Summary
SLC37A4 encodes an endoplasmic reticulum (ER)-localized multitransmembrane protein required for transporting glucose-6-phosphate (Glc-6P) into the ER. Once transported into the ER, Glc-6P is subsequently hydrolyzed by tissue-specific phosphatases to glucose and inorganic phosphate during times of glucose depletion. Pathogenic variants in SLC37A4 cause an established recessive disorder known as glycogen storage disorder 1b characterized by liver and kidney dysfunction with neutropenia. We report seven individuals who presented with liver dysfunction multifactorial coagulation deficiency and cardiac issues and were heterozygous for the same variant, c.1267C>T (p.Arg423∗), in SLC37A4; the affected individuals were from four unrelated families. Serum samples from affected individuals showed profound accumulation of both high mannose and hybrid type N-glycans, while N-glycans in fibroblasts and undifferentiated iPSC were normal. Due to the liver-specific nature of this disorder, we generated a CRISPR base-edited hepatoma cell line harboring the c.1267C>T (p.Arg423∗) variant. These cells replicated the secreted abnormalities seen in serum N-glycosylation, and a portion of the mutant protein appears to relocate to a distinct, non-Golgi compartment, possibly ER exit sites. These cells also show a gene dosage-dependent alteration in the Golgi morphology and reduced intraluminal pH that may account for the altered glycosylation. In summary, we identify a recurrent mutation in SLC37A4 that causes a dominantly inherited congenital disorder of glycosylation characterized by coagulopathy and liver dysfunction with abnormal serum N-glycans.
Keywords: glycosylation, congenital disordes of glycosylation, exome sequencing, coagulopathy, Golgi pH
Introduction
Congenital disorders of glycosylation (CDGs) are a clinically and biochemically heterogenous group of disorders characterized by abnormal lipid or protein glycosylation.1,2 To date, more than 140 types of CDGs have been identified across at least seven different glycosylation pathways.2 The vast majority of CDGs follow an autosomal recessive or X-linked inheritance pattern, although disorders due to de novo or dominant variants have been observed with increased frequency.3 In several instances, a single gene has been shown to cause both dominantly and recessively inherited disorders that present with unique phenotypes as is the case for DHDDS (MIM: 608172), NUS1 (MIM: 610463), POFUT1 (MIM: 607491), and COG4 (MIM: 606976).4, 5, 6, 7, 8
SLC37A4 (MIM: 602671) encodes for a multitransmembrane domain glucose-6-phosphate (Glc-6P) transporter that is localized to the endoplasmic reticulum (ER).9,10 In gluconeogenic organs, such as the liver and kidney, its primary function is to transport Glc-6P into the ER during times of glucose depletion, thus regulating glucose homeostasis.10 SLC37A4 does this in cooperation with various tissue-specific phosphatases (G6PC, G6PC2, and G6PC3) to hydrolyze Glc-6P to glucose and inorganic phosphate (Pi), which exit the ER. Glucose is subsequently transported out of the cell to maintain glucose homeostasis.10,11 SLC37A4 also participates in the ER-localized pentose phosphate pathway (PPP) by providing Glc-6P to hexose-6-phosphate dehydrogenase (H6PD), which catalyzes the first two reactions of ER-localized PPP generating nicotinamide adenine dinucleotide phosphate (NADPH).12
Pathogenic variants in SLC37A4 cause autosomal recessive glycogen storage disorder 1b (GSD1b) (MIM: 232220), which is characterized by an inability to properly metabolize glycogen.9,13 This results in the accumulation of glycogen in gluconeogenic organs such as the liver and kidneys, ultimately leading to organ dysfunction and disease.9,13 Neutropenia is frequently seen in affected individuals as well. Pathogenic variants in the tissue-specific phosphatases have also been identified. GSD1a (also known as von Gierke disease) (MIM: 232200) is caused by mutations in G6PC (MIM: 613742), causing similar accumulation of glycogen in the liver and kidney.14,15 Pathogenic variants in G6PC3 (MIM: 611045) cause autosomal recessive severe congenital Neutropenia 4 and Dursun syndrome (MIM: 612541).16,17 Cortisone reductase deficiency 1 (MIM: 604931) is caused by pathogenic variants in H6PD (MIM: 138090).18
We report the molecular, biochemical, and clinical characterization of seven individuals who clinically presented with a coagulopathy affecting several coagulation factors and were heterozygous for a c.1267C>T (p.Arg423∗) variant in SLC37A4; the affected individuals were from four unrelated families. All affected individuals were identified by coagulation abnormalities and subsequently found to have abnormal N-glycosylation of serum glycoproteins, specifically the accumulation of high mannose and hybrid type N-glycans. Introduction of the c.1267C>T (p.Arg423∗) variant into Huh7 hepatocellular carcinoma cells revealed clear gene dosage-dependent morphological and functional deficiencies in the Golgi apparatus function that recapitulate the abnormal N-glycans seen in our affected individuals.
Material and methods
Subjects
Families included in our SLC37A4-CDG research study provided written consent under an approved Sanford Burnham Prebys Medical Discovery Institute IRB protocol or an approved IRB through each family’s primary medical physician. The only inclusion criteria for this study was the presence of the c.1267C>T (p.Arg423∗) variant in SLC37A4. Blood samples were obtained for all individuals. Primary fibroblasts were obtained for two affected individuals (P6 and P7) and grown from a skin biopsy obtained by the subjects’ physician.
Genomic analysis
Exome sequencing (ES) and genome sequencing (GS) with analysis were carried out by two independent groups with a shared heterozygous c.1267C>T (p.Arg423∗) variant in SLC37A4 identified in all affected individuals. Sanger sequencing of all seven affected individuals and unaffected family members was used to confirm presence of the variant and to assess segregation. Primers to amplify exon 10 of SLC37A4 are available upon request.
Cell culture
Fibroblasts from apparently healthy controls GM-00038, GM-05565, and GM-09503 (Coriell Institute); proband primary fibroblasts (P6 and P7); and the hepatocellular carcinoma line Huh7 were grown (unless otherwise stated) in complete 1 g/L (5.5 mM) glucose containing Dulbecco’s modified Eagle’s medium (Corning) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich LOT# H176268), 100 U/mL penicillin and 100 mg/mL streptomycin (GIBCO), and 2 mM L-Glutamine (GIBCO). Induced pluripotent stem cells (iPSCs) were established from affected individual fibroblasts as described previously.19 The wild-type control iPSC line K3 was utilized.20 Human iPSCs were generated via lentivirus transduction of pluripotency factors as described previously.21 Human hepatocytes from individual P7-derived cells were generated by a previously reported method.19
Glycosylation studies
Individuals P1, P2, P4, P5, and P6, all had carbohydrate-deficient transferrin (CDT) analysis performed with capillary zone electrophoresis (CZE), while P7 had both electrospray ionization mass spectrometry (ESI-MS) and liquid chromatography-mass spectrometry (LC-MS) as previously described.22, 23, 24 Apolipoprotein C-III (apoC-III) mucin core 1 O-glycosylation was analyzed with 2D electrophoresis as previously described.25 Profiles of total serum N-glycans were obtained by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) following N-glycan cleavage by peptide N-glycosidase F (PNGase F) and permethylation as previously described.26 Analysis of iPSCs, derived hepatocytes, and Huh7-edited cell line-secreted and cell-associated N-glycans was performed by multi-dimensional nanospray ionization mass spectrometry (NSI-MSn) as previously described.27 High performance liquid chromatography (HPLC) analysis of N-glycans secreted by CRISPR-edited Huh7 cells was done following labeling with procainamide (ProA) reagent as described with modification available upon request.28
CRISPR-edited Huh7 cell line
The guide RNA (gRNA) sequencing GATGGGCCGAGTGTCCAAGA targeting exon 10 of SLC37A4 was cloned into pGuide-EF1a-GFP (OriGene - GE100044). This vector does not express Cas9, only the gRNA to SLC37A4 and turboGFP (tGFP), which allows for sorting positively transfected cells. Both pGuide-R423X-SLC37A4 and pCMV-BE3 (Addgene plasmid 73021) were co-transfected together into Huh7 cells and allowed to grow for 48 h. Subsequently, tGFP positive cells were fluorescence-activated cell sorting (FACS) sorted with a FACSAria IIu instrument (BD Biosciences) into 96-well plates to generate isogenic clones, which were expanded and screened by Sanger sequencing.
Cellular fractionations
Subcellular fractionations were performed as previously described with some modifications.29,30 Briefly, cells from one 150 mm plate were washed twice with Dulbecco’s phosphate-buffered saline (DPBS) and then homogenized in buffer (10 mM HEPES-KOH, 25 mM KCl, 250 mM sucrose, and 1 mM EDTA, pH 7.4, protease inhibitor cocktail [Thermo Fisher Scientific]) by passing through 27-gauge needle 15 times. A post-nuclear supernatant (PNS) was separated by centrifugation at 1,000g for 15 min at 4°C and then subjected to 15,000g spin for 20 min at 4°C. The resulting pellet was resuspended in 5% Nycodenz, applied to a 10%–24% discontinuous Nycodenz gradient (Fisher Scientific), and centrifuged at 100,000g in an SW40 rotor for 18 h at 4°C. Fractions (1 mL) were collected, trichloroacetic acid (TCA) precipitated, dissolved in 2× loading buffer, and analyzed by SDS-PAGE.
Immunofluorescence
Cells were seeded on Millicell EZ 8-well glass slides (Sigma Aldrich) and, 1 day later, washed twice with DPBS and fixed for 15 min with 4% PFA at room temperature. Next, cells were washed three times with DPBS and blocked with 1% BSA solution in DPBS containing 0.1% saponin. After 1 h of blocking at room temperature, respective primary antibodies (Table S1), diluted in blocking solution, were added and slides were incubated overnight at 4°C. The next day, cells were washed three times with blocking solution and respective secondary antibodies (Table S1) diluted in blocking solution. After 1-h incubation at room temperature, DAPI (Thermo Fisher Scientific) was added directly to the solution, so its final dilution was 1:500. 20 min later, cells were washed twice with blocking buffer, twice with DPBS, and twice with water. Cover slides were stuck to the slides via Immu-Mount mounting medium (Thermo Fisher Scientific). Cells were analyzed with LSM 710 Zeiss confocal microscope and EC Plan-Neofluar 40×/1.30 Oil DIC M27 objective. For immunostaining of iPSC-derived hepatocytes, differentiated hepatocyte spheroids were seeded in Matrigel-coated slide chambers and grown for 48 h to allow a monolayer of cells to migrate from the spheroid prior to fixation and staining as described above.
Golgi pH measurements
Golgi pH measurements were performed as described by Galenkamp et al.31 Briefly, Huh7 or fibroblast cells were seeded on 6-well plates and, after 24 h, transfected with GalT-mCherry-eGFP construct via FuGENE HD (Promega) according to the manufacturer’s instructions. 24 h after transfection, cells were seeded on CELLview glass bottom cell culture dishes (Greiner Bio-One). 48 h later, cells were washed twice with DPBS and media exchanged for phenol red-free DMEM 4.5 g/L glucose media (Thermo Fisher Scientific). For pH determinations, cells were incubated for 15 min with one of the calibration buffers: 30 mM ammonium formate (pH 4.9), 30 mM 2-Morpholinoethanesulfonic acid (MES) (pH 5.4), 30 mM MES (pH 5.9), 30 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 6.5), 30 mM MOPS (pH 7.0), supplemented with 130 mM KCl, 1 mM MgCl2, 10 μM Nigericin, 10 μM Valinomycin, and 10 μM Monensin. Imaging was performed on living cells at room temperature and at atmospheric CO2 via Zeiss LSM 710 laser scanning confocal microscope and C-Apochromat 40×/1.20 W Korr FCS M27 water immersion objective. eGFP and mCherry signals were collected simultaneously by 488 nm and 594 nm excitation and ratios were determined by region of interest selection of fluorescence via ImageJ software (NIH).
Transport assay
SLC37A4 transport assays were performed as previously described.13
Results
Clinical summary for seven individuals presenting with liver dysfunction
A cohort of seven individuals, from four unrelated non-consanguineous families, with an undefined type II CDG (Figure 1A) were found to have a strikingly similar clinical phenotype involving liver dysfunction with abnormal coagulation factor activities. All individuals had an abnormal CDT and apolipoprotein C-III (apoC-III) pattern.
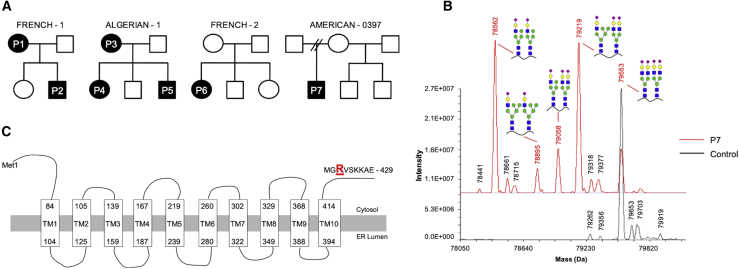
Figure 1.
Identification of a recurrent SLC37A4 mutation in four unrelated families
(A) Pedigrees showing segregation of the SLC37A4 c.1267C>T (p.Arg423∗) mutation in seven affected individuals from four unrelated families.
(B) LC-MS of serum transferrin from control and P7 serum with deconvoluted masses of intact serum TF from full scans showing the appearance of several peaks corresponding to distinctive peaks containing hybrid N-glycans.
(C) Schematic of human SLC37A4 showing the p.Arg423∗ localizing to the cytoplasmic tail (UniProt: O43826-1).
P1 is a 35-year-old female of French origin with a history of elevated aspartate aminotransferase (AST) level (91 U/L; normal value < 35 U/L) but normal alanine aminotransferase (ALT) on routine blood analysis. A complete blood count, as well as prothrombin time (PT) and activated partial thromboplastin time (aPTT), was normal. However, clotting factor II (F2) was unexpectedly decreased (42%), while factor V (F5), factor VII (F7), factor VIII (F8), factor IX (F9), factor X (F10), factor XI (F11), factor XII (F12), and fibrinogen (Fg) were within a normal range (Table 1). Medically assisted reproduction led to a non-hemorrhagic spontaneous abortion followed by two successful pregnancies. During this 4-year period, antithrombin (SERPINC1) and F2 levels were often markedly decreased (SERPINC1, 23% to 52%, mean: 45%; F2, 29% to 67%, mean: 38%) with low levels of protein S (PROS1) (45% to 53%). AST levels were consistently elevated (62 U/L to 91 U/L, mean: 73 U/L) (Table 1). The unexplained fluctuating coagulation abnormalities seen in P1 led to CDG screening, which showed a marked CDG-II transferrin CDT pattern coupled with altered apoC-III glycoforms (Table 1; Figures S1 and S2).
Table 1.
Clinical summary of seven individuals carrying the c.1267C>T (p.Arg423∗) variant in SLC37A4
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | Marquardt et al.32 | Wilson et al.33 | Current cohort summary | |
|---|---|---|---|---|---|---|---|---|---|---|
| Family | French-1 | French-1 | Algerian-1 | Algerian-1 | Algerian-1 | French-2 | American-0397 | – | – | – |
| Gender | female | male | female | Female | male | female | male | female | female | – |
| Year of birth | 1985 | 2018 | 1973 | 2002 | 2007 | 2010 | 2009 | N/A | 1997 | – |
| Age of diagnosis | 29 years | 1 month | 42 years | 11 years | 4 months | 8 years | 5 years | 10 weeks | 36 days | – |
| Variant | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | c.1267C>T (p.Arg423∗) | – |
| Inheritance | unknown | maternally inherited | unknown | maternally inherited | maternally inherited | de novo | de novo | de novo | de novo | – |
| Method of detection | ES | Sanger | ES | ES | ES | Sanger | ES/GS | ES | ES | – |
| CDT results | abnormal-type II | abnormal-type II | abnormal-type II | abnormal-type II | abnormal-type II | abnormal-type II | abnormal-type II | abnormal-type II | abnormal-type II | 7/7 |
| apoC-III | altered O-glycan | altered O-glycan | altered O-glycan | altered O-glycan | altered O-glycan | altered O-glycan | altered O- glycan | N/A | N/A | 7/7 |
| Serum N-glycans | high man, hybrid | high man, hybrid | high man, hybrid | high man, hybrid | high man, hybrid | high man, hybrid | high man, hybrid | high man, hybrid | high man, hybrid | 7/7 |
| Cardiac abnormalities | none reported | perimembranous ventricular septal defect | none reported | none reported | tetralogy of Fallot | ventricular septal defect | none reported | none reported | none reported | 3/7 |
| Skeletal abnormalities | no | no | scoliosis | scoliosis | kyphoscoliosis | no | no | no | scoliosis | 3/7 |
| Ankyloglossia | no | no | no | no | yes | no | no | yes | no | 1/7 |
| AST (ref. 7–40 U/L) | 65 U/L (H) | N/A | 50 U/L (H) | 77 U/L (H) | 91 U/L (H) | 147 U/L (H) | 80 U/L (H) | 228 U/L (H) | 522 U/L (H) | 6/6 |
| F2 (ref. 60%–140%) | 31 (L) | 22 (L) | 57 (L) | 30 (L) | 20 (L) | 18 (L) | 27 (L) | 5.5 (L) | N/A | 7/7 |
| F5 (ref. 60%–140%) | 51 (L) | 52 (L) | 64 | 40 (L) | 39 (L) | 50 (L) | 29 (L) | 38.5 (L) | N/A | 6/7 |
| Fg (ref. 1.5–3.5 g/L) | 1.7 g/L | 1.7 g/L | 2.8 g/L | 1.8 g/L | 1.3 g/L (L) | 1.7 g/L | 1.5 g/L | 0.4 g/L (L) | 0.1 g/L (L) | 1/7 |
| F8 (ref. 60%–150%) | 117 | N/A | 165 | 130 | 144 | 109 | 85 | N/A | N/A | 0/6 |
| F9 (ref. 60%–140%) | 63 | N/A | 85 | 58 (L) | 42 (L) | 55 (L) | 44 (L) | N/A | N/A | 4/6 |
| F11 (ref. 60%–140%) | 59 (L) | N/A | 55 (L) | 31 (L) | 34 (L) | 22 (L) | 33 (L) | N/A | normal | 6/6 |
| SERPINC1 (ref. 80%–120%) | 28 (L) | 34 (L) | 60 (L) | 37 (L) | 32 (L) | 19 (L) | 32 (L) | 0 (L) | normal | 7/7 |
| PROC (ref. 50%–120%) | 110 | 49 | 97 | 72 | 73 | 59 | 94 | N/A | normal | 0/7 |
| PROS1 (ref. 60%–120%) | 41 (L) | 48 (L) | 70 | 43 (L) | 81 | 35 (L) | 35 (L) | N/A | normal | 5/7 |
Reference ranges: ALT, ref.; AST, ref. 7–40 U/L; F2, ref. 60%–140% activity; F5, ref. 60%–140% activity; Fg (g/L), ref. 1.5–3.5 g/L; F8, ref. 60%–150% activity; F9, ref. 60%–140% activity; F11, ref. 60%–140% activity; SERPINC1, ref. 80%–120% activity; PROC, ref. 50%–120% activity; PROS1, ref. 60%–120% activity. Abbreviations are as follows: L, low; H, high; N/A, not available.
P2 is the second child of P1 (Figure 1A). At around 1 month of age, his pre-surgical testing for perimembranous ventricular septal defects (VSDs) showed similar coagulation abnormalities and CDT pattern as his mother. No bleeding episodes occurred during this corrective surgery. At 6 months of age, his coagulation parameters were globally altered with F2 (22%), F5 (52%), PROS1 (48%), SERPINC1 (34%), and protein C (PROC) (49%) (Table 1), however, there were no bleeding or thrombotic episodes. He, like his mother, also showed the same CDG-II CDT and altered apoC-III glycoform profile (Table 1; Figures S1 and S2).
P3 is an adult female of Algerian origin (Figure 1A). At 42 years of age, moderate deficiencies in both clotting factors and coagulation inhibitors were noted, including F2 (57%), F11 (55%), and SERPINC1 (60%), with normal PT, aPTT, Fg, PROC, and PROS1 (Table 1). She had neither bleeding nor thrombotic symptomatology. Besides progressive deafness, strabismus treated by surgery, and delayed diagnosed scoliosis, she had a normal clinical examination. Complete blood count was normal, but an elevated AST (50 U/L) was noted (Table 1). CDT and apoC-III analyses were unable to be performed for P3.
P4 is a female and the first child of P3 (Figure 1A). P4 has had multiple operations under fresh frozen plasma (FFP) and tranexamic acid for repair of a severe lip/palate cleft with bifid uvula. At 13 years of age, she showed altered PT (56%) and aPTT ratio (1.49) with normal F8 and F9. An overall decrease was seen in both the procoagulant factors F2 (30%) and F5 (40%) and anticoagulant factors SERPINC1 (37%) and PROS1 (43%) (Table 1). A complete blood count was normal, as were liver ALT and bilirubin levels. However, AST was elevated (77 U/L) (Table 1). CDG screening tests showed the same CDG II CDT pattern and abnormal apoC-III patterns (Table 1; Figures S1 and S2).
P5 is a male sibling of P4 and the third child of P3 (Figure 1A). At 4 months of age, PT and aPTT ratios were 61% and 1.72, respectively, with markedly decreased F2 (20%) and F11 (30%) with normal F5, Fg, F8, and F9 (Table 1). 1 month later, examination showed decreased SERPINC1 (32%) but normal PROC and PROS1. P5 had a tetralogy of Fallot surgically repaired at the age of 6 months, and at 2 years of age, he had a tongue-tie surgery (under FFP), both without complications. At 5 and 8 years of age, coagulation defects were seen in F2 (17% and 20%) and F5 (41% and 39%). At 8 years of age, AST level was elevated (91 U/L) with normal ALT (Table 1). There was a tendency for easily bruising (in contrast with his sister P4) and he had impaired wound healing. However, no bleeding disorders were reported. CDG screening tests showed a CDG II CDT and abnormal apoC-III pattern (Table 1; Figures S1 and S2).
P6 is a female of French origin (Figure 1A) who, at 8 years of age, showed a coagulation profile with altered PT (62%) and aPTT (ratio 1.86) with markedly decreased activities of F2 (18%) and F11 (22%) and decreased F5 (50%), F7 (44%), and F10 (48%). Coagulation inhibitors SERPINC1 and PROS1 were markedly decreased (19% and 35%) with normal PROC (Table 1). A complete blood count was normal, and no bleeding disorders were reported. She has a VSD under clinical supervision. A vesicoureteral reflux and strabismus were noted. Apparently healthy parents and two younger siblings had normal CDT patterns, while P6 had the same abnormal CDG II CDT and apoC-III patterns (Table 1; Figure S1 and S2).
P7 is a male of Irish-American origin (Figure 1A), born to healthy non-consanguineous parents, who displayed a long history of easy bruising and bleeding. Prior to a tonsillectomy and adenoidectomy, coagulation test revealed abnormalities in multiple coagulation factors, including F2 (27%), F5 (29%), F11 (37%), and PROS1 (35%). Liver function tests showed elevated liver enzyme AST (80 U/L) (Table 1). A liver biopsy determined no fibrosis or gross abnormalities. However, due to the history of elevated AST and abnormal coagulation factors, CDT screening was performed with both LC-MS and ESI-MS that showed accumulation of multiple high mannose and/or hybrid type N-glycans (Figure 1B; Table 1). An abnormal apoC-III pattern was also seen for P7 (data not shown).
In total, from our cohort, 7/7 (100%) individuals showed decreases in both F2 and SERPINC1 activities, 6/7 (86%) showed reduced F5 activity, 6/6 (100%) reduced F11 activity, 5/7 (71%) reduced protein S, and 4/6 (67%) reduced F9. AST levels were elevated in 6/6 (100%) individuals tested, while ALT was unaffected in all. (Table 1). F8 was within the reference range for all seven individuals, most likely because it is produced by non-hepatocyte endothelial cells. No neurological deficiencies were noted in the seven individuals. For all seven affected individuals, no candidate gene was identified through either clinical or glycan phenotyping. Recently, two more individuals were reported to carry the c.1267C>T (p.Arg423∗) variant in SLC37A4 and presented with strikingly similar biochemical and clinical phenotypes (Table 1).32,33 A less frequently shared feature included ankyloglossia, which was treated in both affected individuals (Table 1).
Glycosylation studies of serum showing accumulation of high mannose and hybrid type N-glycans
P1, P2, P4, P5, and P6 showed abnormalities consistent with a type II CDG by CZE (Figure S1). In P7, LC-MS showed the accumulation of several glycans absent from control samples. On the basis of the assigned masses for each of these peaks, we predicted they represented transferrin (TF) containing high mannose and/or hybrid type N-glycans (Figure 1B).
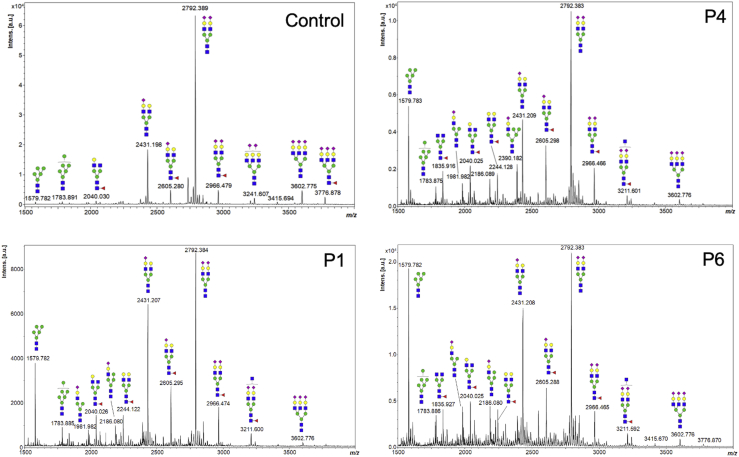
We used MALDI-TOF MS to analyze N-glycans from total serum glycoproteins and found that all seven individuals accumulated multiple species of both oligo and high mannose (e.g., peaks at m/z 1,579.8 and 1,783.9) and hybrid type N-glycans (e.g., peaks at m/z 1,981.9, 2,186.1, and 2,390.2) (Figures 2 and S3). These changes suggested alterations in the cis and medial Golgi compartments. Accumulation of truncated N-glycans with deficiencies in galactosylation (e.g., peaks at m/z 1,835.9 and 2,040.0) were also observed and suggests a defect in the trans-Golgi compartment (Figures 2 and S3).
Figure 2.
N-glycan abnormalities in serum from affected individuals
MALDI-TOF MS spectra of serum protein-derived N-glycans from unrelated individuals (P1, P4, and P6) are abnormal. Specifically, both high mannose (peaks at m/z 1,579.8 and 1,783.9) and hybrid type N-glycans (peaks at m/z 1,981.9, 2,186.1, and 2,390.2) increases were seen in positive-ion mode as sodiated forms. Green circles, mannose; yellow circles, galactose; blue squares, N-acetyl glucosamine; red triangles, fucose; purple diamonds, sialic acid.
A similar N-glycan phenotype occurs in cell lines defective in Golgi α-Mannosidase II (MAN2A1) or cells treated with the α-mannosidase II inhibitor swainsonine.34 On the basis of this similarity, individual P7 was Sanger sequenced for the entire coding, intronic, and promotor regions of both MAN2A1 (MIM: 154582) and its paralog MAN2A2 (MIM: 600988). However, no likely pathogenic variants were identified in either gene.
Identification of a recurrent c.1267C>T (p.Arg423∗) variant in SLC37A4
ES was performed on individuals P1, P3, P4, and P5. ES and GS were performed on P7. Sanger sequencing was used for P2 and P6. All seven individuals were found to share a heterozygous and likely pathogenic variant c.1267C>T (p.Arg423∗) in SLC37A4 (Table 1; Figure 1C). The variant in P6 and P7 was found to be de novo, while it was maternally inherited for P2, P4, and P5 (Table 1). Inheritance is unclear for both adults P1 and P3, and unaffected family members did not carry the variant. This variant was not found in several public databases, including gnomAD v.2.1.1 (>125,000 individuals), DiscovEHR (>50,000 individuals), and Geno2MP (>18,000 individuals). It is important to note the c.1267C>T variant is observed twice in Geno2MP, but both are from our individual P7 because he had both ES and GS. The variant was also absent from ClinVar.
Generating CRISPR-edited Huh7 cells carrying the c.1267C>T (p.Arg423∗) variant
Given the clear N-glycan abnormalities seen in serum samples from all seven affected individuals, we next sought to analyze N-glycans from fibroblasts. HPLC analyses of N-glycans from two affected individuals (P6 and P7) did not reveal any meaningful changes when compared against three healthy control fibroblast lines (data not shown). This was not completely unexpected given the liver-specific phenotype seen in the seven affected individuals. We also attempted to replicate the biochemical phenotype by either transiently or stably expressing the mutant transporter in Huh7 or HEK293 cells. In both cell types, we failed to see measurable changes in the N-glycans (data not shown). However, it is critical to note we observed a significant rapid and progressive cell death from overexpressing either the wild-type or the mutant transporter by using either a strong (CMV) or a weaker (EF1) promoter. As an alternative approach, we generated iPSCs from P7 and carried out several successful experiments (see below). However, technical issues with the generation of iPSC-derived hepatocytes in sufficient quantities for experiments other than immunostaining led us to take a different approach.
Therefore, we CRISPR modified the hepatocarcinoma cell line, Huh7, and generated both heterozygous and homozygous c.1267C>T (p.Arg423∗) clones. Genotyping showed several heterozygous clones with varying ratios of wild-type to mutant alleles. For example, Sanger sequencing showed clone C49 had a wild-type-to-mutant ratio of 40:60, while C71 was exactly 50:50, which matches affected individuals (Figure S4). We identified a single homozygous clone (C21) and multiple clones (C8, C9, and C10) that underwent treatment without producing an edit. These were used as Huh7 control lines because they were biochemically indistinguishable from the parental Huh7 cell line. In some cases, a silent variant c.1266C>T (p.G422G) also occurred (Figure S4).
High mannose and hybrid type N-glycans in glycoproteins secreted from p.Arg423∗-edited Huh7 cells
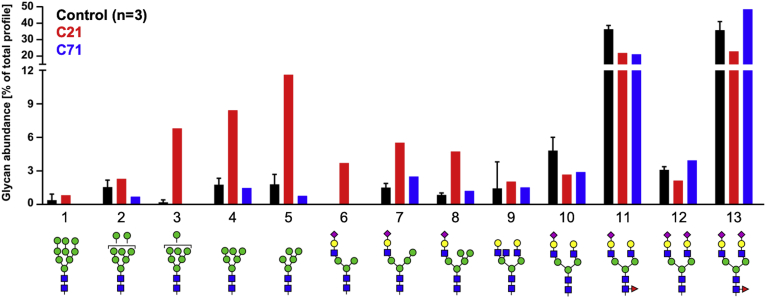
The hallmark of this disorder is the abnormal dramatic accumulation of high mannose and hybrid type N-glycans on liver-derived serum glycoproteins. We wanted to determine whether our CRISPR-edited Huh7 cells could replicate that N-glycan profile. Using NSI-MSn, we analyzed the N-glycans released from glycoproteins secreted by three isogenic controls (C8, C9, and C10) to clones carrying either a heterozygous (C71) or a homozygous (C21) c.1267C>T (p.Arg423∗) variant in SLC37A4. C21 best replicated the N-glycan profile of the affected individuals, showing a substantial increase in the amount of high mannose (glycans 3–5) and hybrid type (glycans 6–8) N-glycans when compared with the three controls (Figure 3). C71 did not show a difference (Figure 3). HPLC analysis confirmed the observed N-glycan changes seen in C21 (Figure S5).
Figure 3.
Characterization of N-glycans from p.Arg423∗ base-edited Huh7 cells
N-glycans released from secreted glycoproteins by PNGase F digestion showing the accumulation of both high mannose and hybrid type glycans in C21 with glycan abundances deduced from NSI-MSn measurements.
Glycan analysis of cell associated material from undifferentiated P7 iPSCs was identical to the control (K3), however, when P7 iPSCs were differentiated to hepatocytes, they replicated the accumulation of several, but not all, serum N-glycans seen in affected individuals (Figure S6). Specifically, hybrid type (glycans 10–12) N-glycans were the most significantly affected, while high/oligo mannose (glycans 1–5) were not.
Mislocalized SLC37A4-encoded transporter in p.Arg423∗-edited Huh7 cells
The c.1267C>T (p.Arg423∗) in SLC37A4 is predicted to delete the last seven amino acids from the cytoplasmic tail (423 - RVSKKAE - 429aa) (UniProt: O43826-1) (Figure 1C). This region includes a KKXX ER retrieval motif typically found in and required to maintain ER localization of resident proteins. However, the deleted region also has a possible Golgi retention sequence KXD/E motif (Figure 1C).35,36 To better understand the consequences of losing these motifs, we used subcellular fractionation and confocal microscopy to compare wild-type or mutant transporter in our Huh7-edited cell lines.
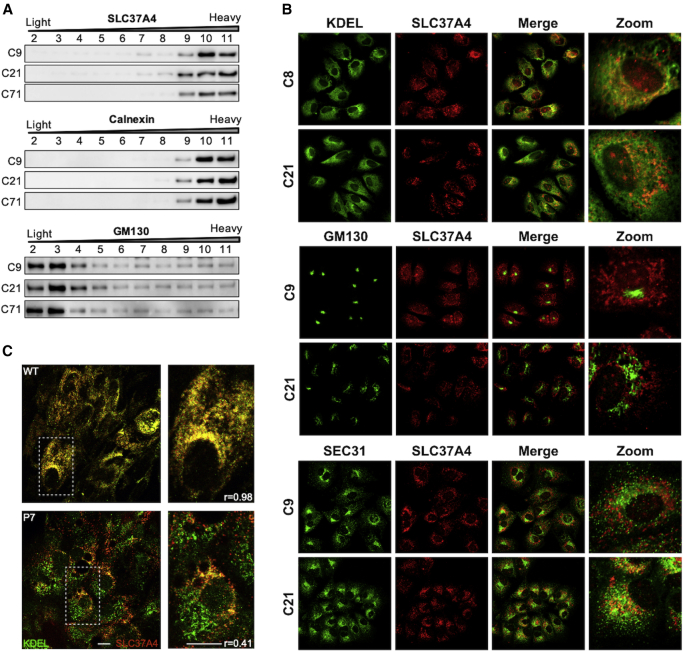
Using a Nycodenz density gradient to fractionate subcellular organelles29,30 of C9 control and C21 and C71, we clearly separate markers for ER (Calnexin) and cis Golgi (GM130) and found that neither the wild-type nor the mutant transporter was present in the Golgi fractions but enriched in the ER fractions (Figure 4A).
Figure 4.
Localization of mutant SLC37A4
(A) Subcellular fractionations of extracts from control C9 and edited lines C21 and C71 showing SLC37A4 protein is absent from the GM130 Golgi-containing fractions, but with similar fractionation pattern as the ER marker, calnexin. Subcellular fractionations were performed with three biological replicates via the Nycodenz gradient method, and representative images are shown.
(B) Immunofluorescence staining of Huh7 control and edited cells showing localization of SLC37A4 with the ER marker, KDEL (upper panel), the Golgi marker, GM130 (middle panel), and the ER exit site marker, SEC31 (lower panel).
(C) Immunofluorescence staining for the ER marker, KDEL, and SLC37A4 in iPSC-derived hepatocytes from control and P7. Control (upper panel) showed strong colocalization (r = 0.98), while P7 (lower panel) showed reduced colocalization (r = 0.41).
Next, we used high-resolution confocal microscopy to colocalize SLC37A4 with either the ER marker (anti-KDEL) or the Golgi marker (anti-GM130) in our mutant Huh7 lines. When compared with the two control Huh7 lines (C8 and C9), we did not see mislocalization of mutant SLC37A4 to the Golgi (Figure 4B). Instead, the majority remained in the ER-containing fractions. However, iPSC-derived differentiated hepatocytes showed an altered distribution of SLC37A4 within the ER. Analysis showed the KDEL-SLC37A4 overlap in control hepatocytes was substantial (R = 0.98), while in iPSC-derived P7 hepatocytes, this co-localization was far less pronounced (R = 0.41; R, Pearson correlation coefficient) (Figure 4C). It is not fully clear where this portion of mislocalized mutant transporter is residing within the early secretory pathway or whether the defect has caused alterations in the structure of the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) compartment or mislocalization of the ER and ER exit site markers. In the Huh7 C21, we did see an increased colocalization of the ER exit site marker, SEC31, with SLC37A4, which was absent from controls (Figure 4B).
These data provide clear evidence that the mutant transporter is not localized to the Golgi, contradicting the claims of a previous report based on overexpression of the mutant transporter in HepG2 cells.32 It is unclear where the mutant transporter resides, but it is most likely an undefined intermediate sub-compartment between the total ER and the cis-Golgi. The mutant transporter did not, however, colocalize with the ERGIC marker protein ERGIC-53 (Figure S7).
Abnormal Golgi morphology and pH in p.Arg423∗-edited Huh7 cells
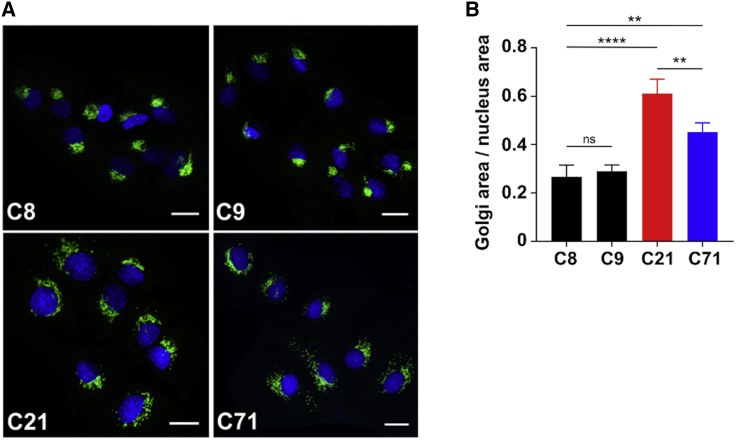
While SLC37A4 was clearly not located in the Golgi, its redistribution correlated with a striking dose-dependent abnormal appearance in the Golgi morphology in both mutant clones (C21 and C71). Immunofluorescence staining with the Golgi-associated marker, GM130, showed a compact Golgi architecture in all three controls (C8, C9, and C10), while in mutant clones C21 and C71, the structure was reminiscent of Golgi fragmentation (Figure 5A). By measuring the ratio between the Golgi area versus the area of the nucleus, we were able to quantify this difference (Figure 5B). The abnormal distribution of cis-Golgi marker GM130 was also seen in the medial (Stx5 and Giantin) and trans-Golgi network (TGN46) markers that showed the same abnormal morphology (Figure S8). Because abnormal trafficking is a hallmark of many CDG Golgi defects, we examined the effects of brefeldin A (BFA)-induced retrograde transport in control and mutant cells but saw no apparent difference (Figure S9).
Figure 5.
Abnormal Golgi structure and function in p.Arg423∗ base-edited Huh7 cells
(A) Immunofluorescence staining of Huh7 control (C8 and C9) and edited cells (C21 and C71) with the Golgi marker, GM130, showing abnormalities Golgi morphology/area. Scale bar represents 20 µm.
(B) Golgi area was quantified and then normalized to the area of the nucleus to provide a ratio showing both C21 (homozygous clone) and C71 (heterozygous clone) had significantly increased Golgi area. The effect is significantly more pronounced when both alleles are mutated as they are in C21. In three separate biological replicates, conducted within monthly intervals, each time with freshly thawed cells, n = 8 cells were analyzed, and the mean was taken. The graph represents an average of the means acquired over three different biological measurements. Total cells measured N = 24 over three biological replicates. Statistical significance ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 was calculated via one-way ANOVA.
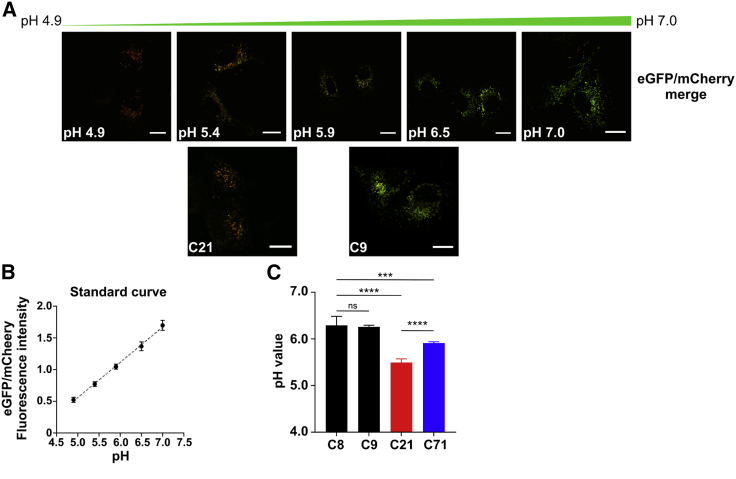
Because intra-Golgi pH is also critical for Golgi homeostasis, we also considered the possibility that an active SLC37A4 relocated to an inappropriate early compartment could indirectly increase the concentration of Glc-6P and/or Pi, leading to an altered intraluminal Golgi pH downstream. To test this, we transiently expressed a Golgi localized B4GALT1 protein fused to both a pH-sensitive eGFP and a pH-indifferent mCherry. Reduced Golgi pH lowers fluorescence of the modified eGFP, while the mCherry is unaffected (Figure 6A). Using this method, we determined the Golgi pH in two controls (C8 and C9) to be 6.23 and 6.21, respectively; C21 was approximately 5.51, 0.7 units lower than that of the controls (Figures 6A–6C), while the heterozygous C71 clone was intermediate at approximately 5.89. This method measures the entire Golgi and cannot discern the pH within each individual cis, medial, or trans compartment.31
Figure 6.
Calibration and quantification of the Golgi pH in Huh7 control and edited cells
(A) Calibration buffers were used to create a pH response curve for the GalT-mCherry-eGFP construct and allowed for the calculation of pH in Huh7 cells. Scale bar represents 20 µm.
(B) Standard curve using calibration buffers to determine estimated luminal pH values.
(C) Quantification of the Golgi luminal pH values in Huh7 controls (C8 and C9) and edited (C21, a homozygous clone, and C71, a heterozygous clone) cells showing acidification of the Golgi upon introduction of the c.1267C>T (p.Arg423∗) mutation. The effect is significantly more pronounced when both alleles are mutated. Data were acquired in four (C8 and C71) or six (C9 and C21) biological replicates, conducted on different weeks, with cells freshly transfected with GalT-mCherry-eGFP construct. In each biological replicate, 10–15 cells were analyzed, and the mean was taken. The graph represents an average of the means acquired over the different biological measurements. Statistical significance ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 was calculated via one-way ANOVA.
SLC37A4 transport activity in CRISPR-edited Huh7 cells
We previously mentioned that autosomal recessive loss-of-function variants in SLC37A4, which are primarily due to reduced Glc-6P transport activity, result in GSD-Ib. We show the p.Arg423∗ does not affect Glc-6P transport activity of SLC37A4 (Figure S10), which is consistent with previous findings showing that the last twelve amino acids of SLC37A4 are not required for protein stability or transport activity.37 It also means that the mislocalized or redistributed mutants are fully capable of transporting and possibly accumulating Glc-6P.
Discussion
We define a dominantly inherited metabolic disorder in seven individuals as a result of a p.Arg423∗ variant in SLC37A4, resulting in a liver-specific CDG characterized by altered serum protein N-glycosylation and a decreased series of coagulation proteins. These proteins include clotting factors (F2, F5, F9, and F11) and inhibitors (SERPINC1, PROS1, and PROC). Interestingly, three of our seven affected individuals also presented with cardiac abnormalities, which is also seen in G6PC3 deficiency.38,39
The p.Arg423∗ variant is predicted to delete a conserved C-terminal ER retrieval signal, which if removed, could result in mislocalization of a portion of the protein because affected individuals are heterozygous for the variant. It is known that varying-sized deletions within the C-terminal tail of SLC37A4 (as early as Arg418) do not impair its transport activity or stability, although localization was not addressed.37 The C-terminal cytosolic tail of SLC37A4 contains a potential COP1-dependent Golgi retention signal KXD/E motif, suggesting possible inability to relocate a portion of the fully active protein to the Golgi. However, it should be noted that SLC37A4 has never been shown to be localized to the Golgi under physiological conditions.
Affected individuals who are compound heterozygous or homozygous for a p.Arg415∗ variant, which deletes the entire retrieval signal, have classical GSD-Ib. Biochemical studies show this variant reduces transport activity because of dramatically decreased transporter protein stability.37 Glycosylation studies in one individual carrying the p.Arg415∗ variant were described as normal.32 However, a lack of defective glycosylation could be due to the fact the p.Arg415∗ mutant is unstable and protein is lost. Therefore, the p.Arg415∗ mutant is unable to exert similar effects as the p.Arg423∗ variant, whose expression is not decreased (Figure S11).
The consequences of these differences in SLC37A4 protein expression highlight a key issue seen in both our study and the one reported by Marquardt et al.32 In both studies, overexpression of either wild-type or mutant transporters was highly toxic, making interpretations from sick or dying cells unreliable.
We initially approached this problem and the lack of a measurable phenotype in fibroblasts by generating iPSCs from one individual (P7). While both control and P7 undifferentiated iPSCs showed no N-glycan abnormalities, we were able to show an abnormal N-glycan phenotype in hepatocytes derived from P7 (Figure S6). Because conversion of iPSCs to hepatocytes is lengthy and incomplete, we sought a more practical and reproducible approach to studying the c.1267C>T (p.Arg423∗) variant in SLC37A4 and opted to CRISPR-base edit Huh7 cells. We were able to generate several isogenic clones carrying variable ratios of wild-type to mutant transporter. This was very likely due to the presence of multiple gene copies of the transporter. These c.1267C>T (p.Arg423∗) SLC37A4-modified Huh7 cell lines maintained stability of the mutant protein at comparable levels to controls (Figure S11). Fibroblasts from affected individuals also express similar levels of SLC37A4 protein to controls, but because they are not gluconeogenic cells, they do not exhibit an abnormal N-glycan phenotype. (Figure S11).
The CRISPR-edited Huh7 cells were found to have a gene dose-dependent abnormal Golgi morphology. There was a clear hierarchy of mutant SLC37A4 dose-dependent phenotypes: reduced Golgi pH → altered Golgi morphology → altered N-glycosylation of secreted and cellular proteins, possibly reflecting a cause-and-effect relationship.
We show mutant protein does not relocate to the Golgi, rather, it maintains activity and a portion of it localizes to an undefined region within the early secretory apparatus. Although several different approaches failed to show localization of the mutant transporter in the Golgi, we do show a portion localized with both an ER-marker (anti-KDEL) and a non-KDEL-containing compartment. This pattern was also seen in P7 hepatocyte-differentiated iPSCs (Figure 4C). We hypothesize the mutant transporter with its normal activity alters the localized environment within this undefined compartment and subsequently causes downstream changes via vesicular trafficking (Figure 7). These changes result in lower Golgi pH, which we hypothesize is sufficient to affect the activity or optimal organization of one or more Golgi-localized glycan maturation enzymes, also altering homeostasis and Golgi architecture (Figure 7). Glycosylation disorders impairing maintenance of luminal pH homeostasis have been reported (e.g., ATP6V0A2-CDG [MIM: 219200], ATP6V1A-CDG [MIM: 617403], ATP6V1E1-CDG [MIM: 617402], ATP6AP1-CDG [MIM: 300972], ATP6AP2-CDG [MIM: 301045], and SLC9A7-CDG [MIM: 301024]), and those disorders often display clear N-glycan and O-glycan defects involving galactosylation and sialylation but not the disturbance in earlier processing steps we observed.40, 41, 42, 43, 44 While the cause of this pH change in our mutant lines remains unclear, we speculate that it is due to abnormal accumulation of Glc-6P or Glc+Pi, which would normally be transported back into the cytoplasm.
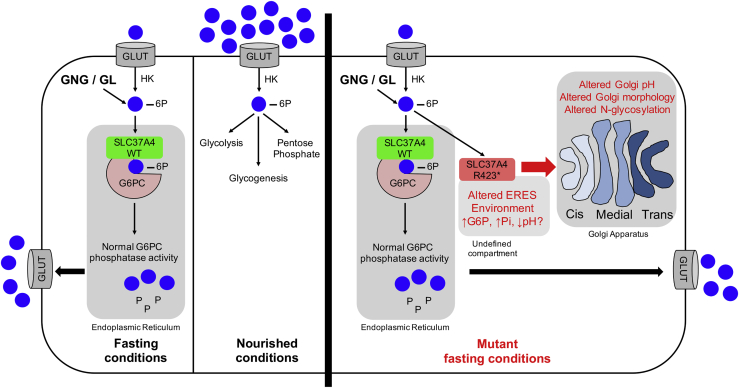
Figure 7.
Overview showing function of SLC37A4 in hepatocytes
A schematic showing the function of wild-type SLC37A4 in hepatocytes during nourished or fasting conditions. Under nourished conditions, exogenous Glc provides ample Glc-6P for glycolysis, glycogenesis, and pentose phosphate pathways. Under fasting conditions, hepatocytes must generate Glc-6P from glycogenolysis (GL) and gluconeogenesis (GNG) for these pathways and also normalize plasma Glc. SLC37A4 imports Glc-6P into the ER and G6PC releases Pi+Glc so both can be returned to the cytoplasm and Glc to the circulation. Mutant SLC37A4 (p.Arg423∗) is fully active and maintains normal glucose homeostasis under fasting conditions, but a portion of the active transporter becomes mislocalized to an undefined, spatially restricted pre-Golgi/post-ER compartment, possibly ERES. Glc-6P and/or Pi accumulates there, leading to a dose-dependent reduction of Golgi pH and Golgi architecture/homeostasis. Reduced pH is propagated in subsequent Golgi compartments, altering the activity and/or localization of multiple N- and O-glycan-modifying enzymes. Because Mn and Mg are critical co-factors for many of these reactions, reduced pH could affect their solubility and availability.
The dominantly inherited disorder presented here is characterized by liver dysfunction, coagulation deficiencies, and profound abnormalities in N-glycosylation of serum-specific proteins; the recessive form GSD-Ib only overlaps with the liver dysfunction. GSD-Ib does not present with the N-glycosylation deficiency seen in liver-specific factors, however neutrophils from affected individuals do show hypogalactosylation.45 It is intriguing that both disorders do show hypogalactosylation, albeit in different tissue types, and suggests dysregulation of glucose homeostasis could be contributing to the hypogalactosylation. In GSD-Ib and G6PC3 deficiency, accumulation of 1,5-anhydroglucitol-6-phosphate significantly alters glucose homeostasis and causes the observed neutropenia and neutrophil dysfunction.38,39
Here, we present a dominantly inherited disorder of glycosylation due to a heterozygous c.1267C>T (p.Arg423∗) variant in SLC37A4. Affected individuals present with a multifactorial coagulation defect characterized by abnormal serum glycoproteins specifically harboring high mannose and hybrid type N-glycans. Hepatoma cells base edited to carry the p.Arg423∗variant recapitulate the N-glycan abnormality and additionally show a profoundly dysfunctional Golgi apparatus characterized by gene dosage-dependent hierarchy of reduced luminal pH, altered morphology, and disrupted homeostasis.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We would like to thank all the families for their continued support and for providing valuable biological specimens and the SBP Medical Discovery Institute FACS core. This work was supported by The Rocket Fund and R01DK99551 (H.H.F.) and the European Union’s Horizon 2020 research and innovation program under the ERA-NET cofund action no. 643578 (A.B., N.S., S.V.B., and T.D.). We thank Koen Galenkamp for providing technical assistance, plasmids, and reagents for pH measurements; Kazuhiro Aoki and Mayumi Ishihara for assistance with N-glycan analysis and annotation; and Jamie Smolin for her assistance. Exome sequencing for P1, P3, P4, and P5 was supported by a grant from Fondation Maladies Rares (FMR) as part of the high-throughput sequencing and rare diseases 2016 program WES-20160717. This work was also supported by Commissariat à l’Energie Atomique et aux Energies Alternatives and the MetaboHUB infrastructure (ANR-11-INBS-0010) (F.F.).
Published: May 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.04.013.
Data and code availability
The accession number for the c.1267C>T (p.Arg423∗) variant in SLC37A4 is GenBank: NM_001164277.1. The UniProt ID is UniProt: O43826-1.
Web resources
Combined Annotation Dependent Depletion (CADD), https://cadd.gs.washington.edu/
DiscovEHR, http://www.discovehrshare.com/
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org/
Supplemental information
References
- 1.Freeze H.H., Chong J.X., Bamshad M.J., Ng B.G. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 2014;94:161–175. doi: 10.1016/j.ajhg.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francisco R., Marques-da-Silva D., Brasil S., Pascoal C., Dos Reis Ferreira V., Morava E., Jaeken J. The challenge of CDG diagnosis. Mol. Genet. Metab. 2019;126:1–5. doi: 10.1016/j.ymgme.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira C.R., Altassan R., Marques-Da-Silva D., Francisco R., Jaeken J., Morava E. Recognizable phenotypes in CDG. J. Inherit. Metab. Dis. 2018;41:541–553. doi: 10.1007/s10545-018-0156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., Deciphering Developmental Disorders Study High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M., Cheng R., Liang J., Yan H., Zhang H., Yang L., Li C., Jiao Q., Lu Z., He J. Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease. Am. J. Hum. Genet. 2013;92:895–903. doi: 10.1016/j.ajhg.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi H., Wong D., Schneider M., Freeze H.H., Takeuchi M., Berardinelli S.J., Ito A., Lee H., Nelson S.F., Haltiwanger R.S. Variant in human POFUT1 reduces enzymatic activity and likely causes a recessive microcephaly, global developmental delay with cardiac and vascular features. Glycobiology. 2018;28:276–283. doi: 10.1093/glycob/cwy014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira C.R., Xia Z.J., Clément A., Parry D.A., Davids M., Taylan F., Sharma P., Turgeon C.T., Blanco-Sánchez B., Ng B.G., Undiagnosed Diseases Network. Scottish Genome Partnership A Recurrent De Novo Heterozygous COG4 Substitution Leads to Saul-Wilson Syndrome, Disrupted Vesicular Trafficking, and Altered Proteoglycan Glycosylation. Am. J. Hum. Genet. 2018;103:553–567. doi: 10.1016/j.ajhg.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynders E., Foulquier F., Leão Teles E., Quelhas D., Morelle W., Rabouille C., Annaert W., Matthijs G. Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum. Mol. Genet. 2009;18:3244–3256. doi: 10.1093/hmg/ddp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veiga-da-Cunha M., Gerin I., Chen Y.T., de Barsy T., de Lonlay P., Dionisi-Vici C., Fenske C.D., Lee P.J., Leonard J.V., Maire I. A gene on chromosome 11q23 coding for a putative glucose- 6-phosphate translocase is mutated in glycogen-storage disease types Ib and Ic. Am. J. Hum. Genet. 1998;63:976–983. doi: 10.1086/302068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou J.Y., Jun H.S., Mansfield B.C. Glycogen storage disease type I and G6Pase-β deficiency: etiology and therapy. Nat. Rev. Endocrinol. 2010;6:676–688. doi: 10.1038/nrendo.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou J.Y., Mansfield B.C. The SLC37 family of sugar-phosphate/phosphate exchangers. Curr. Top. Membr. 2014;73:357–382. doi: 10.1016/B978-0-12-800223-0.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitt K.N., Walker E.A., Stewart P.M. Minireview: hexose-6-phosphate dehydrogenase and redox control of 11beta-hydroxysteroid dehydrogenase type 1 activity. Endocrinology. 2005;146:2539–2543. doi: 10.1210/en.2005-0117. [DOI] [PubMed] [Google Scholar]
- 13.Hiraiwa H., Pan C.J., Lin B., Moses S.W., Chou J.Y. Inactivation of the glucose 6-phosphate transporter causes glycogen storage disease type 1b. J. Biol. Chem. 1999;274:5532–5536. doi: 10.1074/jbc.274.9.5532. [DOI] [PubMed] [Google Scholar]
- 14.Burchell A., Jung R.T., Lang C.C., Bennet W., Shepherd A.N. Diagnosis of type 1a and type 1c glycogen storage diseases in adults. Lancet. 1987;1:1059–1062. doi: 10.1016/s0140-6736(87)90484-3. [DOI] [PubMed] [Google Scholar]
- 15.Lei K.J., Chen Y.T., Chen H., Wong L.J., Liu J.L., McConkie-Rosell A., Van Hove J.L., Ou H.C., Yeh N.J., Pan L.Y. Genetic basis of glycogen storage disease type 1a: prevalent mutations at the glucose-6-phosphatase locus. Am. J. Hum. Genet. 1995;57:766–771. [PMC free article] [PubMed] [Google Scholar]
- 16.Boztug K., Appaswamy G., Ashikov A., Schäffer A.A., Salzer U., Diestelhorst J., Germeshausen M., Brandes G., Lee-Gossler J., Noyan F. A syndrome with congenital neutropenia and mutations in G6PC3. N. Engl. J. Med. 2009;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banka S., Newman W.G., Ozgül R.K., Dursun A. Mutations in the G6PC3 gene cause Dursun syndrome. Am. J. Med. Genet. A. 2010;152A:2609–2611. doi: 10.1002/ajmg.a.33615. [DOI] [PubMed] [Google Scholar]
- 18.Draper N., Walker E.A., Bujalska I.J., Tomlinson J.W., Chalder S.M., Arlt W., Lavery G.G., Bedendo O., Ray D.W., Laing I. Mutations in the genes encoding 11beta-hydroxysteroid dehydrogenase type 1 and hexose-6-phosphate dehydrogenase interact to cause cortisone reductase deficiency. Nat. Genet. 2003;34:434–439. doi: 10.1038/ng1214. [DOI] [PubMed] [Google Scholar]
- 19.Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colunga T., Hayworth M., Kress S., Reynolds D.M., Chen L., Nazor K.L., Baur J., Singh A.M., Loring J.F., Metzger M. Human Pluripotent Stem Cell-Derived Multipotent Vascular Progenitors of the Mesothelium Lineage Have Utility in Tissue Engineering and Repair. Cell Rep. 2019;26:2566–2579.e10. doi: 10.1016/j.celrep.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A.M., Zhang L., Avery J., Yin A., Du Y., Wang H., Li Z., Fu H., Yin H., Dalton S. Human beige adipocytes for drug discovery and cell therapy in metabolic diseases. Nat. Commun. 2020;11:2758. doi: 10.1038/s41467-020-16340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carchon H.A., Chevigné R., Falmagne J.B., Jaeken J. Diagnosis of congenital disorders of glycosylation by capillary zone electrophoresis of serum transferrin. Clin. Chem. 2004;50:101–111. doi: 10.1373/clinchem.2003.021568. [DOI] [PubMed] [Google Scholar]
- 23.Lacey J.M., Bergen H.R., Magera M.J., Naylor S., O’Brien J.F. Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin. Chem. 2001;47:513–518. [PubMed] [Google Scholar]
- 24.Bengtson P., Ng B.G., Jaeken J., Matthijs G., Freeze H.H., Eklund E.A. Serum transferrin carrying the xeno-tetrasaccharide NeuAc-Gal-GlcNAc2 is a biomarker of ALG1-CDG. J. Inherit. Metab. Dis. 2016;39:107–114. doi: 10.1007/s10545-015-9884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen-Nicolaÿ S., Boursier C., Rio M., Lefeber D.J., Pilon A., Seta N., Bruneel A. MALDI-TOF MS applied to apoC-III glycoforms of patients with congenital disorders affecting O-glycosylation. Comparison with two-dimensional electrophoresis. Proteomics Clin. Appl. 2015;9:787–793. doi: 10.1002/prca.201400187. [DOI] [PubMed] [Google Scholar]
- 26.Bruneel A., Cholet S., Drouin-Garraud V., Jacquemont M.L., Cano A., Mégarbané A., Ruel C., Cheillan D., Dupré T., Vuillaumier-Barrot S. Complementarity of electrophoretic, mass spectrometric, and gene sequencing techniques for the diagnosis and characterization of congenital disorders of glycosylation. Electrophoresis. 2018;39:3123–3132. doi: 10.1002/elps.201800021. [DOI] [PubMed] [Google Scholar]
- 27.Mehta N., Porterfield M., Struwe W.B., Heiss C., Azadi P., Rudd P.M., Tiemeyer M., Aoki K. Mass Spectrometric Quantification of N-Linked Glycans by Reference to Exogenous Standards. J. Proteome Res. 2016;15:2969–2980. doi: 10.1021/acs.jproteome.6b00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keser T., Pavić T., Lauc G., Gornik O. Comparison of 2-Aminobenzamide, Procainamide and RapiFluor-MS as Derivatizing Agents for High-Throughput HILIC-UPLC-FLR-MS N-glycan Analysis. Front Chem. 2018;6:324. doi: 10.3389/fchem.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puglielli L., Konopka G., Pack-Chung E., Ingano L.A., Berezovska O., Hyman B.T., Chang T.Y., Tanzi R.E., Kovacs D.M. Acyl-coenzyme A: cholesterol acyltransferase modulates the generation of the amyloid beta-peptide. Nat. Cell Biol. 2001;3:905–912. doi: 10.1038/ncb1001-905. [DOI] [PubMed] [Google Scholar]
- 30.Ko M.H., Puglielli L. Two endoplasmic reticulum (ER)/ER Golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. J. Biol. Chem. 2009;284:2482–2492. doi: 10.1074/jbc.M804901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galenkamp K.M.O., Sosicka P., Jung M., Recouvreux M.V., Zhang Y., Moldenhauer M.R., Brandi G., Freeze H.H., Commisso C. Golgi Acidification by NHE7 Regulates Cytosolic pH Homeostasis in Pancreatic Cancer Cells. Cancer Discov. 2020;10:822–835. doi: 10.1158/2159-8290.CD-19-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquardt T., Bzduch V., Hogrebe M., Rust S., Reunert J., Grüneberg M., Park J., Callewaert N., Lachmann R., Wada Y., Engel T. SLC37A4-CDG: Mislocalization of the glucose-6-phosphate transporter to the Golgi causes a new congenital disorder of glycosylation. Mol. Genet. Metab. Rep. 2020;25:100636. doi: 10.1016/j.ymgmr.2020.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson M.P., Quelhas D., Leão-Teles E., Sturiale L., Rymen D., Keldermans L., Race V., Souche E., Rodrigues E., Campos T. SLC37A4-CDG: Second patient. JIMD Rep. 2021;58:122–128. doi: 10.1002/jmd2.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crispin M., Chang V.T., Harvey D.J., Dwek R.A., Evans E.J., Stuart D.I., Jones E.Y., Lord J.M., Spooner R.A., Davis S.J. A human embryonic kidney 293T cell line mutated at the Golgi alpha-mannosidase II locus. J. Biol. Chem. 2009;284:21684–21695. doi: 10.1074/jbc.M109.006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao C., Cai Y., Wang Y., Kang B.H., Aniento F., Robinson D.G., Jiang L. Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci. 2014;19:508–515. doi: 10.1016/j.tplants.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Jackson L.P., Lewis M., Kent H.M., Edeling M.A., Evans P.R., Duden R., Owen D.J. Molecular basis for recognition of dilysine trafficking motifs by COPI. Dev. Cell. 2012;23:1255–1262. doi: 10.1016/j.devcel.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L.Y., Lin B., Pan C.J., Hiraiwa H., Chou J.Y. Structural requirements for the stability and microsomal transport activity of the human glucose 6-phosphate transporter. J. Biol. Chem. 2000;275:34280–34286. doi: 10.1074/jbc.M006439200. [DOI] [PubMed] [Google Scholar]
- 38.Veiga-da-Cunha M., Chevalier N., Stephenne X., Defour J.P., Paczia N., Ferster A., Achouri Y., Dewulf J.P., Linster C.L., Bommer G.T., Van Schaftingen E. Failure to eliminate a phosphorylated glucose analog leads to neutropenia in patients with G6PT and G6PC3 deficiency. Proc. Natl. Acad. Sci. USA. 2019;116:1241–1250. doi: 10.1073/pnas.1816143116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wortmann S.B., Van Hove J.L.K., Derks T.G.J., Chevalier N., Knight V., Koller A., Oussoren E., Mayr J.A., van Spronsen F.J., Lagler F.B. Treating neutropenia and neutrophil dysfunction in glycogen storage disease type Ib with an SGLT2 inhibitor. Blood. 2020;136:1033–1043. doi: 10.1182/blood.2019004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornak U., Reynders E., Dimopoulou A., van Reeuwijk J., Fischer B., Rajab A., Budde B., Nürnberg P., Foulquier F., Lefeber D., ARCL Debré-type Study Group Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 2008;40:32–34. doi: 10.1038/ng.2007.45. [DOI] [PubMed] [Google Scholar]
- 41.Van Damme T., Gardeitchik T., Mohamed M., Guerrero-Castillo S., Freisinger P., Guillemyn B., Kariminejad A., Dalloyaux D., van Kraaij S., Lefeber D.J. Mutations in ATP6V1E1 or ATP6V1A Cause Autosomal-Recessive Cutis Laxa. Am. J. Hum. Genet. 2017;100:216–227. doi: 10.1016/j.ajhg.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jansen E.J., Timal S., Ryan M., Ashikov A., van Scherpenzeel M., Graham L.A., Mandel H., Hoischen A., Iancu T.C., Raymond K. ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat. Commun. 2016;7:11600. doi: 10.1038/ncomms11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rujano M.A., Cannata Serio M., Panasyuk G., Péanne R., Reunert J., Rymen D., Hauser V., Park J.H., Freisinger P., Souche E. Mutations in the X-linked ATP6AP2 cause a glycosylation disorder with autophagic defects. J. Exp. Med. 2017;214:3707–3729. doi: 10.1084/jem.20170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khayat W., Hackett A., Shaw M., Ilie A., Dudding-Byth T., Kalscheuer V.M., Christie L., Corbett M.A., Juusola J., Friend K.L. A recurrent missense variant in SLC9A7 causes nonsyndromic X-linked intellectual disability with alteration of Golgi acidification and aberrant glycosylation. Hum. Mol. Genet. 2019;28:598–614. doi: 10.1093/hmg/ddy371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letkemann R., Wittkowski H., Antonopoulos A., Podskabi T., Haslam S.M., Föll D., Dell A., Marquardt T. Partial correction of neutrophil dysfunction by oral galactose therapy in glycogen storage disease type Ib. Int. Immunopharmacol. 2017;44:216–225. doi: 10.1016/j.intimp.2017.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the c.1267C>T (p.Arg423∗) variant in SLC37A4 is GenBank: NM_001164277.1. The UniProt ID is UniProt: O43826-1.