Abstract
Influenza A virus (IAV) poses a significant threat to human health, which calls for the development of efficient detection methods. The present study constructed a fluorescence resonance energy transfer (FRET) system based on novel fluorescent probes and graphene oxide (GO) for detecting H5N1 IAV hemagglutinin (HA). Here, we synthesized small (sub-20 nm) sandwich-structured upconversion nanoparticles (UCNPs) (SWUCNPs for short) with a high energy transfer efficiency, which allows for controlling the emitter in a thin shell. The π–π stacking interaction between the aptamer and GO shortens the distance between the fluorescent probe and the receptor, thereby realizing fluorescence resonance energy transfer (FRET). When HA is present, the aptamer enables changes in their conformations and move away from GO surface. Fluorescence signals display a linear relationship between HA quantitation in the range of 0.1–15 ng mL–1 and a limit of detection (LOD) of 60.9 pg mL–1. The aptasensor was also applicable in human serum samples with a linear range from 0.2 to 12 ng mL–1 and a limit of detection of 114.7 pg mL–1. This strategy suggested the promising prospect of the aptasensor in clinical applications because of the excellent sensing performance and sensitivity. This strategy may be promising for vitro diagnostics and provides new insights into the functioning of the SWUCNPs system.
1. Introduction
Influenza A viruses (IAVs) belong to the family of Orthomyxoviridae and are spherical in shape of about 80–120 nm diameter.1 The subtypes of influenza A viruses are further classified according to the antigenicity of surface glycoproteins hemagglutinin (HA) and neuraminidase (NA).2 HA exists as trimeric spikes on the viral membrane, mediating the attachment of influenza virus to host cells containing sialic acid and facilitating viral infection.3 The highly conserved areas of HA are crucial for viral function and replication, which is suitable as the detection target for H5N1 diagnosis.4 The infection of influenza A virus has to represent a grave threat to public health as well as the global economy. Since 2003, the avian influenza A (H5N1) virus has caused 455 deaths of 861 people infected.5 Rapid and sensitive detection for the diagnosis of H5N1 infection are essential for disease control. At present, the traditional methods of detection and diagnosis include viral culturing, serology, and viral nucleic acid detection techniques.6 Viral culturing requires professionally trained technicians, as well as complex and expensive equipment.7 Enzyme-linked immunosorbent assay (ELISA) is an antibody-based conventional assay method; however, ELISA has a relatively low sensitivity and complicated operation steps.8 The polymerase chain reaction (PCR)-based viral nucleic acid detection requires professionally trained personnel, and this detection method is prone to cross-contamination and false-positive results. Nonspecific amplification is actually a real problem that influences the accuracy of PCR methods.9
Fluorescence resonance energy transfer (FRET) is distance-dependent energy transfer between two fluorophores and shows particular advantages of a separation-free step.10,11 Lanthanide-doped upconversion nanoparticles (UCNPs) can be excited with near-infrared (NIR) light,12 avoiding the influence of biological background fluorescence. Its advantages are significant Stokes shift, narrow emission spectrum, long lifetime, etc. Compared with downconversion conventional fluorescent probes, UCNPs reduce the background of autofluorescence and enhances light stability, significantly improving the signal-to-noise ratio in biological detection based on fluorescent labels.13−18 Consequently, UCNPs hold great promise for biodetection based on fluorescence labeling. It is well known that the efficiency of FRET is highly dependent on the distance between the donor and the acceptor, and nonradiative energy transfer is usually effective within a distance of 10 nm.19 The small-size UCNP doped with lanthanide ions is used to shorten the distance between the donor and the acceptor. However, the small size of UCNP will reduce the efficiency of upconversion luminescence (UCL), which is susceptible to quenching from surroundings.20−22 To effectively improve the energy transmission efficiency, we adopt a layer-by-layer seed-mediated shell growth strategy to fabricate a sandwich-structured UCNPs.23 This strategy allows the emission center to be located in the sandwich layer close to the surface of the UCNPs, which enables most emitting ions to be confined in a thin layer near, thereby ensuring improved signal intensity. As such, it would be able to achieve higher detection sensitivity of HA.
It has been proven that graphene oxide (GO) is an oxidized single-atom-thick, two-dimensional carbon material.24 A few reports have indicated that GO can bind with single-stranded DNA (such as aptamer) via a π–π stacking interaction between the carbon cycle and single-stranded DNA. As a fluorescent acceptor, GO has a high solubility in an aqueous solution, a large specific surface area, and a highly efficient quenching ability, making it suitable as a high-efficiency quencher of UCNP.25,26 This FRET design eliminates the need for a covalent label and shortens the total assay time, which inspired the design of novel FRET sensors based on GO and the aptamer.27,28
Aptamers are single-stranded DNA or RNA, which are selected through the systematic evolution of ligands by the exponential enrichment (SELEX) technique.29,30 Compared with antibodies, aptamers are much smaller in size and have thermal and chemical stability and flexible modification. The properties of high affinity and specificity in binding targets make aptamers effective molecular elements for biomedical detection.31 Heretofore, aptamer-based sensors have been widely used in the homogeneous determination of biomolecules.32−34
To date, various strategies based on the aptamer for influenza A virus A (IAV) detection have been developed. However, few HA detection methods have been developed based on UCNPs and GO. This study aimed to test a new type of UCNPs to establish a highly selective and sensitive fluorescent probe for in vitro assays. In this paper, combining the advantage of GO and sandwich-structured UCNPs (SWUCNPs) and NaGdF4@NaYF4:Yb,Er@NaYF4 UCNPs as energy donors and GO as energy receptors are designed as a FRET system for the sensitive detection of H5N1 HA. The HA aptamer-modified SWUCNPs could be brought in close proximity to the GO surface based on the π–π stacking interaction between the aptamer and GO, and then, FRET-induced fluorescence quenching can be observed. In the presence of HA of the H5N1 virus, the formation of the aptamer–HA complex restrained the interaction between the aptamer and GO, which inhibited the FRET process and resulted in the restoration of fluorescence. Based on the SWUCNPs as the energy donor by conjugating the aptamers, a high-sensitivity platform for FRET-based applications was designed. The proposed concept and strategy provides a versatile platform for probing the upconversion emission-based aptasensor for practical applications.
2. Results and Discussion
2.1. Principle of the SWUCNP-FRET-Based Aptasensor for HA of the H5N1 Virus
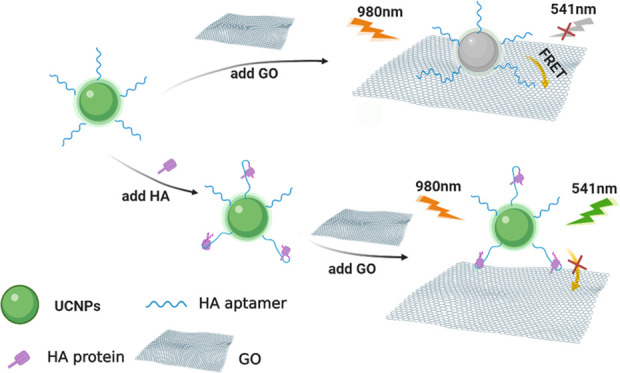
The aptasensor strategy for HA detection was based on FRET from modified SWUCNPs to GO, as briefly depicted in Figure 1. The amino-modified H5N1 HA aptamers were attached to the polyacrylic acid (PAA)-functionalized SWUCNPs by condensation, which can be employed as probes for sensitive and selective detection of HA. Moreover, a system of this type can be implemented by employing a single mixing step. In the absence of H5N1 HA, the distance between SWUCNPs and GO was shortened to <10 nm because the aptamer-modified UCNPs (UCNP-Apt for short, 0.05 mg mL–1) can be brought in close proximity to the GO surface based on the strong π–π stacking effect. Meanwhile, the resonance energy transfer process enables quenching of upconversion luminescence due to their highly overlapped spectrum. In contrast, in the presence of HA, aptamers preferentially bind to HA, which leads to changes in the G-quadruplex structure,2,35−37 thereby promoting the separation of the donor and the acceptor, and the upconversion fluorescence is regained.38 Substitution of UCNPs with FAM dye shows the feasibility of using the H5N1 HA aptamer as the recognition probes(Figure S1).Aptamer binding triggers a conformational change, restoring the prequenched fluorescence signal.
Figure 1.
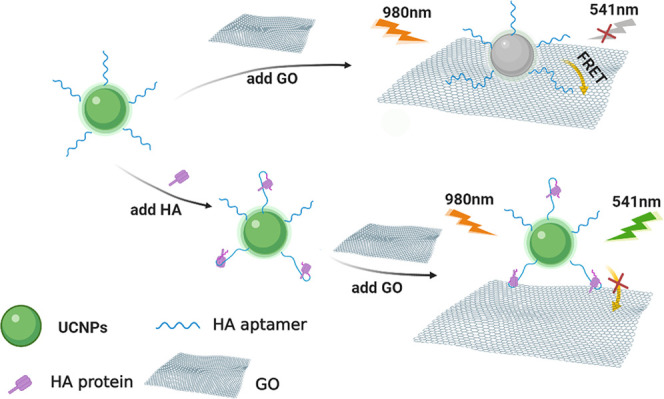
Schematic illustration to detect HA of influenza based on the FRET method.
2.2. Characterization of the Sandwich-Structured UCNPs
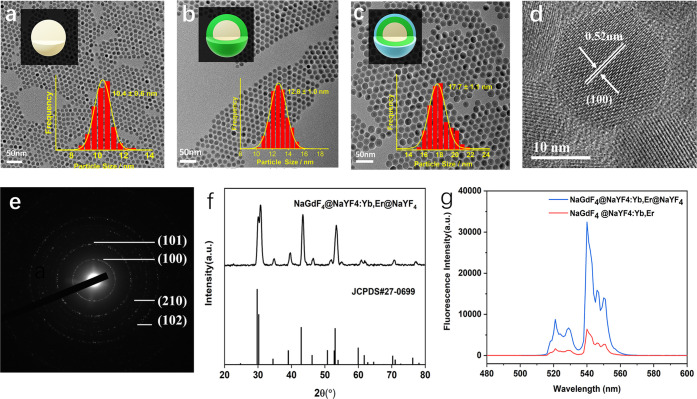
To protect the emitters from environmental quenching and improve quantum yield and brightness, we synthesized the sandwich-structured NaGdF4@NaYF4:Yb,Er@NaYF4 with an average diameter of ∼17.7 nm (Figure 2c) using a layer-by-layer seed-mediated shell growth strategy. The inert shell thickness was a compromise between passivating the structure and therefore improving the quantum yield and brightness while maintaining a small nanoparticle size. NaGdF4 nanocrystals were first synthesized as the core and subsequently coated with the NaYF4:Yb,Er shell through epitaxial growth. To protect the emitting ions from environmental quenching, another NaYF4 shell was further deposited on the surface of the inner shell to form the SWUCNPs. Transmission electron microscopy (TEM) images of the as-synthesized nanoparticles (Figure 2a–c) illustrate the size evolution of the materials, from the ∼10.4 nm NaYF4 cores to the ∼12.8 nm NaGdF4@NaYF4:Yb,Er core–shell particles and further to the ∼17.7 nm NaGdF4@NaYF4:Yb,Er@NaYF4 sandwich structure. According to the size histograms obtained from the large-area TEM images, the size distributions of all of these materials are quite narrow (σ < 4%), indicating the fine control of the material morphology by the growth pathway. Legible lattice fringes of 0.52 nm in the high-resolution TEM (HRTEM) image (Figure 2d) are correlated with the (100) plane of hexagonal NaYF4, which is also verified by the selected area electron diffraction (SAED) pattern result (Figure 2e). The X-ray diffraction (XRD) patterns (Figure 2f) indicate that the NaGdF4@NaYF4:Yb,Er@NaYF4 sandwich structure was hexagonal in-phase (JCPDS 27-0699). All of the results demonstrate the successful synthesis of SWUCNPs. As displayed in Figure 2g, the protecting layer significantly strengthens the fluorescence intensity in comparison to the core–inner shell material.
Figure 2.
TEM images of (a) NaGdF4 core, (b) NaGdF4@NaYF4:Yb,Er, core–inner shell structure, (c) NaGdF4@NaYF4:Yb,Er@NaYF4 sandwich structure. (d) High-resolution TEM image and (e) SAED pattern of SWUCNPs. (f) XRD patterns of the obtained nanoparticles. (g) Emission spectra of NaGdF4@NaYF4:Yb,Er@NaYF4 and NaGdF4@NaYF4:Yb,Er UCNPs.
2.3. Characterization of PAA-Coated UCNPs and GO
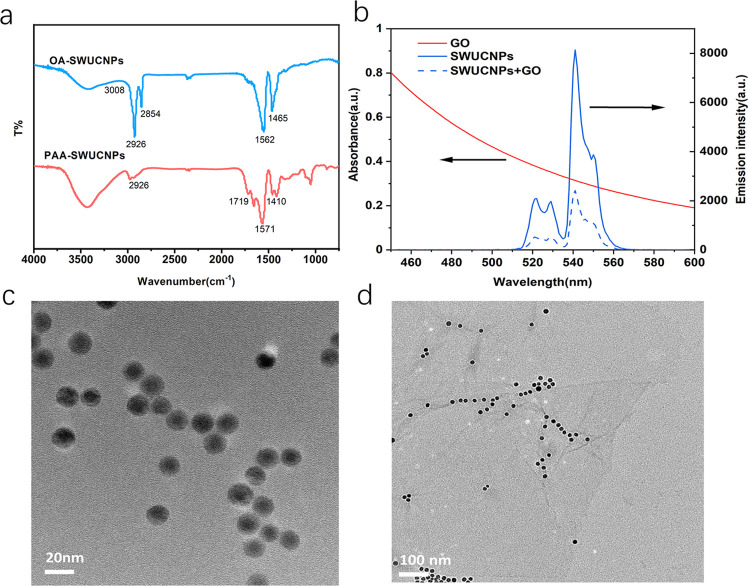
A modified two-step ligand-exchange route, NOBF4-PAA treatment, was also adopted to transfer the oleic acid (OA)-capped UCNPs to water. To realize the above design, PAA-coated upconverting nanoparticles were first prepared by a modified two-step ligand-exchange route, NOBF4-PAA treatment, to enhance the dispersibility in water. The carboxyl group of PAA on the surface of UCNPs could further conjugate with other biomolecules such as aptamers. The existence of PAA molecules on the surface of UCNPs is confirmed by Fourier transform infrared (FT-IR) spectra (Figure 3a); in the case of OA-UCNPs, the peaks at 2926 (C–H stretching vibration), 1562 (C=O stretching vibration), and 1465 cm–1 (C–O stretching vibration) demonstrate the presence of OA molecules. After the two-step ligand-exchange route, the bands centered at 1719 and 1571 cm–1 suggest an increased quantity of −COOH groups on the particle surface. Furthermore, the shoulder band at 2926 cm–1 associated with the characteristic absorption of OA is also obviously weakened, demonstrating that OA molecules had been efficiently substituted by PAA. After ligand exchange, the TEM image of PAA-capped SWUCNPs (Figure 3c) shows that the nanoparticles were well dispersed in water and demonstrated good stability and uniform size. To explore the possible mechanisms behind energy transfer between UCNP and GO, we sought to examine the upconversion emission spectrum of UCNPs and the UV–visible absorption spectrum of GO (Figure 3b). The as-synthesized SWUCNPs exhibited maximum fluorescence at 541 nm. Moreover, there is a good overlap between the emission and absorption spectra of GO (approximately 300–700 nm), thus facilitating energy transfer from UCNPs to GO.
Figure 3.
(a) FT-IR spectra of SWUCNPs capped with OA molecules and PAA molecules. (b) Upconversion fluorescence spectra of SWUCNPs and the UV–vis absorption spectra of GO. (c) TEM images of PAA-capped SWUCNPs and (d) complex of GO (0.1 mg mL–1) and UCNP-Apt (0.05 mg mL–1).
2.4. Characterization of the UCNP-Aptamer Nanoprobe
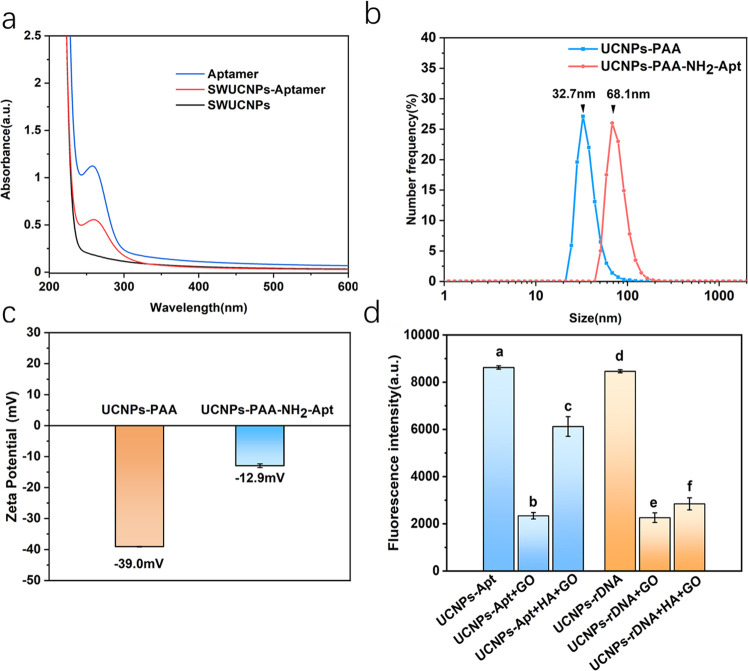
As shown in Figure 4a, there was one peak at 260 nm in the UV–vis spectrum of the aptamer. A new absorption peak appeared at 260 nm for UCNP-PAA after aptamer conjugation through 3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (sulfo-NHS) activation, verifying the successful conjugation. We also analyzed the sizes of UCNP-PAA and the UCNP-PAA-NH2 aptamer by dynamic light scattering (DLS). After covalent coupling, the particle size of nanoparticles turned 32.7 to 68.1 nm (Figure 4b). From Figure 4c, It could be seen that PAA-modified UCNPs were negatively charged. After the reaction, the ζ-potential changed from −39 to −13mV, indicating that UCNPs conjugated with the aptamer through coupling reaction. After incubation with GO, the complexes were formed by a π–π stacking effect between the interaction of aptamer and the sp2 atoms of GO. As depicted in Figure 3d, there are intermolecular π–π stacking interactions between UCNP-Apt (0.05 ng mL–1) and GO sheets. Since the aptamer affinity to the protein is much higher than that of the aptamer and GO (Figure S2), the conformational change of the aptamer makes a difference in the nanoparticle density.
Figure 4.
(a) UV–vis absorption spectra of UCNP and UCNP-Apt conjugates. (b) DLS and (c) ζ-potential of UCNP-PAA and UCNP-PAA-NH2-aptamer. (d) Fluorescence intensity at 541 nm of UCNP-Apt (a–c) and UCNP-rDNA (d–f) under different circumstances. The concentrations of UCNP-Apt, GO, and HA were 0.05 mg mL–1, 150 μg mL–1, and 10 ng mL–1, respectively. Error bars represented the standard deviation (SD) of three parallel tests.
To verify the specific recognition of aptamers and HA, a control experiment was also performed using random ssDNA (rDNA) instead of the H5N1 aptamer (Figure 4d). The results displayed that UCNP-rDNA could not be released from GO by HA in this experimental condition. The results of UV–vis absorption, TEM, DLS, and ζ-potential proved that UCNPs conjugated with the aptamer through a coupling reaction and UCNP-Apt could recognize HA.
2.5. Optimization of the FRET System
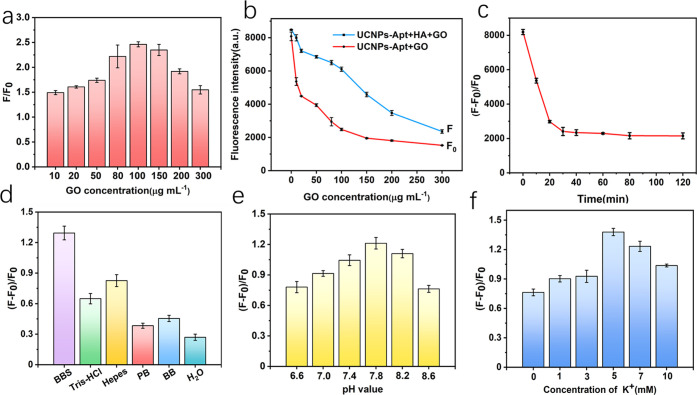
To investigate the energy transfer between the donor–acceptor pair, we fixed the concentration of UCNP-Apt (0.05 mg mL–1) and changed the concentration of GO, wherein F0 and F represent the upconversion fluorescence intensities of the UCNP-Apt–GO complex in the absence and presence of HA, respectively. As we have seen from Figure 5a, the quenching efficiency was strongly dependent on the GO concentration. Excessive GO concentration might yield unspecific HA binding, which is not conducive to the improvement of sensitivity. We tend to choose 100 μg mL–1 as a balanced pattern. Figure 5b shows the effect of different concentrations of GO on the fluorescence intensity of UCNP-Apt (0.05 mg mL–1) in the presence (Figure 5b, F) and absence (Figure 5b, F0) of HA. To preclude the nonspecific adsorption and make the fluorescent aptasensor more sensitive, a GO concentration of 100 μg mL–1 was selected for subsequent experiments. To ensure reaching the quenching equilibrium and getting stable fluorescence signals (Figure 5c), the UCNP-Apt (0.05 mg mL–1) displayed a slow decrease in the luminescence intensity after 30 min of incubation with HA (10 ng mL–1) at 37 °C. Thus, the optimal incubation time was determined to be 30 min. The recovery of the fluorescence signal depends on whether the aptamer can maintain a specific configuration. Different buffers, pH values, and metal ions often affect correct aptamer folding, so we optimized the detection conditions of the FRET system based on aptamers. Figure 5d showed the results of buffer tests, which demonstrate that borate-buffered saline (BBS) is the most suitable for FRET detection. The folding of the aptamer is affected by the surrounding buffer conditions (such as pH, ionic strength, or the presence of specific ions). We speculate that the salt content and pH of BBS are not easily affected by storage time and temperature, which is conducive to the stability of the aptamer configuration. Simultaneously, the composition of the buffer solution does not coordinate with the rare-earth ions, which ensures the stability of the probe function. Then, the effect of pH on upconversion fluorescence was studied. As shown in Figure 5e, during the range of 6.6–8.6 BBS, the response reached the maximum when the pH was 7.8. Thus, BBS with a pH of 7.8 was used as the buffer in the following study. In addition, the G-quadruplex secondary structure of the aptamer was normally stable under monovalent cation (K+) conditions, as shown in Figure 5f; KCl concentrations (0–10 mM) in borate buffer were investigated, and KCl at 5 mM in borate buffer was selected as the working ionic strength condition.
Figure 5.
(a) Fluorescence intensity rate (F/F0) of UCNP-Apt (0.05 mg mL–1) by 10 ng mL–1 HA with various concentrations of graphene oxide (excitation wavelength 980 nm). (b) Effect of GO concentration on the fluorescence intensity of UCNP-Apt in the presence (F) and absence (F0) of 10 ng mL–1 HA. (c) Time dependence of the fluorescence quenching degree with UCNP-Apt (0.05 mg mL–1) and 100 μg mL–1 GO. All experiments were performed in borate buffer (10 mM, 5 mM KCl, pH 7.8) under excitation at 980 nm. (d) Quenching using various reaction buffers (the concentration of buffer solutions is 20 mM with the same pH). (e) pH dependence of the fluorescence intensity. (f) Quenching efficiency with various concentrations of KCl in borate buffer.
2.6. HA of H5N1 Virus Detection with the FRET System
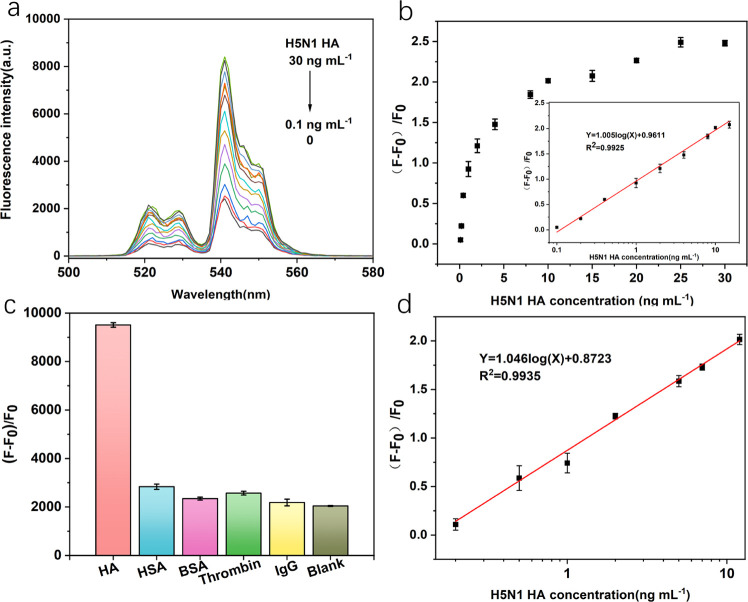
Under the optimum conditions described above, the FRET system was established for quantitative analysis. When HA interacts with the UCNP-Apt system, the aptamer preferentially bound to target HA, which leads to changes in the formation of aptamers. Therefore, the π–π stacking interaction between the aptamer and GO was weakened and parts of luminescence of UCNPs were reserved. In this condition, the energy donor and acceptor have separated from each other. Therefore, the FRET process was inhibited and the fluorescence of UCNPs recovered accordingly. As shown in Figure 6a, increasing the concentration of HA resulted in increasing degrees of fluorescence restoration. Under optimal conditions, the logarithm of HA concentration was proportional to the relative intensity [(F – F0)/F0]. Figure 6b shows that with the increasing number of HA from 0 to 30 ng mL–1, a significant increase in the intensity at 541 nm was observed accordingly. Besides, a linear relationship was obtained between the increasing fluorescence intensity of UCNPs and the logarithm of HA concentration in the range from 0.1 to 15 ng mL–1 (Figure 6b). The regression equation could be represented by Y = 1.005 log(X) + 0.9611, with a reliable correlation coefficient R2 = 0.9925. In particular, the limit of detection (LOD) was estimated to be 60.9 pg mL–1 (S/N = 3). The result demonstrates that this proposed UCNP-FRET system was a promising approach for the highly sensitive detection of H5N1 HA because it was a homogeneous and straightforward procedure without any separation or repeated reagent addition.
Figure 6.
(a) Upconversion emission spectra of the UCNP aptasensor in the presence of various concentrations of H5N1 HA (0, 0.1, 0.2, 0.4, 1, 2, 4, 8, 10, 15, 20, 25, 30 ng mL–1) under optimal experimental conditions, where F0 represents the fluorescence intensity of the UCNP-Apt–GO complex (0.05 mg mL–1 UCNP-Apt and 0.1 mg mL–1 GO) and F is the fluorescence intensity of the complex plus different concentrations of HA. (b) Relationship between relative fluorescence intensity (F – F0)/F0 at 541 nm against the H5N1 HA concentration. Inset: linear part of the plot ranging from 0.01 to 15 ng mL–1 in borate buffer. (c) Specific selectivity evaluation of the proposed method for HA (20 ng mL–1) against other proteins (100 ng mL–1). (d) Relationship curve of relative fluorescence intensity for added standard HA from human serum samples. It shows the linear response with HA concentrations from 0.2 to 12 ng mL–1, under the experimental conditions the same as an aqueous buffer. Error bars: SD, n = 3.
2.7. Specificity for H5N1 HA Detection
To assess the specificity of the proposed aptasensor for HA detection, some abundant proteins in blood or analogous proteins including human serum albumin (HSA), bovine serum albumin (BSA), thrombin, and immunoglobulin G (IgG) were introduced individually into the aptasensor instead of HA in the borate buffer. Different proteins were taken in a concentration of 100 ng mL–1, 5 folds higher than that of HA (20 ng mL–1) under identical experimental conditions. As shown in Figure 6c, none of these species caused a significant recovery of the fluorescence intensity of UNCPs and the fluorescence intensity of HA was eight times higher than that of other controls. Thus, the results above vividly indicated that the designed fluorescent aptamer sensor is highly specific to HA.
2.8. Detection of the HA Protein in Human Serum
Serum is a known complicated biological matrix containing various biomolecules that tend to increase autofluorescence and scattering light in optical sensing. To investigate the potential application of this method, tests for HA in human serum were performed by spiking the samples with a series of predetermined amounts of HA. Figure 6d shows the linear relationship between the fluorescence changes (F – F0)/F0 and the logarithm of HA concentration. For detection in the serum sample, the linear relationship was observed in the range from 0.2 to 12 ng mL–1, which was a little different from that in aqueous buffer. The linear equation was Y = 1.046 log(X) + 0.8723 with a LOD as low as 114.7 pg mL–1 (S/N = 3, n = 9). Figure S3 shows the corresponding fluorescence spectrum. Meanwhile, the applicability of the sensor for real sample analysis was investigated by employing the sensor to analyze these serum samples. The recovery test of different concentrations of HA in serum samples was determined using this sensor. The recovery of HA in three spiked serum samples and the relative standard deviation (RSD) levels (n = 3) are presented in Table 1. The excellent recoveries from 95.26 to 105.12% indicate that the designed method possesses reliable accuracy for H5N1 HA detection. This finding indicates that the approach has the potential for practical applications in the early diagnosis of the HA protein. As shown in Figure S3, the HA concentration measured by the detection platform is consistent with the ELISA method (R2 = 0.996). To verify the stability of the UCNPs, we measured the fluorescence intensity within 15 days and compared the interbatch accuracy in samples. As shown in Figure S4, the results are in line with this expectation. To further investigate the stability of this method, we monitored the interassay variation of five serum samples. The interassay accuracy varied between 95.37 and 114.50%, while the precision ranged between 2.21 and 9.78% RSD (Table S1). A comparison between the present HA of the H5N1 virus biosensor and other biosensors reported is summarized in Table S2. It indicated that the developed SWUCNP-GO aptasensor has acceptable sensitivity and advantages in selectivity and simplicity of preparation. Remarkably, the detection limit of other HA aptamer-based fluorescence biosensors was 33 times higher than that of our method. Because of their near-infrared (NIR) excitation nature that minimizes autofluorescence from serum samples and interference from scattered light, this aptasensor shows reliable stability and sensitivity in serum detection. The SWUCNP-based aptamer platform combines the advantages of small size and reversible secondary structure conformation of the aptamer, which provides favorable conditions for constructing a highly sensitive homogenous immunoassay based on the FRET system.
Table 1. Detection of the HA Protein in Serum Samples of This Method.
| samples | volume of addition (ng mL–1) | found ± SD (ng mL–1) | recovery (%) | RSD (%) n = 3 |
|---|---|---|---|---|
| 1 | 2 | 2.10 ± 0.023 | 105.12 | 1.09 |
| 5 | 5.02 ± 0.42 | 100.46 | 8.36 | |
| 10 | 10.17 ± 0.73 | 101.72 | 7.18 | |
| 2 | 1.83 ± 0.012 | 95.26 | 0.66 | |
| 2 | 5 | 5.23 ± 0.52 | 104.54 | 9.94 |
| 10 | 10.41 ± 0.33 | 104.11 | 3.17 | |
| 2 | 1.90 ± 0.12 | 94.95 | 6.31 | |
| 3 | 5 | 4.90 ± 0.39 | 97.92 | 7.96 |
| 10 | 10.16 ± 0.63 | 101.66 | 6.20 |
3. Conclusions
Aptamer-based targeting offers a novel approach to construct convenient, ultrasensitive, specific, and stable platforms for bioassays. Due to the sandwich-structured UCNPs and the introduction of GO, the SWUCNP-GO aptasensors exhibited high FRET efficiency. The virtue of the SWUCNPs improved the signal-to-noise ratio and avoided background luminescence through a process of generating visible light by a NIR versatile platform. There was a good linear relationship between the fluorescence signal and the HA concentrations of 0.1–15 ng mL–1 with a low limit of detection of 60.9 pg mL–1, and a selective method was applicable for HA quantification in human serum, which has practical application value in real sample detection. In summary, the sensitivity of detection is due to the low background luminescence of SWUCNPs and high specificity of the aptamer toward the HA protein of IAV.
4. Materials and Methods
4.1. Materials
2-(N-morpholino)-ethanesulfonic acid (MES) hydrate, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES), Tris, 3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), and N-hydroxysulfosuccinimide (sulfo-NHS) sodium salt were purchased from Sigma-Aldrich Chemical (St. Louis, MO). Water (DNase and RNase free), HEPES, and Tris were from Beijing Solarbio Science & Technology Co., Ltd. (China). Boric acid (H3BO3), sodium tetraborate decahydrate (Na2B4O7·10H2O), sodium chloride (NaCl), and potassium chloride (KCl) were obtained from Beijing Chemical Works (Beijing, China). Influenza A H5N1 HA protein was purchased from Sino Biological Inc. (Beijing, China). Bovine serum albumin (BSA) was from Sigma (St. Louis, MO), human serum albumin (HSA) was purchased from Linc-bio Science Co., Ltd. (Shanghai, China), and immunoglobin G (IgG) was purchased from Yuduo Biotech Co., Ltd. (He-nan, China). All chemicals were of analytical reagent grade and obtained from a commercial source unless mentioned otherwise. Healthy human serum samples were provided by the Chinese Academy of Inspection and Quarantine. The composition of the buffer used for the experiment was reported as BBS (100 mmol L–1 H3BO3, 20 mmol L–1 Na2B4O7·10H2O, 5 mM KCl, pH 7.8). NaCl, KCl, MgCl2, and other common metal salts used were obtained from Beijing Chemical Works (Beijing, China). All chemicals were commercial products and used as received without further purification. Milli-Q water (Z18.2 MΩ) was used to prepare all stock solutions and buffer solutions. Amine-modified H5N1 HA aptamer39 (5′-NH2- TTGGGGTTATTTGGGAGGGCGGGGGTT-3′) was supplied by Sangon Biotechnology Co., Ltd. (Shanghai, China).
4.2. Synthesis of Oleic Acid-Stabilized SWUCNPs
Oleic acid (OA)-stabilized NaGdF4@NaYF4:Ln@NaYF4 SWUCNPs were synthesized according to the seed-mediated method developed previously.40
In a typical procedure, 1 mmol CF3COONa and 1 mmol (CF3COO)3Gd were added to a 100 mL three-necked flask containing 10 mmol OA, 10 mmol OM, and 20 mmol ODE. The slurry was heated to 110 °C to remove residual water and oxygen. Then, the mixture was heated to 320 °C and kept for 15 min in a N2 atmosphere. After that, an excess amount of ethanol was added to precipitate the nanoparticles. Nanoparticles were dispersed in 10 mL of cyclohexane. Five millilters of the as-prepared nanoparticle colloidal solution (nominal 0.5 mmol) was added to precursors (0.5 mmol CF3COONa, 0.5 mmol Gd(CF3COO)3, and solvents (20 mmol OA and 20 mmol ODE)). Reaction conditions and after-treatments are the same as aforementioned. The resulting NaGdF4 nanoparticles were stocked in 10 mL of cyclohexane.
Five millilters of the hexagonal NaGdF4 solution, used as seeds for epitaxial growth, was added to the precursors of the shell (1 mmol CF3COONa and 0.78 mmol Y(CF3COO)3, 0.2 mmol (CF3COO)3Yb, and 0.02 mmol (CF3COO)3Er) and solvents (20 mmol OA and 20 mmol ODE). Reaction conditions and after-treatments were the same as those in the first step to get NaGdF4@NaYF4:Yb,Er nanoparticles.
Ten millilters of the as-prepared NaGdF4@NaYF4:Yb,Er colloidal solution (nominal 0.25 mmol) was added to a 100 mL three-necked flask containing precursors (1 mmol CF3COONa, 1 mmol Y(CF3COO)3) and solvents (20 mmol OA and 20 mmol ODE). Reaction conditions and after-treatments were the same as aforementioned. The final products were dispersed in 5 mL of cyclohexane for further characterizations.
4.3. Preparation of Water-Dispersible Nanoparticles
Typically, 1 mL of colloidal solutions of oleate-capped nanoparticles was dispersed in a mixture of cyclohexane and N,N-dimethylformamide (DMF) (10 mL, volume ratio 1:1). Then, 30 mg of NOBF4 was added to the system with vigorous stirring for 30 min.41 The oleate ligands left in the cyclohexane phase, while the oleate-free nanoparticles transferred to the DMF phase. Subsequently, the nanoparticles were collected by centrifugation (12 000 rpm, 15 min) after adding an excess amount of toluene. The nanoparticles were redispersed in 10 mL of DMF containing 20 mg of polyacrylic acid (PAA, Mw = 1800, 50% saponification). The mixture was stirred overnight to render PAA modification. Finally, the excess amount of acetone was added to the solution to precipitate PAA-modified nanoparticles. The hydrophilic nanoparticles were collected by centrifugation (12 000 rpm, 15 min) and dispersed in water for further experiments.
4.4. Surface Modification and Bioconjugation of PAA-UCNPs with Aptamers
The amine-modified H5N1 HA aptamer was covalently conjugated to PAA-SWUCPs following the standard EDC–NHS conjugation protocol. Briefly, 2 mg of PAA-UCNPs was added to 2 mL of MES buffer solution (10 mM, pH 5.6), and the aqueous solution was sonicated for 10 min. Then, 0.4 mg of EDC and 0. 6 mg of sulfo-NHS were added to the solution to activate the carboxyl groups on PAA. The mixture was incubated at room temperature with gentle shaking for 15 min.
After centrifugation, the activated PAA-SWUCNPs were washed with ultrapure water three times and then dispersed in 2 mL of borate buffer (10 mM, pH 7.8, 5 mM KCl) containing 2 nmol amino-modified HA aptamer. The reaction lasted overnight at room temperature with slow shaking. To block the excess NHS, 5 mg of Tris was added to the reaction mixture. The SWUCNP-aptamer was obtained by centrifugation at 8000 rpm, washed with ultrapure water three times, redispersed in 2 mL of borate buffer solution (10 mM, pH 7.8, 5 mM KCl), and preserved at 4 °C to keep its activity.
4.5. HA Detection in Aqueous Solution and Human Serum
For the sake of determining a suitable concentration of GO to proceed the subsequent fluorescence recovery experiments, different amounts of GO were separately incubated with a fixed amount of UCNP-aptamer (0.05 mg mL–1) in borate buffer (10 mM, pH 7.8, 5 mM KCl) for 25 min at room temperature. Afterward, upconversion fluorescence measurements were carried out. In a typical FRET assay process, various concentrations of HA were first mixed with the SWUCNP-aptamer (0.05 mg mL–1) conjugates in 10 mM borate buffer (pH 7.8, 5 mM KCl) and the mixture was incubated at room temperature for 30 min. Afterward, graphene oxide was added into the above mixtures with an ultimate concentration of 0.01 mg mL–1 followed by incubation for 25 min. Finally, the upconversion luminescence spectra of the final mixture were then measured using an FLS fluorescence spectrometer with an external 980 nm laser. To verify the specificity of the above FRET aptasensor toward HA, a series of interfering biomolecules including HSA, thrombin, BSA, and IgG were added to the SWUCNP-aptamer/GO system in place of HA following the same experimental procedures. For the detection in serum samples, newly obtained serum 20 times diluted with borate buffer was used as the assay medium and then the same assay procedure as that in the aqueous solution was followed. Furthermore, the standard addition method was adopted to determine the concentration of HA in practical serum samples.
4.6. Instruments and Characterizations
The morphologies and sizes of UCNPs and GO were characterized by transmission electron microscopy (TEM) (JEM-2010, Japan). Ambient X-ray diffraction (XRD) patterns were recorded on a PANalytical X’Pert3 powder diffractometer using Cu Kα radiation (λ = 1.5406 Å). The UV–vis absorption measurements were carried out using a UV–vis spectrophotometer (UV3600Plus, Shimadzu Scientific Instruments Inc.). A Fourier transform infrared (FT-IR) absorption spectrometer (Tensor 27, Bruker) was used to characterize the PAA-SWUCNPs. The ζ-potential measurements were performed using a Zetasizer Nano ZS instrument (Malvern Instruments, U.K.). The upconversion fluorescence spectra were measured at 541 nm using an FLS980 steady-state and time-resolved fluorescence spectrometer (Edinburgh Instruments) with an external 980 nm laser source.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFA0205102) and the Seed Foundation of Tianjin University (Grant No. 2020XY-0078).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c01491.
Feasibility of the FRET system, TEM image of UCNP-Apt on the GO sheet in the presence of HA, fluorescence spectrum in human serum samples, relationship between commercial standard ELISA kits and this method, between-group repeated measures HA, stability of probes, and comparison of this work with other methods (PDF)
Author Contributions
The manuscript was written through the contribution of all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Anderson C. E.; Holstein C. A.; Strauch E.-M.; Bennett S.; Chevalier A.; Nelson J.; Fu E.; Baker D.; Yager P. Rapid Diagnostic Assay for Intact Influenza Virus Using a High Affinity Hemagglutinin Binding Protein. Anal. Chem. 2017, 89, 6608–6615. 10.1021/acs.analchem.7b00769. [DOI] [PubMed] [Google Scholar]
- Zavyalova E.; Kopylov A. Aptamers to Hemagglutinin: A Novel Tool for Influenza Virus Recognition and Neutralization. Curr. Pharm. Des. 2016, 22, 4835–4853. 10.2174/1381612822666160203142513. [DOI] [PubMed] [Google Scholar]
- Hideshima S.; Hinou H.; Ebihara D.; Sato R.; Kuroiwa S.; Nakanishi T.; Nishimura S.-I.; Osaka T. Attomolar Detection of Influenza A Virus Hemagglutinin Human H1 and Avian H5 Using Glycan-Blotted Field Effect Transistor Biosensor. Anal. Chem. 2013, 85, 5641–5644. 10.1021/ac401085c. [DOI] [PubMed] [Google Scholar]
- Chávez Ramos K.; Nishiyama K.; Maeki M.; Ishida A.; Tani H.; Kasama T.; Baba Y.; Tokeshi M. Rapid, Sensitive, and Selective Detection of H5 Hemagglutinin from Avian Influenza Virus Using an Immunowall Device. ACS Omega 2019, 4, 16683–16688. 10.1021/acsomega.9b02788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO. https://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/ (accessed Dec 9, 2020).
- Gopinath S. C. B.; Tang T.-H.; Chen Y.; Citartan M.; Tominaga J.; Lakshmipriya T. Sensing strategies for influenza surveillance. Biosens. Bioelectron. 2014, 61, 357–369. 10.1016/j.bios.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Eisfeld A. J.; Neumann G.; Kawaoka Y. Influenza A virus isolation, culture and identification. Nat. Protocols 2014, 9, 2663–2681. 10.1038/nprot.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y.; Cheng Q. J. A. Detection of influenza virus: traditional approaches and development of biosensors. Anal. Bioanal. Chem. 2005, 381, 156–164. 10.1007/s00216-004-2927-0. [DOI] [PubMed] [Google Scholar]
- Miarka M.; Horban A.; Maliszewska H.; Bilinski P.; Prus-Kowalczuk W. A clinical utility of a strip test for influenza A/B and comparison with detection by RT PCR. Acta Biochim. Pol. 2014, 61, 485–487. 10.18388/abp.2014_1868. [DOI] [PubMed] [Google Scholar]
- Kikuchi K.; Takakusa H.; Nagano T. Recent advances in the design of small molecule-based FRET sensors for cell biology. TrAC, Trends Anal. Chem. 2004, 23, 407–415. 10.1016/S0165-9936(04)00608-9. [DOI] [Google Scholar]
- Wang Y.; Si B.; Lu S.; Liu E.; Hu X.; Fan J. Near-infrared excitation of CdTe quantum dots based on fluorescence resonance energy transfer and their use as fluorescent sensors. Sens. Actuators, B 2017, 246, 127–135. 10.1016/j.snb.2017.02.069. [DOI] [Google Scholar]
- Dong H.; Sun L.-D.; Yan C.-H. Upconversion emission studies of single particles. Nano Today 2020, 35, 100956 10.1016/j.nantod.2020.100956. [DOI] [Google Scholar]
- Xu M.; Zhuang J.; Jiang X.; Liu X.; Tang D. A three-dimensional DNA walker amplified FRET sensor for detection of telomerase activity based on the MnO2 nanosheet-upconversion nanoparticle sensing platform. Chem. Commun. 2019, 55, 9857–9860. 10.1039/C9CC05387E. [DOI] [PubMed] [Google Scholar]
- Luo Z.; Qi Q.; Zhang L.; Zeng R.; Su L.; Tang D. Branched Polyethylenimine-Modified Upconversion Nanohybrid-Mediated Photoelectrochemical Immunoassay with Synergistic Effect of Dual-Purpose Copper Ions. Anal. Chem. 2019, 91, 4149–4156. 10.1021/acs.analchem.8b05959. [DOI] [PubMed] [Google Scholar]
- Qiu Z.; Shu J.; Tang D. NaYF4:Yb,Er Upconversion Nanotransducer with in Situ Fabrication of Ag2S for Near-Infrared Light Responsive Photoelectrochemical Biosensor. Anal. Chem. 2018, 90, 12214–12220. 10.1021/acs.analchem.8b03446. [DOI] [PubMed] [Google Scholar]
- Luo Z.; Zhang L.; Zeng R.; Su L.; Tang D. Near-Infrared Light-Excited Core-Core-Shell UCNP@Au@CdS Upconversion Nanospheres for Ultrasensitive Photoelectrochemical Enzyme Immunoassay. Anal. Chem. 2018, 90, 9568–9575. 10.1021/acs.analchem.8b02421. [DOI] [PubMed] [Google Scholar]
- Qiu Z.; Shu J.; Tang D. Near-Infrared-to-Ultraviolet Light-Mediated Photoelectrochemical Aptasensing Platform for Cancer Biomarker Based on Core Shell NaYF4:Yb,Tm@TiO2 Upconversion Microrods. Anal. Chem. 2018, 90, 1021–1028. 10.1021/acs.analchem.7b04479. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Shikha S.; Liu J.; Zhang J.; Mei Q.; Zhang Y. Upconversion Nanoprobes: Recent Advances in Sensing Applications. Anal. Chem. 2019, 91, 548–568. 10.1021/acs.analchem.8b04049. [DOI] [PubMed] [Google Scholar]
- Marin R.; Labrador-Páez L.; Skripka A.; Haro-González P.; Benayas A.; Canton P.; Jaque D.; Vetrone F. J. A. P. Upconverting Nanoparticle to Quantum Dot Förster Resonance Energy Transfer: Increasing the Efficiency through Donor Design. ACS Photonics 2018, 5, 2261–2270. 10.1021/acsphotonics.8b00112. [DOI] [Google Scholar]
- Wang F.; Han Y.; Lim C. S.; Lu Y.; Wang J.; Xu J.; Chen H.; Zhang C.; Hong M.; Liu X. Simultaneous phase and size control of upconversion nanocrystals through lanthanide doping. Nature 2010, 463, 1061–1065. 10.1038/nature08777. [DOI] [PubMed] [Google Scholar]
- Wang F.; Wang J.; Liu X. Direct Evidence of a Surface Quenching Effect on Size-Dependent Luminescence of Upconversion Nanoparticles. Angew. Chem., Int. Ed. 2010, 49, 7456–7460. 10.1002/anie.201003959. [DOI] [PubMed] [Google Scholar]
- Siefe C.; Mehlenbacher R. D.; Peng C. S.; Zhang Y.; Fischer S.; Lay A.; McLellan C. A.; Alivisatos A. P.; Chu S.; Dionne J. A. Sub-20 nm Core-Shell-Shell Nanoparticles for Bright Upconversion and Enhanced Forster Resonant Energy Transfer. J. Am. Chem. Soc. 2019, 141, 16997–17005. 10.1021/jacs.9b09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Lv S.; Wang Y.; Chen S.; Liu Z. Construction of LRET-Based Nanoprobe Using Upconversion Nanoparticles with Confined Emitters and Bared Surface as Luminophore. J. Am. Chem. Soc. 2015, 137, 3421–3427. 10.1021/jacs.5b01504. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Li Z.; Hu D.; Lin C.-T.; Li J.; Lin Y. Aptamer/Graphene Oxide Nanocomplex for in Situ Molecular Probing in Living Cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- Zeng R.; Luo Z.; Zhang L.; Tang D. Platinum Nanozyme-Catalyzed Gas Generation for Pressure-Based Bioassay Using Polyaniline Nanowires-Functionalized Graphene Oxide Framework. Anal. Chem. 2018, 90, 12299–12306. 10.1021/acs.analchem.8b03889. [DOI] [PubMed] [Google Scholar]
- Shu J.; Qiu Z.; Tang D. Self-Referenced Smartphone Imaging for Visual Screening of H2S Using CuxO-Polypyrrole Conductive Aerogel Doped with Graphene Oxide Framework. Anal. Chem. 2018, 90, 9691–9694. 10.1021/acs.analchem.8b03011. [DOI] [PubMed] [Google Scholar]
- Giust D.; Lucio M. I.; El-Sagheer A. H.; Brown T.; Williams L. E.; Muskens O. L.; Kanaras A. G. Graphene Oxide-Upconversion Nanoparticle Based Portable Sensors for Assessing Nutritional Deficiencies in Crops. ACS Nano 2018, 12, 6273–6279. 10.1021/acsnano.8b03261. [DOI] [PubMed] [Google Scholar]
- Niu J.; Hu X.; Ouyang W.; Chen Y.; Liu S.; Han J.; Liu L. Femtomolar Detection of Lipopolysaccharide in Injectables and Serum Samples Using Aptamer-Coupled Reduced Graphene Oxide in a Continuous Injection-Electrostacking Biochip. Anal. Chem. 2019, 91, 2360–2367. 10.1021/acs.analchem.8b05106. [DOI] [PubMed] [Google Scholar]
- Darmostuk M.; Rimpelova S.; Gbelcova H.; Ruml T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Fang X.; Tan W. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc. Chem. Res. 2010, 43, 48–57. 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Shikha S.; Liu J.; Zhang J.; Mei Q.; Zhang Y. J. A. c. Upconversion Nanoprobes: Recent Advances in Sensing Applications. Anal. Chem. 2019, 91, 548–568. 10.1021/acs.analchem.8b04049. [DOI] [PubMed] [Google Scholar]
- Tang D.; Tang J.; Li Q.; Su B.; Chen G. Ultrasensitive Aptamer-Based Multiplexed Electrochemical Detection by Coupling Distinguishable Signal Tags with Catalytic Recycling of DNase I. Anal. Chem. 2011, 83, 7255–7259. 10.1021/ac201891w. [DOI] [PubMed] [Google Scholar]
- Liu B.; Cui Y.; Tang D.; Yang H.; Chen G. Au(III)-assisted core-shell iron oxide@poly(o-phenylenediamine) nanostructures for ultrasensitive electrochemical aptasensors based on DNase I-catalyzed target recycling. Chem. Commun. 2012, 48, 2624–2626. 10.1039/c2cc17790k. [DOI] [PubMed] [Google Scholar]
- Sun A.-L.; Zhang Y.-F.; Sun G.-P.; Wang X.-N.; Tang D. Homogeneous electrochemical detection of ochratoxin A in foodstuff using aptamer-graphene oxide nanosheets and DNase I-based target recycling reaction. Biosens. Bioelectron. 2017, 89, 659–665. 10.1016/j.bios.2015.12.032. [DOI] [PubMed] [Google Scholar]
- Shiratori I.; Akitomi J.; Boltz D. A.; Horii K.; Furuichi M.; Waga I. Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. 10.1016/j.bbrc.2013.11.041. [DOI] [PubMed] [Google Scholar]
- Pang Y.; Rong Z.; Wang J.; Xiao R.; Wang S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF). Biosens. Bioelectron. 2015, 66, 527–532. 10.1016/j.bios.2014.10.052. [DOI] [PubMed] [Google Scholar]
- Diba F. S.; Kim S.; Lee H. J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015, 72, 355–361. 10.1016/j.bios.2015.05.020. [DOI] [PubMed] [Google Scholar]
- Hermann T.; Patel D. J. S. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Shiratori I.; Akitomi J.; Boltz D.; Horii K.; Furuichi M.; Waga I. J. B. communications, b. r., Selection of DNA aptamers that bind to influenza A viruses with high affinity and broad subtype specificity. Biochem. Biophys. Res. Commun. 2014, 443, 37–41. 10.1016/j.bbrc.2013.11.041. [DOI] [PubMed] [Google Scholar]
- Dong H.; Sun L. D.; Feng W.; Gu Y.; Li F.; Yan C. H. J. A. N. Versatile Spectral and Lifetime Multiplexing Nanoplatform with Excitation Orthogonalized Upconversion Luminescence. ACS Nano 2017, 11, 3289–3297. 10.1021/acsnano.7b00559. [DOI] [PubMed] [Google Scholar]
- Dong A.; Ye X.; Chen J.; Kang Y.; Gordon T.; Kikkawa J. M.; Murray C. J. J. o. t. A. C. S., A generalized ligand-exchange strategy enabling sequential surface functionalization of colloidal nanocrystals. J. Am. Chem. Soc. 2011, 133, 998–1006. 10.1021/ja108948z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.