ABSTRACT
Francisella tularensis is a facultative, intracellular, Gram-negative bacterium that causes a fatal disease known as tularemia. Due to its extremely high virulence, ease of spread by aerosolization, and potential to be used as a bioterror agent, F. tularensis is classified by the CDC as a tier 1 category A select agent. Previous studies have demonstrated the roles of the inflammasome sensors absent in melanoma 2 (AIM2) and NLRP3 in the generation of innate immune responses to F. tularensis infection. However, contributions of both the AIM2 and NLRP3 to the development of vaccine-induced adaptive immune responses against F. tularensis are not known. This study determined the contributions of Aim2 and Nlrp3 inflammasome sensors to vaccine-induced immune responses in a mouse model of respiratory tularemia. We developed a model to vaccinate Aim2- and Nlrp3-deficient (Aim2−/− and Nlrp3−/−) mice using the emrA1 mutant of the F. tularensis live vaccine strain (LVS). The results demonstrate that the innate immune responses in Aim2−/− and Nlrp3−/− mice vaccinated with the emrA1 mutant differ from those of their wild-type counterparts. However, despite these differences in the innate immune responses, both Aim2−/− and Nlrp3−/− mice are fully protected against an intranasal lethal challenge dose of F. tularensis LVS. Moreover, the lack of both Aim2 and Nlrp3 inflammasome sensors does not affect the production of vaccination-induced antibody and cell-mediated responses. Overall, this study reports a novel finding that both Aim2 and Nlrp3 are dispensable for vaccination-induced immunity against respiratory tularemia caused by F. tularensis.
KEYWORDS: AIM2, Francisella tularensis, NLRP3, adaptive immunity, immunization, inflammasome
INTRODUCTION
Francisella tularensis is a nonmotile, facultative, intracellular, Gram-negative bacterium that causes a fatal disease known as tularemia. Strains of F. tularensis subsp. tularensis (also known as type A strains) are remarkably virulent. Due to its extremely high virulence, ease of spread by aerosolization, and potential to be used as a bioterror agent, F. tularensis is classified by the CDC as a tier 1 category A select agent (1). Strains of F. tularensis subsp. holarctica (also known as type B strains), although responsible for causing illness in healthy individuals throughout the Northern Hemisphere, are relatively less virulent than type A strains (2). F. tularensis can cause infection in a broad range of hosts and can survive in contaminated water, soil, or vegetation. F. tularensis is transmitted to humans or between animals through bites of infected arthropod vectors, direct contact with infected animal tissues, consumption of contaminated food or water, and inhalation of infective aerosols (3). Clinical presentation includes ulceroglandular, glandular, oculoglandular, oropharyngeal, and pneumonic forms of tularemia and depends on the route of infection, virulence, and dose of the infecting subspecies (1, 4). The other two subspecies of F. tularensis, F. tularensis subsp. novicida and F. tularensis subsp. mediasiatica, do not cause disease in immunocompetent individuals. The live vaccine strain (LVS), derived from type B F. tularensis subsp. holarctica, was developed in the former Soviet Union and subsequently gifted to the United States (5). F. tularensis LVS shows a high degree of sequence homology and a very similar life cycle to F. tularensis type A strains. However, F. tularensis LVS is attenuated for virulence in humans and therefore is used as a surrogate for highly virulent type A strains in research laboratories to study the virulence mechanisms of F. tularensis.
Toll-like receptors (TLRs) are the first identified class of pattern recognition receptors (PRRs) located extra- and intracellularly. They are primarily responsible for detecting pathogen-associated molecular patterns (PAMPs) to initiate innate immune responses (6). The cytosolic sensors that recognize PAMPs are grouped according to their structural features into the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), NOD-like receptors (NLRs), and absent in melanoma 2-like receptors (ALRs). NLRs are subdivided based on the structure of their N-terminal recruitment domains by the presence of either the CARD domain (NLRCs), the pyrin domain (PYD) (NLRPs), or both (7). Among the NLR family members, NLRP3 is the most studied NLR. NLRP3 recognizes a wide variety of PAMPs/damage-associated molecular patterns (DAMPs), including bacterial and viral nucleic acids, bacterial toxins such as nigericin, environmental nanoparticles such as silica and alum, and DAMPs such as ATP, crystals, and β-amyloid aggregates (8). The AIM2 receptor from the ALR family is an interferon (IFN)-inducible protein that consists of a C-terminal HIN-200 domain, which directly binds in a sequence-independent fashion to cytosolic double-stranded DNA (dsDNA) and an N-terminal pyrin motif for ASC recruitment and interaction. AIM2 recognizes bacterial and viral DNAs during infection and recognizes mislocalized self-DNA inadvertently released into the cytosol during inflammatory and autoimmune diseases (9, 10).
The released DNA is sensed by AIM2, which leads to the recruitment of the adaptor ASC and procaspase1. The assembly of this multiprotein complex, known as the inflammasome, cleaves procaspase1 into active caspase1. Active caspase1 acts as a biological scissor and cleaves immature pro-interleukin-1β (IL-1β) and pro-IL-18 into their active forms. Active caspase1 also cleaves the pore-forming protein gasdermin D (GSDMD). The cleaved N-terminal domain of GSDMD forms pores in the cell membrane. These pores allow the release of mature IL-1β and IL-18 from the cell and electrolyte imbalance such as K+ efflux that eventually leads to the lytic form of cell death, pyroptosis (11, 12). These mechanisms and signaling pathways leading to AIM2 activation have been elucidated using a human avirulent F. novicida strain. On the other hand, previous studies have shown that in F. tularensis LVS-infected macrophages, the activation of the inflammasome is delayed, and the induction of IL-1β is very low (13, 14). The inability of the type A F. tularensis SchuS4 strain to induce inflammation early during infection is like F. tularensis LVS, with a weaker induction of inflammasome activation (15). However, the role of Nlrp3 in innate immune responses to F. tularensis is not fully established. Nlrp3 has been shown to mediate an inflammasome-independent function in response to F. tularensis infection that is detrimental to the host (16).
Several studies provide strong evidence for a significant role played by inflammasome-dependent cytokines in shaping adaptive immune responses (13, 17–21). The inflammasome-mediated cytokine IL-1β is required for T helper type 17 (Th17) differentiation, while IL-18 amplifies the production of interferon gamma (IFN-γ), a Th1-polarizing cytokine (22–24). However, contributions of both the AIM2 and NLRP3 inflammasomes to the development of vaccine-induced adaptive immune responses against F. tularensis are not known.
EmrA1 encoded by the FTL_0687 gene of F. tularensis is a transmembrane component of the multidrug efflux pumps belonging to the Emr type major facilitator superfamily of transporters. The emrA1 mutant of F. tularensis LVS is sensitive to oxidative stress and antibiotics and is attenuated for intramacrophage growth and virulence in mice (25). The oxidant sensitivity of the emrA1 mutant is due to its failure to secrete the antioxidant enzymes superoxide dismutase B (SodB) and catalase (KatG). In a previous study, we tested the vaccine potential of the emrA1 mutant in the prevention of respiratory tularemia. We reported that the emrA1 mutant could be safely administered at a very high dose via the intranasal route without any adverse effects on vaccinated mice. Moreover, emrA1 mutant-vaccinated mice are protected against 1,000 to 10,000 100% lethal doses (LD100s) of F. tularensis LVS, and the emrA1 mutant provides partial protection against respiratory challenge with the virulent F. tularensis SchuS4 strain in vaccinated C57BL/6 mice (26). Based on these features, in this study, we used the emrA1 mutant to immunize Aim2−/− and NlrP3−/− mice to determine the contributions of Aim2 and Nlrp3 inflammasomes to vaccine-induced immune responses against respiratory tularemia caused by F. tularensis LVS.
RESULTS
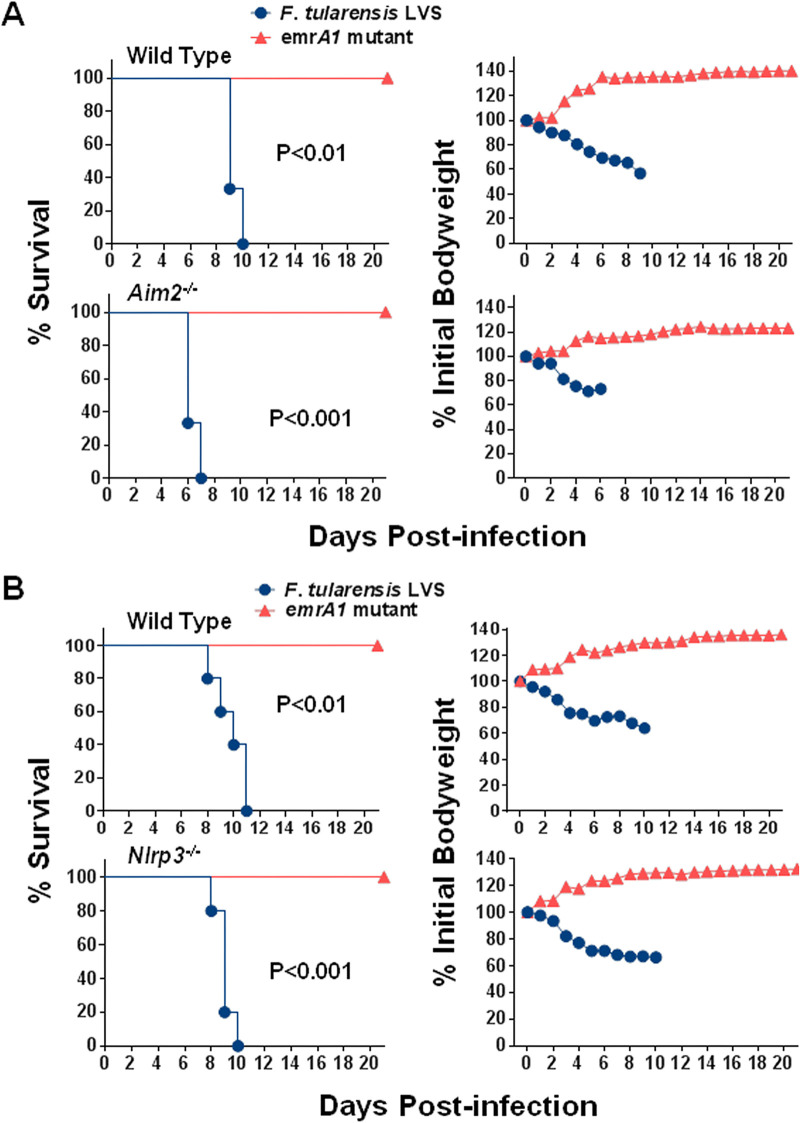
Similar to wild-type mice, the emrA1 mutant is attenuated for virulence in Aim2−/− and Nlrp3−/− mice.
Both Aim2−/− and Nlrp3−/− mice vary in their susceptibilities to primary respiratory infection caused by wild-type F. tularensis LVS, with the former being more susceptible and the latter being more resistant than their respective wild-type mice (16). We took advantage of an attenuated mutant in the emrA1 gene (FTL_0687) of F. tularensis LVS, previously characterized by our group (25). The emrA1 mutant not only is highly attenuated for virulence in mice but, when used as a vaccine, also protects against a very high lethal challenge dose of wild-type F. tularensis LVS (26).
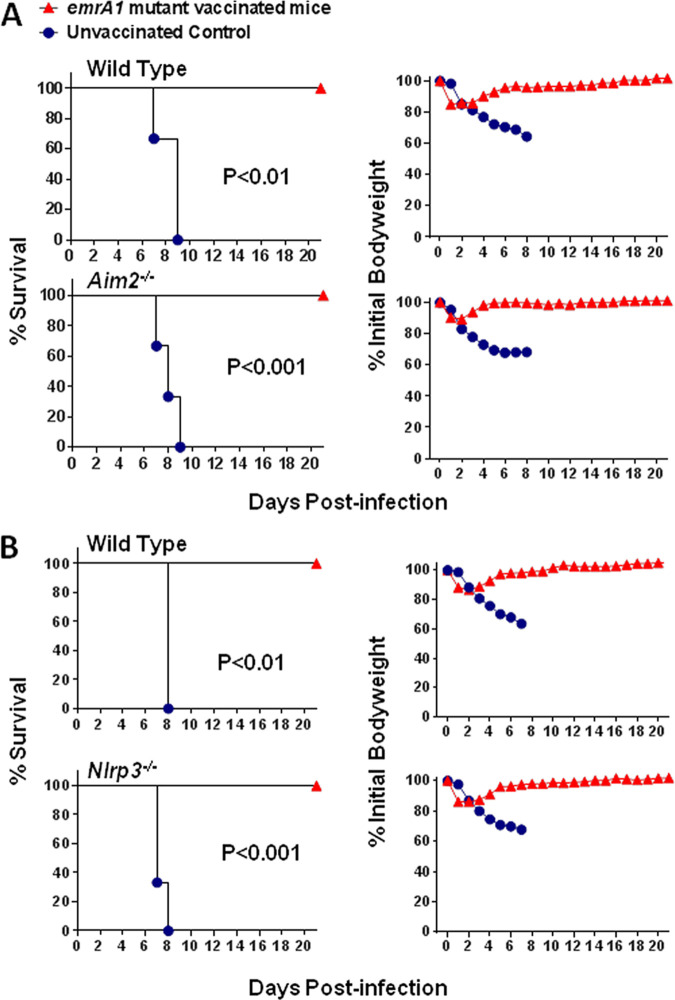
Wild-type, Aim2−/−, and Nlrp3−/− mice infected intranasally with 1 × 106 CFU of either the emrA1 mutant or wild-type F. tularensis LVS were monitored for morbidity by recording body weights daily and mortality. It was observed that 100% of both wild-type and Aim2−/− mice infected with 1 × 106 CFU of the emrA1 mutant survived the infection, with no difference in the body weight loss patterns. One hundred percent of both wild-type and Aim2−/− mice in the control group infected with 1 × 106 CFU of F. tularensis LVS succumbed to infection. However, the Aim2−/− mice were more susceptible to infection than to wild-type F. tularensis, as indicated by the significantly reduced median time to death (MTD) compared to that of the wild-type mice (6 versus 9 days). The weight loss patterns were comparable (Fig. 1A). Like the Aim2−/− mice, all the Nlrp3−/− mice infected intranasally with 1 × 106 CFU of the emrA1 mutant survived the infection. All the corresponding wild-type mice similarly survived, and no differences were observed in body weights. One hundred percent of the wild-type and Nlrp3−/− mice in the control group infected with F. tularensis LVS succumbed to infection. However, unlike Aim2−/− mice, no differences in the MTD were observed (Fig. 1B). These results demonstrated that both Aim2 and Nlrp3 are dispensable for the clearance of the emrA1 mutant. Furthermore, these results also showed that the emrA1 mutant could be used to vaccinate both Aim2−/− and Nlrp3−/− mice to study their contributions to shaping vaccine-induced immunity.
FIG 1.
As for wild-type mice, the emrA1 mutant is attenuated for virulence in Aim2−/− and Nlrp3−/− mice. Wild-type and Aim2−/− mice (n = 5 mice/group) (A) and wild-type and Nlrp3−/− mice (n = 5 mice/group) (B) were infected intranasally with 1 × 106 CFU of either the emrA1 mutant or F. tularensis LVS. Mice were observed for morbidity and mortality for 21 days. The survival results are plotted as Kaplan-Meier survival curves, and the body weights are expressed as percentages of the initial body weight. The P values were determined by the log rank test.
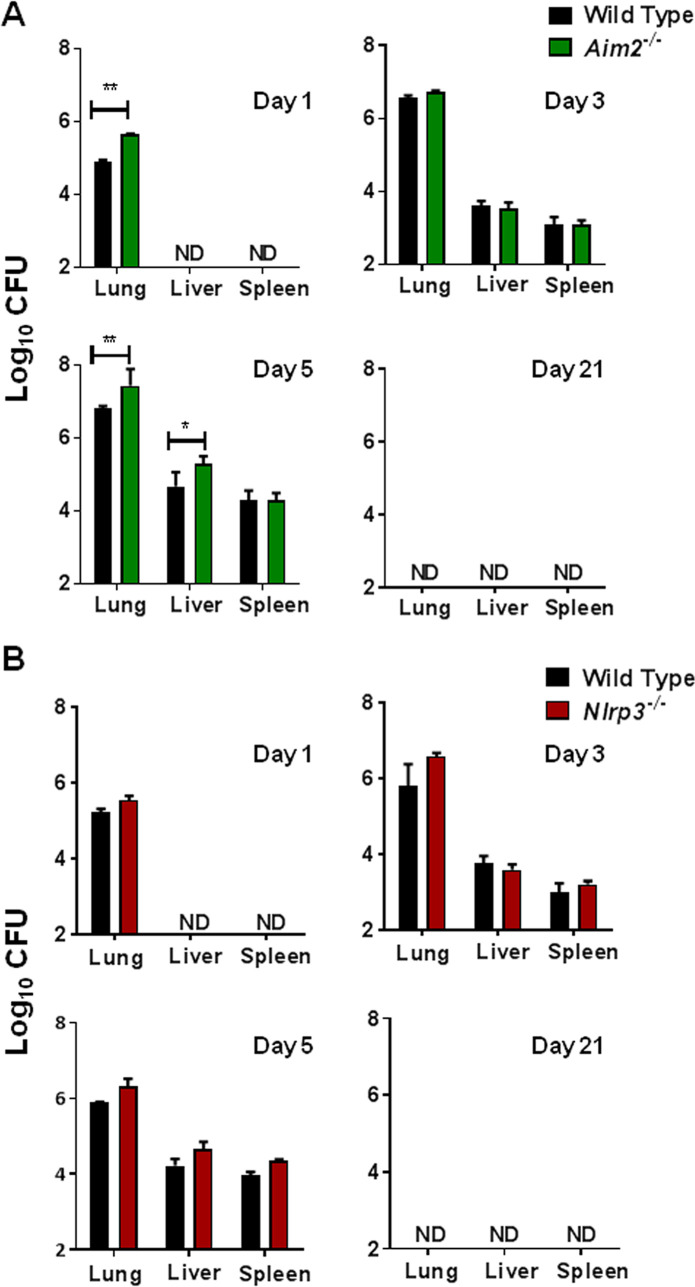
Both Aim2−/− and Nlrp3−/− mice infected with the emrA1 mutant clear the bacteria completely.
It was observed that Aim2−/− and Nlrp3−/− mice survived similarly to wild-type mice following intranasal infection with the emrA1 mutant. Next, we performed a kinetic experiment to determine how long the emrA1 mutant persists in the infected mice and if both Aim2−/− and Nlrp3−/− mice clear the bacteria from their organs completely. Wild-type, Aim2−/−, and Nlrp3−/− mice were infected intranasally with 1 × 106 CFU of the emrA1 mutant. The lungs, livers, and spleens of mice were harvested on days 1, 3, 5, and 21 postinfection and evaluated for their bacterial burdens. The numbers of emrA1 mutant bacteria increased steadily in the lungs, livers, and spleens of the infected mice as the infection progressed through days 1 to 5. On days 1 and 5 postinfection, Aim2−/− mice infected with the emrA1 mutant exhibited significantly higher bacterial numbers in their lungs than the wild-type mice (5.649 ± 0.03 versus 4.917 ± 0.014 and 7.44 ± 0.18 versus 6.83 ± 0.02 log10 CFU, respectively). Both the wild-type and Aim2−/− mice exhibited similar bacterial burdens on day 3 postinfection. Livers of Aim2−/− mice also harbored significantly more bacteria on day 5 postinfection than those observed for the wild-type mice (5.3 ± 0.08 versus 4.67 ± 0.15 log10 CFU). Irrespective of these differences in the bacterial numbers between the wild-type and Aim2−/− mice on days 3 and 5, the emrA1 mutant was cleared completely, and no bacteria were recovered from the lungs, livers, and spleens of both wild-type and Aim2−/− mice on day 21 postinfection (Fig. 2A). Wild-type and Nlrp3−/− mice were also infected intranasally with 1 × 106 CFU of the emrA1 mutant, their organs were collected, and bacterial burdens were quantitated. However, unlike Aim2−/− mice, both wild-type and Nlrp3−/− mice had similar bacterial burdens at all time points tested (Fig. 2B). Collectively, these results demonstrate that the emrA1 mutant replicates for a limited period in the wild-type, Aim2−/−, and Nlrp3−/− mice before being cleared completely. These results also corroborate the survival results and demonstrate that both Aim2 and Nlrp3 are dispensable for the clearance of the emrA1 mutant.
FIG 2.
Aim2−/− and Nlrp3−/− mice infected with the emrA1 mutant clear the bacteria completely. Wild-type and Aim2−/− mice (A) and wild-type and Nlrp3−/− mice (B) (n = 3 per group/time point) infected intranasally with 1 × 106 CFU of the emrA1 mutant were sacrificed at the indicated time points. Lungs, livers, and spleens were collected, homogenized, and plated on MH chocolate agar plates to enumerate the bacteria. Results are expressed as log10 CFU. The P values were determined using one-way ANOVA. *, P < 0.05; **, P < 0.01. ND, not detected. The data shown are representative of results from two independent experiments.
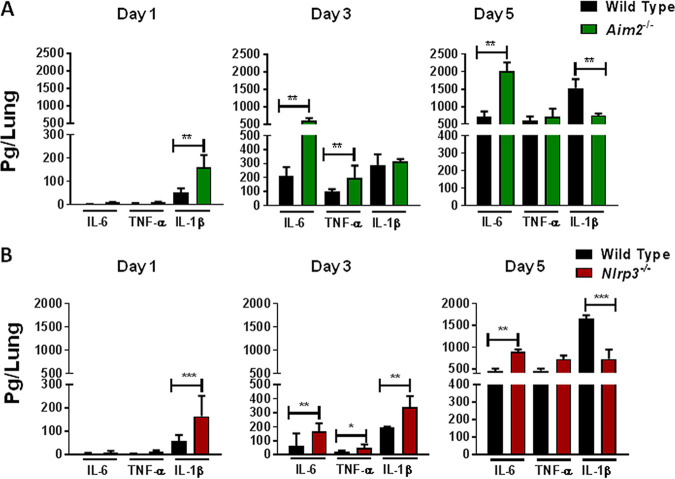
The cytokine responses in Aim2−/− and Nlrp3−/− mice differ from those of wild-type mice following infection with the emrA1 mutant.
Since both Aim2 and Nlrp3 inflammasomes are involved in innate immune responses, we next investigated the type of innate immune responses generated in Aim2−/− and Nlrp3−/− mice following infection with the emrA1 mutant. Wild-type, Aim2−/−, and Nlrp3−/− mice were infected intranasally with the emrA1 mutant, and the levels of proinflammatory cytokines such as IL-1β, IL-6, and tumor necrosis factor alpha (TNF-α) were determined in the lung homogenates. On day 1 postinfection, the lungs of Aim2−/− mice had undetectable levels of IL-6 and TNF-α but significantly higher IL-1β levels (160.08 ± 30.9 pg/ml) than the wild-type mice (51.6 ± 11.10 pg/ml). Significantly elevated IL-6 levels were observed in the lungs of Aim2−/− mice compared to the wild-type mice on days 3 and 5 postinfection, while differences in TNF-α levels were observed only on day 3 postinfection (Fig. 3A). On the other hand, significantly lower IL-1β levels were observed in the lungs of the Aim2−/− mice than in the wild-type mice on day 5 postinfection. These results demonstrate different profiles of proinflammatory cytokines in the Aim2−/− mice compared to the wild-type mice.
FIG 3.
The cytokine responses in Aim2−/− and Nlrp3−/− mice differ from those in wild-type mice following infection with the emrA1 mutant. Wild-type and Aim2−/− mice (A) and wild-type and Nlrp3−/− mice (B) (n = 3 per group/time point) infected intranasally with 1 × 106 CFU of the emrA1 mutant were sacrificed at the indicated time points. Lungs were collected, homogenized, and assayed for the levels of IL-6, TNF-α, and IL-1β. Results are expressed as picograms per lung (means ± SEM). The P values were determined using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The data shown are representative of results from two independent experiments.
When observed in the Nlrp3−/− mice, significantly higher levels of IL-6 and TNF-α were found in the lungs of the Nlrp3−/− mice than in the wild-type mice on days 3 and 5 postinfection. The IL-1β levels were higher in the lungs of Nlrp3−/− mice than in the wild-type mice on days 1 and 3 postinfection. In contrast, the trends were reversed, and significantly higher levels of IL-1β were observed in the lungs of the wild-type mice than in the Nlrp3−/− mice on day 5 postinfection (Fig. 3B). Collectively, these results demonstrate that the profile of proinflammatory cytokines produced in the lungs of Aim2−/− and Nlrp3−/− mice following infection with the emrA1 mutant differs from that observed in the wild-type mice.
Immunization with the emrA1 mutant protects both Aim2−/− and Nlrp3−/− mice against lethal intranasal challenge with F. tularensis LVS.
Wild-type, Aim2−/−, and Nlrp3−/− mice immunized intranasally with 1 × 106 CFU of the emrA1 mutant were challenged intranasally with 2 × 107 CFU (2,000 LD100s) of F. tularensis LVS on day 28 postimmunization. All the unvaccinated wild-type and Aim2−/− mice succumbed to infection by day 9 postchallenge, while 100% of the vaccinated mice survived the challenge. Also, there was minimal weight loss for the first 2 to 4 days postchallenge, after which all mice regained their prechallenge body weights (Fig. 4A). Similar results were observed for the emrA1 mutant-immunized wild-type and Nlrp3−/− mice (Fig. 4B). Collectively, these results demonstrate that both Aim2 and Nlrp3 are dispensable for the generation of vaccination-induced protective immunity against respiratory tularemia caused by a very high challenge dose of F. tularensis LVS.
FIG 4.
Immunization with the emrA1 mutant protects both Aim2−/− and Nlrp3−/− mice against lethal intranasal challenge with F. tularensis LVS. Wild-type and Aim2−/− mice (n = 5 mice/group) (A) and wild-type and Nlrp3−/− mice (n = 5 mice/group) (B) vaccinated intranasally with 1 × 106 CFU of the emrA1 mutant were challenged intranasally with 2 × 107 CFU of F. tularensis LVS on day 28 after primary vaccination. Unvaccinated mice were used as controls. Mice were observed for 21 days for morbidity and mortality. The survival results are plotted as Kaplan-Meier survival curves, and body weight is expressed as a percentage of the initial body weight. P values were determined by the log rank test. The data shown are representative of results from two independent experiments.
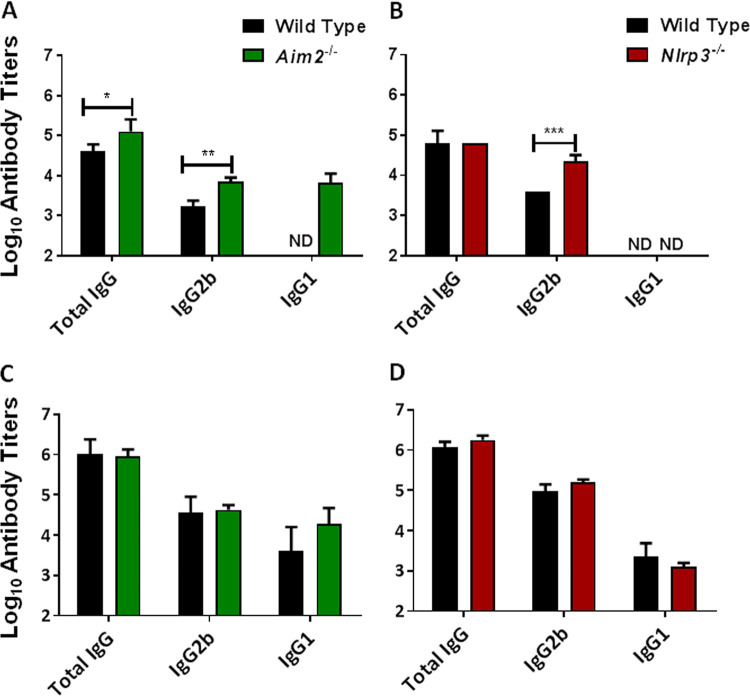
Aim2 and Nlrp3 are dispensable for immunoglobulin isotype responses following immunization and challenge.
Protection against F. tularensis requires both cellular and humoral immune responses. Our previous results demonstrated 100% survival for wild-type, Aim2−/−, and Nlrp3−/− mice immunized with the emrA1 mutant and challenged with F. tularensis LVS. We next evaluated the antibody responses in wild-type, Aim2−/−, and Nlrp3−/− mice by measuring the levels of F. tularensis-specific IgG, IgG2b, and IgG1 prechallenge and postchallenge antibody titers. Mice immunized with the emrA1 mutant were bled to collect serum before the challenge (day 28 postimmunization) and 28 days after the challenge. Sera collected from naive unvaccinated mice were used as controls. Our results show that Francisella-specific prechallenge total IgG and IgG2b titers were significantly higher in the immunized Aim2−/− mice than those observed in the immunized wild-type mice. Interestingly, IgG1 antibodies were detected only in immunized Aim2−/− mice but were not detected in the wild-type mice (Fig. 5A).
FIG 5.
Aim2 and Nlrp3 are dispensable for antibody responses following immunization and challenge. Wild-type and Aim2−/− mice (n = 5 mice/group) (A and C) and wild-type and Nlrp3−/− mice (n = 5 mice/group) (B and D) were immunized intranasally with 1 × 106 CFU of the emrA1 mutant. The serum samples were collected on day 28 postimmunization (A and B) or 28 days postchallenge (C and D). The results are shown as endpoint titers of anti-Francisella-specific antibodies as determined by an ELISA and are represented as log10 values ± SEM. Serum from naive mice was used as a control. The P values were determined using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ND, not detected. The data shown are representative of results from two independent experiments.
In Nlrp3−/− mice, the Francisella-specific prechallenge total IgG antibody titers were similar to those observed for the wild-type mice. However, significantly higher levels of IgG2b antibodies were detected in Nlrp3−/− mice than in the wild-type mice. No prechallenge Francisella-specific IgG1 antibodies were detected in either the wild-type or the Nlrp3−/− mice (Fig. 5B). Collectively, these results demonstrate that prechallenge antibody profiles of immunized Aim2−/− and Nlrp3−/− mice differ from those of immunized wild-type mice.
We also determined the Francisella-specific antibody levels in the serum on day 28 after challenge with 2 × 107 CFU of F. tularensis LVS. The titers of all three antibody types tested were 10- to 100-fold higher postchallenge in the wild-type, Aim2−/−, and Nlrp3−/− mice than their prechallenge antibody titers. However, the Francisella-specific total IgG, IgG2b, and IgG1 antibody titers were similar in the wild-type and Aim2−/− mice (Fig. 5C). Similarly, no differences in the postchallenge total IgG, IgG2b, and IgG1 antibody titers were observed in the wild-type and Nlrp3−/− mice (Fig. 5D). However, unlike the prechallenge IgG1 profile, Francisella-specific IgG1 antibodies were detected in the wild-type as well as Aim2−/− and Nlrp3−/− mice postchallenge. These results demonstrate that although the prechallenge antibody profiles of Aim2−/− and Nlrp3−/− mice differ from those of their wild-type counterparts following immunization with the emrA1 mutant, the postchallenge antibody profiles of both the Aim2−/− and Nlrp3−/− mice are identical to those of the wild-type mice. These results also indicate that both Aim2 and Nlrp3 are dispensable for vaccination-induced protective antibody responses against intranasal F. tularensis challenge.
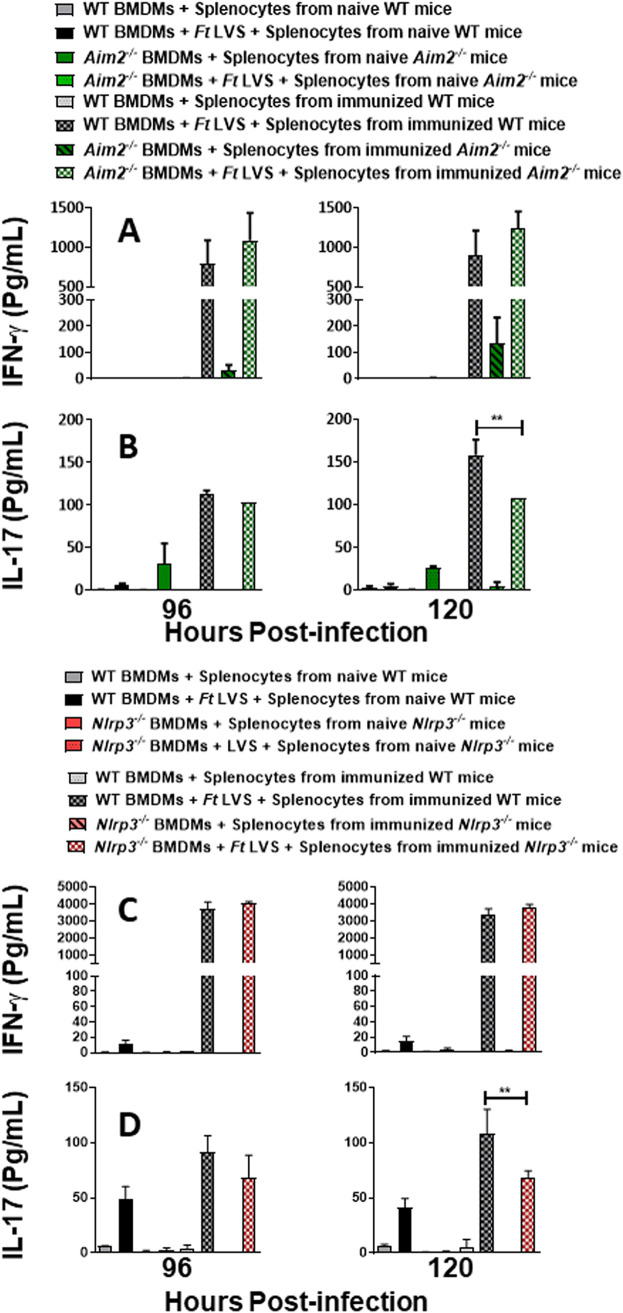
The immune responses responsible for producing IL-17 but not IFN-γ differ between wild-type and Aim2−/− or Nlrp3−/− mice immunized with the emrA1 mutant.
We next determined the cell-mediated immune responses in the wild-type, Aim2−/−, and Nlrp3−/− mice immunized with the emrA1 mutant at 28 days postimmunization. We employed an in vitro splenocyte overlay assay, which can assess adaptive T-cell immunity. This assay evaluates the ability of the splenocytes from immunized mice to produce the T-cell-mediated cytokines IFN-γ and IL-17 in response to the antigens presented by bone marrow-derived macrophages (BMDMs) infected with F. tularensis LVS. Moreover, this assay can also quantitate the bacterial load in infected BMDMs in the presence of an ongoing T-cell response (27–30). To perform this assay, wild-type, Aim2−/−, and Nlrp3−/− mice were vaccinated intranasally with 1 × 106 CFU of the emrA1 mutant. On day 28 postimmunization, splenocytes from immunized and naive mice were harvested. BMDMs derived from wild-type, Aim2−/−, and Nlrp3−/− mice were either left uninfected or infected with F. tularensis LVS at a multiplicity of infection (MOI) of 100, and splenocytes isolated from naive or immunized mice were then added to these cultures. The BMDMs were lysed 24 and 48 h after the addition of splenocytes to enumerate intracellular bacterial replication. The bacteria replicated at a significantly higher rate in Aim2−/− than in wild-type BMDMs in the presence of naive as well as immune splenocytes from their wild-type controls at 24 h postinfection. However, Aim2−/− BMDMs cocultured with splenocytes from immunized Aim2−/− mice controlled intracellular bacterial growth similarly to their wild-type counterparts after 48 h of infection (Fig. 6A). Although the bacterial numbers recovered from the Aim2−/− BMDMs cocultured with splenocytes isolated from naive or immunized Aim2−/− mice were higher than those of the wild-type controls at 48 h postinfection, statistical significance was not achieved (Fig. 6A).
FIG 6.
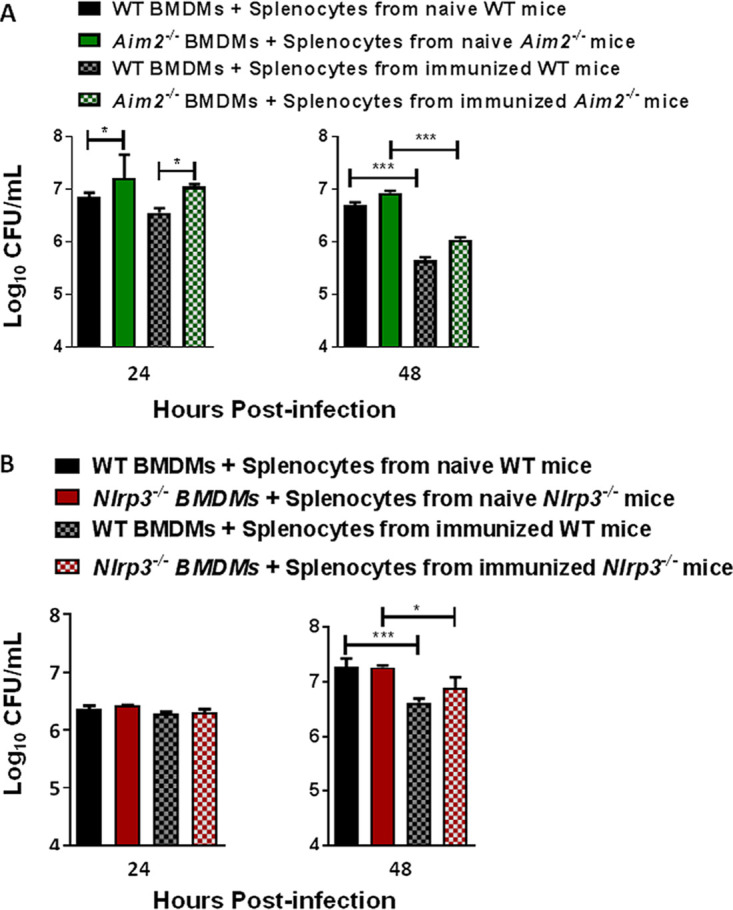
F. tularensis-infected Aim2−/− and Nlrp3−/− BMDMs clear bacteria similarly to wild-type BMDMs when cocultured with splenocytes derived from emrA1 mutant-immunized mice. Wild-type (WT) and Aim2−/− mice (A) and wild-type and Nlrp3−/− mice (B) (n = 6 mice/group) were immunized intranasally with 1 × 106 CFU of the emrA1 mutant. On day 28 postimmunization, mice were sacrificed, and splenocytes were isolated. The splenocytes obtained from naive mice served as controls. The coculture assays were performed as described in Materials and Methods. BMDMs were lysed and plated to enumerate intracellular bacteria at 24 h and 48 h postinfection. Results are expressed as log10 CFU per milliliter. The P values were determined using one-way ANOVA. *, P < 0.05; ***, P < 0.001.
BMDMs from wild-type and Nlrp3−/− mice infected with F. tularensis LVS were also cocultured with splenocytes isolated from naive and immunized mice. It was observed that bacteria replicated similarly in both wild-type and Nlrp3−/− macrophages in the presence of splenocytes derived from naive or immunized mice at 24 h postinfection. The addition of splenocytes from immunized wild-type and Nlrp3−/− mice controlled intracellular bacterial replication at 48 h postinfection. However, no differences were observed in bacterial numbers recovered from the wild-type or Nlrp3−/− BMDMs cocultured with splenocytes isolated from immunized wild-type or Nlrp3−/− mice (Fig. 6B). Collectively, these results demonstrate that the splenocytes derived from the immunized Aim2−/− and Nlrp3−/− mice contribute to bacterial clearance as efficiently as their respective wild-type counterparts at 48 h postinfection.
To further evaluate the ability of T cells in the splenocyte populations from immunized wild-type, Aim2−/−, and Nlrp3−/− mice to induce T-cell-derived cytokines, supernatants from the coculture assay were analyzed for the levels of IFN-γ and IL-17, representing Th1 and Th17 effector functions, respectively. Higher levels of IFN-γ were observed only in cocultures performed with splenocytes from immunized wild-type and Aim2−/− mice at 96 and 120 h postinfection. However, these levels did not achieve statistical significance (Fig. 7A). No differences in IL-17 levels were observed in the wild-type and Aim2−/− BMDMs cocultured with splenocytes derived from immunized wild-type and Aim2−/− mice at 96 h postinfection. However, significantly higher IL-17 levels were observed in coculture assays performed with the wild-type BMDMs and splenocytes derived from the immunized wild-type mice than in their corresponding Aim2−/− counterparts at 120 h postinfection (Fig. 7B).
FIG 7.
The immune responses responsible for producing IL-17 but not IFN-γ differ between wild-type and Aim2−/− or Nlrp3−/− mice immunized with the emrA1 mutant. Wild-type (WT) and Aim2−/− mice (A and B) and wild-type and Nlrp3−/− mice (C and D) (n = 6 mice/group) were immunized intranasally with 1 × 106 CFU of the emrA1 mutant. On day 28 postimmunization, mice were sacrificed, and the splenocytes were isolated. The splenocytes obtained from naive mice served as controls. BMDMs isolated from wild-type, Aim2−/−, or Nlrp3−/− mice were infected with F. tularensis LVS (FtLVS) (MOI of 100) and then overlaid with splenocytes from immunized or naive mice. After 96 and 120 h, culture supernatants were collected and analyzed for the quantification of IFN-γ (A and C) and IL-17 (B and D) by a cytometric bead array. Results are expressed as picograms per milliliter (means ± SEM) from triplicate samples. The P values were determined using one-way ANOVA. **, P < 0.01.
As observed for Aim2−/− mice, no differences in IFN-γ levels were observed in coculture assays performed with the wild-type BMDMs and splenocytes derived from the immunized wild-type mice and their corresponding Nlrp3−/− counterparts at 96 and 120 h postinfection (Fig. 7C). Similarly, significantly higher levels of IL-17 were observed in the wild-type BMDMs and splenocytes derived from the immunized wild-type mice than in their corresponding Nlrp3−/− counterparts only at 120 h postinfection (Fig. 7D). Collectively, these results demonstrate that there are no differences in cell-mediated responses responsible for the production of IFN-γ between wild-type, Aim2−/−, and Nlrp3−/− mice. However, cell-mediated IL-17 responses vary significantly between the immunized wild-type and Aim2−/− or Nlrp3−/− mice. Collectively, these results demonstrate that Th17-mediated cytokines, but not the IFN-γ recall responses, are partially impaired in mice lacking Aim2 or Nlrp3.
DISCUSSION
The activation of Aim2 and Nlrp3 inflammasomes results in the secretion of the proinflammatory cytokines IL-1β and IL-18. Both these cytokines serve as essential innate antimicrobial host defenses by activating neutrophils and macrophages to phagocytose and kill the invading pathogens. IL-1β enhances antigen-driven responses in CD4 and CD8 T cells to promote the expansion and activation of antigen-specific Th1, Th2, Th17, and granzyme B-positive CD8 T cells (31). IL-1β is also required for CD4-positive (CD4+) T cells to become fully functional memory T cells (32). IL-18, first described as an IFN-γ-inducing factor, is required for the activation of NK cells, Th1 cells, and cytotoxic T cells; the expansion of CD4+ IFN-γ+ and CD4+ IL-17+ T cells; and protection against mucosal bacterial pathogens (33–35). Thus, both IL-1β and IL-18 serve to bridge the innate and adaptive immune responses.
Several previous studies have shown the significance of Aim2 inflammasome-mediated innate immune responses in protection against F. novicida infection (36–40). The production of IL-1β is completely dependent on the Aim2 inflammasome in F. novicida-infected macrophages. Although the role of Aim2 against virulent type A and type B Francisella strains, including LVS, is not fully established, both Aim2 and Nlrp3 inflammasomes are required for the production of IL-1β and IL-18 (16). Studies have also reported that the latter Francisella strains suppress Aim2 inflammasome-mediated innate immune responses (13, 15). An inflammasome-independent detrimental role of Nlrp3 during Francisella infection has been demonstrated, and it has been shown that Nlrp3−/− mice are more resistant than wild-type mice to respiratory infection caused by F. tularensis (16). In this study, we determined the contributions of Aim2 and Nlrp3 inflammasomes to vaccine-induced immune responses against respiratory tularemia caused by F. tularensis LVS.
Our results show that no differences in susceptibilities to the intranasal administration of a very high dose of the emrA1 mutant were observed in wild-type, Aim2−/−, and Nlrp3−/− mice. No adverse effects or morbidity was observed in the emrA1 mutant-vaccinated mice. The emrA1 mutant replicated in the infected mice for a short period and was cleared thereafter. These results indicated that the emrA1 mutant is a suitable vaccine candidate to uniformly vaccinate the three mouse strains with various susceptibilities to wild-type F. tularensis LVS.
Despite several differences in proinflammatory cytokine responses induced between the wild-type and Aim2−/− or Nlrp3−/− mice, they all were successful in clearing the primary infection caused by a very high dose of the emrA1 mutant. The higher IL-6 and TNF-α levels in Aim2−/− mice could be due to the higher bacterial load present in the lungs of these mice on days 3 and 5 postinfection. It has been reported that IL-1β and IL-18 levels are markedly reduced but not ablated in Aim2−/− and Nlrp3−/− mice compared to wild-type mice infected with F. tularensis LVS (16). Interestingly, the IL-1β profiles revealed that despite the lack of Aim2 or Nlrp3, significantly higher (days 1 and 3 postinfection) (Fig. 3A and B) or substantial (day 5 postinfection) (Fig. 3A and B) levels of IL-1β were observed in the lung homogenates of Aim2−/− and Nlrp3−/− mice infected with the emrA1 mutant. It has been reported previously that inflammasome-independent mechanisms such as proteinase-3, elastase, and cathepsin G can process pro-IL-1β and pro-IL-18 into active IL-1β and IL-18 (41). It is probable that in the absence of either Aim2 or Nlrp3, the elevated IL-1β levels could be the result of these compensatory inflammasome-independent mechanisms or that the loss of one inflammasome component is compensated for by the other. It has been suggested that Aim2 negatively regulates the Nlrp3 response, resulting in higher levels of IL-1β in Aim2-deficient macrophages (16). Collectively, the elevated IL-1β production in Aim2−/− or Nlrp3−/− mice along with other proinflammatory cytokines observed in the present study are thus sufficient to carry out the innate microbicidal functions resulting in the complete clearance of the emrA1 mutant.
The activation of the inflammasome, with the associated induction of IL-1β and IL-18, is required for the generation of antigen-specific adaptive immune responses. The improved protection provided by a recombinant Mycobacterium bovis BCG vaccine against Mycobacterium tuberculosis and the activation of the Aim2 inflammasome for an efficient antigen-specific adaptive immune response in mice vaccinated with viral DNA vaccines provide a proof of concept for these notions (17, 42). CD8α+ dendritic cells can sense bacterial flagellin to activate the NCLR4 inflammasome with the resultant production of IL-1β and IL-18. IL-18 mediates the production of IFN-γ by memory CD8+ T cells, which provide a mechanistic framework for the induction of inflammasome-mediated but antigen-independent effector functions of memory T-cell responses during bacterial infections (43). Our results demonstrate that both Aim2 and Nlrp3 are dispensable for vaccination-induced protective immune responses in mice, especially when tested against a very high challenge dose of F. tularensis LVS. The Aim2−/− and Nlrp3−/− mice survived similarly to the wild-type mice and after initial weight loss for the first few days postchallenge regained their original body weight. When sacrificed at the end of the experiment, the surviving mice of all three strains did not carry any bacteria in their organs, indicating that vaccination, irrespective of their genotypes, renders sterilizing immunity against F. tularensis LVS.
Higher levels of IgG2b and IgG1 antibodies are protective against F. tularensis infection (44–47). The antibody profiles varied between the emrA1 mutant-vaccinated wild-type and Aim2−/− or Nlrp3−/− mice, with higher levels of total IgG, IgG2b, and IgG1 in Aim2−/− mice and IgG2b antibodies in Nlrp3−/− mice. Intriguingly, as we also observed in our previous study (26), no detectable IgG1 antibodies were observed in the wild-type and Nlrp3−/− mice following immunization with the emrA1 mutant. The reason for the total absence of IgG1 antibodies in the latter strains of mice remains unknown. However, the antibody profiles were identical in the wild-type and Aim2−/− or Nlrp3−/− mice postchallenge. A previous study reported that the levels of Francisella-specific IgM antibodies do not differ between wild-type and Nlrp3−/− mice (16). Collectively, our results convincingly demonstrate that both Aim2 and Nlrp3 are dispensable for the generation of strong antibody responses following immunization with the emrA1 mutant and challenge with lethal doses of F. tularensis.
It has been reported that IFN-γ and IL-17 are induced immediately following challenge with F. tularensis (48, 49). IFN-γ is required for protection during both primary and secondary infection as well as the priming and effector phases of respiratory infection caused by F. tularensis (50–52). IFN-γ plays an essential role in the clearance of Francisella infection by stimulating alveolar macrophages and recruiting neutrophils to the site of infection (53, 54). Similarly, IL-17 produced by Th17-polarized T cells is required to clear primary F. tularensis infection (55–57). Furthermore, the IL-17-dependent induction of IFN-γ controls F. tularensis infection. The induction of both IFN-γ and IL-17 serves as a correlate of immune protection in vaccinated mice, and the production of IFN-γ and IL-17 in an ex vivo assay with splenocytes from immunized mice is an indicator of protective efficacy and the ability of the vaccine to induce a memory recall response against F. tularensis challenge (29, 30, 58, 59). The cell-mediated recall responses were evaluated on day 28 after primary immunization. Similar IFN-γ responses were observed in ex vivo assays using splenocytes from emrA1 mutant-immunized wild-type, Aim2−/−, and Nlrp3−/− mice. On the other hand, the IL-17 recall responses were significantly diminished in both Aim2−/− and Nlrp3−/− mice. However, despite these subtle alterations in the recall responses, the overall abilities of the splenocytes from immunized Aim2−/− or Nlrp3−/− mice to effectively clear the bacteria from infected BMDMs in ex vivo coculture assays as well as to counter the lethal F. tularensis LVS challenge dose in vivo remained similar to those of the wild-type mice.
To conclude, the roles of both Aim2 and Nlrp3 inflammasomes in the generation of innate immune responses against primary F. tularensis infection have been amply demonstrated. However, their contributions to shaping the vaccine-induced adaptive response have not been studied to date. The results from this study demonstrate that both Aim2 and Nlrp3 are dispensable for vaccine-induced immunity against F. tularensis. The overall results point to the fact that alternate mechanisms responsible for IL-1β processing may compensate for the loss of Aim2 or Nlrp3 or that one inflammasome component compensates for the loss of the other component. Future studies using Aim2−/− Nlrp3−/− double-knockout mice would be required to determine the alternate compensatory mechanisms and assign the collective roles of these two inflammasome components in vaccine-induced responses against F. tularensis.
MATERIALS AND METHODS
Mice.
Aim2−/− [B6.129P2-Aim2Gt(CSG445)Byg/J; stock number 013144] and Nlrp3−/− (B6.129S6-Nlrp3tm2Bhk/J; stock number 021302) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred in the Animal Resource Facility of New York Medical College. Corresponding wild-type C57BL6/J (000664 C57BL/6J) mice were purchased from The Jackson Laboratory before the experiments. All mice were 6 to 8 weeks old when used in the experiments. All mice were housed in a pathogen-free environment in the Animal Resource Facility of New York Medical College, and all procedures were conducted according to approved institutional animal care and use committee (IACUC) protocols.
Bacterial strains.
F. tularensis subsp. holarctica LVS obtained from BEI Resources, Manassas, VA, was used in this study. The emrA1 mutant of F. tularensis LVS previously developed in our laboratory was used (25). All work with these strains was conducted under biosafety level 2 containment conditions.
The bacterial stocks were prepared by growing bacteria on Mueller-Hinton (MH) chocolate agar plates (BD Life Sciences) at 37°C with 5% CO2. After 48 h of growth, individual colonies were inoculated into MH broth (MHB; BD Biosciences, San Jose, CA) supplemented with 10% glucose, 0.021% anhydrous calcium chloride, 0.000138% hydrous magnesium chloride, 2.5% ferric pyrophosphate, and 2.5% IsoVitaleX (BD Biosciences, San Jose, CA). These bacterial suspensions were grown at 37°C overnight with constant shaking at 175 rpm (MaxQ 4000; Thermo Scientific). Next, 0.5-ml aliquots of mid-log-phase bacteria grown in MHB (optical density at 600 nm [OD600] of 0.2) were collected in 1.5-ml cryovials, snap-frozen in liquid nitrogen, and then stored at −80°C. Before use in an experiment, a vial of the previously frozen stock was thawed in a 37°C water bath and then plated on MH chocolate agar at 37°C with 5% CO2 for 48 h. For culturing the emrA1 mutant, MH chocolate agar or MHB supplemented with kanamycin (10 μg/ml) was used.
Bone marrow-derived macrophages.
Bone marrow-derived macrophages (BMDMs) were derived from wild-type C57BL6/J, Aim2−/−, and Nlrp3−/− mice using standard isolation protocols. Femurs from mice were flushed using a syringe filled with 5 ml ice-cold Dulbecco’s modified Eagle’s medium (DMEM) into a 50-ml tube (2 mice/tube). The single-cell bone marrow suspension was centrifuged at 1,000 rpm for 10 min. The supernatant was decanted carefully, and the pellet was suspended in 5 ml complete BMDM medium, transferred into a T25 flask, and incubated overnight at 37°C in the presence of 5% CO2. The following day, the nonadherent cells were transferred to a 90-mm petri dish, BMDM medium was then added, and the plates were incubated at 37°C in the presence of 5% CO2 for 7 days, allowing the transformation of monocytes into adherent macrophages. Once confluence was attained, cells were collected and either used directly in experiments or frozen for future use in sterile fetal bovine serum (FBS) with 10% dimethyl sulfoxide (DMSO) (1 ml/vial) in liquid nitrogen.
Infection of mice with F. tularensis LVS and the emrA1 mutant and assessments of bacterial burdens and cytokines.
Wild-type, Aim2−/−, and Nlrp3−/− mice deeply anesthetized with a cocktail of ketamine and xylazine were infected intranasally with 1 × 106 CFU of wild-type F. tularensis LVS or the emrA1 mutant. The mice were observed for survival for 21 days and monitored for any morbidity by recording the body weights daily. To determine the kinetics of clearance of the emrA1 mutant, mice (3 mice/group/time point) were infected with 1 × 106 CFU of the emrA1 mutant. Lungs, livers, and spleens of mice were harvested aseptically on days 1, 3, 5, and 21 postinfection and homogenized using a bead beater. Homogenates were diluted 10-fold in phosphate-buffered saline (PBS), plated on MH chocolate agar plates in duplicate, and incubated at 37°C with 5% CO2 for 48 h to enumerate the bacterial colonies. The results were expressed as log10 CFU in each organ. The organ homogenates were also centrifuged at 1,000 rpm for 10 min to pellet the tissue debris, and supernatants were collected and stored at −80°C to be used for cytokine analysis. Levels of cytokines such as IL-1β, IL-6, and TNF-α in the lung homogenates were determined using cytometric bead array (CBA) analysis and expressed as picograms per milliliter (means ± standard errors of the means [SEM]).
Immunization and challenge experiments.
Wild-type, Aim2−/−, and Nlrp3−/− mice deeply anesthetized with a cocktail of ketamine and xylazine were immunized intranasally with a single dose of 1 × 106 CFU of the emrA1 mutant in 20 μl PBS (10 μl/naris). The immunized mice on day 28 of the primary immunization were challenged intranasally with 2 × 107 CFU of F. tularensis LVS. The mice were observed for morbidity by recording the body weights daily and for mortality for 28 days postchallenge. The survival results were expressed as survival curves, the data were analyzed using the log rank test, and a P value of <0.05 was considered statistically significant. Mice were bled before challenge on day 28 postimmunization and on day 28 postchallenge to harvest serum. Francisella-specific total IgG, IgG1, and IgG2b antibody levels were determined in the harvested serum by an enzyme-linked immunosorbent assay (ELISA) as described previously (26, 29, 30, 47).
In vitro coculture assay.
Spleens were isolated from naive wild-type, Aim2−/−, and Nlrp3−/− mice, and the mice were immunized intranasally with 1 × 106 CFU of the emrA1 mutant on day 28 postimmunization (n = 5/group). The splenocytes were collected, and the numbers of viable splenocytes were counted using a hemocytometer as described previously (29, 30).
BMDMs (5 × 105) from wild-type, Aim2−/−, and Nlrp3−/− mice were seeded in 12-well plates and infected with F. tularensis LVS (MOI of 100). After 4 h, the medium was removed and replaced by BMDM medium containing splenocytes from either naive or immunized mice at a ratio of 1:1 and incubated at 37°C with 5% CO2 for up to 120 h. The cells in coculture were lysed after 24 and 48 h, diluted 10-fold, and plated on MH chocolate agar plates to quantitate bacterial numbers. The culture supernatants were collected 24, 48, 72, 96, and 120 h after the addition of the splenocytes and analyzed for IL-17 and IFN-γ levels by CBA analyses and ELISAs as described previously (29, 30). Uninfected cells and splenocytes collected from naive, wild-type, Aim2−/−, and Nlrp3−/− mice were used as controls.
Statistical analysis.
All data were statistically analyzed using the InStat program (GraphPad Software). The data were analyzed by one-way analysis of variance (ANOVA). Results were expressed as means ± SEM or standard deviations (SD), and differences between the experimental groups were considered statistically significant at a P value of <0.05. The survival data were expressed as Kaplan-Meier survival curves, and statistical significance for survival results was evaluated by analyzing the mean time to death by the log rank test.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R56AI101109 and R21AI151277 (C.S.B.) and R15AI107698 (M.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No financial conflicts of interest exist regarding the contents of the manuscript and its authors.
Contributor Information
Meenakshi Malik, Email: Meenakshi.Malik@acphs.edu.
Chandra Shekhar Bakshi, Email: Shekhar_Bakshi@nymc.edu.
Guy H. Palmer, Washington State University
REFERENCES
- 1.Ellis J, Oyston PCF, Green M, Titball RW. 2002. Tularemia. Clin Microbiol Rev 15:631–646. 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pechous RD, McCarthy TR, Zahrt TC. 2009. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol Mol Biol Rev 73:684–711. 10.1128/MMBR.00028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Tonat K, Working Group on Civilian Biodefense . 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763–2773. 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 4.Oyston PCF, Sjöstedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2:967–978. 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 5.Eigelsbach HT, Downs CM. 1961. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J Immunol 87:415–425. [PubMed] [Google Scholar]
- 6.Janeway CA, Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54:1–13. 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Kim YK, Shin JS, Nahm MH. 2016. NOD-like receptors in infection, immunity, and diseases. Yonsei Med J 57:5–14. 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis BK, Wen H, Ting JP-Y. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 29:707–735. 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518. 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458:509–513. 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526:666–671. 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 12.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526:660–665. 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 13.Dotson RJ, Rabadi SM, Westcott EL, Bradley S, Catlett SV, Banik S, Harton JA, Bakshi CS, Malik M. 2013. Repression of inflammasome by Francisella tularensis during early stages of infection. J Biol Chem 288:23844–23857. 10.1074/jbc.M113.490086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosio CM. 2011. The subversion of the immune system by Francisella tularensis. Front Microbiol 2:9. 10.3389/fmicb.2011.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crane DD, Bauler TJ, Wehrly TD, Bosio CM. 2014. Mitochondrial ROS potentiates indirect activation of the AIM2 inflammasome. Front Microbiol 5:438. 10.3389/fmicb.2014.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Periasamy S, Le HT, Duffy EB, Chin H, Harton JA. 2016. Inflammasome-independent NLRP3 restriction of a protective early neutrophil response to pulmonary tularemia. PLoS Pathog 12:e1006059. 10.1371/journal.ppat.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suschak JJ, Wang S, Fitzgerald KA, Lu S. 2015. Identification of Aim2 as a sensor for DNA vaccines. J Immunol 194:630–636. 10.4049/jimmunol.1402530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nordlander S, Pott J, Maloy KJ. 2014. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal Immunol 7:775–785. 10.1038/mi.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritz JH, Le Bourhis L, Sellge G, Magalhaes JG, Fsihi H, Kufer TA, Collins C, Viala J, Ferrero RL, Girardin SE, Philpott DJ. 2007. Nod1-mediated innate immune recognition of peptidoglycan contributes to the onset of adaptive immunity. Immunity 26:445–459. 10.1016/j.immuni.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Mencacci A, Bacci A, Cenci E, Montagnoli C, Fiorucci S, Casagrande A, Flavell RA, Bistoni F, Romani L. 2000. Interleukin 18 restores defective Th1 immunity to Candida albicans in caspase 1-deficient mice. Infect Immun 68:5126–5131. 10.1128/iai.68.9.5126-5131.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell JR, Yadav R, Rossi RJ, Ruby CE, Weinberg AD, Aguila HL, Vella AT. 2006. IL-18 bridges innate and adaptive immunity through IFN-gamma and the CD134 pathway. J Immunol 177:234–245. 10.4049/jimmunol.177.1.234. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi K, Tsutsui H, Yoshimoto T. 2010. Importance of IL-18-induced super Th1 cells for the development of allergic inflammation. Allergol Int 59:137–141. 10.2332/allergolint.10-RAI-0208. [DOI] [PubMed] [Google Scholar]
- 23.Chai D, Yue Y, Xu W, Dong C, Xiong S. 2014. Mucosal co-immunization with AIM2 enhances protective SIgA response and increases prophylactic efficacy of chitosan-DNA vaccine against coxsackievirus B3-induced myocarditis. Hum Vaccin Immunother 10:1284–1294. 10.4161/hv.28333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markel G, Bar-Haim E, Zahavy E, Cohen H, Cohen O, Shafferman A, Velan B. 2010. The involvement of IL-17A in the murine response to sub-lethal inhalational infection with Francisella tularensis. PLoS One 5:e11176. 10.1371/journal.pone.0011176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Z, Banik S, Rane H, Mora VT, Rabadi SM, Doyle CR, Thanassi DG, Bakshi CS, Malik M, Rane H, Banik S, Doyle CR, Bakshi CS, Rabadi SM, Mora VT, Thanassi DG, Ma Z. 2014. EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol Microbiol 91:976–995. 10.1111/mmi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suresh RV, Ma Z, Sunagar R, Bhatty V, Banik S, Catlett SV, Gosselin EJ, Malik M, Bakshi CS. 2015. Preclinical testing of a vaccine candidate against tularemia. PLoS One 10:e0124326. 10.1371/journal.pone.0124326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Pascalis R, Chou AY, Ryden P, Kennett NJ, Sjostedt A, Elkins KL. 2014. Models derived from in vitro analyses of spleen, liver, and lung leukocyte functions predict vaccine efficacy against the Francisella tularensis live vaccine strain (LVS). mBio 5:e00936-13. 10.1128/mBio.00936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Pascalis R, Hahn A, Brook HM, Ryden P, Donart N, Mittereder L, Frey B, Wu TH, Elkins KL. 2018. A panel of correlates predicts vaccine-induced protection of rats against respiratory challenge with virulent Francisella tularensis. PLoS One 13:e0198140. 10.1371/journal.pone.0198140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansour AA, Banik S, Suresh RV, Kaur H, Malik M, McCormick AA, Bakshi CS. 2018. An improved tobacco mosaic virus (TMV)-conjugated multiantigen subunit vaccine against respiratory tularemia. Front Microbiol 9:1195. 10.3389/fmicb.2018.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahawar M, Rabadi SM, Banik S, Catlett SV, Metzger DW, Malik M, Bakshi CS. 2013. Identification of a live attenuated vaccine candidate for tularemia prophylaxis. PLoS One 8:e61539. 10.1371/journal.pone.0061539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, Shapira I, Dinarello CA, Paul WE. 2009. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci U S A 106:7119–7124. 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schenten D, Nish SA, Yu S, Yan X, Lee HK, Brodsky I, Pasman L, Yordy B, Wunderlich FT, Brüning JC, Zhao H, Medzhitov R. 2014. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity 40:78–90. 10.1016/j.immuni.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamura H, Tsutsi H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, Torigoe K, Okura T, Nukada Y, Hattori K. 1995. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature 378:88–91. 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 34.Garlanda C, Dinarello CA, Mantovani A. 2013. The interleukin-1 family: back to the future. Immunity 39:1003–1018. 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakanishi K. 2018. Unique action of interleukin-18 on T cells and other immune cells. Front Immunol 9:763. 10.3389/fimmu.2018.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belhocine K, Monack DM. 2012. Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells. Cell Microbiol 14:71–80. 10.1111/j.1462-5822.2011.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juruj C, Lelogeais V, Pierini R, Perret M, Py BF, Jamilloux Y, Broz P, Ader F, Faure M, Henry T. 2013. Caspase-1 activity affects AIM2 speck formation/stability through a negative feedback loop. Front Cell Infect Microbiol 3:14. 10.3389/fcimb.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng K, Broz P, Jones J, Joubert LM, Monack D. 2011. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol 13:1586–1600. 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Rühl S, Dussurgey S, Dick MS, Kistner A, Rigard M, Degrandi D, Pfeffer K, Yamamoto M, Henry T, Broz P. 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol 16:476–484. 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Q, Man SM, Karki R, Malireddi RKS, Kanneganti T-D. 2018. Detrimental type I interferon signaling dominates protective AIM2 inflammasome responses during Francisella novicida infection. Cell Rep 22:3168–3174. 10.1016/j.celrep.2018.02.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greten FR, Arkan MC, Bollrath J, Hsu L-C, Goode J, Miething C, Göktuna SI, Neuenhahn M, Fierer J, Paxian S, Van Rooijen N, Xu Y, O’Cain T, Jaffee BB, Busch DH, Duyster J, Schmid RM, Eckmann L, Karin M. 2007. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell 130:918–931. 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gröschel MI, Sayes F, Shin SJ, Frigui W, Pawlik A, Orgeur M, Canetti R, Honoré N, Simeone R, van der Werf TS, Bitter W, Cho S-N, Majlessi L, Brosch R. 2017. Recombinant BCG expressing ESX-1 of Mycobacterium marinum combines low virulence with cytosolic immune signaling and improved TB protection. Cell Rep 18:2752–2765. 10.1016/j.celrep.2017.02.057. [DOI] [PubMed] [Google Scholar]
- 43.Kupz A, Guarda G, Gebhardt T, Sander LE, Short KR, Diavatopoulos DA, Wijburg OLC, Cao H, Waithman JC, Chen W, Fernandez-Ruiz D, Whitney PG, Heath WR, Curtiss R, Tschopp J, Strugnell RA, Bedoui S. 2012. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8+ T cells. Nat Immunol 13:162–169. 10.1038/ni.2195. [DOI] [PubMed] [Google Scholar]
- 44.Mara-Koosham G, Hutt JA, Lyons CR, Wu TH. 2011. Antibodies contribute to effective vaccination against respiratory infection by type A Francisella tularensis strains. Infect Immun 79:1770–1778. 10.1128/IAI.00605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia Q, Bowen R, Sahakian J, Dillon BJ, Horwitz MA. 2013. A heterologous prime-boost vaccination strategy comprising the Francisella tularensis live vaccine strain capB mutant and recombinant attenuated Listeria monocytogenes expressing F. tularensis IglC induces potent protective immunity in mice against virulent F. tularensis aerosol challenge. Infect Immun 81:1550–1561. 10.1128/IAI.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakshi CS, Malik M, Mahawar M, Kirimanjeswara GS, Hazlett KRO, Palmer LE, Furie MB, Singh R, Melendez JA, Sellati TJ, Metzger DW. 2008. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine 26:5276–5288. 10.1016/j.vaccine.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banik S, Mansour AA, Suresh RV, Wykoff-Clary S, Malik M, McCormick AA, Bakshi CS. 2015. Development of a multivalent subunit vaccine against tularemia using tobacco mosaic virus (TMV) based delivery system. PLoS One 10:e0130858. 10.1371/journal.pone.0130858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Y, Ritchea S, Logar A, Slight S, Messmer M, Rangel-Moreno J, Guglani L, Alcorn JF, Strawbridge H, Park SM, Onishi R, Nyugen N, Walter MJ, Pociask D, Randall TD, Gaffen SL, Iwakura Y, Kolls JK, Khader SA. 2009. Interleukin-17 is required for T helper 1 cell immunity and host resistance to the intracellular pathogen Francisella tularensis. Immunity 31:799–810. 10.1016/j.immuni.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khader SA, Gopal R. 2010. IL-17 in protective immunity to intracellular pathogens. Virulence 1:423–427. 10.4161/viru.1.5.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts LM, Tuladhar S, Steele SP, Riebe KJ, Chen C, Cumming RI, Seay S, Frothingham R, Sempowski GD, Kawula TH, Frelinger JA. 2014. Identification of early interactions between Francisella and the host. Infect Immun 82:2504–2510. 10.1128/IAI.01654-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirimanjeswara GS, Olmos S, Bakshi CS, Metzger DW. 2008. Humoral and cell-mediated immunity to the intracellular pathogen Francisella tularensis. Immunol Rev 225:244–255. 10.1111/j.1600-065X.2008.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cowley SC, Meierovics AI, Frelinger JA, Iwakura Y, Elkins KL. 2010. Lung CD4− CD8− double-negative T cells are prominent producers of IL-17A and IFN-γ during primary respiratory murine infection with Francisella tularensis live vaccine strain. J Immunol 184:5791–5801. 10.4049/jimmunol.1000362. [DOI] [PubMed] [Google Scholar]
- 53.Steiner DJ, Furuya Y, Jordan MB, Metzger DW. 2017. Protective role for macrophages in respiratory Francisella tularensis infection. Infect Immun 85:e00064-17. 10.1128/IAI.00064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polsinelli T, Meltzer MS, Fortier AH. 1994. Nitric oxide-independent killing of Francisella tularensis by IFN-gamma-stimulated murine alveolar macrophages. J Immunol 153:1238–1245. [PubMed] [Google Scholar]
- 55.Anthony LSD, Ghadirian E, Nestel FP, Kongshavn PAL. 1989. The requirement for gamma interferon in resistance of mice to experimental tularemia. Microb Pathog 7:421–428. 10.1016/0882-4010(89)90022-3. [DOI] [PubMed] [Google Scholar]
- 56.Karttunen R, Surcel HM, Andersson G, Ekre HPT, Herva E. 1991. Francisella tularensis-induced in vitro gamma interferon, tumor necrosis factor alpha, and interleukin 2 responses appear within 2 weeks of tularemia vaccination in human beings. J Clin Microbiol 29:753–756. 10.1128/JCM.29.4.753-756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. 2007. Interleukins 1β and 6 but not transforming growth factor-β are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol 8:942–949. 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 58.Paranavitana C, Zelazowska E, DaSilva L, Pittman PR, Nikolich M. 2010. Th17 cytokines in recall responses against Francisella tularensis in humans. J Interferon Cytokine Res 30:471–476. 10.1089/jir.2009.0108. [DOI] [PubMed] [Google Scholar]
- 59.De Pascalis R, Taylor BC, Elkins KL. 2008. Diverse myeloid and lymphoid cell subpopulations produce gamma interferon during early innate immune responses to Francisella tularensis live vaccine strain. Infect Immun 76:4311–4321. 10.1128/IAI.00514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]