Abstract
Sugarcane (Saccharum officinarum L.) is a cash crop grown commercially for its higher amounts of sucrose, stored within the mature internodes of the stem. Numerous studies have been done for the resistance development against biotic and abiotic stresses to save the sucrose yields. Quality and yield of sugarcane production is always threatened by the damages of cane borers and weeds. In current study two problems were better addressed through the genetic modification of sugarcane for provision of resistance against insects and weedicide via the expression of two modified cane borer resistant CEMB-Cry1Ac (1.8 kb), CEMB-Cry2A (1.9 kb) and one glyphosate tolerant CEMB-GTGene (1.4 kb) genes, driven by maize Ubiquitin Promoter and nos terminator. Insect Bio-toxicity assays were carried out for the assessment of Cry proteins through mortality percent of shoot borer Chilo infuscatellus at 2nd instar larvae stage. During V0, V1 and V2 generations young leaves from the transgenic sugarcane plants were collected at plant age of 20, 40, 60, 80 days and fed to the Chilo infuscatellus larvae. Up to 100% mortality of Chilo infuscatellus from 80 days old transgenic plants of V2 generation indicated that these transgenic plants were highly resistant against shoot borer and the gene expression level is sufficient to provide complete resistance against target pests. Glyphosate spray assay was carried out for complete removal of weeds. In V1-generation, 70–76% transgenic sugarcane plants were found tolerant against glyphosate spray (3000 mL/ha) under field conditions. While in V2-generation, the replicates of five selected lines 4L/2, 5L/5, 6L/5, L8/4, and L9/6 were found 100% tolerant against 3000 mL/ha glyphosate spray. It is evident from current study that CEMB-GTGene, CEMB-Cry1Ac and CEMB-Cry2A genes expression in sugarcane variety CPF-246 showed an efficient resistance against cane borers (Chilo infuscatellus) and was also highly tolerant against glyphosate spray. The selected transgenic sugarcane lines showed sustainable resistance against cane borer and glyphosate spray can be further exploited at farmer’s field level after fulfilling the biosafety requirements to boost the sugarcane production in the country.
Subject terms: Biotechnology, Plant biotechnology
Introduction
Sugarcane (Saccharum officinarum L.) is significantly prominent among the essential food cash crops of the world1–3 which has been cultivated in 58 countries around the globe. The projected area of the world under sugarcane cultivation was 26.9 million hectares4. This grass of genus Saccharum meets 80% of world sugar requirements though most popular sweetener chemically known as sucrose5,6. Sucrose deposited in stalk’s internodes7. About two billion metric ton stalks are crushed in sugar mills to get the sucrose juice that produces 2/3 world sugar production8. Sugarcane is more than just a sugar crop among the cash crops of the world9,10. Sugarcane yields a variety of products from fiber to biofuel, chemicals and industrial products such as alcohol, furfural, dextran, chipboard, paper, beverages, confectionery, plastics, paints, synthetics, pharmaceutical products, industrial enzymes, insecticides and detergents1,11,12.
In Pakistan, sugarcane has been cultivated on an area of 1.2171 million hectares and contributes 3.4% value added in agriculture sector amounting to nearly 0.7% of the GDP. Pakistan boosted the overall sugarcane production to 73.6 million tons of the commodities13. This gives Pakistan a reputation as being one of the top five sugarcane producers across the world. It occupies an important position in the national economy of Pakistan, by employing more than 9 million Pakistanis, significantly helping out driving the export economy of Pakistan14 and proving itself a backbone of second largest agroindustry of Pakistan by providing raw material to 84 sugar mills for sugar production2. Pakistan exports sugar to the Tajikistan, Afghanistan, and other central Asian countries. Sugar industry is entirely dependent on the availability of sugarcane for sugar production in Pakistan15.
Globally, pests along with diseases cause 37% loss to the agriculture production from which 13% is just because of insects16,17. Approximately 1300, insect pests attack on sugarcane crop around the world. In Pakistan 61 insect species have been accounted for attacking sugarcane crop18. The borers species reported from Pakistan included Chilo infuscatellus and Chilo auricilius (stem borers) with 15–36% yield loss, Scirpophaga excerptalis and Scirpophaga novella (top borers) with 10–15% loss, Emmalocera depressella (root borer) with 10–20% crop infestation, Acigona steniella (Gurdaspur borer) with 20% usual yield loss and Pyrilla perpusilla (sugarcane leaf hoper/Walker) with 25% crop loss respectively. Due to 80% leaf infestation and 15–25% yield reduction, Aleurolobus barodensis (white fly) is also considered among the most deleterious insects. Saccharicoccus sacchari (Mealy bug) and Cavelerious excavatus (black bug) are minor pests of sugarcane who attack the crop throughout the year19. Extensive and excessive use of insecticides/pesticides is increasing every year to control yield losses from insect’s damage.
Cry genes from Bacillus thuringiensis (Bt.) produces crystalline protein that defend transgenic plants against Lepidopteran, Dipterans and Coleopteran20–22. Bt. crops now contain Cry genes such as Cry1Ab, Cry1Ac, Cry1Ac + Cry1F, Cry2A, Cry1Ac + Cry2A significantly efficient in controlling Lepidopteran insects23,24. The Bt. transformed crops were observed with minimum insecticidal sprays throughout the season while non-transgenic plants were treated with five to twelve pesticidal sprays25. Glyphosate (N-phosphonomethyl glycine) is a broad spectrum and most effective herbicide to control weeds. Glyphosate usually kills the plants through the blockage of EPSPS26 enzyme. Incorporation of this soil bacterium gene produces a glyphosate tolerant form of EPSPS enzyme; its binding affinity for glyphosate confers the increased tolerance against glyphosate in transgenic plants27–31. Its distinctive target (EPSPS enzyme), non-selective nature for the plants/weeds, great potential to sink inside the tissues of the plants, and minor toxicity effects to the atmosphere and the human beings has rendered it to its current status32,33. The objectives of our study was that by controlling these cane borers and weeds, improved self sufficiency in sugar production is achievable to execute the compelling/demanding requirement of the increasing population for the sweetener utility.
Materials and methods
Construction of plant expression vectors
The CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene are patented by the Centre of Excellence in Molecular Biology (CEMB), University of the Punjab Lahore, Pakistan (Patent No. K140649-aroA). It have been confirmed that the experimental research and field studies on sugarcane plants, including the collection of plant material, complied with relevant institutional, national, and international guidelines and legislation with appropriate permissions from Centre of Excellence in Molecular Biology (CEMB), University of the Punjab Lahore, and Ayub Agricultural Research Institute Faisalabad, Pakistan for collection of plant specimens. These genes were constructed with maize Ubiquitin promoter and Nos terminator independently in separate cassettes (Supplementary Figure 1). Chemically synthesized genes were confirmed via restriction digestion as well as through protein expression studies as was done (Rao et al.34; Qamar et al.35).
Transient expression of transgene (Gus) to determine the efficacy of plasmid constructs
Electroporation
Plasmids constructs pCEMB-SC12 and pCEMB-SGTG were transformed into Agrobacteriurm tumefaciens competent cells (LBA4404) with Bio-Rad Electroporator (Model 1652078) using standard procedure as done by Puspito et al.27. Transformed cells were spread on YEP medium plates with kanamycin (50 mg/L) and incubated at 30 °C for overnight. Positive transformants were confirmed by colony PCR that was performed using gene specific primers.
Agroinfiltration of tobacco leaves
Agrobacterium-mediated transient transformation of tobacco was conducted on fully expanded leaves attached to 8–10 weeks-old intact plants. Bacterial suspension prepared as reported by Bhaskar et al.36 was infiltrated into intercellular spaces of intact leaves using a 1 ml syringe. Tagged agroinfiltrated leaves were left for 72 h in the greenhouse at 28 °C with 16 h light period.
Histochemical detection of GUS activity
The agroinfiltrated leaves were sacrificed for histochemical detection of GUS activity that was used as a reporter for expression of transgenes. They were incubated with substrate solution (0.08% w/v 5-bromo-4-chloro-3-indolyl-b-Dglucuronic acid (X-Gluc; sigma) in 100 mM Na2HPO4/KH2PO4 pH 7.0, 0.01% w/v Chloramphenicol, 20% Triton X-100, 20% Methanol) at 37 °C for 16 h. The GUS expression was detected in the form of blue precipitates in agroinfiltrated leaves under fluorescent microscope that were produced as a result of enzyme mediated hydrolysis of X-Gluc.
Genetic transformation and regeneration of transformants
Four sugarcane cultivars SPF-213, SPF-234, HSF-240 and CPF-246 were received from Agricultural Research Institute (AARI), Faisalabad, Pakistan. Genetic transformation of these lines was done using the biolistic transformation method. Tungsten particles were used as microprojectiles and were prepared as described Qamar et al.35. The sugarcane transformation was proceeded by following the protocol described by Nasir et al.37.
Molecular characterization of transgenic lines
GUS assay was performed on young shoots to detect expression of transgene during early events of transgenesis. Subsequently potential transgenic lines (V0 as well as V1 generation) were further screened for integration and expression of foreign genes by gene specific PCR, dipstick assay, southern blotting and enzyme linked immunosorbent assay (ELISA). After molecular characterization, qualitative assessment of transgenic plants of V0, V1 and V2 generation was performed via biotoxicity assay and Glyphosate spray assay.
Screening through polymerase chain reaction (PCR)
Genomic DNA was extracted from fresh leaves of transgenic lines using the method described by Edward et al.38. After qualitative as well as quantitative assessment of DNA, PCR reactions were set up using CEMB-CrylAc, CEMB-Cry2A and CEMB-GTGene specific primers. DNA from non-transformed plant was included as a negative control while plant expression constructs were included as positive controls. Each PCR reaction was carried out in a total volume of 20 µL having 100 ng of template DNA, 50 pm of each forward and reverse primers, 200 mM dNTPs and 1–2 units of Taq DNA polymerase. Success and specificity of reaction was checked by running 15 µL of each reaction on 1% agarose gel. Primer sequences are given in Supplementary material (Table 1).
Table 1.
Varietal responses of sugarcane cultivars for tissue culturing.
| Sr. no | Varieties | Callus induction (%) | Regenerated callus (%) | Shoot multiplication (%) |
|---|---|---|---|---|
| 1 | SPF-213 | 80 | 65 | 80 |
| 2 | HSF-234 | 81 | 80 | 90 |
| 3 | CPF-240 | 68 | 62 | 81 |
| 4 | CPF-246 | 98 | 99 | 100 |
Southern blotting
Southern blot analysis was performed on PCR positive transformed lines to confirm stable integration. High quality genomic DNA was isolated from leaf tissues using plant phytopure DNA extraction kit (Amersham) following the manufacturer’s instructions. Approximately 20 µg of genomic DNA from each transgenic line along with control plants was double digested with KpnI & SacI for CEMB-GTGene, KpnI and HindIII for CEMB-Cry1Ac and CEMB-Cry2A. After complete digestion, each digestion mixture was resolved on 1% agarose gel at 15 V for 18 h. and transferred onto the nylon membrane (Hi-bond) using standard capillary transfer39. DIG labeled probe of each gene was prepared using Decanucleotide Biotin labeling Kit (Cat # Ko561) according to the manufacturer’s instructions and then hybridization and detection was done using Cat# K0562.
Dipstick assay
Total protein was extracted from fresh leaves of transgenic plants and then quantified as described by Qamar et al.35. The GTGene, Cry1Ac and Cry2A expression was detected with EnviroLogix® dipstick kits (Cat # AS011LSS; Cat # AS003 # CTLSS and Cat # AS005 LSS) following the manufacturer’s instructions. Dipsticks were coated with monoclonal antibodies IgG of purified EPSPS, Cry1Ac and Cry2A respectively. The antibody coated dipsticks were dipped in transgenic plants protein samples and incubated at room temperature for 15 min.
Enzyme linked immunosorbant assay (ELISA)
Protein expressions of CEMB-Cry1Ac, CEMB-Cry2A, and CEMB-GTGene were quantified in sugarcane positive lines through ELISA. Total crude protein from fresh leaves was used in ELISA. Total crude protein was extracted using protein extraction buffer (10% Glycerol, 40 mM EDTA (pH 7.5), 100 mM NaCl, 10 mM Tris, 100 mM NH4Cl, 20 mM DTT, 2 mM PMSF) and then quantified by Bradford assay40. ELISA was performed according to the instruction manual of Envirologix Kit (Portland USA; Cat # 051) and its results were quantified using Micro Plate ELISA Reader Model ELx800. Serial dilutions of the calibrator were used to generate standard curve by plotting OD450 of each dilution on Y-axis and corresponding concentration on X-axis. The concentrations of trans-proteins were determined by finding their OD450 values on the curve and relating them to the corresponding concentration on X-axis.
Leaf biotoxicity assay
Leaf biotoxicity assay was used to demonstrate toxic effect of Cry1Ac and Cry2A encoded endotoxins on larvae of shoot borer chilo infuscatellus. Leaves from transgenic as well as control plants at the age of 20, 40, 60 and 80 days were used for the biotoxicity assays. Leaves from each plant were used to feed three 2nd instar larvae in a petri dish. Each experiment was performed in triplicates. After 3 days, rate of mortality was measured with reference to control.
Glyphosate spray assay
Glyphosate spray (Roundup Ready Dupont Staple LX Plus USA) with active ingredient [(Pyrithiobac sodium, Sodium-2-Chloro-6(4,6 dimethoxy pyrimidine-2-yl) thio]benzoate) assay was used for physiological evaluation of CEMB-GTGene (modified CP4 EPSPS) encoded herbicide resistant protein. Glyphosate was sprayed at the optimized rate of 1200 mL/80 L H2O/acre on the sugarcane transgenic plants of V0 as well as V1 and V2 generations during field study. Same concentration of spray was also used on non-transgenic plants along with weeds present in the sugarcane field.
Statistical analysis
Analysis of variance (ANOVA), least significant difference test (LSD), and Dunnett’s test were performed for comparison of significantly different mean values of different lines between the control plants and transgenic lines.
Results
Restriction digestion for the confirmation of sugarcane codon optimized synthetic BT. genes (pUC-57 vector)
The synthetic genes received in pUC-57 vector and confirmed through the Restriction Digestion and protein expressions analyses. Before cloning in plant expression vector (pCAMBIA-1301), CEMB-Cry1Ac, CEMB-Cry2A genes were analyzed through the restriction digestion by using Sph1 sites that released the fragments of 1.8 Kb and 1.9 Kb respectively. Restriction digestion with Sph-I also confirmed a fragment of 2.1 Kb for the Ubiquitin promoter. The undigested transformed vector (pUC-57) also shown in the (Supplementary Figure 2; Supplementary Tables 2 and 3).
Confirmation of His-tagged protein expressions (CEMB-Cry1Ac + CEMB-Cry2A, CEMB-GTGene) through Western Blot analysis
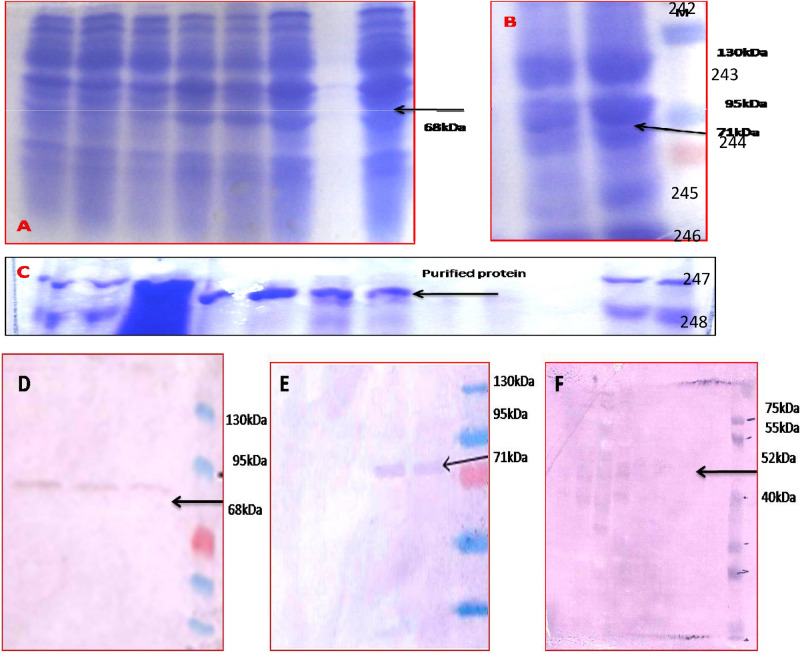
For the confirmation of protein expressions, pET-30 protein expression vector was constructed with CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene modified synthetic genes with KpnI, HindIII and SacI restriction sites. Physical mapping is drawn in the (Fig. 1A–C). The constructed plasmid vectors were transformed in Rosetta cells (E. coli) and analyzed on 10% SDS–Polyacrylamide gel (PAGE) stained with Coomassie blue. His-tagged proteins of CEMB-Cry1Ac (68 kDa), CEMB-Cry2A (71 kDa) and CEMB-GTG (52 kDa) were confirmed through Western Blot analysis (Fig. 1D–F) on nitrocellulose membranes (HyBond C, Amersham) through His-specific primary antibodies and AP-conjugated (Alkaline Phosphatase) secondary antibodies. For color development BCIP/NBT substrate tablets were used.
Figure 1.
Protein expressions analyzed through SDS-PAGE and confirmed by Western Blot analysis. (A,B) SDS-PAGE of induced crude (1 mM-IPTG) proteins of CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene genes. (C) Purified proteins for Western Blot analysis. (D) Western Blot Analysis for CEMB-Cry1Ac protein, Lanes 1–3: CEMB-Cry1Ac protein (68 kDa), Lane M. Pre-strained protein ladder (Fermentas). (E) Western Blot Analysis for CEMB-Cry2Ac protein, Lanes 1–2: CEMB-Cry1Ac protein (71 kDa), Lane M: Pre-strained protein ladder (Fermentas). (F) Western Blot Analysis for CEMB-GTG protein, Lanes 1–4 CEMB-GTGene protein (54 kDa), Lane M. Pre-strained protein ladder (Fermentas).
Cloning of CEMB-Cry1Ac, CEMB-Cry2A, CEMB-GTGene in plant expression vector (pCAMBIA 1301)
After successful protein expressions of three genes through pET-30 vector, for transformation, two final expression plasmids pCEMB-SC12 and pCEMB-SGTG were constructed to tailor the sugarcane with glyphosate tolerance, insect resistance and GUS as a reporter. The three synthetic genes were expressed from Maize Ubiquitin-1 for constitutive expression and NOS terminator for the stability of mRNA. CEMB-Cry1Ac + CEMB-Cry2A and CEMB-GTG expression cassettes were cloned in plant expression vector (pCAMBIA-1301) with the help of KpnI, SacI, SphI, and HindIII diagnostic sites. Restriction analysis of the final plant expression constructs confirmed the cloning of CEMB-Cry1Ac (4 kb), CEMB-Cry2A (4 kb) and CEMB-GTGene (1.4) (Supplementary Figure 3A,B). After confirmation, maxi-prep of the plasmids was done (Supplementary Figure 3C).
Transient expression of transgene (GUS) to determine the efficacy of plasmid constructs
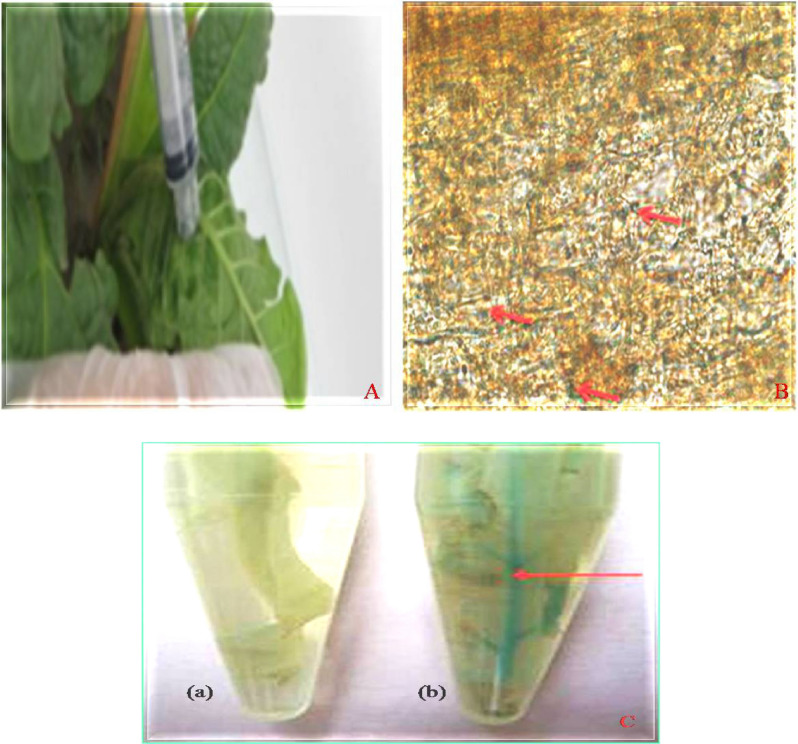
For the transient expression of GUS trasngnene in tobacco leaves, first of all these plant expression plasmids constructs pCEMB-SC12 and pCEMB-SGTG were introduced into Agrobacterium tumefaciens (LBA4404) by electroporation. The positive colonies of these constructs from test cultures were screened through colony PCR (Supplementary Figure 4A–C). The screened positive colonies were used for Agro-infiltration. The leaves were analyzed after 72 h. Under fluorescent micro scope (Olympus Szx7) bluish green color confirmed the GUS-transient expression and expression efficiency of the constructs (Fig. 2A–C).
Figure 2.
Through syringe method Agro-infiltrated Gus expression was efficiently observed in tobacco leaves. (A) Agroinfiltrated tobacco leaves. (B) Bluish-green color under fluorescent microscope. (C) Visual GUS expression in Tobacco leaf (a): Non-Agroinfiltrated tobacco leaves (negative control) (b): Agroinfiltrated tobacco leaves predicting bluish-green color.
Selection of sugarcane variety for transformation and field study
In this study, SPF-213, SPF-234, HSF-240 and CPF-246 sugarcane varieties were selected on the basis of their cane yield (t/ha) and sugar recovery (%) for sugarcane tissue culturing and transformation (Supplementary Figure 5), however, after optimization of glyphosate spray assays the best responded variety was selected for further field studies.
Tissue culture response of sugarcane cultivars
Diverse responses under tissue culture and regeneration of four varieties were noted. Four combinations for best callus induction media were used to obtain maximum embryogenic calli from these varieties. The MS Media 4.43 g/L, sucrose 30 g/L and phytagel 3 g/L were constant ingredients of every media combination. The maximum regeneration ability of SPF-234 (90%) was observed on media (RM-4) containing Kinetein (3 mg/L), BAP (3 mg/L), charcoal (1 g/L), Myo-inositol (200 mg/L). The maximum shoot multiplication percentage of regenerated calli for CPF-246 (100%), SPF-234 (90%), SPF-213 (90%), and HSF-240 (81%), were recorded on media (SMM-2) consisting of Kinetin (2 mg/L), BAP (2 mg/L), GA3 (2 mg/L), IAA (1 mg/L) and charcoal (1 g/L). The sugarcane plants after multiplication were shifted on rooting media. The half strength media with NAA (1 mg/L) was used for rooting. These, one and a half month old rooted plants were acclimatized in the soil pots under green house conditions. From these selected sugarcane varieties CPF-246 revealed maximum regeneration and shoot multiplication (Supplementary Figure 6; Supplementary Tables 4 and 5).
Biolistic transformation in sugarcane and selection of transgenic plants
Tungsten particles coated with pCEMB-SC12 and CEMB-GTGene plasmid constructs were used for Biolistic transformation. The CEMB-CrylAc + CEMB-Cry2A and CEMB-GTG genes driven by Maize Ubiquitin-1 promoter with kanamycin as selection marker, were successfully transformed into four sugarcane varieties SPF-213, SPF-234, HSF-240, and CPF-246 by using CEMB homemade gene gun (Fig. 3). A total of 400 calli were used for the transformation process (Table 2). The transformed calli and regenerated transgenic sugarcane plants with these three genes were screened first on selection medium with only Kanamycine (50 mg/L), the response of resistant calli of four varieties SPF-213, SPF-234, HSF-240, and CPF-246 was as 70%, 74%, 45% and 91% respectively, while on double selection medium (kanamycine and glyphosate) 34% of SPF-213, 40% of SPF-234, 29% of HSF-240 and 81% of CPF-246 transformed calli survived (Supplementary Table 1). After shoot multiplication 1000 plantlets for each variety were obtained. From molecular analyses, PCR with glyphosate gene specific primers was carried out for this initial stage screening of these multiplied transgenic putative plants. From this screening survived plant shifted on rooting media were as 656, 680, 205, and 900 of CPF-213, CPF-234, HSF-240 and CPF-246 respectively. From PCR, Southern Blot, Dipstick and ELISA analyses 14 plants of CPF-213, 15 of CPF-234, 11 of HSF-240, and 30 of CPF-246 were positive for CEMB-Cry1Ac and 11 plants of CPF-213, 13 of CPF-234, 10 of HSF-240, and 15 of CPF-246 were positive for CEMB-Cry2A genes. For glyphosate tolerant gene, 27 plants of CPF-213, 39 of CPF-234, 15 of HSF-240, and 66 of CPF-246 were resulted positive through these analyses. Through these molecular analyses, 11 plants of CPF-213, 13 of CPF-234, 10 of HSF-240, and 15 of CPF-246 were considered three genes positive for protein expressions (Table 2). Stable putative transgenic sugarcane plants containing CEMB-Cry1Ac, CEMB-GTGene and CEMB-GTG genes were selected for further generation’s study. Finally, 15 lines of CPF-246 were selected for further generation’s studies under field conditions. These lines were further analyzed through molecular techniques to check the consistent trans-protein expression of the transformed genes in the next generations. For CPF-246, 66 transgenic plantlets were obtained after three screenings of glyphosate spray assays (2250 mL/80 L/hectare, 2750 mL/80 L/hectare, 3000 mL/80 L/hectare). Overall transformation efficiency of CPF-246 with respect to three genes protein expressions was 1.5%.
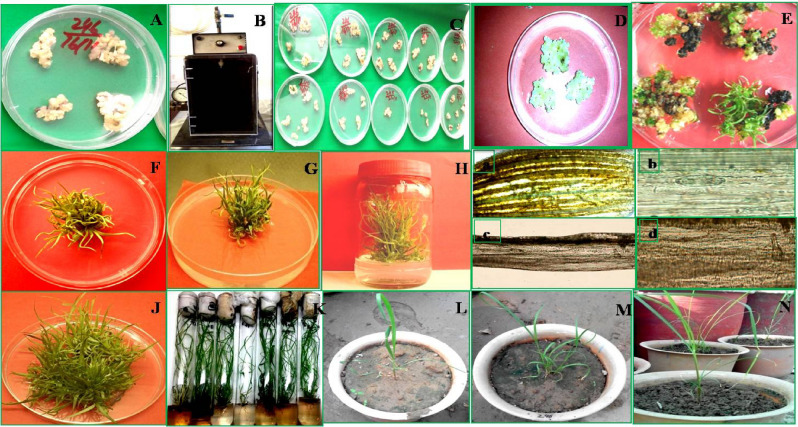
Figure 3.
Schematic presentation of all the steps involved in genetic modification of sugarcane. (A) Callus for Bombardment. (B) Homemade Biolistic machine. (C) Bombarded Callus after bombardment with DNA-coated tungsten particles. (D) Bombarded callus shifted on selection media with Kanamycime (50 mg/L) after 2 days. (E,F) Transformed callus regenerated on double selection (Kanamycine 50 mg/L + Glyphosate 40 mM) media, (G,H) Regenerated sugarcane plantlets on glyphosate selection media (45 mM), shifting on shoot multiplication media with Kanamycime (50 mg/L) and glyphosate (50 mM) selections. (I) Gus Assay for transgenic plant screening (abcd). (J) Transgenic plants for rooting. (K) Shifting on rooting media without any selection drug. (L,M) Acclimatization: Transgenic sugarcane plantlets in soil pots under green house conditions.
Table 2.
Biolistic transformation in sugarcane and selection of transgenic plants.
| Variety | Total no of calli bombarded | No of calli survived on kanamycine selection (%) | No of calli survived on kanamycine + glyphosate selection (%) | No of calli regenerated on selection medium (%) | Regenerated plantlets on selection media | Plantlets after GUS assay screening multiplied in the tubes | PCR (GTGene) screening and in the process of acclimatization | In tubes on rooting media | Shifted to green house in soil pots | Survived (one and half month old plants) after spray 900 mL/ 80 L/acre | In field | 2-month old plants after glyphosate spray 1100 mL/80 L | Spray tolerant (1200 mL/80 L/acr) plants further analyzed for three genes through PCR, Southern Blot, DIPSTICK and ELISA | Positive for CEMB-Cry1Ac | Positive for CEMB-Cry2A | Positive for CEMB-GTGene | Three genes positive plants (transgenic plants survived in field after glyphosate spray (1200 mL/80 L/acre) | Transformation efficiency (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPF-213 | 100 | 70 | 34 | 21 | 500 | 1000 | 800 | 752 | 656 | 200 | 122 | 54 | 27 | 14 | 11 | 27 | 11 | 1.1 |
| SPF-234 | 100 | 74 | 40 | 32 | 600 | 1000 | 700 | 680 | 525 | 250 | 141 | 90 | 39 | 15 | 13 | 39 | 13 | 1.3 |
| HSF-240 | 100 | 45 | 29 | 13 | 300 | 1000 | 320 | 205 | 125 | 75 | 60 | 40 | 15 | 11 | 10 | 15 | 10 | 1 |
| CPF-246 | 100 | 91 | 81 | 48 | 950 | 1000 | 966 | 900 | 900 | 900 | 751 | 300 | 66 | 30 | 15 | 66 | 15 | 1.5 |
| Total | 147 | 71 | 58 | 147 | 49 | |||||||||||||
Screening of sugarcane putative transgenic plants (CPF-246) with glyphosate resistance
For the optimization of glyphosate resistance four selection pressures were given to the transformed callus as well as regenerated transgenic and control plantlets (Fig. 4). After, 48 h of bombardment the transformed calli were transferred on kanamycine (50 mg/L) selection media. Following one week, survived calli were shifted to the regeneration media (RM-1 and RM-2) containing Kanamycine 50 mg/L and Glyphosate 40 mM. After 2-weeks resistant transgenic plants were further multiplied on shoot multiplication media (SMM-2) contained glyphosate concentration (50 mM).
Figure 4.
Sugarcane putative transgenic plantlets (CPF-246) on glyphosate selection medium. (A) Control (Non-Transgenic plants) Kanamycine (50 mg/L). (B) Kanamycine (50 mg/L). (C) Kanamycine (50 mg/L) + Glyphosate (40 mM). (D) Kanamycine (50 mg/L) + Glyphosate (45 mM). (E) Kanamycine (50 mg/L) + Glyphosate (50 mM). (Fa) Resistant Plant on Kanamycine (50 mg/L) + Glyphosate (50 mM). (Fb) Resistant transgenic sugarcane pants multiplied on Kanamycin (50 mg/L) and Glyphosate (50 mM) media.
GUS transient expression for the screening of transgenic sugarcane plants
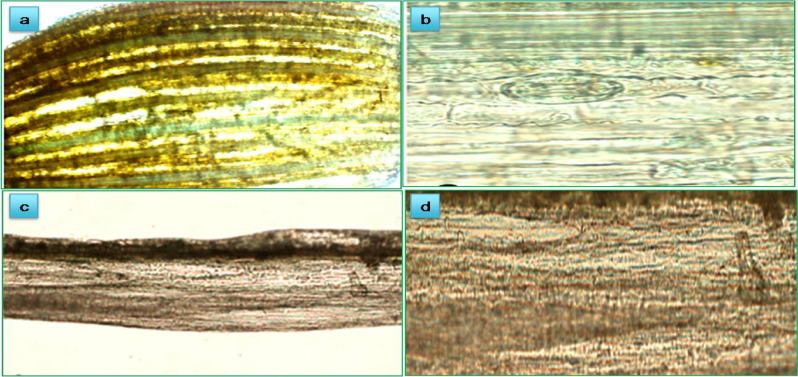
Plasmid constructs pCEMB-SC12 and pCEMB-SGTG was transformed in the sugarcane calli through Biolistic delivery system. These calli containing three transgenes and were regenerated on double selection pressure Kanamycine (50 mg/L) and Glyphosate (45 mM). Young regenerating plants from the screened resistant calli were further selected through the GUS assay. The transgenic expression of GUS assay of reporter gene was done for early detection of transgenic events. After visual observation, GUS expression in leaves and stems is further confirmed by a fluorescent microscope, cells were showing uniform expression through its evenly distributed bluish green color in cross sections, depicting the success of whole transformation process (Fig. 5).
Figure 5.
GUS Assay of transgenic sugarcane plants by using fluorescent microscope. (a,b) Young transgenic leaf. (c,d) Immature transgenic stem portion.
Molecular analyses of putative transgenic sugarcane plants at v0 generation
The putative transgenic sugarcane plants containing CEMB-Cry1Ac + CEMB-Cry2A and CEMB-GTGene were subjected to molecular analyses i.e. PCR, Southern DNA Hybridization Dipstick assay and ELISA.
PCR screening of three genes CEMB-Cry1AC, CEMB-Cry2A and CEMB-GTG positive putative transgenic plants (V0)
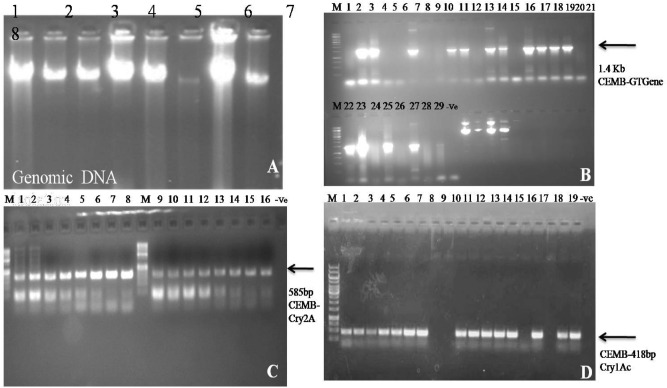
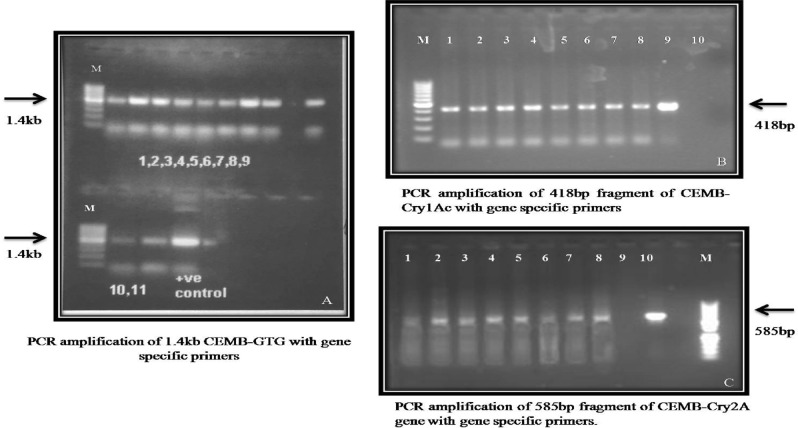
The glyphosate tolerant 15 putative transgenic plants of CPF-246 were designated as CPF-246 (2L/6), CPF-246 (2L/8),V- (3L/3), CPF-246 (3L/4), CPF-246 (4L/7), CPF-246 (4L/8), CPF-246 (5L/1), CPF-246 (5L/5), CPF-246 (6L/2), CPF-246 (6L/5), CPF-246 (7L/2), CPF-246 (7L/7), CPF-246 (L8/4),and CPF-246(L9L/6).These putative transgenic plants were positive for CEMB-Cry1Ac, CEMB-Cry2A, and GTGene genes as confirmed through PCR by using gene specific primers during screening process. The amplified products of 1.4 kb for CEMB-GTGene, 418 bp for CEMB-Cry1Ac and 580 bp for CEMB-Cry2A confirmed the integration of three genes in the genome so referred these plants for further molecular analyses. No amplification was found in non transgenic plants (Fig. 6A–D).
Figure 6.
PCR screened positive plants of CPF-246 plant containing CEMB-GTGene, CEMB-Cry2A and CEMB-Cry1Ac. (A) Genomic DNA isolated from putative sugarcane plants. (B) Lane M: 1 kb DNA Ladder, Lane 1–28: Putative transgenic sugarcane plants with CEMB-GTGene Lane 29: Positive control, Lane −ve: Non-transgenic plant (control plant). (C) Lane M: 1 kb DNA Ladder, Lane 1: Positive control, Lane 2–16: Putative transgenic sugarcane plants with CEMB-Cry1Ac Lane −ve: Non-transgenic plant (control plant). (D) Lane M: 1 kb DNA Ladder, Lane 1: Positive control Ladder, Lane 2–19: Screening of Putative transgenic sugarcane plants with CEMB-Cry2Ac, Lane −ve: Non-transgenic plant (control plant).
Southern Blot analysis
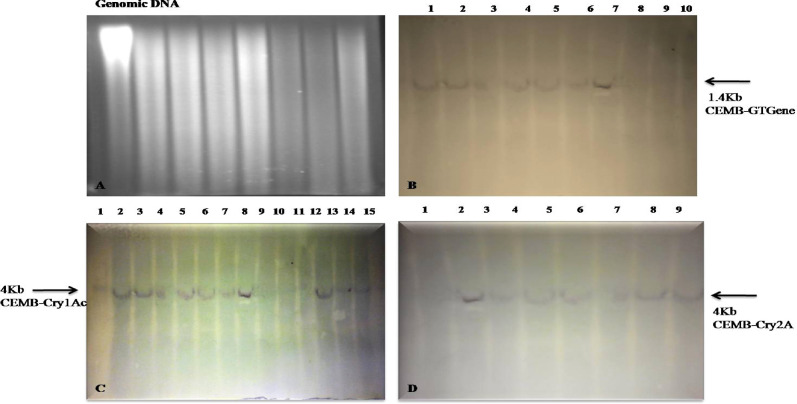
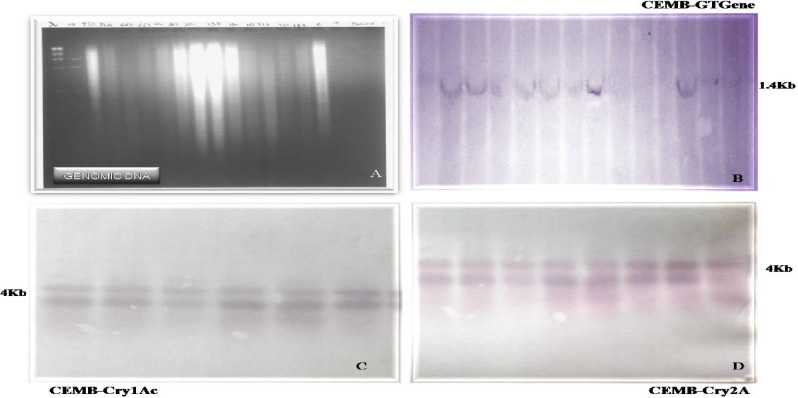
The stable integration of three genes CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene in the putative transgenic sugarcane plants was confirmed by the Southern Blot analysis through probe binding and its succeeding detection with the substrate. The PCR positive putative transgenic plants CPF-246 (2L/6), CPF-246 (2L/8), V- (3L/3), CPF-246 (3L/4), CPF-246 (4L/7), CPF-246 (4L/8), CPF-246 (5L/1), CPF-246 (5L/5), CPF-246 (6L/2), CPF-246 (6L/5), CPF-246 (7L/2), CPF-246 (7L/7), CPF-246 (L8/4), and CPF-246 (L9L/6) were analyzed for gene integration. Their respective probes highlighted the integration of the transgenes when the plant genomic DNA was digested with enzymes, KpnI-SacI for CEMB-GTGene, KpnI and HindIII for CEMB-Cry1Ac and CEMB-Cry2A and hybridized with probes. This hybridization with the specific probes confirmed the stable integrity of all three transgenes in the genome of these 15 sugarcane plants. The CEMB-GTGene with 1.4 kb, CEMB-Cry1Ac with 4 kb and CEMB-Cry2A with 4 kb fragments (full cassettes) were revealed by Southern Blot analysis (Fig. 7A–D).
Figure 7.
Southern Blot analysis of the transgenic sugarcane plants (CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene) in V0-generation. (A) Denaturing of genomic DNA. (B) Lane 1: Positive control, Lane 2–14: Southern DNA hybridization of transgenic sugarcane plants with CEMB-GTGene probe. (C) Lane 1: Positive control, Lane 2–15: Southern DNA hybridization of transgenic sugarcane plants with CEMB-Cry1Ac probe. (D) Lane 1: Positive control, Lane 2–9: Southern DNA hybridization of transgenic sugarcane plants with CEMB-Cry2A probe.
Dipstick assay
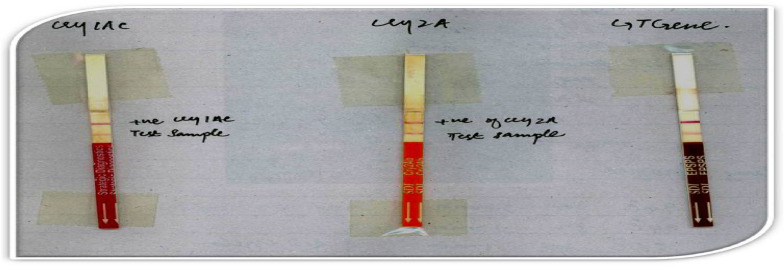
All the 15 putative transgenic plants CPF-246 (2L/6), CPF-246 (2L/8),V- (3L/3), CPF-246 (3L/4), CPF-246 (4L/7), CPF-246 (4L/8), CPF-246 (5L/1), CPF-246 (5L/5), CPF-246 (6L/2), CPF-246 (6L/5), CPF-246 (7L/2), CPF-246 (7L/7), CPF-246 (L8/4), and CPF-246 (L9L/6) were positive for Dipstick assay that confirmed the protein expressions of the CEMB-Cry1Ac,CEMB-Cry2A and CEMB-GTGene transgenes into the genome of putative transgenic sugarcane plants (Fig. 8).
Figure 8.
ELISA-Dipstick assay for the expression confirmation of three trans-proteins CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene in V0-generation. Lane 1: ELISA-Dipstick Positive for CEMB-Cry1Ac transgenic plants. Lane 2: ELISA-Dipstick Positive for CEMB-Cry2A transgenic plants. Lane 3: ELISA-Dipstick Positive for CEMB-GTGene transgenic plants.
Enzyme linked immuno-sorbent assay (ELISA) in V0-generation
The ultimate objective of the sugarcane transformation experiments was to get maximum expression of these transgenes (CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene) in sugarcane. Enzyme linked Immuno-sorbant Assay was done to quantify the proteins for CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene. The total crude protein was extracted from under study plant leaves (Supplementary Figure 7; Supplementary Table 6). The proteins from putative transgenic sugarcane plants were coated in the ELISA plates. The detections of the respective transprotein from transformed plants were confirmed after specific antibodies with the appearance of yellow color. Quantification of CEMB-Bt. and CEMB-GTGene proteins of 15 clones was done through ELISA, maximum 0.475 µg/g protein of CEMB-Cry1Ac from fresh leaves protein was acquired, 0.567 µg/g CEMB-Cry2A of fresh leaves protein was obtained, while for CEMB-GTGene maximum 0.486 µg/g of total leaf protein was acquired as represented in (Supplementary Table 7; Supplementary Figures 8–10). Based on ELISA quantification, comparison between proteins of CEMB-Cry1Ac CEMB-Cry2A and CEMB-GTGene was also made between transgenic sugarcane plants of V0-generation (Supplementary Figures 10, 11).
Leaf bio-assay assay of the transgenic sugarcane
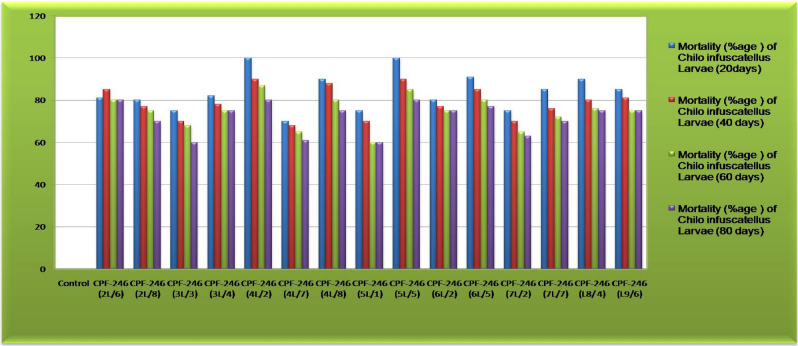
To interpret the efficacy of the CEMB-CrylAc, CEMB-Cry2A and CEMB-GTGene transproteins in transgenic sugarcane plant, immature leaf portions were fed to the larvae of Chilo infuscattellus at the plant age of 20, 40, 60 and 80 days in the Petri plates. The calculated mortality of the Chilo infuscattellus larvae ranged from 60 to 100% against transgenic sugarcane leaves, whereas in non-transgenic plants, all insects were active, alived and showed increased weight. The results showed that the transgenic sugarcane plants have such enough quantities of CEMB-Bt.toxins that can kill the cane borers even at initial stages of the plant growth (Figs. 9, 10, 11; Table 3).
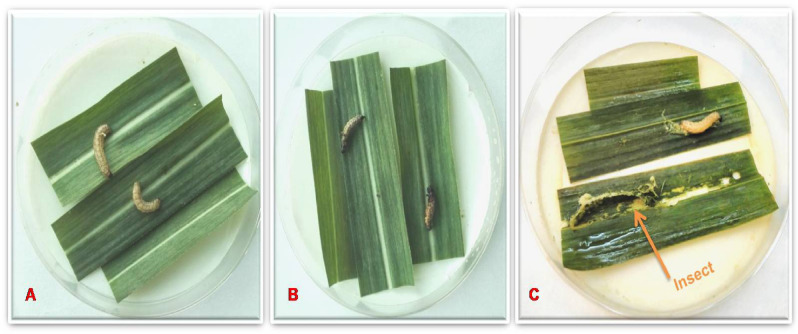
Figure 9.
Leaf bioassay of transgenic plant leaves and control sugarcane plants. Plate (A) Chilo infuscatellus from CEMB-insectray Lab. Plate (B) Non-transgenic sugarcane and transgenic plant leaves after 20 days. Plate (C) Chilo infuscatellus with transgenic sugarcane leaves. Plate (D,E) Dead cane borer, 3rd day of infestation. Plate (F) Chilo infuscatellus is alive and healthy with control; non transgenic sugarcane leaves.
Figure 10.
Leaf bioassay of 80 days old transgenic and non transgenic sugarcane. Plate (A) Transgenic sugarcane leaves killed Chilo infuscatellus larvae. Plate (B) Non-transgenic sugarcane plant. Chilo infustcatellus, alive and actively growing.
Figure 11.
Graphical presentation of morality percentages of Chilo infuscatellus against putative transgenic sugarcane plants at the plant age of 20, 40, 60 and 80 days in V0-generation.
Table 3.
Leaf bio-toxicity assay of transgenic sugarcane plants from V0-generation.
| Sugarcane plants | Leaf bio-assay, mortality percentage of Chilo infuscatellus larvae at different ages of transgenic plants (days) | |||
|---|---|---|---|---|
| 20 days | 40 days | 60 days | 80 days | |
| Control | 0 | 0 | 0 | 0 |
| CPF-246 (2L/6) | 81 | 85 | 80 | 80 |
| CPF-246 (2L/8) | 80 | 77 | 75 | 70 |
| CPF-246 (3L/3) | 75 | 70 | 68 | 60 |
| CPF-246 (3L/4) | 82 | 78 | 75 | 75 |
| CPF-246 (4L/2) | 100 | 90 | 87 | 80 |
| CPF-246 (4L/7) | 70 | 68 | 65 | 60 |
| CPF-246 (4L/8) | 90 | 88 | 80 | 75 |
| CPF-246 (5L/1) | 75 | 70 | 60 | 60 |
| CPF-246 (5L/5) | 100 | 90 | 85 | 80 |
| CPF-246 (6L/2) | 80 | 77 | 75 | 75 |
| CPF-246 (6L/5) | 91 | 85 | 80 | 77 |
| CPF-246 (7L/2) | 75 | 70 | 65 | 63 |
| CPF-246 (7L/7) | 85 | 76 | 72 | 70 |
| CPF-246 (L8/4) | 90 | 80 | 76 | 75 |
| CPF-246 (L9/6) | 85 | 81 | 75 | 75 |
Numbers in brackets are representing the event nos.
Glyphosate spray assay in V0-generation
Spray assays were carried out to optimize the tolerance level of transgenic sugarcane plant against glyphosate under open environmental conditions in V0-generation (1200 mL/80 L/acre). Finally, Roundup, sprayed at the proportion of 1200 ml/acre on the field plants. After 5–7 days, the weed/herbs and control plants that first shown necrotic patches, dead tissues ultimately resulted into complete withering of plant (100% mortality) (Fig. 12). The sugarcane transgenic plants CPF-246 (2L/6), CPF-246 (2L/8), V- (3L/3), CPF-246 (3L/4), CPF-246 (4L/7), CPF-246 (4L/8), CPF-246 (5L/1), CPF-246 (5L/5), CPF-246 (6L/2), CPF-246 (6L/5), CPF-246 (7L/2), CPF-246 (7L/7), CPF-246 (L8/4), and CPF-246 (L9L/6) were tolerant for glyphosate spray at the rate of 1200 mL/80 L/acre (Supplementary material Table 8).
Figure 12.
Glyphosate herbicide spray assay (1200 ml/acre) of sugarcane transgenic plants and Weed/non-transgenic (V0-genearation). (a) Transgenic sugarcane field plants before spray. (b) Non-transgenic plant and weeds died after glyphosate spray (1200 ml/80 L/acre), while survived transgenic sugarcane plants in the field after 5–7 days of spray.
Inheritance of CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene genes in sugarcane V1-generaton
These 15 putative transgenic sugarcane plants of CPPF-246, resistant against cane borers and tolerant for glyphosate herbicide spray assay (1200 ml/acre) were selected further for generation’s advancement (Table 4) and evaluation by molecular analyses like PCR with amplified products 418 for CEMB-Cry1Ac, 585 bp for CEMB-Cry2A and 1.4 kb for CEMB-GTGene (Fig. 15a–c), Southern Blot analysis (Fig. 16a–d), Dipstick assay and ELISA (Supplementary Figures 12–16) in V1-generation. Insect Bioassay and Glyphosate Herbicide Spray assay were also carried out (Supplementary material Table 9). Like Dunnett and Tamhane41, statistical analyses such as Analysis of Variance, least significant difference test (LSD) and Dunnett’s test were also applied for the evaluation of data from transgenic sugarcane plants in V1-generation. Fifteen lines each with 10 replicates (nodes) (15 × 10), total 150 transgenic plants in V1-generation were field studied. Initially randomly selected plants during field study were analyzed from V1-generation. All lines were highly cane borer (Chilo infuscatellus) resistant and efficiently glyphosate tolerant.
Table 4.
Summary of molecular analysis of randomly selected transgenic sugarcane plants (V1-generation).
| Transgenic lines of sugarcane (CPF-246) | PCR | Southern Blot | Dipstick assay | ELISA (µg/g) | Glyphosate spray assay (1200 mL/acre) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTG | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTG | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTG | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTG | ||
| CPF-246 (2L/6-3) 1 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.490 | 0.411 | 0.451 | Tolerant |
| CPF-246 (2L/8-5) 2 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.410 | 0.480 | 0.432 | Tolerant |
| CPF-246 (3L/3-8) 3 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.510 | 0.441 | 0.421 | Tolerant |
| CPF-246 (3L/4-4) 4 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.464 | 0.542 | 0.434 | Tolerant |
| CPF-246 (4L/2-7) 5 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.672 | 0.642 | 0.569 | Tolerant |
| CPF-246 (4L/7-1) 6 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.456 | 0.467 | 0.459 | Tolerant |
| CPF-246 (4L/8-4) 7 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.552 | 0.560 | 0.454 | Tolerant |
| CPF-246 (5L/1-7) 8 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.345 | 0.423 | 0.458 | Tolerant |
| CPF-246 (5L/5-2) 9 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.678 | 0.599 | 0.578 | Tolerant |
| CPF-246 (6L/2-10) 10 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.458 | 0.523 | 0.455 | Tolerant |
| CPF-246 (6L/5-3) 11 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.687 | 0.611 | 0.589 | Tolerant |
| CPF-246 (7L/2-1) 12 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.489 | 0.478 | 0.458 | Tolerant |
| CPF-246 (7L/7-7) 13 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.521 | 0.455 | 0.404 | Tolerant |
| CPF-246 (L8/4-10) 14 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.642 | 0.648 | 0.551 | Tolerant |
| CPF-246 (L9/6-9) 15 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.652 | 0.622 | 0.564 | Tolerant |
Bold values indicate higher protein concentration
Figure 15.
PCR amplified transgenic plants of CPF-246 for CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene. (A) Lane M: 1 kb DNA Ladder, Lane 1–11: Putative transgenic sugarcane plants with CEMB-GTGene (1.4 kb) Lane + ve: Positive control Lane 13: Non-transgenic plant (control plant). (B) Lane M: 1 kb DNA Ladder Lane 1–8: Putative transgenic sugarcane plants with CEMB-Cry1Ac (418 bp) Lane 9: Positive control Lane 10: Non-transgenic plant (control plant). (C) Lane M: 1 kb DNA Ladder Lane 1–11: 1–8 Putative transgenic sugarcane plants with CEMB-Cry2A (585 bp) Lane 9: Non-transgenic plant (control plant), Lane 10: Positive control.
Figure 16.
Southern Blot analysis of the transgenic sugarcane plants (CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene) in V1-generation. (A) Denaturing genomic DNA. (B) Southern DNA hybridization of transgenic sugarcane plants with CEMB-GTGene probe. (C) Southern DNA hybridization of transgenic sugarcane plants with CEMB-Cry1Ac probe. (D) Southern DNA hybridization of transgenic sugarcane plants with CEMB-Cry2A probe.
Inheritance of CEMB-Cry1Ac, CEMB-Cry2Ac and CEMB-GTGene genes in sugarcane lines of V2-generation
The transgenic sugarcane (CPF-246) lines 4L/2, 5L/5, 6L/5, L8/4, and L9/6 were screened from V1-generation (Fig. 13) on the basis of three selection criteria, (1) overall transgenes expressions (CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene, (2) leaf Bio-toxicity (mortality %age) and (3) Glyphosate herbicide spray assays, for the generation advancement as V2-generation in the field (Fig. 14). Five from 15 V1-generation free of any kind insect attack (Stem, Root and Top borers in the field) and completely tolerant to glyphosate spray were screened as V2-generation for further molecular analyses and field expression studies. The highest CEMB-Cry1Ac and CEMB-Cry2A expression was found 0.680 (µg/g) and 0.656 (µg/g) respectively in sugarcane transgenic line (4L/2-9) of V2-generation (Tables 5, 6, 7, 8, 9). The highest dose of CEMB-GTGene 0.595 (µg/g) was noted in line L6/5-4 from V2-generation. All the transgenic sugarcane lines (Table 10) showed 100% cane borer (Chilo infuscatellus) mortality and efficiently complete tolerance against glyphosate application 3000 mL/ha (Figs. 15, 16, Table 10).
Figure 13.
Glyphosate tolerant and cane borer resistant lines of V1-generation during the field studies.
Figure 14.
Five sugarcane transgenic lines from 15 V1-generation free of any kind of insect attack and tolerant to glyphosate spray 3000 mL/ha) were screened as V2-generation.
Table 5.
Molecular analysis of CPF-246 (L4/2-9) transgenic sugarcane plants from V2-generation.
| Serial no | PCR | Southern Blot analysis | Dipstick assay | ELISA (µg/g) | Glyphosate spray assay (1200 m/acre) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transgenic lines of sugarcane CPF-246 | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | |
| CPF-246 (4L/2-9) 1 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.675 | 0.645 | 0.565 | Tolerant |
| CPF-246 (4L/2-9) 2 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.677 | 0.641 | 0.561 | Tolerant |
| CPF-246 (4L/2-9) 3 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.598 | 0.596 | 0.551 | Tolerant |
| CPF-246 (4L/2-9) 4 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.645 | 0.644 | 0.560 | Tolerant |
| CPF-246 (4L/2-9) 5 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.678 | 0.656 | 0.562 | Tolerant |
| CPF-246 (4L/2-9) 6 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.670 | 0.651 | 0.567 | Tolerant |
| CPF-246 (4L/2-9) 7 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.671 | 0.655 | 0.570 | Tolerant |
| CPF-246 (4L/2-9) 8 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.612 | 0.634 | 0.559 | Tolerant |
| CPF-246 (4L/2-9) 9 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.615 | 0.629 | 0.545 | Tolerant |
| CPF-246 (4L/2-9) 10 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.679 | 0.642 | 0.568 | Tolerant |
Table 6.
Molecular analysis of CPF-246 (L5/5-8) transgenic sugarcane plants from V2-generation.
| Serial no | PCR | Southern Blot analysis | Dipstick asaay | ELISA (µg/g) | Glyphosate spray assay (1200 m/acre) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transgenic lines of sugarcane CPF-246 | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | |
| CPF-246 (5L/5-8) 1 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.680 | 0.598 | 0.577 | Tolerant |
| CPF-246 (5L/5-8) 2 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.670 | 0.590 | 0.560 | Tolerant |
| CPF-246 (5L/5-8) 3 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.676 | 0.595 | 0.575 | Tolerant |
| CPF-246 (5L/5-8) 4 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.678 | 0.601 | 0.572 | Tolerant |
| CPF-246 (5L/5-8) 5 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.674 | 0.596 | 0.580 | Tolerant |
| CPF-246 (5L/5-8) 6 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.670 | 0.590 | 0.578 | Tolerant |
| CPF-246 (5L/5-8) 7 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.665 | 0.580 | 0.581 | Tolerant |
| CPF-246 (5L/5-8) 8 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.673 | 0.598 | 0.572 | Tolerant |
| CPF-246 (5L/5-8) 9 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.675 | 0.591 | 0.570 | Tolerant |
| CPF-246 (5L/5-8) 10 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.678 | 0.588 | 0.575 | Tolerant |
Table 7.
Molecular analysis of CPF-246 (L6/5-4) transgenic sugarcane plants from V2-generation.
| Serial no | PCR | Southern Blot analysis | Dipstick assay | ELISA (µg/g) | Glyphosate spray assay (1200 m/acre) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transgenic lines of sugarcane CPF-246 | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | |
| CPF-246 (6L/5-4) 1 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.685 | 0.605 | 0.589 | Tolerant |
| CPF-246 (6L/5-4) 2 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.680 | 0.598 | 0.590 | Tolerant |
| CPF-246 (6L/5-4) 3 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.681 | 0.596 | 0.585 | Tolerant |
| CPF-246 (6L/5-4) 4 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.680 | 0.601 | 0.591 | Tolerant |
| CPF-246 (6L/5-4) 5 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.682 | 0.591 | 0.595 | Tolerant |
| CPF-246 (6L/5-4) 6 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.679 | 0.595 | 0.586 | Tolerant |
| CPF-246 (6L/5-4) 7 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.680 | 0.599 | 0.591 | Tolerant |
| CPF-246 (6L/5-4) 8 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.677 | 0.596 | 0.594 | Tolerant |
| CPF-246 (6L/5-4) 9 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.621 | 0.610 | 0.588 | Tolerant |
| CPF-246 (6L/5-4) 10 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.678 | 0.611 | 0.585 | Tolerant |
Table 8.
Molecular analysis of CPF-246 (L8/4-3) transgenic sugarcane plants from V2-generation.
| Serial no | PCR | Southern Blot analysis | Dipstick assay | ELISA (µg/g) | Glyphosate spray assay (1200 ml/acre) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transgenic lines of sugarcane CPF-246 | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | |
| CPF-246 (L8/4-3) 1 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.645 | 0.640 | 0.551 | Tolerant |
| CPF-246 (L8/4-3) 2 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.640 | 0.644 | 0.542 | Tolerant |
| CPF-246 (L8/4-3) 3 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.641 | 0.642 | 0.534 | Tolerant |
| CPF-246 (L8/4-3) 4 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.640 | 0.622 | 0.538 | Tolerant |
| CPF-246 (L8/4-3) 5 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.630 | 0.635 | 0.525 | Tolerant |
| CPF-246 (L8/4-3) 6 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.638 | 0.635 | 0.535 | Tolerant |
| CPF-246 (L8/4-3) 7 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.642 | 0.632 | 0.552 | Tolerant |
| CPF-246 (L8/4-3) 8 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.635 | 0.638 | 0.551 | Tolerant |
| CPF-246 (L8/4-3) 9 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.635 | 0.640 | 0.540 | Tolerant |
| CPF-246 (L8/4) 10 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.641 | 0.646 | 0.550 | Tolerant |
Table 9.
Molecular analysis of CPF-246 (L9/6-10) transgenic sugarcane plants from V2-generation.
| Serial no | PCR | SOUTHERN | DIPSTICK | ELISA (µg/g) | Glyphosate spray assay (1200 m/acre) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transgenic lines of sugarcane CPF-246 | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | CEMB-Cry1Ac | CEMB-Cry2A | CEMB-GTGene | |
| CPF-246 (L9/6-10) 1 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.648 | 0.620 | 0.565 | Tolerant |
| CPF-246 (L9/6-10) 2 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.646 | 0.611 | 0.561 | Tolerant |
| CPF-246 (L9/6-10) 3 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.644 | 0.603 | 0.558 | Tolerant |
| CPF-246 (L9/6-10) 4 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.650 | 0.599 | 0.555 | Tolerant |
| CPF-246 (L9/6-10) 5 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.640 | 0.598 | 0.560 | Tolerant |
| CPF-246 (L9/6-10) 6 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.645 | 0.599 | 0.559 | Tolerant |
| CPF-246 (L9/6-10) 7 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.641 | 0.610 | 0.556 | Tolerant |
| CPF-246 (L9/6-10) 8 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.639 | 0.596 | 0.567 | Tolerant |
| CPF-246 (L9/6-10) 9 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.633 | 0.613 | 0.560 | Tolerant |
| CPF-246 (L9/6-10) 10 | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | +VE | 0.640 | 0.615 | 0.562 | Tolerant |
Table 10.
Level of pest control and dose assessment of CEMB-Cry1Ac, CEMB Cry2A (µg/g) and CEMB-GTGene (µg/g) from five transgenic sugarcane lines in V2-generation.
| Serial no | Level of pest control and dose assessment at the plant age of 80 days | Glyphosate spray assay (3000 mL/hectare) | ELISA CEMB-GTGene dose assessment (µg/g) |
||
|---|---|---|---|---|---|
| Transgenic lines of sugarcane CPF-246 | Mortality (%age) | ELISA | |||
| CEMB-Cry1Ac (µg/g) | CEMB-Cry2A (µg/g) | ||||
| 1-CPF-246 (4L/2-9) 5 | 100% | 0.678 | 0.656 | Tolerant | 0.562 |
| 2-CPF-246 (5L/5-8) 4 | 100% | 0.678 | 0.601 | Tolerant | 0.572 |
| 3-CPF-246 (5L/5-8) 8 | 100% | 0.673 | 0.598 | Tolerant | 0.572 |
| 4-CPF-246 (6L/5-4) 2 | 100% | 0.680 | 0.598 | Tolerant | 0.590 |
| 5-CPF-246 (6L/5-4) 4 | 100% | 0.680 | 0.601 | Tolerant | 0.591 |
| 6-CPF-246 (6L/5-4) 5 | 100% | 0.682 | 0.591 | Tolerant | 0.595 |
| 7-CPF-246 (6L/5-4) 7 | 100% | 0.680 | 0.599 | Tolerant | 0.591 |
| 8-CPF-246 (6L/5-4) 8 | 100% | 0.677 | 0.596 | Tolerant | 0.594 |
| 9-CPF-246 (L8/4-3) 3 | 100% | 0.641 | 0.642 | Tolerant | 0.534 |
| 10-CPF-246 (L8/4-3) 9 | 100% | 0.635 | 0.640 | Tolerant | 0.540 |
| 11-CPF-246 (L8/4-3) 10 | 100% | 0.641 | 0.646 | Tolerant | 0.550 |
| 12-CPF-246 (L9/6-10) 2 | 100% | 0.646 | 0.611 | Tolerant | 0.561 |
| 13-CPF-246 (L9/6-10) 4 | 100% | 0.650 | 0.599 | Tolerant | 0.555 |
| 14-CPF-246 (L9/6-10) 8 | 100% | 0.639 | 0.596 | Tolerant | 0.567 |
| 15-CPF-246 (L9/6-10) 10 | 100% | 0.640 | 0.615 | Tolerant | 0.562 |
Dipstick assay
Dipstick assay confirmed the protein expression of three genes CEMB-Cry1Ac, CEMB-Cry2Ac and CEMB-GTGene in the lines of V1-generation. The bright dark color bands on the dipstick showed higher protein expressions in these lines CPF-246(4L/2-7) CPF-246(5L/5-2), CPF-246(6L/5-3), CPF-246(L8/4-10), and CPF-246 (L9/6-9) of sugarcane transgenic plants that verified the stability or stable expression of these transgenes into the genome of V1-geneartion.
Enzyme linked immuno sorbent assay (ELISA)
PCR and Dipstick assay confirmed transgenic sugarcane plants were further evaluated by ELISA. ELISA was performed to check and quantify the level of protein expressions from CEMB-Cry1A, CEMB-Cry2A and CEMB-GTGene transgenes in the transgenic sugarcane leaves of V1-generation. Initially at randomly selected transgenic sugarcane (CPF-246) lines (2L/6)-3, (2L/6)-5, (3L/3)-8, (3L/4)-4, (4L/2)-7, (4L/7)-1, (4L/8)-4, (5L/1)-7, (5L/5)-2, (6L/2)-10, (6 6L/5)-3, (7L/2)-1, (7L/7)-7, (L8/4)-10, and (L9/6)-9 from field were analyzed for expression level of CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene genes. ELISA results demonstrated through plate reader Model-ELx800 confirmed that expressions of CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene proteins was much higher as compared to negative control and non transgenic sugarcane plants. Accordingly, V1-generation plants that were selected randomly showed almost stable quantities of these trans-proteins in plants (Table 4; Supplementary Figures 8–15). With reference to overall CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene proteins expression from all the transgenic lines, (4L/2)-7, (5L/5)-2, (6L/5)-3, (L8/4)-10, and (L9/6)-9 lines were distinct with respect to their quantified transproteins (Supplementary Figures 16, 17). These lines were highly insect resistant and glyphosate tolerant in their respective bioassays from V1 and V2-generations.
Leaf bio-toxicity assay of transgenic sugarcane plants in V1-generation
For further evaluation, these sugarcane transgenic lines were subjected to leaf bioassay against cane borer Chilo infuscatellus larvae (Fig. 17). Larvae of three different instars from Chilo infuscatellus were permitted, in three replicates, to feed on non-transgenic and transgenic plant leaves. The time intervals of 20, 40, 60 and 80 days were same as in-V0-generation after plantation in field soil. After 3 days of leaf bioassay, data was composed by counting the alive and dead insect larvae alongside assessing the leaf damages done by these larvae during this experimental time period (Supplementary material Table 9). Up to 70 to 100 (%) mortality of Chilo infuscatellus larvae was resulted from the expressed toxins of these transgenic sugarcane plants, according to variation in the toxins quantity which were observed from these lines during the course of time. Moreover, in control plates, the non transgenic leaves were totally damaged, eaten up and all the larvae were alived and healthy. Different statistical analyses such as Analysis of Variance (ANOVA), Least Significant Difference test (LSD) and Dunnett’s test were applied for toxicity-significance-analysis of transgenic sugarcane lines in comparison with non transgenic plant leaves. ANOVA showed that all the sugarcane transgenic lines significantly differ from the non transgenic plants and Dunnett’s (Supplementary material Table 9) results demonstrated that five (05) transgenic lines from V1-geneartion of CPF-246 are considerably differ from non-transgenic plants, as well as from other transgenic sugarcane lines (within the group) i.e. 4L/2, 5L/5,6L/5, L8/4 and L9/6 lines with respect to their toxicity expression against Chilo Infuscatellus insects.
Figure 17.
Leaf bioassay of transgenic sugarcane plant leaves and control plants in V1-generation. (A) Transgenic young leaf with Chilo infuscatellus. (B) Dead insects Chilo infuscatellus. (C) Control (non-transgenic plants) Chilo infuscatellus alived and healthy.
Discussion
High yielding is the ultimate objective of the crop production including sugarcane1,35,42,43. The major constraints to sugarcane are cane borer insects, weeds, drought stress and different viruses2,5,44. The present study was aimed to control the cane borers and weeds through the genetic modification of sugarcane with codon optimized insect resistant (CEMB-Cry1Ac + CEMB-Cry2A) and glyphosate tolerant genes (CEMB-GTGene).
The agro-climatic environment of the Pakistan is relatively encouraging for the sugarcane cultivation. In spite of worldwide area wise fifth position, Pakistan stands at fifteenth position in sugar production which is lowest yield per unit area. In monocots, a stable reproducible transformation method has been very important45,46 and in sugarcane44,47. In the current study, an effort was made for the development of an efficient procedure for its regeneration from calli and inbuilt resistance/tolerance in sugarcane transgenic plants against cane borers and glyphosate herbicide. For this purpose, four local elite varieties CPF-213, CPF-234, HSF-240, and CPF-246 were selected for establishing tissue culture experiments. Young immature leaves were used as explants for calli induction. Immature leaves were found excellent explants for embryogenic callus formation48, which is compulsory for genetic modification of sugarcane and also strengthen the genetic transformation procedure in the sugarcane49. The embryogenic calli of all the four varieties were obtained on a callus formation media containing 2, 4-D50,51. This media was supplemented with casein for the enhancement of embryogenic potential of sugarcane calli52,53. The calli were regenerated into micro-shoots, and after the multiplication of these micro-shoots, root intiation was started. These four varieties gave different responses for tissue culturing with reference to callogenesis and regeneration due to genotypic variability among cultivars37,54. It was also observed that increased concentration of 2,4-D, more than 4 mg/L, in callus induction media significantly adversely affected the regeneration efficiency of the calli. On increased concentrations, complete loss of the regeneration potential of these calli was noted. These results were consistent with the study by Chengalrayan et al.55, who described that fresh biomass of the calli reduced on adding upto 6 mg/L of 2,4-D in the media.
All the four varieties were critically judged/screened through tissue culture response2,50, and through transformation efficiency on the basis of calli survived (%) and calli regenerated (%) on selection media and finally through glyphosate field spray assays. After through screening, CPF-246 variety was selected for the next generation field studies. The results of tissue culture studies proved CPF-246 (callus induction 90%, regeneration 99% and multiplication 100%) as an excellent variety. Bakhsh et al.56 also selected varieties for genetic modification by means of regeneration response. Crop genetic modification through the introduction of insecticidal (Cry1Ac, Cry2A) and glyphosate tolerant (GTG) genes is more advantageous over conventional breeding techniques57. The transgenic sugarcane provides resistance against the cane borer pests with Bt. genes5,58. More than 100 Bt. genes have been sequenced which were highly dissimilar in amino-acid sequence and in their few biochemical properties as well. Studies proved that in these toxins’ combination, Cry1Ac with Cry2A is better against borer insects or lepidopteran insects59,60. The introduction of double Bt. genes Cry1Ac and Cry2A also represents the gene pyramiding which helps in creating multiple traits in one variety61.
Presently, a broad spectrum herbicide, glyphosate is the most commonly used herbicide. Glyphosate is a non-selective- herbicide that can stop the growth of all plants along with a wide range of herbs and weeds62. Glyphosate inhibits the synthesis of 5-enolpyuvyl-3-phosphoshikimate (EPSPS) during the shikimate metabolic pathway and subsequently stops the synthesizing of three very important amino acid tyrosine, tryptophan and phenylalanine63. Many crops for herbicide resistance has been developed formerly, including the non-glyphosate herbicide-tolerant transgenics with 2,4-D, Dicamba, HPPD inhibitors and bar transgenes64,65. However, all these crops were resistant against specific kind of herbicide, while transgenics with cp4EPSPS are not confined with single kind of herbicid27,66. Up till now, nine glyphosate-resistant crops have been introduced, comprising soybean, wheat, canola, polish canola, corn, alfalfa, sugar beet, creeping bent-grass and cotton17,37.
The modified sequences of CEMB for Bt. (CEMB-Cry1Ac + CEMB-Cry2A) and glyphosate tolerant (cp4EPSPS) genes were used for this transformation study. The nuclear encoded chimeric genes were used in preliminary efforts for the expression of Bt. toxins but the resulted expression level was very low67. It was supposed that Bt. genes are AT-rich in comparison with plant genes and therefore led to the consideration that the reasons for low expression can be termination of pre-mature transcription, abnormal mRNA splicing, instable mRNA or incompetent codon usage68. Synthetic Bt. toxin genes were created (designed), constructed and cloned to neglect all these undesirable aspects, and the resulted expression of these transgenes in plants was significantly enhanced69,70. This jointly with experimenting a variety of promoters and other sequences, led to a considerable development of insect borer resistance and improvement in expressing the toxin levels 0.8% of the overall leaf protein71.
In the current study we have constructed and introduced three codon optimized synthetic genes named as CEMB-Cry1Ac + CEMB-Cry2A and CEMB-GTGene in the nuclear genome of sugarcane. The expression of these synthetic genes is under the control of maize Ubiquition-1 promoter37. Simple and minimal plasmid cassettes were constructed for maximum protein expression72,73. A study was made on comparison between expression of Bt. and GTG genes through two different promoters CAMV35S and Maize Ubiquitin-1 (data not included). The resulted expression under Ubiquitin promoter confirmed higher toxin levels70. So maize Ubiquitin promoter was used to enhance the expression of these transgenes and improved increased resistance against cane bores and tolerance for glyphosate herbicide. After transformation a number of transgenic sugarcane plants were regenerated from transformed calli using this plant expression constructs (pCEMB-SC12) for CEMB-Cry1Ac + CEMB-Cry2A and pCEMB-SGTG for glyphosate tolerant gene (CEMB-GTGene).
In this present study the stability and inheritance of three transgenes up to three vegetative generations (V0, V1, and V2) verified that this technique of transformation can be used efficiently2. From 100 transformed calli, 81% was survived on the selection pressure (Kanamycine 50 mg/L) while 48% transformed calli were regenerated on double selection medium (Kanamycine 50 mg/L + glyphosate 50 mM). From 1000 resistant multiplied plants (CPF-246), 15 plants were positive for CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene, resulting transformation efficiency 1.5% after optimized glyphosate spray assay (1200 mL/80 L/acre). In monocots, the transformation efficiency was reported from 1 to 5%30 while co-integration of combine two genes was 85% when a combination of 14 diverse (pUC- based) plasmids was transformed74. Joyce et al.52 reported 0.8–4.8% transformation efficiency in sugarcane.
From these, 15 putative transgenic plants positive for three genes CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene were obtained. Molecular analyses of putative transgenic plants were carried out, first for their screening and then for their stable integration and expression level of transformed transgenes. Transgenic plants obtained on the basis of selection (kanamycine 50 mg/L) were considered as putative plants. The GUS assay revealed that activity of this gene is observable in all parts of the plants. The leaf and stem segments from the GUS positive plants were examined under fluorescent microscope and uniformly distributed expression was observed. The linked genes with 100% co-integration frequency are already reported75. Chen et al.74 also described the successful co-transformation with 14 different transgenes in rice. PCR analysis was carried out for the screening of putative transgenic plants selected on Kanamycine and glyphosate selection media. At V0-generation, the fifteen (15) screened plants were positive for CEMB-Cry1AC + CEMB-Cry2A and CEMB-GTGene in PCR amplification through gene specific primers.
The integration studies of these three transgenes in the genome of the sugarcane were confirmed by Southern DNA hybridization. Both single and double restriction digestion of the plasmids pCEMB-SC12 and pCEMB-SGTG was performed to digest the full cassettes and for having an estimation of the copy numbers. In current study 1–2 copy numbers were found. There are many reports for 1–5 copy numbers of transgenes introduced into crop plants by gene gun method in monocots76,77. For V1-generation, most of the time there was a co-relation between Cry1Ac and Cry2A banding pattern and intensity of the hybridizing bands, indicative of similar copy numbers of the transgenes. This recommends that the integrated plasmids, in general, integrate as a whole unit77. There were no rearrangements detections for these transgenes, Southern DNA hybridization confirming the integration of intact transgenes copies. Komari78 reported 1–6 copies of integrated genes while majority of the transgenics consist of 1–2 copies.
Transprotein expression levels were confirmed by the ELISA and Dipstick assay. The results of Dipstick-ELISA assay or immuno-Strip assay and quantification through DAS-ELISA confirmed that all of the 15 clones (V0) were consisting of CEMB-Bt. and CEMB-GTGene genes, although the expression levels of these transgenes were variable79. There might be many reasons for this, such as variability in transgene copy number80, transgene insertion locations in the genome8 or heterozygosity of the sugarcane transgenic lines, internal as well as external environment81. Koncz et al.82 and Zambryski et al.83 also reported that in transgenic plants, foreign DNA can integrate randomly on the chromosomal sites in the cells of plants through non-homologous recombination. The highest transprotein expression of CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene were observed as 0.475 µg/g, 0.567 µg/g, and 0.486 µg/g respectively. Overall ELISA based comparison CPF-246-(5L/5), CPF-246-(6L/5) were highest in their three expressions and CPF-246 (2L/8) and CPF-246 (5L/1) lines were lowest. The clones of V0-generation multiplied with 150 replicates (V1-generation) in the field showed positive results for PCR amplification through gene specific primers. Fifteen plants which were randomly selected from V1 and V2-generation showed stability for the integration and in protein expressions of CEMB-Cry1Ac, CEMB-Cry2Ac and CEMB-GTGene. The highest transprotein expressions of CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene were observed as 0.687 µg/g, 0.611 µg/g, and 0.589 µg/g respectively. Overall ELISA based comparison CPF-246-(5L/5), CPF-246-(6L/5) were highest in their three expressions and CPF-246-(2L/8) and CPF-246 (5L/1) lines were lowest. The stable integration of these transgenes was confirmed through Southern Blot analysis. These results showed similarity to those achieved by Bashir et al.84, for transgenic Basmati rice. These results were similar to those obtained by Kiani et al.85 while studying the expression of Cry1Ac protein through ELISA in transgenic cotton plants. Deng et al.86 observed similar quantified values in the transgenic lines of rice. In V2-generation, screened five transgenic sugarcane lines as 50 clones were multiplied in the field. Molecular analyses of three transgenes confirmed its stable trans-protein expression. The clones of these five lines were positive for all the three transgenes by PCR amplification, Southern Blotting, Dipstick assay and ELISA. All the sugarcane clones in this (3rd) generation (V2) were stable in their expressions87. The glyphosate tolerant gene expression was stable with respect to their ELISA protein quantification values and as well as by spray assays. The results showed similarity to those found by Leibbrandt et al.88 and Manickavasagam et al.89. Inheritance of integrated genes was studied stable in these three generations. The transprotein of integrated genes in V0, V1, and V2-generations further verified through insect (Chilo infuscatellus) bioassay.
Bio-toxicity leaf assay
Leaf bio-assay was carried out to check the efficacy of transgenes (CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene) proteins against Chilo infuscatellus (2nd instar larvae) shoot borer at four different time intervals. The expressional level of toxins from these genes is significantly imprtant as it should be in such sufficient quantitative level at the infestation time that can protect the crop against attack of shoot borers of Lepidopteran. In V0-generation, with the passage of time the toxin levels of CEMB-CrylAc and CEMB-Cry2A were decreased and percentage of cane borer damage increased, as the toxin level dropped from 0.475 to 0.252 µg/g for CEMB-Cry1Ac and 0.467 to 0.278 µg/g for CEMB-Cry2A, but all the transgenic lines showed significant insect mortality (60–100%) in comparison with control plants90. In V1-generation 15 lines with 150 clones were analyzed for leaf bioassay, each transgenic line was significantly differed from non-transgenic plants and performed better against in leaf bio-toxicity assays. Five lines were stable in their transproteins (CEMB-Cry1Ac and CEMB-Cry2A) expression and were also free of any kind of insect attack in the field where as control plants were totally damaged. In bioassay less damage to transgenic plant and complete consumption of non-transgenic plants verified strong defense of transgenic sugarcane against cane borer (Chilo infuscatellus). Similar results were published against borers5,8,91.
Analysis of Variance (ANOVA), the Least Significant Difference Test (LSD) and Dunnett’s Test confirmed the significant differences for Chilo infuscatellus mortality (%) between the control and transgenic sugarcane plants. ANOVA and Dunnett’s test verified highly significant difference at 5% level of significance for transgenic sugarcane line with respect to Chilo infuscattelus mortality. In V2-generation insect mortality was highly stable (90%-100). LSD analysis illustrated that transgenic leaf samples from CPF-246 (6L/5-4) at 20-days produced significantly high Chilo infuscatellus larvae mortality percentage (88%) compared with transgenic line (5L/1-9), which showed lowest mortality percentage value (66%). In general this data revealed the significant leaf damage and insect mortality (%) differences in control and transgenic plants respectively and also supported the conclusions of Riaz et al.2; Weng et al.8; Manikandan58.
Success of transformation studies depends upon the integration of the desired genes in the genome of the desired plants in addition to its inheritance to the next generation plants. Inheritance of stably integrated genes was studied up to three vegetative generations. Results acquired from PCR, Southern Blot analysis, Dipstick and ELISA (V0-generation) clearly confirmed the amplification and integration of three genes in the genome of sugarcane plants. Mainly the Southern Blot analysis of V1-generation indicated that two copies of transformed genes were present in the genome of sugarcane plants. ELISA analysis, on the other hand also confirmed the actively integrated genes were efficiently transcribed into protein as toxin. The transprotein of integrated genes in V0, V1 and V2 generation were verified through insect (Chilo infuscatellus) bioassay.
Herbicide spray assay
Weeds affect by limiting the available nutrients to the major cash crops92. When glyphosate (3000 mL/ha) sprayed on the transgenic sugarcane plants, after 15 days 160 out of 300 plants was tolerant to glyphosate stress. The rest of the field plants showed necrotic symptoms, expired tissues and weeds were totally dead after this spray application. In the V1-generation, 75% of plants were alived and tolerant for glyphosate (3000 mL/ha) while 25 percent non tolerant plants were observed dead in some lines. The low expression may be due to inner or outer environmental conditions. In the V2-generation, ten clone plants from each of five transgenic (CPF-246) lines 4L/2, 5L/5, 6L/5, L8/4, and L9/6 were tested. Such comparable results were formerly reported by Joyce et al.52. In V2-generation all the lines with their clone replicates were tolerant to glyphosate spray (3000 ml/80 L/hectare). Finally, CPF-246 produced five tolerant lines with CEMB glyphosate tolerant gene. These results are in accordance with the previous studies37,93–95. It can be concluded that CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTG genes successfully introduced into the genome of the sugarcane CPF-246. Stable expression and insecticidal activity of transgenic plants was confirmed in V0, V1 and V2-generations. Molecular analyses confirmed the expression level of toxin in the transgenic lines of V0, V1 and V2 generations. Transformation of sugarcane with two Bt. and GTGene will open new directions for the development of a high yielding cane borer resistant and glyphosate tolerant sugarcane. These transgenic lines can be further improved with other high sugar yielding characters. The end product can be a transgenic variety with a number of beneficial traits. There is also an international inclination towards economically safe, ecologically and environmentally friendly methods which can enhance the defense mechanism of the crop against pest pathogens96,97.
Biological insect control methods by using naturally available bacteria, fungi, and viruses received limited acceptance from the users, as their capability to defend plants has commonly been substandard to results achieved by chemical ways. Insecticides have been relatively successful in increasing crop production by minimize the losses rooted by insects, but the use of potentially dangerous chemical sprays and pesticides is disfavored in several countries and even a few compounds deregistered, secondly chemical sprays are not acceptable in sugarcane. The most effectual way of achieving high crop production is to set up the desired resistance into the crop plant by genetic transformation. Now it’s possible to create plants that are resistant to insect borers or having multiple novel disease resistant genes which are modified according to the crop genome and adapted for specific soil or environmental conditions.
Conclusion
The five transgenic sugarcane lines (4L/2, 5L/5, 6L/5, L8/4, L9/6) from CPF-246 variety harboring codon optimized CEMB-Cry1Ac, CEMB-Cry2A and CEMB-GTGene have revealed excellent potential up to 100% mortality of Chilo infuscattellus larvae (Lepidopteran) at the plant age of 80 days along with complete weed removal on 3000 mL/ha glyphosate tolerance level. This study concludes that if approved by the biosafety committee, this transgenic sugarcane can be a good starting material for the farmer’s community for the cost effective control of insects and weeds. Further studies are recommended to increase the stable expression of Bt. toxins up to the maturity, which can be achieved by further modification of the gene constructs. It has also been reported that glyphosate crops with tolerance level up to 5000 mL/ha might be more useful to control all types of sugarcane weeds in all the agroclimatic regions of the country. To achieve the above goals new vegetative genes such as Vip3A and Vip3B from Bt. in combination with CEMB-Cry1Ac and CEMB-Cry2A can be more effective, with different enhancers, promoters etc. can be developed for further strengthening/safeguarding this industrial cash crop from the insects and weeds.
Supplementary Information
Acknowledgements
The authors acknowledge the Higher Education Commission (HEC), Pakistan for providing IRSIP grant to Z.Q for research visit to Canada, Prof. Dr. Mounir G. Abouhaidar, Department of Cell and Systems Biology, University of Toronto, Prof. Dr. Kathleen L. Hefferon, Department of Microbiology, Cornell University Ithaca, NY, USA, and also thankful to Punjab Agriculture Research Board (PARB), Pakistan for the award of research project.
Author contributions
Z.Q. and I.A.N. designed this research study. Z.Q.: All the research work and designed experiments were performed by this author. Z.Q. wrote the draft manuscript and prepared all the figures. I.A.N: The project was planned by this author. M.G.A. and K.L.H. helped in protein expression experiments, A.Q.R.: this author helped in experimental design and statistical analyses, A.L., Q.A., S.A. and B.R. contributed in manuscript editing, formatting and reviewed the article. A.A.S.: This author helped technically and proofread the manuscript. All authors reviewed the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91985-8.
References
- 1.Yao W, et al. Field performance of transgenic sugarcane lines resistant to sugarcane mosaic virus. Front. Plant Sci. 2017;8:104. doi: 10.3389/fpls.2017.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raza S, et al. Regeneration in sugarcane via somatic embryogenesis and genomic instability in regenerated plants. J. Crop Sci. Biotechnol. 2012;2(15):131–136. doi: 10.1007/s12892-011-0111-6. [DOI] [Google Scholar]
- 3.Khan MF, et al. Genetic modification of Saccharum officinarum for herbicide tolerance. Cytol. Genet. 2019;53(3):239–249. doi: 10.3103/S0095452719030101. [DOI] [Google Scholar]
- 4.Mayavan S, et al. Agrobacterium-mediated in planta genetic transformation of sugarcane setts. Plant Cell Rep. 2015;34:1835–1848. doi: 10.1007/s00299-015-1831-8. [DOI] [PubMed] [Google Scholar]
- 5.Gao S, et al. Transgenic sugarcane with a Cry1Ac gene exhibited better phenotypic traits and enhanced resistance against sugarcane borer. PLoS ONE. 2016;11(4):1–16. doi: 10.1371/journal.pone.0153929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li YR, Yang LT. Sugarcane agriculture and sugar industry in China. Sugar Technol. 2015;17(1):1–8. doi: 10.1007/s12355-014-0342-1. [DOI] [Google Scholar]
- 7.Wekesa R, Justus MO, Bernard AN, Leonard SW. Sugarcane in vitro culture technology: Opportunities for Kenya’s sugar industry. Afr. J. Biotechnol. 2015;47(14):3170–3178. [Google Scholar]
- 8.Weng LX, et al. Transgenic sugarcane plants expressing high levels of modified Cry1Ac provide effective control against stem borers in field trials. Transgenic Res. 2011;20(4):759–772. doi: 10.1007/s11248-010-9456-8. [DOI] [PubMed] [Google Scholar]
- 9.Jung JH, et al. RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 2012;10(9):1067–1076. doi: 10.1111/j.1467-7652.2012.00734.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, et al. Establishment and application of a loop-mediated isothermal amplification (LAMP) system for detection of Cry1Ac transgenic sugarcane. Sci. Rep. 2014;9:4. doi: 10.1038/srep04912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghavi S, et al. Development of a novel sequential pretreatment strategy for the production of bioethanol from sugarcane trash. Bioresour. Technol. 2015;199:202–210. doi: 10.1016/j.biortech.2015.08.062. [DOI] [PubMed] [Google Scholar]
- 12.Poovaiah CR, et al. Sugarcane transgenics expressing MYB transcription factors show improved glucose release. Biotechnol. Biofuels. 2016;9:143. doi: 10.1186/s13068-016-0559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Economic Survey of Pakistan. Pakistan Bureau of Statistics. 17–33 (Govt of Pakistan, Ministry of Finance, Agriculture, 2016–17).
- 14.Sattar M, Mehmood SS, Khan MR, Ahmad S. Influence of egg parasitoid Trichogramma chilonisishii on sugarcane stem borer (chilo infuscatellus snellen) in Pakistan. Pak. J Zool. 2016;48(4):989–994. [Google Scholar]
- 15.Shahid MR, et al. Effectiveness of Trichogramma chilonis (Ishii) (Hymenoptera: Trichogrammatidae) against sugarcane stem borer (Chiloinfuscatellus Snellen) (Lepidoptera: Pyrallidae) Pak. Entomol. 2007;29:141–146. [Google Scholar]
- 16.Gatehouse AMR, Boulter D, Hilder VA. Potential of plant-derived genes in the genetic manipulation of crops for insect resistance. In: Gatehouse AMR, Hilder VA, Boulter D, editors. Plant Genetic Manipulation for Crop Protection. Wallingford: CAB International; 1992. pp. 155–181. [Google Scholar]
- 17.Awan MF, et al. Transgenic cotton: Harboring Broad Term Resistance against Insect and Weeds through Incorporation of CEMB Double Bt and cp4EPSPS Genes. Pak. J. Agric. Sci. 2016;53(3):501–505. [Google Scholar]
- 18.Hashmi, A.A. Insect pest of sugarcane. Insect pest management cereal and cash crops. Pak. Agric. Res. Council. 261–285 (1994).
- 19.Bhatti IB, et al. Incidence and intensity of borer complex infestation on different sugarcane genotypes under agro-climatic conditions of Thatta. Pak. J. Sci. 2008;6(34):103–106. [Google Scholar]
- 20.Bravo A, Gill SS, Soberón M. Bacillus thuringiensis mechanisms and use. Compr. Mol. Insect Sci. 2005;6:175–205. doi: 10.1016/B0-44-451924-6/00081-8. [DOI] [Google Scholar]
- 21.Parker MW, Feil SC. Pore-forming protein toxins: From structure to function. Progress Biophys. Mol. Biol. 2005;88(1):91–142. doi: 10.1016/j.pbiomolbio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, et al. Preliminary comparing the toxicities of the hybrid Cry1Acs fused with different heterogenous genes provided guidance for the fusion expression of Cry proteins. World J. Microbiol. Biotechnol. 2012;28(1):397–400. doi: 10.1007/s11274-011-0825-0. [DOI] [PubMed] [Google Scholar]
- 23.Bakhsh A, et al. Insect resistance and risk assessment studies in advance lines of Bt. cotton Harboring Cry1Ac and Cry2A genes. American-Eurasian J. Agric. Environ. Sci. 2009;1(6):01–11. [Google Scholar]
- 24.Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensisCry and Cyt. toxins and their potential for insect control. Toxicon. 2007;49(4):423–435. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azam S, et al. Dissemination of Bt. cotton in cotton growing belt of Pakistan. Adv. Life Sci. 2013;1(1):18–26. [Google Scholar]
- 26.Pline-Srnic W. Physiological mechanisms of glyphosate resistance. Weed Technol. 2006;20(2):290–300. doi: 10.1614/WT-04-131R.1. [DOI] [Google Scholar]
- 27.Puspito AN, et al. Transformation and evaluation of Cry1Ac+Cry2A and GTGene in Gossypium hirsutum L. Front. Plant Sci. 2015;6:943. doi: 10.3389/fpls.2015.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qamar Z, et al. An overview of genetic transformation of glyphosate resistant gene in Zea mays. Nat. Sci. 2015;13:80–90. [Google Scholar]
- 29.Qamar Z, et al. Transformation and transgenic expression studies of glyphosate and cane borer resistance genes in sugarcane (Saccharum officinarum L.) Mol. Plant Breed. 2015;12:1–17. [Google Scholar]
- 30.Aaliya K, et al. Transformation, evaluation of GTGene and multivariate genetic analysis for morphophysiological and yield attributing traits in Zea mays. Genetika. 2016;48:423–433. doi: 10.2298/GENSR1601423A. [DOI] [Google Scholar]
- 31.Javied M, et al. Agrobacterium-mediated transformation of cotton (Gossypium hirsutum L.) using dmo gene for enhanced tolerance against dicamba pesticide. Biol. Clin. Sci. Res. J. 2021;2021(1):e009. doi: 10.47264/bcsrj0201009. [DOI] [Google Scholar]
- 32.Paganelli A. Glyphosate-based herbicides produce teratogenic effects on vertebrates by impairing retinoic acid signaling. Chem. Res. Toxicol. 2010;23(10):1586–1595. doi: 10.1021/tx1001749. [DOI] [PubMed] [Google Scholar]
- 33.Tesfamariam T, et al. Glyphosate in the rhizosphere—Role of waiting times and different glyphosate binding forms in soils for phytotoxicity to non-target plants. Eur. J. Agron. 2009;31(3):126–132. doi: 10.1016/j.eja.2009.03.007. [DOI] [Google Scholar]
- 34.Rao AQ, et al. Overexpression of the phytochrome B gene from Arabidopsis thaliana increases plant growth and yield of cotton (Gossypium hirsutum) J. Zhejiang Univ. Sci. B. 2011;12:326–334. doi: 10.1631/jzus.B1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qamar Z, Riaz S, Nasir IA, Ali Q, Husnain T. Transformation and evaluation of different transgenic lines for glyphosate tolerance and cane borer resistance genes in sugarcane (Saccharum officinarum L.) Cytol. Genet. 2017;51(5):401–412. doi: 10.3103/S0095452717050085. [DOI] [Google Scholar]
- 36.Bhaskar PB, et al. Agrobacterium-mediated transient gene expression and silencing: A rapid tool for functional gene assay in potato. PLoS ONE. 2009;4(6):e5812. doi: 10.1371/journal.pone.0005812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasir IA, et al. Herbicide-tolerant sugarcane (Saccharum officinarum L.) plants: An unconventional method of weed removal. Turkish J. Biol. 2014;38(4):439–449. doi: 10.3906/biy-1306-81. [DOI] [Google Scholar]
- 38.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19(6):1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975;98(3):503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1–2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Dunnett, C. W. & Tamhane, A. C. A step-up multiple test procedure. J. Am. Stat. Assoc.87(417), 162–170 (1992).
- 42.Ali A, et al. Genetically modified foods: Engineered tomato with extra advantages. Adv. Life Sci. 2014;3(1):139–152. [Google Scholar]
- 43.Su Y, et al. Early Selection for smut resistance in sugarcane using pathogen proliferation and changes in physiological and biochemical indices. Front. Plant Sci. 2016;7:1133. doi: 10.3389/fpls.2016.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiebaut F, et al. Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics. 2012;13(1):290. doi: 10.1186/1471-2164-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta SK, Datta K, Potrykus I. Embryogenesis and plant regeneration from microspores of both Indica and Japonica rice (Oryza sativa) Plant Sci. 1990;67:83–88. doi: 10.1016/0168-9452(90)90053-Q. [DOI] [Google Scholar]
- 46.Lai KL, Liu LF. Increased plant regeneration frequency in water-stressed rice tissue cultures. Jpn. J. Crop Sci. 1988;57:553–557. doi: 10.1626/jcs.57.553. [DOI] [Google Scholar]
- 47.McQualter RB, et al. Production and evaluation of transgenic sugarcane containing a Fiji disease virus (FDV) genome segment S9-derived synthetic resistance gene. Crop Pasture Sci. 2004;2(55):139–145. doi: 10.1071/AR03131. [DOI] [Google Scholar]
- 48.Snyman SJ. Refining the application of direct embryogenesis in sugarcane: Effect of the developmental phase of leaf disc explants and the timing of DNA transfer on transformation efficiency. Plant Cell Rep. 2006;25:1016–1023. doi: 10.1007/s00299-006-0148-z. [DOI] [PubMed] [Google Scholar]
- 49.Fitch MMM, et al. Elimination of sugarcane yellow leaf virus from infected sugarcane plants by meristem tip culture visualized by tissue blot immunoassay. Plant Pathol. 2001;50:676–680. doi: 10.1046/j.1365-3059.2001.00639.x. [DOI] [Google Scholar]
- 50.Ali K, et al. Ideal in-vitro culture and selection conditions for sugarcane genetic transformation Pakistan. J. Agric. Sci. 2015;52(1):43–49. [Google Scholar]
- 51.Nawaz M, et al. Improving in vitro leaf disk regeneration system of sugarcane (Saccharum officinarum L.) with concurrent shoot/root induction from somatic embryos. Turkish J. Biol. 2013;37:726–732. doi: 10.3906/biy-1212-10. [DOI] [Google Scholar]
- 52.Joyce P, et al. Field performance of transgenic sugarcane produced using Agrobacterium and biolistics methods. Plant Biotechnol. J. 2014;12:411–424. doi: 10.1111/pbi.12148. [DOI] [PubMed] [Google Scholar]
- 53.Akram A, Arshad K, Hafeez M. Cloning and expression of universal stress protein 2 (USP2) gene in Escherichia coli. Biol. Clin. Sci. Res. J. 2021;2021(1):e002. doi: 10.47264/bcsrj0201002. [DOI] [Google Scholar]
- 54.Joshi R, Shukla A, Sairam RK. In vitro screening of rice genotypes for drought tolerance using polyethylene glycol. Acta Physiol. Plant. 2011;33:2209–2217. doi: 10.1007/s11738-011-0760-6. [DOI] [Google Scholar]
- 55.Chengalrayan K, Abouzid A, Gallo-Meagher M. In vitro regeneration of plants from sugarcane seed-derived callus. In vitro cellular and developmental biology. Plant. 2005;41:477–482. [Google Scholar]
- 56.Bakhsh A, Rao AQ, Shahid AA, Husnain T. Spatio temporal expression pattern of an insecticidal gene (Cry2A) intransgenic cotton lines. Notulae Sci. Biol. 2012;4(4):115–119. doi: 10.15835/nsb448217. [DOI] [Google Scholar]
- 57.Qamar Z, et al. Trackable CEMB-klean cotton transgenic technology: Affordable climate neutral agribiotech industrialization for developing countries. Adv. Life Sci. 2019;6(3):131–138. [Google Scholar]
- 58.Manikandan R, Balakrishnan N, Sudhakar D, Udayasuriyan V. Development of leaf folder resistant transgenic rice expressing cry2AX1 gene driven by green tissue-specific rbcS promoter. World J. Microbiol. Biotechnol. 2016;3(32):37. doi: 10.1007/s11274-015-2006-z. [DOI] [PubMed] [Google Scholar]
- 59.Gahan LJ, et al. Genetic basis of resistance to Cry1Ac and Cry2Aa in Heliothis virescens (Lepidoptera: Noctuidae) J. Econ. Entomol. 2005;98:1357–1368. doi: 10.1603/0022-0493-98.4.1357. [DOI] [PubMed] [Google Scholar]
- 60.Riaz N, et al. Development of Indica Basmati rice harbouring two insecticidal genes for sustainable resistance against lepidopteron insects. S. Afr. J. Bot. 2006;72:217–233. doi: 10.1016/j.sajb.2005.07.005. [DOI] [Google Scholar]
- 61.Rashid B, Zafar S, Husnain T, Riazuddin S. Transformation and inheritance of Bt. genes in Gossypium hirsutum. J. Plant Biol. 2008;51:248–254. doi: 10.1007/BF03036123. [DOI] [Google Scholar]
- 62.Nawaz A, Haseeb A, Malik H, Ali Q, Malik A. Genetic association among morphological traits of zea mays seedlings under salt stress. Biol. Clin. Sci. Res. J. 2020;2020(1):e021. doi: 10.47264/bcsrj0101021. [DOI] [Google Scholar]
- 63.Castle LA. Discovery and directed evolution of a glyphosate tolerance gene. Science. 2004;304(5674):1151–1154. doi: 10.1126/science.1096770. [DOI] [PubMed] [Google Scholar]
- 64.Aasim M, Khawar KM, Ozcan S. Production of herbicide resistant cowpea (Vigna unguiculata L.) transformed with the bar gene. Turkish J Biol. 2013;37:472–478. doi: 10.3906/biy-1207-44. [DOI] [Google Scholar]
- 65.Herman PL, et al. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6 gene isolation, characterization, and heterologous expression. J. Biol. Chem. 2005;280(26):24759–24767. doi: 10.1016/j.abb.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 66.Latif A, et al. Herbicide-resistant cotton (Gossypium hirsutum) plants: An alternative way of manual weed removal. BMC Res. Notes. 2015;8:453. doi: 10.1186/s13104-015-1397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, L.H., Weng, L.X. & Jiang, Z.D. Sugarcane. Biotechnology in Agriculture and Forestry Transgenic Crops V. (eds Pua, E. C. & Davey, M. R.) (Springer, 2007).
- 68.Vinogradov AE. DNA helix: The importance of being GC-rich. Nucleic Acids Res. 2003;31(7):1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee KR, et al. Molecular characterization of lepidopteran pest-resistant transgenic rice events expressing synthetic Cry1Ac. Plant Biotechnol. Rep. 2009;3(4):317–324. doi: 10.1007/s11816-009-0105-8. [DOI] [Google Scholar]
- 70.Wang J, Jiang J, Oard JH. Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.) Plant Sci. 2000;156(2):201–211. doi: 10.1016/s0168-9452. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Oard JH. Rice ubiquitin promoters: Deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 2003;22(2):129–134. doi: 10.1007/s00299-003-0657-y. [DOI] [PubMed] [Google Scholar]
- 72.Jackson MA, Anderson DJ, Birch RG. Comparison of Agrobacterium and particle bombardment using whole plasmid or minimal cassette for production of high-expressing, low-copy transgenic plants. Transgenic Res. 2013;22(1):143–151. doi: 10.1007/s11248-012-9639-6. [DOI] [PubMed] [Google Scholar]
- 73.Kim JY, Gallo M, Altpeter F. Analysis of transgene integration and expression following biolistic transfer of different quantities of minimal expression cassette into sugarcane (Saccharum spp. hybrids) Plant Cell Tissue Organ Culture. 2012;108(2):297–302. doi: 10.1007/s11240-011-0043-3. [DOI] [Google Scholar]
- 74.Chen YF, et al. Correlation between appearance of embryogenic cells and the IAA levels in rice somatic cell culture. Acta Botanica Sinica. 1998;40:474–477. [Google Scholar]
- 75.Hadi MZ, McMullen MD, Finer JJ. Transformation of 12 different plasmids into soybean via particle bombardment. Plant Cell Rep. 1996;15(7):500–505. doi: 10.1007/BF00232982. [DOI] [PubMed] [Google Scholar]
- 76.Goto F, Toki S, Uchimiya H. Inheritance of a co-transferred foreign gene in the progenies of transgenic rice plants. Transgenic Res. 1993;5(2):300–305. doi: 10.1007/BF01968842. [DOI] [Google Scholar]
- 77.Kohli A, et al. Transgene organization in rice engineered through direct DNA transfer supports a two-phase integration mechanism mediated by the establishment of integration hot spots. Proc. Natl. Acad. Sci. USA. 1998;12(95):7203–7208. doi: 10.1073/pnas.95.12.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Komari T, et al. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 1996;10(1):165–174. doi: 10.1046/j.1365-313x.1996.10010165.x. [DOI] [PubMed] [Google Scholar]
- 79.Dong HZ, Li WJ. Variability of endotoxin expression in Bt. transgenic cotton. J. Agron. Crop Sci. 2007;193(1):21–29. doi: 10.1111/j.1439-037X. [DOI] [Google Scholar]
- 80.Ingelbrecht IL, Irvine JE, Mirkov TE. Post-transcriptional gene silencing in transgenic sugarcane. Dissection of homology-dependent virus resistance in a monocot that has a complex polyploid genome. Plant Physiol. 1999;4(119):1187–1198. doi: 10.1104/pp.119.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tassy C, et al. Biolistic transformation of wheat: Increased production of plants with simple insertions and heritable transgene expression. Plant Cell Tissue Organ Cult. 2014;119(1):171–181. doi: 10.1007/s11240-014-0524-2. [DOI] [Google Scholar]
- 82.Koncz, C. et al. Homology recognition during T-DNA integration into the plant genome. In Homologous Recombination and Gene Silencing in Plants. 167–189 ISBN: 978-94-010-4478-3 (1994).
- 83.Zambryski P. Basic processes underlying Agrobacterium-mediated DNA transfer to plant cells. Annu. Rev. Genet. 1998;22(1):1–30. doi: 10.1146/annurev.ge.22.120188.000245. [DOI] [PubMed] [Google Scholar]
- 84.Bashir K, Husnain T, Riazuddin S. Response of transgenic rice expressing two Bt. genes to nontarget insects. Int. Rice Res. Notes. 2004;2(29):15–16. [Google Scholar]
- 85.Kiani S, et al. Chloroplast-targeted expression of recombinant crystal-protein gene in cotton: An unconventional combat with resistant pests. J. Biotechnol. 2013;166(3):88–96. doi: 10.1016/j.jbiotec.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 86.Deng LH, Weng LS, Xiao GY. Optimization of Epsps gene and development of double herbicide tolerant transgenic PGMS rice. J. Agric. Sci. Technol. 2014;16(1):217–228. [Google Scholar]
- 87.Basnayake SW, et al. Field performance of transgenic sugarcane expressing isomaltulose synthase. Plant Biotechnol. J. 2012;2:217–225. doi: 10.1111/j.1467-7652.2011.00655.x. [DOI] [PubMed] [Google Scholar]
- 88.Leibbrandt NB, Snyman SJ. Stability of gene expression and agronomic performance of a transgenic herbicide-resistant sugarcane line in South Africa. Crop Sci. 2003;43(2):671–677. doi: 10.2135/cropsci2003.0671. [DOI] [Google Scholar]
- 89.Manickavasagam M, et al. Agrobacterium mediated genetic transformation and development of herbicide-resistant sugarcane (Saccharum species hybrids) using axillary buds. Plant Cell Rep. 2004;3(23):134–143. doi: 10.1007/s00299-004-0794-y. [DOI] [PubMed] [Google Scholar]
- 90.Kim S, et al. Inheritance and field performance of transgenic Korean Bt. rice lines resistant to rice yellow stem borer. Euphytica. 2008;164(3):829–839. doi: 10.1007/s10681-008-9739-9. [DOI] [Google Scholar]
- 91.James, C. Global Status of Commercialized Biotech/GM Crops (International Service for the Acquisition of Agri-Biotech Applications (ISAAA), 2010).
- 92.Manickavasagam M, Ganapathi A. Direct somatic embryogenesis and plant regeneration from leaf explants of sugarcane. Indian J. Exp. Biol. 1998;36:832–835. [Google Scholar]
- 93.Park SH, et al. Cross-protection and selectable marker genes in plant transformation. In Vitro Cell. Dev. Biol. Plant. 1998;34(2):117–121. doi: 10.1007/BF02822775. [DOI] [Google Scholar]
- 94.Tahir T, Ali Q, Rashid M, Malik A. The journey of CRISPR-Cas9 from bacterial defense mechanism to a gene editing tool in both animals and plants. Biol. Clin. Sci. Res. J. 2020;2020:e017. doi: 10.47264/bcsrj0101017m. [DOI] [Google Scholar]
- 95.Ahmad M, Ali Q, Hafeez M, Malik A. Improvement for biotic and abiotic stress tolerance in crop plants. Biol. Clin. Sci. Res. J. 2021;2021(1):e004. doi: 10.47264/bcsrj0201004. [DOI] [Google Scholar]
- 96.Bashir M, Ali Q, Rashid M, Malika A. CRISPR/CAS9 in genome editing: A nature gifted molecular tool. Biol. Clin. Sci. Res. J. 2020;2020(1):e018. doi: 10.47264/bcsrj0201018. [DOI] [Google Scholar]
- 97.Ali M, Rafique F, Ali Q, Malik A. Genetic modification for salt and drought tolerance in plants through SODERF3. Biol. Clin. Sci. Res. J. 2020;2020(1):e022. doi: 10.47264/bcsrj0101022. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.