Abstract
Universally conserved residues (UCRs) are invariable amino acids evolutionarily conserved among members of a protein family across diverse kingdoms of life. UCRs are considered important for stability and/or function of protein families, but it has not been experimentally examined systematically. Cryptochromes are photoreceptors in plants or light-independent components of the circadian clocks in mammals. We experimentally analyzed 51 UCRs of Arabidopsis cryptochrome 2 (CRY2) that are universally conserved in eukaryotic cryptochromes from Arabidopsis to human. Surprisingly, we found that UCRs required for stable protein expression of CRY2 in plants are not similarly required for stable protein expression of human hCRY1 in human cells. Moreover, 74% of the stably expressed CRY2 proteins mutated in UCRs retained wild-type-like activities for at least one photoresponses analyzed. Our finding suggests that the evolutionary mechanisms underlying conservation of UCRs or that distinguish UCRs from non-UCRs determining the same functions of individual cryptochromes remain to be investigated.
Keywords: cryptochrome, blue-light receptor, universally conserved residues, Arabidopsis
Introduction
Cryptochrome is one of the most ancient and common photoreceptors found in nature (Ahmad and Cashmore 1993; Lin et al. 1995; Cashmore 2003; Lin and Shalitin 2003; Sancar 2003). Cryptochromes are homologous to DNA photolyases that repair DNA lesions resulting from ultraviolet light (Ahmad and Cashmore 1993; Lin et al. 1995). Cryptochromes do not repair DNA, instead they act as blue-light receptors to regulate photomorphogenic development in plants or transcriptional regulators to control circadian clock in plants and animals (Cashmore et al. 1999; Sancar 2003). Cryptochromes are composed of two domains, the universally conserved N-terminal PHR (Photolyase-Homologous Region) domain and the unstructured and poorly conserved CCE (Cryptochrome C-terminal Extension) domain (Sancar 2003). The PHR domain of cryptochromes contains universally conserved residues (UCRs), which are invariable amino acids of members of a protein family from distantly related phylogenetic lineages (Mirny and Shakhnovich 1999). It is intuitive that UCRs must be essential to the overall structure integrity of the proteins, such that mutations are prevented from accumulating during evolution of the protein family (Valencia et al. 1991; Landau et al. 2005). And it is commonly hypothesized that UCRs determine the common structure elements that are universally important to the stability and functions of individual members of the protein family. Mutations altered in functionally important UCRs of cryptochromes from diverse lineages, such as Arabidopsis (Ahmad and Cashmore 1993; Guo et al. 1998; Li et al. 2011; Gu et al. 2012; Gao et al. 2015; Taslimi et al. 2016), Drosophila (Stanewsky et al. 1998), and mammals (McCarthy et al. 2009; Ode et al. 2017; Rosensweig et al. 2018), have been reported. However, the structural and functional importance of UCRs has not been systematically investigated for signaling proteins. One technical difficulty to experimentally test the above hypothesis appears to lie in how to measure the specific activities or the protein abundance-adjusted physiological activities of signaling proteins in vivo. We developed a method to systematically analyze the functional importance of most (∼90%) UCRs of Arabidopsis CRY2 in vivo. Our results are consistent with the notion that the UCRs are important for the function of CRY2, because every mutation altered the UCRs of CRY2 impaired at least one activity of CRY2 tested. However, we found that most (∼94%) stably expressed CRY2 proteins mutated in UCRs remained photophysiologically or photobiochemically active in one or more photoresponses examined. We also found that the UCRs required for stable protein expression of Arabidopsis CRY2 are not similarly required for stable protein expression of a human cryptochrome 1 (hCRY1). We further demonstrate that the specific functions of Arabidopsis and mouse cryptochromes are determined by both UCRs and non-UCRs of the respective cryptochromes, arguing it is important to elucidate the evolutionary mechanisms to distinguish UCRs from non-UCRs of cryptochromes or other protein families.
Results
The Double and Triple Mutants of the Universally Conserved Trp-Triad Residues of CRY2 Remained Photobiologically Active In Vivo
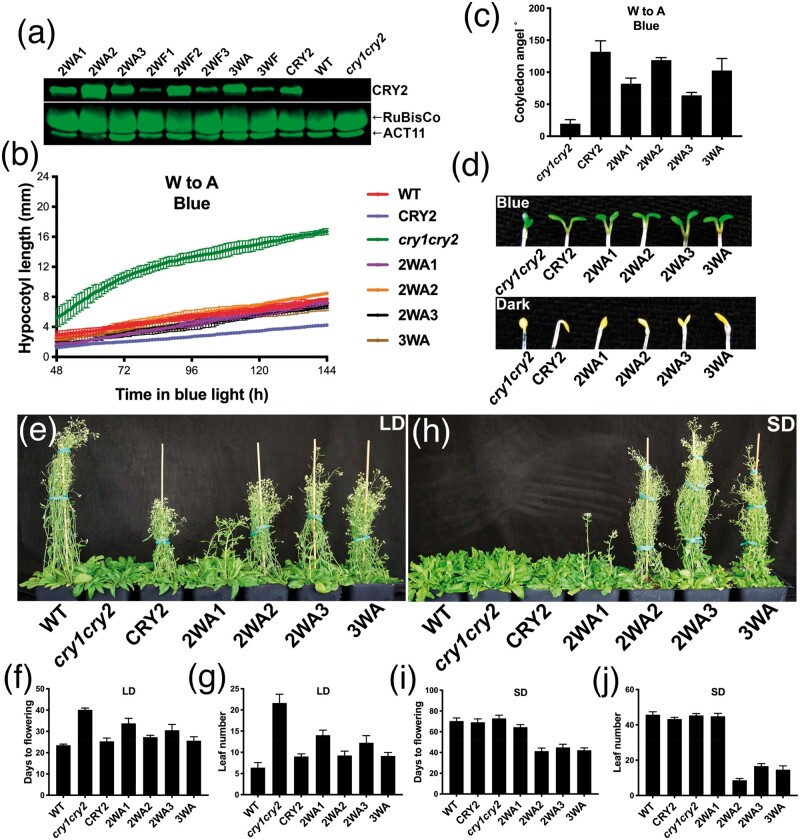
All members of the photolyase/cryptochrome proteins contain three universally conserved tryptophan residues, referred to as Trp-triad (Aubert et al. 2000). Trp-triad are known to be critical to the photoreduction of cryptochromes in vitro, whereby the photon-absorbing chromophore FAD (Flavin Adenine Dinucleotide) is reduced (Li et al. 1991; Lin et al. 1995; Aubert et al. 2000; Chaves et al. 2011). The Trp-triad-dependent photoreduction has been hypothesized to be the photobiochemical mechanism underlying the function of the photolyase/cryptochrome proteins (Aubert et al. 2000; Zeugner et al. 2005; Banerjee et al. 2007; Langenbacher et al. 2009; Müller and Carell 2009; Chaves et al. 2011; Solov’yov et al. 2012; Engelhard et al. 2014; Müller et al. 2015; Ahmad 2016). According to this hypothesis, photolyases and cryptochromes become biochemically and physiologically active upon electron transfer to FAD through the Trp-triad residues. However, this hypothesis has been challenged by several genetics studies, whereby mutations of the Trp-triad residues abolish photoreduction of the mutant proteins in vitro without abolishing their physiological activities in vivo (Gegear et al. 2010; Li et al. 2011; Gao et al. 2015). For example, we have previously reported that mutations altered in any one of the Trp-triad residues of Arabidopsis CRY1 or CRY2 completely abolished FAD photoreduction in vitro, but the mutant proteins remain photophysiologically active in vivo (Li et al. 2011; Gao et al. 2015). It has been proposed that some small molecules in the cell, such as ATP, could bind to cryptochromes and rescue both the photoreduction activity and the physiological activities of a Trp-triad mutant (Engelhard et al. 2014). However, a nonuniversally conserved tryptophan residue has been reported recently for an alternative electron transport pathway governing activities of the Drosophila dCRY mutations (Lin et al. 2018), and ATP fails to rescue the photoreduction activity of every Trp-triad single mutant of Arabidopsis CRY1 (Gao et al. 2015). Therefore, we further investigated the Trp-triad hypothesis by asking whether the double and triple mutants of CRY2 altered in two or all three of the Trp-triad residues (supplementary table S1, Supplementary Material online) might abolish the activity of CRY2 in vivo. In this experiment, the wild-type CRY2 and the W-to-A (redox inactive) or W-to-F (redox inactive but structurally more similar to W) double or triple Trp-triad mutants of CRY2 were constitutively expressed as the green fluorescent protein (GFP)-fusion proteins in the cry1cry2 mutant background. Although a transgenic study using the CRY2 native promoter would be the optimal, we used the constitutive promoter to be consistent with that used in the previous studies (Li et al. 2011; Gao et al. 2015). We have previously shown that the GFP-CRY2 fusion protein is active in all photophysiological and photobiochemical responses tested (Gegear et al. 2010; Li et al. 2011; Gao et al. 2015). The transgenic plants were analyzed for the CRY2 protein abundance and three best known photophysiological activities of CRY2, including blue-light inhibition of hypocotyl elongation, blue-light stimulation of cotyledon unfolding, and promotion of floral initiation (fig. 1 and supplementary fig. S1 and table S1, Supplementary Material online). It is interesting that double or triple mutations of the trp-triad residues often eliminate the gain-of-function hyperactivity of the single mutations affecting residues W374 and W397 (Li et al. 2011). For example, plants expressing the single trp-triad mutants W374A or W397A exhibited constitutive hypocotyl inhibition and floral acceleration phenotype (Li et al. 2011), whereas the double mutant 2WA3 (W374A and W397A) exhibited blue-light-dependent hypocotyl inhibition but constitutive floral acceleration phenotype (fig. 1 and supplementary fig. S1 and table S1, Supplementary Material online). This observation suggests that the hyperactivities of the single mutants may result from structural changes, which may or may not be suppressed by the additional mutations. Remarkably, all double and triple Trp-triad mutants of CRY2 (supplementary table S1, Supplementary Material online), including those expressed at the levels markedly lower than that of the control GFP-CRY2, were able to rescue, to various extent, the defective phenotypes of the cry1cry2 mutant parent, suggesting that all double and triple Trp-triad mutants of CRY2 remained physiologically active (fig. 1 and supplementary fig. S1, Supplementary Material online). For example, a continuous imaging-based kinetic analysis clearly demonstrated that the three double mutants altered in any two of the three Trp-triad residues (2WA1 or CRY2W321A, W374A, 2WA2 or CRY2W321A, W397A, and 2WA3 or CRYsW374A, W397A) and the triple mutant (3WA or CRY2W321A, W374A, W397A) altered in all three Trp-triad residues were active in mediating blue-light inhibition of hypocotyl elongation, such that they all rescued the blue-light-specific long-hypocotyl phenotype of the cry1cry2 parent (fig. 1b). Moreover, transgenic cry1cry2 plants expressing the triple mutant (3WA) protein at the level comparable to that of the wild-type GFP-CRY2 control (fig. 1a) fully rescued the parental late-flowering phenotype (fig. 1e–g). These results confirm that the universally conserved Trp-triad residues are not essential to photophysiological functions of CRY2 in vivo, although they are essential to the photoreduction of CRY2 in vitro (Li et al. 2011).
Fig. 1.
Analyses of double and triple mutants of the Trp-triad residues of CRY2. (a) Immunoblots showing expression of double and triple Trp-triad mutants of the GFP-CRY2 fusion protein. (b) Kinetics analysis of elongation of seedlings germinated and grown under the blue light (15 µmol m−2 s−1). Seedlings are imaged 48 h after germination at the frequency of one image per hour for another 96 h (n = 3). (c) Angles between the two cotyledons were measured from the images taken at 114 h after germination in B (n = 3). (d) The cotyledon unfolding phenotype of 6-day-old seedlings grown in blue light (20 µmol m−2 s−1) (upper) or darkness (lower). (e–j) Images of 40 (e)- or 60 (h)-day-old plants grown in LD (16-h day/8-h night) or SD (8-h day/16-h night). Days to flowering (f, i) and rosette leaf number (g, j) at flowering are shown (n ≥ 8). The wild-type (WT) and transgenic plants constitutively expressing the “wild-type” GFP-CRY2 or the double (2WA1, 2WA2, 2WA3) or triple (3WA) mutants of GFP-CRY2 fusion proteins in the cry1cry2 mutant background are indicated (see supplementary table S1, Supplementary Material online, for more detailed information). Bars in b, c, f, g, i and j indicates SD of the mean.
The UCRs Required for Stable Protein Expression of Arabidopsis CRY2 Are Not Equally Required for Stable Protein Expression of Human hCRY1
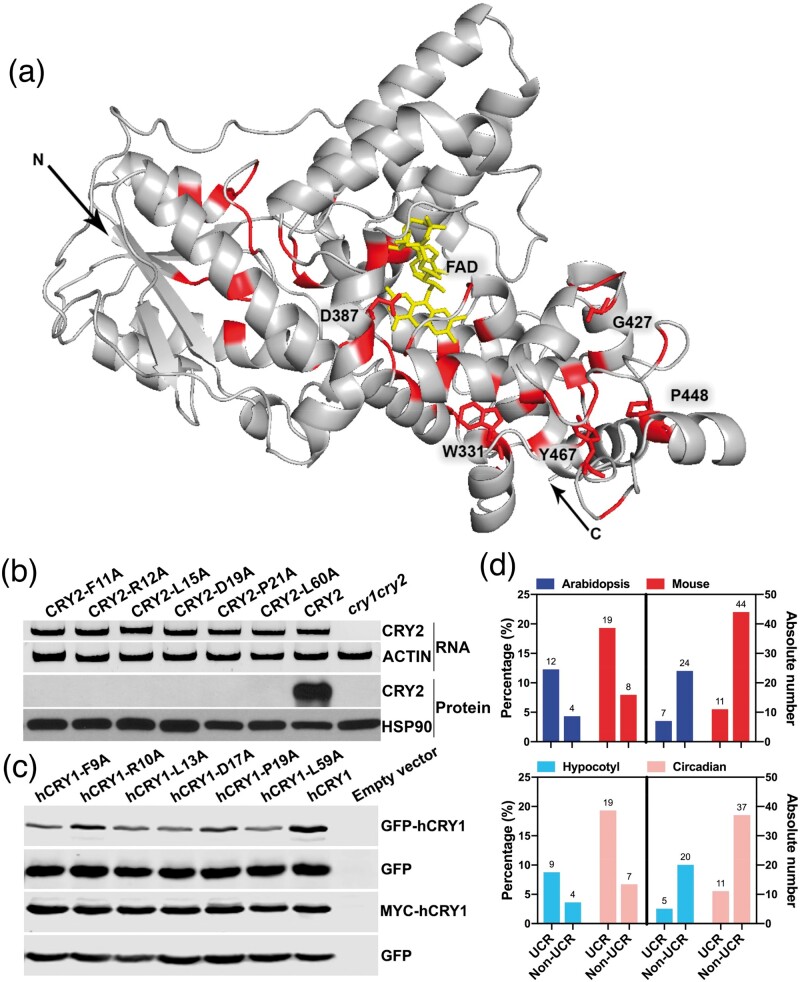
The observation that none of the three universally conserved Trp-triad residues of CRY2 are universally required for all three physiological functions examined seems counterintuitive, because they are UCRs that are commonly considered to be important to preserve integrity of the appropriate conformation of members of a protein family. We hypothesize that the structure elements commonly preserved for members of a protein family may not be required for all functions of individual members under all experimental conditions tested, but such “partial functional requirement” is sufficient to prevent accumulation of any mutations in nature during evolution. Cryptochromes are ancient proteins evolutionarily conserved in all major lineages including plants and human (Sancar 2003), a systematic analysis of mutations of all or most UCRs of cryptochromes, such as Arabidopsis CRY2, would allow us to test this hypothesis. Based on multiple sequence alignment analyses, we identified 57 UCRs of Arabidopsis CRY2, which are defined for the present study as the invariable amino acids that are conserved in the same position of cryptochromes of Arabidopsis and human (fig. 2a and supplementary fig S2 and tables S2–S4, Supplementary Material online). Those 57 residues are also universally conserved among an arbitrarily selected group of 24 cryptochromes from 3 plant families, 1 algae family, and 4 animal families (supplementary fig S2 and table S2, Supplementary Material online).
Fig. 2.
Analyses of UCRs and non-UCRs of plant and mammalian cryptochromes. (a) Structure model of the PHR domain of Arabidopsis CRY2. FAD (yellow), UCRs (red), the N-terminus, the C-terminus (arrows), and 5 UCRs discussed in the text are indicated. (b) Reverse transcription polymerase chain reaction (RT-PCR) (upper two panels) and immunoblots (lower two panels) showing representative Arabidopsis CRY2 UCR mutants that stably express mRNA but not recombinant protein in transgenic plants of the indicated constructs. (c) Immunoblot showing stable protein expression of hCRY1 UCR mutant proteins, each being altered at the residue equivalent to the corresponding Arabidopsis CRY2 UCR mutants shown in (b). Samples were prepared from whole cell lysates of HEK293T cells cotransfected by two plasmids: The sample plasmid encoding the indicated GFP-hCRY1 (upper two panels) or MYC-hCRY1 (lower two panels) protein mutated at the indicated UCR and the control plasmid encoding GFP as the transfection and immunoblot controls. (d) A comparison of the functions of UCR and non-UCR. The functionally defective Arabidopsis (blue) and mouse (red) cryptochromes previously examined based on protein functions (top, all functions, bottom, specific functions) but not sequence conservations were assigned to the UCR or Non-UCR groups according to supplementary fig. S2, Supplementary Material online. Right panels: absolute incidences of mutation independently reported. Left panels: percentage of the functionally defective mutations of the total number of the respective group of residues (UCR or non-UCRs) (see supplementary table S5, Supplementary Material online, for details).
None of the 57 UCRs of CRY2 are alanine, so we changed each of them individually to alanine by site-directed mutagenesis (Zhu et al. 2007) and prepared transgenic plants each constitutively expressing one site-specific CRY2 mutant as GFP-CRY2 fusion protein in the cry1cry2 mutant background. Among the 57 UCRs of CRY2, we successfully obtained transgenic lines expressing 51 UCR mutants (fig. 2a and supplementary fig. S2 and tables S3 and S4, Supplementary Material online). Among these, stable protein expression in plants were observed for 61% (31/51) CRY2 UCR mutants examined, whereas 39% (20/51) failed to stably express detectable amount of mutant proteins in all five independent transgenic lines of each mutant examined. Because all 20 CRY2 UCR mutant genes expressed mRNAs in plants at the level comparable to that of the “wild-type” GFP-CRY2 control (fig. 2b and supplementary fig. S3 A–C and table S4, Supplementary Material online), these UCRs are most likely required for translation or protein stability of Arabidopsis CRY2 in vivo. To test whether these UCRs are similarly required for the stable protein expression of another cryptochrome, we analyzed protein expression of human cryptochrome 1 (hCRY1) mutated in the equivalent UCRs. Interestingly, we found that all the hCRY1 UCR mutant proteins tested expressed at the levels comparable to that of the “wild-type” hCRY1 control in HEK293 (human embryonic kidney) cells (fig. 2c and supplementary fig. S3 D and E, Supplementary Material online), which is in stark contrast to their Arabidopsis counterparts that expressed mRNA but not protein in plant cells (fig. 2b). The mouse mutant mCRY1F257A, which is an UCR mutant equivalent to Arabidopsis CRY2F253A that failed to stably express the protein in plants (supplementary fig. S3 A and B), has also been reported by others to stably express the mutant protein in both HEK293 and MEF (Mouse Embryonic Fibroblasts) cell lines (51). We concluded that the UCRs of cryptochromes are not universally required for stable protein expression of cryptochromes.
UCRs Are Not Universally Required for Photophysiological Activities of CRY2
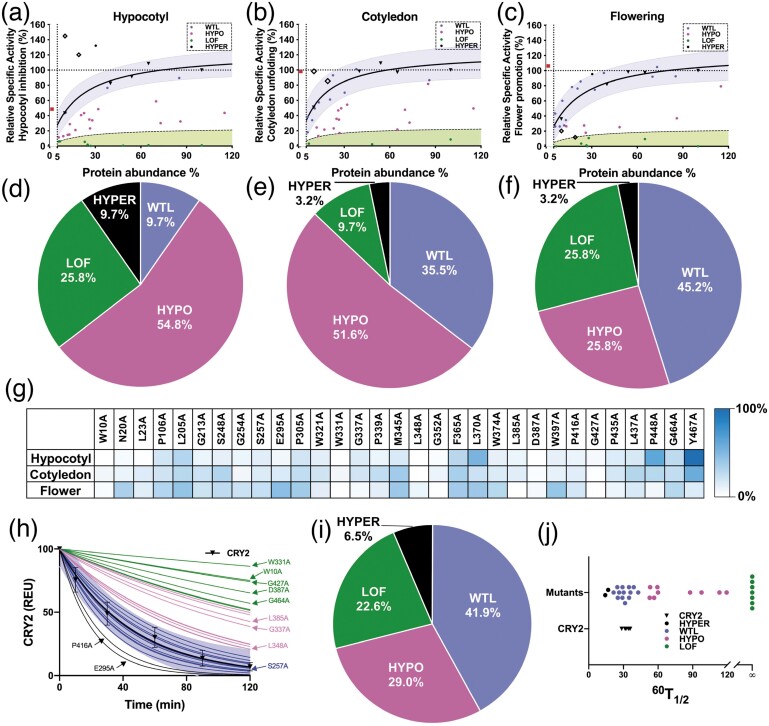
CRY2 is nuclear protein that undergoes blue-light-dependent dimerization (Wang et al. 2016), phosphorylation (Liu et al. 2017), photobody formation (Yu et al. 2009; Zuo et al. 2012), ubiquitination, and degradation (Yu et al. 2007) in nucleus. We examined the subcellular localization of the stably expressed CRY2 UCR mutant proteins and found all of them still locate in the nucleus (supplementary fig. S4, Supplementary Material online). We next tested the photophysiological activities of the 31 CRY2 UCR mutant proteins that are stably expressed in plants. Like enzymes, the cellular concentration of signaling proteins, such as photoreceptors, is expected to determine the total activity of the photoreceptor and photoresponsiveness of the plants (Li et al. 2011). Because it is technically difficult, if not impossible, to obtain different mutant lines that express the identical amount of different CRY2 mutant proteins by either reverse genetics or forward genetics methods, we determined the relative specific-activity of individual CRY2 mutants, based on the standard curves constructed using transgenic lines expressing the “wild-type” GFP-CRY2 protein at different levels (fig. 3). Specifically, we first screened and selected five transgenic lines, referred to as L1, L2, L3, L4, and L5, that express the “wild-type” GFP-CRY2 fusion protein at gradually increased levels, with L5 expressing GFP-CRY2 at the highest level. We quantified the relative abundance of the GFP-CRY2 protein in those five control lines by the quantitative fluorescence immunoblot assay using an Odyssey imager (LI-COR Biotechnology, Lincoln, NE 68504). We next analyzed photomorphogenic phenotypes of the transgenic lines L1–L5, including blue-light inhibition of hypocotyl elongation, blue-light stimulation of cotyledon unfolding, and photoperiodic promotion of flowering (fig. 3a–c). We then constructed the standard curves, whereby the relative protein abundances of the “wild-type” GFP-CRY2 protein in the L1, L2, L3, L4, and L5 transgenic lines were plotted against the relative light responsiveness of the respective transgenic lines. In the three standard curves shown in figure 3a–c, individual light responses of plants are used as the proxy of the relative photophysiological activities of the “wild-type” GFP-CRY2 expressed at the levels measured in the transgenic lines L1–L5, with both the protein abundance and CRY2 activity of the L5 line set as 100%. As expected, the “wild-type” GFP-CRY2 protein shows strong correlations between the protein abundances and the photophysiological activities inferred from the photomorphogenic phenotypes of the lines L1–L5, including blue-light inhibition of hypocotyl elongation (r = 0.9636, P < 0.01), blue-light stimulation of cotyledon unfolding (r = 0.8585, P < 0.01), and CRY2 promotion of floral initiation (r = 0.9701, P < 0.001) (supplementary fig. S5 A–C, Supplementary Material online). This correlation is better observed at relatively lower levels of the CRY2 protein (fig. 3a–c), which is consistent with the expectation that the total activity of CRY2 is apparently saturable. All three standard curves showed saturation of activities of GFP-CRY2 at the approximate protein abundance between that of the transgenic lines L4 and L5, indicating the appropriate sensitivity ranges for our analyses. The “wild-type” GFP-CRY2 protein has been considered similar to the endogenous CRY2, but its activities have never been quantitatively compared with that of the endogenous CRY2 (Yu et al. 2009; Li et al. 2011). We plot the relative abundance and activities of the endogenous CRY2 to the standard curves of GFP-CRY2 and compared the relative specific-activity of the two proteins. Figure 3a–c shows that the endogenous CRY2 protein expresses at the level ∼40% that of the “wild-type” GFP-CRY2 of the transgenic line L1, which expressed the lowest level of GFP-CRY2 among the standard lines L1–L5 (fig. 3a–c and supplementary table S3, Supplementary Material online). However, the endogenous CRY2 has the similar photophysiological activity (∼100%) mediating blue-light inhibition of hypocotyl growth as that of GFP-CRY2 of the L1 line (fig. 3a and supplementary table S3, Supplementary Material online), demonstrating that the relative specific-activity, or the protein abundance-adjusted photophysiological activity, of the “wild-type” GFP-CRY2 is about 40% (40/100) that of the endogenous CRY2 mediating this photoresponse. Similarly, we estimated that the relative specific-activity of GFP-CRY2 mediating blue-light promotion of cotyledon unfolding or CRY2 promotion of floral initiation are ∼20% (40/200) or 16% (40/250) that of the endogenous CRY2 (fig. 3b and c and supplementary table S3, Supplementary Material online). These results suggest that the endogenous CRY2 is ∼2.5–6.25-fold more active than that of the “wild-type” GFP-CRY2. The lower relative specific-activity of GFP-CRY2 in comparison to that of the endogenous CRY2, which is likely caused by the structure disturbances of GFP fusion, minimizes a potential bias in our study due to saturation of activities of the overexpressed GFP-CRY2 mutant proteins tested.
Fig. 3.
Systematic analyses of the activities of the CRY2 UCR mutant proteins. (a–c) Using the standard curve approach to determine the relative specific photophysiological activities of the CRY2 UCR mutants. Blue shades represent regions within 95% prediction bands of the standard curves. Green shades indicate regions lower than 20% of the standard curve (black dashed lines). Round dots, red squires, and hollow squires indicate CRY2 UCR mutants, endogenous CRY2, CRY2P448A, and CRY2Y467A (further analyzed in fig. 4), respectively. (d–f) Classification of CRY2 UCR mutants for hypocotyl inhibition (d), and cotyledon unfolding (e), floral promotion (f), respectively. (g) Heat map showing normalized specific activities of all CRY2 UCR mutants. (h–j) The blue-light-induced photobiochemical activity of the CRY2 UCR mutant proteins. Six-day-old etiolated seedlings were transferred to blue light (60 µmol m−2 s−1) for the indicated time before sample collection for immunoblot analysis. The levels of CRY2 in different samples were quantified by fluorescent immunoblots (Odyssey CLx Infrared Imaging System, LI-COR Inc), normalized to signals of the dark samples of the respective genotypes, and presented as REU (Relative Expression Unit). (h) Blue shades represent regions within 95% prediction bands of the standard proteolytic time-course of the “wild-type” GFP-CRY2. Color of thin lines match with color in (i). SD of the mean (n ≥ 6) are shown. (i) Classification of the photobiochemical activities of the CRY2 UCR mutant proteins. (j) Distribution of 60 T 1/2.
We next estimated the relative specific-activity of the individual CRY2 mutants expressed as the GFP-CRY2 fusion proteins, assuming that the GFP fusion has similar effects on the photophysiological activities of the mutant GFP-CRY2 proteins as that on the “wild-type” GFP-CRY2. In this experiment, the protein abundance of the mutant GFP-CRY2 and the light responsiveness of the respective transgenic lines were measured as described above and plotted to the standard curves of the “wild-type” GFP-CRY2 (fig. 3a–c). The relative specific-activity of all mutants was classified according to their relative positions in the individual standard curves of the “wild-type” GFP-CRY2 proteins. The photophysiological activities of the 31 CRY2 UCR mutant proteins are classified as hypermorph (HYPER) for those positioned higher than the upper limits of the 95% prediction bands of the standard curves (fig. 3a–f), wild-type-like (WTL) for those positioned within 95% prediction bands of the standard curves, hypomorph (HYPO) for those positioned below the lower limits of the 95% prediction bands of the standard curves but higher than the upper limits of the 20% prediction bands of the standard curves, or loss-of-function (LOF) for those positioned below 20% prediction bands of the standard curves.
All 31 stably expressed CRY2 mutant proteins exhibited defects in at least one of the three photophysiological activities examined. However, only two mutants, D387A and G427A, lost all three photophysiological activities examined, and they also lost the blue-light-induced homodimerization activity (supplementary fig. S6 A, Supplementary Material online). Fifty-eight percent (18/31) stably expressed CRY2 mutant proteins exhibited the WTL activity mediating at least one of the three photophysiological activities measured (fig. 3 and supplementary table S3, Supplementary Material online). Figure 3d–f shows that among the 31 stably expressed CRY2 mutants analyzed, 9.7%, 35.5%, or 45.2% are classified as WTL that exhibited the relative specific-activity similar to that of the “wild-type” GFP-CRY2 for the blue-light inhibition of hypocotyl elongation, blue-light stimulation of cotyledon unfolding, and promotion of flowering, respectively (fig. 3d–f). It is interesting that about four times as many CRY2 mutants showed WTL activity promoting floral initiation (45.2%) or cotyledon unfolding (35.5%) in comparison to that of the mutants retained WTL activity inhibiting hypocotyl elongation (9.7%). One possible interpretation of this observation is that different structural elements determined by different UCRs of CRY2, albeit their universal conservativity, contribute to different physiological activities of the photoreceptor, such that individual structure disturbance resulting from different mutations manifest differently in different activities of CRY2. In summary, our results demonstrate that among the 51 UCRs of CRY2 analyzed, 20 may determine protein stability, 2 are universally required for all functions of CRY2, whereas 29 are not universally required for all activities of CRY2 examined under the experimental conditions used.
UCRs Are Not Universally Required for the Photobiochemical Activity of CRY2
It is intuitive that UCRs of a protein family should be universally important for the members of the respective protein family, otherwise mutations would have accumulated at those positions throughout evolution in at least some members of some lineages. It is somewhat unexpected that more than half (29/51) of the UCRs of CRY2 affect only a subset of the functions of CRY2. We next analyzed how mutations of the UCRs affecting the photobiochemical activity of the CRY2 protein. CRY2 undergoes blue-light-dependent phosphorylation, ubiquitination, and degradation, presumably leading to the negative feedback regulation of CRY2 activity and plant photosensitivity (Shalitin et al. 2002; Yu et al. 2007). Since different transgenic lines express different CRY2 mutants at different levels, we first examined whether the difference in protein abundance might affect the blue-light-induced degradation of the “wild-type” GFP-CRY2 protein in three transgenic lines expressing the “wild-type” GFP-CRY2 protein at different levels (supplementary fig. S7 A–D, Supplementary Material online). Figure S7 D, Supplementary Material online, shows that the “wild-type” GFP-CRY2 protein expressed at different levels have the similar half-life (60 T 1/2, see fig. 3j) of ∼30–40 min when etiolated seedlings were exposed to blue light of the indicated fluence rate, which is also similar to the half-life of the endogenous CRY2 reported previously (supplementary fig. S7 A–D, Supplementary Material online) (Yu et al. 2007). These results suggest that the rate of blue-light-induced proteolysis of CRY2 is not significantly affected by fusion to GFP or the absolute abundance of CRY2 protein, at least within the ranges of protein abundance and blue-light intensities examined. We next analyzed the half-life of all 31 stably expressed GFP-CRY2 mutant proteins in etiolated seedlings exposed to blue light. Similar to the photophysiological activities of CRY2, we classified the blue-light-induced proteolysis activity of the CRY2 UCR mutant proteins into four groups for the experimental conditions of 2-h blue-light treatment at the fluence rate of 60 µmol m−2 s−1 (figs. 3h–j and 4a and supplementary fig. S7 E and F and table S3, Supplementary Material online), including hypermorphic (HYPER, e.g., P416A) for those degraded faster than the “wild-type” GFP-CRY2 in response to blue light; hypomorphic (HYPO, e.g., L348A) for those degraded slower than the “wild-type” GFP-CRY2 but achieved at least 50% of degradation within 2 h of blue-light exposure; LOF (e.g., D387A) for those failed to achieve 50% of degradation within 2 h of blue-light exposure; and WTL (e.g., S257A) for those degraded at comparable rates of the “wild-type” GFP-CRY2.
The two mutations, D387A (CRY2D387A) and G427A (CRY2G427A) that lost all their photophysiological activities tested, also showed no photobiochemical activity in light-dependent proteolysis (supplementary table S3, Supplementary Material online). D387A is a known chromophore-less mutant that fails to bind to the FAD chromophore (Stanewsky et al. 1998; Liu et al. 2008), which is presumably required for all light-dependent activities of any cryptochrome family proteins. G427A is located far away from the FAD-binding pocket in the modeled structure of CRY2, and it is universally conserved in the photolyase/cryptochrome of eukaryotes (fig. 2a and supplementary fig. S2 and table S3, Supplementary Material online). Why the mutation at G427 has the same detrimental effect to all CRY2 activities examined as that of the FAD-less D387A mutant remains unclear. Also analogous to the photophysiological activity, a large number (41.9%, 13/31) of the stably expressible CRY2 UCR mutant proteins (e.g., S257A) analyzed exhibited the half-life (60 T 1/2) similar to that of “wild-type” GFP-CRY2 (fig. 3i and supplementary fig. S7 E and F and table S3, Supplementary Material online). This result is consistent with the observation that UCRs are not required for all activities of CRY2. Moreover, the altered activity of light-induced proteolysis of most CRY2 mutants, except D387A and G427A, do not correlate with the other three photophysiological activities examined (supplementary figs. S5D–F and S5B, Supplementary Material online). For example, the CRY2P416A mutant, which showed WTL activity in promoting flowering and lower activity in mediating blue-light inhibition of hypocotyl elongation or blue-light stimulation of cotyledon unfolding, degrades faster than the “wild-type” GFP-CRY2 in response to blue light (supplementary table S3, Supplementary Material online). In contrast, the CRY2L370A mutant, which is classified as hypomorphic for the blue-light-induced proteolysis, appears WTL or hypermorphic for all the three photophysiological activities examined (supplementary table S3, Supplementary Material online). Taken together, our analyses of the photophysiological and photobiochemical activities of the CRY2 UCR mutants may be interpreted by a hypothesis that the structure elements determined by UCRs are required for specific functions but not universally required for all functions of members of a protein family.
Neighboring UCRs Have Similar Effects on the CRY2 Function and Regulation
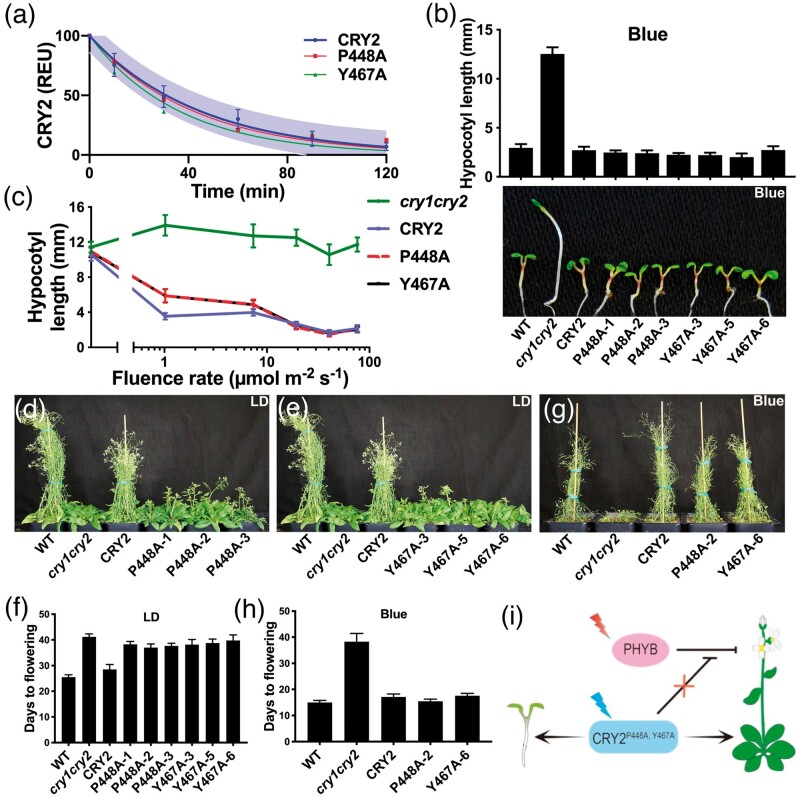
We tested the above hypothesis by examination of whether neighboring UCRs, which are presumably associated with the same structure elements, may determine the same functions of CRY2. We analyzed the functional effects of two CRY2 UCR mutants altered in the residues P448 and Y467. Because these two UCRs appear to locate in the close vicinity of each other (∼2.4 Å for the closest atoms) (fig. 2a) and therefore likely associated with the same structural element. We analyzed effects of the mutations of P448 and Y467 on different functions of CRY2 under different experimental conditions. First, the P448A (GFP-CRY2P448A) and Y467A (GFP-CRY2Y467A) mutants exhibited WTL activity in blue-light-induced proteolysis in etiolated seedlings exposed to blue light (fig. 4a and supplementary table S3, Supplementary Material online). Second, both mutants exhibited similarly complex fluence rate-dependent activity mediating the blue-light inhibition of hypocotyl growth (fig. 4b and c). When grown under blue light with the fluence rate lower than 15–20 µmol m−2 s−1, both mutants developed hypocotyls slightly longer than that of the GFP-CRY2 control (fig. 4c). However, seedlings expressing either mutants developed hypocotyls indistinguishable from that of the GFP-CRY2 control when grown under blue light with the fluence rate higher than 15–20 µmol m−2 s−1 (fig. 4b and c). Because seedlings expressing the P448A and Y467A mutant proteins at the similar level, which are 10%–20% that of the GFP-CRY2 control (supplementary table S3, Supplementary Material online), these two mutants appear to have higher relative specific-activity than that of the “wild-type” GFP-CRY2 protein mediating light inhibition of hypocotyl elongation in at least high light. Third, the P448A and Y467A mutants exhibited similar wavelength-dependent activities promoting floral initiation (fig. 4d–i). The cry2 mutant is known to exhibit delayed flowering in white light or blue-plus-red light but normal flowering time when grown under monochromatic blue light, whereas the cry1cry2 double mutant exhibits delayed flowering in both white light and monochromatic blue light (Guo et al. 1998; Mockler et al. 1999). These wavelength-dependent flowering-time phenotypes have been interpreted to result from two different modes of actions of CRY2 in promoting floral initiation: a phytochrome B (phyB)-dependent pathway and a phyB-independent pathway (Guo et al. 1998; Mockler et al. 1999; Valverde et al. 2004; Zuo et al. 2011). In the phyB-dependent pathway, CRY2 exerts blue-light-dependent inhibition of the red light-dependent suppression of flowering by phyB, such that this function of CRY2 is dependent on both blue light and red light. In the phyB-independent pathway, CRY2 acts redundantly with CRY1 to promote floral initiation directly, such that this function of CRY2 is dependent on blue light but not red light (Mockler et al. 1999). Plants expressing the P448A and Y467A mutants flowered later than the wild-type plants when they were grown in white light composed of both blue and red wavelengths, suggesting both mutants are impaired in the phyB-dependent activity of CRY2 (fig. 4d–f and supplementary table S3, Supplementary Material online). However, plants expressing the “wild-type” GFP-CRY2, P448A, or Y467A mutant proteins all rescued the later-flowering phenotype of cry1cry2 when grown in continuous blue light (fig. 4g and h), indicating that neither mutant is compromised in their activity mediating phyB-independent promotion of floral initiation. Therefore, the structure elements determined by the neighboring P448 and Y467 residues are required for the phyB-dependent but not phyB-independent activity of CRY2 (fig. 4i). Taken together, these results are consistent with the hypothesis that the structure elements determined by UCRs are required for specific functions of CRY2 but not universally required for all functions of CRY2.
Fig. 4.
Functional analyses of neighboring UCRs of CRY2. (a) Blue-light-dependent proteolysis of the “wild-type” GFP-CRY2, GFP-CRY2P448A, and GFP-CRY2Y467A UCR mutant proteins. The assay was carried out as described in figure 3h. (b) Representative 6-day-old seedlings of the indicated genotypes grown in continuous blue light (20 µmol m−2 s−1). Bars indicate SD of the mean (n ≥ 20) are shown. (c) Hypocotyl lengths of 6-day-old seedlings grown in dark or continuous blue light with fluence rates of 1–80 µmol m−2 s−1. Bar indicates SD of the mean (n ≥ 20). (d–e) Plants of indicated genotypes grown in LD photoperiods (16-h day/8-h night) for 40 days. (f) Days to flowering of the indicated genotypes grown in LD. Bars indicates SD of the mean (n ≥ 8). (g) Plants of indicated genotypes grown in continuous blue light (70–80 µmol m−2 s−1) for 35 days. (h) Days to flowering of the indicated genotypes grown in LD. Bars indicate SD of the mean (n ≥ 8). (i) A hypothetic model depicting CRY2P448A, Y467-mediated regulation of de-etiolation and flowering time.
Discussion
UCRs are commonly considered critical for the structural integrity common to all members of a protein family. To our knowledge, this notion has not been systematically tested experimentally. In the present study, we analyzed the in vivo relative specific activities of UCR mutations of the Arabidopsis blue-light receptor CRY2, using a standard curve-based quantitative approach. Our result that all UCR mutations of CRY2 exhibited at least a minor impairment in at least one of the four physiological or biochemical activities examined is consistent with the expectation that UCRs are evolutionarily conserved for functional reasons. On the other hand, it is interesting that none of the UCRs required for stable protein expression of Arabidopsis CRY2 in plants is required for stable protein expression of human hCRY1 in a human cell line (fig. 2). Moreover, 74% (23/31) of the stably expressible CRY2 mutant proteins exhibited the WTL activity mediating at least one of the four photophysiological and photobiochemical responses examined (fig. 3 and supplementary table S3, Supplementary Material online). These results and our follow-up analyses (fig. 4) demonstrate that UCRs of CRY2 are not universally required for protein stability or all functions of cryptochromes. UCRs of closely related cryptochromes may also exert different effects on the same function of the closely related family members. For example, the Arabidopsis CRY2 UCR mutant, CRY2W331A, exhibits the LOF phenotype for the blue-light inhibition of hypocotyl growth response in the present study (supplementary table S3, Supplementary Material online), whereas the CRY2W331A equivalent mutant of Arabidopsis CRY1, CRY1W334A, exhibited WTL activity for the same photoresponse in our previous study (Gao et al. 2015). The mechanisms underlying different functions of the equivalent residues in different cryptochromes may include different post-translational protein modifications. This may be illustrated by the comparison of protein phosphorylation and functions of the Arabidopsis CRY2S257A mutant examined in the present study with that of the corresponding mouse mCRY1S261A mutant reported recently (Ode et al. 2017). Arabidopsis CRY2 or mouse mCRY1 are phosphorylated in at least 24 or 27 residues, respectively (Liu et al. 2017; Ode et al. 2017). None of the phosphorylated residues of Arabidopsis CRY2 is universally conserved. But 2 of the 27 phosphosites of mouse mCRY1, S252 and S261 that correspond to the unphosphorylated S248 and S257 of Arabidopsis CRY2, are universally conserved (supplementary fig. S2 and table S3, Supplementary Material online). The Arabidopsis CRY2S248A or CRY2S257A mutants, which are not expected to directly impair CRY2 phosphorylation (13), fully or partially rescued three photophysiological phenotypes of the cry1cry2 mutant plants, respectively (fig. 3 and supplementary online table S3, Supplementary Material online). In contrast, the mCRY1S261A mutant that directly impaired phosphorylation of mCRY1 failed to rescue the arrhythmic phenotype of the mCry1mCry2 knockout mice (Ode et al. 2017). On the other hand, the mCRY1S252D mutant altered in the other phosphorylated UCR exhibits short period and lower amplitude (Ode et al. 2017). These results demonstrate that UCRs of different cryptochromes can differentially affect protein phosphorylation to impact the functions of respective cryptochromes differently.
Results of our studies demonstrate that the structure elements determined by UCRs common to different members of a protein family are not universally required for the protein stability, post-translational modification, and functions of the individual family members of cryptochromes. We hypothesize that common structure elements associated with the UCRs may evolve to play different subsets of functions in different members of a protein family, and those diverse subsets of functions all contribute to the long-term fitness of the host organisms, sanctioning their universal conservation in evolution. It is conceivable that cryptochromes of different evolutionary origins, which are believed to evolve independently from the ancestral DNA photolyases, may adopt the same folds for different but functionally essential purposes specific to individual members of a protein family. For example, plant cryptochromes may rely on certain UCRs for the light-dependent homodimerization whereas metazoan CRYs might rely on the similar UCRs for light-independent interaction with transcription activators. However, how those UCRs associated with different functions of different members of the protein family are universally conserved remains to be elucidated. Moreover, UCRs are apparently not the only structural elements that are essential for function. Mutations of many non-UCRs are also known to affect the same function impacted by UCRs of cryptochromes in both Arabidopsis and mouse, although functionally defective UCR mutants appear in higher percentages of total UCRs than that of the functionally defective non-UCR mutants among the total non-UCRs in both types of cryptochromes (fig. 2d and supplementary table S5, Supplementary Material online). Therefore, how are the UCRs distinguished from non-UCRs during evolution need to be further investigated to better understand the evolutionary history and the structure–function relationship of the cryptochrome family of proteins that play important functions in plant development and human health.
Materials and Methods
Multiple Sequence Alignment and Structure Simulation
Species, residues forming PHR domain of each protein used for multiple sequence alignment, and NCBI protein accession numbers (supplementary fig. S2, Supplementary Material online) were listed in supplementary table S2, Supplementary Material online, respectively. Multiple sequence alignment was conducted using T-Coffee (Notredame et al. 2000). The resulting Clustalw_aln files were uploaded onto ESPript 3.0 (Robert and Gouet 2014) to generate a black and white version of the alignment, and then manually edited in Adobe Photoshop CC 2017 to add blue shades at desired positions.
The CRY2 structure was simulated using SWISS-MODEL (Waterhouse et al. 2018) from CRY2 full-length protein sequence based on crystal structure of CRY1 (PDB: 1U3C) (Brautigam et al. 2004).
Plasmid Construction and Plant Materials
All Arabidopsis plant lines used in this study were in Columbia (Col) background. The wild-type plants used in this study are rdr6-11 (Peragine et al. 2004). The Ti plasmid pFGFP (Liu et al. 2017) was modified from pCambia3301. The coding sequence (CDS) of wild-type or site-specific mutants of CRY2 was Polymerase Chain Reaction (PCR) amplified and incorporated into the BamHI site of pFGFP using In-Fusion Cloning Kit (Clontech, Mountain View, CA). The resulting constructs were PACTIN2::FLAG-EGFP-CRY2::T35S and were introduced into the cry1cry2rdr6 plants by standard floral dip method (Clough 2005). The cry1cry2rdr6 lines were acquired by crossing cry1-304 (Mockler et al. 1999), cry2-1 (Guo et al. 1998), and rdr6-11 (which suppresses gene silencing) (Peragine et al. 2004). The transgenic T1, T2, and T3 populations were screened and maintained on compound soil subirrigated with the Basta solution (Clough 2005).
The pQCMV-EGFP plasmid was modified from pEGFP-N1 vectors (Clontech) by 1) inserting DNA sequences for a Kozak motif and a flexible protein linker (PAPAP) (gccaccATGgctACTAGTgccCCTAGGgctCCAGCTCCAGCTCCA) between SacI site (GAGCTC, “GAG” remained in the new pQCMV-EGFP plasmid, whereas “CTC” was removed) and start codon of EGFP (Wang et al. 2016); 2) inserting DNA sequences for a flexible protein linker (PAPAP) and multiple cloning site containing KpnI and SacI recognition site (tccggaCCAGCTCCAGCTCCAgctGGTACCgctGAGCTCgct) right before the stop codon (TAA) of EGFP. The CDS of wild-type or site-specific mutants (F9A, R10A, L13A, D17A, P19A, L59) of human hCRY1 was PCR amplified and cloned into the KpnI site of pQCMV-EGFP. The resulting constructs were PCMV::EGFP-hCRY1::TSV40.
The pCMV plasmid was described previously (Liu et al. 2017). The CDS of wild-type or site-specific mutants (F9A, R10A, L13A, R14, D17A, P19A, L59, D110A, L132A, F257A, R293A, D341A, H354A, R358A, F381A, D389A, D423A, P424A, P440A, W448A) of human hCRY1 was PCR amplified and cloned into the BamHI site of pCMV plasmid. The resulting constructs were PCMV::MYC-hCRY1::Tβ-globin.
Plant Growth Conditions and Physiological Analyses
For hypocotyl inhibition assays in darkness or blue light that were not used for growth kinetics analysis, seeds were sterilized and sown onto fresh-made MS Agar (0.8%) plates, subjected to 4 °C cold treatment in darkness for 4 days, exposed to white light at room temperature for 24 h, and then put into indicated light conditions at room temperature for 5 days. The resulting seedlings were sandwiched between two plastic sheets (one transparent, the other black), scanned and measured by Fiji (NIH).
For image-based hypocotyl growth kinetics analyses (Wang et al. 2017), seeds were sterilized and sown onto MS Agar (0.8%) plates, subjected to 4 °C cold treatment for 4 days and exposed to white light at room temperature for 24 h. The imbibed seeds were then transferred onto 100 mm × 100 mm squire MS Agar (0.8%) plates with grids. The plates were placed vertically under blue light (15 µmol m−2 s−1) and images were captured each hour for the next 7 days, by using a CCD camera (Jinghang JHSM500B) equipped with a prime macrolens. Image acquisition was controlled by a custom-designed software. Images captured between 48 and 144 h postexposure of blue light were manually measured by using Fiji (NIH) to get hypocotyl length. Three seedlings were measured for each genotype. Absolute activities (AAs) were acquired as reciprocal of the slope of a linear regression of growth kinetics of 48–96 h. Relative specific activity of hypocotyl inhibition of CRY2 (fig. 2a) was calculated by the following formula:
| (1) |
For cotyledon unfolding assay in darkness, seeds were sterilized and sown onto MS Agar (0.8%) plates, subjected to 4 °C cold treatment for 4 days and exposed to white light at room temperature for 24 h, and then put into corresponding light conditions for 5 days before analysis. Seedlings were carefully sandwiched between adhesive sides of transparent tapes without disturbing cotyledon unfolding angles and then taped onto black paper for scanning. More than 20 seedlings were measured for each genotype. For cotyledon unfolding assay in blue light, plants were similarly prepared as in hypocotyl growth kinetics analyses. Cotyledon unfolding angles were measured by drawing lines between the shoot apical meristem and tips of cotyledons in Fiji (NIH) as previously described (Neff and Chory 1998). Three seedlings were measured for each genotype. Cotyledon unfolding activities of CRY2 shown in figure 2b were calculated by the following formula:
| (2) |
For measuring flowering time in blue light, seeds were sown in soil, subjected to 4 °C cold treatment for 4 days, exposed to white light at room temperature for 24 h, and then put into blue light (70–80 µmol m−2 s−1) as previously described (Mockler et al. 1999). For measuring flowering time in the long-day (16-h day/8-h night) or short-day (8-h day/16-h night) period, seeds were sown in soil, subjected to 4 °C cold treatment for 4 days, and then moved into corresponding light conditions. Days to flowering and leaf number were counted daily. The day when there was at least 1 cm of inflorescence with visible floral meristem at top was regarded as day of flowering. Only rosette leaves were counted. Days to flowering (Days) in the long-day period were used to calculate floral promotion activities shown in figure 2c. The floral initiation activities were calculated by the following formula:
| (3) |
Standard curves were created by fitting data points of wild-type GFP-CRY2 L1–L5 into the hyperbola nonlinear regression model in GraphPad Prism version 8.0.2 (159) for Mac OS X (GraphPad Software, San Diego, California, www.graphpad.com, last accessed September 28, 2019). Ninety-five percent prediction curve was automatically calculated by GraphPad Prism.
Normalized activities used in figure 3g were calculated by the following formula:
| (4) |
where relative specific activityGFP-CRY2 was determined case by case: First, protein abundance of the respective mutants was identified. The protein abundance was then introduced into the formulas of standard curves to get the respective Relative specific activityGFP-CRY2.
Long-day (16-h day/8-h night) and short-day (8-h day/16-h night) photoperiod-treated plants were grown in walk-in growth chambers at 22 °C, 65% relative humidity under cool white fluorescent tubes. Light-emitting diode was used to obtain monochromatic blue light (peak 450 nm; half-bandwidth of 20 nm).
Immunoblot and Blue-Light-Induced Proteolysis of Plant Samples
To prepare protein extracts, plant materials were dipped into liquid N2 and homogenized by TissueLyser (QIAGEN). The resulting plant tissue powders were added 0.8× volume of powder of protein extraction buffer (120 mM Tris–HCl pH 6.8; 100 mM ethylenediaminetetraacetic acid pH 8.0, 4% w/v SDS, 10% v/v 2-mercaptoethanol, 5% glycerol, and 0.01% Bromophonol Blue), boiled for 8 min, and then centrifuged with table top centrifuges at top speed for 10 min. The resulting protein extract supernatant was separated by home-made 10% (for checking protein abundance) or 8% (for assaying proteolysis) sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to Pure Nitrocellulose Blotting Membranes (BioTrace NT, Pall Life Sciences) using wet electroblotting system (Bio-Rad, Hercules, CA). Ponceau S Red solution (0.1% w/v Ponceau S; 5% v/v acetic acid) was used to stain transferred membranes to gauge transferring efficiency. The membranes were then cut horizontally along ∼70 kDa for separate incubation with primary and secondary antibodies. For immunoblot signals captured by the Odyssey CLx Infrared Imaging System (LI-COR Inc, Lincoln, NE), membranes were blocked with 0.5% Casein in PBS (Phosphate-buffered saline, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HCO3, 1.8 mM K2HCO3) solution, blotted with primary antibodies in 0.5% Casein in PBST (PBS with 0.3% Tween-20) solution, and then blotted with fluorescent secondary antibodies (ThermoFisher, A11357, A11369) in 0.5% Casein in PBST solution. Images captured by Odyssey CLx System (LI-COR) were processed with Image Studio Lite software (LI-COR) and organized with Adobe Photoshop CC 2017. Primary antibodies used in this study were rabbit-anti-CRY2 (1:3,000, home-made) (Guo et al. 1998), mouse-anti-ACTIN11 (1:3,000, 26F7, Ab-mart, Inc., Berkeley Heights, NJ) and rabbit-anti-HSP90 (1:3,000, sc-33755, Santa Cruz Biotechnology, Inc., Dallas, TX). Secondary antibodies used here were goat-anti-rabbit IgG (1:15,000, A11369, Thermo Fischer Scientific, Inc., Waltham, MA) and goat-anti-mouse IgG (1:15,000, A11357, Thermo Fischer Scientific), both conjugated to Alexa Fluro 790. For immunoblot signals captured by exposure to X-ray film, membranes were blocked with 5% skimmed milk in PBST solution, blotted with primary antibodies in PBST solution, and then blotted with secondary antibodies in PBST solution. After blotted with secondary antibodies, the membranes were incubated in the home-made ECL solution (Solution A: 100 mM Tris–HCl pH 8.5; 0.2 mM coumaric acid; Solution B: 100 mM Tris–HCl pH 8.5; 1.25 mM luminol; Right before use, mix 3-ml Solution A with 3-ml Solution B and add 2 µl 30% H2O2) and exposed to X-ray films. The resulting films were scanned and organized by Adobe Photoshop CC 2017. Primary antibodies were the same as above. Secondary antibodies used here were donkey-anti-rabbit IgG (1:10,000, NA9340-1ML, GE Healthcare, Chicago, IL) and sheep-anti-mouse IgG (1: 10,000, NA9310-1ML, GE Healthcare), both conjugated to horse radish peroxidase (HRP).
To quantify protein abundance of CRY2, fluorescent signals captured by Odyssey CLx System (LI-COR) were quantified by an internal method of Image Studio Lite software (LI-COR). The resulting signals were used to calculate protein abundance by the following formula:
| (5) |
For the 20 CRY2 mutants that are transcribed but fail to accumulate the mutant protein in plants (fig. 2c and supplementary fig. S3 A–C, Supplementary Material online), the results were verified in at least 6 independent transgenic lines except GFP-CRY2D112A (D112A), GFP-CRY2F253A (F253A), and GFP-CRY2W449A (W449A). Expression of proteins was detectable in D112A, F253A, and W449A lines with abundance lower than 5% of that of the L5 of wild-type GFP-CRY2 line and were thus categorized into the “lack of protein” group.
Blue-light-induced proteolysis curves were plotted to degradation curve:
| (6) |
where Y (%) is percentage of initial signal, e is Euler’s number, k (%/min) is rate of degradation, and t (min) is independent variable time. Half-life (60 t 1/2) was calculated by the formula:
| (7) |
The blue-light-dependent proteolysis activities were calculated by the following formula:
| (8) |
Human Cell Culture, Transfection, Protein Expression, and Coimmunoprecipitation Assay
Human embryonic kidney (HEK) 293 T cells were routinely cultured in Dulbecco’s modified Eagle’s medium (10-013-CM, Corning, New York) supplemented with 10% (v/v) Fetal Bovine Serum (FBS), 100 IU penicillin and 100 mg/l streptomycin, in humidified 5% (v/v) CO2 incubator at 37 °C.
For protein expression assays, HEK293T were seeded at a density of ∼3 × 105 cells per well of a 6-well plate and transfected using a calcium phosphate precipitation protocol as previously described (Wang et al. 2016). 2.5 μg of wild-type or mutant PCMV::EGFP-hCRY1::TSV40, or PCMV::MYC-hCRY1::Tβ-globin plasmids were cotransfected with 2 μg of PCMV::EGFP::TSV40 plasmids (control). Cells were harvested 36–48 h after transfection and lysed in three volumes of 1% Brij buffer (1% Brij-35, 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid and 1× protease inhibitor cocktail). The cells were kept on ice for 30 min, followed by centrifugation at 14,000 × g for 10 min at 4 °C. The supernatant was mixed with equal volume of 2× SDS-PAGE Sample Buffer and heated at 100 °C for 3 min. The protein samples were separated by 10% SDS-PAGE and analyzed by immunoblot using the Odyssey CLx Imaging System (LI-COR) as described above. The primary antibodies used in this assay were rabbit-anti-FLAG (1:3,000, F7425, Sigma-Aldrich Corp., St. Louis, MO). The secondary antibodies were as listed above. The expression levels of wild-type and mutant hCRY1 were normalized against the expression of GFP, and converted to relative expression units (REU) by dividing with the mean (n = 3) of wild-type hCRY1 expression level.
Coimmunoprecipitation was conducted as described in Wang et al. (2016). Basically, 36 h after transfection, the HEK293T cells expressing the indicated proteins were treated with blue light (100 µmol m−2 s−1) or darkness for 120 min. After light treatment, the cells were harvested and washed with PBS, and then lysed with 1% Brij buffer. FLAG-affinity beads (F2426, Sigma) were added to cell lysate, and incubated with gentle rocking at 4 °C in darkness for 2 h. After incubation, the beads were washed with 1% Brij buffer for three times. The proteins were eluted by competition with 30 µl of 500 µg/ml of 3× FLAG peptide with shaking of 1,400 rpm at 4 °C for 30 min. The eluted proteins were then analyzed by fluorescent immunoblot.
Fluorescence Microscopy
Seeds were sterilized and sown onto fresh-made MS Agar (0.8%) plates, subjected to 4 °C cold treatment in darkness for 1 day, and then put into the long-day (16-h day/8-h night) period of white light at room temperature for 2 days. The resulting seedlings were directly imaged using a Zeiss LSM 700 confocal microscope.
Supplementary Material
Acknowledgments
The authors thank Dr Xu Wang for growth kinetics image analyses and helpful discussions. This work was supported in part by National Institute of Health (GM56265 to C.L.), UCLA SOL LESHIN fund, UCLA-FAFU Joint Research Center on Plant Proteomics, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, UCLA Whitcome Pre-doctoral Training Program (H.L.), and China Scholarship Council (H.L.).
Author Contributions
H.L. performed experiments, processed and analyzed data, and wrote and edited the manuscript. T.S. performed experiments, processed data, and helped write the manuscript. W.H. and Q.W. provided research material. C.L. supervised the project, secured funding, helped with data analysis, and wrote and edited the manuscript.
References
- Ahmad M. 2016. Photocycle and signaling mechanisms of plant cryptochromes. Curr Opin Plant Biol. 33:108–115. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR. 1993. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nat Lett. 366(6451):162–166. [DOI] [PubMed] [Google Scholar]
- Aubert C, Vos MH, Mathis P, Eker AP, Brettel K. 2000. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature 405(6786):586–590. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R, Batschauer A. 2007. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem. 282(20):14916–14922. [DOI] [PubMed] [Google Scholar]
- Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. 2004. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci U S A. 101(33):12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR. 2003. Cryptochromes: enabling plants and animals to determine circadian time. Cell 114(5):537–543. [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. 1999. Cryptochromes: blue light receptors for plants and animals. Science 284(5415):760–765. [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen L-O, van der Horst GTJ, Batschauer A, Ahmad M. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 62(1):335–364. [DOI] [PubMed] [Google Scholar]
- Clough SJ. 2005. Floral dip: agrobacterium-mediated germ line transformation. Transgenic Plants: Methods and Protocols. Humana Press, 91–101. [DOI] [PubMed] [Google Scholar]
- Engelhard C, Wang X, Robles D, Moldt J, Essen L-O, Batschauer A, Bittl R, Ahmad M. 2014. Cellular metabolites enhance the light sensitivity of Arabidopsis cryptochrome through alternate electron transfer pathways. Plant Cell 26(11):4519–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang X, Zhang M, Bian M, Deng W, Zuo Z, Yang Z, Zhong D, Lin C. 2015. Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proc Natl Acad Sci U S A. 112(29):9135–9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegear RJ, Foley LE, Casselman A, Reppert SM. 2010. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463(7282):804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu N-N, Zhang Y-C, Yang H-Q. 2012. Substitution of a conserved glycine in the PHR domain of Arabidopsis CRYPTOCHROME 1 confers a constitutive light response. Mol Plant 5(1):85–97. [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. 1998. Regulation of flowering time by Arabidopsis photoreceptors. Science 279(5355):1360–1363. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33(Web Server):W299–W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbacher T, Immeln D, Dick B, Kottke T. 2009. Microsecond light-induced proton transfer to flavin in the blue light sensor plant cryptochrome. J Am Chem Soc. 131(40):14274–14280. [DOI] [PubMed] [Google Scholar]
- Li X, Wang Q, Yu X, Liu H, Yang H, Zhao C, Liu X, Tan C, Klejnot J, Zhong D, et al. 2011. Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc Natl Acad Sci U S A. 108(51):20844–20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Heelis PF, Sancar A. 1991. Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry 30(25):6322–6329. [DOI] [PubMed] [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. 1995. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269(5226):968–970. [DOI] [PubMed] [Google Scholar]
- Lin C, Shalitin D. 2003. Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 54(1):469–496. [DOI] [PubMed] [Google Scholar]
- Lin C, Top D, Manahan CC, Young MW, Crane BR. 2018. Circadian clock activity of cryptochrome relies on tryptophan-mediated photoreduction. Proc Natl Acad Sci U S A. 115(15):3822–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322(5907):1535–1539. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Deng W, Wang X, Piao M, Cai D, Li Y, Barshop WD, Yu X, Zhou T, et al. 2017. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat Commun. 8:15234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EV, Baggs JE, Geskes JM, Hogenesch JB, Green CB. 2009. Generation of a novel allelic series of cryptochrome mutants via mutagenesis reveals residues involved in protein-protein interaction and CRY2-specific repression. Mol Cell Biol. 29(20):5465–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Shakhnovich EI. 1999. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J Mol Biol. 291(1):177–196. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. 1999. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126(10):2073–2082. [DOI] [PubMed] [Google Scholar]
- Müller M, Carell T. 2009. Structural biology of DNA photolyases and cryptochromes. Curr Opin Struct Biol. 19(3):277–285. [DOI] [PubMed] [Google Scholar]
- Müller P, Bouly J-P, Hitomi K, Balland V, Getzoff ED, Ritz T, Brettel K. 2015. ATP binding turns plant cryptochrome into an efficient natural photoswitch. Sci Rep. 4(1):5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J. 1998. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118(1):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 302(1):205–217. [DOI] [PubMed] [Google Scholar]
- Ode KL, Ukai H, Susaki EA, Narumi R, Matsumoto K, Hara J, Koide N, Abe T, Kanemaki MT, Kiyonari H, et al. 2017. Knockout-rescue embryonic stem cell-derived mouse reveals circadian-period control by quality and quantity of CRY1. Mol Cell 65(1):176–190. [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18(19):2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42(W1):W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosensweig C, Reynolds KA, Gao P, Laothamatas I, Shan Y, Ranganathan R, Takahashi JS, Green CB. 2018. An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat Commun. 9(1):1138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 103(6):2203–2238. [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C. 2002. Regulation of Arabidopsis cryptochrome 2 by blue-light- dependent phosphorylation. Nature 417(6890):763–767. [DOI] [PubMed] [Google Scholar]
- Solov’yov IA, Domratcheva T, Moughal Shahi AR, Schulten K. 2012. Decrypting cryptochrome: revealing the molecular identity of the photoactivation reaction. J Am Chem Soc. 134(43):18046–18052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95(5):681–692. [DOI] [PubMed] [Google Scholar]
- Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. 2016. Optimized second-generation CRY2–CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol. 12(6):425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A, Chardin P, Wittinghofer A, Sander C. 1991. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry 30(19):4637–4648. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. 2004. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303(5660):1003–1006. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, Yang L, Liu Q, Liu W, Han Y-J, et al. 2016. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354(6310):343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Q, Han Y-J, Liu Q, Gu L, Yang Z, Su J, Liu B, Zuo Z, He W, et al. 2017. A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 92(3):426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1):W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C. 2007. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19(10):3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. 2009. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21(1):118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeugner A, Byrdin M, Bouly J-P, Bakrim N, Giovani B, Brettel K, Ahmad M. 2005. Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem. 280(20):19437–19440. [DOI] [PubMed] [Google Scholar]
- Zhu B, Cai G, Hall EO, Freeman GJ. 2007. In-FusionTM assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43(3):354–359. [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. 2011. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 21(10):841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo ZC, Meng YY, Yu XH, Zhang ZL, Feng DS, Sun SF, Liu B, Lin CT. 2012. A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol Plant 5(3):726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.