Abstract
Reverse‐Transcription Polymerase Chain Reaction (RT‐PCR) method is currently the gold standard method for detection of viral strains in human samples, but this technique is very expensive, take time and often leads to misdiagnosis. The recent outbreak of COVID‐19 has led scientists to explore other options such as the use of artificial intelligence driven tools as an alternative or a confirmatory approach for detection of viral pneumonia. In this paper, we utilized a Convolutional Neural Network (CNN) approach to detect viral pneumonia in x‐ray images using a pretrained AlexNet model thereby adopting a transfer learning approach. The dataset used for the study was obtained in the form of optical Coherence Tomography and chest X‐ray images made available by Kermany et al. (2018, https://doi.org/10.17632/rscbjbr9sj.3) with a total number of 5853 pneumonia (positive) and normal (negative) images. To evaluate the average efficiency of the model, the dataset was split into on 50:50, 60:40, 70:30, 80:20 and 90:10 for training and testing respectively. To evaluate the performance of the model, 10 K Cross‐validation was carried out. The performance of the model using overall dataset was compared with the means of cross‐validation and the currents state of arts. The classification model has shown high performance in terms of accuracy, sensitivity and specificity. 70:30 split performed better compare to other splits with accuracy of 98.73%, sensitivity of 98.59% and specificity of 99.84%.
Keywords: artificial intelligence, CNN, COVID‐19, pretrained AlexNet, viral pneumonia
Abbreviations
- AI
Artificial Intelligence
- AUC
Area Under the Curve
- BN
Batch Normalization
- CAD
Computer Assisted Diagnosis
- CNN
Convolutional Neural Network
- CONV
Convolution
- CT
Computerized Tomography
- CXR
Chest X‐ray
- DL
Deep Learning
- FCL
Fully Connected Layers
- FM
Feature Map
- FN
False Negative
- FP
False Positive
- GPU
Graphical Processing Unit
- MERS
Middle East Respiratory Syndrome
- ML
Machine Learning
- MRI
Magnetic Resonance Imaging
- RAM
Random Access
- ReLu
Rectified Linear Unit (ReLu)
- RT‐PCR
Reverse‐Transcription Polymerase Chain Reaction
- SAR‐CoV‐1 and 2
Severe Acute Respiratory Syndrome Coronavirus 1 and 2
- SVM
Support Vector Machine
- TL
Transfer Learning
- TN
Transfer Learning
- TP
True Positive
- WHO
World Health Organization
1. INTRODUCTION
Pneumonia is a disease caused by different types of pathogens, which include viruses, bacteria and fungi. Different species that causes pneumonia are shown in Table 1. According to World Health Organization (2018), over 4 million premature deaths occur as a result of diseases related to household air pollution including pneumonia and Tuberculosis. More than 150 million people were estimated to be infected with pneumonia annually and the disease is more prevalence in children less than 5 years of age. Globally, pneumonia is among the top diseases that affect children and account for 15% of mortality of infants and children below 5 years leading to over 1.4 million death in 2018 and 2.56 million in 2017. Even though the prevalence of the disease is common between children, it can also affect all age range. The cases of pneumonia are predominant in underdeveloped countries with poor healthcare sectors, lack of medical personnel and resources for diagnosis and treatment (Gilani et al., 2012; Stephen et al., 2019).
TABLE 1.
Classification of pneumonia based on pathogens
| Pathogen | Species |
|---|---|
| Viruses | Influenza virus, Severe Acute Respiratory Syndrome Coronavirus (SAR‐CoV‐1 and 2), Middle East Respiratory Syndrome (MERS) Coronavirus, Adenovirus, Enteroviruses, Hantavirus etc. |
| Bacteria | Legionella pneumophila, Streptococcus pneumoniae, Mycoplasma pneumoniae, Chlamydophila pneumoniae etc. |
| Fungi | Aspergillus spp, Histoplasmosis, Pneumocystis jirovecii, Coccidioidomycosis, Mucoromycetes, Cryptococcus etc. |
COVID‐19 is among diseases cause by virus from the Coronaviridae family. Several strains of this family have caused global concerns in the past such as Middle East respiratory syndrome coronavirus (MERS‐CoV) in 2012 and severe acute respiratory syndrome coronavirus (SARS‐CoV) in 2002 (Dowel et al., 2004; Oboho et al., 2015). COVID‐19 was declared pandemic by the World Health Organization (WHO) in mid‐March 2020 as a result of outbreak of a new viral strain, which was first recorded on the eve of January, 2020 in Wuhan China. COVID‐19 has spread to almost every country infecting more than 30 million with over 800 thousand deaths globally as of 08 October, 2020 (WHO, 2020). The major symptoms of COVID‐19 include fever, cough, difficulty in breathing and in severe cases, it can lead to pneumonia, kidney failure and eventually death (Banerjee et al., 2019; Chen et al., 2020). The disease is more severe to patients suffering from other diseases such as impaired immune system disorders, patients placed on a ventilator machine, people who smoke and patients suffering from asthma and other chronic diseases (Kolhar et al., 2020; Rahman et al., 2020; Srivastava et al., 2020).
The use of artificial intelligence and machine learning in healthcare systems is growing exponentially due to its ability in detecting diseases, diagnosing clinical issues, discovering drugs, etc. The use of specific machine learning models has even outperformed both microbiologists and pathologists in diagnosis of specific cases due to their pattern recognition ability (Bakator & Radosav, 2018; Hu et al., 2020; Paules et al., 2020; Wang, Casalino, et al., 2019).
Clinicians employ different approaches to diagnose viral pneumonia such as the use of blood test, chest x‐ray, sputum test and pulse oximetry. The gold standard technique is the use of RT‐PCR for detection of the viral strain and the use of Computerized Tomography (CT) scan images which are interpreted by radiologist. Even though many studies have reported the efficiency of using artificial intelligence models for detection of viral pneumonia, these approaches are limited to the use of CT scan images acquired from patients who visited clinics or in a health care setting. Different strains of viruses are associated with viral pneumonia such as Influenza virus, Respiratory syncytial virus, Human metapneumovirus, adenoviruses, coronaviruses with COVID‐19 as the recent viral strain on the list (Chowdhury et al., 2020; Ruuskanen et al., 2011). The unavailability of test kit and lockdown of cities as a result of the COVID‐19 pandemic are other major challenges. In order to solve these challenges, we utilized pretrained AlexNet Model to classify viral pneumonia and normal (i.e., healthy) CT scan.
The integration of IoT, artificial intelligence and biosensors contributed to the advancement of smart systems that can be used to detect, manage and control diseases. The use of smart sensing tools and monitoring devices designed using chips and sensors improved various aspect of healthcare systems in terms of detection of pathogens that causes disease, monitoring of medication, storage and analysis of vital signals, medical records management and rehabilitation of diseases. The potential of IoT in detection of diseases revolve around the use of AI driven biosensor which collects physiological and other form of data from patient's smartphones and wearable devices and applies AI or ML techniques to detect changes in patient's vital signal patterns (Kanaparthi et al., 2019; Kavakiotis et al., 2017; Paiva et al., 2018).
1.1. Machine learning (ML) and deep learning (DL)
Machine learning is a subset of AI that often uses statistical methods to give computer the ability to learn patterns from data without being explicitly programmed. ML algorithms are categorized into supervised, unsupervised, semi‐supervised (hybrid of supervised and unsupervised) and reinforcement. Supervised ML is the most common approach employ in healthcare system which utilize labelled data for models to learn features for prediction and classification (base on patterns). Supervised ML algorithms include neural networks (NNs), support vector machine (SVM), Decision Tree, Random Forests etc. (Paiva et al., 2018). Unsupervised ML utilize unlabelled data to enable model to learn and predict output based on patterns learn from input data. Clustering and rule mining are the most common algorithms use in unsupervised ML However, reinforcements learning relies on the use of experience acquired by performing a given task (Catthoor & Van Hoof, 2018; Wang, Casalino et al., 2019).
The use of DL as a sub‐branch of artificial intelligence comprises of deeper neural networks that can identify more complex non‐linear patterns in data acquired from medical devices (such as microscope, MRI) and IoT ecosystems (such as sensors, devices implants and monitors) and provide meaningful output for decision making. There are various neural networks architectures that have been developed. Some of the architectures have performed better than others in terms of regression, classification and denoising images. The current architectures based on CNN include AlexNet with eight layers, VGGNet with 19 and 16 layers, Inception module also known as GoogleNet with 22 layers and 9 modules and Residual or ResNet with 152 layers (Russakovsky et al., 2015; Yu et al., 2020).
The principle behind the application of CNN in classification or regression revolves around series of dot products of weight matrices and input matrix. These processes are categorized into two stages known as feature learning and classification (Wang, Sun, et al., 2020). Feature learning is based on the use of convolutional blocks with operations such as convolution, a process of computing input matrix and feature matrix to obtain a convolve map or feature map. Activation operation is the use of activation function such as tanh, sigmoid and Rectified Linear Unit (ReLu) to squash output into zero or within ranges of 0 and 1 or from −1 to 1. The main function of pooling operation is to reduce computation by taking the most important part of the convolve map by either max pooling or average (mean) pooling (Kang et al., 2020; Wang, Muhammad, et al., 2020). The output is obtained after these operations in all the layers (including fully connected layers or global average pooling layers) and the use of classifier such as SoftMax based on probabilities to categorized output.
1.2. Application of artificial intelligence in detection of pathogenic diseases
Artificial intelligence has been applied in different field of medicine for detection of diseases associated with cancer, tuberculosis, diabetic retinopathy, pneumonia such as bacterial pneumonia and viral pneumonia (influenza virus and recently, SAR‐CoV‐2). The most common type of dataset used by medical expert includes microscopic slide images and radiographic images (such as CT and CXR). These diseases are classified using different DL models such as ImageNet models (AlexNet, VGGNet, GoogleNet and ResNet). However, these diseases can also be classified using models designed from scratch or using hybrid models (Bakator & Radosav, 2018; Chowdhury et al., 2020; Kallianos et al., 2019; Wang, Casalino, et al., 2019).
1.3. Challenges
As the number of people suffering from pneumonia (especially the ones caused by Influenza virus and SAR‐CoV‐2) continue to grow rapidly. There is high need for testing kits that can enable massive detection and provide result in a short period of time. Detection of viral pneumonia such as COVID‐19 and non‐COVID‐19 viral pneumonia is very critical for prevention and control. Health expert required sophisticated technology to accurately detect these pathogens. Moreover, detection of individual pathogens using molecular testing is still not up to standard of point of care diagnostics, instead specimens are sent out to specialized or equipped laboratories for RT‐PCR sequencing and diagnosis. Pneumonia as one of the symptoms of COVID‐19 and other Bacterial pneumonia have been a major challenge for medical and healthcare sectors in many underdeveloped countries and remote communities with limited diagnosis tools and treatment approach. Other approach utilized by medical experts is the use of chest X‐ray images which are cheaper, reliable and fast. However, interpretation of the images can sometimes be tedious to qualified radiologists. Therefore, the development of fast, cheap, simple and accurate detection approach for diagnosis and predictions of these diseases are highly required.
1.4. Contribution
Accordingly, our contributions can be summarized as follows.
We utilized Pre‐trained (through transfer learning) AlexNet model for detection of pneumonia in CT Scan images.
We carried out 10 k cross validation to estimate the model will perform on unseen dataset.
We evaluated the performance of the models based on accuracy, sensitivity and specificity for general dataset and mean average of the parameters for 10 K cross validation.
The remaining parts of this article are organized as follows. Section 2 overviews related work on the use of AI for the detection of pneumonia. In Section 3, we introduced the adopted model with dataset description, model training and cross validation. In Section 4, we discuss about the result obtained from training and testing of the model, comparison of general dataset with cross validation and comparison of our models with the state of art. Finally, we include concluding remarks in Section 5.
2. RELATED WORK
Throughout the last decade, scientists have been trying to integrate the application of AI, ML, DL in healthcare system. Researchers have utilized CNN to solve challenges in medicine such as disease detection using classification and segmentation approaches in skin disorders, brain and breast cancer, and in diabetes (retinopathy) diseases. In the field of microbiology, microbiologists, radiologists and computer scientists have been working together to detect microbial diseases such as tuberculosis, malaria and pneumonia using computer aided diagnosis (Kallianos et al., 2019).
X‐ray images are the basic data used for detection of pneumonia using ML approach. This idea is adopted by Stephen et al. (2019). The authors utilized DL approach to classify X‐ray images samples. The research employed a CNN that is built from scratch using Keras open source with TensorFlow to extract distinctive features from positive and negative images. The dataset contains 5856 X‐ray images of normal and pneumonia images collected from pediatric patients between 1 to 5 years old. The dataset was further augmented to yield a greater number of training dataset. The model was tested on different data size (100–300) and the model achieved average accuracy of 94.81%, 93.01% training and validation respectively.
ChestX‐Ray8 a new dataset from Chest X‐ray Database and Benchmarks was utilized by Wang et al. (2019). The datasets contain X‐ray images with total number of 108,948 from 32,717 patients for detection of thoracic diseases. The authors trained the dataset using CNN networks such as AlexNet, VGGNet‐16, GoogleNet and ResNet‐50. The research achieved AUC value of 0.6333 for “pneumonia”. A similar study carried out by Rajpurkar et al. (2017) based on 121‐layer CNN called CHeXNet. This research utilized more than 100 thousand frontal view X‐ray images with 14 different diseases. For detection of pneumonia, the model achieved AUC value of 0.8887 with the model outperforming radiologist.
The use of AI and CT scans for detection of COVID‐19 is provided by Wang, Kang, et al. (2020). 453 CT scan images of confirmed COVID‐19 cases of patient diagnosed with viral pneumonia are utilized as dataset. The images are classified into training, testing and validation. The model achieved validation accuracy of 82.9%, sensitivity of 84% and specificity of 80.5% while the external testing result has shown an accuracy of 73.1%, sensitivity of 74% and specificity of 67%.
Saraiva et al. (2019) classified X‐ray Images of childhood pneumonia using CNN model. The research datasets were made available online by Kermany, Zhang, et al. (2018) which are labelled as Optical Coherence Tomography (OCT) and Chest X‐Ray Images with total number of 5863 images. The model was train base on cross validation (k = 5) and the model achieved 95.30% average accuracy. Recently, Chouhan et al. (2020) utilized transfer learning to classify X‐ray images into positive and negative pneumonia samples. The research employed transfer learning models of Resnet (Inception V3), GoogleNet, DenseNet121 and AlexNet. A total of 5856 normal and pneumonia (bacteria and virus) were used. The models achieved respective training (at different epochs) and testing accuracies with AlexNet (98.97% and 92.86%), DenseNet121 (99.23% and 92.62%), GoogleNet (99.48% and 93.12%) and ResNet (99.48% and 94.23%).
A broader study is reported by Xu et al. (2020). The authors proposed an artificial intelligence technique to screen and distinguish between two different types of viral pneumonia which include COVID‐19, Influenza‐A and healthy cases using patients CT images. 618 CT scans (224 CT samples from 224 patients with Influenza‐A virus, 219 from 110 patients with COVID‐19 and 175 CT samples from healthy people) are utilized as dataset which undergoes image processing before training using 3‐dimensional DL model. The result has shown that the model achieved overall accuracy of 86.7%. Peng et al. (2020) utilized small number of datasets obtained from 32 patient already diagnosed with COVID‐19 using RT‐PCR method. The study utilized four AI‐driven tools and the study has shown AI can be used to improve confirmed diagnosis rate for clinical cases of COVID‐19.
To discriminate between viral and bacterial pneumonia, Rajaraman et al. (2018) employed CNN (VGG‐16, residual and inception CNN) for detection of pneumonia in pediatric chest radiographs by localizing the Region of Interest (ROI). The dataset contains total number of 5856 (which include viral, bacterial pneumonia and normal CXR images). The models achieved 96.2% accuracy for bacterial pneumonia and 93.6% for viral pneumonia. A more sophisticated study is carried out by Zech et al. (2018) who utilized deep NN and split validation approach to detect pneumonia in X‐ray images. The study employed a total number of 158,323 chest radiographs collected from three different institutions. The results have shown higher accuracy and AUC values. The summary of literature review is presented in Table 2.
TABLE 2.
Detection of different types of pneumonia using AI‐driven tools
| Reference | Type of pneumonia | Dataset | Result |
|---|---|---|---|
| Stephen et al., 2019 | Viral pneumonia (strain not specified) | 5856 X‐ray images | Average Ac of 94.81% training and 93.01% for validation |
| Rajpurkar et al., 2017 | Not specified | 108,948 X‐ray images | 0.6333 AUC |
| X. Wang et al., 2017 | Not specified | 100, 000 X‐ray images | 0.8887 AUC |
| Wang, Kang et al. 2020 | Viral pneumonia (COVID‐19) | 453 CT scan images | The model achieved validation AC of 82.9%, SV of 84% and SP of 80.5%, testing AC of 73.1%, SV of 74% and SF of 67%. |
| Saraiva et al., 2019 | Viral pneumonia (strain not specified) | 5863 Chest X‐Ray Images | AC of 95.30% |
| Chouhan et al., 2020 | Viral and Bacterial pneumonia (strains not specified) | 5863 Chest X‐Ray Images | Different models were used |
| Xu et al., 2020 | viral pneumonia (COVID‐19, Influenza‐A) | 618 CT scan Images | Ac of 86.7%. |
| Rajaraman et al., 2018 | Viral and Bacterial pneumonia (strains not specified) | 5856 chest X‐Ray | Ac of 96.2% accuracy for bacterial pneumonia and 93.6% for viral pneumonia |
| Zech et al., 2018 | Viral and Bacterial pneumonia (strains not specified) | 158,323 chest radiographs | Different models were used |
Abbreviations: Ac, Accuracy; AUC, Area under the curve; Sf, Specificity; Sv, Sensitivity.
3. THE PROPOSED APPROACH
In this section, we detailed the proposed approach procedures and its main assumptions. The work flow of the study design is schematically shown in Figure 1. In this study, a pretrained AlexNet model is used for classification of pneumonia from normal Chest X‐ray images. Apart from AlexNet, there are other high performing CNN models such as VGGNet, GoogleNet and ResNet, but due its simplicity, a smaller number of layers, minimum error and computational time restraints, it was utilized, nonetheless.
FIGURE 1.
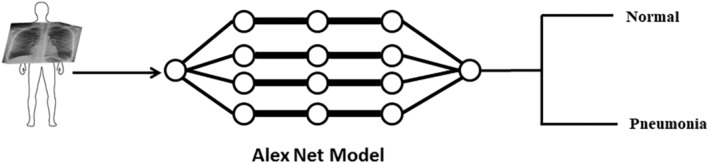
The workflow is represented schematically. CXR images are used to train the network using Pretrained AlexNet model for classification of pneumonia and normal (healthy)
3.1. Dataset
We obtained X‐ray images made available by Kermany, Zhang et al. (2018). The dataset contains three folders (training, validation and testing with a total number of 5856 positive and negative cases. In each folder there is a subfolder with names pneumonia and Normal folders. The dataset description is based on X‐ray images collected from retrospective pediatric patients between the age of 1 to 5 as shown in Figure 2 and describe in Table 3.
FIGURE 2.
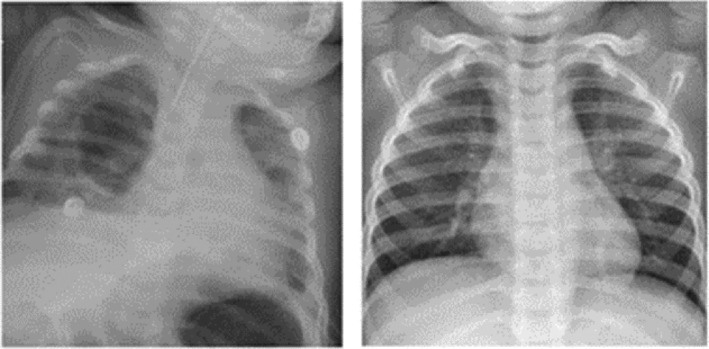
Pediatric CXR scans. Left: Pneumonia. Right: Normal CXR scan
TABLE 3.
Dataset description
| Label | Number |
|---|---|
| Positive | 4273 |
| Negative | 1583 |
| Total | 5856 |
3.1.1. Model training
For training of datasets, we employed Matlab installed on personal computer with window‐64‐bit, 8GB random access memory (RAM), with an intel ® Core i7‐3537U and graphical Processing unit (GPU). 30% of the dataset split as testing dataset are used to evaluate the model performance. Pretrained AlexNet model is employed due to its high accuracy in carrying out feature extraction and image classification. Figure 3 shows the AlexNet architecture employed to classify X‐ray images. AlexNet model contain 5 convolution (CONV) blocks or layer with convolutional filters size of 3×3 without padding and 2×2 window size for max pooling operation. The last 3 layers are 2 fully connected layers (FCL) and output layer. Other terms include Batch Normalization (BN) and Feature Map (FM). SoftMax activation function is utilized in the output layer for classification. Minibatch optimization is a gradient descent that is used to optimize the model. The training is carried out using 20 epochs with 0.0001 as learning rate.
FIGURE 3.
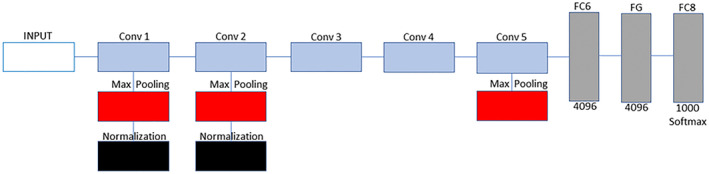
Training of models using AlexNet model. AlexNet model contain 5 convolution (CONV) blocks or layers. The first 2 CONV layers are made up of 3 operations which include convolution, max pooling and normalization. Third and fourth layer are made up of only convolution while fifth layer is made up of convolution and max pooling. The last 3 layers are 2 fully connected layers (FCL) and output layer with SoftMax activation function for classification
3.1.2. Data split
According to literature, scientist recommended the use of 80% for training and 20% for testing. In order to check the performance of different split ratios, we trained the model based on 50:50, 60:40, 70:30, 80:20 and 90:10 for training and testing respectively. The data split for each ratio is presented in Table 4.
TABLE 4.
Data split
| Split | Training | Split | Testing | |||
|---|---|---|---|---|---|---|
| S/No | % | Positive | Negative | % | Positive | Negative |
| 1 | 50 | 2137 | 792 | 50 | 2136 | 791 |
| 2 | 60 | 2564 | 950 | 40 | 1709 | 633 |
| 3 | 70 | 2991 | 1108 | 30 | 1282 | 475 |
| 4 | 80 | 3418 | 1266 | 20 | 855 | 317 |
| 5 | 90 | 3846 | 1425 | 10 | 427 | 158 |
Note: Total number of dataset = 5856, Positive = 4273, Negative = 1583.
3.1.3. Cross validation
Cross validation is a vital method used in machine learning for parameter selection and evaluation of learning performance and prediction. In this study, we utilized K‐fold cross validation approach where the datasets are split into K sets of equal size (i.e., K = 10). In each K sets K−1 is used as training dataset and 1 set is used as validation dataset. Training of the dataset is repeated for K number of times (i.e., n = k) (Fan & Hauser, 2018). The average performance of the training and testing dataset is computed as the evaluation index for the models. This approach is very efficient especially when there are limited number of samples as it takes advantage of the whole dataset. Hence, cross validation dataset classifications are presented in Supplimentary Data S1.
3.1.4. Evaluation and confusion matrix
To evaluate the performance of the trained models, three parameters are employed; accuracy, sensitivity and specificity. Accuracy is termed as the ratio of correctly classified images over total number of images, it is also termed as the sum of sensitivity and specificity. For evaluating the accuracy and loss of a model the following formulas are utilized as shown in Equations (1) and (2).
| (1) |
| (2) |
where N is the overall number of images during training and testing, and n is the number of images and PC is the probability of the correctly classified images.
Confusion matrix is the common approach used for evaluation of model performance based on True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN). TPs is the number of samples that are correctly identified by the model as positive cases or number of cases who actually have pneumonia according to each model. TNs is the number of samples that are correctly identified by the model as negative cases or number of cases who are actually healthy (normal) and classified as negative according to each model. FPs are the number of samples that are incorrectly classified as negative by the model or number of cases that are actually negative (normal or healthy) but classified as pneumonia according to each model. FNs are the number of samples that are incorrectly classified as positive by the model or number of cases that are actually positive (pneumonia) but classified as normal or healthy according to each model as shown in Table 5.
TABLE 5.
Confusion matrix
|
│ Predicted │ |
— Actual — | ||
| True Positive (+) | False Negative (−) | ||
| True Positive | True + | False + | |
| False Negative | False − | True − | |
True Positive rate (Sensitivity) is the proportion of positive image samples that are correctly identified as positive sample (i.e., it shows the percentage of positive samples that are correctly identified as positives). The formula of sensitivity is shown in Equation (3).
| (3) |
False positive rate (FPR) also known as Specificity is the proportion of positive samples that are incorrectly identified as positive samples (i.e., it shows the percentage of negative samples that are incorrectly identified as positives). The formula of sensitivity is shown in Equation (4).
| (4) |
4. RESULT AND DISCUSSION
4.1. General dataset
We trained the models with the entire dataset without cross validation. We utilized 5856 total images which are partition into 50:50, 60:40, 70:30, 80:20 and 90:10 for training and testing. The models were trained in Matlab with 5740 number of iterations, 20 epochs and 0.0001 learning rate.
In terms of 50:50 split, the model achieved training accuracy of 97.98%, testing accuracy of 97.94%, sensitivity of 96.21% and specificity of 99.00%. By increasing the number of training dataset to 60% and reducing testing dataset to 40%, the model achieved training accuracy of 98.94%, testing accuracy of 98.95%, sensitivity of 99.09% and specificity of 98.81%. The difference between training accuracy and testing accuracy achieved by the models (trained on 50:50 and 60:40) are less compare to models trained on 70, 80 and 90%. This is as a result of using same amount or close amount of training and testing splits. Training the model using 70% and testing using 30% (i.e., 70:30) result in training accuracy of 99.19%, testing accuracy of 98.73%, sensitivity of 98.59% and specificity of 99.84%.
In terms of data 80:20 split, the model achieved training accuracy of 99.36%, testing accuracy of 100%, sensitivity of 99.11% and specificity of 99.65%. By increasing the number of training dataset to 90% and reducing testing dataset to 10%, the model achieved training accuracy of 99.86%, testing accuracy of 100%, sensitivity of 99.70% and specificity of 100%. These higher performances are achieved as a result of training the models with large number of datasets and testing using fewer number of datasets (Table 6).
TABLE 6.
General dataset result
| Split | Training accuracy | Testing accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| 50–50 | 97.96 | 97.94 | 96.71 | 99.00 |
| 60–40 | 98.94 | 98.95 | 99.09 | 98.81 |
| 70–30 | 99.19 | 98.73 | 98.59 | 98.84 |
| 80–20 | 99.36 | 100.00 | 99.11 | 99.66 |
| 90–10 | 99.86 | 100.00 | 99.70 | 100.00 |
4.2. Cross validation
The results have shown that training accuracy is greater than testing accuracy in all the k‐folds except 4‐fold where testing accuracy is higher than training accuracy. However, the average result of training accuracy (i.e., 97.70%) is greater than average result of testing accuracy (i.e., 96.04%). The result of sensitivity and specificity varies in the 10‐folds. The average result of sensitivity (97.34%) and specificity (97.79%) indicated that the model has successfully classified both negative and positive images. The result of cross validation is presented in Table 7.
TABLE 7.
Cross validation result for pneumonia
| K fold | Tr(A) | V | Ts(A) | Sv | Sf |
|---|---|---|---|---|---|
| 1 | 98.35 | 0.9835 | 96.67 | 0.9800 | 0.9846 |
| 2 | 96.78 | 0.9678 | 94.71 | 0.9767 | 0.9650 |
| 3 | 97.72 | 0.9772 | 96.55 | 0.9867 | 0.9743 |
| 4 | 97.56 | 0.9756 | 94.71 | 0.9567 | 0.9815 |
| 5 | 97.72 | 0.9772 | 98.16 | 0.9567 | 0.9835 |
| 6 | 97.48 | 0.9748 | 94.14 | 0.9867 | 0.9712 |
| 7 | 96.86 | 0.9686 | 93.45 | 0.9800 | 0.9650 |
| 8 | 98.35 | 0.9835 | 96.21 | 0.9633 | 0.9897 |
| 9 | 98.27 | 0.9827 | 95.63 | 0.9867 | 0.9815 |
| 10 | 97.88 | 0.9788 | 97.13 | 0.9633 | 0.9835 |
| Average |
976.97/10 97.70 |
9.76970/10 0.9770 |
960.36/10 96.04 |
9.37368/10 0.9734 |
9.7798/10 0.9779 |
Abbreviations: Sf, Specificity; Sv, Sensitivity; Tr(A), Training accuracy; Ts(A), Testing accuracy; V, Validation.
4.3. General dataset performance against cross validation
As shown in Table 8, for general dataset we obtained different performance parameters based on training accuracy, testing accuracy, sensitivity and specificity for all the dataset split, while for cross validation we obtained an average performance of 97.70% training accuracy, 96.04% testing accuracy, 97.35% sensitivity and 97.78% specificity. This shows that the average performance of cross validation achieved lower training accuracy, testing accuracy and specificity than general dataset.
TABLE 8.
Comparison between general dataset and cross validation
| Split | Training accuracy | Testing accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| 50–50 | 97.96 | 97.94 | 96.71 | 99.00 |
| 60–40 | 98.94 | 98.95 | 99.09 | 98.81 |
| 70–30 | 99.19 | 98.73 | 98.59 | 98.84 |
| 80–20 | 99.36 | 100.00 | 99.11 | 99.66 |
| 90–10 | 99.86 | 100.00 | 99.70 | 100.00 |
| CV | 97.70 | 96.04 | 97.35 | 97.98 |
Abbreviation: CV, Cross validation.
4.4. Discussion
Radiologist have been relying on radiological images for interpreting pneumonia based on the presence of infiltrates (white spots in the patient's lungs) to identify or interpret the presence of the infection and other complications such as pleural effusions or abscesses. This approach can be very tedious for large images and thus, can lead to misinterpretation. The use of computer aided diagnosis (CAD) which was introduced in 1990s offer a simple, reliable, precise and fast approach of interpreting results related to medical images. CAD approach assist pathologist and radiologist in identifying disease and healthy images while preventing misinterpretation (Matsugu et al., 2003).
The use of CNN to classify and characterize X‐ray images has shown a better accuracy and precision than manual classification by some radiologist. Since the development of deep neural network, scientist have been utilizing different CNN models such as AlexNet, VGGNet 16 and 17, GoogleNet, ResNet and other networks built from scratch to detect pneumonia in x‐ray images. These computer models are developed based on mathematical algorithms to solve problems such as predictions and image classification using probability score.
The results presented in Table 6 has shown that increasing the number of dataset lead to increase in training accuracy. However, our results are in line with the study carried out by Prashanth et al. (2020) based on data splits from 50%–90%. Moreover, 70:30 split is chosen as the best performing model which is “fit” compare to 80:20 and 90:10 which are relatively “overfit” due to testing on small number of datasets. The result obtained from training and testing the performance of the models are presented in Table 6 and Figure 4.
FIGURE 4.
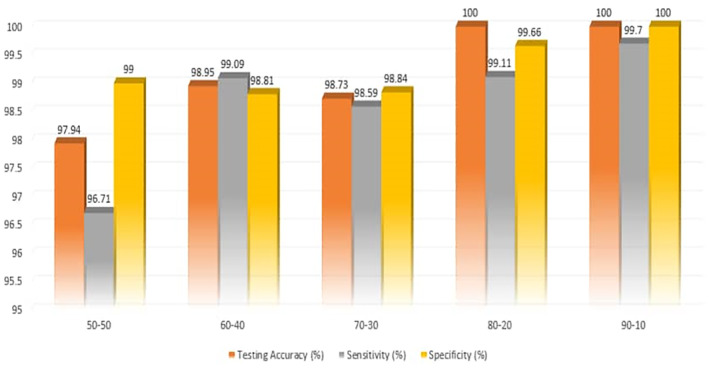
Classification of pneumonia using AlexNet
Comparing our result (based on 70:30 split) with state of art, we obtained a testing accuracy of 98.73% using general dataset and testing accuracy of 97.70% using the average accuracies of cross validation result. Our model has achieved a better accuracy than the study conducted by Stephen et al. (2019) using the same dataset but different model that is built from scratch which achieved average accuracy of 94.81%. Saraiva et al. (2019) utilized the same dataset with our study, the authors split the dataset into 5 K‐folds and achieved 95.30% average accuracy while we split our dataset into 10 k‐folds and achieved average accuracy of 97.70%. Rajaraman et al. (2018) utilized VGG‐16 to classify both bacterial and viral pneumonia. The models achieved 96.2% and 93.6% compare to our model that achieved 98.73% for 70:30 dataset split t using Pretrained AlexNet models. The result presented in Table 9 has shown that using transfer learning yield higher accuracy than building network from scratch as well as using large amount of dataset.
TABLE 9.
Comparison with similar studies from literature
| Rf | No of dataset | Model | A/AUC | Sv | Sf |
|---|---|---|---|---|---|
| 70:30 | 5856 | PA | 98.73 | 98.59 | 98.84 |
| CV | 5856 | PA | 97.35 | 97.35 | 97.78 |
| Stephen et al., 2019 | 5856 | CNN | 94.81 | ‐ | ‐ |
| Chouhan et al., 2020 | 5856 | PA | 92.86 | ‐ | ‐ |
| Saraiva et al., 2019 | 5856 | CNN | 95.30 | ‐ | ‐ |
| Rajaraman et al., 2018 | 5856 | CNN | 92.2, 93.6 | ‐ | ‐ |
| Kanaparthi et al., 2019 | 108,948 | PA | 0.6333 | ‐ | ‐ |
| Rajpurkar et al., 2017 | 100,000 | CHeXNet | 0.8887 | ‐ | ‐ |
Abbreviations: A, Accuracy; AUC, Area under the curve; CV, Cross validation; PA, Pretrained AlexNet; Sf, Specificity; Sv, Sensitivity.
5. CONCLUSION
The recent outbreak of COVID‐19 has caused a global concern leading to over 30 million confirmed cases and more than 800 thousand death. Pneumonia is among the symptoms associated with COVID‐19. However, other pathogens are known to cause pneumonia such as viral pneumonia (caused by Influenza virus) and bacterial pneumonia (caused by Streptococcus pneumoniae). These viruses are mostly diagnosed using bench diagnosis assays which utilize chemical reagent, trained pathologist and radiologist, with longer procedure and heavy workload. To solve these challenges, we utilized a method based on DL and transfer learning approach. We trained our models using 5865 CT scan images based on different splits (50:50, 60:40…90:10) and CV to distinguish between viral pneumonia and healthy patients. For classification of pneumonia using X‐ray images based on 70:30 split, our model achieved testing accuracy of 98.73%, sensitivity of 98.59% and specificity of 99.84% and 96.04% testing accuracy, 97.35% sensitivity and 97.78% specificity using cross validation means. Our result is in line with the notion that CNN models can be used for classifying medical images with higher accuracy and precision. These models can now serve as a confirmation system for diagnosis of viral pneumonia by maximizing miss diagnosis and offer an alternative to relieve the heavy and tedious workload experiencing by radiologist and pathologist in Near East University Hospital.
Some of the limitations of our study include the use of frontal radiographs without augmentation. Normally, frontal images are the types interpreted by radiologist without the need of rotation or colour shift. Another challenge is the lack of sufficient amount of dataset. Thus, with large amount of dataset we can utilize different pretrained architectures such as VGGNet, GoogleNet and ResNet. In the future, this model can be used for classification of COVID‐19 as well as the use of IoT‐enabled system integrated with artificial intelligence for prediction of viral pneumonia. Different image processing techniques can also be applied on the datasets such as annotations and image segmentations. The performance of the models can also be improved by using hybrid models such as combining SVM with pretrained models or models designed from scratch.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Appendix S1: Supporting information
Biographies
Abdullahi Umar Ibrahim PhD Abdullahi Umar Ibrahim was born in Zaria, Kaduna state on the 25th of October 1989. He studied at Nigerian Institute of Science and leather technology Zaria to obtain both OND and HND in Laboratory Science technology. He served at General Hospital Mina Niger state for 1 year. He studied MSc in Bioengineering at Cyprus International university and worked as Student Assistant at the university's information center. He also obtained his Doctoral degree in Biomedical Engineering and currently he is working as a research assistant in the same department. He is also working at Kaduna State University as a laboratory technology. He is also a review many journals from science direct journals and Springer. His research interest is related to CRISPR as genetic engineering tool and application of Artificial Intelligence in Medicine and environment.
Prof. dr. Mehmet Özsöz received his undergraduate degree from Middle East Technical University, Department of Chemical Engineering. He received his doctorate from Ege University Faculty of Pharmacy, Department of Analytical Chemistry. He did postdoctoral work on Electrochemical Biosensors in the United States of America for 2 years. He has 150 scientific articles and 46 h factor and has contributed to more than 50 invited speakers and conferences. He has articles in journals with high impact factor such as ACSLangmiur, ACS Analytical Chemistry, RSC Analyst, Chemical Communications and Biosensors & Bioelectronics.
Sertan Serte completed his undergraduate education at Eastern Mediterranean University, Department of Electrical and Electronics Engineering. He completed his master's degree in the fields of Multimedia, Signal Processing, and Communication at the University of Surrey. He completed his PhD in Electronics in Queen Mary, University of London, in 2015. During his PhD, he worked on facial movement analysis and human‐computer interaction. He is currently working as an Assistant Professor at Near East University, Faculty of Engineering, Department of Electrical and Electronics Engineering.
Prof. Fadi Al‐Turjman received his Ph.D. degree in computer science from Queen's University, Canada, in 2011. He is a Professor with Near East University, North Cyprus. He is a leading authority in the areas of smart / cognitive, wireless and mobile networks' architectures, protocols, deployments, and performance evaluation. His record spans over 200 publications in journals, conferences, patents, books, and book chapters, in addition to numerous keynotes and plenary talks at flagship venues. He has authored / edited more than 20 published books about cognition, security, and wireless sensor networks' deployments in smart environments with Taylor & Francis, and the Springer (Top tier publishers in the area). He was a recipient of several recognitions and best papers' awards at top international conferences. He also received the prestigious Best Research Paper Award from Elsevier COMCOM Journal for the last three years prior to 2018, in addition to the Top Researcher Award for 2018 at Antalya Bilim University, Turkey. He led a number of international symposia and workshops in flag‐ship IEEE ComSoc conferences. He is serving as the Lead Guest Editor in several journals, including the IET Wireless Sensor Systems, Springer EURASIP, MONET, MDPI sensors, Wiley & Hindawi WCM, and the Elsevier COMCOM, SCS, and Internet of Things.
Salahudeen Habeeb Kolapo Habeeb is a PhD student at Cyprus International University In Computer Engineering. He obtained Hid MSC from the same university and is also working as a Research Assistant.
Umar Ibrahim A, Ozsoz M, Serte S, Al‐Turjman F, Habeeb Kolapo S. Convolutional neural network for diagnosis of viral pneumonia and COVID‐19 alike diseases. Expert Systems. 2021;e12705. 10.1111/exsy.12705
Correction added on 5 July 2021, after first online publication: Affiliations have been corrected in this version.
REFERENCES
- Bakator, M. , & Radosav, D. (2018). Deep learning and medical diagnosis: A review of literature. Multimodal Technologies and Interaction, 2(3), 47. 10.3390/mti2030047 [DOI] [Google Scholar]
- Banerjee, A. , Kulcsar, K. , Misra, V. , Frieman, M. , & Mossman, K. (2019). Bats and coronaviruses. Viruses, 11(1), 41. 10.3390/v11010041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catthoor, F. , & Van Hoof, C. (2018). Unsupervised heart‐rate estimation in wearables with Liquid states and a probabilistic readout. Neural Networks, 99, 134–147. 10.1016/j.neunet.2017.12.015 [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Liu, Q. , & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92, 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouhan, V. , Singh, S. K. , Khamparia, A. , Gupta, D. , Tiwari, P. , Moreira, C. , Damaševičius, R. , & de Albuquerque, V. H. C. (2020). A novel transfer learning based approach for pneumonia detection in chest X‐ray images. Applied Sciences, 10(2), 559. 10.3390/app10020559 [DOI] [Google Scholar]
- Chowdhury, M. E. , Rahman, T. , Khandakar, A. , Mazhar, R. , Kadir, M. A. , Mahbub, Z. B. , Islam, K. R. , Salman Khan, M. , Iqbal, A. , Al Emadi, N. , Reaz, M. B. I. , & Islam, M. T . (2020). Can AI help in screening viral and COVID‐19 pneumonia?. arXiv preprint arXiv:2003.13145. Retrieved from 10.1109/ACCESS.2020.3010287 [DOI]
- Dowell, S. F. , Simmerman, J. M. , Erdman, D. D. , Wu, J. S. J. , Chaovavanich, A. , Javadi, M. , & Ho, M. S. (2004). Severe acute respiratory syndrome coronavirus on hospital surfaces. Clinical Infectious Diseases, 39(5), 652–657. 10.1086/422652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , & Hauser, H. (2018). Fast and accurate cnn‐based brushing in scatterplots. Computer Graphics Forum, 37(3), 111–120. 10.1111/cgf.13405 [DOI] [Google Scholar]
- Gilani, Z. , Kwong, Y. D. , Levine, O. S. , Deloria‐Knoll, M. , Scott, J. A. G. , O'Brien, K. L. , & Feikin, D. R. (2012). A literature review and survey of childhood pneumonia etiology studies: 2000–2010. Clinical infectious diseases, 54(Suppl 2), S102–S108. 10.1093/cid/cir1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , Ge, Q. , Jin, L. , & Xiong, M. (2020). Artificial intelligence forecasting of Covid‐19 in China. arXiv preprint arXiv:2002.07112.
- Kallianos, K. , Mongan, J. , Antani, S. , Henry, T. , Taylor, A. , Abuya, J. , & Kohli, M. (2019). How far have we come? Artificial intelligence for chest radiograph interpretation. Clinical Radiology, 74, 338–345. 10.1016/j.crad.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Kanaparthi, S. , Supraja, P. , & Singh, S. G. (2019). Smart, portable, and noninvasive diagnostic biosensors for healthcare. In Advanced biosensors for health care applications (pp. 209–226). Elsevier. Retrieved from 10.1016/B978-0-12-815743-5.00007-X [DOI] [Google Scholar]
- Kang, C. , Yu, X. , Wang, S. H. , Guttery, D. , Pandey, H. , Tian, Y. , & Zhang, Y. (2020). A heuristic neural network structure relying on fuzzy logic for images scoring. IEEE Transactions on Fuzzy Systems, 29, 34–45. 10.1109/TFUZZ.2020.2966163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavakiotis, I. , Tsave, O. , Salifoglou, A. , Maglaveras, N. , Vlahavas, I. , & Chouvarda, I. (2017). Machine learning and data mining methods in diabetes research. Computational and Structural Biotechnology Journal, 15, 104–116. 10.1016/j.csbj.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany, D. , Zhang, K. , & Goldbaum, M. (2018). Large dataset of labeled optical coherence tomography (oct) and chest x‐ray images. Mendeley Data, v3. Retrieved from 10.17632/rscbjbr9sj.3 [DOI]
- Kolhar, M. , Al‐Turjman, F. , Alameen, A. , & Abualhaj, M. (2020). A three layered decentralized IoT biometric architecture for City lockdown during COVID‐19 outbreak. IEEE Access, 8, 163608–163617. 10.1109/ACCESS.2020.3021983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsugu, M. , Mori, K. , Mitari, Y. , & Kaneda, Y. (2003). Subject independent facial expression recognition with robust face detection using a convolutional neural network. Neural Networks, 16(5–6), 555–559. 10.1016/S0893-6080(03)00115-1 [DOI] [PubMed] [Google Scholar]
- Oboho, I. K. , Tomczyk, S. M. , Al‐Asmari, A. M. , Banjar, A. A. , Al‐Mugti, H. , Aloraini, M. S. , Alkhaldi, K. Z. , Almohammadi, E. L. , Alraddadi, B. M. , Gerber, S. I. , Swerdlow, D. L. , Watson, J. T. , & Madani, T. A. (2015). 2014 MERS‐CoV outbreak in Jeddah—A link to health care facilities. New England Journal of Medicine, 372(9), 846–854. 10.1056/NEJMoa1408636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva, J. S. , Cardoso, J. , & Pereira, T. (2018). Supervised learning methods for pathological arterial pulse wave differentiation: A SVM and neural networks approach. International Journal of Medical Informatics, 109, 30–38. 10.1016/j.ijmedinf.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Paules, C. I. , Marston, H. D. , & Fauci, A. S. (2020). Coronavirus infections—More than just the common cold. Journal of the American Medical Association, 323, 707. 10.1001/jama.2020.0757 [DOI] [PubMed] [Google Scholar]
- Prashanth, D. S. , Mehta, R. V. K. , & Sharma, N. (2020). Classification of Handwritten Devanagari Number–An analysis of Pattern Recognition Tool using Neural Network and CNN. Procedia Computer Science, 167, 2445–2457. [Google Scholar]
- Peng, M. , Yang, J. , Shi, Q. , Ying, L. , Zhu, H. , Zhu, G. , Ding, X. , He, Z. , Qin, J. , Wang, J. , Yan, H. , Bi, X. , Shen, B. , Wang, D. , Luo, L. , Zhao, H. , Zhang, C. , Lin, Z. , Hong, L ., … & Li, J . (2020). Artificial intelligence application in COVID‐19 diagnosis and prediction.
- Rahman, M. A. , Zaman, N. , Asyhari, A. T. , Al‐Turjman, F. , Bhuiyan, M. Z. A. , & Zolkipli, M. F. (2020). Data‐driven dynamic clustering framework for mitigating the adverse economic impact of Covid‐19 lockdown practices. Sustainable Cities and Society, 62, 102372. 10.1016/j.scs.2020.102372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman, S. , Candemir, S. , Kim, I. , Thoma, G. , & Antani, S. (2018). Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Applied Sciences, 8(10), 1715. 10.3390/app8101715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurkar, P. , Irvin, J. , Zhu, K. , Yang, B. , Mehta, H. , Duan, T. , Ding, D. , Bagul, A. , Langlotz, C. , Shpanskaya, K. , Lungren, M. P. , & Ng, A. Y . (2017). Chexnet: Radiologist‐level pneumonia detection on chest x‐rays with deep learning. arXiv preprint arXiv:1711.05225.
- Russakovsky, O. , Deng, J. , Su, H. , Krause, J. , Satheesh, S. , Ma, S. , Huang, Z. , Karpathy, A. , Khosla, A. , Bernstein, M. , Berg, A. C. , & Fei‐Fei, L. (2015). Imagenet large scale visual recognition challenge. International Journal of Computer Vision, 115(3), 211–252. 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- Ruuskanen, O. , Lahti, E. , Jennings, L. C. , & Murdoch, D. R. (2011). Viral pneumonia. The Lancet, 377(9773), 1264–1275. 10.1016/S0140-6736(10)61459-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva, A. , Ferreira, N. , Sousa, L. , Carvalho da Costa, N. , Sousa, J. , Santos, D. , & Soares, S. (2019). Classification of images of childhood pneumonia using convolutional neural networks. Paper presented at 6th International Conference on Bioimaging (pp. 112–119).
- Srivastava, V. , Srivastava, S. , Chaudhary, G. , & Al‐Turjman, F. (2020). A systematic approach for the COVID‐19 prediction and parameters estimation. Personal and Ubiquitous Computing, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen, O. , Sain, M. , Maduh, U. J. , & Jeong, D. U. (2019). An efficient deep learning approach to pneumonia classification in healthcare. Journal of Healthcare Engineering, 2019, 1–7. 10.1155/2019/4180949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. H. , Muhammad, K. , Hong, J. , Sangaiah, A. K. , & Zhang, Y. D. (2020). Alcoholism identification via convolutional neural network based on parametric ReLU, dropout, and batch normalization. Neural Computing and Applications, 32(3), 665–680. 10.1007/s00521-018-3924-0 [DOI] [Google Scholar]
- Wang, F. , Casalino, L. P. , & Khullar, D. (2019). Deep learning in medicine—Promise, progress, and challenges. JAMA Internal Medicine, 179(3), 293–294. 10.1001/jamainternmed.2018.7117 [DOI] [PubMed] [Google Scholar]
- Wang, S. , Kang, B. , Ma, J. , Zeng, X. , Xiao, M. , Guo, J. , Cai, M. , Yang, J. , Li, Y. , Meng, X ., & Xu, B. (2020. ). A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID‐19). medRxiv. Retrieved from 10.1101/2020.02.14.20023028 [DOI] [PMC free article] [PubMed]
- Wang, S. , Sun, J. , Mehmood, I. , Pan, C. , Chen, Y. , & Zhang, Y. D. (2020). Cerebral micro‐bleeding identification based on a nine‐layer convolutional neural network with stochastic pooling. Concurrency and Computation: Practice and Experience, 32(1), e5130. 10.1002/cpe.5130 [DOI] [Google Scholar]
- Wang, X. , Peng, Y. , Lu, L. , Lu, Z. , Bagheri, M. , & Summers, R. M. (2017, July). Chestx‐ray8: Hospital‐scale chest x‐ray database and benchmarks on weakly‐supervised classification and localization of common thorax diseases. Paper presented at Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, pp. 2097–2106.
- World Health Organization . (2018). Household Air Pollution and Health [Fact Sheet], Geneva, Switzerland: WHO. Retrieved from http://www.who.int/newa-room/fact-sheets/detail/household-airpollution-and-health
- World Health Organization . (2020). WHO Coronavirus Disease (COVID‐19) Dashboard. Retrieved from https://covid19.who.int/?gclid=EAIaIQobChMI8I_w_Yba6QIVC6h3Ch1hTgS7EAAYASAAEgLR1vD_BwE
- Xu, H. , Zhong, L. , Deng, J. , Peng, J. , Dan, H. , Zeng, X. , Li, T. , & Chen, Q. (2020). High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. International Journal of Oral Science, 12(1), 1–5. 10.1038/s41368-020-0074-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Kang, C. , Guttery, D. S. , Kadry, S. , Chen, Y. , & Zhang, Y. D. (2020). ResNet‐SCDA‐50 for breast abnormality classification. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 18, 94–102. 10.1109/TCBB.2020.2986544 [DOI] [PubMed] [Google Scholar]
- Zech, J. R. , Badgeley, M. A. , Liu, M. , Costa, A. B. , Titano, J. J. , & Oermann, E. K. (2018). Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross‐sectional study. PLoS Medicine, 15(11), e1002683. 10.1371/journal.pmed.1002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information


