Significance
During meiosis, programmed chromosome breakage and subsequent double-stranded DNA (dsDNA) break repair help ensure correct chromosome segregation and promote genetic diversity of the progeny. In budding yeast, which utilizes meiotic recombination pathways conserved in mice and humans, the majority of meiotic crossovers are initiated through the formation of a DNA Holliday junction, which requires the endonuclease activity of the Mlh1-Mlh3 DNA mismatch repair factor to be resolved exclusively into a crossover product. Here, we combined structural biology, biochemical, and genetic analyses to compare the Mlh1-Mlh3 structure and functions with the main mismatch repair endonuclease Mlh1-Pms1. We characterize structural differences around their respective endonuclease sites. We also characterize mutants associated with condensation and filament formation of the Mlh1-Mlh3 heterodimer.
Keywords: biochemistry, structural biology, DNA recombination, DNA repair, genetics
Abstract
In budding yeast, the MutL homolog heterodimer Mlh1-Mlh3 (MutLγ) plays a central role in the formation of meiotic crossovers. It is also involved in the repair of a subset of mismatches besides the main mismatch repair (MMR) endonuclease Mlh1-Pms1 (MutLα). The heterodimer interface and endonuclease sites of MutLγ and MutLα are located in their C-terminal domain (CTD). The molecular basis of MutLγ’s dual roles in MMR and meiosis is not known. To better understand the specificity of MutLγ, we characterized the crystal structure of Saccharomyces cerevisiae MutLγ(CTD). Although MutLγ(CTD) presents overall similarities with MutLα(CTD), it harbors some rearrangement of the surface surrounding the active site, which indicates altered substrate preference. The last amino acids of Mlh1 participate in the Mlh3 endonuclease site as previously reported for Pms1. We characterized mlh1 alleles and showed a critical role of this Mlh1 extreme C terminus both in MMR and in meiotic recombination. We showed that the MutLγ(CTD) preferentially binds Holliday junctions, contrary to MutLα(CTD). We characterized Mlh3 positions on the N-terminal domain (NTD) and CTD that could contribute to the positioning of the NTD close to the CTD in the context of the full-length MutLγ. Finally, crystal packing revealed an assembly of MutLγ(CTD) molecules in filament structures. Mutation at the corresponding interfaces reduced crossover formation, suggesting that these superstructures may contribute to the oligomer formation proposed for MutLγ. This study defines clear divergent features between the MutL homologs and identifies, at the molecular level, their specialization toward MMR or meiotic recombination functions.
During the first meiotic division, in most organisms, each pair of homologous chromosomes (homologs) needs to experience at least one crossover to ensure their accurate segregation and increase genetic diversity of the progeny (1, 2). Crossovers are generated after programmed DNA double-strand break (DSB) formation, and their subsequent repair by homologous recombination (3). Failure to achieve at least one crossover per homolog pair results in aneuploid gametes. These dysfunctions are frequent causes of spontaneous miscarriages and birth defects in humans (1). DSBs are generated by the meiosis-specific Spo11 protein and resected to form 3′ single-stranded tails that are directed to invade and pair with an unbroken homologous template, preferentially on the homolog (3). Invasion intermediates are a substrate for DNA synthesis. After the capture of the second DSB end, a subset of the intermediates is converted into double Holliday junctions (dHJs), which in meiosis are primarily resolved into crossovers (4, 5). The remaining recombination intermediates are repaired as noncrossovers.
MutLγ (Mlh1-Mlh3 in yeast and MLH1-MLH3 in human) is essential for the formation of meiotic crossovers in many organisms. The MutLγ heterodimer possesses, similarly to MutLα (Mlh1-Pms1 in yeast, MLH1-PMS2 in human), a latent endonuclease activity (6–10). It has been proposed that the MutLγ endonuclease activity catalyzes the resolution of the dHJ intermediates and promotes the formation of crossovers (8, 11). In agreement with this, Mlh1 and Mlh3 form foci on pachytene chromosomes in different organisms at future crossover sites (12–14). In yeast mlh3 mutants, the crossover rates are reduced to 50 to 70% of the wild-type level (8, 15–17). These mutants exhibit failure in chromosome disjunction and consequently a decrease of spore viability. In Mlh3-/- mice, males and females present a crossover defect that leads to aneuploidy (18). In agreement with the proposed resolvase role of MutLγ, a mutant of the active site within the conserved DQHAX2EX4E motif of Saccharomyces cerevisiae Mlh3, D523N, results in a loss of activity and confers a phenotype similar to the mlh3∆ mutant with decreased crossover frequencies (8). A mutation in the endonuclease domain of mouse MLH3 leads to infertile males and strongly reduced crossover numbers, despite a correct loading of factors essential for crossover resolution (19). In budding yeast and mammals, MLH1 deletion is also associated with severe dysfunctions in meiosis due to crossover defects, combined with high genetic instability due to its additional central role with Pms1 (as part of the MutLα complex) in mutation avoidance (20).
In budding yeast, the majority of meiotic crossovers are formed by MutLγ, requiring in addition other proteins including the ZMM proteins (Zip1-4, Spo16, Mer3, and Msh4-Msh5), Exo1, and the Sgs1-Top3-Rmi1 complex (11, 21). The ZMM factors are proposed to stabilize the recombination intermediates and protect them from the action of helicases (reviewed in ref. 22). The 5′-3′ exonuclease, Exo1, is important for crossover formation independently of its nuclease activity and likely acts as scaffold factor in particular through its interaction motif with Mlh1 (17). We recently reported that MutLγ-Exo1 associates with recombination intermediates, followed by direct Cdc5 recruitment that triggers MutLγ crossover activity (23). Despite its central role in the formation of crossovers, to date, the MutLγ endonuclease does not show any specific activity on either a single HJ or dHJ DNA substrate in vitro, in contrast to other resolvases or structure-specific endonucleases (24). Recently, it was shown that human MutLγ is part of an ensemble with MutSγ, EXO1, RFC, and PCNA that preferentially cleaves plasmid DNA containing a HJ, although it does not cleave symmetrically across the junction, as would be expected for a canonical resolvase (25, 26).
MutLγ is also an accessory factor of the postreplicative mismatch repair (MMR). In S. cerevisiae, MutLα is the major MutL homolog involved in MMR. It is targeted to DNA mismatches by the MutS homologs and introduces DNA nicks to initiate the excision of the strand containing the mismatch. MutLα contains an endonuclease site formed by three conserved motifs in Pms1 and the last conserved amino acids of Mlh1 (6, 7, 27). Mutation of this active site (e.g., pms1E707K in yeast) is associated with high mutations rates. A minor role of MutLγ has been reported in the repair of a subset of DNA mismatches recognized by Msh2-Msh3 (28, 29). The third MutL heterodimer, Mlh1-Mlh2 (MLH1-PMS1 in mammals) or MutLβ, has no endonuclease motif or activity. In yeast, mlh2Δ cells present only a slight defect in the repair of a subset of frameshift mutations (30–32). Apart from this function in MMR, we recently identified an interaction of MutLβ with Mer3 helicase that limits gene conversion tract lengths (31). A major challenge is to further characterize the molecular basis of the specific interactions and regulations of the three MutL homologs with their DNA substrates and protein partners in MMR and in meiotic recombination.
MutLγ and MutLα present an overall common organization with a N-terminal domain (NTD) bearing an ATPase function, connected through a long linker to a C-terminal domain (CTD). The CTD mediates the dimerization of the complexes (33, 34) and possesses the endonuclease site (6–8). Upon ADP and ATP binding, the NTD undergoes large asymmetric conformational changes that can position it into a close proximity to the CTD (35, 36). The heterodimers MutLα and MutLγ can slide on double-stranded DNA (dsDNA) with linear diffusion modes (37, 38) and control strand excision during MMR (39). However, both heterodimers differ in their DNA-binding properties. MutLα has very low affinity for short dsDNA in physiological salt concentrations and can bind cooperatively to long dsDNA (>200 bp), forming long continuous protein tracts (40). In contrast, MutLγ binds short-branched DNA substrates with a higher affinity (in the nanomolar range) and with a marked preference for HJs (9). MutLγ also binds, albeit more weakly, short and long dsDNA and is proposed to form oligomers on long dsDNA (9, 41). The Pms1 and Mlh3 subunits present a moderate sequence identity precluding correct modeling of MutLγ from the MutLα crystal structures and thus limiting our understanding of the molecular basis of the specificity of MutLα and MutLγ heterodimers in MMR and meiosis, respectively.
Here, we present the crystal structure of the S. cerevisiae MutLγ(CTD), and we compare it with the three-dimensional structure of the S. cerevisiae MutLα(CTD) that we previously reported (27). We reveal differences between the two complexes with regard to the size of the heterodimerization interface, the regulatory domain position, and the shape of the cavity surrounding the endonuclease site. We characterize the role of the last amino acids of Mlh1 using mutant alleles and identify a central role of the last three conserved residues of Mlh1 in vivo for both MMR and meiotic recombination. We analyze the DNA-binding specificities of the MutLα(CTD) and Mut-Lγ(CTD) and compare to the properties of full-length proteins. We then identify positions in Mlh3(NTD) and Mlh3(CTD) that can participate to the positioning of the NTD close to the CTD and characterize the corresponding mutants in meiosis. Finally, we report a filament arrangement of the MutLγ(CTD) in the crystal in agreement with oligomers proposed in previous studies (25, 41), and we characterize an allele mutated in this oligomerization region.
Results
MutLγ(CTD) Presents a Structure Similar to that of MutLα(CTD) with Variations on Functional Sites.
Four crystal structures of the S. cerevisiae MutLγ(CTD) heterodimer are reported here (SI Appendix, Table S1). They differ in either their space group or on the number of zinc atoms present in the active site. Forms called Zn2 and Zn2b contain two zinc atoms in the endonuclease sites (positions A and B) (SI Appendix, Table S1). Forms ZnA and ZnB contain one zinc atom respectively in position A or B. The crystal structure of MutLγ(CTD) presents an overall structural similarity with the one previously reported for MutLα(CTD), although with important local differences (Fig. 1 A and B). The Mlh1(CTD), present in both MutLα and MutLγ heterodimers, superimposes fully in the two heterodimers, both on their dimerization and regulatory subdomains, as well as on the relative position of these two subdomains (rmsd of 0.5Å over 202 Cα). The dimerization domains of Pms1(CTD) and Mlh3(CTD) superimpose also well (rmsd of 0.93Å over 60Cα) (Fig. 1B and SI Appendix, Fig. S1A for a multiple sequence alignment of the MutL homologs).
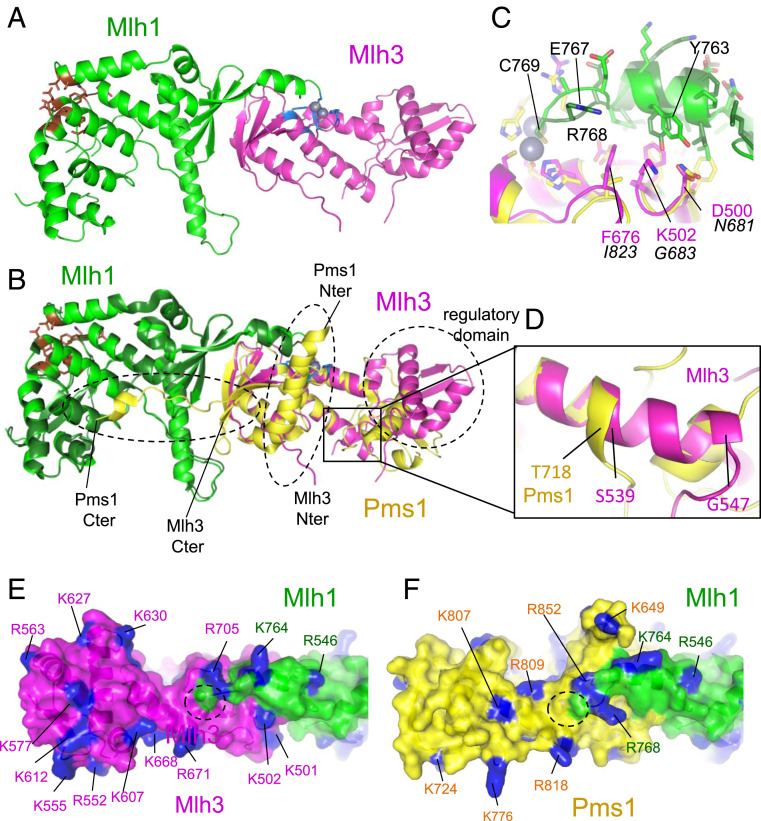
Fig. 1.
Crystal structure of MutLγ(CTD). (A) Overall view of the MutLγ(CTD) heterodimer (Mlh1 and Mlh3 are colored respectively in green and magenta). The endonuclease site is colored in blue with the two zinc atoms in sphere representation and the Exo1 binding site in brown. (B) Superimposition of the MutLα(CTD) on the MutLγ(CTD). Mlh1 and Pms1 in MutLα are colored respectively in dark green and yellow. The three main regions of Pms1(CTD) that differ from Mlh3(CTD) are highlighted with dashed lines: 1) the regulatory domains, 2) the first residues of Pms1 and Mlh3 CTDs (the position of the first residues of the two CTDs are indicated [“Nter”]), and 3) the last residues of both CTDs (“Cter”). (C) The last residue Cys769 of Mlh1 adopts the same position in the endonuclease site in both complexes. The preceding residues of Mlh1 adopt a slightly different conformation when they are in contact with Mlh3 residues (numbers in magenta) or Pms1 ones (numbers in italic). (D) The helix αA of Mlh3 located in the hinge between the regulatory and dimerization domains is two turns longer in Mlh3 than in Pms1 pushing away the regulatory domain of Mlh3 compared to Pms1. The region zoomed corresponds to the black rectangle shown in B. The distribution of the basic residues (colored in blue) around the Mlh3 (E) and Pms1 (F) active sites is different (active site located with dashed lines). The position of the regulatory domain in Mlh3 also differs from the one observed in Pms1 resulting in different shapes of the surface surrounding the endonuclease sites of Mlh1-Mlh3 and Mlh1-Pms1.
The most conserved region in structure between the Mlh3(CTD) and Pms1(CTD) subunits is, as expected, the core dimerization interface mediated by a four-strand β-sheet of each subunit (Fig. 1 A and B). This region is highly conserved in all crystal structures from bacterial MutL to eukaryotic MutL homologs (27, 42–44). A second conserved interface between the MutLα(CTD) and MutLγ(CTD) heterodimers is the contact between the last 10 amino acids of Mlh1 and the surrounding residues from the other subunit (Pms1 or Mlh3). We previously showed that the last 10 residues of the C terminus of Mlh1 tightly interact with Pms1 so that the last residue of Mlh1, Cys-769, is positioned in the endonuclease site of Pms1 (27). We observed a similar interaction between the last residues of Mlh1 and the Mlh3 amino acids involved in the endonuclease site. The superimposition of the two complexes shows that Mlh3 and Pms1 present some sequence variation in this region (Fig. 1C). The main difference is the presence of a Lys in Mlh3 (K502) in place of a Gly in Pms1 (G683) that are both in contact with a Mlh1 Tyr (Y763) in the Cter tail. Despite this variation, the position of the Cter helix is maintained with some reorientation of the Mlh1 Y763 side chain, and the last three amino acids are positioned close to the endonuclease site in both heterodimers, suggesting an evolutionary requirement for the presence of Mlh1 residues in the active sites (Fig. 1C).
The regulatory domains of Pms1(CTD) and Mlh3(CTD) are oriented differently with respect to the rest of the complex (Fig. 1B). The regulatory domain of Mlh3(CTD) is rotated by about 40° compared to the equivalent region of Pms1. This difference comes from the presence of an eight amino acid–longer helix in Mlh3 compared to Pms1 (Fig. 1D and SI Appendix, Fig. S1A). This helix is located in the hinge region between the dimerization domain and the regulatory domain. It is two turns longer in Mlh3 than in Pms1, and it pushes away the regulatory domain in Mlh3 (Fig. 1D). The orientation of the Mlh3 regulatory domain is observed in the two crystal forms, Zn2 and Zn2b. To further characterize the conformation of the MutLγ(CTD) in solution, we performed small angle X-ray scattering (SAXS) analyses. We first compared the scattering curve obtained with MutLγ(CTD) with the theoretical scattering curve calculated with the crystal structure of MutLγ(CTD) (SI Appendix, Fig. S1B). Disordered loops of MutLγ(CTD) were modeled and refined using DADIMODO (see Materials and Methods). The fit between experimental and theoretical scattering curves shows a good agreement [χ2 score of 1.37 using Crysol and the crystal structure refined using DADIMODO (45)]. The fit was performed with a model in which the regulatory domain of Mlh3 is in the orientation observed for the regulatory domain of Pms1 (Fig. 1B). It also shows a good agreement (χ2 score of 1.41), suggesting that in our experimental conditions, SAXS cannot discriminate between both conformations.
The MutLα(CTD) and MutLγ(CTD) present additional differences. We previously characterized an extended structure made by Pms1 C terminus (last 15 amino acids of the subunit) that mediates a large interaction interface with Mlh1. This C terminus of Pms1 includes residues reported as essential for the dimer formation (Fig. 1B, refer to Pms1 Cter) (27, 46). This region is absent in Mlh3, in which the C terminus stops at the beginning of this Pms1 extension close to the dimerization interface (Fig. 1B). Due to the absence of this patch, the surface buried by Mlh3 and Mlh1 is smaller than the one buried by Pms1 and Mlh1 (2,140 Å2 for Mlh1-Mlh3 compared to 3,700 Å2 for Mlh1-Pms1). Finally, another interesting difference between Mlh3(CTD) and Pms1(CTD) concerns their N terminus. We previously characterized two helices at the beginning of the Pms1(CTD) that are located in the vicinity of the endonuclease site (Fig. 1B, refer to Pms1 Nter). The corresponding residues of Mlh3 are not visible in the crystal structure and are probably disordered. This results in important differences between the two heterodimers close to the endonuclease site. The shape of the surface surrounding the endonuclease site is different in Mlh1-Pms1 and Mlh1-Mlh3 with potential impacts on DNA substrate specificity of each complex (Fig. 1 E and F).
The Apo-Form of the Endonuclease Site of MutLγ(CTD) Contains Two Zinc-Binding Sites.
The full-length MutLγ is a latent endonuclease on supercoiled DNA with an activity that is stimulated by Mn2+ cations in the absence of other cofactors (9, 25). The superimposition of the endonuclease sites of the MutLα(CTD) and MutLγ(CTD) shows that the metal binding sites of the two complexes are located at the same positions. These sites are formed by four motifs conserved between Pms1 and Mlh3 and the last residues of Mlh1 (27). In the four crystal structures presented here, the forms Zn2 and Zn2b have two zinc atoms in the active site that occupy positions superimposable with the ones observed in MutLα(CTD) (Fig. 2 A and B and SI Appendix, Fig. S2). The six residues that chelate the two zinc atoms have the same position in the two complexes, including the last residue of Mlh1, Cys769. The 766-FER-768 motif of Mlh1 that precedes Cys769 is less superimposable, suggesting constraints imposed by the zinc atom on the thiol group of Cys769 (Fig. 1C).
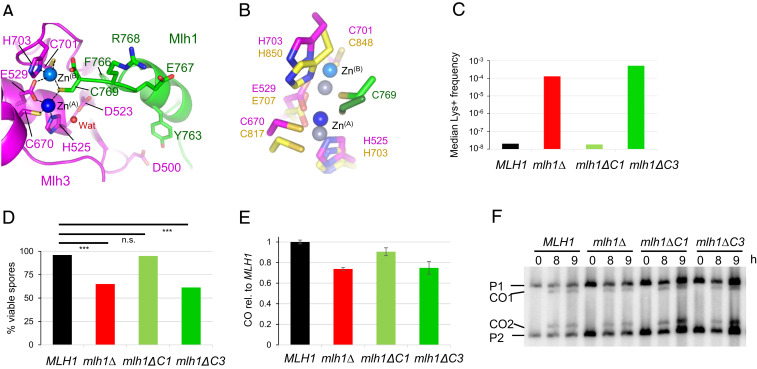
Fig. 2.
Role of the last residues of Mlh1 in the active site of Mlh3 and Pms1. (A) Endonuclease site of MutLγ with two zinc atoms. Mlh1 and Mlh3 are colored respectively in green and magenta. The Zn atom called ZnA is colored in dark blue and the second one, called ZnB, is colored in light blue. Water molecules are colored in red. (B) Superimposition of Pms1 and Mlh3 active sites. ZnA and ZnB in Pms1 are colored in greys. Names of Pms1 residues are in yellow. C769 of Mlh1 in Mlh1-Pms1 complex is in green. (C) Mutation rates measured with a Lys+ reported assay with mlh1 alleles deleted of the last residue (mlh1∆C1) or the last three residues of Mlh1 (mlh1∆C3). The median values from a fluctuation analysis (see Materials and Methods) are plotted. MLH1 and mlh1∆ data were measured in (48). (D) Spore viability of diploid strains bearing the indicated MLH1 genotype at its endogenous locus. ***P < 0.001, Fisher’s exact test. Refer also to SI Appendix, Table S2. (E–F) Crossing over frequency at the HIS4LEU2 hotspot monitored by Southern blot. Graph shows quantification at 8 and 9 h from two independent biological replicates ± range and are expressed relative to levels in MLH1 (same strains as in E).
The two additional crystal structures, ZnA and ZnB, have one Zn atom in position A and in position B, respectively (SI Appendix, Fig. S2). We collected several additional data sets on crystals soaked with ethylenediaminetetraacetic acid (EDTA), Mn2+, and Mg2+ but did not observe active sites with Mn2+ or Mg2+ atoms. We neither observed active sites that do not contain any Zn2+ atom. These observations are similar to those reported for MutLα(CTD) (27). Rogacheva et al. showed that Zn2+ alone is not sufficient to promote the endonuclease activity of MutLγ and that the addition of Zn2+ to reactions containing Mn2+ results in a significant stimulation of the endonuclease activity of MutLγ (10). This suggests that MutLγ endonuclease site has high affinity for the two zinc atoms which may have a structural role to stabilize the structure of the MutLγ active site in the absence of DNA in an inactive apo-form. We propose that upon DNA binding and some local conformational changes, a Mn2+ or Mg2+ atom may replace one of the two Zn2+ and switch the active site into an active state (10, 25, 26).
The Last Three Residues of Mlh1 Are Necessary for Both MMR and Meiotic Recombination.
The last 10 residues of Mlh1 are dispensable for physical interaction with Pms1 or Mlh3 but are necessary for MMR (27, 34, 47). Some discrepancies were observed regarding the ability of a mutant lacking the last Cys769 residue to complement a mlh1Δ mutant (27, 47). In this study, we further investigated the role of the last residues of Mlh1 by introducing deletions of these residues in the MLH1 gene at its endogenous locus. We then performed a mutation rate assay using a LYS2 reporter gene. The strain containing the mlh1∆C1 mutant, deleted for Cys769, presents no increase in mutagenesis and is therefore wild type with respect to MMR (Fig. 2C). In contrast, the mlh1∆C3 mutant, deleted for the last three residues 767-ERC-769, presents a high mutation rate (2.3 × 104-fold compared to wild type), similar to that of the mlh1∆ mutant [Fig. 2C and (48)], indicating a disruption of the MMR function of MutLα.
Strains bearing the same mlh1 alleles were analyzed for their spore viability and crossover frequency to assess their meiotic function. We observed 65% spore viability for the mlh1∆ mutant in agreement with previous studies (31) (Fig. 2D). The mlh1∆C1 mutant strain exhibits no significant difference in spore viability compared to the wild type. Interestingly, the mlh1ΔC3 strain presents a strong reduction (61% spore viability), similar to mlh1Δ mutant. Consistently, crossover frequency at the HIS4LEU2 locus was reduced as mlh1∆ in the mlh1∆C3 mutant but was almost unaffected in the mlh1∆C1 mutant (Fig. 2 E and F). Overall, these genetic data show that the last three residues of Mlh1 are necessary for both MMR repair function of MutLα and meiotic recombination function of MutLγ.
MutLγ(CTD) Contributes to the Specificity of the Heterodimer for HJ Structure Recognition.
A previous study showed that the full-length MutLγ heterodimer preferentially binds to HJs (9). Using a variety of oligonucleotide-based DNA structures and electrophoretic mobility shift assays, MutLγ was shown to interact with strong affinity (Kd ∼16 nM) with HJs and with a fivefold lower affinity with dsDNA. We investigated the binding properties of the MutLγ(CTD) with HJ and dsDNA and compared them with the binding properties of MutLα(CTD) (Fig. 3 A and B and SI Appendix, Fig. S3 A and B). We used a four-way HJ with 25-bp arms and two DNA duplexes of either 50 bp (equivalent of two arms of HJ in number of nucleotides) or 100 bp (same amount of nucleotides as the HJ) (SI Appendix, Fig. S3 C and D). For MutLγ(CTD), we observed a preference for HJ binding compared to dsDNA (50 bp or 100 bp) as reported with the full-length protein, although the affinity was about three orders of magnitude lower than that with the full-length MutLγ(CTD) (9) (Fig. 3A and SI Appendix, Fig. S3C). The interaction of MutLγ(CTD) with HJ gave a discrete band on the electrophoretic mobility shift assay (EMSA) gels whereas the interactions with linear dsDNA resulted in smears, similarly to results obtained with the full-length MutLγ. The smears suggest some dissociation during EMSA (9).
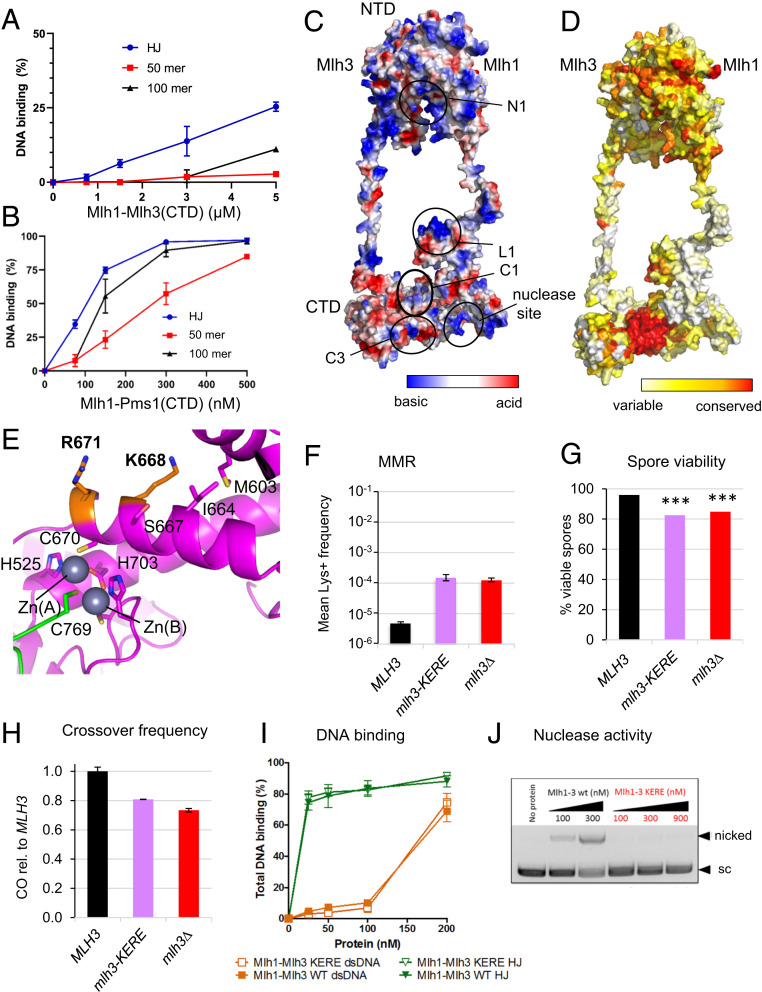
Fig. 3.
DNA-binding properties of MutLγ and identification of a mlh3-KERE separation of function mutant. (A) EMSA of MutLγ(CTD) with a HJ with four arms of 25 bp each, 50 bp–long dsDNA or 100 bp–long dsDNA. The quantification of the gels is from the following number of experiments: HJ (n = 3), 50mer (n = 3), and 100mer (n = 2). Values are the mean ± SEM when n = 3, the mean ± range when n = 2. (B) EMSA of Mlh1-Pms1(CTD) with the same DNA substrates as in A. Values are the mean ± SEM from four independent experiments for each condition. (C and D) Molecular modeling of full-length Mlh1-Mlh3. The surface are colored according (C) to the electrostatic potential and (D) to the conservation rate of the amino acids deduced from multiple sequence alignments of Mlh1 or Mlh3 eukaryotes sequences. The circles represent the five main DNA-binding sites proposed from the literature and the experiments presented in this study. The N1, L1, and C1 sites are respectively in the NTD, linker, and CTD of Mlh1. The C3 is in the CTD of Mlh3. (E) The C3 site contains two basic residues (K668 and R671) that are exposed at the surface of Mlh3(CTD). These residues are close to the C670 position that is involved in the endonuclease site. (F) Mutation rate as measured with the Lys+ reporter assay of the Mlh3-KERE allele compared to wild type and mlh3Δ. Values are the mean of nine independent colonies ± SEM. (G) Spore viability of diploid strains bearing the indicated MLH3 genotype at its endogenous locus. ***P < 0.001, Fisher’s exact test. Refer also to SI Appendix, Table S2. (H) Crossing over frequency at the HIS4LEU2 hotspot monitored by Southern blot. Graph shows quantification from two independent biological replicates ± range and are expressed relative to levels in MLH3. (I) EMSA of wild-type Mlh1-Mlh3 and Mlh1-Mlh3(KERE) mutant with dsDNA and HJ. The quantification of the EMSA is from two independent experiments. Values are the mean ± range. (J) Nuclease activity of wild-type Mlh1-Mlh3 and Mlh1-Mlh3(KERE) mutant on supercoiled pUC19 plasmid DNA.
We performed the same analyses with purified MutLα(CTD) (Fig. 3B and SI Appendix, Fig. S3D). Contrary to MutLγ(CTD), MutLα(CTD) had similar affinity in the submicromolar range for HJ or for 100 bp linear DNA. MutLα(CTD) interactions with HJ gave smears contrary to the discrete band observed with MutLγ(CTD). These results indicate that the CTD of Mlh1-Mlh3 confers some specific recognition activity of the HJ structure that is not present in Mlh1-Pms1. The large difference of affinity of MutLγ(CTD) compared to the full-length heterodimer (micromolar versus nanomolar range) indicates that the NTD and linker regions cooperate with the CTD for specific and tight HJ recognition.
DNA-Binding Sites of the Full-Length MutLγ(CTD).
The crystal structures of NTD and CTD of several bacterial and eukaryotic MutL have been characterized (43). We used these structures, molecular modeling, and multiple sequence alignments to construct a model of the full-length MutLγ heterodimer (see Materials and Methods). Fig. 3C shows a surface representation of the full-length S. cerevisae MutLγ heterodimer. The complex is presented with the linkers of Mlh1 and Mlh3 in an extended structure (the linker between the NTD and CTD is 175 amino acid–long in Mlh1 and 111 amino acid–long in Mlh3). This overall architecture corresponds to one conformation, named semicondensed, previously characterized for the MutLα and the MutLγ heterodimers by atomic force microscopy (AFM) (35, 36). In this model, the complex is 180 Å long in its longer distance. It has a central cavity with a diameter of about 50 Å that can accommodate a dsDNA (20 Å large) as well as a HJ (40 Å large, Protein Data Bank code 5t9j). This ring shape is in agreement with single molecule experiments made with MutLα and MutLγ, which show an efficient linear diffusion of the eukaryotic MutL homologs on long DNA substrates (37, 38).
To identify putative DNA-binding sites of the heterodimer, we used our model to map the residues proposed to be involved in the DNA binding of bacterial and eukaryotic MutL family members (49–51). We analyzed the electrostatic charge at the surface of the complex and the conservation rates (Fig. 3 C and D). From this, we identified five main patches conserved among the MutL complexes: three patches on the Mlh1 subunit (called here N1 in NTD, L1 in linker, and C1 in CTD), one patch on Mlh3 CTD (C3), and the endonuclease site (Fig. 3C). In our model of the semicondensed conformation, the five patches are distant. AFM studies have shown that upon ATP binding, the MutL structures can condense further bringing together the NTD and CTD regions (35, 36). The proposed five patches may thus relocate into close proximity after such conformational changes.
Characterization of a Basic Patch in the C-Terminal Region of Mlh3.
We next investigated the contribution of the C3 patch identified in the C-terminal region of Mlh3 (Fig. 3 C–E). We constructed a strain with the double mutation Mlh3-K668E-R671E (called KERE) introduced at the MLH3 endogenous locus and analyzed the mutation rate with the Lys+ reporter assay. We observed a ∼30-fold higher mutation rate than in the wild-type strain, similar to the one observed in the mlh3∆ strain analyzed in parallel (Fig. 3F). Importantly, we checked that the mutated protein still interacted with Mlh1 in a two-hybrid assay (SI Appendix, Fig. S4A). We then investigated the effect of this double point mutation on spore viability and crossover formation to evaluate its role in meiosis. The spore viability of the mlh3-KERE allele was significantly reduced compared to the wild-type strain to a value of ∼80%, similar to the one observed with mlh3Δ allele (Fig. 3G). We then measured meiotic crossover frequencies at the HIS4LEU2 locus and observed that the mlh3-KERE mutant had a reduced frequency, similar to that of mlh3∆ (Fig. 3H and SI Appendix, Fig. S4B). The mutation on the C3 patch of Mlh3 thus severely affects both MMR and meiotic recombination activities of the MutLγ heterodimer to a level similar to the deletion of the MLH3 gene.
To better understand the molecular mechanism of these impaired activities, we expressed in insect cells and purified the full-length MutLγ heterodimer wild type and KERE mutant (SI Appendix, Fig. S4C). We then compared the DNA-binding activities of the purified MutLγ(KERE) mutant and wild-type complex for dsDNA and HJ substrates. The KERE mutant presented wild-type DNA-binding capacity for both DNA substrates with affinities similar to the ones previously reported (Fig. 3I and SI Appendix, Fig. S4D) (9). In accord, the mutated protein still fully associated in vivo with meiotic recombination hotspots (SI Appendix, Fig. S4E). However, the KERE mutation completely abrogated the endonuclease activity, as measured by nicking of a supercoiled plasmid DNA substrate (Fig. 3J). These results indicate that the KERE mutation does not impair the integrity of the MutLγ heterodimer since it presents wild-type activity for in vivo and in vitro DNA binding but that the C3 patch plays an important role for the nuclease activity. The mutated basic motif K688-R671 surrounds the conserved Cys670 that chelates the zinc and is part of the endonuclease site (Fig. 3E). The double mutant may affect the position of the Cys in the active site and/or may affect the precise positioning of the DNA on the endonuclease site. Additional structural studies in presence of DNA will help to unveil the DNA-binding site of MutLγ and the role of this basic K688-R671 conserved motif.
Characterization of MutLγ Mutants with a Potential Role in the Condensation of the Heterodimer.
The full-length MutLγ heterodimer presents two long linker regions between the NTD and CTD of each subunit as mentioned above. AFM studies of the full-length MutLα and MutLγ previously reported four main states of the heterodimer that are regulated by the presence of ATP and ADP (Fig. 4A, for a schematic representation) (35, 36). In all these conformations, the CTD of the MutL homolog subunits mediate the heterodimer formation through their dimerization. The NTD are observed in different positions. They can be dissociated and far from the CTD (extended state). Alternatively, the NTD can be associated in a position distinct from the CTD (semicondensed state) (as in Fig. 3 C and D). In addition, one or both NTDs may interact with the CTD dimers (one arm state or condensed state, respectively).
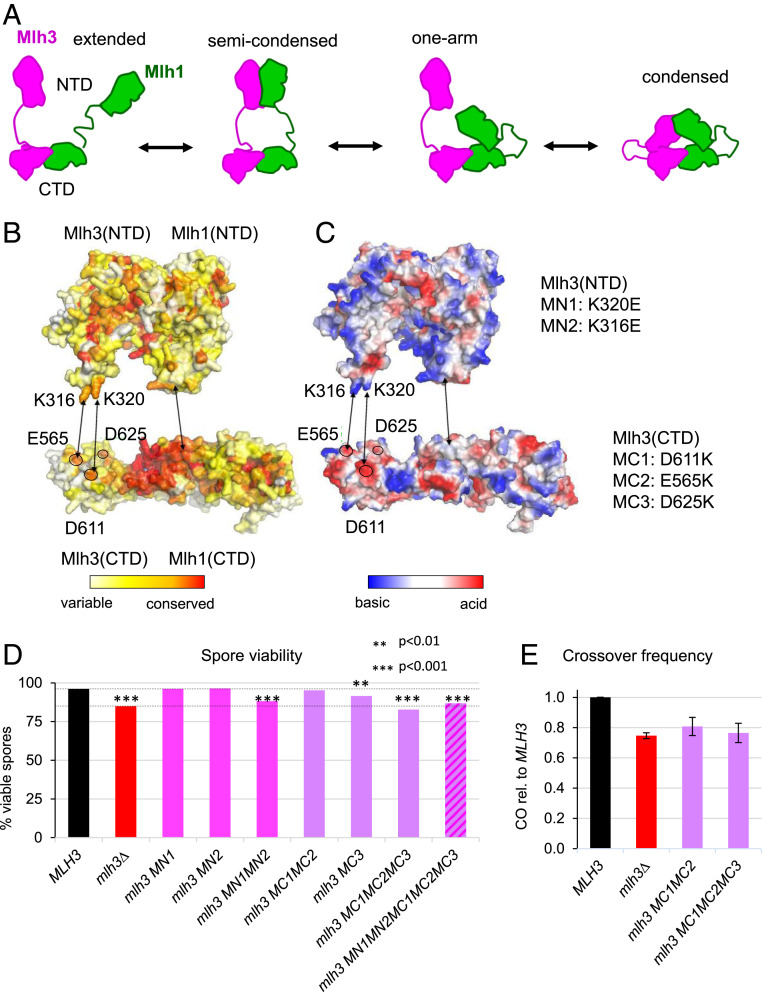
Fig. 4.
Condensation of Mlh1 and Mlh3 through NTD and CTD interactions. (A) Schematic representation of the four conformational states proposed for MutL homologs from AFM studies. (B and C) Surface representation of the NTD and CTD of Mlh1 and Mlh3 colored according to conservation rate of the amino acids (B) and to electrostatic potential (C). Two conserved basic residues in Mlh3-NTD (K316 and K320) and three conserved acid residues in Mlh3-CTD (E565, D611 and D625) are proposed to contribute to the condensation of the full-length heterodimer through their interaction in the condensed state. The five corresponding mutations in Mlh3(NTD) (MN1 and MN2) and in Mlh3(CTD) (MC1, MC2, and MC3) performed in this study are indicated. The structure of Mlh1 and Mlh3 NTDs were modeled from the crystal structures of MutL NTDs. (D) Spore viability of diploid strains bearing the indicated mlh3 genotype at its endogenous locus. The dotted lines indicate MLH3 (WT) and mlh3∆ spore viability levels for comparison. ***P < 0.001, **P < 0.01, Fisher’s exact test. Refer also to SI Appendix, Table S2. (E) Crossing over frequency at the HIS4LEU2 hotspot monitored by Southern blot. Graph shows quantification from three independent biological replicates ± SD, except for mlh3MC1MC2 (two replicates, mean values ± range), and are expressed relative to levels in MLH3 (same strains as in D).
We used our model of the full-length MutLγ to identify residues in Mlh1 or Mlh3 that may contribute to stabilize the condensed states of the heterodimer. We analyzed the electrostatic distribution and the conservation rates on the NTD and CTD surfaces. We identified two charged positions on Mlh3 NTD surface (Lys316 and Lys320) and three charged positions on Mlh3 CTD (Glu565, Asp611, and Asp625) that seem well positioned to participate in the condensing events between Mlh3 NTD and CTD (Fig. 4 B and C).
We constructed 11 mlh3 alleles. They contain mutations with between one to three charge reversions (K in E and D or E in K) on the NTD (mutations called MN1 and MN2) or on the CTD (mutations called MC1, MC2, or MC3) and combinations with mutations on both sides. We measured the effect on spore viability for these 11 alleles. Six alleles have a reduced spore viability (Fig. 4D and SI Appendix, Table S2). Among these, the single mutant D625K (allele mlh3-MC3) has an intermediate spore viability defect, but combination with two other mutations in the vicinity (E565K and D611K, mlh3-MC1MC2), showing no apparent spore viability defect, further reduces spore viability to a value similar to mlh3Δ. Indeed, the mlh3-MC1MC2, although without apparent spore viability defect, has reduced crossover frequencies and has a synergic effect in combination with the D625K mutant, the triple mutant also showing mlh3∆ crossover frequencies (Fig. 4E). Similarly, two single mutations in the NTD (K316E and K320E) have no effect on spore viability on their own, but their combination leads to a null phenotype (allele mlh3-mn1mn2). We thus characterized meiotic recombination-defective Mlh3 alleles with mutations in conserved positions of the NTD and CTD of the MutLγ subunit that present severe spore viability defect outside the dimerization and endonuclease sites which may indicate, according to our model, that the compaction of the MutLγ heterodimer is important for meiotic recombination.
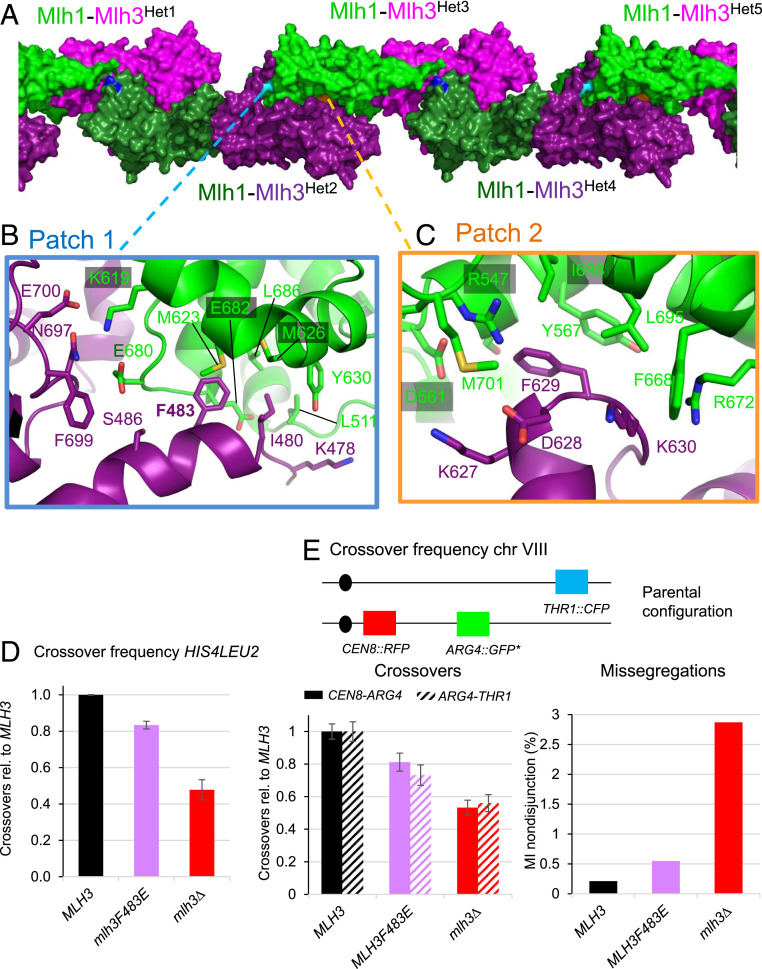
The MutLγ(CTD) Molecules Are Arranged in Filamentous Structures in the Crystal, and a Mutation in the Corresponding Interfaces Lowers Crossover Frequency.
Previous biochemical studies of full-length MutLγ (9, 10, 41) as well as recent biochemical (25, 26) and genetic data (15) showed that it functions as a noncanonical resolvase. While it specifically recognizes HJs and its cleavage activity is stimulated by them, it incises DNA not at the HJ but at some distance away from the HJ (25, 26, 41). It has been proposed that MutLγ can form oligomers that nucleate on the HJ and extend on the adjacent dsDNA arms, where they will be activated by their partners (25, 41).
By examining the crystal packing of the apo-form of the MutLγ(CTD) studied here, we identified regular arrangements of Mlh1-Mlh3 molecules in filament-like structures (Fig. 5A). This oligomerization is mediated by a large, buried surface of the Mlh3 subunit of one heterodimer and the Mlh1 subunit from a second heterodimer. The buried surface at each filament contact is 2,500 Å2, compared to a buried surface of 2,100 Å2 for the canonical heterodimer interface. In these filaments, the N terminus and C terminus of Mlh1 and Mlh3 are accessible, suggesting that this oligomerization can occur with the full-length proteins without steric hindrance.
Fig. 5.
Formation of MutLγ filaments through Mlh3-Cter and Mlh1-Cter interactions. (A) Filament-like structures made by Mlh1-Mlh3 heterodimers in the crystals. The interactions between five heterodimers (Het1 to Het5) in the crystal are represented by alternating the color of the heterodimers with Mlh1-Mlh3 in (green-magenta) or in (dark green-purple). The positions of the three main patches that mediate these structures are shown with the central residues colored respectively in cyan (patch 1), in orange (patch 2), and in blue (patch 3, SI Appendix, Fig. S5A). (B) Patch 1 includes interactions between six Mlh3 residues and nine Mlh1 residues. Most Mlh1 residues belong to the MIP-binding site that is involved in the interaction of Mlh1 with the MIP-motifs observed in Exo1, Ntg2 and Sgs1 (27). The F483 position of Mlh3 is buried in the center of this Mlh1 pocket in a position similar to the aromatic of the MIP motifs of the Mlh1 partners (SI Appendix, Fig. S5 A–C, for the superposition of the MIP motif of Exo1 with the positions of the Mlh3 residues). (C) Patch 2 includes interactions between four Mlh3 residues and eight Mlh1 residues. The Mlh1 side contains the R547 position that interacts in MutLα with a Pms1 C-terminal extension that is not present in Mlh3(CTD). (D) Crossing over frequency at the HIS4LEU2 hotspot monitored by Southern blot. Graph shows quantification at 8 and 9 h from two independent biological replicates ± range and are expressed relative to levels in MLH3. (E) Meiotic crossovers on chromosome VIII. Top: illustration of the fluorescent spore setup (60). Genetic distances are measured in the CEN8-ARG4 and ARG4-THR1 adjacent intervals and expressed relative to the MLH3 values. The MI nondisjunction frequency was measured from the same dataset as for genetic distances, as described previously (60). MLH3: 1898 tetrads; mlh3F483E: 1277 tetrads; and mlh3∆: 1463 tetrads. Error bars: SE.
The interface is constituted by three interfaces. The first interface involves the Mlh3 478-KTITDFS-484 region that interacts with Mlh1 binding site for the Mlh1 Interacting Protein (MIP) motif (including residues E682, M623, and M626) (Fig. 5B, patch 1). The second interface involves the Mlh3 627-KDFKKL-632 region that interacts with Mlh1 positions from helices αA (Y567 to S564), αC (R671 to F668), αD (L695), and strand β2 (R547) (Fig. 5C, patch 2). Finally, a third patch is mediated by Mlh3 residues in the extreme Cter of the Mlh3 CTD close to the 701-CxH-703 motif of the catalytic site and polar residues of Mlh1 (D609, K615, and K619) from helix αB (SI Appendix, Fig. S5A).
In the first patch, the interactions of the Mlh3 residues with Mlh1 are reminiscent of the binding mode of the MIP-motif of Exo1 and Ntg2, suggesting a potential competitive binding between this Mlh3 motif and the MIP-box proteins (SI Appendix, Fig. S5 B–D) (27). The F483 position of Mlh3 is deeply buried in the MIP-site and superposes with the last residue of Exo1 motif (F447). We then analyzed by calorimetry the interaction between peptides of Exo1 and Ntg2 containing their MIP motifs and the MutLγ(CTD). The MutLγ(CTD) binds the Exo1 and Ntg2 peptides with respective Kd of 4.2 ± 0.9 µM and 0.7 ± 0.04 µM, similar to the one measured previously with Mlh1(CTD) or MutLα(CTD) (27). This shows that the heterodimerization of Mlh1 with either Pms1 or Mlh3 has no effect on the MIP-motif binding (SI Appendix, Fig. S5E).
In the second patch, Mlh3 residues interact with a position of Mlh1 (R547) that stabilizes in MutLα(CTD) the interaction with Pms1(CTD) through a salt bridge with Pms1 D670. This position of Pms1 is in its extreme Cter extension, an extension not present in Mlh3. In the heterodimer Mlh1-Mlh3, this region of Mlh1 is thus accessible for interaction with other proteins like another Mlh3 subunit as in the filaments described here (Fig. 5C, patch 2).
We analyzed the potential role of the filaments observed in the crystal by characterizing a mutant of a residue positioned at the core of the main patch, patch 1. We constructed a mlh3F483E allele to weaken the interaction observed between Mlh3(F483) and the MIP-binding site of a proximal Mlh1 from another heterodimer. Interestingly, this mutation significantly reduced meiotic crossover frequency, measured on two different chromosomes: at the HIS4LEU2 locus on chromosome III (Fig. 5D) and in the CEN8-THR1 interval on chromosome VIII (Fig. 5E). Moreover, although spore viability was not affected in the mlh3F483E mutant, meiosis I nondisjunction was elevated, indicative of a specific defect in homolog segregation due to impaired crossovers (Fig. 5E and SI Appendix, Table S2).
Therefore, our results are consistent with the suggestion that interactions between several MutLγ heterodimers are important for MutLγ function in meiotic crossovers.
Discussion
Budding yeast and most eukaryotic genomes encode four proteins (Pms1, Mlh1, Mlh2, and Mlh3) homologous to the bacterial MutL MMR factor (16). They can form three heterodimers with Mlh1 as the common subunit and are involved in many DNA metabolic pathways including MMR and meiotic recombination (52). In humans, these heterodimers are also involved in additional processes including Class Switch Recombination, DNA damage signaling, and ICL repair (52–54). The characterization of the molecular mechanisms of these heterodimers is central to understand the specialization acquired by the respective MutL homologs. The crystal structure of MutLγ(CTD) allows a fine analysis of the main differences with its MutLα counterpart at the endonuclease site and other functional sites.
We previously reported that the core of the dimerization interface involves the packing of a four-strand β-sheet of Mlh1(CTD) with the related one of Pms1(CTD) and six hydrophobic residues that are conserved between Mlh1, Pms1, and bacterial MutL homologs (27). The crystal structure presented here shows that Mlh3 uses the same six conserved hydrophobic residues to form the core of the heterodimer, highlighting strong evolutionary conservation in this heterodimerization interface. A second conserved feature between MutLα and MutLγ is the position of the last residue C769 of Mlh1 that is positioned in the endonuclease site (Fig. 2 A and B). The position of C769 superimposes well in both complexes, although Pms1 and Mlh3 present some sequence differences in this region, suggesting structural and functional constraints on C769. This is in good agreement with the central role identified in vivo in mutation and meiotic recombination assays using an allele deleted for the last three residues of Mlh1.
How do the three subunits Pms1, Mlh2, and Mlh3 compete for interaction with the Mlh1 subunit? Systematic proteomic studies indicate that Mlh1 and Pms1 are the most abundant subunits among yeast MutL homologs (ref. 55 and Saccharomyces Genome Database). The Mlh1-Pms1 heterodimer has the largest buried surface due to the additional interface mediated by the last 12 residues of Pms1, which are absent in Mlh3 and Mlh2. This additional patch contributes to the stability of Mlh1-Pms1, since mutation of a salt bridge in this region (between Mlh1 R547 and Pms1 D870) leads to a destabilization of the heterodimer and causes a strong mutator phenotype in exo1Δ mutant strains (27, 46). The role of this Pms1 C terminus extension was confirmed with a chimeric construct, containing the Mlh3 sequence, followed by the last 12 amino acid of Pms1. This Mlh3 chimera presents a stronger interaction with Mlh1, as evaluated by two-hybrid assays (56). Mlh1 itself can compete with the other MutL homologs since overexpression of Mlh1 was shown to have a mutator phenotype through Mlh1 homodimerization (57). The competition between Pms1, Mlh3, and Mlh2 for Mlh1 may also depend on cell stage–specific posttranslational modifications or expression program. It has indeed been shown that Mlh3 expression increases during meiosis, which may change the balance between the different heterodimers (58).
The structure of MutLγ(CTD) represents the second structure of a eukaryotic endonuclease MutL homolog. Despite moderate sequence identity between the Mlh3 and Pms1 CTD sequences (17% identity, 33% homology), the structures of the active site in its apo-form superimpose well. This result further confirms that the Mlh1 C terminus contributes to the formation of the endonuclease site of the heterodimer. The C terminus of Mlh1 superimposes well in MutLα(CTD) and MutLγ(CTD) despite differences between Pms1 and Mlh3 sequences in this region. We clearly demonstrated the in vivo role of the Mlh1 C terminus by characterizing the severe phenotype in MMR and meiosis of alleles deleted for the last three Mlh1 residues (ERC). The present study also confirms a similar position of two zinc atoms in the MutLα(CTD) and MutLγ(CTD) with or without Mn and Mg cations in the crystallization conditions. These structures reinforce the hypothesis of a structural role for the zinc atoms in absence of DNA.
MutLα and MutLγ present different DNA-binding properties. MutLα interacts weakly with short DNA and cooperatively with longer DNA (40). MutLγ, in contrast, has strong affinities for short DNA substrates, in particular for HJ structures (9), and it can form multimers on longer DNA (41). In a previous study, contacts between DNA and MutLγ were mapped by footprinting on the NTD and in the linker regions (35). Some of these positions correspond to conserved patches on the surface of the full-length heterodimer modeled in this study (Fig. 3D). In addition, we defined a mutant called KERE in the C-terminal region of Mlh3. This allele is affected in its MMR activity and meiotic recombination in a manner similar to an mlh3∆ allele. This mutant is still able to interact with Mlh1 and to bind DNA. We propose that in the KERE mutant, the DNA adopts an alternative position distant from the endonuclease site.
The crystal structure of MutLγ(CTD) allows to position residues mutated in several Mlh3 alleles studied in the literature. In particular, a recent study described 60 alleles of Mlh3 including 25 alleles located in the CTD region (56). Mutations in the Mlh3-conserved residues proposed to participate in the endonuclease site disrupt MMR and meiotic crossovers. This is in agreement with previous studies and with our observation that these residues interact with two zinc atoms in Mlh3 as already reported for Pms1 (27). Three alleles that disrupt both MMR and meiosis activities are remote from the active site (SI Appendix, Fig. S6A). One allele (mlh3-41) corresponds to positions that may stabilize indirectly the loop containing the Cys701 and His703 of the active site. Two alleles (mlh3-48, -49) are located on the last helix of the regulatory domain. The corresponding mutations likely affect the position of the regulatory domain and the electrostatic surface surrounding the endonuclease site. One allele (mlh3-45) is a separation of function mutant (MMR−, crossover+). It corresponds to the mutation K577A-K578A and presents wild-type endonuclease activity and interaction with Mlh1 as measured by two-hybrid (56). These two positions are exposed to the solvent and are located at the periphery of the regulatory domain far from the active site. It will be interesting to further study this mutant to understand why it specifically affects MMR and not meiosis. Noteworthy, the KERE allele reported here was not included in that systematic study since the basic residues are separated by more than one residue; that was a selection criteria in that study (56).
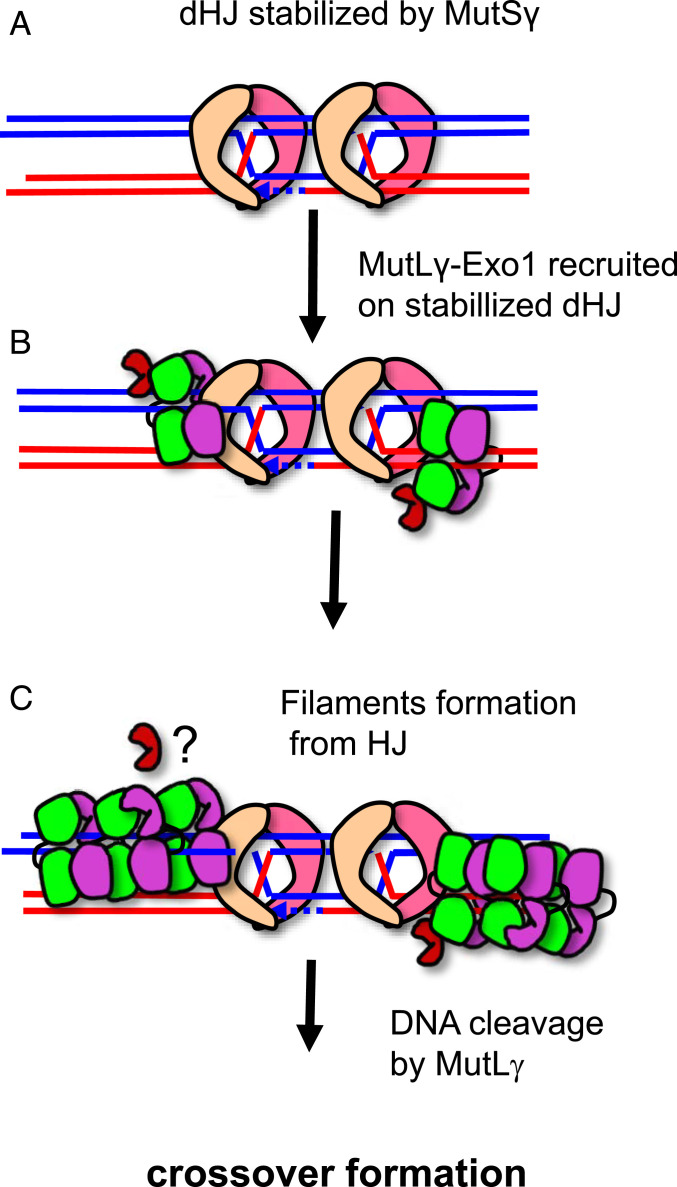
The proposed molecular mechanisms of MutLα in MMR and of MutLγ in meiotic recombination present some interesting parallels suggesting that their function may have diverged while keeping conserved features. Firstly, both heterodimers are recruited by a MutS homolog (Fig. 6A), MutSα or MutSβ bound on mismatch for MutLα and MutSγ bound on HJ for MutLγ (Fig. 6B and SI Appendix, Fig. S6B). Recent studies on MutLα and MutLγ proposed that these heterodimers can condense and oligomerize upon their recruitment by the MutS homologs and that this oligomerization will contribute to the incision of DNA away from the mismatch for MutLα and from the HJ for MutLγ (Fig. 6C and SI Appendix, Fig. S6B) (9, 25, 41, 59). In both pathways, the interaction between Mlh1 and Exo1 through the MIP-motif of Exo1 will be important for the efficiency of the pathway either to resect the newly synthesized strand in MMR or to activate the Mlh1-Mlh3 nuclease in meiotic recombination.
Fig. 6.
Model for MutLγ mechanism in meiotic recombination. In meiotic recombination, MutLγ, in complex with Exo1, is recruited by MutSγ on dHJ intermediates (A–B). Transient MutLγ polymerization is proposed to occur at this step leading to an incision of the DNA away from the dHJ (C). This polymerization is proposed to involve three patches including the MIP-binding site of Mlh1. Further studies should help to evaluate whether competition exists between Exo1 and Mlh3 for the Mlh1 MIP-binding site.
In this study, we further investigate the molecular mechanisms of the condensation and oligomerization of MutLγ. We first identified positions in Mlh3(NTD) and Mlh3(CTD) that may favor contacts between the NTD and CTD upon condensation. We showed that their mutations impact crossover efficiency. Secondly, we observed a filament structure of MutLγ(CTD) in the crystal (Fig. 5). We analyzed an allele with a mutation at the center of the filament interface and showed an impact of this mutation on meiotic recombination. A detailed analysis of the mutant Mlh3-F483E will help in the future to further characterize the impact of this mutation on the MutLγ properties of HJ binding or oligomer formation. We propose that MutLγ may form oligomers that initiate at the HJ and extend away from the junction. The oligomerization may be initiated by the interaction of one Mlh3 subunit with the Mlh1 subunit from a second heterodimer. Potentially, Exo1 and Mlh3 may be in competition for the Mlh1 MIP-binding site (Fig. 6C). Additional biochemical analyses are needed to evaluate this possibility. The precise molecular mechanisms of the condensation and oligomerization of MutLγ will require additional studies in the presence of DNA and full-length proteins. Our results pave the way for such studies by identifying candidate positions that may be involved in these processes.
Our previous structural study of the MutLα(CTD) and the one reported in this study of MutLγ(CTD) allow molecular comparison of the two MutL homologs domains and shed light on their specialization acquired during evolution for the main functions of MutLα in MMR or MutLγ in meiosis. The major challenge is now to further complete this comparison of the two MutL homologs with structural and functional analyses of the full-length proteins in complex with their favorite DNA substrates.
Materials and Methods
Details are provided in SI Appendix.
Preparation of Mlh1-Mlh3 Complexes.
The S. cerevisiae Mlh1-Mlh3 (CTD) that contains Mlh1 residues 485 to 769 and Mlh3 residues 460 to 715 was expressed in Escherichia coli. The full-length S. cerevisiae Mlh1-Mlh3 heterodimer was expressed in insect cells.
DNA Binding and Nucleases Assays.
The 50-bp dsDNA and HJ DNA substrates were prepared as described previously (9). The DNA-binding experiments were performed without added salt. The final NaCl concentration was 43 mM due to the addition of the recombinant proteins. The nuclease assays were carried out as described (9).
Crystal Structures and SAXS Analyses of the MutLγ(CTD) Complex.
Diffraction data were collected at 100 K on protein crystallography beamline Proxima1 (SOLEIL) (SI Appendix, Table S1). SAXS measurements were conducted at the French synchrotron SOLEIL on the SWING beamline. A structural model of the complex between the full-length sequences of yeast Mlh1 and Mlh3 was obtained using the RosettaCM software for homology modeling.
Yeast Manipulations and Strains Construction.
S. cerevisiae haploid strains used for the study of spontaneous mutagenesis are isogenic to E134 (MATα ade5-1 lys2::InsEA14 trp1-289 his7-2 leu2-3,112 ura3-52) that was kindly provided by Erik Johansson (Umea University, Umea, Sweden). All diploid yeast strains are derivatives of the SK1 background. Details are provided in SI Appendix, and yeast strains genotype is provided in SI Appendix, Table S3.
Spontaneous Mutation Rates and Yeast Two-Hybrid Assays.
For data presented in Fig. 2C, the fluctuation tests to determine spontaneous mutation rates were performed in two to five independent experiments of nine independent cultures. Yeast two-hybrid assays were performed exactly as described (31).
Analysis of Crossover Frequencies and Chromatin Immunoprecipitation.
For crossover analysis at the HIS4LEU2 locus, cells were harvested from meiotic time courses at the indicated time point. The data were further normalized to the crossover frequency observed in the wild type. Chromatin immunoprecipitation of Mlh3-Myc8 and qPCR were performed as described (23).
Supplementary Material
Acknowledgments
We are very grateful to Serge Boiteux for his continuous interest and extremely helpful discussions all along the study. J.-B.C. and V.B. are supported by French Agence Nationale de la Recherche (ANR) (Grant ANR-15-CE11-0011 Resolve). J.-B.C. has access to I2BC Platforms supported by the French Infrastructure for Integrated Structural Biology (FRISBI) (Grant ANR-10-INBS-05). J.-B.C. is supported by Institut National du Cancer (INCA) (Grant INCA 2016-159). A.S. was supported by a postdoctoral fellowship from the Fondation ARC pour la Recherche sur la Cancer. J.D. was supported by a doctoral fellowship (Grant Doc4) from the Fondation ARC.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022704118/-/DCSupplemental.
Data Availability
Coordinate data have been deposited in Protein Data Bank (6RMN, 6SHX, 6SNS, and 6SNV).
References
- 1.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zickler D., Kleckner N., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeney S., Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2, 81–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allers T., Lichten M., Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Hunter N., Kleckner N., The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Kadyrov F. A., et al., Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishant K. T., Plys A. J., Alani E., A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179, 747–755 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranjha L., Anand R., Cejka P., The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J. Biol. Chem. 289, 5674–5686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogacheva M. V., et al., Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J. Biol. Chem. 289, 5664–5673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakharyevich K., Tang S., Ma Y., Hunter N., Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barlow A. L., Hultén M. A., Crossing over analysis at pachytene in man. Eur. J. Hum. Genet. 6, 350–358 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Chelysheva L., et al., An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129, 143–153 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Kolas N. K., et al., Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171, 447–458 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsolier-Kergoat M. C., Khan M. M., Schott J., Zhu X., Llorente B., Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Wang T. F., Kleckner N., Hunter N., Functional specificity of MutL homologs in yeast: Evidence for three mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. U.S.A. 96, 13914–13919 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zakharyevich K., et al., Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40, 1001–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipkin S. M., et al., MLH3: A DNA mismatch repair gene associated with mammalian microsatellite instability. Nat. Genet. 24, 27–35 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Toledo M., et al., A mutation in the endonuclease domain of mouse MLH3 reveals novel roles for MutLγ during crossover formation in meiotic prophase I. PLoS Genet. 15, e1008177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edelmann W., et al., Meiotic pachytene arrest in MLH1-deficient mice. Cell 85, 1125–1134 (1996). [DOI] [PubMed] [Google Scholar]
- 21.De Muyt A., et al., BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyatnitskaya A., Borde V., De Muyt A., Crossing and zipping: Molecular duties of the ZMM proteins in meiosis. Chromosoma 128, 181–198 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Sanchez A., et al., Exo1 recruits Cdc5 polo kinase to MutLγ to ensure efficient meiotic crossover formation. Proc. Natl. Acad. Sci. U.S.A. 117, 30577–30588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West S. C., et al., Resolution of recombination intermediates: Mechanisms and regulation. Cold Spring Harb. Symp. Quant. Biol. 80, 103–109 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Cannavo E., et al., Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Kulkarni D. S., et al., PCNA activates the MutLγ endonuclease to promote meiotic crossing over. Nature 586, 623–627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gueneau E., et al., Structure of the MutLα C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat. Struct. Mol. Biol. 20, 461–468 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Flores-Rozas H., Kolodner R. D., The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 95, 12404–12409 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadyrova L. Y., Gujar V., Burdett V., Modrich P. L., Kadyrov F. A., Human MutLγ, the MLH1-MLH3 heterodimer, is an endonuclease that promotes DNA expansion. Proc. Natl. Acad. Sci. U.S.A. 117, 3535–3542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell C. S., et al., Mlh2 is an accessory factor for DNA mismatch repair in Saccharomyces cerevisiae. PLoS Genet. 10, e1004327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duroc Y., et al., Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. eLife 6, e21900 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harfe B. D., Minesinger B. K., Jinks-Robertson S., Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10, 145–148 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Guerrette S., Acharya S., Fishel R., The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J. Biol. Chem. 274, 6336–6341 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Pang Q., Prolla T. A., Liskay R. M., Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol. Cell. Biol. 17, 4465–4473 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claeys Bouuaert C., Keeney S., Distinct DNA-binding surfaces in the ATPase and linker domains of MutLγ determine its substrate specificities and exert separable functions in meiotic recombination and mismatch repair. PLoS Genet. 13, e1006722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sacho E. J., Kadyrov F. A., Modrich P., Kunkel T. A., Erie D. A., Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol. Cell 29, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallardo I. F., et al., High-throughput universal DNA curtain arrays for single-molecule fluorescence imaging. Langmuir 31, 10310–10317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorman J., Plys A. J., Visnapuu M. L., Alani E., Greene E. C., Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 17, 932–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeon Y., et al., Dynamic control of strand excision during human DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 113, 3281–3286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall M. C., Wang H., Erie D. A., Kunkel T. A., High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J. Mol. Biol. 312, 637–647 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Manhart C. M., et al., The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biol. 15, e2001164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukui K., Nishida M., Nakagawa N., Masui R., Kuramitsu S., Bound nucleotide controls the endonuclease activity of mismatch repair enzyme MutL. J. Biol. Chem. 283, 12136–12145 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Guarné A., Charbonnier J. B., Insights from a decade of biophysical studies on MutL: Roles in strand discrimination and mismatch removal. Prog. Biophys. Mol. Biol. 117, 149–156 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Namadurai S., et al., The C-terminal domain of the MutL homolog from Neisseria gonorrhoeae forms an inverted homodimer. PLoS One 5, e13726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudenko O., Thureau A., Perez J., “Evolutionary refinement of the 3D structure of multi-domain protein complexes from small angle X-ray scattering data” in Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO ’19), López-Ibáñez M., Ed. (ACM, New York, NY, USA, 2019), pp. 401–402. [Google Scholar]
- 46.Amin N. S., Nguyen M. N., Oh S., Kolodner R. D., exo1-Dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21, 5142–5155 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith C. E., et al., Dominant mutations in S. cerevisiae PMS1 identify the Mlh1-Pms1 endonuclease active site and an exonuclease 1-independent mismatch repair pathway. PLoS Genet. 9, e1003869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksenova A., et al., Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase ε. PLoS Genet. 6, e1001209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Groothuizen F. S., Sixma T. K., The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst.) 38, 14–23 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Junop M. S., Yang W., Funchain P., Clendenin W., Miller J. H., In vitro and in vivo studies of MutS, MutL and MutH mutants: Correlation of mismatch repair and DNA recombination. DNA Repair (Amst.) 2, 387–405 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Robertson A., Pattishall S. R., Matson S. W., The DNA binding activity of MutL is required for methyl-directed mismatch repair in Escherichia coli. J. Biol. Chem. 281, 8399–8408 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Jiricny J., The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7, 335–346 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Kato N., et al., Sensing and processing of DNA interstrand crosslinks by the mismatch repair pathway. Cell Rep. 21, 1375–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahman M. M., et al., Genetic evidence for the involvement of mismatch repair proteins, PMS2 and MLH3, in a late step of homologous recombination. J. Biol. Chem. 295, 17460–17475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Al-Sweel N., et al., mlh3 mutations in baker’s yeast alter meiotic recombination outcomes by increasing noncrossover events genome-wide. PLoS Genet. 13, e1006974 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shcherbakova P. V., Kunkel T. A., Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 19, 3177–3183 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borde V., et al., Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28, 99–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bradford K. C., et al., Dynamic human MutSα-MutLα complexes compact mismatched DNA. Proc. Natl. Acad. Sci. U.S.A. 117, 16302–16312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thacker D., Lam I., Knop M., Keeney S., Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae. Genetics 189, 423–439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinate data have been deposited in Protein Data Bank (6RMN, 6SHX, 6SNS, and 6SNV).