Abstract
Background
Neonatal sepsis is one of the major public health problems globally, particularly, in developing countries. Klebsiella, Staphylococcus aureus, Coagulase-negative Staphylococcus, and Escherichia coli are the common pathogens for neonatal sepsis in developing countries. However, the pooled estimate of common pathogens causing neonatal sepsis in developing countries is still unknown. Therefore, this study is aimed at computing the pooled proportion of the leading cause of pathogens for neonatal sepsis in developing countries.
Methods
We strictly followed the Preferred Reporting Items for Systemic Reviews and Meta-analysis guidelines to report this systematic review and meta-analysis. PubMed, Cochrane Library, Web of Science, CINAHL, Science Direct, and other search engines such as Google Scholar, Africa Journals Online, and Hinari were used to obtain studies related to the leading cause of pathogens for neonatal sepsis in developing countries. The search was done from October 1 to December 30, 2018, by considering both published and gray literature. Studies were evaluated based on the PRISMA guideline checklist by using their titles, abstracts, and full texts. Studies were extracted using Microsoft Excel spreadsheets, and STATA software version 14 was used to analyze data. Heterogeneity between studies was checked based on Cochran's Q-test and the corresponding I2 statistic test.
Results
The pooled prevalence of the leading cause of pathogens of neonatal sepsis in developing countries were Klebsiella (26.36%), Staphylococcus aureus (23.22%), Coagulase-negative Staphylococcus (23.22%), and Escherichia coli (15.30%). Common pathogens were varied across regions; for instance, pooled isolated Coagulase-negative Staphylococcus was 25.98% in Africa, 16.62% in Asia, and 36.71% in Latin America, and Klebsiella was 29.80% in Africa, 23.21% in Asia, and 22.00% in Latin America. Also, Staphylococcus aureus was 27.87% in Africa and 18.28% in Asia, and Escherichia coli was 22.97% in Asia and 9.43% in Africa.
Conclusions
This study highlights that the more prevalent common isolated pathogens in developing countries were Klebsiella, Staphylococcus aureus, Coagulase-negative Staphylococcus, and Escherichia coli, Klebsiella, and Staphylococcus aureus pathogens were predominantly high in Africa as compared to other Asian and Latin American countries. At the same time, Coagulase-negative Staphylococcus was more prevalent in Latin America compared to other regions. Escherichia coli is more dominant in Asia as compared to Africa and Latin America.
1. Background
Despite the lack of consensuses in definitions and variability between regions, neonatal sepsis is defined as a clinical syndrome of bacteremia with systemic signs and symptoms of infection in the first four weeks of life [1–5].
Neonatal sepsis is a major cause of mortality and morbidity in developing countries [6]. An estimated 3 million newborns suffer from sepsis globally every year [7]. A report showed that three out of every ten deaths were due to neonatal sepsis [8]. Globally, 15% of neonatal mortality was related to sepsis in 2016 [9, 10]. From the total mortality, nearly about 1.6 million deaths occur due to neonatal infections worldwide, and 40% of this death was found in developing countries [11]. Neonatal sepsis remains a significant global problem with little progress made despite major efforts [5] especially in developing countries [12]. This causes an annual economic burden ranging from $10 billion to $469 billion in sub-Saharan countries [13].
The common pathogens of early-onset neonatal sepsis in developed countries were Group B Streptococcus (43-58%), Escherichia coli (E. coli) (18-29%), and other gram-negative bacteria (7-8%). Similarly, in late-onset neonatal sepsis, the common pathogens were Coagulase-negative Staphylococcus (39-54%), E. coli (5-13%), Staphylococcus aureus (6-18%), and Klebsiella (4-9%) [14–16]. Hospital-acquired common pathogens of neonatal sepsis in developing countries were Klebsiella (16-28%), Coagulase-negative Staphylococcus (8-28%), Staphylococcus aureus (8-22%), and E. coli (5-16%). Also, community-acquired common pathogens of neonatal sepsis were Staphylococcus aureus, Klebsiella, E. coli, and Group B Streptococcus which accounted for 13-26%, 14-21%, 8-18%, and 2-8%, respectively [17–19].
E. coli is identified as the second leading cause of early-onset neonatal sepsis and accounted for about 24% of early-onset neonatal sepsis episodes and most (81%) infection seen in preterm newborn babies [20]. In very low birth weight babies, E. coli is responsible for 33.4% of the cases of early-onset neonatal sepsis [21, 22]. Similarly, Staphylococcus aureus and CoNS are more frequent causes of late-onset neonatal sepsis particularly in very low birth weight infants. Also, CoNS was commonly associated with neonatal sepsis in preterm infants, which accounts for 60 to 93% of bloodstream infections [23, 24].
International experience showed that the Gram-positive and Gram-negative microorganisms accounted for 44.5% of Staphylococcus aureus, 31.3% for other staphylococci, and 9.3% for E. coli [25]. Similarly, in developing countries, early-onset neonatal sepsis (EONS) is usually caused by Gram-negative pathogens, i.e., E. coli and Klebsiella, while late-onset neonatal sepsis is mainly caused by Gram-positive organisms like CoNS, Staphylococcus aureus, and S. pneumonia, although the percentage of late-onset sepsis caused by Gram-negative organisms are increasing [26–28]. Klebsiella and E. coli, in particular, are responsible for 61% of neonatal infections, and staphylococci are the most common Gram-positive bacteria for neonatal infection [29].
Moreover, Klebsiella, Staphylococcus aureus, E. coli, Group B Streptococcus, S. pneumonia, and Salmonella sp. have a major contribution for community-acquired neonatal sepsis, whereas hospital-acquired neonatal sepsis caused by Klebsiella, Staphylococcus aureus, E. coli, CoNS, Pseudomonas sp., Enterobacter sp., and Candida sp. were the common pathogens [12]. Staphylococcus aureus, E. coli, and Klebsiella are also the major causes of neonatal sepsis in developing countries [18].
Based on the acquisition of infection, neonatal sepsis can be classified as hospital acquired or community acquired [18, 30]. Hospital-acquired neonatal infection is the most common and severe infection among neonates hospitalized in the hospital [31, 32]. Although there is uncertainty on the source of infection, whether “maternally acquired” or “hospital acquired,” any infection associated with birth in a hospital is considered a hospital-acquired neonatal infection [17]. On the other hand, community-acquired neonatal sepsis is defined as “an infection occurring in nonhospitalized infants between the age of 7-90 days with ≥1 positive blood or CSF cultures with a recognized blood pathogen.” A new infection had to be separated by 48 hours from prior hospitalization discharge [30].
Despite the high burden of neonatal sepsis observed worldwide, there is no clear evidence on the rank of common pathogens leading to neonatal sepsis particularly in developing countries [33]. Gaps were identified on the current knowledge of common pathogens causing neonatal sepsis in low-income countries [34]. Current evidence on the leading cause of neonatal sepsis is varied across developing countries [35]; this may be due to the presence of heterogeneous population and healthcare settings [36]. Based on these regional variations, reporting a pooled analysis stratified by region is essential to synthesize recent evidence. Therefore, the ultimate aim of this systematic review and meta-analysis was to generate updated evidence on common pathogens causing neonatal sepsis in developing countries and to compute a single estimated proportion of common pathogens causing neonatal sepsis in developing countries. This may be important to choose appropriate antibiotics as an empirical treatment in low-income settings
2. Methods
2.1. Eligibility Criteria
We restricted our search to studies published in English. Obtained studies that cover pathogens causing neonatal sepsis in developing countries were carefully assessed whether they fulfilled our criteria or not. We only included observational studies that had full text and information on neonatal sepsis caused by common pathogens such as Klebsiella and/or Coagulase-negative Staphylococcus and/or Staphylococcus aureus and/or E. coli and neonatal sepsis diagnosed according to standard laboratory methods, i.e., blood culture, and supported with clinical presentations to diagnose Coagulase-negative Staphylococcus because of false-positive blood cultures due to contamination. The gold standard for diagnosis of neonatal sepsis, however, remains blood culture [37, 38]. In this study, we included studies from developing countries. For this study, we defined developing countries as “countries in the process of change with economic growth that increases in production, per capita consumption, and income.” Studies were excluded if the age of the study population was beyond the neonatal period (28-day-old infants) and had low sample sizes of less than 60 subjects. Also, we excluded studies that have methodological problems and flaws (lack of clear measurement, incomplete diagnostic criteria in choosing, selection bias, and unclear presentation of study population). Studies with a case-control study design were also excluded from the study.
2.2. Information Sources
Electronic databases and search engines were used to gather data about common pathogens of neonatal sepsis. The search was done from October 1 to December 30, 2018, which considered both published and gray literature.
2.3. Search Strategy
We strictly followed the Preferred Reporting Items for Systemic Reviews and Meta-analysis (PRISMA) flow diagram [39] to report this study. International electronic databases such as PubMed, Cochrane Library, Web of Science, CINAHL, Science Direct, and other searching engines such as Google Scholar, Africa Journals Online, and Hinari Access to Research for Health program were used to obtain studies related to the leading cause of pathogens for neonatal sepsis in developing countries. Our search protocol was developed using the following keywords: neonatal, neonatal sepsis, sepsis, Klebsiella, CoNS, Staphylococcus aureus, developing countries, developing, countries, developing nations, less developed nations, Africa, Latin America, and Asia. These terms were predefined to have an inclusive search strategy that involved all fields within records and searching medical literature using medical subject headings in the National Library of Medicine to control the vocabulary that indexed articles from the MEDLINE and/or PubMed database (Sup. File).
2.4. Study Selection and Data Extraction
Articles identified by the search were imported into EndNote version 7 to screen for duplication. Initially, the studies were selected by DAZ using their titles based on predefined inclusion and exclusion criteria. The coauthors (GD, EW, and MB) checked the consistency of the selected articles. The disparities between these reviewers were resolved by the other coauthors (SE and MM) through discussions. In the second phase, DAZ, GD, and EW screened articles using their abstracts. In the third phase, full-text articles were screened by DAZ and GD. The disagreements among reviewers during the selection process were resolved by discussion with MB, SE, and MM. Finally, all reviewed studies that fulfill the inclusion criteria were saved.
Studies were extracted by considering the preferred reporting items for systematic reviews and meta-analyses guidelines [40] and evaluated based on the PRISMA guideline. Duplicate studies were removed on the first screening process. Then, titles were carefully assessed and articles irrelevant to our objective were removed from the study. The authors DAZ, GD, and EW extracted data using Microsoft Excel spreadsheets. The extracted data comprised of authors' names, publication years, sites (institution or community), types of study (cross-sectional, retrospective), and total sample sizes. Discrepancies among reviewers were resolved by discussion and consensus with reviewers (MM, MB, and SE).
2.5. Quality of the Study
The quality of studies was approved using the Newcastle-Ottawa Scale (NOS) [41]. The NOS is designed to evaluate the qualitative evaluation of observational studies. This examines each study by seven items in three groups: selection, comparability, and outcome. Stars were given to each item. Items of good quality received 3 or 4 stars in the selection domain, 1 or 2 stars in the comparability domain, and 2 or 3 stars in the outcome/exposure domain. Items of fair quality received 2 stars in the selection domain, 1 or 2 stars in the comparability domain, and 2 or 3 stars in the outcome/exposure domain. Items of poor quality received 0 or 1 star in the selection domain, 0 stars in the comparability domain, and 0 or 1 stars in the outcome/exposure domain. In general, each item was scored for a maximum of six scores. Publications which scored 0–2, 3, 4, and 5 were classified as “unsatisfactory,” “satisfactory,” “good,” and “very good,” respectively. Finally, studies that had been categorized as low quality were excluded in the study [41].
2.6. Outcome of Interest
The outcome of interest was to determine the leading cause of pathogens (Klebsiella, CoNS, Staphylococcus aureus, and E. coli) for neonatal sepsis in developing countries. The pooled isolated common pathogens were measured as the number of neonates with sepsis caused by each respective pathogen divided by the number of neonates in a study multiplied by 100.
2.7. Statistical Analysis
Microsoft Excel spreadsheet was used to extract data, and STATA software version 14 was used to analyze data. Laird's random effects model was used to estimate the pooled proportion of common isolated pathogens for neonatal sepsis because a high degree of heterogeneity was observed across studies. Metaregression was employed to identify the source of heterogeneity using the year of publication, study design, and setting, but there was no statistically significant variable. Subgroup analysis was done by study setting to minimize the random variations between the point estimates of the primary studies. A funnel plot was applied to identify the presence of publication bias. Also, we employed Egger's test and Begg's statistical test to identify publication bias. Then, the trim and fill analysis was done to approve the presence of publication bias. A sensitivity analysis was performed to investigate how each study affects the estimated pooled prevalence. Statistical tests were significant if the P value was <0.05.
2.8. Publication Bias, Heterogeneity, and Sensitivity Test
Heterogeneity among studies was checked based on Cochran's Q-test and the corresponding I2 statistic test [42]. Continuous and categorical metaregression analyses were done to determine the sources of heterogeneity. Begg's test and Egger's test were also used to evaluate the publication bias [43].
3. Results
3.1. Description of the Studies
In the initial search, a total of 2157 potentially relevant studies were identified by searching international electronic databases, and 1232 studies were removed as a result of irrelevance and duplicates. Then, 925 studies were assessed in depth, and finally, a total of 52 studies were eligible for all common pathogens (25 studies for CoNS, 39 studies for Klebsiella, 13 studies for E. coli, and 27 studies for Staphylococcus aureus) (Figure 1) and 152217 infants were eligible for the final systematic review and meta-analysis. Studies involved in this systematic review and meta-analysis included 21 studies from different countries in developing regions. About 21 studies were from Africa, 4 studies were from Latin America, and 12 studies were from Asian countries. The publication year included in this study was from 2005 to 2018; the individual study with the largest sample size had 34362 infants, and the individual study with the smallest sample size had 68 infants.
Figure 1.
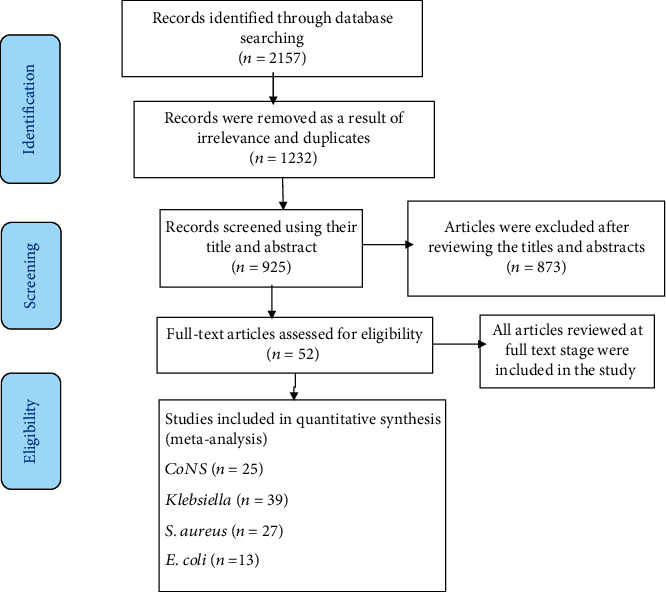
The PRISMA chart to report systematic reviews and meta-analyses of studies.
3.2. Study Characteristics
3.2.1. Coagulase-Negative Staphylococcus
In this study, twenty-five studies were included. All studies were conducted in hospitals. There were eleven studies each from Africa and Asia. The remaining three studies were conducted in Latin America. The largest sample size is 11,790, and the smallest sample size is 68 (Table 1).
Table 1.
Studies characteristics of CoNS in developing countries.
| Authors' names | Years | Countries | Region | Setting | Study design | Sample size |
|---|---|---|---|---|---|---|
| Africa | ||||||
| Arowosegbe et al. [62] | 2017 | Nigeria | Africa | Hospital | Cross-sectional | 180 |
| Kheir et al. [63] | 2014 | Sudan | Africa | Hospital | Cross-sectional | 354 |
| Kabwe et al. [64] | 2016 | Zambia | Africa | Hospital | Cross-sectional | 313 |
| Tsehaynesh et al. [60] | 2017 | Ethiopia | Africa | Hospital | Cross-sectional | 251 |
| Shobowale et al. [65] | 2016 | Nigeria | Africa | Hospital | Cross-sectional | 250 |
| Kumar et al. [66] | 2010 | Kenya | Africa | Hospital | Cross-sectional | 310 |
| El-Din et al. [67] | 2015 | Egypt | Africa | Hospital | Retrospective cohort | 778 |
| Lebea et al. [68] | 2017 | South Africa | Africa | Hospital | Retrospective cohort | 1903 |
| Ballot et al. [69] | 2012 | South Africa | Africa | Hospital | Retrospective cohort | 246 |
| Mhada et al. [70] | 2012 | Tanzania | Africa | Hospital | Cross-sectional | 330 |
| Mkony et al. [71] | 2014 | Tanzania | Africa | Hospital | Cross-sectional | 208 |
| Asia | ||||||
| Thapa et al. [72] | 2014 | Nepal | Asia | Hospital | Cross-sectional | 300 |
| Verma P et al. [73] | 2015 | India | Asia | Hospital | Prospective cohort | 3130 |
| Shah AJ et al. [74] | 2012 | India | Asia | Hospital | Prospective cohort | 190 |
| Ansari et al. [75] | 2015 | Nepal | Asia | Hospital | Cross-sectional | 918 |
| Pokhrel et al. [76] | 2018 | Nepal | Asia | Hospital | Retrospective cohort | 336 |
| Sharma RS et al. [77] | 2016 | India | Asia | Hospital | Retrospective cohort | 98 |
| Sharma M et al. [78] | 2015 | India | Asia | Hospital | Prospective cohort | 200 |
| Sharma CM et al. [79] | 2013 | India | Asia | Hospital | Prospective cohort | 364 |
| Samaga MP [80] | 2016 | India | Asia | Hospital | Prospective cohort | 128 |
| Yusef D et al. [81] | 2017 | Jordan | Asia | Hospital | Retrospective cohort | 68 |
| Sharma P et al. [82] | 2013 | India | Asia | Hospital | Retrospective cohort | 311 |
| Latin America | ||||||
| Dal-Bó et al. [83] | 2015 | Brazil | Latin A | Hospital | Retrospective cohort | 239 |
| Leal et al. [84] | 2012 | Mexico | Latin A | Hospital | Retrospective cohort | 11,790 |
| Bell et al. [85] | 2005 | Jamaica | Latin A | Hospital | Retrospective cohort | 4702 |
3.2.2. Klebsiella
Thirty-nine studies were included, and there were eighteen studies each from Africa and Asia. Three studies were included from Latin America. One study was conducted in a health center, and one study was conducted in a community. The remaining 37 studies were conducted in hospitals. Of the total, 18 studies were conducted using a cross-sectional study design and 21 studies were conducted using a cohort study design. The largest sample had 34362 infants, and the smallest size had 75 infants (Table 2).
Table 2.
Studies characteristics of Klebsiella in developing countries.
| Authors' names | Years | Countries | Setting | Study design | Sample size |
|---|---|---|---|---|---|
| Africa | |||||
| Arowosegbe et al. [62] | 2017 | Nigeria | Hospital | Cross-sectional | 180 |
| Kheir et al. [63] | 2014 | Sudan | Hospital | Cross-sectional | 354 |
| Kabwe et al. [64] | 2016 | Zambia | Hospital | Cross-sectional | 313 |
| Moges et al. [60] | 2017 | Ethiopia | Hospital | Cross-sectional | 251 |
| Peterside et al. [86] | 2015 | Nigeria | Hospital | Retrospective cohort | 233 |
| Shobowale et al. [65] | 2016 | Nigeria | Hospital | Cross-sectional | 250 |
| Kumar et al. [66] | 2010 | Kenya | Hospital | Cross-sectional | 310 |
| Lebea et al. [68] | 2017 | South Africa | Hospital | Retrospective cohort | 1903 |
| Chiabi et al. [87] | 2011 | Cameron | Hospital | Retrospective cohort | 628 |
| Olatunde et al. [88] | 2016 | Nigeria | Hospital | Retrospective cohort | 450 |
| Ballot et al. [69] | 2012 | South Africa | Hospital | Retrospective cohort | 246 |
| Babiker et al. [89] | 2018 | Sudan | Hospital | Cross-sectional | 119 |
| Mhada et al. [70] | 2012 | Tanzania | Hospital | Cross-sectional | 330 |
| Mkony et al. [71] | 2014 | Tanzania | Hospital | Cross-sectional | 208 |
| Kiwanuka et al. [90] | 2013 | Uganda | Hospital | Cross-sectional | 80 |
| John B et al. [91] | 2015 | Uganda | Health center | Cross-sectional | 174 |
| Mugauri H et al. [92] | 2018 | Zimbabwe | Hospital | Prospective cohort | 641 |
| Pius S et al. [93] | 2016 | Nigeria | Hospital | Cross-sectional | 723 |
| Asia | |||||
| Raha et al. [94] | 2014 | Bangladesh | Hospital | Cross-sectional | 720 |
| Hasibuan [95] | 2018 | Indonesia | Hospital | Cross-sectional | 626 |
| Jajoo M et al. [96] | 2015 | India | Hospital | Prospective cohort | 174 |
| Panigrahi et al. [97] | 2017 | India | Community | Prospective cohort | 842 |
| Sundaram et al. [11] | 2009 | India | Hospital | Retrospective cohort | 34362 |
| Agrawal et al. [98] | 2018 | India | Hospital | Cross-sectional | 850 |
| Verma et al. [73] | 2015 | India | Hospital | Prospective cohort | 3130 |
| Shah et al. [74] | 2012 | India | Hospital | Prospective cohort | 190 |
| Dharapur et al. [99] | 2016 | India | Hospital | Cross-sectional | 75 |
| Ansari et al. [75] | 2015 | Nepal | Hospital | Cross-sectional | 918 |
| Pokhrel et al. [76] | 2018 | Nepal | Hospital | Retrospective cohort | 336 |
| Sharma et al. [77] | 2016 | India | Hospital | Retrospective cohort | 98 |
| Sharma M et al. [78] | 2015 | India | Hospital | Retrospective cohort | 200 |
| Sharma CM et al. [79] | 2013 | India | Hospital | Retrospective cohort | 364 |
| Samaga MP [80] | 2016 | India | Hospital | Retrospective cohort | 128 |
| Yusef D et al. [81] | 2017 | Jordan | Hospital | Retrospective cohort | 68 |
| Sharma P et al. [82] | 2013 | India | Hospital | Retrospective cohort | 311 |
| Hasibuan BS [95] | 2018 | Indonesia | Hospital | Cross-sectional | 626 |
| Latin America | |||||
| Boulos et al. [100] | 2017 | Haiti | Hospital | Retrospective cohort | 1292 |
| Leal et al. [84] | 2012 | Mexico | Hospital | Retrospective cohort | 11,790 |
| Bell et al. [85] | 2005 | Jamaica | Hospital | Retrospective cohort | 4702 |
3.2.3. Staphylococcus aureus
A total of twenty-seven studies on the Staphylococcus aureus pathogen were included. Thirteen studies were conducted in Asia, and fourteen studies were conducted in Africa. Two studies were conducted in health centers, and one study was conducted in a community. Fourteen studies were conducted using a cohort study design (Table 3).
Table 3.
Study characteristics of Staphylococcus aureus in developing countries.
| Authors' names | Years | Countries | Region | Setting | Study design | Sample size |
|---|---|---|---|---|---|---|
| Asia | ||||||
| Agrawal A et al. [98] | 2018 | India | Asia | Hospital | Cross-sectional | 850 |
| Hasibuan BS [95] | 2018 | Indonesia | Asia | Hospital | Cross-sectional | 626 |
| Panigrahi P et al. [97] | 2017 | India | Asia | Community | Prospective cohort | 842 |
| Dharapur et al. [99] | 2016 | India | Asia | Hospital | Cross-sectional | 75 |
| Samaga MP [80] | 2016 | India | Asia | Hospital | Prospective cohort | 128 |
| Sharma RS et al. [77] | 2016 | India | Asia | Hospital | Retrospective cohort | 98 |
| Ansari S et al. [75] | 2015 | Nepal | Asia | Hospital | Cross-sectional | 918 |
| Jajoo M et al. [96] | 2015 | India | Asia | Hospital | Prospective cohort | 174 |
| Sharma M et al. [78] | 2015 | India | Asia | Hospital | Prospective cohort | 200 |
| Sharma CM et al. [79] | 2013 | India | Asia | Hospital | Prospective cohort | 364 |
| Sharma P et al. [82] | 2013 | India | Asia | Hospital | Retrospective cohort | 311 |
| Shah AJ [74] | 2012 | India | Asia | Hospital | Retrospective cohort | 190 |
| Sundaram V et al. [11] | 2009 | India | Asia | Hospital | Retrospective cohort | 34362 |
| Africa | ||||||
| Babiker W et al. [89] | 2018 | Sudan | Africa | Hospital | Cross-sectional | 119 |
| Lebea MM et al. [68] | 2017 | South Africa | Africa | Hospital | Retrospective cohort | 1903 |
| Roca A et al. [101] | 2017 | Gambia | Africa | Health Center | Retrospective cohort | 361 |
| Tsehaynesh G et al. [60] | 2017 | Ethiopia | Africa | Hospital | Cross-sectional | 251 |
| Kabwe M et al. [64] | 2016 | Zambia | Africa | Hospital | Cross-sectional | 313 |
| Labi A-K et al. [102] | 2016 | Ghana | Africa | Hospital | Retrospective cohort | 8025 |
| Olatunde OE et al. [88] | 2016 | Nigeria | Africa | Hospital | Prospective cohort | 450 |
| Pius S et al. [93] | 2016 | Nigeria | Africa | Hospital | Cross-sectional | 723 |
| Shobowale OE et al. [65] | 2016 | Nigeria | Africa | Hospital | Cross-sectional | 250 |
| John B et al. [91] | 2015 | Uganda | Africa | Health center | Cross-sectional | 174 |
| Peterside O et al. [86] | 2015 | Nigeria | Africa | Hospital | Retrospective cohort | 233 |
| Kheir et al. [63] | 2014 | Sudan | Africa | Hospital | Cross-sectional | 354 |
| Mkony et al. [71] | 2014 | Tanzania | Africa | Hospital | Cross-sectional | 208 |
| Kiwanuka et al. [90] | 2013 | Uganda | Africa | Hospital | Cross-sectional | 80 |
3.2.4. Escherichia coli
Thirteen studies were included for the E. coli pathogen. All studies were conducted in hospitals. Majority (10/13) of the studies were conducted using a cross-sectional study design (Table 4).
Table 4.
Studies characteristics of E. coli in developing countries.
| Authors' names | Years | Countries | Region | Setting | Study design | Sample size |
|---|---|---|---|---|---|---|
| Singh et al. [103] | 2017 | India | Asia | Hospital | Retrospective cohort | 102 |
| Aku FY et al. [104] | 2016 | Ghana | Africa | Hospital | Cross-sectional | 150 |
| El-Din EMS et al. [67] | 2015 | Egypt | Africa | Hospital | Cross-sectional | 140 |
| Iregbu KC et al. [105] | 2013 | Nigeria | Africa | Hospital | Retrospective cohort | 251 |
| Akindolire AE et al. [106] | 2016 | Nigeria | Africa | Hospital | Cross-sectional | 202 |
| Jajoo M et al. [96] | 2015 | India | Asia | Hospital | Cross-sectional | 440 |
| Afrin M et al. [107] | 2016 | Bangladesh | Asia | Hospital | Cross-sectional | 116 |
| Chiabi A et al. [87] | 2011 | Cameroon | Africa | Hospital | Cross-sectional | 208 |
| Ansari S et al. [75] | 2018 | Nepal | Asia | Hospital | Cross-sectional | 82 |
| Kumar et al. [108] | 2017 | India | Asia | Hospital | Cross-sectional | 175 |
| Mhada TV et al. [70] | 2012 | Tanzania | Africa | Hospital | Cross-sectional | 371 |
| Mkony MF et al. [71] | 2014 | Tanzania | Africa | Hospital | Cross-sectional | 208 |
| Sharma RS et al. [77] | 2016 | India | Asia | Hospital | Retrospective cohort | 98 |
3.3. Leading Pathogens of Neonatal Sepsis
Among the bacterial pathogens causing neonatal sepsis, overall pooled isolation of CoNS was accounted 23.22% (95% CI: 12.15-34.29) (Figure 2) and Klebsiella was the most prevalent causative pathogen for neonatal sepsis that accounted 26.36% (95% CI: 21.19-30.50) (Figure 3). Staphylococcus aureus was 23.22% (95% CI: 18.37-28.07) (Figure 4) and E. coli at 15.30% (95% CI: 9.60-21.01) (Figure 5). Pooled isolation of CoNS across continents was varied, i.e., 25.73% in Africa, 15.59% in Asia, and 36.55% in Latin America. Pooled isolation of Klebsiella was 31.15% in Africa, 22.98% in Asia, and 21.81% in Latin America. Pooled isolation of Staphylococcus aureus was 27.63% in Africa and 18.01% in Asia. Pooled isolation of E. coli was 22.97% in Asia and 9.43% in Africa (Figures 6–9).
Figure 2.
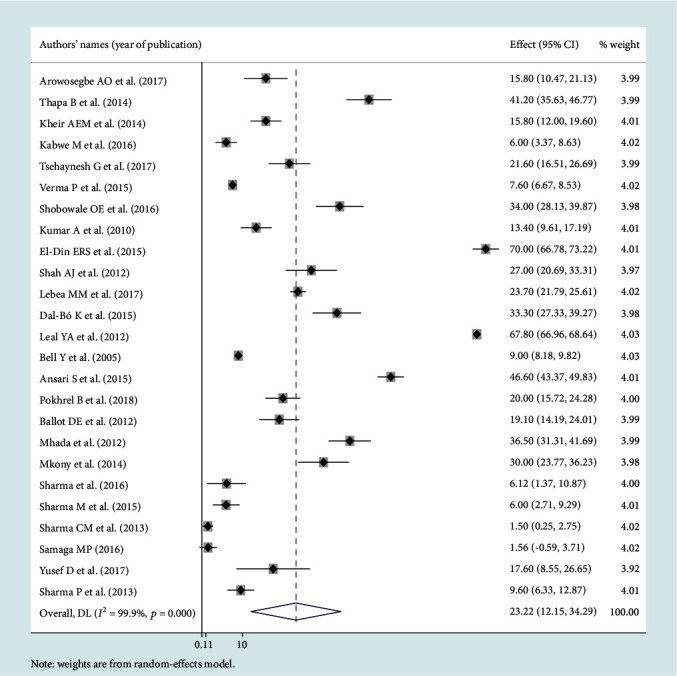
Forest plot for coagulase-negative Staphylococcus pathogen.
Figure 3.
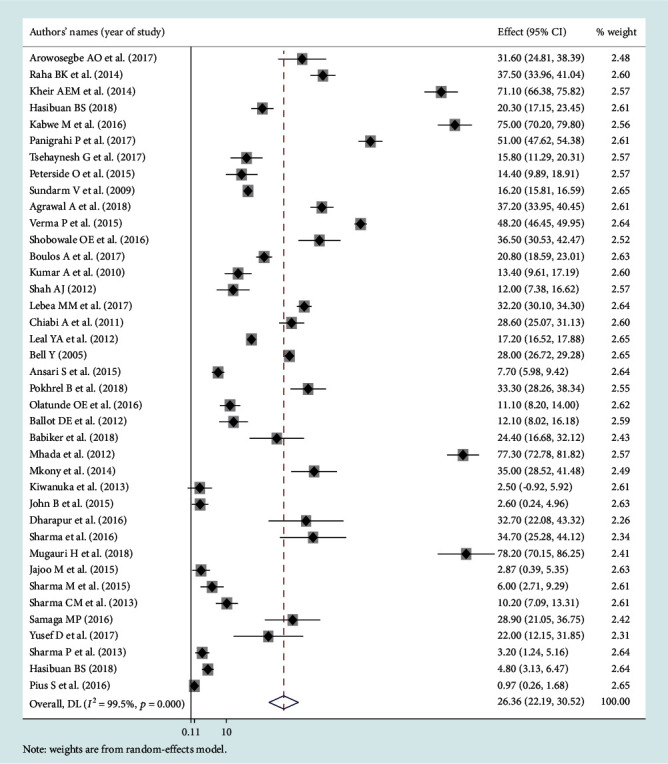
Forest plot for Klebsiella pathogen.
Figure 4.
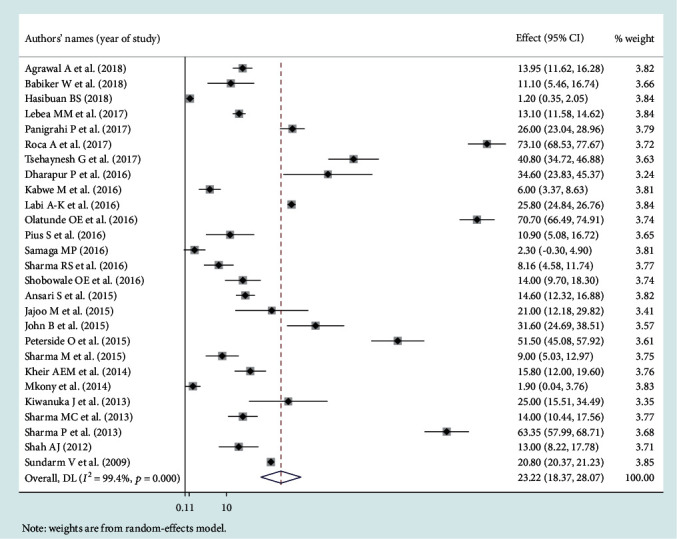
Forest plot for Staphylococcus aureus pathogen.
Figure 5.
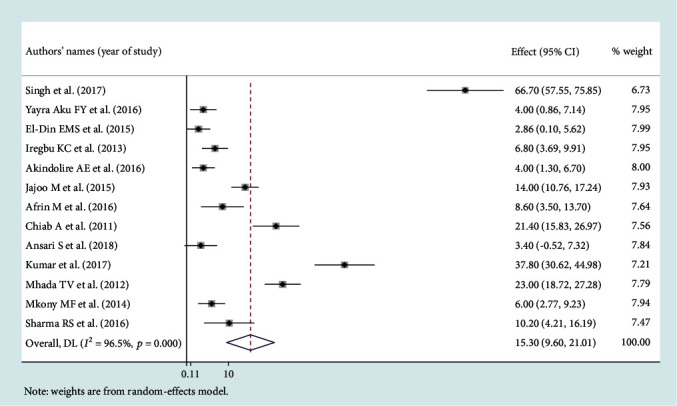
Forest plot for E. coli pathogen.
Figure 6.
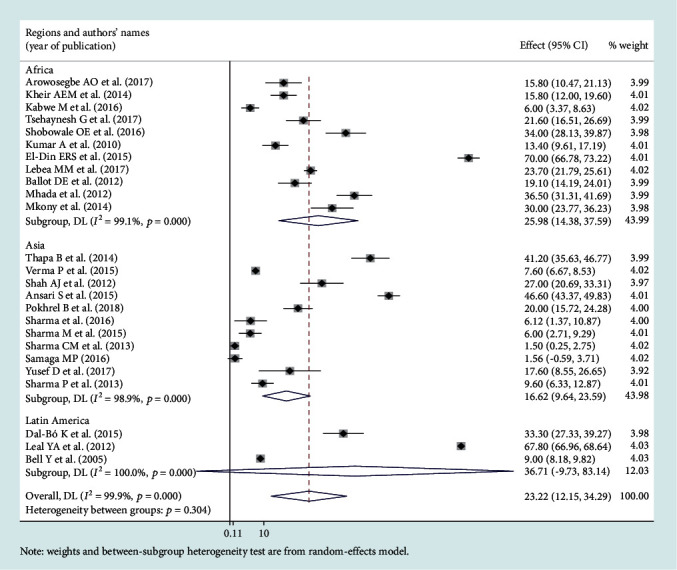
Subgroup analysis for coagulase-negative Staphylococcus by region.
Figure 7.
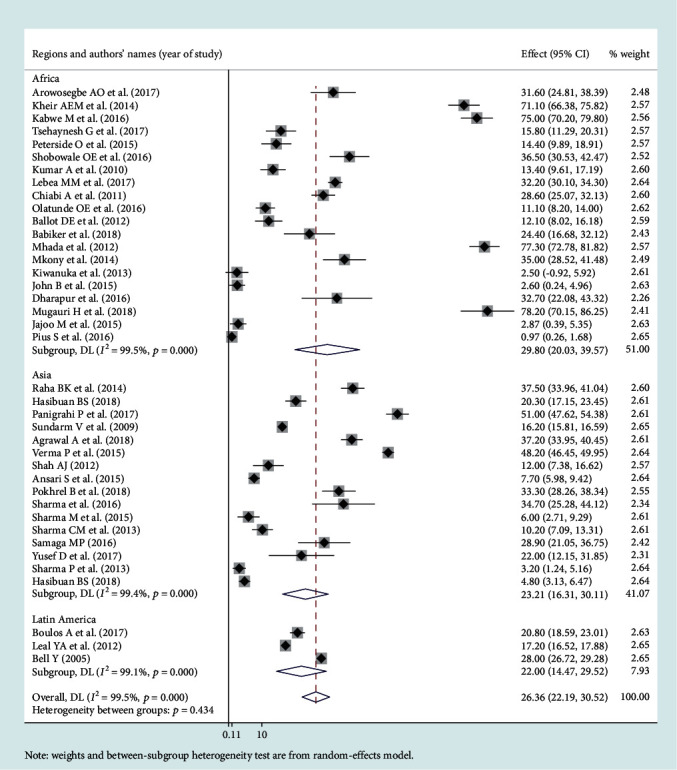
Subgroup analysis for Klebsiella by region.
Figure 8.
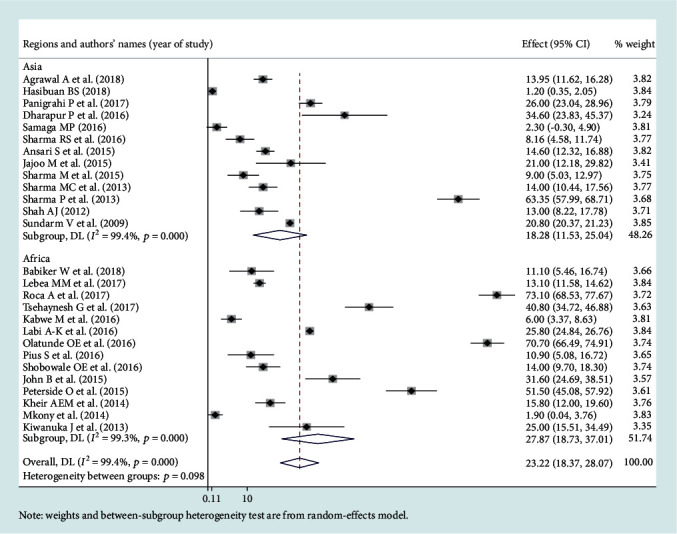
Subgroup analysis for Staphylococcus aureus pathogen by region.
Figure 9.
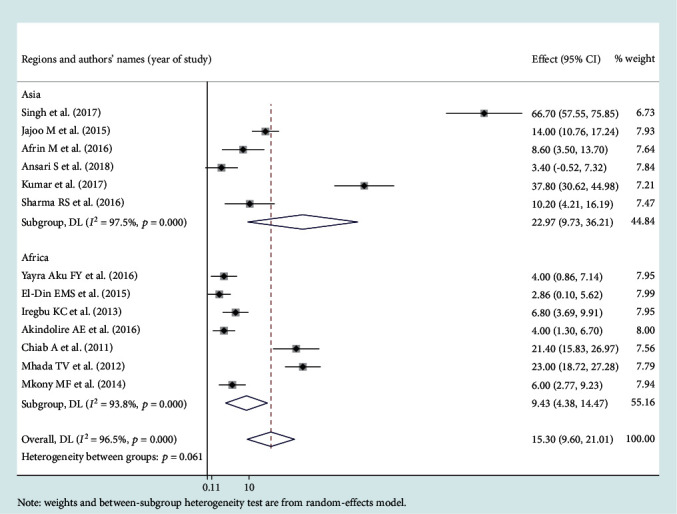
Subgroup analysis for E. coli pathogen by region.
3.4. Level of Heterogeneity
Significant heterogeneity was observed across studies of this systematic review and meta-analysis. The significant level of statistical heterogeneity across studies was assessed using the I2 test, and the presence of heterogeneity was determined through Cochran's Q test. A P < 0.05 was considered statistically significant. The overall I2 was 99.9% and belongs to CoNS, P < 0001. In Africa, Asia, and Latin America, each had overall I2 of 99.1%, 98.9%, and 100%, respectively (Figure 6). In the case of the Klebsiella pathogen, subgroup analysis showed that I2 = 99.5%, 99.4%, and 99.1% in Africa, Asia, and Latin America, respectively (Figure 7). Staphylococcus aureus also showed a significant heterogeneity with an overall I2 of 99. 4%. In subgroup analysis, Africa and Asia had an I2 of 99.3% and 99.4%, respectively (Figure 8). And also, the overall I2 for the E. coli pathogen was 96.5%. In subgroup analysis, Africa and Asia had an I2 of 93.8% and 97.5%, respectively (Figure 9).
3.5. Publication Bias
In the case of CoNS, there was no significant publication bias (P = 0.365), and Egger's test also had no had publication bias (95% CI, -0.7202, 0.877). The funnel plot with pseudo-95% CI using a random effect model was significantly asymmetric. The funnel plots were distributed asymmetrically at which more plots were distributed towards the right side of the midline of the graph (Figure 10). The metatrim test at pseudo-95% CI with a random effects model showed that there was no significant difference from the original pooled prevalence.
Figure 10.
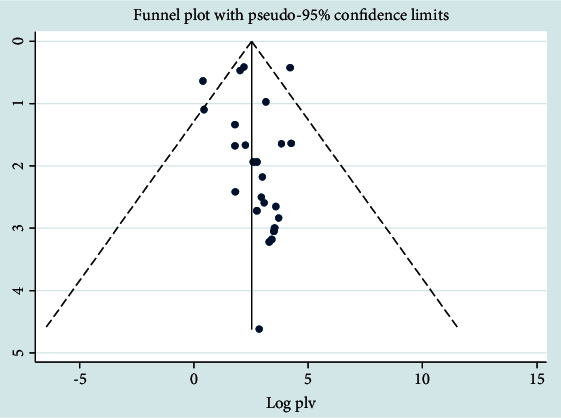
Funnel plot analysis for CoNS pathogen.
In the case of the Klebsiella pathogen, Begg's test and Egger's test showed that there was no significant publication bias across studies (P > 0.05). The number of studies missing from a meta-analysis was estimated using the trim and fill method, and there was a significant asymmetric observation. The funnel and filled funnel plot of the pseudo-95% CI showed that there was a significant asymmetric observation. This showed the presence of publication bias because we observed an asymmetric distribution of plots towards the right side of the midline with a random effects model. The model showed that there was symmetry at the top, but it was missing in the middle and bottom of the graph, and the direction of the effect is towards the right then near the bottom of the plot; we also observed a gap on the left (Figure 11). The metatrim test at pseudo-95% CI with a random effects model showed that there was no significant difference from the original pooled prevalence.
Figure 11.
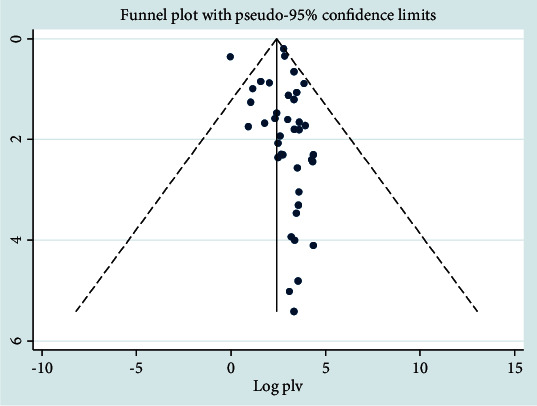
Funnel plot analysis for Klebsiella pathogen.
In the case of Staphylococcus aureus, Begg's test was applied to determine the publication bias, and statistically, there was no publication bias across studies (P > 0.05). Egger's test also showed no publication bias (P > 0.05). The distribution of plots was asymmetrical with the random effects model, and the majority of the plots were distributed towards the right side (Figure 12).
Figure 12.
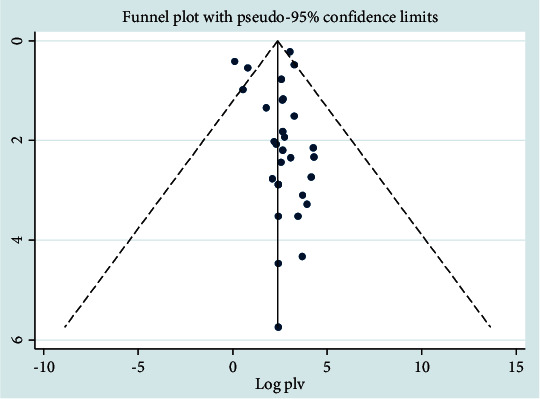
Funnel plot analysis for Staphylococcus aureus pathogen.
In the case of E. coli, the funnel plot showed that there is an asymmetric distribution of plots (Figure 13), and Begg's test showed that there is a significant publication bias. However, Egger's test showed that there was no significant publication bias.
Figure 13.
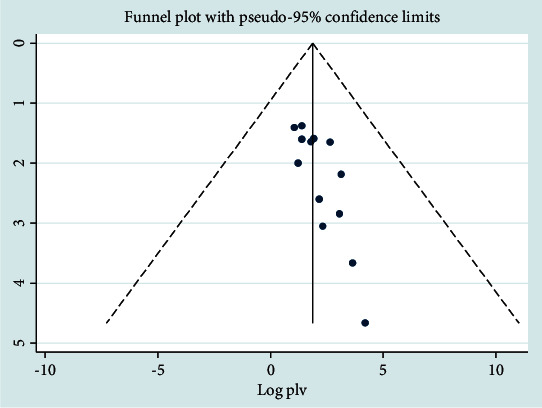
Funnel plot for E. coli pathogen.
3.6. Metaregression Analysis
We conducted a metaregression analysis since there was statistically significant heterogeneity across the studies, with I2 test statistics less than 0.05. This analysis is vital to identify the source of heterogeneity; consequently, a corrective measure during the interpretation of findings was made. In this metaregression analysis, the study that we conducted showed that countries and cross-sectional study designs were found to be a significant source of heterogeneity. However, sample size, publication year, study quality score, and subregion were not found significant for the source of heterogeneity (Table 5).
Table 5.
Metaregression analysis of Coagulase-negative Staphylococcus, Staphylococcus aureus, and E. coli in developing countries.
| Variable | Coefficient | P value |
|---|---|---|
| Coagulase-negative Staphylococcus | ||
| Years of study | -0.17 | 0.826 |
| Sample size | -0.23 | 0.983 |
| Brazil | -35.24 | 0.027 |
| Ethiopia | -70.95 | 0.002 |
| India | -61.44 | 0.000 |
| Jamaica | -59.55 | 0.001 |
| Jordan | -50.94 | 0.005 |
| Kenya | -79.15 | 0.001 |
| Mexico | -0.745 | 0.956 |
| Nepal | -48.54 | 0.005 |
| Nigeria | -67.75 | 0.002 |
| South Africa | -47.81 | 0.001 |
| Sudan | -76.75 | 0.001 |
| Tanzania | 59.23 | 0.004 |
| Zambia | -86.55 | 0.001 |
| Cross-sectional | 24.00 | 0.044 |
| Prospective | 1.45 | 0.813 |
| Staphylococcus aureus | ||
| Years of study | -0.48 | 0.483 |
| Sample size | -0.23 | 0.654 |
| Ethiopia | 49.42 | 0.134 |
| Gambia | 60.00 | 0.046 |
| Ghana | 22.20 | 0.472 |
| India | 18.84 | 0.406 |
| Indonesia | 9.74 | 0.758 |
| Nepal | 23.22 | 0.466 |
| Nigeria | 34.65 | 0.163 |
| Sudan | 21.30 | 0.437 |
| Tanzania | 18.62 | 0.558 |
| Uganda | 34.48 | 0.236 |
| Zambia | 14.62 | 0.644 |
| Cross-sectional | -21.72 | 0.149 |
| Prospective cohort | -9.50 | 0.445 |
| E. coli | ||
| Year of study | -0.6604672 | 0.556 |
| Sample size | 0.022806 | 0.401 |
| Bangladesh | 5.2 | 0.866 |
| Cameroon | 18 | 0.565 |
| Egypt | -0.5400002 | 0.986 |
| Ghana | 0.5999999 | 0.984 |
| India | 28.44278 | 0.273 |
| Nigeria | 1.998955 | 0.940 |
| Tanzania | 11.07911 | 0.679 |
4. Discussion
This review was conducted to estimate the pooled isolated common cause of neonatal sepsis in developing countries. In this study, the first most prevalent bacterial pathogen for neonatal sepsis is Klebsiella pneumonia, and the second most prevalent identified pathogen is Staphylococcus aureus. The third and fourth common pathogens are coagulase-negative staphylococci and Escherichia coli, respectively. Another systematic review and meta-analysis study on causative pathogens of neonatal sepsis in low- and middle-income countries demonstrated that the most prevalent bacterial pathogens for neonatal sepsis were Staphylococcus aureus, E. coli, and Klebsiella [44–46]. However, in this study, common pathogens across regions are varied in its proportion. In Africa, Staphylococcus aureus (27.87%) and Klebsiella (29.80%) were common causes of neonatal sepsis. Coagulase-negative Staphylococcus and E. coli are more common in Latin America and Asia, respectively. Similarly, Staphylococcus aureus and Streptococcuspneumoniae were most prevalent in Africa while Klebsiella was highly prevalent in South-East Asia [44]. Bacterial infection was a leading cause of neonatal mortality in low-income countries, and to date, it is a major cause of morbidity and mortality globally, particularly more common in developing countries as compared to developed countries [35, 47, 48] and little progress was noticed [5].
This systematic review and meta-analysis revealed that Klebsiella pneumonia, Staphylococcus aureus, and CoNS are the common causes of neonatal sepsis in developing countries. Similarly, a large-scale survey across the world showed that neonatal sepsis is more common in developing countries than in developed countries, and the causative pathogens were different with a predominance of Gram-negative bacteria and Staphylococcus aureus [35, 49, 50]. The gram-negative organism Klebsiella [49, 50] and the gram-positive microorganisms Staphylococcus aureus [50, 51] and CoNS [52–54] were the most common pathogens of neonatal sepsis in developing countries.
The present study demonstrated that CoNS is ranked as the third common cause of neonatal sepsis with a pooled prevalence of 23.22%. However, in developed countries, Group B Streptococcus and CoNS were the major organisms implicated in early-onset and late-onset sepsis, respectively [53–56], because the risk factors associated with pathogen-specific sepsis are different based on the pathogen [57].
The present study showed that there are pathogen variations across the study setting. For instance, Klebsiella and Staphylococcus aureus are the most common pathogens of neonatal sepsis in Africa, Coagulase-negative Staphylococcus is the predominant pathogen of neonatal sepsis in Latin America, and E. coli is the most common pathogen in Asia. Likewise, another systematic review showed that Klebsiella and Staphylococcus aureus represented 25% and 18% of neonatal sepsis [18]. Other systematic review findings show that Klebsiella is more prevalent as compared to our study, which accounted for 39% to 70% [19]. On the contrary, a systematic review of community-acquired neonatal sepsis revealed that Staphylococcus aureus represented 14.9% which was lower than that in our study [44] and the reason for this discrepancy may be because the pathogens causing neonatal sepsis in the communities are different from those in the health facilities and the bacterial spectrum of neonatal sepsis varies among healthcare settings and communities [35, 57].
About 70% of the cases of early-onset neonatal sepsis in the developed countries are represented by Streptococcus agalactiae and Escherichia coli [20, 58]. The majority of LOS (70%) in the developed world is due to Gram-positive infections [26, 58], Staphylococcus aureus, Enterococcus spp., and GBS, being most common in very low birth weight and preterm infants [26]. The fact is that in low-income countries, CoNS is responsible for the colonization and development of infection especially in low birth weight babies. Exposures to environmental risks and the timing of exposure, access to healthcare, catheter complications, immune status of the infant, and virulence of the causative agent influence the clinical expression of neonatal sepsis [59–61].
This study found that it is important to choose appropriate antibiotics based on the leading common pathogens especially for clinicians who are working in areas where there is a lack of standard laboratory methods such as blood culture. Also, in this study, it may be important to design appropriate protective measures against known germs associated with neonatal infections. It can play an important role in developing better prevention and treatment policies and programs for policymakers and health planners in low-income settings. Since there is a lack of clarity on variations in the distribution of common pathogens of neonatal sepsis in developing and developed countries [35, 49, 50], it can therefore be used as a starting point for future researchers on this topic.
A limitation of this study is a lack of data to display the community-based neonatal sepsis in African, Asian, and Latin American regions. It has also been indicated by a previous study that data on community-acquired neonatal sepsis are limited [44]. This study is also limited to protocol registration and publication. Future research should be focused on systematic review and meta-analysis of randomized clinical trial studies, and pathogen variations across settings may be the other future research area.
5. Conclusions
This study highlights that Klebsiella is the leading cause of the pathogen, while Staphylococcus aureus and coagulase-negative staphylococcus are the second leading cause of neonatal sepsis. E. coli is ranked as the least common cause of neonatal sepsis. This systematic review shows that these pathogens are highly prevalent in developing countries compared to the developed world. The pooled prevalence of Klebsiella and Staphylococcus aureus pathogens was predominantly high in Africa compared to other Asian and Latin American countries. Coagulase-negative Staphylococcus was also more prevalent in Latin America as compared to other regions. Escherichia coli was also more dominant in Asia compared to Africa and Latin America. Heterogeneity was identified across the regions and within each region. Since most low-income countries have no laboratory access, this finding helps in the selection of antibiotics for the empirical treatment of neonates with a high risk of sepsis. Governments should take preventive and control measures to reduce the burden of infections due to these pathogens.
Abbreviations
- CoNS:
Coagulase-negative Staphylococcus
- E. coli:
Escherichia coli
- EONS:
Early-onset neonatal sepsis
- LOS:
Late-onset sepsis.
Conflicts of Interest
The authors declare that they have no competing interests. Besides, no organization has a competing interest in this work.
Authors' Contributions
DAZ and GD participated in study conception, data extraction, data analysis, and manuscript writing. EWM, MMM, and MBS participated in data analysis and manuscript writing. SE participated in manuscript writing and editing.
Supplementary Materials
This section contains the methods how to search for articles in the PubMed/Medline databases. The search was conducted by searching Medical Literature Using Medical Subject Headings (MeSH) terms. These were neonate OR neonatal OR infant OR newborn AND pathogens OR coagulase-negative staphylococcus OR Staphylococcus aureus OR Klebsiella OR Escherichia coli AND developing countries OR developing country OR countries OR developing OR least developed countries OR least developed country OR less developed countries OR nations OR under developed OR under developed nations OR third world countries OR third world country OR third world nations. This information can be found in the last part of this manuscript appended in the annex.
References
- 1.Sepsis N. Teaching aids: newborn care. Neonatal sepsis, Slide NS-4 Early vs. late.
- 2.Rawat S., Neeraj K., Preeti K., Prashant M. A review on type, etiological factors, definition, clinical features, diagnosis management and prevention of neonatal sepsis. Journal of Scientific and Innovative Research. 2013;2(4):802–813. [Google Scholar]
- 3.Group WYIS. Clinical prediction of serious bacterial infections in young infants in developing countries. The Pediatric Infectious Disease Journal. 1999;18(10):S23–S31. doi: 10.1097/00006454-199910001-00005. [DOI] [PubMed] [Google Scholar]
- 4.Weber M. W., Carlin J. B., Gatchalian S., et al. Predictors of neonatal sepsis in developing countries. The Pediatric Infectious Disease Journal. 2003;22(8):711–717. doi: 10.1097/01.inf.0000078163.80807.88. [DOI] [PubMed] [Google Scholar]
- 5.Wynn J. L. Defining neonatal sepsis. Current Opinion in Pediatrics. 2016;28(2):135–140. doi: 10.1097/MOP.0000000000000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Chavannes-de-Bogis, Switzerland: WHO; 2018. WHO Sepsis Technical Expert Meeting. https://www.who.int. [Google Scholar]
- 7.Fleischmann-Struzek C., Goldfarb D. M., Schlattmann P., Schlapbach L. J., Reinhart K., Kissoon N. The global burden of paediatric and neonatal sepsis: a systematic review. The Lancet Respiratory Medicine. 2018;6(3):223–230. doi: 10.1016/S2213-2600(18)30063-8. [DOI] [PubMed] [Google Scholar]
- 8.Laxminarayan R., Matsoso P., Pant S., et al. Access to effective antimicrobials: a worldwide challenge. The Lancet. 2016;387(10014):168–175. doi: 10.1016/S0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- 9.WHO. Geneva, Switzerland: World Health Organization; 2018. WHO Sepsis Technical Expert Meeting-Meeting Report. [Google Scholar]
- 10.World Health Organization. WHO Recommendations on Intrapartum Care for a Positive Childbirth Experience. World Health Organization; 2018. [PubMed] [Google Scholar]
- 11.Sundaram V., Kumar P., Dutta S., et al. Blood culture confirmed bacterial sepsis in neonates in a North Indian tertiary care center: changes over the last decade. Japanese Journal of Infectious Diseases. 2009;62(1):46–50. [PubMed] [Google Scholar]
- 12.Zea-Vera A., Ochoa T. J. Challenges in the diagnosis and management of neonatal sepsis. Journal of Tropical Pediatrics. 2015;61(1):1–13. doi: 10.1093/tropej/fmu079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranjeva S. L., Warf B. C., Schiff S. J. Economic burden of neonatal sepsis in sub-Saharan Africa. BMJ Global Health. 2018;3(1, article e000347) doi: 10.1136/bmjgh-2017-000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoll B. J., Hansen N., Fanaroff A. A., et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110(2):285–291. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 15.Vergnano S., Menson E., Kennea N., et al. Neonatal infections in England: the NeonIN surveillance network. Archives of Disease in Childhood-Fetal and Neonatal Edition. 2011;96(1):F9–F14. doi: 10.1136/adc.2009.178798. [DOI] [PubMed] [Google Scholar]
- 16.Stoll B. J., Hansen N. I., Sanchez P. J., et al. Early onset neonatal sepsis: the burden of group B streptococcal and E. coli disease continues. Pediatrics. 2011;127(5):817–826. doi: 10.1542/peds.2010-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaidi A. K. M., Huskins W. C., Thaver D., Bhutta Z. A., Abbas Z., Goldmann D. A. Hospital-acquired neonatal infections in developing countries. The Lancet. 2005;365(9465):1175–1188. doi: 10.1016/S0140-6736(05)71881-X. [DOI] [PubMed] [Google Scholar]
- 18.Zaidi A. K., Thaver D., Ali S. A., Khan T. A. Pathogens associated with sepsis in newborns and young infants in developing countries. The Pediatric Infectious Disease Journal. 2009;28(1):S10–S18. doi: 10.1097/INF.0b013e3181958769. [DOI] [PubMed] [Google Scholar]
- 19.Downie L., Armiento R., Subhi R., Kelly J., Clifford V., Duke T. Community-acquired neonatal and infant sepsis in developing countries: efficacy of WHO’s currently recommended antibiotics—systematic review and meta-analysis. Archives of Disease in Childhood. 2013;98(2):146–154. doi: 10.1136/archdischild-2012-302033. [DOI] [PubMed] [Google Scholar]
- 20.Simonsen K. A., Anderson-Berry A. L., Delair S. F., Davies H. D. Early-onset neonatal sepsis. Clinical Microbiology Reviews. 2014;27(1):21–47. doi: 10.1128/CMR.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shane A. L., Stoll B. J. Recent developments and current issues in the epidemiology, diagnosis, and management of bacterial and fungal neonatal sepsis. American Journal of Perinatology. 2013;30(2):131–142. doi: 10.1055/s-0032-1333413. [DOI] [PubMed] [Google Scholar]
- 22.Hornik C. P., Fort P., Clark R. H., et al. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Human Development. 2012;88:S69–S74. doi: 10.1016/S0378-3782(12)70019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.André P., Thébaud B., Guibert M., Audibert F., Lacaze-Masmonteil T., Dehan M. Maternal-fetal staphylococcal infections: a series report. American journal of Perinatology. 2000;17(8):423–428. doi: 10.1055/s-2000-13455. [DOI] [PubMed] [Google Scholar]
- 24.D'Angio C. T., McGowan K. L., Baumgart S., Geme J. S., Harris M. C. Surface colonization with coagulase-negative staphylococci in premature neonates. The Journal of Pediatrics. 1989;114(6):1029–1034. doi: 10.1016/S0022-3476(89)80457-3. [DOI] [PubMed] [Google Scholar]
- 25.Shin Y. J., Ki M., Foxman B. Epidemiology of neonatal sepsis in South Korea. Pediatrics International. 2009;51(2):225–232. doi: 10.1111/j.1442-200X.2008.02685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivanandan S., Soraisham A. S., Swarnam K. Choice and duration of antimicrobial therapy for neonatal sepsis and meningitis. International Journal of Pediatrics. 2011;2011:9. doi: 10.1155/2011/712150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong Y., Speer C. P. Late-onset neonatal sepsis: recent developments. Archives of Disease in Childhood-Fetal and Neonatal Edition. 2015;100(3):F257–F263. doi: 10.1136/archdischild-2014-306213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coetzee M., Mbowane N., de Witt T. Neonatal sepsis: highlighting the principles of diagnosis and management. South African Journal of Child Health. 2017;11(2):99–103. doi: 10.7196/SAJCH.2017.v11i2.1244. [DOI] [Google Scholar]
- 29.Moreno M. T., Vargas S., Poveda R., Sáez-Llorens X. Neonatal sepsis and meningitis in a developing Latin American country. The Pediatric Infectious Disease Journal. 1994;13(6):516–520. doi: 10.1097/00006454-199406000-00010. [DOI] [PubMed] [Google Scholar]
- 30.Bulkowstein S., Ben-Shimol S., Givon-Lavi N., Melamed R., Shany E., Greenberg D. Comparison of early onset sepsis and community-acquired late onset sepsis in infants less than 3 months of age. BMC Pediatrics. 2016;16(1):1–8. doi: 10.1186/s12887-016-0618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emori T. G., Culver D. H., Horan T. C., et al. National nosocomial infections surveillance system (NNIS): description of surveillance methods. American Journal of Infection Control. 1991;19(1):19–35. doi: 10.1016/0196-6553(91)90157-8. [DOI] [PubMed] [Google Scholar]
- 32.Gaynes R. P., Edwards J. R., Jarvis W. R., et al. Nosocomial infections among neonates in high-risk nurseries in the United States. Pediatrics. 1996;98(3):357–361. [PubMed] [Google Scholar]
- 33.Oeser C., Lutsar I., Metsvaht T., Turner M. A., Heath P. T., Sharland M. Clinical trials in neonatal sepsis. Journal of Antimicrobial Chemotherapy. 2013;68(12):2733–2745. doi: 10.1093/jac/dkt297. [DOI] [PubMed] [Google Scholar]
- 34.Seale A. C., Mwaniki M., Newton C. R., Berkley J. A. Maternal and early onset neonatal bacterial sepsis: burden and strategies for prevention in sub-Saharan Africa. The Lancet Infectious Diseases. 2009;9(7):428–438. doi: 10.1016/S1473-3099(09)70172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vergnano S., Sharland M., Kazembe P., Mwansambo C., Heath P. Neonatal sepsis: an international perspective. Archives of Disease in Childhood-Fetal and Neonatal Edition. 2005;90(3):F220–f224. doi: 10.1136/adc.2002.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali M., Salehnejad R., Mansur M. Hospital heterogeneity: what drives the quality of health care. The European Journal of Health Economics. 2018;19(3):385–408. doi: 10.1007/s10198-017-0891-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam P.-Y. I., Bendel C. M. Diagnostics for neonatal sepsis: current approaches and future directions. Pediatric Research. 2017;82(4):574–583. doi: 10.1038/pr.2017.134. [DOI] [PubMed] [Google Scholar]
- 38.Cortese F., Scicchitano P., Gesualdo M., et al. Early and late infections in newborns: where do we stand? A review. Pediatrics & Neonatology. 2016;57(4):265–273. doi: 10.1016/j.pedneo.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Liberati A., Altman D. G., Tetzlaff J., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Medicine. 2009;6(7, article e1000100) doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moher D., Shamseer L., Clarke M., et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Systematic Reviews. 2015;4(1):p. 1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells G., Shea B., O’Connell D., et al. Newcastle-Ottawa quality assessment form for cohort studies. Ottawa Hospital Research Institute. 2014;113:17–18. [Google Scholar]
- 42.Huedo-Medina T. B., Sánchez-Meca J., Marín-Martínez F., Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychological Methods. 2006;11(2):193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 43.Lin L., Chu H. Quantifying publication bias in meta-analysis. Biometrics. 2018;74(3):785–794. doi: 10.1111/biom.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters D., Jawad I., Ahmad A., et al. Aetiology of community-acquired neonatal sepsis in low and middle income countries. Journal of Global Health. 2011;1(2):154–170. [PMC free article] [PubMed] [Google Scholar]
- 45.Akbarian-Rad Z., Riahi S. M., Abdollahi A., et al. Neonatal sepsis in Iran: a systematic review and meta-analysis on national prevalence and causative pathogens. PLoS One. 2020;15(1, article e0227570) doi: 10.1371/journal.pone.0227570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amare D., Mela M., Dessie G. Unfinished agenda of the neonates in developing countries: magnitude of neonatal sepsis: systematic review and meta-analysis. Heliyon. 2019;5(9, article e02519) doi: 10.1016/j.heliyon.2019.e02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seale A. C., Blencowe H., Zaidi A., et al. Neonatal severe bacterial infection impairment estimates in South Asia, sub-Saharan Africa, and Latin America for 2010. Pediatric Research. 2013;74(S1):73–85. doi: 10.1038/pr.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.World Health Organization. World Health Organization; 2018. WHO sepsis technical expert meeting. [Google Scholar]
- 49.Tallur S. S., Kasturi A., Nadgir S. D., Krishna B. Clinico-bacteriological study of neonatal septicemia in Hubli. The Indian Journal of Pediatrics. 2000;67(3):169–174. doi: 10.1007/BF02723654. [DOI] [PubMed] [Google Scholar]
- 50.Karthikeyan G., Premkumar K. Neonatal sepsis: Staphylococcus aureus as the predominant pathogen. The Indian Journal of Pediatrics. 2001;68(8):715–717. doi: 10.1007/BF02752407. [DOI] [PubMed] [Google Scholar]
- 51.Mulholland E. K., Ogunlesi O. O., Adegbola R. A., et al. Etiology of serious infections in young Gambian infants. The Pediatric Infectious Disease Journal. 1999;18(Supplement):S35–S41. doi: 10.1097/00006454-199910001-00007. [DOI] [PubMed] [Google Scholar]
- 52.Pennie R., Malik A. Early onset neonatal septicaemia in a level II nursery. The Medical Journal of Malaysia. 1994;49(1):17–23. [PubMed] [Google Scholar]
- 53.Marchant E. A., Boyce G. K., Sadarangani M., Lavoie P. M. Neonatal sepsis due to coagulase-negative staphylococci. Clinical and Developmental Immunology. 2013;2013:10. doi: 10.1155/2013/586076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J.-y., Chen S.-q., Yan Y.-y., et al. Identification and antimicrobial resistance of pathogens in neonatal septicemia in China--A meta-analysis. International Journal of Infectious Diseases. 2018;71:89–93. doi: 10.1016/j.ijid.2018.04.794. [DOI] [PubMed] [Google Scholar]
- 55.Gheibi S., Fakoor Z., Karamyyar M., et al. Coagulase negative Staphylococcus; the most common cause of neonatal septicemia in Urmia, Iran. Iranian journal of Pediatrics. 2008;18(3):237–243. [Google Scholar]
- 56.Camacho-Gonzalez A., Spearman P. W., Stoll B. J. Neonatal infectious diseases: evaluation of neonatal sepsis. Pediatric Clinics of North America. 2013;60(2):367–389. doi: 10.1016/j.pcl.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo J., Luo Y., Wu Y., Lai W., Mu X. Clinical characteristic and pathogen spectrum of neonatal sepsis in Guangzhou City from June 2011 to June 2017. Medical Science Monitor. 2019;25:2296–2304. doi: 10.12659/MSM.912375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah B. A., Padbury J. F. Neonatal sepsis: an old problem with new insights. Virulence. 2014;5(1):170–178. doi: 10.4161/viru.26906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Oliveira A., Sanches P., Lyra J. C., Bentlin M. R., Rugolo L. M. S. S., Da Cunha M. D. L. R. D. S. Risk factors for infection with coagulase-negative staphylococci in newborns from the neonatal unit of a Brazilian University Hospital. Clinical Medicine Insights: Pediatrics. 2012;6, article CMPed.S7427 doi: 10.4137/cmped.s7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moges F., Eshetie S., Yeshitela B., Abate E. Bacterial etiologic agents causing neonatal sepsis and associated risk factors in Gondar, Northwest Ethiopia. BMC pediatrics. 2017;17(1):p. 137. doi: 10.1186/s12887-017-0892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LShane A. S. P., Stoll B. J. Neonatal sepsis. The Lancet. 2017;390(10104):1770–1780. doi: 10.1016/S0140-6736(17)31002-4. [DOI] [PubMed] [Google Scholar]
- 62.Arowosegbe A. O., Ojo D. A., Dedeke I. O., Shittu O. B., Akingbade O. A. Neonatal sepsis in a Nigerian Tertiary Hospital: clinical features, clinical outcome, aetiology and antibiotic susceptibility pattern. Southern African Journal of Infectious Diseases. 2017;32(4):127–131. doi: 10.4102/sajid.v32i4.37. [DOI] [Google Scholar]
- 63.Kheir A., Khair R. A. Neonatal sepsis; prevalence and outcome in a tertiary neonatal unit in Sudan. Time Journals of Medical Sciences Report and Research. 2014;2:21–25. [Google Scholar]
- 64.Kabwe M., Tembo J., Chilukutu L., et al. Etiology, antibiotic resistance and risk factors for neonatal sepsis in a large referral center in Zambia. The Pediatric Infectious Disease Journal. 2016;35(7):e191–e198. doi: 10.1097/INF.0000000000001154. [DOI] [PubMed] [Google Scholar]
- 65.Shobowale E. O., Ogunsola F., Oduyebo O., Ezeaka V. Aetiology and risk factors for neonatal sepsis at the Lagos University Teaching Hospital, Idi-Araba, Lagos, Nigeria. South African Journal of Child Health. 2016;10(3):147–150. doi: 10.7196/SAJCH.2016.v10i3.965. [DOI] [Google Scholar]
- 66.Kumar R., Musoke R., Macharia W., Revathi G. Validation of c-reactive protein in the early diagnosis of neonatal sepsis in a tertiary care hospital in Kenya. East African Medical Journal. 2010;87(6):255–261. doi: 10.4314/eamj.v87i6.63084. [DOI] [PubMed] [Google Scholar]
- 67.El-Din S., Rabie E. M., El-Sokkary M. M. A., Bassiouny M. R., Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: a study from Egypt. BioMed Research International. 2015;2015:11. doi: 10.1155/2015/509484.509484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lebea M. M., Davies V. Evaluation of culture-proven neonatal sepsis at a tertiary care hospital in Johannesburg, South Africa. South African Journal of Child Health. 2017;11(4):170–173. [Google Scholar]
- 69.Ballot D. E., Nana T., Sriruttan C., Cooper P. A. Bacterial bloodstream infections in neonates in a developing country. ISRN Pediatrics. 2012;2012:6. doi: 10.5402/2012/508512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mhada T. V., Fredrick F., Matee M. I., Massawe A. Neonatal sepsis at Muhimbili National Hospital, Dar es Salaam, Tanzania; aetiology, antimicrobial sensitivity pattern and clinical outcome. BMC Public Health. 2012;12(1):p. 904. doi: 10.1186/1471-2458-12-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mkony M. F., Mizinduko M. M., Massawe A., Matee M. Management of neonatal sepsis at Muhimbili National Hospital in Dar es Salaam: diagnostic accuracy of C-reactive protein and newborn scale of sepsis and antimicrobial resistance pattern of etiological bacteria. BMC Pediatrics. 2014;14(1):p. 293. doi: 10.1186/s12887-014-0293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thapa B., Thapa A., Aryal D. R., et al. Neonatal sepsis as a major cause of morbidity in a tertiary center in Kathmandu. Journal of Nepal Medical Association. 2013;52(192) doi: 10.31729/jnma.2424. [DOI] [PubMed] [Google Scholar]
- 73.Verma P., Berwal P. K., Nagaraj N., Swami S., Jivaji P., Narayan S. Neonatal sepsis: epidemiology, clinical spectrum, recent antimicrobial agents and their antibiotic susceptibility pattern. International Journal of Contemporary Pediatrics. 2017;2(3):176–180. [Google Scholar]
- 74.Shah A. J., Mulla S. A., Revdiwala S. B. Neonatal sepsis: high antibiotic resistance of the bacterial pathogens in a neonatal intensive care unit of a tertiary care hospital. Journal of Clinical Neonatology. 2012;1(2):72–75. doi: 10.4103/2249-4847.96753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ansari S., Nepal H. P., Gautam R., Shrestha S., Neopane P., Chapagain M. L. Neonatal septicemia in Nepal: early-onset versus late-onset. International Journal of Pediatrics. 2015;2015:1–6. doi: 10.1155/2015/379806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pokhrel B., Koirala T., Shah G., Joshi S., Baral P. Bacteriological profile and antibiotic susceptibility of neonatal sepsis in neonatal intensive care unit of a tertiary hospital in Nepal. BMC Pediatrics. 2018;18(1):p. 208. doi: 10.1186/s12887-018-1176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma R. S., Tiwari M., Bansal R. P. Neonatal septicemia: isolates and their sensitivity pattern with emergence of Citrobacter septicemia. International Journal of Research in Medical Sciences. 2016;4(4):1128–1131. [Google Scholar]
- 78.JS S. M., Shree N., Bhagwani D., Kumar M. Neonatal sepsis—a prospective study from a tertiary care hospital of North Delhi. International Journal of Microbiology Research. 2015;7(3):644–647. [Google Scholar]
- 79.Sharma C. M., Agrawal R. P., Sharan H., Kumar B., Sharma D., Bhatia S. S. “Neonatal sepsis”: bacteria & their susceptibility pattern towards antibiotics in neonatal intensive care unit. Journal of Clinical and Diagnostic Research. 2013;7(11):2511–2513. doi: 10.7860/JCDR/2013/6796.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samaga M. P. Prevalence of neonatal septicaemia in a tertiary care hospital in Mandya, Karnataka, India. International Journal of Research in Medical Sciences. 2017;4(7):2812–2816. [Google Scholar]
- 81.Yusef D., Shalakhti T., Awad S., Khasawneh W. Clinical characteristics and epidemiology of sepsis in the neonatal intensive care unit in the era of multi-drug resistant organisms: a retrospective review. Pediatrics & Neonatology. 2018;59(1):35–41. doi: 10.1016/j.pedneo.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 82.Sharma P., Kaur P., Aggarwal A. Staphylococcus aureus—the predominant pathogen in the neonatal ICU of a tertiary care hospital in Amritsar, India. Journal of Clinical and Diagnostic Research. 2013;7(1):66–69. doi: 10.7860/JCDR/2012/4913.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dal-Bó K., Silva R. M., Sakae T. M. Infecção hospitalar em uma unidade de terapia intensiva neonatal do Sul do Brasil. Revista Brasileira de terapia intensiva. 2012;24(4):381–385. doi: 10.1590/S0103-507X2012000400015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leal Y. A., Álvarez-Nemegyei J., Velázquez J. R., et al. Risk factors and prognosis for neonatal sepsis in southeastern Mexico: analysis of a four-year historic cohort follow-up. BMC Pregnancy and Childbirth. 2012;12(1):p. 48. doi: 10.1186/1471-2393-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bell Y., Barton M., Thame M., Nicholson A., Trotman H. Neonatal sepsis in Jamaican neonates. Annals of Tropical Paediatrics. 2005;25(4):293–296. doi: 10.1179/146532805X72449. [DOI] [PubMed] [Google Scholar]
- 86.Peterside O., Pondei K., Akinbami F. O. Bacteriological profile and antibiotic susceptibility pattern of neonatal sepsis at a teaching hospital in Bayelsa State, Nigeria. Tropical Medicine and Health. 2015;43(3):183–190. doi: 10.2149/tmh.2015-03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chiabi A., Djoupomb M., Mah E., et al. The clinical and bacteriogical spectrum of neonatal sepsis in a tertiary hospital in Yaounde, Cameroon. Iranian Journal of Pediatrics. 2011;21(4):441–448. [PMC free article] [PubMed] [Google Scholar]
- 88.Olatunde O., Akinsoji A., Florence D., et al. Neonatal septicaemia in a rural Nigerian hospital: aetiology, presentation and antibiotic sensitivity pattern. British Journal of Medicine and Medical Research. 2016;12(7):1–11. doi: 10.9734/BJMMR/2016/22325. [DOI] [Google Scholar]
- 89.Babiker W., Ahmed A., Babiker T., Ibrahim E., Almugadam B. Prevalence and causes of neonatal sepsis in Soba University Hospital, Sudan. Medical Microbiology Reports. 2018;3:11–13. [Google Scholar]
- 90.Kiwanuka J., Bazira J., Mwanga J., et al. The microbial spectrum of neonatal sepsis in Uganda: recovery of culturable bacteria in mother-infant pairs. PLoS One. 2013;8(8, article e72775) doi: 10.1371/journal.pone.0072775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.John B., David M., Mathias L., Elizabeth N. Risk factors and practices contributing to newborn sepsis in a rural district of Eastern Uganda, August 2013: a cross sectional study. BMC Research Notes. 2015;8(1):p. 339. doi: 10.1186/s13104-015-1308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magwenzi M. T., Gudza-Mugabe M., Mujuru H. A., Dangarembizi-Bwakura M., Robertson V., Aiken A. M. Carriage of antibiotic-resistant Enterobacteriaceae in hospitalised children in tertiary hospitals in Harare, Zimbabwe. Antimicrobial Resistance & Infection Control. 2017;6(1):p. 10. doi: 10.1186/s13756-016-0155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pius S., Bello M., Galadima G., Ibrahim H., Yerima S., Ambe J. Neonatal septicaemia, bacterial isolates and antibiogram sensitivity in Maiduguri North-Eastern Nigeria. Nigerian Postgraduate Medical Journal. 2016;23(3):p. 146. doi: 10.4103/1117-1936.190340. [DOI] [PubMed] [Google Scholar]
- 94.Raha B. K., Baki M. A., Begum T., Nahar N., Jahan N., Begum M. Clinical, bacteriological profile & outcome of neonatal sepsis in a tertiary care hospital. Medicine Today. 2014;26(1):18–21. doi: 10.3329/medtoday.v26i1.21306. [DOI] [Google Scholar]
- 95.Hasibuan B. Comparison of microbial pattern in early and late onset neonatal sepsis in referral center Haji Adam Malik Hospital Medan Indonesia. IOP Conference Series: Earth and Environmental Science; 2018; pp. 1–4. [Google Scholar]
- 96.Jajoo M., Kapoor K., Garg L., Manchanda V., Mittal S. To study the incidence and risk factors of early onset neonatal sepsis in an out born neonatal intensive care unit of India. Journal of Clinical Neonatology. 2015;4(2):p. 91. doi: 10.4103/2249-4847.154106. [DOI] [Google Scholar]
- 97.Panigrahi P., Chandel D. S., Hansen N. I., et al. Neonatal sepsis in rural India: timing, microbiology and antibiotic resistance in a population-based prospective study in the community setting. Journal of Perinatology. 2017;37(8):911–921. doi: 10.1038/jp.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Agrawal A., Awasthi S., Ghanghoriya P., Singh S. Study of current status of bacteriological prevalence and profile in an inborn unit of SNCU in central India. International Journal of Contemporary Pediatrics. 2018;5(3):764–769. doi: 10.18203/2349-3291.ijcp20181436. [DOI] [Google Scholar]
- 99.Dharapur P. M., Kanaki A. R. Antibiotic sensitivity and resistance pattern for neonatal sepsis in Klebsiella and Pseudomonas isolated pathogens in neonatal intensive care unit at tertiary care hospital. International Journal of Basic & Clinical Pharmacology. 2016;5(5):2110–2113. [Google Scholar]
- 100.Boulos A., Rand K., Johnson J. A., Gautier J., Koster M. Neonatal sepsis in Haiti. Journal of Tropical Pediatrics. 2017;63(1):70–73. doi: 10.1093/tropej/fmw077. [DOI] [PubMed] [Google Scholar]
- 101.Roca A., Bojang A., Camara B., et al. Maternal colonization with Staphylococcus aureus and Group B Streptococcus is associated with colonization in newborns. Clinical Microbiology and Infection. 2017;23(12):974–979. doi: 10.1016/j.cmi.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Labi A.-K., Obeng-Nkrumah N., Bjerrum S., Enweronu-Laryea C., Newman M. J. Neonatal bloodstream infections in a Ghanaian Tertiary Hospital: are the current antibiotic recommendations adequate? BMC Infectious Diseases. 2016;16(1):p. 598. doi: 10.1186/s12879-016-1913-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh R., Singh S., Singh S., Sachan D. Prevalence and susceptibility pattern of E. coli in low birth weight neonates of early onset sepsis. International Journal of Pharmaceutical Sciences and Research. 2011;2(12):p. 3122. [Google Scholar]
- 104.Aku F. Y., Akweongo P., Nyarko K., et al. Bacteriological profile and antibiotic susceptibility pattern of common isolates of neonatal sepsis, Ho Municipality, Ghana—2016. Maternal Health, Neonatology and Perinatology. 2018;4(1):p. 2. doi: 10.1186/s40748-017-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iregbu K., Zubair K., Modibbo I., Aigbe A., Sonibare S., Ayoola O. Neonatal infections caused by Escherichia coli at the National Hospital, Abuja: a three-year retrospective study. African Journal of Clinical and Experimental Microbiology. 2013;14(2):95–100. [Google Scholar]
- 106.Akindolire A. E., Tongo O., Dada-Adegbola H., Akinyinka O. Etiology of early onset septicemia among neonates at the University College Hospital, Ibadan, Nigeria. The Journal of Infection in Developing Countries. 2016;10(12):1338–1344. doi: 10.3855/jidc.7830. [DOI] [PubMed] [Google Scholar]
- 107.Afrin M., Siddique M. A., Ahmed A. A., et al. Neonatal septicemia: isolation, identification and antibiotic sensitivity pattern of bacteria in a tertiary hospital in Bangladesh. Faridpur Medical College Journal. 2016;11(2):58–61. doi: 10.3329/fmcj.v11i2.32886. [DOI] [Google Scholar]
- 108.Kumar R., Kumari A., Kumari A., Verma N. Evaluation of perinatal factors in neonatal sepsis at tertiary centre. International Journal of Reproduction, Contraception, Obstetrics and Gynecology. 2017;6(11):4981–4985. doi: 10.18203/2320-1770.ijrcog20175012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This section contains the methods how to search for articles in the PubMed/Medline databases. The search was conducted by searching Medical Literature Using Medical Subject Headings (MeSH) terms. These were neonate OR neonatal OR infant OR newborn AND pathogens OR coagulase-negative staphylococcus OR Staphylococcus aureus OR Klebsiella OR Escherichia coli AND developing countries OR developing country OR countries OR developing OR least developed countries OR least developed country OR less developed countries OR nations OR under developed OR under developed nations OR third world countries OR third world country OR third world nations. This information can be found in the last part of this manuscript appended in the annex.


