Summary
Virtually all genome sequencing efforts in national biobanks, complex and Mendelian disease programs, and medical genetic initiatives are reliant upon short-read whole-genome sequencing (srWGS), which presents challenges for the detection of structural variants (SVs) relative to emerging long-read WGS (lrWGS) technologies. Given this ubiquity of srWGS in large-scale genomics initiatives, we sought to establish expectations for routine SV detection from this data type by comparison with lrWGS assembly, as well as to quantify the genomic properties and added value of SVs uniquely accessible to each technology. Analyses from the Human Genome Structural Variation Consortium (HGSVC) of three families captured ~11,000 SVs per genome from srWGS and ~25,000 SVs per genome from lrWGS assembly. Detection power and precision for SV discovery varied dramatically by genomic context and variant class: 9.7% of the current GRCh38 reference is defined by segmental duplication (SD) and simple repeat (SR), yet 91.4% of deletions that were specifically discovered by lrWGS localized to these regions. Across the remaining 90.3% of reference sequence, we observed extremely high (93.8%) concordance between technologies for deletions in these datasets. In contrast, lrWGS was superior for detection of insertions across all genomic contexts. Given that non-SD/SR sequences encompass 95.9% of currently annotated disease-associated exons, improved sensitivity from lrWGS to discover novel pathogenic deletions in these currently interpretable genomic regions is likely to be incremental. However, these analyses highlight the considerable added value of assembly-based lrWGS to create new catalogs of insertions and transposable elements, as well as disease-associated repeat expansions in genomic sequences that were previously recalcitrant to routine assessment.
Keywords: structural variation, whole-genome sequencing, long-read technology, genome assembly, segmental duplication, simple repeats, copy number variation
Main text
The field of genomics has seen remarkable advances in the accuracy and efficiency of massively parallel sequencing-by-synthesis technologies that generate pairs of short reads from the ends of small 400–800 base pair (bp) fragments (referred to herein as short-read whole-genome sequencing [srWGS]). This technical leap and derivative approaches such as targeted whole-exome capture sequencing (WES) have catalyzed a deluge of gene discoveries for rare diseases and insights into population genetics and genome biology. Correspondingly, srWGS has been adopted by all major human disease and biobank sequencing initiatives, including the NHGRI Centers for Common Disease Genomics (CCDG)1 and Centers for Mendelian Genetics (CMG),2 the Deciphering Developmental Disorders (DDD) project,3 the Trans-Omics for Precision Medicine (TOPMed),4 the All of Us Research Program,5 the NICHD Gabriella Miller Kids First (GMKF) initiative, the UK BioBank,6 and Genomics England,7 to name just a few. As such, a critical step for the field is to establish uniform methods for srWGS data processing and rational benchmarking standards to set expectations for variant detection.
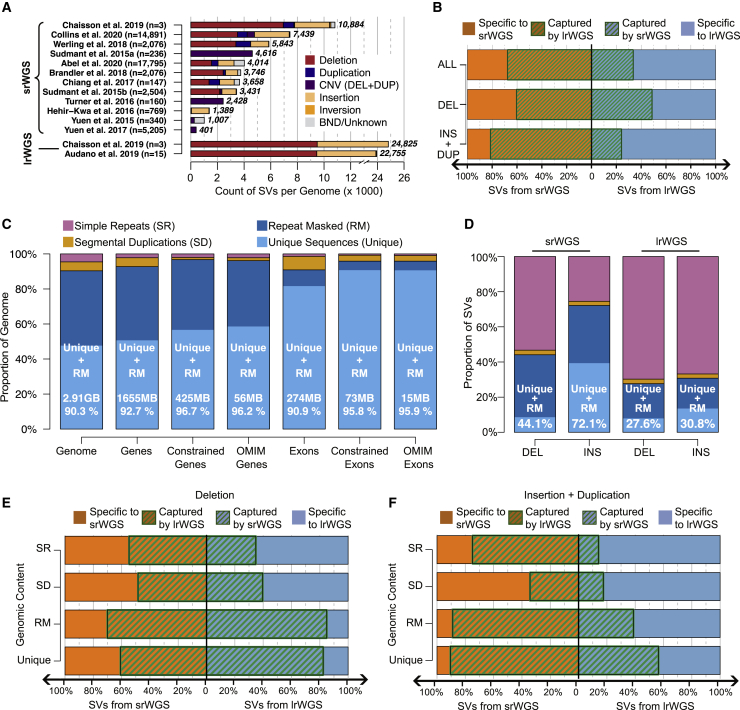
The technical processes of genome alignment and single-nucleotide variant (SNV) detection have been an intensive focus of genomics since the inception of the 1000 Genomes Project8 and more recently updated for cross-institute functional equivalence as part of the NHGRI Genome Sequencing Program and variant detection with the Genome Analysis Toolkit (GATK) best practices.9, 10, 11 However, no comparable standardized methods have been adopted for detection of structural variants (SVs), defined here as genomic alterations greater than 50 bp, from srWGS, and there are limited gold-standard benchmarking approaches for SV discovery. This lack of uniformity has introduced a barrier to establishing reliable estimates of the SV counts and characteristics per genome. Not surprisingly, as shown in Figure 1A, these estimates have varied considerably across studies. The initial discovery effort from the 1000 Genomes Project12,13 revealed the landscape of SVs that could be captured from srWGS with just 4–7× coverage (3,431 SVs per genome). More recent population genetic and human disease studies using deeper (30× or higher) srWGS and diverse analytic methods have varied in estimates of SVs that can be captured via srWGS; these estimates vary from 401 to 10,884 per genome. At present, the most sensitive studies have utilized the integration of multiple SV detection methods from the Genome Aggregation Database (gnomAD) and the Human Genome Structural Variation Consortium (HGSVC) projects (Figure 1A).1,14,15,13, 16, 17, 18, 19, 20
Figure 1.
Comparison of SV callsets from srWGS and lrWGS
(A) The substantial increased yield of lrWGS in SV detection is displayed from the HGSVC14 and the largest Pacific Biosciences (PacBio) lrWGS study published to date15 by comparison with contemporary srWGS studies. As shown, there is wide variability of SV detection across srWGS studies to date that report SVs detected per individual in more than 100 genomes. Parenthetical numbers next to each study label indicate the number of genomes analyzed, and bold numbers next to each bar represent the number of SVs per genome reported by each study.
(B) Overlap of SVs from the HGSVC srWGS and lrWGS callsets across children of the three trio families, partitioned by SV class.
(C) Distribution of repetitive sequences across the genome, genes, and exons. “Constrained” refers to genes and exons with pLI > 0.9,21 and “OMIM genes” includes a curated list of autosomal dominant genes that were defined in both Berg et al.22 and Blekhman et al.23 Gb, gigabase; Mb, megabase. Percentage listed within each bar is the fraction of each group composed of "unique + RM" sequences.
(D) Distribution of SVs from srWGS and lrWGS split by repetitive sequence context. Formatting conventions are the same as in (C).
(E and F) Concordance of deletions (E) and insertions and duplications (F) between srWGS and lrWGS split by repetitive sequence context.
Emerging long-read WGS (lrWGS) technologies, which involve sequencing thousands to millions of contiguous nucleotides from a single strand of DNA, have significantly increased sensitivity for SV discovery in the human genome. The most widely tested lrWGS technologies include single-molecule real-time (SMRT) sequencing from Pacific Biosciences (PacBio)24 and sequencing by ionic current through a nanopore channel (Oxford Nanopore Technologies [ONT]).25 A key advantage of lrWGS is the abundance of reads that span entire SVs, allowing for direct observation of SVs rather than SV detection by inference as required for srWGS. These unique properties of lrWGS are beginning to revolutionize de novo assembly approaches,26,27 and methods are already maturing for telomere-to-telomere assembly of individual human chromosomes.15,28,29 The most recent analyses used the combination of multiple sequencing platforms (e.g., lrWGS, strand-specific sequencing,30 and optical mapping31) in relatively small numbers of genomes to generate assembly-based SV callsets,14,32 which have approximately doubled the number of SVs able to be captured in each genome to ~25,000 as compared with srWGS14,15 (Figure 1A).
These lrWGS studies have thus opened access to SVs in the genome that were traditionally refractory to discovery by srWGS or interpretation in disease association studies, such as repeat expansions and other alterations within repetitive genomic regions and centromeres.33 Unfortunately, the current cost of lrWGS is a significant premium over srWGS. For example, as of this writing the cost for generation of PacBio lrWGS over srWGS for equivalent coverage at leading academic platforms from the HGSVC ranges from 5.9-fold increase for continuous long-read technology to 12-fold increase for circular consensus sequencing HiFi technology. Moreover, the comparatively lower throughput of modern lrWGS platforms renders them impractical for adoption in large-scale population studies on the order of tens to hundreds of thousands of individuals. The largest published assembly-based PacBio study to date has analyzed just 15 genomes,15 and a recent preprint from the HGSVC describes 35 genomes,34 while a published study from Iceland analyzed 3,622 ONT genomes.35 By comparison, millions of genomes have already been sequenced or commissioned via srWGS across international initiatives. Given this predominance of srWGS in the current landscape of genomics research, we present here a series of analyses from the HGSVC to (1) benchmark expectations for the number and class of variants that can be reliably detected from srWGS, (2) predict the genomic features that drive false positive and false negative discoveries for each technology, and (3) establish the scientific and clinical advances offered by state-of-the-art lrWGS assembly as a complementary approach to srWGS.
In this study, we performed a detailed comparison of SVs detected from alignment-based srWGS and assembly-based lrWGS methods on three matched trio families (HG00514, HG00733, and NA19240) from the 1000 Genomes Project, and all results per genome reported here are averages across the three children in these families.14 For srWGS, this initial study applied a highly sensitive ensemble approach to integrate 13 SV detection algorithms (supplemental material and methods) and discovered an average of 10,884 SVs per genome. The emphasis on sensitivity suggests that approximately 11,000 SVs per genome most likely reflects an upper bound on the total number of SVs that can be routinely captured from srWGS with the alignment-based algorithms applied by the HGSVC, as demonstrated in Figure 1A by comparison with other contemporary studies. However, this sensitivity came at the significant cost of specificity: 685 de novo SVs were observed per genome, over 1,000-fold more than our expectation from srWGS based on family studies, population genetic estimators, and molecular validation, therefore representing many SV predictions that are most likely false positives.16 The lrWGS-derived SV callset combined whole-genome phasing with two state-of-the-art genome assembly approaches (Phase-SV and MS-PAC14,26,36) and was supplemented by additional technologies (HiC37 and StrandSeq,38 see Chaisson et al.14). These methods discovered an average of 24,825 haplotype-resolved SVs per genome, or over 2-fold more than the most sensitive srWGS approaches. Surprisingly, although the srWGS and lrWGS callsets were generated on identical samples, only a limited subset of SVs (66.8% of srWGS and 33.5% of lrWGS) overlapped between technologies. Moreover, the mutational class of SVs dramatically impacted concordance: 60.6% of srWGS and 48.7% of lrWGS deletions demonstrated overlap as compared with 81.7% of srWGS and 24.1% of lrWGS insertions (Figure 1B).
We sought to define and quantify the factors contributing to the poor concordance between SVs derived from each technology to improve SV discovery, filtering, and prioritization from srWGS in future large-scale medical and population genetic initiatives. We first explored the role of genomic features such as repetitive sequences that are enriched for SVs via repeat-mediated mechanisms39,40 because short-read alignment has well-documented limitations within these genomic regions.41,42 We annotated all SVs with sequence context based on RepeatMasker43 and segmental duplication44 tracks from the UCSC genome browser.45,46 For simplicity, we consolidated all repetitive sequence annotations into three categories: segmental duplication (SD; 5.1% of the genome), simple repeat (SR; 4.6%), and “repeat masked” (RM; 42.9%), where this RM category referred to all other repetitive sequence not overlapping SD or SR elements. The remaining 47.4% of the genome not overlapping any of these repeat categories was labeled as “unique” sequence, which is a term used for simplicity here, although these regions are not completely devoid of repetitive sequences. The "unique" and RM categories collectively encompass 90.3% of the annotated human reference sequence, 90.9% of all currently annotated protein-coding sequence, 95.8% of all currently annotated coding sequence from evolutionarily constrained genes, and 95.9% of genes currently associated with human disease from the Online Mendelian Inheritance in Man (OMIM; Figure 1C).21,47, 48, 49
As expected, the distribution of SVs was non-uniform and varied by sequence context for each technology (Figure 1D). Most prominently, the enrichment of SV breakpoints in highly repetitive genomic sequences (SD + SR regions) was dramatic and their distribution differed significantly between technologies: despite representing just 9.7% of the reference genome, SD + SR annotated sequences contained at least one breakpoint from 49.8% of all SVs from srWGS and 70.4% of all SVs from lrWGS (p < 2.2e−16 for both technologies, chi-square test, Table S1, see supplemental material and methods for details). This enrichment of SVs in repetitive sequence was also strongly correlated with concordance between srWGS and lrWGS: SVs located in repetitive SD + SR sequences displayed 57.0% concordance among srWGS variants and 22.5% in lrWGS variants, whereas those ratios improved considerably in less repetitive sequences ("unique + RM") to 76.5% in srWGS and 59.9% in lrWGS (Figures 1E and 1F).
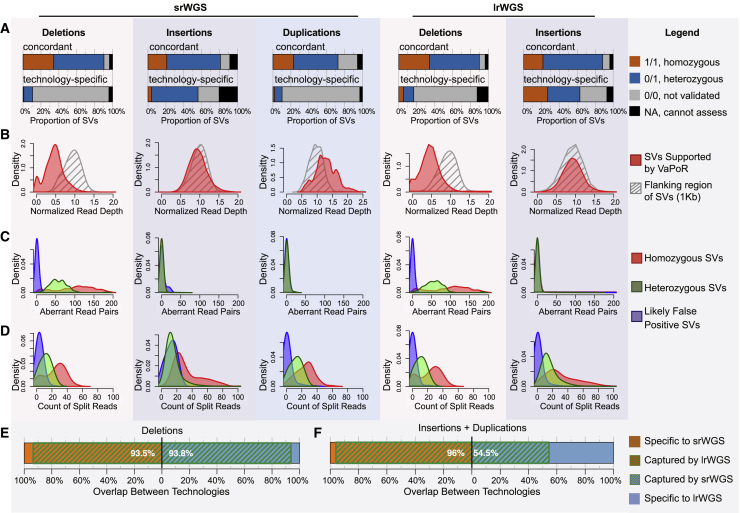
Although the divergent distributions and diminished concordance of SV detection by technology aligned with expectations for SD + SR regions, the paucity of overlap between technologies in "unique + RM" regions was unexpected because breakpoints localized to these regions should not suffer from the same technical confounders of SV discovery in highly repetitive sequences. Therefore, we next sought to decouple and quantify the discordance driven by underlying biological features of the genome from technical noise driven by false positive SVs present in the underlying HGSVC callsets that were optimized for sensitivity as described above. We also reasoned that identifying the covariates that have the greatest influence on false positive SV calls would be valuable in guiding the human genetics community toward principled improvements in SV detection and filtering algorithms. To accomplish this, we developed an in silico SV assessment to improve the precision of srWGS and lrWGS callsets in non-repetitive regions. This procedure re-evaluated the following three pieces of orthogonal information from both lrWGS and srWGS for each SV: (1) supporting evidence from raw lrWGS reads in the parent and offspring genomes for the presence of an SV (VaPoR;50 Figure 2A); (2) copy states based on srWGS normalized read depth within SVs (Figures 2B and S1); (3) discordant paired-end and split reads information at the breakpoint of each predicted SV (Figures 2C, 2D, and S2, Table S2). We considered the SVs with one or more modes of supporting evidence as “high confidence” and explored their overlap on the basis of repeat context for SV calls from different technologies (see supplemental material and methods for further details).
Figure 2.
Methods to recalibrate SVs in "unique + RM" sequences based on read-level alignment signatures
(A) In silico evaluation results from VaPoR on deletions (pink background), insertions (purple background), and duplications (blue background). Duplications and insertions reported by srWGS were both compared against insertions from lrWGS. “Concordant” represents SVs discovered by both lrWGS and srWGS, and “technology-specific” represents SVs specifically discovered from one technology.
(B) Distribution of normalized read depth of srWGS across deletions (pink background), insertions (purple background), and duplications (blue background) that were supported by VaPoR (red) and the 1 kb genomic regions that flank these SVs (gray).
(C and D) Distribution of aberrant srWGS read pairs (C) and split reads (D) around deletions (pink background), insertions (purple background), and duplications (blue background) that were either homozygous (red), heterozygous (green), or false positives (blue). The homozygous, heterozygous, and likely false positive SV sets were selected with the criteria described in the supplemental material and methods.
(E and F) Concordance of deletions (E) and insertions and duplications (F) in "unique + RM" sequences that were supported by the in silico SV refinement procedure. Percentages represent the fraction of total variants shared between srWGS and lrWGS.
We initially applied this in silico SV refinement procedure to deletions, which represent the most interpretable class of SVs for genomics applications. As expected, the in silico confirmation rate—i.e., the proportion of SVs supported by one or more of the evidence classes described above—was high (93.5%) for deletions concordant between technologies in "unique + RM" regions compared to just 13.5% and 33.1% for those that were only discovered by a single technology for srWGS or lrWGS, respectively (Figure S3). After restricting to high-confidence SVs, we observed a substantial improvement in concordance: 93.5% to 93.8% of deletions were shared between srWGS and lrWGS (Figure 2E). Although mutational processes such as somatic SVs or sub-clonal mutations arising in cell culture can contribute to false positive findings, these results implied that most of the discordance between srWGS and lrWGS for SV discovery in the 90.3% of the genome not encompassed by SD + SR sequence was most likely technical in origin. Importantly, it appeared that most of the discordance wasdriven by false positive SV calls that can be pruned by post hoc heuristic filtering.
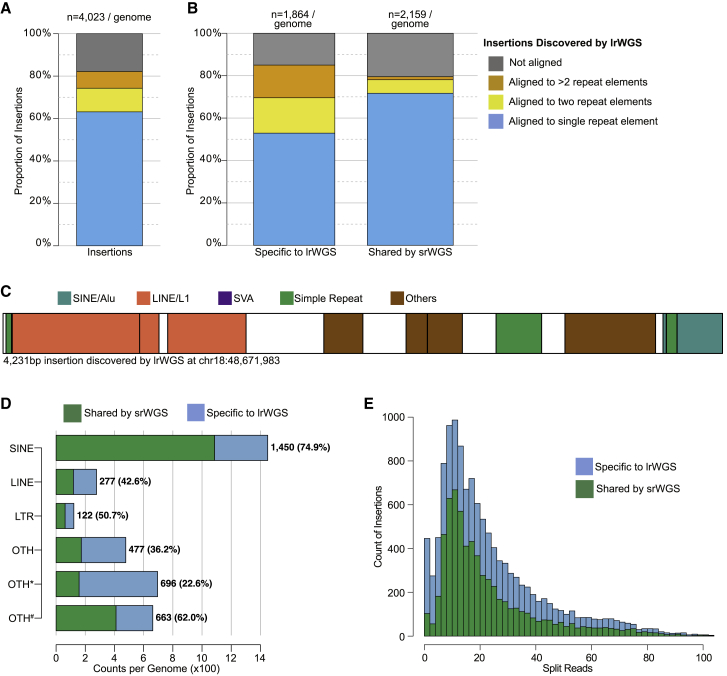
We next explored the impact of post hoc filtering on SVs other than deletions. While duplications and insertions were reported as separate SV classes by srWGS, the lrWGS methods applied by the HGSVC treated both classes as insertions. Given this, we considered all srWGS duplications as insertions for subsequent comparisons. In contrast to the strong concordance between srWGS and lrWGS observed for deletions, 45.5% of high-confidence lrWGS insertions in "unique + RM" regions had no matching SV call from srWGS, while the majority (96.0%) of srWGS insertions and duplications were captured by lrWGS (Figures 2F and S4). To investigate the properties of insertions specifically captured by lrWGS in "unique + RM" sequences, we aligned the assembled sequences of high-confidence insertions against a catalog of known repeat elements.43 Most of these insertions aligned to specific types of repeat elements (61.8%, n = 2,485/genome), such as short interspersed nuclear elements (SINEs, n = 1,494/genome), long interspersed nuclear elements (LINEs, n = 312/genome), and long terminal repeat (LTR, n = 139/genome) retrotransposons (Figures 3A and 3B). Notably, a “chimeric” alignment pattern was observed for 31.7% of the insertions specifically discovered by lrWGS where inserted sequences were aligned to multiple different repeat types (Figures 3B and 3C). These results indicate that the complexity of insertion repeat structure is a major determinant of srWGS sensitivity for insertion SVs, as has been previously demonstrated for certain classes of nested insertions.51 We further observed high variability in the current capabilities of srWGS detection algorithms depending on the type of transposable element insertions when comparing with lrWGS: 74.9% of SINEs, 42.6% of LINEs, and 50.7% of LTRs detected by lrWGS were also discovered by srWGS (Figure 3D). Intriguingly, almost all (95.8%) of the high-confidence lrWGS insertions in "unique + RM" regions that were only discovered by lrWGS nevertheless had some detectable support in the raw srWGS data, indicating that continued development of insertion detection algorithms could substantially improve sensitivity for identification of this variant class from srWGS (Figure 3E). Taken together, these analyses indicate that lrWGS and assembly-based approaches provide substantial improvements over srWGS for insertion discovery, particularly for those events with complex repeat structures.
Figure 3.
Alignment of assembled lrWGS insertion sequences against known repeat elements
(A) Count of lrWGS insertions in "unique + RM" sequences per genome by alignment of inserted sequences to known repeat elements. The number on top of the bar represents the averaged count of high-confidence insertions in "unique + RM" sequences per genome.
(B) Count of lrWGS insertions that are specifically discovered by lrWGS and shared by srWGS, by alignment of inserted sequences to known repeat elements. Formatting conventions are the same as in (A).
(C) An example of an insertion SV assembled by lrWGS, annotated with sequences that align to known repeat element classes. White shading represents sequences not annotated as a known repeat element.
(D) Counts of lrWGS insertions in "unique + RM" sequences per genome by the class of inserted sequence and the proportion that was overlapped by srWGS. “OTH∗” represents insertions aligned to multiple known repeat elements, such as the example shown in (B). “OTH#” stands for insertions that were not aligned to any repeat elements. Numbers in parentheses represent the proportion of insertions that were overlapped by srWGS.
(E) Count of split reads around the lrWGS high-confidence insertions in histogram.
We also examined SVs in highly repetitive SD + SR regions by using the same in silico evaluation framework (Figures S5A–S5D) as described above with the caveat that the orthogonal evaluation of variants in these regions is more challenging and prone to false positives due to alignment artifacts that do not arise in the less repetitive regions of the genome. Similar to the "unique + RM" regions, insertions were poorly captured by srWGS, and only 17.0% overlapped lrWGS insertions, while 74.0% of srWGS insertions were captured by lrWGS (Figure S5F). The high concordance for deletions in "unique + RM" sequences also dissipated in these more repetitive SD + SR regions, as the concordance was 69.6% and 40.4% of high-confidence deletions from srWGS and lrWGS that were shared by the other technology, respectively (Figure S5E).
Finally, we explored the concordance of SV detection for a class of SVs that is strongly enriched for pathogenic variation and appears to be a significant blind spot for long-read assembly technologies: large CNVs captured by depth-based analyses from srWGS. Our initial analyses suggested that lrWGS assembly methods failed to capture all but one of the small number of large (>5 kb) CNVs that could be detected by srWGS read-depth methods in three probands (average size = 14.7 kb). Recognizing the limitation of read-depth analyses to capture large CNVs in a small number of families, we explored CNV calls from 3,202 individuals from our ongoing analyses of 30× srWGS in the 1000 Genomes Project that included all three families used in this study (see HGSVC preprints for complete details).34,52 We found an average of 167 large CNVs per genome that were exclusively detected by depth-based methods, 88.2% of which were not detected by lrWGS assembly. These findings highlight an important blind spot in variant detection from lrWGS assembly in the absence of depth-based analyses and have significant implications for human disease studies because large CNVs have a profound deleterious impact on a spectrum of human diseases.
In conclusion, we demonstrate the strong influence of genomic context on expectations for SV detection from srWGS in genomic studies, as well as estimating the anticipated yields of emerging lrWGS technologies. Initial surveys have implied highly variable outcomes and limited overall concordance in SV detection between the two technologies;14 however, in-depth analyses of these variants emphasize that genome organization, variant type, variant size, and high type I error rates in SV detection from each technology were the predominant features driving discordance. After applying post hoc filters to correct for the relatively high type I error rates for SV detection from this ensemble srWGS approach optimized for sensitivity, and the assembly-based lrWGS approach that was optimized with orthogonal data types, we were able to extrapolate the informative genomic features that influence differences in SV distributions between technologies. The concordance between srWGS and lrWGS was remarkably high for deletions localized to the least-repetitive regions of the genome (93.8%), while almost all lrWGS-specific deletions were localized to repetitive SD + SR regions. We observed poor sensitivity in the detection of large CNVs (>5 kb) via lrWGS assemblies by comparison with srWGS, and this limitation is most likely due to the lack of depth-based lrWGS methods. In contrast, lrWGS showed superior sensitivity for detection of insertions regardless of the genomic context, although most (95.8%) insertions in the least-repetitive genomic regions had detectable alignment signatures in the srWGS data, indicating further improvement in insertion discovery methods for srWGS should continue to bridge this disparity in insertion detection between technologies. Variant types other than deletions and insertions (e.g., inversions, translocations, balanced and complex SVs) were excluded from these analyses because they were not uniformly called by lrWGS assemblies, although we expect future improvement in lrWGS methods to provide novel insights into repeat-mediated mechanisms for these variant classes.
The value added for long-read assembly to discover new disease-associated SVs, or to provide resolution to “unsolved” cases in Mendelian genetics research and clinical diagnostics, is thus a complex calculus. As we note above, srWGS captures virtually all high-quality deletions derived from lrWGS assembly in the regions of the genome that encompass over 95% of currently annotated coding sequence in genes with existing evidence for dominant-acting pathogenic mutations from OMIM. We therefore anticipate that a minority of “unsolved” cases will be explained by novel and readily interpretable deletions that can be captured by lrWGS but remain cryptic to srWGS in known disease-associated genes. However, given that the most highly repetitive regions of the genome have been traditionally inaccessible in human disease studies, it is anticipated that new disease-associated genes and sequences will emerge as functional annotation of these repetitive sequences and duplicated genes continues to improve. Indeed, germline and somatic repeat expansions and contractions are already well established mechanisms of human disease, particularly neurodegenerative disorders.53 As telomere-to-telomere assembly methods continue to mature and eventually reach into centromeres, telomeres, and other highly repetitive regions, the catalog of disease-associated variants will certainly expand beyond what is applied to current clinical interpretation. Moving forward, long-read technologies also offer the opportunity to detect novel transcripts from RNA-seq54 and methylation status from technologies such as ONT, which will further expand the list of disease-associated variants.54, 55, 56
Collectively, we estimate from these analyses that future genomic studies and clinical initiatives using srWGS can expect to capture upward of ten to eleven thousand SVs in each human genome, and current large-scale international initiatives are poised to provide exciting new insights into the 90% of the annotated reference genome that encompasses most known genic sequence. Our analyses also confirmed that assembly-based lrWGS methods will access regions of the genome that were previously intractable to conventional technologies and srWGS. We anticipate that advances in lrWGS technologies, and associated analytic approaches, will provide significant long-term value in expanding the catalog of functional variation associated with insertions, mobile elements, and the most challenging sequence features in the human genome.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
Data and analyses were conducted by the Human Genome Structural Variation Consortium (HGSVC). Analyses, data, and personnel were supported by the following grants from the National Institutes of Health (NIH): U24HG007497, R01MH115957, R03HD099547, UM1HG008895, R01HD081256, R01HD091797, R01HD096326, R01HG002898, R01HG010169, R00DE026824, GRFP2017240332, and F31HG010569. X.Z. was supported in part by the MGH ECOR Fund for Medical Discovery (FMD) Postdoctoral Fellowship. R.L.C. was supported by NHGRI T32HG002295 and NSF GRFP #2017240332. C. Lee was supported in part by the operational funds from The First Affiliated Hospital of Xi’an Jiaotong University. C. Lee is supported in part by the Ewha Womans University research grant of 2019. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Published: March 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.014.
Data and code availability
Resource data used in this paper were generated by Chaisson et al. (2019)14. These data are available under dbVar: nstd152.
Web resources
HGSV integration pipeline, https://github.com/xuefzhao/HGSV_SV_integration_pipe
lrWGS data of HGSVC sample, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/hgsv_sv_discovery/working/20160623_chaisson_pacbio_aligns/
OMIM, https://omim.org/
srWGS data of HGSVC sample, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/hgsv_sv_discovery/data/
Supplemental information
References
- 1.Abel H.J., Larson D.E., Regier A.A., Chiang C., Das I., Kanchi K.L., Layer R.M., Neale B.M., Salerno W.J., Reeves C., NHGRI Centers for Common Disease Genomics Mapping and characterization of structural variation in 17,795 human genomes. Nature. 2020;583:83–89. doi: 10.1038/s41586-020-2371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posey J.E., O’Donnell-Luria A.H., Chong J.X., Harel T., Jhangiani S.N., Coban Akdemir Z.H., Buyske S., Pehlivan D., Carvalho C.M.B., Baxter S., Centers for Mendelian Genomics Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E., All of Us Research Program Investigators The “All of Us” Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rusk N. The UK Biobank. Nat. Methods. 2018;15:1001. doi: 10.1038/s41592-018-0245-2. [DOI] [PubMed] [Google Scholar]
- 7.Turro E., Astle W.J., Megy K., Gräf S., Greene D., Shamardina O., Allen H.L., Sanchis-Juan A., Frontini M., Thys C., NIHR BioResource for the 100,000 Genomes Project Whole-genome sequencing of patients with rare diseases in a national health system. Nature. 2020;583:96–102. doi: 10.1038/s41586-020-2434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regier A.A., Farjoun Y., Larson D.E., Krasheninina O., Kang H.M., Howrigan D.P., Chen B.-J., Kher M., Banks E., Ames D.C. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 2018;9:4038. doi: 10.1038/s41467-018-06159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Audano P.A., Sulovari A., Graves-Lindsay T.A., Cantsilieris S., Sorensen M., Welch A.E., Dougherty M.L., Nelson B.J., Shah A., Dutcher S.K. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell. 2019;176:663–675.e19. doi: 10.1016/j.cell.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werling D.M., Brand H., An J.-Y., Stone M.R., Zhu L., Glessner J.T., Collins R.L., Dong S., Layer R.M., Markenscoff-Papadimitriou E. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat. Genet. 2018;50:727–736. doi: 10.1038/s41588-018-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang C., Scott A.J., Davis J.R., Tsang E.K., Li X., Kim Y., Hadzic T., Damani F.N., Ganel L., Montgomery S.B., GTEx Consortium The impact of structural variation on human gene expression. Nat. Genet. 2017;49:692–699. doi: 10.1038/ng.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandler W.M., Antaki D., Gujral M., Kleiber M.L., Whitney J., Maile M.S., Hong O., Chapman T.R., Tan S., Tandon P. Paternally inherited cis-regulatory structural variants are associated with autism. Science. 2018;360:327–331. doi: 10.1126/science.aan2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., Khera A.V., Lowther C., Gauthier L.D., Wang H., Genome Aggregation Database Production Team. Genome Aggregation Database Consortium A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I., Raja A., Baker C., Hoekzema K., Stessman H.A. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg J.S., Adams M., Nassar N., Bizon C., Lee K., Schmitt C.P., Wilhelmsen K.C., Evans J.P. An informatics approach to analyzing the incidentalome. Genet. Med. 2013;15:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhoads A., Au K.F. PacBio Sequencing and Its Applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain M., Olsen H.E., Paten B., Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pendleton M., Sebra R., Pang A.W.C., Ummat A., Franzen O., Rausch T., Stütz A.M., Stedman W., Anantharaman T., Hastie A. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods. 2015;12:780–786. doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaisson M.J., Huddleston J., Dennis M.Y., Sudmant P.H., Malig M., Hormozdiari F., Antonacci F., Surti U., Sandstrom R., Boitano M. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cretu Stancu M., van Roosmalen M.J., Renkens I., Nieboer M.M., Middelkamp S., de Ligt J., Pregno G., Giachino D., Mandrile G., Espejo Valle-Inclan J. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat. Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., Brooks S., Howe E., Porubsky D., Logsdon G.A. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders A.D., Falconer E., Hills M., Spierings D.C.J., Lansdorp P.M. Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat. Protoc. 2017;12:1151–1176. doi: 10.1038/nprot.2017.029. [DOI] [PubMed] [Google Scholar]
- 31.Chan S., Lam E., Saghbini M., Bocklandt S., Hastie A., Cao H., Holmlin E., Borodkin M. Structural Variation Detection and Analysis Using Bionano Optical Mapping. Methods Mol. Biol. 2018;1833:193–203. doi: 10.1007/978-1-4939-8666-8_16. [DOI] [PubMed] [Google Scholar]
- 32.Zook J.M., Hansen N.F., Olson N.D., Chapman L., Mullikin J.C., Xiao C., Sherry S., Koren S., Phillippy A.M., Boutros P.C. A robust benchmark for detection of germline large deletions and insertions. Nat. Biotechnol. 2020;38:1347–1355. doi: 10.1038/s41587-020-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eichler E.E. Genetic Variation, Comparative Genomics, and the Diagnosis of Disease. N. Engl. J. Med. 2019;381:64–74. doi: 10.1056/NEJMra1809315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M.J. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021;eabf7117 doi: 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyter D., Ingimundardottir H., Oddsson A., Eggertsson H.P., Bjornsson E., Jonsson H., Atlason B.A., Kristmundsdottir S., Mehringer S., Hardarson M.T. Long read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. bioRxiv. 2020 doi: 10.1101/848366. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez O.L., Ritz A., Sharp A.J., Bashir A. MsPAC: a tool for haplotype-phased structural variant detection. Bioinformatics. 2020;36:922–924. doi: 10.1093/bioinformatics/btz618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Berkum N.L., Lieberman-Aiden E., Williams L., Imakaev M., Gnirke A., Mirny L.A., Dekker J., Lander E.S. Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp. 2010;(39):1869. doi: 10.3791/1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders A.D., Hills M., Porubský D., Guryev V., Falconer E., Lansdorp P.M. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 2016;26:1575–1587. doi: 10.1101/gr.201160.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monlong J., Cossette P., Meloche C., Rouleau G., Girard S.L., Bourque G. Human copy number variants are enriched in regions of low mappability. Nucleic Acids Res. 2018;46:7236–7249. doi: 10.1093/nar/gky538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tattini L., D’Aurizio R., Magi A. Detection of Genomic Structural Variants from Next-Generation Sequencing Data. Front. Bioeng. Biotechnol. 2015;3:92. doi: 10.3389/fbioe.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kosugi S., Momozawa Y., Liu X., Terao C., Kubo M., Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samonte R.V., Eichler E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- 45.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhn R.M., Haussler D., Kent W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrovski S., Gussow A.B., Wang Q., Halvorsen M., Han Y., Weir W.H., Allen A.S., Goldstein D.B. The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity. PLoS Genet. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X., Weber A.M., Mills R.E. A recurrence-based approach for validating structural variation using long-read sequencing technology. Gigascience. 2017;6:1–9. doi: 10.1093/gigascience/gix061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou W., Emery S.B., Flasch D.A., Wang Y., Kwan K.Y., Kidd J.M., Moran J.V., Mills R.E. Identification and characterization of occult human-specific LINE-1 insertions using long-read sequencing technology. Nucleic Acids Res. 2020;48:1146–1163. doi: 10.1093/nar/gkz1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrska-Bishop M., Evani U.S., Zhao X., Basile A.O., Abel H.J., Regier A.A., Corvelo A., Clarke W.E., Musunuri R., Nagulapalli K. High coverage whole genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. bioRxiv. 2021 doi: 10.1101/2021.02.06.430068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 54.Uapinyoying P., Goecks J., Knoblach S.M., Panchapakesan K., Bonnemann C.G., Partridge T.A., Jaiswal J.K., Hoffman E.P. A long-read RNA-seq approach to identify novel transcripts of very large genes. Genome Res. 2020;30:885–897. doi: 10.1101/gr.259903.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gigante S., Gouil Q., Lucattini A., Keniry A., Beck T., Tinning M., Gordon L., Woodruff C., Speed T.P., Blewitt M.E., Ritchie M.E. Using long-read sequencing to detect imprinted DNA methylation. Nucleic Acids Res. 2019;47:e46. doi: 10.1093/nar/gkz107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gouil Q., Keniry A. Latest techniques to study DNA methylation. Essays Biochem. 2019;63:639–648. doi: 10.1042/EBC20190027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Resource data used in this paper were generated by Chaisson et al. (2019)14. These data are available under dbVar: nstd152.