Abstract
BACKGROUND
Deciphering the function of the many genes previously classified as uncharacterized open reading frame (ORF) would complete our understanding of a cell’s function and its pathophysiology.
METHODS
Whole-exome sequencing, yeast 2-hybrid and transcriptome analyses, and molecular characterization were performed in this study to uncover the function of the C2orf69 gene.
RESULTS
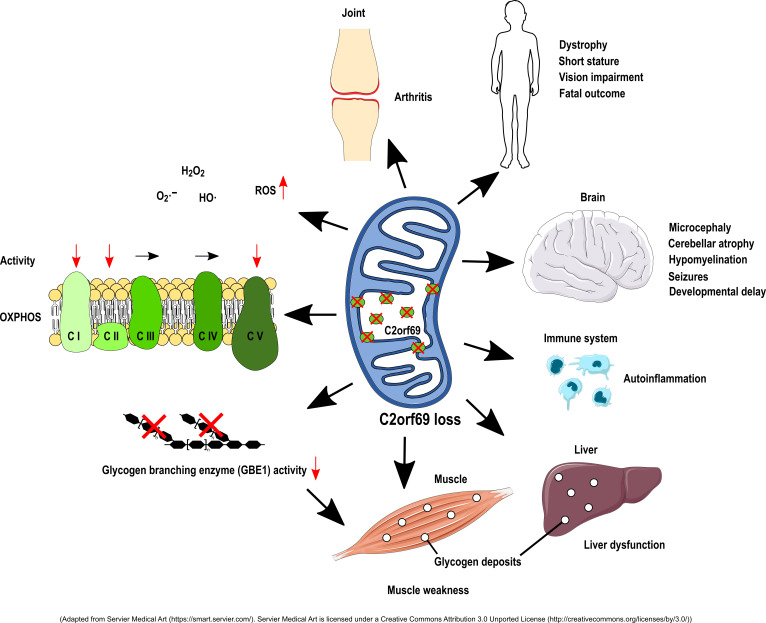
We identified loss-of-function mutations in the uncharacterized C2orf69 gene in 8 individuals with brain abnormalities involving hypomyelination and microcephaly, liver dysfunction, and recurrent autoinflammation. C2orf69 contains an N-terminal signal peptide that is required and sufficient for mitochondrial localization. Consistent with mitochondrial dysfunction, the patients showed signs of respiratory chain defects, and a CRISPR/Cas9-KO cell model of C2orf69 had similar respiratory chain defects. Patient-derived cells revealed alterations in immunological signaling pathways. Deposits of periodic acid–Schiff–positive (PAS-positive) material in tissues from affected individuals, together with decreased glycogen branching enzyme 1 (GBE1) activity, indicated an additional impact of C2orf69 on glycogen metabolism.
CONCLUSIONS
Our study identifies C2orf69 as an important regulator of human mitochondrial function and suggests that this gene has additional influence on other metabolic pathways.
Keywords: Genetics
Keywords: Genetic diseases, Mitochondria, Neurological disorders
Introduction
Inborn errors of metabolism (IEMs) are a genetically heterogeneous group of more than 1000 diseases (1). They result from metabolic defects due to a deficiency of enzymes, membrane transporters, or other functional proteins. IEMs can fall into many categories such as defects in carbohydrate or protein metabolism, fatty acid oxidation, glycogen storage, or dysfunction of peroxisomes, lysosomes, or mitochondria (2). Initially considered rare, IEMs have a significant global prevalence of approximately 1 in 2000 births (3). Given the large number of clinically overlapping disorders and manifold manifestations, IEMs are often difficult to diagnose. Specific signs can be absent, and clinical symptoms range from mild developmental delay to severe metabolic acidosis with periods of remission or sudden death. A large class of IEMs directly affects energy metabolism, especially the function of mitochondria, which are crucial for ATP supply via oxidative phosphorylation (OXPHOS) (4). The mitochondrial proteome consists of approximately 1500 proteins, many of which are still uncharacterized, leaving an incomplete understanding of mitochondrial physiology (5). Here, we identify homozygous loss-of-function mutations in the uncharacterized C2orf69 gene as a cause for a complex multisystem disease and describe an important role for this gene in mitochondrial metabolism.
Results
Mutations in the uncharacterized C2orf69 gene cause a multisystem human disease.
We studied a male index patient (patient I), who was born to consanguineous parents and had severe developmental delay, hepatopathy, and recurrent septic and aseptic inflammation of the joints starting at the age of 2 months, followed by aseptic pericarditis at the age of 9 months. The episodes of inflammation were accompanied by elevated C-reactive protein (CRP) and partially responsive to glucocorticoids. The severe dystrophy made feeding via percutaneous gastrostomy necessary. A brain MRI revealed prominent cerebellar atrophy, a thin corpus callosum, and diffuse hypomyelination (Supplemental Figure 1; Supplemental material available online with this article; https://doi.org/10.1172/JCI143078DS1). The patient had muscle weakness, general dystrophy, short stature, thyroid dysfunction, biventricular cardiac hypertrophy, and osteopenia. An episode of severe epilepsy ultimately made adrenocorticotropic hormone (ACTH) therapy necessary, which also reduced the autoinflammation. The patient died at the age of 2 years and 10 months from multiorgan failure (Tables 1 and 2). Whole-exome sequencing (WES) revealed a potential disease-causing homozygous 1 bp deletion (NM_153689.6:c.298del) in chromosome 2 open reading frame 69 (C2orf69), a hitherto uncharacterized gene (Figure 1A). The variant results in a premature stop codon (p.[Gln100Serfs*18]). Both healthy parents and the 2 unaffected siblings were heterozygous for the C2orf69 deletion. C2orf69 consists of 2 exons and encodes a protein that is highly conserved among vertebrates and Drosophila. According to in silico predictions, the gene product has an N-terminal 24-aa signal peptide (SP) and a protein family domain of unknown function (UPF0565) with phosphorylation, ubiquitination, SUMOylation (small ubiquitin-like modifier), and NEDDylation (neural precursor cell–expressed developmentally downregulated) sites (refs. 6–9 and Figure 1B).
Table 1. Summary of clinical findings for patients I–VIII.
Table 2. Summary of clinical findings for patients I–VIII.
Figure 1. Gene and protein structures of C2orf69.
(A) C2orf69 is composed of 2 exons (boxes E1 and E2). The 3′- and 5′-UTRs are indicated by thick lines and the intron region by an arrowed line. Triangles represent the mutations found in the patients. (B) Domain architecture of the human C2orf69 protein. The protein harbors a 24-aa SP and an uncharacterized UPF0565 protein domain. Circles and squares indicate posttranscriptional modifications and triangles the mutation sites. (C) Pedigrees of families 1–5 show the segregation of the C2orf69 variants. The homozygously mutated allele is shown in black, the WT in white, and heterozygous carriers as a split black-and-white symbol.
Through GeneMatcher (10) and existing collaborations, we identified 7 additional patients from 4 different families who had homozygous loss-of-function mutations in C2orf69 (Figure 1C). Five patients from 3 different families harbored the same homozygous c.298del mutation in C2orf69, whereas 2 patients from another family showed a homozygous 5 bp deletion (c.843_847del), resulting in a premature stop codon (p.[Lys282Glnfs*55]). Neither variant is listed in a public database, and, to our knowledge, no homozygous loss-of-function variants of C2orf69 are known to date. All 8 individuals showed very similar clinical signs, and their brain MRI findings were remarkably consistent (Tables 1 and 2 and Supplemental Figure 1). Different degrees of frontotemporal atrophy, hypomyelination, thin corpus callosum, and the Dandy Walker variant with hypoplasia of the caudal vermis were typical findings on MRI. Patients I, III, V, and VI died at a young age. Interestingly, 4 patients (patients III, IV, V, and VI) were initially suspected to have a glycogen storage disease (GSD), because glycogen branching enzyme 1 (GBE1) activity was absent or reduced in tissues (Supplemental Table 1). In accordance with the possibility of GSD, we observed periodic acid–Schiff–positive (PAS-positive) deposits in myocytes and macrophages of patient III and in the liver of patients III, V, and VI. The autopsy of patient I showed PAS-positive accumulations in hepatocytes, cardiomyocytes, and macrophages (Figure 2, A–F and Supplemental Figure 1D). These deposits were resistant to digestion with amylase (PAS diastase stain) (Figure 2, B, C, E, and F), which led to the assumption of GSD type IV (GSD IV), although no pathogenic variants in the GBE1 gene were present. The muscle biopsy from patient III showed polyglucosan bodies on electron microscopy (EM) (Figure 2, G and H). Disintegration of several mitochondria accompanied by osmiophilic membranous material in the mitochondria was evident in the muscle (Figure 2, I and J).
Figure 2. Histological findings in patients I and III.
Histology for patient I revealed PAS-positive deposits in heart and liver that were still stained positive with PAS diastase stain. (A–C) PAS-stained sections of heart muscle. Original magnification, ×100 (A) and ×400 (B and C). Liver sections at original magnification, ×100 (D) and ×400 (E and F), before and after diastase staining. Arrows in A–F indicate PAS-positive accumulations. (G–J) Electron micrographs of the muscle biopsy from patient III. (G) Muscle fiber containing several polyglucosan bodies. Scale bar: 1 μm. (H) Higher magnification of a polyglucosan body composed of finely granular and filamentous material. Scale bar: 0.4 μm. (I) Degenerating mitochondria undergoing autophagy. Scale bar: 0.5 μm. (J) Disintegrating muscle fiber mitochondria containing osmiophilic membranous material. Scale bar: 0.3 μm.
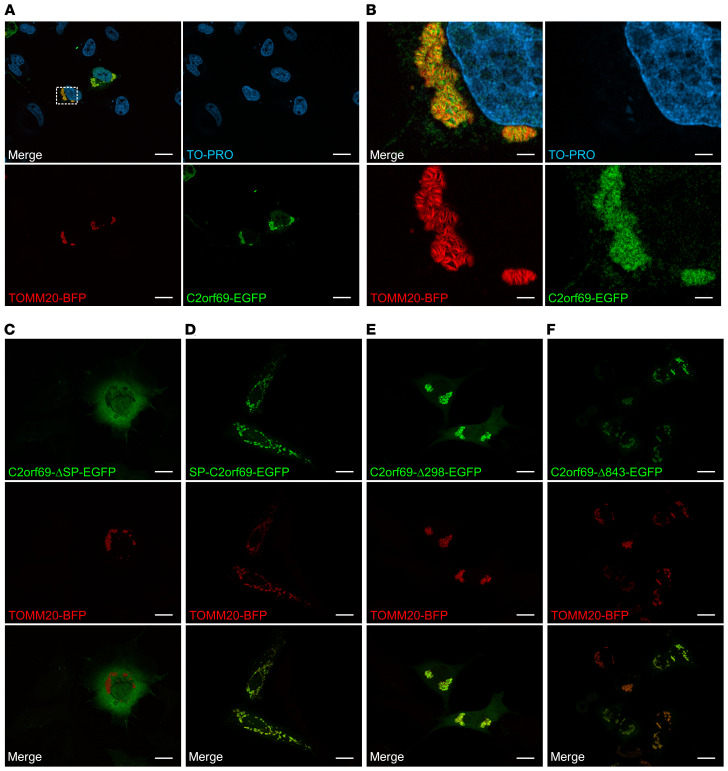
C2orf69 is targeted to mitochondria by a 24-aa motif.
To address the subcellular localization of C2orf69, we cloned the cDNA into EGFP and FLAG expression vectors. Overexpression of N-terminally EGFP-tagged C2orf69 (EGFP-C2orf69) showed homogenous distribution in the cell (Supplemental Figure 3C), whereas the C-terminal EGFP fusion (C2orf69-EGFP) had a distinct and potentially mitochondrial localization (Figure 3A). We observed the same localization for FLAG-tagged (Supplemental Figure 3, A and B) and mCherry-tagged C2orf69 (Supplemental Figure 3, D and E). The mitochondrial localization of C2orf69-EGFP was confirmed by coexpression with the mitochondrial marker TOMM20-BFP. Both proteins showed a robust overlap, but the C2orf69-EGFP signal was close to and not congruent with the outer mitochondrial membrane protein TOMM20, consistent with localization in the mitochondrial matrix (Figure 3B). A mutant lacking the first 24 aa of C2orf69 and a fusion protein of the SP (aa 1–24) attached to a C-terminal EGFP epitope were expressed in cells. Deletion of the SP completely abolished the mitochondrial localization (Figure 3C). However, the SP alone was sufficient to target EGFP to mitochondria (Figure 3D). C-terminally truncated C2orf69 isoforms mimicking the patients’ mutations (C2orf69-Δ298, corresponding to c.298del, and C2orf69-Δ843, corresponding to c.843_847del) were localized in the mitochondria, which is in line with the presence of the SP being sufficient for mitochondrial targeting (Figure 3, E and F). However, Western blot analysis, which showed an absence of C2orf69 proteins in whole-cell lysates of EBV-transformed lymphocytes from patient I, in contrast to control samples from unaffected family members, indicated that the truncated proteins were not stable, as there was no apparent reduction in RNA levels (Supplemental Figure 2, A and C).
Figure 3. C2orf69 shows mitochondrial localization.
COS-7 cells were transfected with the indicated constructs, and nuclei were stained with TO-PRO. Localization of C2orf69‑EGFP (A), C2orf69‑ΔSP‑EGFP (C), SP‑C2orf69‑EGFP (D), C2orf69‑Δ298‑EGFP (E), C2orf69‑Δ843‑EGFP (F), and the mitochondrial marker TOMM20‑BFP. Scale bars: 20 μm. (B) Higher-magnification images of the mitochondria from A. Scale bars: 3.5 μm. Representative images are shown (n = 3).
To further classify the function of C2orf69, we performed next-generation sequencing–based (NGS-based) yeast 2-hybrid (Y2H) screening using full-length human C2orf69 and a normalized human brain cDNA library. We identified 35 putative interaction partners, among which were important mitochondrial proteins (BOLA3, CMPK2, ETFA, KLHDC9, SDHB) (Supplemental Figure 4). Furthermore, RNA-Seq of EBV-transformed lymphocytes from patient I and all family members revealed an upregulation of genes involved in the OXPHOS process. Moreover, we observed an upregulation of immunological pathways such as that for Fc receptor and TNF-α signaling. On the other hand, pathways related to cell proliferation, e.g., those for histone synthesis, RNA polymerase 2–related transcription, and TOR signaling, were downregulated (Supplemental Figure 5).
C2orf69 loss causes respiratory chain defects and accumulation of ROS.
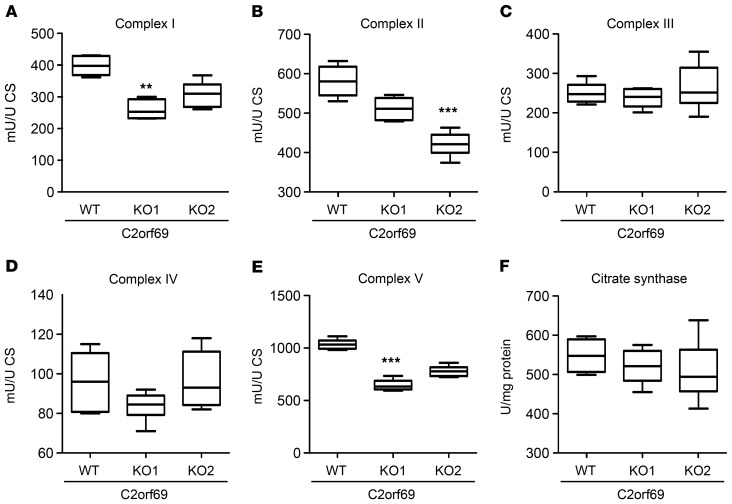
To further address the function of C2orf69, we generated a KO via CRISPR/Cas9-based mutagenesis in HAP1 cells. KO in 2 independent C2orf69-KO clones (KO1 and KO2) was confirmed by Sanger sequencing (Supplemental Figure 6) and Western blotting (Supplemental Figure 2, A and B). Analysis revealed no alterations in mitochondrial mass (Supplemental Figure 7C) or mitochondrial DNA copy numbers (Supplemental Figure 7, D–F) compared with WT cells. However, OXPHOS function analysis showed a greater than 20% reduction in complex I activity in both C2orf69-KO clones. Examination of OXPHOS function in a muscle biopsy from patient III also revealed a strong reduction in complex I activity (Supplemental Table 1). In addition, we detected a reduction in complex II activity, whereas complex III and complex IV activity was not altered in the KO clones. Of note, we found that activity of the ATP synthase (complex V) was impaired in C2orf69-KO cells (Figure 4). Another hallmark of mitochondrial malfunction is an increase in ROS within the cell. Therefore, we analyzed cellular ROS levels by DCFDA staining in C2orf69 WT and -KO cells. DCFDA is oxidized by ROS, which results in a fluorescent form of the reagent. The analysis revealed a significantly higher amount of ROS in C2orf69-KO cells under resting conditions (Supplemental Figure 7, A and B). In summary, the data indicated OXPHOS defects that led to alterations in the activity of complexes I and II, and, most robustly, complex V.
Figure 4. Biochemical studies of mitochondrial respiratory chain complexes in HAP1 cells.
(A–F) Box plots show the mean values of enzymatic activity of complexes I (A), II (B), III (C), IV (D), and V (E) of OXPHOS and citrate synthase (F) measured in HAP1 WT and -KO cell lysates (n = 3). Boxes show the lower and upper interquartiles, horizontal lines indicate the median, and whiskers show the minimum-to-maximum values. **P < 0.01 and ***P < 0.001, by Kruskal-Wallis test. CS, citrate synthase.
Discussion
Understanding the function of yet-uncharacterized genes is a major challenge in biology. C2orf69 is one such protein-coding gene with unknown function. It encodes a highly conserved 385-aa polypeptide without any obvious orthologs or paralogs, indicating an essential and nonredundant function. The first evidence for an important biological role of C2orf69 came from our unbiased sequencing approach in a patient with a complex disease compromising several organs and their function. Using WES, we identified a homozygous frameshift deletion (c.298del) in C2orf69 in an affected individual that segregated with the phenotype in the family. Human genomic databases do not reveal biallelic C2orf69 loss-of-function mutations, further suggesting that loss of both C2orf69 gene copies is deleterious. Our subsequent identification of homozygous loss-of-function mutations in the gene in another 7 individuals with similar clinical symptoms corroborated its relevance in human disease.
Here, we provide multiple lines of evidence that C2orf69 is a mitochondrial protein that affects OXPHOS function. Strong evidence for mitochondrial localization is already provided by in silico prediction tools, which, on the other hand, suggested that C2orf69 is secreted. Our studies showed a predominant mitochondrial localization upon overexpression of C-terminally tagged C2orf69 and a partial overlap with the outer mitochondrial membrane marker TOMM20. We showed that the most N-terminal 24 aa of C2orf69 were both required and sufficient for its targeting to mitochondria and that N-terminal protein tags hindered mitochondrial targeting. N-terminal SPs are characteristic of mitochondrial matrix proteins and only rare or absent in proteins located in other parts of the mitochondria (11). Immunohistochemical confirmation of the localization of endogenous C2orf69 proteins was not possible because of a lack of suitable antibodies. However, other studies using a proteomics approach (ascorbic acid peroxidase mass spectrometry [APEX MS]) showed that endogenous C2orf69 is found in the mitochondrial matrix (12). Moreover, in silico predictions classify C2orf69 as an oxidoreductase of the mitochondrial matrix (13). Nevertheless, we cannot exclude the possibility that the protein is secreted in other cell types or under certain conditions, or, further, that it localizes to cell compartments other than mitochondria.
Y2H screening revealed 35 interaction partners of C2orf69, and several interacting proteins support its role in mitochondrial function: SDHB is a major subunit of complex II of the respiratory chain; BOLA3 is an essential protein in the Fe-S cluster synthesis and involved in the assembly of respiratory chain complexes; ETFA is involved in the electron transfers from mitochondrial β-oxidation to the ubiquinone pool in the inner mitochondrial membrane; KLHDC9 is predicted to localize to mitochondria; and CMPK2 plays an essential role in the mitochondrial nucleotide synthesis salvage pathway. Other interacting proteins could be assigned to the secretion of proteins and the extracellular matrix, indicating a possible role of C2orf69 outside the cell. However, in our secretion assays, we did not observe C2orf69 in cell supernatants. Publicly available high-throughput MS data showed ubiquitination and SUMOylation of C2orf69. Consistent with this, we found 3 proteins involved in ubiquitination and SUMOylation in our Y2H screen.
There is evidence from our HAP1 C2orf69-KO model that mitochondrial complex I, II, and V activity was reduced. The clinical data for patient III also revealed lower activity of complexes I and IV in muscle (complex V activity was not measured). A similar impairment of complexes I and IV is described in patients with Leigh syndrome, which is caused by mutations in NDUFAF5 (C20orf7), an assembly factor of complex I (14). Immortalized lymphocytes from patient I exhibited a transcriptional upregulation of genes associated with OXPHOS compared with control lymphocytes, and together with increased ROS levels in the HAP1-KO cells, these data corroborate mitochondrial impairment upon C2orf69 loss (15, 16). RNA-Seq data showed upregulation mainly of the genes encoded by the mitochondrial genome of complexes I and III–V (Supplemental Figure 8). This constellation is observed in response to increased ROS levels and OXPHOS alterations (17, 18). Moreover the induction of NDUFA4 is remarkable and may be a compensatory effect, because aside from being an essential part of complex I, NDUFA4 acts as an assembly factor for complex IV (19). Further, the disintegration and degeneration of mitochondria observed in patient III by EM underlines the essential role of C2orf69 in mitochondria maintenance.
The clinical symptoms of the patients are in line with a multisystem mitochondrial disorder. Typical signs of mitochondriopathies include myopathy with or without cardiac involvement, deafness, various ophthalmological problems, diabetes mellitus, and, in some, liver failure. Encephalopathy, seizures, cerebellar ataxia, and spasticity indicate an effect on the CNS (20). In accordance with this, patients with C2orf69 mutations have muscular weakness, eye abnormalities, liver dysfunction, epilepsy, and cardiomyopathy as overlapping features. Other common signs of congenital mitochondrial defects such as hypomyelination/delay of myelination, cerebellar atrophy, microcephaly, and short stature are also present. Lactate in blood was transiently elevated in some patients (I, III, and VII), and patient III showed a lactate peak on MR spectroscopy of the brain, however, this was not consistent for all patients.
A remarkable feature of the C2orf69 disorder was the initial suspicion of a GSD in several patients. For example, in patients III, V, and VI, we found a lack of glycogen branching enzyme activity, and we observed PAS-positive and diastase-resistant deposits in tissue biopsies (patients I, III, V, and VI), which are features characteristic of the abnormally branched glycogen found in GSD IV. The presence of polyglucosan bodies seen on EM of muscle from patient III further indicated a glycogen pathology. GSD IV is an autosomal-recessive disorder with reduced or absent GBE1 activity and a continuum of different subtypes with variable outcomes ranging from fatality in newborns to a late-onset neuromuscular phenotype. Early-onset forms show profound hypotonia, respiratory distress, and dilated cardiomyopathy. The progressive hepatic subtype is characterized by developmental delay, hepatomegaly, liver dysfunction, and cirrhosis accompanied by hypotonia and cardiomyopathy (21). Thus, there is a marked clinical overlap between mitochondriopathies and GSD, and the clinical presentation of our patients may in part be a combination of both. Inhibition of glycogen synthase 1 (GYS1) was shown to reduce glycogen deposits in a polyglucosan body disease (APBD) mouse model (22), and thus GYS1 inhibition could be a possible option to mitigate symptoms of the C2orf69 disorder.
A hypothetical link between mitochondrial dysfunction and glycogen accumulation could be that both pathways are involved in energy metabolism. Mitochondria are the main source of ATP, which is provided by metabolizing glucose. On the other hand, glucose is stored in the form of glycogen. The branching of glycogen via GBE1 activity enables rapid synthesis and degradation to supply the cell with glucose under starving conditions. Indeed, several mitochondriopathies show signs of altered glycogen storage. The exact mechanism of the C2orf69-related pathology remains elusive, but there might be a direct or, because of the mitochondrial localization of C2orf69, rather indirect impact on GBE1 activity.
Another remarkable feature of the C2orf69-related disease was the recurrence of inflammatory events, which were associated with an increased number of monocytes (patients I and III). In EBV-immortalized lymphocytes from patient I, several immune system–related pathways appeared to be disturbed. This may be in accordance with a dysregulation of the immune system in these patients, who initially seemed to have an immune deficiency and later developed autoinflammation, as is also seen in other mitochondrial disorders (23, 24). Glucocorticoid therapy was partially successful in patient I; however, upon a reduction of glucocorticoids, the inflammation returned. Remarkably, the antiepileptic therapy with ACTH resulted in a long-lasting suppression of inflammation that was most probably due to the corticotropic effect. Anakinra therapy resulted in a remarkable improvement in patient II, with attenuation of serositis and arthritis and complete normalization of the patient’s laboratory features. Suppression of the overacting immune system, either by glucocorticoids or by targeting the IL-1 signaling pathway, might therefore be considered a therapeutic option for these patients.
KO of C2orf69 in HAP1 cells revealed a reduction in cell proliferation (Supplemental Figure 9). In line with this, RNA-Seq data on family 1 also showed downregulation of several proliferation-related genes (Supplemental Figure 5). This suggests that the attempt to compensate for C2orf69 loss by upregulation of other mitochondrial genes is insufficient and cannot provide enough energy for proliferation. Additionally, Müller et al. identified a cell-cycle genes homology region (CHR) element in the promoter of C2orf69, which can be bound and repressed by the DREAM (dimerization partner, RB-like, E2F, and multivulval class B) complex in response to p53 activation to regulate G2/M cell-cycle genes (25, 26). C2orf69 may therefore also play a direct role in proliferation, but confirmation of this requires further study. Hence, the impaired cell proliferation could be partly responsible for the observed clinical features, e.g., microcephaly and dystrophy.
Taken together, we have identified a disorder with autoinflammation as a core characteristic and clinical overlap with mitochondriopathies and GSDs. We found that the affected gene product, C2orf69, localizes to mitochondria and is involved in respiratory chain function, but simultaneously regulates GBE1 enzyme activity. The exact mechanism that leads to impairment of these 2 energy-producing systems remains a subject for further investigation.
Methods
Materials.
If not stated otherwise, sequencing materials were purchased from Illumina, cell culture reagents and cloning enzymes were purchased from Thermo Fisher Scientific, and primers were purchased from biomers.net.
WES.
WES was performed on DNA from peripheral blood obtained from 3 family members including the index patient and both parents (family 1). Enrichment was performed with Illumina Nextera Rapid Capture Exome, version 1.2, and the respective libraries were sequenced on an Illumina NextSeq 500 sequencer. Alignment and variant calling were performed with SeqMule (version 1.2; ref. 27). The resulting variant files were annotated with KGGSeq (version 1.0, April 14, 2017; ref. 28). Variants with a minor allele frequency above 0.75% in public databases were excluded. Mutations were confirmed by Sanger sequencing.
Exome sequencing for patient II was done as published elsewhere (29). Validation of the C2orf69 mutation was done by Sanger sequencing. For family 4, exome sequencing was performed on peripheral blood DNA from the affected patient VI, the unaffected sister, and both parents. DNA libraries were prepared with the Illumina TruSeq DNA Library Preparation Kit. Agilent SureSelect V4 (Agilent Technologies) was used for exome enrichment. Sequencing was performed on an Illumina HiSeq 2000. Data analysis was performed using Highlander software as previously described (30), and the results were confirmed by Sanger sequencing of the whole family, including patient V.
WES analysis of patients III and VI and patients VII and VIII, respectively, was carried out by the commercial diagnostic providers CeGaT and GeneDx.
Cloning.
The C2orf69 coding sequence (NM_153689.6) was amplified from a human fibroblast cDNA and cloned via BamHI and EcoRI into pEGFP-C1/N3 (Clontech Laboratories) and p3xFlag-CMV-10/14 (MilliporeSigma) vectors. For the deletion and SP mutants, the C2orf_EcoRI_F or C2orf69_d24_EcoRI-F primer and the reverse primer C2orf69_BamHI_del_R, C2orf69_BamHI_del2_R, or C2orf69_BamHI_24AA_R was used to amplify the C2orf69 fragments from the pEGFP-C1-C2orf69 vector and cloned as for the full-length C2orf69 described above.
pmCherry-N1-C2orf69, pEGFP-C1-C2orf69, and pmCherry-N1 were cut by EcoRI and Cfr9I and blunted with Klenow. The resulting fragments were purified from agarose gel and ligated with T4 ligase. pmCherry-C2-C2orf69 was generated by digestion of pEGFP-C1-C2orf69 and pmCherry-C2 with BamHI and EcoRI. The resulting fragments were purified via agarose gel and ligated with T4 ligase.
For the generation of px330-EGFP-C2orf69, sgRNA oligonucleotides (forward, 5′-CACCGCCAGAACCATGAACCCGGG-3′; reverse, 5′-AAACCCCGGGTTCATGGTTCTGGC-3′) were cloned into the pX330-EGFP vector according to the Zhang laboratory protocol available from Addgene.
Transformation for cloning and plasmid preparation were carried out using One Shot Top10 chemically competent E. coli (Thermo Fisher Scientific).
All constructed plasmids were verified by Sanger sequencing. All mutagenesis and sequencing primers and vectors used in this study are listed in Tables 3–5.
Table 3. Primers for mutagenesis.

Table 5. Vectors.
Cells and cell culture.
C2orf69-KO HAP1 cells were generated by CRISPR/Cas9. In brief, HAP1 cells (Horizon Discovery) were seeded at 70% confluence onto 6-well dishes and transfected with the pX330-EGFP-C2orf69 vector. After 48 hours, cells were trypsinized and sorted to identify strong EGFP-positive populations. EGFP-positive cells were seeded onto a 10 cm dish, and colonies were selected and transferred onto a 96-well plate after 3 days. KO of the single clones was confirmed by sequencing and Western blot analysis. The cells were cultured in IMDM supplemented with 5% FCS and 1% penicillin/streptomycin at 37°C in an atmosphere of 5% CO2. B lymphocytes from blood were immortalized by infection with EBV according to the protocol by Tosato et al. (31). HeLa (American Type Culture Collection [ATCC]) and COS-7 (ATCC) cells were cultured in DMEM supplemented with 5% FCS and 1% penicillin/streptomycin at 37°C in an atmosphere of 5% CO2. Transfection of HeLa, COS-7, and HAP1 cells was achieved using PolyJet or GenJet in vitro transfection reagent (tebu-Bio) according to the manufacturer’s protocol.
Histology.
Tissue samples obtained during autopsy were fixed in neutral buffered formalin (4%) followed by paraffin embedment. After sectioning, slides were stained according to standard protocols with hematoxylin (Merck Millipore) and eosin (BioGnost), PAS (Merck Millipore) reaction with hematoxylin as a counterstain, as well as Diastase-PAS stain, similar to the PAS staining with prior digestion with synthetic amylase (Diapath). After coverslipping, the stained slides were scanned with the Hamamatsu Nanozoomer 2.0HT (Hamamatsu Photonics K.K.) whole-slide scanner, and histological images were extracted with NDP.view software (Hamamatsu Photonics K.K.).
Microscopy.
Transfected COS-7 cells were seeded onto coverslips in a 24-well plate and fixed after 24 hours with 4% paraformaldehyde in PBS. TO-PRO-3 Iodide (1:1000; Thermo Fisher Scientific) was used for nucleic acid staining. Images were taken with a Zeiss Observer Z.1 microscope (Carl Zeiss) equipped with an ApoTome.2 and an HXP 120 lamp.
Y2H screening.
A commercially available Y2H screen was performed using Next Interactions with a Clontech human brain (normalized) library (32).
Immunoprecipitation.
HeLa cells were transfected with pEGFP-N1 (Clontech Laboratories), IL-6–EGFP (Addgene), pC2ORF69-EGFP, or pEGFP-C2orf69 in a 6-well plate, and the media were changed 24 hours later. After an additional 6 hours, 500 μL media were collected and centrifuged to eliminate detached cells. The supernatant was incubated with 1 μL anti-EGFP antibody (MAB3580, Merck Millipore) for 2 hours at 4°C with agitation. Protein A/G Bead slurry (20 μL) (Santa Cruz Biotechnology) was added overnight, followed by 3 washes with PBS (MilliporeSigma) plus 0.2% Tween (AppliChem). Beads were resuspended in 30 μL 2× SDS loading buffer, cooked, and loaded onto an SDS gel.
Western blot analysis.
Protein isolation and Western blotting were performed as described previously (33). The following primary antibodies were used for immunodetection: anti-C2orf69 (ab188870, 1:1000, Abcam), anti-GFP (ab6556; 1:2000, Abcam), and anti–α-tubulin (ab15246, 1:2000, Abcam). HRP-conjugated anti-rabbit antibody (sc-2370, 1:10,000; Santa Cruz Biotechnology) was used as a secondary antibody. Detection was done using Clarity Western ECL Substrate (Bio-Rad) and a Fujifilm LAS 3000 system (Fujifilm). The PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) was used for protein molecular weight estimation.
FACS.
Measurements were carried out on a BD FACSCanto II flow cytometer. Analysis of the data was done in FlowJo (BD).
Mitochondrial membrane potential.
Cells were seeded onto 6-well plates to approximately 40% confluence. Twenty-four hours later, the media were replaced with growth media containing 200 nM tetramethylrhodamine ethyl ester (TMRE) (Biomol) and incubated for 20 minutes at 37°C. Cells were harvested with trypsin and measured directly with a FACSCanto II in the phycoerythrin (PE) channel. Normalization to mitochondrial mass was performed by parallel MitoSpy Green staining (1:10,000; BioLegend).
ROS.
Cells were seeded onto 6-well plates to reach approximately 50% confluence at the time of the experiment. The cells were incubated with 25 μM DCFDA (Biomol) for 90 minutes at 37°C, harvested with trypsin, and measured directly with the FACSCanto II in the FITC channel.
Biochemical examination of OXPHOS function.
OXPHOS enzyme complex activity was measured in HAP cells as previously described (34, 35).
Apoptosis.
Cells were seeded onto 6-well plates at 40% confluence, harvested 24 hours later with trypsin, and collected in a reaction tube followed by a 1-hour incubation with CellEvent (2 μM in media; Thermo Fisher Scientific) at 37°C. Counterstaining was performed with 7-aminoactinomycin D (7-AAD) (1:100; BIOZOL Diagnostica Vertrieb), which was in a solution of PBS with 5% FCS. CellEvent and 7-AAD signals were measured by FACS in the FITC or PerCP‑Cy5-5 channel, respectively.
Growth curve.
Five hundred HAP1 WT and -KO cells were seeded onto 24-well plates on day 0 in IMDM supplemented with 2% FCS and 1% penicillin/streptomycin at 37°C in an atmosphere of 5% CO2. The cells were trypsinized and counted on days 2, 4, 6, and 8 in a Neubauer counting chamber.
Quantitative PCR.
DNA was isolated from HAP1 cells with the Quick-DNA Miniprep Plus Kit (Zymo Research Europe) according to the manufacturer’s instructions. DNA (1 ng) was used for quantitative PCR (qPCR) with PowerUp SYBR Green (Thermo Fisher Scientific) according to the manufacturer’s protocol. Denaturation: 2 minutes at 95°C; cycle, 40 times, 5 seconds at 95°C, 10 seconds at 60°C, 30 seconds at 72°C; melting curve, 1 minute at 95°C, 1 minute at 60°C, ramp +0.6°C, and 10 seconds at 95°C. Expression levels were calculated using the ΔΔCt method.
Real-time qPCR.
RNA was isolated from EBV immortalized lymphocytes with peqGOLD TriFast (VWR International) according to the manufacturer’s instructions. RNA (1–5 μg) was used for cDNA synthesis with the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. cDNA (25 ng) was used for qPCR with PowerUp SYBR Green (Thermo Fisher Scientific) according to the manufacturer’s protocol. Denaturation: 2 minutes at 95°C; cycle, 40 times, 5 seconds at 95°C, 10 seconds at 60°C, 30 seconds at 72°C; melting curve: 1 minute at 95°C, 1 minute at 60°C, ramp +0.6°C, and 10 seconds at 95°C. Expression levels were calculated using the ΔΔCt method. The primers used for qPCR are listed in Table 6.
Table 6. qPCR primers.

RNA-Seq.
EBV cells were seeded at 500,000/mL for 24 hours before RNA preparation. RNA was extracted with peqGOLD TriFast (VWR International), and libraries for RNA-Seq were prepared with the Illumina TruSeq Stranded Total RNA kit according to the manufacturer’s protocol. Sequencing was performed on an Illumina NextSeq 500 with a high-output 1 × 75 flow cell. Raw data were demultiplexed, and FASTQ files were generated with bcl2fastq (version 2.20). Data were analyzed and visualized with BioJupies using default parameters (36).
In accordance with data protection requirements, we do not have the option of making the genetic data publicly available at this time. However, the data can be shared upon request and with appropriate legal safeguards.
Statistics.
Results are presented as the mean ± SD from at least 3 independent experiments using GraphPad Prism 5 (GraphPad Software). Statistical significance was evaluated by Kruskal-Wallis test or 2way ANOVA, with a P value of less than 0.05 considered statistically significant.
Study approval.
Written informed consent was obtained from the parents or legal guardians of the study participants after approval from the IRBs at the participating institutions (EK302-16; UK D 4886R; H12-00067, UBC). Consent was obtained according to Declaration of Helsinki principles. Approval EK302-16 was granted by the ethics committee of the Medical Faculty of the RWTH Aachen (Universitaetsklinikum Aachen, Aachen, Germany). Approval UK D 4886R was granted by the ethics committee of the Medical Faculty of the HHU Düsseldorf (Universitaetsklinikum Düsseldorf, Düsseldorf, Germany). Approval H12-00067 was granted by the UBC’s Clinical Research Ethics Office (Vancouver, British Columbia, Canada).
Author contributions
IK and FK designed the study and analyzed and interpreted the data. IK, FK, and EL wrote the manuscript. EL and SG acquired, analyzed, and interpreted the majority of the experimental data. PK, KO, MH, KT, ME, CSP, and NW acquired, analyzed, and interpreted the clinical data for patient I. MB, TE, ME, and RM acquired, analyzed, and interpreted the WES data for patient I. MM analyzed and interpreted the MRI data for all patients. TB acquired, analyzed, and interpreted the histology data for patient I. JW and CJS acquired, analyzed, and interpreted the EM data for patient I. PTO and AB acquired, analyzed, and interpreted the clinical data for patient II. HS and DW acquired, analyzed, and interpreted the WES data for patient II. CH, DC, and SD acquired, analyzed, and interpreted the clinical data for patient III. AH acquired, analyzed, and interpreted the WES data for patient III. FGD, CL, JPD, EW, and EVS acquired, analyzed, and interpreted the clinical and WES data for patients V and VI. RS and CDVK acquired, analyzed, and interpreted the clinical and WES data for patients VII and VIII. LVDH acquired, analyzed, and interpreted the OXPHOS data for the HAP cells. All authors revised the manuscript.
Supplementary Material
Table 4. Sequencing primers.

Acknowledgments
This work was funded by the Deutsche Forschungsgemeinschaft (DFG) (948/32–1 FUGG). Support for this study was provided by the Flow Cytometry Facility and the Genomics Facility, core facilities of the Interdisciplinary Center for Clinical Research (IZKF) Aachen within the Faculty of Medicine at RWTH Aachen University. The study was also supported by a Fund Invest for Scientific Research (FIRS) grant from the CHU de Liège, Belgium. We are grateful to the TIDEX team (Britt Drogemoller, Aisha Ghani, Colin Ross, Maja Tarailo-Graovac, and Wyeth Wassermann) at the UBC’s Centre for Molecular Medicine and Therapeutics (Vancouver, British Columbia, Canada). We also acknowledge the physicians who participated in the patients’ clinical care: Hal Siden, Canuck Place, Michelle Demos, and Lori Tucker (Department of Pediatrics, British Columbia Children’s Hospital).
Version 1. 05/04/2021
In-Press Preview
Version 2. 06/15/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(12):e143078.https://doi.org/10.1172/JCI143078.
Contributor Information
Eva Lausberg, Email: elausberg@ukaachen.de.
Sebastian Gießelmann, Email: sgiesselmann@ukaachen.de.
Joseph P. Dewulf, Email: joseph.dewulf@uclouvain.be.
Elsa Wiame, Email: elsa.wiame@uclouvain.be.
Anja Holz, Email: Anja.Holz@humangenetik-tuebingen.de.
Ramona Salvarinova, Email: rsalvarinova@cw.bc.ca.
Clara D. van Karnebeek, Email: Clara.vanKarnebeek@radboudumc.nl.
Patricia Klemm, Email: Patricia.klemm@rwth-aachen.de.
Kim Ohl, Email: kohl@ukaachen.de.
Michael Mull, Email: mmull@ukaachen.de.
Till Braunschweig, Email: tbraunschweig@ukaachen.de.
Joachim Weis, Email: jweis@ukaachen.de.
Clemens J. Sommer, Email: Clemens.Sommer@unimedizin-mainz.de.
Stephanie Demuth, Email: demuth@genetik-erfurt.de.
Claudia Haase, Email: claudia.haase@helios-kliniken.de.
Claudia Stollbrink-Peschgens, Email: cstollbrink-peschg@ukaachen.de.
François-Guillaume Debray, Email: francois-guillaume.debray@chc.be.
Cecile Libioulle, Email: c.libioulle@chuliege.be.
Daniela Choukair, Email: daniela.choukair@med.uni-heidelberg.de.
Prasad T. Oommen, Email: Prasad.Oommen@med.uni-duesseldorf.de.
Arndt Borkhardt, Email: arndt.borkhardt@med.uni-duesseldorf.de.
Harald Surowy, Email: Harald.Surowy@med.uni-duesseldorf.de.
Dagmar Wieczorek, Email: Dagmar.Wieczorek@med.uni-duesseldorf.de.
Norbert Wagner, Email: nwagner@ukaachen.de.
Robert Meyer, Email: rmeyer@ukaachen.de.
Thomas Eggermann, Email: teggermann@ukaachen.de.
Matthias Begemann, Email: mbegemann@ukaachen.de.
Emile Van Schaftingen, Email: emile.vanschaftingen@uclouvain.be.
Martin Häusler, Email: mhaeusler@ukaachen.de.
Klaus Tenbrock, Email: ktenbrock@ukaachen.de.
Lambert van den Heuvel, Email: Bert.vandenHeuvel@radboudumc.nl.
Miriam Elbracht, Email: mielbracht@ukaachen.de.
Ingo Kurth, Email: ikurth@ukaachen.de.
Florian Kraft, Email: fkraft@ukaachen.de.
References
- 1.Ferreira CR, et al. A proposed nosology of inborn errors of metabolism. Genet Med. 2019;21(1):102–106. doi: 10.1038/s41436-018-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blau N, et al, eds. Physician’s guide to the diagnosis, treatment, and follow-up of inherited metabolic disease. Springer; 2014. [Google Scholar]
- 3.Waters D, et al. Global birth prevalence and mortality from inborn errors of metabolism: a systematic analysis of the evidence. J Glob Health. 2018;8(2):021102. doi: 10.7189/jogh.08.021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman J, Rahman S. Mitochondrial medicine in the omics era. Lancet. 2018;391(10139):2560–2574. doi: 10.1016/S0140-6736(18)30727-X. [DOI] [PubMed] [Google Scholar]
- 5.Stenton SL, et al. The diagnosis of inborn errors of metabolism by an integrative “multi-omics” approach: A perspective encompassing genomics, transcriptomics, and proteomics. J Inherit Metab Dis. 2020;43(1):25–35. doi: 10.1002/jimd.12130. [DOI] [PubMed] [Google Scholar]
- 6.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534(7605):55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mertins P, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 2014;13(7):1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4(179):rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akimov V, et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 2018;25(7):631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 10.Sobreira N, et al. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacinska A, et al. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvo SE, et al. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(d1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khorsandi SE, et al. An in silico argument for mitochondrial microRNA as a determinant of primary non function in liver transplantation. Sci Rep. 2018;8(1):3105. doi: 10.1038/s41598-018-21091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saada A, et al. Combined OXPHOS complex I and IV defect, due to mutated complex I assembly factor C20ORF7. J Inherit Metab Dis. 2012;35(1):125–131. doi: 10.1007/s10545-011-9348-y. [DOI] [PubMed] [Google Scholar]
- 15.Miwa S, et al. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat Commun. 2014;5:3837. doi: 10.1038/ncomms4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mimaki M, et al. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Al-Kafaji G, et al. Increased expression of mitochondrial DNA-encoded genes in human renal mesangial cells in response to high glucose-induced reactive oxygen species. Mol Med Rep. 2016;13(2):1774–1780. doi: 10.3892/mmr.2015.4732. [DOI] [PubMed] [Google Scholar]
- 18.Reinecke F, et al. OXPHOS gene expression and control in mitochondrial disorders. Biochim Biophys Acta. 2009;1792(12):1113–1121. doi: 10.1016/j.bbadis.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Balsa E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16(3):378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Finsterer J, Zarrouk-Mahjoub S. Cerebellar atrophy is common among mitochondrial disorders. Metab Brain Dis. 2018;33(4):987–988. doi: 10.1007/s11011-018-0238-y. [DOI] [PubMed] [Google Scholar]
- 21. Magoulas PL, El-Hattab AW. Glycogen storage disease type IV. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al, eds. GeneReviews. University of Washington; 2013:NBK115333. [PubMed] [Google Scholar]
- 22.Kakhlon O, et al. Guaiacol as a drug candidate for treating adult polyglucosan body disease. JCI Insight. 2018;3(17):e99694. doi: 10.1172/jci.insight.99694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker MA, et al. Predisposition to infection and SIRS in mitochondrial disorders: 8 years’ experience in an academic center. J Allergy Clin Immunol Pract. 2014;2(4):465–468. doi: 10.1016/j.jaip.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Walker MA, et al. Powering the immune system: mitochondria in immune function and deficiency. J Immunol Res. 2014;2014:164309. doi: 10.1155/2014/164309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller GA, et al. The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42(16):10331–10350. doi: 10.1093/nar/gku696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer M, et al. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016;44(1):164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, et al. SeqMule: automated pipeline for analysis of human exome/genome sequencing data. Sci Rep. 2015;5:14283. doi: 10.1038/srep14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MX, et al. Predicting mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2013;9(1):e1003143. doi: 10.1371/journal.pgen.1003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taeubner J, et al. Diagnostic challenges in a child with early onset desmoplastic medulloblastoma and homozygous variants in MSH2 and MSH6. Eur J Hum Genet. 2018;26(3):440–444. doi: 10.1038/s41431-017-0071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewulf JP, et al. SLC13A3 variants cause acute reversible leukoencephalopathy and α-ketoglutarate accumulation. Ann Neurol. 2019;85(3):385–395. doi: 10.1002/ana.25412. [DOI] [PubMed] [Google Scholar]
- 31.Tosato G, Cohen IJ. Generation of Epstein-Barr Virus (EBV)–immortalized B cell lines. Curr Protoc Immunol. 2007;76:7.22.1–7.22.4. doi: 10.1002/0471142735.im0722s76. [DOI] [PubMed] [Google Scholar]
- 32.Suter B, et al. Next-generation sequencing for binary protein-protein interactions. Front Genet. 2015;6:346. doi: 10.3389/fgene.2015.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esmaeili M, et al. The tumor suppressor ING1b is a novel corepressor for the androgen receptor and induces cellular senescence in prostate cancer cells. J Mol Cell Biol. 2016;8(3):207–220. doi: 10.1093/jmcb/mjw007. [DOI] [PubMed] [Google Scholar]
- 34.Janssen AJ, et al. Some practical aspects of providing a diagnostic service for respiratory chain defects. Ann Clin Biochem. 2003;40(pt 1):3–8. doi: 10.1258/000456303321016114. [DOI] [PubMed] [Google Scholar]
- 35.Smeitink J, et al. Human NADH:ubiquinone oxidoreductase. J Bioenerg Biomembr. 2001;33(3):259–266. doi: 10.1023/A:1010743321800. [DOI] [PubMed] [Google Scholar]
- 36.Torre D, et al. Biojupies: automated generation of interactive notebooks for RNA-Seq data analysis in the cloud. Cell Syst. 2018;7(5):556–561. doi: 10.1016/j.cels.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.