Abstract
Functionally similar IDPs (intrinsically disordered proteins) often have little sequence similarity. This is in stark contrast to folded proteins and poses a challenge for the inverse problem, functional classification of IDPs using sequence alignment. The problem is further compounded because of the lack of structure in IDPs, preventing structural alignment as an alternate tool for classification. Recent advances in heteropolymer theory unveiled a powerful set of sequence-patterning metrics bridging molecular interaction with chain conformation. Focusing only on charge patterning, these set of metrics yield a sequence charge decoration matrix (SCDM). SCDMs can potentially identify functionally similar IDPs not apparent from sequence alignment alone. Here, we illustrate how these information-rich “molecular blueprints” encoded in SCDMs can be used for functional classification of IDPs with specific application in three protein families—Ste50, PSC, and RAM—in which electrostatics is known to be important. For both the Ste50 and PSC protein family, the set of metrics appropriately classifies proteins in functional and nonfunctional groups in agreement with experiment. Furthermore, our algorithm groups synthetic variants of the disordered RAM region of the Notch receptor protein—important in gene expression—in reasonable accordance with classification based on experimentally measured binding constants of RAM and transcription factor. Taken together, the novel classification scheme reveals the critical role of a high-dimensional set of metrics—manifest in self-interaction maps and topology—in functional annotation of IDPs even when there is low sequence homology, providing the much-needed alternate to a traditional sequence alignment tool.
Significance
Functional classification of proteins is critical to understand fundamental biology and molecular evolution. Folded proteins can be functionally grouped based on their sequence and/or structural similarity. However, the same does not apply for intrinsically disordered proteins (IDPs) that lack unique folded structure. Sequence alignment often fails to identify functionally similar IDPs because of low sequence similarity. Yet, functional clues must be in the sequence! How do we unlock the code? Progress in theoretical physics of IDPs yielded novel mathematical formulae revealing hidden features of sequences. We applied these information-rich metrics to classify IDPs consistent with experimental data but not possible by sequence alignment. The success of our approach offers a, to our knowledge, new avenue for IDP classification grounded on physicochemical rules.
Introduction
Intrinsically disordered proteins and disordered regions (generally termed IDPs) are ubiquitous and participate in numerous biological functions (1,2): from signaling, chromatin remodeling, and cellular differentiation to the formation of membraneless organelles. However, functional classification of IDPs—in contrast to that of folded proteins—is in its infancy because of two primary challenges. First, IDP sequences of functionally similar proteins have very low sequence similarity (3). Consequently, traditional sequence alignment tools, successful for folded proteins, cannot be used to detect functionally similar IDPs. Second, IDPs do not have a definite native structure; instead, they interconvert among disordered conformations. Thus, functional classification by structure alignment is not possible either.
Despite low sequence similarity, IDPs across different species can perform similar function. An intriguing question emerges: are there hidden molecular blueprints in the apparently diverged set of sequences that are perhaps conserved for function (4)? Support for this idea comes from recent, but limited, experimental studies identifying specific metrics based on charge amino acids encoded in the sequence. Functional similarity of proteins can manifest in metrics as simple as overall charge composition (4,5) or more abstruse, such as contiguous stretches of charge (beyond composition) (6), while sequence alignment fails to detect any similarity. The intricate role of charge decoration is further evidenced when synthetic sequences generated by charge shuffling while maintaining the same charge composition exhibit widely different behavior (7). In a recent approach, it has been noted that conformational fluctuations arising from an all-atom force field may be used to classify IDP sequences (8). A large-scale proteome-wide analysis of IDP function revealed the importance of a set of molecular features extracted from a sequence for functional characterization (4). It is now timely to unravel these cryptic features of the sequence—stemming from specific interaction rules—to identify functionally similar or dissimilar proteins that are not apparent from simple sequence alignment alone.
So, what are these cryptic features and how do we decode them? Intuitive features of sequences have been shown to describe the experimentally measured sizes of IDPs (9, 10, 11, 12). Recent progress in heteropolymer theory unveiled several nonintuitive metrics encoded in sequence that determine IDP conformational ensemble (13, 14, 15, 16, 17, 18). Some of these metrics are a direct outcome of mathematical averaging over the ensemble (14, 15, 16). These closed-form mathematical expressions are functions of sequence decoration and not just the composition. The same set of decoration metrics also plays a critical role in describing the phase-separation propensity of IDP chains (19,20), consistent with the finding that single-chain conformation can dictate multichain physics of phase separation (21,22). Attempts are underway to use sequence features to build predictors of diverse function such as phase separation (23) and formation of fuzzy complexes involving IDPs (24,25), to name a few. Gross conformational features such as radius of gyration and limited proteolysis have been implicated to detect the functional similarities of POLII between human and fly, despite having little sequence similarity (26). Several other studies also hint at a possible role of the conformational ensemble and the associated disorder to tune function (27, 28, 29). These observations convey two main messages: 1) novel sequence-decoration metrics extracted from sequence alone can be used to describe conformational features and 2) conformational features ultimately dictate function. We notice the emergence of a “molecular metric, conformation, function” paradigm in IDPs.
With the possibility of identifying and using hidden molecular metrics to detect functional similarity between two apparently dissimilar sequences, we also recognize the challenge. Two functionally similar proteins can happen to be dissimilar in a given metric. On the other hand, two functionally dissimilar proteins can have the same values of a given metric, although this is less likely. We need a high-dimensional yet finite set of metrics to have enough specificity to detect similarities or dissimilarities between sequences. At the same time, the metrics should be representative of the conformational ensemble because function is expected to depend on conformation. In our recent work, we discovered such a set that dictates the inter-residue distance maps holding the keys to IDP conformations. Specifically, we identified an electrostatic self-interaction matrix that determines the ensemble average distance profiles between two residues i, j in a given chain (30). These information-rich quantities define several metrics of charge patterning that can be organized as a matrix called sequence charge decoration matrix (SCDM). A previously discovered metric, defined as sequence charge decoration or simply SCD (see (14)), is just one element of this matrix. SCDM provides the much-needed high-dimensional yet manageable set of numbers—derived directly from sequence—to quantify IDP similarity or dissimilarity.
IDPs are low-complexity sequences with significant conformational dependence on electrostatics (9, 10, 11,13,31, 32, 33). In this work, as a proof of concept, we focus on the set of IDPs for which electrostatics has been implicated to influence function. We show that SCDM arising from intrachain electrostatic interaction can be used to group functionally similar proteins in two IDP families, Ste50 and PSC, for which experimental data are available (4, 5, 6). Finally, we notice SCDM-based classification of synthetic variants of RAM proteins also moderately correlates with classification using measured binding data (7). We emphasize that the elements of the SCDM were derived within an analytical formalism distinct from molecular simulation. Moreover, SCDMs provide physical insights by depicting electrostatic origin to intrachain conformational profile such as collapse and swelling at different parts of the chain. Intrachain conformational features, in turn, dictate the protein’s accessibility and ability to interact with other biomolecules, often required for function. Thus, similarity or dissimilarity in the patterns of these easily computable molecular features concealed in SCDM is perfectly suited to classify many IDP sequences across multiple species and designed sequences in which electrostatics is critical.
Methods
We need a high-dimensional set of metrics that are functions of sequence to identify similarity or dissimilarity between two IDPs. In a recent publication, we have demonstrated the dependence of inter-residue distance profiles on different interactions in a sequence-specific manner (30). For example, the ensemble average distance between two amino acids i and j depends on the sequence details by an SCDM whose elements SCDMij are defined as
| (1) |
where qm, qn are the charges on the residues at position m and n, respectively, and N is the total number of amino acids in a protein. The origin of SCDM can be understood by noticing that SCDMij values are proportional to (κl = 0)/(i − j), where (κl) is defined in (14). Note that κl = 0 denotes zero salt condition. The division by (i − j) ensures consistency with the previously defined one-dimensional patterning metric SCD (14). Specifically, SCD = SCDMi = N, j = 1. Thus, the SCDM is more descriptive of IDP conformation than the single-metric SCD defined earlier (14).
For classification purposes, SCDMij values were calculated for each i, j pair of amino acids yielding a large set of metrics. Furthermore, they were assigned +1 if the elements were positive (repulsive) or −1 when negative (attractive). Thus, the SCDM was binarized. For the sake of brevity, binary SCDM will be referred to as bSCDM (binarized SCDM) for the rest of this manuscript. To address the issue of different chain lengths, each bSCDM was resized to the dimensions of the longest protein chain using the image rescaling package in OpenCV with an interpolation algorithm. The elements of bSCDM bridge electrostatic interaction with distance maps () describing the chain conformation for different residue pairs (i, j). Thus, the NX(N − 1)/2 dimensional bSCDM provides the map of attractive and repulsive regions within the chain holding the blueprint of IDPs.
Next, bSCDM was converted to a one-dimensional array describing NX(N − 1)/2 features for a given IDP. However, the topology of the matrix information was preserved by properly ordering the elements keeping track of their indices based on i, j. A consolidated protein data matrix was created in which each row contained the one-dimensional ordered array of NX(N − 1)/2 features specific to a protein. If n proteins are to be classified, the protein data matrix will have n rows for each protein. To eliminate possible redundancy, principal component analysis was carried out on this high-dimensional protein data matrix using the SciKitLearn module PCA. The number of principal components were determined to ensure that at least 90% of the variance in the data can be explained. For Ste50, we used the top three components accounting for 92% of the variance; for PSC, we used the top five components with 94.1% of the variance; and for RAM, we used eight components accounting for 91.7% of the variance. Finally, clustering of the proteins in this principal component space was performed using the SciPy hierarchical clustering package with the centroid algorithm. Euclidean distance matrices have been included in the Supporting material to further highlight the insights gained from the clusters presented in dendrograms. Sequences used for all three protein families can be found in Tables S5–S7 for Ste50, RAM, and PSC, respectively.
Results and discussion
Ste50
The first group of proteins chosen were the Ste50 proteins studied by Moses and colleagues (4,5). Ste50 is an intrinsically disordered region (IDR) between two highly conserved folded domains regulating MAPK pathways. We consider the active Ste50 IDR from Lachancea kluyveri with phosphorylations at positions 17 and 81, referred to as LKCharge. For Saccharomyces cerevisiae, we consider two forms of Ste50 IDR: SC5A and SCCharge. SC5A is nonfunctional with alanine-replacing phosphorylatable positions 13, 54, 60, 102, and 106 (in the truncated sequence). The functional form (SCCharge) is phosphorylated at positions 102 and 106 (in the truncated sequence). Phosphorylation is modeled by adding two glutamic acids to mimic the charge of the phosphate group. SCCharge is functionally comparable to the maximally phosphorylated sequence (5), justifying the double phosphorylation in SCCharge. In addition to LKCharge, SC5A, and SCCharge, we included two IDRs, PEX5 and RAD26, that have negligible sequence similarity to Ste50. When wild-type Ste50 was replaced by RAD26, the function was lost, whereas PEX5 retained wild-type function (4). The functional classification of these proteins was determined by measuring cell morphology, viability, MAPK signaling, and/or response to pheromone (4,5).
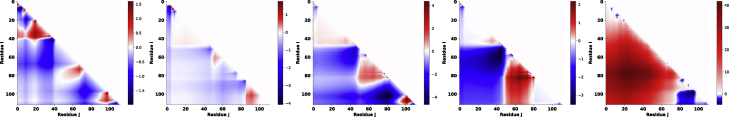
Fig. 1 reveals a visual trend in the topology of the SCDMs. All the functional proteins (LKCharge, PEX5, and SCCharge) have three distinct repulsive regions (red) near the diagonal and rest of the interaction maps are primarily attractive (blue). This is in stark contrast to the nonfunctional proteins (RAD26 and SC5A). Thus, the topology of the intrachain interaction maps quantified by SCDM clearly separates functional and nonfunctional proteins. Electrostatic interaction is expected to induce collapse in the blue regions and swelling in the red regions. The ability to distinguish functional and nonfunctional proteins using these maps highlight important role of single-chain conformational ensemble, including local features, that dictate function.
Figure 1.
Sequence charge decoration matrices (SCDMs) reveal protein specific patterns facilitating functional classification. SCDMs are shown for LKCharge, PEX5, SCCharge, SC5A, and RAD26 (from left to right). The color coding above depicts where the contribution of electrostatics is predicted to be repulsive (red) or attractive (blue). A clear visual pattern of three repulsive clusters near the diagonal is seen to emerge in the functional linkers (LKCharge, PEX5, and SCCharge) that is not present in the nonfunctional linkers (SC5A and RAD26). To see this figure in color, go online.
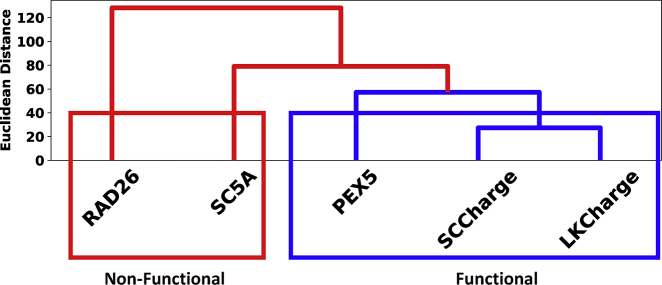
To automate such classification without using visual inspection, we developed a quantitative platform to classify and cluster based on the bSCDMs. As reported in the Methods, the matrix was transformed in binary with attractive interactions assigned a value of −1 and the repulsive interactions a value of +1. This generated sequence-specific patterns in the interaction maps similar to Fig. 1, which we also term bSCDM. We resized these interaction maps to that of the largest protein. Next, we performed principal component analysis to include only dimensions capturing at least 90% of the variance. Once the coordinates along these new dimensions were determined for each protein, they were clustered by their coordinates using a hierarchical agglomerative algorithm. The results of this clustering for Ste50 are shown in Fig. 2.
Figure 2.
Clustering Ste50 using bSCDM matches with functional classification. Clustering using bSCDM groups functional proteins PEX5, SCCharge, and LKCharge (blue) in one cluster and places the two proteins SC5A and RAD26, found nonfunctional in experiments, outside of that cluster (red, left). To see this figure in color, go online.
PEX5, SCCharge, and LKCharge are all classified together on the right (see blue cluster). RAD26 and SC5A are distinct from this initial cluster, shown in red. This is consistent with the experimental readout that identifies PEX5, SCCharge, and LKCharge as functional and RAD26 and SC5A as nonfunctional. The order and proximity of the proteins within the cluster determined by the distance map (see Table S1) provide further insights. The closest two proteins are SCCharge and LKCharge, which are the two functional orthologs of Ste50 linker sequences. PEX5 is the next addition to the cluster, which is not orthologous to Ste50 linker yet retains the normal function (4). Notably, the next addition is SC5A, the nonfunctional form of the Ste50 sequence from S. cerevisiae, which is clustered outside of the functional group but closer to the functional group compared to RAD26. Finally, RAD26 is the furthest from all the other proteins, consistent with the observation that RAD26 is nonfunctional and is not a member of the orthologous set.
As a control, we used three additional classification schemes. First, we performed clustering by using charge content only (see Fig. S4). We notice SC5A, a nonfunctional protein, is clustered closely to PEX5, a functional protein, contradicting experimental observation. This highlights the importance of the sequence specificity captured by bSCDM to classify function and not just composition metrics. We carried out further control by shuffling the elements of bSCDM in random order to test whether the exact topology of the matrix (i.e., the order in which the elements in the matrix appear) is critical for proper classification (see Supporting materials and methods). Classification using randomized bSCDM clusters nonfunctional SC5A and functional SCCharge together, in disagreement with experiment (see Fig. S6). The third control scheme uses the charge-product metric, in which each i, j element of the matrix is calculated simply by qiqj (see Supporting materials and methods). Fig. S9 shows that clustering using the charge-product method does not agree with the functional classification. The failure of the charge-product metric shows that the important contribution of the neighboring charges and their conformation—both embedded in SCDM—is critical to correctly classify “functional” and “nonfunctional” proteins. Ultimately, all three control studies reveal a more nuanced role of sequence charge decoration in grouping functionally similar proteins. Additionally, clustering these proteins by their SCD (one element of SCDM, specifically SCDMN, 1) does not distinguish between functional and nonfunctional proteins. However, our method cannot a priori determine which of the two groups is functional.
PSC-CTR
Next, we considered polycomb repressive complex 1 PSC, a set of highly charged and highly disordered proteins (6). PSC binds to DNA with nanomolar affinity to inhibit chromatin formation and is essential for viability in Drosophila melanogaster. Moreover, it has been found that the C-terminal disordered region of PSC (termed as PSC-CTR) is necessary and sufficient for the function of PSC inhibiting chromatin structure. Beh et al. (6) identified and studied sets of PSC-CTR from different metazoan species and classified them as “inhibitory” or “noninhibitory” based on the 50% inhibition point for the respective protein. Two of the identified PSC-CTR proteins, Daphnia pulex PSC2 (also termed D. pulex2) and D. pulex PSC1 (also termed D. pulex1), inhibit chromatin formation less well (“noninhibitory”) than all the other members of the set. The sequence feature discriminating these two proteins from the rest of the proteins are contiguous stretches of negative charges (6). The scrambled versions (D. pulex PSC1 Act1 and D. pulex PSC1 Act2) of the wild-type sequence D. pulex PSC1 were generated by reducing the contiguous negative charges and increasing the binding affinity to DNA, classifying them as “inhibitory” (6).
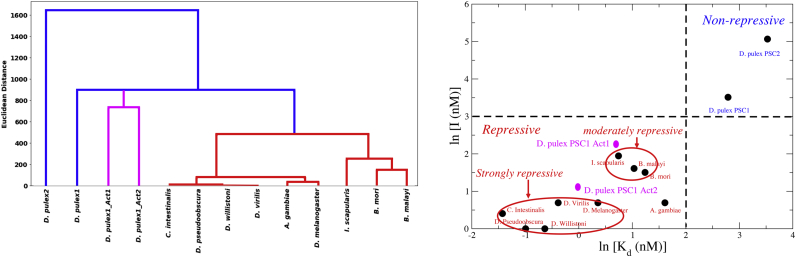
We tested the discriminatory power of our theoretical machinery using bSCDM to cluster PSC-CTR sequences (Fig. 3, left panel). We excluded Helobdella sp. and Lottia gigantea from the original list because of less than 75% disorder in these sequences predicted by the IUPRED server. Theoretical classification compares well with grouping based on dissociation constant (Kd) and 50% inhibition point (denoted as I for this manuscript) measured experimentally (see right panel in Fig. 3). First, we note that D. pulex PSC2 clusters on its own in both the theoretical method and clustering using experimental data (top right in Fig. 3, right panel). Next, our theoretical scheme classifies D. pulex PSC1 separately from all the other wild-type sequences, consistent with the observation that Kd and I of D. pulex PSC1 are much greater than all the “inhibitory or repressive” proteins. A significant difference in Kd and I between D. pulex PSC1 and D. pulex PSC2 is also consistent with our modeling that separates the two proteins. Apart from success in broad grouping, bSCDM captures finer differences among “inhibitory” PSC-CTRs. For example, Drosophila virilis, Ciona intestinalis, D. willistoni, D. pseudoobscura, and D. melanogaster all form their own subcluster in the theoretical method. The same set of proteins can also be grouped together by defining a “strongly repressive” group characterized by Kd less than 2 nM and I between 1 and 2 nM (right panel in Fig. 3). Next, we note that Bombyx mori, Brugia malayi, and Ixodes scapularis are members of their own subcluster using a bSCDM-based classification scheme. The same proteins can also be classified as “moderately repressive” with boundary defined as 2.1 nM < Kd < 3.5 nM and 4.5 nM < I < 7 nM. It is important to discuss Anopheles gambiae within the repressive class; it has unusually high Kd = 5 but low I = 2, in contrast to all the other proteins, which tend to have high (low) Kd associated with high (low) I-values. Thus, A. gambiae is expected to be subclassified on its own among the repressive proteins; theoretical classification, however, fails to capture this finer classification.
Figure 3.
Clustering using bSCDM for PSC proteins closely resembles clustering using experimentally measured Kd and I. The left panel shows the clustering using bSCDM for the PSC proteins, and the right panel shows clustering using Kd (x axis) and the 50% inhibition point I (y axis). With the exception of very few outliers, it can be seen that clustering by bSCDMs is in agreement with clustering using experimental data. To see this figure in color, go online.
Next, we consider the two synthetic sequences D. pulex1 Act1 (also called D. pulex PSC1 Act1) and D. pulex1 Act2 (also called D. pulex PSC1 Act2)—both in magenta—that are clustered together but separate from the parent sequence D. pulex PSC1 using our theoretical algorithm. The separation is consistent with experimental classification of D. pulex1 Act1 and D. pulex1 Act2 as “repressive” compared to “nonrepressive” D. pulex PSC1. At a finer resolution, our algorithm, however, differs from the experimental data (see magenta points in Fig. 3, right panel) that show D. pulex1 Act2 is closer to the subgroup D. virilis, C. intestinalis, D. willistoni, D. pseudoobscura, and D. melanogaster. Closer inspection at the Kd and I data somewhat alleviates this concern. We note that although D. pulex1 Act2 has low Kd = 0.97, similar to the proteins in the “strongly repressive” group defined above, the value of I = 3 nM falls just outside the range of 1 < I < 2 nM loosely associated with the “strongly repressive” group. Furthermore, the distance matrix from the theoretical classification scheme (see Table S2) places D. pulex1 Act2 closer to the wild-type “repressive” sequences compared to D. pulex1 Act1. This relative ordering between D. pulex1 Act2 and D. pulex1 Act1 is in agreement with the observation that D. pulex1 Act2 has a lower Kd and I compared to D. pulex1 Act1 and hence is considered more “repressive.” Fig. S1 shows theoretical clustering using only the first two principal components (PC1 and PC2) capturing 74% of the total variance. Using only two PCs as two axes provides an easy visual interpretation similar to experimental data shown in Fig. 3 (right panel). The repressive PSCs (in red) cluster together and are far from nonrepressive proteins D. pulex PSC2 and D. pulex PSC1. In agreement with experiment, D. pulex1 Act2 appears to be closer to the wild-type repressive group compared to D. pulex1 Act1. Subclustering between repressive groups, in major agreement with the data, is also visible in the PC1-PC2 space.
In addition to the quantitative analysis provided above, the topology maps using SCDMs (Fig. S2) provide important insights. We note that proteins with the strongest binding (low Kd) have an entirely repulsive (red) contribution from electrostatics to IDP conformation. Marginal increases in dissociation (i.e., higher Kd seen in A. gambiae, B. mori, and B. malayi) visually correspond to increasing regions of attractive electrostatics (blue regions). The weakest-binding proteins, D. pulex PSC2 and D. pulex PSC1, in contrast, have significant regions of attractive (blue regions) electrostatics contribution to intrachain conformation. Interestingly, the two synthetic sequences D. pulex1 Act1 and D. pulex1 Act2 have a more central region of repulsion (red), facilitating binding to DNA. The wild-type sequence D. pulex PSC1 has a region where repulsive electrostatics causes local swelling (small red in the top left) that favorably interacts with distal parts of the chain reflected in blue islands. These blue islands create favorable nonlocal intrachain contacts that compete with DNA binding, explaining the lower binding to DNA in D. pulex PSC2 and D. pulex PSC1. The removal of long stretches of negative charges in the two synthetic sequences (D. pulex1 Act1, and D. pulex1 Act2) relieves the small-scale local repulsion (red corners) that, in turn, disrupts distal contacts (blue). This is reflected in the reduction of blue patches and appearance of large red regions in the synthetic sequences. These changes in electrostatics contribution to chain conformation prevent the formation of nonlocal contacts (self-collapse), promoting binding to DNA. Thus, the topography of the interaction maps can provide valuable insights to local and nonlocal interactions that can repress or promote DNA binding.
Despite minor deviations in subclasses noted above, it is encouraging that our algorithm similarly classifies “repressive” proteins identified by Beh et al. (6). Moreover, our method delineates subtle differences within subgroups of “repressive” proteins subclassified as 1) “strongly repressive” and 2) “moderately repressive.” Minor deviations noted for the wild-type A. gambiae and synthetic sequences D. pulex1 Act1 and D. pulex1 Act2 could potentially be due to nonelectrostatic effects on the binding of the PSC-CTR to DNA. These differences may also arise from nondisordered structure forming in regions critical to binding, neglected in our formalism.
As before, we carried out a control study to classify proteins using sequence composition. Fig. S5 shows that when clustering is done based on charge composition only, D. melanogaster, D. pseudoobscura, D. virilis, and D. wilistoni are clustered together, consistent with data. However, major outliers are evident; for example C. intestinalis and I. scapularis are classified far away from the other “repressive” proteins. Moreover, classification based on composition will not discriminate D. pulex1 from D. pulex1 Act1 and D. pulex1 Act2, failing to explain the data. Next, we randomized elements of bSCDM and performed clustering (see Fig. S7). This method also fails to capture major features of the data. For example, D. pulex2 is grouped with the moderately repressive proteins B. mori, B. malayi, and I. scapularis, inconsistent with data. Further analysis using our charge-product method again disagrees with the data, failing to distinguish between strongly repressive, moderately repressive, and nonrepressive (see Fig. S10). These findings reiterate the observation that subtle features of bSCDMs are important for accurate classification of IDPs.
RAM
Finally, we considered the disordered RAM region of the Notch receptor protein (7). The intrinsically disordered RAM region and the folded ANK domain together regulate binding to the transcription factor CSL. Unlike previous examples with PSC and Ste50, RAM has a specific motif that binds to CSL. Sherry et al. generated synthetic sequences of RAM by charge scrambling, with some (RAM 2, 5, 7, and 8) maintaining the noncharged residue positions intact and others (RAM 1, 3, 4, 6, 9, 10, 11, 12, and 13) shuffling the entire sequence, excluding the conserved motif (7). RAM sequences provide an ideal case to test the ability of bSCDM to classify protein sequences that have the same charge composition but different decoration.
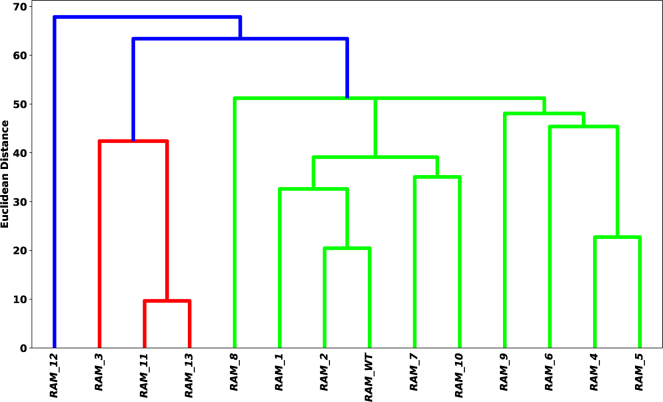
Fig. 4 shows classification using bSCDM produces three major classes: class 1, with RAM 12 only; class 2, consisting of RAM 3, 11, and 13; and class 3, containing RAM 1, 2, 4, 5, 6, 7, 8, 9, 10, and the wild-type (WT). Interestingly, this categorization compares well with a broad classification based on experimentally measured Kd (in nanomolar) values, reported in the Supporting material (see Table S4, color coded by theoretically assigned cluster). The rough classification using Kd also identifies three clusters: RAM 12 as the weakest binder (Kd ≈ 100); RAM 10, 11, and 13 clustered together as the moderate binder (40 > Kd > 29); and the rest of the RAM permutations (RAM 1, 2, 3, 4, 5, 6, 7, 8, 9, and WT) grouped as the strong binder (23 > Kd > 9). This classification is primarily in accordance with our model, with the exception of RAM 3 and RAM 10. It is important to note both RAM 3 and RAM 10 have all amino acids shuffled, excluding in the original binding motif, in addition to charges. Despite RAM 10 not being classified with RAM 11 and 13 in our theoretical model, the distance matrix reveals RAM 10 is closest to RAM 7 but is also relatively close to RAM 11 (see Table S3). Similarly, RAM 13 has its nearest neighbors in the following order: sequence 11, 3, and 10. These observations show although RAM 10 is not directly clustered with sequence 11 and 13 in the dendrogram, they are closer to RAM 10 when compared with many other RAM sequences.
Figure 4.
Clustering of RAM sequences using bSCDM majorly agrees with experimental data. The dendrogram showing clustering of RAM IDR using bSCDMs shows three major groupings, in good agreement with the experimental data using Kd. RAM 3 and 10 are two outliers (see text). To see this figure in color, go online.
In addition to the quantitative analysis and automated clustering, qualitative insights can be gleaned from color-coded SCDMs (see Fig. S3). We note that RAM 12 has distinct topology from all the other sequences with primarily blue regions. Next, RAM 3, 11, and 13 all have a fairly large blue island in the middle when compared to all other sequences, explaining the clustering of RAM 3, 11, and 13 seen in the dendrogram. The blue island in RAM 10 is also visually similar to RAM 11 and RAM 13, consistent with the distance matrix-based similarity noted above. These color maps again highlight the importance of intrachain interaction profiles and topologies in determining IDP binding affinity with other macromolecule, CSL in this case.
The overall agreement between theoretical and experimental categorization indicates Kd is greatly influenced by electrostatic interaction, in accordance with previous studies (7). Sherry et al. found that the hydrodynamic radius of the RAM sequences strongly depends on two different charge segregation metrics (7), albeit much less detailed than the high-dimensional SCDM. However, it is important to recognize the possible role of nonelectrostatic interactions, given there are specific binding motifs (noncharge) that have been disrupted in the designed sequences (RAM 1, 3, 4, 6, 9, 10, 11, 12, and 13). The outliers such as RAM 3 and RAM 10, noted above, may have influence from nonelectrostatic effects not captured in our theory. These effects could either be affecting the strength of the binding of RAMANK to CSL or altering the access that CSL has to the conserved binding motif present in RAM.
The role of the ANK folded domain on the conformation of RAM is also neglected in our model. We note the binding data with Kd-values (discussed above) correspond to the full RAMANK sequence of the RAM permutation to CSL, not the binding of the RAM region alone. This data set was used because limited binding data were available for truncated (without the flanking ANK domain) version of the RAM. Although transcriptional activation data were available, we only compared classification using Kd because the intrachain conformational map is expected to directly influence binding with other partners. Furthermore, lack of correlation between transcriptional activity and Kd shows the possible role of other factors, including in vivo effects, controlling transcription.
For RAM, we did not use sequence composition as a control, unlike Ste50 and PSC, because all the permutants would have the same composition. Thus, as a control we classified sequences by shuffling the elements of bSCDM (see Fig. S8). There are two main clusters, one containing RAM 3 and 12, and all others are assigned in the second cluster. Although this correctly assigns RAM 12 outside of all the other RAM sequences, RAM 3 is not placed correctly. Moreover, the second cluster consists of two subclusters, one containing RAM 2, 4, 5, 6, 7, and 10 and the other containing RAM 1, WT, 8, 9, 11, and 13. Experimental data, however, show RAM 10, 11, and 13 should be clustered together. Clustering based on the charge product (Fig. S11) again shows few to no trends that agree with experimental data. 10 out of 14 proteins appear to be virtually unclassifiable. Clustering these proteins based upon their SCD also does not capture the same effects as clustering by bSCDM. These results further support our previous observation that the nuanced topology of the intrachain interaction maps—quantified by bSCDMs—is key to detect functional similarities in IDPs.
Cohan et al. provided an alternate approach using ensemble entropy to cluster RAM sequences (8). The information-entropy-based classification of Cohan creates four primary clusters in comparison to three using bSCDM and binding (Kd) data. In their classification, cluster 1 contains RAM 11 and 13; cluster 2 contains RAM 7, 10, and 12; cluster 3 contains RAM 2, 3, 5, WT, 6, 8, and 9; and cluster 4 consists of RAM 1 and 4. Similar to our bSCDM-based classification, information entropy classifies RAM 11 and 13 together without RAM 10, in contradiction to the Kd-based grouping. However, RAM 12 is classified close to many other proteins, whereas experimental data (and our classification using bSCDM) show it should be clustered on its own. We also note that RAM 1 and 4, which should be classified very close to WT, are classified as far away as possible. Overall, we conclude that our algorithm using bSCDM, despite the outliers of RAM 10 and 3, clusters proteins in reasonable accordance with Kd data.
Conclusions
We devised a high-dimensional intrachain interaction matrix containing a set of sequence-patterning metrics that mathematically projects protein sequences on a smaller yet meaningful space. This set of sequence-decoration metrics reveals the hidden relation between interaction and chain conformation. In the space of these metrics, we can classify proteins that strongly correlate with experimental classification based on function. We specifically used an interaction matrix arising from electrostatics and defined it as SCDM. We show the success of bSCDM-based clustering in three protein families in which electrostatics is known to be important for function. All these protein families have available experimental data to test our proposed method. For the Ste50 family, proteins are correctly classified as functional or nonfunctional. Likewise, for the PSC-CTR family, our algorithm correctly discriminates between repressive and nonrepressive for wild-type and synthetic sequences. Moreover, for PSC-CTR families our algorithm can depict finer subclassifications such as “strong” and “moderate” repressive in agreement with experimental data based on binding affinity and inhibition concentration. Even for the challenging cases in which functional readout can vary continuously without sharp demarcation between subclasses such as synthetic RAM sequences—classified using binding affinity—our algorithm shows moderate success. The emerging theme is that protein self-interaction captured by the patterns in the bSCDM can also serve as an indicator of interaction with other biomolecules important for function. Consequently, similarity (dissimilarity) in these patterns of bSCDM can be used to detect proteins that are functionally similar (or dissimilar). The success of this approach is further evident by the control study, in which disrupting these patterns by shuffling the decoration matrix failed to cluster proteins in accordance with data. It is important to note the algorithm only identifies proteins that are similar or dissimilar but cannot a priori determine which cluster will be functional. These results demonstrate power of mathematical metrics, derived on physical principles, to classify IDPs that typically evade traditional sequence and structure alignment tools successful in modeling folded proteins.
Acknowledgments
We acknowledge support from the National Institutes of Health (R15GM128162-01A1 and R01GM138901) and Knoebel Institute for Health Aging at the University of Denver.
Editor: Monika Fuxreiter.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2020.11.2282.
Author contributions
J.H. and K.G. designed research. J.H. performed research. J.H. and K.G. analyzed data and wrote the manuscript.
Supporting material
References
- 1.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 2.Dunker A.K., Silman I., Sussman J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Lange J., Wyrwicz L.S., Vriend G. KMAD: knowledge-based multiple sequence alignment for intrinsically disordered proteins. Bioinformatics. 2016;32:932–936. doi: 10.1093/bioinformatics/btv663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarin T., Strome B., Moses A.M. Proteome-wide signatures of function in highly diverged intrinsically disordered regions. eLife. 2019;8:46883. doi: 10.7554/eLife.46883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zarin T., Tsai C.N., Moses A.M. Selection maintains signaling function of a highly diverged intrinsically disordered region. Proc. Natl. Acad. Sci. USA. 2017;114:E1450–E1459. doi: 10.1073/pnas.1614787114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beh L.Y., Colwell L.J., Francis N.J. A core subunit of Polycomb repressive complex 1 is broadly conserved in function but not primary sequence. Proc. Natl. Acad. Sci. USA. 2012;109:E1063–E1071. doi: 10.1073/pnas.1118678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherry K.P., Das R.K., Barrick D. Control of transcriptional activity by design of charge patterning in the intrinsically disordered RAM region of the Notch receptor. Proc. Natl. Acad. Sci. USA. 2017;114:E9243–E9252. doi: 10.1073/pnas.1706083114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohan M.C., Ruff K.M., Pappu R.V. Information theoretic measures for quantifying sequence-ensemble relationships of intrinsically disordered proteins. Protein Eng. Des. Sel. 2019;32:191–202. doi: 10.1093/protein/gzz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsh J.A., Forman-Kay J.D. Sequence determinants of compaction in intrinsically disordered proteins. Biophys. J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller-Späth S., Soranno A., Schuler B. From the cover: charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao A.H., Crick S.L., Pappu R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holehouse A.S., Das R.K., Pappu R.V. CIDER: resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das R.K., Pappu R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawle L., Ghosh K. A theoretical method to compute sequence dependent configurational properties in charged polymers and proteins. J. Chem. Phys. 2015;143:085101. doi: 10.1063/1.4929391. [DOI] [PubMed] [Google Scholar]
- 15.Firman T., Ghosh K. Sequence charge decoration dictates coil-globule transition in intrinsically disordered proteins. J. Chem. Phys. 2018;148:123305. doi: 10.1063/1.5005821. [DOI] [PubMed] [Google Scholar]
- 16.Huihui J., Firman T., Ghosh K. Modulating charge patterning and ionic strength as a strategy to induce conformational changes in intrinsically disordered proteins. J. Chem. Phys. 2018;149:085101. doi: 10.1063/1.5037727. [DOI] [PubMed] [Google Scholar]
- 17.Samanta H.S., Chakraborty D., Thirumalai D. Charge fluctuation effects on the shape of flexible polyampholytes with applications to intrinsically disordered proteins. J. Chem. Phys. 2018;149:163323. doi: 10.1063/1.5035428. [DOI] [PubMed] [Google Scholar]
- 18.Zheng W., Dignon G., Mittal J. Hydropathy patterning complements charge patterning to describe conformational preferences of disordered proteins. J. Phys. Chem. Lett. 2020;11:3408–3415. doi: 10.1021/acs.jpclett.0c00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y.H., Chan H.S. Phase separation and single-chain compactness of charged disordered proteins are strongly correlated. Biophys. J. 2017;112:2043–2046. doi: 10.1016/j.bpj.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y.H., Brady J.P., Ghosh K. A unified analytical theory of heteropolymers for sequence-specific phase behaviors of polyelectrolytes and polyampholytes. J. Chem. Phys. 2020;152:045102. doi: 10.1063/1.5139661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignon G.L., Zheng W., Mittal J. Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput. Biol. 2018;14:e1005941. doi: 10.1371/journal.pcbi.1005941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng X., Holehouse A.S., Pappu R.V. Connecting coil-to-globule transitions to full phase diagrams for intrinsically disordered proteins. Biophys. J. 2020;119:402–418. doi: 10.1016/j.bpj.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernon R.M., Forman-Kay J.D. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019;58:88–96. doi: 10.1016/j.sbi.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Amin A.N., Lin Y.H., Chan H.S. Analytical theory for sequence-specific binary fuzzy complexes of charged intrinsically disordered proteins. J. Phys. Chem. B. 2020;124:6709–6720. doi: 10.1021/acs.jpcb.0c04575. [DOI] [PubMed] [Google Scholar]
- 25.Miskei M., Horvath A., Fuxreiter M. Sequence-based prediction of fuzzy protein interactions. J. Mol. Biol. 2020;432:2289–2303. doi: 10.1016/j.jmb.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Portz B., Lu F., Gilmour D.S. Structural heterogeneity in the intrinsically disordered RNA polymerase II C-terminal domain. Nat. Commun. 2017;8:15231. doi: 10.1038/ncomms15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flock T., Weatheritt R.J., Babu M.M. Controlling entropy to tune the functions of intrinsically disordered regions. Curr. Opin. Struct. Biol. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Motlagh H.N., Wrabl J.O., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohan M.C., Eddelbuettel A.M.P., Pappu R.V. Dissecting the functional contributions of the intrinsically disordered C-terminal tail of Bacillus subtilis FtsZ. J. Mol. Biol. 2020;432:3205–3221. doi: 10.1016/j.jmb.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huihui J., Ghosh K. An analytical theory to describe sequence-specific inter-residue distance profiles for polyampholytes and intrinsically disordered proteins. J. Chem. Phys. 2020;152:161102. doi: 10.1063/5.0004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann H., Soranno A., Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sizemore S.M., Cope S.M., Vaiana S.M. Slow internal dynamics and charge expansion in the disordered protein CGRP: a comparison with Amylin. Biophys. J. 2015;109:1038–1048. doi: 10.1016/j.bpj.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das R.K., Ruff K.M., Pappu R.V. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2015;32:102–112. doi: 10.1016/j.sbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.