ABSTRACT
Multidrug efflux systems belonging to the resistance-nodulation-cell division (RND) family are ubiquitous in Gram-negative bacteria and are critical for antimicrobial resistance. This realization has led to efforts to develop efflux pump inhibitors (EPI) for use as adjuvants for antibiotic treatment of resistant organisms. However, the functions of RND transporters extend beyond antimicrobial resistance to include physiological functions that are critical for pathogenesis, suggesting that EPIs could also be used as antivirulence therapeutics. This was documented in the enteric pathogen Vibrio cholerae, in which EPIs were shown to attenuate the production of the critical virulence factors cholera toxin (CT) and the toxin-coregulated pilus (TCP). In this study, we investigated the antivirulence mechanism of action of the EPI phenyl-arginine-β-naphthylamide (PAβN) on V. cholerae. Using bioassays, we documented that PAβN inhibited virulence factor production in three epidemic V. cholerae isolates. Transcriptional reporter studies and mutant analysis indicated that PAβN initiated a ToxR-dependent regulatory circuit to activate leuO expression and that LeuO repressed the expression of the critical virulence activator aphA to attenuate CT and TCP production. The antivirulence activity of PAβN was found to be dependent on the ToxR periplasmic sensing domain (PPD), suggesting that a feedback mechanism was involved in its activity. Collectively, the data indicated that PAβN inhibited V. cholerae virulence factor production by activating a ToxR-dependent metabolic feedback mechanism to repress the expression of the ToxR virulence regulon. This suggests that efflux pump inhibitors could be used as antivirulence therapeutics for the treatment of cholera and perhaps that of other Gram-negative pathogens.
KEYWORDS: efflux pump inhibitors, virulence inhibition, RND, efflux, cholera, pathogenesis, virulence
INTRODUCTION
Vibrio cholerae is an enteric Gram-negative bacterium that causes the severe diarrheal disease cholera (1, 2). Cholera is an epidemic disease that affects 2 to 3 million people each year, particularly in places with poor sanitation. V. cholerae is a common inhabitant of aquatic ecosystems across the globe and can be found in both fresh (3, 4) and brackish waters (5), from which people acquire cholera through the consumption of contaminated food or water. Following ingestion, V. cholerae passes through the gastric acid barrier of the stomach before colonizing the crypts of the small intestine to cause disease. As this pathogen must traverse a wide variety of environments to colonize the small intestine and cause disease in its human host, its pathogenicity hinges on its ability to coordinate the expression of multiple virulence genes in response to environmental stimuli. This process is orchestrated by the ToxR virulence regulon.
The ToxR virulence regulon is a hierarchical regulatory cascade that activates the expression of critical virulence genes following host entry that code for the production of cholera toxin (CT) and the toxin-coregulated pilus (TCP) (6). The regulon is named after the first regulator identified, ToxR, which is a membrane-associated transcription factor with a periplasmic sensing domain (PPD) that is thought to modulate ToxR activity and the expression of the regulon in response to environmental cues. ToxR functions in conjunction with another membrane protein, ToxS, which stabilizes the ToxR PPD in response to environmental cues (7–9). CT is an enterotoxin that is responsible for the secretory diarrhea that is the hallmark of cholera, and the TCP is a type IV pilus that is essential for intestinal colonization. The ToxR regulon consists of five main transcription factors that respond to environmental cues in the host to activate virulence gene expression (10). AphA and AphB are cytoplasmic transcription factors that respond to quorum sensing molecules and oxygen, respectively. Together, they bind to the tcpP promoter to activate its expression. TcpP then binds with the toxT promoter to activate its expression. ToxT then directly activates the expression of the genes responsible for CT and TCP production. Several other regulatory genes function to modulate the expression of the ToxR regulon in response to environmental cues. This includes the LysR family transcriptional regulator leuO and the two-component regulator ompR. The expression of leuO is positively regulated by ToxR in response to bile salts and is predominantly expressed at high cell density during growth in vitro (11, 12). LeuO negatively regulates the ToxR regulon by repressing aphA transcription. OmpR is activated in response to membrane intercalating agents and represses the ToxR regulon by repressing aphB transcription (13).
The ability of V. cholerae to colonize the human gastrointestinal tract is not only dependent on virulence factor production but also requires the expression of efflux systems belonging to the resistance-nodulation-cell division (RND) family. RND efflux systems are ubiquitous tripartite transporters that are found in Gram-negative bacteria (14). V. cholerae encodes six RND transporters that share TolC as their outer membrane pore protein (15). These systems have been shown to be essential for intestinal colonization and for the intrinsic resistance of V. cholerae to antibiotics, detergents, bile acids, and antimicrobial peptides (16). In addition, the RND efflux systems were also required to produce CT and the TCP (16). The absence of RND-mediated efflux was shown to result in increased leuO transcription, leading to repression of the ToxR regulon and attenuated CT and TCP production (11). These studies suggested that impaired efflux resulted in the initiation of a metabolite feedback loop via the periplasmic sensing domain of ToxR to effect virulence repression (11). Collectively, these findings indicate that RND-mediated efflux has dual functions in V. cholerae pathogenesis, i.e., providing resistance to antimicrobial compounds in the host and modulating virulence gene expression.
The observation that RND-mediated efflux was required for V. cholerae virulence gene expression suggested that efflux could be a target for the development of antivirulence therapeutics. Indeed, we previously found that the RND efflux pump inhibitors (EPIs) phenylalanine-arginine-β-naphthylamide (PAβN) and 1-(1-naphthylmethyl)-piperazine (NMP) attenuated V. cholerae CT and TCP production (17). However, the mechanism by which EPIs attenuated virulence factor production was unresolved. In this work, we used PAβN to investigate the mechanism of EPI inhibition of V. cholerae virulence factor production. We found that PAβN activated leuO expression by a process that was dependent on the presence of the periplasmic sensing domain of ToxR. LeuO then repressed aphA transcription, leading to downregulation of the ToxR regulon and attenuated CT and TCP production. The data supported a model where PAβN activated an efflux-dependent regulatory feedback circuit by inhibiting the efflux of native substrates of the RND transporters to repress virulence gene expression. Our results suggest that the use of EPIs could extend beyond antibiotic potentiation and be used as antivirulence therapeutics for treatment of cholera and, likely, that of other bacterial diseases.
RESULTS
PAβN inhibits virulence factor production in pandemic V. cholerae strains.
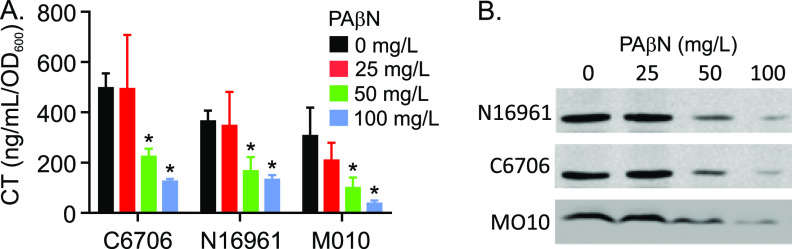
A previous study suggested that EPIs inhibited virulence factor production in V. cholerae O1 El Tor strain N16961. To confirm and extend this finding, we tested the effect of PAβN on CT and TCP production in three epidemic strains of V. cholerae, O1 El Tor strains N16961 and C6706 and O139 strain MO10. The three V. cholerae strains were cultured under virulence gene-inducing conditions (i.e., AKI conditions) in the presence of 0, 25, 50 or 100 mg/liter of PAβN. The MIC for PAβN in V. cholerae was previously shown to be 300 mg/liter (17). Consistent with this, PAβN at the test concentrations did not affect the growth of any of the V. cholerae strains. The following day, culture aliquots were collected, normalized by optical density, and assayed for CT production and TcpA production as described in Materials and Methods. The results showed a PAβN concentration-dependent inhibition of both CT (Fig. 1A) and TcpA (Fig. 1B) production in each strain beginning at 50 mg/liter PAβN. The PAβN-dependent decrease in CT and TcpA production in N16961 confirmed the results of a previous study (17). The observation that PAβN exhibited similar virulence-repressing activity in strains C6706 and MO10 showed that the virulence inhibitor activity of PAβN was not strain specific and likely extends to all currently circulating epidemic and pandemic V. cholerae strains.
FIG 1.
PAβN inhibits virulence factor production in V. cholerae. The indicated V. cholerae strains were cultured for 15 h under AKI conditions in AKI medium containing the indicated concentrations of PAβN. Cholera toxin (CT) production in each culture was then determined by GM1 enzyme-limited immunosorbent assay (ELISA) (A), and toxin-coregulated pilus (TCP) production was assessed by TcpA Western blotting (B). The results are representative of at least three independent experiments. Statistical significance was determined by analysis of variance (ANOVA) with the Tukey-Kramer multiple-comparison test. *, P ≤ 0.05 relative to the no-PAβN control.
PAβN inhibits ToxR regulon expression.
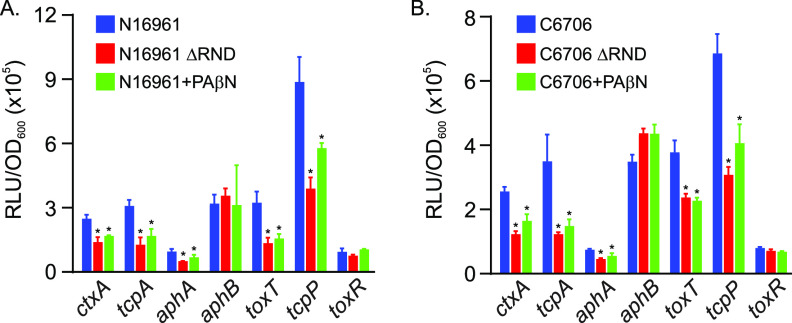
The results described above confirmed that PAβN inhibited V. cholerae virulence factor production, but the mechanism by which PAβN affected this process was undetermined. The production of CT and TCP in V. cholerae is positively regulated by the ToxR regulon. We therefore tested if PAβN was affecting the expression of the primary regulatory genes in the ToxR regulon. We cultured V. cholerae strains N16961 (Fig. 2A) and C6706 (Fig. 2B), bearing lux-based transcriptional reporters for the primary regulatory genes (i.e., aphA, aphB, toxT, tcpP, and toxR), and CT and TCP (i.e., ctxA and tcpA) under AKI conditions in AKI broth plus and minus PAβN (75 mg/liter) for 6 h, at which time we assessed gene expression as relative light units (RLU) divided by the optical density at 600 nm (OD600). An RND-deficient V. cholerae strain (ΔRND) cultured in AKI broth without PAβN was included as a control. The results showed that PAβN inhibited the expression of ctxA and tcpA (Fig. 2A) in both N16961 and C6706, confirming the CT enzyme-limited immunosorbent assay (ELISA) and TcpA Western blotting results presented above. The expression of aphB and toxR was not decreased in the ΔRND mutant or in PaβN-treated N16961 or C6706, confirming previous reports (11, 16). In contrast, the expression of aphA, tcpP, and toxT was repressed by PAβN in both N16961 and C6706 to a level that was similar to what was observed in the ΔRND mutant. As aphA is one of the most upstream regulators in the ToxR regulon, and it directly activates tcpP transcription and indirectly activates toxT (via tcpP), this result indicates that PAβN inhibition of aphA transcription was likely responsible for downregulation of the ToxR regulon and the attenuated CT and TCP production. The fact that the effects of PAβN on the ToxR regulon were conserved between N16961 and C6706 further confirms that the antivirulence activity of PAβN is not strain specific.
FIG 2.
Effect of PAβN on the expression of genes in the ToxR virulence regulon. The indicated V. cholerae N16961 JB58 (A) or C6706 (B) strains bearing plasmids with lux-based promoter fusions for the indicated genes were cultured under AKI conditions in the presence or absence of PAβN (75 mg/liter) for 5 h, at which time gene expression was quantified as relative light units (RLU) divided by the optical density at 600 nm (OD600). RND efflux pump-negative mutants (ΔRND) for both strains were included as a comparator. The results represent the average RLU/OD and standard deviation of three independent experiments. Statistical significance was determined by ANOVA with the Tukey-Kramer multiple-comparison test. *, P ≤ 0.05 relative to the wild type (WT) cultured in AKI broth.
PAβN represses virulence factor production via ToxR-dependent activation of leuO.
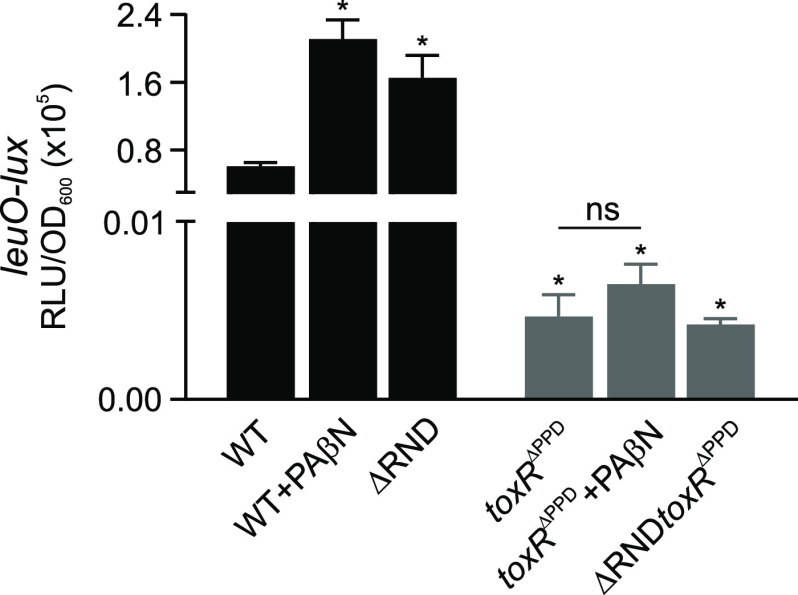
We then set out to further determine the mechanism by which PAβN repressed aphA transcription. The expression of aphA is negatively regulated by the main quorum sensing (QS) regulator HapR, with maximal expression occurring at low cell density (28). However, the fact that N16961, like most pandemic strains, contains a frameshift mutation in hapR and is QS negative suggested that another mechanism must be involved in aphA repression (18, 29). ToxR also indirectly regulates aphA via leuO. ToxR activates leuO transcription in response to environmental cues, including V. cholerae metabolites such as indole, cyclo(phe-pro), and malate (11, 21, 30). LeuO can directly bind to the aphA promoter to repress its expression (21). We therefore tested if PAβN was repressing aphA through leuO by repeating the above-described experiments using N16961 bearing a leuO-lux reporter. The results showed a PAβN-dependent increase in leuO expression (Fig. 3, black bars) in the wild type (WT). The increase in leuO expression in the PaβN-treated cells was similar to what was observed in an isogenic RND-deficient mutant (ΔRND), confirming that treatment of WT with PAβN phenocopied a ΔRND mutant for leuO expression.
FIG 3.
PAβN activates leuO expression. V. cholerae strain N16961 and isogenic mutants lacking the ToxR periplasmic domain (ΔPPD) and all six RND efflux pump proteins (ΔRND) bearing a leuO-lux promoter fusion reporter were cultured under AKI conditions in AKI medium in the presence or absence of PAβN (75 mg/liter) for 5 h, at which time gene expression was quantified as relative light units (RLU) divided by the optical density at 600 nm. The results represent the average RLU/OD and standard deviation of three independent experiments. Statistical relevance to the WT was calculated using a one-way ANOVA with Dunnett’s post hoc test. Statistical significance for the effects of PAβN on leuO expression in toxRΔPPD and toxRΔPPD was determined using Student’s t test. *, P ≤ 0.05 relative to the WT. NS, not significant.
Previous studies documented that ToxR activation of leuO expression in response to environmental cues was dependent on the presence of the ToxR periplasmic domain (PPD) (11, 21, 30). We therefore tested if the PAβN-dependent induction of leuO was also dependent on the ToxR PPD. We cultured leuO-lux reporter bearing derivatives of N16961 and the ΔRND mutant that expressed a truncated ToxR allele that lacked the periplasmic sensing domain (toxRΔPPD). The toxRΔPPD allele was previously shown to support virulence factor production and porin regulation (11, 21). The results showed that leuO was not expressed in the strains expressing the toxRΔPPD allele and that the addition of PAβN to the toxRΔPPD-expressing mutants did not significantly affect leuO expression (Fig. 3, gray bars). This confirmed that the PAβN induction of leuO was dependent on the presence of the periplasmic sensing domain of ToxR and suggested that leuO may modulate the PAβN-dependent repression of virulence factor production.
PAβN antivirulence activity is mediated by LeuO.
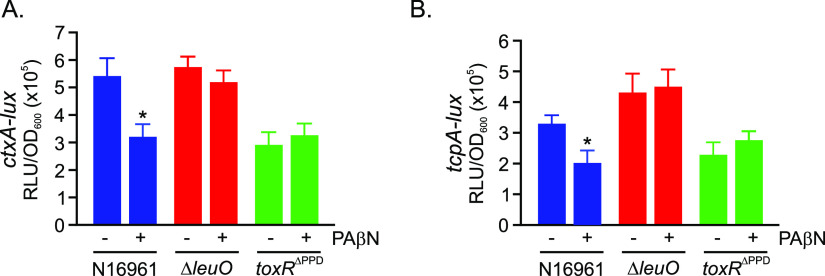
The results described above indicated that PAβN indirectly activated leuO via ToxR. As leuO was previously reported to repress aphA (21), and aphA expression was reduced by the addition of PAβN (Fig. 2), we hypothesized that PAβN-dependent virulence repression was mediated by leuO. If this was true, then deletion of leuO should abrogate the antivirulence activity of PAβN. To test this, we cultured N16961 and isogenic leuO and toxRΔPPD mutants under AKI conditions in the presence and absence of PAβN and then assayed for CT and TcpA production. The results showed that leuO deletion restored near-WT-level production of CT and TcpA in the cultures grown in the presence of PAβN (Fig. 4A and B). In contrast, PAβN did not have any effect on ctxA or tcpA expression in N16961 expressing the truncated toxRΔPPD allele, which is consistent with the inability of this mutant to activate leuO transcription. Collectively, these results, combined with the above-described data, suggested that the antivirulence activity of PAβN resulted from the activation of a ToxR-dependent signaling cascaded that resulted in the increased expression of leuO, with LeuO then repressing aphA transcription to downregulate the ToxR regulon to attenuate CT and TcpA production (Fig. 5).
FIG 4.
Deletion of leuO restores virulence factor production. The indicated Vibrio cholerae N16961 strains bearing lux-based promoter fusions for (A) ctxA and (B) tcpA were cultured under AKI conditions in the presence or absence of PAβN (75 mg/liter) for 5 h, at which time gene expression was quantified as relative light units (RLU) divided by the optical density at 600 nm. The results represent the average RLU/OD and standard deviation of three independent experiments. Statistical analysis was determined using Student’s t test. *, P ≤ 0.05.
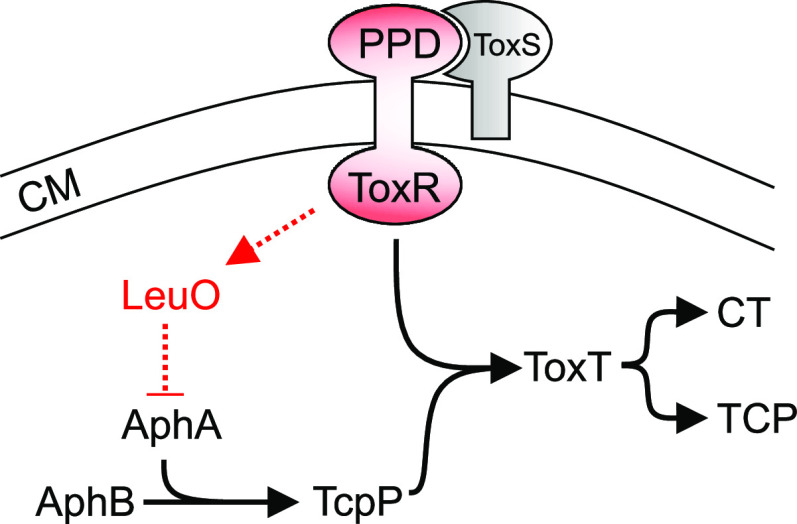
FIG 5.
Model for the antivirulence activity of PAβN in V. cholerae. Exposure of V. cholerae to PAβN results in the activation of ToxR by a process that is dependent upon its periplasmic signaling domain (PPD). Activation of ToxR results in increased leuO transcription. LeuO then represses aphA transcription, leading to downregulation of the ToxR virulence regulon and attenuated CT and TCP production. PAβN, phenyl-arginine-β-naphthylamide; CM, cytoplasmic membrane; CT, cholera toxin; TCP, toxin-coregulated pilus.
DISCUSSION
Here, we documented that exposure of V. cholerae to PAβN resulted in the activation of a negative regulatory circuit that attenuated production of the two most critical V. cholerae virulence factors, CT and the TCP. In this regulatory cascade, PAβN initiated signaling through the periplasmic signaling domain of ToxR (Fig. 5), which led to the activation of leuO transcription. LeuO then repressed the expression of aphA. As AphA is one of the most upstream regulators in the ToxR regulon, its repression results in downregulation of the ToxR regulon and attenuated CT and TCP production. The fact that PAβN activation of this regulatory circuit was dependent upon the periplasmic sensing domain of ToxR is consistent with a previous study suggesting that impaired RND-mediated efflux leads to intracellular metabolite accumulation to initiate adaptive responses via periplasmic sensors such as ToxR and other two-component regulatory systems (11, 30–33). The fact that production of CT and TCP are essential for the development of cholera suggests that EPIs could be used to treat cholera. These results also have potential implications for the application of EPIs to other Gram-negative pathogens in which RND-mediated efflux is critical for virulence.
While the present studies document that PAβN activated a putative metabolite-dependent negative regulatory circuit to suppress V. cholerae virulence gene expression, the environmental cue(s) that initiated this regulatory cascade remains unresolved. The dependence of this regulatory circuit on the ToxR PPD suggests that the activating stimuli is localized to the periplasm, a conclusion that is consistent with the function of RND transporters in capturing substrates from the periplasmic compartment. This conclusion is further supported by multiple studies showing that the ToxR PPD senses and responds to extracellular chemical cues, which include indole (30), cyclic di-peptides (21), malate (11), and bile salts (12). It is noteworthy that all of these compounds have been shown to induce the ToxR-dependent transcription of leuO and that indole, malate, and bile salts are substrates of the RND transporters. Based on this, we propose that treatment of V. cholerae with PAβN resulted in the periplasmic accumulation of metabolites. Once the metabolites reached a critical concentration, they interact with the ToxR PPD to activate leuO expression to attenuate virulence factor production. The fact that PAβN treatment of wild-type V. cholerae phenocopies an RND efflux pump null mutant provides additional support for this model (16). Furthermore, metabolite efflux was proposed to explain dysregulated virulence gene expression in a V. cholerae classical biotype strain lacking tolC (the outer membrane pore protein for the V. cholerae RND systems) (34, 35). The identity of the specific metabolite (or metabolites) responsible for activating the ToxR-dependent regulatory circuit remain unknown and will require additional studies.
The studies described here suggest that EPIs could also be used as antivirulence therapeutics in cholera. However, the contributions of RND efflux to virulence is not limited to V. cholerae. RND systems have been shown to be important for virulence in a large number of pathogens, including pathogens that are rapidly becoming refractory to antibiotics (e.g., Acinetobacter, Burkholderia, Escherichia, Klebsiella, Moraxella, Mycobacterium, Pseudomonas, Salmonella, and Stenotrophomonas) (14, 36–38) and for which new therapeutic approaches are needed. While the mechanism linking RND efflux to virulence in these pathogens is largely unknown, the metabolite feedback circuits described in V. cholerae appear to be conserved in other Gram-negative bacteria. For example, AcrAB in Escherichia coli has been shown to be feedback regulated in response to at least four metabolic pathways, namely, enterobactin, cysteine and purine biosynthesis, and gluconeogenesis (39). Consistent with this, additional studies have linked TolC to metabolite feedback in E. coli (40). In Salmonella, mutation of the AcrD efflux pump resulted in dramatic transcriptional changes that were consistent with metabolite accumulation initiating feedback mechanisms (41). Furthermore, RND systems have been linked to efflux of quorum sensing molecules and siderophores in multiple organisms with impaired efflux, affecting feedback responses for both classes of molecules (32, 42–46). Although not well studied, there is accumulating evidence to suggest that EPIs negatively impact virulence in other organisms. For example, EPIs were shown to inhibit quorum sensing and virulence factor production in Pseudomonas aeruginosa (47). EPIs were also shown in a cell culture model to inhibit P. aeruginosa invasion (48), and EPIs were also shown affect biofilm production in Klebsiella pneumoniae. P. aeruginosa, and E. coli (49). Thus, we conclude that EPIs may have utility beyond antibiotic potentiation and suggest that EPIs represent potential antivirulence therapeutics for Gram-negative pathogens and warrant further studies. If efficacious, EPIs could help to combat the global pandemic of antimicrobial resistance that threatens human health.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
V. cholerae O1 El Tor strains N16961 (18) and C6706 (19) and serogroup O139 strain MO10 (20) were obtained from our laboratory collection. N16961 JB58 (ΔlacZ Smr), JB58 toxRΔPPD, ΔRND (ΔvexB ΔvexD ΔvexF ΔvexH ΔvexK ΔvexM), and ΔRND toxRΔPPD mutants have been previously described (11, 12, 21). N16961 JB58 was used as the WT in this study. Escherichia coli strain EC100pir (Epicentre; Madison, WI) was used as a host for DNA cloning experiments. E. coli and V. cholerae strains were routinely grown in lysogeny broth (LB) or on LB agar at 37°C. Induction of the ToxR virulence regulon was accomplished by culturing V. cholerae strains under AKI conditions as follows. Overnight LB broth cultures of the test strains were individually diluted (10−3) into 10 ml of AKI broth (15 g Bacto peptone, 4 g Difco yeast extract and 5 g of NaCl per liter [pH 7.4]) in 150- × 15-mm glass test tubes (22). The inoculated test tubes were then incubated statically for 4 h at 37°C or until the OD600 reached ≥0.08 before the cultures were transferred to 125-ml Erlenmeyer flasks and incubated with shaking at 37°C for 1 h for the lux reporter assays or overnight for CT and TCP quantification. Carbenicillin and streptomycin were used at 100 μg/ml as necessary.
Chemicals and reagents.
Chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO). PAβN stock solutions were made in ultrapure water and filter sterilized before being aliquoted and stored at −20°C until needed. Enzymes for cloning were purchased from New England Biolabs (Beverly, MA), and oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and designed based on the V. cholerae C6706 genome (23).
Strain and plasmid construction.
The promoter probe vector pBBR-lux (24), which codes for chloramphenicol resistance, was used to generate transcriptional reporter plasmids for the V. cholerae ToxR regulon genes as follows. We amplified the ampicillin resistance gene from pBAD18 (25) using the AmpR-F (gcccgcctgatgaatgctcatccgggaattc-TGACGGATGGCCTTTTTGCGTTTCT) and AmpR-R (ctcaccgtctttcattgccatacgggaattc-TACAGGGCGCGTAAATCAATCTAAAG) PCR primers. These primers were tailed with homology (lowercase letters) to the DNA flanking the EcoRI site in pBBR-lux that intersected the chloramphenicol resistance gene to render the resulting plasmids ampicillin (Amp) resistant and chloramphenicol sensitive. The resulting PCR amplicon was then recombined by Gibson cloning into EcoRI-restricted pBBR-lux or pBBR-lux that already contained ToxR regulon promoter reporters to generate the following plasmids (26, 27): pTB17 (toxT-lux), pTB18 (pBBR-lux-Ampr; empty vector control), pTB19 (aphA-lux), pTB20 (tcpP-lux), pTB21 (toxR-lux), pTB22 (tcpA-lux), pTB23 (ctxA-lux), and pTB25 (aphB-lux). pTB32 (leuO-lux) was constructed by PCR amplifying the leuO promoters from pXB266 (21) using the pTL61T-F-SacI (ATGAGCTCGTTTGACAGCTTATCATCGGAGCTC) and pTL61T-R-BamHI (TTGGATCCGTCGGGATCGCTAGTTAGTTAGG) PCR primers. The resulting PCR amplicon was restricted with SacI and BamHI and cloned into similarly restricted pTB18 to generate pTB31 (leuO-lux). The reporter plasmids were sequence verified prior to use.
Virulence factor production.
Virulence factor production was assessed in all strains following overnight growth under AKI conditions in AKI medium (22). All cultures were normalized by optical density prior to assessing CT and TCP production. AKI culture supernatants were collected to quantify CT production using a GM1 enzyme-linked immunosorbent assay as previously described, with purified CT being used as a standard for quantitation (11). Cell pellets from the overnight AKI cultures were used to assess TCP production by Western immunoblotting for TcpA as previously described (11).
Transcriptional reporter assays.
V. cholerae strains bearing the indicated reporter plasmids were cultured under AKI growth conditions in AKI medium, with PAβN being added as indicated. Triplicate culture aliquots (200 μl) were collected at the indicated time points and transferred to the wells of a white microtiter plate with a clear bottom. Luminescence production and the optical density at 600 nm were then determined using a BioTek Synergy HT plate reader, with the results being reported as relative light units (RLU) divided by the optical density. The reported results are the average and standard deviation of at least three independent experiments.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health under award number R01AI132460.
The content is solely the responsibility of the authors.
Contributor Information
James E. Bina, Email: JBina@pitt.edu.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.Kaper JB, Morris JG, Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. 10.1128/CMR.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233. 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 3.Bina RF, Bina JE, Weng Y. 2020. Genome sequence of Vibrio cholerae strain RFB16, isolated from North Park Lake in Allegheny County, Pennsylvania. Microbiol Resour Announc 9:e00111-20. 10.1128/MRA.00111-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daboul J, Weghorst L, DeAngelis C, Plecha SC, Saul-McBeth J, Matson JS. 2020. Characterization of Vibrio cholerae isolates from freshwater sources in northwest Ohio. PLoS One 15:e0238438. 10.1371/journal.pone.0238438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty S, Nair GB, Shinoda S. 1997. Pathogenic vibrios in the natural aquatic environment. Rev Environ Health 12:63–80. 10.1515/reveh.1997.12.2.63. [DOI] [PubMed] [Google Scholar]
- 6.Childers BM, Klose KE. 2007. Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol 2:335–344. 10.2217/17460913.2.3.335. [DOI] [PubMed] [Google Scholar]
- 7.Midgett CR, Almagro-Moreno S, Pellegrini M, Taylor RK, Skorupski K, Kull FJ. 2017. Bile salts and alkaline pH reciprocally modulate the interaction between the periplasmic domains of Vibrio cholerae ToxR and ToxS. Mol Microbiol 105:258–272. 10.1111/mmi.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller VL, DiRita VJ, Mekalanos JJ. 1989. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol 171:1288–1293. 10.1128/JB.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almagro-Moreno S, Root MZ, Taylor RK. 2015. Role of ToxS in the proteolytic cascade of virulence regulator ToxR in Vibrio cholerae. Mol Microbiol 98:963–976. 10.1111/mmi.13170. [DOI] [PubMed] [Google Scholar]
- 10.Peterson KM, Gellings PS. 2018. Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants. Pathog Dis 76:ftx126. 10.1093/femspd/ftx126. [DOI] [PubMed] [Google Scholar]
- 11.Bina XR, Howard MF, Taylor-Mulneix DL, Ante VM, Kunkle DE, Bina JE. 2018. The Vibrio cholerae RND efflux systems impact virulence factor production and adaptive responses via periplasmic sensor proteins. PLoS Pathog 14:e1006804. 10.1371/journal.ppat.1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. 2015. Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J Bacteriol 197:3499–3510. 10.1128/JB.00419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunkle DE, Bina XR, Bina JE. 2020. Vibrio cholerae OmpR contributes to virulence repression and fitness at alkaline pH. Infect Immun 88. 10.1128/IAI.00141-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colclough AL, Alav I, Whittle EE, Pugh HL, Darby EM, Legood SW, McNeil HE, Blair JM. 2020. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol 15:143–157. 10.2217/fmb-2019-0235. [DOI] [PubMed] [Google Scholar]
- 15.Bina JE, Mekalanos JJ. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun 69:4681–4685. 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. IAI 76:3595–3605. 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bina XR, Philippart JA, Bina JE. 2008. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J Antimicrob Chemother 63:103–108. 10.1093/jac/dkn466. [DOI] [PubMed] [Google Scholar]
- 18.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thelin KH, Taylor RK. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun 64:2853–2856. 10.1128/IAI.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldor MK, Mekalanos JJ. 1994. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun 62:72–78. 10.1128/IAI.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bina XR, Taylor DL, Vikram A, Ante VM, Bina JE. 2013. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). mBio 4:e00366-13. 10.1128/mBio.00366-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwanaga M, Yamamoto K. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J Clin Microbiol 22:405–408. 10.1128/JCM.22.3.405-408.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng Y, Bina XR, Bina JE. 2021. Complete genome sequence of Vibrio cholerae O1 El Tor strain C6706. Microbiol Resour Announc 10:e01301-20. 10.1128/MRA.01301-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer BK, Bassler BL. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 104:11145–11149. 10.1073/pnas.0703860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. 10.1128/JB.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Yang M, Peterfreund GL, Tsou AM, Selamoglu N, Daldal F, Zhong Z, Kan B, Zhu J. 2011. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci U S A 108:810–815. 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Stern AM, Liu Z, Kan B, Zhu J. 2010. Virulence regulator AphB enhances toxR transcription in Vibrio cholerae. BMC Microbiol 10:3. 10.1186/1471-2180-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacikova G, Skorupski K. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol 46:1135–1147. 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 29.Joelsson A, Liu Z, Zhu J. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect Immun 74:1141–1147. 10.1128/IAI.74.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard MF, Bina XR, Bina JE. 2019. Indole inhibits ToxR regulon expression in Vibrio cholerae. Infect Immun 87:e00776-18. 10.1128/IAI.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor DL, Ante VM, Bina XR, Howard MF, Bina JE. 2015. Substrate-dependent activation of the Vibrio cholerae vexAB RND efflux system requires vexR. PLoS One 10:e0117890. 10.1371/journal.pone.0117890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkle DE, Bina XR, Bina JE. 2017. The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 8:e00126-17. 10.1128/mBio.00126-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunkle DE, Bina TF, Bina XR, Bina JE. 2020. Vibrio cholerae OmpR represses the ToxR regulon in response to membrane intercalating agents that are prevalent in the human gastrointestinal tract. Infect Immun 88:e00912-19. 10.1128/IAI.00912-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minato Y, Siefken RL, Hase CC. 2011. TolC affects virulence gene expression in Vibrio cholerae. J Bacteriol 193:5850–5852. 10.1128/JB.05222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minato Y, Fassio SR, Wolfe AJ, Hase CC. 2013. Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Microbiology (Reading) ) 159:792–802. 10.1099/mic.0.064865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tipton KA, Farokhyfar M, Rather PN. 2017. Multiple roles for a novel RND-type efflux system in Acinetobacter baumannii AB5075. Microbiologyopen 6:e00418. 10.1002/mbo3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcalde-Rico M, Hernando-Amado S, Blanco P, Martinez JL. 2016. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 7:1483. 10.3389/fmicb.2016.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Ortega C, Olivares J, Martinez JL. 2013. RND multidrug efflux pumps: what are they good for? Front Microbiol 4:7. 10.3389/fmicb.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz C, Levy SB. 2014. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J Antimicrob Chemother 69:390–399. 10.1093/jac/dkt352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol 191:5283–5292. 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buckner MM, Blair JM, La Ragione RM, Newcombe J, Dwyer DJ, Ivens A, Piddock LJ. 2016. Beyond antimicrobial resistance: evidence for a distinct role of the AcrD efflux pump in Salmonella biology. mBio 7:e01916-16. 10.1128/mBio.01916-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horiyama T, Nishino K. 2014. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 9:e108642. 10.1371/journal.pone.0108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamarche MG, Deziel E. 2011. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS One 6:e24310. 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawano H, Miyamoto K, Yasunobe M, Murata M, Myojin T, Tsuchiya T, Tanabe T, Funahashi T, Sato T, Azuma T, Mino Y, Tsujibo H. 2014. The RND protein is involved in the vulnibactin export system in Vibrio vulnificus M2799. Microb Pathog 75:59–67. 10.1016/j.micpath.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Pearson JP, Van Delden C, Iglewski BH. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210. 10.1128/JB.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. 2009. Assessment of three resistance-nodulation-cell division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol 9:200. 10.1186/1471-2180-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Shaer S, Shaaban M, Barwa R, Hassan R. 2016. Control of quorum sensing and virulence factors of Pseudomonas aeruginosa using phenylalanine arginyl beta-naphthylamide. J Med Microbiol 65:1194–1204. 10.1099/jmm.0.000327. [DOI] [PubMed] [Google Scholar]
- 48.Hirakata Y, Kondo A, Hoshino K, Yano H, Arai K, Hirotani A, Kunishima H, Yamamoto N, Hatta M, Kitagawa M, Kohno S, Kaku M. 2009. Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa. Int J Antimicrob Agents 34:343–346. 10.1016/j.ijantimicag.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Kvist M, Hancock V, Klemm P. 2008. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol 74:7376–7382. 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]