Abstract
Over the two last decades, coronaviruses have affected human life in different ways, especially in terms of health and economy. Due to the profound effects of novel coronaviruses, growing tides of research are emerging in various research fields. This paper employs a co-word analysis approach to map the intellectual structure of the coronavirus literature for a better understanding of how coronavirus research and the disease itself have developed during the target timeframe. A strategic diagram has been drawn to depict the coronavirus domain’s structure and development. A detailed picture of coronavirus literature has been extracted from a huge number of papers to provide a quick overview of the coronavirus literature. The main themes of past coronavirus-related publications are (a) “Antibody-Virus Interactions,” (b) “Emerging Infectious Diseases,” (c) “Protein Structure-based Drug Design and Antiviral Drug Discovery,” (d) “Coronavirus Detection Methods,” (e) “Viral Pathogenesis and Immunity,” and (f) “Animal Coronaviruses.” The emerging infectious diseases are mostly related to fatal diseases (such as Middle East respiratory syndrome, severe acute respiratory syndrome, and COVID-19) and animal coronaviruses (including porcine, turkey, feline, canine, equine, and bovine coronaviruses and infectious bronchitis virus), which are capable of placing animal-dependent industries such as the swine and poultry industries under strong economic pressure. Although considerable research into coronavirus has been done, this unique field has not yet matured sufficiently. Therefore, “Antibody-virus Interactions,” “Emerging Infectious Diseases,” and “Coronavirus Detection Methods” hold interesting, promising research gaps to be both explored and filled in the future.
Keywords: COVID-19, Coronavirus, Co-word analysis, Text mining, Science mapping, Emerging infectious diseases
Introduction
Over the last few decades, highly contagious fatal diseases have increased dramatically. Humans and animals alike have been struck down, sometimes resulting in economic crisis. According to the literature, coronavirus fields are recognized as degenerative, incurable diseases causing various health problems. Coronaviruses are highly infectious diseases and may lead to health threats including enteric, respiratory, hepatic, and neurological diseases, so they are extremely demanding to control and even to treat, and they can spread rapidly among communities on a large scale (Gorbalenya et al., 2004). In addition, through adaptive evolution owing to a high mutation rate and genetic recombination, distinct, unique coronaviruses have arisen that have damaging effects on humans and animals. Hence, the adaptive evolution feature has resulted in a novel coronavirus emergence (May et al., 2004).
Coronaviruses have caused primarily enzootic infections in birds and mammals; however, research outputs have recently revealed that the virus can easily affect humans and pass from person to person, leading to a dramatic increase in cases. Generally, coronaviruses have caused respiratory diseases for humans and enteric diseases for animals (Banerjee et al., 2019). In 2002 SARS‐CoV infected 8098 Chinese residents, leading to 774 deaths (CDC. Severe Acute Respiratory Syndrome, 2018). Besides, 2260 people were affected by MERS-CoV in 2012, resulting in 803 deaths in 27 countries (WHO. the Middle East Respiratory Syndrome Coronavirus, 2018). Animal coronaviruses have also contributed substantially to economic problems and animal loss over the last few decades. For instance, infectious bronchitis virus (IBV) took a toll of around $3,567.40 per 1000 birds in Brazilian poultry farms (Colvero et al., 2015). Furthermore, porcine epidemic diarrhea virus (PEDV) dealt a 3.1% loss to the U.S. pig-farming industry (Schulz & Tonsor, 2015). Coronaviruses have also had a devastating effect on the world economy and human health, which has convinced many researchers to focus on them. Thus, many researchers and practitioners have attempted to either mitigate or overcome the issue using different modern methods reported in a great number of publications on a daily basis, giving rise to a body of work that is extremely complicated to collect and analyze.
Additionally, the novel coronavirus could be interrelated with previous types of coronavirus fields, so a comprehensive analysis of studies in the areas already explored might uncover hidden patterns and useful knowledge that could shed light on the causes of the disease and efficient treatments. Unfortunately, traditional methodologies, such as reviewing and surveying the literature, are neither effective nor efficient means of going about this work; the traditional literature review can neither investigate all material carefully nor provide the researcher with detailed, accurate information on hidden patterns and complex relationships. Furthermore, no work has yet been done to provide researchers with a conceptual map in this critical field of research. Therefore, the co-word analysis approach is utilized here to cope with the problems traditional methods cannot handle and to address the main questions listed below.
How has the literature of coronavirus fields evolved? What journals actively publish coronavirus-related research?
What are the main themes and subthemes running throughout coronavirus research?
How are coronavirus themes related to each other?
What themes are peripheral and undeveloped?
The key aims of this paper are (a) to provide a comprehensive overview of a huge number of coronavirus papers dating to the period between 2000 and 2020 and indexed in Clarivate Analytics’ Web of Science Core Collection (WoS); (b) to examine the current state of the field and promising research directions. The paper is structured into four sections. Following the introduction, the theoretical background and coronavirus-related literature are surveyed and presented, and then the methodology is described. Next, primary data analyses and principal outputs are reported, including the most prolific, reputable journals and themes as well as the relationships among the keywords, and the strategic diagram is presented. Lastly, a discussion and conclusion are provided.
Literature review
Coronaviruses and related diseases
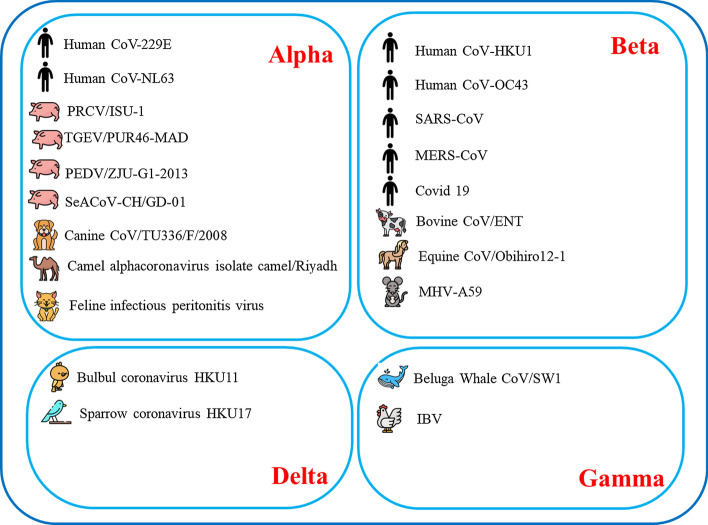
Coronaviruses are important pathogens for humans and other vertebrates. They can infect the hepatic, respiratory, gastrointestinal, and central nervous system of humans, livestock, mice, bats, birds, and other wild animals (Chen & Guo, 2016; Ge et al., 2013; Wang et al., 2006). Coronaviridae, with its two main subfamilies Letovirinae and Orthocoronavirinae, is the largest family in the order Nidovirales. Orthocoronavirinae consists mainly of four major genera: alpha-coronavirus (α‐coronavirus), beta-coronavirus (β‐coronavirus), gamma-coronavirus (γ‐coronavirus), and delta-coronavirus. Both α‐coronavirus and β‐coronavirus are found in mammals; γ‐coronavirus and delta-coronavirus, however, have evolved to infect birds (Woo et al., 2010).
Until late 2019, only six coronaviruses (CoVs) negatively affecting the respiratory system had been identified as serious threats to public health. Mild upper respiratory disease and, in a few cases, severe infection are associated with HKU1, HCoV‐229E, HCoV‐NL63, and HCoV‐OC43. Additionally, severe respiratory syndrome has often been seen in SARS-CoV- and MERS-CoV-related cases infecting the lower respiratory tract (Fehr & Perlman, 2015; Su et al., 2016). Besides human health issues caused by the virus, some viruses may have influenced livestock, bats, birds, mice, and other families of wild animals (Zhou et al., 2018). Different types of coronaviruses with their groups are briefly illustrated in Fig. 1.
Fig. 1.
Coronavirus types
Over the last few months, a novel virus, COVID-19, has emerged from Wuhan, China. The world population has been severely affected by this virus, which has created life-threatening conditions. Due to the highly dangerous nature of the virus for humans, the world economy has been hurt significantly (Maria et al., 2020).
Theoretical background
Bibliometrics has proven itself an inestimable tool for understanding how areas of scientific specialization are organized and how they evolve over time. One of the objectives of bibliometrics is science mapping. Science mapping is a generic process for analyzing and visualizing domains of knowledge, based on extracting and manipulating a certain corpus of scientific literature. The techniques it involves are drawn primarily from computer science, metric studies of science, data and information visualization, visual analytics, and network science. The realm to map may be a scientific discipline, a field of research, topic areas or scientific specialties, or an individual researcher’s career. The spatial representations in which science maps consist are created by preprocessing and processing aggregate data containing intellectual contributions retrieved from the literature stored in bibliographic databases like the Web of Science, Scopus, PubMed, Lens, Dimensions, and Microsoft Academic. The bibliographic records contain a number of variables or bibliographic entities, such as publication titles, authors, journals, years, places, institutions, keywords, abstracts, and bibliographic references. By working with these variables and using techniques and methods of citation analysis, concept or topic extraction, bibliometric metrics, algorithms, clustering techniques, visualization techniques, and python libraries or programs, maps of domains of knowledge or science can be designed and built (Thijs, 2019).
The theoretical rationality behind science maps has been addressed by many authors. When a science map is created, it reveals the links between different areas of knowledge, as well as the processes whereby knowledge is disseminated and the direction in which knowledge travels between areas. Science maps afford a means of analyzing the current knowledge within a domain from a bird’s-eye view, facilitating the systematic, synthetic review of the literature and sociological or historical analyses. They help to determine the present consensus in regard to a concept, by itself or in combination with other concepts, and to trace variations in research discourse. They are instruments that aid in the detection of potential partners who might be interested in working on related subjects, based on representations of co-authorship maps. One classic subject in this subspecialty is the creation of maps that successfully represent emerging research fronts, or key scientific topics and assets, based on an analysis of the scientific and technological literature (Xu et al., 2020). The most common questions about research fronts and scientific topics include the methodological aspects and techniques involved in their creation (i.e., similarity calculation, clustering). Design and visualization are currently implemented in the form of “global” maps, “local” maps, or hybrid maps. “Global” maps represent the whole or substantially whole structure of a bibliographic database, while “local” maps select micro-areas of interest or focus on a particular field or specialty. If the research fronts or topics in a given specific subject are projected as a layer of information superimposed on top of a base constructed in the form of a “global” science map, then the product is a hybrid map (Chen, 2019; Boyack & Klavans, 2019).
In this paper, a co-word analysis technique is applied to map the intellectual structure of the “CoronaVirus” field using 13,518 papers collected from a mainstream scholarly database, the Web of Science Core Collection (WoS). The method is recognized as an effective solution for mapping the strength of association between information sections in textual contexts (Viedma-Del-Jesus et al., 2011). In other words, co-word analysis is considered a quantitative method for mapping the relationships and interrelationships among concepts in different textual scientific works (Zhang et al., 2017). The main idea of co-word analysis is to describe documents based on keyword co-occurrence (Callon et al., 1991). Utilizing co-word analysis enables researchers to draw a detailed picture of a specific domain using words’ co-occurrence in different types of bibliometric units, such as book chapters, journal and conference papers, and books (Liu et al., 2012). Co-word analysis is generally used to convert massive networks of keyword information into smaller, more intelligible networks of related concepts with graph theory (Cobo et al., 2011). The co-word network consists of nodes that represent the keywords and links between them. Given a network of keywords, a compound of community detection, network analysis, and strategic diagrams are depicted to detect interesting trends and patterns within a complex keyword interaction network and reveal the conceptual structure of a research field (Callon et al., 1991). The method has been widely utilized in various areas like urbanization (Zhang et al., 2017), supply chain management (Shiau et al., 2015), library and information science (Olmeda-Gómez et al., 2017), the Internet of Things (Yan et al., 2015), marketing (Z. Wang et al., 2015), social media (Gan & Wang, 2015), scientometrics (Ravikumar et al., 2015), non-biomedical modalities (Nguyen, 2019), medical tourism research (Hoz-Correa et al., 2018), child-computer interaction (Giannakos et al., 2020), energy (Wu & Leu, 2014), open data (Corrales-Garay et al., 2019), recommendation systems (Hu & Zhang, 2015), gender differences (Dehdarirad et al., 2014), and epidemiology (Baziyad et al., 2020). Some papers have also utilized co-word analysis in coronavirus research areas.
Bibliometric methods have already been used in regard to some types of coronaviruses, such as COVID-19 (Lou et al., 2020; Pal, 2021; Yang et al., 2020), SARS (Chiu et al., 2004), MERS (Zyoud, 2016) or all three (Haghani & Bliemer, 2020). Papers on COVID-19 have focused on limited features outlining correlations in existing research on the novel COVID-19. However, the latest papers on COVID-19, SARS, and MERS are still restricted to certain areas, and they help practitioners move efficiently forward in a limited direction only. Hence, the scientific work contains major flaws, because the relationships among earlier coronavirus families and other viruses are not sufficiently investigated.
Methodology
Method and data collection
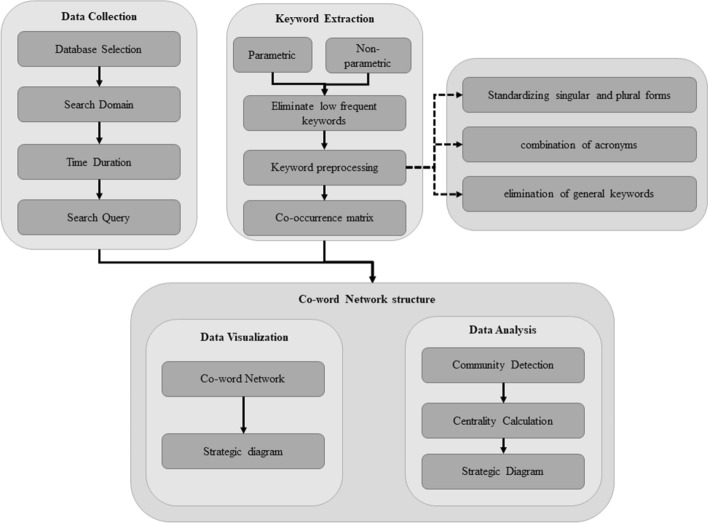
This paper employs the co-word analysis approach for visualizing and analyzing the CoronaVirus-19-related research work published between 2000 and 2020, using the WoS. A comprehensive framework for designing a co-word-based intellectual map as applied in this paper is shown in Fig. 2.
Fig. 2.
A comprehensive framework of intellectual structure based on co-word analysis
Data collection
Data collection is categorized into four main stages, namely: (a) database selection, (b) search query, (c) search domain selection, and (d) time duration selection. In this study, the WoS database was set as the key source for data extraction. Although Scopus and the WoS are both among the leading databases researchers use, a wide range of co-word analysis papers has utilized the WoS. In addition, the WoS has a large number of journals indexed and complete and contains consistently formatted citation information for its entries (Khasseh et al., 2017). For these reasons the WoS database was selected.
To identify the keywords to use in the publication search process, the literature on the topic was inspected and experts’ advice was taken. This paper mostly concentrates on recent important human coronaviruses, such as COVID-19, MERS, and SARS. The keywords eventually settled on are "coronavirus," "COVID-19," "SARS-CoV," and "MERS-CoV," because COVID-19, SARS-CoV, and MERS-CoV are the main types of coronaviruses causing respiratory diseases in humans and enteric diseases in animals. To find all relevant publications, other combinations and the full names of the acronyms were added, including "severe acute respiratory syndrome," "Middle East respiratory syndrome," and "2019-nCoV." Boolean operators were also used to make the searches more detailed and to target the literature with an appropriate degree of specificity.
The search query was performed based on Advanced Search. A careful, structured search for document keywords, titles, and abstracts (Topic search format) was conducted on the WoS. A Topic Search query (TS) searches simultaneously by titles, abstracts, author keywords, and Keywords Plus. Thus, the query strings used in this study are the following:
TS = (("COVID-19" OR “Coronavirus” OR "Corona virus" OR "2019-nCoV" OR "SARS-CoV" OR “MERS-CoV" OR “Severe Acute Respiratory Syndrome" OR “Middle East Respiratory Syndrome")).
All document types (including journal articles, review and conference papers, letters, book chapters, proceedings papers, short surveys, and editorials) were classified as the main input for this study. Other publication types (such as meeting abstracts and news items that do not represent sound publications) were not taken into account. The database contained different languages, including English, Spanish, Russian, Chinese, Japanese, and Korean, but only the search terms in the English language were retrieved. The search query looked for English COVID-19-oriented publications published from January 1, 2000, to January 1, 2020. The search, which was updated on April 1, 2020, returned 13,518 items.
The records were downloaded from the WoS database over the Internet connection provided by the central library at the University of Ottawa. Full records of the dataset were retrieved from the WoS in the CSV Excel format. The databases in which the search was formulated were mostly Science Citation Index Expanded (SCI-EXPANDED), Social Sciences Citation Index (SSCI), Conference Proceedings Citation Index-Science (CPCI-S), Emerging Sources Citation Index (ESCI), and Arts and Humanities Citation Index (AHCI). The retrieved information included the authors, document type, citation information, publication name, research areas, funding agency, bibliographic information, publication year, title, and keywords. Extracting the dataset posed challenges due to the WoS restriction of a maximum of 500 documents per export. Thus, a strategy was adopted to extract 13,518 documents. The list of selected studies was first sorted by the most recently published, and then 500 items were extracted in one export. This strategy was repeatedly employed until all remaining documents were extracted. Around 28 exports were done to extract all documents in the CSV Excel format from the WoS.
Keyword extraction
Co-word analysis research is based on the keywords extracted from textual data such as papers and blogs. There is, then, a close direct correlation between the quality of the keyword extraction process and the quality of the results. Also, keywords with a frequency of at least 10 were chosen to cooperate in the co-word network according to Hosseini et al. (2021), and the collected keywords were filtered through a number of criteria including (1) standardization of singular and plural keyword forms (e.g., “pig,” “pigs”); (2) combination of acronyms (e.g., “MERS-CoV,” “Middle East respiratory syndrome coronavirus”); (3) elimination of general keywords (e.g., “corona,” “coronavirus”).
After the final list of keywords was found, the co-occurrence matrix (symmetrical matrix) of keywords was calculated using Python 3.6, and the visualization process and community detection were conducted by Gephi 0.9.2 (Bastian et al., 2009).
Co-word network
Co-word analysis generally finds clusters of keywords that appear often in conceptually counterpart papers. The co-word network is calculated based on the normalized co-occurrence matrix. A co-occurrence matrix is also obtained from keywords in input papers. For example, keywords A and B appeared in 25 papers simultaneously; consequently, the weight of their co-occurrence is equal to 25. Finally, the matrix was normalized depending on the Jaccard index. The Jaccard index is a measurement for analyzing similarities between two objects calculated by the following formulation (Jaccard, 1902):
| 1 |
where Cij is the number of co-occurrences of keywords i and j. Also, Ci and Cj are the numbers of occurrences of keywords i and j, respectively (Jaccard, 1902). The normalized co-occurrence matrix is shown in Table 1 for three selected major keywords.
Table 1.
An example of a normalized co-occurrence matrix
| COVID-19 | SARS-CoV-2 | 2019-nCoV | MERS | SARS | |
|---|---|---|---|---|---|
| COVID-19 | 1 | 0.4419 | 0.1791 | 0.0039 | 0.0145 |
| SARS-CoV-2 | 0.4419 | 1 | 0.1731 | 0.0013 | 0.0140 |
| 2019-nCoV | 0.1791 | 0.1731 | 1 | 0.0053 | 0.0113 |
| MERS | 0.0039 | 0.0013 | 0.0053 | 1 | 0.0433 |
| SARS | 0.0145 | 0.0140 | 0.0113 | 0.0433 | 1 |
After the co-occurrence matrix is calculated, the co-word network is presented. Each of the keywords in the co-occurrence matrix plays the role of a node in the co-word network, and each normalized weight of the matrix is considered as an edge weight in the co-word graph. For instance, the importance of 0.4419 is the edge weight between the keywords “COVID-19” and “SARS-CoV-2” in the co-word graph in Table 1.
Identification of themes
Next, a community detection algorithm (a type of clustering method frequently used for graph-based datasets such as co-occurrence data) known as the Louvain method (Blondel et al., 2008) was utilized to map each community. ForceAtlas2 was employed for the layout algorithm (Jacomy et al., 2014). Some manual changes and modifications were also made to node movements to improve coordination. Community detection and the layout algorithm visualization were performed using Gephi 0.9.2. The findings can be seen in the result section. Quantitative validation was done by calculating the modularity values.
The modularity of a network measures the extent to which the network can be broken down into multiple components or modules. This metric provides a reference for the overall clarity of a given decomposition of the network (Newman, 2004).
Usually, a graph with a modularity of between 0.3 and 0.7 is considered a strong cluster in which each community has strong internal relations with its vertices (dense connections within communities or clusters) while having weak relationships with other communities (Xie & Szymanski, 2011). The co-word network’s modularity was measured and found to be 0.378, which lies in the desired range. According to Newman (2004), modularity is found by the following formula:
| 2 |
where and are the nodes’ degrees and m is the total number of edges. is the number of edges between vertices and . The quantities are the elements of the so-called adjacency matrix. is the maximum number of edges being between vertices and .
Strategic map
Another analysis method is the strategic diagram of cluster density and centrality. The density of each cluster indicates the internal cohesion of a particular theme, and the centrality of each cluster shows how close each cluster comes to being a component of the central field. Each theme begins at low centrality and density, in the Chaos/Unstructured quadrant. As it gains in centrality, the theme shifts into the Bandwagon quadrant. When the theme’s internal cohesion becomes mature, the theme moves into the Mainstream quadrant. Finally, the theme moves to the Ivory Tower quadrant when it loses its centrality (Giannakos et al., 2020). A schematic strategic diagram is illustrated in Fig. 3.
Fig. 3.
Strategic diagram of density and centrality
Many papers in the field of co-word analysis have used keyword clustering and strategic diagramming for data mapping and data analysis. This paper employed keyword clustering and a strategic diagram for CoronaVirus-19-related research. Centrality and density were calculated using Eq. 3 and Eq. 4, respectively (given below). Node centrality equals the total number of direct connections related to a node. Greater centrality for a specific keyword shows that the keyword is used more frequently in research. Also, network density (D) is the proportion between the actual amount of links and the maximum possible number of links that can be obtained from the network.
| 3 |
where is the centrality of community , denotes nodes in community , the list of other existing communities except community is given by , and is existing nodes in all communities. Also, is a binary variable that is equal to 1 if an edge has existed between node and node , and otherwise it is equal to 0. Lastly, is considered an edge weight between node and node .
| 4 |
where states the density of community wherein is the total number of edges in and is the total number of nodes in .
Result
Top keywords and journals
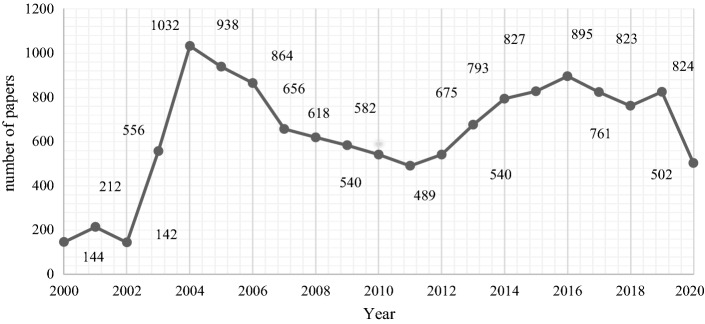
The evolution of the papers published on the topic per year from 2000 to 2020 is illustrated in Fig. 4. Coronavirus research grew significantly between 2003 and 2004 due to the emergence of SARS in 2002 and 2003. The maximum number of coronavirus research publications (1032 publications) was reached in 2004. Between 2004 and 2011, the number of publications gradually declined; nevertheless, academic publications rose steadily once again with the emergence of MERS in 2012. The trend in publications can be expected to continue rising in 2020 because of the COVID-19 pandemic.
Fig. 4.
Quantification of papers on coronavirus topics (between 2000 and 2020, updated on April 1, 2020)
After careful examination, 491 keywords were selected for the co-word network. Thirty highly frequent keywords are listed in Table 2.
Table 2.
Highly frequent keywords in 13,518 papers
| No. | Keyword | Occurrences | No | Keyword | Occurrences |
|---|---|---|---|---|---|
| 1 | Respiratory disease | 2630 | 16 | Antibody | 762 |
| 2 | Spike | 1429 | 17 | Enteritis | 733 |
| 3 | Cats | 1349 | 18 | Emerging | 706 |
| 4 | Human | 1334 | 19 | AR-PCR | 701 |
| 5 | Pathogen | 1329 | 20 | Receptor | 695 |
| 6 | PCR | 1238 | 21 | Protease | 677 |
| 7 | RNA | 1215 | 22 | Response | 661 |
| 8 | E-protein | 1165 | 23 | Pneumonia | 619 |
| 9 | Diarrhea | 995 | 24 | Control | 606 |
| 10 | Middle East | 920 | 25 | Influenza | 598 |
| 11 | Spike protein | 888 | 26 | Replication | 596 |
| 12 | Epidemic | 843 | 27 | Vaccines | 585 |
| 13 | Antivirals | 830 | 28 | Nucleocapsid | 584 |
| 14 | Epidemiology | 791 | 29 | Recombinant | 549 |
| 15 | Respiratory tract | 780 | 30 | Diagnosis | 526 |
Table 3 lists the journals that have published more than 100 papers on the topic of coronavirus over the last two decades. Journal of Virology accounts for 6.76% of the most active publications on the subject. Emerging Infectious Diseases and Virology are ranked second and third, respectively, with 2.34% and 2.26% of the publications.
Table 3.
Leading journals in terms of number of coronavirus-related publications (2000–2020)
| Journal | # papers | % |
|---|---|---|
| Journal of virology | 907 | 6.7621 |
| Emerging infectious diseases | 314 | 2.341 |
| Virology | 303 | 2.259 |
| PLoS ONE | 261 | 1.9459 |
| Virus Research | 201 | 1.4985 |
| Journal of general virology | 193 | 1.4389 |
| Viruses-Basel | 177 | 1.3196 |
| Archives of virology | 162 | 1.2078 |
| Antiviral research | 154 | 1.1481 |
| Journal of virological methods | 152 | 1.1332 |
| Veterinary microbiology | 147 | 1.0960 |
| Journal of medical virology | 138 | 1.0289 |
| Journal of infectious diseases | 122 | 0.9096 |
| Journal of clinical virology | 121 | 0.9021 |
| Vaccine | 118 | 0.8797 |
| Journal of clinical microbiology | 117 | 0.8723 |
| Proceedings of the National Academy of Sciences of the United States of America | 117 | 0.8723 |
| Clinical infectious diseases | 113 | 0.8425 |
| Virology Journal | 110 | 0.8201 |
| Lancet | 108 | 0.8052 |
| Total | 13,518 | 100 |
Different areas of coronaviruses
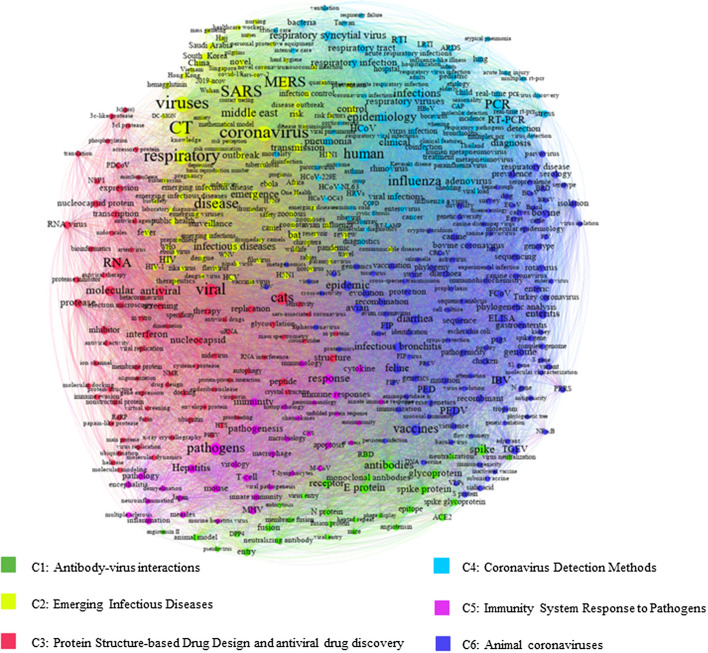
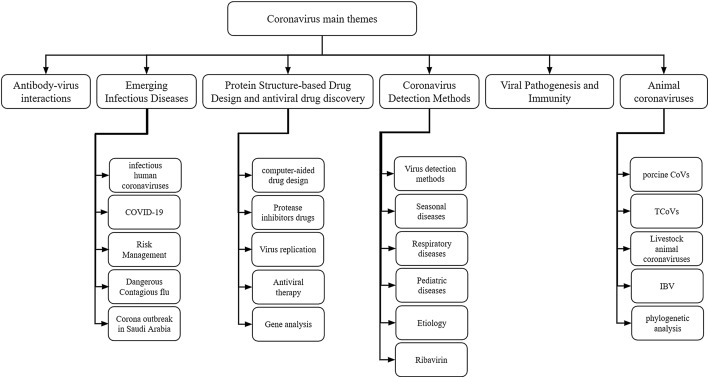
The Louvain algorithm was applied to the co-word matrix (Lambiotte et al., 2008), and six main clusters were manually labeled based on pathologists’ recommendations. The clusters were “Antibody-virus Interactions,” “Emerging Infectious Diseases,” “Protein Structure-based Drug Design and Antiviral Drug Discovery,” “Coronavirus Detection Methods,” “Viral Pathogenesis and Immunity,” and “Animal Coronaviruses.” The whole structure of the co-word network is shown in Fig. 5. Each cluster regarded as a theme in coronaviruses and concepts related to similar communities contains a set of keywords and is represented by a unique color. More frequent keywords are displayed in bigger fonts.
Fig. 5.
The co-word network of coronaviruses
As explained before, the Louvain algorithm was applied to clusters with more than 50 nodes to learn more about the distinctive communities shown in the figure and interpreted in the sections below. The modularity figures were found for “Emerging Infectious Diseases,” “Protein Structure-based Drug Design and Antiviral Drug Discovery,” “Coronavirus Detection Methods,” and “Animal Coronaviruses:” approximately 0.421, 0.436, 0.308, and 0.383, respectively, all within the desired range. The themes of “Antibody-virus Interactions” and “Viral Pathogenesis and Immunity” had smaller scales than the others, so no further detail was required. Community detection was therefore not applied to these themes. The results of the modularity criteria are given in Table 4.
Table 4.
Modularity measurement of the communities yielded by the community detection algorithm
| Id | Cluster | No. sub-communities | Modularity |
|---|---|---|---|
| C1 | “Antibody-virus interactions” | – | – |
| C2 | “Emerging infectious diseases” | 7 | 0.421 |
| C3 | “Protein structure-based Drug design and antiviral drug discovery” | 7 | 0.436 |
| C4 | “Coronavirus Detection methods” | 6 | 0.308 |
| C5 | “Viral Pathogenesis and immunity” | – | – |
| C6 | “Animal coronaviruses” | 6 | 0.383 |
The main themes and sub-themes of coronavirus-related research over the two past decades are depicted in Fig. 6, including six main general themes. The results obtained from the map of each community and the structure of each theme and sub-theme were validated using pathologist’s expert opinions. Additionally, the experts verified both the research findings and future research directions.
Fig. 6.
The main themes of coronavirus
Antibody-virus interactions C1
Antibodies play four major roles in connection with pathogenic viruses: antibody-dependent cell-mediated virus inhibition (ADCVI), virus neutralization, phagocytosis, and antibody-dependent cellular cytotoxicity (ADCC) (Forthal & Moog, 2009). The “Antibody” theme, shown in green in Fig. 7, mostly focuses on antibody-virus interactions in a host cell, especially from the perspective of neutralizing antibodies. Neutralizing antibodies are the antibodies that bind to cell-free viruses, thus blocking the viruses; as a result, they eliminate and interrupt virus replication and prevent cell infection (Gorny et al., 2005; Greenspan & Cavacini, 2019; Ringe & Bhattacharya, 2013; Tuen et al., 2005). Furthermore, the theme also covers distinctive virus components associated with viral entry in host cells, including membrane envelope (E) proteins and spike peptides or glycoproteins (S) (David Cavanagh, 1995). Spike protein, a viral membrane protein, plays a vital role in viral entry: It binds to a receptor in the target cell, such as angiotensin-converting enzyme 2 (ACE2) (Hamming et al., 2004)), thus providing opportunities for virus-cell fusion (Bosch et al., 2003). A coronavirus structure and the ACE2 viral receptor on the host cell surface are illustrated in Fig. 8 as represented by Oguh et al. (2020). As Fig. 7 shows, the major nodes in the co-occurrence network correspond to neutralizing agents, such as antibodies and monoclonal antibodies, and coronavirus structures, such as E protein, spike, glycoprotein, and receptor. In the light of the literature review and the strong relationships between the network’s nodes, the specific community can be mapped onto “Antibody-virus Interactions.”
Fig. 7.
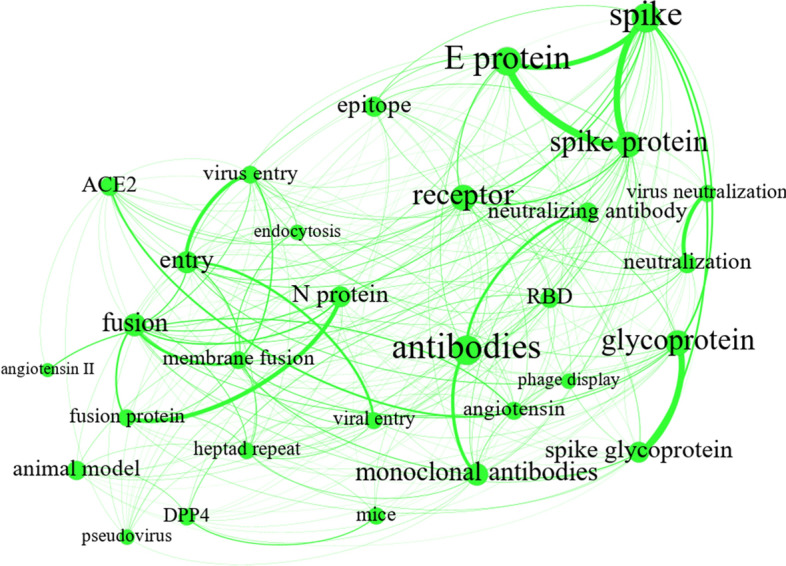
“Antibody-virus interactions C1”
Fig. 8.
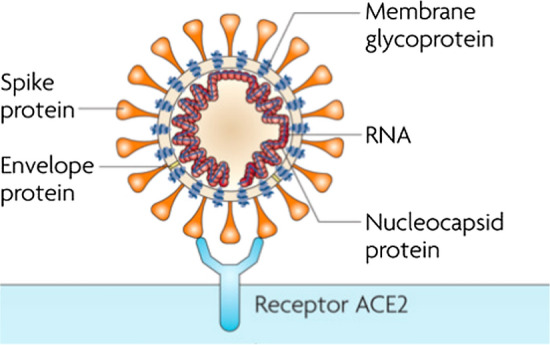
Illustration of the coronavirus structure and the ACE2 viral receptor
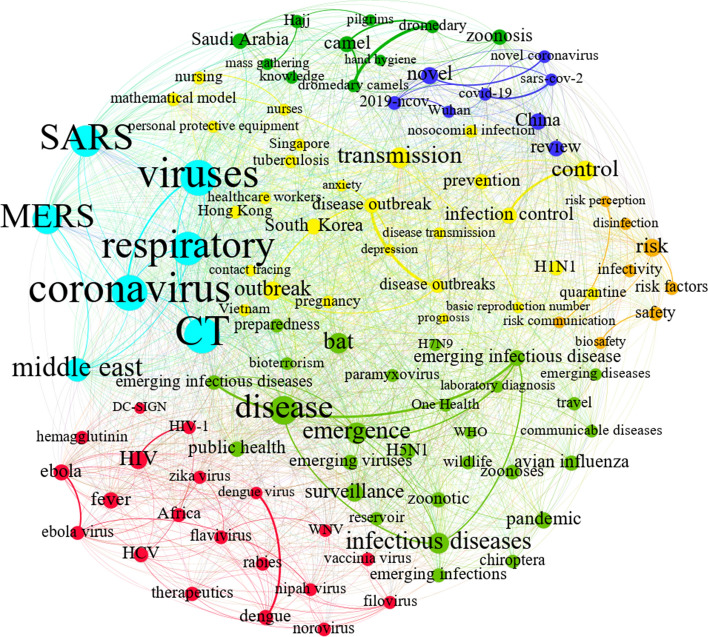
Emerging infectious diseases C2
This theme (yellow color in Fig. 9) divides emerging infectious diseases into two groups: (a) non-coronaviruses and (b) coronaviruses. The first group describes several types of viral infectious diseases (not including the coronavirus family) in both humans and animals, such as contagious flu (H5N1, H7N9) (light green in Fig. 9), hepatitis C, HIV, and flavivirus. Fever is reported as a common symptom of most of these viruses, shown by the “fever” node (red in Fig. 9). The other parts of the graph, however, mention emerging fatal coronaviruses, namely, MERS, SARS (light blue in Fig. 9), and COVID-19 (dark blue in Fig. 9), and transmission, disease outbreaks, risk management, and infection control (yellow and orange in Fig. 9).
Fig. 9.
“Emerging infectious diseases C2”
Due to the large size of this theme, the Louvain algorithm was also applied to extract sub-themes, as depicted in Fig. 9. Based on existing nodes and their relations inside the light blue sub-community, this sub-community can be identified by titling it as highly infectious human coronaviruses, especially MERS and SARS. Also, Fig. 9 shows that the dark blue sub-community can be mapped onto COVID-19 due to its nodes’ interrelation in this sub-community (e.g., “SARS-CoV-2,” “novel coronavirus,” and “2019-ncov”).
After the sub-communities were investigated and various components of the community were classified, an explanation of how the map was generated was found. Nodes related to cities and countries appear in Fig. 9, proving that outbreaks of novel human coronaviruses were reported in China (the origin of COVID-19 and the SARS virus outbreak), Saudi Arabia (the origin of the MERS outbreaks), Vietnam, Hong Kong, South Korea, and Singapore. All these countries (except Saudi Arabia) are located near China, so Asian nations, particularly China, are clarified as the main sources of the SARS, MERS, and COVID-19 outbreaks/infections (Peeri et al., 2020). Other nodes, such as “quarantine,” “infection control,” “prevention,” “mathematical modeling,” and “risk management,” can be helpful for surveying the outputs of community mapping. They will therefore be discussed below.
Risk management, as a leading issue of emerging human coronaviruses, has three primary controversial pillars: risk factors, perception, and communication. A personal intellectual judgment about disease characteristics and risk severity which can instigate behavior designed to prevent the spread of disease is what is considered risk perception (Ferrer & Klein, 2015). When experts who have combatted previous coronavirus outbreaks share information about their experience, making it accessible to the public, they enable researchers and practitioners to make better decisions about how to prevent the disease (Krewski et al., 2011). Furthermore, medical treatment and care can be investigated and then applied by researchers. For example, quarantine, which has been detected as a major effective means of virus transmission prevention, has recently received considerable attention from mathematical modeling standpoints. Indeed, mathematical models are providing managers with useful information for scheduling and planning the quarantine process, leading to outbreak reductions with an efficient use of effort (Mandal et al., 2020). Mathematical modeling not only can promote the optimization of quarantine processes, but also can be utilized for calculating the potential reproduction number of different disease types (Fraser et al., 2004). Mathematical modeling can also reveal the patterns of virus spread and the time when a peak is likely to be reached. Note that in quarantine periods policymakers are faced with two key factors of outbreaks, namely traveling and gathering.
In general, the historical origins of various virus types and the protective methods used to restrict and immunize humans against infection are connected to emerging infectious diseases through the map (Fig. 9), based on a similarity between emerging novel coronaviruses. This insight uncovers useful hidden knowledge and also helps researchers channel their considerable efforts in the right directions.
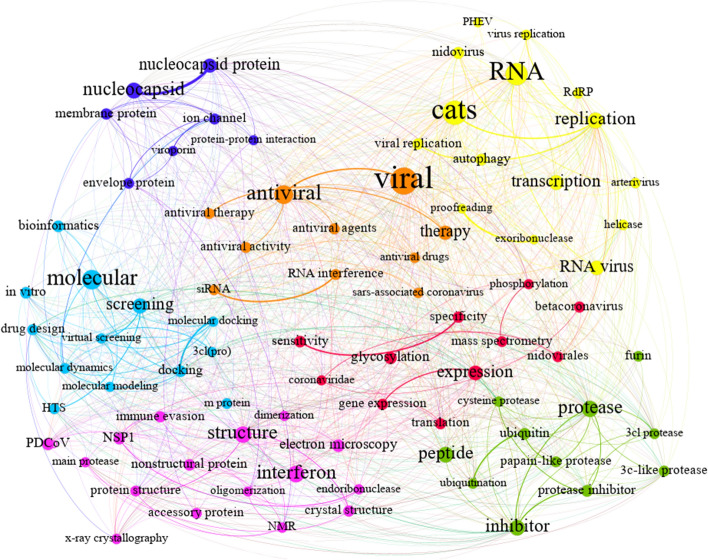
Protein structure-based drug design and antiviral drug discovery C3
Some of the nodes in Fig. 10, such as “antiviral,” “molecular,” “RNA,” “protease,” “inhibitor,” and “interferon,” are larger than others due to their centralized position as calculated in the co-occurrence network. Our survey of biological resources has proven that nodes and their links have the utmost importance in antiviral drug development. Based on the given description, this theme can be mapped onto “Protein Structure-based Drug Design and Antiviral Drug Discovery.” Nowadays, computer-aided drug design (CADD) plays a key role in how pharmaceutical companies discover drugs (Muegge et al., 2017; Hillisch et al., 2015). Antiviral drugs are classified into two main classes, namely, ligand-based (two-dimensional or 2D) drugs and protein structure-based (three-dimensional or 3D) drugs. Protein structure-based drug design approaches include various methods, from docking to molecular dynamics (Śledź & Caflisch, 2018). Further details about the theme are provided below.
Fig. 10.
“Protein structure-based drug design and antiviral drug discovery C3”
As can be observed in Fig. 10, the third theme was split into different sub-communities, and their maps are listed below in order of importance. According to the co-word network, there are three types of molecular modeling methods employed in coronavirus-associated research: molecular docking, molecular dynamics, and virtual screening (light blue in Fig. 10). The most important nodes of the community are “molecular” and its related nodes, such as “molecular modeling” and “screening,” illustrated in the light-blue sub-theme.
The function of molecular docking has been increasingly enhanced in the last three decades due to the growing tides of computing power and computer availability and also growing accessibility to databases of small protein molecules (Berman et al., 2000; Hendlich, 1998). Moreover, molecular dynamics provide a dynamic behavioral model on a microscopic scale for different types of systems, such as solids, liquids, gases, surfaces, and clusters (Tuckerman & Martyna, 2000), and for drug design (Ganesan et al., 2017). In addition, virtual screening, as a computational technique of drug discovery, is utilized to find the most similar structures binding to a targeted drug, typically a protein receptor or enzyme (Rester, 2008; Rollinger et al., 2008). Some other keywords of this theme indirectly depend on the coronavirus life cycle (CLC), which is important for drug discovery. Some stages of the CLC (namely entry, replication, and transcription) are given in major nodes in Fig. 10. Thus, the stages of the CLC as leading nodes in the theme are surveyed as follows.
The cycle generally has two steps, namely a) entry, replication, and transcription and b) virion assembly and release (da Costa et al., 2020). The sub-community, however, mostly focuses on entry, replication, and transcription. Viral infection begins with the virus binding to the receptor-binding domains. This binding is a fusion of the virion and the cell membrane. It happens on the cell surface or within the endosome, and it culminates with the release of the nucleocapsid into the cytoplasm (dark blue in Fig. 10). Viral gene translation, in turn, is done to produce polyproteins (red in Fig. 10). The polyproteins are then processed by the viral proteases (green in Fig. 10) to make the viral proteins. Finally, a series of sub-genomic mRNA is released by discontinuous transcription that is required for the expression of the structural protein that leads to virus replication (yellow in Fig. 10).
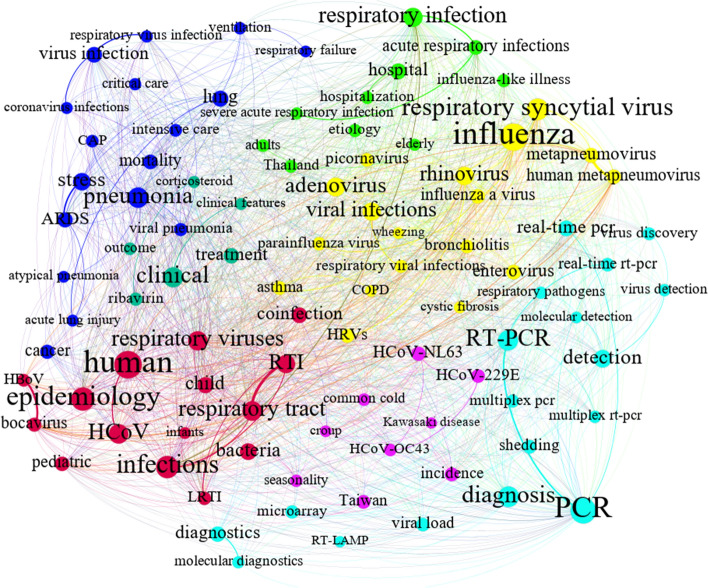
Coronavirus detection methods C4
The major keywords applied in the theme in Fig. 11 are “PCR,” “LAMP,” “diagnosis,” and “detection.” For mapping and extracting useful information through the Louvain algorithm, distinctive sub-communities were derived from this community. One of the most important concepts examined in the community has to do with coronavirus detection and diagnosis. Some of the leading detection and diagnosis approaches are therefore discussed below to clarify the concepts involved and evaluate the results of the community mapping endeavor.
Fig. 11.
“Coronavirus detection methods C4”
Three significant methods for detecting coronavirus appear in the graph (light blue in Fig. 11). One of them is polymerase chain reaction (PCR), a prevalent assay taken as a reliable technique for coronavirus detection due to its applicability and high sensitivity. This is particularly so for the new versions of PCR assays, such as real-time PCR and real-time reverse transcriptase-PCR (Balboni et al., 2012; Shen et al., 2020; Uhlenhaut et al., 2012). Two other coronavirus detection approaches, namely, loop-mediated isothermal amplification (LAMP) (Enosawa et al., 2003) and the microarray-based method (Chen et al., 2010), are identified as cost-effective, efficient, applicable means of virus detection. However, according to the co-word network, the PCR approaches have been used more than LAMP and microarray methods. For example, multiplex RT-PCR, which has been considered an appropriate method for diagnosing respiratory human CoVs, could be further adapted to novel CoVs (Emery et al., 2004; Gaunt et al., 2010). Misdiagnosis may be an issue in cases of co-infection and divergent viral respiratory infections; other viral infections mostly cause pneumonia, including respiratory syncytial virus, human metapneumovirus, parainfluenza virus, and adenovirus (yellow in Fig. 11). Pediatric diseases with mild respiratory symptoms in children (red in Fig. 11) and HCoVs (purple in Fig. 11) have also been studied alongside coronavirus detection.
Viral pathogenesis and immunity C5
As can be seen in Fig. 12, the keywords of this theme are associated with viral pathogenesis and coronavirus immunology. Viral pathogenesis is a mechanism for finding the viruses that cause disease in interactions between viruses and their hosts (Heise, 2014). As such, it appears in association with keywords such as “pathogens,” “viral pathogenesis,” “pathology,” and “histopathology.” With the keywords in the community, the concept of immune system response can be mapped onto pathogens. Keywords such as “MHV,” “MuCoV,” and “ER stress” were subsequently investigated further to explore the outputs of the map.
Fig. 12.
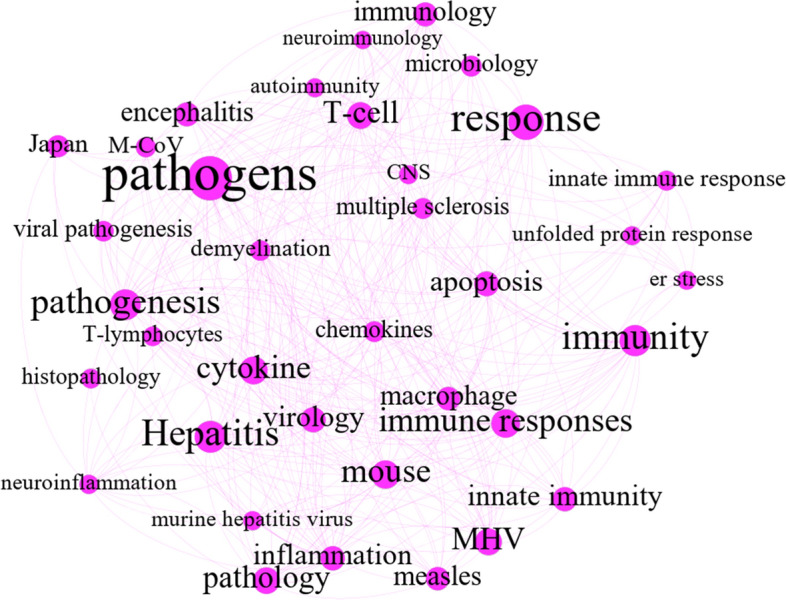
“Viral pathogenesis and immunity C5”
The main studies in the theme concern mouse hepatitis virus (MHV) and murine coronavirus (MuCoV), which provide animal model systems for examining viral tropism and pathogenesis in various tissue organs, including the CNS (for instance, surveying demyelination of the CNS in multiple sclerosis) (Weiss & Leibowitz, 2011), lung, and liver. Other keywords describe immune system responses, including immune-related cells (T-lymphocyte and macrophage), innate immune response, immunology, unfolded protein response, and ER stress. The community is illustrated separately in Fig. 12 to visualize the keyword correlations. By releasing diverse inflammatory mediators such as chemokines, cytokines, and proteases, macrophages recognize and engulf antigens of the pathogens through T lymphocytes (T-cells) (Arango Duque & Descoteaux, 2014; Gordon & Plüddemann, 2013; Kassab et al., 2019). T-cells are stimulated by the antigens presented by macrophages and then release lymphokines to signal other cells. Also, T-cells can react properly to antigens through the recognition of antigens (Filiano et al., 2017; Park & Mitchell, 2012).
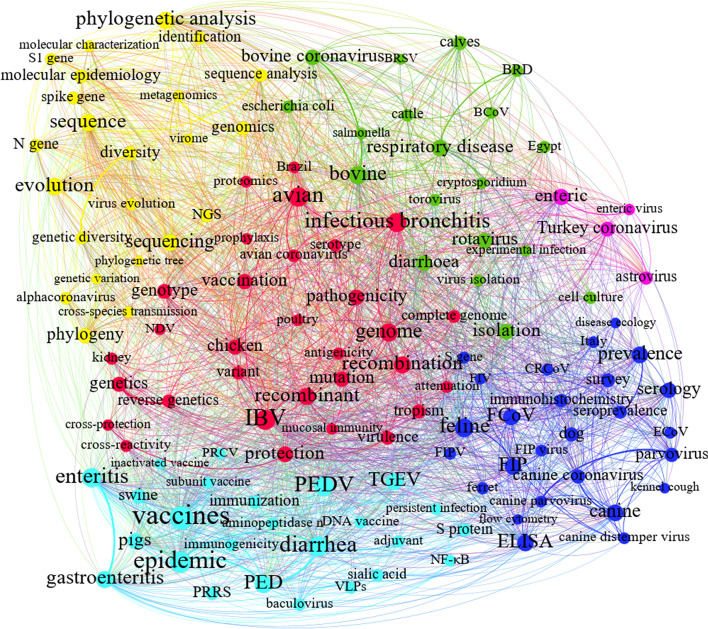
Animal coronaviruses C6
The main reason why this community is mapped this way can be outlined by probing its nodes and relationships in Fig. 13. Keywords in this community concentrate on three main animal coronaviruses, namely, a) alpha coronaviruses, such as porcine CoVs, feline coronavirus (FCoVs), and canine coronavirus (CCoV); b) beta coronaviruses, including equine coronaviruses (ECoV) and bovine coronaviruses (BCoV); and c) gamma coronaviruses, like avian infectious bronchitis virus (IBV) and turkey coronavirus (TCoV). The yellow sub-graph displays a phylogenetic analysis of different animal coronavirus types. For further investigation and classification of animal coronaviruses, the Louvain algorithm is employed to survey the sub-communities in Fig. 13. One leading virus group is listed and described below.
Fig. 13.
“Animal coronaviruses C6”
The three porcine coronaviruses (light blue in Fig. 13) are transmissible gastroenteritis coronavirus (TGEV), porcine epidemic diarrhea virus (PEDV), which causes enteric infections, and porcine respiratory coronavirus (PRCV), which leads to respiratory disorders. Considering how the porcine CoVs have emerged and re-emerged since 2013, they have been considered a major threat to swine industries the world over (Leyi Wang & Zhang, 2016). FCoV (dark blue in Fig. 13) evolved from the fecal–oral route, resulting in feline infectious peritonitis (FIP), identified as a systematic infectious disease in both domestic cats and their wild peers (Hartmann, 2005). CRCoV (dark blue in Fig. 13) is closely related to feline and porcine strains. It produces gastroenteritis in dogs (de Cássia Nasser Cubel Garcia, 2016). ECoV (dark blue in Fig. 13) is associated with the most common symptoms in equine species (e.g., fever, anorexia, diarrhea, leukopenia, lethargy) (Miszczak et al., 2016). BCoV (green in Fig. 13) is responsible for neonatal calf diarrhea, respiratory disorders, and winter dysentery in cattle (Alfieri et al., 2016). The agent of infectious bronchitis, accompanied by tracheal ciliostasis, nasal discharge, snicking, and rales, is also labeled as “IBV” (red in Fig. 13). Additionally, IBV has harmful effects on kidneys, enteric surfaces, and oviducts, and it impairs egg production and quality (Ambali & Jones, 1990; Cavanagh, 2005; Cook & Mockett, 1995). A highly epidemic enteric disease in turkey peers which has caused severe damage for turkey-related industries has evolved from TCoV (purple in Fig. 13).
Strategic diagram
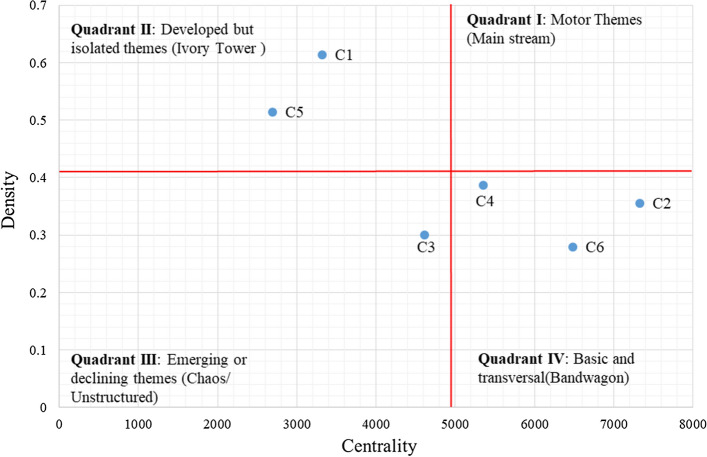
The strategic diagram of co-word-related centrality and density is described in this section. The density and centrality of each community are given in Table 5.
Table 5.
The strategic diagram measurement
| Q | ID | Top keywords | Centrality | No. nodes | No. edges | Density |
|---|---|---|---|---|---|---|
| QI: Motor | – | – | – | – | – | – |
| QII: Ivory towers | C1 | Spike, spike protein, E protein, glycoprotein, entry, fusion, spike glycoprotein, membrane fusion, antibodies, and receptor | 3321 | 30 | 267 | 0.613793 |
| C5 | Demyelination, immunology, inflammation, response, pathogenesis, immune responses, multiple sclerosis, neuroimmunology, neuroinflammation, and hepatitis | 2694 | 36 | 324 | 0.514286 | |
| QIII: Emerging/Declining | C3 | Replication, protease, inhibitor, docking, antiviral, structure, nucleocapsid, molecular docking, and nucleocapsid protein | 6408 | 83 | 1036 | 0.3044 |
| QIV: Basic/Transversal, Bandwagon | C2 | Emerging infectious disease, infectious diseases, emerging infectious diseases, emergence, disease, control, Middle East, outbreak, dromedary, and Ebola | 7331 | 107 | 2013 | 0.354963 |
| C4 | RTI, respiratory tract, metapneumovirus, human metapneumovirus, rhinovirus, infections, respiratory syncytial virus, respiratory viruses, bocavirus, and human | 5355 | 93 | 1652 | 0.386162 | |
| C6 | Diarrhea, PED, enteritis, epidemic, infectious bronchitis, canine, gastroenteritis, bovine, phylogenetic analysis, and enteric | 6481 | 128 | 2274 | 0.279774 |
For the clustering results, the strategic diagram of the six communities was drawn in Fig. 14 using the centrality and density measures. Moreover, Fig. 14 shows that no motor theme has existed in coronavirus research work.
Fig. 14.
Strategic diagram
According to Fig. 9, the two clusters placed in QII (Ivory Towers), “Antibody-virus Interactions C1” and “Viral Pathogenesis and Immunity C5”, are well structured internally but have rather weak external relations. Combining these themes with the other four themes drives them into QI. In other words, a further effort is required to help place this theme in more effective subject areas, shown in Fig. 14.
Also, theme 3 (“Protein Structure-based Drug Design and Antiviral Drug Discovery C3”) falls into the Chaos/Unstructured quadrant, as either an emerging theme or a disappearing theme. Thus, greater concentration on C3 can move it to QII, reflecting greater development as illustrated in Fig. 14.
Finally, three main communities (“Emerging Infectious Diseases C2”, “Coronavirus Detection Methods C4” and “Animal Coronaviruses C6”) are in QIV (Bandwagon). They are all central, albeit distinctive from each other. Thus, these themes should be also studied further so they can be moved into QI.
Generally, there remains a paucity of concentration in C2, C4, and C6. Owing to their low density, these areas are more interesting to focus on. Consequently, each of these three areas can be considered a potential subject for further improvement. C1 and C5 are already sufficiently structured, and they have less research potential than the other clusters. Even so, they can be combined with other themes to create novelty values because of the low centrality of the other clusters. Furthermore, due to the diversity and novelty of the coronavirus, no themes lie in QI. Hence, interested researchers have a golden opportunity for further contribution and achievement in that sense.
Discussion
The recent trend in keyword extraction has shown just how important past studies of coronaviruses are. Coronavirus-related research surged with the emergence of SARS and MERS in 2003–2004 and 2012–2016. Therefore, it is predicted that COVID-19 studies will rise noticeably, appearing primarily in Journal of Virology, Emerging Infectious Diseases, and PLoS ONE.
This paper employed a co-word analysis methodology to provide a comprehensive review of coronavirus field research published in the WOS between 2000 and 2020, updated on April 1, 2020. Six main themes emerged from our analysis (Fig. 3). They are named (a) “Antibody-virus Interactions C1,” (b) “Emerging Infectious Diseases C2” (c) “Protein Structure-based Drug Design and Antiviral Drug Discovery C3” (d) “Coronavirus Detection Methods C4” (e) “Viral Pathogenesis and Immunity C5” and (f) “Animal Coronaviruses C6” Our research findings substantiate the original definition of the coronavirus field summarized below.
Antibody-virus interactions C1
This theme mostly focuses on interactions between antibodies and viruses, particularly in terms of the neutralization process. Also, this theme lies in QII, which indicates strong density and low centrality. In other words, the keywords of this theme are frequently researched and discovered together. On the other hand, it has only weak relationships with the other five themes. Weak relationships indicate golden opportunities for researchers to discover new dimensions of antibody-virus attractions in coronaviruses.
From bioassay perspectives, neutralization assays are not only highly sensitive and quite specific but also time consuming and prohibitively expensive because of their labor-intensive nature and cell culture prerequisites (Atmar, 2014). To bring down time and costs, mathematical modeling and optimization could be applied; mathematical modeling could pave the way for setting arbitrary parameters during neutralization processes to yield the desired results promptly at a lower cost.
Emerging infectious diseases C2
The second theme explains the virology of coronaviruses. It has significant centrality and low density. Accordingly, more work is needed to develop the theme more thoroughly: Coronavirus and non-coronavirus cases in this cluster should be simultaneously subjected to greater investigation and closer examination. Indeed, the body of work done so far has not been integrated well enough to create novelties. Research into HIV, Ebola, dengue fever, Nipah virus, Zika virus, West Nile virus (WNV), HCV, and so on has provided inspiration in the past for non-coronavirus-related achievement and can contribute further to different types of coronavirus-preventive actions. Comparative analysis of non-coronaviruses and coronaviruses is recognized as containing a wide gap; this gap can be closed with multidisciplinary efforts. Utilizing different ideas and methodologies drawn from non-coronavirus work, then, will culminate in an impressive success in the coronavirus research area.
Drug repurposing is also one of the major triggers suggested for devising effective coronavirus drugs. Drug repurposing is an effective strategy, as it involves developing products based on current drugs and thus requires less effort and lower cost than randomized clinical trials and de-novo drug discovery (Cheng, 2019; Cheng et al., 2016). For example, IFN-α, one of the antivirals most widely used against hepatitis, could be useful to inhibit SARS-CoV reproduction in vitro. Furthermore, the HIV medication lopinavir/ritonavir could play a role in SARS-CoV treatment, as could darunavir (a second-generation HIV-1 protease inhibitor) (Lisheng Wang et al., 2020). Consequently, drug-repurposing strategies are crucial for discovering effective drugs and antibodies of coronaviruses.
Protein structure-based drug design and antiviral drug discovery C3
This theme largely concerns protein structure-based drug design, especially using computer-aided drug design methods. According to the strategic diagram, it lies in QIII and has both low density and low centrality. In other words, it is still in its infancy. As mentioned, computer-aided drug design methods fall into two main classes, namely, ligan-based drug design and protein structure-based drug design. In comparison to the ligan-based drug design methods, the protein structure-based methods account for a major part of computer-aided coronavirus drug design, while Wilson and Lill (2011) state that combining ligan-based and protein-based design methods is essential for promoting the efficiency and reliability of computer drug design. Moreover, ligan-based drug design is employed when limited information about the target protein structure and structure-based methods is available (Wilson & Lill, 2011). Therefore, it can be helpful for detecting drugs to use against emerging coronaviruses, especially those that are in the early stages of outbreak.
Coronavirus detection methods C4
The fifth theme defines coronavirus detection methods that have strong relationships with other coronaviruses; nevertheless, it has not been properly developed in terms of methodologies and technologies. Some advanced technologies, such as the Internet of Things (IoT) and artificial intelligence (AI), and various smart devices can be employed to produce novelties for real-time virus detection. For instance, Mohammed et al. (2020) introduced an IoT-based system for discovering people suspected of being infected by COVID-19. In the proposed system, the suspect zone is scanned by an infrared camera to detect high body temperatures. When high-temperature bodies are found, an optical camera captures the suspected faces of the high-temperature persons, and their location (found via a geographic information system) is sent to health officials. Salman et al. (2020) developed an algorithm based on deep learning methods for detecting COVID-19 from X-ray images.
Viral pathogenesis and immunity C5
This theme investigates pathogenesis and the immunology of coronaviruses. It lies in the Ivory Tower quadrant of the strategic diagram. The theme has high density, which means it could develop well internally. Besides, it has low centrality, which suggests it should be combined with other themes to develop externally and yield novelties. The following viral pathogenesis research questions are proposed (Heise, 2014):
Which viral infections lead to diseases?
Does a virus cause disease or not?
What form of the disease is taken?
How severe will the disease be?
In short, interactions of coronavirus pathogens and immune system responses are presented in biological terms in this theme. Although pathogenesis studies culminate with a deeper understanding of different types of coronaviruses, investigating effective factors for preventive measures against the emergence of new diseases may significantly prevent novel coronavirus outbreaks even worse than the MERS, SARS, and COVID-19 outbreaks. Methods for finding such effective factors and measuring their effectiveness will come from pathogenesis-related research.
Animal coronaviruses C6
The sixth theme describes the pathology of major animal coronaviruses. While the animal coronavirus community is well connected to other coronavirus fields, the community is not very dense; thus, it can be moved to the Motor Theme quadrant with a little effort. As such, it should be studied more from the viewpoints of pathology and immunology. Additionally, animal coronavirus outbreaks have affected livestock considerably and consequently damaged the economy. Animal health measures can effectively block animal coronavirus outbreaks. Wearable technologies are classified among the leading concepts for improving animal health through time-monitoring systems.
Indeed, biosensors and wearable technologies have become very important in animal health management systems and have been proved to be helpful in poultry farming and dairy cattle raising. One of the main achievements of these technologies is that they provide real-time disease diagnosis, which mitigates profit losses. Sensors can assess and measure different biomedical parameters and provide reliable data that can reduce human error. Also, these technologies can be customized to meet the needs of various types of animals, pets, and livestock (Harrop et al., 2016). Accordingly, designing integrated systems to trace alpha coronavirus symptoms can restrict outbreaks of the virus, eventually resulting in economic recovery.
Conclusions, limitations, and future research
The present research aimed to map the intellectual structure of the coronavirus field from 2000 to 2020 using co-word analysis of publications in the WoS Core Collection database. The results confirmed that research in the coronavirus field is still in its early stage, which consists of a wide range of heterogeneous, interdisciplinary knowledge domains. The study also laid the groundwork for future research in the subject areas. To achieve that goal, this paper explored a knowledge map of the main CoV themes, including six dominant communities, namely (a) “Antibody-virus Interactions C1,” (b) “Emerging Infectious Diseases C2,” (c) “Protein Structure-based Drug Design and Antiviral Drug Discovery C3,” (d) “Coronavirus Detection Methods C4,” (e) “Viral Pathogenesis and Immunity C5,” and (f) “Animal Coronaviruses C6.” Furthermore, a strategic diagram was created showing the communities’ current situation. According to the strategic diagram, none of the communities lie in Quadrant I. This means that there is no domain in the coronavirus field that is both central and mature. As a result, many research gaps still need to be bridged by further research. “Antibody-virus Interactions C1” and “Viral Pathogenesis and Immunity C5” lie in the Ivory Towers quadrant, which means they should be combined with other themes to create novelties in CoV research. “Protein Structure-based Drug Design and Antiviral Drug Discovery C3” is in QIII; this indicates that it needs more concentration before it can develop further. “Emerging Infectious Diseases C2”, “Coronavirus Detection Methods C4” and “Animal Coronaviruses C6” are situated in QIV; they are more central but low in density, traits that mark potential future areas of research.
Although this paper provides a good intellectual structure of the subject, it still has great room for further improvement before it becomes a comprehensive work in the coronavirus field. The major limitations of this study are listed as follows:
Time-consuming
Co-word analysis requires preprocessing and feature selection, which are highly time-consuming tasks, especially when either the number of features (selected words) is large or powerful hardware is not available. Big Data-based co-word analysis could pave the way for both more research papers and better keyword selection, yielding more accurate, comprehensive results.
The future trend
We applied co-word analysis to develop an intellectual map of coronavirus research. However, we did not consider the predictive aspect of research. Consequently, prediction methods, such as link prediction, could be employed to provide a broad overview of future research.
Acknowledgements
We gratefully thank Marzieh Houshvar (medical laboratory scientist) for providing us with her constructive comments on the results and helping us move forward.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Contributor Information
Aliakbar Pourhatami, Email: a.pourhatami@modares.ac.ir.
Mohammad Kaviyani-Charati, Email: Kaviyani_ie@aut.ac.ir.
Bahareh Kargar, Email: kargar.iust@gmail.com.
Hamed Baziyad, Email: hamed_baziyad@modares.ac.ir.
Maryam Kargar, Email: mkargar16@yahoo.com.
Carlos Olmeda-Gómez, Email: olmeda@bib.uc3m.es.
References
- Alfieri, A. A., Alfieri, A. F., & Takiuchi, E. (2016). Detection of Bovine Coronavirus by Conventional Reverse Transcription Polymerase Chain Reaction. In: Animal Coronaviruses, Leyi Wang (ed.); pp. 101–113. Humana Press, New York, NY. 10.1007/978-1-4939-3414-0_9
- Ambali AG, Jones RC. Early pathogenesis in chicks of infection with an enterotropic strain of infectious bronchitis virus. Avian Diseases. 1990;34(4):809–817. doi: 10.2307/1591367. [DOI] [PubMed] [Google Scholar]
- Arango Duque G, Descoteaux A. Macrophage cytokines: Involvement in immunity and infectious diseases. Frontiers in Immunology. 2014;5:491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar, R. L. (2014). Immunological Detection and Characterization. In: Viral Infections of Humans: Epidemiology and Control, R. A. Kaslow, L. R. Stanberry, & J. W. Le Duc (eds.); pp. 47–62. Springer US. 10.1007/978-1-4899-7448-8_3
- Balboni A, Gallina L, Palladini A, Prosperi S, Battilani M. A real-time PCR assay for bat SARS-like coronavirus detection and its application to italian greater horseshoe bat faecal sample surveys. The Scientific World Journal. 2012 doi: 10.1100/2012/989514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):41. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian, M., Heymann, S., & Jacomy, M. (2009). Gephi: An Open Source Software for Exploring and Manipulating Networks. Proceedings of the International AAAI Conference on Web and Social Media, 3(1). https://ojs.aaai.org/index.php/ICWSM/article/view/13937
- Baziyad, H., Shirazi, S., Hosseini, S., & Norouzi, R. (2019). Mapping the intellectual structure of epidemiology with use of co-word analysis. Journal of Biostatistics and Epidemiology. 10.18502/jbe.v5i3.3618
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008(10):P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. Journal of Virology. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyack, K.W. & Klavans, R. (2019). Creation an analysis of large-scale bibliometric networks. In: Springer Handbook of Science and Technology Indicators, W. Glanzel, H.F. Moed, U. Schmoch, M. Thelwall (eds.); pp. 187–212. Springer Handbooks. Springer, Cham. 10.1007/978-3-030-02511-3_8
- Callon M, Courtial JP, Laville F. Co-word analysis as a tool for describing the network of interactions between basic and technological research: The case of polymer chemistry. Scientometrics. 1991;22(1):155–205. doi: 10.1007/BF02019280. [DOI] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathology. 2005;34(6):439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh, David. (1995). The Coronavirus Surface Glycoprotein. In: The Coronaviridae The viruses. S. G. Siddell (ed.); pp. 73–113. Springer US. 10.1007/978-1-4899-1531-3_5
- CDC. Severe Acute Respiratory Syndrome. (n.d.). Retrieved December 20, 2018, from https://www.cdc.gov/sars/about/fs sars.html
- Chen C, Song M. Visualizing a field of research: A methodology of systematic scientometric reviews. PLoS ONE. 2019;14(10):e0223994. doi: 10.1371/journal.pone.0223994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Li J, Deng Z, Xiong W, Wang Q, Hu Y. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a Microarray Hybridization Assay. Intervirology. 2010;53(2):95–104. doi: 10.1159/000264199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Guo D. Molecular mechanisms of coronavirus RNA capping and methylation. Virologica Sinica. 2016;31(1):3–11. doi: 10.1007/s12250-016-3726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. (2019) In Silico Oncology Drug Repositioning and Polypharmacology. In: Cancer Bioinformatics. Methods in Molecular Biology, vol 1878, A. Krasnitz (eds). Humana Press, New York, NY. 10.1007/978-1-4939-8868-6_15 [DOI] [PubMed]
- Cheng F, Murray JL, Rubin DH. Drug repurposing: New treatments for Zika virus infection? Trends in Molecular Medicine. 2016;22(11):919–921. doi: 10.1016/j.molmed.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Chiu W-T, Huang J-S, Ho Y-S. Bibliometric analysis of severe acute respiratory syndrome-related research in the beginning stage. Scientometrics. 2004;61(1):69–77. doi: 10.1023/B:SCIE.0000037363.49623.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobo MJ, López-Herrera AG, Herrera-Viedma E, Herrera F. Science mapping software tools: Review, analysis, and cooperative study among tools. Journal of the American Society for Information Science and Technology. 2011;62(7):1382–1402. doi: 10.1002/asi.21525. [DOI] [Google Scholar]
- Colvero LP, Villarreal LYB, Torres CA, Brañdo PE. Assessing the economic burden of avian infectious bronchitis on poultry farms in Brazil. Review of Science and Technology. 2015;34(3):993–999. doi: 10.20506/rst.34.3.2411. [DOI] [PubMed] [Google Scholar]
- Cook, J. K. A., & Mockett, A. P. A. (1995). Epidemiology of Infectious Bronchitis Virus. In: The Coronaviridae. S. G. Siddell (ed.); pp. 317–335. Springer US. 10.1007/978-1-4899-1531-3_15
- Corrales-Garay D, Ortiz-de-Urbina-Criado M, Mora-Valentín E-M. Knowledge areas, themes and future research on open data: A co-word analysis. Government Information Quarterly. 2019;36(1):77–87. doi: 10.1016/j.giq.2018.10.008. [DOI] [Google Scholar]
- da Costa VG, Moreli ML, Saivish MV. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Archives of Virology. 2020;165(7):1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cássia Nasser Cubel Garcia, R. (2016). Molecular Characterization of Canine Coronavirus. In Animal Coronaviruses, Leyi Wang (ed.); pp. 189–198). Springer New York. 10.1007/978-1-4939-3414-0_17
- Dehdarirad T, Villarroya A, Barrios M. Research trends in gender differences in higher education and science: A co-word analysis. Scientometrics. 2014;101(1):273–290. doi: 10.1007/s11192-014-1327-2. [DOI] [Google Scholar]
- Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, Meyer RF, Tong S, Cook BT, Holloway BP, McCaustland KA, Rota PA, Bankamp B, Lowe LE, Ksiazek TG, Bellini WJ, Anderson LJ. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerging Infectious Diseases. 2004;10(2):311–316. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enosawa M, Kageyama S, Sawai K, Watanabe K, Notomi T, Onoe S, Mori Y, Yokomizo Y. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp paratuberculosis. Journal of Clinical Microbiology. 2003;41(9):4359–4365. doi: 10.1128/JCM.41.9.4359-4365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An Overview of Their Replication and Pathogenesis. In: Coronaviruses: Methods and Protocols, H. J. Maier, E. Bickerton, & P. Britton (eds.); pp. 1–23). Springer New York. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed]
- Ferrer RA, Klein WMP. Risk perceptions and health behavior. Current Opinion in Psychology. 2015;5:85–89. doi: 10.1016/j.copsyc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano AJ, Gadani SP, Kipnis J. How and why do T cells and their derived cytokines affect the injured and healthy brain? Nature Reviews Neuroscience. 2017;18(6):375–384. doi: 10.1038/nrn.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthal DN, Moog C. Fc receptor-mediated antiviral antibodies. Current Opinion in HIV and AIDS. 2009;4(5):388–393. doi: 10.1097/COH.0b013e32832f0a89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Riley S, Anderson RM, Ferguson NM. Factors that make an infectious disease outbreak controllable. Proceedings of the National Academy of Sciences. 2004;101(16):6146–6151. doi: 10.1073/pnas.0307506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan C, Wang W. Research characteristics and status on social media in China: A bibliometric and co-word analysis. Scientometrics. 2015;105(2):1167–1182. doi: 10.1007/s11192-015-1723-2. [DOI] [Google Scholar]
- Ganesan A, Coote ML, Barakat K. Molecular dynamics-driven drug discovery: Leaping forward with confidence. Drug Discovery Today. 2017;22(2):249–269. doi: 10.1016/j.drudis.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Gaunt, E. R., Hardie, A., Claas, E. C. J., Simmonds, P., & Templeton, K. E. (2010). Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected over 3 Years Using a Novel Multiplex Real-Time PCR Method. Journal of Clinical Microbiology, 48(8), 2940 LP – 2947. 10.1128/JCM.00636-10 [DOI] [PMC free article] [PubMed]
- Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakos M, Papamitsiou Z, Markopoulos P, Read J, Hourcade JP. Mapping child–computer interaction research through co-word analysis. International Journal of Child-Computer Interaction. 2020;23–24:100165. doi: 10.1016/j.ijcci.2020.100165. [DOI] [Google Scholar]
- Gorbalenya AE, Snijder EJ, Spaan WJM. Severe acute respiratory syndrome coronavirus phylogeny: Toward consensus. Journal of Virology. 2004;78(15):7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Plüddemann A. Role of macrophages in autoimmunity. The Autoimmune Diseases: Fifth Edition; 2013. [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang X-H, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus Type 1 virus particles. Journal of Virology. 2005;79(8):5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan, N. S., & Cavacini, L. A. (2019). 15 - Immunoglobulin Function. In: Clinical Immunology. R. R. Rich, T. A. Fleisher, W. T. Shearer, H. W. Schroeder, A. J. Frew, & C. M. Weyand (eds.); pp. 223–233.e1). 10.1016/B978-0-7020-6896-6.00015-6
- Haghani M, Bliemer M. Covid-19 pandemic and the unprecedent mobilisation of scholarly efforts prompted by a health crisis: Scientometric comparisons across SARS, MERS and 2019-nCOV literature. Scientometrics. 2020;125(3):2695–2726. doi: 10.1007/s11192-020-03706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrop, P., Napier, E., Das, R., & Tsao, N. (2016). Wearable technology for animals 2017–2027: technologies, markets, forecasts. IDTechEx Research.
- Hartmann K. Feline infectious peritonitis. Veterinary Clinics: Small Animal Practice. 2005;35(1):39–79. doi: 10.1016/j.cvsm.2004.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise, M. T. (2014). Viral Pathogenesis. Reference Module in Biomedical Sciences, B978–0–12–801238–3.00079–9. 10.1016/B978-0-12-801238-3.00079-9
- Hendlich M. Databases for protein-ligand complexes. Acta Crystallographica Section D. 1998;54(6):1178–1182. doi: 10.1107/S0907444998007124. [DOI] [PubMed] [Google Scholar]
- Hillisch A, Heinrich N, Wild H. Computational chemistry in the pharmaceutical industry: from childhood to adolescence. ChemMedChem. 2015;10(12):1958–1962. doi: 10.1002/cmdc.201500346. [DOI] [PubMed] [Google Scholar]
- Hosseini S, Baziyad H, Norouzi R, Jabbedari Khiabani S, Gidófalvi G, Albadvi A, Alimohammadi A, Seyedabrishami S. Mapping the intellectual structure of GIS-T field (2008–2019): A dynamic co-word analysis. Scientometrics. 2021 doi: 10.1007/s11192-020-03840-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Hoz-Correa A, Muñoz-Leiva F, Bakucz M. Past themes and future trends in medical tourism research: A co-word analysis. Tourism Management. 2018;65:200–211. doi: 10.1016/j.tourman.2017.10.001. [DOI] [Google Scholar]
- Hu J, Zhang Y. Research patterns and trends of recommendation system in China using co-word analysis. Information Processing & Management. 2015;51(4):329–339. doi: 10.1016/j.ipm.2015.02.002. [DOI] [Google Scholar]
- Jaccard P. Distribution comparée de la flore alpine dans quelques régions des Alpes occidentales et orientales. Bulletin De La Murithienne. 1902;31:81–92. [Google Scholar]
- Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE. 2014;9(6):e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassab, C., Kerrigan, B. P., Caruso, H., Al Enazy, S., & Heimberger, A. B. (2019). Immunomodulatory Methods. In: Nervous System Drug Delivery. Principles and Practice. R. R. Lonser, M. Sarntinoranont, & K. Bankiewicz (eds.); pp. 297–334). Academic Press. 10.1016/B978-0-12-813997-4.00015-3
- Khasseh AA, Soheili F, Moghaddam HS, Chelak AM. Intellectual structure of knowledge in iMetrics: A co-word analysis. Information Processing & Management. 2017;53(3):705–720. doi: 10.1016/j.ipm.2017.02.001. [DOI] [Google Scholar]
- Krewski, D., Turner, M. C., & Tyshenko, M. G. (2011). Risk Management in Environmental Health Decision. In: Encyclopedia of Environmental Health. J. O. Nriagu (ed.); pp. 868–877). Elsevier. 10.1016/B978-0-444-52272-6.00621-8
- Lambiotte, R., Delvenne, J.-C., & Barahona, M. (2008). Laplacian dynamics and multiscale modular structure in networks. ArXiv Preprint.
- Liu G-Y, Hu J-M, Wang H-L. A co-word analysis of digital library field in China. Scientometrics. 2012;91(1):203–217. doi: 10.1007/s11192-011-0586-4. [DOI] [Google Scholar]
- Lou J, Tian S-J, Niu S-M, Kang X-Q, Lian H-X, Zhang L-X, Zhang J-J. Coronavirus disease 2019: A bibliometric analysis and review. European Review for Medical and Pharmacological Sciences. 2020;24(6):3411–3421. doi: 10.26355/eurrev_202003_20712. [DOI] [PubMed] [Google Scholar]
- Mandal S, Bhatnagar T, Arinaminpathy N, Agarwal A, Chowdhury A, Murhekar M, Gangakhedkar RR, Sarkar S. Prudent public health intervention strategies to control the coronavirus disease 2019 transmission in India: A mathematical model-based approach. The Indian Journal of Medical Research. 2020;151(2–3):190. doi: 10.4103/ijmr.IJMR_504_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maria N, Zaid A, Catrin S, Ahmed K, Ahmed A-J, Christos I, Maliha A, Riaz A. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. International Journal of Surgery. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RM, McLean AR, Pattison J, Weiss RA, Holmes EC, Rambaut A. Viral evolution and the emergence of SARS coronavirus. Philosophical Transactions of the Royal Society of London Series b: Biological Sciences. 2004;359(1447):1059–1065. doi: 10.1098/rstb.2004.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miszczak, F., Kin, N., Tesson, V., & Vabret, A. (2016). Real-Time RT-PCR Detection of Equine Coronavirus BT - Animal Coronaviruses (Leyi Wang (Ed.); pp. 93–100). Springer New York. 10.1007/978-1-4939-3414-0_8
- Mohammed MN, Syamsudin H, Al-Zubaidi S, Sairah A, Ramli R, Yusuf E. Novel COVID-19 detection and diagnosis system using IOT based smart helmet. International Journal of Psychosocial Rehabilitation. 2020;24(7):2296–2303. [Google Scholar]
- Muegge I, Bergner A, Kriegl JM. Computer-aided drug design at Boehringer Ingelheim. Journal of Computer-Aided Molecular Design. 2017;31(3):275–285. doi: 10.1007/s10822-016-9975-3. [DOI] [PubMed] [Google Scholar]
- Newman MEJ. Fast algorithm for detecting community structure in networks. Physical Review E. 2004;69(6):66133. doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- Nguyen D. Mapping knowledge domains of non-biomedical modalities: A large-scale co-word analysis of literature 1987–2017. Social Science & Medicine. 2019;233:1–12. doi: 10.1016/j.socscimed.2019.05.044. [DOI] [PubMed] [Google Scholar]
- Oguh, C. E., Obiwulu, E. N. O., Oniwon, W. O., Okekeaji, U., Ugwu, C. V, Umezinwa, O. J., & Osuji, C. A. (2020). Structure and Function of COVID-19 Encode Proteins in the Transcription and Replication Mechanism with Its Preventive Measures and Propose Efficacy Treatments: A Critical Systematic Review. Asian Journal of Immunology, 15–29.
- Olmeda-Gómez C, Ovalle-Perandones M-A, Perianes-Rodríguez A. Co-word analysis and thematic landscapes in Spanish information science literature, 1985–2014. Scientometrics. 2017;113(1):195–217. doi: 10.1007/s11192-017-2486-8. [DOI] [Google Scholar]
- Pal JK. Visualizing the knowledge outburst in global research on COVID-19. Scientometrics. 2021 doi: 10.1007/s11192-021-03912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Mitchell MS. What happens in biological therapy and immunotherapy. Cureus. 2012 doi: 10.7759/cureus. [DOI] [Google Scholar]
- Peeri NC, Shrestha N, Rahman MS, Zaki R, Tan Z, Bibi S, Baghbanzadeh M, Aghamohammadi N, Zhang W, Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? International Journal of Epidemiology. 2020;49(3):717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar S, Agrahari A, Singh SN. Mapping the intellectual structure of scientometrics: A co-word analysis of the journal Scientometrics (2005–2010) Scientometrics. 2015;102(1):929–955. doi: 10.1007/s11192-014-1402-8. [DOI] [Google Scholar]
- Rester U. From virtuality to reality - virtual screening in lead discovery and lead optimization: A medicinal chemistry perspective. Current Opinion in Drug Discovery & Development. 2008;11(4):559–568. [PubMed] [Google Scholar]
- Ringe R, Bhattacharya J. Preventive and therapeutic applications of neutralizing antibodies to Human Immunodeficiency virus type 1 (HIV-1) Therapeutic Advances in Vaccines. 2013;1(2):67–80. doi: 10.1177/2051013613494534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollinger, J. M., Stuppner, H., & Langer, T. (2008). Virtual screening for the discovery of bioactive natural products. In: Natural Compounds as Drugs Volume I Progress in Drug Research, vol 65.F. Petersen & R. Amstutz (eds.); pp. 211–249). Birkhäuser Basel. 10.1007/978-3-7643-8117-2_6 [DOI] [PMC free article] [PubMed]
- Salman, F. M., Abu-Naser, S. S., Alajrami, E., & Abu-Nasser, Bassem S.; Alashqar, B. A. M. (2020). COVID-19 detection using artificial intelligence. http://dstore.alazhar.edu.ps/xmlui/handle/123456789/587
- Schulz LL, Tonsor GT. Assessment of the economic impacts of porcine epidemic diarrhea virus in the United States. Journal of Animal Science. 2015;93(11):5111–5118. doi: 10.2527/jas.2015-9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Zhou Y, Ye J, Abdullah AL-maskri AA, Kang Y, Zeng S, Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. Journal of Pharmaceutical Analysis. 2020;10(2):97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau W-L, Dwivedi YK, Tsai C-H. Supply chain management: exploring the intellectual structure. Scientometrics. 2015;105(1):215–230. doi: 10.1007/s11192-015-1680-9. [DOI] [Google Scholar]
- Śledź P, Caflisch A. Protein structure-based drug design: from docking to molecular duynamics. Current opinion in structural biology. 2018;48:93–102. doi: 10.1016/j.sbi.2017.10.010. [DOI] [PubMed] [Google Scholar]
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends in Microbiology. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs, B. (2019). Science mapping and the identification of topics: Theoretical and methodological considerations. In: Springer Handbook of Science and Technology Indicators, W. Glanzel, H.F. Moed, U. Schmoch, M. Thelwall (eds.), pp. 213–233. Springer Handbooks. Springer, Cham. 10.1007/978-3-030-02511-3_9
- Tuckerman ME, Martyna GJ. Understanding modern molecular dynamics: Techniques and applications. The Journal of Physical Chemistry B. 2000;104(2):159–178. doi: 10.1021/jp992433y. [DOI] [Google Scholar]
- Tuen M, Visciano ML, Chien PC, Jr, Cohen S, Chen P, Robinson J, He Y, Pinter A, Gorny MK, Hioe CE. Characterization of antibodies that inhibit HIV gp120 antigen processing and presentation. European Journal of Immunology. 2005;35(9):2541–2551. doi: 10.1002/eji.200425859. [DOI] [PubMed] [Google Scholar]
- Uhlenhaut C, Cohen JI, Pavletic S, Illei G, Gea-Banacloche JC, Abu-Asab M, Krogmann T, Gubareva L, McClenahan S, Krause PR. Use of a novel virus detection assay to identify coronavirus HKU1 in the lungs of a hematopoietic stem cell transplant recipient with fatal pneumonia. Transplant Infectious Disease. 2012;14(1):79–85. doi: 10.1111/j.1399-3062.2011.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viedma-Del-Jesus MI, Perakakis P, Muñoz MÁ, López-Herrera AG, Vila J. Sketching the first 45 years of the journal Psychophysiology (1964–2008): A co-word-based analysis. Psychophysiology. 2011;48(8):1029–1036. doi: 10.1111/j.1469-8986.2011.01171.x. [DOI] [PubMed] [Google Scholar]
- Wang L-F, Shi Z, Zhang S, Field H, Daszak P, Eaton BT. Review of bats and SARS. Emerging Infectious Diseases. 2006;12(12):1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Leyi, & Zhang, Y. (2016). Animal Coronaviruses: A Brief Introduction. In: Animal Coronaviruses. Leyi Wang (ed.); pp. 3–11). Springer Protocols Handbooks. New York. 10.1007/978-1-4939-3414-0_1
- Wang, Lisheng, Wang, Y., Ye, D., & Liu, Q. (2020). Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. International Journal of Antimicrobial Agents, 55(6), 105948. 10.1016/j.ijantimicag.2020.105948 [DOI] [PMC free article] [PubMed]
- Wang Z, Zhao H, Wang Y. Social networks in marketing research 2001–2014: A co-word analysis. Scientometrics. 2015;105(1):65–82. doi: 10.1007/s11192-015-1672-9. [DOI] [Google Scholar]
- Weiss, S. R., & Leibowitz, J. L. (2011). Coronavirus Pathogenesis. In: Advances in Virus Research. K. Maramorosch, A. J. Shatkin, & F. A. Murphy (eds.); Vol. 81, pp. 85–164. Academic Press. 10.1016/B978-0-12-385885-6.00009-2 [DOI] [PMC free article] [PubMed]
- WHO. Middle East Respiratory Syndrome Coronavirus. (n.d.). Retrieved November 10, 2018, from http://www.who.int/ emergencies/mers-cov/en
- Wilson GL, Lill MA. Integrating structure-based and ligand-based approaches for computational drug design. Future Medicinal Chemistry. 2011;3(6):735–750. doi: 10.4155/fmc.11.18. [DOI] [PubMed] [Google Scholar]
- Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-C, Leu H-J. Examining the trends of technological development in hydrogen energy using patent co-word map analysis. International Journal of Hydrogen Energy. 2014;39(33):19262–19269. doi: 10.1016/j.ijhydene.2014.05.006. [DOI] [Google Scholar]
- Xie J, Szymanski BK. Community detection using a neighborhood strength driven label propagation algorithm. IEEE Network Science Workshop. 2011;2011:188–195. doi: 10.1109/NSW.2011.6004645. [DOI] [Google Scholar]
- Xu S, Hao L, An X, Pang H, Li T. Review of emerging research topics with key-route main path analysis. Scientometrics. 2020;122(1):607–624. doi: 10.1007/s11192-019-03288-5. [DOI] [Google Scholar]
- Yan B-N, Lee T-S, Lee T-P. Mapping the intellectual structure of the Internet of Things (IoT) field (2000–2014): A co-word analysis. Scientometrics. 2015;105(2):1285–1300. doi: 10.1007/s11192-015-1740-1. [DOI] [Google Scholar]
- Yang K-L, Jin X-Y, Gao Y, Xie J, Liu M, Zhang J-H, Tian J-H. Bibliometric analysis of researches on traditional Chinese medicine for coronavirus disease 2019 (COVID-19) Integrative Medicine Research. 2020;9(3):100490. doi: 10.1016/j.imr.2020.100490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-R, Li Y, Liu J-S, Chen Y-D, Chai L-H. A dynamic co-word network-related approach on the evolution of China’s urbanization research. Scientometrics. 2017;111(3):1623–1642. doi: 10.1007/s11192-017-2314-1. [DOI] [Google Scholar]
- Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, Zheng X-S, Li B, Li J-M, Guo H, Pei G-Q, An X-P, Chen J-W, Zhou L, Mai K-J, Ma J-Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyoud SH. Global research trends of Middle East respiratory syndrome coronavirus: A bibliometric analysis. BMC Infectious Diseases. 2016;16(1):255. doi: 10.1186/s12879-016-1600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]