Abstract
Background
The COVID-19 pandemic caused by the severe acute SARS-CoV-2 virus has undeniably highlighted the importance of reliable nucleic acid quantification. Digital PCR (dPCR) is capable of the absolute quantification of nucleic acids.
Method
By using the droplet dPCR (QX200) and the digital real-time PCR (LOAA), the copy numbers were compared via multiple assays for three distinct targerts; EGFR DNA, SARS-CoV-2 and HIV-1 RNA.
Results
The droplet dPCR and digital real-time PCR showed similar copy numbers for both DNA and RNA quantification. When the limit of detection (LOD) and limit of quantitation (LOQ) of each method were estimated for DNA and RNA targets, the digital real-time PCR showed a higher sensitivity and precision especially with low copy number targets.
Conclusion
The breath of nucleic acid testing in diagnostic applications continues to expand. In this study we applied common diagnostic targets to a novel digital real-time PCR methodology. It performed comparably to the established dPCR method with distinctive advantages and disadvantages for implementing in laboratories. These rapidly developing dPCR systems can be applied to benefit the accurate and sensitive nucleic acid testing for various clinical areas.
Keywords: Digital PCR, EGFR, SARS-CoV-2, HIV-1, Digital real-time PCR
1. Introduction
The incidence of infectious diseases due to the transmission of novel viruses, including the coronavirus disease 2019 (COVID-19) pandemic, is increasing. Diagnostic testing for such infections is typically based on the detection of viral genetic materials such as DNA and RNA. Molecular genetic testing for cancer diagnosis has likewise become important due to the development of targeted therapeutics and liquid biopsies. Accordingly, accurate and rapid methods for DNA and RNA quantification are currently in high demand. Among a number of related methods, quantitative PCR (or real-time PCR, qPCR) has been the most commonly adopted in testing laboratories.
Recently, digital PCR (dPCR) methods have reached technical maturity in commercial instrumentation and are therefore starting to become popular in both research and testing laboratory settings [1]. Unlike conventional qPCR approaches in quantifying sequence-specific DNA, dPCR does not require a calibration curve for target quantification [2], and achieves the absolute quantitation of the target sequence. The main distinction of dPCR is that each PCR reaction is first divided into at least 10,000 partitions. In these individual and parallel partitions, some contain the target molecule, while others contain none. After amplification through thermal cycling, those partitions with the amplified product are designated positive, and those with no amplified product are designated negative. The ratio of positive partitions to the total number of partitions is statistically analyzed using the Poisson distribution to calculate the copy number of the target. The volume of partitions is used to calculate the copy number concentration of a specific target in the reaction. Overall, compared to qPCR, dPCR provides comparatively reproducible results with high sensitivity and precision [3].
One of the key technologies in dPCR instrumentation is generation of the stable nanoliter-sized partitions. Two partitioning principles have been applied in dPCR instruments: droplet- and chip-based. The QX200 and LOAA are representatives of each partitioning methods, respectively. The most wildly used QX200 adopts the droplet-based end-point PCR method. The QX200 can read up to two channels in fluorescein (FAM) and hexachloro-fluorescein (HEX) spectra from approximately 20,000 oil droplets. Employing the other emerging approach, the LOAA system physically separates the partitions on a two-dimensional semiconductor chip in a cartridge (Fig. 1 A). As a digital real-time PCR approach, the LOAA shows amplification curves of each partition over cycles, with the fluorescent intensities for each channel displayed on a scatter chart (Fig. 1A). The LOAA also reads two channels: FAM and FRET (fluorescence resonance energy transfer)-based FAM-Cy5, referred to as FRET probe in this study. The second channel utilizes the FRET phenomena using the same light source as FAM for excitation. In this work, these two distinct dPCR platforms are compared for the quantification of epidermal growth factor receptor (EGFR) DNA variants, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA, and human immunodeficiency virus type 1 (HIV-1) RNA molecules.
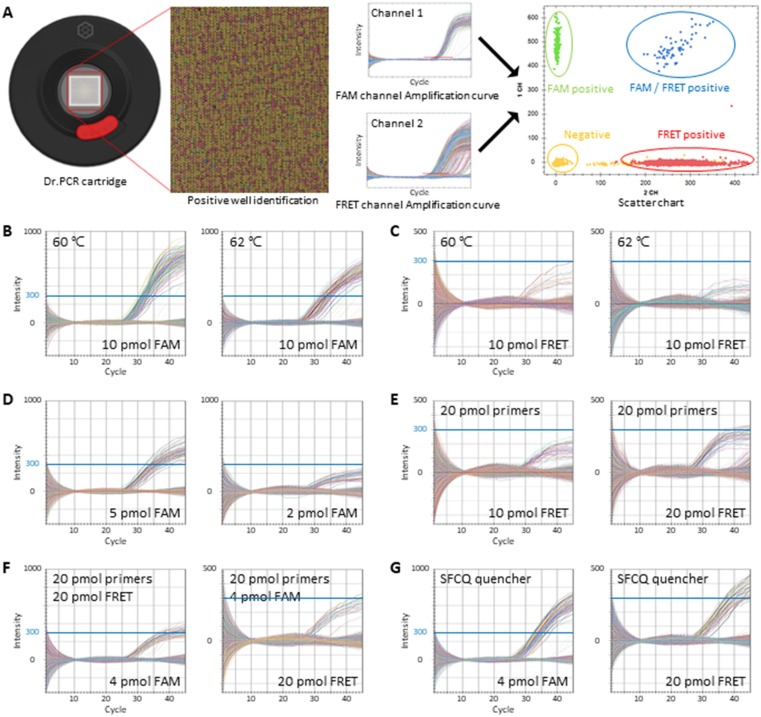
Fig. 1.
Optimization of the digital real-time PCR platform LOAA. A: Images of the LOAA cartridge and wells within it (left), and representative results including real-time amplification curves and a two-dimensional scatter chart of FAM (channel 1) and FRET (channel 2) signals (right). B–G: Digital real-time PCR amplification curves using a set of assays distinguishing the T790M mutation (FAM) from the wild-type (FRET). FAM channel (B and D) and FRET channel (C and E) with indicated conditions. Digital real-time PCR amplification curves using the following optimal concentrations of primers and probes for two assays (different quenchers): forward primer: reverse primer: FAM probe: FRET probe as 20:20:4:20 pmol per reaction (F and G). Blue lines indicate a fixed fluorescence intensity of 300.
EGFR is a transmembrane protein that is a receptor for members of the epidermal growth factor family (EGF family) of extracellular protein ligands [4]. Mutations affecting EGFR expression or activity often result in cancer [5]. Among many EGFR mutations with clinical significance, T790M and L858R mutations are two of the most commonly found in long-term cancer patients with acquired resistance [6]. Detection of these EGFR mutations is part of the diagnostic routine for cancer treatment using EGFR tyrosine kinase inhibitors (i.e., gefitinib and erlotinib) [7]. In particular, non-invasive tests allow for the monitoring of disease progression in patients with the T790M mutation [8]; however, high sensitivity is required due to the low concentrations of the liquid biopsy samples. Various methods including dPCR have therefore been devised to detect the T790M mutation more sensitively [9].
The SARS-CoV-2 virus that originated in Wuhan, China, in December 2019 [10] causes COVID-19, often leading to severe lung damage and mortality [11], [12], [13]. As the COVID-19 pandemic has negatively affected every sector of the globe, early and accurate diagnosis has become the key to success in preventing the spread of virus. Detection of SARS-CoV-2 RNA through real-time reverse transcription PCR (RT-qPCR) is the gold standard for the diagnosis of SARS-CoV-2 infections. Within its genome, E, RdRP, and N genes are most often targeted for SARS-CoV-2 diagnosis [14]. Despite being widely adopted in molecular testing laboratories, the qPCR technique has been repeatedly challenged over its sensitivity and precision [15]. It is still unclear if viral copy concentrations at early or late infection stages can be measured at the requisite levels of sensitivity and accuracy by qPCR [16], [17]. Therefore, considering the high demand for the measurement of accurate copies of SARS-CoV-2 RNA, dPCR, offering the high sensitivity and precision required in the field, should be explored.
HIV causes acquired immunodeficiency syndrome (AIDS) [18], with HIV-1 being the most common subtype [19], [20], [21]. For HIV detection, PCR tests are required for early diagnosis, drug resistance testing, and dose determination [22]. Recent guidelines suggest that the initiation of treatment when HIV RNA reaches above 55 copies/μL is considered a suitable time to prevent immunological destruction and disease progression [23], [24]. This points to the importance of accurately measuring the number of copies of HIV-1.
The recent COVID-19 pandemic caused by the SARS-CoV-2 virus has highlighted the importance of reliable nucleic acid quantification methods. Furthermore, accurate nucleic acid measurement is essential for the development of molecular diagnostics and therapeutic research in diverse clinical areas, such as liquid biopsies for cancer. Here, two distinct dPCR methods, droplet dPCR (QX200) and digital real-time PCR (LOAA), were compared in measuring the copy numbers of EGFR mutations, SARS-CoV-2, and HIV-1. To determine their sensitivities in the given settings, limit of detection (LOD) and limit of quantitation (LOQ) values for each target were estimated.
2. Methods
2.1. Sample preparation
A549 and NCI-H1975 (ATCC) cells were grown in RPMI 1640 media (Gibco, Carlsbad, CA) supplemented with 10% FBS (Gibco) at 37 °C, 5% CO2. 1 × 106 of these cells were prepared as pellets to extract genomic DNA using a Blood & Cell Culture DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The extracted genomic DNA was eluted with TE buffer (pH 8.0) and the concentration was measured using a NanoDrop (XP205, METTLER TOLEDO, Columbus, OH). SARS-CoV-2 reference material 111-10-506, batch 1 (Korea Research Institute of Standards and Science, Daejeon, Republic of Korea), and WHO international standard for HIV-1 RNA (National Institute for Biological Standards and Control, Hertfordshire, UK) were used as RNA templates.
2.2. QX200 (Droplet dPCR)
Hydrolysis probe-based assays were designed for each target, namely EGFR, SARS-CoV-2, and HIV-1 (Table 1 ). Experiments were conducted with a QX200 Droplet Digital PCR system (Bio-Rad, Hercules, CA). For DNA quantification, PCR reactions were prepared in a final volume of 20 μL that contained 10 μL of Supermix for Probes (no dUTP; Bio-Rad) and 3 μL of the assay for EGFR (10 pmol forward primer, 10 pmol reverse primer, 5 pmol FAM probe, and 5 pmol HEX probe per reaction). In the RNA experiments, 5 μL of supermix, 2 μL of reverse transcriptase, 1 μL of 300 mM dithiothreitol (DTT) from a One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad), and 5 μL assays for SARS-CoV-2 or HIV-1 (20 pmol forward primer and reverse primer plus 10 pmol FAM probe per reaction) were used. For each assay, a non-template control (NTC) reaction was included. A QX200 Droplet Generator (Manual DG, Bio-Rad) or Automated Droplet Generator (Auto DG, Bio-Rad) was used to generate the droplets. PCR was performed in a Veriti 96-well Thermal Cycler (Applied Biosystems, Waltham, MA). For DNA quantification, the reactions were conducted under the conditions of 10 min at 95 °C, and 45 cycles of 30 s at 95 °C and 1 min at 58 °C. In the RNA experiments, the reactions were conducted under the conditions of 60 min at 42 °C, 10 min at 95 °C, 70 cycles of 30 s at 95 °C and 2 min 30 s at 59 °C, and 10 min at 98 °C. After amplification, the plate was loaded onto the QX200 Droplet Reader (Bio-Rad) and analyzed using QuantaSoft Software version 1.7.4. (Bio-Rad). All of the thresholds were set up manually to allow the distinction between positive and negative droplets. Only the reactions with more than 10,000 accepted droplets were used for analysis.
Table 1.
Sequences of the hydrolysis probes and primers for the QX200 and LOAA
| Target | Probe sequences | Forward/Reverse Primers | |
|---|---|---|---|
| EGFR | T790M | MT 5′-[6-FAM]-CATCATGCAGCTCATGCCC-[BHQ1]- 3′ | 5′-CATCTGCCTCACCTCCAC-3′ |
| MT 5′-[6-FAM]-CATCATGCAGCTCATGCCC-[SFCQ]- 3′ | 5′- ACCAGTTGAGCAGGTACTGG-3′ | ||
| WT 5′-[HEX]-CATCACGCAGCTCATGCC-[SFCQ]- 3′ | |||
| WT 5′-[SFC620]-AAA-[FAM]-CATCACGCAGCTCATGCC-[SFCQ]- 3′ | |||
| L858R | MT 5′-[6-FAM]-GATTTTGGGCGGGCCAAACT-[SFCQ]- 3′ | 5′- CCAGGAACGTACTGGTGAAA-3′ | |
| WT 5′-[HEX]-GATTTTGGGCTGGCCAAACT-[SFCQ]-3′ | 5′- TACTTTGCCTCCTTCTGCAT-3′ | ||
| WT 5′-[SFC620]-AAA-[FAM]-GATTTTGGGCTGGCCAAACT-[SFCQ]-3′ | |||
| SARS-CoV-2 | RdRP-1* | 5′-[6-FAM] - CAGGTGGAACCTCATCAGGAGATGC - [BHQ1]-3′ | 5′-GTGARATGGTCATGTGTGGCGG-3′ |
| 5′-CARATGTTAAASACACTATTAGCATA-3′ | |||
| RdRP-2 | 5′-[6-FAM]-CCGTAGCTGGTGTCTCTATCTGT-[SFCQ]-3′ | 5′-TGCAAAGAATAGAGCTCGCA-3′ | |
| 5′-CTCCTCTAGTGGCGGCTATT-3′ | |||
| E | 5′-[6-FAM]-TCTTGCTTTCGTGGTATTCTTGCT-[SFCQ]-3′ | 5′-CGGAAGAGACAGGTACGTTAA-3′ | |
| 5′-GCAGTAAGGATGGCTAGTGT-3′ | |||
| N | 5′-[6-FAM]-CACCAATAGCAGTCCAGATGACC-[SFCQ]-3′ | 5′-ACTCAACATGGCAAGGAAGA-3′ | |
| 5′-GCTCTTCGGTAGTAGCCAAT-3′ | |||
| HIV-1 | A assay | 5′-[6-FAM]-CCAGGGATGGATGGCCCAAAAGTTAAACAATG-[BHQ1]-3′ | 5′- CCAATWAGTCCTATTGAAACTGTKCCAGT-3′ |
| 5′-RYTAATGCTTTTATTTTYTCTTCTGTCAATG-3′ | |||
| B assay | 5′-[6-FAM]- AGAT/ZENTM/CTGGCCTTCCTACAAGGGAAG-[IBFQ]-3′ | 5′- ATGGGTGCGAGAGCGTCA-3′ | |
| 5′-GTTGGCTCTGGTCTGCTCTG-3′ | |||
FAM, fluorescein; HEX, hexachloro-fluorescein; MT, mutant allele; WT, wild-type allele; *, assay from Charité Virology, Germany [14].
2.3. LOAA (Digital real-time PCR)
In the DNA quantification experiments, except for the optimization process, total 30 μL reaction mixtures contained 10 μL 3X Dr. PCR Master mix (Optolane, Seongnam, Republic of Korea) and 6.4 μL T790M or L858R assays (20 pmol forward primer, 20 pmol reverse primer, 4 pmol FAM probe, and 20 pmol FRET probe per reaction). In the RNA experiments, 10 μL 3X Dr. PCR One-step dRT-PCR Mixture (Optolane) and 5 μL assays (20 pmol forward primer, 20 pmol reverse primer, and 10 pmol FAM probe per reaction) were used. The reaction mixture was loaded into the cartridge and spread evenly within the chip using a LOAA POSTMAN sample loader (Optolane). The PCR reactions were performed with the LOAA (Optolane). The reactions for EGFR were conducted under the conditions of 3 min at 50 °C, 10 min at 95 °C, and 45 cycles of 15 s at 95 °C and 1 min at 60 °C. The reactions for SARS-CoV-2 were conducted under the conditions of 10 min at 50 °C, 15 min at 95 °C, and 40 cycles of 10 s at 95 °C and 10 s at 57 °C, and those for HIV-1 were 10 min at 50 °C, 10 min at 95 °C, and 50 cycles of 10 s at 95 °C and 30 s at 57 °C. After amplification, results were analyzed using Dr. PCR Analyzer version 1.3.24. (Optolane), with an exception for Fig. 1 where a beta version of Analyzer was used.
2.4. Statistical analyses
Experiments were independently repeated at least in triplicates, otherwise as indicated in the corresponding figures, and were analyzed with Welch’s t-test (two-tailed) Microsoft Excel 2016 (Microsoft, Redmond, WA). Error bars in the graphical data represent the mean ± standard deviation. Statistical significance was claimed when the p-value was lower than 0.05. The coefficient of variation (CV) was calculated for more than ten replicates, according to standard deviation (SD) / average copy number. The lambda values were calculated according to the following formula [25],
where λ (lambda) is the average number of target molecules per partition, P is the number of partitions containing amplified product, and R is the number of partitions analyzed. The LOD and LOQ were calculated at approximate levels according to the formula [26], [27].
3. Results
3.1. Optimization for digital real-time PCR
As dPCR relies on the successful amplification of target DNA during thermal cycles, factors affecting amplification efficiency need to be thoroughly optimized. These factors include the cycling conditions for the given assay and the concentration and ratio of primers and hydrolysis probes [28], [29]. In order to find out the optimal conditions specifically for the recently developed LOAA, the EGFR T790M mutation and wild-type copy number concentrations in 1 ng of genomic DNA from the NCI-H1975 cancer cell line were subjected to measurement. This cell line carries the T790M (c.2369C > T) and L858R (c.2573 T > G) EGFR variants as well as the wild-type allele, which are all amplified within the genome. The FAM probe targets the EGFR T790M mutant allele, and the FRET probe targets the wild-type counterpart.
First, the effect of the temperature during annealing and extension in the LOAA was tested. As the melting temperature of the primers is around 58 °C whereas that of the hydrolysis probes is approximately 62 °C, the assays with the QX200 were previously optimized to produce stable and reliable results with the annealing and extension temperature between 58 and 60 °C (manuscript in preparation). Similarly, temperatures of 58, 60, and 62 °C with the LOAA were tested. At 58 and 60 °C, the plateau FAM fluorescent levels were comparable (Supplementary Fig. S1A); however the FAM intensity was decreased when using 62 °C (Fig. 1B). Similar trends were observed with the FRET probe (Fig. 1C). These results suggest that the optimal extension and annealing temperature for the given EGFR assays is between 58 and 60 °C.
While the FAM channel exists in both the QX200 and LOAA, the second channel is different: QX200 reads HEX or VIC, whereas LOAA adopts an uncommon FRET channel that is co-excited by FAM and then transferred by Cy5 (or SFC620) fluorophore. Probably for this reason, the fluorescent signal from the FRET channel was significantly lower compared to the FAM when the same amount of probes were used (Fig. 1B, C). In order to obtain results with a robust amplification, the ratio of primers and probes in the reactions was adjusted. The fluorescence intensity of the amplification curve changed according to the amount of probe added in the reaction. In the case of FAM, the intensity was relatively low at 2 pmol in 30 μL reaction, but it became sufficiently high at 4 or 5 pmol (Fig. 1D, F-G). For the FRET signal, both probes and each primer at 20 pmol were required to obtain a comparable intensity (Fig. 1E). Based on these results, the optimal concentrations were set as following: 20 pmol for each primer, 4 pmol for the FAM probe and 20 pmol for the FRET probe per reaction (Fig. 1F). During the course of our experiments, an alternative quencher (SFCQ) was discovered that yielded an even higher FAM fluorescent signal (Fig. 1G). For the rest of this study handling EGFR targets, the optimized settings identified in this section were used.
3.2. Comparing copy number values of EGFR variants
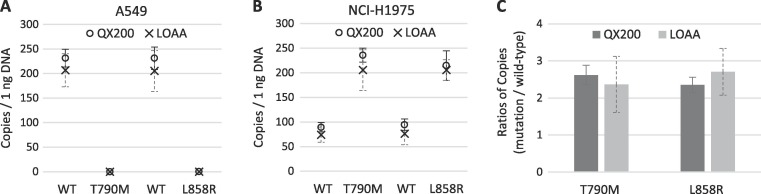
In order to compare the two dPCR platforms for DNA measurement, the copy number concentrations of the EGFR variants were analyzed. Templates were prepared from two cancer cell lines: NCI-H1975 and A549. Unlike NCI-H1975, A549 carries only the wild-type EGFR allele [30], [31]. As expected, none were detected in the A549 genomic DNA using T790M and L858R mutation assays (Fig. 2 A). With the same assays using A549 genomic DNA, the corresponding wild-type copy numbers from LOAA and QX200 were comparable (Fig. 2A). When genomic DNA from
Fig. 2.
Results of EGFR copy number concentration measured by droplet digital PCR (QX200) and digital real-time PCR (LOAA) using 1 ng of genomic DNA isolated from cancer cell lines A549 and NCI-H1975. A and B: Graphs comparing copy numbers of T790M and L858R EGFR mutations and the corresponding wild-type copies in A549 (A) and NCI-H1975 (B). Circles indicate the copy number from the QX200 and crosses indicate the copy number from the LOAA. C: Copy number ratio of each EGFR mutation over wild-type in NCI-H1975 genomic DNA by instrument. Error bars indicate the standard deviation (SD) at each data point with the mean of replicated measurements (n ≥ 5). Significant difference is tested by t-test (p > 0.05, not significant).
NCI-H1975 was analyzed with the T790M and L858R assays, LOAA and QX200 copy number results were similar for both wild-type and variants (Fig. 2B). The QX200 tended to show slightly higher copy numbers than the LOAA, but there was no significant difference in repeated experiments (Fig. 2A,B). The ratio of EGFR mutations to wild-type allele measured by the two systems also showed similar results (Fig. 2C). These results suggest that the QX200 and LOAA show a comparable performance in copy number measurement for EGFR variants.
3.3. Linearity and sensitivity for EGFR variant quantification
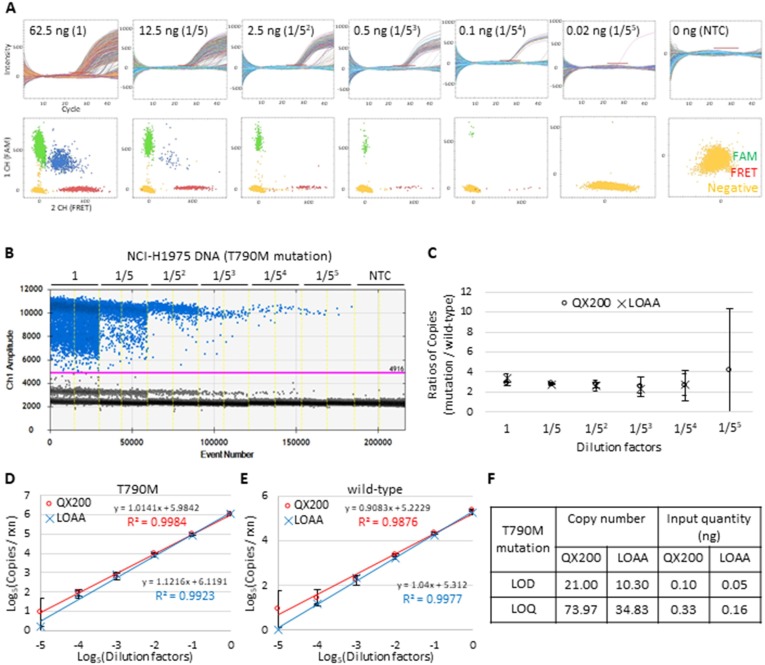
Next, serially diluted NCI-H1975 genomic DNA (62.5, 12.5, 2.5, 0.5, 0.1, 0.02 ng) was used to test the accuracy of DNA measurement with the QX200 and LOAA. Real-time amplification curves and scattered plots from the LOAA clearly showed a decrease in positive partitions throughout the dilution series for the T790M mutation (Fig. 3 A) and wild-type allele (Supplementary Fig. S1B). A similar trend was found in the one-dimensional amplitude graphs from the QX200 (Fig. 3B and Supplementary Fig. S1C). It is worth noting that essentially no signal was detected in the NTC by either QX200 or LOAA. The copy number ratios of the T790M mutation to wild-type allele derived from the QX200 (circles) and LOAA (crosses) results were then calculated (Fig. 3C). At relatively high concentrations, the ratio remained constant around 2.6; however, the value began to significantly deviate at low concentrations (Fig. 3C). It should be noted that the FRET signal from the LOAA produced zero copies in our experiments at the lowest concentration (data point omitted in Fig. 3C). Both QX200 and LOAA results from these dilution series showed excellent linearity (R2 > 0.987) for the T790M mutation (Fig. 3D) and the wild-type allele (Fig. 3E). These results suggest that both QX200 and LOAA produce relatively stable copy number concentrations using as little as 0.1 ng (dilution factor 1/54) of human genomic DNA.
Fig. 3.
Linearity assessment of T790M mutation quantification using the LOAA and QX200. A: LOAA results for the T790M mutation showing amplification curves for each partition over cycles (upper) and end-point two-dimensional scatter plots (lower). The red bar indicates the position of the threshold. B: Representative QX200 results showing end-point one-dimensional amplitude graphs. The ordinate scales indicate the fluorescent amplitude. The pink line indicates the threshold, above which are positive droplets (blue) containing at least one copy of the target DNA and below which are negative droplets (gray) without any target DNA. C: Ratios of copy number concentrations of the T790M mutation over wild-type in serially diluted templates. Significant difference is tested by t-test (p > 0.05, not significant). Error bars indicate the SD at each data point. D and E: Log-scale graphs showing the linear correlation between the input DNA amount and copy number concentrations of T790M mutation (D) and wild-type allele (E) from the QX200 (red) and LOAA (blue). Error bars indicate the SD with the mean of replicated measurements (n ≥ 5). F: LOD and LOQ values for T790M quantification using the QX200 and LOAA (n > 10 per sample).
To further investigate the analytical sensitivity of the QX200 and LOAA for DNA measurement, the LOD and LOQ were obtained for each platform. LOD and LOQ are parameters that describe the minimum concentration of a target necessary for detection and quantitation, respectively [32], [33]. Measurements of the NCI-H1975 genomic DNA dilution series to detect the T790M mutation were repeated (n > 10); each data point and corresponding SD and CV is plotted and summarized in Supplementary Fig. S2A. LOD and LOQ values were calculated using the SD of each y-intercept and the slope of the average data (Supplementary Fig. S2B) [26], [34], [35], [36]. The LOD and LOQ for our EGFR T790M assay using the QX200 were 21 and 74 copies per reaction, which is 0.10 ng and 0.33 ng of NCI-H1975 genomic DNA, respectively (Fig. 3F). On the other hand, the LOD and LOQ of the LOAA were approximately 50% lower than those of the QX200 (Fig. 3F). The QX200 and LOAA showed similar precision with the high-concentration template in our experiments (2.5 ng, Supplementary Fig. S2A). When a lower amount of template was used, the LOAA results showed a better precision (Supplementary Fig. S2A). In conclusion, for the EGFR T790M mutation assay, the LOAA has higher sensitivity and better precision for targets with an extremely low concentration.
3.4. SARS-CoV-2 and HIV-1 RNA quantification
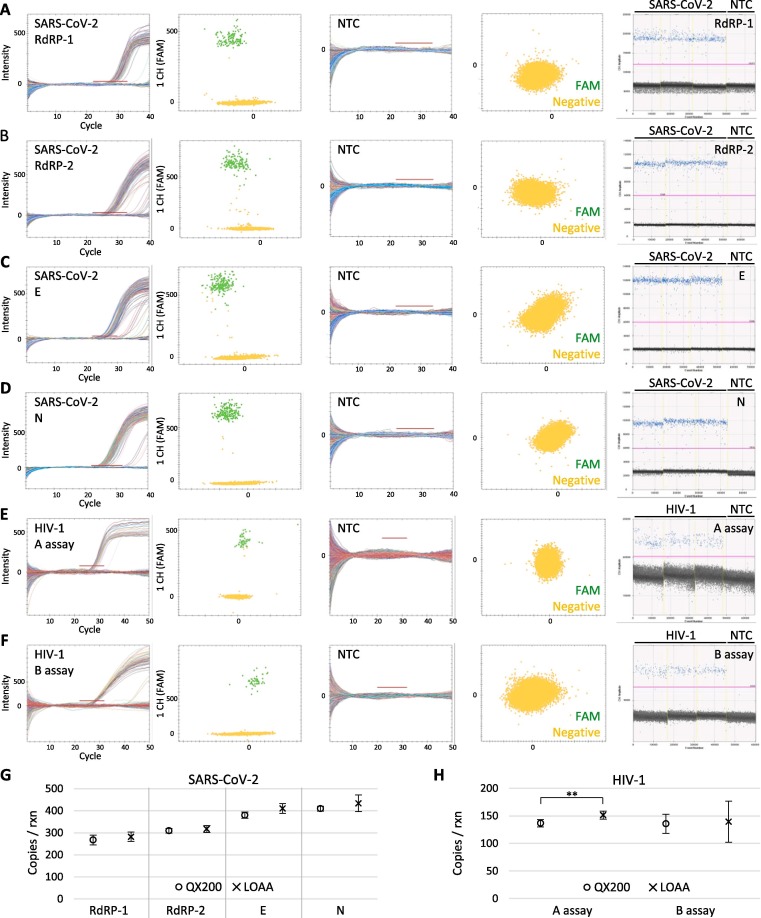
Like the DNA measurements, RNA copy number quantification using the QX200 and LOAA showed a comparable performance with SARS-CoV-2 and HIV-1. In order to accurately compare the RNA copy number values from the two platforms, SARS-CoV-2 reference material, containing most of the coding region of the virus, was utilized. This reference material was measured with the one-step reverse transcription dPCR protocol in each instrument using multiple assays targeting the RdRP (RdRP-1, RdRP-2), N, and E genes. Both platforms performed well in all the assays, showing clear separations and no NTC signals (Fig. 4 A-D). The measured values for the RdRP, E, and N genes showed good agreement between the QX200 and LOAA (Fig. 4G), which fit within the expected intervals according to the reference value of the material (manuscript in preparation). In comparing the HIV-1 RNA copy number values, an assay-dependent conclusion was obtained. Unlike with the B assay where similar values were shown by the QX200 and LOAA, the two dPCR platforms produced copy number values with a statistically significant difference in the A assay (Fig. 4E-F,H). However, this difference is unlikely to be common as a number of other assays in this study proved their similarities. In conclusion, both the QX200 and LOAA are capable of producing comparable copy number values for RNA in addition to DNA.
Fig. 4.
Quantification of SARS-CoV-2 and HIV RNA. A–D: Representative SARS-CoV-2 amplification curves and scatter plots from LOAA (left four panels: positive and NTC) and one-dimensional plots from QX200 (rightmost) using four different assays: RdRP-1 (A), RdRP-2 (B), E (C), and N (D). E and F: Representative HIV-1 amplification curves and scatter plots from LOAA (left four panels: positive and NTC) and one-dimensional plots from QX200 (rightmost) using two different assays: A assay (E) and B assay (F). The red bars and pink lines indicate the set thresholds. G and H: Graphs comparing copy number results for SARS-CoV-2 (G) and HIV-1 (H) using the QX200 (circles) and LOAA (crosses). Error bars indicate the SD with the mean of replicated measurements (n ≥ 3). Significant difference analyzed by t-test is labeled (**p < 0.01) and otherwise not significant p > 0.05.
3.5. Linearity and sensitivity assessment for SARS-CoV-2 and HIV-1 RNA
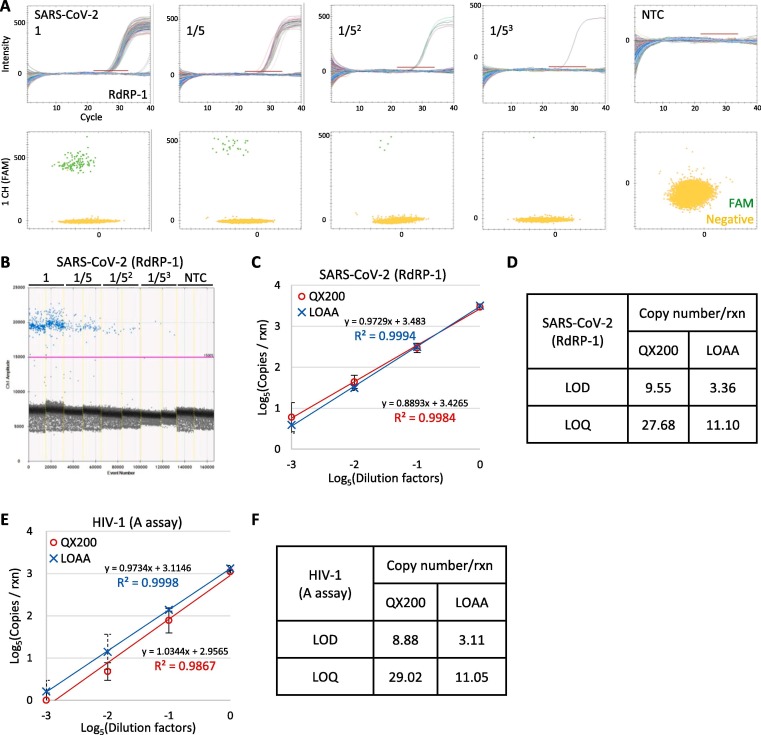
The precision and sensitivity of the QX200 and LOAA were tested for RNA measurements using a similar experimental scheme as for DNA (Fig. 3). It was evident that the numbers of FAM positive partitions targeting the SARS-CoV-2 RdRP gene were reduced (Fig. 5 A,B) as the dilution factors increases. The dilution series results showed an excellent linearity for both QX200 and LOAA (Fig. 5C). Consistent with the EGFR T790M results, the LOAA results had better precision, showing smaller CVs than the QX200 (Supplementary Fig. S3A). The experiments were then expanded to estimate the LOD and LOQ for RdRP copy number measurement (n = 11, Supplementary Fig. S3B). The LOD was about 9.55 and 3.36 copies per reaction and the LOQ was 27.68 and 11.10 copies per reaction for the QX200 and LOAA, respectively (Fig. 5D). Similar results were obtained in the HIV-1 RNA quantification, yielding a comparable linearity (Fig. 5E, and Supplementary Fig. S4, S5). The calculated LOD and LOQ for HIV-1 were strikingly similar to those for SARS-CoV-2 (Fig. 5F), suggesting that the analytical sensitivity of QX200 and LOAA RNA measurement by one-step reverse transcription is consistent across multiple targets.
Fig. 5.
Linearity assessment of RNA quantification. A: LOAA results showing amplification curves for each partition over cycles (upper) and scatter plots (lower). Red bars indicate the set threshold. B: Representative QX200 results showing an end-point one-dimensional amplitude plot. The pink line indicates the set threshold. C: Log-scale graph showing the linear correlation between the input SARS-CoV-2 RNA amount and the copy number concentrations from QX200 (red) and LOAA (blue). D: LOD and LOQ values for SARS-COV-2 quantification using the QX200 and LOAA (RdRP-1 assay). E: Log-scale graph showing the linear correlation between input HIV-1 RNA amount and copy number concentrations from QX200 (red) and LOAA (blue) using A assay. F: LOD and LOQ values for HIV-1 quantification using the QX200 and LOAA (A assay). Error bars indicate the SD with the mean of replicated measurements (n > 10 per sample).
4. Discussions
Accurate nucleic acid measurement has become of great interest in cancer genetic diagnosis, such as in liquid biopsies, as well as in infectious disease testing, such as for COVID-19. As interest in dPCR continues to grow, various dPCR methods are being actively developed and adapted for laboratory medicine. Here, two distinct dPCR platforms were compared: the QX200, a previously established droplet-based dPCR instrument, and the LOAA, a more recently developed digital real-time PCR instrument. Using these two instruments, the copy numbers of EGFR mutations and matching wild-type alleles (T790M, L858R), SARS-CoV-2, and HIV-1 were measured. Respective LOD and LOQ values were also determined to compare analytical sensitivity.
From this study, the following conclusions were reached. First, the overall performance of the two platforms in DNA and RNA measurement was strikingly comparable. They produced comparable copy number concentrations of the EGFR mutations and wild-type alleles, multiple SARS-CoV-2 diagnostic targets, and HIV-1 genes (Fig. 2, Fig. 4). Second, at least for the described assays, the estimated LOD and LOQ copy number values were lower for RNA than for DNA in both the QX200 and LOAA (Fig. 3, Fig. 5). Third, compared to the QX200, the LOAA seemed to offer a higher analytical sensitivity for both DNA and RNA targets tested (Fig. 3, Fig. 5). Further investigation with extended targets and assays is needed to confirm these findings, as multiple aspects of dPCR affect its final results, such as the composition of reagents, assay designs, selection of targets, and PCR cycling conditions [37].
Apart from the aforementioned similarities, the QX200 and LOAA have a number of practical differences. Multiple samples (up to 96) can be run and analyzed simultaneously using the QX200, while only one cartridge housing one reaction is run on the LOAA. Unless equipped with multiple LOAA systems, it is rather inconvenient to test a number of samples in parallel on this platform. Particularly when handling unstable materials like RNA at a low concentration, the limited number of samples allowed in LOAA unavoidably leads to higher variations in copy number values (Fig. 4G,H). Otherwise, compared to the QX200, the hands-on time to set up a reaction is shorter with the LOAA, as it generates partitions by simply pressing the cartridge and then placing it to start the thermal cycling. This convenience together with the shorter thermal cycling time represents the key advantage of the LOAA. In sum, the LOAA is favorable when a small number of samples needs to be measured with a relatively short turn-around time, while the QX200 offers a practical advantage when replicates of multiple samples need to be measured in parallel.
The number of accepted partitions in the QX200 and LOAA showed a notable trend. An important consideration, the number of total accepted partitions is an essential component for target copy number estimation and determination of the dynamic range of a measurement [38]. The LOAA produced about 16,000 accepted wells regardless of the target types and the reagent used (Supplementary Fig. S6); on the other hand, for the QX200 in either Manual or Auto DG setting, the number of accepted partitions differed depending on the target and the reagent (Supplementary Fig. S6). This observation likely resulted from the differences in the partitioning principles of the two instruments. The LOAA partitions are more resilient to changes in chemical composition of reaction, which may stem from the fact that LOAA’s partitions are (1) physically separated and (2) not dependent on a stable interface between oil and reagent, unlike the QX200 droplets. In addition, the lambda values (average copy number per partition) with a given template concentration were higher with the QX200 than with the LOAA, suggesting that the partition volume used to calculate the copy number concentration differs in these two platforms (Supplementary Fig. S6).
Accurate DNA and RNA quantification is in high current demand, as recently highlighted by the explosion in molecular testing for the SARS-CoV-2 virus. It is encouraging to discover that a new instrument enabling digital real-time PCR analyses shows at least comparable performance with an established one. The increasing awareness of the sensitive and precise nucleic acid quantification that dPCR can achieve is clear; in the future, the continuously developing dPCR technologies will make true absolute nucleic acid quantification a reality in the biomedical field. At present though, its current high running costs impede wide applications in clinical laboratories and research institutes. Further improvements in dPCR instruments will benefit accurate and reproducible molecular diagnosis covering various laboratory medicine areas, such as pathogen measurement and identification, somatic mutation detection in cancer, and non-invasive prenatal testing.
5. Conclusion
The overall measurement results for DNA and RNA copy numbers using the two different dPCR methods (droplet dPCR and digital real-time PCR) were strikingly comparable. Our combined results suggest that the novel digital real-time PCR method can show a higher sensitivity and precision especially with low copy number targets. We project that these rapidly developing dPCR methods and their implementation will benefit the nucleic acid testing performance in laboratory medicine.
CRediT authorship contribution statement
Sang-Soo Lee: Conceptualization, Methodology, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization, Funding acquisition. Jae-Hyeong Park: Validation, Investigation. Young-Kyung Bae: Conceptualization, Methodology, Resources, Data curation, Writing - original draft, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We acknowledge the members of Biomolecular Measurement Team at KRISS, especially Drs. Da-Hye Lee and In-Chul Yang, for their insightful discussions and support for this work. We would like to thank Drs. Kyung-Ok Cho and Jong-Hoon Won at KAIST for their continued encouragement.
Funding
This work was supported by the National Research Council of Science and Technology given to Korea Research Institute of Standards and Science (GP2020-0003/20011003) and Young Scientists Fellowship to Sang-Soo Lee (IP2019-0039/19601001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2021.06.016.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Bogožalec Košir A., Demšar T., Štebih D., Žel J., Milavec M. Digital PCR as an effective tool for GMO quantification in complex matrices. Food Chem. 2019;294:73–78. doi: 10.1016/j.foodchem.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 2.Ricchi M., Bertasio C., Boniotti M.B., Vicari N., Russo S., Tilola M., Bellotti M.A., Bertasi B. Comparison among the quantification of bacterial pathogens by qPCR, dPCR, and cultural methods. Front. Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devonshire A.S., Honeyborne I., Gutteridge A., Whale A.S., Nixon G., Wilson P., Jones G., McHugh T.D., Foy C.A., Huggett J.F. Highly Reproducible Absolute Quantification of Mycobacterium tuberculosis Complex by Digital PCR. Anal. Chem. 2015 doi: 10.1021/ac5041617. [DOI] [PubMed] [Google Scholar]
- 4.Herbst R.S. Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys. 2004 doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., Berezov A., Wang Q., Zhang G., Drebin J., Murali R., Greene M.I. ErbB receptors: From oncogenes to targeted cancer therapies. J. Clin. Invest. 2007 doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma S.V., Bell D.W., Settleman J., Haber D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007 doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi K., Uchida J., Nishino K., Kumagai T., Okuyama T., Okami J., Higashiyama M., Kodama K., Imamura F., Kato K. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin. Cancer Res. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 8.Kosaka T., Yatabe Y., Endoh H., Yoshida K., Hida T., Tsuboi M., Tada H., Kuwano H., Mitsudomi T. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin. Cancer Res. 2006 doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 9.Thress K.S., Brant R., Carr T.H., Dearden S., Jenkins S., Brown H., Hammett T., Cantarini M., Barrett J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015 doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Phelan A.L., Katz R., Gostin L.O. The Novel Coronavirus Originating in Wuhan, China: Challenges for Global Health Governance. JAMA - J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 11.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:1–8. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.G.A. E, B.S. C, B.R. S, de Groot Raoul J, D. Christian, G.A. A, H.B. L, L. Chris, L.A. M, N.B. W, P. Dmitry, P. Stanley, P.L.L. M, S.D. V, S.I. A, S. Isabel, Z. John, The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat. Microbiol. (2020). [DOI] [PMC free article] [PubMed]
- 13.Ren L.L., Wang Y.M., Wu Z.Q., Xiang Z.C., Guo L., Xu T., Jiang Y.Z., Xiong Y., Li Y.J., Li X.W., Li H., Fan G.H., Gu X.Y., Xiao Y., Gao H., Xu J.Y., Yang F., Wang X.M., Wu C., Chen L., Liu Y.W., Liu B., Yang J., Wang X.R., Dong J., Li L., Huang C.L., Zhao J.P., Hu Y., Cheng Z.S., Liu L.L., Qian Z.H., Qin C., Jin Q., Cao B., Wang J.W. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. (Engl). 2020 doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Chen Y., Yang J., Han Y., Shi J., Zhan S., Peng R., Li R. External Quality Assessment for Molecular Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Clinical Laboratories. J. Mol. Diagnostics. 2021;23:19–28. doi: 10.1016/j.jmoldx.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J., Han D., Zhang R., Li J., Zhang R. Molecular and Serological Assays for SARS-CoV-2: Insights from Genome and Clinical Characteristics. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barré-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vézinet-Brun F., Rouzioux C., Rozenbaum W., Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;80-:). doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 19.Wei X., Ghosh S.K., Taylor M.E., Johnson V.A., Emini E.A., Deutsch P., Lifson J.D., Bonhoeffer S., Nowak M.A., Hahn B.H., Saag M.S., Shaw G.M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995 doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 20.Russell K.L., Carcamo C., Watts D.M., Sanchez J., Gotuzzo E., Euler A., Blanco J.C., Galeano A., Alava A., Mullins J.I., Holmes K.K., Carr J.K. Emerging genetic diversity of HIV-1 in South America. AIDS. 2000 doi: 10.1097/00002030-200008180-00014. [DOI] [PubMed] [Google Scholar]
- 21.Junqueira D.M., de Medeiros R.M., Matte M.C.C., Araújo L.A.L., Chies J.A.B., Ashton-Prolla P., de Matos Almeida S.E. Reviewing the history of HIV-1: Spread of subtype B in the Americas. PLoS ONE. 2011 doi: 10.1371/journal.pone.0027489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiebig E.W., Wright D.J., Rawal B.D., Garrett P.E., Schumacher R.T., Peddada L., Heldebrant C., Smith R., Conrad A., Kleinman S.H., Busch M.P. Dynamics of HIV viremia and antibody seroconversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS. 2003 doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 23.Fauci A.S., Bartlett J.G., Goosby E.P., Smith M.D., Kaiser H.J., Chang S.W., Anderson J., Armstead R., Baker A.C., Barr D., Bozzette S., Cox S., Delaney M., Gordin F., Greaves W., Harrington M., Henning J.J., Hirsch M.S., Jacobs J., Marlink R., Maxwell C., Mellors J.W., Nash D.B., Perryman S., Schooley R.T., Sherer R., Spector S.A., Torres G., Volberding P., Brady B.A., Daniels E.M., Feigal D., Feinberg M., Gayle H.D., Graydon T.R., Kaplan J., Macher A., Martin R.F., Masur H., Mofenson L., Murray J., O’Neill J., Perez L.C., Riseberg R., Shekar S., Stanley S.K., Whitescarver J. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. Ann. Intern. Med. 1998 doi: 10.7326/0003-4819-128-12_part_2-199806151-00003. [DOI] [Google Scholar]
- 24.Yerly S., Kaiser L., Perneger T.V., Cone R.W., Opravil M., Chave J.P., Furrer H., Hirschel B., Perrin L. Time of initiation of antiretroviral therapy: Impact on HIV-1 viraemia. AIDS. 2000 doi: 10.1097/00002030-200002180-00006. [DOI] [PubMed] [Google Scholar]
- 25.Dube S., Qin J., Ramakrishnan R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE. 2008 doi: 10.1371/journal.pone.0002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taleuzzaman M. Limit of Blank (LOB), Limit of Detection (LOD), and Limit of Quantification (LOQ) Org. Med. Chem IJ. 2018;7 doi: 10.19080/OMCIJ.2018.07.555722. [DOI] [Google Scholar]
- 27.Marsin S.M., Ling S.L., Zalilah N., Dadan H., Wan I.W.A., Abu N.A. Comparison of Signal-to-Noise, Blank Determination, and Linear Regression Methods for the Estimation of Detection and Quantification Limits for Volatile Organic Compounds by Gas Chromatography. J. Aoac Int. 2019 [PubMed] [Google Scholar]
- 28.Taylor S.C., Carbonneau J., Shelton D.N., Boivin G. Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations. J. Virol. Methods. 2015 doi: 10.1016/j.jviromet.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Gerdes L., Iwobi A., Busch U., Pecoraro S. Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomol. Detect. Quantif. 2016;7:9–20. doi: 10.1016/j.bdq.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han H.S., Lim S.N., An J.Y., Lee K.M., Choe K.H., Lee K.H., Kim S.T., Son S.M., Choi S.Y., Lee H.C., Lee O.J. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J. Thorac. Oncol. 2012 doi: 10.1097/JTO.0b013e31823c4c1b. [DOI] [PubMed] [Google Scholar]
- 31.Das A.K., Sato M., Story M.D., Peyton M., Graves R., Redpath S., Girard L., Gazdar A.F., Shay J.W., Minna J.D., Nirodi C.S. Non-small cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res. 2006 doi: 10.1158/0008-5472.CAN-06-2627. [DOI] [PubMed] [Google Scholar]
- 32.B. Magnusson and U. Örnemark, Eurachem Guide: The Fitness for Purpose of Analytical Methods, in: Eur. J. Pharmacol., 2014.
- 33.B. Magnusson, U. Örnemark, Eurachem Guide: A Laboratory Guide to Method Validation and Related Topics, 2014.
- 34.M. Ribani, C.H. Collins, C.B.G. Bottoli, Validation of chromatographic methods: Evaluation of detection and quantification limits in the determination of impurities in omeprazole, J. Chromatogr. A. (2007). https://doi.org/10.1016/j.chroma.2006.12.080. [DOI] [PubMed]
- 35.Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008 [PMC free article] [PubMed] [Google Scholar]
- 36.Saadati N., Abdullah M.P., Zakaria Z., Sany S.B.T., Rezayi M., Hassonizadeh H. Limit of detection and limit of quantification development procedures for organochlorine pesticides analysis in water and sediment matrices. Chem. Cent. J. 2013;7:1–10. doi: 10.1186/1752-153X-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svec D., Tichopad A., Novosadova V., Pfaffl M.W., Kubista M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015 doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majumdar N., Wessel T., Marks J. Digital PCR modeling for maximal sensitivity, dynamic range and measurement precision. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0118833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.