Abstract
Background
Carbapenem resistance in Gram-negative bacteria is an ongoing public health problem of global dimensions leaving very few treatment options for infected patients.
Objectives
To study the dissemination of plasmid-borne carbapenemase genes in Gram-negative bacteria from a diagnostic centre in Tamil Nadu, India.
Methods
A total of 151 non-repetitive isolates belonging to 10 genera were collected between January 2015 and December 2016 from a diagnostic centre in Tamil Nadu. The isolates included Escherichia coli (n = 57), Klebsiella pneumoniae (n = 45), Pseudomonas aeruginosa (n = 10), Salmonella Typhi (n = 8), Enterobacter cloacae (n = 8), Acinetobacter baumannii (n = 7), Serratia marcescens (n = 5), Achromobacter xylosoxidans (n = 5), Proteus mirabilis (n = 5), Klebsiella oxytoca (n = 5) and Elizabethkingia meningoseptica (n = 1).
Results
Of the 151 isolates, 71% (n = 107) and 68% (n = 103) were found to be resistant to meropenem and imipenem, respectively. The most prevalent β-lactamase gene was blaNDM-1 (n = 22), followed by blaOXA-181 (n = 21), blaGES-1 (n = 11), blaOXA-51 (n = 9), blaGES-9 (n = 8), blaOXA-23 (n = 7) and blaIMP-1 (n = 3). We also observed blaOXA-23 in E. coli (n = 4), and three K. pneumoniae were positive for both, blaOXA-23 and blaOXA-51. Plasmid incompatibility (inc/rep) typing results showed that the resistance genes (n = 11) were present in the isolates carrying plasmid-types IncX, IncA/C, IncFIA-FIB and IncFIIA. The plasmid-borne resistance genes in E. coli and K. pneumoniae were transferred to susceptible E. coli AB1157.
Conclusions
This study highlights the prevalence of carbapenem resistance and the acquisition of plasmid-borne carbapenemase genes in Gram-negative bacteria isolated at this centre.
Introduction
Antibiotic resistance is an emerging global health problem due to the injudicious use of antibiotics.1 It is considered as a major clinical and public health problem because of the limited treatment options available to treat infections caused by antibiotic-resistant bacteria. The increasing bacterial resistance rates to most available antibiotics, including penicillin, cephalosporins, carbapenems, and colistin pose a serious threat.1 The WHO recently listed carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa and ESBL-producing Enterobacteriaceae as pathogens of critical importance.2 Gram-negative bacteria (GNB), especially Enterobacteriaceae, have developed resistance to a broad-spectrum of antibiotics responsible for significant mortality around the globe.3 Carbapenems are considered as one of the last resort antibiotics against infections caused by multidrug-resistant GNB.4 The emergence of carbapenem resistance in Enterobacteriaceae is a major clinical problem, particularly for patients with complex infections, especially when they are immunocompromised or suffering from multiple diseases.5 Pathogens that are resistant to carbapenems often show high levels of resistance to other commonly used antibiotics. This not only leads to high mortality rates, but often the patient’s time in the hospital is prolonged and medical expenses accumulate, placing an emotional, economic and financial burden on families, especially in resource-limited countries.6
The assessment of the worldwide rise in antibiotic resistance has become very difficult due to the increasing rates of multidrug resistance shown by pathogens and the lack of harmonized surveillance systems.7 Moreover, the coexistence of carbapenem resistance genes with other genes such as plasmid-mediated AmpC or plasmid-mediated quinolone resistance has resulted in an increased acquisition of resistance, causing community- and hospital-acquired infections.8,9 The carbapenem-hydrolysing oxacillinases (CHDL) are the major source of carbapenem resistance in A. baumannii. The first report of OXA-23-type β-lactamase in A. baumannii was in 1985 in Edinburgh, UK.10 Recently, OXA-23 was also reported in members of the Enterobacteriaceae family.11–14 The OXA-51-like β-lactamase was first reported by the same laboratory in Edinburgh from isolates collected from three hospitals in Buenos Aires, Argentina. At present, more than 150 variants of OXA-51 have been reported worldwide.15 These intrinsic enzymes (OXA-51-like) in A. baumannii are naturally chromosome-borne, but in rare cases are also reported to be encoded on plasmids.16 Previously, we reported the distribution of colistin resistance in the study region, and investigated the importance of integrons in disseminating antibiotic resistance.17,18 In the present study, dissemination of carbapenem resistance among Gram-negative bacteria was evaluated, and the role of plasmid transfer in developing carbapenem resistance was also explored in further detail.
Materials and methods
Ethics approval
Ethics approval was from the Institutional Ethical Committee for studies on Human subjects (IECH), ref. no. VIT/IECH/004/Jan2015.
Isolate collection and classification
During January 2015 and December 2016, a total of 151 Gram-negative bacterial isolates were collected from Hi-Tech diagnostic centre in Chennai, Tamil Nadu, India. Bacteria were isolated from urine, blood, pus, bronchial secretion, CSF, pulmonary secretion and bile fluid. The collected isolates were received at the Antibiotic Resistance and Phage Therapy Laboratory, VIT, Vellore, for further analyses. Bacterial identification was carried out using the VITEK identification system (bioMérieux) and 16S rRNA gene nucleotide sequence analysis using universal primers 27 F and 1492 R.18 DNA was extracted from all the isolates using a boiling lysis method. Briefly, overnight-grown bacterial cultures were centrifuged at 8000 g for 10 min, and the bacterial pellet was resuspended in 100 μL of sterile distilled water. The cells were boiled at 100°C for 10 min and the mixture was centrifuged at 2000 g for 2 min. The supernatant was extracted and used as a source of template for PCR. The PCR products were sequenced and identified to the species level using the BLASTN tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch).
Antibiotic susceptibility testing and MICs
Antibiotic resistance profiling was performed using the disc diffusion method according to CLSI guidelines.19 The antibiotic discs used for this study were gentamicin (10 μg), co-amoxiclav (30 μg), cefotaxime (30 μg), ertapenem (10 μg), amikacin (30 μg), meropenem (10 μg), colistin (10 μg) and cefepime (30 μg). Briefly, on the Muller-Hinton (MH) agar plate, a lawn culture of bacteria was prepared by adjusting the bacterial culture to 0.5 McFarland turbidity standards. The antibiotic discs were placed on the bacterial lawn and the MH plates were incubated at 37°C for 18 h. Based on the zone of inhibition, the results were interpreted as susceptible, intermediate or resistant. MICs were determined by the broth microdilution method for meropenem and imipenem, as described previously.18 Briefly, in the 96-well microtitre plate, 100 μL of cation-adjusted MH broth was added to each well. Meropenem or imipenem was added at concentrations ranging from 0.06 to 128 mg/L in columns 1 to 11, whereas column 12 served as growth control. The bacterial culture at 5 × 105 dilutions from the overnight grown cells was added and the plates were incubated at 37°C for 20 h. Escherichia coli ATCC 25922 was used as a control strain and the results were interpreted according to CLSI guidelines.19
Molecular analysis of resistance-related genes
The isolates were screened for the presence of the carbapenem resistance genes blaNDM, blaOXA-48-like, blaKPC, blaIMP and blaVIM.18 A second multiplex PCR was also performed for blaDIM, blaBIC, blaGIM, blaSIM and blaAIM.20 The blaOXA-1, blaOXA-4, blaOXA-30, blaGES-1-9 and blaGES-11 were screened as described earlier.21 The blaOXA-23-like, blaOXA-24-like, blaOXA-51-like and blaOXA-58-like were screened for according to Woodford et al.22 The primers and PCR conditions used for analyses are given in Tables S1 to S5 (available as Supplementary data at JAC-AMR Online). The PCR amplicons of the resistance genes were sequenced and genes were confirmed using NCBI BLASTN program (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch).
Plasmid isolation and plasmid incompatibility grouping
Plasmid isolation was performed for all the isolates harbouring resistance genes. The isolation of plasmid DNA was performed using HiPurA Plasmid DNA Miniprep Purification Kit (Himedia, India). Chromosomal DNA contamination was checked using the 16S rRNA primers as described earlier.23 The purified plasmid DNA was used for screening β-lactamase genes. Plasmid incompatibility (inc/rep) typing (FIA, FIB, FIC, HI1, HI2, I1-Ig, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA replicons) was performed using multiplex PCR following the primers and PCR conditions as described by Carattoli et al.24 The primers and PCR conditions used for analysis are given in Table S6.
Conjugation studies
Representative carbapenem-resistant isolates harbouring plasmid-borne resistance were tested for conjugation using the broth-mating method.18 Briefly, the donor strain (strains carrying resistance genes) and the recipient strain (E. coli AB1157, Strr) were grown overnight in MH broth at 37°C and mixed in 9:1 ratio each of donor and recipient. The cells were kept undisturbed for 6 h at 37°C and plated onto antibiotic-containing medium. The isolates which grew on both meropenem and streptomycin were considered as transconjugants. All the donor strains were tested for streptomycin resistance and MIC values (<2 mg/L) were found to be suitable for the assay. The transconjugants were confirmed for the presence of respective carbapenem resistance genes using PCR. The list of isolates used for conjugation studies is given in Table S7.
Results
Bacterial identification
In this cross-sectional study, a total of 151 non-duplicate, Gram-negative bacteria belonging to 10 genera were studied which included E. coli (n = 57, 37.7%), Klebsiella pneumoniae (n = 40, 26.4%), Klebsiella oxytoca (n = 5, 3.3%), P. aeruginosa (n = 10, 6.6%), Salmonella Typhi (n = 8, 5.2%), Enterobacter cloacae (n = 8, 5.2%), A. baumannii (n = 7, 4.6%), Serratia marcescens (n = 5, 3.3%), Achromobacter xylosoxidans (n = 5, 3.3%), Proteus mirabilis (n = 5, 3.3%) and Elizabethkingia meningoseptica (n = 1, 0.6%). Most of the isolates were isolated from urine (37%; 56/151) and blood (28%; 42/151) and from other sources such as pus (7%), bronchial secretion (2%), CSF (1%), pulmonary secretion (1%), bile fluid (5%) or from sources that were not documented (19%).
Antibiotic susceptibility studies
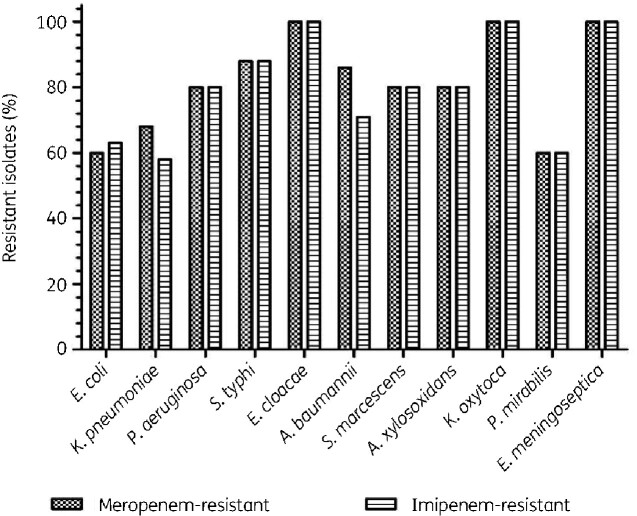
Table 1 summarizes the antibiotic susceptibility pattern of all the isolates tested against eight different antibiotics. Meropenem MICs showed that 107/151 (71%) isolates were resistant (Figure 1), whereas 128 (84.7%) isolates were meropenem-resistant when analysed by the disc diffusion method. For imipenem, 68% (n = 103) were resistant by microbroth dilution method whereas 83% (n = 125) were resistant according to the disc diffusion method. MIC50 and MIC90 values for meropenem were 4 mg/L and 16 mg/L, respectively, and for imipenem the MIC50 was 4 mg/L and the MIC90 was 16 mg/L.
Table 1.
Antibiotic susceptibility testing employing the disc diffusion method and the prevalence of MDR isolates among 151 Gram-negative bacteria isolated from clinical samples
| Bacteria/antibiotic | No. of resistant isolates (%) |
Total MDR isolates (n = 151) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| GEN | AMC | IPM | ETP | AMK | MEM | CST | FEP | ||
| E. coli (n = 57) | 51 (89) | 45 (79) | 46 (81) | 38 (67) | 49 (86) | 43 (75) | 35 (61) | 45 (79) | 54 (95) |
| K. pneumoniae (n = 40) | 33 (83) | 31 (78) | 32 (80) | 28 (70) | 36 (90) | 32 (80) | 29 (73) | 30 (75) | 32 (80) |
| P. aeruginosa (n = 10) | 10 (100) | 10 (100) | 9 (90) | 6 (60) | 8 (80) | 10 (100) | 7 (70) | 8 (80) | 10 (100) |
| S. Typhi (n = 8) | 6 (75) | 7 (88) | 5 (63) | 5 (63) | 6 (75) | 7 (88) | 5 (63) | 7 (88) | 7 (88) |
| E. cloacae (n = 8) | 7 (88) | 8 (100) | 7 (88) | 6 (75) | 8 (100) | 8 (100) | 6 (75) | 7 (88) | 8 (100) |
| A. baumannii (n = 7) | 7 (100) | 6 (86) | 7 (100) | 6 (86) | 7 (100) | 7 (100) | 5 (71) | 7 (100) | 7 (100) |
| S. marcescens (n = 5) | 5 (100) | 5 (100) | 5 (100) | 3 (60) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 5 (100) |
| A. xylosoxidans (n = 5) | 5 (100) | 5 (100) | 4 (80) | 2 (40) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 5 (100) |
| K. oxytoca (n = 5) | 4 (80) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 5 (100) |
| P. mirabilis (n = 5) | 5 (100) | 5 (100) | 4 (80) | 4 (80) | 5 (100) | 5 (100) | 4 (80) | 5 (100) | 5 (100) |
| E. meningoseptica (n = 1) | 1 (100) | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
Values represent the number of resistant isolates, % is listed in brackets. Isolates were defined as MDR only when the isolates are resistant to three or more antibiotics. Abbreviations: GEN, gentamicin; AMC, co-amoxiclav; IPM, imipenem; ETP, ertapenem; AMK, amikacin; MEM, meropenem; CST, colistin; FEP, cefepime.
Figure 1.
The distribution of Gram-negative bacteria and comparison of imipenem and meropenem resistance.
Distribution of carbapenemase resistance genes
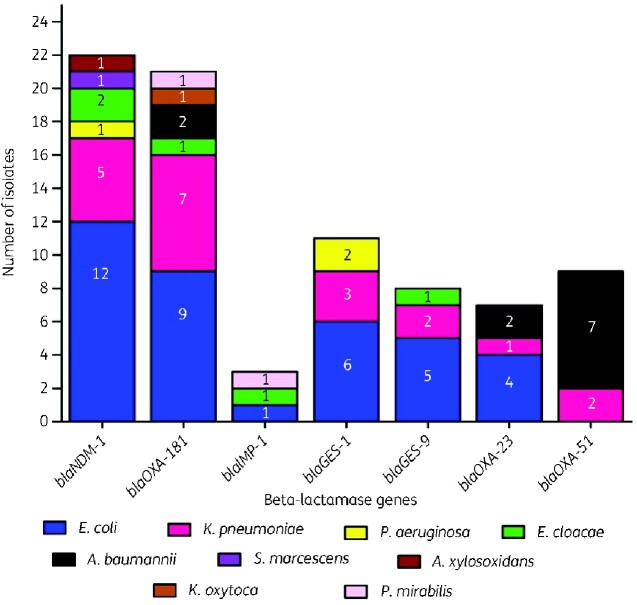
Of the 57 E. coli, 32 isolates carried carbapenemases (blaNDM, blaOXA-48-like, blaGES-1, blaGES-9, blaOXA-23-like and blaIMP) and five E. coli isolates carried more than one of the carbapenem resistance genes (Figure 2). Among the K. pneumoniae strains, 19/40 carried the studied genes (blaNDM, blaOXA-48-like, blaGES-1, blaGES-9, blaOXA-23-like, blaOXA-51-like), and one isolate was positive for both blaNDM and blaOXA-48-like. Carbapenem resistance genes were detected in 70/151 by PCR, and 10 isolates had more than one gene type. The most prevalent resistance genes were blaNDM (n = 22), blaOXA-48-like (n = 21), blaGES-1 (n = 11), blaGES-9 (n = 8), blaOXA-23-like (n = 7), blaOXA-51-like (n = 9) and blaIMP (n = 3). None of the β-lactamase genes blaKPC, blaVIM, blaBIC, blaGIM, blaDIM, blaSIM or blaAIM were detected in the isolates. Sequencing of genes showed that all the amplified blaNDM genes were blaNDM-1, blaOXA-48-like genes were blaOXA-181, and blaIMP genes were blaIMP-1.
Figure 2.
The distribution of carbapenemase genes among Gram-negative bacteria isolated from the clinical samples. A total of 20 resistance genes were studied that include blaNDM, blaOXA-48-like, blaKPC, blaIMP, blaVIM, blaDIM, blaBIC, blaGIM, blaSIM, blaAIMblaOXA-1, blaOXA-4, blaOXA-30, blaGES-1-9, blaGES-11, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like. The genes blaKPC, blaVIM, blaDIM, blaBIC, blaGIM, blaSIM, blaAIM, blaOXA-1, blaOXA-4, blaOXA-30, blaGES-11, blaOXA-24-like and blaOXA-58-like were not observed in any of the isolates.
Plasmid incompatibility typing and conjugation
Plasmid DNA was isolated from 70 isolates that carried resistance genes (Table 2). In total, of the 151 isolates studied, 70 isolates carried resistance genes, of which 11 were plasmid-borne and 59 were chromosomal. Of the 37 E. coli isolates, 32 isolates carried resistance genes, of which six were plasmid-encoded. Among the 40 K. pneumoniae strains, only 19 isolates carried resistance genes, of which three were encoded on plasmids. In E. cloacae, one isolate carried blaNDM-1 on a plasmid and one P. mirabilis carried plasmid-borne blaIMP-1. Plasmid incompatibility/replicon (inc/rep) typing results showed that the plasmids belonged to inc/rep types: IncX, IncA/C, IncFIA-FIB and IncFIIA (Table 2). E. coli isolates that carried IncX (EC10), IncA/C (EC21) and IncFIA-FIB (EC29) -type plasmids harboured blaNDM-1 genes. E. coli strains carrying IncFIIA (EC39) and IncFIA-FIB (EC29) harboured blaOXA-181 genes and IncFIA-FIB (EC47) type plasmids carried blaGES-1/9 genes. K. pneumoniae isolates carrying IncFIA-FIB (KP10) -type plasmids carried blaNDM-1 genes, and IncA/C (KP31 and KP39) carried blaGES-1, blaOXA-23/51-like genes. One E. cloacae isolate with IncFIIA (EL3)-type plasmid harboured blaNDM-1 gene and one P. mirabilis isolate carrying IncFIA-FIB (PM5)-type plasmid had the blaIMP-1 gene.
Table 2.
Distribution of resistance genes, plasmid incompatibility grouping and transconjugation studies on Gram-negative isolates that were harbouring resistance genes
| Isolate | Source | MIC (mg/L) |
Resistance gene | Plasmid inc/rep typing | Conjugative plasmid | |
|---|---|---|---|---|---|---|
| Meropenem | Imipenem | |||||
| E. coli EC1 | Urine | 16 | 8 | bla NDM-1 | – | – |
| E. coli EC2 | Urine | 0.25 | 0.12 | ND | – | – |
| E. coli EC3 | Blood | 1 | 1 | ND | – | – |
| E. coli EC4 | Pus | 32 | 32 | bla NDM-1 | – | – |
| E. coli EC5 | Urine | 8 | 16 | bla NDM-1 | – | – |
| E. coli EC6 | Pus | 2 | 0.5 | ND | – | – |
| E. coli EC7 | Urine | 8 | 8 | bla NDM-1 | – | – |
| E. coli EC8 | Urine | 1 | 2 | ND | – | – |
| E. coli EC9 | Urine | 0.25 | 0.5 | ND | – | – |
| E. coli EC10 | Blood | 32 | 8 | bla NDM-1 | IncX | + |
| E. coli EC11 | Unknown | 0.5 | 0.25 | ND | – | – |
| E. coli EC12 | Urine | 64 | 32 | bla NDM-1 | – | – |
| E. coli EC13 | Unknown | 0.5 | 1 | ND | – | – |
| E. coli EC14 | Unknown | 2 | 2 | ND | – | – |
| E. coli EC15 | Urine | 32 | 32 | ND | – | – |
| E. coli EC16 | Urine | 0.5 | 1 | ND | – | – |
| E. coli EC17 | Urine | 8 | 4 | bla NDM-1 | – | – |
| E. coli EC18 | Urine | 0.25 | 0.25 | ND | – | – |
| E. coli EC19 | Unknown | 4 | 4 | ND | – | – |
| E. coli EC20 | Urine | 0.12 | 0.25 | ND | – | – |
| E. coli EC21 | Blood | 16 | 8 | bla NDM-1 | IncA/C | + |
| E. coli EC22 | Unknown | >128 | >128 | bla NDM-1 | – | – |
| E. coli EC23 | Urine | 64 | 8 | bla NDM-1 | – | – |
| E. coli EC24 | Unknown | 1 | 1 | ND | – | – |
| E. coli EC25 | Urine | >128 | 128 | bla NDM-1 | – | – |
| E. coli EC26 | Urine | 2 | 8 | ND | – | – |
| E. coli EC27 | Urine | 0.25 | 0.5 | ND | – | – |
| E. coli EC28 | Bile fluid | 1 | 1 | ND | – | – |
| E. coli EC29 | Unknown | 8 | 32 | bla NDM-1, blaOXA-181 | IncFIA-FIB | + |
| E. coli EC30 | Urine | 4 | 4 | bla OXA-181 | – | – |
| E. coli EC31 | Unknown | 16 | 32 | bla OXA-181 | – | – |
| E. coli EC32 | Blood | 2 | 2 | ND | – | – |
| E. coli EC33 | Blood | 8 | 8 | bla OXA-181 | – | – |
| E. coli EC34 | Bile fluid | 16 | 8 | bla OXA-181 | – | – |
| E. coli EC35 | Urine | 0.5 | 0.25 | ND | – | – |
| E. coli EC36 | Urine | 32 | 16 | bla OXA-181 | – | – |
| E. coli EC37 | Bile fluid | 2 | 8 | ND | – | – |
| E. coli EC38 | Urine | 1 | 0.5 | ND | – | – |
| E. coli EC39 | Blood | 64 | >128 | bla OXA-181 | IncFIIA | + |
| E. coli EC40 | Blood | 8 | 8 | bla OXA-181 | – | – |
| E. coli EC41 | Blood | 16 | 16 | bla OXA-181 | – | – |
| E. coli EC42 | Blood | 32 | 16 | bla IMP-1 | – | – |
| E. coli EC43 | Urine | 0.5 | 1 | ND | – | – |
| E. coli EC44 | Pus | 16 | 64 | bla GES-1 | IncFIA-FIB | + |
| E. coli EC45 | Unknown | 32 | 32 | bla GES-1 | – | – |
| E. coli EC46 | Urine | 16 | 8 | bla GES-1 | – | – |
| E. coli EC47 | Blood | >128 | >128 | bla GES-1, blaGES-9 | IncFIA-FIB | + |
| E. coli EC48 | Pus | 0.06 | 0.06 | ND | – | – |
| E. coli EC49 | Blood | 64 | 64 | bla GES-1, blaGES-9 | – | – |
| E. coli EC50 | Pus | 16 | 32 | bla GES-1, blaGES-9 | – | – |
| E. coli EC51 | Bile fluid | 4 | 4 | bla GES-9 | – | – |
| E. coli EC52 | Unknown | >128 | 64 | bla GES-9, blaOXA-23 | – | – |
| E. coli EC53 | Blood | 64 | >128 | bla OXA-23 | – | – |
| E. coli EC54 | Urine | 8 | 8 | bla OXA-23 | – | – |
| E. coli EC55 | Unknown | 1 | 0.5 | ND | – | – |
| E. coli EC56 | Urine | 32 | 64 | bla OXA-23 | – | – |
| E. coli EC57 | Blood | 0.5 | 0.5 | ND | – | – |
| K. pneumoniae KP1 | Urine | 1 | 1 | ND | – | – |
| K. pneumoniae KP2 | Urine | 0.5 | 0.25 | ND | – | – |
| K. pneumoniae KP3 | Blood | >128 | 128 | bla NDM-1 | – | – |
| K. pneumoniae KP4 | Bile fluid | 2 | 1 | ND | – | – |
| K. pneumoniae KP5 | Urine | 8 | 32 | ND | – | – |
| K. pneumoniae KP6 | Blood | 0.25 | 0.12 | ND | – | – |
| K. pneumoniae KP7 | Blood | 8 | 16 | bla NDM-1 | – | – |
| K. pneumoniae KP8 | Blood | 0.5 | 0.25 | ND | – | – |
| K. pneumoniae KP9 | Urine | 32 | 32 | bla NDM-1 | – | – |
| K. pneumoniae KP10 | Blood | 64 | >128 | bla NDM-1 | IncFIA-FIB | + |
| K. pneumoniae KP11 | Unknown | 8 | 8 | bla NDM-1 | – | – |
| K. pneumoniae KP12 | Bile fluid | 1 | 1 | ND | – | – |
| K. pneumoniae KP13 | Urine | 32 | 64 | ND | – | – |
| K. pneumoniae KP14 | Urine | 0.06 | 0.12 | ND | – | – |
| K. pneumoniae KP15 | Pulmonary secretion | 8 | 4 | ND | – | – |
| K. pneumoniae KP16 | Urine | 2 | 2 | ND | – | – |
| K. pneumoniae KP17 | Blood | 16 | 16 | bla OXA-181 | – | – |
| K. pneumoniae KP18 | Unknown | 0.5 | 0.25 | ND | – | – |
| K. pneumoniae KP19 | Blood | 32 | 8 | bla OXA-181 | – | – |
| K. pneumoniae KP20 | Unknown | 128 | 64 | bla OXA-181 | – | – |
| K. pneumoniae KP21 | Unknown | 8 | 8 | bla OXA-181 | – | – |
| K. pneumoniae KP22 | Unknown | 16 | 2 | ND | – | – |
| K. pneumoniae KP23 | Blood | 16 | 8 | ND | – | – |
| K. pneumoniae KP24 | Blood | 0.25 | 0.25 | ND | – | – |
| K. pneumoniae KP25 | Unknown | 32 | 64 | bla OXA-181 | – | – |
| K. pneumoniae KP26 | Blood | 8 | 2 | ND | – | – |
| K. pneumoniae KP27 | Unknown | 64 | >128 | bla OXA-181 | – | – |
| K. pneumoniae KP28 | Blood | 32 | 8 | bla OXA-181 | – | – |
| K. pneumoniae KP29 | Unknown | 4 | 0.5 | ND | – | – |
| K. pneumoniae KP30 | Urine | 1 | 2 | ND | – | – |
| K. pneumoniae KP31 | Blood | 16 | 4 | bla GES-1 | IncA/C | – |
| K. pneumoniae KP32 | Unknown | 32 | 32 | bla GES-1 | – | – |
| K. pneumoniae KP33 | Urine | 128 | >128 | bla GES-1 | – | – |
| K. pneumoniae KP34 | Blood | 8 | 1 | ND | – | – |
| K. pneumoniae KP35 | Unknown | 0.5 | 1 | ND | – | – |
| K. pneumoniae KP36 | Urine | 64 | 16 | bla GES-9 | – | – |
| K. pneumoniae KP37 | Bile fluid | 64 | 64 | bla GES-9 | – | – |
| K. pneumoniae KP38 | Blood | 0.5 | 2 | ND | – | – |
| K. pneumoniae KP39 | Urine | >128 | >128 | bla OXA-23, blaOXA-51 | IncA/C | – |
| K. pneumoniae KP40 | Urine | 32 | 8 | bla OXA-51 | – | – |
| P. aeruginosa PA1 | Pus | 8 | 16 | bla NDM-1 | – | – |
| P. aeruginosa PA2 | Pus | 1 | 0.5 | ND | – | – |
| P. aeruginosa PA3 | Pus | 16 | 32 | ND | – | – |
| P. aeruginosa PA4 | Bronchial secretion | 0.25 | 0.25 | ND | – | – |
| P. aeruginosa PA5 | Urine | >128 | 128 | bla GES-1 | – | – |
| P. aeruginosa PA6 | Unknown | 8 | 4 | ND | – | – |
| P. aeruginosa PA7 | Blood | 32 | 32 | bla GES-1 | – | – |
| P. aeruginosa PA8 | Pus | 64 | 128 | ND | – | – |
| P. aeruginosa PA10 | Pus | >128 | 64 | ND | – | – |
| S. Typhi ST1 | Blood | 8 | 64 | ND | – | – |
| S. Typhi ST2 | Unknown | 32 | 32 | ND | – | – |
| S. Typhi ST3 | Urine | 0.5 | 1 | ND | – | – |
| S. Typhi ST4 | Blood | 32 | 64 | ND | – | – |
| S. Typhi ST5 | Urine | 128 | 64 | ND | – | – |
| S. Typhi ST6 | Blood | 16 | 8 | ND | – | – |
| S. Typhi ST7 | Blood | 4 | 4 | ND | – | – |
| S. Typhi ST8 | Unknown | 8 | 32 | ND | – | – |
| E. cloacae EL1 | Urine | 64 | >128 | ND | – | – |
| E. cloacae EL2 | Blood | 16 | 8 | bla NDM-1 | – | – |
| E. cloacae EL3 | Urine | 4 | 64 | bla NDM-1 | IncFIIA | – |
| E. cloacae EL4 | Bronchial secretion | 32 | 128 | ND | – | – |
| E. cloacae EL5 | Blood | 32 | 32 | bla OXA-181 | – | – |
| E. cloacae EL6 | Urine | 16 | 8 | – | – | – |
| E. cloacae EL7 | Urine | 128 | 128 | bla IMP-1 | – | – |
| E. cloacae EL8 | Urine | 32 | 8 | bla GES-9 | – | – |
| A. baumannii AB1 | CSF | 8 | 8 | bla OXA-181, bla OXA-51 | – | – |
| A. baumannii AB2 | Urine | 16 | 64 | bla OXA-181, bla OXA-51 | – | – |
| A. baumannii AB3 | Unknown | 0.5 | 1 | bla OXA-51 | – | – |
| A. baumannii AB4 | Pus | 8 | 64 | bla OXA-23, bla OXA-51 | – | – |
| A. baumannii AB5 | Blood | 32 | 32 | bla OXA-23, bla OXA-51 | – | – |
| A. baumannii AB6 | Urine | 32 | >128 | bla OXA-51 | – | – |
| A. baumannii AB7 | Urine | 16 | 2 | bla OXA-51 | – | – |
| S. marcescens SM1 | Bronchial secretion | 8 | 4 | ND | – | – |
| S. marcescens SM2 | Blood | 32 | 64 | bla NDM-1 | – | – |
| S. marcescens SM3 | Unknown | 128 | 64 | ND | – | – |
| S. marcescens SM4 | Urine | 2 | 2 | ND | – | – |
| S. marcescens SM5 | Unknown | 32 | 8 | ND | – | – |
| A. xylosoxidans AY1 | Unknown | 4 | 8 | ND | – | – |
| A. xylosoxidans AY2 | Blood | 128 | 128 | ND | – | – |
| A. xylosoxidans AY3 | Urine | 32 | 32 | ND | – | – |
| A. xylosoxidans AY4 | Urine | 1 | 0.5 | ND | – | – |
| A. xylosoxidans AY5 | Urine | 64 | 128 | bla NDM-1 | – | – |
| K. oxytoca KO1 | Blood | 32 | 128 | ND | – | – |
| K. oxytoca KO2 | Urine | 8 | 16 | ND | – | – |
| K. oxytoca KO3 | Blood | 32 | 32 | ND | – | – |
| K. oxytoca KO4 | Blood | 128 | 128 | bla OXA-181 | – | – |
| K. oxytoca KO5 | Urine | 8 | 2 | ND | – | – |
| P. mirabilis PM1 | Unknown | 1 | 2 | ND | – | – |
| P. mirabilis PM2 | Blood | 128 | 128 | bla OXA-181 | – | – |
| P. mirabilis PM3 | Urine | 8 | 8 | ND | – | – |
| P. mirabilis PM4 | Blood | 0.06 | 0.25 | ND | – | – |
| P. mirabilis PM5 | Urine | 64 | 32 | bla IMP-1 | IncFIA-FIB | – |
| E. meningoseptica EM1 | CSF | 64 | 64 | ND | – | – |
ND, not detected; ‘–’ denotes absence; ‘+’ denotes conjugation positive; bold text indicates the isolates were carrying resistance genes on conjugative plasmids. The resistance breakpoint (CLSI) for both meropenem and imipenem is MIC ≥ 4 mg/L.
In total, 11 carbapenem-resistant isolates harbouring plasmid-encoded resistance were subjected to conjugation studies (Table 2). The six E. coli isolates EC10, 21, 29, 39, 44, and 47 were found to facilitate the transfer of plasmid-mediated resistance to susceptible E. coli AB1157. Inter-generic transfer of NDM-1 was observed in one K. pneumoniae isolate (KP10) (Table 2).
Discussion
In India, carbapenem-resistant Gram-negative bacteria have been reported as becoming more frequent.17,18 In this study, the distribution of carbapenem-resistant isolates in 10 genera of Gram-negative bacteria isolated at a diagnostic centre in Tamil Nadu, India, has been investigated. Previous studies describe the increasing prevalence of ESBL and MBL producers among Gram-negative bacteria in India.25–28
In this study, experiments determining MIC values show that 107/151 (71%) isolates were resistant to meropenem, correlating with the observation made by the disc diffusion method (n = 128). All the 70 isolates harbouring carbapenem resistance genes were resistant according to the results of both the methods (MIC and disc diffusion). As carbapenems are one of the last-resort antibiotics available to treat infections caused by Gram-negative bacteria, the prevalence of carbapenem resistance is of worldwide concern. Our previous studies had reported the dissemination of carbapenem-resistant bacteria and carbapenem resistance genes among Gram-negative bacteria in Tamil Nadu.17,18 Here, we report the prevalence (71%) of carbapenem-resistant isolates among 10 genera of Gram-negative bacteria. β-Lactamase genes such as blaNDM-1 (n = 22), blaOXA-181 (n = 21), blaGES-1 (n = 11), blaGES-9 (n = 8), blaOXA-23 (n = 7), blaOXA-51 (n = 9) and blaIMP-1 (n = 3) were found in 70 isolates (with 10 isolates carrying more than one gene type), in contrast to our earlier study which reported a lower prevalence (27%) of blaNDM-1 and blaOXA-181 genes among carbapenem-resistant isolates.18 The coexistence of blaNDM-1 and blaOXA-181 in E. coli is a reason for major concern from the healthcare perspective. All the A. baumannii isolates (n = 7) were found to have either OXA-23 or OXA-181 along with OXA-51 intrinsic β-lactamase.29 Earlier reports from India showed the presence of OXA-23 and OXA-51 in carbapenem-resistant Acinetobacter causing serious healthcare problems.12 Enterobacteriaceae carried OXA-48-like genes, which are carbapenem-hydrolysing class D β-lactamases.13,30 The unusual occurrence of blaOXA-23 in E. coli, and plasmid-encoded blaOXA-23 and blaOXA-51 in K. pneumoniae are very important findings of this study, as only very few earlier studies have reported the presence of the blaOXA-23 gene in E. coli.31,32 OXA-23-like genes in Enterobacteriaceae may be embedded within a transposon but were not characterized in this study. The resistance reports on E. meningoseptica are very rare in India,31,32 and in this study it was found that one isolate of E. meningoseptica was resistant to imipenem and meropenem. Although earlier studies showed the presence of carbapenemase genes in E. meningoseptica, in this study no carbapenem resistance genes were found.
Carbapenem resistance among Gram-negative bacteria is becoming very common in Tamil Nadu, India. The isolates producing carbapenemases are mostly MDR and the rapid spread of carbapenem resistance genes is highly concerning. These resistance genes are located adjacent to mobile genetic elements (integrons and transposons), which facilitates the easy transposition between replicons.33 The extrachromosomal plasmids are the primary carriers of antibiotic resistance genes and can spread horizontally between strains or species. The recent molecular and genomic surveillance studies are also focused to track the clonally evolving lineages, besides plasmids being the primary focus.34 The most common plasmid replicon types for carbapenem resistance genes are IncF, IncA/C2, IncX3, IncL/M and IncH.35 In this study, blaNDM-1 was found in the isolates that carried IncX, IncA/C, IncFIA-FIB and IncFIIA; blaOXA-181 in IncA/C, IncFIA-FIB and IncFIIA; blaGES-1/9 in IncFIA-FIB and IncA/C; blaIMP-1 in IncFIA-FIB; and blaOXA-23/51 in IncA/C. The presence of plasmid-encoded blaOXA-23/51 is an important finding, considering the rapid spread of carbapenem resistance among Gram-negative bacteria. Interestingly, the isolates we investigated (such as P. aeruginosa, Salmonella Typhi, A. baumannii, S. marcescens, A. xylosoxidans, K. oxytoca, and E. meningoseptica) do not carry any plasmids harbouring resistance genes. This clearly showed that the β-lactamase or carbapenemase genes were confined to certain strains and present in the different replicon types (plasmids) in the study region. Earlier, the blaNDM IncFII plasmids were reported from India,35 and IncFIA-FIB plasmids carrying carbapenem resistance genes such as blaNDM were described in samples collected from river and sewage treatment plants in India.7,35 This study also showed that some isolates with plasmids were carrying more than one resistance gene, an alarming public health threat. Conjugative plasmids are known to spread their resistance among the bacteria of the same or of different genera. This study showed that all the six E. coli isolates carrying plasmid-encoded resistance genes (blaNDM-1, blaOXA-181, blaGES-1, and blaGES-9) were conjugative and one K. pneumoniae plasmid (IncFIA-FIB with blaNDM-1) was transferable, illustrating how resistance genes rapidly spread in clinically relevant bacteria.
We acknowledge several limitations of our study. First, the clinical samples or isolates were collected randomly from the diagnostic centre, which receives clinical samples from multiple hospitals (both in- and out-patient) in the study region. Second, the presence of insertion sequence (IS) elements was not studied. Finally, the transfer of resistance genes between the bacteria was studied using simple conjugation experiments but we did not confirm the results using Southern hybridization or sequencing techniques.
Conclusions
The increasing frequency of antibiotic resistance in bacteria is a major healthcare problem. This study highlights the distribution of carbapenem-resistant isolates in the region we studied, with the emphasis on the existence of blaNDM-1, blaOXA-181, blaIMP-1, blaGES-1, blaGES-9, blaOXA-23-like, and blaOXA-51-like among the clinical pathogens. The unusual presence of an E. coli strain carrying blaOXA-23, and K. pneumoniae isolates carrying blaOXA-23 and blaOXA-51 require targeted antibiotic resistance surveillance programmes. The development of alternative therapeutic options should be undertaken immediately to be able to combat the problem of resistance, especially to treat carbapenem-resistant infections in the future. Our study shows that conjugative plasmids are a major contributor to the transfer of resistance in pathogens leading to further dissemination of resistance genes. A One-Health approach is necessary to combat the problem of resistance both at the local and international level.
Supplementary Material
Acknowledgements
We thank Vellore Institute of Technology (VIT) for providing partial funding as ‘VIT Seed Grant’ and Council of Scientific and Industrial Research (CSIR) for providing financial assistance to P.M. in the form of senior research fellowship (SRF) to support this research work. A preprint of this article has been published in BioRxiv and Research Square (Manohar et al.).
Funding
This study was supported by internal funding.
Transparency declarations
None to declare. All the datasets are presented in the main manuscript. The raw datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author contributions
Authors P.M. and R.N. collected the isolates from the clinical samples. Authors P.M. and R.N. undertook the laboratory work, R.N. and B.S.L. interpreted the data, and P.M. and R.N. wrote the initial manuscript. Authors S.L., R.N. and B.S.L. revised and finalized manuscript. All the authors read and approved the manuscript.
Supplementary data
Tables S1 to S7 are available as Supplementary data at JAC Online.
References
- 1. Roca I, Akova M, Baquero F. et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect 2015; 6: 22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO. WHO publishes list of bacteria for which new antibiotics are urgently needed. 2017. http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
- 3. Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T 2015; 40: 277. [PMC free article] [PubMed] [Google Scholar]
- 4. McKenna M. Antibiotic resistance: the last resort. Nat News 2013; 499: 394. [DOI] [PubMed] [Google Scholar]
- 5. Ben‐David D, Kordevani R, Keller N. et al. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect 2012; 18: 54–60. [DOI] [PubMed] [Google Scholar]
- 6. Xu Y, Gu B, Huang M. et al. Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000-2012 in Asia. J Thorac Dis 2015; 7: 376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akiba M, Sekizuka T, Yamashita A. et al. Distribution and relationships of antimicrobial resistance determinants among extended-spectrum-cephalosporin-resistant or carbapenem-resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrob Agents Chemother 2016; 60: 2972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandran SP, Sarkar S, Diwan V. et al. Detection of virulence genes in ESBL producing, quinolone resistant commensal Escherichia coli from rural Indian children. J Infect Dev Ctries 2017; 11: 387–92. [DOI] [PubMed] [Google Scholar]
- 9. Manohar P, Shanthini T, Marathe N. et al. Genetic characteristics of plasmid-mediated extended-spectrum β-lactamases (CTX-M) and its coexistence with carbapenemases (NDM-1) in clinical Gram negative bacteria. Ind J Biotechnol 2017; 16: 189–94. [Google Scholar]
- 10. Scaife W, Young HK, Paton RH. et al. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother 1995; 36: 585–6. [DOI] [PubMed] [Google Scholar]
- 11. Teixeira AB, Martins AF, Barin J. et al. First report of carbapenem-resistant Acinetobacter nosocomialis isolates harboring ISAba1-bla OXA-23 genes in Latin America. J Clin Microbiol 2013; 51: 2739–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. La MV, Jureen R, Lin RT. et al. Unusual detection of an Acinetobacter class D carbapenemase gene, bla OXA-23, in a clinical Escherichia coli isolate. J Clin Microbiol 2014; 52: 3822–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paul D, Ingti B, Bhattacharjee D. et al. An unusual occurrence of plasmid-mediated blaOXA-23 carbapenemase in clinical isolates of Escherichia coli from India. Int J Antimicrob Agents 2017; 49: 642–5. [DOI] [PubMed] [Google Scholar]
- 14. Österblad M, Karah N, Halkilahti J. et al. Rare detection of the Acinetobacter class D carbapenemase blaOXA-23 gene in Proteus mirabilis. Antimicrob Agents Chemother 2016; 60: 3243–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown S, Young HK, Amyes SGB.. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect 2005; 11: 15–23. [DOI] [PubMed] [Google Scholar]
- 16. Chen TL, Lee YT, Kuo SC. et al. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother 2010; 54: 4575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nachimuthu R, Subramani R, Maray S. et al. Characterization of carbapenem-resistant Gram-negative bacteria from Tamil Nadu. J Chemother 2016; 28: 371–4. [DOI] [PubMed] [Google Scholar]
- 18. Manohar P, Shanthini T, Ayyanar R. et al. The distribution of carbapenem-and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol 2017; 66: 874–83. [DOI] [PubMed] [Google Scholar]
- 19. CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Sixth Edition: M100. 2017.
- 20. Gheorghe I, Czobor I, Chifiriuc MC. et al. Molecular screening of carbapenemase-producing Gram-negative strains in Romanian intensive care units during a one year survey. J Med Microbiol 2014; 63: 1303–10. [DOI] [PubMed] [Google Scholar]
- 21. Dallenne C, Da Costa A, Decré D. et al. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 2010; 65: 490–5. [DOI] [PubMed] [Google Scholar]
- 22. Woodford N, Ellington MJ, Coelho JM. et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006; 27: 351–3. [DOI] [PubMed] [Google Scholar]
- 23. Lin L, Ling BD, Li XZ.. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii–Acinetobacter calcoaceticus complex. Int J Antimicrob Agents 2009; 33: 27–32. [DOI] [PubMed] [Google Scholar]
- 24. Carattoli A, Bertini A, Villa L. et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 2005; 63: 219–28. [DOI] [PubMed] [Google Scholar]
- 25. Oberoi L, Singh N, Sharma P. et al. ESBL, MBL and AmpC β lactamases producing superbugs–Havoc in the Intensive Care Units of Punjab India. J Clin Diagn Res 2013; 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Azim A, Dwivedi M, Rao PB. et al. Epidemiology of bacterial colonization at intensive care unit admission with emphasis on extended-spectrum β-lactamase-and metallo-β-lactamase-producing Gram-negative bacteria–an Indian experience. J Med Microbiol 2010; 59: 955–60. [DOI] [PubMed] [Google Scholar]
- 27. Nordmann P, Naas T, Poirel L.. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis 2011; 17: 1791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khurana S, Mathur P, Kapil A. et al. Molecular epidemiology of β-lactamase producing nosocomial Gram-negative pathogens from North and South Indian hospitals. J Med Microbiol 2017; 66: 999–1004. [DOI] [PubMed] [Google Scholar]
- 29. Durante-Mangoni E, Zarrilli R.. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol 2011; 6: 407–22. [DOI] [PubMed] [Google Scholar]
- 30. Bali NK, Fomda BA, Bashir H. et al. Emergence of carbapenem-resistant Acinetobacter in a temperate north Indian State. Br J Biomed Sci 2013; 70: 156–60. [DOI] [PubMed] [Google Scholar]
- 31. Shinha T, Ahuja R.. Bacteremia due to Elizabethkingia meningoseptica. IDCases 2015; 2: 13–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhat KS, Priya R, Krishnan L. et al. Elizabethkingia meningoseptica bacteremia in a neonate: A case report and mini-review of the literature. J Curr Res Sci Med 2016; 2: 42. [Google Scholar]
- 33. Partridge SR, Iredell JR.. Genetic contexts of blaNDM-1. Antimicrob Agents Chemother 2012; 56: 6065–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. David S, Cohen V, Reuter S. et al. ; ESCMID Study Group for Epidemiological Markers (ESGEM). Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci Usa 2020; 117: 25043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sugawara Y, Akeda Y, Sakamoto N. et al. Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS One 2017; 12: e0184720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.