ABSTRACT
Francisella tularensis causes the deadly zoonotic disease tularemia in humans and is able to infect a broad range of organisms including arthropods, which are thought to play a major role in Francisella transmission. However, while mammalian in vitro and in vivo infection models are widely used to investigate Francisella pathogenicity, a detailed characterization of the major Francisella virulence factor, a noncanonical type VI secretion system (T6SS), in an arthropod in vivo infection model is missing. Here, we use Galleria mellonella larvae to analyze the role of the Francisella T6SS and its corresponding effectors in F. tularensis subsp. novicida virulence. We report that G. mellonella larvae killing depends on the functional T6SS and infectious dose. In contrast to other mammalian in vivo infection models, even one of the T6SS effectors PdpC, PdpD, or OpiA is sufficient to kill G. mellonella larvae, while sheath recycling by ClpB is dispensable. We further demonstrate that treatment by polyethylene glycol (PEG) activates Francisella T6SS in liquid culture and that this is independent of the response regulator PmrA. PEG-activated IglC secretion is dependent on T6SS structural component PdpB but independent of putative effectors PdpC, PdpD, AnmK, OpiB1, OpiB2, and OpiB3. The results of larvae infection and secretion assay suggest that AnmK, a putative T6SS component with unknown function, interferes with OpiA-mediated toxicity but not with general T6SS activity. We establish that the easy-to-use G. mellonella larvae infection model provides new insights into the function of T6SS and pathogenesis of Francisella.
KEYWORDS: Francisella tularensis subsp. novicida, Galleria mellonella, tularemia, in vivo infection model, type VI secretion system activation and effectors, polyethylene glycol, T6SS
INTRODUCTION
Francisella tularensis is the causative agent of the deadly zoonotic disease called tularemia (1). The most virulent subspecies Francisella tularensis subsp. tularensis is considered a tier 1 select agent due to high infectivity in humans (50% lethal dose, <10 CFU) and a high mortality rate if left untreated (up to 60%) (1, 2). In Europe, the less virulent Francisella tularensis subsp. holarctica is most prevalent (1). The closely related Francisella tularensis subsp. novicida is often used as a model organism to study Francisella pathogenicity, as it has a high infectivity in mice but not in humans (2).
Francisella virulence depends on the Francisella pathogenicity island (FPI) (3). Interestingly, F. tularensis subsp. novicida encodes one FPI, while the more virulent subspecies F. tularensis subsp. tularensis and F. tularensis subsp. holarctica both possess two identical FPIs (4). The FPI encodes a noncanonical type VI secretion system (T6SS) (see Fig. S1 in the supplemental material) required for intracellular survival (4–7). T6SS is a contractile nanomachine that can translocate effector proteins into bacterial and eukaryotic cells (8). The T6SS consists of the following three subcomplexes: a membrane complex spanning the bacterial cell envelope; a baseplate complex harboring the spike and effectors; and a contractile, cytosolic sheath with inner tube (8). Contraction of the cytosolic sheath propels the inner tube with the spike and effectors into a target cell (9–11). Dynamics of sheath assembly and contraction can be visualized by live-cell fluorescence microscopy and serves as a readout for a functional T6SS (5, 12, 13).
For F. tularensis subsp. novicida, six secreted T6SS effectors were identified as follows: PdpC, PdpD, OpiA, and OpiB1, OpiB2, and OpiB3 (OpiB1–3) (14). While PdpC and PdpD are required for phagosomal escape, their exact mode of action remains elusive (15, 16). Interestingly, OpiA and OpiB1–3 are encoded outside the FPI at different genomic sites (14). OpiA was shown to be a bacterial phosphatidylinositol 3-kinase delaying phagosomal maturation (17). Conversely, the function of the three almost identical OpiB proteins is unclear (14). The FPI encodes additional components PdpE and AnmK, which are dispensable for T6SS assembly and dynamics, and thus may be putative effectors (5). However, the corresponding deletion mutants were indistinguishable from the parental strains in various infection models (5, 15, 18, 19).
Strikingly, Francisella is able to infect and survive in a wide range of hosts ranging from amoeba and insects to mammals (20–24). Although the primary niche of Francisella is phagocytic cells, such as macrophages, Francisella is able to infect a broad range of cells, including nonphagocytic cells such as HeLa cells, Drosophila melanogaster cells, or erythrocytes (6, 25–28). Furthermore, there is clear evidence that tularemia is transmitted either by aerosols, infected animals, or by arthropod vectors such as ticks (21, 29, 30). Particularly, the broad range of arthropods that are susceptible for Francisella infections suggests that arthropods may play a role in maintaining Francisella in the environment (24).
An increasingly used arthropod in vivo infection model for studying host-pathogen interactions as well as for antimicrobial drug testing is Galleria mellonella larva (31). G. mellonella larvae combine several advantages for research, such as low maintenance costs and few ethical problems, compared to mammalian in vivo infection models (32). Moreover, G. mellonella larvae contain a complex innate immune system, including phagocytic cells called hemocytes and a humoral response (31). A part of the humoral response is a melanization process required for encapsulation of pathogens (33). Melanization results in a color change of the larvae from a healthy yellow into different shades of brown and black depending on the strength of the immune response (34). Recently, the complete G. mellonella genome was sequenced, facilitating genetic manipulations in the future (35).
G. mellonella larvae were already used as an in vivo infection model for Francisella. However, these studies focused on initial characterization of inoculum concentrations and infection conditions for robust killing of G. mellonella larvae by various Francisella species (36–39). Crucially, in-depth characterization of the major Francisella virulence factor, the noncanonical Francisella T6SS, and its role in killing of G. mellonella larvae is lacking.
Here, we show that virulence of F. tularensis subsp. novicida in G. mellonella larvae depends on a functional T6SS. However, ClpB-mediated T6SS sheath recycling is less important than reported previously in mice and bone marrow-derived macrophages (BMDMs). In addition, the main T6SS effectors PdpC and PdpD were dispensable for killing G. mellonella larvae. In contrast to mammalian in vivo infection models, individual effectors PdpC, PdpD, or OpiA were sufficient to kill G. mellonella larvae in a manner comparable to the parental strain. We demonstrate that Francisella T6SS can be activated in vitro by polyethylene glycol (PEG) in a PmrA-independent manner and use this to show that AnmK affects OpiA-mediated killing of G. mellonella larvae without altering T6SS activity or IglC secretion. In summary, our results suggest that G. mellonella larvae serve as a suitable model for testing roles of uncharacterized Francisella genes in infection.
RESULTS
T6SS is required for efficient killing of larvae by F. tularensis subsp. novicida.
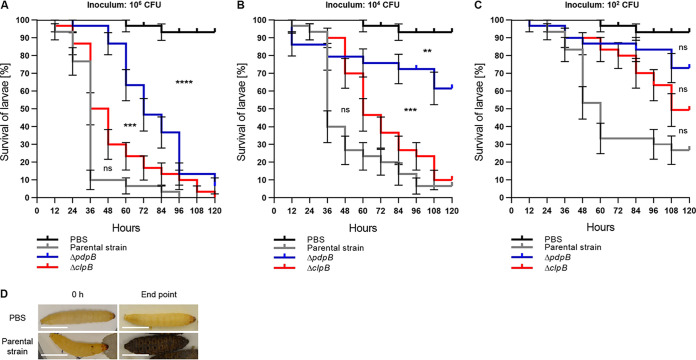
In order to characterize G. mellonella larvae as an in vivo infection model for Francisella, we first tested if F. tularensis subsp. novicida establishes infection in a T6SS-dependent manner. We used F. tularensis subsp. novicida U112 iglA-sfGFP (10) as the parental strain, which has T6SS sheath component IglA labeled with superfolder green fluorescent protein (sfGFP) and, thus, allows monitoring of T6SS dynamics by live-cell fluorescence microscopy (Fig. 1A; see also Movies S1 and S2 in the supplemental material) (5). We compared survival of G. mellonella larvae infected with the parental strain and that of a T6SS-negative control, in which pdpB, part of the T6SS membrane complex, was deleted (Fig. 1A; see also Fig. S1 and Movies S1 and S2 in the supplemental material) (5). G. mellonella survival was monitored for three different calculated inocula (106 CFU, 104 CFU, and 102 CFU per injection) and compared to a phosphate-buffered saline (PBS) control (Fig. 2A to C). Fifty percent of larvae infected with the parental strain were dead after 36 to 60 h, depending on the infection dose, while significantly more larvae infected with the T6SS-negative strain remained alive (Fig. 2A to C). PBS-treated G. mellonella larvae survived over 120 h (Fig. 2A to C). The infection dose of 104 CFU killed 93% of larvae when infected with the parental strain; however, less than 40% of larvae died when infected with the T6SS-negative strain after 120 h (Fig. 2B). Moreover, the parental F. tularensis subsp. novicida strain was able to robustly trigger an immune response in G. mellonella larvae, indicated by the melanization process and darkening of the larvae (Fig. 2D). In general, the killing rate for each strain and infection dose was reproducible over three independent infection experiments (see Fig. S2 in the supplemental material). While our data show that the Francisella T6SS is a major virulence factor in G. mellonella larvae, at higher infection doses, larvae infected with the T6SS-negative strain were also killed, suggesting that additional virulence factors play a role during infection.
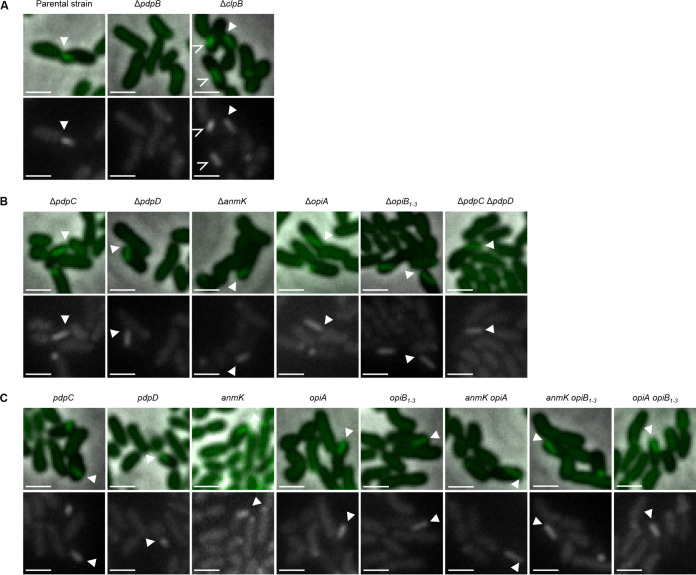
FIG 1.
T6SS assembly in F. tularensis subsp. novicida is independent of PdpC, PdpD, AnmK, OpiA, and OpiB1–3. All F. tularensis subsp. novicida mutants used in this study exhibit a functional T6SS. Filled arrows point to examples of assembled T6SS. Upper images are a merge of phase contrast and GFP channel. The lower images show GFP channel only. The 3.3- by 3.3-μm fields of view are shown. Scale bars represent 1 μm. (A) Assembly of IglA-sfGFP containing T6SS sheath in F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain) and the ΔclpB mutant. Empty arrows point to sfGFP aggregates in the F. tularensis subsp. novicida U112 iglA-sfGFP ΔclpB strain. No T6SS assembly was observed in the F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpB strain (T6SS-negative control). (B) Assembly of IglA-sfGFP containing T6SS sheath in F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpC, ΔpdpD, ΔanmK, ΔopiA, ΔopiB1–3, and ΔpdpC ΔpdpD strains. (C) Assembly of IglA-sfGFP containing T6SS sheath in F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpD ΔanmK ΔopiA ΔopiB1–3 (pdpC), ΔpdpC ΔanmK ΔopiA ΔopiB1–3 (pdpD), ΔpdpC ΔpdpD ΔopiA ΔopiB1–3 (anmK), ΔpdpC ΔpdpD ΔanmK ΔopiB1–3 (opiA), ΔpdpC ΔpdpD ΔanmK ΔopiA (opiB1–3), ΔpdpC ΔpdpD ΔopiB1–3 (anmK opiA), ΔpdpC ΔpdpD ΔopiA (anmK opiB1–3), and ΔpdpC ΔpdpD ΔanmK (opiA opiB1–3) strains.
FIG 2.
F. tularensis subsp. novicida kills G. mellonella larvae in a T6SS- and concentration-dependent manner. (A to C) Survival curves represent three individual experiments over 5 days pooled together (n0 total = 30; n0 = 10 per experiment). State of G. mellonella larvae was monitored every 12 h. Pupating larvae were censored (vertical dashes). Error bars indicate standard error. Black survival curves, PBS-treated G. mellonella larvae; gray survival curves, G. mellonella infected with F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain); blue survival curves, G. mellonella larvae infected with F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpB strain (T6SS-negative control); red survival curves, G. mellonella infected with F. tularensis subsp. novicida U112 iglA-sfGFP ΔclpB strain. Individual curves were compared with log rank (Mantel-Cox) test. P values above a Bonferroni-corrected threshold were considered nonsignificant (ns). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Following curves were compared. Parental strain versus ΔclpB mutant, ΔclpB mutant versus ΔpdpB mutant, and ΔpdpB mutant versus PBS control. Calculated infection inocula are as follows: 106 CFU (A), 104 CFU (B), and 102 CFU (C). (D) Representative examples of G. mellonella larvae morphology directly after PBS treatment and infection with F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain) at an infection dose of 104 CFU and after 120 h or 96 h, respectively.
ClpB and effectors PdpD and PdpC are less important for establishing infection in G. mellonella larvae than in mammalian infection models.
ClpB-mediated refolding of the T6SS sheath is essential for Francisella virulence in BMDMs and mice (5, 40). To test the role of ClpB in G. mellonella, we infected the larvae with a ΔclpB mutant (Fig. 1A; see also Fig. S1 and Movies S1 and S2). Surprisingly, a ΔclpB mutant killed G. mellonella larvae almost as efficiently as the parental strain (Fig. 2A to C). An average delay in killing of approximately 12 to 24 h was observed for the ΔclpB mutant for all infection doses, suggesting that while ClpB contributes to infection, it is largely dispensable.
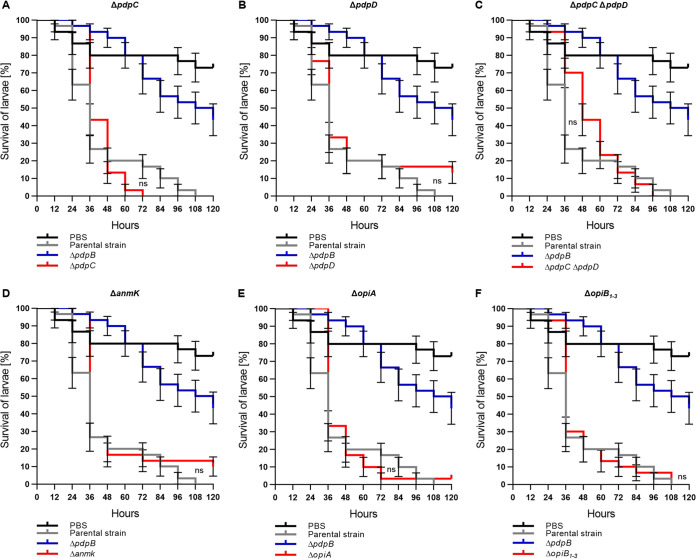
Next, we focused on the role of the FPI components, which are not required for T6SS assembly (5) (Fig. 1B; see also Fig. S1 and Movies S1 and S2). First, we tested the role of T6SS effectors PdpC and PdpD, which are secreted in a T6SS-dependent manner and have a major role in phagosomal escape in BMDMs and mice (5, 14–16). Since the infection with 104 CFU (Fig. 2B) resulted in the biggest survival difference between the parental strain and a T6SS-negative strain, we used this dose for all remaining infections. Surprisingly, single in-frame deletions of pdpC and pdpD had no effect on Francisella virulence in larvae (Fig. 3A and B). Even a ΔpdpC ΔpdpD double mutant, which is avirulent in BMDM and mice (5), killed the G. mellonella larvae as efficiently as the parental strain (Fig. 3C). We further analyzed the contribution of AnmK, a FPI component of unknown function (4) and the four secreted effectors OpiA and OpiB1–3, which are located outside of the FPI (14, 17) (Fig. 1B; see also Fig. S1 and Movies S1 and S2). Single deletion of either of the genes encoding these proteins had no effect on Francisella virulence in larvae (Fig. 3D to F; see also Fig. S3 in the supplemental material), suggesting that T6SS effectors play redundant roles in killing of G. mellonella.
FIG 3.
PdpC and PdpD are dispensable for T6SS-dependent Francisella virulence in G. mellonella. Survival curves represent three individual experiments over 5 days pooled together (n0 total = 30; n0 = 10 per experiment). State of G. mellonella larvae was monitored every 12 h. Pupating larvae were censored (vertical dashes). Error bars indicate standard error. Individual curves were compared with log rank (Mantel-Cox) test. P values above a Bonferroni-corrected threshold were considered nonsignificant (ns). Parental strain versus mutant curves were compared. Black survival curves, PBS-treated G. mellonella larvae; gray survival curves, G. mellonella infected with F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain); blue survival curves, G. mellonella larvae infected with F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpB strain (T6SS-negative control); red survival curves, G. mellonella infected with F. tularensis subsp. novicida U112 iglA-sfGFP (A) ΔpdpC mutant; (B) ΔpdpD mutant; (C) ΔpdpC ΔpdpD mutant; (D) ΔanmK mutant; (E) ΔopiA mutant; (F) ΔopiB1–3 mutant.
PdpC, PdpD, and OpiA alone are sufficient for killing of larvae.
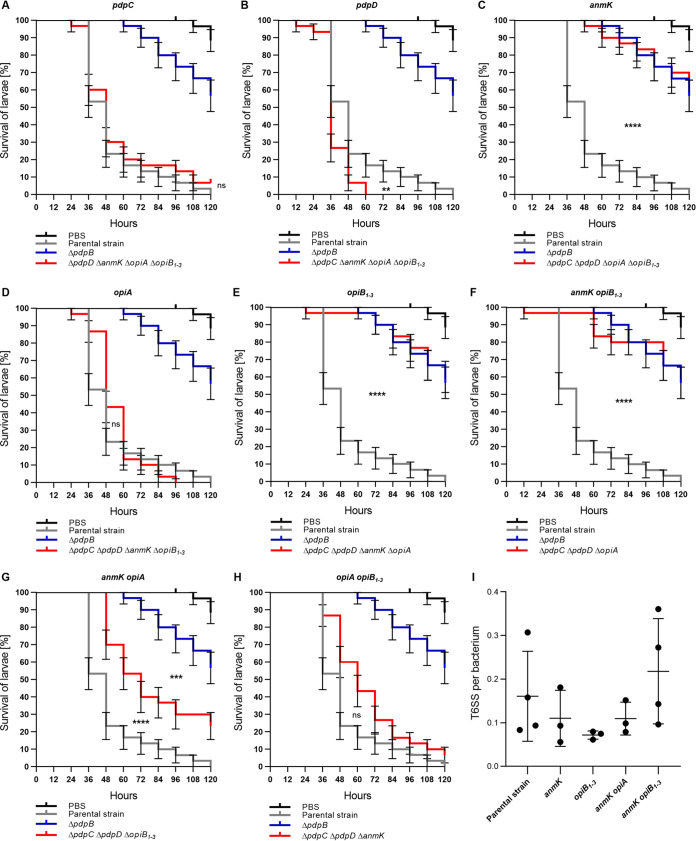
Since both PdpC and PdpD are dispensable for Francisella virulence in G. mellonella larvae, we hypothesized that either other effectors, such as OpiA and OpiB1–3, may compensate for the loss of PdpC and PdpD or that Francisella secretes additional T6SS effectors. To distinguish between these two possibilities, we first assessed if the previously identified T6SS effectors are individually sufficient to kill G. mellonella. We prepared strains where we deleted genes encoding all but one of the known or suspected effectors (PdpC, PdpD, AnmK, OpiA, and OpiB1–3) (see Fig. S1). Interestingly, the strains with PdpC, PdpD, and OpiA alone were as efficient in killing of larvae as the parental strain (Fig. 4A, B, and D; see also Fig. S4 in the supplemental material). A strain expressing only PdpD killed larvae even faster than the parental strain (Fig. 4B). In contrast, F. tularensis subsp. novicida strains with pdpC, pdpD, and opiA deleted (only anmK and/or opiB1–3 present) killed larvae at the same rate as the T6SS-negative strain (ΔpdpB mutant) (Fig. 4C, E, and F; see also Fig. S4). Interestingly, killing of G. mellonella larvae was significantly delayed in a strain with both anmK and opiA present compared to a strain having only opiA (Fig. 4G; see also Fig. S4). In contrast, the presence of opiB1–3 had no significant effect on OpiA-mediated killing of larvae (Fig. 4H; see also Fig. S4). Importantly, all of these strains assembled T6SS with a frequency and dynamics comparable to those of the parental strain (Fig. 1C and Fig. 4I; see also Movie S1 and S2). In summary, these results suggest that PdpC, PdpD, or OpiA are individually sufficient to kill G. mellonella larvae and that AnmK specifically reduces OpiA-mediated killing.
FIG 4.
PdpC, PdpD, and OpiA are sufficient for T6SS-dependent Francisella virulence in G. mellonella. Survival curves represent three individual experiments over 5 days pooled together (n0 total = 30; n0 = 10 per experiment). State of G. mellonella larvae was monitored every 12 h. Pupating larvae were censored (vertical dashes). Error bars indicate standard error. Individual curves were compared with log rank (Mantel-Cox) test. P values above a Bonferroni-corrected threshold were considered nonsignificant (ns). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Parental strain versus mutant curves were compared to each other. For panel G, mutant versus ΔpdpB strain curve was also compared. Black survival curves, PBS-treated G. mellonella larvae; gray survival curves, G. mellonella infected with F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain); blue survival curves, G. mellonella larvae infected with F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpB strain (T6SS-negative control); red survival curves, G. mellonella infected with F. tularensis subsp. novicida U112 iglA-sfGFP ΔpdpD ΔanmK ΔopiA ΔopiB1–3 mutant (pdpC) (A), ΔpdpC ΔanmK ΔopiA ΔopiB1–3 mutant (pdpD) (B), ΔpdpC ΔpdpD ΔopiA ΔopiB1–3 mutant (anmK) (C), ΔpdpC ΔpdpD ΔanmK ΔopiB1–3 mutant (opiA) (D), ΔpdpC ΔpdpD ΔanmK ΔopiA mutant (opiB1–3) (E), ΔpdpC ΔpdpD ΔopiA mutant (anmK opiB1–3) (F), ΔpdpC ΔpdpD ΔopiB1–3 mutant (anmK opiA) (G), and ΔpdpC ΔpdpD ΔanmK mutant (opiA opiB1–3) (H). (I) Quantification of T6SS sheaths per bacterium within 5 min for F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain), ΔpdpC ΔpdpD ΔopiA ΔopiB1–3 mutant (anmK), ΔpdpC ΔpdpD ΔanmK ΔopiA mutant (opiB1–3), ΔpdpC ΔpdpD ΔopiB1–3 mutant (anmK opiA), and ΔpdpC ΔpdpD ΔopiA mutant (anmK opiB1–3). At least three biological replicates with at least 3,200 bacteria each were analyzed per strain. Mean with standard deviation is shown. No significant differences in means were detected with Tukey’s multiple-comparison test and 95% confidence level.
Polyethylene glycol activates Francisella T6SS assembly and IglC secretion.
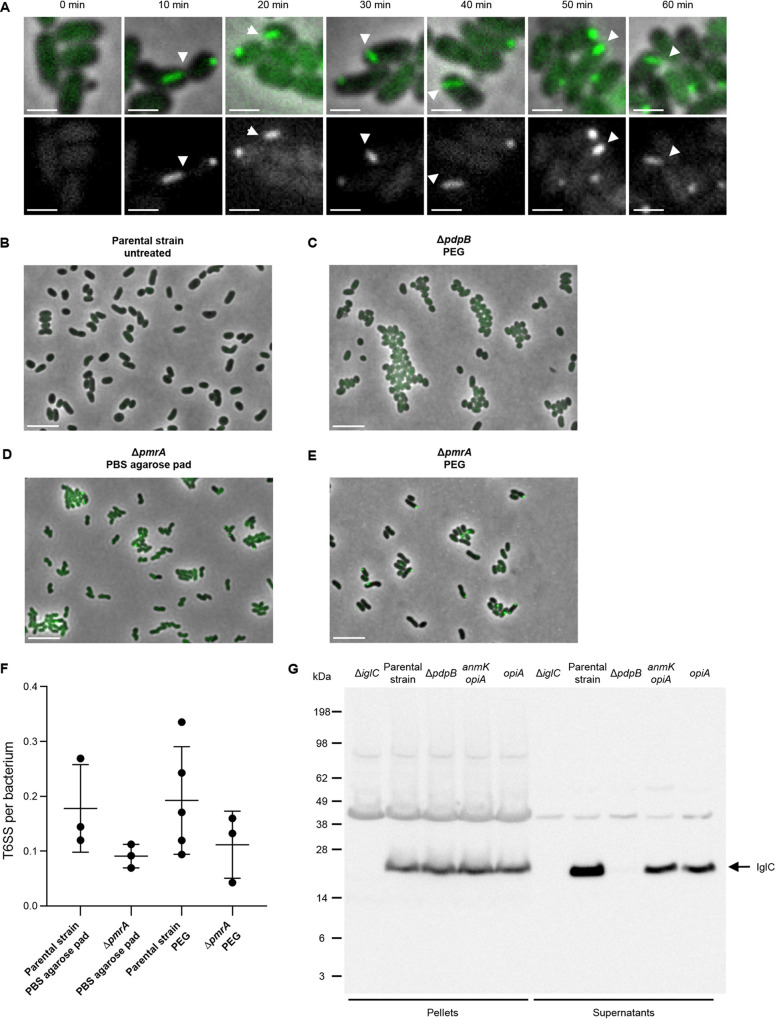
Previous work identified activation of T6SS expression and assembly by 5% KCl treatment (10) or by 30 to 60 min of incubation on PBS-agarose pads (5). Inspired by the observation that T6SS in Vibrio fischeri is activated by increasing the viscosity of the medium (41), we tested if similar conditions could activate F. tularensis subsp. novicida T6SS. We show that treatment of an exponentially growing culture of F. tularensis subsp. novicida U112 iglA-sfGFP by 10% polyethylene glycol 4000 (PEG) induces assembly of IglA-sfGFP into dynamic structures in less than 20 min (Fig. 5A). In contrast, no such structures were detected in untreated cells (Fig. 5B) or PEG-treated T6SS-negative mutant (ΔpdpB mutant) cells (Fig. 5C).
FIG 5.
PEG activates Francisella T6SS in liquid culture. (A) Examples of assembled T6SS (IglA-sfGFP) in F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain) during PEG treatment. Filled arrows point to examples of assembled T6SS. Upper images are a merge of phase contrast and GFP channel. The lower images show GFP channel only. The 3.3- by 3.3-μm fields of views are shown. Scale bars represent 1 μm. (B) No T6SS assemblies (IglA-sfGFP) were observed in untreated F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain) and in the ΔpdpB mutant (T6SS-negative control) (C) after PEG treatment for 60 min. (D) T6SS activation in F. tularensis subsp. novicida U112 iglA-sfGFP ΔpmrA mutant on PBS agarose pad after 60 min incubation. (E) T6SS assemblies in F. tularensis subsp. novicida U112 iglA-sfGFP ΔpmrA mutant after PEG treatment for 60 min. (B to E) Merge of phase contrast and GFP channel and 39- by 26-μm fields of view are shown. Scale bars represent 5 μm. (F) Quantification of T6SS sheaths per bacterium within 5 min. At least three biological replicates with at least 750 bacteria each were analyzed per strain and condition. Mean with standard deviation is shown. No significant differences in means were detected with Tukey’s multiple-comparison test and 95% confidence level. (G) Levels of inner tube protein IglC was assessed in bacterial pellets and concentrated supernatants of F. tularensis subsp. novicida U112 iglA-sfGFP (parental strain), ΔiglC mutant (negative control for α-IglC antibody), ΔpdpB mutant (T6SS-negative control), ΔpdpC ΔpdpD ΔopiB1–3 mutant (anmK opiA), and ΔpdpC ΔpdpD ΔanmK ΔopiB1–3 mutant (opiA) after PEG treatment for 1 h. Arrow points to IglC bands (theoretical size, 22.1 kDa). An exposure time of 1 min was used for developing the immunoblot.
In Francisella, the orphan response regulator PmrA is required for regulation of FPI expression and intracellular replication upon environmental cues (42–44). Thus, we hypothesized that PmrA could be involved in agarose pad-dependent and/or PEG-dependent Francisella T6SS activation. However, deletion of pmrA did not significantly change T6SS activity compared to the parental strain with either of the two activation methods (Fig. 5D to F). Moreover, we observed that T6SS dynamics and number of T6SS assemblies per cell upon PEG treatment were comparable to what was observed upon starvation on PBS-agarose pads (Fig. 5F).
The advantage of PEG treatment is that it activates T6SS assembly in liquid culture similarly to the previously used 5% KCl treatment (10, 14). Therefore, we tested if PEG treatment also results in T6SS-dependent IglC secretion. Indeed, IglC was secreted by T6SS-positive parental strain while no IglC was detected in the supernatant of the T6SS-negative mutant (ΔpdpB mutant) (Fig. 5G). We also used this IglC secretion assay to rule out that the observed AnmK-dependent modulation of F. tularensis subsp. novicida infection is due to its role in T6SS-mediated secretion. Importantly, both strains containing only opiA or opiA and anmK secreted IglC at comparable levels, albeit at a slightly lower level than that of the parental strain (Fig. 5G). This suggests that a general defect in T6SS function is an unlikely explanation for the observed AnmK-dependent decrease in virulence toward G. mellonella (Fig. 4G).
DISCUSSION
Since Francisella is able to infect a broad range of arthropods, it is important to understand Francisella pathogenicity in suitable infection models. Here, we characterized the contribution of the noncanonical T6SS and its known effectors to F. tularensis subsp. novicida pathogenicity in an in vivo arthropod infection model, namely, G. mellonella larvae. Our data show that F. tularensis subsp. novicida robustly kills G. mellonella larvae in a T6SS- and dose-dependent manner (Fig. 2; see also Fig. S2 in the supplemental material). These findings are in agreement with reports for other established in vivo infection models (4, 5, 45). Moreover, we could replicate the different dose-dependent killing dynamics with the parental strain reported previously (39). Interestingly, even a T6SS-negative strain killed some G. mellonella larvae, especially at high infection doses (Fig. 2; see also Fig. S2), suggesting that other virulence factors contribute to Francisella virulence in G. mellonella larvae. Indeed, Francisella encodes a variety of other bacterial virulence factors, such as type II secretion systems, type IV pili, outer membrane vesicles, nutritional virulence factors, as well as mechanisms to avoid oxidative stress and immune recognition (46–54).
We observed striking differences in the importance of individual T6SS components for F. tularensis subsp. novicida virulence in G. mellonella larvae compared to other mammalian in vivo infection models such as mice. First, T6SS sheath recycling and thus repeated T6SS firing is less important for Francisella pathogenicity in G. mellonella larvae than in BMDMs and mice (5). A ΔclpB mutant killed G. mellonella larvae slower but to the same extent as the parental strain (Fig. 2; see also Fig. S2). In contrast, a ΔclpB mutant was attenuated in Drosophila melanogaster, another arthropod in vivo infection model (25). It is important to note that the ΔclpB mutant is likely able to secrete a limited number of effectors because assembly and contraction of the T6SS sheath is independent of ClpB (5). Thus, one explanation for the observed difference between G. mellonella larvae and other in vivo infection models could be that G. mellonella cells are more sensitive to T6SS effectors or less capable of inhibiting the bacteria, and thus less effector translocation is sufficient for Francisella survival. Indeed, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica ΔclpB mutants are reported to replicate to higher numbers in J774A.1 cells than in bone marrow-derived macrophages, suggesting that some cell types may be more sensitive to T6SS effectors than others (40). Interestingly, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica ΔclpB mutants were less attenuated in mice than the F. tularensis subsp. novicida ΔclpB mutant (5, 40). However, both F. tularensis subsp. tularensis and F. tularensis subsp. holarctica encode two T6SS (4) and thus are potentially capable of secreting more effectors even with impaired T6SS compared to F. tularensis subsp. novicida. In summary, the general sensitivity to T6SS effectors as well as the number of translocation events may at least partially explain the variety of Francisella ΔclpB mutant phenotypes in different infection models.
Another striking difference in G. mellonella larvae compared to mice and other mammalian infection models is that Francisella virulence did not solely depend on T6SS effectors PdpC and PdpD (Fig. 3A to C; see also Fig. S3 in the supplemental material) (4, 5, 15, 16, 29, 55, 56). These results suggest that Francisella may manipulate different host cell components in insects and in mammal infection models or that arthropods are more sensitive to other T6SS effectors, such as OpiA. In agreement, single interruptions of pdpC and pdpD by transposons had no effect on Francisella virulence in Drosophila melanogaster or in a cell line derived from Anopheles gambiae (25, 57).
Interestingly, individual PdpC, PdpD, or OpiA effectors were sufficient to mediate Francisella virulence in G. mellonella larvae (Fig. 4A, B, and D; see also Fig. S4 in the supplemental material), which explains why deletion of pdpC and pdpD results in no change in virulence (Fig. 3C). In agreement, redundant functions for PdpC and OpiA were previously proposed (17). A strain with pdpC, anmK, opiA, and opiB1–3 deleted and only left with pdpD was even significantly faster in killing G. mellonella larvae than the parental strain (Fig. 4B). It remains to be determined if this is due to increased translocation rate of PdpD in the absence of other effectors.
While we cannot rule out that F. tularensis subsp. novicida encodes additional yet unidentified T6SS effectors, deletion of pdpC, pdpD, and opiA resulted in an attenuated phenotype in G. mellonella larvae comparable to that of a T6SS-negative mutant (Fig. 4F), and mutants with only anmK or opiB1–3 were also severely attenuated (Fig. 4C and E). Therefore, we conclude that PdpC, PdpD, and OpiA are the most important effectors for Francisella virulence in G. mellonella larvae.
Surprisingly, OpiA-mediated toxicity was affected by AnmK while general T6SS-dependent secretion was comparable to that of a fully virulent single opiA mutant (Fig. 4G and 5G; see also Fig. S4). Previously, no function of AnmK was observed in mice or macrophages (5, 15, 19, 25, 58). In contrast to OpiA, AnmK was never shown to be secreted (14). AnmK is predicted to contain an anhydro-N-acetylmuramic acid kinase domain, which is normally involved in peptidoglycan recycling (59), while OpiA was found to be a phosphatidylinositol 3-kinase delaying phagosomal maturation (17). It is possible that AnmK is a T6SS effector, which potentially competes with OpiA for secretion by T6SS. Another possibility is that either AnmK modulates OpiA expression levels or AnmK directly regulates OpiA function. Intriguingly, anmK is missing in F. tularensis subsp. holarctica and is expressed in two separate open reading frames in F. tularensis subsp. tularensis (15, 60). Surprisingly, the addition of opiB1–3 to anmK and opiA background (ΔpdpD ΔpdpC mutant) reverts the intermediate phenotype to parental strain-like killing of G. mellonella larvae (Fig. 3C). This shows that further studies are necessary to fully understand the role of these proteins in infection.
Several different environmental signals, such as biotin, iron limitation, pH changes, oxidative stress, or starvation, were identified to increase FPI transcription or IglC production (50, 61–64). Nevertheless, our understanding of what triggers T6SS assembly remains limited. Here, we show that PEG, next to KCl and incubation on PBS agarose pads, activates T6SS assembly in F. tularensis subsp. novicida (Fig. 5A) (5, 10). It is still unclear which physiological signal is mimicked by incubation on PBS agarose pads and 10% PEG treatment. However, we show that orphan response regulator PmrA is dispensable for both of the two T6SS activation methods (Fig. 5D to F). The demonstration that PEG activates Francisella T6SS expands the toolbox for Francisella T6SS research, as it allows robust T6SS activation that is compatible with downstream analyses without exposing cells to high KCl concentration, which could potentially stress the cells.
In summary, we demonstrate that G. mellonella larvae are an easy to handle and robust in vivo infection model for studying Francisella virulence and its T6SS. Moreover, this model makes it possible to uncover new functions and interactions between T6SS components as shown for AnmK and OpiA. Further investigations about why some effectors are more toxic in one infection model than another will lead to a more detailed understanding of the mode of action of different effectors. Intriguingly, a well-characterized arthropod in vivo model might help to study Francisella traits important for persistence in the environment and in potential reservoir hosts.
MATERIALS AND METHODS
Bacterial strains.
F. tularensis subsp. novicida U112 and derivative strains were grown in brain heart infusion (BHI) broth with aeration or on BHI agar plates at 37°C. The medium was supplemented with 0.1% l-cysteine (Acros Organics) and 100 μg/ml ampicillin (AppliChem) for overnight cultures and plates. Escherichia coli DH5α λpir and derivative strains were aerobically grown in Luria broth (LB) or on agar plates supplemented with 50 μg/ml kanamycin at 37°C. All strains are listed in Table S1 in the supplemental material.
Bacterial mutagenesis.
F. tularensis subsp. novicida in-frame deletion mutants were created with suicide vector pDMK3 (66) as reported previously (see Table S2 in the supplemental material) (5, 67). In brief, pDMK3 containing a DNA sequence of interest, including homology arms (750 bp each), was introduced into a donor E. coli strain from Harms and Dehio (68) and conjugated into F. tularensis subsp. novicida. For conjugation, liquid cultures of recipient F. tularensis subsp. novicida and donor E. coli strains were grown until an optical density at 600 nm (OD600) of 1 was reached. Day cultures were washed once in LB and 1 ml of both donor and recipient strain culture was concentrated and mixed together. Conjugation took place on an LB agar plate supplemented with 300 μM 2,6-diaminopimelic acid at 25°C overnight. Then, the mixture was transferred on Mueller-Hinton agar plates supplemented with 0.1% l-cysteine, 0.1% d-glucose (Millipore), 0.1% fetal calf serum (BioConcept), 100 μg/ml ampicillin, and 15 μg/ml kanamycin to select for recipients containing the suicide vector. After incubation at 37°C for 2 days, colonies were restreaked on BHI agar plates supplemented with 0.1% l-cysteine, 100 μg/ml ampicillin, and 15 μg/ml kanamycin. Negative selection was carried out on LB agar plates supplemented with 0.1% l-cysteine, 5% sucrose, and 100 μg/ml ampicillin, which were incubated at room temperature for several days. All plasmids and remaining peptides of in-frame deletions are listed in Table S2. All cloning products were sequenced, and sites of homologous recombination were verified by PCR.
Galleria mellonella infections.
Weight and aged defined Galleria mellonella larvae from TruLarv (BioSystems Technology) were used for all infection experiments. For each experiment and condition, 10 randomly chosen larvae were infected as previously described (69). F. tularensis subsp. novicida strains were prepared as follows. Day cultures of bacterial strains from plates were inoculated at an OD600 of 0.2 and grown without antibiotics as described above for 3 h. Then, cultures were washed once with Dulbecco’s phosphate saline buffer without CaCl2 and MgCl2 (PBS; Sigma), and OD600 was adjusted to 1 in PBS. Ten-fold dilutions in PBS were carried out. Ten microliters of the 108, 106, or 104 CFU/ml dilution (106, 104, or 102 CFU per injection) was used for injection into the second left proleg with a Hamilton syringe (10-μl volume, 26s ga bevel tip, needle length of 51 mm; Sigma-Aldrich). All infected larvae per condition were placed in one petri dish (Greiner Bio-One) and incubated at 37°C for 5 days. Survival was scored manually every 24 h. Death was defined as no movement of legs, head, or body. Pupated larvae were considered alive as long as they exhibited any movement but were censored and not considered for calculating the percentage of surviving larvae. As control for proper handling, each experiment included larvae injected with PBS.
Petri dishes with 10 dead G. mellonella larvae and after 5 days all remaining G. mellonella larvae were incubated at −20°C overnight before disposal.
Plating of inoculum.
The prepared 10-fold dilution series of F. tularensis subsp. novicida strains was also used to determine the actual inoculum concentration. A total of 100 μl of the calculated 103 CFU/ml dilution was plated on Mueller-Hinton agar plates supplemented with 0.1% l-cysteine, 0.1% d-glucose (Millipore), 0.1% fetal calf serum (BioConcept), and 100 μg/ml ampicillin. The plates were incubated for 2 days at 37°C, and colonies were counted afterward.
T6SS-dependent secretion assay.
Overnight cultures were washed twice with PBS and then resuspended in BHI and used for inoculation of day cultures without antibiotics at an OD600 of 0.2. After 3.5 h, the OD600 was adjusted to 2, and the bacterial cultures were washed twice with PBS and resuspended in 1 ml of BHI without l-cysteine. Then, 1 ml of 20% polyethylene glycol 4000 (Sigma-Aldrich) in BHI without l-cysteine was added so that a final PEG 4000 concentration of 10% was achieved. The cultures were incubated at 37°C shaking for 1 h. Afterward, 1 ml of the PEG 4000-treated samples was centrifuged at 16,000 × g for 1.5 min. A total of 0.9 ml of supernatant was used for trichloroacetic acid (TCA)/acetone protein precipitation. In brief, 100 μl of 100% TCA (wt/vol) (Sigma-Aldrich) was added to the harvested supernatants, followed by incubation at 4°C for 10 min with mixing in between. After centrifugation at 18,000 × g and 4°C for 5 min, the precipitated proteins were washed twice with cold acetone (Merk Millipore) and left to dry at room temperature. Then, the precipitated proteins were resuspended in 40 μl 1× lithium dodecyl sulfate (LDS) buffer (Thermo Fisher). The remaining bacterial cells were resuspended in 100 μl PBS, boiled at 95°C for 10 min, and sonicated afterwards. Thirty microliters of these samples were mixed with 10 μl 4× LDS buffer.
SDS-PAGE and Western blotting.
Samples prepared for the T6SS-dependent secretion assay (see above) were supplemented with 4 μl of 1 M dithiothreitol (DTT; Roche) and incubated at 72°C for 10 min. Then, 20 μl of the samples was loaded on 10% polyacrylamide gels, and proteins were separated by gel electrophoresis. For immunodetection, proteins were transferred to a nitrocellulose membrane (25 V for 45 min). After blocking of the nitrocellulose membrane in 5% milk in Tris‐buffered saline containing Tween 0.1% (TBST) at room temperature for 2 h and three washing steps with TBST for 5 min each, the nitrocellulose membrane was incubated with the primary antibody at room temperature for 2 h. Primary α-IglC antibody (polyclonal antibody raised in rabbit; Genescript) was used at a final concentration of 1 μg/ml in 5% milk in TBST. Incubation for 1 h with secondary antibody α-rabbit conjugated to horseradish peroxidase (Jackson ImmunoResearch) at a final concentration of 30 ng/ml in 5% milk in TBST followed. LumiGLO chemiluminescent substrate (KPL) was used for detection of peroxidase on a gel imager (GE ImageQuant LAS 4000). Exposure time is given in the figure legend.
Live-cell fluorescence imaging.
Microscope set up was described previously (5, 11, 70). The Nikon Ti-E inverted motorized microscope was equipped with a Perfect Focus system and a Plan Apo 1003 Oil Ph3 DM (NA 1.4) objective lens. Fluorescence was excited and filtrated with a SPECTRA X light engine (Lumencor) along with an ET-GFP (Chroma number 49002) filter set. The exposure time for each channel was set to 150 ms. Images were collected with a scientific complementary metal oxide semiconductor (sCMOS) camera pco.edge 4.2 with a pixel size of 65 nm (PCO) and VisiView software (Visitron). For imaging, day cultures of F. tularensis subsp. novicida parental and mutant strains were inoculated from plate at an OD600 of 0.2 without antibiotics. At an OD600 of 1, the cultures were concentrated in phosphate saline buffer to an OD600 of 10. A total of 1.5 μl of the concentrated cultures was then spotted on a pad consisting of 1% agarose in phosphate saline buffer. The agarose pad was covered with a cover slip and incubated at 37°C for 1 h before imaging at 30°C and 95% humidity (T-unit; Okolab). To monitor T6SS activation through PEG 4000 treatment (see T6SS-dependent secretion assay), 1.5 μl of liquid culture was spotted on a pad consisting of 1% agarose in BHI, covered by a coverslip, and imaged immediately. Images were collected every 30 s for 5 min.
Image analysis.
Image analysis was carried out with Fiji software (71) as previously described (5, 70, 72). Images in the same subfigure were set to the same contrast values for comparison of fluorescent signal intensities. For quantification of T6SS assemblies per bacterium within 5 min, the “temporal color code” function was used together with the “Cell Counter” plugin.
Statistical analysis.
Three infection experiments with independent G. mellonella larvae batches were performed. Mutants of a given set were tested in the same infection experiments. Pooled and single survival plots were calculated with Prism 8 (GraphPad Software). For more clarity, the graphs contain only data of the indicated mutant and the controls (G. mellonella larvae treated with PBS and infected with parental strain and T6SS) of the whole experiment. Thus, for a given set of mutants, the controls are the same for individual graphs. Standard errors were calculated for pooled survival plots. The log rank (Mantel-Cox) test in combination with Bonferroni corrected threshold (significance level, 0.05; number of comparisons, 6) was used to determine if compared curves are significantly different. P values are given in the figure legends.
Number of T6SS assemblies per bacterium was quantified in biological replicates. The smallest number of analyzed bacteria for a data set is given in the figure legend. Means with standard deviation were calculated. To test for significant differences in means, the Tukey’s multiple-comparison test with a confidence level of 95% in Prism 8 (GraphPad software) was used.
ACKNOWLEDGMENTS
We thank M. A. Horwitz (UCLA) for providing the strain F. tularensis subsp. novicida U112 iglA-sfGFP, D. M. Monack (Stanford University) for the conjugation plasmid pDMK3, A. Harms and C. Dehio (Biozentrum, University of Basel) for the E. coli conjugation strain, and C. Kemmer (BioVersys, Basel) for initial advice for how to handle G. mellonella larvae.
This work was supported by Swiss National Science Foundation (SNSF) grant BSSGI0_155778 and the University of Basel. M. Brodmann and S. T. Schnider were supported by the “Biozentrum PhD Fellowships” program.
We declare no competing financial interests.
M. Brodmann and M. Basler designed experiments as well as analyzed and interpreted the results. M. Brodmann generated strains and acquired all data except for Fig. 4I and Fig. 5E and F. S. T. Schnider acquired and analyzed data for Fig. 4I and Fig. 5E and F. M. Brodmann and M. Basler wrote the manuscript, and all authors approved it.
Footnotes
Supplemental material is available online only.
Contributor Information
Marek Basler, Email: marek.basler@unibas.ch.
Denise Monack, Stanford University.
REFERENCES
- 1.Oyston PCF, Sjostedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2:967–978. 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 2.Kingry LC, Petersen JM. 2014. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol 4:35. 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KKM, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol 186:6430–6436. 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bröms JE, Sjöstedt A, Lavander M. 2010. The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol 1:136. 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodmann M, Dreier RF, Broz P, Basler M. 2017. Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat Commun 8:15853. 10.1038/ncomms15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong A, Celli J. 2010. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol 1:138. 10.3389/fmicb.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker JR, Chong A, Wehrly TD, Yu J-J, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. 2009. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol 74:1459–1470. 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Brodmann M, Basler M. 2019. Assembly and subcellular localization of bacterial type VI secretion systems. Annu Rev Microbiol 73:621–638. 10.1146/annurev-micro-020518-115420. [DOI] [PubMed] [Google Scholar]
- 9.Brackmann M, Wang J, Basler M. 2018. Type VI secretion system sheath inter-subunit interactions modulate its contraction. EMBO Rep 19:225–233. 10.15252/embr.201744416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens DL, Ge P, Lee B-Y, Horwitz MA, Zhou ZH. 2015. Atomic structure of T6SS reveals interlaced array essential to function. Cell 160:940–951. 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudryashev M, Wang RY-R, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. 2015. Structure of the type VI secretion system contractile sheath. Cell 160:952–962. 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basler M, Mekalanos JJ. 2012. Type 6 secretion dynamics within and between bacterial cells. Science 337:815. 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basler M, Pilhofer M, Henderson P, Jensen JG, Mekalanos J. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eshraghi A, Kim J, Walls AC, Ledvina HE, Miller CN, Ramsey KM, Whitney JC, Radey MC, Peterson SB, Ruhland BR, Tran BQ, Goo YA, Goodlett DR, Dove SL, Celli J, Veesler D, Mougous JD. 2016. Secreted effectors encoded within and outside of the Francisella Pathogenicity island promote intramacrophage growth. Cell Host Microbe 20:573–583. 10.1016/j.chom.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludu JS, de Bruin OM, Duplantis BN, Schmerk CL, Chou AY, Elkins KL, Nano FE. 2008. The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J Bacteriol 190:4584–4595. 10.1128/JB.00198-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uda A, Sekizuka T, Tanabayashi K, Fujita O, Kuroda M, Hotta A, Sugiura N, Sharma N, Morikawa S, Yamada A. 2014. Role of pathogenicity determinant protein C (PdpC) in determining the virulence of the Francisella tularensis subspecies tularensis SCHU. PLoS One 9:e89075. 10.1371/journal.pone.0089075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledvina HE, Kelly KA, Eshraghi A, Plemel RL, Peterson SB, Lee B, Steele S, Adler M, Kawula TH, Merz AJ, Skerrett SJ, Celli J, Mougous JD. 2018. A phosphatidylinositol 3-kinase effector alters phagosomal maturation to promote intracellular growth of Francisella. Cell Host Microbe 24:285–295. 10.1016/j.chom.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bröms JE, Lavander M, Meyer L, Sjöstedt A. 2011. IglG and IglI of the Francisella pathogenicity island are important virulence determinants of Francisella tularensis LVS. Infect Immun 79:3683–3696. 10.1128/IAI.01344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. 2011. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology (Reading) 157:3483–3491. 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abd H, Johansson T, Golovliov I, SandströM G, Forsman M. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl Environ Microbiol 69:600–606. 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keim P, Johansson A, Wagner DM. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci 1105:30–66. 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 22.Lampe EO, Brenz Y, Herrmann L, Repnik U, Griffiths G, Zingmark C, Sjöstedt A, Winther-Larsen HC, Hagedorn M. 2016. Dissection of Francisella-host cell interactions in Dictyostelium discoideum. Appl Environ Microbiol 82:1586–1598. 10.1128/AEM.02950-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santic M, Ozanic M, Semic V, Pavokovic G, Mrvcic V, Kwaik YA. 2011. Intra-vacuolar proliferation of F. novicida within H. Vermiformis. Front Microbiol 2:78. 10.3389/fmicb.2011.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Telford SR, III, Goethert HK. 2020. Ecology of Francisella tularensis. Annu Rev Entomol 65:351–372. 10.1146/annurev-ento-011019-025134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ÅHlund MK, Rydén P, SjöStedt A, StöVen S. 2010. Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster. Infect Immun 78:3118–3128. 10.1128/IAI.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elkins KL, Cowley SC, Bosio CM. 2007. Innate and adaptive immunity to Francisella. Ann N Y Acad Sci 1105:284–324. 10.1196/annals.1409.014. [DOI] [PubMed] [Google Scholar]
- 27.Jones BD, Faron M, Rasmussen JA, Fletcher JR. 2014. Uncovering the components of the Francisella tularensis virulence stealth strategy. Front Cell Infect Microbiol 4:32. 10.3389/fcimb.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt DM, Barnes R, Rogerson T, Haught A, Mazzella LK, Ford M, Gilson T, Birch JW-M, Sjöstedt A, Reed DS, Franks JM, Stolz DB, Denvir J, Fan J, Rekulapally S, Primerano DA, Horzempa J. 2017. The role and mechanism of erythrocyte invasion by Francisella tularensis. Front Cell Infect Microbiol 7:173. 10.3389/fcimb.2017.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozanic M, Marecic V, Abu Kwaik Y, Santic M. 2015. The divergent intracellular lifestyle of Francisella tularensis in evolutionarily distinct host cells. PLoS Pathog 11:e1005208. 10.1371/journal.ppat.1005208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittwer M, Altpeter E, Pilo P, Gygli SM, Beuret C, Foucault F, Ackermann-Gäumann R, Karrer U, Jacob D, Grunow R, Schürch N. 2018. Population genomics of Francisella tularensis subsp. holarctica and its implication on the eco-epidemiology of tularemia in Switzerland. Front Cell Infect Microbiol 8:89. 10.3389/fcimb.2018.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai CJ-Y, Loh JMS, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramarao N, Nielsen-Leroux C, Lereclus D. 2012. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J Vis Exp 70:e4392. 10.3791/4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H. 2009. Regulation and function of the melanization reaction in Drosophila. Fly (Austin) 3:105–111. 10.4161/fly.3.1.7747. [DOI] [PubMed] [Google Scholar]
- 34.Cerenius L, Söderhäll K. 2004. The prophenoloxidase-activating system in invertebrates. Immunol Rev 198:116–126. 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 35.Lange A, Beier S, Huson DH, Parusel R, Iglauer F, Frick J-S. 2018. Genome sequence of Galleria mellonella (greater wax moth). Genome Announc 6:e01220-17. 10.1128/genomeA.01220-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmad S, Hunter L, Qin A, Mann BJ, van Hoek ML. 2010. Azithromycin effectiveness against intracellular infections of Francisella. BMC Microbiol 10:123. 10.1186/1471-2180-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aperis G, Fuchs BB, Anderson CA, Warner JE, Calderwood SB, Mylonakis E. 2007. Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect 9:729–734. 10.1016/j.micinf.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Propst CN, Pylypko SL, Blower RJ, Ahmad S, Mansoor M, van Hoek ML. 2016. Francisella philomiragia infection and lethality in mammalian tissue culture cell models, Galleria mellonella, and BALB/c mice. Front Microbiol 7:696. 10.3389/fmicb.2016.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thelaus J, Lundmark E, Lindgren P, Sjödin A, Forsman M. 2018. Galleria mellonella reveals niche differences between highly pathogenic and closely related strains of Francisella spp. Front Cell Infect Microbiol 8:188. 10.3389/fcimb.2018.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam A, Golovliov I, Javed E, Sjöstedt A. 2018. ClpB mutants of Francisella tularensis subspecies holarctica and tularensis are defective for type VI secretion and intracellular replication. Sci Rep 8:11324. 10.1038/s41598-018-29745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speare L, Smith S, Salvato F, Kleiner M, Septer AN. 2020. Environmental viscosity modulates interbacterial killing during habitat transition. mBio 11:e03060-19. 10.1128/mBio.03060-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell BL, Mohapatra NP, Gunn JS. 2010. Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect Immun 78:2189–2198. 10.1128/IAI.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohapatra NP, Soni S, Bell BL, Warren R, Ernst RK, Muszynski A, Carlson RW, Gunn JS. 2007. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun 75:3305–3314. 10.1128/IAI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey KM, Dove SL. 2016. A response regulator promotes Francisella tularensis intramacrophage growth by repressing an anti-virulence factor. Mol Microbiol 101:688–700. 10.1111/mmi.13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rick Lyons C, Wu TH. 2007. Animal models of Francisella tularensis infection. Ann N Y Acad Sci 1105:238–265. 10.1196/annals.1409.003. [DOI] [PubMed] [Google Scholar]
- 46.Forsberg A, Guina T. 2007. Type II secretion and type IV pili of Francisella. Ann N Y Acad Sci 1105:187–201. 10.1196/annals.1409.016. [DOI] [PubMed] [Google Scholar]
- 47.Hoang KV, Chen CG, Koopman J, Moshiri J, Adcox HE, Gunn JS. 2016. Identification of genes required for secretion of the Francisella oxidative burst-inhibiting acid phosphatase AcpA. Front Microbiol 7:605. 10.3389/fmicb.2016.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones CL, Sampson TR, Nakaya HI, Pulendran B, Weiss DS. 2012. Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell Microbiol 14:1531–1543. 10.1111/j.1462-5822.2012.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanistanon D, Powell DA, Hajjar AM, Pelletier MR, Cohen IE, Way SS, Skerrett SJ, Wang X, Raetz CRH, Ernst RK. 2012. Role of Francisella lipid A phosphate modification in virulence and long-term protective immune responses. Infect Immun 80:943–951. 10.1128/IAI.06109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lenco J, Pavkova I, Hubalek M, Stulik J. 2005. Insights into the oxidative stress response in Francisella tularensis LVS and its mutant ΔiglC1+2 by proteomics analysis. FEMS Microbiol Lett 246:47–54. 10.1016/j.femsle.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 51.Meibom KL, Charbit A. 2010. Francisella tularensis metabolism and its relation to virulence. Front Microbiol 1:140. 10.3389/fmicb.2010.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sampath V, McCaig WD, Thanassi DG. 2018. Amino acid deprivation and central carbon metabolism regulate the production of outer membrane vesicles and tubes by Francisella. Mol Microbiol 107:523–541. 10.1111/mmi.13897. [DOI] [PubMed] [Google Scholar]
- 53.Zarrella TM, Singh A, Bitsaktsis C, Rahman T, Sahay B, Feustel PJ, Gosselin EJ, Sellati TJ, Hazlett KRO. 2011. Host-adaptation of Francisella tularensis alters the bacterium’s surface-carbohydrates to hinder effectors of innate and adaptive immunity. PLoS One 6:e22335. 10.1371/journal.pone.0022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zogaj X, Chakraborty S, Liu J, Thanassi DG, Klose KE. 2008. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology (Reading) 154:2139–2150. 10.1099/mic.0.2008/018077-0. [DOI] [PubMed] [Google Scholar]
- 55.Lindgren M, Bröms JE, Meyer L, Golovliov I, Sjöstedt A. 2013. The Francisella tularensis LVS ΔpdpC mutant exhibits a unique phenotype during intracellular infection. BMC Microbiol 13:20. 10.1186/1471-2180-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long ME, Lindemann SR, Rasmussen JA, Jones BD, Allen L-AH. 2013. Disruption of Francisella tularensis Schu S4 iglI, iglJ, and pdpC genes results in attenuation for growth in human macrophages and in vivo virulence in mice and reveals a unique phenotype for pdpC. Infect Immun 81:850–861. 10.1128/IAI.00822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Read A, Vogl SJ, Hueffer K, Gallagher LA, Happ GM. 2008. Francisella genes required for replication in mosquito cells. J Med Entomol 45:1108–1116. 10.1093/jmedent/45.6.1108. [DOI] [PubMed] [Google Scholar]
- 58.Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, Brittnacher MJ, Manoil C, Skerett SJ, Salama NR. 2009. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun 77:232–244. 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nano FE, Schmerk C. 2007. The Francisella pathogenicity island. Ann N Y Acad Sci 1105:122–137. 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 61.Napier BA, Meyer L, Bina JE, Miller MA, Sjostedt A, Weiss DS. 2012. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc Natl Acad Sci U S A 109:18084–18089. 10.1073/pnas.1206411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. 2008. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun 76:2671–2677. 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murch AL, Skipp PJ, Roach PL, Oyston PCF. 2017. Whole genome transcriptomics reveals global effects including up-regulation of Francisella pathogenicity island gene expression during active stringent response in the highly virulent Francisella tularensis subsp. tularensis SCHU S4. Microbiology (Reading) 163:1664–1679. 10.1099/mic.0.000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng K, Blick RJ, Liu W, Hansen EJ. 2006. Identification of Francisella tularensis genes affected by iron limitation. Infect Immun 74:4224–4236. 10.1128/IAI.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reference deleted. [Google Scholar]
- 66.Lindgren H, Shen H, Zingmark C, Golovliov I, Conlan W, SjöStedt A. 2007. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect Immun 75:1303–1309. 10.1128/IAI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brodmann M, Heilig R, Broz P, Basler M. 2018. Mobilizable plasmids for tunable gene expression in Francisella novicida. Front Cell Infect Microbiol 8:284. 10.3389/fcimb.2018.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harms A, Segers FHID, Quebatte M, Mistl C, Manfredi P, Körner J, Chomel BB, Kosoy M, Maruyama S, Engel P, Dehio C. 2017. Evolutionary dynamics of pathoadaptation revealed by three independent acquisitions of the VirB/D4 type IV secretion system in Bartonella. Genome Biol Evol 9:761–776. 10.1093/gbe/evx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harding CR, Schroeder GN, Collins JW, Frankel G. 2013. Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J Vis Exp 81:e50964. 10.3791/50964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vettiger A, Basler M. 2016. Type VI secretion system substrates are transferred and reused among sister cells. Cell 167:99–110. 10.1016/j.cell.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 71.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4; Movie S1 and S2 legends; Tables S1 and S2. Download IAI.00579-20-s0001.pdf, PDF file, 1.39 MB (1.4MB, pdf)
Movie S1. Download IAI.00579-20-s0002.mp4, MP4 file, 6.17 MB (6.2MB, mp4)
Movie S2. Download IAI.00579-20-s0003.mp4, MP4 file, 17.1 MB (17.2MB, mp4)