Abstract
Despite huge efforts towards understanding the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pathogenesis, little is known about the long‐term consequences of the disease. Here, we critically review existing literature about oncogenesis as a potential long‐term effect of SARS‐CoV‐2 infection. Like other viral infections, SARS‐CoV‐2 may promote cancer onset by inhibiting tumor suppressor genes. We conclude that, although unlikely, such hypothesis cannot be excluded a priori and we delineate an experimental approach to address it. Also see the video abstract here: https://youtu.be/TBUTDSLR7vY
Keywords: cancer, COVID‐19, oncogenic viruses, SARS‐CoV‐2
Several human viruses possess the ability to transform normal cells into cancerous cells. The potential for some severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) proteins to be oncogenic suggests a link between SARS‐CoV‐2 infection and cancer. In this essay, we review existing literature in favor or against the role of SARS‐CoV‐2 in cancer development.
INTRODUCTION
To date, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and the ongoing outbreak of the novel coronavirus disease 2019 (known as COVID‐19) has rapidly become one of the most important global health problems—infecting more than 150 million people worldwide and causing over 3 million deaths. Besides acute symptoms, there are rising concerns about long‐term effects of the viral infection. Convalescent patients, with no detectable viral load, report a variety of long‐lasting symptoms that puzzle scientists and clinicians.[ 1 , 2 ] These long‐lasting symptoms are worrisome from a public health perspective, as people experiencing them may need medical care for months or years.
A recent report proposed that among long‐term effects linked to SARS‐CoV‐2 infection there may be an oncogenic potential.[ 3 ] Here, we further discuss this hypothesis by comparing SARS‐CoV‐2 with other oncogenic viruses and evaluate potential molecular mechanisms that may link the viral infection with cancer. We conclude that although a link between SARS‐CoV‐2 and tumorigenesis is unlikely, it cannot be excluded due to the incomplete understanding of some aspects of the viral biology. To address this possibility, we believe more data are needed and propose a few experimental strategies to close this gap.
CANCER AND VIRUSES
Cancer can arise through a series of stochastic mutations in the human genome. Events that increase the rate of genetic mutation also correlate with cancer incidence. For example, the exposition of melanocytes to ultraviolet radiation induces mutations that will eventually lead to the development of melanoma.[ 4 ] Among the events that can induce cancer, viral infections are of particular interest. The relationship between viruses and cancer represents one of the most remarkable observations in modern biology. Pioneering work in poultry identified a causal link between viral infections and some forms of cancer [ 5 , 6 ] and paved the way to understanding the oncogenic potential of viruses. Such oncogenic potential was later described in other animal viruses, including human viruses.[ 7 , 8 ]
Today, the link between viral infection and cancer is well established and recognized as one of the most pressing public health problems. The mechanism through which a viral infection degenerates into cancer can vary from a sustained inflammatory reaction to a suppression of the immune system to an active reprogramming of the host cell. Broadly speaking, a virus can directly trigger cell transformation in the following ways: by providing an external oncogene, by over‐activating human oncogenes, and/or by inhibiting tumor suppressors.[ 9 ] A textbook example of virus‐mediated inhibition of oncosuppressor is the human papilloma virus (HPV). HPV infected cells express two viral proteins named E6 and E7 that bind and inhibit the two tumor suppressors p53 and pRB. HPV E7 protein contains a LXCXE motif and mutation or deletion of this short sequence abolishes the interaction between E7 and pRB.[ 10 ] In addition, the oncogenic potential of different HPV strains seems to correlate with the amount of E6 and E7 expression and with their affinity to their targets.[ 11 , 12 ]
Overall, it is not uncommon for a virus to spark cell transformation and cancer. Since Sars‐CoV‐2 is a novel virus with uncertainty about its long‐term effects, we asked whether Sars‐CoV‐2 infection could promote cancer onset.
ONCOGENIC POTENTIAL FOR SARS‐CoV‐2
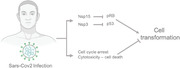
Multiple mechanisms could link Sars‐CoV‐2 infection with cancer onset. The most important one being a potential inhibition of oncosuppressors in Sars‐CoV‐2‐infected cells.
SARS‐CoV, the virus responsible for the 2002 pandemic of the severe acute respiratory syndrome (SARS), expresses the endoribonuclease non‐structural protein 15 (Nsp15) that interacts with pRB tumor suppressor through a LXCXE motif.[ 13 ] The interaction between Nsp15 and pRB induces its nuclear export and ubiquitination, targeting pRB for proteasomal degradation. NIH‐3T cells expressing Nsp15 lose contact inhibition and display an increased proliferative potential compared to wild‐type cells—indicating a role of Nsp15 in cell transformation.[ 13 ] Altogether, Nsp15 expression has the potential to inactivate pRB, to induce cell transformation and eventually cancer. Nsp15 nuclease is present and conserved in the SARS‐CoV‐2 genome, sharing 88.7% sequence similarity with its orthologue in SARS‐CoV. In addition, the pRB interacting motif LXCXE is conserved both in terms of sequence and primary structure location. Although no experimental evidence exists to date, an interaction between Nsp15 and the tumor suppressor pRB seems highly likely. In this instance, a cell infected with SARS‐CoV‐2 would experience Nsp15‐mediated pRB inhibition.
In addition to pRB inhibition, Sars‐CoV‐1 could also increase p53 degradation via non‐structural protein 3 (Nsp3) protein. The papain‐like protease of Nsp3 interacts and stabilizes the E3 ubiquitin ligase RCHY1 (ring‐finger and CHY zinc‐finger domain‐containing).[ 14 ] One of the main targets of RCHY1 is the tumor suppressor p53.[ 15 , 16 ] Stabilization of RCHY1 results in decreased levels of p53 and a weaker G1 checkpoint. Similar to Nsp15, Nsp3 is highly conserved between SARS‐CoV‐1 and SARS‐CoV‐2 (76% sequence similarity) suggesting that Nsp3 from SARS‐CoV‐2 has the potential to reduce p53 levels increasing the likelihood of cell transformation.
Both p53 and pRB are widely recognized as important tumor suppressor genes. p53 is the most frequently mutated gene in human cancer[ 17 ] and pRB is often lost in human malignancies (reviewed in [18, 19]). In addition, germ‐line mutation of either gene results in genetic diseases associated with increased cancer risk.[ 20 , 21 ] The ability of SARS‐CoV‐2′s proteins to inhibit both p53 and pRB suggests that SARS‐CoV‐2 may have an oncogenic potential through a mechanism similar to HPV (see above).
SARS‐CoV‐2 AND CANCER: ARGUMENTS AGAINST
In the previous section, we described how some proteins of SARS‐CoV‐2 may exert oncogenic potential by inhibiting tumor suppressor genes. However, the relationship between a virus and its host is a complex phenomenon, involving several regulators from both the virus and the host. Although the expression of certain proteins may have an oncogenic effect, this does not mean that the viral infection necessarily degenerates into cancer.
Well‐known oncogenic viruses typically establish long‐term relationships with their host through some forms of latency or persistence. Two infamous examples are HPV, which follows the differentiation of keratinocytes as well as herpes simplex virus—which remains latent into neural ganglia. Unlike these oncogenic viruses, most Sars‐CoV‐2 infections resolve in a limited amount of time. A recent work speculated about the possibility of viral latency in the testis[ 22 ]; however, Pan and colleagues found no evidence of Sars‐CoV‐2 genome in the semen of convalescent males.[ 23 ] While the persistence of Sars‐CoV‐2 RNA in fecal samples may indicate a potential gastrointestinal reservoir, no direct evidence has been reported. Furthermore, RNA persistence in fecal samples is limited to a period of months which is much shorter than the infections caused by other oncogenic viruses.[ 24 , 25 , 26 ] Other reports of “chronic” SARS‐CoV‐2 infections or “reactivation” of the virus can be more easily explained by the worsening of low‐grade infections rather than by the existence of a viral reservoir. The fact that SARS‐CoV‐2 does not seem to establish a long‐lasting infection argues against its role in cancer onset.
If Sars‐CoV‐2 does not establish a stable relationship with its host, the only possibility for cell transformation relies on the ability of infected cells to escape immune surveillance and the cytopathic effect of the virus. This possibility is particularly attractive considering the link between pRB haploinsufficiency and chromosomal instability. The loss of a single pRB copy induces chromosomal instability[ 27 ] and creates an optimal environment for cell transformation, even in the case of an acute and transient event.[ 28 ] Although Nsp15‐mediated pRB inhibition and genomic instability are likely to self‐resolve with virus clearance, the mutations that originated through genomic instability are irreversible. Some of these mutations may have long‐term effects and increase susceptibility to cancer even years after the primary infection. However, SARS‐CoV‐2 infection results in extensive tissue damage and cell death in cultured cells and organoids[ 29 , 30 , 31 ]; this indicates that the virus tends to kill its host and severely reduces the likelihood for cell transformation. Some lines of evidence suggest that alternative outcomes of viral infection exist. Recent results indicate that the egression strategy of β‐coronaviruses does not cause major alterations of the plasma membrane or increases in cell death 18 h post‐infection.[ 32 , 33 ] In addition, SARS‐CoV‐2 infects but does not replicate in at least some cell types such as monocytes and monocyte‐derived macrophages.[ 34 ] The reduced cytotoxicity and the existence of “abortive” viral cycle indicate that cell death may not be the only possible outcome of SARS‐CoV‐2 infections; in this context, the host would experience pRB and p53 inhibition without the cytopathic effect of the virus.
Considering a scenario where some cells survive the infection, the chances of cell transformation are linked to an active cell cycle machinery. While SARS‐CoV‐2 seems to promote cell cycle progression by inhibiting pRB and counteracting the G1 checkpoint, other data suggest a cell cycle arrest in the infected cells. β‐Coronaviruses display multiple mechanisms to block cell cycle progression in the host cell (reviewed in [35]). For example, SARS‐CoV‐1 inhibits pRB phosphorylation through 3a and 7b proteins and inhibits cyclin‐cyclin‐dependent kinases activity through the N‐protein, inducing a cell cycle arrest.[ 36 , 37 , 38 ] Similarly, the coronavirus infectious bronchitis virus induces a G2/M arrest in cells lacking p53[ 39 ] and SARS‐CoV‐2 infected cells also arrest in G2/M.[ 40 ] Moreover, cells infected by SARS‐CoV‐2 activate the pro‐apoptotic regulator caspase 8 and undergo programmed cell death.[ 41 ] Overall, even postulating the cell survival following SARS‐CoV‐2 infection, the chances of cell transformation are severely reduced by the virus‐induced cell cycle arrest and by the activation of the apoptosis cascade.
A final piece of evidence arguing against oncogenic potential of SARS‐CoV‐2 comes from epidemiological data on SARS‐CoV‐1. Extensive follow‐up studies on the long‐term symptoms of SARS‐CoV‐1 have not reported increased cancer incidence thus far[ 42 , 43 , 44 ] arguing against an increased susceptibility of cancer in SARS‐CoV‐1‐infected individuals. It must be noted that only a subset of the 8096 total reported cases of SARS‐CoV‐1 were analyzed to epidemiologically investigate the long‐term effects of the syndrome. In addition, it is plausible that increased incidence of cancer has not yet arisen in the relatively short period of time between the 2002–2004 SARS outbreak and the time of the study.
In conclusion, although cells infected by SARS‐CoV‐2 may experience downregulation of the tumor suppressors pRB and p53, the virus‐mediated cell cycle arrest and the cytopathic effect argue against a direct oncogenic potential of SARS‐CoV‐2. However, one cannot exclude the possibility of cell transformation in cell types where the virus undergoes abortive cycles and in conditions where the cytopathic effects are reduced.
SARS‐CoV‐2 AND CANCER: AN EXPERIMENTAL ROADMAP
In previous sections, we speculated about the mechanisms that could link SARS‐CoV‐2 with cancer and reviewed the evidence for and against such a hypothesis. In this section, we consider how to address any outstanding questions experimentally.
Histology is a classic technique to detect early signs of cell transformation in biopsies and autopsies. Thus far, post‐mortem histological analysis of SARS‐CoV‐2 focused on patients that have died from the infection without specifically searching for cancer markers.[ 45 , 46 ] In the future, it would be interesting to study individuals that have recovered from the disease and died of independent causes at different time after the infection. This analysis should focus primarily on tissues with high trophism for SARS‐CoV‐2, such as the lungs, as these are the prime suspect location for cancer to potentially develop.
In parallel with the histological analysis, important evidence on the oncogenic potential of SARS‐CoV‐2 could come from epidemiological studies on patients that have recovered from COVID‐19 syndrome. The unprecedented detail and quality of epidemiological data collected during the COVID‐19 pandemic, along with recent advancement in data analysis, will be invaluable tools for evaluating the long‐term effects of the infection.
Both histological and epidemiological data could show a correlation between COVID‐19 and cancer, but a more direct approach is needed to demonstrate causation. In this context, the cellular effects of the virus on cell physiology remain only partially understood. One should assess the long‐term effects of SARS‐CoV‐2 on cell cultures and organoids using functional assays such as proliferation curves, soft agar assay, and cell cycle profile analysis. A replication‐defective or attenuated version of the virus is an important tool to differentiate the viral effects on cell physiology from its cytotoxicity.
Although cells and organoids enable investigations of biological phenomena at high resolution, they often fall short in capturing the complexity of an organism. For this reason, it is crucial to assess the long‐term effects of mild SARS‐CoV‐2 infection in laboratory animals. The effects of SARS‐CoV‐2 infection can be modeled in several animals.[ 47 , 48 ] The development of mice expressing human angiotensin‐converting enzyme 2 (hACE2 – K18‐hACE2 mice) receptors[ 49 , 50 , 51 ] and the availability of cancer mouse models[ 52 ] makes Mus musculus a promising model to study the effect of SARS‐CoV‐2 infection and cancer development. It would be interesting to cross p53−/+[ 53 , 54 ] with K18‐hACE2 mice to investigate the effect of tumor development in SARS‐CoV‐2‐infected mice compared to non‐infected mice.
CONCLUSIONS
The novel coronavirus, SARS‐CoV‐2, emerged for the first time in China in December 2019.
It is characterized by modest mortality (Infection Fatality Rate, circa 0.6) and high infectiousness that can cause a severe respiratory syndrome known as COVID‐19.[ 55 ] There is a growing body of evidence that indicates long‐term effects well after primary SARS‐CoV‐2 infections have resolved. Among these long‐term effects, we have speculated about the oncogenic potential of SARS‐CoV‐2.
The viral proteins Nsp15 or Nsp3 are likely to have pro‐oncogenic effects by inhibiting the two important tumor suppressors, pRB and p53. On the other hand, the transient nature of the infection, the virus mediated cell cycle arrest, and the cytopathic effect argue against the possibility of cell transformation. The most significant contribution of the virus to cancer could be indirect and related to the extensive and irreversible lung fibrosis that characterizes severe COVID‐19 pneumonia.[ 56 , 57 , 58 , 59 ]
Nevertheless, the effects of SARS‐CoV‐2 infection on a single cell remain unclear and may be dramatically different depending on the cell type and the initial viral load. In conclusion, the role of SARS‐CoV‐2 in cell transformation is unlikely but cannot be excluded in specific contexts. Additional studies on the long‐term effects of the virus are needed to shed light on such possibility.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
We would like to thank Prof. Patrick Meraldi (University of Geneva, Geneva) and Prof. Jonathon Pines (The Institute for Cancer Research, London) for their support. We are grateful to Matteo Bosso (Institute of Molecular Virology, Ulm University Hospital) for critically reading the manuscript. A.S. is supported by IGE3 grant and L.C. is supported by the SNF fellowship P2GEP3‐187775.
Stingi, A. , & Cirillo, L . (2021). SARS‐CoV‐2 infection and cancer. BioEssays, 43, e2000289. 10.1002/bies.202000289
Aureliano Stingi and Luca Cirillo contributed equally to this study.
Contributor Information
Aureliano Stingi, Email: Aureliano.Stingi@unige.ch.
Luca Cirillo, Email: Luca.Cirillo@icr.ac.uk.
DATA AVAILABILITY STATEMENT
Data sharing not applicable – no new data generated.
REFERENCES
- 1. Gao, L. , Ge, M. Q. , Hung, I. F. N. , Ip, M. S. M. , Ip, P. , Lau, K. K. , Lau, L. K. W. , Leung, W. K. , Li, X. , Luo, H. , Man, K. K. C. , Ng, V. W. S. , Wan, E. Y. F. , Wing, Y. K. , Wong, C. S. M. , Wong, K. H. T. , Wong, I. C. K. , Chan, A. Y. L. , Chan, E. W. , … Chui, C. S. L. (2020). Short‐ and potential long‐term adverse health outcomes of COVID‐19: A rapid review. Emerging Microbes & Infections, 9(1), 2190–2199. 10.1080/22221751.2020.1825914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yelin, D. , Wirtheim, E. , Vetter, P. , Kalil, A. C. , Bruchfeld, J. , Runold, M. , Guaraldi, G. , Mussini, C. , Gudiol, C. , Pujol, M. , Bandera, A. , Scudeller, L. , Paul, M. , Kaiser, L. , & Leibovici, L. (2020). Long‐term consequences of COVID‐19: Research needs. The Lancet Infectious Diseases, 20(10), 1115–1117. 10.1016/S1473-3099(20)30701-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alpalhão, M. , Ferreira, J. A. , & Filipe, P. (2020). Persistent SARS‐CoV‐2 infection and the risk for cancer. Medical Hypotheses, 143(May), 109882. 10.1016/j.mehy.2020.109882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arisi, M. , Zane, C. , Caravello, S. , Rovati, C. , Zanca, A. , Venturini, M. , & Calzavara‐Pinton, P. (2018). Sun exposure and melanoma, certainties and weaknesses of the present knowledge. Frontiers in Medicine, 5, 235. 10.3389/fmed.2018.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellermann, V. , & Bang, O. (1909). Experimentelle Leukämie bei Hühnern. II. Zeitschrift für Hygiene und Infektionskrankheiten, 63(1), 231–272. 10.1007/BF02227892 [DOI] [Google Scholar]
- 6. Rous, P. (1910). A transmissible avian neoplasm. (Sarcoma of the common fowl.). The Journal of Experimental Medicine, 12(5), 696–705. 10.1084/jem.12.5.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burkitt, D. (1958). A sarcoma involving the jaws in African children. The British Journal of Surgery, 46(197), 218–223. 10.1002/bjs.18004619704 [DOI] [PubMed] [Google Scholar]
- 8. Epstein, M. A. , Henle, G. , Achong, B. G. , & Barr, Y. M. (1965). Morphological and biological studies on a virus in cultured lymphoblasts from Burkitt's lymphoma. The Journal of Experimental Medicine, 121(5), 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mesri, E. A. , Feitelson, M. A. , & Munger, K. (2014). Human viral oncogenesis: A cancer hallmarks analysis. Cell Host & Microbe, 15(3), 266–282. 10.1016/j.chom.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim, H.‐Y. , Ahn, B.‐Y. , & Cho, Y. (2001). Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. The EMBO Journal, 20(1–2), 295–304. 10.1093/emboj/20.1.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Araldi, R. P. , Sant'Ana, T. A. , Módolo, D. G. , de Melo, T. C. , Spadacci‐Morena, D. D. , de Cassia Stocco, R. , Cerutti, J. M. , & Souza, E. B. (2018). The human papillomavirus (HPV)‐related cancer biology: An overview. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 106, 1537–1556. 10.1016/j.biopha.2018.06.149 [DOI] [PubMed] [Google Scholar]
- 12. Brianti, P. , De Flammineis, E. , & Mercuri, S. R. (2017). Review of HPV‐related diseases and cancers. The New Microbiologica, 40(2), 80–85. [PubMed] [Google Scholar]
- 13. Bhardwaj, K. , Liu, P. , Leibowitz, J. L. , & Kao, C. C. (2012). The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. Journal of Virology, 86(8), 4294–4304. 10.1128/JVI.07012-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma‐Lauer, Y. , Carbajo‐Lozoya, J. , Hein, M. Y. , Müller, M. A. , Deng, W. , Lei, J. , Meyer, B. , Kusov, Y. , von Brunn, B. , Bairad, D. R. , Hünten, S. , Drosten, C. , Hermeking, H. , Leonhardt, H. , Mann, M. , Hilgenfeld, R. , & von Brunn, A. (2016). P53 down‐regulates SARS coronavirus replication and is targeted by the SARS‐unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proceedings of the National Academy of Sciences of the United States of America, 113(35), E5192–E5201. 10.1073/pnas.1603435113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leng, R. P. , Lin, Y. , Ma, W. , Wu, H. , Lemmers, B. , Chung, S. , Parant, J. M. , Lozano, G. , Hakem, R. , & Benchimol, S. (2003). Pirh2, a p53‐induced ubiquitin‐protein ligase, promotes p53 degradation. Cell, 112(6), 779–791. 10.1016/s0092-8674(03)00193-4 [DOI] [PubMed] [Google Scholar]
- 16. Sheng, Y. , Laister, R. C. , Lemak, A. , Wu, B. , Tai, E. , Duan, S. , Lukin, J. , Sunnerhagen, M. , Srisailam, S. , Karra, M. , Benchimol, S. , & Arrowsmith, C. H. (2008). Molecular basis of Pirh2‐mediated p53 ubiquitylation. Nature Structural & Molecular Biology, 15(12), 1334–1342. 10.1038/nsmb.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Surget, S. , Khoury, M. P. , & Bourdon, J.‐C. (2013). Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets and Therapy, 7, 57–68. 10.2147/OTT.S53876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burkhart, D. L. , & Sage, J. (2008). Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nature Reviews. Cancer, 8(9), 671–682. 10.1038/nrc2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanahan, D. , & Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell, 144(5), 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 20. McBride, K. A. , Ballinger, M. L. , Killick, E. , Kirk, J. , Tattersall, M. H. N. , Eeles, R. A. , Thomas, D. M. , & Mitchell, G. (2014). Li‐Fraumeni syndrome: Cancer risk assessment and clinical management. Nature Reviews Clinical Oncology, 11(5), 260–271. 10.1038/nrclinonc.2014.41 [DOI] [PubMed] [Google Scholar]
- 21. Thériault, B. L. , Dimaras, H. , Gallie, B. L. , & Corson, T. W. (2014). The genomic landscape of retinoblastoma: A review. Clinical & Experimental Ophthalmology, 42(1), 33–52. 10.1111/ceo.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. López‐Romero, R. , Nambo‐Lucio, M. J. , Salcedo‐Carrillo, E. , Hernández‐Cueto, M. , L. Á., & Salcedo‐Vargas, M. (2020). The big challenge of SARS‐CoV‐2 latency: Testes as reservoir. Gaceta Medica De Mexico, 156(4), 328–333. 10.24875/GMM.20000295 [DOI] [PubMed] [Google Scholar]
- 23. Pan, F. , Xiao, X. , Guo, J. , Song, Y. , Li, H. , Patel, D. P. , Spivak, A. M. , Alukal, J. P. , Zhang, X. , Xiong, C. , Li, P. S. , & Hotaling, J. M. (2020). No evidence of severe acute respiratory syndrome‐coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertility and Sterility, 113(6), 1135–1139. 10.1016/j.fertnstert.2020.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo, M. , Tao, W. , Flavell, R. A. , & Zhu, S. (2021). Potential intestinal infection and faecal‐oral transmission of SARS‐CoV‐2. Nature Reviews. Gastroenterology & Hepatology, 18(4), 269–283. 10.1038/s41575-021-00416-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalkeri, R. , Goebel, S. , & Sharma, G. D. (2020). SARS‐CoV‐2 shedding from asymptomatic patients: Contribution of potential extrapulmonary tissue reservoirs. The American Journal of Tropical Medicine and Hygiene, 103(1), 18–21. 10.4269/ajtmh.20-0279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xing, Y.‐H. , Ni, W. , Wu, Q. , Li, W.‐J. , Li, G.‐J. , Wang, W.‐D. , Tong, J.‐N. , Song, X.‐F. , Wing‐Kin Wong, G. , & Xing, Q.‐S. (2020). Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. Journal of Microbiology, Immunology and Infection, 53(3), 473–480. 10.1016/j.jmii.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coschi, C. H. , Ishak, C. A. , Gallo, D. , Marshall, A. , Talluri, S. , Wang, J. , Cecchini, M. J. , Martens, A. L. , Percy, V. , Welch, I. , Boutros, P. C. , Brown, G. W. , & Dick, F. A. (2014). Haploinsufficiency of an RB – E2F1 – Condensin II complex leads to aberrant replication and aneuploidy. 4, 840–853. 10.1158/2159-8290.CD-14-0215 [DOI] [PubMed]
- 28. Manning, A. L. , Longworth, M. S. , & Dyson, N. J. (2010). Loss of pRB causes centromere dysfunction and chromosomal instability. Genes and Development, 24(13), 1364–1376. 10.1101/gad.1917310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mason, R. J. (2020). Pathogenesis of COVID‐19 from a cell biology perspective. European Respiratory Journal, 55(4), 2000607. 10.1183/13993003.00607-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou, J. , Li, C. , Liu, X. , Chiu, M. C. , Zhao, X. , Wang, D. , Wei, Y. , Lee, A. , Zhang, A. J. , Chu, H. , Cai, J.‐P. , Yip, C. C.‐Y. , Chan, I. H.‐Y. , Wong, K. K.‐Y. , Tsang, O. T.‐Y. , Chan, K.‐H. , Chan, J. F.‐W. , To, K. K.‐W. , Chen, H. , & Yuen, K. Y. (2020). Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nature Medicine, 26(7), 1077–1083. 10.1038/s41591-020-0912-6 [DOI] [PubMed] [Google Scholar]
- 31. Zhu, N. , Wang, W. , Liu, Z. , Liang, C. , Wang, W. , Ye, F. , Huang, B. , Zhao, L. , Wang, H. , Zhou, W. , Deng, Y. , Mao, L. , Su, C. , Qiang, G. , Jiang, T. , Zhao, J. , Wu, G. , Song, J. , & Tan, W. (2020). Morphogenesis and cytopathic effect of SARS‐CoV‐2 infection in human airway epithelial cells. Nature Communications, 11(1), 3910. 10.1038/s41467-020-17796-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghosh, S. , Dellibovi‐Ragheb, T. A. , Kerviel, A. , Pak, E. , Qiu, Q. , Fisher, M. , Takvorian, P. M. , Bleck, C. , Hsu, V. W. , Fehr, A. R. , Perlman, S. , Achar, S. R. , Straus, M. R. , Whittaker, G. R. , de Haan, C. A. M. , Kehrl, J. , & Altan‐Bonnet, G. , & Altan‐Bonnet, N . (2020). β‐Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell, 183(6), 1520–1535.e14. 10.1016/j.cell.2020.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. V'kovski, P. , Kratzel, A. , Steiner, S. , Stalder, H. , & Thiel, V. (2021). Coronavirus biology and replication: Implications for SARS‐CoV‐2. Nature Reviews. Microbiology, 19(3), 155–170. 10.1038/s41579-020-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boumaza, A. , Gay, L. , Mezouar, S. , Bestion, E. , Diallo, A. B. , Michel, M. , Desnues, B. , Raoult, D. , La Scola, B. , Halfon, P. , Vitte, J. , Olive, D. , & Mege, J.‐L. (2021). Monocytes and macrophages, targets of SARS‐CoV‐2: The clue for Covid‐19 immunoparalysis. The Journal of Infectious Diseases, jiab044. 10.1093/infdis/jiab044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su, M. , Chen, Y. , Qi, S. , Shi, D. , Feng, L. , & Sun, D. (2020). A mini‐review on cell cycle regulation of coronavirus infection. Frontiers in Veterinary Science, 7, 943. 10.3389/fvets.2020.586826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Surjit, M. , Liu, B. , Chow, V. T. K. , & Lal, S. K. (2006). The nucleocapsid protein of severe acute respiratory syndrome‐coronavirus inhibits the activity of cyclin‐cyclin‐dependent kinase complex and blocks S phase progression in mammalian cells. The Journal of Biological Chemistry, 281(16), 10669–10681. 10.1074/jbc.M509233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuan, X. , Yao, Z. , Wu, J. , Zhou, Y. , Shan, Y. , Dong, B. , Zhao, Z. , Hua, P. , Chen, J. , & Cong, Y. (2007). G1 phase cell cycle arrest induced by SARS‐CoV 3a protein via the cyclin D3/pRb pathway. American Journal of Respiratory Cell and Molecular Biology, 37(1), 9–19. 10.1165/rcmb.2005-0345RC [DOI] [PubMed] [Google Scholar]
- 38. Yuan, X. , Wu, J. , Shan, Y. , Yao, Z. , Dong, B. , Chen, B. , Zhao, Z. , Wang, S. , Chen, J. , & Cong, Y. (2006). SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology, 346(1), 74–85. 10.1016/j.virol.2005.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li, F. Q. , Tam, J. P. , & Liu, D. X. (2007). Cell cycle arrest and apoptosis induced by the coronavirus infectious bronchitis virus in the absence of p53. Virology, 365(2), 435–445. 10.1016/j.virol.2007.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bouhaddou, M. , Memon, D. , Meyer, B. , White, K. M. , Rezelj, V. V. , Correa Marrero, M. , Polacco, B. J. , Melnyk, J. E. , Ulferts, S. , Kaake, R. M. , Batra, J. , Richards, A. L. , Stevenson, E. , Gordon, D. E. , Rojc, A. , Obernier, K. , Fabius, J. M. , Soucheray, M. , Miorin, L. , …, & Krogan, N. J. (2020). The global phosphorylation landscape of SARS‐CoV‐2 infection. Cell, 182(3), 685–712.e19. 10.1016/j.cell.2020.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li, S. , Zhang, Y. , Guan, Z. , Li, H. , Ye, M. , Chen, X. , Shen, J. , Zhou, Y. , Shi, Z.‐L. , Zhou, P. , & Peng, K. (2020). SARS‐CoV‐2 triggers inflammatory responses and cell death through caspase‐8 activation. Signal Transduction and Targeted Therapy, 5(1), 235. 10.1038/s41392-020-00334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lam, M. H. ‐B. (2009). Mental morbidities and chronic fatigue in severe acute respiratory syndrome survivors: Long‐term follow‐up. Archives of Internal Medicine, 169(22), 2142–2147. 10.1001/archinternmed.2009.384 [DOI] [PubMed] [Google Scholar]
- 43. Ngai, J. C. , Ko, F. W. , Ng, S. S. , To, K.‐W. , Tong, M. , & Hui, D. S. (2010). The long‐term impact of severe acute respiratory syndrome on pulmonary function, exercise capacity and health status. Respirology, 15(3), 543–550. 10.1111/j.1440-1843.2010.01720.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang, P. , Li, J. , Liu, H. , Han, N. , Ju, J. , Kou, Y. , Chen, L. , Jiang, M. , Pan, F. , Zheng, Y. , Gao, Z. , & Jiang, B. (2020). Long‐term bone and lung consequences associated with hospital‐acquired severe acute respiratory syndrome: A 15‐year follow‐up from a prospective cohort study. Bone Research, 8(1), 1–8. 10.1038/s41413-020-0084-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanley, B. , Naresh, K. N. , Roufosse, C. , Nicholson, A. G. , Weir, J. , Cooke, G. S. , Thursz, M. , Manousou, P. , Corbett, R. , Goldin, R. , Al‐Sarraj, S. , Abdolrasouli, A. , Swann, O. C. , Baillon, L. , Penn, R. , Barclay, W. S. , Viola, P. , & Osborn, M. (2020). Histopathological findings and viral tropism in UK patients with severe fatal COVID‐19: A post‐mortem study. The Lancet Microbe, 1(6), e245–e253. 10.1016/S2666-5247(20)30115-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pernazza, A. , Mancini, M. , Rullo, E. , Bassi, M. , De Giacomo, T. , Rocca, C. D. , & d'Amati, G. (2020). Early histologic findings of pulmonary SARS‐CoV‐2 infection detected in a surgical specimen. Virchows Archiv: An International Journal of Pathology, 477(5), 743–748. 10.1007/s00428-020-02829-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johansen, M. D. , Irving, A. , Montagutelli, X. , Tate, M. D. , Rudloff, I. , Nold, M. F. , Hansbro, N. G. , Kim, R. Y. , Donovan, C. , Liu, G. , Faiz, A. , Short, K. R. , Lyons, J. G. , McCaughan, G. W. , Gorrell, M. D. , Cole, A. , Moreno, C. , Couteur, D. , Hesselson, D. , … Hansbro, P. M. (2020). Animal and translational models of SARS‐CoV‐2 infection and COVID‐19. Mucosal Immunology, 13(6), 877–891. 10.1038/s41385-020-00340-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Muñoz‐Fontela, C. , Dowling, W. E. , Funnell, S. G. P. , Gsell, P.‐S. , Riveros‐Balta, A. X. , Albrecht, R. A. , Andersen, H. , Baric, R. S. , Carroll, M. W. , Cavaleri, M. , Qin, C. , Crozier, I. , Dallmeier, K. , de Waal, L. , de Wit, E. , Delang, L. , Dohm, E. , Duprex, W. P. , Falzarano, D. , … Barouch, D. H. (2020). Animal models for COVID‐19. Nature, 586(7830), 509–515. 10.1038/s41586-020-2787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hassan, A. O. , Case, J. B. , Winkler, E. S. , Thackray, L. B. , Kafai, N. M. , Bailey, A. L. , McCune, B. T. , Fox, J. M. , Chen, R. E. , Alsoussi, W. B. , Turner, J. S. , Schmitz, A. J. , Lei, T. , Shrihari, S. , Keeler, S. P. , Fremont, D. H. , Greco, S. , McCray, P. B. , Perlman, S. , …, & Diamond, M. S. (2020). A SARS‐CoV‐2 infection model in mice demonstrates protection by neutralizing antibodies. Cell, 182(3), 744–753.e4. 10.1016/j.cell.2020.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rathnasinghe, R. , Strohmeier, S. , Amanat, F. , Gillespie, V. L. , Krammer, F. , García‐Sastre, A. , Coughlan, L. , Schotsaert, M. , & Uccellini, M. (2020). Comparison of transgenic and adenovirus hACE2 mouse models for SARS‐CoV‐2 infection. bioRxiv. 10.1101/2020.07.06.190066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao, J. , Li, K. , Wohlford‐Lenane, C. , Agnihothram, S. S. , Fett, C. , Zhao, J. , Gale, M. J. , Baric, R. S. , Enjuanes, L. , Gallagher, T. , McCray, P. B. , & Perlman, S. (2014). Rapid generation of a mouse model for Middle East respiratory syndrome. Proceedings of the National Academy of Sciences of the United States of America, 111(13), 4970–4975. 10.1073/pnas.1323279111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lampreht Tratar, U. , Horvat, S. , & Cemazar, M. (2018). Transgenic mouse models in cancer research. Frontiers in Oncology, 8, 268. 10.3389/fonc.2018.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harvey, M. , McArthur, M. J. , Montgomery, C. A. , Butel, J. S. , Bradley, A. , & Donehower, L. A. (1993). Spontaneous and carcinogen‐induced tumorigenesis in p53‐deficient mice. Nature Genetics, 5(3), 225–229. 10.1038/ng1193-225 [DOI] [PubMed] [Google Scholar]
- 54. Kemp, C. J. , Wheldon, T. , & Balmain, A. (1994). P53‐deficient mice are extremely susceptible to radiation‐induced tumorigenesis. Nature Genetics, 8(1), 66–69. 10.1038/ng0994-66 [DOI] [PubMed] [Google Scholar]
- 55. Meyerowitz‐Katz, G. , & Merone, L. (2020). A systematic review and meta‐analysis of published research data on COVID‐19 infection‐fatality rates. International Journal of Infectious Diseases, 101, 138–148. 10.1016/j.ijid.2020.09.1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. George, P. M. , Wells, A. U. , & Jenkins, R. G. (2020). Pulmonary fibrosis and COVID‐19: The potential role for antifibrotic therapy. The Lancet Respiratory Medicine, 8(8), 807–815. 10.1016/S2213-2600(20)30225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lechowicz, K. , Drożdżal, S. , Machaj, F. , Rosik, J. , Szostak, B. , Zegan‐Barańska, M. , Biernawska, J. , Dabrowski, W. , Rotter, I. , & Kotfis, K. (2020). COVID‐19: The potential treatment of pulmonary fibrosis associated with SARS‐CoV‐2 infection. Journal of Clinical Medicine, 9(6), 1917. 10.3390/jcm9061917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Spagnolo, P. , Balestro, E. , Aliberti, S. , Cocconcelli, E. , Biondini, D. , Casa, G. D. , Sverzellati, N. , & Maher, T. M. (2020). Pulmonary fibrosis secondary to COVID‐19: A call to arms? The Lancet Respiratory Medicine, 8(8), 750–752. 10.1016/S2213-2600(20)30222-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tao, S.‐L. , Wang, X. , Feng, Y. , Kang, P. , Li, Q. , Sun, T. , Tan, Q. , & Deng, B. (2020). Is the presence of lung injury in COVID‐19 an independent risk factor for secondary lung cancer? Medical Hypotheses, 143, 110074. 10.1016/j.mehy.2020.110074 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable – no new data generated.


