Abstract
Motivation
Although genome-wide association studies (GWASs) have identified thousands of variants for various traits, the causal variants and the mechanisms underlying the significant loci are largely unknown. In this study, we aim to predict non-coding variants that may functionally affect translation initiation through long-range chromatin interaction.
Results
By incorporating the Hi-C data, we propose a novel and powerful deep learning model of artificial intelligence to classify interacting and non-interacting fragment pairs and predict the functional effects of sequence alteration of single nucleotide on chromatin interaction and thus on gene expression. The changes in chromatin interaction probability between the reference sequence and the altered sequence reflect the degree of functional impact for the variant. The model was effective and efficient with the classification of interacting and non-interacting fragment pairs. The predicted causal SNPs that had a larger impact on chromatin interaction were more likely to be identified by GWAS and eQTL analyses. We demonstrate that an integrative approach combining artificial intelligence—deep learning with high throughput experimental evidence of chromatin interaction leads to prioritizing the functional variants in disease- and phenotype-related loci and thus will greatly expedite uncover of the biological mechanism underlying the association identified in genomic studies.
Availability and implementation
Source code used in data preparing and model training is available at the GitHub website (https://github.com/biocai/DeepHiC).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Genome-wide association studies (GWASs) are designed to study the associations between genetic variants and diseases. GWASs facilitate studies in complex-trait genetics and the biology of diseases. Thousands of variants which are assumed to tag one or a few causal variants have been identified for various diseases and traits (Welter et al., 2013). However, most of these variants (93%) are located in the non-coding regions including introns, long terminal repeats and intergenic regions (Maurano et al., 2012). The major challenge of GWAS lies in interpreting the involvement of these non-coding variants in the etiology of diseases.
Many methods have been proposed to tackle this problem by combining other omics data. Summary data-based Mendelian randomization integrates independent GWAS summary statistics data with expression quantitative trait loci (eQTL) data to identify potential functional genes (Zhu et al., 2016). Similarly, a Bayesian analysis approach, COLOC, colocalizes GWAS and eQTL signals at known GWAS risk loci (Giambartolomei et al., 2014) and combines the DNA methylation quantitative trait loci (mQTL) signals (Giambartolomei et al., 2018). Regulatory information, including CHIP-seq peaks, DNase I hypersensitivity peaks, DNase I footprints, ATAC-seq, conserved motifs, eQTL and transcription factor (TF) binding sites, was used to suggest functional hypotheses for variants associated with diseases (Boyle et al., 2012; Bryois et al., 2018). Other studies used chromatin interaction information to explain GWAS significant variants through predicting the target genes (Chen and Tian, 2016; Lu et al., 2013).
DNA looping is a widely held model that posits that enhancers can be brought proximately to the promoter of the target genes by bending DNA structure via transcription factor binding and mediation of cohesin and mediators (Mossing and Record, 1986). This process of DNA looping facilitates the regulation of gene expression. The structure of chromatin interaction can be captured at the level of a single locus (3C, 4C), a set of loci (5C, ChIA-PET and Capture-C) and genome-wide (Hi-C) (Ay and Noble, 2015). The mutations on enhancers may disturb normal cell activity and influence pathogenesis of diseases, for example, aniridia (Kleinjan, 2001) and Hirschsprung’s disease (Emison et al., 2005).
Previous studies have used chromatin interaction information to understand the mechanisms underlying the non-coding variants. For example, rs11610206, located on the enhancer of VDR, is associated with Alzheimer’s disease (Yu et al., 2011). There is an interaction between the enhancer region and the VDR gene region. This SNP influences the function of the enhancer, which then affects the expression of VDR and leads to Alzheimer’s disease (Lu et al., 2013). The hypothesis is that the non-coding variant may disturb the interaction between these two regions.
Prioritizing candidate variants and elucidating the function of non-coding variants is challenging. Many computational tools have been developed to assess the functional impact of non-coding variants (Nishizaki and Boyle, 2017). The general framework is to build predictive models that learn the rules of combining genome sequences, multiple genomic annotations, functional attributes and evolutionary features to differentiate disease-related non-coding variants from neutral ones (Liu et al., 2019). Deep learning (an artificial intelligence approach) is a state-of-the-art technology that has been widely used in genomics (Zou et al., 2019). Several studies combine deep learning with regulatory information to predict chromatin effects of sequence alterations with single-nucleotide sensitivity (Wang et al., 2018; Zhou and Troyanskaya, 2015). For example, Deep learning-based Functional impact of non-coding variants evaluator (DeFine) combines a deep learning approach with large-scale TF ChIP-seq data to predict the TF binding intensities to given DNA sequences (Wang et al., 2018). The changes in TF binding intensities between the reference sequence and the alternative sequence reflect the functional impact of the variant on TF binding (Wang et al., 2018). This suggests that well-trained deep learning models can be used to reveal, illuminate and prioritize potential functional variants.
In this study, combining the Hi-C data, we for the first time develop a deep learning model (DeepHiC) to predict the effects of sequence alteration of single nucleotide on chromatin interaction. Further, we demonstrate that it is useful to determine whether a non-coding variant has a functional impact and identify the potential target gene affected by this variant.
2 Materials and methods
2.1 Hi-C data
Although Capture-C enables us to obtain cis interactions at hundreds of selected loci at high resolution (Hughes et al., 2014), Hi-C which captures the genome-wide interactions is more suitable to study features of interacted sequences. A previous study suggested that local chromatin interaction domains and topological domains are stable across different cell types (Dixon et al., 2012). Hi-C data generated from three representative cell lines (a human embryonic stem cell line, a human lymphoblastoid cell line and a human erythroleukemic cell line) were used in this study. Hi-C data generated from human embryonic stem cells (hESC) at a resolution of 5 kb were downloaded from the Gene Expression Omnibus (GEO) database (accession number GSE52457) (Dixon et al., 2015). Hi-C data generated from GM12878 (a human lymphoblastoid cell line) and K562 (a human erythroleukemic cell line) at a resolution of 5 kb were downloaded from the GEO database (accession number GSE63525) (Rao et al., 2014). In the original paper, the authors aggregated the results of nine biological replicate experiments of GM12878 cell line to generate a Hi-C map that reached the resolution of 950 bp. To match the resolution in other cell lines, we only used two replication results of GM12878 cell line (accession numbers GSM1551584 and GSM1551585). All the raw sequencing data of the two replications of these three cell lines were downloaded.
2.2 Whole genome sequencing data and processing
The whole genome sequencing reads of hESC were downloaded from the GEO database (accession number GSE69471) (Mertes et al., 2016). The whole genome sequencing reads for the GM12878 cell lines were downloaded from ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/NA12878/NIST_NA12878_HG001_HiSeq_300x/RMNISTHS_30xdownsample.bam. The K562 whole-genome sequencing reads were downloaded from https://www.ncbi.nlm.nih.gov/sra/SRX118400. The raw short reads were first cleaned to remove adaptor sequences and truncate low quality reads using Trimmomatic (Bolger et al., 2014). Burrows–Wheeler Aligner (BWA) was used to map all the cleaned short reads to the reference genome with default parameters (Li and Durbin, 2009). The Variant Call Format (VCF) file was generated using GATK after removing polymerase chain reaction duplicates (Van der Auwera et al., 2013). The reference bases at variation sites were replaced to generate cell type-specific genome sequences using GATK (Van der Auwera et al., 2013). The cell type-specific genome sequences were used in the following steps to correct the cell type-specific variants (Wang et al., 2018).
2.3 Processing Hi-C data
We analyzed the raw sequencing reads of the two replications of hESC using HOMER (Heinz et al., 2010). The pair-end reads were separately aligned to the cell type-specific genome using Subread (Liao et al., 2013). HOMER taking SAM file as input only removes reads if their ends form a self-ligation with adjacent restriction sites and removes reads from the selected bins (for examples, 10 kb) that contain more than 5× the average number of reads (Heinz et al., 2010). HOMER can identify significant interactions by searching for pairs of loci that have a greater number of Hi-C reads than expected by chance (Heinz et al., 2010). We used HOMER to identify significant interactions for each replication using different bin sizes (10, 40 and 100 kb). We also used HOMER to run principal component analysis (PCA) of Hi-C data. The first principal component (PC1) was used to classify each region of the chromosome into active (‘A’) and inactive (‘B’) compartments. Compartment A is gene-rich and has relatively high GC content, while compartment B is gene desert.
2.4 Data preparation
Since the sequence data produced by Hi-C are noisy, Hi-C may capture random chromatin interactions. The significant interactions presented in both replications with FDR < 0.05 were considered as positive, and the other bin pairs as negative. Since the interactions between the same compartment were overestimated, we balanced the number of interaction pairs within compartment A, the number of interaction pairs within compartment B and the number of interaction pairs between different compartments. The distribution of distances between selected positive interaction pairs was shown in Supplementary Figure S1. For each positive interaction pairs in the positive set, a negative pair with matched GC% content and compartment was included in the negative set. Because there was only a small fraction of inter-chromosomal chromatin interaction pairs which was too rare to train a deep learning model, we did not consider inter-chromosomal chromatin interactions any further (Supplementary Fig. S2). Finally, we generated 95 849 intra-chromosomal interactions of 10 kb bin pairs, 654 156 intra-chromosomal interactions of 40 kb bin pairs and 63 015 intra-chromosomal interactions of 100 kb bin pairs. The sequences of 10, 40 and 100 kb bins generated from the hESC genome were extracted from the hESC genome sequences using SAMtools (Li et al., 2009). As DNA is a double helix, both the forward sequence and the reverse sequence were considered. Nucleotides A, T, C and G are encoded as [1,0,0,0], [0,1,0,0], [0,0,1,0] and [0,0,0,1] (Zou et al., 2019). The sequences of each fragment pair were merged and converted to a one-hot matrix with 10 000 rows for 10 kb bins (40 000 rows for 40 kb bins and 100 000 rows for 100 kb) and 16 columns encoding 4 nucleotides. The matrix can be viewed as a gray image and used as the input to feed to the deep learning model. We compared with the models taking different length of bin pairs (10–100 kb) as input.
2.5 Overview of the DeepHiC deep learning model
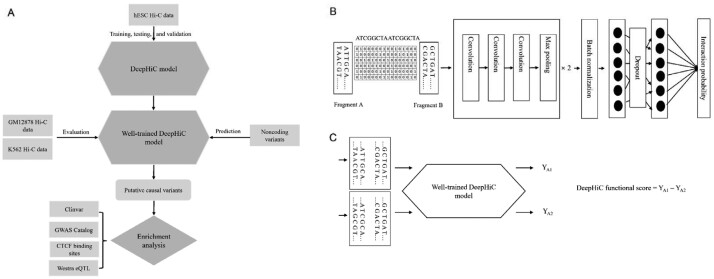
DeepHiC employed the convolutional neural network (CNN) to understand the sequence feature that characterizes real interactions. The overall architecture of DeepHiC is shown inFigure 1B. The CNN model consisted of convolution layers, pooling layers, fully connected layers and a SoftMax layer. The dropout layer with a probability of 0.5 was added between two fully connected layers to improve the generalization capability of the model and avoid overfitting (Srivastava et al., 2014).
Fig. 1.
Overview of DeepHiC method. (A) The workflow of this study. (B) The overall architecture of DeepHiC. The sequences of each fragment pair were merged and converted to a one-hot matrix with 10 000 rows for 10 kb bins. The matrix can be viewed as a gray image and used as the input to feed to the CNN model. (C) DeepHiC functional score was defined as the difference between the two predicted interaction probabilities (YA1 calculated using the reference allele A1 and YA2 calculated using the other allele A2)
2.6 Training of DeepHiC
The whole dataset generated from hESC Hi-C data was separated into training, validation and testing with a ratio of 3:1:1. The validation dataset was used in the grid search progress to determine the hyper-parameters in the deep learning model during training. The following hyper-parameters were applied: (batch size was 32, the filter was a 24-by-1 matrix, the number of filters in each convolution layer were 64, 64, 64, 128, 128 and 128, the number nodes in the full connected layers was 2048, learning rate was 0.0001). In the supervised training step, CNN learned features that help to differentiate interacting fragment pairs from randomly selected fragment pairs. We trained the deep learning model with a mini-batch stochastic gradient descent algorithm (Adam) (Kingma and Ba, 2015). During each mini-batch training, the parameters in the model were updated based on a gradient calculated using backpropagation. Training was run for 30 epochs. During each epoch of training, the loss in the validation set was calculated and monitored. When the loss in the validation dataset did not decrease in five epochs, the training was stopped, and the model weights from the epoch with the smallest loss in the validation dataset were saved. The training and testing procedures were implemented on an Ubuntu 18.04 computer with a NVIDIA GTX 2080Ti 11 Gb GPU. We used python library Keras 2.3 with Tensorflow backend (https://keras.io) for data preprocessing and CNN model training and testing.
2.7 Prediction
There currently remains no database of causal SNPs affecting the target gene through chromatin interaction. We assumed those SNPs changing the interaction state as putatively causal SNPs. In the inference step, the trained model predicted the interaction probability of the paired fragments with one altered fragment sequence centered at the variant site (Fig. 1B). The difference between the two predicted interaction probabilities (YA1 calculated using the reference allele A1 and YA2 calculated using the other allele A2) was used to assess the functional impact of the SNP which was defined as DeepHiC functional score. Non-coding variant dataset was collected from the 1000 Genomes Project phase 3, which comprises 8 251 605 non-coding SNPs. In total, 2 844 552 SNPs from the non-coding variant dataset were located in the positive bin pairs in the positive set. We then measured the scores of each non-coding SNP in the positively interacted bin pairs. To estimate the statistical significance of interaction probability difference caused by the alteration of a single nucleotide, FastPval which computed the empirical P-value by a two-stage ranking strategy (Li et al., 2010) was used to calculate the P-value of each absolute DeepHiC functional score. The direction of the DeepHiC functional score was used to show which allele was helpful with the interaction (e.g. positive DeepHiC functional score meant that the reference allele was helpful with the interaction).
2.8 Enrichment analysis
To check whether the putatively causal SNPs predicted by DeepHiC were enriched in ClinVar, GWAS, eQTL datasets and CTCF binding sites, we performed enrichment analysis. ClinVar aggregates information about genomic variation and the relationships among human variations and phenotypes (Landrum et al., 2014). Functional non-coding variants from ClinVar database (released on 03/09/2019) were employed in this study. We collected GWAS SNPs identified by previous studies from the NHGRI-EBI GWAS Catalog (Welter et al., 2014), publicly available at https://www.ebi.ac.uk/gwas/, and downloaded on May 16, 2019. We limited our study to the most significant associations, eliminating SNPs with P-values larger than the genome-wide significance (P-value = 5 × ). Westra eQTLs dataset which were performed in peripheral blood samples of 5311 individuals were used in this study (Westra et al., 2013). Since CTCF plays a critical role in chromatin interaction, we also evaluated whether the putatively causal SNPs were enriched in CTCF binding sites with the data downloaded from https://github.com/gkichaev/PAINTOR_V3.0/wiki/2b.-Overlapping-annotations.
3 Results
3.1 Chromatin interaction state was accurately predicted by deep learning model
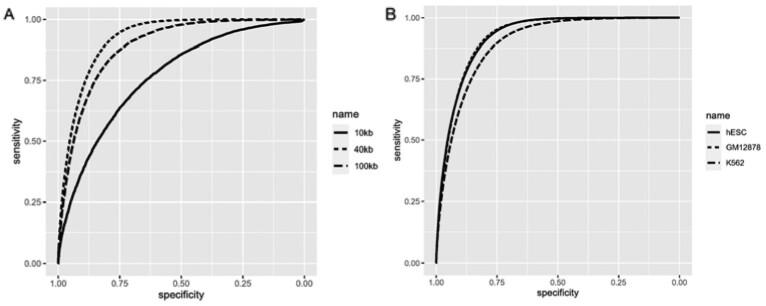
We built deep learning models to predict the interaction probability of bin pairs. The architecture of the deep learning model is shown in Figure 1B. The DeepHiC models took sequences from bin pairs of different sizes generated from hESC as input. The receiver operating characteristic (ROC) curves were generated to select the best predictive model. The performance of different models on the testing dataset was evaluated by comparing the area under the curve (AUC). The model trained on 40 kb bin pairs (Fig. 2A) generated the highest AUC value (0.922) and was used in the following study. The model trained on 100 kb bin pairs generated an AUC of 0.889 while the model taking 10 kb bin pairs as input achieved an AUC of 0.765 (Fig. 2A). Detailed results were shown in Table 1.
Fig. 2.
The performance of different DeepHiC models. (A) DeepHiC models were trained with sequences from bin pairs with length of 10, 40 and 100 kb, respectively. (B) The performance of the best DeepHiC model trained on bin pairs with 40 kb length was evaluated on independent datasets generated from GM12878 cells and K562 cells
Table 1.
Performance of DeepHiC models
| Length of bin pairs | AUC | Accuracy | Specificity | Sensitivity | F1 score |
|---|---|---|---|---|---|
| 10 kb | 0.77 | 0.70 | 0.71 | 0.68 | 0.71 |
| 40 kb | 0.92 | 0.85 | 0.78 | 0.92 | 0.86 |
| 100 kb | 0.89 | 0.81 | 0.83 | 0.78 | 0.80 |
To assess the generalization capability of the CNN model, we further evaluated the performance on independent datasets generated from GM12878 and K562 cell lines. The raw sequencing pair-end reads were separately aligned to the genome of GM12878 or K562 cell line, respectively. HOMER was used to identify significant interactions at the resolution of 40 kb. GM12878 cell line shared 93.37% significant interactions with hESC cell line while K562 cell line shared 75.96% significant interactions (Supplementary Fig. S3). The positive set and the negative set were selected based on the same strategy of the hESC cell line. The CNN model yielded accuracies of 0.86 (AUC = 0.923) on the dataset generated from the GM12878 cell line and 0.82 (AUC = 0.897) on the dataset generated from the K562 cell line (Fig. 2B). The performances were similar or dropped only slightly compared with the performance on the testing dataset which was generated from the hESC cell line.
3.2 Putatively causal SNPs were enriched in GWAS and eQTL datasets
Based on the CNN model, to assess the functional impact of the non-coding variants, DeepHiC functional score was defined as the change of interaction probability of the altered fragment pairs (Fig. 1C). The altered fragment sequence was centered at the non-coding variant site. We investigated the utility of DeepHiC functional scores for discriminating disease-related non-coding variants from neutral ones. After computing the functional scores of 2 844 552 non-coding SNPs (including 26 243 694 different interactions), we get the distribution of interaction probability difference (Supplementary Fig. S4). FastPval was used to generate the empirical P-value of each SNP by the two-stage ranking strategy (Li et al., 2010). To decrease the false-positive rate accumulated by multiple testing, SNPs with an absolute DeepHiC functional score >0.031 (P-value < 0.001) as suggested by FastPval were considered as putatively causal SNPs.
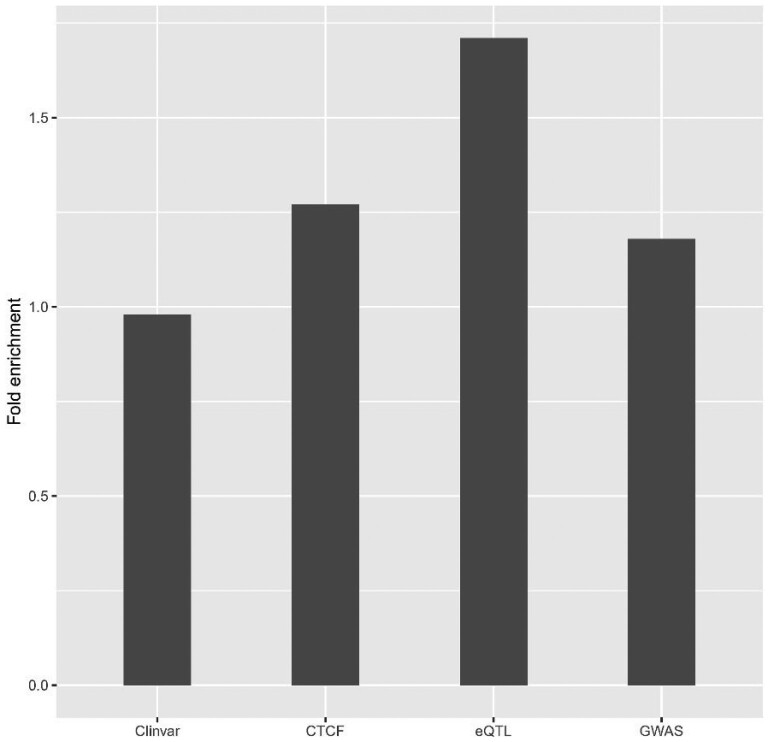
We further checked whether the putatively causal SNPs were enriched in ClinVar, GWAS, eQTL datasets and CTCF binding sites. After comparing the consistency with the non-coding SNPs collected from the 1000 Genome Project, 3933, 22 584 and 51 138 SNPs were selected from ClinVar, GWAS datasets and CTCF binding sites, respectively. Since one SNP may interact with different regions in the genome, the largest score of each SNP was used in the enrichment analyses of SNPs from ClinVar, GWAS datasets and CTCF binding sites. The enrichment analyses showed that the putatively causal SNPs were significantly enriched in the GWAS dataset with a fold change of 1.18 (Fisher’s exact test P-value = 4.9 × , Fig. 3) using 2 844 552 SNPs with the largest score as background. For each phenotype in the GWAS dataset, the putatively causal SNPs were significantly enriched in body mass index, educational attainment, systolic blood pressure and mean corpuscular hemoglobin (Supplementary Fig. S4). We observed a 1.27-fold enrichment for CTCF binding (Fisher’s exact test P-value = 5.6 × ) and no enrichment for ClinVar dataset (0.98, Fisher’s exact test P-value = 1, Fig. 3). The paired fragments harboring the eQTL and the TSS of the target gene were input into the DeepHiC model to generate the interaction probability (YA1 and YA2). If there was an interaction between both fragments, the eQTL with an absolute DeepHiC functional score > 0.031 was considered to influence the target expression by changing the interaction probability between the two fragments. In total, 13 853 eQTL-gene pairs were included in the enrichment analysis. Finally, the enrichment analyses showed that the putatively causal SNPs were significantly enriched in the Westra eQTLs (with a fold change of 1.71, Fisher’s exact test P-value = 6.3 × ) compared with the totally 26 243 694 different interactions. We also identified 31 putatively causal eQTLs (Supplementary Table S1).
Fig. 3.
The putatively causal SNPs predicted by DeepHiC functional scores were significantly enriched in GWAS, eQTL datasets and CTCF binding sites
3.3 DeepHiC was capable of prioritizing disease-related functional non-coding variants
We further explored whether DeepHiC could help to identify disease-related functional non-coding variants from a difficult credible set. We employed a set of eQTLs (rs9533090, rs9594738, rs8001611, rs9533094 and rs9533095) correlated with TNFSF11, which has been identified for osteoporosis (Estrada et al., 2012; Rivadeneira et al., 2009; Zhang et al., 2014). RANKL encoded by TNFSF11 is a key factor for osteoclast differentiation and activation (Wittrant et al., 2004). All of these SNPs are located in a super-enhancer region to regulate the expression of RANKL via long-range chromatin interaction (Zhu et al., 2018). We used DeepHiC to prioritize these SNPs (Table 2). The results showed that DeepHiC predicted rs9533090 with the highest functional score (0.024, P-value = 2.9 × ). This result was consistent with one recent study regarding rs9533090-C as a functional SNP to recruit transcription factor NFIC and increase RANKL expression (Zhu et al., 2018).
Table 2.
The DeepHiC functional scores of eQTLs correlated with TNFSF11
| SNPs | Allele 1 | Allele 2 | DeepHiC functional score | P-value |
|---|---|---|---|---|
| rs9533090 | C | T | 0.024 | 2.9 × 10−3 |
| rs9594738 | C | T | −0.0014 | 0.40 |
| r8001611 | T | C | −0.0013 | 0.42 |
| rs9533094 | A | G | −0.0095 | 0.043 |
| rs9533095 | G | T | −0.0038 | 0.18 |
rs7756521 was a causal variant significantly associated with control of HIV infection (Jin et al., 2018). It may affect the DDR1 expression through chromatin interaction (Jin et al., 2018). The variant was located in a DNase I hypersensitive site (DHS) active in chimpanzees, macaques and humans when the variant position was the T allele, since only the T allele was observed in other non-human primates (Jin et al., 2018). The DeepHiC model showed that this SNP had a strong impact on chromatin interaction (function score = 0.0096, P-value = 0.042). The sequence with the T allele generated a higher interaction probability, thus consistent with earlier study (Jin et al., 2018) in that T allele increased the expression of DDR1.
4 Discussion
In this study, we predicted chromatin interaction based on DNA sequences using a deep learning approach in artificial intelligence. The high accuracy of the DeepHiC model allowed us to quantify the effect of variants on chromatin interaction probability. The performance of DeepHiC model trained on 40 kb bin pairs was better than others. We used PCA to classify each region of the chromosome into A/B compartment. When small resolution was used, the classification results may not accurately reflect the general chromosome structure (Heinz et al., 2010). Then it would have an effect on selection of positive and negative interactions. A previous study suggested that the local chromatin interaction domains, topological domains, are stable across different cell types (Dixon et al., 2012). We used cell type-specific genome sequences to generate interaction sequences. Although the interactions can be the same in different cell lines, the interaction sequences were different. The validation rates were higher than the overlapping rates suggested that the performance of the DeepHiC model trained on data from hESC was successfully validated in the data generated from the GM12878 and K562 cells. The DeepHiC model successfully predicted the functional SNPs that are identified by previous studies using Hi-C data from other cell lines (Jin et al., 2018; Zhu et al., 2018). All of these results suggest that the DeepHiC model captured the common features of chromatin interactions across different cell types. The DeepHiC model can be applied to predict chromatin interactions and potential functional variants affecting translation initiation through chromatin interaction in other cell lines.
Previous studies suggested that algorithmic incorporation of functional and evolutionary scores might resolve true causal variants (Nariai and Greenwald, 2017; Trynka et al., 2015). Two previous deep learning methods (DeFine and DeepSEA) combine the deep learning functional score with the evolutionary conservation scores to prioritize functional variants, and the evolutional conservation helped to improve the prediction accuracy. However, the prediction accuracies in GWAS and eQTL dataset were still modest (AUC ranged from 0.549 to 0.652). This may be due to the fact that the positive variants in GWAS and eQTL datasets were disease- and trait-associated and may not be causal variants. Therefore, we used enrichment analysis to show that the DeepHiC functional scores were helpful with prioritizing the non-coding variants for further functional studies.
We extended DeepHiC to prioritize functional SNPs on the basis of the predicted interaction probability. The putatively causal SNPs predicted by DeepHiC functional scores were significantly enriched in GWAS, eQTL datasets and CTCF binding sites, although no enrichment was observed for pathogenic non-coding variants from ClinVar annotations. The pathogenic non-coding variants implicated in heritable diseases are expected to have stronger functional impact compared with those non-coding variants that underlie complex diseases and traits (Liu et al., 2019). One possible cause is that these non-coding variants from ClinVar annotations may not influence traits through changing the state of chromatin interaction, but rather by transcription factor binding (Wang et al., 2018; Zhou and Troyanskaya, 2015). eQTLs were expected to be mildly correlated with pathogenicity (Liu et al., 2019). Although the enrichment score was relatively small, it can be caused by the fact that many eQTLs in high LD were associated with a target gene, but only one or a limited few of them were causal.
Previous methods including those based on evolution often assigned the variant with one score (Nariai and Greenwald, 2017; Trynka et al., 2015). However, like the eQTL study, the variant may influence the expression of several target genes with different effect sizes. In the present study, the variant may have several scores because the region that harbors the variant may interact with several different fragments. Therefore, inclusion of one evolution conservation score for each SNP may not be appropriate in this study.
During the training and testing, we corrected the sequences by incorporating the cell-type-specific genomic variants to reflect the actual genome sequences in hESC rather than using the reference genome sequences. When testing the performance in the data from GM12878 cells and K562 cells, the sequences were also corrected to reflect the actual genome sequences in GM12878 or K562 cell line. This step is very important in sequence-based models to train the model with the actual sequences in each cell line, especially for models with single-base resolution (Wang et al., 2018).
Elucidating the function of non-coding variants is difficult, since the non-coding variant may affect a number of biological activities, including splicing, transcription, post-transcription regulation, translation initiation/elongation and post-translational modification (Sauna and Kimchi-Sarfaty, 2011). It has been suggested that a single method cannot fully understand the genetic disorders caused by the non-coding variants. Our proposed method, DeepHiC, tried to predict variants affecting translation initiation through chromatin interaction. We demonstrate that an integrative approach combining artificial intelligence—deep learning with experimental evidence of chromatin interaction leads to prioritizing the functional variants in disease- and phenotype-related loci and generates the biological mechanism underlying the association.
Funding
This study was funded by grants from Natural Science Foundation of China [81570807, 30900810, 31271344 and 31071097] to Hunan Normal University. H.-W.D. and X.-H.M. were partially supported by grants [to H.-W.D.] from National Institutes of Health [R01 AR069055, U19 AG055373, R01 MH104680, R01 AR059781 and P20 GM109036], and the Edward G. Schlieder Endowment [to H.-W.D.].
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Xiang-He Meng, Centers of System Biology, Data Information and Reproductive Health, School of Basic Medical Science, Central South University, Changsha, Hunan 410008, China; Tulane Center for Biomedical Informatics and Genomics, Deming Department of Medicine, School of Medicine, Tulane University, New Orleans, LA 70112, USA; Centers of System Biology, Data Information and Reproductive Health, Laboratory of Molecular and Statistical Genetics, College of Life Sciences, Hunan Normal University, Changsha, Hunan 410081, China.
Hong-Mei Xiao, Centers of System Biology, Data Information and Reproductive Health, School of Basic Medical Science, Central South University, Changsha, Hunan 410008, China.
Hong-Wen Deng, Centers of System Biology, Data Information and Reproductive Health, School of Basic Medical Science, Central South University, Changsha, Hunan 410008, China; Tulane Center for Biomedical Informatics and Genomics, Deming Department of Medicine, School of Medicine, Tulane University, New Orleans, LA 70112, USA; Centers of System Biology, Data Information and Reproductive Health, Laboratory of Molecular and Statistical Genetics, College of Life Sciences, Hunan Normal University, Changsha, Hunan 410081, China.
References
- Ay F., Noble W.S. (2015) Analysis methods for studying the 3D architecture of the genome. Genome Biol., 16, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M. et al. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A.P. et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res., 22, 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryois J. et al. (2018) Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nature Communications, 9, 3121–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Tian W. (2016) Explaining the disease phenotype of intergenic SNP through predicted long range regulation. Nucleic Acids Research, 44, 8641–8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R. et al. (2015) Chromatin architecture reorganization during stem cell differentiation. Nature, 518, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R. et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emison E.S. et al. (2005) A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature, 434, 857–863. [DOI] [PubMed] [Google Scholar]
- Estrada K. et al. (2012) Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nature Genetics, 44, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C. et al. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genetics, 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C. et al. ; The CommonMind Consortium. (2018) A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics, 34, 2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S. et al. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.R. et al. (2014) Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature Genetics, 46, 205–212. [DOI] [PubMed] [Google Scholar]
- Jin Y. et al. (2018) Evolution of DNAase I Hypersensitive Sites in MHC Regulatory Regions of Primates. Genetics, 209, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KingmaKingma et.al D.P. 2014. Adam: a method for stochastic optimization. Preprint at arXiv. . 1412.6980. [Google Scholar]
- Kleinjan D.A. (2001) Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet., 10, 2049–2059. [DOI] [PubMed] [Google Scholar]
- Landrum M.J. et al. (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res., 42, D980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. ; 1000 Genome Project Data Processing Subgroup. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.J. et al. (2010) FastPval: a fast and memory efficient program to calculate very low P-values from empirical distribution. Bioinformatics, 26, 2897–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. et al. (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res., 41, e108–e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. et al. (2019) Biological relevance of computationally predicted pathogenicity of noncoding variants. Nat. Commun., 10, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. et al. (2013) Combining Hi-C data with phylogenetic correlation to predict the target genes of distal regulatory elements in human genome. Nucleic Acids Res., 41, 10391–10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M.T. et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science, 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertes F. et al. (2016) Combined sequencing of mRNA and DNA from human embryonic stem cells. Genomics Data, 8, 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossing M.C., Record M.T. Jr. (1986) Upstream operators enhance repression of the lac promoter. Science, 233, 889–892. [DOI] [PubMed] [Google Scholar]
- Nariai N., Greenwald W.W. (2017) Efficient prioritization of multiple causal eQTL variants via sparse polygenic modeling. Genetics, 207, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaki S.S., Boyle A.P. (2017) Mining the unknown: assigning function to noncoding single nucleotide polymorphisms. Trends Genet. TIG, 33, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira F. et al. (2009) Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet., 41, 1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna Z.E., Kimchi-Sarfaty C. (2011) Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet., 12, 683–691. [DOI] [PubMed] [Google Scholar]
- Srivastava N. et al. (2014) Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res., 15, 1929–1958. [Google Scholar]
- Trynka G. et al. (2015) Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet., 97, 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G.A. et al. (2013) From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinf., 43, 11.10.11–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. (2018) DeFine: deep convolutional neural networks accurately quantify intensities of transcription factor-DNA binding and facilitate evaluation of functional non-coding variants. Nucleic Acids Res., 46, e69–e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra H.J. et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittrant Y. et al. (2004) RANKL/RANK/OPG: new therapeutic targets in bone tumours and associated osteolysis. Biochim. Biophys. Acta, 1704, 49–57. [DOI] [PubMed] [Google Scholar]
- Yu J.T. et al. (2011) Genetic association of rs11610206 SNP on chromosome 12q13 with late-onset Alzheimer's disease in a Han Chinese population. Clin. Chim. Acta Int. J. Clin. Chem., 412, 148–151. [DOI] [PubMed] [Google Scholar]
- Zhang L. et al. (2014) Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum. Mol. Genet., 23, 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Troyanskaya O.G. (2015) Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods, 12, 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D.L. et al. (2018) Multiple functional variants at 13q14 risk locus for osteoporosis regulate RANKL expression through long-range super-enhancer. J. Bone Miner. Res Off. J. Am. Soc. Bone Miner. Res., 33, 1335–1346. [DOI] [PubMed] [Google Scholar]
- Zhu Z. et al. (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet., 48, 481–487. [DOI] [PubMed] [Google Scholar]
- Zou J. et al. (2019) A primer on deep learning in genomics. Nat. Genet., 51, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.