, we chose to study a bacterial pLGIC (ELIC) expressed in Escherichia coli’s inner membrane. We obtained a three-dimensional reconstruction of unliganded ELIC (2.5-Å resolution) that shows clear evidence for two types of tightly bound lipid at the protein–bulk-membrane interface. One of them was consistent with a “regular” diacylated phospholipid, in the cytoplasmic leaflet, whereas the other one was consistent with the tetra-acylated structure of cardiolipin, in the periplasmic leaflet. Upon reconstitution in E. coli polar-lipid bilayers, ELIC retained the functional properties characteristic of members of this superfamily, and thus, the fitted atomic model is expected to represent the (long-debated) unliganded-closed, “resting” conformation of this ion channel. Notably, the addition of cardiolipin to phosphatidylcholine membranes restored the ion-channel activity that is largely lost in phosphatidylcholine-only bilayers.
The extent to which the lipid environment contributes to the conformational free-energy landscape of membrane proteins and the mechanism underlying this effect remain outstanding questions (e.g., refs. 1 and 2). In the particular case of the pentameric ligand-gated ion channel (pLGIC) superfamily, an effect of the membrane composition on function was first proposed for the muscle-type nicotinic acetylcholine receptor (AChR) from the electric organ of Torpedo fish several decades ago on the basis of ligand-binding and ion-flux studies (3–10). Since then, the lipid dependence of ion-channel function has become a recurring theme in the literature for the entire superfamily (e.g., ref. 11).
A variety of experimental approaches (such as NMR spectroscopy, mass spectrometry, and X-ray crystallography) have revealed that a fraction of membrane phospholipids often associate tightly and in a stoichiometric manner to integral membrane proteins (e.g., refs. 12–14). In the last few years, cryogenic electron microscopy (cryo-EM) single-particle analysis of lipid nanodisc–embedded proteins (e.g., ref. 15) has emerged as a powerful tool to elucidate this aspect of membrane biology. Moreover, the discovery of amphipathic copolymers that can “carve” nanometer-sized fragments of membrane (16) has paved the way to structural studies of membrane proteins embedded in their native environments in the complete absence of detergent-solubilization steps.
It is intriguing to ponder that structural phospholipids may contribute to the overall effect of the membrane on function by binding to proteins with conformation-specific affinities, much like signaling phospholipids do. Some experimental evidence for this notion has been presented for phosphatidylglycerol in the bacterial KcsA K+ channel (17) and for cardiolipin in the calcium uniporter (18) and the ADP/ATP carrier (19), both from yeast mitochondria, for example. In the case of pLGICs, unambiguous experimental evidence for or against this idea has been difficult to obtain, in part because of the challenging process of identifying tightly bound lipids in the first place. Indeed, on the one hand, natively bound phospholipids may be removed or dislocated by detergent-solubilization steps. On the other hand—even if the use of detergents were omitted and fragments of pLGIC-containing native membrane were purified, instead—the identification of such lipids would not be a foregone conclusion because most of them seem to bind to the periphery of the channel, where the quality of single-particle cryo-EM three-dimensional (3D) reconstructions tends to be poorer (20, 21).
As a step toward gaining a better understanding of the lipid dependence of pLGIC function, we set out to apply single-particle cryo-EM to nanometer-sized fragments of native membrane (“nanodiscs”) containing single copies of protein. In an attempt to reduce the heterogeneity of the lipid composition of these discs, we reasoned that a bacterial expression system could be advantageous because the channel would be inserted in a single type of membrane—the inner membrane of a gram-negative bacterium, for example—as opposed to the several membranes of the secretory pathway of eukaryotic cells (22, 23). Because the bacterial pLGIC ELIC (from the gram-negative bacterium Erwinia chrysanthemi) has been shown to be sensitive to the chemical composition of the lipid bilayer (24–26), we chose it as the subject of this work; we expressed it heterologously in Escherichia coli cells.
We obtained a 3D reconstruction of unliganded ELIC in native E. coli membranes at a global resolution [gold-standard, 0.143 Fourier shell correlation (FSC)-threshold criterion (27, 28)] of 2.5 Å. To the extent that ELIC displayed electrophysiological properties characteristic of pLGICs upon being reconstituted in E. coli polar-lipid bilayers, we reasoned that the atomic model fitted to this density map must represent the unliganded-closed, “resting” conformation. At the protein–membrane interface, two distinct groups of density features consistent with phospholipid acyl chains could be clearly observed. Whereas one of them was consistent with a “regular” diacylated phospholipid, the other one was consistent with the tetra-acylated structure of cardiolipin. Because of its uniqueness, here, we focused on the latter. Notably, the addition of physiological levels of cardiolipin to phosphatidylcholine membranes restored the ion-channel activity that is largely lost in phosphatidylcholine-only bilayers.
Results
Structure and Function of ELIC in E. coli Membranes.
Reasoning that the interactions between membrane proteins and tightly bound phospholipids may be weakened by detergent solubilization, we decided to image ELIC in fragments of native bacterial inner membrane. To achieve this, we overexpressed wild-type ELIC in E. coli cells, converted the native inner membranes into disk-shaped entities using a styrene–maleic acid (SMA) copolymer (16), purified discs containing single copies of ELIC (“ELIC–SMA”), and imaged the resulting nanodiscs using single-particle cryo-EM—being also a γ proteobacterium, the lipid composition of E. coli’s inner membrane is expected to mimic closely that of E. chrysanthemi (29). Micrographs of unliganded protein were collected using a Titan Krios microscope (ThermoFisher Scientific) equipped with a Gatan K2 direct electron detector, and the 3D reconstruction was obtained using cryoSPARC version 2.15 (30, 31). The global resolution (half-map–half-map correlation; 0.143 threshold) was 2.5 Å (SI Appendix, Fig. S1 and Table S1), and the local resolution varied within a narrow range (SI Appendix, Fig. S2). The structural interpretability of the map was enhanced by adjusting the Fourier-coefficient amplitudes of the sum of experimental half maps in a local-resolution–dependent manner using a combination of MonoRes (32) and LocalDeblur (33). This “local-sharpening” step turned out to be crucial for the identification of density features consistent with the presence of bound phospholipids (SI Appendix, Fig. S3). Attempts to generate two or more density maps from our dataset [using cryoSPARC version 2.15 (30, 31)], indicative of two or more conformations of the protein–lipid complex, led us to conclude that, when unliganded, ELIC seems to adopt a single major conformation.
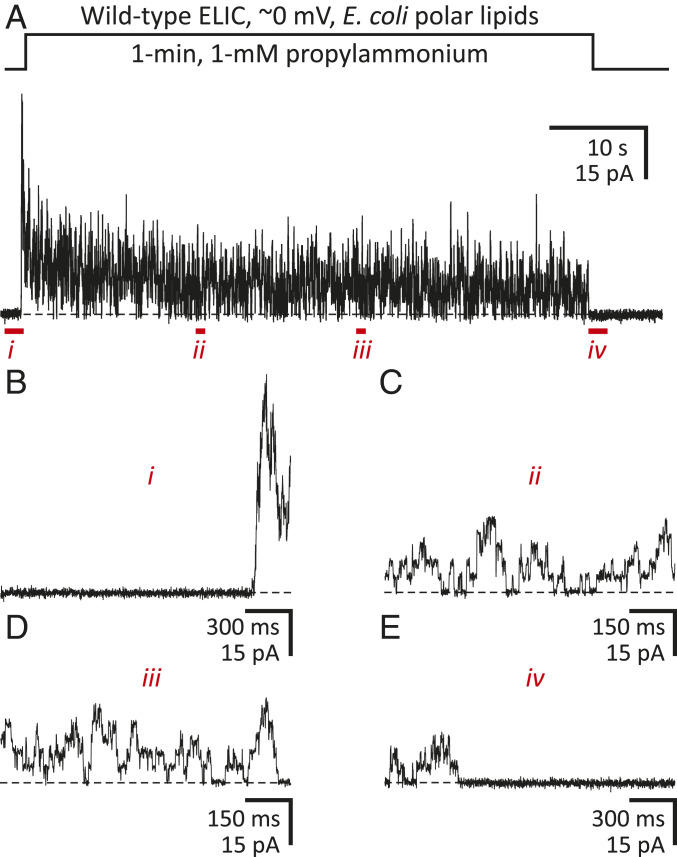
To learn about the functional properties of the channel when embedded in E. coli membranes—and thus, to be able to make inferences as to the functional state of our structural model—we purified detergent-solubilized ELIC, reconstituted it into cell-sized liposomes prepared from a mixture of E. coli polar lipids (Avanti Polar Lipids), and recorded currents elicited in response to the extracellular application of agonist (1 mM propylammonium). In excised patches of liposomal membrane, ELIC proved to be fully functional, activating and desensitizing upon exposure to propylammonium, and deactivating to a zero-current level upon agonist washout (Fig. 1). Thus, assuming that the properties of bilayers formed by this lipid extract from E. coli (67% phosphatidylethanolamine, 23% phosphatidylglycerol, and 10% cardiolipin, by weight) do not deviate much from those of the inner membrane [75% phosphatidylethanolamine, 15 to 20% phosphatidylglycerol, and 5 to 10% cardiolipin, by weight (34, 35)], and that electrically silent patches represent patches of membrane either without channels or with channels inserted in the opposite orientation, it seems reasonable to expect that unliganded ELIC embedded in native nanodiscs adopts the closed, resting conformation.
Fig. 1.
ELIC function in E. coli polar-lipid membranes. Outward currents recorded from inside-out patches of membrane excised from E. coli polar-lipid liposomes reconstituted with wild-type ELIC. (A) Response of a representative excised patch of membrane to a 1-min pulse of 1 mM propylammonium. Openings are upward deflections. The distribution of ions across the membrane was asymmetrical (∼150 mM/5 mM K+, inside/outside). The command potential was zero, and the +2.7-mV liquid-junction potential between the pipette and bath solutions was offset. As a result, the membrane potential was close to zero, thus approximating the conditions used for cryo-EM image acquisition. The black dashed lines denote the zero-current baseline. (B–E) Details of A at expanded time scales.
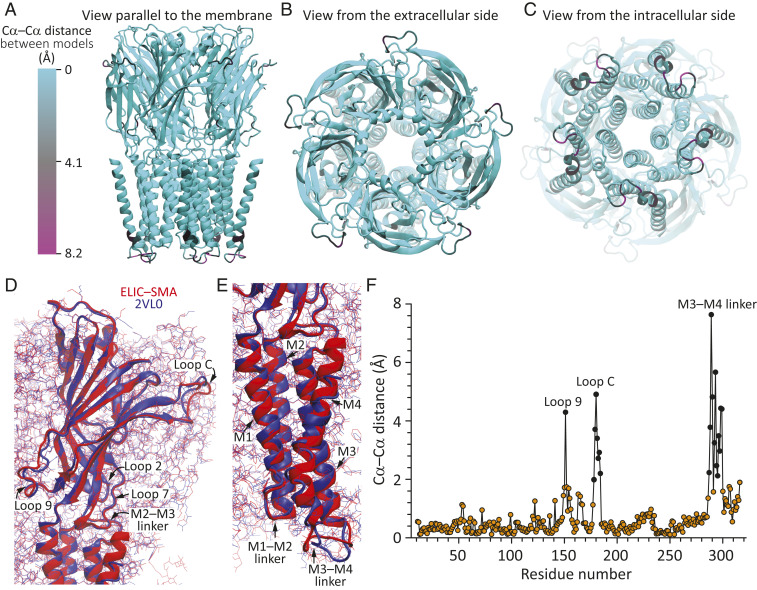
An atomic model was built from Pro-11 to Thr-321 (SI Appendix, Fig. S4); only the first 10 and the last residue(s) of the protein could not be built with confidence. Global superposition of this model of unliganded ELIC–SMA (excluding the phospholipids) to the atomic model of detergent-solubilized unliganded ELIC obtained from X-ray crystallography [Protein Data Bank (PDB) ID: 2VL0 (36)] revealed that both models are very similar (Fig. 2; Cα RMSD = 1.2 Å). A residue-to-residue comparison of Cα atom coordinates revealed that the largest differences (>2.0 Å) between the two atomic models occur at the level of a few residues (178 to 184) in loop C (the distance between these stretches’ geometrical centers being as small as 0.7 Å, however), one residue in loop 9 (Asn-151), and the M3–M4 linker (Fig. 2F). Otherwise, the similarity between the two models is remarkable, particularly in flexible regions such as loops 2 and 7, and the M1–M2 and M2–M3 linkers (Fig. 2 D and E). Comparison of pore-radius profiles revealed very small differences (SI Appendix, Fig. S5) that resulted from minor discrepancies in the dihedral angles of the modeled rotamer side chains; at the level of backbone atoms, the two sets of five pore-lining M2 α-helices overlap nearly completely (SI Appendix, Fig. S6 A–C). Thus, the long-standing question as to whether the X-ray crystal structure of detergent-solubilized ELIC represents the unliganded-closed conformation of this channel (e.g., refs. 38 and 39) seems to be finally settled: it definitely does. Two notable differences between the transmembrane pores of unliganded-closed ELIC and the unliganded-closed conformation of most other pLGICs are the location of the narrowest constriction (the “activation gate”)—at position 16ʹ in ELIC rather than at the more common position 9ʹ—and a wider intracellular mouth. Both differences result from a combination of different amino acid sequences and different rotation and tilt angles of the M2 α-helices, as exemplified by a superposition of unliganded ELIC–SMA to the atomic model of unliganded α-GluCl [PDB ID: 4TNV (40)] (SI Appendix, Fig. S6 D–F).
Fig. 2.
Cryo-EM structure of unliganded ELIC in SMA nanodiscs. Global superposition of two atomic models of unliganded ELIC: reconstituted in SMA nanodiscs and detergent solubilized [PDB ID: 2VL0 (36)]. The two models were superposed “globally” in such a way as to minimize the Cα–Cα distance between identically numbered residues in the stretch ranging from Pro-11 (the first residue in both models) to Ile-317 (the last residue modeled in detergent-solubilized ELIC) of all five subunits and omitting Gly-164 (which was not included in the 2VL0 model). (A–C) Different views of the ELIC–SMA model displayed in ribbon representation and colored according to Cα–Cα distance from the 2VL0 model. The color code is the same for all three panels. (D and E) View (parallel to the plane of the membrane) of the extracellular and transmembrane domains, respectively. For clarity, only one subunit is displayed in ribbon representation; all others are displayed as lines. The color code is the same for both panels. (F) Cα–Cα distances as a function of residue number. Distances ≥2 Å are denoted with black symbols. Including all superposed residues, the mean and RMSD values of the Cα–Cα distance were 0.5 and 1.2 Å, respectively. The molecular images were prepared with Visual Molecular Dynamics (37).
We have previously generated an atomic model of unliganded ELIC embedded in 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC)-only nanodiscs [“ELIC–POPC” (26)]. In addition to the different lipid composition, ELIC–POPC nanodiscs differed from native ones in that ELIC was detergent solubilized prior to its incorporation into lipid discs, and that a membrane-scaffolding protein [MSP1E3D1 (41)], rather than an SMA copolymer, was used to shield the phospholipid acyl chains from the surrounding aqueous solution. The POPC-only model was built into a moderate-resolution (4.1 Å) 3D reconstruction whose quality was particularly poor at the level of the transmembrane region. In an attempt to improve the quality of this map, here, we reprocessed the micrographs using cryoSPARC version 2.15 (30, 31). The global resolution of the new reconstruction increased to 3.3 Å (SI Appendix, Fig. S7A and Table S1), and the structural interpretability of the map was much improved (SI Appendix, Fig. S7B). Global superposition of the corresponding structural model to that of unliganded ELIC–SMA revealed that the two models are, essentially, identical (SI Appendix, Fig. S7 C–E). Thus, whether unliganded ELIC is embedded in a membrane that fully supports ion-channel function (E. coli inner membrane), one that does very poorly [POPC (26)], or whether unliganded ELIC is solubilized and crystallized (36) does not seem to make a difference to its structure.
Structure and Function of Tightly Bound Cardiolipin.
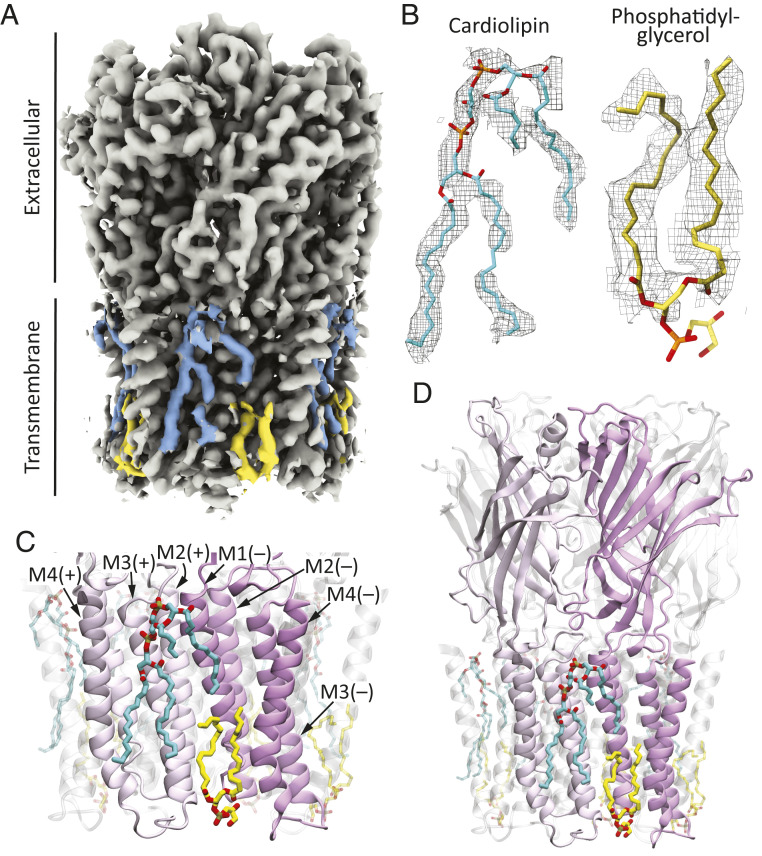
The obtained 3D reconstruction of unliganded ELIC–SMA showed density features consistent with firmly bound phospholipid molecules at the interface between the protein and the bulk membrane (Fig. 3A). Although the unequivocal identification of the type of polar head group was not possible, the quality of the reconstruction was high enough to suggest the presence of two phospholipid molecules per subunit: a diacylated phospholipid with its head group at the membrane–cytoplasm interface and a more-complex structure that we attributed to a tetra-acylated phospholipid with its head group close to the membrane–periplasm interface. We modeled the former as phosphatidylglycerol and the latter as cardiolipin (Fig. 3B). Cardiolipin may be regarded as two phosphatidate moieties esterified to the primary carbons of a single glycerol molecule, and thus, cardiolipin contains two ionizable groups that, at least when exposed to bulk solution, are expected to be negatively charged (43). The density we attributed to phosphatidylglycerol is almost undoubtedly the same density modeled as phosphatidylethanolamine by Ulens and coworkers [PDB ID: 6HJX (24)] in a recent crystal structure of detergent-solubilized ELIC; our data does not allow us to distinguish between the two. As for the density we attributed to cardiolipin, a detergent molecule was modeled in this site in the aforementioned crystal structure.
Fig. 3.
Protein–membrane interface of unliganded ELIC. (A) Unfiltered sum of experimental half maps sharpened locally. The density features modeled as cardiolipin (cyan) and phosphatidylglycerol (yellow) are highlighted. (B) Atomic models of firmly bound phospholipids and corresponding densities (display level = 0.7). (C) Atomic model of the protein–membrane interface. The phospholipids are displayed in stick representation, and the protein, in ribbon. The two front subunits are colored in different shades of magenta to highlight the binding sites of cardiolipin and phosphatidylglycerol in our unliganded ELIC–SMA model. At every subunit–subunit interface, the individual subunits are referred to as “principal” (“+”) or “complementary” (“–”). (D) A lower-magnification view of the model shown in (C) including the entire protein. The molecular images were prepared with Visual Molecular Dynamics (37) and Chimera X (42).
In our atomic model of unliganded ELIC–SMA, cardiolipin binds to the membrane-facing side of transmembrane α-helix M3 (and to a smaller extent, M4) of each subunit and of M1 of the adjacent subunit in the counterclockwise direction, as seen from the periplasmic side (that is, the “principal” and “complementary” subunits, respectively; Fig. 3 C and D). One of the two phosphatidate “halves” of cardiolipin binds to M1 and the other one to M3 (and M4). This site overlaps with the binding site of the allosteric agonist ivermectin in, for example, the invertebrate pLGIC α-GluCl [PDB ID: 3RIF (44)] (SI Appendix, Fig. S8). Unlike ivermectin, however, which intercalates between pairs of adjacent M1 and M3 α-helices, cardiolipin binds “flat” against these two transmembrane segments facing the bulk of the membrane in a manner that resembles the binding pose of cardiolipin to, for example, bacterial photosynthetic reaction centers [e.g., PDB ID: 1QOV (45)] or mitochondrial ATP-binding cassette transporters [e.g., PDB ID: 3ZDQ (46)]. Cardiolipin has also been observed to bind in-between transmembrane α-helices, in an “ivermectin-like” manner, to other membrane proteins (e.g., refs. 47 and 48). ELIC-bound cardiolipin adopts a nonbilayer conformation. Certainly, one of the two 1,2-acylated glycerol moieties is “buried” well below the line expected to be occupied by the outer leaflet glycerol groups, and two acyl chains nearly reach the opposite side of the membrane, whereas the other two acyl chains curve away from an axis perpendicular to the plane of the bilayer (Fig. 3 C and D). Distorted conformations such as this one have been observed for lipids in structural models of several other membrane proteins (SI Appendix, Fig. S9), notably including the muscle-type AChR [PDB ID 6UWZ (49)] (SI Appendix, Fig. S10), and they likely result from the need of the lipids to adapt closely to the irregular surface presented by the protein in order to prevent leaks.
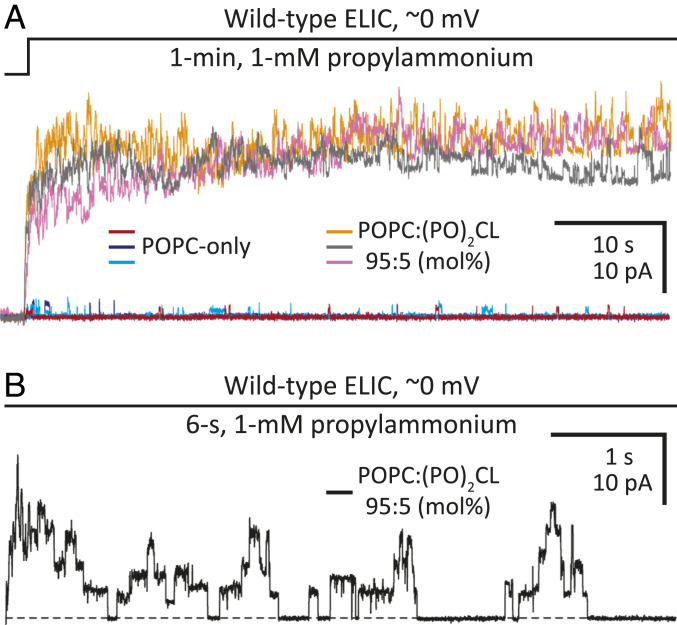
To evaluate the importance of cardiolipin to ion-channel function, we reconstituted detergent-solubilized ELIC in POPC liposomes containing physiological levels of cardiolipin. Although POPC-only membranes supported channel activity very poorly [Fig. 4 (26)], the addition of palmitoyl-oleoyl cardiolipin (1′,3′-bis[1-palmitoyl-2-oleoyl-sn-glycero-3-phospho]-glycerol; (PO)2CL) to form a 95:5 POPC:(PO)2CL (mol%) binary mixture (∼90:10 by weight) increased the channel’s open probability considerably (Fig. 4), strongly suggesting a functional role for this unique acidic phospholipid. As was the case also for POPC-only bilayers, obtaining long-lasting, electrically tight patch-clamp seals with this lipid mixture was far more challenging than it was with membranes of more-complex composition (such as asolectin or E. coli polar lipids), but clear currents, characteristic of ELIC, could nevertheless be recorded. The use of a lipid mixture containing only a low proportion of cardiolipin and lacking other acidic phospholipids was deemed necessary to isolate the effect of physiological levels of cardiolipin on channel function.
Fig. 4.
ELIC function in cardiolipin-free and cardiolipin-containing POPC bilayers. Outward currents recorded from inside-out patches of membrane excised from liposomes of the indicated composition reconstituted with wild-type ELIC. The distribution of ions across the membrane was asymmetrical (∼150 mM/5 mM K+, inside/outside). The command potential was zero, and the +2.7-mV liquid-junction potential between the pipette and bath solutions was offset. As a result, the membrane potential was close to zero, thus approximating the conditions used for cryo-EM image acquisition. (A) Three representative responses to the application of 1 mM propylammonium for each type of bilayer. Openings are upward deflections. In each case, ion-channel activity is shown for 1 min. (B) A 6-s stretch of ELIC activity during the application of 1 mM propylammonium to a patch of cardiolipin-containing POPC bilayer. The overall ion-channel activity of this recording was lower than that in the traces shown in (A), and thus, single-channel openings, with the highly characteristic subconductance behavior of ELIC, could be clearly observed. A black dashed line denotes the zero-current baseline. As expected from a negatively charged lipid, the addition of cardiolipin to POPC bilayers increased the single-channel conductance of this cation-selective channel.
Structure and Function of the Cardiolipin-Binding Sites.
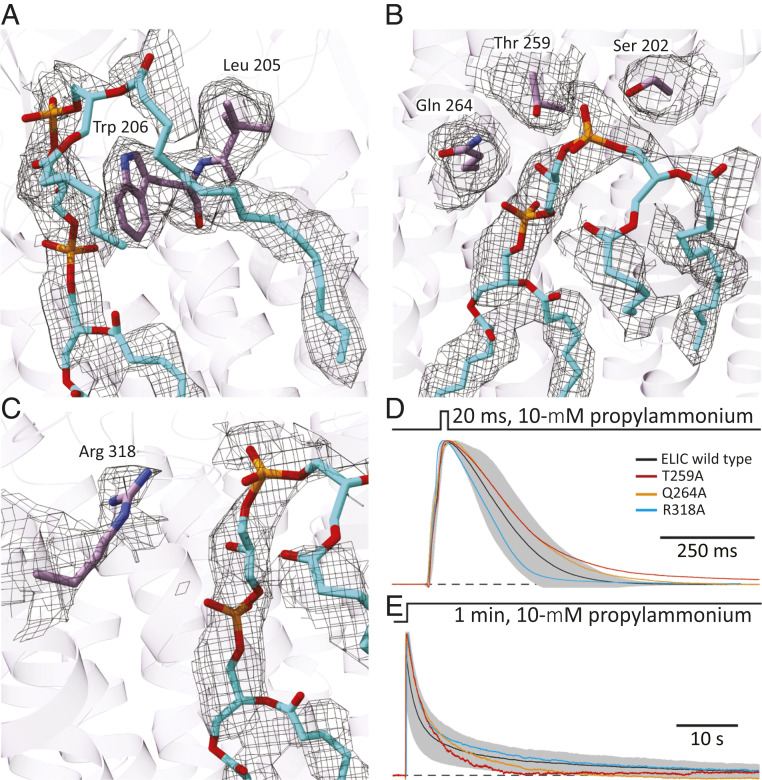
Several residues line the cardiolipin-binding site (defined here as residues with atoms within 5 Å of any atom of bound cardiolipin), and a subset of these (Ser-202, Leu-205, and Trp-206, in M1; Thr-259, in the M2–M3 linker; Gln-264, in M3; and Arg-318, in M4) are close to the phosphate–glycerol–phosphate polar head group (Fig. 5 A–C). The hydroxyl groups of Ser-202 and Thr-259 and one of the guanidinium nitrogen atoms of Arg-318 are close to the oxygen atoms of the “superficial” phosphate and to the hydroxyl group of the “central” head group glycerol. The amide group of Gln-264 is close to the oxygen atoms of the “buried” phosphate and to the hydroxyl group of the head group glycerol; and Leu-205 and Trp-206 form a structure akin to a “coat-hanging hook” on which the superficial phosphate and the superficial glycerol moieties seem to rest. Similar arrangements of arginines and aromatic residues close to the phosphate–glycerol–phosphate polar head group of cardiolipin have been observed in structural models of other membrane proteins (e.g., ref. 50) (SI Appendix, Fig. S11).
Fig. 5.
Cardiolipin-binding site in unliganded ELIC. (A–C) Atomic model of cardiolipin and protein atoms within 5 Å of the phosphate–glycerol–phosphate polar head group. The corresponding densities are also displayed (display level = 0.7). Ser-202, Leu-205, and Trp-206 are in transmembrane α-helix M1; Thr-259, in the M2–M3 linker; Gln-264, in M3; and Arg-318, in M4. Phospholipid and amino-acid atoms are shown in stick representation with carbons colored cyan and purple, respectively. The molecular images were prepared with Chimera X (42). (D and E) Outward currents recorded in the whole-cell configuration from HEK-293 cells transiently transfected with wild-type ELIC or one of the indicated alanine mutants in response to short and long applications of saturating (10 mM) propylammonium. For wild-type ELIC, the mean (black solid line) ± 1 SD (gray error bars) of responses recorded from eight (short pulses) or nine (long pulses) different whole-cell experiments are displayed. For the mutants, the displayed traces are currents recorded from representative individual cells. The distribution of ions across the membrane was asymmetrical (∼150 mM/5 mM K+, inside/outside). The command potential was zero, and the −1.6-mV liquid-junction potential between the pipette and bath solutions was offset. As a result, the membrane potential was close to zero, thus approximating the conditions used for cryo-EM image acquisition. The black dashed lines denote the zero-current baseline. The mutants S202A and L205A + W206A, both in M1, did not elicit measurable currents despite displaying cell-surface expression levels that were comparable to those of the other three mutants.
As a first step toward assessing the relevance of these cardiolipin-binding site residues to ion-channel function, we mutated them to alanine, expressed the mutant constructs in Human embryonic kidney- (HEK-) 293 cells, and recorded whole-cell currents. Although the plasma membrane of eukaryotic cells is not expected to contain cardiolipin [(22), but see refs. 48, 51, and 52], other acidic phospholipids are likely to substitute for it, at least to some degree. Owing to the robustness with which the expression and function of wild-type ELIC and its mutants can be studied in HEK-293 cells, we chose them as the expression system to perform this initial characterization of the cardiolipin-binding sites. In an attempt to mimic the situation of lipid discs (used to determine structure), currents were recorded at approximately zero membrane potential, and thus, an ion-concentration gradient was imposed across the membrane to allow currents to flow. Although the currents recorded from the T259A, Q264A, and R318A mutants were rather wild-type–like (Fig. 5 D and E), no currents could be recorded from the S202A single mutant or the L205A + W206A double mutant on exposure to a saturating concentration of agonist (10 mM propylammonium). To estimate the effect of these mutations on expression, we engineered them in the background of an α-bungarotoxin (α-BgTx)-binding variant of ELIC (ELICα-BgTx) that we designed by replacing the sequence of loop C (I173RYDHLSSVQPNQN) with that of the 13-mer peptide WRYYESSLEPYPD (53). From the amount of 125I-toxin bound to the cell surface of transiently transfected HEK-293 cells, we found that the level of plasma-membrane expression of the mutants relative to the background construct (mutant-to-ELICα-BgTx ratios) are: 0.34 (S202A), 0.47 (L205A + W206A), 0.30 (T259A), 0.24 (Q264A), and 0.43 (R318A). Assuming that the effect of the engineered cardiolipin-binding site mutations on cell-surface expression is independent of that of the loop C–replacement mutation, we inferred that the former reduce expression by a factor of ∼2 to 4 relative to wild-type ELIC. Thus, because robust currents could be recorded from the T259A, Q264A, and R318A mutants (Fig. 5 D and E), our failure to detect currents from the S202A mutant and the L205A + W206A double mutant must have resulted from a severe loss-of-function effect rather than from poor expression. Further experiments will have to be conducted to ascertain the extent to which the detrimental effect of these mutations on function results from an altered affinity of the channel for lipids or from the loss of interactions between the truncated side chains and the rest of the protein.
Discussion
Here, we presented the structure of unliganded ELIC embedded in fragments of E. coli inner membrane purified using an SMA copolymer. Also using an SMA copolymer, several cryo-EM reconstructions of the zebrafish α1 glycine receptor embedded in fragments of insect cell membranes have recently been reported [resolution range: 2.9 to 4.0 Å; (54)]. Lipid molecules, however, were not included in these atomic models, presumably because the structural interpretability of the corresponding density maps was low at the level of the (peripheral) lipid–protein interface. Using a completely unrelated membrane-purification strategy, Unwin and coworkers imaged the native muscle-type AChR from the electric organ of Torpedo fish also in fragments of membrane prepared in the complete absence of detergent-solubilization steps (e.g., ref. 55). Although the limited resolution of the corresponding reconstructions hindered the fitting of structural models, the most recent applications of this approach led to the identification of stoichiometric cholesterol binding to specific transmembrane segments of the receptor (e.g., ref. 56). To the extent that the success of the latter method hinges on the formation of well-ordered two-dimensional (2D) crystals of the imaged protein, however, the applicability of this approach to pLGIC-containing native membranes is likely to be restricted to the electric-organ AChR.
ELIC remained fully functional in membrane patches excised from E. coli polar-lipid liposomes, and thus, it seems reasonable to assume that ELIC function was also retained in SMA-delimited discs of E. coli inner membrane. Hence, we propose that our model of unliganded ELIC represents the resting, low-affinity–closed-pore state of the channel; it seems as though no other interpretation would be warranted. This is the same conformation that has been repeatedly observed in crystal structures of ELIC and whose functional assignment had, justifiably, remained uncertain for several years. We do not think that this result could have been predicted without the corresponding experimental data, however, which is why we consider that obtaining a 3D reconstruction of unliganded ELIC in fragments of native membrane was absolutely necessary to settle this long-standing question.
In some structures of pLGICs (e.g., refs. 15, 24, 39, 49, and 57), the peptide bond between the residues aligned with the highly conserved Phe-119, and the universally conserved Pro-120 of ELIC, in loop-7, has been modeled in the cis conformation. In our case, the quality of the reconstruction at the level of this loop was not high enough to allow the unambiguous distinction between the two alternative conformations, and thus we modeled it as (the much more common) trans; with our density map, modeling this peptide bond as cis did not seem justified. Doing so, however, resulted in an atomic model that differs from the one we are proposing, essentially, only in the orientation of the backbone carbonyl group of Phe-119, with its oxygen atom pointing toward the membrane (cis) rather than the extracellular side (trans). The likely coexistence of both conformational isomers should be borne in mind in studies that address the interactions between the backbone atoms of loop 7 and the rest of the protein.
The high quality of the 3D reconstruction of unliganded ELIC–SMA (2.5-Å resolution) allowed us to model two different phospholipid molecules bound to each subunit: a diacylated phospholipid in the inner leaflet and a cardiolipin molecule in the outer leaflet. A variety of bound lipids (or small fragments thereof) have also been modeled at the protein–membrane interface of nanodisc-reconstituted, detergent-purified animal pLGICs (e.g., refs. 15, 49, 54, and 58), but whether these lipids adopt native binding poses or even whether they represent natively bound lipids is currently unknown. Moreover, none of these lipids is cardiolipin. Quite notably, cardiolipin’s binding site to ELIC overlaps extensively with that of the allosteric agonist ivermectin to its clinically relevant target, the invertebrate pLGIC α-GluCl [PDB ID: 3RIF (44)]. Cardiolipin is a larger molecule than any other membrane phospholipid, and when bound to ELIC, it covers an extensive area contributed by different transmembrane α-helices from different subunits. Hence, cardiolipin seems to be particularly well-poised to bind with different affinities to the different conformational states of the channel, and thus, in a good position to shape its conformational free-energy landscape. It seems as though the affinity of diacylated phospholipids, with their smaller “footprints,” would be less sensitive to the channel’s conformational rearrangements.
The levels of cardiolipin in the inner membrane of E. coli can be experimentally lowered to zero with only minor detrimental effects on the logarithmic phase of growth curves (59, 60). This finding indicates that some of the roles played by cardiolipin in bacteria may be fulfilled by other acidic lipids—most likely, phosphatidylglycerol—at least to a degree that keeps cells dividing at an approximately normal rate. Assuming that no cardiolipin at all is present in the plasma membrane of mammalian cells [(22); but see refs. 51 and 52)]—not even when a bacterial cardiolipin-binding protein is heterologously expressed—a similar phenomenon may underlie the observation that ELIC retains function when expressed in HEK-293 cells; other plasma-membrane acidic phospholipids may replace cardiolipin. However, the mere observation of currents that activate, desensitize, and deactivate upon exposure of ELIC-expressing HEK-293 cells to step changes in agonist concentration does not necessarily mean that cardiolipin can be substituted by other membrane components without physiological consequences. Indeed, the biological role of an ion channel (or any other protein) in the context of a cell often depends on fine details of its behavior that may be missed in experiments. The fact that the identity and in vivo time course of the naturally occurring agonist acting on ELIC remain unknown prevented us from probing the function of ELIC in a more physiologically meaningful manner. For example, we applied long, desensitizing pulses of saturating agonist, but it is currently unknown whether, under in vivo conditions, this slowly desensitizing channel is exposed to concentrations of agonist high enough and for durations long enough for it to desensitize at all. Moreover, it may also be that cardiolipin modulates some aspects of the “life cycle” of ELIC in the bacterial inner membrane, a fundamental facet of ion-channel biology that would go undetected in electrophysiological experiments and that cannot be assessed sensibly using heterologous (over)expression systems.
It is unclear what is required from a specimen to allow a clear visualization of tightly bound lipids. Although the global resolution of the reconstruction must be undoubtedly crucial, we surmise that a high binding-site occupancy and a rigid binding pose must also contribute favorably. For example, in the recent 1.7-Å–resolution (the highest, thus far, for a pLGIC) 3D reconstruction of the homomeric β3 γ-aminobutyric-acid receptor [β3 GABAAR; PDB ID: 7A5V (15)]—incorporated into lipid nanodiscs upon detergent solubilization from HEK-293 cell membranes—the density attributed to lipids is not clearer than that in our 2.5-Å reconstruction, even upon our processing of the sum of β3-GABAAR experimental half maps using the same local-sharpening procedure as that used here for ELIC–SMA. It is likely that our use of a membrane preparation with a more homogeneous chemical composition (bacterial inner membranes versus a mixture of eukaryotic plasma and intracellular membranes) and the complete omission of detergent-solubilization steps may have helped in our case.
The structural characterization of the lipid—protein interface in different conformational states will shed light on conformation-specific interactions, and thus, on the possible role of structural membrane lipids as allosteric ligands of pLGICs. Because one cannot predict the extent to which the lipid–protein interface is disrupted upon the use of detergents, detergent-free purification strategies emerge as the most sensible approach for tackling this type of question. Obtaining 3D reconstructions of the required high quality for different protein states arises, now, as the next challenge.
Materials and Methods
Purification of ELIC-Containing SMA Nanodiscs.
ELIC was overexpressed in E. coli (BL21-Gold; Agilent Technologies), cells were disrupted, and the membrane fraction was isolated, as previously described (61). Membrane pellets were resuspended and extracted for 2 h at 4 °C in a buffer containing 500 mM NaCl, 25 mM KCl, 0.6% (wt/vol) SMA-3000 HNA copolymer (Total Petrochemicals & Refining), and 50 mM Tris/HCl, pH = 8.0. Insoluble material was separated from SMA-solubilized membranes by centrifugation (1 h at 200,000 × g), and the supernatant was combined with Ni-NTA resin (ThermoFisher Scientific) pre-equilibrated in purification buffer (150 mM NaCl, 20 mM Tris/HCl, pH = 8.0). After overnight incubation, the slurry was transferred to a 10-mL Poly-Prep chromatography column (Bio-Rad), washed with 10 column volumes of purification buffer, and incubated with Tobacco Etch Virus (TEV) protease for 8 to 10 h to remove the expression and purification tags. The column was then washed with 10 column volumes of purification buffer, and ELIC-containing SMA nanodiscs were eluted with purification buffer containing 30 mM imidazole (ELIC interacts with the affinity resin, likely, through its M3–M4-linker dihistidine motif). The eluate was concentrated and subjected to size-exclusion chromatography using a Superdex 200 10/300 GL column (Cytiva). The eluted SMA nanodiscs were concentrated using an Amicon ultra-centrifuge filter (MilliporeSigma) to a final value of 10 μM for cryo-EM imaging.
Purification of Detergent-Solubilized ELIC and Reconstitution in Liposomes.
Detergent-solubilized ELIC was purified and reconstituted into cell-sized multilamellar liposomes using the “dehydration–rehydration” method (62, 63) as previously described (26). Dried E. coli polar lipids (Avanti Polar Lipids; catalog no.: 100600) were dispersed in water, and the corresponding protein-reconstituted liposomes were dehydrated first, followed by rehydration in a solution consisting of 150 mM NaCl and 10 mM sodium phosphate, pH = 8.0. The dried mixture of phosphatidylcholine (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine; Avanti Polar Lipids; catalog no.: 850457) and cardiolipin (1′,3′-bis[1-palmitoyl-2-oleoyl-sn-glycero-3-phospho]-glycerol; Avanti Polar Lipids; catalog no.: 710341), at a 95:5 ratio (mol%), was dispersed in water, and the corresponding dehydrated protein-reconstituted liposomes were rehydrated in a solution consisting of 5 mM KCl, 1 mM CaCl2, 250 mM mannitol, and 5 mM Hepes/KOH, pH = 7.4. In all cases, the protein-to-lipid ratio was 1:20 (by weight).
Complementary DNA Clones, Mutagenesis, and Heterologous Expression.
Complementary DNA (cDNA) coding the bacterial pLGIC ELIC (accession no.: E0SJQ4) in pcDNA3.1 was obtained as described previously (61). Mutations were engineered using the QuikChange kit (Agilent Technologies), and the sequences of the resulting cDNAs were verified by dideoxy sequencing of the entire coding region (ACGT). Wild-type and mutant ELIC were heterologously expressed in transiently transfected adherent HEK-293 cells using a calcium-phosphate precipitation method (187.5 ng DNA/cm2). HEK-293 cells were grown at 37 °C and 5% CO2 in Dulbecco’s modified Eagle’s Medium (ThermoFisher Scientific) containing 10% (wt/vol) fetal bovine serum (ThermoFisher Scientific) and 1% (wt/vol) penicillin-streptomycin (Corning). Transfections proceeded for 16 to 18 h after which the cell-culture medium containing the DNA precipitate was replaced by fresh medium. Electrophysiological recordings were performed ∼28 h later.
Electrophysiology.
Currents from ELIC-incorporated cell-sized proteoliposomes and from transiently transfected HEK-293 cells were recorded in the inside-out and whole-cell patch-clamp configurations, respectively, at ∼22 °C using an Axopatch 200B amplifier (Molecular Devices) with an effective bandwidth of 5 kHz. Currents were digitized at 10 kHz and analyzed using pCLAMP 11.1 software (Molecular Devices). In the whole-cell configuration, series-resistance compensation was used and set to ∼80%; throughout the paper, we assumed that this compensation zeroed the series-resistance error. The reference Ag/AgCl wire was connected to the extracellular solution through an agar bridge containing 200 mM KCl. Agonist-concentration jumps were applied using a piece of double-barreled glass “θ-tubing” [Siskiyou (64, 65)]. The flow of solution through the θ-tube was controlled using a gravity-fed system (ALA BPS-8; ALA Scientific Instruments), and the movement of the θ-tube was achieved using a piezo-electric arm (Burleigh-LSS-3100; discontinued) controlled by pCLAMP 11.1 software and a Digidata 1322A interface (Molecular Devices). Signals from this interface were low-pass filtered (900 C; Frequency Devices) at a cutoff frequency of 25 Hz prior to their arrival at the piezoelectric arm to reduce ringing in the θ-tube motion. In the whole-cell configuration, cells remained attached to a piece of poly-l-lysine– (MilliporeSigma) coated glass (Warner Instruments) or glycol-modified polyethylene terephthalate (Thermo Fisher Scientific) coverslip throughout the experiment. In this configuration, the perfusion system achieved a solution-exchange time of ∼4 ms for the t10-90% and ∼10 ms for the t90-10%, as estimated from changes in the liquid-junction current measured with an open-tip patch pipette. In the inside-out configuration, these values were ∼0.4 ms for the t10-90% and ∼0.8 ms for the t90-10%. Although slower than pressurized perfusion (64, 65), we chose to use a gravity-fed system here because it was gentler on the seals and because it was not our intention to perform a detailed characterization of the kinetic aspects of channel function. In the inside-out configuration, the pipette solution was 142 mM KCl, 5.4 mM NaCl, 1.8 mM CaCl2, 1.7 mM MgCl2, and 10 mM Hepes/KOH, pH = 7.4. In the whole-cell configuration, the pipette solution was 110 mM KCl, 40 mM KF, and 5 mM Hepes/KOH, pH = 7.4. For both configurations, the bath solution was 5 mM KCl, 250 mM mannitol, and 5 mM Hepes/KOH, pH = 7.4. The two extracellular solutions applied through the θ-tube barrels were bath solution with and without 1 or 10 mM propylammonium chloride/propylamine. At pH = 7.4, the concentration of propylammonium cation (pKa ≅ 10.71) in a 1-mM solution of the salt/base mixture can be calculated to be 0.9995 mM and that in a 10-mM solution of the mixture, 9.995 mM.
Cell-Surface Expression.
The number of ELICα-BgTx channels and its mutants on the plasma membrane of transfected HEK-293 cells was estimated from the amount of [125I]α-BgTx (PerkinElmer) bound upon incubation with ∼30-nM toxin for 4 h at 4 °C in suspension. The low temperature during the incubation was intended to minimize the uptake of toxin through endocytosis. Once the incubation was completed, the cells were pelleted by centrifugation, and the pellets were resuspended and washed with ice-cold phosphate-buffered saline solution. The pelleting–washing steps were repeated a second time, and the final (third) pellet was resuspended and solubilized in a solution consisting of 0.1 N NaOH and 1% (wt/vol) sodium dodecyl sulfate at 65 °C for 30 min. The radioactivity associated with each cell pellet (counted in a γ-counter) was normalized by the mass of protein (estimated using the bicinchoninic acid [ThermoFisher Scientific] assay). To account for the nonspecific binding of toxin, controls consisting of HEK-293 cells transfected with cDNA coding for pLGIC subunits that do not form α-BgTx–binding sites were run in parallel; we used a 1:1:1 (by weight) mixture of β1, δ, and ε AChR subunit-coding cDNAs from mouse.
Grid Preparation.
Samples were thawed on ice and centrifuged at 21,000 × g for 10 min at 4 °C. Aliquots were applied on freshly glow-discharged quantifoil holey carbon grids (R1.2/1.3 200 Au; Electron Microscopy Sciences) for 30 s, with the carbon side facing up. Grids were incubated at 4 °C for 30 s in the 95%-humidity controlled chamber of a Leica EM GP2 Automatic Plunge Freezer with 1.5-s blotting time before plunge freezing in liquid nitrogen–cooled liquid ethane.
Cryo-EM Data Acquisition.
Grids were clipped and transferred to a Titan Krios microscope (ThermoFisher Scientific) operated at 300 kV and equipped with a Gatan K2 Counting imaging system. Movies were collected using Leginon (66) at a dose rate of 7.06 e−Å−2s−1 with a total exposure of 10 s, for an accumulated dose of 70.58 e−Å−2. Intermediate frames were recorded every 0.20 s for a total of 50 frames per micrograph. A total of 5,627 images were collected at a nominal defocus range between −0.3 and 3.3 μm (SI Appendix, Table S1).
Cryo-EM Data Processing.
The summed, aligned, and dose-weighted micrographs were processed using cryoSPARC version 2 (30, 31), and contrast-transfer function estimation was performed with cryoSPARC version 2’s Patch CTF. Initial particle picking was done manually, and the picked particles were subjected to 2D classification for template generation. Classes representing different particle orientations were selected and used as templates for automatic picking. Picked particles were subjected to several rounds of 2D classification to eliminate bad particles. After the initial rounds of classifications, an ab initio model was generated and refined with homogeneous refinement, followed by the final reconstruction of the map using the non–uniform-refinement option (C5 symmetry) in Local Refinement. Local resolution was estimated with MonoRes (32), and map sharpening was performed locally on the basis of the MonoRes-generated local-resolution map using LocalDeblur (33), both in Xmipp/Scipion 2.0 (67).
Model Building and Refinement.
Structure factors obtained from the EM maps (using Phenix.map_to_structure_factors) and the primary sequence of ELIC were used as input in BALBES (68), and the output coordinate file was used for model building and refinement. The manual adjustment of the model inside the map and the subsequent refinement were performed using Coot (69) and phenix_real_space_refine (70, 71) iteratively. Model building and refinement cycles were performed until no further improvement in cross-correlation (70, 71) or Molprobity scores (72) was observed. As validation tools, we also computed EM ringer scores (73), and map-versus-model FSC-threshold crossings (27) (SI Appendix, Figs. S1D and S7B). The corresponding values are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank N. Godellas for assistance with cell culture and molecular biology, Luis Cuello for advice on protein reconstitution in liposomes, and Z. Zhao and E. Tajkhorshid for discussions. Part of this work was performed at the National Center for CryoEM Access and Training and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and New York State Assembly. This work was supported by a grant from the NIH (R01-NS042169 to C.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100164118/-/DCSupplemental.
Data Availability
The cryoelectron microscopy maps and atomic coordinates reported in this paper have been deposited in the Electron Microscopy Databank, https://www.ebi.ac.uk/pdbe/emdb (ID codes EMD-23207 for unliganded ELIC in SMA nanodiscs, and EMD-23208 for unliganded ELIC in POPC-only nanodiscs at 3.3-Å resolution) and Protein Data Bank, https://www.wwpdb.org (ID codes 7L6Q for unliganded ELIC in SMA nanodiscs and 7L6U for unliganded ELIC in POPC-only nanodiscs at 3.3-Å resolution).
References
- 1.Camara-Artigas A., Brune D., Allen J. P., Interactions between lipids and bacterial reaction centers determined by protein crystallography. Proc. Natl. Acad. Sci. U.S.A. 99, 11055–11060 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wenz T., et al., Role of phospholipids in respiratory cytochrome bc(1) complex catalysis and supercomplex formation. Biochim. Biophys. Acta 1787, 609–616 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Franklin G. I., Potter L. T., Studies of the binding of -bungarotoxin to membrane-bound and detergent-dispersed acetylcholine receptors from Torpedo electric tissue. FEBS Lett. 28, 101–106 (1972). [DOI] [PubMed] [Google Scholar]
- 4.Cohen J. B., Weber M., Changeux J. P., Effects of local anesthetics and calcium on the interaction of cholinergic ligands with the nicotinic receptor protein from Torpedo marmorata. Mol. Pharmacol. 10, 904–932 (1974). [PubMed] [Google Scholar]
- 5.Chang H. W., Bock E., Structural stabilization of isolated acetylcholine receptor: Specific interaction with phospholipids. Biochemistry 18, 172–179 (1979). [DOI] [PubMed] [Google Scholar]
- 6.Dalziel A. W., Rollins E. S., McNamee M. G., The effect of cholesterol on agonist-induced flux in reconstituted acetylcholine receptor vesicles. FEBS Lett. 122, 193–196 (1980). [DOI] [PubMed] [Google Scholar]
- 7.Heidmann T., Sobel A., Changeux J. P., Conservation of the kinetic and allosteric properties of the acetylcholine receptor in its Na cholate soluble 9 S form : Effect of lipids. Biochem. Biophys. Res. Commun. 93, 127–133 (1980). [DOI] [PubMed] [Google Scholar]
- 8.Kilian P. L., et al., Reconstitution of acetylcholine receptor from Torpedo Californica with highly purified phospholipids: Effect of alpha-tocopherol, phylloquinone, and other terpenoid quinones. Biochem. Biophys. Res. Commun. 93, 409–414 (1980). [DOI] [PubMed] [Google Scholar]
- 9.Criado M., Eibl H., Barrantes F. J., Functional properties of the acetylcholine receptor incorporated in model lipid membranes. Differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J. Biol. Chem. 259, 9188–9198 (1984). [PubMed] [Google Scholar]
- 10.Fong T. M., McNamee M. G., Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry 25, 830–840 (1986). [DOI] [PubMed] [Google Scholar]
- 11.Thompson M. J., Baenziger J. E., Structural basis for the modulation of pentameric ligand-gated ion channel function by lipids. Biochim. Biophys. Acta Biomembr. 1862, 183304 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Fyfe P. K., et al., Probing the interface between membrane proteins and membrane lipids by X-ray crystallography. Trends Biochem. Sci. 26, 106–112 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Lee A. G., Lipid-protein interactions in biological membranes: A structural perspective. Biochim. Biophys. Acta 1612, 1–40 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Palsdottir H., Hunte C., Lipids in membrane protein structures. Biochim. Biophys. Acta 1666, 2–18 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Nakane T., et al., Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overduin M., Esmaili M., Memtein: The fundamental unit of membrane-protein structure and function. Chem. Phys. Lipids 218, 73–84 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Marius P., et al., Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys. J. 94, 1689–1698 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S., et al., An essential role for cardiolipin in the stability and function of the mitochondrial calcium uniporter. Proc. Natl. Acad. Sci. U.S.A. 117, 16383–16390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senoo N., et al., Cardiolipin, conformation, and respiratory complex-dependent oligomerization of the major mitochondrial ADP/ATP carrier in yeast. Sci. Adv. 6, eabb0780 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakobi A. J., Wilmanns M., Sachse C., Model-based local density sharpening of cryo-EM maps. eLife 6, e27131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilas J. L., Tagare H. D., Vargas J., Carazo J. M., Sorzano C. O. S., Measuring local-directional resolution and local anisotropy in cryo-EM maps. Nat. Commun. 11, 55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zambrano F., Fleischer S., Fleischer B., Lipid composition of the Golgi apparatus of rat kidney and liver in comparison with other subcellular organelles. Biochim. Biophys. Acta 380, 357–369 (1975). [DOI] [PubMed] [Google Scholar]
- 23.van Meer G., Voelker D. R., Feigenson G. W., Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hénault C. M., et al., A lipid site shapes the agonist response of a pentameric ligand-gated ion channel. Nat. Chem. Biol. 15, 1156–1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong A., et al., Direct binding of phosphatidylglycerol at specific sites modulates desensitization of a ligand-gated ion channel. eLife 8, e50766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P., et al., Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer. Proc. Natl. Acad. Sci. U.S.A. 117, 1788–1798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Chen S., et al., High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sohlenkamp C., Geiger O., Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Punjani A., Zhang H., Fleet D. J., Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Vilas J. L., et al., MonoRes: Automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ramírez-Aportela E., et al., Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanemasa Y., Akamatsu Y., Nojima S., Composition and turnover of the phospholipids in Escherichia coli. Biochim. Biophys. Acta 144, 382–390 (1967). [PubMed] [Google Scholar]
- 35.Cronan J. E., Bacterial membrane lipids: Where do we stand? Annu. Rev. Microbiol. 57, 203–224 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Hilf R. J. C., Dutzler R., X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Gutierrez G., Grosman C., Bridging the gap between structural models of nicotinic receptor superfamily ion channels and their corresponding functional states. J. Mol. Biol. 403, 693–705 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu H., Howard R. J., Bastolla U., Lindahl E., Delarue M., Structural basis for allosteric transitions of a multidomain pentameric ligand-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 117, 13437–13446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Althoff T., Hibbs R. E., Banerjee S., Gouaux E., X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 512, 333–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie T. K., et al., Chapter 11–Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen E. F., et al., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olofsson G., Sparr E., Ionization constants pKa of cardiolipin. PLoS One 8, e73040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hibbs R. E., Gouaux E., Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McAuley K. E., et al., Structural details of an interaction between cardiolipin and an integral membrane protein. Proc. Natl. Acad. Sci. U.S.A. 96, 14706–14711 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shintre C. A., et al., Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. U.S.A. 110, 9710–9715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinzawa-Itoh K., et al., Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pye V. E., et al., A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat. Struct. Mol. Biol. 27, 78–83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahman M. M., et al., Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins. Neuron 106, 952–962.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grba D. N., Hirst J., Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation. Nat. Struct. Mol. Biol. 27, 892–900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorice M., et al., Cardiolipin on the surface of apoptotic cells as a possible trigger for antiphospholipids antibodies. Clin. Exp. Immunol. 122, 277–284 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sorice M., et al., Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ. 11, 1133–1145 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Kasher R., et al., Design and synthesis of peptides that bind alpha-bungarotoxin with high affinity. Chem. Biol. 8, 147–155 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Yu J., et al., Mechanism of gating and partial agonist action in the glycine receptor. Cell 184, 957–968.e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyazawa A., Fujiyoshi Y., Unwin N., Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Unwin N., Protein-lipid architecture of a cholinergic postsynaptic membrane. IUCrJ 7, 852–859 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bocquet N., et al., X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Kumar A., et al., Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs. Nat. Commun. 11, 3752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishijima S., et al., Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J. Bacteriol. 170, 775–780 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan B. K., et al., Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 16504–16509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez-Gutierrez G., et al., Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc. Natl. Acad. Sci. U.S.A. 109, 6331–6336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller B. U., Hedrich R., Vaz W. L., Criado M., Single channel recordings of reconstituted ion channel proteins: An improved technique. Pflugers Arch. 411, 94–100 (1988). [DOI] [PubMed] [Google Scholar]
- 63.Cuello L. G., Cortes D. M., Perozo E., The gating cycle of a K+ channel at atomic resolution. eLife 6, e28032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elenes S., Ni Y., Cymes G. D., Grosman C., Desensitization contributes to the synaptic response of gain-of-function mutants of the muscle nicotinic receptor. J. Gen. Physiol. 128, 615–627 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elenes S., Decker M., Cymes G. D., Grosman C., Decremental response to high-frequency trains of acetylcholine pulses but unaltered fractional Ca2+ currents in a panel of “slow-channel syndrome” nicotinic receptor mutants. J. Gen. Physiol. 133, 151–169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suloway C., et al., Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 67.de la Rosa-Trevín J. M., et al., Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Long F., Vagin A. A., Young P., Murshudov G. N., BALBES: A molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Afonine P. V., et al., New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis I. W., et al., MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barad B. A., et al., EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryoelectron microscopy maps and atomic coordinates reported in this paper have been deposited in the Electron Microscopy Databank, https://www.ebi.ac.uk/pdbe/emdb (ID codes EMD-23207 for unliganded ELIC in SMA nanodiscs, and EMD-23208 for unliganded ELIC in POPC-only nanodiscs at 3.3-Å resolution) and Protein Data Bank, https://www.wwpdb.org (ID codes 7L6Q for unliganded ELIC in SMA nanodiscs and 7L6U for unliganded ELIC in POPC-only nanodiscs at 3.3-Å resolution).