Abstract
Meniscus injuries are common and a major cause of long-term joint degeneration and disability. Current treatment options are limited, so novel regenerative therapies or tissue engineering strategies are urgently needed. Development of new therapies is hindered by a lack of knowledge regarding the cellular biology of the meniscus and lack of well-established methods for studying meniscus cells in vitro. The goals of this study were to 1) establish baseline expression profiles and dedifferentiation patterns of inner and outer zone primary meniscus cells, and 2) evaluate the utility of poly(ethylene glycol) diacrylate (PEGDA) and gelatin methacrylate (GelMA) polymer hydrogels to reverse dedifferentiation trends for long-term meniscus cell culture. Using RT-qPCR, we measured expression levels of putative meniscus phenotype marker genes in freshly isolated meniscus tissue, tissue explant culture, and monolayer culture of inner and outer zone meniscus cells from porcine knees to establish baseline dedifferentiation characteristics, then compared these expression levels to PEGDA/GelMA embedded passaged meniscus cells. COL1A1 showed robust upregulation, while CHAD, CILP, and COMP showed downregulation with monolayer culture. Expression levels of COL2A1, ACAN, and SOX9 were surprisingly similar between inner and outer zone tissue, and were found to be less sensitive as markers of dedifferentiation. When embedded in PEGDA/GelMA hydrogels, expression levels of meniscus cell phenotype genes were significantly modulated by varying the ratio of polymer components, allowing these materials to be tuned for phenotype restoration, meniscus cell culture, and tissue engineering applications.
Keywords: fibrochondrocyte, collagen, aggrecan, cartilage, osteoarthritis
Introduction
Meniscus injuries are highly prevalent, affecting millions of people each year1. Loss of meniscal function has been linked to early onset osteoarthritis within 10-20 years2. However, due to poor healing potential, especially in the inner zone of the meniscus, partial meniscectomy remains the most common surgical treatment for meniscus injuries3. For these reasons, novel ways to treat meniscus injuries and/or augment surgical repair are greatly needed. To this end, efforts at tissue engineering and biological stimulation of repair have been increasingly investigated in recent years. However, these efforts are hindered by a lack of knowledge regarding the cellular biology of the meniscus and lack of well-established methods for studying meniscus cells in vitro.
Only a few studies can be found in the literature describing expression level phenotypic changes of meniscus cells during monolayer expansion4-9. Of the studies that do exist, even fewer report on differences between inner and outer zone meniscus cells7-9 and comparisons with fresh tissue10, and there is a lack of data comparing tissue explant culture to isolated cells. Inner and outer regions of meniscus are significantly different in terms of vascularity and extracellular matrix (ECM) composition11. In addition, the cells in these regions have different phenotypic characteristics based on morphology both in situ12 and when cultured in monolayer13. Based on these morphological characteristics and differences in their regional ECM composition, outer zone cells are often described as “fibroblast-like,” while inner zone cells are described as “chondrocyte-like”11. The genes most commonly used as phenotype markers for studies of meniscus cell dedifferentiation are commonly associated with the articular chondrocyte phenotype, including type I collagen (COL1A1), type II collagen (COL2A1), aggrecan (ACAN), and SOX9. However, COL2A1 and ACAN may not be ideal for evaluation of meniscus cells, as they are expressed at significantly lower levels in meniscus tissue8. While some studies have shown a strong downregulation of COL2A1 with meniscus monolayer culture4-6, in a recent study utilizing microarray to compare inner zone meniscus tissue to monolayer cultured cells, of the chondrocyte phenotype genes only COL1A1 and SOX9 were found among the most significantly differentially regulated genes10. Furthermore, COL2A1 and ACAN were notably absent from the top 20 down-regulated transcripts. Interestingly, the authors identified and validated several less studied genes as potential phenotype markers of inner zone cells, which may be more relevant to meniscus cells, including chondroadherin (CHAD), cartilage intermediate layer protein (CILP), and cartilage oligomeric matrix protein (COMP). In order to address these gaps in knowledge, we have characterized phenotypic changes during monolayer culture in primary meniscus cell gene expression of both traditionally used phenotype marker genes, as well as less-studied targets identified in meniscus cells, and compared this data to fresh tissue and tissue culture expression levels. In addition, we investigated the potential of hydrogel materials for meniscus cell culture and phenotype restoration by directly comparing expression profiles of embedded meniscus cells and meniscus tissue in culture.
Attempts to reverse dedifferentiation patterns have not yet yielded a widely accepted method for maintaining meniscus cell phenotype and thus each lab utilizes their own biomaterials or techniques4-6, and due to the lack of well-defined expression profiles of meniscus cells, it is difficult to evaluate the efficacy of various cell culture techniques. A number of methods have been investigated, such as culture of cells on a variety of modified or biomimetic surfaces4; 5, three-dimensional (3D) culture supported by biomaterials5; 14, microaggregate culture6, and varying the oxygen tension of culture conditions5; 6; 15. It is increasingly appreciated that the physical 3D environment can control cellular phenotype, and biomaterials can be engineered for this purpose16. In particular, substrate stiffness, the availability of cellular adhesion motifs, and matrix degradability are parameters that have been shown to modulate cellular phenotype16. We investigated the utility of poly(ethylene glycol) diacrylate (PEGDA) and gelatin methacrylate (GelMA) hydrogels as substrates for 3D meniscus cell culture. PEGDA is a synthetic photocrosslinkable polymer of poly(ethylene glycol) subunits. The stiffness of PEGDA is tunable and the polymer is generally described as “biologically inert” because it contains no cellular adhesion motifs17; 18. Nevertheless, PEGDA has been successfully used to embed many different cell types for various applications, usually with high cell viability19-21. GelMA is a photocrosslinkable polymer derived from denatured collagen. Therefore, it retains native ECM cellular adhesion motifs and is cell-degradable, but lacks the tunable mechanical properties of PEGDA18; 22. We hypothesized that a hybrid of these materials could provide a physical 3D environment capable of reversing the dedifferentiation trends of primary meniscus cells in monolayer culture.
The goals of this study were 1) to establish baseline expression profiles and dedifferentiation patterns of inner and outer zone primary meniscus cells, and 2) to evaluate the utility of PEGDA/GelMA polymer hydrogels to reverse dedifferentiation trends for long-term meniscus cell culture.
Materials and Methods:
Tissue harvest and culture:
Skeletally mature 2-3 year old female porcine knee joints were obtained from a local abattoir. Tissue explants were harvested from the inner and outer regions of medial menisci (N=12), using 5 mm diameter biopsy punches, and trimmed to approximately 2 mm thick. Samples were either immediately frozen in liquid nitrogen (“fresh tissue”, n=6), or maintained in complete culture media composed of Dulbecco’s Modified Eagle Medium, high glucose (DMEM-HG; Catalog # 11995073; Gibco, Carlsbad, CA), 10% fetal bovine serum (Product # 35-010-CV; Corning, Oneonta, NY), 1x non-essential amino acids (Catalog # 11140050; Gibco), 1x antibiotic/antimycotic (Catalog # 15240062; Gibco), 10 mM HEPES (Cat # 15630080; Gibco), 40 μg/mL L-proline (SKU # H54409; Sigma-Aldrich, St. Louis, MO), and 50 μg/mL ascorbic acid (SKU # A8960; Sigma-Aldrich) for three days, then frozen in liquid nitrogen (“tissue explant”, n=6).
RNA extraction:
Explants were pulverized in TRIzol (Catalog # 15596026; Life Technologies, Carlsbad, CA) under liquid nitrogen using a freezer mill (Model 6875; SPEX SamplePrep, Metuchen, NJ). Then RNA was extracted by TRIzol/choloroform separation following the manufacturer’s protocol and column purified (Catalog # 48300; Norgen Biotek Corp., Thorold, Canada).
Cell isolation and monolayer culture:
Meniscus cells were isolated from the outer or inner zones (approximately outer one-third and inner two-thirds respectively) of the medial meniscus (N=6) by sequential digestion of minced tissue with 0.5% (w/v) pronase (SKU 53702; EMD Millipore Corp., Temecula, CA) digestion for one hour and 0.2% (w/v) collagenase type I (Catalog # LS004197; Worthington Biochemical Corp., Lakewood, NJ) digestion for 16 hours in DMEM-HG with 10x antibiotic/antimycotic and 10% FBS23. Isolated cells were seeded at 5 million cells per 10 cm tissue culture plate and maintained in complete culture media. Media was replaced every 2-3 days and cells were passaged at 90% confluence (5-6 days for passage 1 and then 2 days for subsequent passages). RNA was extracted by column purification (Catalog # 48300; Norgen Biotek Corp.) at the time of the first, second, and third passage (p1, p2, and p3).
Reverse transcription quantitative PCR (RT-qPCR):
RNA samples were reverse-transcribed using SuperScript VILO cDNA Synthesis Kit (Catalog # 11754050; Life Technologies). Then qPCR was performed using PowerUP SYBR Green master mix (Catalog # A25776; Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania) and the StepOne Plus real-time PCR system (Model 4376374; Applied Biosystems, Foster City, CA). Relative fold-change was determined by the 2−ΔΔCt method24 using 18S as a reference gene. Results are presented as fold-change relative to the mean of the indicated group.
Hydrogel preparation:
PEGDA and GelMA precursors were synthesized as previously described21; 25. Polymer precursors were prepared as 10% (w/v) solution and mixed at 8:2, 6:4, 5:5, and 4:6 ratios to yield final concentrations of 10% PEGDA (10P), 8%PEGDA/2%GelMA (8P2G), 6%PEGDA/4%GelMA (6P4G), 5%PEGDA/5%GelMA (5P5G), and 4% PEGDA/6% GelMA (4P6G) respectively. Lithium phenyl(2,4,6-trimethylbenzoyl) phosphinate (LAP; SKU 900889; Sigma-Aldrich) photoinitiator was prepared as a 75 mg/mL aqueous solution and added to the precursor solutions prior to photopolymerization to yield a final concentration of 0.75 mg/mL. The precursor solutions containing LAP were then added to a custom cylindrical mold (4 mm diameter, 2 mm height) and photopolymerized by exposure to UV light at 365 nm for 1 minute.
Equilibrium modulus:
The equilibrium modulus of hydrogel constructs (n=8/group) was determined by stress relaxation testing at 5, 10, 15, and 20% strain in unconfined compression. Construct height and width were measured using a precision camera system (iMetrum; Bristol, United Kingdom), and then constructs were submerged in a PBS bath for testing. An Electroforce 3220 load frame (TA Instruments; Newcastle, DE) and 250g load cell (Model 31; Honeywell, Golden Valley, MN) were used. A preload of 2 g was applied for 15 minutes, and then samples were compressed at a rate of 0.01 mm/s to each strain level and allowed to equilibrate for 30 minutes. The equilibrium modulus was calculated as the slope of the stress/strain curve at the four strain levels tested.
Cell Encapsulation:
Monolayer expanded porcine meniscus cells pooled from three subjects were trypsinized at p2, counted, centrifuged, and resuspended in PEGDA/GelMA solution at a density of 10 million cells/mL. Constructs were maintained in complete culture media, with media replaced every 2-3 days. A subset of samples (n=3/group) were collected after 48 hours in culture for viability measurements and then frozen in PBS for subsequent digestion and quantification of DNA content. The remaining samples were collected after two weeks in culture, and immediately processed for RNA extraction (n=3/group) or frozen in PBS for DNA content analysis (n=3/group) or histological staining (see Supplemental Figure S-2). RNA was extracted by manual pestle homogenization of hydrogel constructs and column purification (Norgen). RT-qPCR was performed as described above.
Viability:
Viability was assessed with the Live/Dead Viability/Cytotoxicity kit (Catalog # L3224; Molecular Probes, Eugene, OR), according to the manufacturer’s instructions. Constructs were stained with 4 μM calcein AM and 4 μM ethidium homodimer-1 in PBS and imaged with a fluorescent microscope (Model IX83; Olympus, Tokyo, Japan). Live and dead cells were quantified using CellSens Dimension software (Olympus).
DNA content:
Hydrogel constructs collected and frozen at 48 hrs post-embedding or after 2 weeks in culture were manually homogenized and digested in papain (SKU P4762; Sigma-Aldrich) solution (125 μg/mL in phosphate buffered EDTA solution) at 65°C for 16 hrs23; 26-29. DNA concentration was measured by Quant-iT Picogreen dye (Catalog # P11496; Molecular Probes). Total DNA was calculated and divided by construct wet weight to obtain a DNA/wet weight for each sample (n=3/group). The DNA/wet weight values at two weeks were divided by the average DNA/wet weight of the 48-hour samples to obtain the fold-change in DNA content.
Statistical analyses:
Expression data were log-transformed to correct for skewness inherent to relative-expression data and meet statistical model assumptions. For dedifferentiation RT-qPCR results, one-way ANOVA followed by Bonferroni corrected post-hoc analyses were performed in order to select relevant comparisons: all inner zone samples to each other, all outer zone samples to each other, and fresh inner to fresh outer tissue. Hydrogel gene expression, DNA content, and mechanical testing data were analyzed by one-way ANOVA followed by Tukey corrected post-hoc analyses. Alpha was set a prioi as < 0.05.
Results:
Meniscus Cell Dedifferentiation:
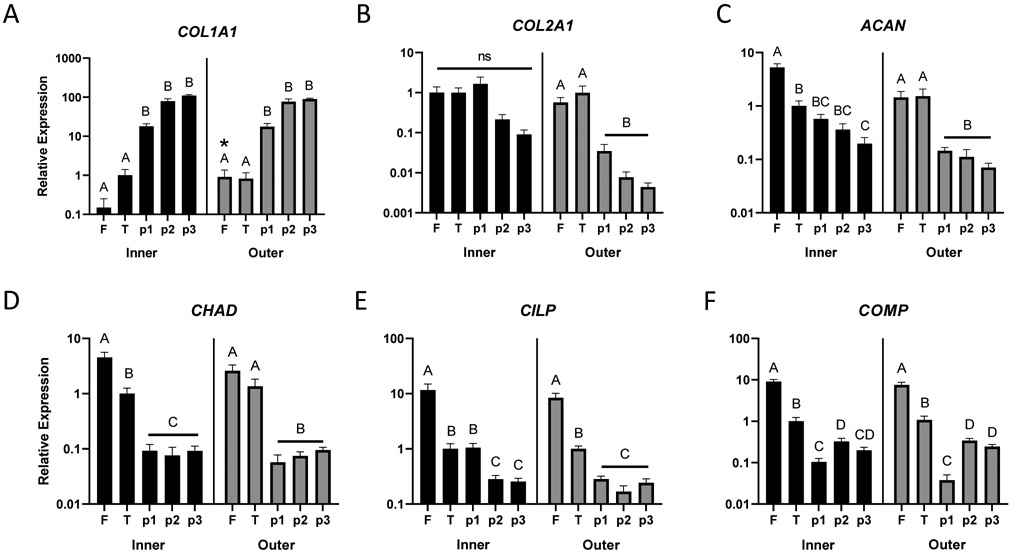
COL1A1 showed a robust increase in expression with monolayer passage in both inner and outer zone cells, increasing approximately 18-fold at passage 1 and 80 to 100-fold at subsequent passages, relative to tissue explant culture (Figure 1A). Outer zone fresh tissue expression of COL1A1 was significantly higher (p=0.02) than inner zone, although this 6-fold difference was modest compared to the changes observed with monolayer culture. Lower mean expression of COL2A1 was observed in later passage monolayer cells, but these differences were surprisingly not significant for inner zone cells (Figure 1B). COL2A1 expression of outer zone monolayer cells at all passages was significantly downregulated compared to fresh tissue or tissue explant culture (p<0.01). There was no difference detected between inner and outer zone fresh tissue expression of COL2A1. ACAN expression was downregulated in monolayer culture for both inner and outer zone cells compared to fresh tissue (Figure 1C, p<0.01). Expression in outer zone cells at all passages and inner zone cells at passage 3 was significantly downregulated compared to the corresponding tissue explant culture (p<0.05). While outer zone fresh tissue ACAN expression appeared lower than inner zone expression, no significant difference was detected (p=0.07). Expression of SOX9 (Supplemental Figure S-1) was significantly downregulated in tissue explant culture and monolayer compared to fresh tissue (p<0.001), but there were no apparent expression differences between tissue explant culture and passage 3 monolayer cells or between inner and outer zone fresh tissue.
Figure 1:
Changes in expression of (A) type I collagen (COL1A1), (B) type II collagen (COL2A1), (C) aggrecan (ACAN), (D) chondroadherin (CHAD), (E) cartilage intermediate layer protein (CILP), and (F) cartilage oligomeric matrix protein (COMP) from freshly isolated tissue, F; tissue explant culture, T; and isolated cells grown in monolayer culture at first, second, and third passage (p1, p2, p3) measured by RT-qPCR. Data are expressed as mean fold-change (+ SEM) relative to inner zone tissue explant culture (n=6/group). Within each zone (inner or outer), groups not sharing a letter are significantly different. *: significantly different from fresh inner (p<0.05, Bonferroni post-hoc).
CHAD (Figure 1D), CILP (Figure 1E), and COMP (Figure 1F) all showed significant decreases in expression in both inner and outer zone monolayer cultured cells compared to fresh tissue (p<0.001) and no significant differences were detected between inner and outer zone fresh tissue. In almost all cases, these genes were significantly differentially expressed between fresh tissue, tissue explant, and monolayer culture (p<0.05), except for CILP (inner zone tissue explant versus passage 1) and outer zone CHAD (fresh tissue versus tissue explant).
Hydrogels for Meniscus Cell Embedding:
Hydrogels for meniscus cell culture were tested for 1) mechanical properties (equilibrium modulus), 2) viability of embedded meniscus cells, and 3) expression level of the phenotype marker genes described above. The equilibrium modulus ranged from 27 kPa for the 5P5G hydrogels to 40 kPa for 10P hydrogels, and only 10P and 5P5G hydrogels had significantly different equilibrium moduli (Figure 2, p<0.05).
Figure 2:
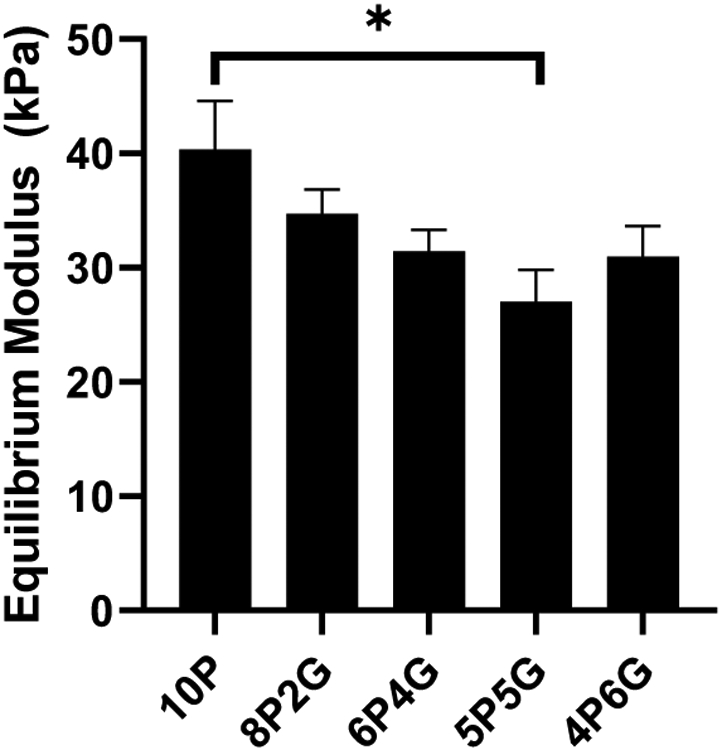
Equilibrium modulus of hydrogel materials. 10P: 10% PEGDA; 8P2G: 8% PEGDA/2%GelMA; 6P4G: 6% PEGDA/4% GelMA; 5P5G: 5% PEGDA/5% GelMA; 4P6G: 4% PEGDA/6% GelMA. Data are expressed as mean equilibrium modulus + SEM (n=8/group). *: p < 0.05 for 10P compared to 5P5G, Tukey post-hoc.
Viability of the cells embedded within the hydrogels varied by gel composition (Figure 3A and Supplemental Figure S-3). The percent cell viability indicated that higher PEGDA content negatively affected cell viability, with 10P hydrogels demonstrating decreased cell viability at 48 hours post-embedding compared to all other materials for inner zone cells (p<0.05), and as compared to 6P4G and 4P6G for outer zone cells (p<0.05). As shown by the fold-change in DNA content (Figure 3B), the cellular content of majority-PEGDA hydrogels declined over two weeks in culture (fold-change <1). Conversely, 5P5G hydrogels maintained about the same overall level of cellularity during culture (fold-change ~1) and 4P6G hydrogels promoted cellular proliferation (fold change >1). These differences were significant when comparing majority-PEGDA hydrogels (10P, 8P2G, and 6P4G) to 5P5G or 4P6G for inner zone cells (p<0.05), or when comparing majority-PEGDA hydrogels to 4P6G for outer zone cells (p<0.05).
Figure 3:
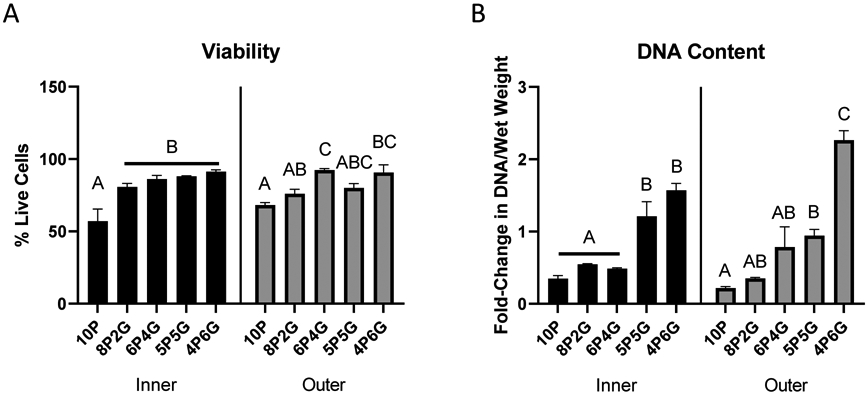
(A) Meniscus cell viability at 48 hours post-embedding. Data are expressed as mean percent live cells + SEM (n=3/group). (B) Hydrogel DNA content. Data are expressed as mean fold change (+ SEM) during culture (n=3/group). Within each zone (inner or outer), groups not sharing a letter are significantly different (p<0.05, Tukey post-hoc). 10P: 10% PEGDA; 8P2G: 8% PEGDA/2%GelMA; 6P4G: 6% PEGDA/4% GelMA; 5P5G: 5% PEGDA/5% GelMA; 4P6G: 4% PEGDA/6% GelMA.
Embedding meniscus cells in hydrogels of varying PEGDA/GelMA composition had striking effects on meniscus cell gene expression (Figure 4). COL1A1 expression was increased in hydrogels with a higher GelMA content. Only 10P and 8P2G for inner zone cells (Figure 4A, p<0.05), and 10P, 8P2G, and 6P4G for outer zone cells (Figure 4G, p<0.05) significantly suppressed COL1A1 expression compared to p3 monolayer cells. All hydrogel materials, except 8P2G and 6P4G with outer zone cells, demonstrated significantly higher COL1A1 expression than tissue explant culture (p<0.05). Higher-GelMA content hydrogels (5P5G and 4P6G for inner zone cells (Figure 4B) and 4P6G for outer zone cells (Figure 4H)) induced expression levels of COL2A1 similar to tissue explant culture and significantly higher than p3 monolayer culture (p<0.001). Similarly, ACAN expression was not significantly different from tissue explant culture for either inner (Figure 4C) or outer zone cells (Figure 4I) embedded in 5P5G or 4P6G hydrogels. For inner zone cells, 10P and 4P6G embedding induced CHAD expression significantly higher than monolayer (p<0.05), but 10P was still significantly lower than tissue explant culture (Figure 4D, p<0.05). For outer zone cells, none of the hydrogels increased CHAD expression compared to monolayer culture (Figure 4J). On the other hand, CILP expression was supported by all hydrogel materials for both inner (Figure 4E) and outer zone cells (Figure 4K); for every group, CILP expression was significantly higher than monolayer culture (p<0.05) and not significantly different than tissue explant culture. COMP expression trended in the opposite direction from the other ECM component genes, showing explant level expression in high-PEGDA hydrogels and decreasing expression as GelMA composition was increased (Figures 4F and 4L); however, for the inner zone cells (Figure 4F), none of the hydrogel constructs were significantly different from tissue explant culture expression levels.
Figure 4:
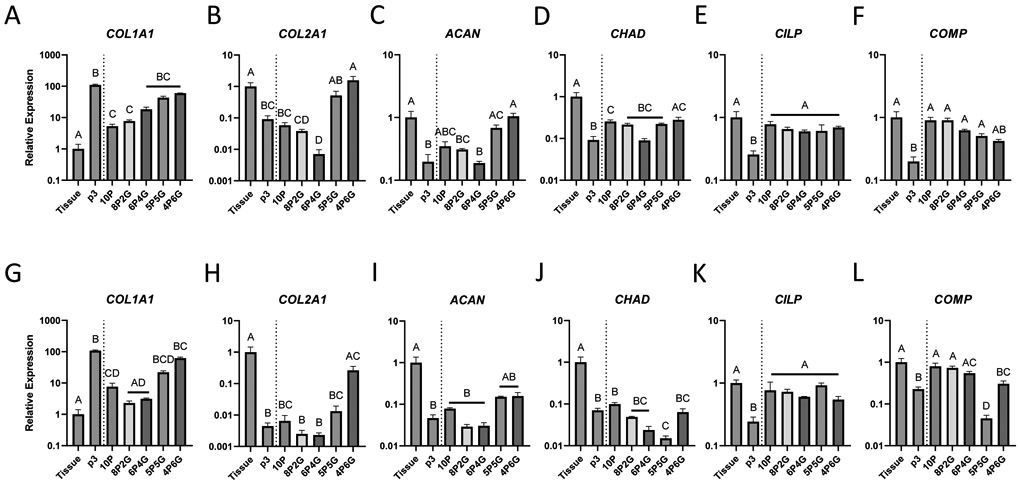
Effect of hydrogel embedding on inner zone (A-F) and outer zone (G-L) meniscus cell gene expression of (A, G) type I collagen (COL1A1), (B, H) type II collagen (COL2A1), (C, I) aggrecan (ACAN), (D, J) chondroadherin (CHAD), (E, K) cartilage intermediate layer protein (CILP), and (F, L) cartilage oligomeric matrix protein (COMP). Expression was measured by RT-qPCR on samples from tissue explant culture (Tissue), isolated cells grown in monolayer to passage 3 (p3), or hydrogel embedded meniscus cells (10P: 10% PEGDA; 8P2G: 8% PEGDA/2%GelMA; 6P4G: 6% PEGDA/4% GelMA; 5P5G: 5% PEGDA/5% GelMA; 4P6G: 4% PEGDA/6% GelMA). Data are expressed as mean fold-change (+ SEM) relative to tissue explant culture (n≥3/group). Groups not sharing a letter: p<0.05, Tukey post-hoc.
Discussion
Although it is generally accepted that meniscus cells dedifferentiate in monolayer culture, our results expand the understanding of meniscus cell dedifferentiation by providing comparisons between inner and outer zone cells and between fresh tissue and explant tissue culture, for a number of genes generally assumed to be dysregulated in meniscus cell dedifferentiation. In addition, we report decreased expression during monolayer culture of CHAD, CILP, and COMP, which were recently identified as potential markers of meniscus cell dedifferentiation10. Overall, COL1A1, CHAD, CILP, and COMP show robust and consistent changes in expression with monolayer culture, which make them useful markers of dedifferentiation in meniscus cells. Mean expression levels of COL2A1 and ACAN did decrease with passage number, but were less sensitive as markers of dedifferentiation for inner zone cells: downregulation of COL2A1 was not found to be significant, and ACAN expression was not significantly different from tissue explant culture until passage 3. In this study, we were able to directly compare the gene expression levels of meniscus tissue explant culture and monolayer culture to the expression profile of meniscus cells in different concentrations of hydrogel biomaterials. Our findings show that hydrogel embedding modulates the dedifferentiation process, and depending on the embedding material, is capable of restoring phenotype marker gene expression to levels similar to tissue explant culture. Of the materials tested, higher GelMA containing hydrogels were the most promising for meniscus cell culture, restoring expression levels of most phenotype markers to tissue explant culture levels and promoting expression of the major ECM components.
One widely accepted definition of articular chondrocyte dedifferentiation is an increase in collagen type I expression concomitant with decreases in collagen type II, aggrecan, and SOX930; 31. However, our results showed that of these genes, only COL1A1 showed robust changes in expression level for monolayer cultured meniscus cells of both inner and outer zone. While COL2A1 and ACAN expression did tend to decrease with passage, changes in relative expression were smaller than those reported in articular chondrocytes30; 31. Furthermore, in this study outer zone cells showed more robust changes in the chondrogenic gene COL2A1 during monolayer culture. SOX9, considered an indicator of chondrocyte dedifferentiation30; 32; 33, only showed differences between fresh tissue and all cultured conditions, including tissue explant culture. Therefore, we did not interrogate SOX9 further, as we focused primarily on genes that showed changes in expression between tissue explant culture and monolayer culture, as we considered the tissue explant to be the best comparator for other culture methods. Genes identified as markers of meniscus cell dedifferentiation by microarray of inner zone meniscus cells and tissue (CHAD, CILP, and COMP)10 displayed similar expression patterns between inner and outer zone tissue and decreased markedly in monolayer cultured cells in this study. Our findings match previous reports of COMP expression4; 7; however, to our knowledge CHAD and CILP have not previously been utilized as phenotype markers for both inner and outer zone meniscus cells.
Meniscus cells are often described as “chondrocyte-like” in the inner zone, and “fibroblast-like” in the outer zone11. However, of all the genes measured in this study, only COL1A1 expression was significantly different in fresh tissue between inner and outer zone cells. ACAN was nearly significant (p=0.07), while all other genes had similar expression levels between fresh tissue from the inner and outer zones. Both in our results and in the literature8; 9, significant changes in COL1A1 expression are observed between inner and outer zone tissue, while differences in expression of chondrogenic phenotype genes, such as COL2A1, ACAN, and SOX9 are relatively small or absent. Previous studies have found significant differences between inner and outer zone expression of COL2A1 and ACAN8; 9. However, the difference in COL2A1 expression reported by Upton et al. was 5.7±3.4 fold higher in inner versus outer tissue, whereas the reported change in COL1A1 expression was >10,000 fold9. This study also reported a similar fold-change in ACAN expression to our results (4.6±1.6-fold and 3.6±0.6-fold, respectively). Son and Levenston reported dramatic increases in COL2A1 expression in the inner third of the meniscus and found ACAN expression to be significantly higher in inner regions but still far lower than in cartilage8. Subdividing the tissue into thirds rather than inner two-thirds and outer third, as performed in the current study, may have increased the sensitivity to measure differences in COL2A1. However, Son and Levenston utilized tissue from two-week old calves8, which may also explain the observed differences, as patterns of COL2A1 and other ECM gene expression are likely to vary with age and health status of the tissue34-37. Therefore, although COL2A1 and ACAN expression appear to vary spatially across the meniscus, the use of these genes as phenotype markers may be complicated by their expression being modulated by a number of patient variables, including age and health. Taken together, these results demonstrate that characterizing meniscus cells as chondrocyte-like or fibroblast-like is insufficient. Indeed, a recent study utilizing single-cell RNA-sequencing identified seven unique cell types in meniscus. Markers of several of the identified cell types, including the major population defined as “fibrochondrocytes”, did not appear to be differently expressed between inner and outer zones38, which highlights the limitations of the currently accepted descriptions and zones of meniscus tissue.
One challenge in studying the meniscus is the difficulty in defining the precise location of inner versus outer zone tissue. Since there is no clear boundary to distinguish the zones, the selection of tissue for cell isolation or explant culture is subjective. In this study, we defined the outer zone tissue as the outer one-third and the inner zone tissue as the inner two-thirds of the meniscus. This method was consistent between samples and was utilized in previous studies of meniscus regional cellular characterization5; 6; 10. It is possible that further subdivision of meniscus regions may have yielded further significant differences in gene expression, as observed by Son and Levenston8. Ultimately, the true spatial variability and distribution of cell types throughout the meniscus remain largely unknown and cannot be established by the bulk-RNA preparation methods used in this paper. Therefore, further characterization of spatial variation in the meniscus cell transcriptome is necessary to understand the distribution and phenotypic characteristics of meniscus-resident cells.
In this study, we used porcine meniscus cells. However, comparisons to the literature suggest that the gene expression patterns revealed in this study generally show concordance across species, with porcine9, bovine4; 8, and human5 menisci showing similar gene expression trends in monolayer culture. Furthermore, the identification of CHAD, CILP, and COMP as potential markers of meniscus cell dedifferentiation by microarray was performed using human tissue, and our results confirmed loss of expression of these genes in porcine tissue, supporting the utility of porcine tissue for studies of meniscus cell phenotyping.
As with dedifferentiation, redifferentiation of articular chondrocytes has been studied more than redifferentiation of meniscus cells. Methods widely accepted for redifferentiation of chondrocytes include 3D culture in agarose or alginate hydrogels and/or treatment with various growth factors, such as TGF-β33; 39-42. In our experience, treating meniscus cells with TGF-β or agarose embedding results in large increases in type II collagen expression (unpublished data), which is by no means a return to baseline, or “redifferentiation” of meniscus cells. Although some have proposed using a ratio of collagen II/collagen I as an indicator of dedifferentiation6, as shown by our results, changes related to dedifferentiation would be driven almost entirely by changes in type I collagen. Furthermore, redifferentiation strategies that increase type II collagen expression could restore the ratio but would fail to restore the baseline phenotype.
The lack of established culture methods to maintain the differentiated meniscus cell phenotype may be largely due to the fact that most currently available methods are attuned to driving a chondrogenic phenotype, as well as the lack of clearly defined markers of the meniscus cell phenotype to use as benchmarks. It is known that cell shape, substrate stiffness, and availability of cellular adhesion sites have profound effects on the phenotype of cells in culture, and indeed these properties have been shown to be effective in modulating the phenotype of chondrocytes21; 43. Therefore, we tested the hypothesis that varying mixtures of PEGDA, which is a synthetic polymer often described as “biologically inert” and GelMA, which is a collagen-derived polymer retaining native cellular adhesion motifs, would alter the phenotype of embedded meniscus cells, and that the right ratio would provide an environment that returns phenotypic markers to tissue explant culture levels. In this study, gene expression was measured after two weeks in hydrogel embedded culture. This was chosen to give adequate time for the reversal of monolayer-induced dedifferentiation, which was observed over a period of 10 days during monolayer culture (time from cell isolation to passage 3). Additionally, prior work has shown that two weeks is sufficient time for hydrogel-embedded chondrocytes to produce a pericellular matrix44; 45, which is important for cells to sense and respond to their physical environment, and therefore may be important for phenotype maintenance mediated by material embedding.
The varying PEGDA/GelMA ratio hydrogels had markedly different effects on the meniscus phenotype markers measured in this study. Higher GelMA content favored expression of major ECM genes COL1A1, COL2A1, and ACAN. For inner zone cells, COL2A1 and ACAN returned to expression levels similar to tissue explant culture in 5P5G and 4P6G materials. Unfortunately, higher GelMA containing gels appeared to be less effective in returning COL1A1 expression to baseline levels. COMP, which is also a major ECM component in both cartilage and meniscus, showed an opposite trend, with expression decreasing with increasing GelMA concentration. Outer zone cells displayed similar overall trends in expression to inner zone cells, but the materials tested seemed less effective at returning their expression levels to tissue explant culture levels, suggesting that other materials or approaches may be necessary for culture of outer zone cells.
Mechanical testing of the hydrogel materials revealed only modest differences in stiffness, suggesting that the effect of cellular adhesion sites or some other property of the GelMA/cell interaction is driving phenotypic changes rather than differences in stiffness. Overall, the stiffness of these hydrogels is similar to materials used to encapsulate chondrocytes for mechanobiology and tissue engineering research46, so they should be suitable for experiments of mechanical stimulation similar to those performed on chondrocyte-embedded agarose constructs. Viability and DNA content measurements showed that higher GelMA content hydrogels supported greater cell survival and 4P6G hydrogels promoted cellular proliferation. Overall, these materials were partially successful in halting or reversing the dedifferentiation trends in the phenotype markers measured in this study. 5P5G and 4P6G materials returned most of the phenotype markers to near-tissue explant culture levels for inner zone meniscus cells; however, these materials did not significantly reduce COL1A1 expression relative to p3 monolayer culture. For tissue engineering applications, 4P6G hydrogels seem to promote maximum ECM component expression and cellular proliferation. However, COL1A1 expression seems to increase and COMP expression decreases with increasing GelMA concentration. Therefore, higher GelMA ratios may further diverge from the meniscus cell phenotype. By directly comparing expression levels to tissue explant culture, we have shown that it is possible to reverse dedifferentiation trends by embedding meniscus cells in biomaterial hydrogels. However, much like previous attempts4; 5; 15, returning COL1A1 expression to baseline remains a challenge. Additional work is still needed to further define meniscus cell phenotype, factors driving dedifferentiation, and methods to maintain a native meniscus cell phenotype in vitro for studies of meniscus cell biology and tissue engineering.
Overall, we identified COL1A1, CHAD, CILP, and COMP as useful markers of meniscus cell dedifferentiation. Our findings provide new details on the meniscus cell dedifferentiation process, including comparisons between inner and outer zone tissue, tissue explant culture, and isolated cells. However, there remains a dearth of knowledge regarding the phenotypes of meniscus cells, including clear definitions of inner and outer zone cell identities, and whether multiple subpopulations exist beyond these major delineations. Attempts to reverse or halt meniscus cell dedifferentiation by embedding meniscus cells in PEGDA/GelMA hydrogels are promising, exhibiting a partial reversal of the dedifferentiation phenotype and returning several marker genes to near tissue explant culture levels. Our findings fill important gaps in knowledge that lay the foundation for future studies of meniscus cell biology and meniscus tissue engineering.
Supplementary Material
Acknowledgements: funding and conflicts of interest
Supported in part by NIH grants AR065527, AR074800, and AR073221, an Orthopaedic Research and Education Foundation grant with funding provided by the Musculoskeletal Transplant Foundation, and the Urbaniak Sports Science Institute. We thank Nivedita Sangaj for technical assistance in hydrogel preparation. The authors have no conflicts of interest to disclose in presenting this work.
References
- 1.Nielsen AB, Yde J. 1991. Epidemiology of acute knee injuries: a prospective hospital investigation. The Journal of trauma 31:1644–1648. [DOI] [PubMed] [Google Scholar]
- 2.Lohmander LS, Englund PM, Dahl LL, et al. 2007. The long-term consequence of anterior cruciate ligament and meniscus injuries: osteoarthritis. Am J Sports Med 35:1756–1769. [DOI] [PubMed] [Google Scholar]
- 3.Abrams GD, Frank RM, Gupta AK, et al. 2013. Trends in Meniscus Repair and Meniscectomy in the United States, 2005-2011. The American Journal of Sports Medicine 41:2333–2339. [DOI] [PubMed] [Google Scholar]
- 4.Gunja NJ, Athanasiou KA. 2007. Passage and reversal effects on gene expression of bovine meniscal fibrochondrocytes. Arthritis Res Ther 9:R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan GK, Dinnes DL, Myers PT, et al. 2011. Effects of biomimetic surfaces and oxygen tension on redifferentiation of passaged human fibrochondrocytes in 2D and 3D cultures. Biomaterials 32:5600–5614. [DOI] [PubMed] [Google Scholar]
- 6.Berneel E, Philips C, Declercq H, et al. 2016. Redifferentiation of High-Throughput Generated Fibrochondrocyte Micro-Aggregates: Impact of Low Oxygen Tension. Cells, tissues, organs 202:369–381. [DOI] [PubMed] [Google Scholar]
- 7.Croutze R, Jomha N, Uludag H, et al. 2013. Matrix forming characteristics of inner and outer human meniscus cells on 3D collagen scaffolds under normal and low oxygen tensions. BMC musculoskeletal disorders 14:353–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son M, Levenston ME. 2012. Discrimination of meniscal cell phenotypes using gene expression profiles. Eur Cell Mater 23:195–208. [DOI] [PubMed] [Google Scholar]
- 9.Upton ML, Chen J, Setton LA. 2006. Region-specific constitutive gene expression in the adult porcine meniscus. Journal of Orthopaedic Research 24:1562–1570. [DOI] [PubMed] [Google Scholar]
- 10.Grogan SP, Duffy SF, Pauli C, et al. 2018. Gene expression profiles of the meniscus avascular phenotype: A guide for meniscus tissue engineering. J Orthop Res 36:1947–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makris EA, Hadidi P, Athanasiou KA. 2011. The knee meniscus: structure-function, pathophysiology, current repair techniques, and prospects for regeneration. Biomaterials 32:7411–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellio Le Graverand MP, Ou Y, Schield-Yee T, et al. 2001. The cells of the rabbit meniscus: their arrangement, interrelationship, morphological variations and cytoarchitecture. J Anat 198:525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauck RL, Martinez-Diaz GJ, Yuan X, et al. 2007. Regional multilineage differentiation potential of meniscal fibrochondrocytes: Implications for meniscus repair. The Anatomical Record 290:48–58. [DOI] [PubMed] [Google Scholar]
- 14.Verdonk PC, Forsyth RG, Wang J, et al. 2005. Characterisation of human knee meniscus cell phenotype. Osteoarthritis Cartilage 13:548–560. [DOI] [PubMed] [Google Scholar]
- 15.Adesida AB, Mulet-Sierra A, Laouar L, et al. 2012. Oxygen tension is a determinant of the matrix-forming phenotype of cultured human meniscal fibrochondrocytes. PLoS One 7:e39339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Eyckmans J, Chen CS. 2017. Designer biomaterials for mechanobiology. Nature Materials 16:1164–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, He P, Lin L, et al. 2012. Biomimetic poly(ethylene glycol)-based hydrogels as scaffolds for inducing endothelial adhesion and capillary-like network formation. Biomacromolecules 13:706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Ma M, Wang J, et al. 2018. Development of a Photo-Crosslinking, Biodegradable GelMA/PEGDA Hydrogel for Guided Bone Regeneration Materials. Materials (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazzoccoli JP, Feke DL, Baskaran H, et al. 2010. Mechanical and cell viability properties of crosslinked low- and high-molecular weight poly(ethylene glycol) diacrylate blends. Journal of biomedical materials research Part A 93:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairbanks BD, Schwartz MP, Bowman CN, et al. 2009. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials 30:6702–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, Sangaj N, Razafiarison T, et al. 2011. Influence of physical properties of biomaterials on cellular behavior. Pharm Res 28:1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichol JW, Koshy ST, Bae H, et al. 2010. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 31:5536–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyons LP, Hidalgo Perea S, Weinberg JB, et al. 2019. Meniscus-Derived Matrix Bioscaffolds: Effects of Concentration and Cross-Linking on Meniscus Cellular Responses and Tissue Repair. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. 2001. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- 25.Aung A, Theprungsirikul J, Lim HL, et al. 2016. Chemotaxis-driven assembly of endothelial barrier in a tumor-on-a-chip platform. Lab on a chip 16:1886–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins AT, Hatcher CC, Kim SY, et al. 2019. Selective Enzymatic Digestion of Proteoglycans and Collagens Alters Cartilage T1rho and T2 Relaxation Times. Ann Biomed Eng 47:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatcher CC, Collins AT, Kim SY, et al. 2017. Relationship between T1rho magnetic resonance imaging, synovial fluid biomarkers, and the biochemical and biomechanical properties of cartilage. J Biomech 55:18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hidalgo Perea S, Lyons LP, Nishimuta JF, et al. 2019. Evaluation of culture conditions for in vitro meniscus repair model systems using bone marrow-derived mesenchymal stem cells. Connective tissue research:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruprecht JC, Waanders TD, Rowland CR, et al. 2019. Meniscus-Derived Matrix Scaffolds Promote the Integrative Repair of Meniscal Defects. Sci Rep 9:8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng T, Maddox NC, Wong AW, et al. 2012. Comparison of gene expression patterns in articular cartilage and dedifferentiated articular chondrocytes. Journal of Orthopaedic Research 30:234–245. [DOI] [PubMed] [Google Scholar]
- 31.Hamada T, Sakai T, Hiraiwa H, et al. 2013. Surface markers and gene expression to characterize the differentiation of monolayer expanded human articular chondrocytes. Nagoya journal of medical science 75:101–111. [PMC free article] [PubMed] [Google Scholar]
- 32.Lefebvre V, Huang W, Harley VR, et al. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol 17:2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao Z, Lu Z, Wu H, et al. 2018. Collagen, agarose, alginate, and Matrigel hydrogels as cell substrates for culture of chondrocytes in vitro: A comparative study. Journal of Cellular Biochemistry 119:7924–7933. [DOI] [PubMed] [Google Scholar]
- 34.Di Giancamillo A, Deponti D, Addis A, et al. 2014. Meniscus maturation in the swine model: changes occurring along with anterior to posterior and medial to lateral aspect during growth. Journal of cellular and molecular medicine 18:1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreinest M, Reisig G, Ströbel P, et al. 2016. Analysis of Gene Expression and Ultrastructure of Stifle Menisci from Juvenile and Adult Pigs. Comparative medicine 66:30–40. [PMC free article] [PubMed] [Google Scholar]
- 36.Le Graverand M-PH, Reno C, Hart DA. 1999. Gene expression in menisci from the knees of skeletally immature and mature female rabbits. Journal of Orthopaedic Research 17:738–744. [DOI] [PubMed] [Google Scholar]
- 37.Rai MF, Sandell LJ, Cheverud JM, et al. 2013. Relationship of age and body mass index to the expression of obesity and osteoarthritis-related genes in human meniscus. International journal of obesity (2005) 37:1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H, Wen X, Li H, et al. 2020. Single-cell RNA-seq analysis identifies meniscus progenitors and reveals the progression of meniscus degeneration. Annals of the Rheumatic Diseases 79:408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aurich M, Hofmann GO, Best N, et al. 2018. Induced Redifferentiation of Human Chondrocytes from Articular Cartilage Lesion in Alginate Bead Culture After Monolayer Dedifferentiation: An Alternative Cell Source for Cell-Based Therapies? Tissue Eng Part A 24:275–286. [DOI] [PubMed] [Google Scholar]
- 40.Caron MMJ, Emans PJ, Coolsen MME, et al. 2012. Redifferentiation of dedifferentiated human articular chondrocytes: comparison of 2D and 3D cultures. Osteoarthritis and Cartilage 20:1170–1178. [DOI] [PubMed] [Google Scholar]
- 41.Jeyakumar V, Niculescu-Morzsa E, Bauer C, et al. 2019. Redifferentiation of Articular Chondrocytes by Hyperacute Serum and Platelet Rich Plasma in Collagen Type I Hydrogels. Int J Mol Sci 20:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong E, Reddi AH. 2013. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs−221/−222, −140, and −143/145 expression. Tissue Eng Part A 19:1015–1022. [DOI] [PubMed] [Google Scholar]
- 43.Schuh E, Hofmann S, Stok K, et al. 2012. Chondrocyte redifferentiation in 3D: The effect of adhesion site density and substrate elasticity. Journal of Biomedical Materials Research Part A 100A:38–47. [DOI] [PubMed] [Google Scholar]
- 44.Jeon JE, Schrobback K, Meinert C, et al. 2013. Effect of Preculture and Loading on Expression of Matrix Molecules, Matrix Metalloproteinases, and Cytokines by Expanded Osteoarthritic Chondrocytes. Arthritis & Rheumatism 65:2356–2367. [DOI] [PubMed] [Google Scholar]
- 45.O’Conor CJ, Leddy HA, Benefield HC, et al. 2014. TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proceedings of the National Academy of Sciences 111:1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buckley CT, Thorpe SD, O'Brien FJ, et al. 2009. The effect of concentration, thermal history and cell seeding density on the initial mechanical properties of agarose hydrogels. Journal of the mechanical behavior of biomedical materials 2:512–521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.