Abstract
Neisseria gonorrhoeae , the bacterium responsible for the sexually transmitted disease gonorrhoea, has shown an extraordinary ability to develop antimicrobial resistance (AMR) to multiple classes of antimicrobials. With no available vaccine, managing N. gonorrhoeae infections demands effective preventive measures, antibiotic treatment and epidemiological surveillance. The latter two are progressively being supported by the generation of whole-genome sequencing (WGS) data on behalf of national and international surveillance programmes. In this context, this study aims to perform N. gonorrhoeae clustering into genogroups based on WGS data, for enhanced prospective laboratory surveillance. Particularly, it aims to identify the major circulating WGS-genogroups in Europe and to establish a relationship between these and AMR. Ultimately, it enriches public databases by contributing with WGS data from Portuguese isolates spanning 15 years of surveillance. A total of 3791 carefully inspected N. gonorrhoeae genomes from isolates collected across Europe were analysed using a gene-by-gene approach (i.e. using cgMLST). Analysis of cluster composition and stability allowed the classification of isolates into a two-step hierarchical genogroup level determined by two allelic distance thresholds revealing cluster stability. Genogroup clustering in general agreed with available N. gonorrhoeae typing methods [i.e. MLST (multilocus sequence typing), NG-MAST ( N. gonorrhoeae multi-antigen sequence typing) and PubMLST core-genome groups], highlighting the predominant genogroups circulating in Europe, and revealed that the vast majority of the genogroups present a dominant AMR profile. Additionally, a non-static gene-by-gene approach combined with a more discriminatory threshold for potential epidemiological linkage enabled us to match data with previous reports on outbreaks or transmission chains. In conclusion, this genogroup assignment allows a comprehensive analysis of N. gonorrhoeae genetic diversity and the identification of the WGS-based genogroups circulating in Europe, while facilitating the assessment (and continuous monitoring) of their frequency, geographical dispersion and potential association with specific AMR signatures. This strategy may benefit public-health actions through the prioritization of genogroups to be controlled, the identification of emerging resistance carriage, and the potential facilitation of data sharing and communication.
Keywords: antimicrobial resistance, molecular epidemiology, Neisseria gonorrhoeae, whole-genome sequencing
Data Summary
All Neisseria gonorrhoeae reads generated in this study have been deposited in the European Nucleotide Archive (ENA) (BioProject accession no. PRJEB36482; www.ebi.ac.uk/ena/data/view/PRJEB36482). ENA sample accession numbers are included in Table S1 (available with the online version of this article). The read datasets used were downloaded from the ENA (BioProject accession numbers PRJEB14933, PRJEB2124, PRJEB23008, PRJEB26560, PRJEB9227, PRJNA275092, PRJNA348107, PRJNA473385, PRJNA315363) and all sample accession numbers are included in Table S2. Supplementary material for this study was deposited in ZENODO and is available at https://doi.org/10.5281/zenodo.3946223.
Impact Statement.
National and international surveillance programmes are increasingly promoting the application of genomic data to track the main circulating lineages of Neisseria gonorrhoeae , and the emergence and spread of antimicrobial resistance (AMR). Using data from 3791 isolates collected in Europe, we report a comprehensive whole-genome-sequencing-based genogroup assignment for N. gonorrhoeae , based on the identification of the maximum discriminatory genetic thresholds reflecting cluster stability. The genogroups defined at these levels, which represent the main circulating lineages, were correlated with other typing techniques and further linked to specific AMR signatures. Insights on the clustering at a higher discriminatory level further supported the suitability of gene-by-gene typing strategies to detect/track outbreaks and transmission chains as a means to promote and/or support epidemiological investigation. This study enhances N. gonorrhoeae surveillance by promoting the prospective monitoring of genogroup frequency and geographical spread, towards more oriented public-health actions to control the spread of N. gonorrhoeae AMR.
Introduction
The disease burden of gonorrhoea has increased in the last decade and remains a major public-health concern worldwide. From 2012 up to 2016, the World Health Organization observed an increase from 78.3 to 87.0 million new cases per year of gonorrhoea worldwide [1, 2], with the European Centre for Disease Prevention and Control reporting around 90 000 notified cases in 2016 across 27 European Union/European Economic Area (EU/EEA) countries [3]. This sexually transmitted disease, caused by the bacterium Neisseria gonorrhoeae , usually affects the urogenital tract, causing urethritis in men and cervicitis in women, resulting in significant morbidity, but can also affect the anal canal and oropharyngeal tract causing proctitis and pharyngitis, respectively. Linked to disease burden, treatment strategies are being continuously challenged due to N. gonorrhoeae ’s ability to acquire antimicrobial resistance (AMR) to multiple classes of antimicrobials, including β-lactams, tetracyclines, macrolides and quinolones, after the acquisition of genes or mutations conferring resistance [4–7]. During the last decade, there has been a steady rise in minimum inhibitory concentrations (MICs) for cephalosporins, with resistant strains and clinical failures being described [8–14]. A worldwide increase in azithromycin resistance is also being reported [15–19], with resistant strains being associated with recent outbreaks [20–22].
With these last lines of treatment seemingly failing, and the fear that gonorrhoea might become untreatable, reaching a ‘superbug’ status, the World Health Organization, the European Centre for Disease Prevention and Control, and the Centers for Disease Control and Prevention (CDC) have issued action plans since 2012, raising the awareness of health-care professionals to define and monitor treatment failures of gonorrhoea worldwide [5]. In the absence of a vaccine, the management of N. gonorrhoeae mainly relies on preventive measures, an effective antibiotic treatment and epidemiological surveillance. As such, whole-genome sequencing (WGS) data are now being generated in order to monitor global N. gonorrhoeae epidemiology and AMR trends, with several countries already performing WGS on behalf of national or international surveillance programmes [23]. Nevertheless, during the last few years, N. gonorrhoeae molecular epidemiology studies, using WGS data, have been mostly focused on the reporting of circulating genogroups defined by typing strategies relying on just a few genomic loci, namely the traditional two gene N. gonorrhoeae multi-antigen sequence typing (NG-MAST) scheme, the seven-loci-based multilocus sequence typing (MLST), and the seven-loci N. gonorrhoeae sequence typing for antimicrobial resistance (NG-STAR) [24]. This has been performed, for instance, to group isolates based on the most prevalent sequence types (STs) [6] or to cluster isolates based on allelic differences (ADs) of both NG-MAST genes [25, 26]. Therefore, these approaches do not take advantage of the full potential of using whole-genome data for typing as well demonstrated by the recent publication of insightful WGS-based studies relevant for the understanding of N. gonorrhoeae evolution [27], population structure [28] and transmission of AMR determinants [29]. Nevertheless, N. gonorrhoeae exhibits a non-clonal population structure [30], due to frequent intraspecies horizontal gene transfer, leading to the need for comprehensive identification and tracking of circulating lineages or genetic groups for this pathogen [27, 28]. Achieving this goal has been a recurring challenge in molecular epidemiology due to the lack of approaches to quantitatively evaluate cluster stability and, therefore, identify genetic thresholds that define stable clusters representing main circulating lineages. In this context, the present study, focused on the European panorama (>3000 isolates from 21 countries), aims to perform WGS-based N. gonorrhoeae genogroup clustering for prospective WGS-based surveillance. Particularly, we aimed to identify the major circulating WGS-genogroups in Europe, assess their geographical spread and search for potential relationships between particular genogroups and specific AMR signatures. To achieve this, we used a large dataset of European gonococcal sequences available in public datasets, now enriched with 600 additional isolates spanning fifteen years of N. gonorrhoeae surveillance in Portugal.
Methods
Selection and characterization of Portuguese isolates
N. gonorrhoeae isolates analysed in the present study are part of the ongoing activity of the Portuguese National Laboratory Network for Neisseria gonorrhoeae Collection (PTGonoNet) hosted at the National Reference Laboratory for Sexually Transmitted Infections of the Portuguese National Institute of Health (NRL) [31]. Six hundred Portuguese isolates (herein designated as ‘PT isolates’) collected from 2003 up to 2017 from distinct specimens, spread across the country, and presenting different antimicrobial-susceptibility profiles were selected for analysis. Regarding antimicrobial-susceptibility testing, MICs for azithromycin, benzylpenicillin, cefixime, ceftriaxone, ciprofloxacin, gentamicin, spectinomycin and tetracycline were determined by Etest (bioMérieux), as previously described [31]. For 117 isolates from 2006 up to 2010, no MIC values were available for some antibiotics, as antibiotic susceptibility was determined by agar dilution breakpoint technique at the time by Public Health England, which provided solely qualitative results [32] (Table S1). All but 65 isolates were tested for β-lactamase production using the chromogenic reagent Nitrocefin (Oxoid), according to the manufacturer’s instructions. Isolate antibiotic resistance was classified according to the current European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint definitions [33]. From the 600 studied PT isolates, 491 isolates were subjected to WGS at the Portuguese National Institute of Health, and 109 isolates collected in 2013 were externally sequenced on behalf of the NRL participation in EURO-GASP (European Gonococcal Antimicrobial Surveillance Programme) [6]. Briefly, DNA was extracted from each isolate using the NucliSens easyMAG platform (bioMérieux) for total nucleic acid extraction, according to the manufacturer’s instructions. DNA was then subjected to Nextera XT library preparation (Illumina) prior to paired-end sequencing (2×250 bp or 2×150 bp) on either a MiSeq or a NextSeq 550 instrument (Illumina), according to the manufacturer’s instructions. Coded isolate designations, available anonymized metadata, antibiotic-susceptibility data and WGS details for all isolates are reported in Table S1.
Additional dataset for the European context and genome assembly
In order to obtain the current genetic diversity of N. gonorrhoeae circulating in European countries and to integrate the novel PT genomes in this global scenario, ultimately strengthening the analyses conducted in the present study, we took advantage of the public availability of 3263 genomes obtained from strains isolated in Europe. As such, WGS read datasets were download from the European Nucleotide Archive (ENA) from the following BioProjects: PRJEB14933 [20]; PRJEB2124 [34]; PRJEB23008 [35]; PRJEB26560 [12]; PRJEB9227 [6]; PRJNA275092 [36]; PRJNA348107 [37]; PRJNA473385 [38]; PRJNA315363 [39]. Available data for the isolates used, including sample accession numbers, are detailed in Table S2.
All genome sequences were assembled using the INNUca v4.0.1 pipeline (https://github.com/B-UMMI/INNUca), an integrative bioinformatics pipeline for read quality analysis and de novo genome assembly [40]. Briefly, read quality analysis and improvement is performed using FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic v0.36 [41] (with sample-specific read trimming criteria determined automatically based on FastQC report), respectively. Genomes were assembled with SPAdes v3.11 [42] and subsequently polished using Pilon v1.18 [43], with Quality Assurance/Quality Control statistics being monitored and reported throughout the analysis. For all isolates, assembly statistics including final genome assembly sizes, number of contigs and mean depth of coverage values are reported in Tables S1 and S2. Only 28 out of the 3861 genomes (2 PT and 26 publicly available genomes) were excluded after assembly, due to a genome size larger than expected, mean depth of coverage below 15× or the detection of contamination.
In silico typing and AMR prediction
For all validated assemblies, in silico seven loci MLST prediction was performed using mlst v2.4 software (https://github.com/tseemann/mlst), which is integrated within the INNUca pipeline. NG-MAST was performed using the ngmaster v0.4 software [44], with novel alleles and STs being assigned after submission to the international database of the NG-MAST website (http://www.ng-mast.net/). Additionally, rplF fine-typing [45] was performed in order to confirm Neisseria species, upon query to the PubMLST Neisseria database (http://pubmlst.org). Identification of AMR determinants in silico was performed using ariba v2.12.2 with the NG database [46], with all known variants analysed reported in Tables S1 and S2. Additional variants not classified in the ariba database as ‘known variants’ were also inspected [7, 47], namely: A39T and R44H in mtrR, 91K in parC and −35Adel in the mtrR promoter. We also performed isolate typing with NG-STAR [24], upon query to the PubMLST Neisseria database, for interpretation purposes. The results of this study were oriented towards the results provided by ariba, as it provides the ability to inspect more loci and, more importantly, variants that are carried heterogeneously.
Gene-by-gene analysis
Gene-by-gene analysis was performed by taking advantage of the publicly available panel of 1649 loci from the PubMLST Neisseria database ( N. gonorrhoeae cgMLST v1.0 [28], available at http://pubmlst.org), which was prepared in the present study for the chewBBACA core suite v2.0.11 [48] using the PrepExternalSchema module. This was a quality-control step to ensure that all loci are coding sequences as required by chewBBACA. Allele calling was performed on all assemblies using chewBBACA, with minimum blast score ratio to 0.6 and size threshold adjusted to 0.3 (i.e. alleles with size variation of 30 % to be tagged), and a training file generated by Prodigal v2.6.3 using the reference genome NCCP11945 (RefSeq accession number NC_011035). After inspection, the scheme was additionally curated by removing 16 loci that were flagged as ‘repeated loci’ after allele calling and another 39 loci that were only successfully called in less than 1 % of the genomes (Table S3). Exact and inferred matches were used to construct an allelic profile matrix, where other allelic classifications (see https://github.com/B-UMMI/chewBBACA/wiki) were assumed as ‘missing’ loci. Assemblies with less than 1545 loci called (~97 %) in the scheme were removed, which occurred only for 44 assemblies (19 PT and 25 publicly available) under analysis (detailed in Tables S1 and S2). Of note, the application of a more stringent cut-off was meant to increase confidence in the cluster stability analysis, although a lower cut-off (namely a ‘traditional’ 95% loci called) can be applied with this adapted scheme.
WGS-based genogroup classification
For WGS-based surveillance purposes, N. gonorrhoeae isolates were classified into two-step hierarchical WGS-based genogroups. For this, we used the maximum-shared loci of the PubMLST cgMLST scheme at 100 % by all 3791 validated genomes (upon assembly- and cgMLST-based exclusion), which resulted in 822 loci [herein designated as the maximum-shared cgMLST (MScgMLST) scheme; Table S4]. From the allelic profile matrix, the goeBURST algorithm [49, 50] implemented in the PHYLOViZ software [51] was used to generate clusters at all possible allelic distance thresholds [here expressed as the number of ADs over the total number of loci under analysis]. Cluster concordance was then assessed by the neighbourhood adjusted Wallace coefficient (nAWC), where all clusters partitioning at adjacent allelic distance thresholds (i.e. cut-off for ADs n and n+1) were compared using the comparing_partitions.py script (https://github.com/jacarrico/ComparingPartitions) [52, 53], as previously described [40, 54]. Following this, we searched for consecutive goeBURST thresholds over which cluster congruence is high (i.e. nAWC plateaus reflecting cluster stability) as a mean to define thresholds (or threshold ranges) useful for longitudinal surveillance, i.e. stable genogroups that represent major circulating lineages [40, 54]. Two stability phases, defined as the first and second intervals at which five or more consecutive thresholds yielded nAWC above 0.99, were used to cluster isolates into low- and high-level WGS-based genogroups, respectively. Each isolate was then classified into a specific genogroup designated as ‘GX.Y’, where X and Y are non-redundant arbitrary three digit numbers associated, respectively, to the high- and low-level WGS-based genogroup clustering, i.e. constituting two-step hierarchical genogroup definition directly associated with the two determined allelic distance thresholds. We further identified the goeBURST threshold ranges with the highest typing concordance with MLST and NG-MAST classifications using the adjusted Rand index (ARI) and adjusted Wallace coefficient (AWC) [52, 53]. Using the same rationale, cluster composition at both threshold levels defining WGS-based genogroups were compared with cluster composition defined by NG-MAST and MLST, where the Shannon index (SI) was used to measure cluster entropy with each partition. This was also performed to compare cluster composition between the MScgMLST scheme and the maximum shared loci obtained when using the cgMLST typing directly on the PubMLST website (i.e. 537 loci), with comparison only performed for the 1977 isolates where allelic profiles were available.
N. gonorrhoea e genogroups and genetic clustering analysis
A gene-by-gene allelic profile matrix was used to construct a minimum spanning tree (MST) using the goeBURST algorithm [50] implemented in the PHYLOViZ online web-based tool [51] based on 100 % shared loci between all isolates [49]. To take advantage of the maximum number of loci from the scheme (which may be key for discriminating potential epidemiological clusters), we used PHYLOViZ online 2.0 beta version (http://online2.phyloviz.net/), which allows maximizing the shared genome in a dynamic manner [40, 55]. As such, for each subset of isolates under comparison, the maximum number of shared loci between them is automatically used for MST construction. AD thresholds for cluster inspection are expressed as percentages of AD over the total number of shared loci under comparison for every analysis of each subset of isolates at any given level. As there is no established cut-off range for N. gonorrhoeae outbreak detection and contact tracing, we applied a conservative approach to investigate (and characterize at AMR level) sub-clusters with potential epidemiological links. In summary, after generating a sub-MST for each low-level genogroup, we applied a cut-off of 1.5 % AD, which represents half of the mean percentage of AD observed within each low-level genogroup of the full dataset.
Data availability
All reads generated for the present study were deposited in the ENA under the study accession number PRJEB36482 (individual run accession numbers are detailed in Table S1). The PubMLST N. gonorrhoeae cgMLST v1 scheme adapted for chewBBACA [48], the MScgMLST scheme, as well as allelic profile matrix for both schemes are available at https://doi.org/10.5281/zenodo.3946223. Additionally, a .json file also has been made available for direct upload into the GrapeTree visualization software [56] in order to interactively explore data/metadata.
Results
N. gonorrhoeae WGS-based genogroups
In the present study, 3791 N . gonorrhoeae genomes from isolates collected across Europe (21 EU/EEA countries) were analysed using the PubMLST N. gonorrhoeae cgMLST scheme. This scheme, enrolling 1594 loci, yielded 3623 unique allelic profiles (including loci not called). In order to determine WGS-based genogroups for long-term and large-scale surveillance, a shorter scheme was used, based on the loci shared (i.e. called) by all isolates. This scheme comprising 822 loci (Table S4) yielded 2422 unique allelic profiles (see Supplementary Material). After clustering using goeBURST, cluster composition was compared at all possible allelic distance thresholds using the nAWC (Fig. S1). Results showed that the two earliest cluster stability points are at 40 ADs (4.87 %; low-level) and at 79 ADs (9.61 %; high-level). Subsequently, we defined WGS-based genogroups at two-step hierarchical clustering levels taking into account these two thresholds. This assignment allows a comprehensive analysis of N. gonorrhoeae genetic diversity and the identification of the WGS-based genogroups circulating in Europe, while facilitating the assessment (and continuous monitoring) of their frequency, geographical dispersion and potential association with specific AMR signatures. As such, we sought to assess the weight of each genogroup (at both high- and low-levels) in the whole dataset. The low-level stability point (i.e. more discriminatory) yielded 321 clusters, of which 208 are represented by one or two isolates and 38 by more than ten isolates, while the high-level cut-off yielded 180 clusters, of which 106 are composed by one or two isolates and 35 by more than ten isolates. A total of 35 high-level (Table 1) and 38 low-level genogroups (Table 2) composed of at least ten isolates comprise 91.6 and 84.4 % of the whole dataset analysed, respectively. Thus, we opted to perform all downstream analysis focusing particularly on these more represented genogroups. The mean allelic diversity within low-level genogroups composed of at least 10 isolates was of ~3 % (AD=25), ranging from 0.2 (AD=2) up to 8.5 % (AD=70) (Fig. S2), while for high-level genogroups it was ~5 % (AD=41), ranging from 0.2 up to 10.7 % (AD=88). When the number of shared loci was maximized within each genogroup (mean of 1.8-fold increase for all genogroups), the mean allelic diversity for low-level was observed at 3.5 %, ranging from 0.2 up to 9.9 % (AD=81), and 5.3 % for high-level, ranging from 0.2 up to 12.4 % (AD=102). These results suggest that diversity within each WGS-based genogroups remains similar when the number of loci under comparison is increased (i.e. when the number of shared loci approaches the entire cgMLST scheme), and that the typing resolution (i.e. degree of genetic relatedness between isolates) is not compromised by using the MScgMLST scheme. For instance, for genogroup G001.002, composed by 491 isolates, when the number of shared loci under analysis increased from 822 to 1408, the mean AD only increased from 4.12 to 4.76 %.
Table 1.
Summary description of the high-level WGS-based genogroups enrolling at least ten isolates
|
High-level WGS-based genogroup |
Most common MLST ST (%) |
2nd most common MLST ST (%) |
Most common NG-MAST ST (%) |
2nd most common NG-MAST ST (%) |
Most common NG-STAR ST (%) |
2nd most common NG-STAR ST (%) |
No. of enrolled isolates |
No. of enrolled countries |
Isolate collection date interval |
No. of enrolled countries with at least five isolates |
|---|---|---|---|---|---|---|---|---|---|---|
|
G001 |
9363 (38.2) |
1580 (18.6) |
2992 (40.1) |
9768 (7.0) |
63 (42.6) |
301 (8.0) |
817 |
20 |
2003–2017 |
15 |
|
G002 |
1901 (82.8) |
1579 (6.3) |
1407 (48.8) |
2212 (3.0) |
90 (72.5) |
89 (2.3) |
574 |
20 |
2004–2017 |
19 |
|
G005 |
1901 (98.5) |
11992 (0.4) |
225 (67.2) |
5967 (3.6) |
26 (70.4) |
310 (6.9) |
274 |
15 |
2004–2017 |
5 |
|
G004 |
7363 (97.6) |
1587 (1.4) |
2400 (55.9) |
6360 (14.2) |
158 (74.9) |
419 (9.0) |
211 |
17 |
2010–2017 |
8 |
|
G003 |
11990 (60.4) |
1594 (35.6) |
51 (35.1) |
25 (34.2) |
20 (59.4) |
722 (9.4) |
202 |
10 |
2005–2017 |
4 |
|
G006 |
1579 (100.0) |
– |
21 (32.0) |
1034 (19.3) |
139 (94.7) |
856 (2.0) |
150 |
13 |
2004–2017 |
7 |
|
G007 |
1918 (99.3) |
10305 (0.7) |
12 (57.3) |
44 (32.9) |
239 (89.5) |
1510 (10.5) |
143 |
1 |
1995–2014 |
1 |
|
G010 |
1584 (86.6) |
11172 (9.0) |
26 (62.7) |
4528 (11.2) |
178 (89.6) |
410 (3.0) |
134 |
7 |
2002–2016 |
3 |
|
G012 |
8122 (100.0) |
– |
292 (75.2) |
210 (21.7) |
299 (76.7) |
85 (19.4) |
129 |
6 |
2003–2015 |
3 |
|
G009 |
7822 (95.0) |
11985 (1.7) |
4995 (67.2) |
10421 (19.3) |
416 (89.1) |
72 (2.5) |
119 |
11 |
2012–2017 |
3 |
|
G008 |
1588 (76.7) |
11247 (13.3) |
3785 (11.1) |
1582 (8.9) |
247 (18.9) |
870 (13.3) |
90 |
16 |
2005–2017 |
6 |
|
G011 |
11 516 (95.0) |
11 980 (5.0) |
1780 (58.8) |
5793 (16.3) |
56 (67.5) |
55 (27.5) |
80 |
6 |
2008–2016 |
2 |
|
G014 |
8143 (96.7) |
11968 (1.6) |
5624 (45.9) |
9918 (34.4) |
426 (42.6) |
436 (36.1) |
61 |
13 |
2012–2017 |
4 |
|
G025 |
1893 (100.0) |
– |
8517 (92.9) |
5993 (3.6) |
142 (100.0) |
– |
56 |
3 |
2011–2016 |
1 |
|
G013 |
1582 (72.2) |
11727 (13.0) |
147 (24.1) |
2997 (9.3) |
186 (72.2) |
896 (7.4) |
54 |
9 |
2003–2014 |
3 |
|
G018 |
1596 (100.0) |
– |
384 (40.5) |
190 (19.0) |
955 (66.7) |
307 (26.2) |
42 |
4 |
2003–2014 |
1 |
|
G019 |
1588 (100.0) |
– |
1479 (40.5) |
19083 (40.5) |
271 (48.6) |
433 (48.6) |
37 |
5 |
2005–2017 |
2 |
|
G015 |
8135 (97.1) |
11997 (2.9) |
987 (55.9) |
5120 (17.6) |
729 (94.1) |
New (2.9) |
34 |
10 |
2013–2017 |
3 |
|
G016 |
1599 (68.8) |
~1599 (31.3) |
645 (62.5) |
11461 (25.0) |
520 (100.0) |
– |
32 |
4 |
2013–2017 |
3 |
|
G017 |
8163 (58.6) |
11975 (31.0) |
2 (65.5) |
226 (10.3) |
84 (86.2) |
424 (6.9) |
29 |
7 |
2005–2015 |
2 |
|
G020 |
7826 (57.1) |
7359 (42.9) |
2487 (57.1) |
4186 (19.0) |
992 (57.1) |
231 (38.1) |
21 |
6 |
2008–2016 |
1 |
|
G021 |
8156 (100.0) |
– |
5441 (68.4) |
13489 (10.5) |
442 (84.2) |
982 (5.3) |
19 |
8 |
2013–2017 |
1 |
|
G022 |
1892 (100.0) |
– |
6129 (36.8) |
387 (26.3) |
563 (94.7) |
867 (5.3) |
19 |
5 |
2008–2013 |
1 |
|
G027 |
7827 (86.7) |
13489 (6.7) |
2318 (20.0) |
8845 (13.3) |
38 (46.7) |
1225 (13.3) |
15 |
4 |
2013–2017 |
1 |
|
G039 |
8776 (100.0) |
– |
1285 (100.0) |
– |
950 (100.0) |
– |
15 |
4 |
2012–2014 |
1 |
|
G023 |
11986 (100.0) |
– |
8465 (85.7) |
26 (7.1) |
162 (85.7) |
432 (7.1) |
14 |
2 |
2012–2015 |
1 |
|
G029 |
1588 (100.0) |
– |
10801 (42.9) |
11575 (42.9) |
969 (50.0) |
970 (21.4) |
14 |
2 |
2013 |
2 |
|
G024 |
10932 (76.9) |
11967 (15.4) |
5004 (15.4) |
4234 (15.4) |
169 (46.2) |
388 (30.8) |
13 |
5 |
2008–2014 |
1 |
|
G031 |
11 956 (91.7) |
8130 (8.3) |
New (83.3) |
7414 (16.7) |
2005 (83.3) |
949(16.7) |
12 |
3 |
2013–2015 |
1 |
|
G026 |
7367 (91.7) |
1579 (8.3) |
40 (16.7) |
18167 (16.7) |
New (33.3) |
733 (25.0) |
12 |
2 |
2003–2009 |
1 |
|
G030 |
1590 (100.0) |
– |
684 (45.5) |
5519 (18.2) |
1559 (45.5) |
190 (27.3) |
11 |
2 |
2004–2013 |
1 |
|
G041 |
11177 (100.0) |
– |
1993 (100.0) |
– |
568 (100.0) |
– |
11 |
1 |
2013 |
1 |
|
G032 |
1585 (100.0) |
– |
471 (60.0) |
752 (20.0) |
153 (80.0) |
913 (20.0) |
10 |
3 |
2004–2013 |
1 |
|
G028 |
8112 (100.0) |
– |
8149 (40.0) |
5560 (20.0) |
352 (60.0) |
468 (40.0) |
10 |
2 |
2008–2017 |
1 |
|
G036 |
8114 (100.0) |
– |
4 (40.0) |
~69 (30.0) |
46 (100.0) |
– |
10 |
1 |
2004–2011 |
1 |
Table 2.
Summary description of the low-level WGS-based genogroups enrolling at least ten isolates
|
Low-level WGS-based genogroup |
Most common MLST ST (%) |
2nd most common MLST ST (%) |
Most common NG-MAST ST (%) |
2nd most common NG-MAST ST (%) |
Most common NG-STAR ST (%) |
2nd most common NG-STAR ST (%) |
No. of enrolled isolates |
No. of enrolled countries |
Isolate collection date interval |
No. of enrolled countries with at least five isolates |
|---|---|---|---|---|---|---|---|---|---|---|
|
G002.001 |
1901 (83.9) |
1579 (6.6) |
1407 (55.8) |
2212 (3.4) |
90 (82.7) |
951 (2.6) |
502 |
20 |
2007–2017 |
18 |
|
G001.002 |
9363 (52.7) |
11428 (19.0) |
2992 (58.5) |
3935 (8.8) |
63 (69.7) |
67 (7.7) |
491 |
19 |
2009–2017 |
14 |
|
G005.004 |
1901 (99.0) |
11992 (0.4) |
225 (67.4) |
5967 (3.7) |
26 (70.7) |
310 (7.0) |
273 |
15 |
2004–2014 |
5 |
|
G004.003 |
7363 (97.6) |
1587 (1.4) |
2400 (56.2) |
6360 (14.3) |
158 (79.5) |
419 (8.6) |
210 |
17 |
2010–2017 |
8 |
|
G006.007 |
1579 (100.0) |
– |
21 (32.0) |
1034 (19.3) |
139 (94.7) |
856 (2.0) |
150 |
13 |
2004–2017 |
7 |
|
G001.005 |
1580 (77.0) |
8126 (19.6) |
9768 (38.5) |
359 (18.2) |
1996 (34.5) |
192 (27.7) |
148 |
7 |
2003–2017 |
4 |
|
G007.008 |
1918 (99.2) |
10305 (0.8) |
12 (62.1) |
44 (35.6) |
239 (88.6) |
1510 (11.4) |
132 |
1 |
1995–2000 |
1 |
|
G003.006 |
11 990 (93.1) |
1594 (4.6) |
25 (46.6) |
51 (43.5) |
20 (65.6) |
722 (14.5) |
131 |
5 |
2008–2017 |
1 |
|
G012.013 |
8122 (100.0) |
– |
292 (75.2) |
210 (21.7) |
299 (76.7) |
85 (19.4) |
129 |
6 |
2003–2015 |
3 |
|
G009.009 |
7822 (95.0) |
11985 (1.7) |
4995 (67.2) |
10421 (19.3) |
416 (89.1) |
72 (2.5) |
119 |
11 |
2012–2017 |
3 |
|
G010.010 |
1584 (88.1) |
11172 (10.2) |
26 (70.3) |
4528 (12.7) |
178 (95.8) |
423 (1.7) |
118 |
5 |
2002–2015 |
2 |
|
G011.011 |
11516 (95.0) |
11980 (5.0) |
1780 (58.8) |
5793 (16.3) |
56 (67.5) |
902 (3.8) |
80 |
6 |
2008–2016 |
2 |
|
G008.012 |
1588 (80.0) |
11247 (20.0) |
3785 (16.7) |
1582 (13.3) |
247 (23.3) |
567 (13.3) |
60 |
16 |
2005–2017 |
3 |
|
G001.015 |
1580 (59.0) |
11999 (39.3) |
995 (64.3) |
1313 (10.7) |
301 (42.9) |
245 (42.9) |
56 |
6 |
2005–2014 |
2 |
|
G025.030 |
1893 (100.0) |
– |
8517 (100.0) |
– |
142 (100.0) |
– |
52 |
3 |
2013–2016 |
1 |
|
G001.024 |
11864 (98.0) |
12521(2.0) |
2992 (78.4) |
~5230 (9.8) |
439 (86.3) |
453 (9.8) |
51 |
3 |
2013–2015 |
1 |
|
G001.014 |
9363 (100.0) |
– |
7445 (76.6) |
11500 (6.4) |
301 (70.2) |
965 (10.6) |
47 |
5 |
2013–2017 |
1 |
|
G019.022 |
1588 (100.0) |
– |
1479 (41.7) |
19083 (41.7) |
433 (50.0) |
271 (47.2) |
36 |
5 |
2005–2014 |
2 |
|
G015.016 |
8135 (97.1) |
11997 (2.9) |
987 (55.9) |
5120 (17.6) |
729 (94.1) |
~729 (2.9) |
34 |
10 |
2013–2017 |
3 |
|
G002.017 |
1901 (91.2) |
11107(8.8) |
3150 (14.7) |
2018 (14.7) |
127 (23.5) |
150 (17.6) |
34 |
7 |
2004–2017 |
2 |
|
G016.018 |
1599 (68.8) |
~1599 (31.3) |
645 (62.5) |
11461 (25.0) |
520 (100.0) |
– |
32 |
4 |
2013–2017 |
3 |
|
G014.020 |
8143 (96.7) |
11971 (3.3) |
5624 (93.3) |
1691 (3.3) |
426 (86.7) |
890 (6.7) |
30 |
9 |
2012–2017 |
3 |
|
G013.021 |
1582 (100.0) |
– |
147 (36.7) |
2997 (16.7) |
186 (66.7) |
752 (10) |
30 |
5 |
2003–2013 |
3 |
|
G018.025 |
1596 (100.0) |
– |
384 (56.7) |
190 (26.7) |
955 (93.3) |
307 (6.7) |
30 |
3 |
2003–2014 |
1 |
|
G017.019 |
8163 (55.6) |
11975 (33.3) |
2 (70.4) |
226 (7.4) |
84 (85.2) |
424 (7.4) |
27 |
7 |
2005–2015 |
2 |
|
G003.023 |
1594 (100.0) |
– |
51 (63.6) |
8148 (22.7) |
851 (77.3) |
2014 (13.6) |
22 |
2 |
2005–2017 |
1 |
|
G014.038 |
8143 (100.0) |
– |
9918 (95.5) |
15909 (4.5) |
436 (100.0) |
– |
22 |
2 |
2014–2017 |
1 |
|
G003.036 |
1594 (100.0) |
– |
10800 (68.4) |
11042 (31.6) |
20 (100.0) |
– |
19 |
3 |
2013 |
2 |
|
G021.029 |
8156 (100.0) |
– |
5441 (72.2) |
13489 (11.1) |
442 (88.9) |
New (5.6) |
18 |
7 |
2013–2017 |
1 |
|
G008.026 |
1588 (100.0) |
– |
3307 (43.8) |
9171 (12.5) |
249 (37.5) |
53 (25) |
16 |
6 |
2009–2013 |
1 |
|
G039.037 |
8776 (100.0) |
– |
1285 (100.0) |
– |
950 (100.0) |
– |
15 |
4 |
2012–2014 |
1 |
|
G023.027 |
11986 (100.0) |
– |
8465 (85.7) |
26 (7.1) |
162 (85.7) |
432 (7.1) |
14 |
2 |
2012–2015 |
1 |
|
G029.032 |
1588 (100.0) |
– |
10801 (42.9) |
11575 (42.9) |
969 (50.0) |
970 (21.4) |
14 |
2 |
2013 |
2 |
|
G013.028 |
11727 (53.8) |
7363 (46.2) |
1466 (30.8) |
4333 (23.1) |
186 (84.6) |
1347 (7.7) |
13 |
1 |
2005–2009 |
1 |
|
G020.031 |
7826 (100.0) |
– |
2487 (100.0) |
– |
992 (100.0) |
– |
12 |
3 |
2008–2013 |
1 |
|
G041.040 |
11177 (100.0) |
– |
1993 (100.0) |
– |
568 (100.0) |
– |
11 |
1 |
2013 |
1 |
|
G031.044 |
11956 (90.0) |
8130 (10.0) |
New (100.0) |
– |
2005 (100.0) |
– |
10 |
1 |
2014–2015 |
1 |
|
G036.034 |
8114 (100.0) |
– |
4 (40.0) |
~69 (30.0) |
46 (100.0) |
– |
10 |
1 |
2004–2011 |
1 |
From traditional to WGS-based typing
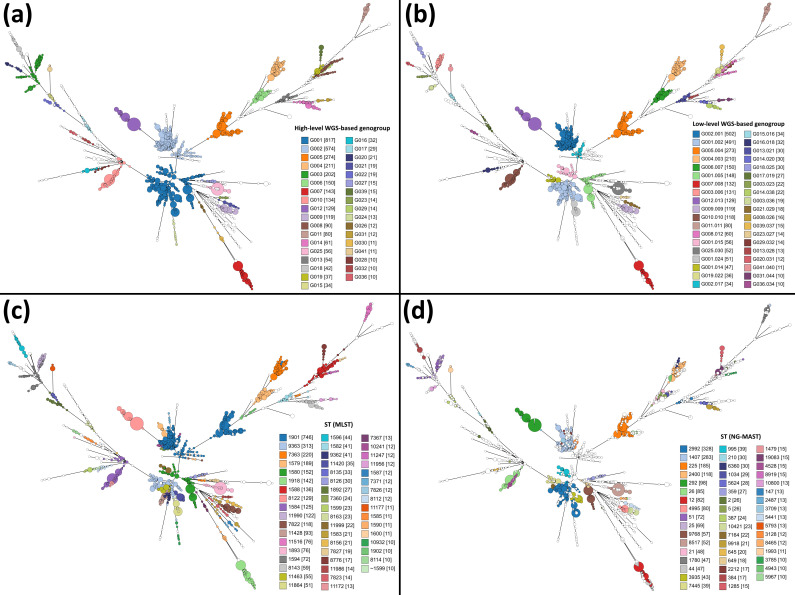
We then compared the relationship between the novel defined WGS-genogroups and the isolates’ classification based on traditional typing methods for N. gonorrhoeae (Fig. 1), namely MLST (which yielded ~180 profiles; SI=5.136) and NG-MAST (which yielded ~800 profiles; SI 7.431). Cluster congruence analysis between MLST types and WGS-genogroups showed that overall cluster congruence is slightly higher with the low-level WGS-genogroups than with the high-level (ARI=0.584 and 0.524, respectively). Nevertheless, the greater cluster agreement between MLST and WGS-genogroups was observed at the high level, as we observed a 64.2 % probability of isolates belonging to the same MLST type to also belong to the same high-level genogroup, and a probability of 54.6 % when evaluating the low-level (based on AWC analysis). The goeBURST threshold range that displayed simultaneously higher concordance and agreement with MLST was between 15.9 and 24.9 % AD (i.e. 131 and 205 ADs), where the highest AWCs were observed (between 0.660 and 0.663) with ARI values above 0.520 (up to 0.529) (Fig. S3). Regarding NG-MAST typing, we observed that the goeBURST threshold range that displayed higher congruence and agreement was between 4.4 and 6.8 % AD (i.e. 36 and 56 ADs). When compared with MLST analysis, this lower threshold range could be explained by a higher number of profiles observed in NG-MAST clustering. Nevertheless, although the results showed that the level of entropy in NG-MAST clustering is higher than with the other methods, the probability that a set of strains evidencing the same NG-MAST also belong to the same high- and low-level genogroups was very high (AWC of 0.979 and 0.887, respectively). This overall trend is especially relevant for abundant genogroups (Fig. 1, Tables 1 and 2), as there is a strong correspondence between particular MLST and NG-MAST types and high- and low-level genogroups. For example, all 52 isolates from genogroup G025.030 are from MLST ST1893 and NG-MAST ST8517. In contrast, isolates from genogroup G006.007 are from MLST ST1579, but present 25 distinct ST in NG-MAST typing (Table 2). Additionally, comparison with PubMLST website results for 1977 isolates (discriminated in Table S2) showed that cluster congruence was high with the novel defined WGS-genogroups, as ARI values were equal to 0.815 and 0.931 when cluster composition was compared at the high and low-level thresholds, respectively. Furthermore, we also compared the recently proposed core-genome groups [28], defined at 400 locus differences threshold (Ng_cgc_400) using the PubMLST cgMLST scheme, for 1830 isolates for which data were available (discriminated in Table S2) and were present in our dataset (Fig. S4). We observed the Ng_cgc_400 groups have a less discriminatory resolution than the two-stable-level WGS-based genogroup defined in the present study (Fig. S4b). More importantly, results showed that when the same cut-off of ~24 % (i.e. 400 ADs in the 1649 loci PubMLST scheme and 199 ADs in the 822 loci MScgMLST scheme) is applied to our data, there is a remarkable agreement between isolate grouping (Fig. S4a), reinforcing that our threshold might be scaled to the entire scheme while keeping the robustness of the clustering. In summary, the novel defined genogroups correlated well with MLST and NG-MAST classification, which largely facilitates backwards compatibility in the transition to WGS-based typing, as associations between ST and AMR profiles have been previously reported [6].
Fig. 1.
Comparison of WGS-based genogroups, defined at two levels, and both traditional typing methods for N. gonorrhoeae . The MSTs enrol all 3791 N . gonorrhoeae isolates based on the MScgMLST scheme (822 loci). Nodes, corresponding to a unique allelic profile, are coloured according to their corresponding (a) high-level WGS-based genogroup, (b) low-level WGS-based genogroup, (c) ST of the traditional seven loci MLST scheme and (d) ST of the two loci NG-MAST scheme. Numbers in parenthesis refer to the number of isolates comprising each genogroup or ST. The MSTs were generated using GrapeTree v1.5.0 software [56].
Distribution of N. gonorrhoeae genogroups by European country
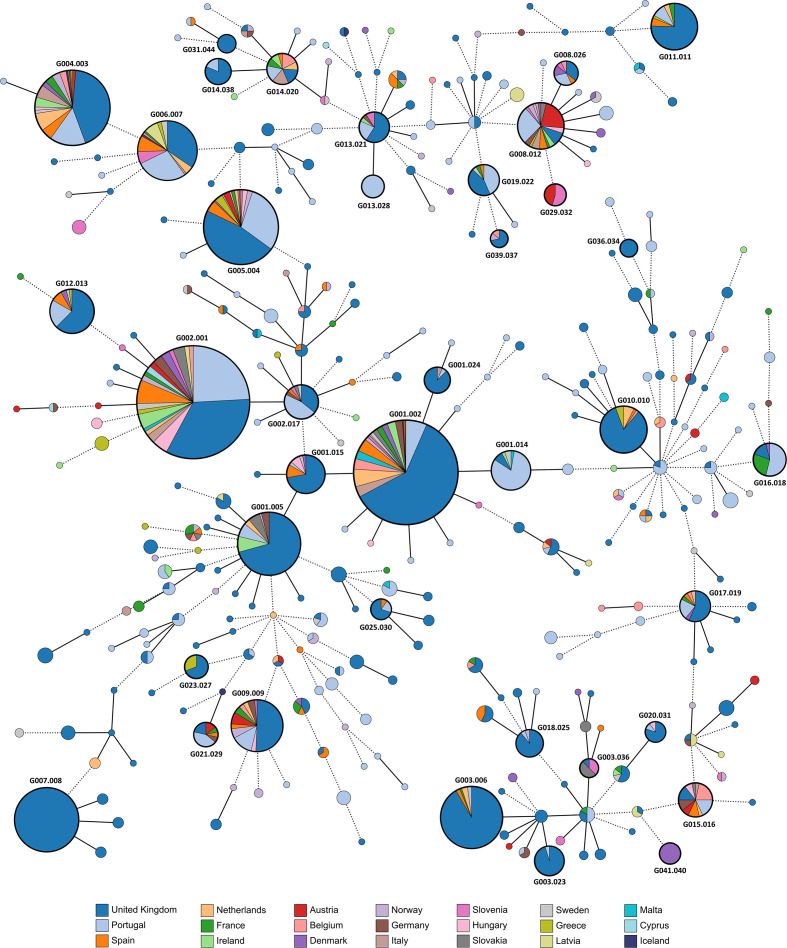
All novel PT isolates were integrated within a dataset of publicly available N. gonorrhoeae genomes, in order to assess their genomic diversity and phylogenetic relationships within the European circulating strains. As it stands, the dataset is highly represented by isolates from the UK (n=2263) and Portugal (n=579) in comparison with other European countries, some of which have less than ten isolates sequenced at the date of the analysis (e.g. Iceland and Cyprus). Global phylogenetic analysis, based on the MScgMLST scheme, revealed that the PT isolates presented high genetic diversity, being dispersed within the European genomic panorama (Fig. 2). PT isolates belonged to 115 distinct WGS-based genogroups, of which 32 were mainly (>50 %) constituted by PT isolates (e.g. G001.014, G016.018 and G013.028) and 60 contained a single isolate. This is a reflection of N. gonorrhoeae ’s global genetic diversity, as a total of 161 low-level genogroups were represented by a single isolate, and 81 genogroups were composed by isolates from a single country (Fig. S5), for example, G007.008 (UK), G013.028 (PT), G041.40 (Denmark), G031.044 (UK), G036.034 (UK), G046.056 (Greece), G053.086 (Slovenia), G170.243 (Latvia) and G002.050 (PT). Results showed that the more geographically widespread and abundant high-level genogroups were G001 and G002, enrolling strains from 20 countries, followed by G004, G008 and G005 enrolling isolates from 17, 16 and 15 countries, respectively (Table 1, Fig. 2). Even when fragmented into low-level genogroups, these were still within the more prevalent genogroups. In fact, 11 low-level genogroups were represented by isolates of at least six countries, of which three were composed by at least five isolates, namely G002.001 (NG-MAST ST1407), G001.002 (ST2992), G005.004 (ST225), G004.003 (ST2400), G006.007 (ST21), G001.005 (ST9768), G012.013 (ST292), G009.009 (ST4995), G008.012 (ST3785), G015.016 (ST977) and G014.020 (ST5624), with isolates being detected with time intervals of 5 up to 14 years apart (Table 2).
Fig. 2.
Phylogeny of 3791 N . gonorrhoeae isolates from Europe, based on a gene-by-gene approach using the MScgMLST scheme. The MST was constructed based on allelic diversity found among the 822 genes shared by 100 % of the isolates. All nodes (which represent a unique allelic profile) presenting an allelic distance below 40, corresponding to the low-level genogroup threshold, have been collapsed for visualization purposes. Nodes are coloured according to different countries of origin. Straight and dotted lines reflect nodes linked with the allelic distances below and above the threshold applied for high-level WGS-based genogroup definition (allelic distance 79), respectively. Low-level WGS-based genogroups comprised more than ten isolates are highlighted by thicker black circles. The MST was generated using GrapeTree v1.5.0 software [56].
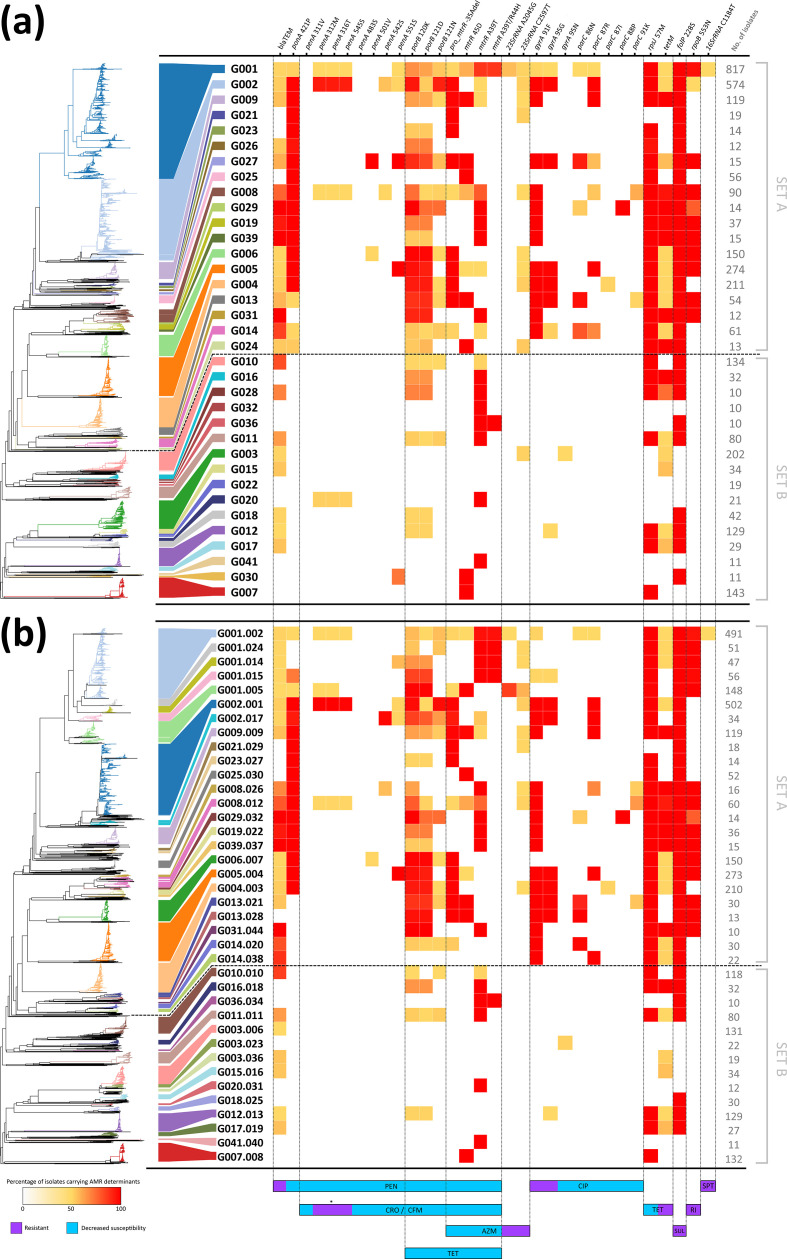
WGS-based genogroups carrying antibiotic-resistance determinants
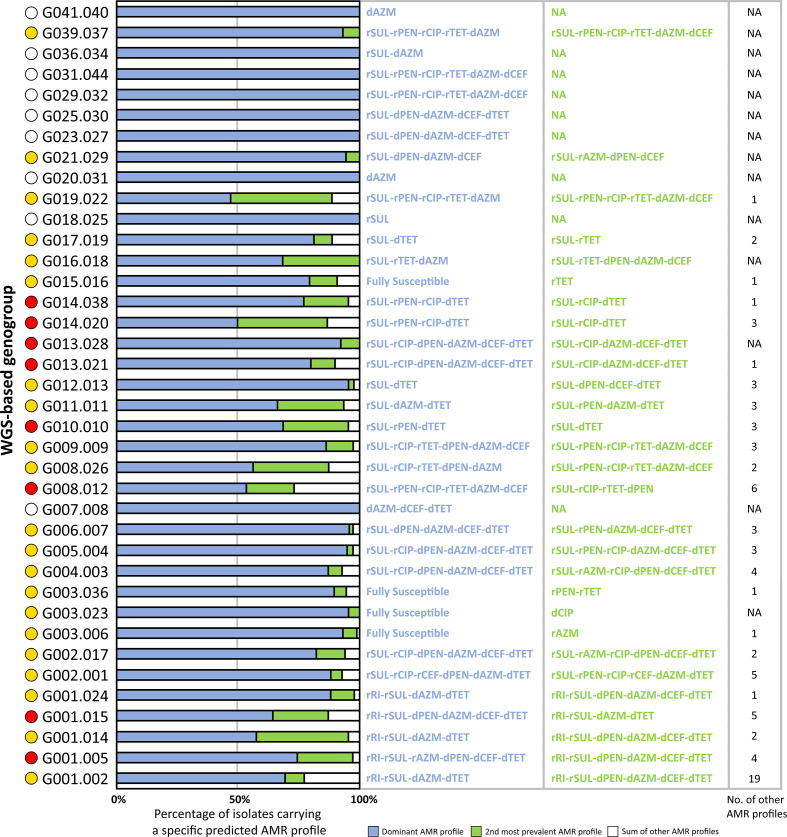
We analysed 32 genetic determinants known to be involved in decreased susceptibility and resistance to antimicrobials in N. gonorrhoeae (namely for penicillin, tetracycline, ciprofloxacin, azithromycin, cephalosporins, spectinomycin, sulphonamides and rifampicin), in order to assess their relationship with WGS-based genogroups at high- and low-levels (Fig. 3). Overall, results showed two distinct sets of isolates displaying a contrasting pattern of AMR, i.e. one with isolates carrying several genetic determinants related with decreased susceptibility and resistance (set A) and another with more susceptible isolates (set B). In fact, 15 out of 19 high-level genogroups from set A contained isolates independently resistant to at least four distinct antimicrobial drugs, contrasting with only 1 out of 16 high-level genogroups from set B (Fig. 3a). The same trend was observed at the low-level, with 19 out of 24 genogroups from set A and only 1 out of 14 genogroups from set B (Fig. 3b). In order to understand whether these resistant markers were carried together by the same isolates (i.e. multidrug resistance), we analysed the most dominant AMR profiles observed within each low-level genogroup (Fig. 4), as well as the dominant NG-STAR ST (Table 2). We observed that for nine genogroups all isolates displayed the same specific AMR profile. Furthermore, results showed that there were 21 genogroups where the most dominant AMR profile was not the one exhibiting the largest repertoire of AMR determinants. In contrast, for eight genogroups, the dominant AMR profile was the one associated with more AMR determinants. Several scenarios can justify both observations, but sampling bias hampers a more in depth analysis of the evolutionary and epidemiological trajectory of the involved genogroups.
Fig. 3.
Heatmap distribution and occurrence of the genetic determinants involved in AMR by high-level (a) and low-level (b) WGS-based genogroups. Genetic determinants are ordered by the affected antimicrobial drug class/antibiotic, with resistance or decreased susceptibility effect described at the bottom. Heatmap colour range correlates with the percentage of isolates carrying each genetic determinant within a given WGS-based genogroup. The numbers of isolates within each genogroup are presented on the right of each panel. The contextual neighbour-joining phylogenetic tree at the left side of each panel was generated based on the MScgMLST allelic profiles using GrapeTree v1.5.0 software [56]. The asterisk indicates that the combination of these three mutations has been proposed as potentially inducing resistance to cephalosporins [8]. AZM, Azithromycin; CFM, cefixime; CIP, ciprofloxacin; CRO, ceftriaxone; PEN, penicillins; RI, rifampicin; SPT, spectinomycin, SUL, sulphonamides; TET, tetracycline.
Fig. 4.
Major predicted AMR profiles observed within each low-level genogroup. Yellow circles indicate genogroups where the dominant AMR profile exhibits less genetic determinants associated with resistance than the second observed profile, for multiple classes of antimicrobials. Red circles indicate genogroups where the dominant AMR profile exhibits more genetic determinants associated with resistance for multiple classes of antimicrobials. White circles indicate genogroups with a unique AMR profile. AZM, Azithromycin; CEF, cephalosporins; CIP, ciprofloxacin; NA, not applicable; PEN, penicillins; RI, rifampicin; SUL, sulphonamides; TET, tetracycline. dXXX indicates decreased susceptibility to the named antibiotic, while rXXX indicates resistance to the named antibiotic.
Regarding specific genetic alterations, for both genogroup levels, resistance to ciprofloxacin was only observed in set A (with exception of one set B G012 resistant isolate from 2013), with all isolates from five different high-level genogroups simultaneously harbouring the resistance-mediating 91F and 95G amino acids in DNA-gyrase subunit A (encoded by gyrA; NG-STAR allele 1), and eight others displaying at least one of them. The same trend was observed for five mutations in the topoisomerase IV subunit C-encoding gene parC associated with decreased susceptibility to quinolones, which were exclusive of set A isolates (albeit dissimilarly spread across distinct high-level genogroups) (Fig. 3a). At the low-level (Fig. 3b), 15 genogroups were almost fully composed by ciprofloxacin-resistant isolates, with two others having only a few resistant isolates (G001.002 with 15 isolates from 2013 to 2015 and G001.0015 with one isolate from 2013). Amino acid alteration 553 N in the RNA polymerase subunit B (rpoB), conferring resistance to rifampicin [47], was exclusively observed in set A. Likewise, the azithromycin resistance-associated mutation C2597T in 23S rRNA was mostly observed in nine genogroups of set A. Furthermore, only isolates from high-level genogroup G001 (Fig. 3a) carried the mutation A2045G (NG-STAR allele 1), conferring high-level resistance to azithromycin. More importantly, this seemed to be exclusive to low-level genogroups G001.005 and G001.002 (Fig. 3b), with isolates having a fixed mutation (i.e. present in all 23S rRNA copies) or carrying it heterogeneously. Spectinomycin-resistance-associated mutation in 16S rRNA was only observed in one isolate from Sweden (G001.002) carrying it heterogeneously. Furthermore, resistance-associated markers for sulphonamides (dihydropteroate synthase-encoding gene folP), tetracycline (presence of the tetM-carrying conjugative plasmid) and penicillin (presence of the blaTEM plasmid, associated with β-lactamase production) were widespread across most of the genogroups. All markers seemed to be more frequent in genogroups from set A. For instance, all of them carried the mutation 228S in folP, while blaTEM and tetM were not found in only three and four high-level genogroups, respectively (Fig. 3a). Regarding the putative resistance to cephalosporins, results showed that most isolates from G002.001 (494 out of 502, ranging from 2007 to 2017) simultaneously possessed the three mutations (i.e. 312M, 316T and 545S) in the penicillin-binding protein 2-encoding gene (penA) that are known to potentially mediate resistance [6, 57], 97.0 % of which precisely carry the mosaic penA-XXXIV allele (NG-STAR allele 266, mostly associated with ST90) [58]. Besides G002.001 isolates, this mutational profile was only detected in 24 other isolates in the whole dataset (from 2013 to 2016, representing eight different genogroups). The whole penA-XXXIV allele was observed in 12 out these 24 isolates, which is suggestive of acquisition by recombination. Of note, other mutations in penA (inducing decreased susceptibility to penicillins and cephalosporins) were only observed in set A isolates. Concerning genetic markers associated with decreased susceptibility to penicillin, the mutation 421P in the penicillin-binding protein 1-encoding gene (ponA) was exclusively present in set A genogroups (Fig. 3); this mutation was carried by >90 % of isolates from 14 distinct low-level genogroups. Three mutations associated with decreased susceptibility to penicillins, cephalosporins and tetracycline, targeting the major porin-encoding gene porB (type porB1b), were observed in 1590 isolates from set A, contrasting with only 26 isolates from set B. Likewise, genetic alterations in the MtrR transcriptional regulator or its promoter, disrupting the MtrCDE efflux of substrate antimicrobials, were also mainly observed in set A. Of note, mutation A39T in mtrR was observed in most genogroups, but the combination of mutations A39T and R44H, associated with decreased susceptibility [59, 60], was only observed in G001 and G036. Finally, it is noteworthy that our data suggests that genogroups G007, G022, G032 and G041 were susceptible to all antimicrobials.
Association between N. gonorrhoeae genetic clustering and epidemiological data
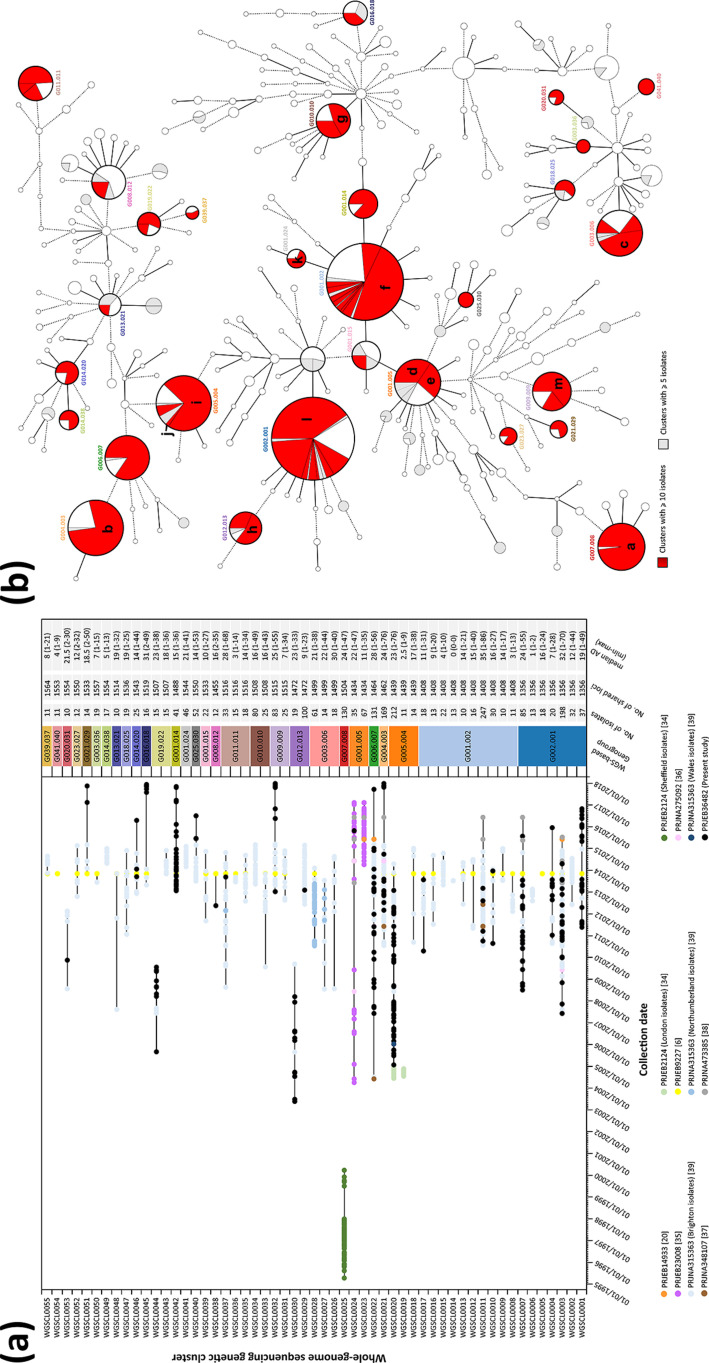
In order to analyse genetic clusters with the potential to be epidemiologically linked, we applied a conservative threshold of 1.5 % AD within each low-level genogroup, after sub-MST generation maximizing the shared loci between isolates at this level. As such, we observed a total of 315 genetic clusters spread out across 141 low-level genogroups. Results are presented for all clusters composed of more than ten isolates (Fig. 5a), with the phylogenetic distribution of all identified clusters with ≥5 isolates presented in Fig. 5(b) (full detailed data described in Tables S1 and S2). Expectedly, the more prevalent low-level genogroups presented the highest number of genetic clusters, namely G001.002, G002.001, G005.004 and G001.005 (Fig. 5b). Nevertheless, we also observed cases where a single large close-related genetic cluster was detected within a given genogroup (e.g. G023.027, G021.029, G041.040 and G001.014; see Fig. 5b). In another perspective, this analysis also highlighted genogroups disseminated at multi-country level with potential regional transmission chains. For instance, G015.016, which is composed by isolates from ten different countries, was fragmented into eight smaller genetic clusters with only two clusters presenting isolates from distinct countries. We also examined each cluster in a time scale by each isolate’s collection date (Fig. 5a), which allowed us to discriminate between clusters that potentially circulated within a specific time interval (and have not since been detected) from clusters that potentially emerged and others that are still circulating, even though this is reliant on the dataset and time period analysed. For example, WGSCL0025 (G007.008, mean AD 1.6 %) that enrols isolates spanning 5 years (1995 up to 2000) seems not to have been detected since, which relates to a described outbreak in Sheffield, UK [34]. This was also the case for WGSCL0044 (G019.022, mean AD 1.5 %) with isolates from 2005 up to 2009 and WGSCL0030 (G012.013, mean AD 1.6 %) with isolates from 2003 up to 2009 (Fig. 5a), although these did not have a described epidemiological link as they contained PT isolates and isolates from the UK [34]. In contrast, WGSCL0024 (G001.005, mean AD 1.5 %) and WGSCL0022 (G006.007, mean AD 1.9 %) were composed by isolates that have been consistently detected from 2004 up to 2017 (the latest year within the dataset). Regarding clusters enriched by PT isolates, this could be observed for instance in WGSCL0020 (G005.004, mean AD 1.6 %) with isolates from 2004 up to 2014, in WGSCL0042 (G001.014, mean AD 1.0 %), a more recent cluster dating from 2013 up to 2017, and WGSCL0001, from 2011 up to 2017. However, we believe that more genomic data is needed in order to reinforce these observations as they rely on the analysed database; for instance, data from the 2013 EURO-GASP survey [6] are a sub-sample restricted to the latter months of the year. Nevertheless, a relationship between the observed genetic clusters at this threshold and published data was clearly observed (Fig. 5). Eleven distinct subsets that were phylogenetically related (i.e. epidemiological verified outbreaks or verified transmission chains) were consistent with the obtained genetic clusters at a threshold of 1.5 %. This was the case for: WGSCL0028 (G003.006, mean AD 1.4 %), an outbreak in North-East England [39]; WGSCL0023 (G001.005, mean AD 0.8 %), an outbreak of high-resistance azithromycin isolates in Leeds [20] linked to isolates from other published data [35]; and WGSCL0020, an outbreak in London [34], which could be linked to 49 PT isolates, 19 isolates from the EURO-GASP 2013 survey [6], 51 isolates from Brighton [39] and 2 isolates from The Netherlands [37]. Of note, one PT isolate (NGPT15194) could be linked to the described high-resistance azithromycin N. gonorrhoeae isolates in Ireland and the UK (WGSCL0024), with sustained transmission [35].
Fig. 5.
Analysis of N. gonorrhoeae WGS-based genetic clusters at low-resolution level potentially concordant with epidemiological link. (a) Genetic cluster isolates’ distribution by collection date, with detailed data of each cluster presented on the right. Each isolate colour refers to a specific study, and black lines link the earliest and latest isolate detected. Numbers in parentheses in the figure key refer to the study reference. (b) MST (also described in Fig. 2) of all isolates analysed in the present study highlighting WGS genetic clusters identified at a conservative threshold of 1.5 % AD. Nodes (which represent a unique allelic profile) presenting an allelic distance below 40, corresponding to the low-level genogroup threshold, have been collapsed for visualization purposes. The letters within (b) represent the following: a, WGSCL0025 Sheffield outbreak described in [34]; b, WGSCL0021 including cluster ST2400 described in [39] and the ST2400 MSM-associated isolates described in [6]; c. WGSCL0028 North-East England outbreak described in [39]; d, WGSCL0024 clade 2 described in [35]; e, WGSCL0023 Leeds outbreak [20] plus clade 1 and 3 described in [35]; f, WGSCL0011 large cluster ST2992 described in [39]; g, WGSCL0034 cluster ST26 described in [39]; h, WGSCL0029 cluster ST292 described in [39]; i and j, WGSCL0019 and WGSCL0020 London outbreaks described in [34]; k, WGSCL0041 small cluster ST2992 described in [39]; l, WGSCL0003 cluster ST1407, which includes linked isolates from Brighton and London described in [39], and isolates described as cephalosporin resistant in [6]; m, WGSCL0032, the ST4995 MSM-associated isolates described in [6].
Discussion
In the present study, we aimed to perform a comprehensive N. gonorrhoeae genogrouping, based on WGS data, for prospective WGS-based laboratory surveillance. Focused on the European panorama, we quantitatively evaluated cluster stability to identify the major circulating WGS-genogroups, assessed their geographical spread, and searched for potential relationships between particular genogroups and specific AMR signatures. To achieve this, we relied on a recently described method (nAWC) [40, 54] to unprecedently analyse cluster-partitioning concordance at all possible allelic distance thresholds for gonococci. Two cluster-stability points could be identified, which enabled the classification of isolates into hierarchical WGS-based genogroups. This assignment facilitates the assessment (and continuous monitoring) of the frequency, geographical dispersion and potential association with specific AMR signatures of N. gonorrhoeae circulating genogroups, while providing a comprehensive analysis of the genetic diversity of this bacterium. Our approach resulted in the identification of 180 high-level and 321 low-level genogroups, several of which were composed by a single isolate or are represented by isolates from a single country. Both reflected the global genetic diversity and spread of N. gonorrhoeae , and showed the more prevalent and widespread strains circulating in Europe (Fig. 2). At least 5 high-level genogroups (Table 1) were found in more than 15 European countries, and 11 low-level genogroups (Table 2) were found in at least 6 countries (of which 3 are composed by at least 5 isolates). In an opposite scenario, we observed 81 genogroups that were composed by isolates from a single country (Fig. S5). Although N. gonorrhoeae infections lack a geographical structure, as revealed by its intercontinental spread [39, 61], our results also sustained the existence of N. gonorrhoeae transmission chains, likely confined at country/regional level. Nevertheless, as WGS is becoming more accessible and the several gonococcal antimicrobial surveillance programmes are starting to generate substantial genomic data, we cannot discard that the inclusion of genomes from other continents may impact the genetic diversity landscape observed in this Europe-oriented study. This impact could be reflected in either the addition of entirely new genogroups or, for instance, the potential merger of genogroups with poorly represented genetic diversity (i.e. represented here by one or two isolates). In fact, several studies have pointed-out a contrasting genetic diversity in countries from other continents [27], such as Kenya [62], Canada [25, 26], Japan [63], New Zealand [64], Australia [65] or the USA [66]. Comparison of typing data showed that the novel defined genogroups were mostly populated by a dominant MLST or NG-MAST ST (being especially relevant for typing comparability), which largely facilitates backwards compatibility and data communication in the transition to WGS-based typing (Fig. 1, Table 2). This observation is essential, as previous surveillance studies report particular NG-MAST genogroups or ST that not only need to be monitored but also display relationships with AMR profiles that cannot be dismissed [6]. Additionally, our study also showed that the quantitatively identified thresholds (4.97 and 9.61 % AD; 40 and 79 ADs in 822 loci, respectively), reflecting the earliest cluster stability (i.e. stable points with maximum resolution), provide a higher discriminatory resolution than the recently described PubMLST core-genome grouping threshold (400 ADs in 1649 loci) [28] (Fig. S4). The observed huge cluster agreement at the same threshold reinforces the likelihood that the robustness of the clustering will be kept if our high- and low-level thresholds are scaled to the entire PubMLST loci scheme.
Here, we observed that several WGS-based genogroups seem to be associated with distinct profiles of AMR (Figs 3 and 4). In fact, one such genogroup was NG-MAST G1407 (identified as genogroup G002.001 in the present study), which has been detected worldwide [27, 66–71] and is one of the major types found across Europe [6, 72]. Most isolates from this genogroup have been shown to be associated with multidrug resistance, presenting resistance or an increased MIC (or both) to cefixime, ceftriaxone, azithromycin and ciprofloxacin [6, 72, 73]. Concordantly, genetic alterations with potential association with cephalosporin resistance [6, 73], i.e. simultaneous carriage of mutations 312M, 316T and 545S in penA (mainly represented by the penA-XXXIV allele [58]), were mostly observed in isolates from this genogroup. Although, we detected potential horizontal acquisition of this specific penA allele by isolates with different genome backgrounds (eight other genogroups). However, it is worth noting that all 102 PT isolates belonging to G002.001 (including 87 harbouring the penA-XXXIV allele) had low MIC values for both cefixime and ceftriaxone, whether or not belonging to NG-MAST 1407; thus, challenging the association between these mutations in NG-MAST G1407 and resistance to cephalosporins. This also highlights the fact that some mechanisms of resistance in N. gonorrhoeae are still not fully disclosed, and further research is needed to understand the linkage between resistance and the simultaneous carriage of genetic AMR determinants, namely the role of epistatic interactions on the level of antimicrobial susceptibility (i.e. differential MIC values). For instance, the first cephalosporin-resistant isolate detected in Portugal (end of 2019) revealed a single alteration (542S) in penA, while presenting MICs of 0.19 and 0.38 mg l−1 to ceftriaxone and cefixime, respectively [14]. Another genogroup that warrants particular attention is G001.005, where the carriage of the azithromycin-resistance-associated mutation A2045G was mostly observed. This genogroup enrolled the isolates associated with high-level azithromycin resistance that have been linked to an outbreak in the UK [20, 35]. Of note, we found a high-level azithromycin-resistant PT isolate that was closely related to other resistant isolates from Ireland [36, 38] and the UK [35] (WGCL0024), suggesting its potential spread.
Overall, two distinct sets of genogroups displaying a contrasting pattern of AMR could be observed, i.e. one set with isolates carrying several genetic determinants related with decreased susceptibility and resistance, and another set mostly involving susceptible isolates (Fig. 3). For the vast majority of genogroups, we observed a dominant AMR profile (of note, for nine genogroups, a single AMR profile was observed) and a lower fraction of one or more additional AMR profiles (Fig. 4). Since a predominant circulating genogroup may not be the one carrying an alarming AMR profile, the combination of these results may help prioritize which genogroups need to be subjected to more close surveillance, management and control at both country and continent levels. Our results showed that, for some genogroups, the less frequently observed AMR profile was the one carrying a larger arsenal of genetic determinants for resistance, suggesting the potential emergence of novel resistant isolates within a WGS subtype. Of note, we detected genogroups where the dominant profile was the one associated with a larger repertoire of AMR determinants. This suggests a progressive accumulation of AMR determinants within these genogroups due to intensive antibiotic-driven selective pressures, although other scenarios cannot be discarded and warrant investigation (such as the presence of naturally resistant lineages). This information may be crucial for surveillance towards the identification of evolving strains whose emergence needs to be alerted to public-health authorities in order to control a local or cross-border spread in early stages.
The strategy described in the present study benefits from the comprehensive definition of WGS subtypes for long-term surveillance (with backwards compatibility) and a conservative threshold range that can be used for contact tracing and outbreak investigation. While for long-term surveillance, the goal is usually to continuously record the bacterial types that are circulating in a particular geographical area, for outbreak investigation a genomic analysis with higher resolution is required, potentiating the identification of shared genomic signatures to infer transmission [40, 61]. Defining clusters composed of likely related isolates by using specific thresholds ranges is an important step in the application of any subtyping scheme, but optimizing these thresholds/parameters has been a recurring challenge in the field of molecular epidemiology [40, 74]. In our approach, we cannot discard that adjusting these thresholds and maximizing the dataset may affect cluster composition and stability, even though our results are highly concordant with previous observations (Figs 2 and 5). Nonetheless, this study provides a comprehensive genome-scale snapshot of the genetic diversity of circulating N. gonorrhoeae strains in Europe. Results suggest that the applied conservative threshold for potential epidemiological linkage may be suitable to highlight clusters for fine-tuned analysis that could promote an epidemiological investigation, as cluster composition at this level seems to match with previous reports on outbreaks or transmission chains (Fig. 5). Additionally, the possibility to expand the number of loci under analysis in the scheme not only allows the performance of a dynamic analysis of epidemiological clusters, but also increases the confidence in results, as the link between strains is strengthened when AD decreases or remains unaltered after generating a sub-MST with a higher number of shared loci (Fig. 5). This concept of dynamically reconstructing phylogenies for sub-sets of strains (by maximizing the number of shared loci) and the bioinformatics tools needed for its operationalization were set-up recently for foodborne pathogens [40] and were applied to respiratory pathogens [75], showing that they facilitate the use of WGS in routine surveillance and, consequently, epidemiological investigations.
Ultimately, in the frame of the demanding short-term transition from traditional genotyping methods, the current comparative study is an important step towards the implementation of a WGS-based laboratory workflow for N. gonorrhoeae surveillance in the NRL. The laboratory surveillance and isolates collection built up on behalf of PTGonoNet, hosted at the NRL [31], has already strengthened our contribution to the EURO-GASP [6]. This has been achieved through the enrichment of the current geo-temporal and genomic diversity of N. gonorrhoeae , by adding WGS data of isolates spanning 15 years of N. gonorrhoeae surveillance in Portugal. Together with the centralization of the AMR phenotypic characterization and molecular typing at the Portuguese National Institute of Health, and a strong articulation with the public-health authorities, the framework described should constitute the driving force towards a faster and robust prospective surveillance of cases, from antibiotic-resistance prediction to transmission-chain detection. Future studies based on this approach will be crucial to consolidate the benefits of this technological transition for public health through the prioritization of genogroups to be monitored, the identification of emerging resistance carriage, and the potential facilitation of data sharing and communication.
Supplementary Data
Funding information
M.P. was supported by the Portuguese Science and Technology Foundation (FCT) through grant SFRH/BD/109264/2015. This work was partially funded by the GenomePT project (POCI-01-0145-FEDER-022184), supported by COMPETE 2020 – Operational Programme for Competitiveness and Internationalisation (POCI), Lisboa Portugal Regional Operational Programme (Lisboa2020), Algarve Portugal Regional Operational Programme (CRESC Algarve2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF), and by the Portuguese Science and Technology Foundation (FCT).
Acknowledgements
The authors would like to acknowledge all participants of the Portuguese National Laboratory Network for Neisseria gonorrhoeae Collection (PTGonoNet – http://www.insa.min-saude.pt/category/areas-deatuacao/doencas-infeciosas/rede-nacional-de-vigilancia-laboratorial-de-estirpes-de-n-gonorrhoeae/).
Author contributions
M. P. and J. C. R. were responsible for isolate AMR assays. L. V. supervised the next-generation sequencing procedures. J. I. carried out the next-generation sequencing procedures. M. P. did the bioinformatics analysis and wrote the article. M. P. and V. B. analysed and interpreted the data. All authors revised and approved the article. V. B., M. J. B. and J. P. G. jointly supervised the study.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All patient data associated with each N. gonorrhoeae isolate were fully anonymized.
Footnotes
Abbreviations: AD, allelic difference; ARI, adjusted Rand index; AWC, adjusted Wallace coefficient; ENA, European Nucleotide Archive; EURO-GASP, European Gonococcal Antimicrobial Surveillance Programme; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; MScgMLST, maximum-shared cgMLST; MST, minimum spanning tree; nAWC, neighbourhood adjusted Wallace coefficient; NG-MAST, Neisseria gonorrhoeae multi-antigen sequence typing; NG-STAR, Neisseria gonorrhoeae sequence typing for antimicrobial resistance; NRL, National Reference Laboratory for Sexually Transmitted Infections of the Portuguese National Institute of Health; SI, Shannon index; ST, sequence type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables and five supplementary figures are available with the online version of this article.
References
- 1.Newman L, Rowley J, Vander Hoorn S, Wijesooriya NS, Unemo M, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One. 2015;10:e0143304. doi: 10.1371/journal.pone.0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Report on Global Sexually Transmitted Infection Surveillance, 2018. Geneva: WHO; 2018. [Google Scholar]
- 3.European Centre for Disease Prevention and Control Gonococcal Antimicrobial Susceptibility Surveillance in Europe – Results Summary 2017. Stockholm: ECDC; 2019. [Google Scholar]
- 4.Unemo M, Golparian D, Eyre DW. Antimicrobial resistance in Neisseria gonorrhoeae and treatment of gonorrhea. Methods Mol Biol. 2019;1997:37–58. doi: 10.1007/978-1-4939-9496-0_3. [DOI] [PubMed] [Google Scholar]
- 5.Goire N, Lahra MM, Chen M, Donovan B, Fairley CK, et al. Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nat Rev Microbiol. 2014;12:223–229. doi: 10.1038/nrmicro3217. [DOI] [PubMed] [Google Scholar]
- 6.Harris SR, Cole MJ, Spiteri G, Sánchez-Busó L, Golparian D, et al. Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect Dis. 2018;18:758–768. doi: 10.1016/S1473-3099(18)30225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyre DW, De Silva D, Cole K, Peters J, Cole MJ, et al. WGS to predict antibiotic MICs for Neisseria gonorrhoeae . J Antimicrob Chemother. 2017;72:1937–1947. doi: 10.1093/jac/dkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole MJ, Spiteri G, Chisholm SA, Hoffmann S, Ison CA, et al. Emerging cephalosporin and multidrug-resistant gonorrhoea in Europe. Euro Surveill. 2014;19:20955. doi: 10.2807/1560-7917.ES2014.19.45.20955. [DOI] [PubMed] [Google Scholar]
- 9.Yu R-X, Yin Y, Wang G-Q, Chen S-C, Zheng B-J, et al. Worldwide susceptibility rates of Neisseria gonorrhoeae isolates to cefixime and cefpodoxime: a systematic review and meta-analysis. PLoS One. 2014;9:e87849. doi: 10.1371/journal.pone.0087849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, et al. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother. 2011;55:3538–3545. doi: 10.1128/AAC.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cámara J, Serra J, Ayats J, Bastida T, Carnicer-Pont D, et al. Molecular characterization of two high-level ceftriaxone-resistant Neisseria gonorrhoeae isolates detected in Catalonia, Spain. J Antimicrob Chemother. 2012;67:1858–1860. doi: 10.1093/jac/dks162. [DOI] [PubMed] [Google Scholar]
- 12.Eyre DW, Sanderson ND, Lord E, Regisford-Reimmer N, Chau K, et al. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Euro Surveill. 2018;23:1800323. doi: 10.2807/1560-7917.ES.2018.23.27.1800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poncin T, Fouere S, Braille A, Camelena F, Agsous M, et al. Multidrug-resistant Neisseria gonorrhoeae failing treatment with ceftriaxone and doxycycline in France, November 2017. Euro Surveill. 2018;23:1800264. doi: 10.2807/1560-7917.ES.2018.23.21.1800264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto M, Matias R, Rodrigues JC, Duarte S, Vieira L, et al. Cephalosporin-resistant Neisseria gonorrhoeae isolated in Portugal, 2019. Sex Transm Dis. 2020;47:e54–e56. doi: 10.1097/OLQ.0000000000001218. [DOI] [PubMed] [Google Scholar]
- 15.Day MJ, Spiteri G, Jacobsson S, Woodford N, Amato-Gauci AJ, et al. Stably high azithromycin resistance and decreasing ceftriaxone susceptibility in Neisseria gonorrhoeae in 25 European countries, 2016. BMC Infect Dis. 2018;18:609. doi: 10.1186/s12879-018-3528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thakur SD, Levett PN, Horsman GB, Dillon JR. High levels of susceptibility to new and older antibiotics in Neisseria gonorrhoeae isolates from Saskatchewan (2003-15): time to consider point-of-care or molecular testing for precision treatment? J Antimicrob Chemother. 2018;73:118–125. doi: 10.1093/jac/dkx333. [DOI] [PubMed] [Google Scholar]
- 17.Liang J-Y, Cao W-L, Li X-D, Bi C, Yang R-D, et al. Azithromycin-resistant Neisseria gonorrhoeae isolates in Guangzhou, China (2009-2013): coevolution with decreased susceptibilities to ceftriaxone and genetic characteristics. BMC Infect Dis. 2016;16:152. doi: 10.1186/s12879-016-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz AR, Komeya AY, Kirkcaldy RD, Whelen AC, Soge OO, et al. Cluster of Neisseria gonorrhoeae isolates with high-level azithromycin resistance and decreased ceftriaxone susceptibility, Hawaii, 2016. Clin Infect Dis. 2017;65:918–923. doi: 10.1093/cid/cix485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole MJ, Spiteri G, Jacobsson S, Woodford N, Tripodo F, et al. Overall low extended-spectrum cephalosporin resistance but high azithromycin resistance in Neisseria gonorrhoeae in 24 European countries, 2015. BMC Infect Dis. 2017;17:617. doi: 10.1186/s12879-017-2707-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chisholm SA, Wilson J, Alexander S, Tripodo F, Al-Shahib A, et al. An outbreak of high-level azithromycin resistant Neisseria gonorrhoeae in England. Sex Transm Infect. 2016;92:365–367. doi: 10.1136/sextrans-2015-052312. [DOI] [PubMed] [Google Scholar]
- 21.Smolarchuk C, Wensley A, Padfield S, Fifer H, Lee A, et al. Persistence of an outbreak of gonorrhoea with high-level resistance to azithromycin in England, November 2014‒May 2018. Euro Surveill. 2018;23:1800287. doi: 10.2807/1560-7917.ES.2018.23.23.1800287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahra MM, Ward A, Trembizki E, Hermanson J, Clements E, et al. Treatment guidelines after an outbreak of azithromycin-resistant Neisseria gonorrhoeae in South Australia. Lancet Infect Dis. 2017;17:133–134. doi: 10.1016/S1473-3099(17)30007-5. [DOI] [PubMed] [Google Scholar]
- 23.Weston EJ, Wi T, Papp J. Strengthening global surveillance for antimicrobial drug–resistant Neisseria gonorrhoeae through the enhanced gonococcal antimicrobial surveillance program. Emerg Infect Dis. 2017;23:S47–S52. doi: 10.3201/eid2313.170443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demczuk W, Sidhu S, Unemo M, Whiley DM, Allen VG, et al. Neisseria gonorrhoeae sequence typing for antimicrobial resistance, a novel antimicrobial resistance multilocus typing scheme for tracking global dissemination of N. gonorrhoeae strains. J Clin Microbiol. 2017;55:1454–1468. doi: 10.1128/JCM.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demczuk W, Lynch T, Martin I, Van Domselaar G, Graham M, et al. Whole-genome phylogenomic heterogeneity of Neisseria gonorrhoeae isolates with decreased cephalosporin susceptibility collected in Canada between 1989 and 2013. J Clin Microbiol. 2015;53:191–200. doi: 10.1128/JCM.02589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demczuk W, Martin I, Peterson S, Bharat A, Van Domselaar G, et al. Genomic epidemiology and molecular resistance mechanisms of azithromycin-resistant Neisseria gonorrhoeae in Canada from 1997 to 2014. J Clin Microbiol. 2016;54:1304–1313. doi: 10.1128/JCM.03195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Busó L, Golparian D, Corander J, Grad YH, Ohnishi M, et al. The impact of antimicrobials on gonococcal evolution. Nat Microbiol. 2019;4:1941–1950. doi: 10.1038/s41564-019-0501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison OB, Cehovin A, Skett J, Jolley KA, Massari P, et al. Neisseria gonorrhoeae population genomics: use of the gonococcal core genome to improve surveillance of antimicrobial resistance. J Infect Dis. 2020;222:1816–1825. doi: 10.1093/infdis/jiaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez-Busó L, Harris SR. Using genomics to understand antimicrobial resistance and transmission in Neisseria gonorrhoeae . Microb Genom. 2019;5:e000239. doi: 10.1099/mgen.0.000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Rourke M, Stevens E. Genetic structure of Neisseria gonorrhoeae populations: a non-clonal pathogen. J Gen Microbiol. 1993;139:2603–2611. doi: 10.1099/00221287-139-11-2603. [DOI] [PubMed] [Google Scholar]
- 31.Pinto M, Rodrigues JC, Matias R, Água-Doce I, Cordeiro D, et al. Fifteen years of a nationwide culture collection of Neisseria gonorrhoeae antimicrobial resistance in Portugal. Eur J Clin Microbiol Infect Dis. 2020;39:1761–1770. doi: 10.1007/s10096-020-03907-7. [DOI] [PubMed] [Google Scholar]
- 32.European Centre for Disease Prevention and Control Gonococcal Antimicrobial Susceptibility Surveillance in Europe 2015. Stockholm: ECDC; 2017. [Google Scholar]
- 33.European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 9.0. Basel: EUCAST; 2019. [Google Scholar]
- 34.Didelot X, Dordel J, Whittles LK, Collins C, Bilek N, et al. Genomic analysis and comparison of two gonorrhea outbreaks. mBio. 2016;7:e00525-16. doi: 10.1128/mBio.00525-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fifer H, Cole M, Hughes G, Padfield S, Smolarchuk C, et al. Sustained transmission of high-level azithromycin-resistant Neisseria gonorrhoeae in England: an observational study. Lancet Infect Dis. 2018;18:573–581. doi: 10.1016/S1473-3099(18)30122-1. [DOI] [PubMed] [Google Scholar]
- 36.Mac Aogáin M, Fennelly N, Walsh A, Lynagh Y, Bekaert M, et al. Fourteen draft genome sequences for the first reported cases of azithromycin-resistant Neisseria gonorrhoeae in Ireland. Genome Announc. 2017;5:e00403-17. doi: 10.1128/genomeA.00403-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wind CM, de Vries E, Schim van der Loeff MF, van Rooijen MS, van Dam AP, et al. Decreased azithromycin susceptibility of Neisseria gonorrhoeae isolates in patients recently treated with azithromycin. Clin Infect Dis. 2017;65:37–45. doi: 10.1093/cid/cix249. [DOI] [PubMed] [Google Scholar]
- 38.Ryan L, Golparian D, Fennelly N, Rose L, Walsh P, et al. Antimicrobial resistance and molecular epidemiology using whole-genome sequencing of Neisseria gonorrhoeae in Ireland, 2014-2016: focus on extended-spectrum cephalosporins and azithromycin. Eur J Clin Microbiol Infect Dis. 2018;37:1661–1672. doi: 10.1007/s10096-018-3296-5. [DOI] [PubMed] [Google Scholar]
- 39.De Silva D, Peters J, Cole K, Cole MJ, Cresswell F, et al. Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis. 2016;16:1295–1303. doi: 10.1016/S1473-3099(16)30157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llarena A‐K, Ribeiro‐Gonçalves BF, Nuno Silva D, Halkilahti J, Machado MP, et al. INNUENDO: a cross‐sectoral platform for the integration of genomics in the surveillance of food‐borne pathogens. EFSA Supporting Publications. 2018;15:EN‐1498. doi: 10.2903/sp.efsa.2018.EN-1498. [DOI] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwong JC, Gonçalves da Silva A, Dyet K, Williamson DA, Stinear TP, et al. NGMASTER: in silico multi-antigen sequence typing for Neisseria gonorrhoeae . Microb Genom. 2016;2:e000076. doi: 10.1099/mgen.0.000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett JS, Watkins ER, Jolley KA, Harrison OB, Maiden MC. Identifying Neisseria species by use of the 50S ribosomal protein L6 (rplF) gene. J Clin Microbiol. 2014;52:1375–1381. doi: 10.1128/JCM.03529-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unemo M, Golparian D, Sánchez-Busó L, Grad Y, Jacobsson S, et al. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J Antimicrob Chemother. 2016;71:3096–3108. doi: 10.1093/jac/dkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, et al. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018;4:e000166. doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Francisco AP, Bugalho M, Ramirez M, Carriço JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, et al. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics. 2012;13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribeiro-Gonçalves B, Francisco AP, Vaz C, Ramirez M, Carriço JA. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016;44:W246–W251. doi: 10.1093/nar/gkw359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, et al. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes . J Clin Microbiol. 2006;44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Severiano A, Pinto FR, Ramirez M, Carriço JA. Adjusted Wallace coefficient as a measure of congruence between typing methods. J Clin Microbiol. 2011;49:3997–4000. doi: 10.1128/JCM.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barker DOR, Carriço JA, Kruczkiewicz P, Palma F, Rossi M, et al. Rapid identification of stable clusters in bacterial populations using the adjusted Wallace coefficient. bioRxiv. 2018:299347 [Google Scholar]
- 55.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, et al. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng X, Klausner JD. Six penA codons accurately and reliably predict cefixime-decreased susceptibility in Neisseria gonorrhoeae . J Infect Dis. 2020;221:851–852. doi: 10.1093/infdis/jiz504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unemo M, Golparian D, Nicholas R, Ohnishi M, Gallay A, et al. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Chemother. 2012;56:1273–1280. doi: 10.1128/AAC.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belkacem A, Jacquier H, Goubard A, Mougari F, La Ruche G, et al. Molecular epidemiology and mechanisms of resistance of azithromycin-resistant Neisseria gonorrhoeae isolated in France during 2013-14. J Antimicrob Chemother. 2016;71:2471–2478. doi: 10.1093/jac/dkw182. [DOI] [PubMed] [Google Scholar]
- 60.Calado J, Castro R, Lopes Ângela, Campos MJ, Rocha M, et al. Antimicrobial resistance and molecular characteristics of Neisseria gonorrhoeae isolates from men who have sex with men. Int J Infect Dis. 2019;79:116–122. doi: 10.1016/j.ijid.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 61.Mortimer TD, Grad YH. Applications of genomics to slow the spread of multidrug-resistant Neisseria gonorrhoeae . Ann N Y Acad Sci. 2019;1435:93–109. doi: 10.1111/nyas.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cehovin A, Harrison OB, Lewis SB, Ward PN, Ngetsa C, et al. Identification of novel Neisseria gonorrhoeae lineages harboring resistance plasmids in coastal Kenya. J Infect Dis. 2018;218:801–808. doi: 10.1093/infdis/jiy240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yahara K, Nakayama S-I, Shimuta K, Lee K-I, Morita M, et al. Genomic surveillance of Neisseria gonorrhoeae to investigate the distribution and evolution of antimicrobial-resistance determinants and lineages. Microb Genom. 2018;4:e000205. doi: 10.1099/mgen.0.000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee RS, Seemann T, Heffernan H, Kwong JC, Gonçalves da Silva A, et al. Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J Antimicrob Chemother. 2018;73:353–364. doi: 10.1093/jac/dkx405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williamson DA, Chow EPF, Gorrie CL, Seemann T, Ingle DJ, et al. Bridging of Neisseria gonorrhoeae lineages across sexual networks in the HIV pre-exposure prophylaxis era. Nat Commun. 2019;10:3988. doi: 10.1038/s41467-019-12053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, et al. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis. 2014;14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin I, Sawatzky P, Liu G, Allen V, Lefebvre B, et al. Antimicrobial susceptibilities and distribution of sequence types of Neisseria gonorrhoeae isolates in Canada: 2010. Can J Microbiol. 2013;59:671–678. doi: 10.1139/cjm-2013-0357. [DOI] [PubMed] [Google Scholar]
- 68.Gose S, Nguyen D, Lowenberg D, Samuel M, Bauer H, et al. Neisseria gonorrhoeae and extended-spectrum cephalosporins in California: surveillance and molecular detection of mosaic penA. BMC Infect Dis. 2013;13:570. doi: 10.1186/1471-2334-13-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shimuta K, Unemo M, Nakayama S, Morita-Ishihara T, Dorin M, et al. Antimicrobial resistance and molecular typing of Neisseria gonorrhoeae isolates in Kyoto and Osaka, Japan, 2010 to 2012: intensified surveillance after identification of the first strain (H041) with high-level ceftriaxone resistance. Antimicrob Agents Chemother. 2013;57:5225–5232. doi: 10.1128/AAC.01295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gianecini RA, Golparian D, Zittermann S, Litvik A, Gonzalez S, et al. Genome-based epidemiology and antimicrobial resistance determinants of Neisseria gonorrhoeae isolates with decreased susceptibility and resistance to extended-spectrum cephalosporins in Argentina in 2011-16. J Antimicrob Chemother. 2019;74:1551–1559. doi: 10.1093/jac/dkz054. [DOI] [PubMed] [Google Scholar]
- 71.Chen SC, Yin YP, Dai XQ, Unemo M, Chen XS. First nationwide study regarding ceftriaxone resistance and molecular epidemiology of Neisseria gonorrhoeae in China. J Antimicrob Chemother. 2016;71:92–99. doi: 10.1093/jac/dkv321. [DOI] [PubMed] [Google Scholar]
- 72.Chisholm SA, Unemo M, Quaye N, Johansson E, Cole MJ, et al. Molecular epidemiological typing within the European gonococcal antimicrobial resistance surveillance programme reveals predominance of a multidrug-resistant clone. Euro Surveill. 2013;18:20358. [PubMed] [Google Scholar]
- 73.Młynarczyk-Bonikowska B, Majewska A, Malejczyk M, Młynarczyk G, Majewski S. Multiresistant Neisseria gonorrhoeae: a new threat in second decade of the XXI century. Med Microbiol Immunol. 2020;209:95–108. doi: 10.1007/s00430-019-00651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 75.Macedo R, Pinto M, Borges V, Nunes A, Oliveira O, et al. Evaluation of a gene-by-gene approach for prospective whole-genome sequencing-based surveillance of multidrug resistant Mycobacterium tuberculosis . Tuberculosis. 2019;115:81–88. doi: 10.1016/j.tube.2019.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reads generated for the present study were deposited in the ENA under the study accession number PRJEB36482 (individual run accession numbers are detailed in Table S1). The PubMLST N. gonorrhoeae cgMLST v1 scheme adapted for chewBBACA [48], the MScgMLST scheme, as well as allelic profile matrix for both schemes are available at https://doi.org/10.5281/zenodo.3946223. Additionally, a .json file also has been made available for direct upload into the GrapeTree visualization software [56] in order to interactively explore data/metadata.