Summary
The human genetic dissection of clinical phenotypes is complicated by genetic heterogeneity. Gene burden approaches that detect genetic signals in case-control studies are underpowered in genetically heterogeneous cohorts. We therefore developed a genome-wide computational method, network-based heterogeneity clustering (NHC), to detect physiological homogeneity in the midst of genetic heterogeneity. Simulation studies showed our method to be capable of systematically converging genes in biological proximity on the background biological interaction network, and capturing gene clusters harboring presumably deleterious variants, in an efficient and unbiased manner. We applied NHC to whole-exome sequencing data from a cohort of 122 individuals with herpes simplex encephalitis (HSE), including 13 individuals with previously published monogenic inborn errors of TLR3-dependent IFN-α/β immunity. The top gene cluster identified by our approach successfully detected and prioritized all causal variants of five TLR3 pathway genes in the 13 previously reported individuals. This approach also suggested candidate variants of three reported genes and four candidate genes from the same pathway in another ten previously unstudied individuals. TLR3 responsiveness was impaired in dermal fibroblasts from four of the five individuals tested, suggesting that the variants detected were causal for HSE. NHC is, therefore, an effective and unbiased approach for unraveling genetic heterogeneity by detecting physiological homogeneity.
Keywords: genetic heterogeneity, physiological homogeneity, incomplete penetrance, herpes simplex encephalitis, network biology, next-generation sequencing, cohort analysis, gene clustering, software
Introduction
The germline genetic component of many human diseases displays substantial heterogeneity, including both locus and allelic heterogeneity.1, 2, 3 Variants of genes from the same or related pathways can underlie the same or similar disorders.1,2 Along with genetic heterogeneity, incomplete penetrance is another obstacle hindering the identification of disease-causing variants, which may be clinically silent, even in relatives of an index case. For example, a study of 589,306 genomes identified 13 healthy adults with genotypes of eight severe Mendelian diseases.4 Severe infectious diseases may display both genetic heterogeneity and incomplete penetrance. Most of the infectious diseases studied to date have been shown to be genetically heterogeneous and can follow a “monogenic non-Mendelian” pattern,2 as shown by the finding that 15 human infections caused by viruses, bacteria, fungi, or parasites were monogenic, often with incomplete penetrance, in at least one affected individual.2 When two or more susceptibility genes were found, their products were often biologically connected.2 This may reflect the nature of the search leading to their discovery, based on an anchor gene, but, alternatively, genetic heterogeneity may underlie physiological homogeneity in many or even most individuals with a given condition. A landscape of genetic heterogeneity underlying physiological homogeneity is emerging, and may apply to a sizeable proportion of individuals with various severe infectious diseases.1,2
Next-generation sequencing (NGS) technologies have greatly accelerated the discovery of genetic lesions underlying human diseases.2 Most of the methods currently widely used in searches for pathogenic variants in NGS data are usually one-dimensional. They sequentially filter the variants by quality control (QC) metrics and population minor allele frequency (MAF), prioritize the variants according to their annotated molecular consequences and predicted deleteriousness, apply burden tests to detect enrichment in particular genes and variants relative to a control group, and finally propose risk alleles on the basis of statistical significance.3 These methods assume genetic homogeneity, in at least a proportion of individuals with the condition studied. They cannot capture genetic lesions in multiple genes of close biological relevance, which may be collectively, but not individually, significant. The notion of “network medicine” may provide a framework for tackling this challenge, as human inherited conditions, rare or common, are rarely consequences of abnormalities in a single gene, and more commonly result from the perturbation of a functionally related molecular network.5 In this light, the detection of physiological homogeneity could facilitate the development of a multidimensional approach for intersecting and leveraging NGS data with biological networks and pathways, to detect the nexus of heterogeneous genetic defects in biological proximity within a cohort of individuals with the same disease.
The application of network/pathway-based methodologies has progressed in multiple studies for rare diseases6,7 and cancer genomics,8,9 and for genome-wide association studies (GWASs) of complex diseases and traits.10,11 These studies have proposed different network concept for the detection of disease-associated genes from pathways, protein-protein interaction (PPI) networks, or gene co-expression networks, providing many examples of the power of network medicine. However, these methodologies have not been presented as practical tools suitable for implementation in clustering analyses on NGS data for case-control studies, and none was designed to identify clusters of gene candidates at the individual level. Meanwhile, there are several methods that could search for variants in physiologically related genes from NGS data, but they need to predefine gene sets before the analysis.12 We therefore aimed to develop a practical genome-wide computational approach connecting deleterious genetic heterogeneity with physiological homogeneity, by integrating NGS data, population genetics, predictions of mutation deleteriousness, biological interaction networks, pathway information, gene ontology annotations and statistics, in order to identify significant disease-specific genetic signals at the gene cluster level in an unbiased, efficient, and systematic manner.6,13,14 We developed the network-based heterogeneity clustering (NHC) approach for this purpose.
Material and methods
Samples
In this study, we used three sets of samples to develop and validate our NHC method. A group of 893 European individuals with severe infectious diseases, including viral, bacterial, and fungal infections, from our in-house Human Genetics of Infectious Diseases (HGID) database, was used for simulation studies. A group of 122 European individuals with herpes simplex virus-1 (HSV-1) encephalitis (HSE) from our HGID database was used for the application of our method to the detection of HSE-causing signals. These individuals of European origin were identified by principal component analysis (PCA) on whole-exome sequencing (WES) data. We also used 490 healthy European individuals from the 1000 Genomes Project (1KGP) as control subjects.15 We focused on the European population in this study, as there is evidence to suggest that ethnic homogeneity between affected individuals and control subjects is important in variant filtration and case-control enrichment tests.3 Figure S1 shows the PCA plot of the first two principal components (PCs) for all the European individuals used in this study, together with all the individuals identified globally for whom data were deposited in our HGID database. The inclusion of all the human subjects studied here was approved by the appropriate institutional review board.
Variant detection and filtration
We performed exome capture with SureSelect Human All Exon V4+UTRs on our in-house WES data, aligned reads to the human reference genome (hg19) with the maximum exact matches algorithm in the Burrows-Wheeler Aligner (BWA),16 and used the Genome Analysis Software Kit (GATK) v3.3 best practice pipeline to process the data.17 We filtered the variants on the basis of multiple QC metrics:18 depth of coverage (DP) ≥ 7, mapping quality (MQ) ≥ 60, variant quality (VARQUAL) ≥ 45, and minor read ratio (MRR) < 0.2. We annotated the variants with SnpEff19 and focused on the missense or loss-of-function (MisLoF) variants annotated as “missense,” “frameshift,” “stop-gained,” “stop-lost,” “start-lost,” “splice-acceptor,” or “splice-donor.”
For a given disease, knowledge of its prevalence and mode of inheritance facilitate the estimation of the compatible MAF cutoff for the disease, and the removal of variants above the MAF cutoff,20 based on the observed MAF in the gnomAD database.21 The common false-positive variants in our in-house HGID database of individuals with infectious diseases were also used to filter the variants further.22 We focused on potentially deleterious variants, using the gene damage index (GDI),23 combined annotation dependent depletion score (CADD),24 and mutation significance cutoff (MSC).25 GDI is an indicator of the biological indispensability (low GDI) or redundancy (high GDI) of a given gene.23 CADD is a composite score representing the deleteriousness of a given variant.24 MSC95 is the lower boundary of the 95% confidence interval of CADD scores for all reported disease-causing mutations of a given gene.25 In this study, we used GDI ≤ 10, and CADD ≥ 10 or CADD ≥ MSC95 as the mutation deleteriousness cutoff for shortlisting the presumably deleterious variants in each individual. The combination of these filtration parameters, population genetics, and predictions of mutation deleteriousness has already been successfully applied to the discovery of a number of disease-causing mutations.3,20,26
Biological network construction
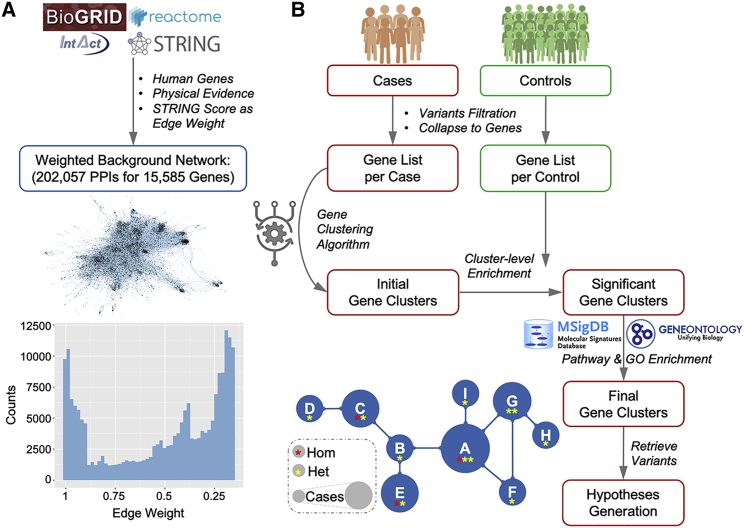
We obtained 1,760,357 PPIs from the BioGRID database,27 1,063,382 PPIs from the IntAct database,28 and 89,955 PPIs from the REACTOME database.29 Following restriction to human genes and the requirement for evidence of physical interactions, we retained 363,547 PPIs, 65,573 PPIs, and 18,119 PPIs, respectively, from these databases. These three PPI datasets were then merged into a set of 420,785 unique PPIs for 18,892 human genes. We also obtained the STRING v.11 database,30 which provides a composite score to approximate the probability of an interaction based on multiple evidence (e.g., experimental, co-expression, text-mining, etc.). We cross-referenced the STRING database with the PPI dataset and assigned the STRING score as the edge weight of each interaction, to represent the biological proximity between two genes. Finally, we constructed an edge-weighted background biological network of 202,057 PPIs for 15,585 human genes (Figure 1A). All the PPIs mentioned above were processed by removing self-interacting genes and duplicated gene pairs.
Figure 1.
The general framework of the NHC approach
(A) Data collection and processing to construct the edge-weighted background biological network based on protein-protein interactions, the graphic visualization of the network, and its edge-weight distribution.
(B) Flowchart of NHC detection of gene clusters harboring genetic heterogeneity with close biological relevance.
NHC gene clustering algorithm
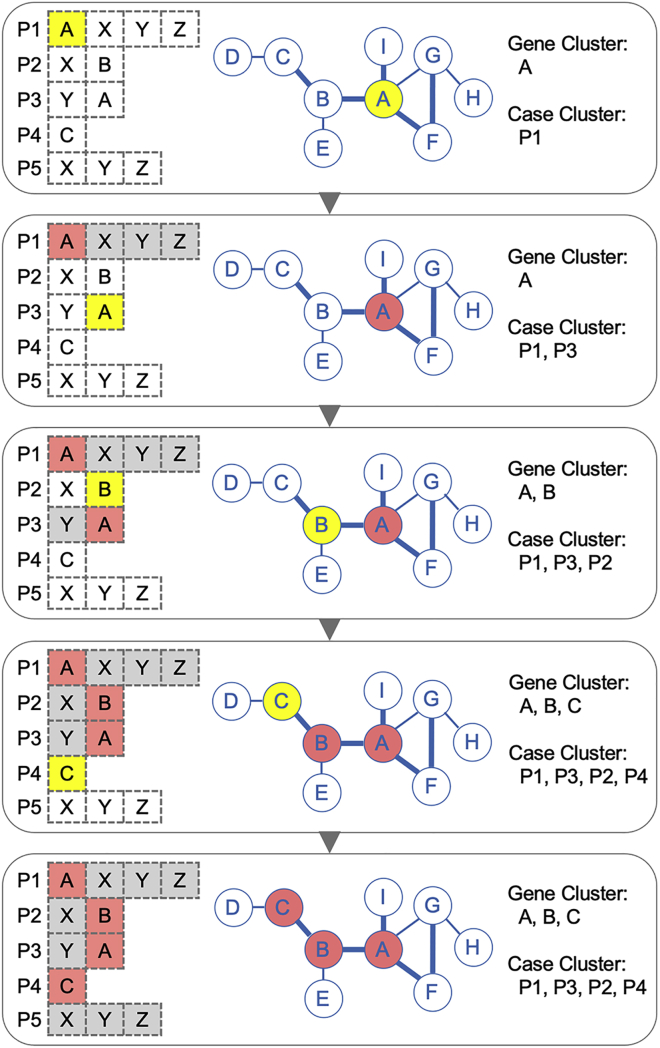
We collapsed the potentially deleterious variants into the genes harboring them, and the gene clustering algorithm was designed to traverse all genes in all individuals in the background network to converge genes that are biologically close. As illustrated in Figure 2, the algorithm is initialized with a list of genes per individual and an edge-weighted background network. The algorithm starts from one gene in one individual and searches for the next closest gene above the edge weight cutoff in the rest of the individuals. We used a stringent STRING score ≥ 0.99 to cluster the genes of the greatest biological relevance. At each step in the clustering iteration, the newly identified gene and individual are grouped into the existing gene cluster and case cluster. The clustering algorithm continues to cluster the genes with the highest degree of biological proximity, by traversing all genes in all individuals iteratively in the background network, until all individuals have been visited or no more genes in the unvisited individuals are beyond the edge-weight threshold for gene clustering. A full round of clustering yields one gene cluster and its corresponding case cluster as output.
Figure 2.
Illustration of the gene-clustering algorithm
The list of genes harboring qualifying variants in five affected individuals is shown on the left, the center shows the background network in which edge thickness represents the edge weight and the bold edge is chosen as the clustering cutoff, and the gene cluster and corresponding case cluster generated are shown on the right (yellow indicates a new gene and a new case identified in each step of clustering; red indicates the genes and cases already in the cluster; white indicates the genes available for clustering; and gray indicates the genes closed for clustering).
As previously reported, disease-associated genes are usually identified by avoiding hub genes, which are highly connected with other molecules.5 During gene clustering, the algorithm skips the hub genes, to prevent the formation of giant gene clusters due to the high connectivity of the hub genes in the background network. The connectivity of each gene is defined by the number of its interacting genes with a STRING score above 0.9 (a high-confidence PPI cutoff defined by the STRING database) in the background network. Here, we considered hub genes to be those with a connectivity ≥ 50 (constituting ∼2% of the total genes in the background network). In other words, we skip the genes with more than 50 high-confidence PPIs in the gene clustering process. In the code provided, this parameter can be modified to include all genes or to exclude more hub genes, as appropriate for the analysis of the cohort concerned and specified by the user.
Once a round of clustering is completed, the algorithm starts again from another gene in the same individual, to converge another gene cluster and its corresponding case cluster. Once all the genes of this individual have been used as the starting point for clustering, the algorithm moves on to the next individual. This process is repeated for each gene of each individual, to generate a number of gene clusters, each displaying internal biological closeness. The algorithm then iteratively merges two gene clusters if one cluster is a superset/subset of the other, or if the two clusters with the greatest overlap have more than 50% (common genes/union genes) of their genes in common, thereby reducing the number of gene clusters, such that these clusters are externally more different from each other. Given a number of n cases with a mean of m qualifying genes per case, the computational complexity of this algorithm is O(n∙m∙log(n∙m)).
NHC-boost gene clustering algorithm
For dealing with large cohorts of cases, we implemented a boosted version of the algorithm (NHC-boost), following the same concept as the original algorithm, but traversing each gene of a specific case only once. If a given gene from a specific case has been assigned to one cluster, it is not traversed or clustered again in the remaining clustering iterations. This modification may slightly decrease the size of the gene clusters, but significantly increases the computation efficiency, by reducing computational complexity form the original O(n∙m∙log(n∙m)) to the boosted O(n∙m).
Cluster-level enrichment
We performed a PC-adjusted cluster-level enrichment test to determine the gene clusters statistically enriched in the case cohort relative to the control cohort, and also to account for the ethnicity-related genetic heterogeneity in the HGID and 1KGP databases (Figure S1). We performed PCA on all individuals, using the high-quality common variants. We extracted the first five PCs for each individual, on which we performed logistic regression analysis with the glm function in R to determine the proportion of carriers between affected individuals and control subjects for each gene cluster. We used the p value indicating the statistical significance of each gene cluster in affected individuals versus control subjects, and took a p value ≤ 0.01 as the threshold for cluster-level significance. This p value cutoff could be customized for different applications, as the number of gene clusters and the size of each gene cluster may vary considerably with the use of different parameters for variant filtration, different edge-weight cutoffs for gene clustering, and different cohort sizes.
Pathway and gene ontology enrichment
We collected a total of 1,720 pathways (187 KEGG pathways and 1,533 REACTOME pathways) from the MSigDB database,31 and performed Fisher’s exact tests to assess the statistical significance of each gene cluster versus each pathway. We used a p value ≤ 1e−5 as the cutoff for pathway enrichment, which is more stringent than the adjusted p value (0.05/1720 = 2.9e−5). We used the most enriched pathway (the pathway with the lowest p value) as a surrogate for the primary physiological nature of each gene cluster. We also collected 8,992 biological process and 2,812 molecular function annotations from the Gene Ontology (GO) database32 that contain more than one gene. We performed Fisher’s exact tests to evaluate the statistical significance of each gene cluster versus each biological process and molecular function with a p value cutoff at 1e−5. Enrichment in gene ontology terms can suggest the biological nature of small gene clusters not displaying significant enrichment at pathway level, but functionally important and potentially disease related.
Control-embedded pathways
As a means of understanding the gene clusters harboring the background variations and their enriched pathways within the control cohort of 490 healthy individuals, we ran our method within control subjects. We randomly picked 100 individuals, in which gene clusters were identified and subjected to enrichment analysis versus the remaining 390 control subjects, for 100 simulations. We identified the gene clusters with cluster-level p values ≤ 0.01 and with at least one pathway enriched with a p value ≤ 1e−5. We extracted the most enriched pathway for each of these gene clusters, determined the number of occurrences of these pathways, and defined the pathways occurring in more than 5 of the 100 simulations as relatively common pathways in control subjects. We thus obtained a list of 58 control-embedded pathways (Table S1), emerging from the background variations. We used these pathways to classify the gene clusters generated from the case cohort.
Gene cluster output
The final output of NHC is the significant gene clusters converged from the genes carrying qualifying variants in the case cohort, with cluster-level p values ≤ 0.01 versus the control cohort, and at least one pathway or one gene ontology term enriched with a p values ≤ 1e−5. Gene clusters for which the most enriched pathway is not a control-embedded pathway are prioritized as class I clusters, with all other gene clusters classified as class II clusters. Gene clusters are first categorized by these two classes, and then ranked by their cluster-level significance. The previously identified qualifying variants are then retrieved for each cluster. Finally, multiple levels of information are summarized for each gene cluster: the list of genes, the individuals carrying variants of these genes, presumably deleterious homozygous, and heterozygous variants of these genes, cluster-level significance, the list of enriched pathways, the most enriched pathway indicating the physiological nature of the cluster, and the list of enriched gene ontology terms. VCF-format files containing the presumably deleterious variants are generated for each cluster for further variant investigations,20,33,34 and network-format files are also generated for each cluster for network-based computations35,36 and visualizations.37
Simulation studies
We performed two simulation studies to gain more insight into the workings of our method. The first was designed to test the null hypothesis that a cohort of individuals with multiple disease phenotypes should not yield biologically and statistically significant gene clusters associated with multiple diseases. We sampled case cohorts from a group of 893 European individuals with different severe infectious diseases in the HGID database, and we used 490 healthy individuals from 1KGP as the control. We took the same variant filtration criteria as in the HSE study. We performed 100 independent tests of our method on a cohort of 100 cases selected at random, and we identified the class I gene clusters (p values ≤ 0.01) in all simulations to analyze the significant gene clusters generated from this null hypothesis test.
The second simulation study was designed to detect the simulated disease signals from varying numbers of simulated individuals with varying numbers of mutated genes in a predefined pathway. We used the same data as in the first simulation study. We chose a development-related Hippo pathway, due to the high intrinsic likelihood of these individuals carrying deleterious signals from immune/infection-related pathways. According to the REACTOME database, the Hippo signaling pathway has a total of 20 genes, from which we selected eight (LATS1, MOB1A, MOB1B, SAV1, STK3, STK4, WWTR1, and YAP1), for which we created artificial stop-gained mutations at the 10th codon in their canonical transcripts (Figure S2, Table S2). If the nucleotides encoding the 10th amino acid span the splice site, the mutation was instead created at the 9th codon, to prevent potentially aberrant splicing. We confirmed these stop-gained mutations with SeqTailor.33 We randomly selected 100 individuals for 100 iterations and randomly assigned the artificial mutations of the eight genes independently to subgroups of 5, 10, 20, or 30 individuals, with 25 simulations for each subgroup size. As mutation assignment was a random process, it was possible for any given mutation to be assigned to more than one individual. In each simulation, we identified the gene cluster displaying the highest enrichment in the Hippo pathway as the Hippo cluster, and we checked whether these Hippo clusters included all the simulated genes and were highly ranked, as a demonstration that the simulated disease signal was effectively captured and prioritized.
SKAT-O test
For comparison with the NHC approach, we implemented a pathway-informed SKAT-O38 analysis of the HSE cohort. We used the same qualifying variants and genes as were obtained in the application of the NHC method in the HSE study. We first ran the SKAT-O test38 at the variant and gene levels in the HSE cohort, using a p value cutoff ≤ 0.01 for significant variant and gene hits. We also scripted a pathway-informed SKAT-O test, by providing 1,720 predefined pathways (187 KEGG and 1,533 REACTOME pathways) before running the SKAT-O test on the HSE cohort, using a p value cutoff ≤ 0.001 to identify significant pathway hits. SKAT-O tests were performed with the default settings. The p value cutoffs used here could be customized for different applications.
Study of TLR3 responsiveness in human fibroblasts
SV40-transformed fibroblasts were plated at a density of 105 cells per well in a 24-well plate and incubated overnight. The cells were left unstimulated or were stimulated with polyinosinic-polycytidylic acid (poly(I:C); Amersham) at a concentration of 25 μg/mL, and the culture medium was harvested after 24 h of stimulation. Cytokine concentrations in the culture medium were determined by ELISA. For the determination of IFN-λ, culture supernatants were incubated for 2 h in plates coated with 1 μg/mL monoclonal anti-human IL29 antibody (R&D). A biotinylated monoclonal secondary antibody directed against human IL29 (R&D) and streptavidin peroxidase were added, together with TMB (3,3′,5,5′-tetramethylbenzidine). The signal at 450 nm was then read with a plate fluorescence reader. The concentration of IL-6 in the cell culture supernatant was determined with an ELISA kit from R&D Systems.
Results
NHC, a network-based approach for detecting physiological homogeneity
NHC is designed to study NGS data from a cohort of individuals with the same disease as the input and to output significantly enriched gene clusters harboring presumably deleterious variants in these individuals as the causal candidates underlying the disease studied. Figure 1 illustrates the general framework of our approach for distilling large-scale biological data with interpretable parameters and converging genes into gene clusters of close biological relevance. The main framework of NHC is composed of the following steps.
-
(1)
A large-scale network of biological relevance is first established, by integrating the human PPIs from the BioGRID,27 IntAct,28 and REACTOME29 databases, and weighting the PPIs by scores from the STRING database30 to represent the level of biological relevance between genes. We thus obtained an edge-weighted background biological network of 202,057 PPIs for 15,585 human genes (Figure 1A).
-
(2)
Users then define a cohort of affected individuals and a cohort of control subjects with their genomic variant data and filter the variants by multiple parameters: QC metrics, MisLoF molecular consequences, population MAF compatible with the prevalence of the disease based on the gnomAD database,21 and predictions of mutation deleteriousness (e.g., CADD,24 MSC25). This variant filtration shortlists the variants most likely to confer a risk of the disease. These qualifying variants, which are presumed to be deleterious, are then collapsed into genes for each individual. Different analyses could be performed based on the different definitions of variant selection.
-
(3)
All the genes carrying qualifying variants are then traversed in all individuals in the edge-weighted background PPI network, and genes in biological proximity are iteratively converged into gene clusters (Figure 2). The algorithm starts from one gene in one individual, and iteratively searches for the closest gene above the edge-weight cutoff in the remaining individuals. Each round of clustering stops when all the individuals have been visited or no other gene in the unvisited individuals is above the edge-weight cutoff. The algorithm continues until every gene of every individual has been used once as the starting point for clustering. A number of gene clusters are obtained, each of which contains genes that are biologically close. The algorithm then iteratively merges two clusters, if one is a superset/subset of the other or if the two clusters with the greatest overlap have more than 50% of their genes in common, to obtain a smaller number of gene clusters that are externally more different from each other.
-
(4)
The statistical significance of each gene cluster in cases versus controls is then determined by PC-adjusted cluster-level enrichment. We used a stringent p value ≤ 0.01 as the cutoff to retain significant gene clusters in the case cohort. This cutoff is based on the average of ∼100 clusters formed in the simulation study under the null hypothesis.
-
(5)
KEGG + REACTOME pathway enrichment analysis is then performed on each gene cluster, retaining all pathways displaying enrichment with a p value ≤ 1e−5, a stringent cutoff adjusted by the number of pathways. The pathway with the greatest enrichment (the lowest p value) is considered to correspond to the biological nature of each gene cluster. Gene ontology enrichment analysis is also performed for biological processes and molecular functions on each gene cluster with a p value cutoff at 1e−5. Gene clusters for which no pathway or gene ontology is enriched are removed.
-
(6)
Steps 3–5 are then performed within the control cohort, to identify control-embedded pathways. A subgroup of controls is selected at random, in which gene clusters are identified and subjected to enrichment analysis versus the rest of the controls, for 100 simulations. The most enriched pathways of these gene clusters will be populated from all simulations. The frequently enriched pathways are considered to be control-embedded pathways.
-
(7)
The gene clusters from the case cohort are classified into classes I and II. If a gene cluster does not display the highest level of enrichment in a control-embedded pathway, it is considered as class I and likely to be more relevant to the disease studied. Otherwise, the gene cluster is considered as class II. Within each class, the gene clusters are ranked by cluster-level significance versus controls, as determined in step 4.
-
(8)
The presumably deleterious variants harbored in each gene cluster are retrieved, and the detected genetic heterogeneous signals are presented at the network, gene, variant, and individual levels.
Simulation study (I): Null hypothesis test
We conducted the first simulation study for a null hypothesis test to assess the assumption that a randomly assembled case cohort with multiple disease phenotypes does not display biologically relevant gene clusters significantly associated with a mixture of diseases. As our method is capable of detecting gene clusters for a portion of individuals displaying physiological homogeneity within a cohort displaying physiological heterogeneity, some disease signals might emerge, with a low discovery rate. We used 893 European individuals with viral, bacterial, and fungal infections from our HGID database. We randomly selected 100 of these individuals, to assemble a case cohort of different clinical phenotypes for 100 independent replicates and used 490 healthy European individuals from 1KGP as the control cohort in all simulations. The variant filtration criteria were as defined in material and methods. On average, each simulation yielded a total of 106 gene clusters (minimum: 90, maximum: 127), after merging the gene clusters from the initial output. By taking a cluster-level p value ≤ 0.01 and focusing on class I gene clusters, we found that 62/100 simulations detected no significant class I gene clusters. One significant class I gene cluster was identified in 30 of the remaining 38 simulations. The top gene clusters in these 38 simulations had p values of 0.00003 to 0.00744 (median: 0.002) and cluster sizes of 3 to 46 genes (median: 9) (Figure S3, Table S3). This null hypothesis simulation gave us a baseline for NHC detection in cohorts of individuals for whom no particular role of any specific pathway was expected to be detected.
Simulation study (II): Alternative hypothesis test
We conducted a second simulation study under the alternative hypothesis, to test NHC for the detection of simulated disease signals, in cohorts with different numbers of simulated individuals carrying mutations of genes from a predefined pathway. We used the same data as in simulation study I, with the random selection of 100 individuals, from whom we randomly selected a subgroup of 5, 10, 20, or 30 individuals to be assigned any of the artificial stop-gained mutations of eight genes of the Hippo signaling pathway (Figure S2, Table S2). We ran a total of 100 simulations, with 25 simulations per subgroup size. Mutations were assigned at random, permitting a given mutation to be assigned to more than one case. We considered the identification of a gene cluster most enriched in the Hippo pathway (Hippo cluster) to constitute the capture of the simulated disease signal.
We found that the Hippo cluster was detected with a p values ≤ 0.01 in all simulations with at least ten selected individuals (Figure S4, Table S4). When only five individuals were selected, the Hippo cluster was detected in 22/25 (88%) of the simulations, with the other three Hippo clusters close to the threshold for cluster-level significance (p values < 0.03). Prioritization of the Hippo cluster may have been affected by the smaller number of simulated genes and the smaller number of carriers, resulting in a sparsely populated cluster with a relatively lower level of significance versus controls. When 20 or 30 individuals were selected, the Hippo cluster was always identified as the top-ranked gene cluster. Thus, NHC was found to be capable of detecting and prioritizing the simulated disease signal with varying mutated genes and varying number of carriers.
Application to an HSE cohort (I): Detecting HSE-specific gene clusters
A good example of genetic heterogeneity underlying severe infectious diseases is provided by HSE, the most common and devastating viral encephalitis in Western countries, with an incidence of about 2–4 affected individuals per 1,000,000 people per year.39,40 Our previous studies have shown that HSE was caused by various monogenic inborn errors of TLR3-dependent IFN-α/β immunity in 13 HSE-affected individuals with causal rare variants of five genes of the TLR3 signaling pathway:39, 40, 41, 42, 43, 44, 45, 46, 47 TLR3 (MIM:48 603029), UNC93B1 (MIM: 608204), TICAM1 (TRIF) (MIM: 607601), TRAF3 (MIM: 601896), and TBK1 (MIM: 604834) (Table 1). Another group identified a case of HSE in a context of IRF3 (MIM: 603734) deficiency,49 which also affects the TLR3 signaling pathway. It is therefore becoming increasingly clear that HSE is caused by a collection of monogenic inborn errors of immunity displaying genetic heterogeneity and incomplete clinical penetrance at individual level but physiological homogeneity at cohort level.50,51 We evaluated the efficacy of our method on data from real individuals, by applying it to the analysis of WES data from a cohort of 122 European individuals with HSE (including the 13 previously published cases) from the HGID database, along with a control cohort of 490 healthy European individuals from 1KGP.15 We used multiple criteria to filter the variants: sequencing QC (DP ≥ 7, MQ ≥ 60, VARQUAL ≥ 45, MRR < 0.2), population MAF compatible with HSE prevalence (MAF ≤ 0.001 for heterozygous variants and MAF ≤ 0.03 for homozygous variants according to gnomAD21), predictions of mutation deleteriousness (CADD ≥ 10 or CADD ≥ MSC95),24,25 and MisLoF variants. We obtained a total of 14,729 rare and potentially deleterious variants from the 122 cases, which were collapsed into 7,512 genes, with a mean of 126 genes per case.
Table 1.
Published HSE-causing single-gene inborn errors of TLR3-dependent IFN-α/β immunity
| Gene | # Cases | Variant | Accession | Consequence | Zygosity | MAF (gnomAD v2.1.1) |
|---|---|---|---|---|---|---|
| TLR3 | 1 | g.187003919T>C | rs768091235 | missense (p.Leu360Pro) | het | 2.39e−05 |
| TLR3 | 3 | g.187004500C>T | rs121434431 | missense (p.Pro554Ser) | het | 4.06e−04 |
| TLR3 | 1 | g.187005068G>A | rs1280549921 | missense (p.Gly743Asp) | het | 3.98e−06 |
| TLR3 | 1 | g.187005076G>T | rs1554064929 | stop-gained (p.Glu746∗) | het | N/A |
| TLR3 | 1 | g.187005272G>T | rs1244010954 | missense (p.Arg811Ile) | het | N/A |
| TLR3 | 1 | g.187005912G>A | rs199768900 | missense (p.Arg867Gln) | hom | 6.48e−04 |
| TBK1 | 1 | g.64854030A>C | rs1010930015 | missense (p.Asp50Ala) | het | 4.20e−06 |
| TBK1 | 1 | g.64860798G>C | rs1555202947 | missense (p.Gly159Ala) | het | N/A |
| TICAM1 | 1 | g.4817833C>T | rs146550489 | missense (p.Ser186Leu) | het | 3.07e−04 |
| TICAM1 | 1 | g.4817969C>T | rs387907307 | stop-gained (p.Arg141∗) | hom | 4.01e−06 |
| TRAF3 | 1 | g.103342015C>T | rs143813189 | missense (p.Arg118Trp) | het | 1.44e−03 |
| UNC93B1 | 1 | g.67764121_67764124del | rs759883057 | indel-frameshift (p.Phe345fs) | hom | 2.55e−05 |
| UNC93B1 | 1 | g.67765829G>A | rs780094017 | missense (p.Gly261Ser) | hom | 4.01e−06 |
NHC generated an initial output of 225 gene clusters, which were then merged by combining the clusters with more than 50% overlap, to yield 143 gene clusters. Eight gene clusters were significantly enriched in affected individuals, with a cluster-level p values ≤ 0.01. Pathway enrichment analysis led to the identification of five gene clusters with significant pathways (Table 2), which were then classified into three class I and two class II gene clusters, based on the control-embedded pathways. These gene clusters were internally biologically close, externally different (no common genes between the five clusters), statistically significant from control subjects, functionally annotated with biological pathways and gene ontology terms, and carried presumably deleterious mutations. Each gene cluster was associated with physiological homogeneity in a proportion of HSE-affected individuals.
Table 2.
The five significant gene clusters, p value ≤ 0.01, detected by the NHC method in the HSE cohort of 122 individuals
| Cluster | #Genes | #Var Hom | Het | #Cases | Cluster p value | #Pathways | Top Pathway | #BP | Top BP | #MF | Top MF |
|---|---|---|---|---|---|---|---|---|---|---|
| Class I | ||||||||||
| #1 | 32 | 4 | 59 | 49 | 0.00125 | 17 | KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY (3.209e−15) | 12 | GO:0035666:TRIF-dependent toll-like receptor signaling pathway (5.015e−16) | 0 | – |
| #2 | 5 | 0 | 14 | 13 | 0.00274 | 11 | REACTOME_HDR_THROUGH_SINGLE_STRAND_ANNEALING_SSA (9.676e−10) | 4 | GO:1901796:regulation of signal transduction by p53 class mediator (2.108e−07) | 0 | – |
| #3 | 6 | 0 | 15 | 15 | 0.00564 | 3 | REACTOME_PEROXISOMAL_PROTEIN_IMPORT (7.262e−11) | 2 | GO:0006625:protein targeting to peroxisome (2.81e−11) | 0 | – |
| Class II | ||||||||||
| #4 | 30 | 1 | 48 | 44 | 0.00092 | 2 | REACTOME_MITOCHONDRIAL_TRANSLATION (1.121e−57) | 4 | GO:0070125:mitochondrial translational elongation (7.605e−64) | 2 | GO:0003735:structural constituent of ribosome (5.791e−42) |
| #5 | 10 | 0 | 24 | 22 | 0.00188 | 7 | REACTOME_PROCESSING_OF_INTRONLESS_PRE MRNAS (2.658e−17) | 6 | GO:0006378:mRNA polyadenylation (2.224e−16) | 0 | – |
Hom, homozygous; Het, heterozygous; BP, biological process; MF, molecular function.
Application to an HSE cohort (II): Genes harboring the known HSE-causing mutations in the top-ranked cluster
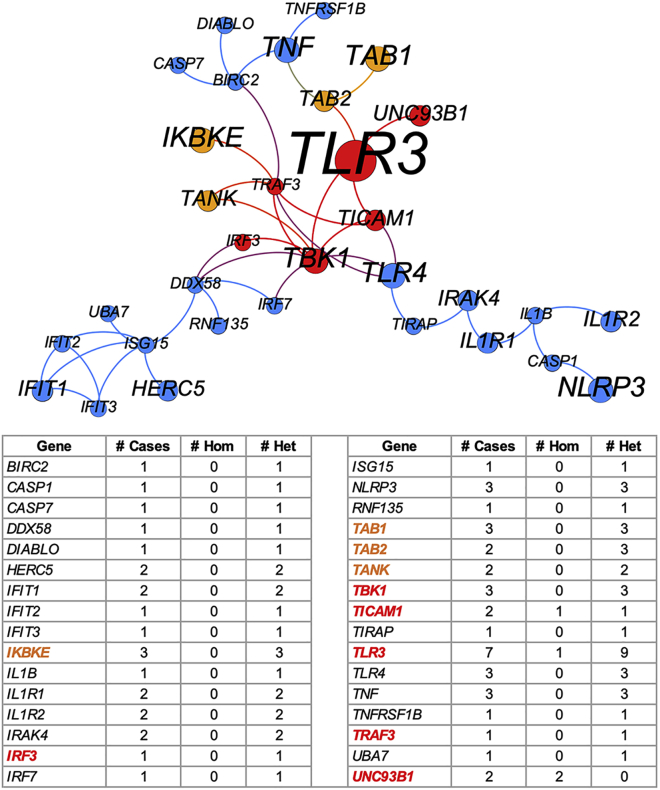
We found that the NHC method successfully detected, converged, and prioritized all five genes harboring HSE-causing mutations reported by us, with 13 deleterious variants in 13 affected individuals in the top-ranked class I gene cluster (15 variants in total, as 2 individuals carry 2 variants each). This gene cluster contained a total of 32 genes, harboring 4 homozygous and 59 heterozygous variants that are presumably deleterious, from 49 HSE-affected individuals, with a cluster-level p value of 0.00125 versus the control cohort (Table 2). This gene cluster was most enriched in the KEGG Toll-like receptor signaling pathway (p value = 3.2e−15) and in another 16 significantly enriched pathways with p values ≤ 1e−5 (Table S5), all of which were closely related to TLR or interferon signaling. This gene cluster was also enriched in 12 biological processes with p values ≤ 1e−5 and was most enriched in the GO:0035666 TRIF-dependent toll-like receptor signaling pathway (p value = 5.0e−16). No molecular function was significantly enriched in this gene cluster. Figure 3 visualizes this top-ranked gene cluster as a network, in which the known HSE genes are gathered at the center of the cluster.
Figure 3.
Network visualization of the top-ranked gene cluster most enriched in the TLR3 signaling pathway
Each node is a gene, and its size is proportional to the number of affected individuals carrying the presumably deleterious variants of the gene. The red nodes are the six reported genes, and the orange nodes are four candidate genes. Each edge represents an interaction between two genes that is above the edge-weight cutoff in the background network. The number of affected individuals carrying each gene and the number of homozygous and heterozygous variants harboring in each gene are provided.
Application to an HSE cohort (III): Detection of candidate genes in the top-ranked cluster and experimental validation
In addition to the previously published HSE-causing mutations of 5 TLR3 pathway genes in 13 affected individuals, this top-ranked cluster captured another three heterozygous variants of three of these genes (TLR3, IRF3, and TBK1) in three individuals that had not previously been studied (Table 3). It also identified another 33 affected indviduals carrying 45 potentially deleterious variants of 26 genes closely related to TLR3 signaling (Figure 3). This top-ranked gene cluster presented candidate genes for testing in these previously unstudied HSE-affected individuals. Based on literature review for the genes concerned and their population genetics data,20,21 we selected IKBKE (MIM: 605048), TAB1 (MIM: 602615), TAB2 (MIM: 605101), and TANK (MIM: 603893) as four candidate genes potentially able to cause HSE due to their involvement in TLR3-IFN signaling. TAB2 is thought to be recruited by TLRs for signal transduction, but this remains a matter of debate, because TAB2-deficient mice do not display TLR signaling abnormalities.52 TANK binds TBK1 and IKBKE to regulate type I interferon induction in antiviral innate immunity, and TBK1 activation is dependent on the integrity of TBK1/TANK interaction.53
Table 3.
Candidate HSE-causing variants in seven genes, three reported genes and four candidate genes, from 10 unstudied HSE-affected individuals, identified from the top-ranked gene cluster
| Case | Gene | Reported gene? (Y/N) | Variant | Accession | Consequence | Zygosity |
Family segregation (F, father; M, mother) |
MAF | CADD | Impaired TLR3 response (Y/N) |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | IKBKE | N | g.206666660C>T | rs782760912 | missense (p.Pro665Leu) | het | N/A | 3.18e−05 | 12.04 | Y |
| TAB2 | N | g.149699800C>T | N/A | missense (p.Pro250Leu) | het | N/A | N/A | 7.39 | ||
| TAB2 | N | g.149700172C>A | rs774198686 | missense (p.Pro374Gln) | het | N/A | 3.98e−06 | 26.2 | ||
| P2 | TAB2 | N | g.149700624A>G | rs142662439 | missense (p.Met525Val) | het | N/A | 2.40e−05 | 7.3 | N |
| P3 | TANK | N | g.162061198G>T | rs187514019 | missense (p.Gly74Val, p.Arg394Gly) | comp. het |
F: G74V het M: R394G het | 1.61e−03 | 10.31 | Y |
| P4 | TANK | N | g.162061198G>T | rs187514019 | missense (p.Gly74Val) | het | F: WT M: het |
1.61e−03 | 10.31 | Y |
| P5 | TLR3 | Y | g.186997949C>A | rs143307508 | missense (p.Thr59Asn) | het | F: WT M: het |
1.83e−04 | 23.9 | Y |
| X1 | IRF3 | Y | g.50164059G>A | rs561346823 | missense (p.Arg342Gln) | het | N/A | 1.15e−04 | 11.02 | N/A |
| X2 | IKBKE | N | g.206650100T>C | N/A | missense (p.Ile207Thr) | het | F: WT M: het |
N/A | 26.3 | N/A |
| X3 | TAB1 | N | g.39811120C>T | rs143512143 | missense (p.Pro48Leu) | het | N/A | 2.80e−04 | 28.7 | N/A |
| X4 | TAB1 | N | g.39811629G>A | rs767710748 | missense (p.Glu99Lys) | het | N/A | 4.01e−05 | 24.7 | N/A |
| X5 | TBK1 | Y | g.64883900G>C | N/A | essential splicing |
het | N/A | N/A | 24.1 | N/A |
P1–P5 are HSE-affected individuals for whom SV40 fibroblasts were available, whereas no SV40 fibroblasts were available for testing for X1–X5.
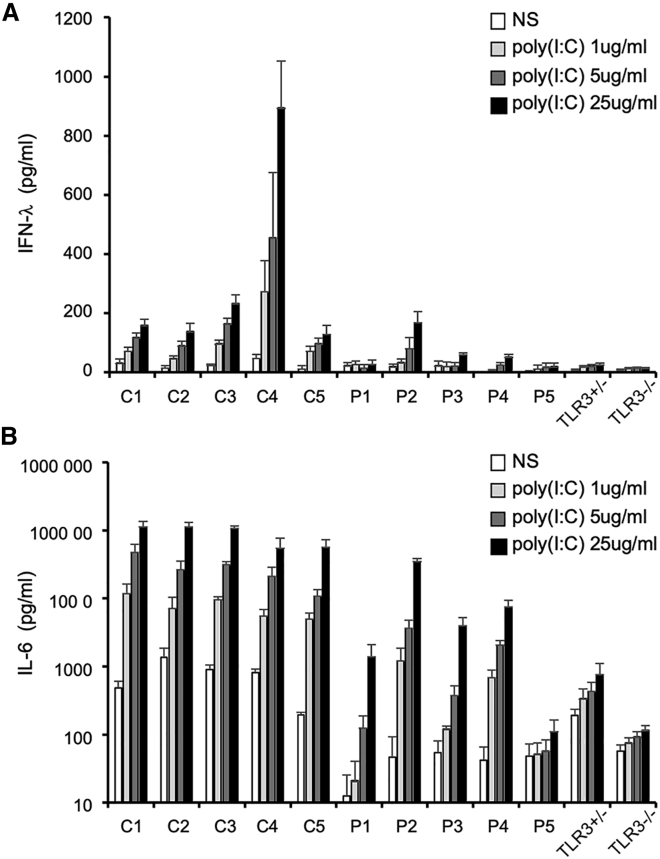
We identified eight presumably deleterious rare variants of these four candidate genes in seven unstudied HSE-affected individuals. Together with the three variants of three reported genes, we thus obtained a total of 11 variants of 7 genes from 10 affected individuals (Table 3). All these variants have a MAF < 0.001 and affect highly conserved residues in the corresponding molecules. The familial segregation of these variants suggested that they may underlie HSE through single-gene autosomal-dominant or digenic modes of inheritance. Dermal fibroblasts are a surrogate cell type for investigating TLR3 responsiveness.41,42 SV40-transformed fibroblast cells (SV40-fibroblasts) are available for five of the ten affected individuals and were used to analyze their responses to TLR3 stimulation. Four of the five affected individuals for whom SV40 fibroblasts were available (with variants of TAB2/IKBKE, TANK, and TLR3) displayed impaired IFN-λ and IL-6 production following simulation with various doses of poly(I:C), suggesting impaired TLR3 signaling due to these genetic variants (Figure 4). The current data suggest that there is a causal relationship between genotype and phenotype, but further experimental validation is required.
Figure 4.
TLR3 responsiveness in the fibroblasts of affected individuals and control subjects in terms of IFN-λ and IL-6 production
IFN-λ and IL-6 production in SV40 fibroblasts left non-stimulated (NS) or treated with poly(I:C) for 24 h, as measured by ELISA. C1-C5 are healthy control subjects. P1-P5 are HSE-affected individuals with TLR3 pathway gene mutations. SV40 fibroblasts from two previously published TLR3+/− and TLR3−/− cases are used as negative controls.
Application to an HSE cohort (IV): Testing on individuals of unknown etiology
We further tested our method on the HSE cohort excluding the 13 published cases with known disease etiologies. We tested the remaining 109 HSE cases with 490 healthy controls and yielded six gene clusters (p value ≤ 0.01): four class I and two class II clusters (Table S6). The top-ranked gene cluster, with a p value of 0.00176, remained the most strongly enrichment in the TLR signaling pathway, confirming the importance of this pathway in the etiology of HSE. This top-ranked gene cluster contained 27 genes in 37 cases, and the 27 genes identified were exactly the same as those in the previous top-ranked gene cluster generated from the full HSE cohort with exclusion of the five genes of published HSE-causing mutations. The other gene clusters may suggest other possible HSE-causing mechanisms. One class I cluster with a p value of 0.01023 just above the cutoff was identified as potentially interesting. It contained six genes from 24 cases and was most enriched in the KEGG regulation of autophagy pathway (p value = 3.1e−6). Some autophagy genes have been shown to inhibit viral replication by inducing type-I interferon production.54
Application to an HSE cohort (V): Comparison with a burden test on the HSE cohort
We compared the performance of our method with that of SKAT-O38 for detecting disease signals in the HSE cohort. SKAT-O is a genome-wide test of association between rare variants and phenotypes, for identifying the significant genes and variants in a given cohort. It combines the strengths of the burden test and variance-component tests and has been shown to outperform other tests for detecting disease signals when prior knowledge of disease etiology is limited.3 We ran SKAT-O on the same HSE cohort, with the same variant filtration parameters, at the gene and variant levels. It shortlisted 67 genes and 22 variants as significant hits, with p values ≤ 0.01. These hits captured one gene (TLR3) and one TLR3 mutation (Figure S5). These findings demonstrate that NHC exceeds SKAT-O for detecting known etiology of HSE, mainly due to the ability of NHC to link the genes with biological relevance and to measure the significance of the enrichment at cluster level, and also due to the genetic heterogeneity of HSE, implying that the causal genes/mutations would not be of genome-wide significance individually.
We further scripted a pathway-informed SKAT-O test, by providing 1,720 predefined pathways before running the SKAT-O on the HSE cohort. Taking a p value ≤ 0.001 as the pathway level significance cutoff, we obtained an output of five pathways (Table S7). The first three pathways were TLR-related or interferon-related: REACTOME trafficking and processing of endosomal TLR (p value = 0.0002), REACTOME TICAM1-dependent activation of IRF3 and IRF7 (p value = 0.0006), and REACTOME ZBP1-mediated induction of type I IFNs (p value = 0.0007), due to the relatively strong intrinsic TLR signals in the HSE cohort. However, none of these three pathways covered all five genes that harbor known HSE-causing mutations, and they did not include the candidate genes. The three previously most enriched pathways in our NHC top-ranked gene cluster (Table S5): KEGG toll-like receptor signaling pathway, REACTOME toll-like receptor cascades, and REACTOME innate immune system were ranked 127th, 9th, and 331st, respectively. The pathway-informed SKAT-O test is gene set biased and assesses all the qualifying genes in a pathway for enrichment in a supervised manner. Our results show that it can detect some of the reported genes that harbor known HSE-causing mutations but lacks power to identify candidate genes that are not part of a predefined pathway. By contrast, NHC is not gene set-biased and focuses on the cluster of closely interacting qualifying genes for pathway enrichment in an unsupervised manner.
Robustness of gene clustering algorithm and computation time
We assessed the robustness and stability of the gene clustering algorithm by randomly shuffling the choice of gene and individual as the starting point for gene clustering in the HSE cohort 100 times. We obtained exactly the same gene clusters, with exactly the same rankings in all tests. This finding demonstrates the robustness of NHC for delivering a stable result. NHC took ∼40 min to analyze data for 122 HSE-affected individuals versus 490 control subjects, on a desktop computer with 20 CPUs and 128 GB RAM. With the fixed variant filtration criteria, it took ∼15 min to analyze a cohort of 50 individuals, and ∼8 h for a cohort of 500 individuals, based on the testing of randomly assembled cohorts. If the variant filtration criteria were relaxed (i.e., more qualifying variants and genes) or the cohort size was increased (i.e., more individuals to transverse to converge gene clusters), the computation time would increase significantly. We therefore provide an alternative version, “NHC-boost,” which required ∼5 min to analyze the HSE cohort, outputting the same number of gene clusters with slightly fewer genes than the original NHC output, in cluster #1 (28 versus 32 genes), cluster #4 (26 versus 30 genes), and cluster #5 (9 versus 10 genes) (Table S8). The top-ranked cluster from NHC-boost converged 28 genes, including all reported genes and candidate genes, but missing four peripheral genes (IFIT2, RNF135, TNFRSF1B, UBA7) (see Figure 3 for visualization) from the 32 genes output by the original NHC. In a test of 500 affected individuals, NHC-boost greatly decreased the computation time, from ∼8 h to ∼50 min.
Discussion
We show here that NHC is a biological network-based genome-wide computational approach that can unravel the genetic heterogeneity underlying the physiological homogeneity of a subset of individuals in a given condition. As proof-of-concept, application of our method to the HSE cohort successfully captured and prioritized all previously published HSE-causing genetic variants in the TLR3 pathway in its top-ranked gene cluster,41, 42, 43, 44, 45, 46, 47 also suggesting candidate genes with products involved in this pathway, in a systematic, efficient, and unbiased manner. These genes and variants would have been identified by NHC, despite never having been reported to underlie HSE. NHC also identified other gene clusters functionally enriched in other pathways, the relationships to HSE and TLR3 of which require further investigation. Moreover, an individual could have multiple deleterious variants from the same or different gene clusters, suggesting possible digenic or oligogenic genetic lesions with similar or different molecular mechanisms leading to the disease phenotype of the individual concerned.55
NHC is of particular interest among the computational methods for analyzing NGS data, as it can detect disease signals from a cohort of affected individuals by accepting genetic heterogeneity and assessing physiological homogeneity. NHC method is suitable for diseases that have a homogeneous clinical phenotype and are likely caused in a substantial number of individuals (e.g., ≥5, as suggested in simulation study II) by rare/uncommon variants (e.g., MAF < 0.01) with strong individual effects and located in physiologically related genes. NHC could be widely applied to rare diseases with a smaller sample size, and to more common complex diseases with a larger sample size, as most human diseases can result from the disturbance of a functionally related molecular network, at least in a core group of individuals. Although the concept of network medicine has long been proposed and applied,5 NHC provides a widely practical and unbiased solution, introducing the network concept into discoveries of the genetic basis of diseases, to deal with the problems that have frustrated traditional approaches, because these methods search for disease signals based on genetic homogeneity or based on predefined gene sets. Given that no disease-causing mutation has yet been detected in vast numbers of individuals, this method has the potential to make a substantial contribution to the discovery of the genetic determinants underlying many other diseases, including life-threatening COVID-19.56,57
We focused here on European populations, but studies of individuals from different ancestries may affect the performance of NHC in terms of gene discovery and statistical significance, without affecting the retention of genetically heterogeneous disease-associated clusters. For such studies, we recommend first testing NHC on the entire cohort with the same cutoffs applied for variant filtration, and then trying to set different MAF cutoffs for different populations if the disease prevalence is different. The use of a set of control subjects representative of the ethnic make-up of the cohort of affected individuals may be helpful. If one population accounts for a large proportion of the total cohort, NHC should be tested on this subset of the cohort displaying ethnicity homogeneity, as defined by PCA. In our NHC code, we provide an option allowing users to provide PC tables for running PC-adjusted enrichment, to adjust for ethnic diversity.
This approach is also subject to several limitations. The gene clustering algorithm is dependent on the scale and quality of the background PPI network. We collected data from multiple reliable databases and used multiple parameters to enhance its representation of the physiological interaction space. However, the PPIs available to date are far from complete and may include false positives. Our method is also limited by a maximum of 15,585 searchable genes for linking genetic heterogeneity, as our background network contains 202,057 PPIs for 15,585 human protein-coding genes. This method cannot detect non-coding RNA genes, such as the reported HSE-causing mutations in snoRNA31.58 Another limitation is the definition of the hub genes to be removed during clustering. If these genes are not removed, giant clusters are formed, with enrichment in very general pathways (e.g., cancer pathways, metabolic pathways). The removal of too many hub genes leads to a risk of missing potentially promising candidate genes and to the formation of small and scattered gene clusters with lower levels of enrichment. The definition of this parameter also depends on the disease studied. We think that a dedicated study is required to gain greater insight into the hub genes for different diseases. In the NHC method, this parameter can be customized by the user.
We intend to update our tool with the latest data available. Future improvement of NHC method will include: (1) the employment of tissue-specific biological networks, with transcriptomic databases such as GTEx;12,59 (2) the consideration of genes with products that are functionally complementary but do not interact physically; (3) the implementation of alternative graph theory algorithms to test computational performance further with experimental evidence;6,14 and (4) parallel programming of our approach to reduce computing time for large cohorts. These are some of the approaches that could be followed to further improve the detection of physiological homogeneity in the midst of genetic heterogeneity, for various human diseases, whether rare or common, and infectious or otherwise.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We warmly thank the clinicians, affected individuals, and their families for participating in this study. We thank Y. Nemirovskaya, D. Papandrea, and M. Woollett for administrative support. We thank P. Bastard, B. Bigio, B. Boisson, D.X. Gao, G. Kerner, W.T. Lei, M. Ogishi, and F. Rapaport for valuable discussions. We thank Z. Yang for the artwork. The Laboratory of Human Genetics of Infectious Diseases is supported by the Howard Hughes Medical Institute, the Rockefeller University, the St. Giles Foundation, the National Institutes of Health (NIH) (R01AI088364), the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award (CTSA) program (UL1 TR001866), the Yale Center for Mendelian Genomics and the GSP Coordinating Center funded by the National Human Genome Research Institute (NHGRI) (UM1HG006504 and U24HG008956), the French National Research Agency (ANR) under the “Investments for the Future” program (ANR-10-IAHU-01), ANR grants (ANR-14-CE14-0008-01 and ANR-18-CE15-0020-02), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798), Institut National de la Santé et de la Recherche Médicale (INSERM), and the University of Paris. This study was also supported by NIH grant number R01DK123530-01 and The Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, United States.
Published: May 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.023.
Contributor Information
Peng Zhang, Email: pzhang@rockefeller.edu.
Jean-Laurent Casanova, Email: casanova@rockefeller.edu.
Data and code availability
NHC program is written in python and publicly accessible under a CC BY-NC-ND 4.0 license. Its gene clustering code works at the gene level and converges genes carrying qualifying variants into gene clusters with pathway and gene ontology enrichment. We provide the code for (1) case-control studies and (2) case-only studies, for both the original NHC version and the NHC-boost version. All the examples described here are case-control studies, but we are aware that a control cohort may not always be available. We therefore also provide a case-only code for such situations. This method is designed to detect gene clusters harboring deleterious mutations from a cohort of affected individuals, but it is difficult to provide the code starting from variant-level processing, as variant data formats and variant filtration criteria vary considerably between laboratories and between studies. We therefore leave variant-level processing to users, who will need to prepare the gene list carrying the qualifying variants for all individuals in the cohort to use the code.
Web resources
Supplemental information
References
- 1.McClellan J., King M.C. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Casanova J.L., Abel L. The human genetic determinism of life-threatening infectious diseases: genetic heterogeneity and physiological homogeneity? Hum. Genet. 2020;139:681–694. doi: 10.1007/s00439-020-02184-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Povysil G., Petrovski S., Hostyk J., Aggarwal V., Allen A.S., Goldstein D.B. Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat. Rev. Genet. 2019;20:747–759. doi: 10.1038/s41576-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 4.Chen R., Shi L., Hakenberg J., Naughton B., Sklar P., Zhang J., Zhou H., Tian L., Prakash O., Lemire M. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat. Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 5.Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P., Itan Y. Biological Network Approaches and Applications in Rare Disease Studies. Genes (Basel) 2019;10:10. doi: 10.3390/genes10100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taroni J.N., Greene C.S., Martyanov V., Wood T.A., Christmann R.B., Farber H.W., Lafyatis R.A., Denton C.P., Hinchcliff M.E., Pioli P.A. A novel multi-network approach reveals tissue-specific cellular modulators of fibrosis in systemic sclerosis. Genome Med. 2017;9:27. doi: 10.1186/s13073-017-0417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leiserson M.D., Vandin F., Wu H.T., Dobson J.R., Eldridge J.V., Thomas J.L., Papoutsaki A., Kim Y., Niu B., McLellan M. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engin H.B., Kreisberg J.F., Carter H. Structure-Based Analysis Reveals Cancer Missense Mutations Target Protein Interaction Interfaces. PLoS ONE. 2016;11:e0152929. doi: 10.1371/journal.pone.0152929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun R., Buetow K. Pathways of distinction analysis: a new technique for multi-SNP analysis of GWAS data. PLoS Genet. 2011;7:e1002101. doi: 10.1371/journal.pgen.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia P., Zhao Z. Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum. Genet. 2014;133:125–138. doi: 10.1007/s00439-013-1377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M., Gelfman S., McCarthy J., Harms M.B., Moreno C.A.M., Goldstein D.B., Allen A.S. Incorporating external information to improve sparse signal detection in rare-variant gene-set-based analyses. Genet. Epidemiol. 2020;44:330–338. doi: 10.1002/gepi.22283. [DOI] [PubMed] [Google Scholar]
- 13.Eckhardt M., Hultquist J.F., Kaake R.M., Hüttenhain R., Krogan N.J. A systems approach to infectious disease. Nat. Rev. Genet. 2020;21:339–354. doi: 10.1038/s41576-020-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P., Tao L., Zeng X., Qin C., Chen S., Zhu F., Li Z., Jiang Y., Chen W., Chen Y.Z. A protein network descriptor server and its use in studying protein, disease, metabolic and drug targeted networks. Brief. Bioinform. 2017;18:1057–1070. doi: 10.1093/bib/bbw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belkadi A., Bolze A., Itan Y., Cobat A., Vincent Q.B., Antipenko A., Shang L., Boisson B., Casanova J.L., Abel L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA. 2015;112:5473–5478. doi: 10.1073/pnas.1418631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P., Bigio B., Rapaport F., Zhang S.Y., Casanova J.L., Abel L., Boisson B., Itan Y. PopViz: a webserver for visualizing minor allele frequencies and damage prediction scores of human genetic variations. Bioinformatics. 2018;34:4307–4309. doi: 10.1093/bioinformatics/bty536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maffucci P., Bigio B., Rapaport F., Cobat A., Borghesi A., Lopez M., Patin E., Bolze A., Shang L., Bendavid M. Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc. Natl. Acad. Sci. USA. 2019;116:950–959. doi: 10.1073/pnas.1808403116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itan Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA. 2015;112:13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itan Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods. 2016;13:109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevim Bayrak C., Itan Y. Identifying disease-causing mutations in genomes of single patients by computational approaches. Hum. Genet. 2020;139:769–776. doi: 10.1007/s00439-020-02179-7. [DOI] [PubMed] [Google Scholar]
- 27.Oughtred R., Stark C., Breitkreutz B.J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P., Boisson B., Stenson P.D., Cooper D.N., Casanova J.L., Abel L., Itan Y. SeqTailor: a user-friendly webserver for the extraction of DNA or protein sequences from next-generation sequencing data. Nucleic Acids Res. 2019;47(W1):W623–W631. doi: 10.1093/nar/gkz326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Hagberg A.A., Schult D.A., Swart P.J. Proceedings of the 7th Python in Science Conference (SciPy2008), T.V. Gäel Varoquaux and Jarrod Millman. 2008. Exploring network structure, dynamics, and function using NetworkX; pp. 11–15. Pasadena, CA, USA. [Google Scholar]
- 36.Zhang P., Tao L., Zeng X., Qin C., Chen S.Y., Zhu F., Yang S.Y., Li Z.R., Chen W.P., Chen Y.Z. PROFEAT Update: A Protein Features Web Server with Added Facility to Compute Network Descriptors for Studying Omics-Derived Networks. J. Mol. Biol. 2017;429:416–425. doi: 10.1016/j.jmb.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S., Emond M.J., Bamshad M.J., Barnes K.C., Rieder M.J., Nickerson D.A., Christiani D.C., Wurfel M.M., Lin X., NHLBI GO Exome Sequencing Project—ESP Lung Project Team Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S.Y. Herpes simplex virus encephalitis of childhood: inborn errors of central nervous system cell-intrinsic immunity. Hum. Genet. 2020;139:911–918. doi: 10.1007/s00439-020-02127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stahl J.P., Mailles A. Herpes simplex virus encephalitis update. Curr. Opin. Infect. Dis. 2019;32:239–243. doi: 10.1097/QCO.0000000000000554. [DOI] [PubMed] [Google Scholar]
- 41.Casrouge A., Zhang S.Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 43.Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y., Audry M., Ciancanelli M., Alsina L., Azevedo J., Herman M., Anguiano E., Sancho-Shimizu V., Lorenzo L., Pauwels E. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J. Exp. Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herman M., Ciancanelli M., Ou Y.H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim H.K., Seppänen M., Hautala T., Ciancanelli M.J., Itan Y., Lafaille F.G., Dell W., Lorenzo L., Byun M., Pauwels E. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology. 2014;83:1888–1897. doi: 10.1212/WNL.0000000000000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47(D1):D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen L.L., Mørk N., Reinert L.S., Kofod-Olsen E., Narita R., Jørgensen S.E., Skipper K.A., Höning K., Gad H.H., Østergaard L. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 2015;212:1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S.Y., Casanova J.L. Inborn errors underlying herpes simplex encephalitis: From TLR3 to IRF3. J. Exp. Med. 2015;212:1342–1343. doi: 10.1084/jem.2129insight4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casanova J.L. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc. Natl. Acad. Sci. USA. 2015;112:E7128–E7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goncalves A., Bürckstümmer T., Dixit E., Scheicher R., Górna M.W., Karayel E., Sugar C., Stukalov A., Berg T., Kralovics R. Functional dissection of the TBK1 molecular network. PLoS ONE. 2011;6:e23971. doi: 10.1371/journal.pone.0023971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du J.L., Ma P., Wang C., Zeng Y., Xue Y.J., Yang X.C., Wan X.M., Chang F.F., Zhao T.Y., Jia X.Y. ATG13 restricts viral replication by induction of type I interferon. J. Cell. Mol. Med. 2019;23:6508–6511. doi: 10.1111/jcmm.14483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerner G., Bouaziz M., Cobat A., Bigio B., Timberlake A.T., Bustamante J., Lifton R.P., Casanova J.L., Abel L. A genome-wide case-only test for the detection of digenic inheritance in human exomes. Proc. Natl. Acad. Sci. USA. 2020;117:19367–19375. doi: 10.1073/pnas.1920650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casanova J.L., Su H.C., COVID Human Genetic Effort A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194–1199. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., COVID-STORM Clinicians. COVID Clinicians. Imagine COVID Group. French COVID Cohort Study Group. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort. NIAID-USUHS/TAGC COVID Immunity Group Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:370. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lafaille F.G., Harschnitz O., Lee Y.S., Zhang P., Hasek M.L., Kerner G., Itan Y., Ewaleifoh O., Rapaport F., Carlile T.M. Human SNORA31 variations impair cortical neuron-intrinsic immunity to HSV-1 and underlie herpes simplex encephalitis. Nat. Med. 2019;25:1873–1884. doi: 10.1038/s41591-019-0672-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NHC program is written in python and publicly accessible under a CC BY-NC-ND 4.0 license. Its gene clustering code works at the gene level and converges genes carrying qualifying variants into gene clusters with pathway and gene ontology enrichment. We provide the code for (1) case-control studies and (2) case-only studies, for both the original NHC version and the NHC-boost version. All the examples described here are case-control studies, but we are aware that a control cohort may not always be available. We therefore also provide a case-only code for such situations. This method is designed to detect gene clusters harboring deleterious mutations from a cohort of affected individuals, but it is difficult to provide the code starting from variant-level processing, as variant data formats and variant filtration criteria vary considerably between laboratories and between studies. We therefore leave variant-level processing to users, who will need to prepare the gene list carrying the qualifying variants for all individuals in the cohort to use the code.