Abstract
Periodontitis is an infection-driven inflammatory disease, which is characterized by gingival inflammation and bone loss. Periodontitis is associated with various systemic diseases, including cardiovascular, respiratory, musculoskeletal, and reproductive system related abnormalities. Recent theory attributes the pathogenesis of periodontitis to oral microbial dysbiosis, in which Porphyromonas gingivalis acts as a critical agent by disrupting host immune homeostasis. Lipopolysaccharide, proteases, fimbriae, and some other virulence factors are among the strategies exploited by P. gingivalis to promote the bacterial colonization and facilitate the outgrowth of the surrounding microbial community. Virulence factors promote the coaggregation of P. gingivalis with other bacteria and the formation of dental biofilm. These virulence factors also modulate a variety of host immune components and subvert the immune response to evade bacterial clearance or induce an inflammatory environment. In this chapter, our focus is to discuss the virulence factors of periodontal pathogens, especially P. gingivalis, and their roles in regulating immune responses during periodontitis progression.
Keywords: P. gingivalis, gingipain, fimbria, LPS, capsule, inflammation, periodontal disease, T cell
1. Periodontitis
Periodontitis is one of the most prevalent inflammatory diseases. In US, 47% of adults, with 70% of the adult population aged 65 years and older, are afflicted with periodontitis (Eke et al., 2015; Eke, Dye, Wei, Thornton-Evans, & Genco, 2012; Papapanou, 2012). Worldwide, 538 million people suffer from severe periodontal disease and 276 million of whom lost their teeth. The figures are expected to rise as population grows and ages (Kassebaum et al., 2017). In 1999, the annual expenditure on the therapy of periodontal disease was estimated to exceed $14 billion in the USA (Brown, Johns, & Wall, 2002). Apart from being the major factor causing loss of teeth, periodontitis is associated with various systemic diseases like atherosclerosis, diabetes, rheumatoid arthritis and adverse pregnancy outcomes like low birth weight (Khader, Dauod, El-Qaderi, Alkafajei, & Batayha, 2006; Kinane, Stathopoulou, & Papapanou, 2017; Madianos, Bobetsis, & Offenbacher, 2013). Periodontal disease may exacerbate these diseases through increased pro-inflammatory mediators or direct infiltration of periodontal pathogens in the circulation (Khader et al., 2006).
2. Periodontal Pathogens and their virulence factors
Periodontitis is induced by microbial biofilm and caused by host-mediated inflammation, which leads to collateral tissue damage and clinical attachment loss (Hajishengallis, Darveau, & Curtis, 2012). Oral microbiota contains the microorganisms found in the oral cavity, which are estimated to be more than 700 different species with distinct subspecies (Aas, Paster, Stokes, Olsen, & Dewhirst, 2005; Dewhirst et al., 2010). A balance between the resident microbiota and host immune response maintains a state of homeostasis, while the disruption of this balance contributes to oral diseases like periodontal disease (Lamont, Koo, & Hajishengallis, 2018). Periodontitis is one typical disease of biofilm-associated infection (Bergstrom, 2006; Hajishengallis & Lamont, 2012; Socransky, Haffajee, Cugini, Smith, & Kent, 1998). Oral bacterial biofilms are among the first to be discovered in human and have been studied extensively. From the time that oral bacterial biofilm was first discovered by James Leon Williams in 1897 as a mass of bacteria adhering to the tooth surface, various periodontal pathogens, including Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Tannerella forsythia (formerly Bacteroides forsythus), Porphyromonas gingivalis, Prevotella intermedia and Treponema denticola, have been extensively investigated (Aruni, Dou, Mishra, & Fletcher, 2015). These bacteria and their pathogenic roles in periodontitis have been explained in various models, including red complex, specific and non-specific microorganisms, keystone pathogen, and polymicrobial synergy and dysbiosis models (Hajishengallis & Lamont, 2012).
Red complex model recognizes bacteria including P. gingivalis, T. Denticola and T. Forsythia, popularly known as the “red complex” in gingival plaques, as the major pathogens that are responsible for periodontitis progression (Socransky et al., 1998). The “red complex model” laid solid foundation for the concept that periodontitis is a multi-microbe disease, and leads to other newly emerged models following the new technology development and new discoveries. Most recently, the etiology of periodontitis has shifted in favor of the theory of polymicrobial synergy and dysbiosis (the ‘PSD model’) (Hajishengallis, Darveau, et al., 2012; Hajishengallis & Lamont, 2012), which proposes that disease is initiated by a synergistic polymicrobial community, in which different members fulfill distinct roles that result in a combined effect on the oral microbial dysbiosis and host immune disruption. Dysbiosis or imbalance in the ecosystem of microorganisms further leads to change in the host-microbe crosstalk that causes destructive inflammation, bone loss, and periodontitis (Abusleme et al., 2013).
During the formation of the pathogenic community, ‘keystone pathogens’ modulate the immune response to impair host immune surveillance and tip the balance from homeostasis to dysbiosis. Keystone pathogens can also promote the pathogenesis through the enhancement of direct interaction among the oral pathogens. Virulence factors are integral molecules expressed by an organism at various stages of its life cycle with capacity to damage the host. Formation of the pro-inflammatory environment and induction of oral microbiome dysbiosis require diverse virulence factors expressed by the bacteria, such as proteolytic enzymes, capsule, lipopolysaccharide, fimbriae, and certain other surface structures/ligands (How, Song, & Chan, 2016)
Our and others’ reports showed even in relatively low numbers, P. gingivalis can interfere with host immune responses and disrupt the symbiosis between the local oral bacteria (Hajishengallis, 2014). It acts as the keystone pathogen in the dysbiosis of oral microbiome and periodontitis development. In this chapter, we will discuss various periodontal pathogens, but focus on P. gingivalis and its virulence factors for their important roles in host immune response modulation, which are critical for the pathogen colonization, oral microbial dysbiosis, and periodontitis pathogenesis.
3. P. gingivalis and its virulence factors
P. gingivalis is a Gram-negative, anaerobic, rod-shaped bacteria forming black colonies on blood agar. It was detected in 85.75% subgingival plaque of chronic periodontitis patients (How et al., 2016). Prevalence of P. gingivalis is related to the severity of periodontal disease and has been identified by researchers as one of the major causative agents in the periodontitis pathogenesis. Recently, the keystone pathogen theory postulates that even at low abundance, P. gingivalis is capable of inducing chronic periodontitis by remodeling the commensal bacterial community to promote a state of dysbiosis, which leads to disease (Olsen, Lambris, & Hajishengallis, 2017). P. gingivalis has been widely investigated, partially benefited from the well-characterized genomics of different P. gingivalis strains and the accessibility of a great variety of mutants (Hajishengallis, Darveau, et al., 2012; Naito et al., 2008; Nelson et al., 2003). P. gingivalis is known to produce a wide array of virulence factors that could cause tissue destruction on their own or act through other mediators to induce inflammation (Hajishengallis, 2014).
P. gingivalis and Streptococcus gordonii are able to interact to form communities and subsequent colonization of the dental plaque. P. gingivalis benefits from its interaction and coaggregation in the subgingival plaque for its destructive effects on periodontal tissues (Kuboniwa et al., 2017). P. gingivalis can efficiently modify the host immune response and create an environment favorable to its own and other pathogens’ continued persistence (Hajishengallis & Lambris, 2011). Virulence factors of P. gingivalis play important roles in the coaggregation, biofilm formation, and oral microbial dysbiosis. They can either directly act on other bacteria or induce an optimal environment. One example of direct interaction is that P. gingivalis binds to GAPDH and surface proteins on the surface of S. gordonii, which ultimately favor the formation of P. gingivalis biofilms on the tooth surface (Maeda, Nagata, Yamamoto, et al., 2004).
4. P. gingivalis LPS
4.1. Structure of P. gingivalis LPS
Lipopolysaccharide (LPS) is a pathogen associated molecular pattern (PAMP) or more recently identified as microbe-associated molecular pattern (MAMP) (Medzhitov, 2001). LPS is also a major component of the cell wall (i.e. outer cell membrane) of Gram-negative bacteria, including periodontal pathogen P. gingivalis (R.P. Darveau, Arbabi, Garcia, Bainbridge, & Maier, 2002). LPS was well known for its toxicity and the ability to cause unwanted host inflammation, which gave it the name endotoxin. LPS from E. coli was found to be able to facilitate the host inflammatory response by stimulating Toll-like receptor 4 (TLR4) (Lien et al., 2000; Poltorak et al., 1998). P. gingivalis LPS, like those of E. coli and other Gram-negative bacteria, comprises the outer leaflet of the bacteria and is composed of lipid A, core oligosaccharide that forms the backbone of LPS, and O-specific polysaccharide (Schromm et al., 2000). Acting as the core factor of LPS in the induction of immune responses, Lipid A moiety confers toxicity and is structurally composed of a phosphorylated β (1–6) D-glucosamine disaccharide backbone and multiacyl chains acylated by fatty acids at specific positions on the backbone (Dixon & Darveau, 2005). LPS from different bacterial species are structurally different, depending on the fatty acid acyl chain composition of lipid A. The structure difference in LPS may explain the distinct mechanisms through which the host cells recognize bacterial species.
P. gingivalis LPS plays a strong pathogenic role in periodontal tissues. The virulence properties of P. gingivalis LPS is determined by its lipid A component. P. gingivalis LPS contains multiple forms of lipid A (R. P. Darveau et al., 2004), the chemical structure of which was determined to be a glucosamine beta-(1–6) disaccharide 1-monophosphate acylated by 3-hydroxy-15-methylhexadecanoic acid and 3-hexadecanoyloxy-15-methylhexadecanoic acid at the 2- and 2’-positions, respectively (Ogawa, 1993). Host cells respond to P. gingivalis LPS lipid A and produce inflammatory responses in gingival tissues, thereby creat a favorable environment for pathogens to sustain and ultimately cause periodontal disease progression (Herath et al., 2013).
While the basic chemical composition of P. gingivalis LPS is similar to that of E. coli endotoxin, P. gingivalis lipid A structure exhibits different variations of acylation, tetra-acylation and penta-acylation. As a consequence, P. gingivalis LPS activates distinctive signaling pathways and initiates differential immune responses. Based on its molecular weight, P. gingivalis LPS with penta-acylated lipid A is named P. gingivalis LPS1690, for its molecular weight of 1690 Da. Similarly, P. gingivalis LPS with tetra-acylated lipid A is called LPS1435/1449 (Curtis et al., 2011). Basic mechanism for the production of distinctive P. gingivalis LPS remains controversial, which can be attributed to different P. gingivalis strains and environmental factors, such as the hemin levels, phosphate availability, and incubation temperatures (R.P. Darveau et al., 2002) (Rangarajan, Aduse-Opoku, Paramonov, Hashim, & Curtis, 2017).
4.2. Immune responses triggered by P. gingivalis LPS
P. gingivalis LPS-induced TLR-specific immune regulatory roles have been extensively investigated. While E. coli-LPS can activate TLR4 but not TLR2 (O. Takeuchi et al., 1999), the TLR activations that triggered by P. gingivalis LPS are much more complicated (Coats et al., 2009; R. Liu, Desta, Raptis, Darveau, & Graves, 2008). The leading signaling pathway activated by P. gingivalis LPS is controversial. Some reports showed that TLR4 exerts a dominant function, while others demonstrated that TLR2 is the major receptor (P.-L. Wang & Ohura, 2002). Reports proved that TLR2 is required for alveolar bone loss caused by P. gingivalis infection in animal models. P. gingivalis stimulation led to the up-regulation of TLR2 expression and pro-inflammatory cytokine production in vitro (Papadopoulos et al., 2013). TLR2 is also critical for P. gingivalis LPS-activated nitric oxide, TNF-α, and IL-6 production by macrophages (Holden et al., 2014). The signaling pathways downstream of TLRs are also controversial. Some reports showed that NF-ĸB pathway is critical in TLR-mediated pro-inflammatory signaling and the production of cytokines, chemokines, and adhesion molecules (R. P. Darveau et al., 2004). NF-ĸB pathway is also important in alveolar bone loss. Its activation by LPS from non-periodontal pathogen E. coli, through TLR4 is enough to cause periodontitis (Li et al., 2014). On the other hand, other pathways including the mitogen-activated protein kinase (MAPK) pathway are also reported to be involved in inflammation and bone loss (Bainbridge & Darveau, 2001).
Recent discovery revealed that the binding and activation of specific TLRs by LPS are determined by the structure of different types lipid A, which is the core component and effector of the LPS molecule. Even small changes in lipid A structure can influence the immune responses (Dixon & Darveau, 2005). Lipid A heterogeneity in P. gingivalis LPS led to significantly different innate immune response and pro‐inflammatory cytokine production (Herath et al., 2011). When binding to TLR4, penta-acylated P. gingivalis LPS1690 is similar to classical hexa-acylated E. coli LPS in certain ways (Bozkurt, Hakki, Hakki, Durak, & Kantarci, 2017). For example, both P. gingivalis LPS1690 and E. coli LPS elevated the LBP protein expression in human oral keratinocytes. P. gingivalis LPS1690 acts differently from traditional E. coli LPS in several ways. While P. gingivalis LPS1690 activated NF-κB and p38/MAPK pathways, E. coli LPS regulated through NF-κB, p38/MPAK and JNK signaling pathways (Ding, Wang, Darveau, & Jin, 2013). P. gingivalis LPS1690 may also bind to TLR2. Through TLR2 activation, P. gingivalis LPS1690 significantly enhanced the transcription of NF-κB in mouse cementoblasts. Comparing to P. gingivalis LPS1690, tetra-acylated LPS1435/1449 stimulation relied even less on TLR4, and more on TLR2 (Nemoto, Darveau, Foster, Nogueira-Filho, & Somerman, 2006). Indeed, LPS1435/1449 acts as a TLR4 antagonist and TLR2 agonist (Herath et al., 2013; Herath et al., 2011). Stimulation with either P. gingivalis LPS1690 or LPS1435/1449 elevated TLR2 expression on the cell surfaces of hGFs, but LPS1435/1449 induced higher TLR2 expression than LPS1690. In contrast, P. gingivalis LPS1690, but not LPS1435/1449, significantly increased TLR4 and MD-2 expression in hGFs. P. gingivalis LPS1690 activated mainly through NF-κB, while P. gingivalis LPS1435/1449 primarily induced the p38/MAPK and ERK1/2 signaling pathways.
The structure of lipid A in LPS also determines its binding to a newly found intracellular LPS receptor, caspase-11. E. coli LPS can also bind to mouse caspase-11 (homologue of human caspase-4), induce caspase-11 oligomerization, and activate the caspases (Kayagaki et al., 2013). On the other hand, underacylated lipid IVa and tetra-acylated LPS, such as Rhodobacter sphaeroides LPS (LPS-RS), cannot induce caspase-11 oligomerization and activation, despite that they can bind to caspase-11 (Shi et al., 2014). Tetra-acylated P. gingivalis lipid A was found to evade activation of the noncanonical inflammasome, which enhanced the intracellular survival of P. gingivalis in macrophages. In contrast, P. gingivalis LPS1690, which contains TLR4 agonist lipid A, induced noncanonical inflammasome activation and led to the production of IL-1β and deprived the survival capability of P. gingivalis in macrophages (Slocum et al., 2014). P. gingivalis might exploit its TLR4 antagonistic lipid A to evade host clearance by interfering with the activation of TLR4 and caspase-11. Indeed, the lipid A phosphatase, which is responsible for lipid A alterations, was shown to be critical for P. gingivalis colonization and even the formation of oral microbiome, indicating the importance of the lipid structure in the processes (Zenobia et al., 2014).
5. Fimbriae
5.1. Types and compositions of fimbriae
P. gingivalis develops fimbriae, or pili, which are thin filamentous surface appendages that protrude from the outer membrane. P. gingivalis fimbriae have been among the focuses of research in periodontal pathogens and periodontitis pathogenesis. Fimbriae enhance the biofilm formation, bacterial motility, bacterial adhesion to host cells, and bacterial invasion into the cells. P. gingivalis expresses two forms of fimbriae, the long fimbriae (also called major fimbriae) comprised of FimA subunits, and the short fimbriae (or minor fimbriae) with Mfa1 subunit proteins. Both FimA and Mfa1 are critical for host immune responses and pathogenesis of periodontitis, even though their amino acid composition and antigenic properties are different (Amano, 2010). With optimal staining, fimbriae can be observed under microscopes, while different P. gingivalis strains showed variance in surface morphology and capability to adhere to host cells (Zheng, Wu, & Xie, 2011).
Long fimbriae are 5 nm wide and extend up to 3 μm long, with sizes varying from 41 to 49 kDa (Dickinson, Kubiniec, Yoshimura, & Genco, 1988; Handley & Tipler, 1986; Lee, Sojar, Bedi, & Genco, 1991). FimA is the shaft subunit, with accessory proteins FimC, FimD, and FimE as minor components of the long fimbriae (Enersen, Nakano, & Amano, 2013; Watanabe et al., 1996). FimE is required for assembly of FimC and FimD onto the distal tip of FimA fibers (Nishiyama et al., 2007), while FimB has been suggested as fimbria anchors and regulators of fimbrial length (Nagano, Hasegawa, Murakami, Nishiyama, & Yoshimura, 2010). Deficiency in FimB expression was reported to lead to the formation of abnormal longer fimbriae, which easily shed from the surface (Nagano et al., 2010).
FimA proteins vary in sizes and amino terminals. Fimbriae can be classified into six types based on nucleotide sequences of the fimA gene (type I, Ib, II, III, IV, and V) (Fujiwara, Morishima, Takahashi, & Hamada, 1993). Strains 381, ATCC 33277, and HG565 are type I strains and abundantly fimbriated that showed significant adhesive capability to host tissues and cells. Strains A7A1, SA2, BH18/10, OMZ314, and OMZ409 belong to type II, while strain BH6/26 is a type III P. gingivalis. Type IV strains, such as W50 and W83, are poorly fimbriated. HNA99 belongs to type V (Enersen, Olsen, Kvalheim, & Caugant, 2008). When tested for the fimA genotypes of P. gingivalis, type II and I are the most widely distributed ones in plaque samples from periodontitis patients (Enersen et al., 2008; Nagano, Hasegawa, Iijima, Kikuchi, & Mitani, 2018).
Short fimbriae, composed of mainly Mfa1 structural subunit proteins, differ from 60 to 500 nm in their length (Enersen et al., 2013). Similar to long fimbriae, short fimbriae also contain accessory proteins, which are named Mfa2–5 (Hasegawa et al., 2016; Hasegawa et al., 2009). Mfa2 is an anchor subunit as well as an assembly and elongation terminator (Hasegawa et al., 2009). Mfa3 is localized at the distal tip of the short fimbriae, implying its role as a ligand to host cell receptors (Hasegawa et al., 2016; Hasegawa et al., 2013). Interestingly, Mfa3, Mfa4, and Mfa5 work closely together, by which the absence of any one of these three accessory proteins would lead to lack of all three in the mature fimbriae (Hasegawa et al., 2016; Hasegawa et al., 2013).
5.2. Roles of fimbriae in biofilm formation
As we mentioned earlier, fimbriae play important roles in many of the adhesive properties of P. gingivalis. P. gingivalis fimbriae bind to host tissues and cells through a wide variety of host molecules and oral substrates, which include proline-rich proteins and glycoproteins, statherins, fibrinogen, fibronectin, and lactoferrin (Amano, 2003). Besides their adhesive function to host tissues and cells, fimbriae are also important for the interaction of P. gingivalis with other oral bacteria (Lamont & Jenkinson, 2000), which is indispensible of biofilm formation. Biofilm formation is a complex process, during which the early colonizers first attach to the tooth surface, followed by the attachment of later colonizers (Kuboniwa & Lamont, 2010; Whittaker, Klier, & Kolenbrander, 1996). Fimbriae have been reported to mediate the aggregation of P. gingivalis with other organisms including Actinomyces viscosus (Goulbourne & Ellen, 1991), T. denticola (Hashimoto, Ogawa, Asai, Takai, & Ogawa, 2003), S. gordonii (Maeda, Nagata, Nonaka, et al., 2004), and Streptococcus oralis (Maeda, Nagata, Yamamoto, et al., 2004). The molecules bound by the P. gingivalis fimbriae have been under investigation. Some manifested molecules include the dentilisin of T. denticola and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of S. gordonii and S. oralis (Hashimoto et al., 2003; Maeda, Nagata, Yamamoto, et al., 2004; Y. Park et al., 2005). Interestingly, P. gingivalis long fimbriae can also bind to human GAPDH, which imply one pathway in host cell invasion and induction of host responses (Sojar & Genco, 2005). Aside from the major components, the minor components of long fimbriae are also critical for the adherence capability of P. gingivalis. It was reported that the mutants lack of fimC-, fimD-, and fimE were less efficient to bind to GAPDH of S. oralis as well as fibronectin and type I collagen (Maeda, Nagata, Yamamoto, et al., 2004; Nishiyama et al., 2007).
On the other hand, the roles and working mechanisms of short fimbriae in biofilm formation are likely different from those of long fimbriae. For instance, P. gingivalis short fimbriae can bind to SspA and SspB proteins of S. gordonii (Lamont, Hsiao, & Gil, 1994). Ssp proteins are members of the antigen I/II family of streptococcal surface proteins. Interestingly, while the antigen I/II proteins are highly conserved among the streptococcal species, P. gingivalis can only adhere to SspA/B proteins of S. gordonii, but not antigen I/II from S. mutans. Research showed that the P. gingivalis Mfa1 specially recognized the amino acid residues 1167–1250 at the C-terminal region of the SspB protein, which was named SspB adherence region (BAR) (Y. Park et al., 2005).
5.3. Host immune responses induced by fimbriae
Compared with wild type P. gingivalis, the mutant lacking FimA induced less severe bone loss in an animal periodontitis model (Malek et al., 1994). In addition to the fimbriae’s roles in biofilm formation, their effects on host immune responses have also been well investigated (Enersen et al., 2013; Pathirana, O’Brien-Simpson, & Reynolds, 2010). The fimbriae can be recognized by the receptors on epithelial, endothelial, and immune cells, which leads to the activation of the cells and the production of cytokines and adhesion molecules (Amano, 2003).
Long fimbriae activated NF-ĸB via TLR2 and induce the production of pro-inflammatory cytokines, such as tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-8, and IL-6 (Hajishengallis et al., 2006). CD14 is an essential TLR2 co-receptor for P. gingivalis FimA (Eskan, Hajishengallis, & Kinane, 2007). Gingival epithelial cells do not express CD14, therefore respond poorly to P. gingivalis FimA. In contrast, monocytes express CD14 and respond to FimA (Eskan et al., 2007). P. gingivalis fimbriae can induce differential immune responses in different host cells, which might be a strategy exploited by P. gingivalis to optimize its colonization and infection.
Long fimbriae of P. gingivalis are also able to exploit complement pathway to subvert the host immune clearance. Our research showed that fimbriae interacted with complement receptor 3 (CR3; CD11b/CD18) in monocytes/macrophages, resulting in extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation and inhibition of LPS-induced IL-12 production. In vivo results proved that CR3 blockage enhanced IL-12 production and P. gingivalis clearance. Furthermore, CR3 antagonist protected the mice from P. gingivalis-induced periodontitis (Hajishengallis, Shakhatreh, Wang, & Liang, 2007). Since IL-12 is involved in oral pathogen clearance, fimbriae activated CR3 signaling cross talk with P. gingivalis-activated TLR pathways, and inhibit IL-12 production to promote the survival of P. gingivalis. Later, our research illustrated that the activation of CXCR4 with P. gingivalis interacted with TLR2 signaling. In this process, binding of long fimbriae to CXCR4 and TLR2 led to cAMP-dependent protein kinase A activation, which inhibited host immune clearance of bacteria (Hajishengallis, Wang, Liang, Triantafilou, & Triantafilou, 2008). Another crosstalk pathway is between CXCR4 and CR3, in which P. gingivalis fimbriae utilize CXCR4 to induce PI3K-dependent activation of CR3 (Hajishengallis, McIntosh, Nishiyama, & Yoshimura, 2012). Both CR3 and CXCR4 exploitation require FimA and FimCDE to assemble fully functional long fimbriae (Pierce et al., 2009; Min Wang et al., 2007).
Research on the effect of short fimbriae is relatively limited. Infection of FimA (long fimbriae) knockout mutant, mfa1 (short fimbriae) knockout, and double mutant P. gingivalis in an animal model showed that both long fimbriae and short fimbriae contributed to alveolar bone loss in periodontitis (Umemoto & Hamada, 2003). Short fimbriae, similar to long fimbriae, interacted with TLR2 and CD14 and induced the cytokine production in both human monocyte cell lines and mouse macrophages (Hamada, Watanabe, Arai, Hiramine, & Umemoto, 2002; Hiramine, Watanabe, Hamada, & Umemoto, 2003). In dendritic cells, Mfa1 interacts with DC-SIGN (Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin), which stimulates the engulfment of P. gingivalis by dendritic cell but promotes the evasion of antibacterial autophagy and lysosome fusion, and leads to the intracellular persistence of P. gingivalis in myeloid dendritic cells (El-Awady et al., 2015).
6. Gingipain
Gingipain is a family of cysteine proteinase, which is also known as “trypsin-like” enzyme (Bostanci & Belibasakis, 2012). Gingipains include arginine-specific gingipains (Rgp, including RgpA and RgpB), which hydrolyze peptide bonds at the carbonyl groups of arginine residue, and lysine-specific gingipain (Kpg), which cleave polypeptides at the C-terminal after lysine residue. Structures of the gingipains are highly conserved among different P. gingivalis strains. Kgp is composed of a catalytic domain that specifically recognize Lys-Xaa peptide bond and an immunoglobulin‐like domain, which are followed by a C-terminal that contains hemagglutinin‐adhesin domain. RgpA’s structure is similar to Kgp, with an Arg-Xaa recognizing catalytic domain, an immunoglobulin‐like domain, and C-terminal hemagglutinin‐adhesin domain. On the other hand, RgpB possesses the catalytic domain and immunoglobulin‐like domain that similar to RgpA and Kgp, but lacks the COOH-terminal hemagglutinin domain (Guo, Nguyen, & Potempa, 2010). Newly Translated products of gingipain genes are inactive zymogens to avoid the unwanted proteolytic activity inside the cell and require further post-translational processing to produce mature functional gingipains. For example, pro‐RgpA and pro‐Kgp need to go through proteolysis to liberate the hemagglutinin‐adhesin domains. In different P. gingivalis strains, the mature gingipains can either be transported and docked on the bacterial outer membrane or released into extracellular environment.
Gingipains are accountable for 85% of the extracellular proteolytic activities of P. gingivalis and are important in the pathogenesis of periodontitis (de Diego et al., 2014). They cause dysregulated immune responses and inflammation through their involvement in the activation of the host matrix metalloproteinases, inactivation of immune inhibitors, degradation of immune factors, and cleavage of immune cell receptors (Guo et al., 2010; Imamura, Travis, & Potempa, 2003; J. Potempa, Banbula, & Travis, 2000). Inhibitors of gingipains can effectively suppress the pathogenicity of P. gingivalis and are studied for their potential therapeutic functions (Kadowaki et al., 2004).
6.1. Roles of gingipains in biofilm formation
Gingipains enhance the interactions of P. gingivalis with other periodontal pathogens, including T. forsythia, T. denticola, and A. actinomycetemcomitans, to facilitate their survival and biofilm formation (Bao et al., 2014; Haraguchi, Miura, Fujise, Hamachi, & Nishimura, 2014; Ito, Ishihara, Shoji, Nakayama, & Okuda, 2010). Gingipains can be directly involved in coaggregation with other bacteria or adhesion to host tissues and cells. P. gingivalis mutants without all three gingipains (rgpA- rgpB- kpg-) exhibit no coaggregative activity (N. Abe et al., 2004; Ito et al., 2010). Reports showed that the hemagglutinin‐adhesin domains of RgpA and Kgp are responsible for the coaggregation (N. Abe et al., 2004; Kamaguchi et al., 2003), which was proved by the observation that P. gingivalis mutants deficient in hemagglutinin domain-bearing gingipains (i.e. RgpA and Kgp) and hemagglutinin hagA genes were deprived of their coaggregative capability as well (N. Abe et al., 2004). Furthermore, gingipains are involved in the fimbria-mediated adherence process through the modification and maturation of pro-fimbrilin into fimbriae. RgpB is vital for the processing and maturation of FimA (Kristoffersen, Solli, Nguyen, & Enersen, 2015), while both RgpA and RgpB are required for the proteolytic processing and polymerization of Mfa1 to produce short fimbriae (Lee et al., 2018). Besides their ability to coaggregate with other bacteria, gingipains participate in P. gingivalis adhesion to host cells like gingival epithelial cells and gingival fibroblasts, which also expedite their survival and infection (Andrian, Grenier, & Rouabhia, 2004; Grenier et al., 2003; Sakanaka, Takeuchi, Kuboniwa, & Amano, 2016).
Gingipains can also provide nutrients for the bacteria and help their proliferation. They degrade fibrinogen/fibrin, which contribute to the gingival tissue breakdown, inhibition of blood coagulation, and increased bleeding (Ally et al., 2003). Gingipain Kgp cleaves host heme proteins, such as hemoglobin, haptoglobin, and hemopexin, to provide P. gingivalis with heme for its growth (Sroka, Sztukowska, Potempa, Travis, & Genco, 2001).
6.2. Immune regulation by gingipains
Gingipains provide P. gingivalis the abilities to evade host immune responses and clearance. Some of the mechanisms involved the degradation of extracellular matrix components, such as collagens, cytokines, immunoglobulins, and complement factors (Curtis, Aduse-Opoku, & Rangarajan, 2001). Gingipains degrade the junctional adhesion molecules of the gingival epithelial cells to break down epithelial barrier and increase the LPS and proteoglycan penetration (H. Takeuchi et al., 2019). Gingipains are also involved in regulating the immune responses and the production of immune mediators in various cells. Defensins are antimicrobial peptides in innate immune processes that play an important role in pathogen clearance. In humans, defensins are classified into α-defensins (human neutrophil peptides) and ß‐defensins (hBDs) (Dommisch & Jepsen, 2015). Gingipains are reported to degrade both neutrophil-derived α-defensins and ß‐defensins (Carlisle, Srikantha, & Brogden, 2009; Maisetta, Brancatisano, Esin, Campa, & Batoni, 2011), which might protect the bacteria from being eradicated. Gingipains reduce the expression of innate immune receptors CD14 on macrophage cell surface, which results in decreased response to bacterial infection (Wilensky, Tzach-Nahman, Potempa, Shapira, & Nussbaum, 2015). In human monocytes, chemical inhibitor of gingipains reduced the IL-1β production, indicated its participation in the regulation of inflammatory (Hamedi et al., 2009).
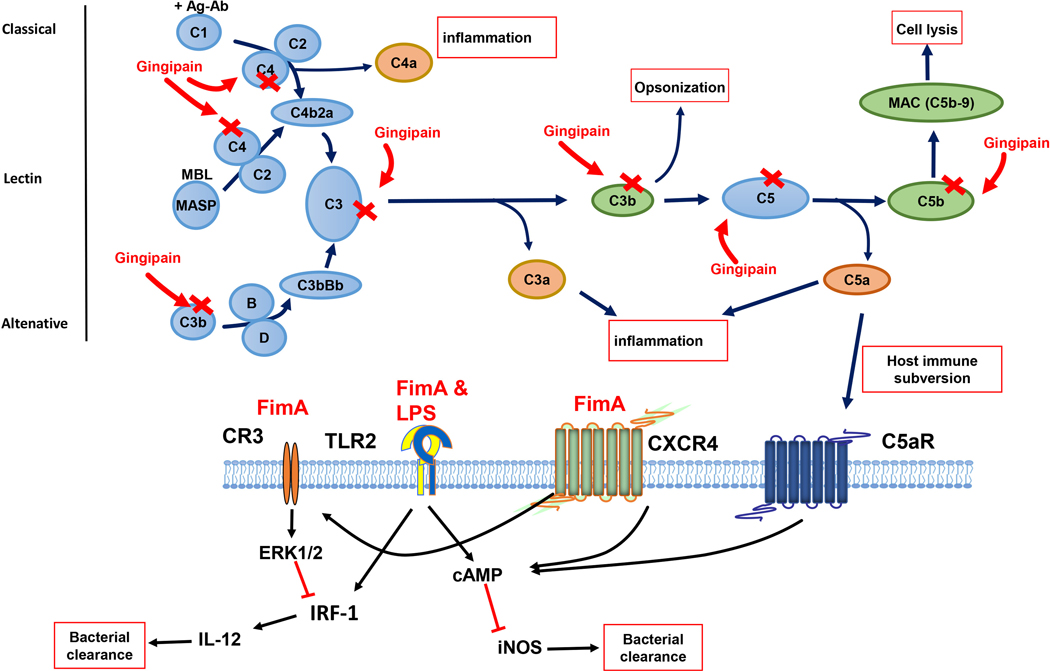
Recently, complement pathways regained the interests of dental research in that they are critical for P. gingivalis to cause microbial dysbiosis in oral cavity (Hajishengallis et al., 2011). Complement system is composed of many proteins and protein fragments, which work through a cascade of reactions to (1) generate opsonization factors to promote phagocytosis of microbes and damaged host cells, (2) promote inflammation, and (3) lyse and destroy foreign cells by forming the membrane attack complex (MAC). The system can be activated through classical pathway, lectin pathway, or alternative pathway. All three pathways converge at the central stage of C3 activation. C3 is cleaved by C3 convertase to generate C3b and C3a. C3b can act as an opsonization factor to promote phagocytosis, while C3a is an important anaphylatoxin. More importantly, C3b is required for complement cascade to cleaves C5 into C5b and C5a. C5b works together with C6, C7, C8, and C9 to form membrane attack complex (MAC), which generates a pore on cell membrane and leads to the lysis of foreign cells. The “side products” of this process, anaphylatoxins C3a, C4a, C5a, are important pro-inflammatory factors. Gingipains disrupt the activation of complement system and host-microbe homeostasis in the periodontal tissue, which further cause periodontitis. Gingipains degrade components C3, C4, and C5 to inhibit complement activation and the formation of MAC, therefore inhibit the bacterial clearance (Popadiak, Potempa, Riesbeck, & Blom, 2007; M. Potempa et al., 2008). Degradation of C3 and C3b molecules by gingipains could also contribute to decreased bacterial opsonization to reduce antibody-mediated bacterial clearance (Wingrove et al., 1992). Rpgs are more potent in degrading C3, C4, and C5 than Kpg is (Popadiak et al., 2007; M. Potempa et al., 2008). Disruption of complement pathway by gingipains not only inhibits bacterial clearance, but also elevates inflammation. For example, Rgps cleave complement component C5 molecule and degrade C5b to block the formation of MAC. At the same time, anaphylatoxin C5a is released and result in higher inflammation (Liang et al., 2011; Schenkein, Fletcher, Bodnar, & Macrina, 1995; Wingrove et al., 1992). On the other hand, Kpg, but not Rpg, can cleave the C5a receptor (C5aR) on neutrophils, which might be a mechanism to curb the recruitment and anti-bacterial action of neutrophils (Jagels, Ember, et al., 1996; Jagels, Travis, Potempa, Pike, & Hugli, 1996).
Moreover, complement systems are involved in a network of cross-interactions with other signaling pathways (Ricklin, Hajishengallis, Yang, & Lambris, 2010) (Hajishengallis, Abe, Maekawa, Hajishengallis, & Lambris, 2013). Indeed, P. gingivalis fimbriae exploit the crosstalks between complement receptor CR3, chemokine receptor CXCR4, and TLR signaling pathways to evade bacterial clearance (Maekawa et al., 2014). Additional complement-TLR crosstalk has been discovered to act through the C5a receptor (C5aR) and C3a receptor (C3aR) signaling, which involve mitogen-activated protein kinases (MAPKs), leading to extracellular signal-regulated kinase (ERK1/2) and the c-Jun N-terminal kinase (JNK) activation (X. Zhang et al., 2007). The production of C5a from C5 by Rgps facilitates the crosstalk (Schenkein et al., 1995; Wingrove et al., 1992). While the complement–TLR crosstalk might intend to serve as a protective mechanism against bacterial infection for the host, enhanced production of TNF-α, IL-1β and IL-6 could upregulate inflammation and cause a detrimental effect in inflammatory diseases (X. Zhang et al., 2007). C5aR and C3aR can be potential therapeutical targets since reports showed that blockade of C5a receptor (C5aR) and C3a receptor (C3aR) signaling decrease inflammation and ameliorate the periodontitis (T. Abe et al., 2012; Hajishengallis et al., 2011; Ricklin et al., 2010).
7. Capsules of P. gingivalis
The bacterial capsule is an outer envelope structure that lies outside bacterial cell. Capsule usually consists of polysaccharides and contains water to keep the bacterial cells from desiccated. It is important for the bacteria to survive in less-than-optimal environment. Capsule also acts as a protective mechanism in that encapsulated bacteria are more resistant to phagocytosis and intracellular killing. There are many bacterial strains that possess capsules, which protect the bacteria from host immune clearance. One well-known example in textbooks is Streptococcus pneumonia, whose capsule was verified as important virulence factors in early 20th century, which showed that the encapsulated pneumococci were virulent while the non-encapsulated variants were avirulent (Griffith, 1928).
The capsule of P. gingivalis is also known as K-antigen (Bostanci & Belibasakis, 2012; Holt, Kesavalu, Walker, & Genco, 1999; Schifferle, Reddy, Zambon, Genco, & Levine, 1989). The gene loci of P. gingivalis capsule have been identified and characterized (Aduse-Opoku et al., 2006). The chemical composition of P. gingivalis capsules varies, by which different P. gingivalis strains can be categorized into different K-serotypes, while some P. gingivalis strains are not encapsulated at all (Laine, Appelmelk, & van Winkelhoff, 1997). For example, P. gingivalis strains ATCC 33277 and W83 have been extensively used in our and others’ previous work (Chen et al., 2004; Hajishengallis, 2009; Hajishengallis et al., 2011; Hajishengallis et al., 2008; Liang et al., 2011; Nishikawa & Duncan, 2010; Pierce et al., 2009; Popadiak et al., 2007; Scheres & Crielaard, 2013; M. Wang et al., 2010; Min Wang et al., 2007). Although they both show strong capability to regulate host immune responses and induce periodontitis, ATCC 33277 is not encapsulated and W83 possesses K1 serotype polysaccharide capsules (Brunner et al., 2010; Sheets, Potempa, Travis, Casiano, & Fletcher, 2005; Sheets, Potempa, Travis, Fletcher, & Casiano, 2006). Similar to its functions in other bacteria, P. gingivalis capsule protects bacteria from the phagocytosis by host leukocytes, while non-encapsulated P. gingivalis strains are less resistant to phagocytosis and killing by macrophages and dentritic cells (Laine et al., 1997; Sundqvist, Figdor, Hanstrom, Sorlin, & Sandstrom, 1991). Encapsulated P. gingivalis strain showed much higher survival rate than the non-encapsulated strain (A. Singh et al., 2011). Report showed that different serotype of capsules exhibited differential adhesion capabilities. Overall, the adhesion capacity of encapsulated P. gingivalis to gingival epithelial cells was lower than the non-encapsulated ones (Dierickx et al., 2003). The roles of capsules in the decreased adhesion ability might contribute to its protective mechanism from phagocytosis by host cells. Capsules of P. gingivalis can also bind to other periodotopathogens to facilitate the co-aggregation of the bacteria and biofilm formation. The binding of P. gingivalis to other bacteria is K serotype specific. For example, K5 and K6, but not K1 capsules, can bind to F. nucleatum PK 1594 (Rosen & Sela, 2006).
Capsule was shown to induce host immune responses. Different P. gingivalis capsular serotypes induced differential cytokine expression in dendritic cells (Vernal et al., 2009) and chemokine production in macrophages (d’Empaire, Baer, & Gibson, 2006). In both cases, W83 strain of K1 serotype was most potent in stimulating chemokine and cytokine production (d’Empaire et al., 2006; Vernal et al., 2009). When the role of capsule in a specific strain was tested, a wild-type encapsulated strain W83 exhibited weaker ability to induce cytokine production in human gingival fibroblasts than its noncapsulated mutant (Brunner et al., 2010). It is possible that the P. gingivalis capsule is a weaker immunity inducer than other virulence factors. P. gingivalis capsule antagonizes the strong stimuli and promotes bacterial evasion of host immune responses, therefore promotes bacterial survival in hosts.
Another report demonstrated that encapsulated strain W50 was more resistant to bactericidal effect of antimicrobial peptide, such as defensin HBD3, than its nonencapsulated mutant (Igboin, Tordoff, Moeschberger, Griffen, & Leys, 2011).
In a mouse abscess model, the encapsulated P. gingivalis exhibited increased virulence than nonencapsulated strains in forming abscesses. The encapsulated strains caused invasive and spreading abscesses, while nonencapsulated strains caused non-invasive and localized abscesses (Katz, Ward, & Michalek, 1996; Laine & van Winkelhoff, 1998)(A. Singh et al., 2011). Furthermore, when animals were subcutaneously injected with non-encapsulated P. gingivalis strain 33277 or strain A7A1–28 of K3 serotype, P. gingivalis 33277-challenged animals produced lower levels of serum immunoglobulin G (IgG) and salivary IgA than A7A1‐28‐challenged ones, implying that capsules may help to induce adaptive immunity and antibody production (Katz et al., 1996). In summary, various reports proved that capsule affected the virulence of P. gingivalis. These results showed that the observed differential virulence among the different P. gingivalis strains could be at least partially attributed to the inequality in capsular structure and function, while the detailed mechanism of capsule’s involvement in the responses needs further investigation and clarification.
8. T cell Immunity induced by P. gingivalis and its virulence factors
Bacteria and their virulence factors play important roles in the pathogen interactions and inflammation. Dysregulated host factors like innate immune cells, cytokines, and other soluble factors, which are affected by the virulence factors, caused periodontitis progression. T cells are involved in various inflammatory diseases and are critical in host immune responses. Almost a half-century ago, Ivanyi and Lehner first reported that oral bacteria stimulated lymphocyte activation in mild periodontal disease patients, while this activation was suppressed in severe periodontitis patients (Ivanyi & Lehner, 1970). Later reports showed that T cells from periodontitis patients exhibited a reduced ability to proliferate, suggesting an impaired T cell response during periodontitis (Cole, Seymour, & Powell, 1987). The suppression of T cell activation can be induced by various periodontal pathogens (Shenker & Datar, 1995; Stashenko, Resmini, Haffajee, & Socransky, 1983). Gingipain Rpgs, which down-regulate IL-2 production and attenuate T-cell proliferation, might be used by P. gingivalis to evade host adaptive immune system (Khalaf & Bengtsson, 2012). Gingipains can also degrade T cell receptors, such as CD4 and CD8, and down-regulate T cell activation (Hajishengallis et al., 2013; Kitamura, Matono, Aida, Hirofuji, & Maeda, 2002). Interestingly, when tested for mixed lymphocyte reactions (MLR), not all the periodontitis patients showed impaired T cell reactions. For the patients who showed lower MLR responses, the T cells were comprised of lower percentage of naïve T cells. After periodontal therapy in these patients, the MLR responses and naïve T cells went back to normality (Kimura, Fujimoto, & Okada, 1991). While these findings pointed to a protective function of T cells, recent research demonstrated that the roles of T cell immunity in periodontitis are complicated, partially due to the existence of divergent T cell subsets.
Baker and colleagues found that P. gingivalis infection induced bone loss at a lower level in the severe combined immunodeficient (SCID) mice, which lack B and T lymphocytes, than immunocompetent mice (P. J. Baker, Evans, & Roopenian, 1994). Later, Baker et al. further demonstrated that MHC-II deficient mice, which cannot induce CD4+ T cell development and responses, were resistant to alveolar bone loss in a P. gingivalis-induced periodontitis model. On the other hand, MHC-I deficient mice that lack CD8+ T cells showed no change in bone loss (Pamela J. Baker et al., 1999). In another report, adoptive transfer of human CD4+ T cells, but not CD8+ T cells, were essential for A. actinomycetemcomitans infection-induced alveolar bone destruction in NOD/SCID mice (Teng et al., 2000). After the initial infection of periodontal pathogens, the bacterial antigens will be processed and presented to CD4+ helper T cells by professional antigen-presenting cells, such as dendritic cells, macrophages, and B cells. These findings imply that CD4+ T cells are the major T cell contributors to bone destruction in periodontitis. Therefore, our following discussion has been focused on the CD4+ T-helper cells (Th) and the effects of P. gingivalis and its virulence factors on Th cell responses. CD4+ T cells are divided into different subsets Th1, Th2, Th17, Treg, and some other subsets, based on their distinct cytokines and specific transcription factors (Ahlers & Belyakov, 2010). Differentiation and activation of these CD4+ T cell subsets from naïve cells are affected by T cell receptor activation, co-stimulatory factors, and cytokine milieu.
8.1. Th1/Th2 cells
From its establishment, Th1/Th2 paradigm classified the differentiated CD4+ T cells into Th1 and Th2 cells and has prevailed for around two decades (Mosmann, Cherwinski, Bond, Giedlin, & Coffman, 1986). Th1 cells express transcription factors T-bet and STAT-4. They preferably produced cytokines IL-12 and IFN-γ and their differentiation is driven by IL-12. On the other hand, Th2 cells express GATA3 and STAT6, and produce cytokines IL-4, IL-5, IL-6, IL-10, and IL-13 (Jankovic & Feng, 2015). Th1 and Th2 cells respond to different stimulations and are associated with different outcomes in infectious and inflammatory diseases. Th1/Th2 paradigm has been investigated to identify the roles of both cell subsets in periodontitis. By testing the T cell subset cytokine profiles, some studies have shown that the Th1 cells are associated with stable lesions while Th2 cells with progressive lesions (Aoyagi, Sugawara-Aoyagi, Yamazaki, & Hara, 1995; Tokoro, Matsuki, Yamamoto, Suzuki, & Hara, 1997; Wassenaar, Reinhardus, Thepen, Abraham-Inpijn, & Kievits, 1995), suggesting that Th2 cells are associated with more severe disease. Meanwhile, other studies demonstrated that Th1 cells were predominant in the gingival tissues from periodontitis patients and were associated with inflammation and periodontitis severity (Takeichi et al., 2000; Ukai, Mori, Onoyama, & Hara, 2001). Interestingly, there are also papers showed that Th1 and Th2 responses were up-regulated to a similarly higher level during periodontitis (Berglundh, Liljenberg, & Lindhe, 2002).
Th cell differentiation during periodontitis can be at least partially attributed to periodontal pathogens and their virulence factors, with controversy on which specific Th subsets are preferably promoted. Tests on P. gingivalis-responsive T-cell lines from periodontitis and healthy or gingivitis subjects implied that P. gingivalis facilitated Th1 differentiation (Gemmell, Grieco, Cullinan, Westerman, & Seymour, 1999). In vitro test suggested that fimbriae might induce the IFN-γ production by dendritic cells and provide Th1-biased environment (Ravi Jotwani & Cutler, 2004). On the other hand, there are reports showing that P. gingivalis and its LPS induce Th2 responses (Aoyagi et al., 1995; R. Jotwani, Pulendran, Agrawal, & Cutler, 2003). P. gingivalis FimA acts through TLR2 and produce IL-10, which suppresses Th1 cell differentiation and function (Gaddis, Maynard, Weaver, Michalek, & Katz, 2013). The capability of P. gingivalis to induce Th cell was explored to develop potential vaccine.
In an animal model for periodontitis, IFN-γ- and IL-6-deficient mice were resistant to P. gingivalis-induced alveolar bone loss, implying that Th1 and Th2 cells play destructive effects during periodontitis (Pamela J. Baker et al., 1999). Later reports showed that transfer of Th1 or Th1 cytokine IFN-γ caused higher alveolar bone loss in the mice injected with A. actinomycetemcomitans or its virulence factors, which indicated the detrimental effects of Th1 cells in periodontitis (Kawai et al., 2000). Interestingly, deficiency in Th1/Th2 cytokines, such as IFN-γ, IL-12p40, IL-4, or IL-10, can lead to more severe alveolar bone loss in aged mice (Alayan, Ivanovski, & Farah, 2007). Since IFN-γ and IL-12 are important in bacterial clearance, and IL-4 and IL-10 are famous for their anti-inflammatory functions, the seemingly contradictory reports might be explained by that the balance between pathogens and different host immune components is required to maintain the periodontal health.
8.2. Th17 cells
Pathogenesis of periodontitis had not been fully explained through the Th1/Th2 paradigm. The discovery of more Th subsets in recent years would help us to further our understanding of the pathogenesis of periodontitis. Among these new subsets are Th17 cells, which express transcription factors RORγt and produce cytokines IL-17 and IL-21. Th17 development and maintenance require cytokines TGF-β, IL-6, IL-21, and IL-23. Th17 is important in protecting against extracellular pathogens and fungi as well as promoting inflammation (O’Connor, Zenewicz, & Flavell, 2010). Recent researches link Th17 to various autoimmune and inflammatory diseases (R. P. Singh et al., 2014; Stadhouders, Lubberts, & Hendriks, 2018; van Bruggen & Ouyang, 2014). Th17 specific cytokine IL-17 and other related cytokines have been found in the gingival tissues or gingival crevicular fluid from periodontitis patients (C. R. Cardoso et al., 2009; Dutzan et al., 2012; Honda et al., 2008; Lester, Bain, Johnson, & Serio, 2007; Moutsopoulos et al., 2012; Ohyama et al., 2009; Vernal et al., 2005). Increased infiltration of Th17 cells in periodontal lesions further proved the association of Th17 with periodontitis (Adibrad et al., 2012; Okui, Aoki, Ito, Honda, & Yamazaki, 2012).
It has been shown that P. gingivalis regulate Th17 development and function (Moutsopoulos et al., 2012; Oda, Yoshie, & Yamazaki, 2003). P. gingivalis, especially the encapsulated strain W83, induced Th17 in periodontal lesions (Moutsopoulos et al., 2012). Capsules of different serotypes seem to possess differential effects on Th subset polarization. After priming DCs with the P. gingivalis strains of different K-serotypes, strains W83 of serotype K1 and HG184 of K2 induced a Th1/Th17 pattern of immune response, while K3, K4 and K5 strains promoted a Th2 response (Vernal, Diaz-Guerra, Silva, Sanz, & Garcia-Sanz, 2014). Moreover, P. gingivalis LPS can promote an environment for Th17 development by inducing the production of IL-17 and IL-23, a cytokine critical for Th17 expansion and maintenance, in human periodontal ligament cells (Y. D. Park et al., 2012). Besides producing pro-inflammatory cytokine IL-17, Th17 lymphocytes also promote bone loss by inducing the expression of RANKL, which is required for osteoclastogenesis (Sato et al., 2006; Stadhouders et al., 2018).
The role of IL-17 in periodontitis is not determined yet and under intense investigation. There are reports showing that IL-17 receptor signaling is protective in periodontitis (Jeffrey J. Yu, Ruddy, Conti, Boonanantanasarn, & Gaffen, 2008; J. J. Yu et al., 2007), and the ones showing that it is detrimental in the disease as well (Eskan et al., 2012). The reason of the inconsistency might be that different animal models were used.
8.3. Treg cells
Regulatory T cell (Treg) represents a special suppressive lymphocyte subset that down-regulates the activation, proliferation, and effector functions of a wide range of immune cells. Tregs specifically express transcription factor Foxp3 and secrete cytokines TGF-β and IL-10. Treg cells also express some cell surface molecules at higher level than naïve helper T cells, such as CD25, CTLA-4, GITR, and etc. Treg cells are critical in the maintenance of host immune homeostasis by controlling the direction and intensity of both adaptive and innate immunity and modulating various host immune responses, including inflammation (Chaudhry & Rudensky, 2013; Shevach, 2018).
Numerous publications on human patients demonstrated that Treg cells were enriched in periodontitis lesions (Cristina Ribeiro Cardoso et al., 2008; Dutzan, Gamonal, Silva, Sanz, & Vernal, 2009; Nakajima et al., 2005). Interestingly, there was also a finding showing reduced Foxp3+CD25+ cell number in periodontitis lesions (Ernst et al., 2007). It is still unclear what caused the inconsistency. In a series of reports with animal models for periodontitis, the association of Tregs with periodontitis and the role of Tregs in the disease have been further illustrated. Inhibition of Treg functions with blocking antibody anti-GITR treatment enhanced A. actinomycetemcomitans-induced inflammation and bone loss (Garlet et al., 2010), while recruitment of Foxp3+ Tregs into gingival tissue inhibited pro-inflammatory cytokine expression and A. actinomycetemcomitans-induced alveolar bone resorption in mice (Glowacki et al., 2013). Because all-trans retinoic acid (atRA) promotes the differentiation of Treg cells and sustains Treg stability (Z. M. Liu, Wang, Ma, & Guo Zheng, 2015), atRA was orally administrated to induce an increase of Treg cells and decrease in Th17 cells in cervical lymph nodes, which were accompanied by the inhibition of P. gingivalis-induced bone loss (L. Wang, Wang, Jin, Gao, & Lin, 2014).
The roles of Treg-associated molecules have also been tested for their roles in periodontitis. TGF-β and IL-10 are famous anti-inflammatory cytokines and major Treg cytokines. TGF-β and IL-10 were expressed higher in periodontitis tissues (Cristina Ribeiro Cardoso et al., 2008; Nakajima et al., 2005; Yamazaki et al., 1997), while their expression levels were negatively correlated to the severity of the disease (Dutzan et al., 2009; Dutzan et al., 2012). P. gingivalis infection caused more severe alveolar bone loss in IL-10-deficient mice, (Sasaki et al., 2004), and the IL-10 administration suppressed alveolar bone loss, indicating the protective role of IL-10 (Q. Zhang et al., 2014; X. Zhang & Teng, 2006). These findings all support the idea that Tregs play a key role in alleviating periodontitis, while the impairment of Treg function may cause exacerbated tissue damage. Indeed, some reported that during periodontitis, Foxp3+CD4+ T cells harvested from periodontitis lesions lost their suppressive function (Okui et al., 2008). Studies have shown that Tregs can convert into IL-17-producing cells in a pro-inflammatory environment (Ayyoub et al., 2009; Koenen et al., 2008; Leveque et al., 2009). Tregs that express RORγt and IL-17 were recently found and believed to play an role in the phenotypic plasticity in T cell lineages, transiting between inflammatory and regulatory function responding to outside stimulation (Nyirenda et al., 2011; Valmori, Raffin, Raimbaud, & Ayyoub, 2010). On the other hand, there are other reports showed that IL-17+Foxp3+ or RORγt+Foxp3+ cells are an independent stable Treg lineage, and are potent in regulating Th17-related diseases (Kluger et al., 2016; B. H. Yang et al., 2016). IL-17+Foxp3+ cells have been detected and seemed to be involved in periodontitis, whose roles are not fully elucidated either (Okui et al., 2012). Since these cells possess both pro-inflammatory and suppressive properties, their roles, protective or destructive, in inflammatory diseases are still unclear (Blatner et al., 2012; Kryczek et al., 2011; Tartar et al., 2010; Voo et al., 2009).
Later in vitro and in vivo research confirmed the ability of P. gingivalis to induce Treg development (Kim et al., 2010; Kobayashi et al., 2011). Interestingly, the effect of P. gingivalis on Treg development varies under different conditions. For example, we found that P. gingivalis induced fewer Treg cells in pregnant mice than non-pregnant mice. Similarly, infection with P. gingivalis, especially the ones with type II FimA, reduced Tregs in atherosclerotic patients, implying that type II FimA might be associated with Treg dysregulation (J. Yang et al., 2014).
Discussion
In this chapter, we have introduced periodontal pathogens and their virulence factors. We focused on P. gingivalis because it has been widely investigated and regarded as the keystone pathogen for periodontitis. Despite of its role as a “keystone” pathogen in periodontitis, P. gingivalis is not a lone criminal in the progression of periodontitis. Other pathogens and their virulence factors also play critical roles in the pathogenesis of periodontitis, which have been reviewed in other articles (de Andrade, Almeida-da-Silva, & Coutinho-Silva, 2019). P. gingivalis uses many methods to cooperate with other pathogens and evade immune clearance in periodontitis. It develops strategies, including their virulence factors, to overcome and exploit host immune pathways to facilitate its own and other pathogens’ colonization and survival. The virulence factors are required for the interactions between P. gingivalis and other bacteria. Fimbriae and capsules are able to adhere to the molecules on other bacteria and host tissues and cells to promote the formation of biofilm. The virulence factors can also act on the host cells to inhibit bacterial clearance, promote bacterial invasion, and cause inflammation and tissue damage (Fig. 1). They are critical in the manipulation and exploitation of host immune responses by P. gingivalis to lead to dysbiosis in oral cavity and periodontitis progression. T cells are major components of host immunity. Previous studies confirmed the importance of Th cells in periodontitis pathogenesis. P. gingivalis and its virulence factors induce biased Th differentiation and function, while the roles of the T cells in the periodontitis pathogenesis remains controversial.
Fig. 1.
P. gingivalis virulence factors subvert immune responses to evade bacterial clearance and promote inflammation. P. gingivalis gingipains degrade C3, C4, and C5 to block complement cascade. Besides, anaphylatoxins C3a, C4a, and C5a are generated to promote inflammation. Degradation of C3b prevents opsonization and phagocytosis, and C5b cleavage prevents the formation of membrane attack complex (MAC). Furthermore, C5a crosstalks with TLR2/1 activation and enhances the cAMP responses, which inhibits iNOS production and bacterial killing. Fimbriae promote the crosstalk between CXCR4 and TLR and further enhance cAMP responses. Fimbriae also induce CXCR4-dependent activation of CR3. The interaction between CR3 and P. gingivalis requires fimbriae and induces ERK1/2 signaling to down-regulate IL-12 production, which results in impaired bacterial clearance.
Great progress has been achieved to understand the complex inter-microbial interactions and host-pathogen interactions during the disease. Previous studies demonstrate that oral bacteria and the host immune system need to maintain a delicate balance. Disruption of the balance would cause dysbiosis in oral cavity, unwanted tissue damage, and diseases. Further exploration on the immune responses induced by the periodontal pathogens and their virulence factors will help us to better understand the pathogenesis of periodontitis and pave the road for its future therapy.
Acknowledgements
S.L. is supported by grants KSEF-3283-RDE-018, KSEF-3835-RDE-020, and NIH DE025388. H.W. is supported by DE026727 from National Institute of Dental and Craniofacial Research, NIH, USA.
Footnotes
Conflicts of Interest Statement
The authors report no conflicts of interest related to this work.
References:
- Aas JA, Paster BJ, Stokes LN, Olsen I, & Dewhirst FE (2005). Defining the normal bacterial flora of the oral cavity. J Clin Microbiol, 43(11), 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe N, Baba A, Takii R, Nakayama K, Kamaguchi A, Shibata Y, . . . Yamamoto K. (2004). Roles of Arg- and Lys-gingipains in coaggregation of Porphyromonas gingivalis: identification of its responsible molecules in translation products of rgpA, kgp, and hagA genes. Biol Chem, 385(11), 1041–1047. doi: 10.1515/BC.2004.135 [DOI] [PubMed] [Google Scholar]
- Abe T, Hosur KB, Hajishengallis E, Reis ES, Ricklin D, Lambris JD, & Hajishengallis G. (2012). Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J Immunol, 189(11), 5442–5448. doi: 10.4049/jimmunol.1202339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, . . . Diaz PI (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J, 7(5), 1016–1025. doi: 10.1038/ismej.2012.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibrad M, Deyhimi P, Ganjalikhani Hakemi M, Behfarnia P, Shahabuei M, & Rafiee L. (2012). Signs of the presence of Th17 cells in chronic periodontal disease. J Periodontal Res, 47(4), 525–531. doi: 10.1111/j.1600-0765.2011.01464.x [DOI] [PubMed] [Google Scholar]
- Aduse-Opoku J, Slaney JM, Hashim A, Gallagher A, Gallagher RP, Rangarajan M, . . . Curtis MA (2006). Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect Immun, 74(1), 449–460. doi: 10.1128/IAI.74.1.449-460.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlers JD, & Belyakov IM (2010). Molecular pathways regulating CD4(+) T cell differentiation, anergy and memory with implications for vaccines. Trends Mol Med, 16(10), 478–491. doi: 10.1016/j.molmed.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Alayan J, Ivanovski S, & Farah CS (2007). Alveolar bone loss in T helper 1/T helper 2 cytokine-deficient mice. J. Periodont. Res, 42(2), 97–103. doi:doi: 10.1111/j.1600-0765.2006.00920.x [DOI] [PubMed] [Google Scholar]
- Ally N, Whisstock JC, Sieprawska-Lupa M, Potempa J, Le Bonniec BF, Travis J, & Pike RN (2003). Characterization of the specificity of arginine-specific gingipains from Porphyromonas gingivalis reveals active site differences between different forms of the enzymes. Biochemistry, 42(40), 11693–11700. doi: 10.1021/bi0349726 [DOI] [PubMed] [Google Scholar]
- Amano A. (2003). Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J. Periodontol, 74(1), 90–96. [DOI] [PubMed] [Google Scholar]
- Amano A. (2010). Host-parasite interactions in periodontitis: microbial pathogenicity and innate immunity. Periodontol 2000, 54(1), 9–14. doi: 10.1111/j.1600-0757.2010.00376.x [DOI] [PubMed] [Google Scholar]
- Andrian E, Grenier D, & Rouabhia M. (2004). In vitro models of tissue penetration and destruction by Porphyromonas gingivalis. Infect Immun, 72(8), 4689–4698. doi: 10.1128/IAI.72.8.4689-4698.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi T, Sugawara-Aoyagi M, Yamazaki K, & Hara K. (1995). Interleukin 4 (IL-4) and IL-6-producing memory T-cells in peripheral blood and gingival tissue in periodontitis patients with high serum antibody titers to Porphyromonas gingivalis. Oral Microbiol Immunol, 10(5), 304–310. [DOI] [PubMed] [Google Scholar]
- Aruni AW, Dou Y, Mishra A, & Fletcher HM (2015). The Biofilm Community-Rebels with a Cause. Curr Oral Health Rep, 2(1), 48–56. doi: 10.1007/s40496-014-0044-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyoub M, Deknuydt F, Raimbaud I, Dousset C, Leveque L, Bioley G, & Valmori D. (2009). Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the T(H)17 lineage-specific transcription factor RORgamma t. Proc Natl Acad Sci U S A, 106(21), 8635–8640. doi: 10.1073/pnas.0900621106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge BW, & Darveau RP (2001). Porphyromonas gingivalis lipopolysaccharide: an unusual pattern recognition receptor ligand for the innate host defense system. Acta Odontol Scand, 59(3), 131–138. [DOI] [PubMed] [Google Scholar]
- Baker PJ, Dixon M, Evans RT, Dufour L, Johnson E, & Roopenian DC (1999). CD4+ T Cells and the Proinflammatory Cytokines Gamma Interferon and Interleukin-6 Contribute to Alveolar Bone Loss in Mice. Infect. Immun, 67(6), 2804–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Evans RT, & Roopenian DC (1994). Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol, 39(12), 1035–1040. [DOI] [PubMed] [Google Scholar]
- Bao K, Belibasakis GN, Thurnheer T, Aduse-Opoku J, Curtis MA, & Bostanci N. (2014). Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol, 14, 258. doi: 10.1186/s12866-014-0258-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglundh T, Liljenberg B, & Lindhe J. (2002). Some cytokine profiles of T-helper cells in lesions of advanced periodontitis. J Clin Periodontol, 29(8), 705–709. doi: 10.1034/j.1600-051x.2002.290807.x [DOI] [PubMed] [Google Scholar]
- Bergstrom J. (2006). Periodontitis and smoking: an evidence-based appraisal. J Evid Based Dent Pract, 6(1), 33–41. doi: 10.1016/j.jebdp.2005.12.018 [DOI] [PubMed] [Google Scholar]
- Blatner NR, Mulcahy MF, Dennis KL, Scholtens D, Bentrem DJ, Phillips JD, . . . Khazaie K. (2012). Expression of RORgammat marks a pathogenic regulatory T cell subset in human colon cancer. Sci Transl Med, 4(164), 164ra159. doi: 10.1126/scitranslmed.3004566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanci N, & Belibasakis GN (2012). Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett, 333(1), 1–9. doi: 10.1111/j.1574-6968.2012.02579.x [DOI] [PubMed] [Google Scholar]
- Bozkurt SB, Hakki SS, Hakki EE, Durak Y, & Kantarci A. (2017). Porphyromonas gingivalis Lipopolysaccharide Induces a Pro-inflammatory Human Gingival Fibroblast Phenotype. Inflammation, 40(1), 144–153. doi: 10.1007/s10753-016-0463-7 [DOI] [PubMed] [Google Scholar]
- Brown LJ, Johns BA, & Wall TP (2002). The economics of periodontal diseases. Periodontology 2000, 29, 223–234. [DOI] [PubMed] [Google Scholar]
- Brunner J, Scheres N, El Idrissi NB, Deng DM, Laine ML, van Winkelhoff AJ, & Crielaard W. (2010). The capsule of Porphyromonas gingivalis reduces the immune response of human gingival fibroblasts. BMC microbiology, 10, 5. doi: 10.1186/1471-2180-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso CR, Garlet GP, Crippa GE, Rosa AL, Junior WM, Rossi MA, & Silva JS (2009). Evidence of the presence of T helper type 17 cells in chronic lesions of human periodontal disease. Oral microbiology and immunology, 24(1), 1–6. doi: 10.1111/j.1399-302X.2008.00463.x [DOI] [PubMed] [Google Scholar]
- Cardoso CR, Garlet GP, Moreira AP, Junior WM, Rossi MA, & Silva JS (2008). Characterization of CD4+CD25+ natural regulatory T cells in the inflammatory infiltrate of human chronic periodontitis. J Leukoc Biol, 84(1), 311–318. doi: 10.1189/jlb.0108014 [DOI] [PubMed] [Google Scholar]
- Carlisle MD, Srikantha RN, & Brogden KA (2009). Degradation of human alpha- and beta-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J Innate Immun, 1(2), 118–122. doi: 10.1159/000181015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, & Rudensky AY (2013). Control of inflammation by integration of environmental cues by regulatory T cells. J Clin Invest, 123(3), 939–944. doi: 10.1172/JCI57175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hosogi Y, Nishikawa K, Abbey K, Fleischmann RD, Walling J, & Duncan MJ (2004). Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. Journal of Bacteriology, 186(16), 5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, . . . Darveau RP (2009). Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4’-phosphatase activities. Cell Microbiol, 11(11), 1587–1599. doi: 10.1111/j.1462-5822.2009.01349.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KL, Seymour GJ, & Powell RN (1987). Phenotypic and functional analysis of T cells extracted from chronically inflamed human periodontal tissues. J Periodontol, 58(8), 569–573. doi: 10.1902/jop.1987.58.8.569 [DOI] [PubMed] [Google Scholar]
- Curtis MA, Aduse-Opoku J, & Rangarajan M. (2001). Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med, 12(3), 192–216. [DOI] [PubMed] [Google Scholar]
- Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, . . . Marsh PD (2011). Temperature-dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defenses. Infect Immun, 79(3), 1187–1193. doi: 10.1128/IAI.00900-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Empaire G, Baer MT, & Gibson FC 3rd. (2006). The K1 serotype capsular polysaccharide of Porphyromonas gingivalis elicits chemokine production from murine macrophages that facilitates cell migration. Infect Immun, 74(11), 6236–6243. doi: 10.1128/IAI.00519-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP, Arbabi S, Garcia I, Bainbridge B, & Maier RV (2002). Porphyromonas gingivalis lipopolysaccharide is both agonist and antagonist for p38 mitogen-activated protein kinase activation. Infect. Immun, 70, 1867–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, . . . Hajjar AM (2004). Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun, 72(9), 5041–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrade KQ, Almeida-da-Silva CLC, & Coutinho-Silva R. (2019). Immunological Pathways Triggered by Porphyromonas gingivalis and Fusobacterium nucleatum: Therapeutic Possibilities? Mediators Inflamm, 2019, 7241312. doi: 10.1155/2019/7241312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Diego I, Veillard F, Sztukowska MN, Guevara T, Potempa B, Pomowski A, . . . Gomis-Ruth FX (2014). Structure and mechanism of cysteine peptidase gingipain K (Kgp), a major virulence factor of Porphyromonas gingivalis in periodontitis. J Biol Chem, 289(46), 32291–32302. doi: 10.1074/jbc.M114.602052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, . . . Wade WG (2010). The human oral microbiome. Journal of Bacteriology, 192(19), 5002–5017. doi: 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DP, Kubiniec MA, Yoshimura F, & Genco RJ (1988). Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J. Bacteriol, 170(4), 1658–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierickx K, Pauwels M, Laine ML, Van Eldere J, Cassiman JJ, van Winkelhoff AJ, . . . Quirynen M. (2003). Adhesion of Porphyromonas gingivalis serotypes to pocket epithelium. J Periodontol, 74(6), 844–848. doi: 10.1902/jop.2003.74.6.844 [DOI] [PubMed] [Google Scholar]
- Ding PH, Wang CY, Darveau RP, & Jin L. (2013). Porphyromonas gingivalis LPS stimulates the expression of LPS-binding protein in human oral keratinocytes in vitro. Innate Immun, 19(1), 66–75. doi: 10.1177/1753425912450348 [DOI] [PubMed] [Google Scholar]
- Dixon DR, & Darveau RP (2005). Lipopolysaccharide heterogeneity: innate host responses to bacterial modification of lipid a structure. J Dent Res, 84(7), 584–595. [DOI] [PubMed] [Google Scholar]
- Dommisch H, & Jepsen S. (2015). Diverse functions of defensins and other antimicrobial peptides in periodontal tissues. Periodontol 2000, 69(1), 96–110. doi: 10.1111/prd.12093 [DOI] [PubMed] [Google Scholar]
- Dutzan N, Gamonal J, Silva A, Sanz M, & Vernal R. (2009). Over-expression of forkhead box P3 and its association with receptor activator of nuclear factor-kappa B ligand, interleukin (IL) −17, IL-10 and transforming growth factor-beta during the progression of chronic periodontitis. J Clin Periodontol, 36(5), 396–403. [DOI] [PubMed] [Google Scholar]
- Dutzan N, Vernal R, Vaque JP, Garcia-Sesnich J, Hernandez M, Abusleme L, . . . Gamonal J. (2012). Interleukin-21 expression and its association with proinflammatory cytokines in untreated chronic periodontitis patients. J Periodontol, 83(7), 948–954. doi: 10.1902/jop.2011.110482 [DOI] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, . . . Genco RJ (2015). Update on Prevalence of Periodontitis in Adults in the United States: NHANES 2009 to 2012. J Periodontol, 86(5), 611–622. doi: 10.1902/jop.2015.140520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Thornton-Evans GO, & Genco RJ (2012). Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res, 91(10), 914–920. doi: 10.1177/0022034512457373 [DOI] [PubMed] [Google Scholar]
- El-Awady AR, Miles B, Scisci E, Kurago ZB, Palani CD, Arce RM, . . . Cutler CW (2015). Porphyromonas gingivalis evasion of autophagy and intracellular killing by human myeloid dendritic cells involves DC-SIGN-TLR2 crosstalk. PLoS Pathog, 10(2), e1004647. doi: 10.1371/journal.ppat.1004647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enersen M, Nakano K, & Amano A. (2013). Porphyromonas gingivalis fimbriae. J Oral Microbiol, 5. doi: 10.3402/jom.v5i0.20265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enersen M, Olsen I, Kvalheim O, & Caugant DA (2008). fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol, 46(1), 31–42. doi: 10.1128/JCM.00986-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst CW, Lee JE, Nakanishi T, Karimbux NY, Rezende TM, Stashenko P, . . . Kawai T. (2007). Diminished forkhead box P3/CD25 double-positive T regulatory cells are associated with the increased nuclear factor-kappaB ligand (RANKL+) T cells in bone resorption lesion of periodontal disease. Clin Exp Immunol, 148(2), 271–280. doi: 10.1111/j.1365-2249.2006.03318.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan MA, Hajishengallis G, & Kinane DF (2007). Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infect. Immun, 75, 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, . . . Hajishengallis G. (2012). The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol, 13(5), 465–473. doi: 10.1038/ni.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Morishima S, Takahashi I, & Hamada S. (1993). Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem Biophys Res Commun, 197(1), 241–247. doi: 10.1006/bbrc.1993.2467 [DOI] [PubMed] [Google Scholar]
- Gaddis DE, Maynard CL, Weaver CT, Michalek SM, & Katz J. (2013). Role of TLR2-dependent IL-10 production in the inhibition of the initial IFN-gamma T cell response to Porphyromonas gingivalis. J Leukoc Biol, 93(1), 21–31. doi: 10.1189/jlb.0512220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlet GP, Cardoso CR, Mariano FS, Claudino M, de Assis GF, Campanelli AP, . . . Silva JS (2010). Regulatory T cells attenuate experimental periodontitis progression in mice. J Clin Periodontol, 37(7), 591–600. doi: 10.1111/j.1600-051X.2010.01586.x [DOI] [PubMed] [Google Scholar]
- Gemmell E, Grieco DA, Cullinan MP, Westerman B, & Seymour GJ (1999). The proportion of interleukin-4, interferon-gamma and interleukin-10-positive cells in Porphyromonas gingivalis--specific T-cell lines established from P. gingivalis-positive subjects. Oral Microbiol Immunol, 14(5), 267–274. doi: 10.1034/j.1399-302x.1999.140501.x [DOI] [PubMed] [Google Scholar]
- Glowacki AJ, Yoshizawa S, Jhunjhunwala S, Vieira AE, Garlet GP, Sfeir C, & Little SR (2013). Prevention of inflammation-mediated bone loss in murine and canine periodontal disease via recruitment of regulatory lymphocytes. Proc Natl Acad Sci U S A, 110(46), 18525–18530. doi: 10.1073/pnas.1302829110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulbourne PA, & Ellen RP (1991). Evidence That Porphyromonas (Bacteroides) Gingivalis Fimbriae Function in Adhesion to Actinomyces-Viscosus. Journal of Bacteriology, 173(17), 5266–5274. doi:DOI 10.1128/jb.173.17.5266-5274.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier D, Roy S, Chandad F, Plamondon P, Yoshioka M, Nakayama K, & Mayrand D. (2003). Effect of inactivation of the Arg- and/or Lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect Immun, 71(8), 4742–4748. doi: 10.1128/iai.71.8.4742-4748.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith F. (1928). The Significance of Pneumococcal Types. J Hyg (Lond), 27(2), 113–159. doi: 10.1017/s0022172400031879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Nguyen KA, & Potempa J. (2010). Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000, 54(1), 15–44. doi: 10.1111/j.1600-0757.2010.00377.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. (2009). Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect, 11(6–7), 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. (2014). Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends in immunology, 35(1), 3–11. doi: 10.1016/j.it.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Abe T, Maekawa T, Hajishengallis E, & Lambris JD (2013). Role of complement in host-microbe homeostasis of the periodontium. Semin Immunol, 25(1), 65–72. doi: 10.1016/j.smim.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, & Curtis MA (2012). The keystone-pathogen hypothesis. Nature reviews. Microbiology, 10(10), 717–725. doi: 10.1038/nrmicro2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, & Lambris JD (2011). Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol, 11(3), 187–200. doi: 10.1038/nri2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, & Lamont RJ (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Molecular oral microbiology, 27(6), 409–419. doi: 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, . . . Curtis MA (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe, 10(5), 497–506. doi: 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, McIntosh ML, Nishiyama SI, & Yoshimura F. (2012). Mechanism and implications of CXCR4-mediated integrin activation by Porphyromonas gingivalis. Molecular oral microbiology. doi: 10.1111/omi.12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Shakhatreh M-AK, Wang M, & Liang S. (2007). Complement receptor 3 blockade Promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J. Immunol, 179(4), 2359–2367. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S-I, Ratti P, Schifferle RE, . . . Yoshimura F. (2006). Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell. Microbiol, 8(10), 1557–1570 [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Wang M, Liang S, Triantafilou M, & Triantafilou K. (2008). Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proceedings of the National Academy of Sciences, 105(36), 13532–13537. doi: 10.1073/pnas.0803852105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Watanabe K, Arai M, Hiramine H, & Umemoto T. (2002). Cytokine production induced by a 67-kDa fimbrial protein from Porphyromonas gingivalis. Oral Microbiol Immunol, 17(3), 197–200. [DOI] [PubMed] [Google Scholar]
- Hamedi M, Belibasakis GN, Cruchley AT, Rangarajan M, Curtis MA, & Bostanci N. (2009). Porphyromonas gingivalis culture supernatants differentially regulate interleukin-1beta and interleukin-18 in human monocytic cells. Cytokine, 45(2), 99–104. doi: 10.1016/j.cyto.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Handley PS, & Tipler LS (1986). An electron microscope survey of the surface structures and hydrophobicity of oral and non-oral species of the bacterial genus Bacteroides. Arch Oral Biol, 31(5), 325–335. doi: 10.1016/0003-9969(86)90047-6 [DOI] [PubMed] [Google Scholar]
- Haraguchi A, Miura M, Fujise O, Hamachi T, & Nishimura F. (2014). Porphyromonas gingivalis gingipain is involved in the detachment and aggregation of Aggregatibacter actinomycetemcomitans biofilm. Mol Oral Microbiol, 29(3), 131–143. doi: 10.1111/omi.12051 [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Iijima Y, Persson K, Nagano K, Yoshida Y, Lamont RJ, . . . Yoshimura F. (2016). Role of Mfa5 in Expression of Mfa1 Fimbriae in Porphyromonas gingivalis. J Dent Res, 95(11), 1291–1297. doi: 10.1177/0022034516655083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Iwami J, Sato K, Park Y, Nishikawa K, Atsumi T, . . . Yoshimura F. (2009). Anchoring and length regulation of Porphyromonas gingivalis Mfa1 fimbriae by the downstream gene product Mfa2. Microbiology, 155(Pt 10), 3333–3347. doi: 10.1099/mic.0.028928-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Nagano K, Ikai R, Izumigawa M, Yoshida Y, Kitai N, . . . Yoshimura F. (2013). Localization and function of the accessory protein Mfa3 in Porphyromonas gingivalis Mfa1 fimbriae. Mol Oral Microbiol, 28(6), 467–480. doi: 10.1111/omi.12040 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Ogawa S, Asai Y, Takai Y, & Ogawa T. (2003). Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol Lett, 226(2), 267–271. doi: 10.1016/S0378-1097(03)00615-3 [DOI] [PubMed] [Google Scholar]
- Herath TD, Wang Y, Seneviratne CJ, Darveau RP, Wang CY, & Jin L. (2013). The expression and regulation of matrix metalloproteinase-3 is critically modulated by Porphyromonas gingivalis lipopolysaccharide with heterogeneous lipid A structures in human gingival fibroblasts. BMC Microbiol, 13, 73. doi: 10.1186/1471-2180-13-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herath TD, Wang Y, Seneviratne CJ, Lu Q, Darveau RP, Wang CY, & Jin L. (2011). Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity differentially modulates the expression of IL-6 and IL-8 in human gingival fibroblasts. J Clin Periodontol, 38(8), 694–701. doi: 10.1111/j.1600-051X.2011.01741.x [DOI] [PubMed] [Google Scholar]
- Hiramine H, Watanabe K, Hamada N, & Umemoto T. (2003). Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol Lett, 229(1), 49–55. doi: 10.1016/S0378-1097(03)00788-2 [DOI] [PubMed] [Google Scholar]
- Holden JA, Attard TJ, Laughton KM, Mansell A, O’Brien-Simpson NM, & Reynolds EC (2014). Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect Immun, 82(10), 4190–4203. doi: 10.1128/IAI.02325-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Kesavalu L, Walker S, & Genco CA (1999). Virulence factors of Porphyromonas gingivalis. Periodontol 2000, 20, 168–238. [DOI] [PubMed] [Google Scholar]
- Honda T, Aoki Y, Takahashi N, Maekawa T, Nakajima T, Ito H, . . . Yamazaki K. (2008). Elevated expression of IL-17 and IL-12 genes in chronic inflammatory periodontal disease. Clin Chim Acta, 395(1–2), 137–141. doi: 10.1016/j.cca.2008.06.003 [DOI] [PubMed] [Google Scholar]
- How KY, Song KP, & Chan KG (2016). Porphyromonas gingivalis: An Overview of Periodontopathic Pathogen below the Gum Line. Front Microbiol, 7, 53. doi: 10.3389/fmicb.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igboin CO, Tordoff KP, Moeschberger ML, Griffen AL, & Leys EJ (2011). Porphyromonas gingivalis-host interactions in a Drosophila melanogaster model. Infect Immun, 79(1), 449–458. doi: 10.1128/IAI.00785-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Travis J, & Potempa J. (2003). The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr Protein Pept Sci, 4(6), 443–450. doi: 10.2174/1389203033487027 [DOI] [PubMed] [Google Scholar]
- Ito R, Ishihara K, Shoji M, Nakayama K, & Okuda K. (2010). Hemagglutinin/Adhesin domains of Porphyromonas gingivalis play key roles in coaggregation with Treponema denticola. FEMS Immunol Med Microbiol, 60(3), 251–260. doi: 10.1111/j.1574-695X.2010.00737.x [DOI] [PubMed] [Google Scholar]
- Ivanyi L, & Lehner T. (1970). Stimulation of lymphocyte transformation by bacterial antigens in patients with periodontal disease. Arch Oral Biol, 15(11), 1089–1096. doi: 10.1016/0003-9969(70)90121-4 [DOI] [PubMed] [Google Scholar]
- Jagels MA, Ember JA, Travis J, Potempa J, Pike R, & Hugli TE (1996). Cleavage of the human C5A receptor by proteinases derived from Porphyromonas gingivalis: cleavage of leukocyte C5a receptor. Adv Exp Med Biol, 389, 155–164. doi: 10.1007/978-1-4613-0335-0_19 [DOI] [PubMed] [Google Scholar]
- Jagels MA, Travis J, Potempa J, Pike R, & Hugli TE (1996). Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infect Immun, 64(6), 1984–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, & Feng CG (2015). CD4(+) T Cell Differentiation in Infection: Amendments to the Th1/Th2 Axiom. Front Immunol, 6, 198. doi: 10.3389/fimmu.2015.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jotwani R, & Cutler CW (2004). Fimbriated Porphyromonas gingivalis Is More Efficient than Fimbria-Deficient P. gingivalis in Entering Human Dendritic Cells In Vitro and Induces an Inflammatory Th1 Effector Response. Infect. Immun, 72(3), 1725–1732. doi: 10.1128/iai.72.3.1725-1732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jotwani R, Pulendran B, Agrawal S, & Cutler CW (2003). Human dendritic cells respond to Porphyromonas gingivalis LPS by promoting a Th2 effector response in vitro. Eur J Immunol, 33(11), 2980–2986. doi: 10.1002/eji.200324392 [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Baba A, Abe N, Takii R, Hashimoto M, Tsukuba T, . . . Yamamoto K. (2004). Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol Pharmacol, 66(6), 1599–1606. doi: 10.1124/mol.104.004366 [DOI] [PubMed] [Google Scholar]
- Kamaguchi A, Ohyama T, Sakai E, Nakamura R, Watanabe T, Baba H, & Nakayama K. (2003). Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia. Microbiology, 149(Pt 5), 1257–1264. [DOI] [PubMed] [Google Scholar]
- Kassebaum NJ, Smith AGC, Bernabe E, Fleming TD, Reynolds AE, Vos T, . . . Collaborators, G. B. D. O. H. (2017). Global, Regional, and National Prevalence, Incidence, and Disability-Adjusted Life Years for Oral Conditions for 195 Countries, 1990–2015: A Systematic Analysis for the Global Burden of Diseases, Injuries, and Risk Factors. J Dent Res, 96(4), 380–387. doi: 10.1177/0022034517693566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Ward DC, & Michalek SM (1996). Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol Immunol, 11(5), 309–318. doi: 10.1111/j.1399-302x.1996.tb00187.x [DOI] [PubMed] [Google Scholar]
- Kawai T, Eisen-Lev R, Seki M, Eastcott JW, Wilson ME, & Taubman MA (2000). Requirement of B7 costimulation for Th1-mediated inflammatory bone resorption in experimental periodontal disease. J. Immunol, 164(4), 2102–2109. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, . . . Dixit VM (2013). Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science, 341(6151), 1246–1249. doi: 10.1126/science.1240248 [DOI] [PubMed] [Google Scholar]
- Khader YS, Dauod AS, El-Qaderi SS, Alkafajei A, & Batayha WQ (2006). Periodontal status of diabetics compared with nondiabetics: a meta-analysis. J Diabetes Complications, 20(1), 59–68. doi: 10.1016/j.jdiacomp.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Khalaf H, & Bengtsson T. (2012). Altered T-cell responses by the periodontal pathogen Porphyromonas gingivalis. PLoS One, 7(9), e45192. doi: 10.1371/journal.pone.0045192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Ko Y, Hong SD, Kim KY, Lee YH, Chae C, & Choi Y. (2010). Presence of Porphyromonas gingivalis and plasma cell dominance in gingival tissues with periodontitis. Oral diseases, 16(4), 375–381. doi: 10.1111/j.1601-0825.2009.01649.x [DOI] [PubMed] [Google Scholar]
- Kimura S, Fujimoto N, & Okada H. (1991). Impaired autologous mixed-lymphocyte reaction of peripheral blood lymphocytes in adult periodontitis. Infect Immun, 59(12), 4418–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinane DF, Stathopoulou PG, & Papapanou PN (2017). Periodontal diseases. Nat Rev Dis Primers, 3, 17038. doi: 10.1038/nrdp.2017.38 [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Matono S, Aida Y, Hirofuji T, & Maeda K. (2002). Gingipains in the culture supernatant of Porphyromonas gingivalis cleave CD4 and CD8 on human T cells. J Periodontal Res, 37(6), 464–468. doi: 10.1034/j.1600-0765.2002.01364.x [DOI] [PubMed] [Google Scholar]
- Kluger MA, Meyer MC, Nosko A, Goerke B, Luig M, Wegscheid C, . . . Steinmetz OM (2016). RORgammat(+)Foxp3(+) Cells are an Independent Bifunctional Regulatory T Cell Lineage and Mediate Crescentic GN. J Am Soc Nephrol, 27(2), 454–465. doi: 10.1681/ASN.2014090880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi R, Kono T, Bolerjack BA, Fukuyama Y, Gilbert RS, Fujihashi K, . . . Yamamoto M. (2011). Induction of IL-10-producing CD4+ T-cells in chronic periodontitis. J Dent Res, 90(5), 653–658. doi: 10.1177/0022034510397838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen HJ, Smeets RL, Vink PM, van Rijssen E, Boots AM, & Joosten I. (2008). Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood, 112(6), 2340–2352. doi: 10.1182/blood-2008-01-133967 [DOI] [PubMed] [Google Scholar]
- Kristoffersen AK, Solli SJ, Nguyen TD, & Enersen M. (2015). Association of the rgpB gingipain genotype to the major fimbriae (fimA) genotype in clinical isolates of the periodontal pathogen Porphyromonas gingivalis. J Oral Microbiol, 7, 29124. doi: 10.3402/jom.v7.29124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Wu K, Zhao E, Wei S, Vatan L, Szeliga W, . . . Zou W. (2011). IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J Immunol, 186(7), 4388–4395. doi: 10.4049/jimmunol.1003251 [DOI] [PubMed] [Google Scholar]
- Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, . . . Lamont RJ (2017). Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol, 2(11), 1493–1499. doi: 10.1038/s41564-017-0021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, & Lamont RJ (2010). Subgingival biofilm formation. Periodontol 2000, 52(1), 38–52. doi: 10.1111/j.1600-0757.2009.00311.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine ML, Appelmelk BJ, & van Winkelhoff AJ (1997). Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients. J Dent Res, 76(12), 1840–1844. doi: 10.1177/00220345970760120601 [DOI] [PubMed] [Google Scholar]
- Laine ML, & van Winkelhoff AJ (1998). Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol, 13(5), 322–325. doi: 10.1111/j.1399-302x.1998.tb00714.x [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Hsiao GW, & Gil S. (1994). Identification of a molecule of Porphyromonas gingivalis that binds to Streptococcus gordonii. Microb Pathog, 17(5), 355–360. doi: 10.1006/mpat.1994.1081 [DOI] [PubMed] [Google Scholar]
- Lamont RJ, & Jenkinson HF (2000). Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol, 15(6), 341–349. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Koo H, & Hajishengallis G. (2018). The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol, 16(12), 745–759. doi: 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Miller DP, Wu L, Casella CR, Hasegawa Y, & Lamont RJ (2018). Maturation of the Mfa1 Fimbriae in the Oral Pathogen Porphyromonas gingivalis. Front Cell Infect Microbiol, 8, 137. doi: 10.3389/fcimb.2018.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Sojar HT, Bedi GS, & Genco RJ (1991). Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect Immun, 59(1), 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester SR, Bain JL, Johnson RB, & Serio FG (2007). Gingival concentrations of interleukin-23 and −17 at healthy sites and at sites of clinical attachment loss. J Periodontol, 78(8), 1545–1550. doi: 10.1902/jop.2007.060458 [DOI] [PubMed] [Google Scholar]
- Leveque L, Deknuydt F, Bioley G, Old LJ, Matsuzaki J, Odunsi K, . . . Valmori D. (2009). Interleukin 2-mediated conversion of ovarian cancer-associated CD4+ regulatory T cells into proinflammatory interleukin 17-producing helper T cells. J Immunother, 32(2), 101–108. doi: 10.1097/CJI.0b013e318195b59e [DOI] [PubMed] [Google Scholar]
- Li C, Li B, Dong Z, Gao L, He X, Liao L, . . . Jin Y. (2014). Lipopolysaccharide differentially affects the osteogenic differentiation of periodontal ligament stem cells and bone marrow mesenchymal stem cells through Toll-like receptor 4 mediated nuclear factor kappaB pathway. Stem Cell Res Ther, 5(3), 67. doi: 10.1186/scrt456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, . . . Hajishengallis G. (2011). The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol, 186(2), 869–877. doi: 10.4049/jimmunol.1003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien E, Means TK, Heine H, Yoshimura A, Kusumoto S, Fukase K, . . . Golenbock DT (2000). Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Invest, 105, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Desta T, Raptis M, Darveau RP, & Graves DT (2008). P. gingivalis and E. coli lipopolysaccharides exhibit different systemic but similar local induction of inflammatory markers. J Periodontol, 79(7), 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZM, Wang KP, Ma J, & Guo Zheng S. (2015). The role of all-trans retinoic acid in the biology of Foxp3+ regulatory T cells. Cell Mol Immunol, 12(5), 553–557. doi: 10.1038/cmi.2014.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madianos PN, Bobetsis YA, & Offenbacher S. (2013). Adverse pregnancy outcomes (APOs) and periodontal disease: pathogenic mechanisms. J Periodontol, 84(4 Suppl), S170–180. doi: 10.1902/jop.2013.1340015 [DOI] [PubMed] [Google Scholar]
- Maeda K, Nagata H, Nonaka A, Kataoka K, Tanaka M, & Shizukuishi S. (2004). Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect, 6(13), 1163–1170. doi: 10.1016/j.micinf.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, & Shizukuishi S. (2004). Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect Immun, 72(3), 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Abe T, Hajishengallis E, Hosur KB, Deangelis RA, Ricklin D, . . . Hajishengallis G. (2014). Genetic and Intervention Studies Implicating Complement C3 as a Major Target for the Treatment of Periodontitis. J Immunol. doi: 10.4049/jimmunol.1400569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisetta G, Brancatisano FL, Esin S, Campa M, & Batoni G. (2011). Gingipains produced by Porphyromonas gingivalis ATCC49417 degrade human-beta-defensin 3 and affect peptide’s antibacterial activity in vitro. Peptides, 32(5), 1073–1077. doi: 10.1016/j.peptides.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, . . . Dyer DW (1994). Inactivation of Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol, 176(4), 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. (2001). Toll-like receptors and innate immunity. Nat. Rev. Immunol, 1(2), 135–145. [DOI] [PubMed] [Google Scholar]
- Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, & Coffman RL (1986). Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol, 136(7), 2348–2357. [PubMed] [Google Scholar]
- Moutsopoulos NM, Kling HM, Angelov N, Jin W, Palmer RJ, Nares S, . . . Wahl SM (2012). Porphyromonas gingivalis promotes Th17 inducing pathways in chronic periodontitis. Journal of autoimmunity, 39(4), 294–303. doi: 10.1016/j.jaut.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano K, Hasegawa Y, Iijima Y, Kikuchi T, & Mitani A. (2018). Distribution of Porphyromonas gingivalis fimA and mfa1 fimbrial genotypes in subgingival plaques. PeerJ, 6, e5581. doi: 10.7717/peerj.5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano K, Hasegawa Y, Murakami Y, Nishiyama S, & Yoshimura F. (2010). FimB regulates FimA fimbriation in Porphyromonas gingivalis. J Dent Res, 89(9), 903–908. doi: 10.1177/0022034510370089 [DOI] [PubMed] [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, . . . Hayashi T. (2008). Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA research : an international journal for rapid publication of reports on genes and genomes, 15(4), 215–225. doi: 10.1093/dnares/dsn013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Ueki-Maruyama K, Oda T, Ohsawa Y, Ito H, Seymour GJ, & Yamazaki K. (2005). Regulatory T-cells infiltrate periodontal disease tissues. J Dent Res, 84(7), 639–643. doi: 10.1177/154405910508400711 [DOI] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, . . . Fraser CM (2003). Complete genome sequence of the oral pathogenic Bacterium porphyromonas gingivalis strain W83. Journal of Bacteriology, 185(18), 5591–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto E, Darveau RP, Foster BL, Nogueira-Filho GR, & Somerman MJ (2006). Regulation of cementoblast function by P. gingivalis lipopolysaccharide via TLR2. J Dent Res, 85(8), 733–738. doi: 10.1177/154405910608500809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, & Duncan MJ (2010). Histidine kinase-mediated production and autoassembly of Porphyromonas gingivalis fimbriae. Journal of Bacteriology, 192(7), 1975–1987. doi: 10.1128/JB.01474-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S-I, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, & Yoshimura F. (2007). Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology, 153, 1916–1925. [DOI] [PubMed] [Google Scholar]
- Nyirenda MH, Sanvito L, Darlington PJ, O’Brien K, Zhang GX, Constantinescu CS, . . . Gran B. (2011). TLR2 stimulation drives human naive and effector regulatory T cells into a Th17-like phenotype with reduced suppressive function. J Immunol, 187(5), 2278–2290. doi: 10.4049/jimmunol.1003715 [DOI] [PubMed] [Google Scholar]
- O’Connor W Jr., Zenewicz LA, & Flavell RA (2010). The dual nature of T(H)17 cells: shifting the focus to function. Nat Immunol, 11(6), 471–476. doi: 10.1038/ni.1882 [DOI] [PubMed] [Google Scholar]
- Oda T, Yoshie H, & Yamazaki K. (2003). Porphyromonas gingivalis antigen preferentially stimulates T cells to express IL-17 but not receptor activator of NF-kappaB ligand in vitro. Oral Microbiol Immunol, 18(1), 30–36. doi: 10.1034/j.1399-302x.2003.180105.x [DOI] [PubMed] [Google Scholar]
- Ogawa T. (1993). Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolysaccharide. FEBS Lett, 332(1–2), 197–201. doi: 10.1016/0014-5793(93)80512-s [DOI] [PubMed] [Google Scholar]
- Ohyama H, Kato-Kogoe N, Kuhara A, Nishimura F, Nakasho K, Yamanegi K, . . . Terada N(2009). The involvement of IL-23 and the Th17 pathway in periodontitis. J Dent Res, 88(7), 633–638. doi: 10.1177/0022034509339889 [DOI] [PubMed] [Google Scholar]
- Okui T, Aoki Y, Ito H, Honda T, & Yamazaki K. (2012). The presence of IL-17+/FOXP3+ double-positive cells in periodontitis. J Dent Res, 91(6), 574–579. doi: 10.1177/0022034512446341 [DOI] [PubMed] [Google Scholar]
- Okui T, Ito H, Honda T, Amanuma R, Yoshie H, & Yamazaki K. (2008). Characterization of CD4+ FOXP3+ T-cell clones established from chronic inflammatory lesions. Oral Microbiol Immunol, 23(1), 49–54. doi: 10.1111/j.1399-302X.2007.00390.x [DOI] [PubMed] [Google Scholar]
- Olsen I, Lambris JD, & Hajishengallis G. (2017). Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. J Oral Microbiol, 9(1), 1340085. doi: 10.1080/20002297.2017.1340085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos G, Weinberg EO, Massari P, Gibson FC 3rd, Wetzler LM, Morgan EF, & Genco CA (2013). Macrophage-specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J Immunol, 190(3), 1148–1157. doi: 10.4049/jimmunol.1202511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapanou PN (2012). The Prevalence of Periodontitis in the US: Forget What You Were Told. J Dent Res, 91(10), 907–908. doi: 10.1177/0022034512458692 [DOI] [PubMed] [Google Scholar]
- Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, . . . Lamont RJ. (2005). Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun, 73(7), 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YD, Kim YS, Jung YM, Lee SI, Lee YM, Bang JB, & Kim EC (2012). Porphyromonas gingivalis lipopolysaccharide regulates interleukin (IL)-17 and IL-23 expression via SIRT1 modulation in human periodontal ligament cells. Cytokine, 60(1), 284–293. doi: 10.1016/j.cyto.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Pathirana RD, O’Brien-Simpson NM, & Reynolds EC (2010). Host immune responses to Porphyromonas gingivalis antigens. Periodontol 2000, 52(1), 218–237. doi: 10.1111/j.1600-0757.2009.00330.x [DOI] [PubMed] [Google Scholar]
- Pierce DL, Nishiyama S, Liang S, Wang M, Triantafilou M, Triantafilou K, . . . Hajishengallis G. (2009). Host adhesive activities and virulence of novel fimbrial proteins of Porphyromonas gingivalis. Infect Immun, 77(8), 3294–3301. doi: 10.1128/IAI.00262-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu M-Y, Huffel CV, Du X, . . . Beutler B. (1998). Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science, 282(5396), 2085–2088. doi: 10.1126/science.282.5396.2085 [DOI] [PubMed] [Google Scholar]
- Popadiak K, Potempa J, Riesbeck K, & Blom AM (2007). Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J. Immunol, 178(11), 7242–7250. [DOI] [PubMed] [Google Scholar]
- Potempa J, Banbula A, & Travis J. (2000). Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000, 24, 153–192. [DOI] [PubMed] [Google Scholar]
- Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, . . . Blom AM (2008). Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol, 181(8), 5537–5544. doi: 10.4049/jimmunol.181.8.5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan M, Aduse-Opoku J, Paramonov NA, Hashim A, & Curtis MA (2017). Hemin binding by Porphyromonas gingivalis strains is dependent on the presence of A-LPS. Mol Oral Microbiol, 32(5), 365–374. doi: 10.1111/omi.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, & Lambris JD (2010). Complement: a key system for immune surveillance and homeostasis. Nat Immunol, 11(9), 785–797. doi: 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen G, & Sela MN (2006). Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol Lett, 256(2), 304–310. doi: 10.1111/j.1574-6968.2006.00131.x [DOI] [PubMed] [Google Scholar]
- Sakanaka A, Takeuchi H, Kuboniwa M, & Amano A. (2016). Dual lifestyle of Porphyromonas gingivalis in biofilm and gingival cells. Microb Pathog, 94, 42–47. doi: 10.1016/j.micpath.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Okamatsu Y, Kawai T, Kent R, Taubman M, & Stashenko P. (2004). The interleukin-10 knockout mouse is highly susceptible to Porphyromonas gingivalis-induced alveolar bone loss. J. Periodontal Res, 39(6), 432–441. [DOI] [PubMed] [Google Scholar]
- Sato K, Suematsu A, Okamoto K, Yamaguchi A, Morishita Y, Kadono Y, . . . Takayanagi H. (2006). Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J. Exp. Med, 203(12), 2673–2682. doi: 10.1084/jem.20061775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkein HA, Fletcher HM, Bodnar M, & Macrina FL (1995). Increased opsonization of a prtH-defective mutant of Porphyromonas gingivalis W83 is caused by reduced degradation of complement-derived opsonins. J Immunol, 154(10), 5331–5337. [PubMed] [Google Scholar]
- Scheres N, & Crielaard W. (2013). Gingival fibroblast responsiveness is differentially affected by Porphyromonas gingivalis: implications for the pathogenesis of periodontitis. Molecular oral microbiology, 28(3), 204–218. doi:Doi 10.1111/Omi.12016 [DOI] [PubMed] [Google Scholar]
- Schifferle RE, Reddy MS, Zambon JJ, Genco RJ, & Levine MJ (1989). Characterization of a polysaccharide antigen from Bacteroides gingivalis. J Immunol, 143(9), 3035–3042. [PubMed] [Google Scholar]
- Schromm AB, Brandenburg K, Loppnow H, Moran AP, Koch MH, Rietschel ET, & Seydel U. (2000). Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur J Biochem, 267(7), 2008–2013. doi: 10.1046/j.1432-1327.2000.01204.x [DOI] [PubMed] [Google Scholar]
- Sheets SM, Potempa J, Travis J, Casiano CA, & Fletcher HM (2005). Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect Immun, 73(3), 1543–1552. doi: 10.1128/IAI.73.3.1543-1552.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets SM, Potempa J, Travis J, Fletcher HM, & Casiano CA (2006). Gingipains from Porphyromonas gingivalis W83 synergistically disrupt endothelial cell adhesion and can induce caspase-independent apoptosis. Infect Immun, 74(10), 5667–5678. doi: 10.1128/IAI.01140-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker BJ, & Datar S. (1995). Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun, 63(12), 4830–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach EM (2018). Foxp3(+) T Regulatory Cells: Still Many Unanswered Questions-A Perspective After 20 Years of Study. Front Immunol, 9, 1048. doi: 10.3389/fimmu.2018.01048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, . . . Shao F. (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature, 514(7521), 187–192. doi: 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- Singh A, Wyant T, Anaya-Bergman C, Aduse-Opoku J, Brunner J, Laine ML, . . . Lewis JP (2011). The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect Immun, 79(11), 4533–4542. doi: 10.1128/IAI.05016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Hasan S, Sharma S, Nagra S, Yamaguchi DT, Wong DT, . . . Hossain A. (2014). Th17 cells in inflammation and autoimmunity. Autoimmun Rev, 13(12), 1174–1181. doi: 10.1016/j.autrev.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Slocum C, Coats SR, Hua N, Kramer C, Papadopoulos G, Weinberg EO, . . . Genco CA (2014). Distinct lipid a moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS Pathog, 10(7), e1004215. doi: 10.1371/journal.ppat.1004215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, & Kent RL Jr. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol, 25(2), 134–144. [DOI] [PubMed] [Google Scholar]
- Sojar HT, & Genco RJ (2005). Identification of glyceraldehyde-3-phosphate dehydrogenase of epithelial cells as a second molecule that binds to Porphyromonas gingivalis fimbriae. FEMS Immunol Med Microbiol, 45(1), 25–30. doi: 10.1016/j.femsim.2005.01.006 [DOI] [PubMed] [Google Scholar]
- Sroka A, Sztukowska M, Potempa J, Travis J, & Genco CA (2001). Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. Journal of Bacteriology, 183(19), 5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Lubberts E, & Hendriks RW (2018). A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J Autoimmun, 87, 1–15. doi: 10.1016/j.jaut.2017.12.007 [DOI] [PubMed] [Google Scholar]
- Stashenko P, Resmini LM, Haffajee AD, & Socransky SS (1983). T cell responses of periodontal disease patients and healthy subjects to oral microorganisms. J Periodontal Res, 18(6), 587–600. doi: 10.1111/j.1600-0765.1983.tb00396.x [DOI] [PubMed] [Google Scholar]
- Sundqvist G, Figdor D, Hanstrom L, Sorlin S, & Sandstrom G. (1991). Phagocytosis and virulence of different strains of Porphyromonas gingivalis. Scand J Dent Res, 99(2), 117–129. doi: 10.1111/j.1600-0722.1991.tb01874.x [DOI] [PubMed] [Google Scholar]
- Takeichi O, Haber J, Kawai T, Smith DJ, Moro I, & Taubman MA (2000). Cytokine profiles of T-lymphocytes from gingival tissues with pathological pocketing. J Dent Res, 79(8), 1548–1555. doi: 10.1177/00220345000790080401 [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Sasaki N, Yamaga S, Kuboniwa M, Matsusaki M, & Amano A. (2019). Porphyromonas gingivalis induces penetration of lipopolysaccharide and peptidoglycan through the gingival epithelium via degradation of junctional adhesion molecule 1. PLoS Pathog, 15(11), e1008124. doi: 10.1371/journal.ppat.1008124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, . . . Akira S. (1999). Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity, 11(4), 443–451. [DOI] [PubMed] [Google Scholar]
- Tartar DM, VanMorlan AM, Wan X, Guloglu FB, Jain R, Haymaker CL, . . . Zaghouani H. (2010). FoxP3+RORgammat+ T helper intermediates display suppressive function against autoimmune diabetes. J Immunol, 184(7), 3377–3385. doi: 10.4049/jimmunol.0903324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YT, Nguyen H, Gao X, Kong YY, Gorczynski RM, Singh B, . . . Penninger JM (2000). Functional human T-cell immunity and osteoprotegerin ligand control alveolar bone destruction in periodontal infection. J Clin Invest, 106(6), R59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokoro Y, Matsuki Y, Yamamoto T, Suzuki T, & Hara K. (1997). Relevance of local Th2-type cytokine mRNA expression in immunocompetent infiltrates in inflamed gingival tissue to periodontal diseases. Clin Exp Immunol, 107(1), 166–174. doi: 10.1046/j.1365-2249.1997.d01-880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai T, Mori Y, Onoyama M, & Hara Y. (2001). Immunohistological study of interferon-gamma- and interleukin-4-bearing cells in human periodontitis gingiva. Arch Oral Biol, 46(10), 901–908. doi: 10.1016/s0003-9969(01)00057-7 [DOI] [PubMed] [Google Scholar]
- Umemoto T, & Hamada N. (2003). Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis. J. Periodontol, 74(1), 119–122. [DOI] [PubMed] [Google Scholar]
- Valmori D, Raffin C, Raimbaud I, & Ayyoub M. (2010). Human RORgammat+ TH17 cells preferentially differentiate from naive FOXP3+Treg in the presence of lineage-specific polarizing factors. Proc Natl Acad Sci U S A, 107(45), 19402–19407. doi: 10.1073/pnas.1008247107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bruggen N, & Ouyang W. (2014). Th17 cells at the crossroads of autoimmunity, inflammation, and atherosclerosis. Immunity, 40(1), 10–12. doi: 10.1016/j.immuni.2013.12.006 [DOI] [PubMed] [Google Scholar]
- Vernal R, Diaz-Guerra E, Silva A, Sanz M, & Garcia-Sanz JA (2014). Distinct human T-lymphocyte responses triggered by Porphyromonas gingivalis capsular serotypes. J Clin Periodontol, 41(1), 19–30. doi: 10.1111/jcpe.12176 [DOI] [PubMed] [Google Scholar]
- Vernal R, Dutzan N, Chaparro A, Puente J, Antonieta Valenzuela M, & Gamonal J. (2005). Levels of interleukin-17 in gingival crevicular fluid and in supernatants of cellular cultures of gingival tissue from patients with chronic periodontitis. J Clin Periodontol, 32(4), 383–389. doi: 10.1111/j.1600-051X.2005.00684.x [DOI] [PubMed] [Google Scholar]
- Vernal R, Leon R, Silva A, van Winkelhoff AJ, Garcia-Sanz JA, & Sanz M. (2009). Differential cytokine expression by human dendritic cells in response to different Porphyromonas gingivalis capsular serotypes. J Clin Periodontol, 36(10), 823–829. doi: 10.1111/j.1600-051X.2009.01462.x [DOI] [PubMed] [Google Scholar]
- Voo KS, Wang YH, Santori FR, Boggiano C, Arima K, Bover L, . . . Liu YJ (2009). Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A, 106(12), 4793–4798. doi: 10.1073/pnas.0900408106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang J, Jin Y, Gao H, & Lin X. (2014). Oral administration of all-trans retinoic acid suppresses experimental periodontitis by modulating the Th17/Treg imbalance. J Periodontol, 85(5), 740–750. doi: 10.1902/jop.2013.130132 [DOI] [PubMed] [Google Scholar]
- Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, . . . Hajishengallis G. (2010). Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal, 3(109), ra11. doi: 10.1126/scisignal.2000697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Shakhatreh M-AK, James D, Liang S, Nishiyama S. i., Yoshimura F, . . . Hajishengallis G. (2007). Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol, 179(4), 2349–2358. [DOI] [PubMed] [Google Scholar]
- Wang P-L, & Ohura K. (2002). Porphyromonas gingivalis lipopolysaccharide signaling in gingival fibroblasts- CD14 and Toll-like receptors. Crit. Rev. Oral Biol. Med, 13, 132–142. [DOI] [PubMed] [Google Scholar]
- Wassenaar A, Reinhardus C, Thepen T, Abraham-Inpijn L, & Kievits F. (1995). Cloning, characterization, and antigen specificity of T-lymphocyte subsets extracted from gingival tissue of chronic adult periodontitis patients. Infect Immun, 63(6), 2147–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Onoe T, Ozeki M, Shimizu Y, Sakayori T, Nakamura H, & Yoshimura F. (1996). Sequence and product analyses of the four genes downstream from the fimbrilin gene (fimA) of the oral anaerobe Porphyromonas gingivalis. Microbiol. Immunol, 40(10), 725–734. [DOI] [PubMed] [Google Scholar]
- Whittaker CJ, Klier CM, & Kolenbrander PE (1996). Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol, 50, 513–552. doi: 10.1146/annurev.micro.50.1.513 [DOI] [PubMed] [Google Scholar]
- Wilensky A, Tzach-Nahman R, Potempa J, Shapira L, & Nussbaum G. (2015). Porphyromonas gingivalis gingipains selectively reduce CD14 expression, leading to macrophage hyporesponsiveness to bacterial infection. J Innate Immun, 7(2), 127–135. doi: 10.1159/000365970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, & Hugli TE (1992). Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem, 267(26), 18902–18907. [PubMed] [Google Scholar]
- Yamazaki K, Nakajima T, Kubota Y, Gemmell E, Seymour GJ, & Hara K. (1997). Cytokine messenger RNA expression in chronic inflammatory periodontal disease. Oral Microbiol Immunol, 12(5), 281–287. doi: 10.1111/j.1399-302x.1997.tb00392.x [DOI] [PubMed] [Google Scholar]
- Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, . . . Lochner M. (2016). Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol, 9(2), 444–457. doi: 10.1038/mi.2015.74 [DOI] [PubMed] [Google Scholar]
- Yang J, Wu J, Liu Y, Huang J, Lu Z, Xie L, . . . Ji Y. (2014). Porphyromonas gingivalis infection reduces regulatory T cells in infected atherosclerosis patients. PLoS One, 9(1), e86599. doi: 10.1371/journal.pone.0086599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JJ, Ruddy MJ, Conti HR, Boonanantanasarn K, & Gaffen SL (2008). The Interleukin-17 Receptor Plays a Gender-Dependent Role in Host Protection against Porphyromonas gingivalis-Induced Periodontal Bone Loss. Infect. Immun, 76(9), 4206–4213. doi: 10.1128/iai.01209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JJ, Ruddy MJ, Wong GC, Sfintescu C, Baker PJ, Smith JB, . . . Gaffen SL. (2007). An essential role for IL-17 in preventing pathogen-initiated bone destruction: recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood, 109(9), 3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenobia C, Hasturk H, Nguyen D, Van Dyke TE, Kantarci A, & Darveau RP (2014). Porphyromonas gingivalis lipid A phosphatase activity is critical for colonization and increasing the commensal load in the rabbit ligature model. Infect Immun, 82(2), 650–659. doi: 10.1128/IAI.01136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen B, Yan F, Guo J, Zhu X, Ma S, & Yang W. (2014). Interleukin-10 inhibits bone resorption: a potential therapeutic strategy in periodontitis and other bone loss diseases. Biomed Res Int, 2014, 284836. doi: 10.1155/2014/284836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kimura Y, Fang C, Zhou L, Sfyroera G, Lambris JD, . . . Song WC (2007). Regulation of Toll-like receptor-mediated inflammatory response by complement in vivo. Blood, 110(1), 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, & Teng YT (2006). Interleukin-10 inhibits gram-negative-microbe-specific human receptor activator of NF-kappaB ligand-positive CD4+-Th1-cell-associated alveolar bone loss in vivo. Infect Immun, 74(8), 4927–4931. doi: 10.1128/IAI.00491-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Wu J, & Xie H. (2011). Differential expression and adherence of Porphyromonas gingivalis FimA genotypes. Mol Oral Microbiol, 26(6), 388–395. doi: 10.1111/j.2041-1014.2011.00626.x [DOI] [PMC free article] [PubMed] [Google Scholar]