Abstract
The lymphatic system is composed of a hierarchical network of fluid absorbing lymphatic capillaries and transporting collecting vessels. Despite distinct functions and morphologies, molecular mechanisms that regulate the identity of the different vessel types are poorly understood. Through transcriptional analysis of murine dermal lymphatic endothelial cells (LECs), we identified Foxp2, a member of the FOXP family of transcription factors implicated in speech development, as a collecting vessel signature gene. FOXP2 expression was induced after initiation of lymph flow in vivo and upon shear stress on primary LECs in vitro. Loss of FOXC2, the major flow‐responsive transcriptional regulator of lymphatic valve formation, abolished FOXP2 induction in vitro and in vivo. Genetic deletion of Foxp2 in mice using the endothelial‐specific Tie2‐Cre or the tamoxifen‐inducible LEC‐specific Prox1‐CreERT2 line resulted in enlarged collecting vessels and defective valves characterized by loss of NFATc1 activity. Our results identify FOXP2 as a new flow‐induced transcriptional regulator of collecting lymphatic vessel morphogenesis and highlight the existence of unique transcription factor codes in the establishment of vessel‐type‐specific endothelial cell identities.
Keywords: lymphatic vessel, shear stress, valve
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Development & Differentiation; Vascular Biology & Angiogenesis
Speech‐linked FOXP2 is a specific marker of collecting lymphatic vessels that regulates lymphatic vessel and valve morphogenesis downstream of flow‐responsive transcriptional regulator FOXC2.
Introduction
The lymphatic vascular system maintains tissue fluid balance and immune homeostasis through a coordinated action of lymphatic capillaries (also known as initial lymphatics) and collecting lymphatic vessels. The blind‐ended lymphatic capillaries are composed of endothelial cells with discontinuous button junctions that allow entry of excess extracellular fluid and immune cells (reviewed in (Potente & Mäkinen, 2017; Oliver et al, 2020; Petrova & Koh, 2020)). In contrast, the lymphatic capillary‐draining collecting vessels have continuous zipper junctions that prevent excess leakage of fluid. Additional unique characteristics of the collecting lymphatic vessels are the smooth muscle coverage that facilitates fluid propulsion through contractions and the existence of bicuspid valves that prevent fluid backflow (Zawieja, 2009). The establishment of a hierarchy of functionally specialized vessel types is critical for normal lymphatic vascular function and, consequently, failure in this process can lead to lymphatic diseases.
During embryonic development, lymphatic vessel formation occurs through transdifferentiation of venous into lymphatic endothelial cells (LECs), with contribution of additional non‐venous sources in certain organs (reviewed in (Ulvmar & Mäkinen, 2016; Kazenwadel & Harvey, 2018)). The key regulator of LEC fate is the homeobox transcription factor PROX1 that is required for the formation of all lymphatic vascular beds (Wigle & Oliver, 1999). PROX1 co‐operates with other transcription factors in regulating the LEC lineage‐specific transcriptome. For example, the venous endothelial cell (EC) fate regulator COUP‐TFII (You et al, 2005) is required for initiation of PROX1 expression and regulates lymphatic vessel formation through heterodimerization with PROX1 (Lee et al, 2009; Yamazaki et al, 2009; Lin et al, 2010; Srinivasan et al, 2010). The subsequent maturation of the primitive lymphatic vascular plexus into functional collecting vessels is controlled by an interplay between mechanical forces, induced by initiation of lymph flow, and another set of transcription factors including GATA2 and FOXC2 (Sabine et al, 2012; Kazenwadel et al, 2015). Oscillatory shear stress, mimicking turbulent flow in the branched network of primitive vessel plexus, induces upregulation of GATA2 and FOXC2 (Sabine et al, 2012; Kazenwadel et al, 2015). These factors subsequently regulate a transcriptional program required for the formation of lymphatic valves. Deficiency of GATA2 or FOXC2, or mutations in their DNA‐binding domains consequently cause abnormal development of lymphatic valves and underlie human hereditary lymphedemas (Petrova et al, 2004; Ostergaard et al, 2011; Kazenwadel et al, 2012, 2015). While the mechanisms that specify and sustain the identity of the phenotypically distinct valve LECs are characterized with increasing detail (Geng et al, 2017), the pathways that specify the functionally different collecting vessels and lymphatic capillaries have not been delineated.
Here, we aimed to determine the mechanism that regulates the functional specification of collecting lymphatic vessels through transcriptional analysis of LECs of distinct vessel subtypes. We identified forkhead box protein P2 (FOXP2), previously implicated in the development of speech and language in humans (Co et al, 2020), as a collecting vessel‐specific transcription factor. Using cultured primary LECs and genetic mouse models, we further show that FOXP2 is transcriptionally induced by flow‐mediated shear stress and regulates collecting lymphatic vessel and valve morphogenesis through co‐operation with the major flow‐responsive FOXC2/NFATc1 signaling pathway. Our results highlight the existence of unique transcription factor codes in the establishment of vessel‐type‐specific LEC identities that may provide an opportunity to exploit for therapeutic restoration of specific vessel functions.
Results
Global transcriptome analysis of endothelial cells of lymphatic vessel subtypes
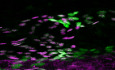
To identify genes regulating the functional specification of collecting lymphatic vessels, we performed transcriptome profiling of dermal ECs isolated from adult mouse ear skin by flow cytometry. Dermal cell suspensions from mice carrying a LEC reporter Prox1‐GFP were first subjected to enrichment for ECs using PECAM1 antibody‐coated magnetic beads, followed by sorting of live blood EC (BEC) and LEC populations based on the expression of the lymphatic markers PDPN, LYVE1, and Prox1‐GFP (Fig 1A). LEC subpopulations were further defined by the level of LYVE1 expression (Mäkinen et al, 2005) and assigned as LYVE1high lymphatic capillaries, LYVE1low/− collecting vessels, and LYVE1intermed pre‐collecting vessels (Fig 1A). Capillary LECs accounted for a majority (60.9 ± 0.2% [n = 3]) of the LEC population (Fig EV1A).
Figure 1. Transcriptome analysis of lymphatic vessels identifies FOXP2 as a collecting vessel‐specific transcription factor.
- Isolation of dermal EC subtypes from the ear skin of 5‐week‐old Prox1‐GFP mice using multicolor flow cytometry. Whole‐mount immunofluorescence for LYVE1 (on the left) defines three Prox1‐GFP + lymphatic vessels subtypes that are sorted using the indicated gating scheme (on the right): LYVE1low/− collecting vessel (green arrow, col); LYVE1intermed pre‐collecting vessel (gray arrow, pre‐col); LYVE1high lymphatic capillary (magenta arrow, cap).
- Top 100 genes upregulated in collecting in comparison to lymphatic capillary LECs. Expression in BECs is shown for comparison. Heat map color coding shows log2 fold change. Red box indicates LEC‐specific/enriched collecting vessel signature genes that include Foxp2 (red arrow).
- Whole‐mount immunofluorescence of ear skin, mesentery, and flank, showing nuclear FOXP2 staining in collecting lymphatic vessels (arrows, col), but not in LYVE1+ lymphatic capillaries (cap) or blood vessels (bv).
- qRT–PCR analysis of Foxp1–4 in murine ECs freshly isolated from P11 mesentery (n = 6 mice, individual data points shown). Data are presented as mean relative expression (normalized to Gapdh) ± SD. Transcript levels for each transcript are presented relative to levels in LECs. P, Student's t‐test. nd, not detected.
Data information: Scale bar: 75 µm (A), 50 µm (C).
Source data are available online for this figure.
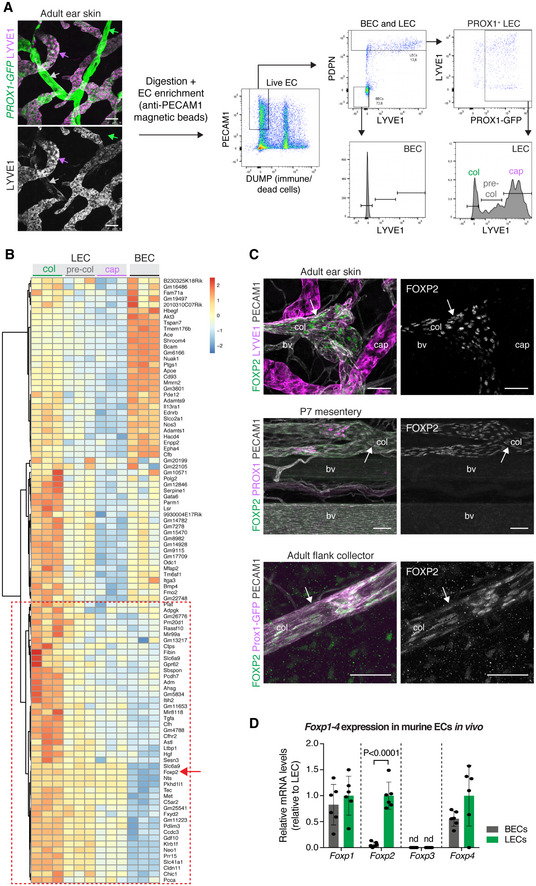
Figure EV1. Transcriptome analysis of dermal endothelial cells.
- Proportion of LEC subpopulations (of the total LEC population) sorted from mouse ear skin based on expression of LYVE1, PDPN, and Prox1‐GFP as shown in Fig 1A (n = 3 samples). Horizontal line indicates mean. Col, collecting vessel; pre‐col, pre‐collecting vessel; cap, lymphatic capillary.
- Heat map showing differential expression of 1,191 genes between dermal LEC and BEC. Color coding shows log2 fold change. Gene ID list is provided in Dataset EV1.
- Intensity bar plots of cell lineage/phenotype genes showing expression in dermal EC populations (each bar represents one sample, n = 3). Y‐axis represents expression level (signal intensity value given in arbitrary units (AU)).
- Intensity bar plot of Foxp2 showing expression in dermal EC populations (n = 3). Y‐axis represents expression level (signal intensity value given in arbitrary units (AU)).
Affymetrix GeneChip analysis of the isolated EC populations revealed 1,191 genes that were differentially expressed (FDR < 0.02, log2 fold change > 1 or < −1) between BECs and (all) LECs, including the established BEC‐LEC lineage markers Flt1, Nrp1, and Prox1 (Fig EV1B and C, Dataset EV1). As expected, Lyve1 was highly expressed in LECs of lymphatic capillaries and pre‐collecting vessels (Fig EV1C). In contrast, the valve‐LEC‐specific Cldn11 (Takeda et al, 2019) was highly expressed in LECs of collecting and pre‐collecting vessels (Fig EV1C). Pdgfrb expression suggested contamination of the BEC, but not LEC population with mural cells (Fig EV1C), which is commonly observed in bulk‐sorted BECs due to close association of the two cell types (Vanlandewijck et al, 2018). No significant contamination by Lum + fibroblasts was observed in any of the EC populations (Fig EV1C). These results indicate a successful generation of a microarray dataset for differential gene expression analysis of LECs of lymphatic capillaries and collecting vessels (Dataset EV2).
The transcription factor FOXP2 is specifically expressed in endothelial cells of collecting lymphatic vessels
Next, we focused on collecting lymphatic vessel enriched genes (Fig 1B, Dataset EV3) that also showed LEC‐specific pattern of expression with no or low expression in the BECs (Fig 1B, boxed). Among these was the gene encoding the transcription factor forkhead box protein P2 (FOXP2; Figs 1B and EV1D) that has previously been studied in the nervous system and implicated in several cognitive functions including development of speech and language in humans ((Lai et al, 2001), reviewed in (Co et al, 2020)), and lung development (Shu et al, 2007). Its role in other organ systems, including the vasculature, is not known.
Selective expression of Foxp2 in collecting lymphatic vessels suggested involvement in establishing vessel‐type‐specific LEC identity. To first validate the transcriptome data, we performed whole‐mount immunofluorescence staining of adult mouse ear skin. Staining with antibodies against FOXP2 revealed nuclear expression in LYVE1− collecting lymphatic vessels, but not in LYVE1+ lymphatic capillaries or in blood vessels (Fig 1C). Uniform expression of FOXP2 was detected along the collecting vessel, including the luminal valves composed of LECs expressing high levels of PROX1 (Fig 1C and Appendix Fig S1A). FOXP2 was also expressed in the mesenteric collecting vessels and large flank collectors (Fig 1C), but not in lymphatic capillaries of the intestinal villi (lacteals) or the diaphragm (Appendix Fig S1B), or in the “lymphatic‐like” Schlemm's canal in the eye (Appendix Fig S1C). qRT–PCR analysis of BECs and LECs freshly isolated from the mesentery of P11 mice by flow cytometry further demonstrated that Foxp2 was the only differentially expressed member of the FOXP family of transcription factors (Fig 1D), which was also supported by the dermal array data (Appendix Fig S1D). The closely related Foxp1 and Foxp4 were expressed in both BECs and LECs, while Foxp3 was not detected (Fig 1D). Taken together, these results identify FOXP2 as a potential new transcriptional regulator of collecting lymphatic vessel identity and morphogenesis.
FOXP2 expression is regulated by flow
To investigate whether FOXP2 is functionally important for collecting lymphatic vessel formation, we first analyzed its expression in the developing vasculature. We studied the mesenteric lymphatic vessels that begin to form at embryonic (E)13 through lymphvasculogenic assembly of LEC progenitors (Stanczuk et al, 2015), and undergo remodeling and valve morphogenesis after the onset of lymph flow at E15‐E16 (Bazigou et al, 2009; Norrmén et al, 2009; Sabine et al, 2015). Whole‐mount immunofluorescence staining of embryonic mesenteries showed no expression of FOXP2 between E15‐E16 (Fig 2A), suggesting that FOXP2 is not involved in the early steps of collecting vessel formation. FOXP2 expression was first observed at E17, and its expression was maintained in the postnatal mesenteric lymphatic vessels (Figs 1C and 2A). FOXP2 expression was not restricted to PROX1high valves but showed a uniform pattern of expression in all LECs of collecting vessels from E17 onwards. A similar pattern of FOXP2 expression was observed in embryonic skin. FOXP2 was not expressed at E14 or E15, when a primary dermal lymphatic capillary plexus forms through vessel sprouting, but was upregulated at E18, when remodeling of lymphatic capillaries into collecting vessels is initiated (Fig EV2).
Figure 2. FOXP2 expression is regulated by flow.
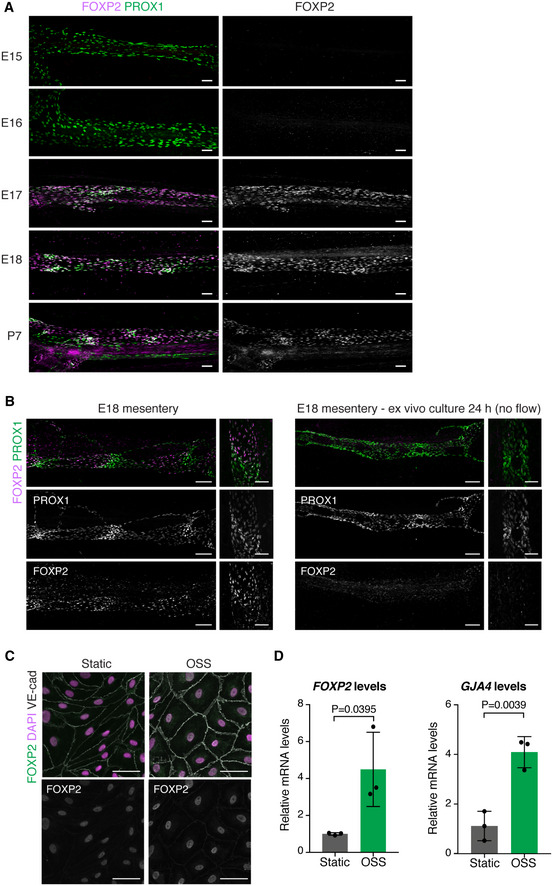
-
AWhole‐mount immunofluorescence of embryonic mesenteries of the indicated developmental stages. Note induction of FOXP2 expression at E17.
-
BWhole‐mount immunofluorescence of E18 mesenteries fixed immediately after dissection (left panel) or after 24 h of ex vivo culture (right panel). Note loss of patterning of PROX1high valve LECs and downregulation of FOXP2 expression in flow‐abrogated vessels after ex vivo culture.
-
C, DImmunofluorescence (C) and qRT–PCR analysis (D) of HDLECs grown under static conditions or exposed to OSS for 48 h (n = 3 independent experiments). Data are presented as mean ± SD. P, Student's t‐test.
Data information: Scale bar: 100 µm (A, B), 50 μm (C).
Source data are available online for this figure.
Figure EV2. FOXP2 expression in the developing dermal vasculature.
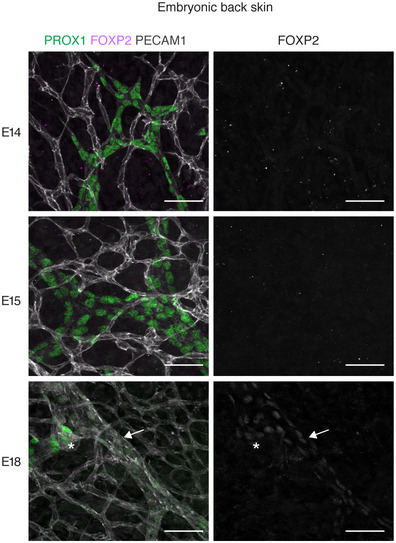
Whole‐mount immunofluorescence of embryonic back skin at the indicated developmental stages. Note expression of FOXP2 (single channel images on the right) in the developing collecting vessel (arrow) including luminal valve (asterisk) at E18. Scale bar: 50 µm.
To investigate if the induction of FOXP2 expression in vivo was related to initiation of flow in mesenteric lymphatic vessels at E15–E16, we first utilized ex vivo culture of E18 mesenteries as a model of abrogated flow (Sabine et al, 2012). As expected, ex vivo culture of vessels for 24 h led to loss of patterned valve regions composed of PROX1high LECs, although PROX1 expression level was not affected (Fig 2B). This coincided with the downregulation of FOXP2 (Fig 2B), suggesting that maintenance of FOXP2 levels/expression is dependent on flow. To directly test if flow regulates FOXP2 expression, we exposed primary human dermal LECs (HDLECs) to different types of fluid shear stress implicated in collecting lymphatic vessel and valve morphogenesis. Oscillatory shear stress (OSS) has previously been used to mimic disturbed flow in the developing lymphatic network and shown to regulate the transcriptional program controlling valve morphogenesis (Sabine et al, 2012, 2015). On the other hand, laminar shear stress (LSS) regulates lymphatic vessel remodeling as well as LEC proliferation and quiescence (Wang et al, 2016; Choi et al, 2017a, 2017b; Geng et al, 2020). Under static conditions, only a weak immunofluorescence signal for FOXP2 protein was detected in HDLECs (Fig 2C). However, exposure to OSS robustly induced FOXP2 protein (Fig 2C) and FOXP2 mRNA (Fig 2D). Notably, this was observed at 48 h but not earlier (Fig EV3A), whereas the OSS‐regulated GATA2 (Fig EV3A) was upregulated already at 24 h, as previously reported (Sweet et al, 2015). LSS led to a more modest upregulation of FOXP2 (Fig EV3B). Increased expression of genes previously shown to be regulated by OSS (GJA4 (Sabine et al, 2012), Fig 2D) or LSS (KLF4 (Choi et al, 2017a), Fig EV3B) in LECs was also observed.
Figure EV3. Regulation of FOXP2 expression by oscillatory and laminar flow.
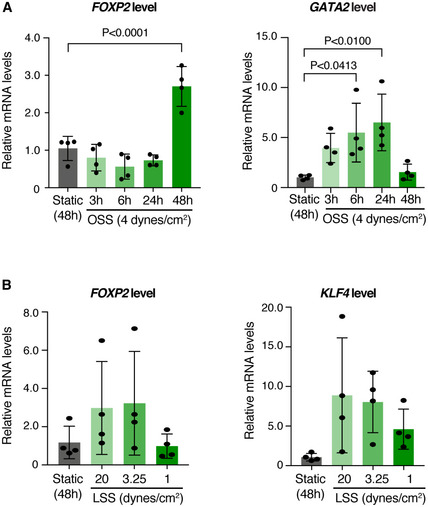
-
A, BqRT–PCR analysis of HDLECs grown under static conditions or exposed to oscillatory shear stress (OSS) for the indicated times (A), or to laminar shear stress (LSS) at the indicated dyn/cm2 for 48 h (n = 4 independent experiments). GATA2 and KLF4 are shown as controls. Data are presented as mean ± SD. P, Student's t‐test.
Source data are available online for this figure.
Taken together, these results demonstrate that FOXP2 expression is induced in vivo after the onset of flow, and regulated in vitro by OSS and, to a lesser extent, laminar flow.
Endothelial‐specific deletion of Foxp2 leads to collecting lymphatic vessel defects
In order to investigate the potential role of FOXP2 in collecting lymphatic vessel development, we genetically deleted it in all ECs using the Tie2‐Cre mice in combination with the floxed Foxp2 allele (Fig 3A). The organization and gross morphology of the blood and lymphatic vessel networks in adult ear or embryonic back skin were not affected by loss of Foxp2 (Appendix Fig S2A–C). Analysis of early postnatal mesenteric vasculature also revealed grossly normal collecting lymphatic vessel morphology in the Foxp2flox / flox; Tie2‐Cre + mutant mice in comparison with littermate controls (Fig 3B). However, staining for lymphatic valve marker integrin‐α9 (Bazigou et al, 2009) revealed shortened valve leaflets in the mutant mice (Fig 3B). Notably, FOXP2 expression was efficiently depleted in the collecting vessels of the mutant mice except for the valves. Unexpectedly, in four out of five mutant mice analyzed the majority of the valves (85 ± 30%, n = 4 mice, 19 vessels) were formed of FOXP2+ cells (Fig 3C). Valve‐specific selection of non‐recombined LECs suggests selective requirement of FOXP2 in their morphogenesis.
Figure 3. Endothelial‐specific deletion of Foxp2 leads to defective collecting vessel and valve morphogenesis.
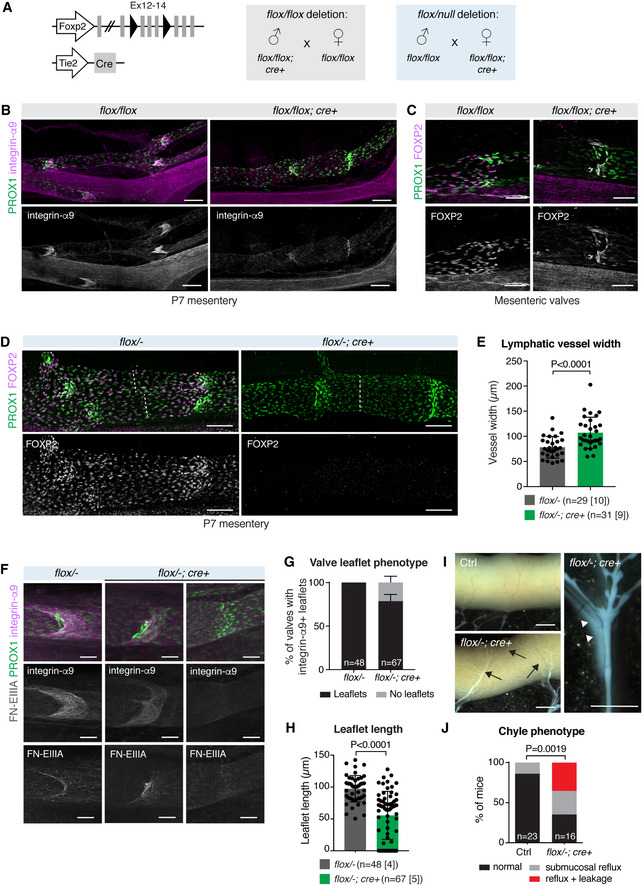
-
AGenetic constructs and breeding strategies used to generate pan‐endothelial Foxp2flox / flox;Tie2‐Cre (gray) or Foxp2flox /− ;Tie2‐Cre (blue) deletion.
-
B, CWhole‐mount immunofluorescence of P7 mesenteries, showing abnormally short integrin‐α9+ lymphatic valve leaflets (B) and selection of non‐recombined FOXP2+ LECs (C) in Foxp2flox / flox;Tie2‐Cre vessels.
-
DWhole‐mount immunofluorescence of P7 mesenteries, showing efficient FOXP2 depletion in Foxp2flox /− ;Tie2‐Cre vessels. Dotted line indicates vessel width.
-
EQuantification of mesenteric collecting lymphatic vessel width in P7 Foxp2flox /− ;Tie2‐Cre and littermate controls (n = vessels [mice], as indicated).
-
FLymphatic valves in P7 mesenteric vessels of control and Foxp2flox /− ;Tie2‐Cre mice. A spectrum of defects in the mutants ranging from shortened integrin‐α9+FN‐EIIIA+ valve leaflets (middle panels) to a complete loss of leaflets (right panels) are shown.
-
G, HQuantification of the proportion of valves with integrin‐α9+ leaflets (G), and leaflet length (H) in P7 Foxp2flox /− ;Tie2‐Cre and littermate controls (n = valves [mice], as indicated). Leaflet length = 0 corresponds to valves with no leaflets.
-
IImages of P7 intestine (on the left) and mesentery (on the right) showing submucosal reflux (arrows) and leakage (arrowheads) in the Foxp2flox /− ;Tie2‐Cre mice.
-
JChyle phenotypes in P7 Foxp2flox /− ;Tie2‐Cre and littermate controls (n = number of mice as indicated).
Data information: In (E, G, H), data are presented as mean ± SD. P, Student's t‐test. In (J), P, Fisher's exact test. Scale bar: 100 µm (B, D), 50 µm (C, F), 500 µm (I).
Source data are available online for this figure.
To maximize gene targeting efficiency, we generated mice carrying the floxed in combination with a null Foxp2 allele (Fig 3A). Efficient FOXP2 depletion in LECs, including the valves, in the Foxp2flox /− ; Tie2‐Cre + mice resulted in a spectrum of valve defects as well as increased collecting lymphatic vessel width (Fig 3D and E). A proportion (~ 25%) of valves composed of PROX1high LECs had no leaflets, as assessed by staining for integrin‐α9 and its ligand fibronectin‐EIIIA (Fig 3F and G). Quantification showed reduced leaflet length in the remaining valves in the mutants compared to littermate controls at this stage (Fig 3F and H). The mutant mice also showed chyle accumulation in the submucosal lymphatic vessel network on the intestinal wall, suggesting backflow from the mesenteric collecting lymphatic vessels, as well as chyle leakage from collecting vessels (Fig 3I and J). These defects did not, however, compromise postnatal survival and growth in the mutant mice, and analysis of adult mesenteries revealed morphologically normal valves in the Foxp2flox /− ; Tie2‐Cre + mice (Appendix Fig S2D). In agreement with the lack of expression of FOXP2 in lacteal LECs (Appendix Fig S1B), we could not observe defects in these vessels in the Foxp2flox /− ; Tie2‐Cre + mice (Appendix Fig S2E). Together, these data demonstrate a critical requirement of Foxp2 for collecting lymphatic vessel and valve morphogenesis.
Loss of FOXP2 leads to reduced expression of known regulators of lymphatic and valve development
FOXP2 is required in the nervous system for several cognitive functions, including general brain development and synaptic plasticity (Kim et al, 2019; Co et al, 2020). Interestingly, validated direct FOXP2 targets in the nervous system include genes encoding known regulators of lymphatic development (NRP2) and valve morphogenesis (EphrinB2 and SEMA3A) (Mäkinen et al, 2005; Xu et al, 2010; Vernes et al, 2011; Bouvrée et al, 2012; Jurisic et al, 2012; Ochsenbein et al, 2014).
Immunofluorescence staining of P6 mesenteric collecting vessels showed marked downregulation of NRP2 in Foxp2flox /− ; Tie2‐Cre + mice compared to littermate controls (Figs 4A and B). In addition, RNAscope‐based whole‐mount in situ hybridization revealed a significant reduction of Efnb2 transcript in Foxp2‐deficient lymphatic vessels (Fig 4C and D). FOXP2 thus regulates genes critical for normal lymphatic vessel and valve development.
Figure 4. Downregulation of FOXP2 targets Nrp2 and Efnb2 in Foxp2‐deficient lymphatic vessels.
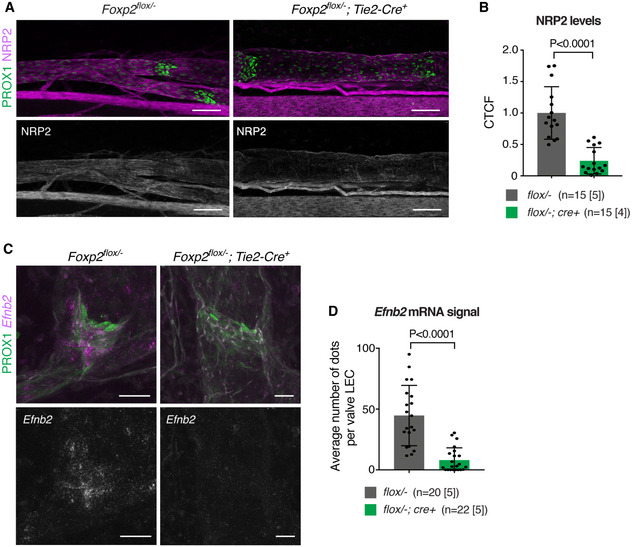
-
A, BWhole‐mount immunofluorescence (A) and quantification of NRP2 staining intensity (B) in P6 mesenteric lymphatic vessels of Foxp2flox /− ;Tie2‐Cre and littermate control mice (n = vessels [mice], as indicated).
-
C, DRNAscope‐based whole‐mount in situ hybridization (C) and quantification of Efnb2 transcript levels (D) in P7 mesenteric lymphatic vessels of Foxp2flox /− ;Tie2‐Cre and littermate control mice (n = vessels [mice], as indicated).
Data information: In (B, D), data are presented as mean ± SD. P, Student's t‐test. Scale bar: 100 µm.
Source data are available online for this figure.
FOXP2 is required LEC‐autonomously for valve formation and maintenance
To study the potential function of FOXP2 in the maintenance of lymphatic valves, we conditionally deleted Foxp2 in LECs using the tamoxifen‐inducible Prox1‐CreERT2 line (Fig 5A). Gene deletion was induced at postnatal day (P)1‐P2. At this stage the majority of mesenteric lymphatic valves have formed, but new valves continue to form during the first postnatal weeks (Sabine et al, 2015). Neonatal LEC‐specific deletion of Foxp2 led to collecting lymphatic vessel and valve defects similar to those observed upon global EC deletion (Fig 5B). At P6, the number of valves (Fig 5C) and the length of valve leaflets (Fig 5D) were reduced in the Foxp2floxflox; Prox1‐CreERT2 mutants in comparison with Cre‐negative littermate controls. However, we observed neither chyle leakage (n = 6 mice analyzed) nor enlargement of collecting vessels (Fig 5E) in the Foxp2floxflox; Prox1‐CreERT2 mice that occurred upon constitutive EC‐specific deletion in the Foxp2flox /− ; Tie2‐Cre mice. This suggests that the Foxp2 loss‐induced collecting vessel defects, including barrier leakage, develop prior to birth or during early neonatal life.
Figure 5. LEC‐autonomous role of FOXP2 in lymphatic valve morphogenesis.
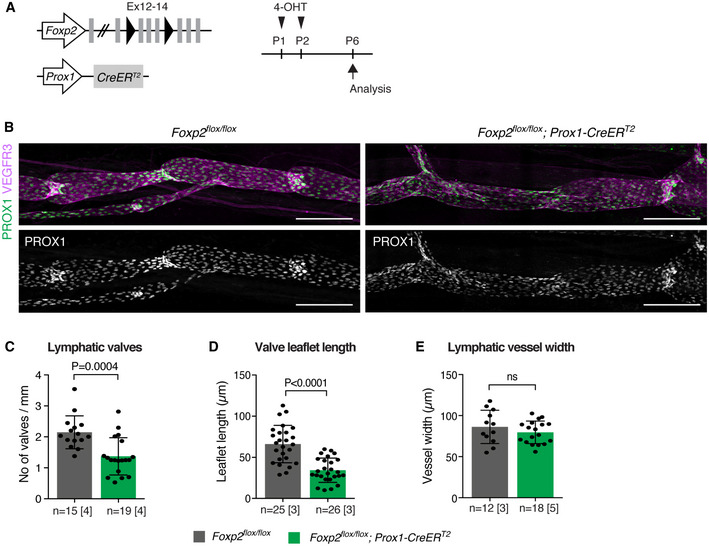
-
AGenetic constructs and 4‐OHT treatment for neonatal LEC‐specific Foxp2 deletion. Ex, exon.
-
BWhole‐mount immunofluorescence of P6 mesenteries, showing valve defects in the mutant vessels.
-
C–EQuantification of lymphatic valve numbers (C), valve leaflet length (D), and vessel width (E) in Foxp2flox / flox;Prox1‐CreERT2 and littermate control mice (n = vessels (C, E) or valves (D) [mice], as indicated).
Data information: In (C–E), data are presented as mean ± SD. P, Student's t‐test. ns = not significant. Scale bar: 200 µm.
Source data are available online for this figure.
Molecular mechanisms that regulate the formation of lymphatic, venous, and lymphovenous valves are shared (Bazigou et al, 2011; Geng et al, 2017). Similar to the lymphatic valve regulators PROX1, integrin‐α9 (Bazigou et al, 2011), and FOXC2 (Lyons et al, 2017), we found that FOXP2 was also expressed in venous valve ECs (Fig EV4A and B). Genetic deletion of Foxp2 using the Tie2‐Cre mice did not, however, lead to apparent defects in the formation of venous valve leaflets (Fig EV4A and B). Developing lymphovenous valves (LVVs) of an E14 embryo were instead negative for FOXP2 staining (Fig EV4C). Embryos with defective LVVs frequently display blood‐filled lymphatic vessels and edema (Geng et al, 2016; Martin‐Almedina et al, 2016). Lack of an overt phenotype in Foxp2flox; Tie2‐Cre embryos suggests presence of functional LVVs in these mice, consistent with the absence of FOXP2 in LVVs.
Figure EV4. Analysis of FOXP2 expression and function in venous and lymphovenous valves.
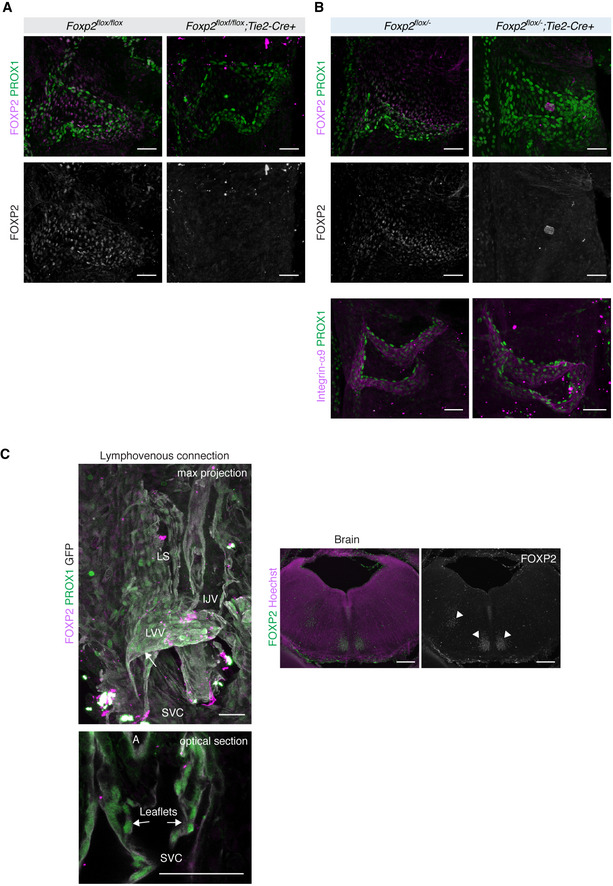
-
A, BWhole‐mount immunofluorescence of venous valves of the proximal femoral vein in P7 Foxp2flox / flox;Tie2‐Cre (A) and Foxp2flox /− ;Tie2‐Cre (B) mice with respective littermate controls. Note expression of FOXP2 in ECs of the valve leaflet but apparently normal PROX1high integrinα9+ valve leaflets in Foxp2‐deficient mice.
-
CLeft: Immunofluorescence of a coronal vibratome section of an E14 Vegfr3‐CreERT2;R26‐mTmG (Martinez‐Corral et al, 2016) embryo showing no expression of FOXP2 in LVV (arrows). FOXP2+ cells are likely (PROX1−) neuronal cells. Maximum intensity projection is shown above, and a single optical section at the level of LVV leaflets below. GFP expression in LECs was induced by intraperitoneal injection of 1 mg of 4‐OHT into pregnant female. Right: Visualization of FOXP2+ neuronal cells confirmed successful staining (arrowheads). LS, lymph sac; IJV, internal jugular vein; SVC, superior vena cava; A, artery.
Data information: Scale bar: 50 µm (A–C, lymphovenous connection), 200 µm (C, brain).
Collectively, these results demonstrate that FOXP2 is required LEC‐autonomously for the development and maintenance of lymphatic, but not venous valves.
FOXP2 is a component of the FOXC2/NFATc1 pathway
Oscillatory shear stress regulates the expression of two key transcription factors controlling collecting vessel and valve morphogenesis: FOXC2 (Sabine et al, 2012) and GATA2 (Kazenwadel et al, 2015). Notably, ChIP‐seq analysis of FOXC2 chromatin occupancy in LECs revealed binding to FOXP2 regulatory regions (Norrmén et al, 2009). To test if FOXC2 regulates FOXP2 expression, we first silenced its expression in HDLECs using siRNA. qRT–PCR analysis of HDLECs showed downregulation of FOXP2 upon FOXC2 silencing (Fig 6A). In contrast, FOXP2 silencing did not affect the expression of FOXC2 or its target GJA4 (encoding CX37; Fig 6A). FOXP2 silencing had also no effect on the expression of GATA2 (Fig EV5A), an upstream regulator of FOXC2 (Kazenwadel et al, 2015).
Figure 6. FOXP2 is a component of the FOXC2/NFATc1 pathway.
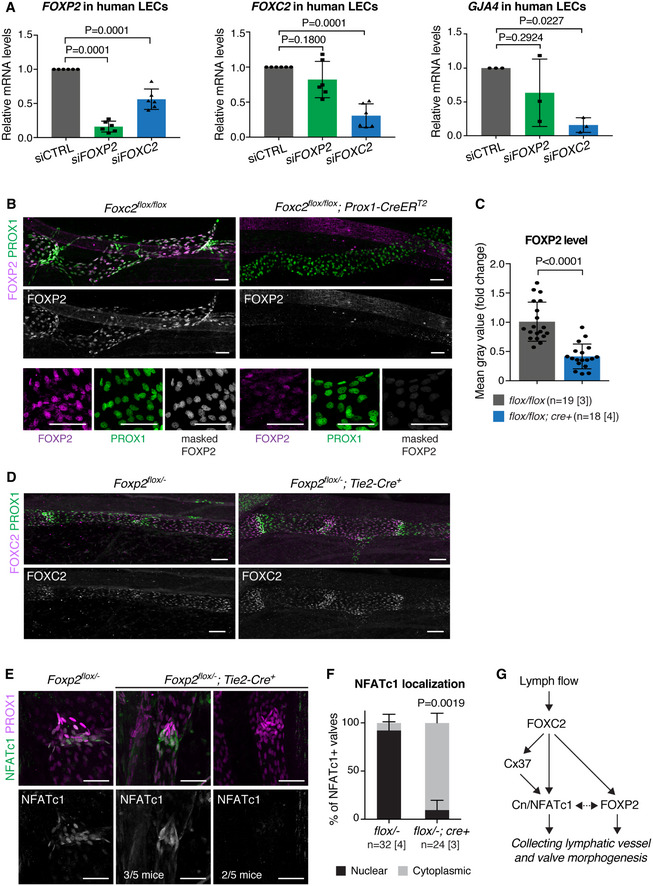
- qRT–PCR analysis of FOXP2, FOXC2, and GJA4 expression in control (siCTRL) and FOXP2 or FOXC2 siRNA‐treated HDLECs (n = 6 (FOXP2, FOXC2) or n = 3 (GJA4) independent experiments).
- Top panels: whole‐mount immunofluorescence of E18 Foxc2flox / flox;Prox1‐CreERT2 and littermate control mesenteries, showing downregulation of FOXP2 in Foxc2‐deficient vessels. Bottom panels: extraction of nuclear FOXP2 staining (gray) using IMARIS surface mask generated based on PROX1 staining (green). Unmasked FOXP2 staining is shown in magenta.
- Quantification of FOXP2 levels in mesenteric lymphatic vessels of Foxc2flox / flox;Prox1‐CreERT2 and control littermates based on nuclear FOXP2 signals as in (B) (n = vessels [mice], as indicated).
- Whole‐mount immunofluorescence of P8 mesenteric lymphatic vessels of Foxp2flox /− ;Tie2‐Cre and littermate control mice showing loss of organized FOXC2high valve regions but unaltered expression of FOXC2 in Foxp2 deficient vessels.
- Whole‐mount immunofluorescence of lymphatic valves in P7 mesenteries of control and Foxp2flox /− ;Tie2‐Cre mice showing NFATc1 subcellular localization. Note predominantly nuclear localization of NFATc1 in control valves (left panels), but cytoplasmic localization (middle panels, three out of five mice) or downregulation of NFATc1 (right panels, two out of five mice) in the mutant valves.
- Quantification of protein localization in NFATc1+ lymphatic valves from (D) (n = valves [mice], as indicated).
- Schematic of the molecular pathway involving FOXC2/FOXP2/NFATc1 in collecting vessel and valve morphogenesis.
Data information: In (A, C, F), data are presented as mean ± SD. P, one‐way ANOVA (A), Student's t‐test (C), Fisher's exact test (F). Scale bar: 50 µm (B, D, E).
Source data are available online for this figure.
Figure EV5. Analysis of FOXC2 pathway components in FOXP2 silenced LECs.
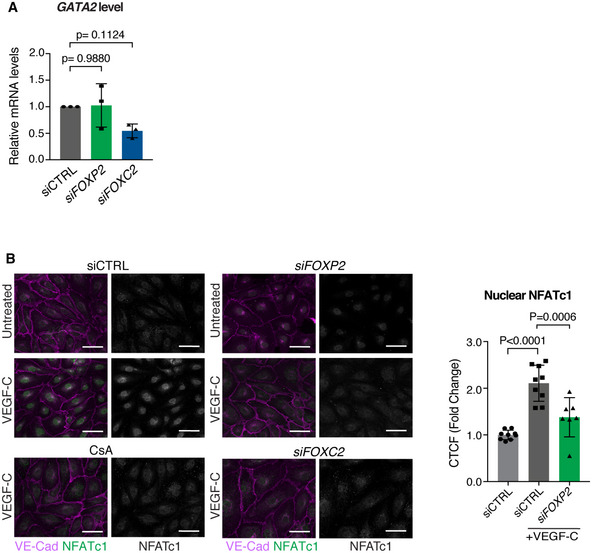
- qRT–PCR analysis of GATA2 expression in control (siCTRL) and FOXP2 or FOXC2 siRNA‐treated HDLECs (n = 3 independent experiments).
- Left: NFATc1 immunofluorescence in control (siCTRL) and FOXP2 or FOXC2 siRNA‐transfected HDLECs under basal conditions (untreated) or after 30‐min stimulation with VEGF‐C (50 ng/ml) in the absence or presence of the calcineurin inhibitor cyclosporin A (CsA). Note nuclear translocation of NFATc1 upon VEGF‐C stimulation, which is abrogated in cells treated with FOXP2 or FOXC2 siRNA, or CsA. Right: Quantification of nuclear NFATc1 in CTRL and FOXP2 siRNA‐treated LECs after VEGF‐C stimulation (n = 7–9 images from two independent experiments).
Data information: In (A, B), data are presented as mean ± SD. P, one‐way ANOVA (A), or Student's t‐test (B). Scale bar: 50 µm (B).
Source data are available online for this figure.
Next, we investigated FOXC2 regulation of Foxp2 in vivo by assessing FOXP2 protein levels in Foxc2‐deficient lymphatic vessels. As previously reported (Sabine et al, 2015), deletion of Foxc2 using the Prox1‐CreERT2 line led to loss of lymphatic valves composed of PROX1high LECs (Fig 6B). To assess the level of nuclear FOXP2 signals in the LECs, we quantified the staining intensity after masking the PROX1 signal (Fig 6B, lower panels). This revealed approximately two‐fold higher FOXP2 immunostaining intensity in control in comparison with Foxc2‐deficient vessels (Fig 6C). Loss of FOXC2 thus leads to downregulation of FOXP2 both in vitro and in vivo. Conversely, whole‐mount immunofluorescence of mesenteric lymphatic vessels of Foxp2flox /− ; Tie2‐Cre mice showed loss of organized FOXC2high valve regions but unaltered expression of FOXC2 in Foxp2‐deficient vessels compared to littermate controls (Fig 6D).
FOXC2 co‐operates with calcineurin/NFATc1 signaling during valve morphogenesis (Norrmén et al, 2009; Sabine et al, 2012), while FOXP2 has been shown to form a co‐operative complex with NFATc2 (Wu et al, 2006). To investigate the potential regulation of NFAT signaling by FOXP2 during lymphatic development, we stained mesenteric lymphatic vessels of Foxp2flox /− ; Tie2‐Cre + and control littermates for NFATc1. In agreement with previous findings (Norrmén et al, 2009; Sabine et al, 2012), increased NFATc1 staining and nuclear localization were observed in the valves of control mice (Fig 6E and F). In contrast, Foxp2‐deficient valves showed predominantly cytoplasmic localization, or overall reduction in NFATc1 levels (Fig 6E and F). FOXP2 silencing also abrogated VEGF‐C‐induced nuclear translocation of NFATc1 in LECs in vitro (Fig EV5B). These results suggest inhibition of NFAT signaling in Foxp2‐deficient LECs.
Collectively, the above results demonstrate that FOXP2 is a downstream effector of the flow‐responsive FOXC2 in the regulation of NFATc1 activity during lymphatic valve formation (Fig. 6G).
Discussion
Coordinated action of the two functionally specialized types of lymphatic vessels, the lymphatic capillaries and collecting lymphatic vessels, ensures removal of excess fluid from the tissues and efficient unidirectional drainage to the venous circulation. Here, we investigated the transcriptional basis of phenotypic identities of specific vessel subtypes and identified the forkhead transcription factor FOXP2 as a signature gene for collecting vessels. Functional analysis of FOXP2 in primary LECs and using genetic loss‐of‐function mouse models further establish it as a new flow‐induced regulator of valve morphogenesis and a component of the FOXC2/NFATc1 pathway.
Cell‐extrinsic factors such as differences in the magnitude and type of fluid shear stress as well as matrix composition and stiffness experienced by different vessel types likely play an important role in the regulation of LEC phenotype and transcription. To identify the transcriptional basis of LEC identities in specific vessel subtypes, we determined differentially expressed genes between ECs sorted from dermal LYVE1+ lymphatic capillaries and LYVE1− collecting vessels. Consistent with the unique collecting vessel features, namely the presence of continuous basement membrane, smooth muscle coverage, and luminal valves, we observed enrichment of genes encoding regulators of EC‐extracellular matrix such as Cd93 and Mmrn2 (Lugano et al, 2018) and EC‐smooth muscle (Nos3 encoding eNOS) communication, as well as the valve LEC signature gene Cldn11 (Takeda et al, 2019). Collecting LEC‐enriched genes also included Foxp2, a member of the FOXP family of transcription factors, which suggests the existence of unique transcription factor codes in defining vessel‐type‐specific LEC identities.
FOXP2 has previously not been implicated in EC biology, but is extensively studied in the nervous system (Co et al, 2020) and also shown to regulate lung development (Shu et al, 2007). Clinically, mutations in FOXP2 are causative of developmental speech and language disorders ((Lai et al, 2001), reviewed in (Co et al, 2020)). Analysis of the developing mesenteric collecting vessels in mouse embryos showed that FOXP2 was not expressed during early stages of vessel formation. It was abruptly upregulated between E16 and E17, after initiation of fluid flow (Sabine et al, 2015), and persisted in the remodeling and mature collecting vessels in vivo. FOXP2 mRNA and protein levels were also increased in primary human LECs in response to oscillatory shear stress that mimics turbulent flow in the branched network of primitive lymphatic vessel plexus, and, to a lesser degree, laminar shear stress. Flow regulation of FOXP2 was further supported by the observations that abrogation of flow in cultured mesenteries ex vivo, or genetic deletion of Foxc2, the major flow‐responsive transcriptional regulator of lymphatic valve formation (Petrova et al, 2004; Sabine et al, 2012) in vivo led to reduction in FOXP2 levels. Several putative FOXC2‐binding sites in FOXP2 identified by ChIP‐chip analysis in human LECs (Norrmén et al, 2009) suggest that FOXP2 is a direct downstream target of FOXC2.
In agreement with the restricted expression of FOXP2 in collecting lymphatic vessels, genetic deletion of Foxp2 in all ECs or specifically in LECs revealed its selective role in the morphogenesis of collecting vessels and valves. Mice with global EC‐specific Foxp2 deletion showed chyle reflux and leakage, as well as a spectrum of valve defects. About 25% of valves in these mice lacked leaflets and showed no expression of integrin‐α9 and its ligand fibronectin‐EIIIA that are critical regulators of leaflet formation (Bazigou et al, 2009). The remaining valves were characterized by shorter integrin‐α9+ leaflets and low fibronectin‐EIIIA deposition. Similarly, early postnatal deletion of Foxp2 specifically in LECs led to reduced valve numbers and leaflet length, but not chyle leakage. The most prominent feature of the phenotype is valve defect, providing an explanation for retrograde lymph flow. Since lack of valves alone, as observed in mice lacking Gja4 (encoding connexin 37), is not sufficient to promote leakage of chyle (Sabine et al, 2012), additional collecting vessels defects must be present in Foxp2 null mice and develop prior to birth. Notably, the phenotype is similar, albeit milder, than that reported in mice lacking Foxc2 (Sabine et al, 2012). Foxp2 deletion did not lead to a complete lack of valve development and thus did not compromise postnatal survival and growth in the mutant mice. This further supports the notion that FOXP2 acts downstream of FOXC2 in regulating lymphatic (valve) development, and other FOXC2 targets can partially compensate for the loss of FOXP2. Although FOXP2 was also expressed in venous valve ECs, we could not observe defects in the formation of these valves in Foxp2 deficient mice. Our assessment of valve morphology was limited to an early developmental stage, and potential effect of Foxp2 deficiency on maturation or long‐term maintenance of venous valves cannot be excluded.
Transcriptional activity of the FOXP proteins is mediated by homo‐ or heterodimerization (Li et al, 2004; Sin et al, 2015). In the nervous system, FOXP2 has dual functionality, acting to repress or activate gene expression, with the different FOXP1/2/4 dimer combinations leading to different transcriptional outcomes (Sin et al, 2015). Interestingly, FOXP2 targets identified in murine brain include several known regulators of lymphatic development, including Nrp2 and Efnb2 (Vernes et al, 2011). Downregulation of NRP2 protein and Efnb2 transcript observed in Foxp2‐deficient lymphatic vessels in vivo suggests conservation of these targets in LECs. NRP2 functions as a co‐receptor for VEGFR3 and regulates lymphangiogenic sprouting (Kärpänen et al, 2006; Xu et al, 2010), but has not been implicated in collecting vessel maturation, and is even absent in valve LECs (Sabine et al, 2012). EphrinB2 has instead been shown to regulate the development and maintenance of lymphatic valves and the integrity of collecting vessel LEC junctions (Mäkinen et al, 2005; Zhang et al, 2015; Frye et al, 2020), and its downregulation may thus contribute to the phenotype observed in Foxp2 mutant mice. Considering the multitude of FOXP interaction partners reported (FOXP1/4, NFAT1, NR2F1, NR2F2, SATB1, SATB2, SOX5, YY1, and ZMYM2) (Stroud et al, 1993; Li et al, 2004; Xu et al, 2010; Sin et al, 2015; Estruch et al, 2018), it is, however, unlikely that FOXP2‐driven cellular functions can be attributed to (a) single downstream target(s) but instead involves a transcriptional program defined by a combinatorial code of transcription factors.
Since the relative levels of FOXP1/2/4 proteins, and thus the different combinations of dimers that may be formed, determine the ability of these factors to act as activators or repressors (Sin et al, 2015), it is interesting to note that Foxp1 and Foxp4 are expressed at comparable levels in the two lymphatic vessel types and are also present in BECs. This suggests that the selective expression of FOXP2 in collecting vessel LECs can profoundly change their transcriptional program, not only via FOXP2 homodimers but also through heterodimerization with other family members. Other FOXP2 interactors of particular interest include the NFAT and NR2F2 (also known as chicken ovalbumin upstream promoter transcription factor II, COUP‐TFII). COUP‐TFII heterodimerizes with PROX1 and regulates LEC specification (Lee et al, 2009; Yamazaki et al, 2009; Aranguren et al, 2013), that does not require FOXP2. However, like FOXP2, COUP‐TFII has been shown to positively regulate NRP2 expression during later stages of lymphatic development (Lin et al, 2010). Co‐operative binding of FOXP2 and FOXP3 to NFAT1 (also known as NFATc2) is characterized structurally by crystallography (Stroud et al, 1993; Wu et al, 2006; Bandukwala et al, 2011), and shown to control regulatory T‐cell function by determining the transcriptional outcome of key downstream targets (Wu et al, 2006). NFAT proteins are dephosphorylated by activated calmodulin‐dependent phosphatase calcineurin, leading to their nuclear translocation. However, in regulatory T cells a fraction of NFAT was constitutively localized in the nucleus where it selectively bound to FOXP3 target genes (Li et al, 2012), indicating calcineurin independent mechanism of NFAT regulation. Interestingly, in the lymphatic vasculature calcineurin/NFATc1 signaling is regulated by flow and FOXC2, and plays an important role in collecting vessel and valve morphogenesis (Norrmén et al, 2009; Sabine et al, 2012). In agreement with previous data (Norrmén et al, 2009), we found that NFATc1 was mainly localized to the nucleus of valve LECs. In contrast, Foxp2‐deficient vessels showed predominantly cytoplasmic NFATc1 localization, but we also observed reduced protein levels in some of the mutant mice. FOXP binding surface is conserved in the NFAT family (Wu et al, 2006), suggesting that within valve LECs FOXP2 may through dimerization control the stability, nuclear localization, and transcriptional activity of NFATc1.
The major lymphangiogenic growth factor VEGF‐C also promotes transient activation of calcineurin/NFAT signaling in vitro (Norrmén et al, 2009) and was recently shown to contribute to lymphatic valve morphogenesis through the regulation of mechanosensitive transcriptional co‐factors YAP and TAZ (Cha et al, 2020). VEGF‐C‐induced nuclear translocation of NFATc1 was abrogated in FOXP2 silenced cells, similar to previously reported upon FOXC2 silencing (Sabine et al, 2012). Although further work is necessary, these results hint at the potential cross‐talk between mechanosensitive and growth factor signaling pathways in lymphatic valve development.
In summary, our study identifies FOXP2 as a new flow‐induced transcriptional regulator of collecting lymphatic vessel and valve development and a critical component of the FOXC2/NFATc1 pathway, thus revealing a unique transcription factor code in determining collecting vessel LEC identity. Pathological conditions associated with vessel damage and reduced fluid flow may through reduced FOXP2 expression contribute to loss of collecting vessel identity and function. Although lymphatic vessel growth can be stimulated by pro‐lymphangiogenic VEGF‐C therapy, remodeling of the initially dysfunctional lymphatic capillary network into functional collecting vessels occurs with a delay (Tammela et al, 2007). Modulation of FOXP2 function to induce a collecting vessel LEC‐specific transcriptional program in the newly formed lymphatic vessels could thereby provide a therapeutic strategy to promote vessel maturation and restoration of collecting vessel function.
Materials and Methods
Mice
Foxp2flox (French et al, 2007), Ti e2‐Cre (Koni et al, 2001), Prox1‐CreERT2 (Bazigou et al, 2011), and Foxc2flox (Sabine et al, 2015) mice were analyzed on a C57BL/6J background. For global EC‐specific deletion using the Tie2‐Cre, two breeding schemes were used; (i) “flox/flox deletion” was induced by breeding Tie2‐Cre + male mice (Foxp2flox / flox;Cre + or Foxp2flox /+ ;Cre +) with Foxp2flox / flox; Cre − females, to avoid inheritance of a null allele when transmitted through the female germ line, and (ii) “flox/null deletion” was induced by breeding Tie2‐Cre + female mice (Foxp2flox / flox;Cre + or Foxp2flox /+;Cre +) with Foxp2flox / flox;Cre − males. For postnatal induction of Cre activity in the Foxp2flox / flox; Prox1‐CreERT2 mice, 4‐hydroxytamoxifen (4‐OHT; Sigma, H7904) was dissolved in ethanol (25 mg/ml) and administered (2 μl/50 μg) by intragastric injection at P1 and P2. For embryonic induction of Cre activity in embryos, pregnant mice were injected with 5 mg of tamoxifen and 5 mg of progesterone dissolved in 100 µl of Cremophor®EL (Sigma, C5135) at E13.5 and E14.5 and embryos were harvested at E18.5 (Foxc2flox / flox; Prox1‐CreERT2), or with 1 mg of 4‐OHT in peanut oil at E15 and E16 and embryos were harvested at E18 (Foxp2flox / flox; Prox1‐CreERT2). The morning of vaginal plug detection was considered as embryonic day (E) 0. All experimental procedures were approved by the Uppsala Animal Experiment Ethics Board (permit numbers C130/15 and 5.8.18‐06383/2020) and performed in compliance with all relevant Swedish regulations, or by the Animal Ethics Committee of Vaud, Switzerland and performed in compliance with all relevant Swiss regulations.
Antibodies
The details of primary antibodies used for immunofluorescence of whole‐mount tissues, cells, and flow cytometry are provided in Table EV1. Secondary antibodies conjugated to Cy3, Alexa Fluor 488, 594, or 647 were obtained from Jackson ImmunoResearch.
Flow cytometry
Each replica for the array analysis was produced from pooled ear skin from 5 to 6 5‐week‐old Prox1‐GFP mice of mixed genders. Ear skins were digested in 10 mg/ml Collagenase IV (Life Technologies) and 0.2 mg/ml DNase I (Roche) in PBS at 37°C for 30 min. Digests were washed and filtered before EC enrichment with CD31/PECAM1 magnetic microbeads (Miltenyi Biotec) on LS columns according to the manufacturer's instructions. Fc receptor binding was blocked by adding anti‐mouse CD16/CD32 (93) (eBioscience), and cells were stained with anti‐CD31/PECAM1 (390) PE‐Cy7, anti‐podoplanin (PDPN) (8.1.1) PE, and anti‐LYVE1 (ALY7) eFluor660. Dump channel included antibodies to exclude immune cells: anti‐CD45 (30‐F11) and anti‐CD11b (M1/70); and red blood cells: TER‐119 (TER‐119); all conjugated to eFluor450 (eBioscience); together with cell‐death stain Sytox blue (Life Technologies). LECs (GFP+PDPNhighPECAM1high) were sorted based on LYVE1 expression into three fractions: LYVE1high lymphatic capillaries, LYVE1low/− collecting vessels, and LYVE1intermed (putative) pre‐collecting vessels. BECs were sorted as GFP−PDPN−LYVE1−PECAM1high cells. Sorting was performed on a FACS Aria III with the FACS Diva software (BD Biosciences) using a 100 µm nozzle, 20–25 pounds per square inch (psi), 4‐way purity sorting modality, and an acquisition rate of 500–2,000 events per second. Three independent sorts were performed and cells (cap LEC: 1,000–2,003 cells, pre‐col LEC: 505–1,097 cells, col LEC: 541–839 cells, and BEC: 1,504–2,023 cells) were collected directly into RLT buffer with added beta‐mercaptoethanol (Sigma) and immediately processed for RNA extraction (RNeasy Micro Kit, Qiagen). FACS data were processed using FlowJo software (BD). Single cells were gated using FSC‐A/SSC‐A followed by FSC‐H/FSC‐W and SSC‐H/SSC‐W.
To obtain BECs and LECs for qRT–PCR analysis, mesenteries of P11 mice were dissected from intestine and mesenteric root lymph node and digested in 2 mg/ml Collagenase type II, 0.2 mg/ml DNase I and 0.2% FBS in PBS at 37°C for 15 min during continuous shaking. After quenching and washing with FACS buffer (PBS, 0.5 % FBS, 2 mM EDTA), Fc receptor binding was blocked with rat anti‐mouse CD16/CD32. Samples were thereafter stained with anti‐podoplanin (clone 8.1.1, APC), anti‐CD31/Pecam1 (390, PE‐Cy7), anti‐CD45 (30‐F11, eFluor450), anti‐CD11b (M1/70, eFluor450), and anti‐TER‐119 (TER‐119, eFluor450). Prior to sorting cells were incubated with 1 μM Sytox blue to label dead cells. Sorting was performed as described above with the differences that an 85 µm nozzle was used and by 4‐way purity sorting modality. After single‐cell gating and exclusion of immune cells and dead cells as above, BECs were sorted as PECAM1+PDPN− cells and LECs as PECAM1+PDPN+ cells.
Microarray expression analysis
RNA concentration and quality were evaluated using the Agilent 2100 Bioanalyzer system (Agilent Technologies Inc, Palo Alto, CA). 500 pg of total RNA was estimated from each sample, and this starting amount was used to generate amplified and biotinylated sense‐strand cDNA from the entire expressed genome according to the GeneChip® WT Pico Reagent Kit User Manual (P/N 703262 Rev 1 Affymetrix Inc., Santa Clara, CA). GeneChip® ST Arrays (GeneChip® Mouse Transcriptome Array 1.0; now called Clariom™ D assays, mouse) were hybridized for 16 h in a 45°C incubator, rotated at 60 rpm, according to the GeneChip® Expression Wash, Stain and Scan Manual (PN 702731 Rev 3, Affymetrix Inc., Santa Clara, CA). The arrays were then washed and stained using the Fluidics Station 450 and finally scanned using the GeneChip® Scanner 3000 7G. The raw data were normalized to gene level using the robust multi‐array average (RMA) method (Li & Wong, 2001; Irizarry et al, 2003), and data were logarithm based two transformed. Statistical test for selecting differentially expressed genes was performed using a method Significance Analysis of Microarrays (SAM) (Tusher et al, 2001) in Bioconductor package siggenes (version: 1.56.0) using R program (version: 3.5.1).
Primary EC culture and flow experiments
Human primary dermal LECs (HDLECs isolated from juvenile foreskin, cat no. C‐12216) were obtained from PromoCell. Cells were grown on bovine fibronectin (Sigma, F1141) coated dishes in complete ECGMV2 medium (PromoCell, C‐22022) and used after three passages. Cells were grown to confluence in 6‐well plates and transfected with AllStars‐negative control siRNA (SI03650318, QIAGEN) or ON‐TARGET plus FOXP2 siRNA‐SMARTpool (M‐010359‐02‐0005, Dharmacon) or siGENOME FOXC2 (2303) siRNA (MQ‐008987‐00‐0002, Dharmacon) at final concentration of 40 nM siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For activation of NFATc1 nuclear translocation, cells were stimulated with VEGF‐C (R&D Systems, 9199‐VC, 50 ng/ml) for 30 min. Calcineurin signaling was inhibited by pretreatment with 200 ng/ml of cyclosporine A (CsA, Merck Millipore, 239835) for 2 h, as described (Norrmén et al, 2009; Sabine et al, 2012).
For flow experiments, LECs at passage 4–5 were seeded on fibronectin‐coated slides (μ‐Slide I0.8 Luer; Ibidi) in complete medium until confluent cell monolayers were formed. Laminar (4 dyn/cm2) or oscillatory (4 dyn/cm2, ¼ Hz) flow was applied in a parallel plate flow‐chamber system (Ibidi, Pump System) for the indicated times. For static conditions, cells were cultured in the μ‐Slide I0.8 Luer without flow but maintained in the same conditions as flow‐induced cells.
Ex vivo culture
Mesenteric lymphatic vessels were dissected at E18 and cultured in complete EC culture medium without VEGF‐A (PromoCell) for 24 h, as described (Sabine et al, 2012).
RNAscope
Detection of Enfb2 mRNA (ACD Bio, Cat no. 477671) in mesenteric lymphatic vessels by whole mount in situ hybridization was performed according to the protocol described by (Gross‐Thebing et al, 2014). Negative and positive controls provided in the kit were used. Quantifications were done using the guideline to quantify RNAscope® Fluorescent Assay Results recommended by ACD (www.acdbio.com). Briefly, mesentery was dissected and fixed in 4% PFA for 2 h at RT followed by methanol dehydration using a series of increasing concentration (25, 50, 75, 2 × 100%) in 0.1% PBT (0.1% Tween‐20 in PBS) for 5 min each. Sample was kept in 100% methanol at −20°C overnight. Sample was then air‐dried for 30 min at RT and exposed to the RNAscope‐based signal amplification (Advanced Cell Diagnostics, ACD) for Efnb2 detection. Following the RNAscope procedure, mesentery was subjected to immunostaining with anti‐PROX1 and anti‐PECAM1 antibodies using the immunofluorescence protocol described above. The total number of dots in the valve region was divided by the number of PROX1high cells to obtain the average number of dots per valve LEC.
Immunofluorescence
Tissue was dissected and fixed in 4% paraformaldehyde (PFA) for 2 h at room temperature (RT) followed by permeabilization in 0.3–0.5% Triton X‐100 in PBS (PBST) for 10 min and blocking in 3% BSA or 5% donkey serum in PBST for 2 h at RT. Immunostaining was performed by adding primary antibodies in blocking buffer at 4°C overnight. Samples were washed in PBST and incubated with fluorochrome‐conjugated secondary antibodies at 1:300 dilution for 2 h at RT. After washing step, samples were mounted in Mowiol.
For analysis of LVVs, 100‐μm coronal vibratome sections of E14 embryos (n = 3) were cut and stained as described above. Single‐plane images of the valve were taken where the valve was clearly visible.
HDLECs were washed with PBS before fixation with 4% PFA for 15 min at RT, washed three times in PBS, and permeabilized in 0.1% Triton X‐100 in PBS for 5 min. After blocking with 1% BSA in PBS for 1 h at RT, cells were incubated with primary antibodies at 1:100 dilution in blocking buffer at 4°C overnight. Cells were washed three times, and fluorochrome‐conjugated secondary antibodies at 1:100 dilution were added for 1 h at RT. Cells were then washed and mounted using Fluoroshield with DAPI mounting medium (Sigma, F6057).
Image acquisition and quantification
Images were acquired using Leica SP8 confocal microscope with HC FLUOTAR L 25×/0.95 W VISIR or HC PL APO 63×/1.30 GLYC CORR CS2 objective and LAS X software. All confocal images represent maximum intensity projection of Z‐stacks of single tile or multiple tile scan images. Stereomicroscope images were acquired with LEICA MZ 16 F stereomicroscope equipped with a Leica DFC420 C camera with PLANAPO 1.6× or PLANAPO 0.63× objective.
Image analysis was performed using ImageJ FIJI or IMARIS. Quantification of number of valves in mesenteric collecting lymphatic vessels and embryonic skin were performed by identifying valves as clusters of LECs expressing high levels of PROX1 and measuring the length of vessel to obtain the average of valves per vessel length (mesentery: n = 4 mice per condition; > 3 vessels per mouse; three independent experiments/litters of mice; skin: n = 2 embryos from 1 litter, 2 tile scan images [3,243.42 µm × 2,201.26 µm]/embryo). Average lymphatic vessel width was determined by measuring the diameter at several points along the vessel and averaging the measurements (n ≥ 3 mice per condition; > 5 measurements per vessel; 3–4 independent experiments). For measurement of valve leaflet length, vessels were stained with integrin‐α9 and PROX1 antibodies (n ≥ 3 mice per condition; at least five images per mouse; three independent experiments). For quantification of FOXP2 expression in the nucleus in Foxc2 mutants, IMARIS software was used to create a 3D surface mask based on PROX1 staining to obtain the pixel intensity of FOXP2 from maximum intensity projection images of Z‐stacks (n = 3 [control mice] or n = 4 [mutant mice]; > 5 images per sample [total n ≥ 150 cells]) and the average pixel intensity value for each was plotted. For quantification of NRP2 levels, pixel intensity was measured by CTCF (corrected total cell fluorescence = integrated density − [area of selected cell × mean fluorescence of background readings], n = 4 each condition; ≥ 10 measurements per mouse; 3 independent experiments). For quantification of nuclear NFATc1 protein levels, DAPI fluorescence was used to identify the nucleus and measure pixel intensity of NFATc1 by CTCF method as described above (n ≥ 3 images per condition; two independent experiments).
Quantitative RT–PCR analysis
Total RNA was extracted from HDLECs subjected to flow or static conditions, and murine BEC and LEC, using RNAeasy Mini kit (QIAGEN). Reverse transcription was performed from 0.5 μg of RNA using SuperScript™ VILO™ Master Mix (Thermo Fisher Scientific) according to the manufacturer's instructions. Gene expression was analyzed by qRT–PCR using TaqMan Gene Expression Assay and StepOnePlus™ Real‐Time PCR System (Applied Biosystems). Relative Gene expression levels were normalized to GAPDH. The following probes were used: Hs00362818_m1 FOXP2, Hs02786624_g1 GAPDH, Hs00270951_s1 FOXC2, Hs00358836_m1 KLF4, Hs00704917_s1 GJA4, Hs00231119_m1 GATA2, Mm00474845_m1 Foxp1, Mm00475030_m1 Foxp2, Mm00475162_m1 Foxp3, Mm00466368_m1 Foxp4.
Statistical analysis
GraphPad Prism was used for graphic representation and statistical analysis of the data. Data between two groups were compared with unpaired two‐tailed Student's t‐test, assuming equal variance. Ordinary one‐way ANOVA Dunnett's multiple comparisons test was used to compare differences between groups (Figs 6A and EV5A), and Fisher's exact test to determine association between two categorical variables (Figs 3J and 6F). Differences were considered statistically significant when P < 0.05. The experiments were not randomized, and no blinding was done in the analysis and quantifications. All quantifications are based on a minimum of three biological replicates and a minimum of two independent experiments/litters, except for experiments shown in Appendix Fig S2B and C (n = 2 embryos from 1 litter) and Fig EV5B (n = 2 biological replicates). Data are presented as mean ± SD.
Author contributions
Conceptualization: MNHV, MHU, TM; Formal analysis: YS, LH; Funding acquisition: CH, TVP, TM; Investigation: MNHV, MHU, AGL, IK; Methodology: MNHV, MHU, TM; Project administration: MNHV, TM; Supervision: CH, TVP, TM; Visualization: MNHV, YS, TM; Writing—original draft: MNHV, TM; Writing—review and editing: MNHV, MHU, AGL, IK, CH, TVP, TM.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Expanded View/Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Amelie Sabine for helpful discussions and advice, Yan Zhang for the immunofluorescence staining in Fig 1A, the BioVis facility (Uppsala University, Sweden) for flow cytometer usage and support, and Henrik Ortsäter, Aissatu Mami Camara, Sofie Sjöberg, and Sofie Lunell Segerqvist for technical assistance. This work was supported by the Swedish Cancer Society (CAN 2016/535), the European Research Council (ERC‐2014‐CoG‐646849), Knut and Alice Wallenberg Foundation (2015.0030 and 2018.0218) and the Swedish Research Council (542‐2014‐3535 and 2020‐02692) to TM, and Swiss National Science Foundation (CRSII5_177191 to TP, 310030_182528 to CH). The computations of transcriptome data were performed by resources provided by the Swedish National Infrastructure for Computing (SNIC) through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) partially funded by the Swedish Research Council through grant agreement no. 2018‐05973, under projects SNIC 2018/8‐62 and SNIC 2020/16‐159.
The EMBO Journal (2021) 40: e107192.
Data availability
RNA microarray data that support the findings of this study have been deposited in GEO (Gene Expression Omnibus) repository with the accession code GSE159842 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159842).
References
- Aranguren XL, Beerens M, Coppiello G, Wiese C, Vandersmissen I, Lo Nigro A, Verfaillie CM, Gessler M, Luttun A (2013) COUP‐TFII orchestrates venous and lymphatic endothelial identity by homo‐ or hetero‐dimerisation with PROX1. J Cell Sci 126: 1164–1175 [DOI] [PubMed] [Google Scholar]
- Bandukwala H, Wu Y, Feuerer M, Chen Y, Barboza B, Ghosh S, Stroud J, Benoist C, Mathis D, Rao A et al (2011) Structure of a domain‐swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity 34: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazigou E, Xie S, Chen C, Weston A, Miura N, Sorokin L, Adams R, Muro AF, Sheppard D, Makinen T (2009) Integrin‐alpha9 is required for fibronectin matrix assembly during lymphatic valve morphogenesis. Dev Cell 17: 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazigou E, Lyons OTA, Smith A, Venn GE, Cope C, Brown NA, Makinen T (2011) Genes regulating lymphangiogenesis control venous valve formation and maintenance in mice. J Clin Invest 121: 2984–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvrée K, Brunet I, del Toro R, Gordon E, Prahst C, Cristofaro B, Mathivet T, Xu Y, Soueid J, Fortuna V et al (2012) Semaphorin3A, Neuropilin‐1, and PlexinA1 are required for lymphatic valve formation. Circ Res 111: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha B, Ho Y‐C, Geng X, Mahamud MR, Chen L, Kim Y, Choi D, Kim TH, Randolph GJ, Cao X et al (2020) YAP and TAZ maintain PROX1 expression in the developing lymphatic and lymphovenous valves in response to VEGF‐C signaling. Development 147: dev195453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Park E, Jung E, Seong YJ, Hong M, Lee S, Burford J, Gyarmati G, Peti‐Peterdi J, Srikanth S et al (2017a) ORAI1 Activates proliferation of lymphatic endothelial cells in response to laminar flow through Krüppel‐like factors 2 and 4. Circ Res 120: 1426–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Park E, Jung E, Seong YJ, Yoo J, Lee E, Hong M, Lee S, Ishida H, Burford J et al (2017b) Laminar flow downregulates Notch activity to promote lymphatic sprouting. J Clin Invest 127: 1225–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co M, Anderson AG, Konopka G (2020) FOXP transcription factors in vertebrate brain development, function, and disorders. Wiley Interdiscip Rev Dev Biol 9: e375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch SB, Graham SA, Quevedo M, Vino A, Dekkers DHW, Deriziotis P, Sollis E, Demmers J, Poot RA, Fisher SE (2018) Proteomic analysis of FOXP proteins reveals interactions between cortical transcription factors associated with neurodevelopmental disorders. Hum Mol Genet 27: 1212–1227 [DOI] [PubMed] [Google Scholar]
- French CA, Groszer M, Preece C, Coupe A‐M, Rajewsky K, Fisher SE (2007) Generation of mice with a conditional Foxp2 null allele. Genesis 45: 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Stritt S, Ortsäter H, Hernandez Vasquez M, Kaakinen M, Vicente A, Wiseman J, Eklund L, Martínez‐Torrecuadrada JL, Vestweber D et al (2020) EphrinB2‐EphB4 signalling provides Rho‐mediated homeostatic control of lymphatic endothelial cell junction integrity. eLife 9: e57732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Cha B, Mahamud MR, Lim K‐C, Silasi‐Mansat R, Uddin MKM, Miura N, Xia L, Simon AM, Engel J et al (2016) Multiple mouse models of primary lymphedema exhibit distinct defects in lymphovenous valve development. Dev Biol 409: 218–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Cha B, Mahamud MR, Srinivasan RS (2017) Intraluminal valves: development, function and disease. Dis Model Mech 10: 1273–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Yanagida K, Akwii RG, Choi D, Chen L, Ho YC, Cha B, Mahamud MR, Berman de Ruiz K, Ichise H et al (2020) S1PR1 regulates the quiescence of lymphatic vessels by inhibiting laminar shear stress‐dependent VEGF‐C signaling. JCI Insight 5: e137652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross‐Thebing T, Paksa A, Raz E (2014) Simultaneous high‐resolution detection of multiple transcripts combined with localization of proteins in whole‐mount embryos. BMC Biol 12: 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer‐Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostat Oxf Engl 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jurisic G, Maby‐El Hajjami H, Karaman S, Ochsenbein AM, Alitalo A, Siddiqui SS, Ochoa Pereira C, Petrova TV, Detmar M (2012) An unexpected role of semaphorin3a‐neuropilin‐1 signaling in lymphatic vessel maturation and valve formation. Circ Res 111: 426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärpänen T, Heckman CA, Keskitalo S, Jeltsch M, Ollila H, Neufeld G, Tamagnone L, Alitalo K (2006) Functional interaction of VEGF‐C and VEGF‐D with neuropilin receptors. FASEB J 20: 1462–1472 [DOI] [PubMed] [Google Scholar]
- Kazenwadel J, Secker GA, Liu YJ, Rosenfeld JA, Wildin RS, Cuellar‐Rodriguez J, Hsu AP, Dyack S, Fernandez CV, Chong C‐E et al (2012) Loss‐of‐function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood 119: 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazenwadel J, Betterman KL, Chong C‐E, Stokes PH, Lee YK, Secker GA, Agalarov Y, Demir CS, Lawrence DM, Sutton DL et al (2015) GATA2 is required for lymphatic vessel valve development and maintenance. J Clin Invest 125: 2979–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazenwadel J, Harvey NL (2018) Lymphatic endothelial progenitor cells: origins and roles in lymphangiogenesis. Curr Opin Immunol 53: 81–87 [DOI] [PubMed] [Google Scholar]
- Kim J‐H, Hwang J, Jung JH, Lee H‐J, Lee DY, Kim S‐H (2019) Molecular networks of FOXP family: dual biologic functions, interplay with other molecules and clinical implications in cancer progression. Mol Cancer 18: 180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA (2001) Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J Exp Med 193: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha‐Khadem F, Monaco AP (2001) A forkhead‐domain gene is mutated in a severe speech and language disorder. Nature 413: 519–523 [DOI] [PubMed] [Google Scholar]
- Lee S, Kang J, Yoo J, Ganesan SK, Cook SC, Aguilar B, Ramu S, Lee J, Hong Y‐K (2009) Prox1 physically and functionally interacts with COUP‐TFII to specify lymphatic endothelial cell fate. Blood 113: 1856–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model‐based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE (2004) Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 24: 809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Shakya A, Guo X, Zhang H, Tantin D, Jensen PE, Chen X (2012) Constitutive nuclear localization of NFAT in Foxp3+ regulatory T cells independent of calcineurin activity. J Immunol 188: 4268–4277 [DOI] [PubMed] [Google Scholar]
- Lin F‐J, Chen X, Qin J, Hong Y‐K, Tsai M‐J, Tsai SY (2010) Direct transcriptional regulation of neuropilin‐2 by COUP‐TFII modulates multiple steps in murine lymphatic vessel development. J Clin Invest 120: 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugano R, Vemuri K, Yu D, Bergqvist M, Smits A, Essand M, Johansson S, Dejana E, Dimberg A (2018) CD93 promotes β1 integrin activation and fibronectin fibrillogenesis during tumor angiogenesis. J Clin Invest 128: 3280–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons O, Saha P, Seet C, Kuchta A, Arnold A, Grover S, Rashbrook V, Sabine A, Vizcay‐Barrena G, Patel A et al (2017) Human venous valve disease caused by mutations in FOXC2 and GJC2. J Exp Med 214: 2437–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen T, Adams RH, Bailey J, Lu Q, Ziemiecki A, Alitalo K, Klein R, Wilkinson GA (2005) PDZ interaction site in ephrinB2 is required for the remodeling of lymphatic vasculature. Genes Dev 19: 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Almedina S, Martinez‐Corral I, Holdhus R, Vicente A, Fotiou E, Lin S, Petersen K, Simpson MA, Hoischen A, Gilissen C et al (2016) EPHB4 kinase‐inactivating mutations cause autosomal dominant lymphatic‐related hydrops fetalis. J Clin Invest 126: 3080–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Corral I, Stanczuk L, Frye M, Ulvmar MH, Diegez‐Hurtado R, Olmeda D, Makinen T, Ortega S (2016) Vegfr3‐CreER (T2) mouse, a new genetic tool for targeting the lymphatic system. Angiogenesis 19: 433–445 [DOI] [PubMed] [Google Scholar]
- Norrmén C, Ivanov KI, Cheng J, Zangger N, Delorenzi M, Jaquet M, Miura N, Puolakkainen P, Horsley V, Hu J et al (2009) FOXC2 controls formation and maturation of lymphatic collecting vessels through cooperation with NFATc1. J Cell Biol 185: 439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein AM, Karaman S, Jurisic G, Detmar M (2014) The role of neuropilin‐1/semaphorin 3A signaling in lymphatic vessel development and maturation. Adv Anat Embryol Cell Biol 214: 143–152 [DOI] [PubMed] [Google Scholar]
- Oliver G, Kipnis J, Randolph GJ, Harvey NL (2020) The lymphatic vasculature in the 21st century: novel functional roles in homeostasis and disease. Cell 182: 270–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P et al (2011) Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet 43: 929–931 [DOI] [PubMed] [Google Scholar]
- Petrova TV, Karpanen T, Norrmén C, Mellor R, Tamakoshi T, Finegold D, Ferrell R, Kerjaschki D, Mortimer P, Ylä‐Herttuala S et al (2004) Defective valves and abnormal mural cell recruitment underlie lymphatic vascular failure in lymphedema distichiasis. Nat Med 10: 974–981 [DOI] [PubMed] [Google Scholar]
- Petrova TV, Koh GY (2020) Biological functions of lymphatic vessels. Science 369: eaax4063 [DOI] [PubMed] [Google Scholar]
- Potente M, Mäkinen T (2017) Vascular heterogeneity and specialization in development and disease. Nat Rev Mol Cell Biol 18: 477–494 [DOI] [PubMed] [Google Scholar]
- Sabine A, Agalarov Y, Maby‐El Hajjami H, Jaquet M, Hägerling R, Pollmann C, Bebber D, Pfenniger A, Miura N, Dormond O et al (2012) Mechanotransduction, PROX1, and FOXC2 cooperate to control connexin37 and calcineurin during lymphatic‐valve formation. Dev Cell 22: 430–445 [DOI] [PubMed] [Google Scholar]
- Sabine A, Bovay E, Demir CS, Kimura W, Jaquet M, Agalarov Y, Zangger N, Scallan JP, Graber W, Gulpinar E et al (2015) FOXC2 and fluid shear stress stabilize postnatal lymphatic vasculature. J Clin Invest 125: 3861–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE (2007) Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 134: 1991–2000 [DOI] [PubMed] [Google Scholar]
- Sin C, Li H, Crawford DA (2015) Transcriptional regulation by FOXP1, FOXP2, and FOXP4 dimerization. J Mol Neurosci 55: 437–448 [DOI] [PubMed] [Google Scholar]
- Srinivasan RS, Geng X, Yang Y, Wang Y, Mukatira S, Studer M, Porto MPR, Lagutin O, Oliver G (2010) The nuclear hormone receptor Coup‐TFII is required for the initiation and early maintenance of Prox1 expression in lymphatic endothelial cells. Genes Dev 24: 696–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczuk L, Martinez‐Corral I, Ulvmar MH, Zhang Y, Laviña B, Fruttiger M, Adams R, Saur D, Betsholtz C, Ortega S et al (2015) cKit lineage hemogenic endothelium‐derived cells contribute to mesenteric lymphatic vessels. Cell Rep 10: 1708–1721 [DOI] [PubMed] [Google Scholar]
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L, Tong H, Chen L (2006) Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14: 159–166 [DOI] [PubMed] [Google Scholar]
- Sweet DT, Jiménez JM, Chang J, Hess PR, Mericko‐Ishizuka P, Fu J, Xia L, Davies PF, Kahn ML (2015) Lymph flow regulates collecting lymphatic vessel maturation in vivo . J Clin Invest 125: 2995–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Hollmén M, Dermadi D, Pan J, Brulois KF, Kaukonen R, Lönnberg T, Boström P, Koskivuo I, Irjala H et al (2019) Single‐cell survey of human lymphatics unveils marked endothelial cell heterogeneity and mechanisms of homing for neutrophils. Immunity 51: 561–572.e5 [DOI] [PubMed] [Google Scholar]
- Tammela T, Saaristo A, Holopainen T, Lyytikkä J, Kotronen A, Pitkonen M, Abo‐Ramadan U, Ylä‐Herttuala S, Petrova TV, Alitalo K (2007) Therapeutic differentiation and maturation of lymphatic vessels after lymph node dissection and transplantation. Nat Med 13: 1458–1466 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvmar MH, Mäkinen T (2016) Heterogeneity in the lymphatic vascular system and its origin. Cardiovasc Res 111: 310–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L et al (2018) A molecular atlas of cell types and zonation in the brain vasculature. Nature 554: 475–480 [DOI] [PubMed] [Google Scholar]
- Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E et al (2011) Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet 7: e1002145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baeyens N, Corti F, Tanaka K, Fang JS, Zhang J, Jin Y, Coon B, Hirschi KK, Schwartz MA et al (2016) Syndecan 4 controls lymphatic vasculature remodeling during mouse embryonic development. Development 143: 4441–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle JT, Oliver G (1999) Prox1 function is required for the development of the murine lymphatic system. Cell 98: 769–778 [DOI] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF et al (2006) FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 126: 375–387 [DOI] [PubMed] [Google Scholar]
- Xu Y, Yuan Li, Mak J, Pardanaud L, Caunt M, Kasman I, Larrivée B, del Toro R, Suchting S, Medvinsky A et al (2010) Neuropilin‐2 mediates VEGF‐C‐induced lymphatic sprouting together with VEGFR3. J Cell Biol 188: 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Yoshimatsu Y, Morishita Y, Miyazono K, Watabe T (2009) COUP‐TFII regulates the functions of Prox1 in lymphatic endothelial cells through direct interaction. Genes Cells 14: 425–434 [DOI] [PubMed] [Google Scholar]
- You L‐R, Lin F‐J, Lee CT, DeMayo FJ, Tsai M‐J, Tsai SY (2005) Suppression of Notch signalling by the COUP‐TFII transcription factor regulates vein identity. Nature 435: 98–104 [DOI] [PubMed] [Google Scholar]
- Zawieja DC (2009) Contractile physiology of lymphatics. Lymphat Res Biol 7: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Brady J, Liang W‐C, Wu Y, Henkemeyer M, Yan M (2015) EphB4 forward signalling regulates lymphatic valve development. Nat Commun 6: 6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Source Data for Expanded View/Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
RNA microarray data that support the findings of this study have been deposited in GEO (Gene Expression Omnibus) repository with the accession code GSE159842 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159842).


