Abstract
Background:
Lung microbiota profiles in patients with early idiopathic pulmonary fibrosis (IPF) have been associated with disease progression; however, the topographic heterogeneity of lung microbiota and their roles in advanced IPF are unknown.
Methods:
We performed a retrospective, case-control study of explanted lung tissue obtained at the time of lung transplantation or rapid autopsy from patients with IPF and other chronic lung diseases (connective tissue disease-associated interstitial lung disease [CTD-ILD], cystic fibrosis [CF], chronic obstructive pulmonary disease [COPD], and donor lungs unsuitable for transplant from Center for Organ Recovery and Education [CORE]). We sampled subpleural tissue and airway-based specimens (bronchial washings and airway tissue) and quantified bacterial load and profiled communities by amplification and sequencing of the 16S rRNA gene.
Findings:
Explants from 62 IPF, 15 CTD-ILD, 20 CF, 20 COPD and 20 CORE patients were included. Airway-based samples had higher bacterial load compared to distal parenchymal tissue. IPF basilar tissue had much lower bacterial load compared to CF and CORE lungs (p<0.001). No microbial community differences were found between parenchymal tissue samples from different IPF lobes. Dirichlet multinomial models revealed an IPF cluster (29%) with distinct composition, high bacterial load and low alpha diversity, exhibiting higher odds for acute exacerbation or death.
Interpretation:
IPF explants had low biomass in the distal parenchyma of all three lobes with higher bacterial load in the airways. The discovery of a distinct subgroup of IPF patients with higher bacterial load and worse clinical outcomes supports investigation of personalized medicine approaches for microbiome-targeted interventions.
Keywords: Idiopathic pulmonary fibrosis, lung microbiome
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a devastating age-associated disease, occurring more frequently in smokers and carriers of host-defense gene mutations.[1, 2] While it is theorized that alveolar injury in a genetically-susceptible host propagates aberrant repair mechanisms resulting in fibrosis, the exact environmental factors provoking lung injury have not been defined.[3] Dysbiosis in the respiratory tract has been proposed as a potential mechanism for precipitation and/or perpetuation of lung injury. The hypothesis that lung microbiota contributes to IPF progression emerged from epidemiologic observations suggestive of host-microbiome interactions in the respiratory tract, such as the finding that immunosuppression increases mortality in IPF, whereas antibiotics may offer survival benefit in treatment-tolerant and adherent IPF patients.[4–6] Three prospective cohort studies in patients with early IPF provided direct evidence for the lung microbiome hypothesis with the use of culture-independent, bacterial DNA sequencing techniques. Bronchoalveolar lavage (BAL) fluid from patients with IPF had higher bacterial burden compared to chronic obstructive pulmonary disease (COPD) patients and healthy controls,[7] and those IPF patients with the highest bacterial burden exhibited worse outcomes.[7–9] Lung microbial profiles were associated with distinct host transcriptome responses.[10, 11] Furthermore, in murine model of fibrosis, lung dysbiosis preceded peak lung injury and was associated with worse survival.[8] Therefore, lung microbiota manipulation with antibiotic therapies has become an attractive target for intervention, currently assessed by ongoing clinical trials.[12, 13]
The role of lung microbiota in later stages of IPF remains unknown. Furthermore, the involvement of microbes across the respiratory tract in such a disease hallmarked by spatial heterogeneity (subpleural predominance and an apicobasal gradient of fibrosis) is poorly understood. The BAL samples used by prior studies capture microbiota from lower generations of the tracheobronchial tree and up to 5% of the alveolar space, but do not provide detailed regional characterization of microbiota. In a previous study from our group (Microbiome in Lung Explants - MiLEs study), we examined distal parenchymal tissue from lung explants of patients with end-stage usual interstitial pneumonia (UIP) at the time of lung transplant or death.[14] Surprisingly, we found exceedingly low bacterial signals by 16S rRNA gene sequencing, in contrast to explant tissue from cystic fibrosis (CF) or donor lungs. These findings suggest that advanced honeycombing may represent not only a physiologic dead-space, but also an area of reduced bacterial load, whereas microbiota may primarily colonize the airways and areas of traction bronchiectasis.[14] The discordance between IPF bacterial burden in BAL versus in distal parenchymal tissue suggests that fibrosis and the resulting honeycombing could be a maladaptive fibrotic response to airway microbiota.[15]
To gain further understanding of the spatial heterogeneity of lung microbiota in IPF, our current study (MiLEs-2) characterized the regional tissue microbiome across the apicobasal axis of UIP as well as the airways in lung explants from patients with IPF. In particular, we sampled up to three different lobes and airway-based samples in IPF explants, as well as similar samples from explants with connective tissue disease-associated interstitial lung disease (CTD-ILD), CF, COPD, and lungs that had been donated but rejected for transplant.
METHODS
Study design
MiLEs-2 is a retrospective, case-control study of explanted lung tissue obtained at the time of lung transplantation or rapid autopsy (less than 6 hours post-mortem) from patients with IPF and diseased controls (CTD-ILD, CF, and COPD). We also obtained control tissue samples from lung donation candidates deemed unsuitable for transplant, via the Center for Organ Recovery and Education (CORE). IPF diagnoses were confirmed according to 2018 clinical practice guidelines, with histopathology results from lung explants (and prior surgical lung biopsies for some cases) reviewed by specialized thoracic pathologists at the University of Pittsburgh Medical Center.[16] We classified patients with IPF as those with an acute exacerbation of IPF (AE-IPF) versus chronic IPF per established criteria.[14, 17] Informed consents for conducting research utilizing the explanted lung specimens were obtained from patients or their designated representatives. The University of Pittsburgh Institutional Review Board and Committee for Oversight of Research and Clinical Training Involving Decedents approved this study.
Sample acquisition and processing
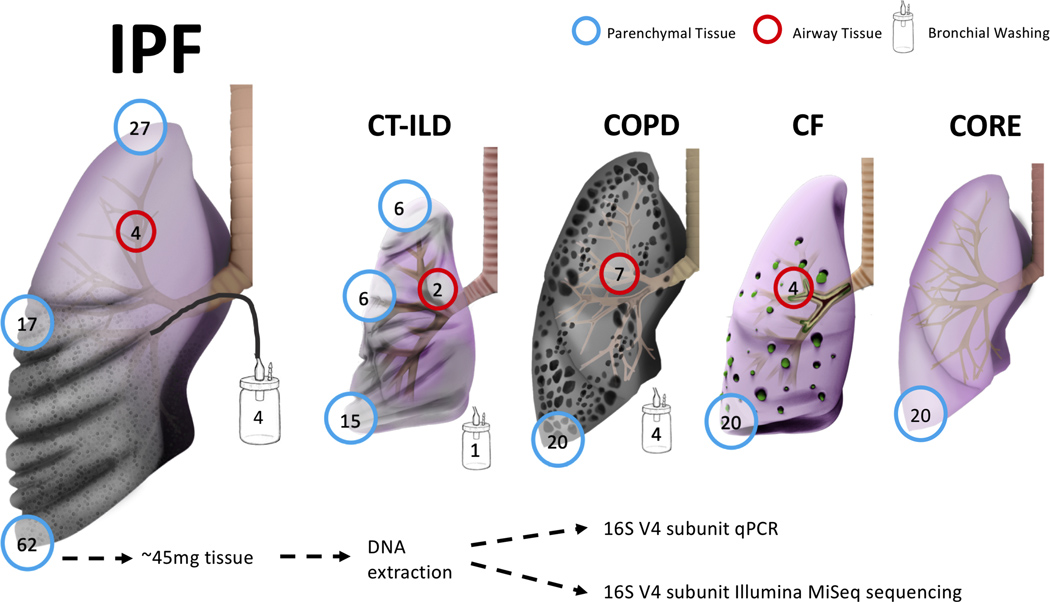
We obtained lung tissue and airway samples in the operating room or autopsy suite per established protocols.[14] We resected a subpleural lower lobe tissue segment, which was further dissected into pieces weighing an average of 45mg under sterile conditions (Figure 1). For a random subset of diseased explants based on logistical feasibility of additional sample collection, we also resected subpleural tissue from the right middle lobe or lingula (for right or left lung explants respectively) and the upper lobe. For a smaller subset of explants, we also collected a bronchial wash specimen (by aspiration of 30mL of phosphate-buffered saline instilled into a bronchial segment using a sterile tube) as well as an airway tissue specimen (from a segmental bronchus) prior to parenchymal tissue sample collection. Samples were frozen in liquid nitrogen and stored at −80°C until processing.
Figure 1.
Explanted lung tissue and bronchial washing samples included in the study, depicted by disease and tissue anatomic location. Numbers in the circles indicate available number of samples from each anatomic site or type. IPF, idiopathic pulmonary fibrosis; CTD-ILD, connective tissue disease-associated interstitial lung disease; COPD, chronic obstructive pulmonary disease; CF, cystic fibrosis; CORE, Center for Organ Recovery and Education.
DNA extraction, 16S rRNA qPCR and pyrosequencing
We extracted genomic DNA and performed polymerase chain reaction (PCR) amplification of the V4 hypervariable region of the 16S rRNA gene.[18] Amplicons of the V4 rRNA bacterial gene subunit were sequenced on the Illumina MiSeq platform and quantified by qPCR.[14] We also quantified the human genomic DNA present in each sample by qPCR of the human Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene. We conducted a series of experiments with lung explant tissue to identify optimal sample type (whole tissue vs. swab) and ruled out the presence of PCR inhibitors (Supplement).
Statistical analysis
From derived 16S sequences, we applied a custom pipeline for Operational Taxonomic Units (OTUs-taxa) classification (Supplement) and performed analyses at genus level. We calculated descriptive statistics of clinical characteristics and performed nonparametric comparisons using the R software (v.3.5.1). Ecological analyses of alpha diversity (Shannon index) and beta diversity (Bray-Curtis index with permutational analysis of variance [permanova] at 1000 permutations) were conducted using the R vegan package and visualized with principal coordinates analyses (PCoA) plots. For comparisons with lower respiratory communities in healthy controls, we utilized 16S sequencing data from a previous study that had analyzed BAL specimens from healthy volunteers.[19] For 30 IPF patients who had survived to lung transplantation and had whole genome sequencing performed in genomic DNA extracted from blood samples, we obtained genotypes for the promoter single nucleotide polymorphism (SNP) rs35705950 of the MUC5B gene. To agnostically examine for distinct clusters of microbial composition (“meta-communities”) in our IPF basilar tissue samples (n=62), we applied unsupervised Dirichlet Multinomial Models (DMM) with Laplace approximations to define the optimal number of clusters in our dataset.[20, 21] We then examined for associations of microbiome variables (bacterial load [log-transformed end-point fluorescence of qPCR assay], alpha diversity, beta-diversity and DMM clusters) with clinical variables (disease classification, diagnosis of AE-IPF, lung transplant vs. death outcome and MUC5B genotypes).
RESULTS
Study population
We analyzed basilar lung explant tissue specimens from 62 IPF patients, 15 CTD-ILD patients, 20 CF patients, 20 COPD patients and 20 CORE lungs. Additional specimens (middle/upper lobe tissue or airway-based samples) were available from a subset of explants (Figure 1). Samples for 32/62 IPF explants (52%) had been previously analyzed by our group.[14] However, we did not use previously generated sequencing data, but performed de-novo experiments with DNA extraction from different tissue specimens available from these 32 IPF subjects to ensure consistency of the methods utilized in our current study. Comparisons of clinical characteristics showed that a higher percentage of IPF patients were male (81%) than within the other three disease groups (Table 1), and that IPF patients had severely decreased forced vital capacity (FVC) (median 41% predicted, interquartile range [IQR] 37.0 %−58.0%) and diffusing capacity for carbon monoxide (DLCO) (median 30% predicted, IQR 22.0%−36.0%), reflecting their end-stage status at the time of lung transplantation (76%) or rapid autopsy (24%). AE-IPF was clinically diagnosed in 35% of patients with IPF and was strongly associated with the finding of diffuse alveolar damage on explant histopathology (odds ratio [OR] = 9.8, 95% confidence interval [CI]: 2.5–44.3, p<0.0001), supporting the accuracy of these patients’ AE-IPF diagnoses, as diffuse alveolar damage with underlying UIP is the anticipated histopathology during an AE-IPF.[17]
Table 1. Clinical data for patients included in the MiLEs-2 cohort.
Data presented as medians (Interquartile range [IQR]) or N (%).
| Variable | IPF | CF | COPD | CTD-ILD |
|---|---|---|---|---|
| N | 62 | 20 | 20 | 15 |
| Age (years), median (IQR) | 64.5 [60.0, 69.0] | 29.0 [23.8, 35.0] | 62.0 [59.0, 65.0] | 63.0 [54.0, 68.5] |
| Male, n (%) | 50 (81%) | 7 (35%) | 9 (45%) | 7 (47%) |
| Ever-smokers, n (%) | 42 (68%) | 0 (0%) | 20 (100%) | 10 (67%) |
| Total pack-years, median (IQR) | 10.2 [0.0, 30.0] | 0.0 [0.0, 0.0] | 57.5 [40.0, 65.5] | 10.0 [0.0, 26.8] |
| FEV1 (L), median (IQR) | 1.5 [1.2, 2.0] | 0.7 [0.6, 0.9] | 0.6 [0.5, 0.7] | 1.4 [1.2, 1.6] |
| FEV1 predicted %, median (IQR) | 53.0 [43.0, 67.0] | 22.0 [18.8, 24.2] | 25.0 [22.0, 30.2] | 52.0 [45.5, 68.0] |
| FVC (L), median (IQR) | 1.8 [1.4, 2.5] | 1.4 [1.1, 1.8] | 2.0 [1.8, 2.3] | 1.7 [1.5, 2.1] |
| FVC predicted %, median (IQR) | 41.0 [37.0, 58.0] | 32.5 [28.0, 41.2] | 58.5 [48.0, 66.0] | 47.0 [39.2, 60.2] |
| DLCO (ml), median (IQR) | 6.4 [4.7, 9.3] | 12.8 [11.8, 14.6] | 6.8 [5.1, 9.9] | 5.1 [4.6, 7.5] |
| DLCO predicted %, median (IQR) | 30.0 [22.0, 36.0] | 58.0 [49.0, 67.0] | 32.5 [26.8, 51.0] | 25.0 [20.5, 28.5] |
| Mean PAP (mmHg), median (IQR) | 24.0 [20.0, 28.0] | 25.0 [21.0, 33.0] | 21.5 [19.0, 27.0] | 24.5 [21.0, 38.8] |
| GERD#, n (%) | 48 (77%) | 19 (95%) | 12 (60%) | 15 (100%) |
| Systemic steroids, n (%) | 15 (24%) | 9 (45%) | 6 (30%) | 9 (60%) |
| Inhaled corticosteroids, n (%) | 11 (18%) | 8 (40%) | 19 (95%) | 5 (33%) |
| Specific therapies: | ||||
| Pirfenidone, n (%) | 10 (16%) | 0 (0%) | 0 (0%) | 1 (7%) |
| Nintedanib, n (%) | 9 (15%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Other immunomodulator, n (%) | 15 (24%) | 0 (0%) | 1 (5.0%) | 9 (60%) |
| BAL + culture or recipient tissue culture*, n (%) | 7 (12%)$ | 20 (100%) | 7 (35%) | 2 (13%)$ |
| Diffuse alveolar damage on explant pathology, n (%) | 21 (36%)$ | 0 (0%) | 0 (0%) | 5 (33%) |
| Antibiotics in past 3 months, n (%) | 25 (40%) | 20 (100%) | 9 (45%) | 9 (60%) |
| Lung transplant, n (%) | 47 (76%) | 20 (100%) | 20 (100%) | 10 (67%) |
GERD definition: based on clinical history, available data from esophagogram or upper endoscopy, or prescription for proton pump inhibitor or histamine receptor 2 blocker.
Systemic steroids refers to patients receiving systemic steroids at the time of the hospital admission resulting in transplant or death.
All patients who underwent lung transplantation had a tissue culture of their resected main stem bronchial tissue, as per the institutional transplant protocol.
Lung transplant, n (%) indicates the number of patients with native lungs sampled at the time of transplant, rather than at autopsy.
Percentage calculated with denominator of all patients with available data (patients with unavailable data for each variable were removed from calculations).
Abbreviations: FEV1: forced expiratory volume in 1 second; FVC: forced vital capacity; DLCO: diffusion capacity of the lungs for carbon monoxide; GERD: gastroesophageal reflux disease; PAP: pulmonary artery pressure.
Airway-based samples have higher bacterial load than corresponding parenchymal tissue
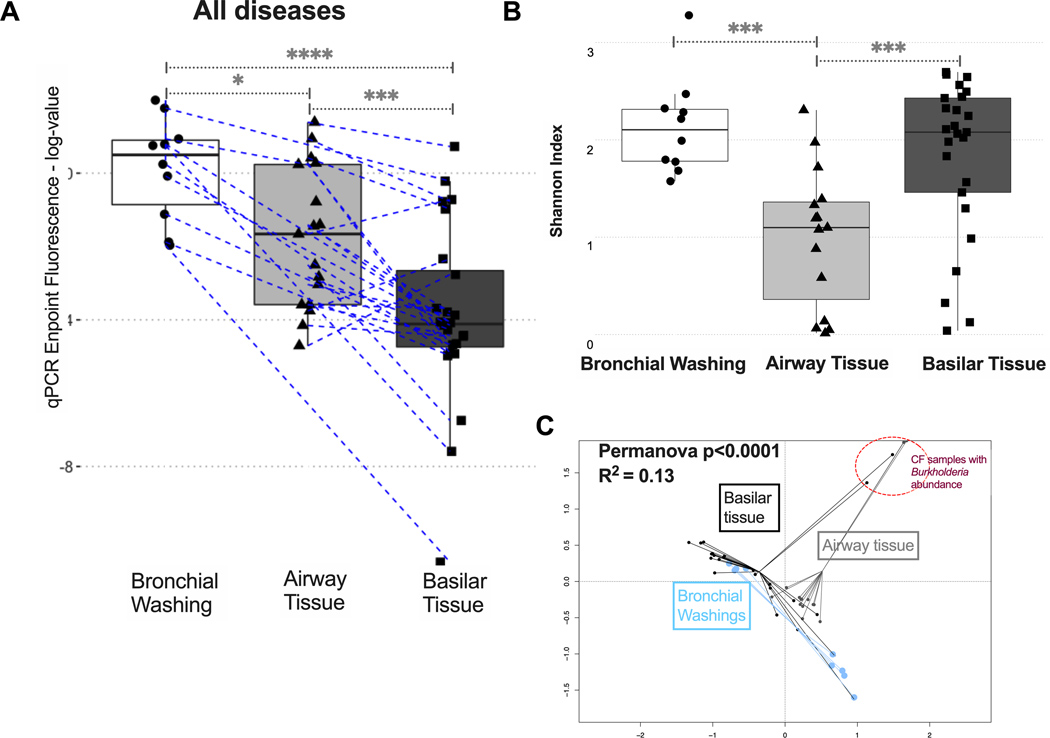
By 16S qPCR across all available samples, bronchial washings had higher bacterial load than corresponding airway tissue, which in turn had higher bacterial load than their counterparts in basilar parenchymal tissue (Figure 2A). This bacterial load gradient from the airways to the distal parenchyma was present within both IPF and COPD lungs (Figure S1). As for the amount of human DNA available in each sample (quantified by GAPDH qPCR), a reverse gradient was observed compared to bacterial DNA load, with tissue samples having much higher human DNA content compared to bronchial washings (p<0.0001) (Figure S2). Sample type was further associated with significant difference in alpha diversity (Shannon index, Figure 2B), with airway tissue samples having the lowest alpha diversity compared to bronchial washing or parenchymal tissue samples, as well as significantly different taxonomic composition by beta-diversity comparisons (Figure 2C). Overall, airway-based samples captured higher microbial biomass with lower human DNA abundance, whereas distal parenchymal tissue (mainly from IPF and COPD explants) had lower microbial biomass and a higher amount of human DNA present.
Figure 2. Airway-based samples have higher bacterial load compared to parenchymal tissue samples.
(A) Bacterial load by qPCR endpoint fluorescence (log-transformed) in bronchial washings, airway tissue and basilar parenchyma tissue samples for all diseased samples available (IPF, CTD-ILD, COPD and CF). Dashed lines connect samples obtained from the same patient. (B) Airway tissue samples had the lowest alpha diversity (Shannon Index) compared to bronchial washings or basilar tissue samples. (C) Significant compositional differences by beta diversity (Bray-Curtis dissimilarity index) comparisons visualized with Principal Coordinates Analysis plot between bronchial washings, airway tissue, and basilar parenchyma tissue samples. Highlighted in a red circle is a cluster of two basilar tissue samples and three airway tissue samples that belonged to five patients with CF with high relative abundance of Burkholderia genera. Pairwise p-values obtained from Wilcoxon tests. *= p<0.05, ***= p<0.001, ****= p<0.0001.
IPF basilar tissue samples have low bacterial load
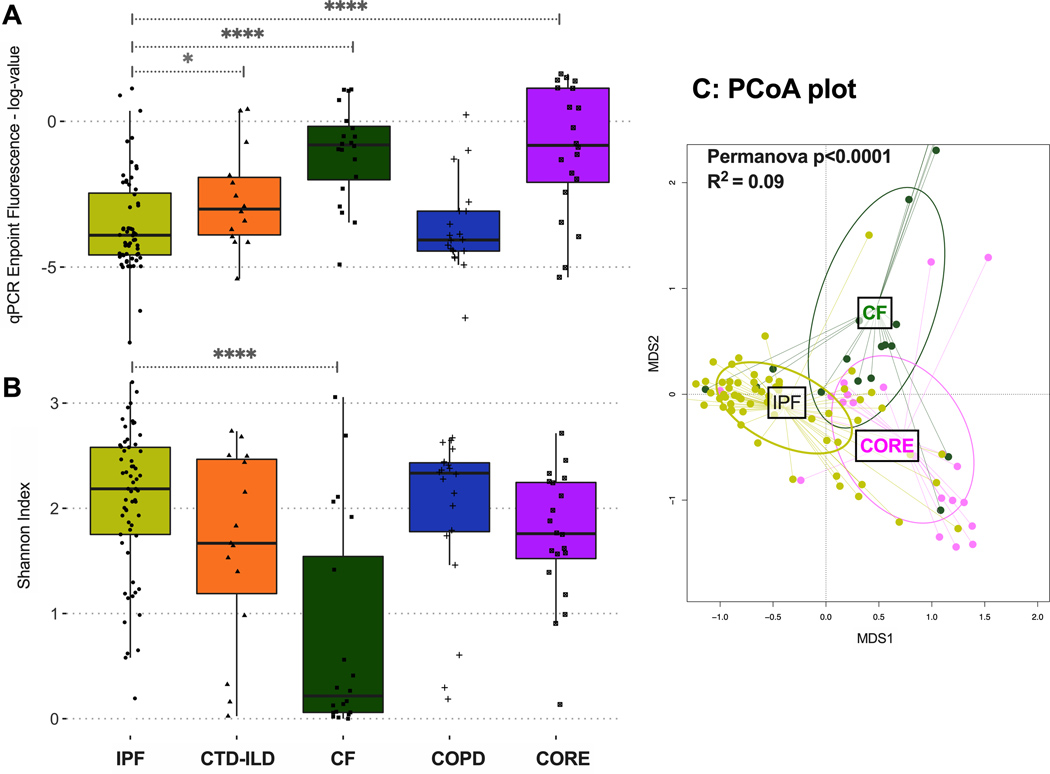
After demonstration of the airway-parenchyma gradient of bacterial load, we then examined for differences in bacterial load and community profiles in basilar tissue parenchymal specimens across the different disease states. By qPCR, IPF and COPD basilar tissue samples had much lower bacterial load (approximately 40-fold less bacterial DNA signal) compared to basilar parenchymal tissue from CF patients or CORE lungs (p<0.0001, Figure 3A). While healthy lung samples are generally expected to have lower biomass, the finding of higher biomass in CORE lungs may reflect differences in the microbial burden between true healthy lungs and those of brain-dead mechanically ventilated organ donors, subjected to the risks of aspiration and undiagnosed secondary pneumonia in CORE patients. Notably, parenchymal samples in IPF and COPD have extensive anatomic destruction and physiologic dead-spaces (due to advanced honeycombing and emphysema, respectively), suggesting the presence of advanced disease microenvironments that are a less hospitable environment for respiratory microbiota. These observations agree with our prior ones in a smaller cohort of end-stage IPF explants.[14] Examination of alpha diversity provided a reciprocal image of bacterial load: IPF samples had much higher Shannon index compared to CF samples (median [IQR]: 2.19[1.75–2.58] vs. 0.22 [0.06–1.54] respectively, p<0.0001, Figure 3B). The pattern of low bacterial burden with high alpha diversity in IPF samples is strongly suggestive of experimental contamination due to low signal/noise ratio.[22] Significant taxonomic composition differences by disease were detected with beta-diversity comparisons (Figure 3C, Figure S3).
Figure 3.
(A) Bacterial burden by qPCR endpoint fluorescence in basilar parenchyma tissue across all diseases and CORE lungs. Significant differences in bacterial burden were identified between IPF and CTD-ILD, IPF and CF, and IPF and CORE basilar tissue. (B) Alpha diversity, as measured by Shannon index, of basilar parenchyma tissue across all diseases and CORE lungs. IPF basilar tissue had significantly higher alpha diversity compared to CF basilar tissue. (C) Beta diversity, as measured by Bray-Curtis dissimilarity in basilar parenchyma tissue of IPF, CF, and CORE lungs, demonstrating significant taxonomic composition differences by disease. P-values are obtained by Wilcoxon tests. *= p<0.05, ***=p<0.001, ****=p<0.0001. IPF, idiopathic pulmonary fibrosis; CTD-ILD, connective tissue disease-associated interstitial lung disease; COPD, chronic obstructive pulmonary disease; CF, cystic fibrosis; CORE, Center for Organ Recovery and Education; MDS1, primary NMDS axis; MDS2, secondary NMDS axis; NMDS, non-metric multidimensional scaling.
To further interpret the low bacterial signal from IPF basilar tissue samples, we examined the specific taxonomic composition of all samples available (Figure S4-9). We first focused on CF samples, which consisted of low diversity communities with a high abundance of typical pathogenic taxa (e.g. Pseudomonas or Burkholderia genera). Among 21 samples from CF patients, 16S sequencing of tissue samples demonstrated dominance by one or two genera that corresponded to the clinically isolated pathogens identified by airway cultures in 80% of samples (Figure S10). This high concordance with clinical isolates established that whenever there is high bacterial load, even basilar tissue samples can reliably capture the bacteria present, despite their smaller biomass compared to corresponding airway-based samples. Importantly, for the majority of IPF (and COPD) parenchymal samples, typical respiratory bacteria (commensal or pathogenic) were not the predominant taxa (Figure S4).
Bacterial load and composition in IPF tissue samples is associated with clinical outcomes.
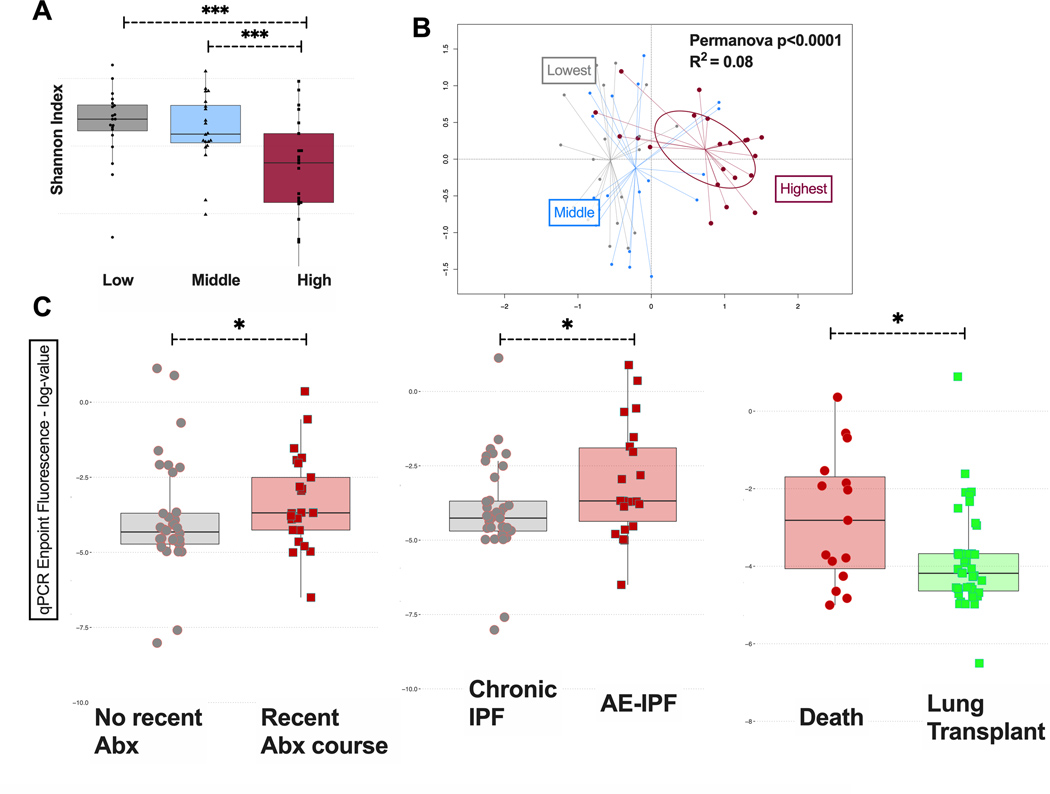
We noted a distinct subgroup of IPF samples with high bacterial load by qPCR, in the range of bacterial loads observed for CF samples. IPF samples within the highest bacterial load tertile had the lowest alpha diversity (Figure 4A) and were taxonomically distinct from samples from the other two tertiles (Figure 4B). Upon retrospective review of associated clinical variables, we found that patients receiving systemic antibiotics within the last three months had higher bacterial load compared to those not receiving antibiotics (p=0.04) (Figure 4C), possibly signaling a recent clinical deterioration that prompted an antibiotic prescription. Patients diagnosed with AE-IPF also had higher bacterial load (p=0.03) as well as those who had not survived to undergo lung transplantation (p=0.02) (Figure 4C).
Figure 4.
(A) Alpha diversity, as measured by Shannon index, of IPF basilar parenchymal tissue stratified by tertiles of bacterial load. IPF samples within the highest bacterial load tertile had the lowest alpha diversity. (B) Beta diversity, as measured by Bray-Curtis dissimilarity, in IPF basilar parenchymal tissue stratified by tertiles of bacterial load. Samples from the highest bacterial load tertile were taxonomically distinct from samples from the other two tertiles (C) Mean bacterial burden by qPCR endpoint fluorescence in basilar parenchyma IPF tissue stratified by clinical outcomes: by whether patients received a recent (within the preceding 90 days) antibiotic prescription, by whether patients had an AE-IPF at the time of lung explantation (transplant or death) and by whether patients died or received a lung transplant. P-values are obtained by Wilcoxon tests. *=p<0.05, ***=p<0.001, ****=p<0.0001. Abx, antibiotics; IPF, idiopathic pulmonary fibrosis; AE-IPF, acute exacerbation of IPF; MDS1, primary NMDS axis; MDS2, secondary NMDS axis; NMDS, non-metric multidimensional scaling.
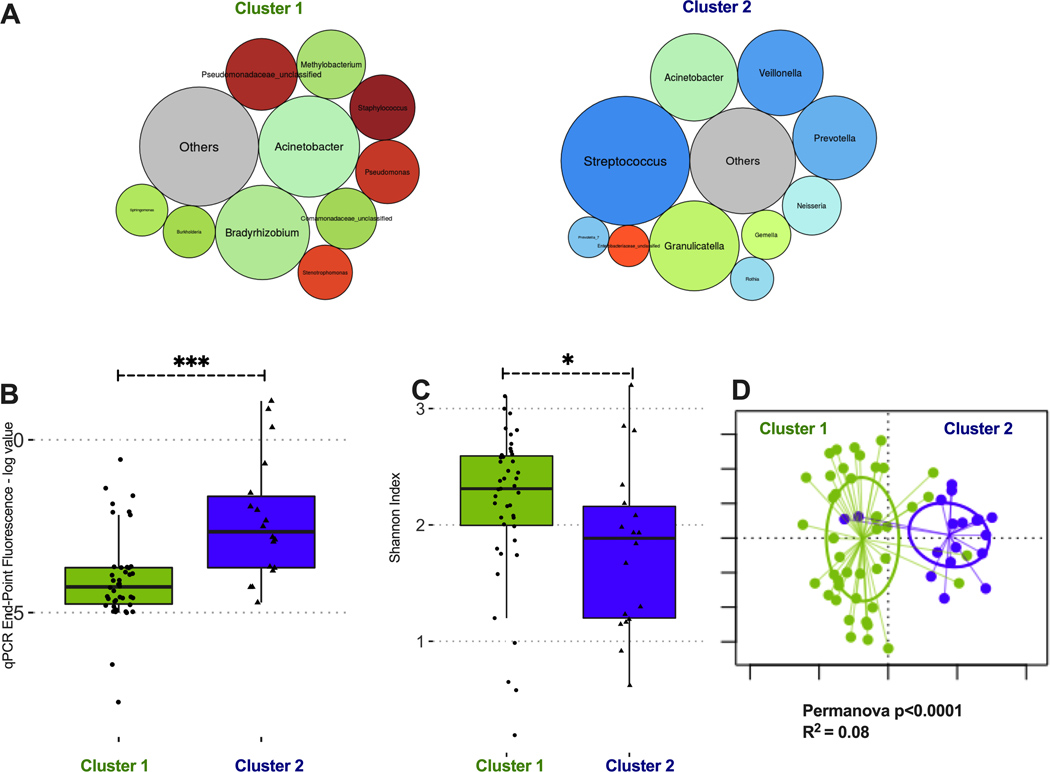
We then agnostically examined for the presence of distinct microbial composition clusters in basilar IPF tissue samples by DMM. Two clusters offered the best model fit. Cluster 1 (n=44, 71% of samples) consisted of communities with low bacterial load, high alpha diversity and abundance of bacteria that are not typical members of the respiratory microbiome (e.g. Bradyrhizobium and Methylobacterium), which likely represent experimental contamination (Figure 5). Cluster 2 (n=18, 29% of samples) demonstrated high abundance of typical members of the microbiome of respiratory tract (Streptococcus, Veillonella or Prevotella genera), and communities with higher bacterial load (p<0.001) and lower alpha diversity (1.89 [1.20–2.16] vs. 2.31 [2.00–2.59], p<0.05) compared to cluster 1 (Figure 5). Membership in cluster 2 was associated with higher odds ratio for diagnosis of AE-IPF (OR=3.3 [0.9–12.2], p=0.04) and recent antibiotic prescription (OR=6.6 [1.8–29.5], p=0.002) and lower odds ratio for survival to lung transplantation (OR=0.16 [0.04–0.66], p=0.007). Cluster 2 membership was not significantly associated with age, last available pulmonary function test results or treatment with antifibrotic therapies. We did not find any association between bacterial load or DMM clusters by MUC5B genotypes in the subset of IPF patients who underwent lung transplantation; however, the limited number of samples with genotyping (n=30) precluded a definitive assessment of microbiome profile differences by MUC5B genotypes.
Figure 5: Dirichlet Multinomial Modeling clustering of IPF basilar tissue sample communities reveals two distinct taxonomic clusters.
(A) Summary relative abundance for top 10 genera in each cluster, visualized as bubble plot (diameter of each circle corresponds to relative abundance of each taxon across all samples in the cluster). Cluster 1 (n=44, 71% of samples) had high abundance for several genera that are not typical members of the respiratory microbiome and may represent procedural contamination (e.g. Bradyrhizobium, Methylobacterium, Comamonadaceae). Cluster 2 (n=18, 29% of samples) had high abundance of typical members of the respiratory microbiome (Streptococcus, Prevotella and Veillonella genera). Genera beyond the top 10 genera demonstrated in these bubble plots were summarized to their overall relative abundance as a single bubble in grey and annotated “Others”. (B) Cluster 2 had significantly higher bacterial load compared to cluster 1 (log-transformed end-point fluorescence of 16S rRNA gene quantitative polymerase chain reaction). (C) Cluster 2 had lower alpha diversity (Shannon index) compared to cluster 1. (D) Principal coordinates analysis for visualization of beta-diversity differences (Bray-Curtis dissimilarity) between Cluster 1 and Cluster 2 samples, demonstrating significant taxonomic compositional differences between clusters. P-values are obtained by Wilcoxon tests. *=p<0.05; ***=p-value <0.001.
Bacterial load and diversity do not differ between apical and basal tissue specimens
We examined for differences of bacterial communities in IPF lungs along the typical apicobasal gradient of fibrosis seen in IPF, by comparing basilar, middle/lingula and upper lobe tissue specimens. Although we did not have available histopathological data to confirm differences in the extent of UIP fibrosis across this apicobasal gradient of tissue samples, we confirmed the presence of more advanced basilar fibrosis in 2/4 cases with available histopathology (Figure S11). Overall, analyses of bacterial load by qPCR revealed no differences between apical and basilar tissue (Figure S12). Upon closer inspection of bacterial distribution, the majority of patient samples exhibited consistently low bacterial signals across all three lobes. Additionally, the taxonomic composition of communities was indistinguishable between each lobe (Permanova p-value for lobe non-significant). However, a distinct subgroup of six IPF lungs with high bacterial load (above 75th percentile) was identified not only in the basilar but also in the middle/lingula and upper lobe samples (Figure S12). In these six cases, similar taxa were highly abundant in samples from all two or three lobes available, further underlying the notion for the presence of a patient-specific rather than a lobe-specific microbiome in IPF (Figure S13). Finally, the higher bacterial load observed for DMM cluster 2 vs. 1 basilar tissue samples was also confirmed for apical and middle lobe/lingular samples, confirming that DMM cluster 2 samples had higher bacterial load and differential taxonomic composition across all lobes (Figure S14).
DISCUSSION
In this study, we identified that basilar lung parenchyma samples have consistently decreased bacterial load compared to airway-based samples. End-stage parenchymal destruction as seen with advanced IPF and COPD results in areas of reduced bacterial load, with the absence of identifiable respiratory bacterial communities in most cases. We also determined that while bacterial communities in IPF do not differ across the apicobasal gradient of fibrosis, a distinct subgroup of IPF patients demonstrated high bacterial load with abundance of typical respiratory microbiota, associated with recent antibiotic exposure as well as worse clinical outcomes including acute exacerbations and death. Amongst the subgroup of IPF patients receiving a lung transplant, no association was detected between MUC5B genotypes and bacterial load, nor with DMM cluster assignment. These novel data generate further hypotheses regarding the potential of personalized microbiome-targeted therapies in IPF.
The spatial and temporal heterogeneity of UIP in IPF classically develop along an apicobasal gradient of fibrosis, with end-stage honeycombing remodeling more of the subpleural lower lobes compared to the upper lobes.[16] Recent studies examining whether BAL-sampled microbiota vary among IPF patients with different radiographic features (e.g. honeycombing or traction bronchiectasis) did not reveal any significant associations.[9, 23] As our findings demonstrate that bacterial load is higher in airway-based samples compared to lung tissue, prior BAL studies cannot inform the bacterial burden within IPF parenchymal tissue with and without honeycombing, rather only its presence within the airways.[23] Additionally, the absence of radiographic honeycombing does not equate the absence of histopathologic honeycombing, as honeycomb cysts <1mm in diameter are generally not detected on high-resolution CT.[24] Similar to the lack of associations with radiographic features, we did not identify significant differences in the microbial communities of apical vs. basilar parenchymal samples. We could not confirm histopathological differences in the extent of UIP between apical and basilar samples due to limited available data. In a small subset of four cases with matched histopathology, two of them demonstrated the classic apicobasal difference. Although we cannot infer the presence of such histopathological apicobasal gradient of fibrosis in our cohort, our findings overall provide evidence of a patient-specific instead of an anatomic lobe-specific microbiome in IPF. If local replication within the lower respiratory tract was a predominant source of origin for the lung microbiome, one might expect to see greater diversity across lobes. Our findings may indicate that microaspiration and dispersion of microbiota in the lower airways and parenchyma is the primary shaping force of microbial communities in IPF.
Although most IPF patients had minimal bacterial burden within the basilar tissue, these regions are not necessarily immune to bacterial harm. Microbiota present in other regions of the lung, including the less advanced upper lobe and/or higher bacterial burden airway, may secrete products able to damage both the immediately adjacent and more distant tissue, resulting in disease progression and possibly acute exacerbation. A recent murine study identified that infection by Staphylococcus species releasing a newly identified pro-apoptic peptide named “corisin,” as well as intratracheal instillation of corisin produces a respiratory illness resembling AE-IPF.[25] BAL samples from human patients with AE-IPF also had significantly elevated corisin compared to those with stable IPF, with all IPF BAL levels elevated compared to healthy controls. Similarly, infection of mice with established fibrosis by Streptococcus pneumoniae has been shown to induce an AE-IPF like reaction via the cytotoxin pneumolysin.[26] These or other yet to be discovered bacterial toxins may be a mechanism via which increased bacterial burden in IPF tissue is associated with worse clinical outcomes.
Our study utilized lung explants for excision of tissue specimens and acquisition of bronchial washings rather than obtaining trans-oral bronchoscopic samples as in prior investigations. Explanted tissue not only allowed us to examine the bacterial burden and communities in patients with advanced IPF that were unable to undergo research bronchoscopy, but also obviated the concern for bronchoscopic contamination by upper airway bacteria. Nonetheless, risk of procedural contamination during sample handling in the operating room or morgue from skin/environmental microbiota remained a possibility, despite the standard precautions of surgical sterility. Previous prospective analyses of IPF lung microbiota have focused on BAL within early-stage IPF patients, which may not be reflective of the microbiota present in those with the most advanced disease. Unlike previous studies,[7, 27] we did not identify an association between Streptococcus or Staphylococcus genus abundance and disease progression within our advanced IPF population. Instead, we utilized an unsupervised clustering approach to identify whether distinct meta-communities of lung microbiota existed in the IPF cohort. With agnostic DMM, we identified that 71% of samples had low bacterial burden and high alpha diversity, with several genera not typically associated with the respiratory tract, suggesting their possible origin from experimental contamination rather than true biologic presence. The remaining 29% of samples exhibited higher bacterial loads with typical respiratory microbiota abundance (Streptococcus, Veillonella, Prevotella genera). Notably, this cluster of patients with high loads of respiratory microbiota was associated with worse clinical outcomes, suggesting that high bacterial burden may impact clinical decline not only in early disease shown in previous studies,[7–9] but also in end-stage IPF. While we did not find an association between Streptococcus and pulmonary function measures marking disease severity, as previously reported, the increased abundance of Streptococcus in cluster 2 patients supports the hypothesis that this species may contribute to disease pathogenesis.[27, 28] Such cluster 2-like patients with higher loads of typical respiratory microbiota may benefit from antimicrobial therapy to address their dysbiosis, while cluster 1-like patients with minimal tissue bacteria could have an inverted risk/benefit calculation dictating avoidance of therapy.
Lung bacterial communities resemble those of the oral cavity in healthy individuals, though certain bacteria are significantly more abundant in the lung.[19] As oral samples were not collected from study patients, we were unable to ascertain the correlation between oral and lung microbiota in these patients. Given the high prevalence of gastroesophageal reflux disease (GERD) in IPF and the subsequent probable repeated micro-aspirations, higher correlation of lung and gut microbiome might be expected in IPF.[29, 30] While much of the association between GERD and IPF may be related to smoking, GERD has been implicated in many other respiratory diseases as well including asthma, COPD, bronchiolitis obliterans, and organizing pneumonia.[31–35] GERD was clinically present in 77% of our IPF patients (as defined by clinical history, esophogram and upper endoscopy results, and prescription for proton-pump inhibitor or H2 blocker), though the rate of silent micro-aspiration could be even higher. Indeed, increased micro-aspiration could account for the higher bacterial burden of BAL fluid in IPF compared to healthy controls previously identified. Explanted lung samples may also be more enriched for those with recent or frequent aspiration if it contributes to a more severe phenotype. Further efforts to treat GERD, and more importantly prevent micro-aspiration events in IPF (rather than acid controlled focused therapy alone), may have a vital role in preventing the dysbiosis associated with worse clinical outcomes. However, our analyses by clinically-defined GERD did not reveal significant differences in microbial communities, similar to previous observations.[9]
Our study was limited by its retrospective design and utilization of a convenience dataset rather than a prospective cohort. As the extent of sampling from each patient varied by feasibility, differences in sample procurement may have influenced our findings. Nonetheless, sampling of basilar subpleural tissue was consistent for all lung explants, regardless of diagnosis. Similarly, comparisons of basilar parenchymal tissue vs. airway-based samples was performed only in explants with both sample types available. Thus, the major findings of our study (lower biomass in IPF basilar tissue compared to CF or CORE lungs, and lower biomass in parenchymal vs. airway-based samples) should not have been influenced by variability in sampling. The limited sample size of our study also restricts the conclusions that can be derived, due to the resulting wide confidence intervals of observed associations and the small number of subjects used for clustering. All IPF samples are from patients with end-stage disease receiving care at a tertiary medical center, and thus may not reflect the general IPF population. Noted associations with clinical outcomes, including the decreased likelihood of receiving a lung transplant and increased incidence of AE-IPF in patients with higher bacterial load, are hypothesis-generating only, given their retrospective nature. How to identify such patients while balancing the potential benefits of targeted antimicrobials with the risks of overtreatment and diagnostic procedures in patients with advanced lung disease remains unclear.
Lung tissue remains a challenging biospecimen for microbiome work due to its low biomass relative to the amount of human DNA present, contamination risks, and readouts at levels near the detection limit of assays used. We utilized standard PCR amplification and sequencing of the V4 subunit of the 16S rRNA gene sequencing, which has well-recognized limitations including sequencing errors, generation of chimeric sequences, and species-specific amplification biases.[36] To mitigate the impact of any sequencing errors on OTU classifications, we limited taxonomic composition analyses at the genus level, and thus there may be concealed species-level variation between communities that was not detectable by our methods.[37] Since our methods were exclusively targeted on bacterial DNA, we could not draw any inferences on the viability of detected bacteria, whereas DNA molecules from viruses and fungi were not within the scope of our study. Such limitations of 16S amplicon sequencing could theoretically be overcome with agnostic, shotgun metagenomic sequencing of all nucleic acids in a sample (DNA and/or RNA).[38] However, the large amounts of human DNA/RNA in lung tissue samples can dominate and overwhelm the sequencing output, not allowing for meaningful microbial nucleic acid detection and analysis.[39] Methods for host DNA depletion are becoming available and may allow for metagenomic sequencing in prospectively collected and real-time processed lung tissue samples before freezing for storage .[40, 41]
In summary, our analysis utilizes the distinct capacity of culture-independent sequencing techniques to discern that end-stage IPF lungs have limited biomass bacterial communities, without evidence of spatial heterogeneity across the apicobasal gradient of fibrosis. A subpopulation of patients with higher bacterial load and taxonomic species dominated by typical respiratory pathogens are more likely to die than undergo lung transplantation and may represent a crucial population towards whom microbiome-targeted interventions ought to be considered. The ongoing development of rapid, culture-independent methods for profiling microbiota holds the promise for personalized medicine approaches in IPF.
Supplementary Material
Key Messages.
• What is the key question?
Bronchoalveolar lavage microbiome profiles in early idiopathic pulmonary fibrosis (IPF) have been associated with disease progression, but the regional heterogeneity of resident microbiota in end-stage IPF has not been defined.
• What is the bottom line?
IPF explants demonstrate higher bacterial load in airway compared to parenchymal samples, but no differences between apical or basilar parenchymal samples. A subgroup of patients with higher bacterial load and respiratory pathogen abundance was associated with worse clinical outcomes.
• Why read on?
Patient-specific heterogeneity in the lung microbiome of IPF supports the need for personalized microbiome-targeted interventions in IPF.
Acknowledgements
The authors would like to acknowledge The Center for Organ Recovery & Education (CORE) as well as organ donors and their families for the generous donation of tissue used in this study.
Funding
National Institutes of Health [K23 HL139987 (GDK); U01 HL098962 (AM); U01 HL137159 (PVB); R01 HL127349 (PVB); K24 HL123342 (AM); R01 HL123766 (MR), RO1 HL126990 (DK), T32 HL007563-31 (EV), CFF RDP to the University of Pittsburgh (MM)], Breathe Pennsylvania Lung Health Research Grant (GDK).
Dr. Bryan J. McVerry is a consultant for The VeraMedica Institute, LLC and receives research funding from Bayer Pharmaceuticals, Inc. Dr. Georgios D. Kitsios has received research funding from Karius, Inc.
Footnotes
Competing interests
The other authors have no conflicts of interest to declare.
Data sharing
All de-identified sequencing data have been submitted to Sequence Read Archive (SRA) database, with BioSample accession numbers of SAMN13906474-13906711.
References
- 1.Allen RJ, Porte J, Braybrooke R, Flores C, Fingerlin TE, Oldham JM, et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med. 2017. November; 5(11):869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013. June; 1(4):309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher TM, Wells AU, Laurent GJ. Idiopathic pulmonary fibrosis: multiple causes and multiple mechanisms? Eur Respir J. 2007. November; 30(5):835–839. [DOI] [PubMed] [Google Scholar]
- 4.Idiopathic Pulmonary Fibrosis Clinical Research N, Raghu G, Anstrom KJ, King TE Jr., Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012. May 24; 366(21):1968–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreuter M, Wuyts W, Renzoni E, Koschel D, Maher TM, Kolb M, et al. Antacid therapy and disease outcomes in idiopathic pulmonary fibrosis: a pooled analysis. Lancet Respir Med. 2016. May; 4(5):381–389. [DOI] [PubMed] [Google Scholar]
- 6.Shulgina L, Cahn AP, Chilvers ER, Parfrey H, Clark AB, Wilson EC, et al. Treating idiopathic pulmonary fibrosis with the addition of co-trimoxazole: a randomised controlled trial. Thorax. 2013. February; 68(2):155–162. [DOI] [PubMed] [Google Scholar]
- 7.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014. October 15; 190(8):906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Dwyer DN, Ashley SL, Gurczynski SJ, Xia M, Wilke C, Falkowski NR, et al. Lung Microbiota Contribute to Pulmonary Inflammation and Disease Progression in Pulmonary Fibrosis. Am J Respir Crit Care Med. 2019. May 1; 199(9):1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Invernizzi R, Barnett J, Rawal B, Nair A, Ghai P, Kingston S, et al. Bacterial burden in the lower airways predicts disease progression in idiopathic pulmonary fibrosis and is independent of radiological disease extent. Eur Respir J. 2020. January 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Ma SF, Espindola MS, Vij R, Oldham JM, Huffnagle GB, et al. Microbes Are Associated with Host Innate Immune Response in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2017. July 15; 196(2):208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molyneaux PL, Willis-Owen SAG, Cox MJ, James P, Cowman S, Loebinger M, et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2017. June 15; 195(12):1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond M, Clark AB, Cahn AP, Chilvers ER, Fraser WD, Livermore DM, et al. The Efficacy and Mechanism Evaluation of Treating Idiopathic Pulmonary fibrosis with the Addition of Co-trimoxazole (EME-TIPAC): study protocol for a randomised controlled trial. Trials. 2018. February 5; 19(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brownell R, Kaminski N, Woodruff PG, Bradford WZ, Richeldi L, Martinez FJ, et al. Precision Medicine: The New Frontier in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2016. June 1; 193(11):1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitsios GD, Rojas M, Kass DJ, Fitch A, Sembrat JC, Qin S, et al. Microbiome in lung explants of idiopathic pulmonary fibrosis: a case-control study in patients with end-stage fibrosis. Thorax. 2018. May; 73(5):481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins G. A big beautiful wall against infection. Thorax. 2018. May; 73(5):485. [DOI] [PubMed] [Google Scholar]
- 16.Raghu G, Remy-Jardin M, Myers JL, Richeldi L, Ryerson CJ, Lederer DJ, et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med. 2018. September 1; 198(5):e44–e68. [DOI] [PubMed] [Google Scholar]
- 17.Collard HR, Moore BB, Flaherty KR, Brown KK, Kaner RJ, King TE, Jr., et al. Acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2007. October 1; 176(7):636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012. August; 6(8):1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013. May 15; 187(10):1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012; 7(2):e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitsios GD, Yang H, Yang L, Qin S, Fitch A, Wang XH, et al. Respiratory Tract Dysbiosis is Associated With Worse Outcomes in Mechanically-Ventilated Patients. Am J Respir Crit Care Med. 2020. July 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014. November 12; 12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson RP, Huffnagle GB, Flaherty KR, White ES, Martinez FJ, Erb-Downward JR, et al. Radiographic Honeycombing and Altered Lung Microbiota in Patients with Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2019. December 15; 200(12):1544–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akira M. Radiographic Differentiation of Advanced Fibrocystic Lung Diseases. Ann Am Thorac Soc. 2017. March; 14(3):432–440. [DOI] [PubMed] [Google Scholar]
- 25.D’Alessandro-Gabazza CN, Kobayashi T, Yasuma T, Toda M, Kim H, Fujimoto H, et al. A Staphylococcus pro-apoptotic peptide induces acute exacerbation of pulmonary fibrosis. Nat Commun. 2020. March 24; 11(1):1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knippenberg S, Ueberberg B, Maus R, Bohling J, Ding N, Tort Tarres M, et al. Streptococcus pneumoniae triggers progression of pulmonary fibrosis through pneumolysin. Thorax. 2015. July; 70(7):636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. 2014. July; 2(7):548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi Y, Saito A, Chiba H, Kuronuma K, Ikeda K, Kobayashi T, et al. Impaired diversity of the lung microbiome predicts progression of idiopathic pulmonary fibrosis. Respir Res. 2018. February 27; 19(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedard Methot D, Leblanc E, Lacasse Y. Meta-analysis of Gastroesophageal Reflux Disease and Idiopathic Pulmonary Fibrosis. Chest. 2019. January; 155(1):33–43. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Bonella F, Li W, Boerner EB, Guo Q, Kong X, et al. Gastroesophageal Reflux Disease in Idiopathic Pulmonary Fibrosis: Uncertainties and Controversies. Respiration. 2018; 96(6):571–587. [DOI] [PubMed] [Google Scholar]
- 31.Harding SM, Guzzo MR, Richter JE. 24-h esophageal pH testing in asthmatics: respiratory symptom correlation with esophageal acid events. Chest. 1999. March; 115(3):654–659. [DOI] [PubMed] [Google Scholar]
- 32.Rascon-Aguilar IE, Pamer M, Wludyka P, Cury J, Coultas D, Lambiase LR, et al. Role of gastroesophageal reflux symptoms in exacerbations of COPD. Chest. 2006. October; 130(4):1096–1101. [DOI] [PubMed] [Google Scholar]
- 33.King BJ, Iyer H, Leidi AA, Carby MR. Gastroesophageal reflux in bronchiolitis obliterans syndrome: a new perspective. J Heart Lung Transplant. 2009. September; 28(9):870–875. [DOI] [PubMed] [Google Scholar]
- 34.Gaillet G, Favelle O, Guilleminault L, de Muret A, Lemarie E, Lecomte T, et al. Gastroesophageal reflux disease is a risk factor for severity of organizing pneumonia. Respiration. 2015; 89(2):119–126. [DOI] [PubMed] [Google Scholar]
- 35.Sedgewick AJ, Buschur K, Shi I, Ramsey JD, Raghu VK, Manatakis DV, et al. Mixed graphical models for integrative causal analysis with application to chronic lung disease diagnosis and prognosis. Bioinformatics. 2019. April 1; 35(7):1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007. May; 69(2):330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, et al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013. December 1; 4(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017. September 12; 35(9):833–844. [DOI] [PubMed] [Google Scholar]
- 39.Cookson W, Cox MJ, Moffatt MF. New opportunities for managing acute and chronic lung infections. Nat Rev Microbiol. 2018. February; 16(2):111–120. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Haidar G, Zia H, Nettles R, Qin S, Wang X, et al. Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir Res. 2019. November 27; 20(1):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charalampous T, Kay GL, Richardson H, Aydin A, Baldan R, Jeanes C, et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019. July; 37(7):783–792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.