This review summarizes and integrates current views on the contribution of inflammatory signals derived from multiple sources to hematopoietic stem cell function and its long-term implications on mutational selection and clonal expansion during aging.
Abstract
Inflammation is an evolutionarily selected defense response to infection or tissue damage that involves activation and consumption of immune cells in order to reestablish and maintain organismal integrity. In this process, hematopoietic stem cells (HSCs) are themselves exposed to inflammatory cues and via proliferation and differentiation, replace mature immune cells in a demand-adapted fashion. Here, we review how major sources of systemic inflammation act on and subsequently shape HSC fate and function. We highlight how lifelong inflammatory exposure contributes to HSC inflamm-aging and selection of premalignant HSC clones. Finally, we explore emerging areas of interest and open questions remaining in the field.
Introduction
The hematopoietic system is responsible for lifelong blood cell production and downstream primary functions, including oxygen transportation (erythrocytes), hemostasis (platelets), and host defense (leukocytes: myeloid and lymphoid cells). Recent estimations suggest that adult bone marrow (BM) produces 2–6 × 1010 cells/kg/d in mice and 5–7 × 1010 cells/kg/d in humans under homeostatic conditions (steady-state hematopoiesis), which can increase several fold upon stress demands, e.g., infection (demand-adapted or “emergency” hematopoiesis; Nombela-Arrieta and Manz, 2017; Sender and Milo, 2021). To replenish high cellular consumption while preserving its function and preventing potentially debilitating conditions derived from hematopoietic failure or excess (e.g., aplasia or leukemia, respectively), the hematopoietic system is tightly regulated and characterized by a hierarchical cellular differentiation system sustained by rare stem cell populations kept in protective BM niches coupled to sensing mechanisms that connect the stress-exposed periphery to the BM tissue.
Hematopoietic stem cells (HSCs; also termed long-term [LT] HSCs) reside at the top of the hematopoietic hierarchy and have two unique properties: self-renewal (ability to generate a daughter cell that retains HSC activity) and multilineage differentiation capacity (ability to produce all blood cell types). HSCs differentiate into multipotent progenitors (MPPs), which together are defined as hematopoietic stem and progenitor cells (HSPCs) and in mice share Lin−Sca-1+ and c-Kit+ marker expression. MPPs subsequently differentiate to increasingly lineage-restricted progenitor cells that undergo terminal differentiation into mature blood cells. MPPs and their progeny are termed hematopoietic progenitor cells (HPCs; Wilson et al., 2008; Manz and Boettcher, 2014). Under steady-state conditions, most adult HSCs are quiescent, residing in the G0 phase of the cell cycle. Highly quiescent HSC fractions have an estimated turnover rate of months (in mice) to years (in humans; Takizawa et al., 2011; Trumpp et al., 2010). Thus, daily hematopoietic production is primarily sustained by highly proliferative HPCs (Sun et al., 2014; Busch et al., 2015). However, HSCs can sense environmental cues and depending on the type, duration, and concentration of the signal(s), initiate demand-adapted hematopoiesis by exiting quiescence, proliferating and differentiating, and/or self-renewing.
Inflammation, generally defined as a protective immune response to infection and tissue damage, is mediated by proinflammatory cytokines and chemokines that are produced by stressed or damaged cells and sensed by effector cells that orchestrate a systemic and/or local response (Medzhitov, 2008). It has now become well recognized that HSCs are key players in systemic inflammatory responses capable of integrating external inflammatory cues into cellular responses and establishing a demand-adapted axis between peripheral stresses and hematopoietic responses in the BM (Takizawa et al., 2012). In recent years, a wealth of new investigations has probed more deeply into how HSC function is impacted by inflammation and how such responses may contribute to HSC aging and oncogenesis (Fig. 1). Here, we summarize current knowledge in the field with an emphasis on highlighting new themes and ambiguities emerging in the current literature.
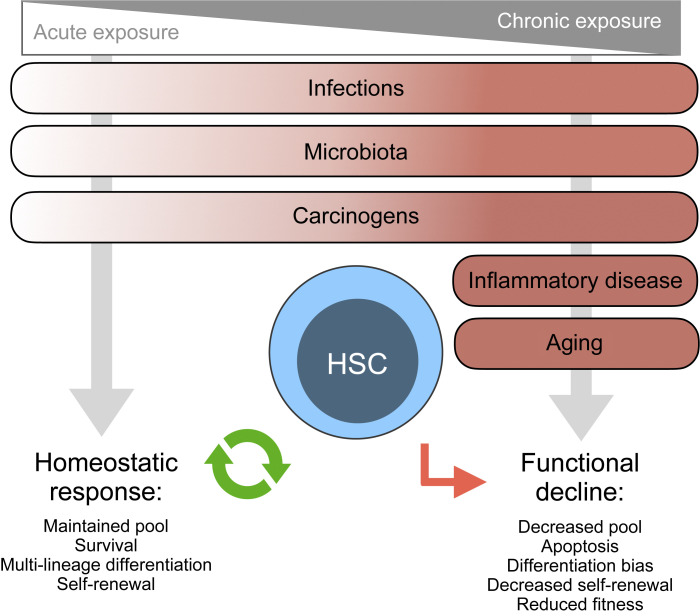
Figure 1.
Causes and consequences of inflammation on HSC functionality. Schematic representation of current understanding in the field on the major causes of systemic inflammation known to impact HSC biology in mouse models (infection, microbiota, carcinogens, inflammatory diseases, aging). Each cause in distributed in a gradient regarding the duration of inflammation: acute to chronic. HSC properties during homeostatic responses (more frequently associated with acute inflammation) or functional decline (more frequently associated with chronic inflammation) are depicted.
Main causes of inflammation and their impact on HSC biology
Infectious diseases
King and Goodell (2011) previously defined four different mechanisms by which infections can influence HSC biology. The first two mechanisms act via direct effects on HSCs: (1) direct infection or (2) direct recognition of a pathogen. The other two mechanisms are indirect: (3) via proinflammatory cytokines released by other cells or (4) through changes in the cellular BM microenvironment. Here, we focus on and dissect mechanism 3, particularly in the context of in vivo mouse infection models. Table 1 summarizes the major sources of infection-derived inflammation (viral, bacterial, protozoan, and fungal infections), their associated inflammatory pathways, and consequent biological impacts on HSCs. Despite the significant variability among the different infection models, there are some conserved features: (a) Infection-derived inflammation leads frequently to HSC functional decline, including reduced repopulating capacity and increased proliferation associated with a myeloid lineage bias; (b) HSC functional decline is most commonly associated with chronic infection, which suggests that prolonged inflammation and the resultant demand may exert cumulative effects on HSC function; (c) inflammatory cytokines can act both directly on HSCs and indirectly, leading to secondary inflammatory signal production by BM niche cells, such as mesenchymal stromal cells (MSCs), endothelial cells, and mature hematopoietic cells. Overall, the studies presented in Table 1 indicate that infection-derived inflammation is a significant source of BM inflammation with the potential to establish long-term functional impairment on HSCs (Binder et al., 1997; de Bruin et al., 2013; Matatall et al., 2014; Schürch et al., 2014; Isringhausen et al., 2020 Preprint; Hirche et al., 2017; Scumpia et al., 2010; Burberry et al., 2014; Shi et al., 2018; Baldridge et al., 2010; Matatall et al., 2016; Florez et al., 2020; Choi et al., 2011; Khan et al., 2020; MacNamara et al., 2011; Smith et al., 2018; Vainieri et al., 2016; Haltalli et al., 2020; Martínez et al., 2018; Mitroulis et al., 2018).
Table 1. Summary of mouse studies describing the contribution of different infection-derived inflammatory sources to HSC biology.
| Source | Inflammatory pathway | Molecular mechanism | Effect on HSCs | Reference |
|---|---|---|---|---|
| Viral | ||||
| Lymphocytic choriomeningitis virus | IFN-α/β–IFNAR | ND | 30-fold reduction on CFU (at infection day 3) | Binder et al. (1997) |
| IFN-γ–IFNGR | IFN-γ–induced SOCS1 inhibits TPO-induced STAT5 phosphorylation and leads to decreased/increased cyclin D1/Cdkn1C | Decreased pool recovery and self-renewal capacity (at infection days 4–12) | de Bruin et al. (2013) | |
| IFN-γ–IFNGR | IFN-γ–induced Cebpb in high IFNGR–expressing myeloid-biased HSCs | Increased proliferation and myeloid bias associated to decreased self-renewal (at infection day 6) | Matatall et al. (2014) | |
| IFN-γ–IFNGR–IL-6 | Cytotoxic CD8+ T cell–produced IFN-γ activates IL-6 production by BM MSCs, leading to reduced Runx-1 and Cebpa expression | Increased myeloid-differentiation bias on HSPCs | Schürch et al. (2014) | |
| IFN-α/β–IFNAR and IFN-γ–IFNGR | Persistent destruction of CARc networks by virus-specific IFN-producing CD8+ T cells—loss of HSC quiescence–enforcing niche | Increased proliferation, decreased pool size, and decreased repopulating capacity | Isringhausen et al., 2020 Preprint | |
| Murine cytomegalovirus | Increased IFN-γ, IL-17, and CCL12 levels | Inflammatory milieu associates in an IFNAR-independent manner with increased Sca-1 and Evi1 gene expression in HSCs | Decreased repopulating capacity and increased myeloid differentiation, after BM viral clearance (at infection day 21) | Hirche et al. (2017) |
| Bacterial | ||||
| Polymicrobial sepsis using cecal ligation and puncture model | ND | HSC expansion associates with reduced BM cellularity but is independent from MyD88, TRIFF, IFNAR, TNF-α, IL-1, IL-6, prostaglandins, oxidative stress, and super antigen signaling | Increased pool size | Scumpia et al. (2010) |
| Escherichia coli | G-CSF/CXCL12 | Dual stimulation of NOD1 and TLR4 leads to increased G-CSF and decreased CXCL12 production in radioresistant endothelial cells | Increased mobilization of BM-expanded HSCs, reduced repopulating capacity, and increased myeloid-differentiation | Burberry et al. (2014) |
| SHH-GLI | ERK1/2–SP1–mediated increased SHH expression in Lin+ BM cells leads to higher GLI levels in HSCs | Proliferation and myeloid differentiation | Shi et al. (2018) | |
| IFN-γ–IFNGR | ND | Increased proliferation and decreased repopulating capacity (4 wk after single bacterial inoculation) | Baldridge et al. (2010) | |
| M. avium | IFN-γ–IFNGR | IFN-γ–induced Batf2 leading to HSPC terminal differentiation | Decreased pool size, repopulating capacity, and increased myeloid differentiation (after six monthly inoculations) | Matatall et al. (2016) |
| IFN-γ–IFNGR | IFN-γ–induced expression of BST2 (noncanonical E-selectin ligand) displaces HSCs from quiescence-enforcing CARc niche to an activating E-selectin–positive vascular niche | Increased proliferation, terminal differentiation, and decreased pool size (after four monthly inoculations) | Florez et al. (2020) | |
| Mycobacterium tuberculosis | TNF-α/IL-6 | TLR2 and MyD88 bacterial sensing dependent | Increased HSPC pool and myeloid differentiation | Choi et al. (2011) |
| IFN-α/β–IFNAR | IFNAR-dependent reprogramming of HSCs leading to dysregulated iron metabolism, depolarized mitochondrial membrane, and necrosis in myeloid progenitors | Decreased pool and reconstitution capacity (up to 1 yr after infection) | Khan et al. (2020) | |
| Ehrlichia muris | IFN-γ–IFNGR | ND | Decreased pool and reconstitution capacity, and increased myeloid differentiation bias (at infection day 8) | MacNamara et al. (2011) |
| IFN-α/β–IFNAR | Direct sensitization of HSPCs to RIPK1-dependent death and increased HSC proliferative arrest | Decreased pool and reconstitution capacity (at infection day 7) | Smith et al. (2018) | |
| Protozoan | ||||
| Plasmodium berghei | ND | ND | Increased proliferation and pool size (at infection days 7–10) | Vainieri et al. (2016) |
| IFN-γ–IFNGR | Loss of BM niche osteoblasts and endothelial cell properties | Increased turnover leading to decreased functionality and transcriptional identity | Haltalli et al. (2020) | |
| Fungal | ||||
| Candida albicans | ND (TNF-α?) | TLR2/MyD88 and dectin-1 fungal sensing dependent | Increased proliferation and pool size | Martínez et al. (2018) |
| IL-1β and GM-CSF/CD131 | β-glucan–mediated IL-1β and GM-CSF production by BM cells leads to increased glycolytic pathways and proliferation in HSCs | Increased proliferation, pool size, and myeloid differentiation bias (at 7 d after exposure) | Mitroulis et al. (2018) |
Microbiota
The intestinal bacterial microbiome is emerging as a key regulator of hematopoiesis (Tada et al., 1996; Deshmukh et al., 2014; Balmer et al., 2014; Khosravi et al., 2014; Lee et al., 2011; Staffas et al., 2018). Concerning the effects of microbiota on HSPC pool size, Balmer et al. (2014) showed that germ-free (GF) mice (mice lacking microbiota) or antibiotic-treated specific pathogen–free (SPF) mice (carrying a full microbiota of >100 bacterial species, but free of known mouse pathogens) have lower steady-state HSPC numbers compared with untreated SPF mice. Interestingly, colonization of GF mice with a defined, low-complexity microbiota led to an intermediate expansion of the HSPC pool to levels still below that of SPF mice. This suggests that HSPC pool size is governed by the strength of microbiota-associated signals and/or the presence of a complex microbial milieu. Later, Josefsdottir et al. (2017) further demonstrated that antibiotic-treated mice show microbiota depletion (but unaltered serum levels of IFNs, IL-6, TNF, G-CSF) and a decrease in HSPC pool size to levels similar to those of Stat1−/− mice. These data suggest that cytokine-dependent STAT1 signaling, stimulated by the microbiome, is required for maintenance of normal BM HSPC numbers. In agreement, Iwamura et al. (2017) showed that NOD1 ligand administration to GF mice activates BM MSCs to produce key hematopoietic factors, including IL-7, Flt3L, stem cell factor, thrombopoietin (TPO), IL-6, and TNF, which restored HSPC pool size to levels equivalent to SPF mice. Furthermore, Lee et al. (2019) revealed that microbiome-derived molecules, including bacterial DNA, reach the BM via systemic blood circulation and are sensed by CX3CR1+ mononuclear cells via a TLR-dependent mechanism, resulting in the production of inflammatory cytokines (TNF-α, IL-1, IL-6) that regulate the HSPC pool size and their differentiation potential toward myeloid lineages. Along these lines, unpublished data from our laboratory suggest that upon aging, the levels of continuously circulating microbiota-derived molecules (e.g., TLR4 and 8 agonists) increase. High abundance of these molecules leads to enhanced myeloid-derived systemic IL-1 production, causing HSC pool expansion and increased myeloid differentiation bias. Overall, these studies suggest that tonic, microbiota-associated inflammatory signaling is essential to maintain HSPC pool size and myelopoiesis in steady-state conditions but can potentially become detrimental in the context of an aged hematopoietic system. Furthermore, whether signaling via multiple pathways (e.g., STAT1, NOD1, TLRs) are all required in concert to effectively govern HSPC pool size and myelopoietic activity or whether requirements for these signaling pathways may reflect differences in microbial communities established in different SPF mouse colonies remains to be rigorously addressed.
Carcinogens
Carcinogens include any substance, radionuclide, or radiation that promotes cancer formation. Long-term BM injury mediated by carcinogens, like ionizing radiation (IR) and chemotherapy (CT), is attributed to direct effects on HSCs as well as on the BM environment. These effects include induction of apoptosis, differentiation, senescence, and/or damage to BM HSC niche (reviewed in Shao et al., 2013). Additionally, IR and CT (doxorubicin, 5-fluorouracil [5-FU], cisplatin, paclitaxel) are known to induce a rapid and dynamic production of multiple proinflammatory cytokines and chemokines, including IL-1α/β, IL-6, IL-8, IFN-α/β, TNF-α, CXCL9, CCL2, CCL3, and GM-CSF, within the first 1–2 wk following exposure (Citrin and Mitchell, 2017; McKelvey et al., 2018; Zhang et al., 2012; Vyas et al., 2014; Pietras et al., 2016). However, the effects of IR- and CT-mediated inflammation are difficult to dissociate from the direct effects of IR and CT on the metabolic and genomic integrity of HSCs themselves (Mohrin et al., 2010). Analysis is further confounded by the complexity of the systemic “storm” of cytokines and damage-associated molecular patterns produced in response to these insults in addition to other effects, including loss of gut integrity, which can release inflammatory microbial products into the bloodstream. These confounders have been partially addressed by transplanting cytokine or chemokine receptor-null HSCs into IR- or CT-treated animals and following the kinetics by which they regenerate the blood system. Interestingly, most primary receptor-null mice do not have a clear HSC phenotype, suggesting a redundant role of cytokines in HSC regulation in steady state. On the other hand, HSCs from these mice often show improved long-term repopulating capacity compared with their WT counterparts upon transplantation into irradiated recipients, including Tnfrsf1a−/− and Tnfrsf1b−/− (Pronk et al., 2011), Ifnar−/− (Essers et al., 2009), and Ifngr−/− (Matatall et al., 2014). This suggests that IR- and CT-mediated inflammation restricts HSC self-renewal and repopulating capacity, particularly in the context of serial transplantation. Besides IR and CT, cigarette smoke is one of the most widespread carcinogens (containing at least 98 hazardous components; Talhout et al., 2011), yet its impact on hematopoiesis may be overlooked. Mice exposed to cigarette smoke exhibited increased extramedullary hematopoiesis, decreased MSCs and HSCs, and increased expression of pro-proliferation genes that lead to the expansion of HSPCs (Khaldoyanidi et al., 2001; Pandit et al., 2006; Siggins et al., 2014). Cigarette smoke extract also inhibits osteogenic differentiation and increases expression of proinflammatory markers (Cyprus et al., 2018). Furthermore, in the context of human hematopoiesis, individual smoking status associates with ASXL1 loss-of-function–driven clonal hematopoiesis (Dawoud et al., 2020). While cigarette smoke elicits a similar hematopoietic phenotype as many inflammatory challenges, the contribution of inflammation and identity of elicited inflammatory factors, such as cytokines in this context, remains elusive. Altogether, similar to infection, carcinogen exposure can induce inflammatory stresses that negatively impact HSC function, particularly in contexts where IR or CT induces myeloablation and subsequent replicative “aging” of HSCs that degrades their function (Beerman et al., 2014; Bernitz et al., 2016). Nonetheless, it should be noted that highly quiescent and drug-resistant “reserve” HSC fractions have been identified (Zhao et al., 2019; Wilson et al., 2008) that retain HSC activity following exposure to CT challenges, such as 5-FU. Hence, not all HSCs respond uniformly to the inflammatory, cytotoxic, and replicative challenges associated with carcinogen exposure. This feature is likely a significant contributor to hematopoietic recovery following induction CT and/or HSC transplantation in the context of hematological malignancy.
Inflammatory diseases
Inflammatory diseases are a diverse group of conditions resulting from dysregulated immune responses that lead to a deleterious state of chronic inflammation and/or periodic inflammatory “flares” that potentially affect HSC function. Broadly speaking, BM failure is rare in such “sterile” inflammatory contexts, although cytopenias, anemia, overproduction of myeloid cells, immunosenescence, and other hematologic comorbidities have been documented in patients who suffer from rheumatoid arthritis (RA) and juvenile idiopathic arthritis, systemic lupus erythematosus (SLE), colitis, and various autoinflammatory conditions such as gout where IL-1 is the primary pathogenic factor (Colmegna and Weyand, 2011; Papadaki et al., 2002; Pascual et al., 2005; Chalayer et al., 2017; Gasche et al., 2004). Many of these diseases have been modeled in the mouse. Using an IL-23–driven chronic intestinal inflammation model, Griseri et al. (2012) showed an IFN-γ–dependent increase in HSC proliferation, leading to an eightfold expansion of the HSC pool. This effect was associated with GM-CSF–dependent myeloid differentiation bias and extramedullary hematopoiesis in colitic mice (Griseri et al., 2012). Chronic granulomatous disease (CGD) is a primary immunodeficiency characterized by dysfunctional microbicidal activity and chronic inflammation, often related to deficiency in the NOX genes. In mice with CGD, increased cycling of HSCs, expansion of HPCs, and impaired long-term engraftment capacity were associated with high concentrations of proinflammatory cytokines, including IL-1β. Interestingly, treatment of WT mice with IL-1β mimicked these effects, while inhibition of IL-1 signaling (using IL-1R1 antagonist Anakinra and anti-inflammatory steroid dexamethasone) in CGD mice reduced HPC numbers but had only minor effects on the repopulating ability of HSCs (Weisser et al., 2016). RA is a debilitating autoimmune disease characterized by chronic inflammation and progressive destruction of joint tissue. Using a collagen-induced arthritis mouse model, Hernandez et al. (2020) identified systemic inflammation (increased serum levels of TNF, IFN-γ, G-CSF) and myeloid overproduction associated with activation of a myeloid differentiation gene program in HSCs. Myeloid overproduction has been characterized in a genetic RA model as well (Oduro et al., 2012). In a similar vein, using an experimental spondyloarthritis mouse model that generates systemic and local inflammation, Regan-Komito et al. (2020) documented an IL-33–GM-CSF axis that promotes HSC myeloid differentiation bias and a GM-CSF–dependent accumulation of granulocyte/macrophage progenitors at extramedullary sites that leads to severe symptoms in spondyloarthritis mice. Likewise, using a genetic mouse model of SLE, Niu et al. (2013) showed an intrinsic and extrinsic (increased serum levels of IL-6, IL-10, TNF-α, and IFN-α/β) inflammatory-dependent expansion and mobilization of HSCs with myeloid lineage differentiation bias. While the cytokine milieus induced by these diseases overlap with those observed in the chronic infection models described above, disruption of HSC repopulating function is generally much less significant. Indeed, HSCs from collagen-induced arthritic mice activate a proliferation arrest gene program associated with maintenance of quiescence and reconstitution capacity, despite ongoing inflammation. Furthermore, treatment with the IL-1R1 antagonist Anakinra reverted myeloid overproduction and associated gene programs in these HSCs (Hernandez et al., 2020), suggesting that disease has an impact on the hematopoietic system, including HSCs themselves, and can be at least partially reversed following inflammatory challenge. Likewise, HSCs from the genetic SLE model retained robust self-renewal capacity and, in fact, exhibited a repopulating advantage over WT HSCs, which resulted from a single nucleotide polymorphism in the cdkn2c gene that causes reduced p18INK4c expression (Niu et al., 2013). Unpublished data from our laboratory using a pristane-induced model of chronic SLE confirm the absence of HSC pool attrition despite ongoing IFN signaling, and HSCs from these mice exhibit only a moderate defect in long-term repopulating activity. These data raise the interesting question as to why some chronic infections may impart a more severe impact on HSC pool size and function than the sterile inflammation models described above, despite inducing similar cytokine milieus. Possibilities include differences in cytokine levels, increased hematopoietic demands caused by active turnover of immune cells during infection, and/or direct effects of pathogens on HSCs (including direct infection or recognition). Overall, chronic inflammatory disease can significantly impact HSC function; key next steps are to assess the extent to which anti-inflammatory therapies, including cytokine blockade approaches currently in clinical use, reverse these phenotypes and to address the impact of recently characterized “immune training” on long-term HSC and blood system function in this context.
Aging
Aging is associated with chronic low-grade inflammation (i.e., a two- to fourfold increase in IL-1, IL-6, TNF, and C-reactive protein serum levels; Vasto et al., 2007), a state often referred to as inflamm-aging. This phenotype, observed in both mice and humans, is attributed to a cumulative lifetime exposure to infectious and noninfectious agents and/or senescence-associated cytokines that leads to a self-sustained, vicious proinflammatory cycle (reviewed in Baylis et al., 2013; Kovtonyuk et al., 2016b; Fulop et al., 2018). Aging also associates with functional changes in both mature and immature hematopoietic cells. Aged HSCs show increased pool size and quiescence, reduced self-renewal capacity, increased myeloid/megakaryocytic-differentiation bias, accumulated DNA damage and proliferative stress marks, increased resistance to apoptosis, epigenetic and transcriptional alterations, loss of autophagy capacity, decreased homing capacity, and increased expression of adhesion molecules (reviewed in Geiger et al., 2013; Kovtonyuk et al., 2016a). The contribution of inflamm-aging to HSC dysfunction has been addressed in multiple studies. Ergen et al. (2012) performed heterochronic BM transplantation and BM cytokine profiling to identify RANTES (Regulated And Normal T cell Expressed and Secreted)/Ccl5 as a driver of HSC myeloid differentiation bias during aging. Using RANTES overexpression and Rantes KO mice, the HSC myeloid bias phenotype was mechanistically linked to increased Gata2 (promyeloid transcription factor), decreased Gata3 and Ikaros (lymphoid-associated transcription factors) expression, and increased mTOR activity (previously implicated in HSC aging; Saxton and Sabatini, 2017). Frisch et al. (2019) showed that impaired neutrophil efferocytosis (specialized phagocytosis that minimizes the inflammatory consequences of apoptosis; Arandjelovic and Ravichandran, 2015) and increased caspase-1 activity in old BM macrophages associates with increased BM levels of IL-1β and with enhanced HSC megakaryocytic differentiation bias (associated with expression of CD41 and CD61). In another study, aged BM stromal cells showed enhanced IL-6 and TGF-β signaling. While IL-6 inhibition led to improved erythroid progenitor function in aged mice, TGF-β neutralization resulted in reversal of age-associated HSC megakaryocytic differentiation bias, increased generation of lymphoid progenitors, and rebalanced HSC lineage output in transplantation assays (Valletta et al., 2020). Pioli et al. (2019) suggested that plasma cells from old mice accumulate in the BM where they display increased TLR-driven activation and produce (together with stromal cells) a positive feedback network of proinflammatory cytokines (including IL-1, TNF-α, and M-CSF) that promote HSC myeloid differentiation bias. In overall agreement, work by Helbling et al. (2019) from our laboratory demonstrated that the BM stromal compartment (particularly CXCL12-abundant reticular cells [CARc]) contributes significantly to IL-1– and IL-6–mediated inflamm-aging. Furthermore, transcriptomic pathways induced during aging were largely recapitulated by in vivo administration of prototypic microbial molecules, thereby supporting an inflammatory basis of age-related alterations to HSC function. Collectively, these studies indicate that inflamm-aging is a key, and at least to some extent reversible, mediator of age-associated HSC myeloid/megakaryocyte differentiation biases, while having as-yet undetermined impacts on other aging-associated HSC impairments. Further disentangling the respective contributions of accumulated replicative history, age-related alterations in BM niche function, and inflamm-aging to impaired HSC function and oncogenesis could provide a basis for improving blood system function and health span in old age.
Role of key cytokines on HSC biology: New concepts and ambiguities
HSCs are exposed to multiple and diverse inflammatory milieus during their lifetime. A large body of work using models of single-cytokine stimulation in mice has demonstrated both unique and overlapping impacts of these factors on HSC function independently of the inflammation source. The most commonly studied factors include type I and II IFNs, TNF-α, IL-1, and G-CSF. In this section, we dissect the direct consequences of in vivo administration of these key cytokines on HSC biology and provide a critical review of insights and limitations emerging from mouse models of acute and chronic cytokine exposure (Fig. 2).
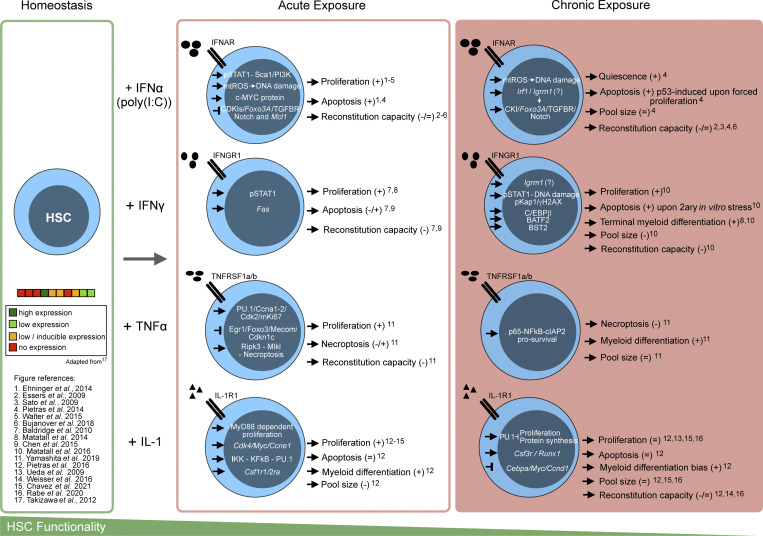
Figure 2.
Effects of acute and chronic inflammatory signaling in HSC biology. Schematic representation of the antagonizing effects of acute or chronic in vivo cytokine exposure on murine HSC functions. Here, acute is defined has exposure of the indicated cytokine in one to three consecutive single daily doses, while chronic represents more than three consecutive daily exposures (in agreement with classic definitions in the field). The left panel depicts HSCs in homeostatic state and the published expression levels of cytokine receptors. Acute and chronic exposures panels depict the pathways and the respective cellular effects of the exposure to the indicated cytokines. The bottom scheme shows loss of HSC functionality from homeostasis to acute to chronic inflammatory exposure. 2ary, secondary.
Type I IFN
Acute exposure of type I IFN inducer polyinosinic:polycytidylic acid (polyI:C) or IFN-α itself can drive proliferation of dormant HSCs through a pathway dependent on the IFN-α/β receptors IFNAR and STAT1 (Essers et al., 2009; Pietras et al., 2014) that associate with increased MYC protein levels (Ehninger et al., 2014), decreased expression of quiescence-enforcing genes, and increased mitochondrial ROS-induced DNA damage (Walter et al., 2015). Interestingly, work by Haas et al. (2015) indicated a stronger type I IFN–induced proliferation in CD41hi HSC-like megakaryocytic progenitors within the phenotypic SLAM (Signaling Lymphocytic Activation Molecule) gate. Notably, this effect is transient, as the compartment returns to a quiescent state under continued IFN-α administration (Pietras et al., 2014). Effects of type I IFNs are negatively regulated by Irf2, as HSC deficiency in this transcription factor exhibits significantly impaired function (Sato et al., 2009). The overall impact of type I IFNs on HSC function remains an area of continued interest. HSCs remain protected from the p53-dependent proapoptotic effects of type I IFNs so long as they remain quiescent; however, forced entry of HSCs into the cell cycle by culture and/or 5-FU induces apoptosis and rapid depletion of HSCs (Pietras et al., 2014). Along these lines, transplantation assays using SLAM cells and/or unfractionated BM have suggested that acute and chronic exposures to type I IFNs impair HSC function (Essers et al., 2009; Pietras et al., 2014). On the other hand, recent work has shown that long-term HSCs, prospectively isolated based on an HSC-specific Fgd5 reporter, do not exhibit impaired reconstitution activity following acute type I IFN exposure (Bujanover et al., 2018). Hence, the effect of type I IFNs on HSC reconstitution activity appears to vary significantly based on the phenotypic definition used to prospectively identify HSCs, calling into question the fidelity of commonly used surface marker definitions such as the SLAM code or Sca-1 expression levels (discussed in King and Goodell, 2011; Kanayama et al., 2020) under nonhomeostatic conditions.
Type II IFN
Acute IFN-γ exposure drives HSC proliferation via IFNGR and STAT1; associates with Fas-mediated apoptosis (Chen et al., 2015); and, like type I IFNs, is associated with reduced repopulation capacity (Baldridge et al., 2010; Matatall et al., 2014). Chronic IFN-γ exposure (via chronic Mycobacterium avium infection) results in increased proliferation stress, potentially resulting from an override of Irgm1-dependent cell-cycle and IFN response suppression (King et al., 2011). Like type I IFNs, IFN-γ primes HSCs to undergo apoptosis upon secondary stress (in vitro culture). Impaired HSC function has also been attributed to enhanced Batf2-dependent terminal myeloid differentiation that depletes the HSC pool and severely impairs self-renewal capacity (Matatall et al., 2016). Notably, surface expression of IFNGR is heterogenous in the phenotypic HSC compartment, and IFNGR+ HSCs exhibit myeloid bias and reduced repopulating activity, further confirming the importance of direct IFNGR signaling in eliciting reduced HSC function (Matatall et al., 2014) as well as the inherent heterogeneity contained within the phenotypic HSC compartment.
TNF-α
The role of TNF-α in HSC function remains an area of significant interest and open questions. Work by Pronk et al. (2011) demonstrated that TNF-α signaling restricts HSC proliferative and reconstitution activity via both TNFRI and TNFRII pathways. On the other hand, recent work by Yamashita and Passegué (2019) showed that the duration of TNF-α stimulation is an essential variable that determines HSC responses. Accordingly, acute TNF-α exposure leads to HSC proliferation and a transient activation of the canonical NF-κB pathway, which maintains HSC survival during proliferation. Interestingly, 48 h later, the initial TNF-α exposure no longer sustains NF-κB pathway activity, resulting in Ripk3-Mlkl–mediated necroptosis associated with reduced HSC repopulating capacity. During chronic TNF-α exposure, there is a sustained p65-NF-κB–mediated prosurvival signaling activity that prevents necroptosis and poises HSCs to undergo myeloid differentiation by inducing Pu.1 expression (Etzrodt et al., 2019). This potentially further contributes to the reestablishment of HSC quiescence by inhibiting cell-cycle activators (Staber et al., 2013), thus protecting HSCs from necroptosis while terminating the regenerative response (Yamashita and Passegué, 2019). Hence, TNF signaling may exert distinct effects based on context and dose. It remains to be addressed whether the capacity of TNF to induce PU.1, which can promote terminal differentiation and restrict HSC cell-cycle activity (see below), may provide insight into the seemingly paradoxical capacity of TNFR signaling to restrict HSC proliferation and reconstitution activity while promoting HSC survival and expansion in other contexts.
IL-1
HSCs exposed to acute IL-1β appear to exhibit Myd88-dependent proliferation and myeloid differentiation through activation of the transcription factor PU.1 (downstream of NF-κB activation) without losing self-renewal capacity (Ueda et al., 2009; Pietras et al., 2016; Weisser et al., 2016). Indeed, short-term or low-dose administration of IL-1α/IL-1β prevents IR- or CT-induced myelosuppression and protects mice with cyclophosphamide-induced neutropenia against sepsis (van der Meer et al., 1988; Damia et al., 1992; Pietras et al., 2016). Likewise, IL-1R1–deficient mice exhibit slowed myeloid recovery following 5-FU administration, further supporting a nonredundant role for IL-1 in hematopoietic regeneration (Pietras et al., 2016). On the other hand, chronic IL-1β exposure results in a profound, yet reversible reduction in long-term repopulating activity in the phenotypic SLAM HSC compartment (Pietras et al., 2016; Weisser et al., 2016). However, more recent work has shown that the long-term impact of chronic IL-1β exposure on HSC reconstitution activity might in fact be minimal if stringent phenotypic definitions (ECPR+/CD34− SLAM cells or Fgd5+ SLAM cells) are used to prospectively isolate HSCs (Rabe et al., 2020). These data suggest that like type I IFNs, the impact of IL-1 signaling on long-term HSC function may be relatively small. Indeed, IL-1 triggers only limited proliferative activity in vivo within the ECPR+/CD34− long-term HSC compartment following acute IL-1 treatment, and quiescence is quickly reestablished during chronic exposure (Rabe et al., 2020; Chavez et al., 2021). Along these lines, IL-1 rapidly induces repression of Myc and numerous protein synthesis genes within the first day of exposure, an effect that is sustained with chronic stimulation. Interestingly, IL-1–induced activation of PU.1 is a proliferation-limiting mechanism, as IL-1 triggers aberrant protein synthesis activity, proliferation, and HSC pool expansion in PU.1-deficient HSC (Chavez et al., 2021). These findings are consistent with previous characterizations of PU.1 as a cell-cycle inhibitor (Staber et al., 2013; Kueh et al., 2013) that facilitates myeloid differentiation by promoting intracellular PU.1 accumulation. Furthermore, recent work has identified IL-1 as a key player in HSC immune training, suggesting that IL-1 and other proinflammatory cytokines can induce heritable, long-term effects on HSC function (Mitroulis et al., 2018). Taken together, these results illustrate an ongoing redefinition of how IL-1 impacts HSC function in contexts such as regeneration versus chronic inflammation and the extent to which prospective HSC isolation strategies can affect experimental results and their interpretation.
G-CSF
G-CSF administration in vivo results in transient HSC proliferation (Wilson et al., 2008), peripheral mobilization, and expansion of the HSC pool within the central but not endosteal BM region (Grassinger et al., 2012). However, chronically G-CSF–treated HSCs have an overall loss of repopulating and self-renewal activity (Kovtonyuk et al., 2016b) that associates with enhanced TLR/MyD88 signaling (Schuettpelz et al., 2014). Interestingly, work from Cain et al. (2011) suggested that transient IL-1–mediated HSPC proliferation may be indirectly regulated by IL-1R1–dependent G-CSF production. These data are consistent with prior BM chimera studies from the same group, showing that alum-induced HSPC proliferation requires IL-1R1 expression in the radioresistant (nonhematopoietic) compartment (Ueda et al., 2009), which aligns with our finding that direct IL-1 signaling triggers a PU.1-mediated proliferation-limiting mechanism in HSCs instead. Interestingly, later work from Bernitz et al. (2017) suggested that like type I IFNs, G-CSF–induced proliferation is limited to CD41hi SLAM cells based on H2B-GFP dilution studies. Hence, as with IL-1 and type I IFNs, the precise functional impacts of G-CSF on HSC function remain to be fully clarified.
Taken as a complete body of work, it is clear that proinflammatory cytokines can exert significant effects on HSCs. Strikingly however, and perhaps contrary to initial published characterizations, most cytokines directly elicit only transient, if any, proliferative activity in true long-term HSCs. Moreover, the mechanisms underlying these activities require revisiting, as with PU.1 and IL-1. Importantly, limitations imposed by established experimental model setups need scrutiny. In particular, standard definitions of inflammatory stimulus duration (i.e., acute and chronic) often lack consistency; do not align with established clinical definitions of acute and chronic disease; and thus, represent a significant source of noise in the field. In addition, time points after inflammatory stimulus at which hematopoietic outcomes are analyzed are of key importance. As perturbed biological systems tend to return to a homeostatic state over time, it is likely that analysis closer to inflammatory stimulation associates with stronger phenotypes. Finally, as discussed above, the phenotypic definition of HSCs needs to be updated and systematized in order to accommodate the growing evidence of molecular heterogeneity within this compartment. In spite of these limitations, the totality of evidence suggests that chronic exposure to individual cytokines exerts fairly mild effects on HSC function. Such results are again in contrast to chronic infection models where the HSC compartment exhibits more profound changes in proliferation and functional decline. Hence, individual proinflammatory cytokines on their own (alongside any secondary signals they induce) may not be sufficient to severely degrade HSC function. On the other hand, they may trigger much more significant impairment of HSC and blood system function in the context of ongoing hematopoietic demand, such as during exposure to combined IFN-1 and 5-FU challenge or complex “cytokine storms.” Furthermore, while the majority of experimental systems address acute and chronic exposures, the impact of serial inflammatory episodes on HSC function is less well studied. This further highlights the fact that HSC responses to inflammatory cytokines are the result of biological processes that are highly dependent on the (patho)physiological context in which the inflammatory signals are produced as well as on cytokine doses and duration of exposure. Care must therefore be taken not to derive broad generalizations about HSC responses to inflammation based on the results of individual studies using specific cytokines and/or experimental models.
Inflammation as a driver of somatic evolution in BM
Chronic inflammation and/or tonic levels of inflammatory signaling sustained over time and combined with aging-associated declines can have deleterious impacts on overall blood system function. This includes multiple HSC properties, such as survival, proliferation, differentiation, self-renewal, and genomic integrity (see previous sections). While HSCs are clearly capable of adapting to inflammatory signals necessary for hematopoietic regeneration and blood system maintenance without incurring dramatic functional costs, the extent to which chronic or even punctuated inflammatory insults degrade HSC function over time and contribute to morbidities, such as BM failure and/or myeloid oncogenesis, remains an area of active investigation. Indeed, there is an association between inflammatory burden, preleukemic states, and full-blown leukemia development (reviewed in Craver et al., 2018). Along these lines, revised models of cancer development, such as the theory of adaptive oncogenesis, implicate inflammation as a crucial driver of hematological and other malignancies by impairing the fitness (defined as capacity to generate progeny) of normal cells, thereby altering the tissue fitness landscape to favor cells harboring oncogenic mutations (Laconi et al., 2020). Here, we summarize multiple studies that have suggested that selective pressure imposed by chronic inflammation on the HSC pool might drive genetic mutations and/or selection/expansion of inflammation-adapted mutant clones, leading to a preleukemic state and, potentially, evolution to leukemia (Fig. 3).
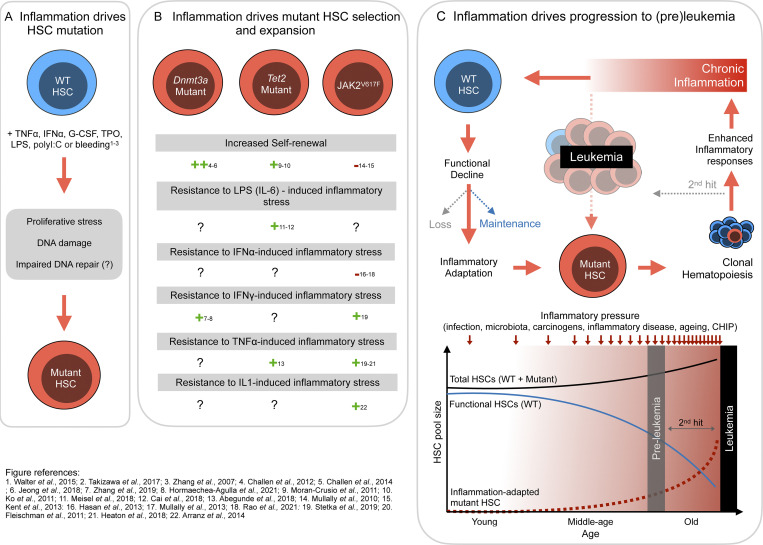
Figure 3.
Proposed mechanisms of HSCs inflammation-driven evolution. Schematic representation of the three main processes by which sustained inflammation drives HSC somatic evolution. (A) Inflammation as a driver of mutation incidence in HSC by inducing increased proliferation stress and DNA damage. Impact of inflammation on DNA repair efficiency is unknown. (B) Inflammatory pressure leads to the selection and expansion of HSC-carrying mutations (in DNMT3A, TET, and JAK2). Depicted are the published adaptations of each mutation to inflammatory stress (++ strongly present, + present, − absent, ? unknown). (C) Speculative role of inflammation on establishment of clonal hematopoiesis and its progression to leukemia. Lifelong inflammatory exposure leads to functional decline of WT HSCs and selects inflammatory-adapted mutant HSCs that expand, leading to a preleukemic stage. Enhanced cytokine production by mutant myeloid cells further increased the systemic inflammatory load, leading to inflammatory-adapted mutant HSC clonal expansion and increasing the risk of secondary hit mutations and potential leukemia transformation.
Inflammation and DNA damage in HSCs
Cellular mutation occurrence probability is defined by the product of the mutation rate per division per DNA base pair (defined as 3 × 10−9) and the cellular division rate (Rozhok et al., 2014, 2016). Therefore, accelerated inflammation-induced HSC cycling can enhance mutational occurrence. Indeed, in vivo cell-cycle induction using IFN-α, G-CSF, TPO, serial bleeding, or chronic polyI:C regimens leads to increased mitochondrial ROS-induced DNA damage in WT HSCs and results in HSC depletion and BM failure in mice with a nonfunctional Fanconi anemia DNA repair pathway (Fanca−/−; Walter et al., 2015). Work from our laboratory further established that HSCs exposed to prolonged bacterial LPS stimulation or Salmonella typhimurium infection exhibit elevated proliferative stress and DNA damage formation mainly via TLR4–TRIF signaling (Takizawa et al., 2017). Importantly, these effects are amplified in genetic contexts associated with defective DNA repair. Zhang et al. (2007) showed that TNF-α–induced senescence in WT HSPCs correlates with the accumulation of ROS-mediated DNA damage and leads to increased chromosomal aberrations (in Lin− cells), which are further enhanced in Fancc−/− mice. This strongly suggests that inflammatory exposure leads to increased mutagenesis in Lin− cells; however, direct evidence in HSCs remains to be established (Fig. 3 A). Along these lines, HSPCs with compound loss of single-stranded DNA–binding proteins Ssb1 and Ssb2 rapidly induce type I IFNs associated with replication stress and depletion of HSC (Shi et al., 2017). Indeed, BM failure syndromes are commonly associated with chronic production of TNF and IFNs (Hara et al., 2004; Chen et al., 2015), reinforcing the pathogenic role of inflammation in this context. Finally, it is worth noting that genotoxin exposure can trigger DNA breaks in quiescent HSCs, which are repaired by nonhomologous end joining, leading to acquisition of chromosomal aberrations (Mohrin et al., 2010). The extent to which inflammation can induce somatic mutations in quiescent HSCs via induction of nonhomologous end joining and other proliferation-independent DNA modification or repair mechanisms remains an important question. Altogether, inflammation functions as both a cause and an effect of DNA damage. Defining the extent to which inflammation directly induces mutations that lead to hematological malignancy, as opposed to serving as a selective mechanism for mutated HSC clones, remains an important frontier of investigation in the field.
Inflammation-adapted mutant HSCs
Inflammatory milieus are associated with hematological malignancies, such as myelodysplastic syndromes (reviewed in Sallman and List, 2019), myeloproliferative neoplasms (reviewed in Lussana and Rambaldi, 2017), chronic myeloid leukemia (reviewed in Hoermann et al., 2015), and acute myeloid leukemia (AML; reviewed in Binder et al., 2018). Again, whether inflammation is a cause or consequence (or both) of these established malignancies remains to be determined. In healthy, young tissue systems, mutated cell clones often exhibit low relative fitness levels and are routinely eliminated via apoptosis, terminal differentiation, immune system surveillance, and/or entry into a senescent state (Laconi et al., 2020). However, the proinflammatory stimuli discussed above are capable of inducing these processes in normal HSCs, thereby reducing their relative fitness. Indeed, as we describe above, IFNs can induce p53-dependent apoptosis and/or cell-cycle arrest, whereas IL-1 and TNF can activate PU.1 and promote terminal differentiation and/or enforce quiescence, altogether limiting proliferative activity. Mutations that render HSCs insensitive to these effects can thus confer a relative fitness advantage in an inflammatory environment, leading to an altered tissue landscape that now selects for such mutations. Indeed, a wide variety of mutations associated with myeloid malignancy interfere with the normal expression and/or function of proinflammatory cytokine targets, such as PU.1, C/EBPα, and p53, and/or lead to increased activation of survival and proliferation pathways, such as Flt3 and Ras (Gerloff et al., 2015; McKenzie et al., 2019; Mueller et al., 2006; Noguera et al., 2016; Vangala et al., 2003; Yang et al., 2012; Kaasinen et al., 2019; Henry et al., 2015). Along these lines, we recently showed that IL-1 rapidly suppresses HSC expression of Myc, a key player regulating relative cell fitness in many tissues, including HSCs (Laurenti et al., 2008; Scognamiglio et al., 2016; Wilson et al., 2004). On the other hand, PU.1-deficient HSCs are largely insensitive to this effect of IL-1, fail to repress Myc, and instead continue to proliferate and expand in the BM (Chavez et al., 2021) during IL-1 exposure. Likewise, Cebpa-deficient MPPs can outcompete their WT counterparts in vivo when exposed to chronic IL-1 (Higa et al., 2021). Strikingly, both PU.1- and Cebpa-deficient HSPCs are not selected for in the absence of the inflammatory signal, providing important evidence that the loss of these factors alone is not sufficient to promote their expansion.
Preleukemic states where founding mutations are present but where the hematopoietic system is still uncompromised (reviewed in Corces-Zimmerman and Majeti, 2014), constitute an ideal system to address how inflammatory pressure acts on a genetically heterogeneous HSC pool to select for clones carrying specific mutations. Clonal hematopoiesis of indeterminate potential (CHIP) is a hematological condition first characterized in humans by the presence of an expanded somatic blood cell clone carrying a mutation in leukemia driver genes (e.g., DNMT3A, TET2, ASXL1, TP53, JAK2, and SF3B1) at a variant allele frequency of at least 2% in the absence of any other hematological abnormalities. CHIP incidence increases with age (and is thus sometimes also termed age-related clonal hematopoeisis, ARCH) and associates with an increased risk of hematological malignancies, cardiovascular disease, and all-cause mortality (Boettcher and Ebert, 2019; Jaiswal et al., 2014; Genovese et al., 2014; Shlush et al., 2014), which is attributed to increased inflammatory responses from mutant immune cells (reviewed in Cook et al., 2020). Indeed, individuals with CHIP have higher serum IL-6, TNF-α, IL-1β, and IL-18 levels than those without (Cook et al., 2019; Bick et al., 2020). Furthermore, enhanced inflammatory responses have been described in DNMT3A (Leoni et al., 2017; Sano et al., 2018a), TET2 (Sano et al., 2018b; Fuster et al., 2017; Zhang et al., 2015; Cull et al., 2017), and JAK2 mutant myeloid cells (Wang et al., 2018). Importantly, multiple studies in mice and humans have suggested that CHIP-mutant HSPCs, contrary to their WT counterparts, have developed molecular mechanisms that potentially allow adaptation to the sustained inflammatory cues present in CHIP individuals (Fig. 3 B). Here, we summarize the main inflammatory adaptation mechanisms observed in DNMT3A,TET2, andJAK2 mutant HSPCs.
DNMT3A
Mutations in DNMT3A, which encodes a DNA methyltransferase enzyme responsible for establishing de novo DNA methylation patterns during stem cell differentiation (Okano et al., 1999), are the most common CHIP mutations and are detectable in Lin−CD34+CD38− HSCs. These mutations are heterozygous, occur in structural and functional domains, and are thought to lead to loss of protein function (Arends et al., 2018). HSCs from Dnmt3a−/− mice show enhanced self-renewal and reduced differentiation efficiency, which results from biased HSC fate decisions (Challen et al., 2012; Jeong et al., 2018; Challen et al., 2014) and has been partially attributed to hypomethylation-mediated increased expression of self-renewal genes (Jeong et al., 2014). In the context of inflammation, a higher frequency of DNMT3A (and PPM1D) mutations in ulcerative colitis patients was recently described, possibly related to higher IFN-γ serum levels in patients with DNMT3A mutations (Zhang et al., 2019). In agreement, it was also shown that chronic mycobacterial infection as well as chronic IFN-γ administration drive the expansion of Dnmt3a−/− HSCs over WT HSCs. Mechanistically, this associated with hypermethylation of prodifferentiation genes (Jun/Fos gene family members and Batf2) in Dnmt3a−/− HSCs, rendering them more resistant to pool exhaustion via infection-mediated terminal differentiation and secondary stress-induced apoptosis (Hormaechea-Agulla et al., 2021).
TET2
The TET2 gene encodes a dioxygenase that promotes DNA demethylation via the conversion of 5-methylcytosine to 5-hydroxymethylcytosine and other oxidized derivatives (Ito et al., 2010). TET2 mutations are the second most common in CHIP, are detectable in Lin−CD34+CD38− HSCs, and are also loss of function (Arends et al., 2018). HSCs from Tet2−/− mice (or with targeted disruption of the catalytic domain) also show increased self-renewal, impaired differentiation, and a predisposition to hematological malignancy development (Moran-Crusio et al., 2011; Ko et al., 2011). In the context of inflammation, Tet2-deficient murine and human HSCs have a strong proliferative and antiapoptotic advantage compared with WT cells when exposed in vitro to TNF-α (Abegunde et al., 2018). Murine Tet2-deficient HSPCs further show enhanced proliferation and myeloid differentiation in response to microbial-induced IL-6 (Meisel et al., 2018). Moreover, Tet2−/− HSPCs maintain a repopulation advantage after a chronic inflammatory insult (LPS), which is related to hyperactivation of the IL-6/Shp2/Stat3/lncRNA–Morrbid axis, resulting in reduced apoptosis compared with WT cells (Cai et al., 2018). Tet2-deficient HSPCs also exhibit a competitive advantage over WT cells during low-grade inflammation in a manner associated with increased TLR-TRAF6 and noncanonical NF-κB activation (Muto et al., 2020), confirming their enhanced fitness in inflammatory contexts.
JAK2
JAK2 is a member of the JAK family of nonreceptor protein tyrosine kinases involved in cytoplasmic signaling components of cytokine receptors. The JAK2V617F variant can be found in 3% of individuals with CHIP (Cordua et al., 2019). JAK2V617F reduces the self-renewal capacity of individual HSCs but leads to expansion of downstream progenitor cells (Mullally et al., 2010; Kent et al., 2013). JAK2V617F HSPCs proliferate and exhaust after IFN-α treatment, losing disease initiation capacity in secondary transplantation (Hasan et al., 2013; Mullally et al., 2013). In agreement, JAK2V617F megakaryocyte-biased CD41hi HSCs were recently shown to exhaust after IFN-α treatment (Rao et al., 2021). On the other hand, JAK2V617F HSCs exhibit increased IL-1β secretion, which induces MSC death, resulting in mutant HSC expansion and accelerated myeloproliferative neoplasm progression (Arranz et al., 2014). Furthermore, JAK2V617F HSPCs exposed to IFN-γ, TNF-α, and TGF-β1 treatment show a DUSP1-dependent proliferation and protection against DNA damage accumulation and inflammatory stress (Stetka et al., 2019). Additionally, TNF-α drives JAK2V617F HSPC expansion (Fleischman et al., 2011) in an autocrine Tnfr2-NF-κB–dependent fashion that downregulates XIAP and MAPK8, leading to increased survival compared with WT progenitors (Heaton et al., 2018). Altogether, these studies provide compelling evidence that inflammatory alterations to the BM fitness landscape can activate a process of somatic evolution that favors oncogenic mutations. Notably, many of these studies rely on generation of BM chimeras using lethal irradiation, which imparts lasting effects on BM integrity, including increased ROS (Rodrigues-Moreira et al., 2017), which could accelerate and/or alter selection processes in the BM. The use of nonconditioned recipients (Wang et al., 2020); less-inflammatory methods of BM conditioning, such as low-dose CT (Henry et al., 2015; Higa et al., 2021); or genetic mosaic approaches may provide more faithful readouts of selective processes triggered by inflammation and assist in the development of novel therapies to address BM oncogenesis.
Inflammation as a driver of CHIP-mutant HSC malignant transformation
CHIP increases the risk of hematologic malignancy (in aggregate) by 0.5–1% per year (Jaiswal et al., 2014). Subsequent studies indicate that the type of mutation, higher number of mutations, and higher mutational variant allele frequencies predict which CHIP-carrying individuals are more likely to progress to AML (Desai et al., 2018; Abelson et al., 2018). Thus, identifying the factors leading to expansion of CHIP mutant clones and to acquisition of secondary mutations is key to understanding leukemia pathogenesis. Based on evidence discussed here, we propose that lifelong chronic inflammation derived from cumulative sources (infection, microbiota, inflammatory diseases, aging, and CHIP) might be a driver of AML progression in CHIP carriers, particularly in the ones carrying inflammation-adapted mutant clones (TET2+/−, DNMT3A+/−, JAK2V617F). Accordingly, selection and expansion of inflammation-adapted CHIP mutant over WT HSPCs will generate an increasingly larger mutant HSC-derived allele fraction in mature cells that further contributes to systemic inflammation. With time, this proinflammatory cycle can facilitate the acquisition of secondary mutations, increasing the potential for leukemia development (Fig. 3 C). Continued effort into developing in vivo models that can report and/or replicate how sequential mutations lead to malignant transformation (Loberg et al., 2019) will provide opportunities for important advancement in this area. Moreover, whether lifelong proinflammatory stress is also a relevant driver of clonal evolution in nonhematopoietic tissues (e.g., skin, colon, liver; Martincorena et al., 2015; Blokzijl et al., 2016; Brunner et al., 2019) remains to be determined.
Concluding remarks
At least three decades’ worth of work and resultant literature have sought to address the functional and mechanistic impacts of inflammatory challenges on HSCs (Fig. 1 and Tables 1 and 2). This process has been greatly accelerated over the past decade by advances in HSC isolation and functional profiling as well as by development of advanced mouse models of disease and infection. Considering these investigations as a whole, it is now clear that in the absence of defects in DNA repair or other HSC maintenance systems, the hematopoietic system (and the HSC pool in particular) is sufficiently robust to withstand inflammatory challenges, respond to them, and restore homeostasis without incurring immediate dramatic functional costs. On the other hand, in extreme circumstances, inflammatory responses can break this homeostatic buffer and compromise the hematopoietic system, as observed in some infection models or in the context of BM failure. Second, models used to study the impact of inflammation on HSCs vary considerably in terms of the content of the inflammatory milieus, the relative quantities of cytokines, damage-associated molecular patterns, metabolites, and other factors; the duration of the inflammatory stimuli; the extent to which supportive niches are disrupted; and, finally, the proliferative demands exerted upon HSCs. Direct comparisons between models and even to human biology (which is analyzed using a substantially different time frame), must be made cautiously and with due consideration of these confounding factors. Third, it has become clear that as methods for prospective isolation of HSCs improve alongside advanced technologies that allow for interrogation of molecular state and clonal dynamics at the single-cell level, we will be required to revisit, refine, and perhaps discard elements of current dogma related to how inflammation shapes the HSC pool and alters its functional properties. Finally, it is also important for the field to consider the cumulative impact on HSCs of a lifetime of inflammatory challenges and exposure to tonic inflammatory signals generated by the microbiome and other environmental factors. The extent to which “normal” levels of inflammation or serial exposures to inflammatory signals contribute to eventual HSC decline in the context of aging remains a fertile area of investigation, with the potential for significant impact on human health span and risk for hematological malignancy. As the field further clarifies the contribution of inflammation to oncogenesis (Fig. 3), significant opportunities will arise to translate this knowledge into potential novel strategies that prevent and/or redirect somatic evolution to reduce the risk of malignancy, cardiovascular disease, and other comorbidities. Taken together, it is clear that the study of inflammation and HSC biology remains an exciting and active topic replete with ambiguities, evolving concepts, and new opportunities to address key problems in human health.
Table 2. Summary of human studies describing the effects of different inflammatory sources and pathways on phenotypically defined HSC.
| Source | Inflammatory pathway | Effect on HSCs | HSC phenotype | Reference |
|---|---|---|---|---|
| Viral | ||||
| HIV patients | Not direct infection | Decreased pool | CD34+CD38− | Marandin et al. (1996) |
| TNF-α–induced Fas | Apoptosis | CD34+ | Isgrò et al. (2004) | |
| IL-18 increase; stem cell factor decrease | Decreased pool | Circulating Lin−CD34+CD45RA+CD10+CD117− | Bordoni et al. (2018) | |
| HIV humanized mice | pDCs (via IFN-α/β?) | Decreased pool and colony formation | CD34+CD38− | Li et al. (2017) |
| Bacterial | ||||
| Sepsis patients | Increased CXCL12 and decrease S1P at day 1 after septic shock | Increased mobilization and proliferation at day 3 after septic shock | Circulating Lin−CD133+CD45+ and CD34+CD38− | Skirecki et al. (2019) |
| ND | Minor increase mobilization at days 4 and 7 after septic shock | Circulating CD34+CD38−CD90+CD45RA− | Wang et al. (2021) | |
| Sepsis humanized mice | Increased TLR4 and CXCR4 expression/notch–Jagged1 signaling | Increased pool and proliferation | CD34+CD38− | Skirecki et al. (2015) |
| Inflammatory disease | Increased proinflammatory milieu (IL-1β, according to mouse model) | Decreased pool | Lin−CD34+CD38− | Weisser et al. (2016) |
| CGD patients | Exhausted after in vitro culture | G-CSF–mobilized CD34+CD38−CD90+ | ||
| Increased TNF-α | Decreased pool and increased apoptosis | CD34+ | Papadaki et al. (2002) | |
| RA patients | ND/premature telomere shortening | Decreased pool | Circulating CD34+ | Colmegna et al. (2008) |
| Impaired proliferative potential | ||||
| Increased TNF-α | Decreased pool and colony formation | CD34+ | Porta et al. (2004) | |
| SLE patients | Increased Fas (via TNF-α?) | Decreased pool and colony formation | CD34+CD38− | Papadaki et al. (2001) |
| Enhanced CD40–CD40L | Decreased pool and increased apoptosis | CD34+ | Pyrovolaki et al. (2009) | |
| ND | Increased pool | CD34+CD38− | Taraldsrud et al. (2009) | |
| ND | Increased pool | CD34+CD38− | Kuranda et al. (2011) | |
| Decrease in myeloid lineage bias in vivo (NSG mice) and in vitro | ||||
| Increased ERK/MAPK and GM-CSF signaling (tanscriptomics) | Increased pool and reduced quiescence | Lin−CD34+CD38−CD90+CD45RA− | Pang et al. (2011) | |
| Aging | ||||
| Elderly individuals | Reduced in vitro plating efficiency and increased myeloid bias | |||
| Reduced in vivo (NSG mice) engraftment and increased myeloid bias | ||||
| ND | Increased pool | CD34+ | Woolthuis et al. (2014) | |
| Impaired reconstitution capacity at 1 yr after autologous stem cell transplantation | ||||
| Increased pool | Lin−CD34+CD123low/−CD90+CD45RA− | Rundberg Nilsson et al. (2016) | ||
| ND | Reduced in vitro plating efficiency and colony size | |||
| Intrinsic megakaryocytic/ erythroid bias (transcriptomics) |
pDC, plasmacytoid dendritic cells.
Acknowledgments
This work was supported by National Institutes of Health grant R01 DK119394 and the Cleo Meador and George Ryland Scott Endowed Chair in Hematology (to E.M. Pietras), a Comprehensive Cancer Center Zurich fellowship (to F. Caiado), and the Swiss National Science Foundation (310030B_166673/1 to M.G. Manz).
Author contributions: F. Caiado, E.M. Pietras, and M.G. Manz researched and wrote the paper.
References
- Abegunde, S.O., Buckstein R., Wells R.A., and Rauh M.J.. 2018. An inflammatory environment containing TNFα favors Tet2-mutant clonal hematopoiesis. Exp. Hematol. 59:60–65. 10.1016/j.exphem.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Abelson, S., Collord G., Ng S.W.K., Weissbrod O., Mendelson Cohen N., Niemeyer E., Barda N., Zuzarte P.C., Heisler L., Sundaravadanam Y., et al. 2018. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 559:400–404. 10.1038/s41586-018-0317-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arandjelovic, S., and Ravichandran K.S.. 2015. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16:907–917. 10.1038/ni.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends, C.M., Galan-Sousa J., Hoyer K., Chan W., Jäger M., Yoshida K., Seemann R., Noerenberg D., Waldhueter N., Fleischer-Notter H., et al. 2018. Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia. 32:1908–1919. 10.1038/s41375-018-0047-7 [DOI] [PubMed] [Google Scholar]
- Arranz, L., Sánchez-Aguilera A., Martín-Pérez D., Isern J., Langa X., Tzankov A., Lundberg P., Muntión S., Tzeng Y.S., Lai D.M., et al. 2014. Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature. 512:78–81. 10.1038/nature13383 [DOI] [PubMed] [Google Scholar]
- Baldridge, M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature. 465:793–797. 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer, M.L., Schürch C.M., Saito Y., Geuking M.B., Li H., Cuenca M., Kovtonyuk L.V., McCoy K.D., Hapfelmeier S., Ochsenbein A.F., et al. 2014. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 193:5273–5283. 10.4049/jimmunol.1400762 [DOI] [PubMed] [Google Scholar]
- Baylis, D., Bartlett D.B., Patel H.P., and Roberts H.C.. 2013. Understanding how we age: insights into inflammaging. Longev. Healthspan. 2:8. 10.1186/2046-2395-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman, I., Seita J., Inlay M.A., Weissman I.L., and Rossi D.J.. 2014. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 15:37–50. 10.1016/j.stem.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernitz, J.M., Kim H.S., MacArthur B., Sieburg H., and Moore K.. 2016. Hematopoietic stem cells count and remember self-renewal divisions. Cell. 167:1296–1309.e10. 10.1016/j.cell.2016.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernitz, J.M., Daniel M.G., Fstkchyan Y.S., and Moore K.. 2017. Granulocyte colony-stimulating factor mobilizes dormant hematopoietic stem cells without proliferation in mice. Blood. 129:1901–1912. 10.1182/blood-2016-11-752923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick, A.G., Weinstock J.S., Nandakumar S.K., Fulco C.P., Bao E.L., Zekavat S.M., Szeto M.D., Liao X., Leventhal M.J., Nasser J., et al. NHLBI Trans-Omics for Precision Medicine Consortium . 2020. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 586:763–768. 10.1038/s41586-020-2819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, D., Fehr J., Hengartner H., and Zinkernagel R.M.. 1997. Virus-induced transient bone marrow aplasia: major role of interferon-α/β during acute infection with the noncytopathic lymphocytic choriomeningitis virus. J. Exp. Med. 185:517–530. 10.1084/jem.185.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, S., Luciano M., and Horejs-Hoeck J.. 2018. The cytokine network in acute myeloid leukemia (AML): A focus on pro- and anti-inflammatory mediators. Cytokine Growth Factor Rev. 43:8–15. 10.1016/j.cytogfr.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Blokzijl, F., de Ligt J., Jager M., Sasselli V., Roerink S., Sasaki N., Huch M., Boymans S., Kuijk E., Prins P., et al. 2016. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 538:260–264. 10.1038/nature19768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher, S., and Ebert B.L.. 2019. Clonal hematopoiesis of indeterminate potential. J. Clin. Oncol. 37:419–422. 10.1200/JCO.2018.79.3588 [DOI] [PubMed] [Google Scholar]
- Bordoni, V., Viola D., Sacchi A., Pinnetti C., Casetti R., Cimini E., Tumino N., Antinori A., Ammassari A., and Agrati C.. 2018. IL-18 and Stem Cell Factor affect hematopoietic progenitor cells in HIV-infected patients treated during primary HIV infection. Cytokine. 103:34–37. 10.1016/j.cyto.2017.12.033 [DOI] [PubMed] [Google Scholar]
- Brunner, S.F., Roberts N.D., Wylie L.A., Moore L., Aitken S.J., Davies S.E., Sanders M.A., Ellis P., Alder C., Hooks Y., et al. 2019. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 574:538–542. 10.1038/s41586-019-1670-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujanover, N., Goldstein O., Greenshpan Y., Turgeman H., Klainberger A., Scharff Y., and Gazit R.. 2018. Identification of immune-activated hematopoietic stem cells. Leukemia. 32:2016–2020. 10.1038/s41375-018-0220-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burberry, A., Zeng M.Y., Ding L., Wicks I., Inohara N., Morrison S.J., and Núñez G.. 2014. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe. 15:779–791. 10.1016/j.chom.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., and Rodewald H.R.. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 518:542–546. 10.1038/nature14242 [DOI] [PubMed] [Google Scholar]
- Cai, Z., Kotzin J.J., Ramdas B., Chen S., Nelanuthala S., Palam L.R., Pandey R., Mali R.S., Liu Y., Kelley M.R., et al. 2018. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 23:833–849.e5. 10.1016/j.stem.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain, D.W., Snowden P.B., Sempowski G.D., and Kelsoe G.. 2011. Inflammation triggers emergency granulopoiesis through a density-dependent feedback mechanism. PLoS One. 6:e19957. 10.1371/journal.pone.0019957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalayer, E., Costedoat-Chalumeau N., Beyne-Rauzy O., Ninet J., Durupt S., Tebib J., Asli B., Lambotte O., Ffrench M., Vasselon C., and Cathébras P.. 2017. Bone marrow involvement in systemic lupus erythematosus. QJM. 110:701–711. 10.1093/qjmed/hcx102 [DOI] [PubMed] [Google Scholar]
- Challen, G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., et al. 2012. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44:23–31. 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen, G.A., Sun D., Mayle A., Jeong M., Luo M., Rodriguez B., Mallaney C., Celik H., Yang L., Xia Z., et al. 2014. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 15:350–364. 10.1016/j.stem.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez, J.S., Rabe J.L., Loeffler D., Higa K.C., Hernandez G., Mills T.S., Ahmed N., Gessner R.L., Ke Z., Idler B.M., et al. 2021. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J. Exp. Med. 218:e20201169. 10.1084/jem.20201169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Feng X., Desierto M.J., Keyvanfar K., and Young N.S.. 2015. IFN-γ-mediated hematopoietic cell destruction in murine models of immune-mediated bone marrow failure. Blood. 126:2621–2631. 10.1182/blood-2015-06-652453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.H., Kim K.K., Kim K.D., Kim H.J., Jo E.K., and Song C.H.. 2011. Effects of mycobacterial infection on proliferation of hematopoietic precursor cells. Microbes Infect. 13:1252–1260. 10.1016/j.micinf.2011.08.001 [DOI] [PubMed] [Google Scholar]
- Citrin, D.E., and Mitchell J.B.. 2017. Mechanisms of normal tissue injury from irradiation. Semin. Radiat. Oncol. 27:316–324. 10.1016/j.semradonc.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmegna, I., and Weyand C.M.. 2011. Haematopoietic stem and progenitor cells in rheumatoid arthritis. Rheumatology (Oxford). 50:252–260. 10.1093/rheumatology/keq298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmegna, I., Diaz-Borjon A., Fujii H., Schaefer L., Goronzy J.J., and Weyand C.M.. 2008. Defective proliferative capacity and accelerated telomeric loss of hematopoietic progenitor cells in rheumatoid arthritis. Arthritis Rheum. 58:990–1000. 10.1002/art.23287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, E.K., Izukawa T., Young S., Rosen G., Jamali M., Zhang L., Johnson D., Bain E., Hilland J., Ferrone C.K., et al. 2019. Comorbid and inflammatory characteristics of genetic subtypes of clonal hematopoiesis. Blood Adv. 3:2482–2486. 10.1182/bloodadvances.2018024729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, E.K., Luo M., and Rauh M.J.. 2020. Clonal hematopoiesis and inflammation: Partners in leukemogenesis and comorbidity. Exp. Hematol. 83:85–94. 10.1016/j.exphem.2020.01.011 [DOI] [PubMed] [Google Scholar]
- Corces-Zimmerman, M.R., and Majeti R.. 2014. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 28:2276–2282. 10.1038/leu.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordua, S., Kjaer L., Skov V., Pallisgaard N., Hasselbalch H.C., and Ellervik C.. 2019. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood. 134:469–479. 10.1182/blood.2019001113 [DOI] [PubMed] [Google Scholar]
- Craver, B.M., El Alaoui K., Scherber R.M., and Fleischman A.G.. 2018. The Critical role of inflammation in the pathogenesis and progression of myeloid malignancies. Cancers (Basel). 10:104. 10.3390/cancers10040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull, A.H., Snetsinger B., Buckstein R., Wells R.A., and Rauh M.J.. 2017. Tet2 restrains inflammatory gene expression in macrophages. Exp. Hematol. 55:56–70.e13. 10.1016/j.exphem.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Cyprus, G.N., Overlin J.W., Hotchkiss K.M., Kandalam S., and Olivares-Navarrete R.. 2018. Cigarette smoke increases pro-inflammatory markers and inhibits osteogenic differentiation in experimental exposure model. Acta Biomater. 76:308–318. 10.1016/j.actbio.2018.06.018 [DOI] [PubMed] [Google Scholar]
- Damia, G., Komschlies K.L., Futami H., Back T., Gruys M.E., Longo D.L., Keller J.R., Ruscetti F.W., and Wiltrout R.H.. 1992. Prevention of acute chemotherapy-induced death in mice by recombinant human interleukin 1: protection from hematological and nonhematological toxicities. Cancer Res. 52:4082–4089. [PubMed] [Google Scholar]
- Dawoud, A.A.Z., Tapper W.J., and Cross N.C.P.. 2020. Clonal myelopoiesis in the UK Biobank cohort: ASXL1 mutations are strongly associated with smoking. Leukemia. 34:2660–2672. 10.1038/s41375-020-0896-8 [DOI] [PubMed] [Google Scholar]
- de Bruin, A.M., Demirel Ö., Hooibrink B., Brandts C.H., and Nolte M.A.. 2013. Interferon-γ impairs proliferation of hematopoietic stem cells in mice. Blood. 121:3578–3585. 10.1182/blood-2012-05-432906 [DOI] [PubMed] [Google Scholar]
- Desai, P., Mencia-Trinchant N., Savenkov O., Simon M.S., Cheang G., Lee S., Samuel M., Ritchie E.K., Guzman M.L., Ballman K.V., et al. 2018. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24:1015–1023. 10.1038/s41591-018-0081-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh, H.S., Liu Y., Menkiti O.R., Mei J., Dai N., O’Leary C.E., Oliver P.M., Kolls J.K., Weiser J.N., and Worthen G.S.. 2014. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20:524–530. 10.1038/nm.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger, A., Boch T., Uckelmann H., Essers M.A., Müdder K., Sleckman B.P., and Trumpp A.. 2014. Posttranscriptional regulation of c-Myc expression in adult murine HSCs during homeostasis and interferon-α-induced stress response. Blood. 123:3909–3913. 10.1182/blood-2013-10-531038 [DOI] [PubMed] [Google Scholar]
- Ergen, A.V., Boles N.C., and Goodell M.A.. 2012. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 119:2500–2509. 10.1182/blood-2011-11-391730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, M.A.G., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Etzrodt, M., Ahmed N., Hoppe P.S., Loeffler D., Skylaki S., Hilsenbeck O., Kokkaliaris K.D., Kaltenbach H.M., Stelling J., Nerlov C., and Schroeder T.. 2019. Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood. 133:816–819. 10.1182/blood-2018-02-832998 [DOI] [PubMed] [Google Scholar]
- Fleischman, A.G., Aichberger K.J., Luty S.B., Bumm T.G., Petersen C.L., Doratotaj S., Vasudevan K.B., LaTocha D.H., Yang F., Press R.D., et al. 2011. TNFα facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood. 118:6392–6398. 10.1182/blood-2011-04-348144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez, M.A., Matatall K.A., Jeong Y., Ortinau L., Shafer P.W., Lynch A.M., Jaksik R., Kimmel M., Park D., and King K.Y.. 2020. Interferon gamma mediates hematopoietic stem cell activation and niche relocalization through BST2. Cell Rep. 33:108530. 10.1016/j.celrep.2020.108530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, B.J., Hoffman C.M., Latchney S.E., LaMere M.W., Myers J., Ashton J., Li A.J., Saunders J. II, Palis J., Perkins A.S., et al. 2019. Aged marrow macrophages expand platelet-biased hematopoietic stem cells via Interleukin1B. JCI Insight. 5:e124213. 10.1172/jci.insight.124213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop, T., Larbi A., Dupuis G., Le Page A., Frost E.H., Cohen A.A., Witkowski J.M., and Franceschi C.. 2018. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front. Immunol. 8:1960. 10.3389/fimmu.2017.01960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster, J.J., MacLauchlan S., Zuriaga M.A., Polackal M.N., Ostriker A.C., Chakraborty R., Wu C.L., Sano S., Muralidharan S., Rius C., et al. 2017. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 355:842–847. 10.1126/science.aag1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasche, C., Lomer M.C.E., Cavill I., and Weiss G.. 2004. Iron, anaemia, and inflammatory bowel diseases. Gut. 53:1190–1197. 10.1136/gut.2003.035758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, H., de Haan G., and Florian M.C.. 2013. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 13:376–389. 10.1038/nri3433 [DOI] [PubMed] [Google Scholar]
- Genovese, G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371:2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerloff, D., Grundler R., Wurm A.A., Bräuer-Hartmann D., Katzerke C., Hartmann J.U., Madan V., Müller-Tidow C., Duyster J., Tenen D.G., et al. 2015. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 29:535–547. 10.1038/leu.2014.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassinger, J., Williams B., Olsen G.H., Haylock D.N., and Nilsson S.K.. 2012. Granulocyte colony stimulating factor expands hematopoietic stem cells within the central but not endosteal bone marrow region. Cytokine. 58:218–225. 10.1016/j.cyto.2012.01.014 [DOI] [PubMed] [Google Scholar]
- Griseri, T., McKenzie B.S., Schiering C., and Powrie F.. 2012. Dysregulated hematopoietic stem and progenitor cell activity promotes interleukin-23-driven chronic intestinal inflammation. Immunity. 37:1116–1129. 10.1016/j.immuni.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast Á.M., Schnell A., Hexel K., et al. 2015. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell. 17:422–434. 10.1016/j.stem.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Haltalli, M.L.R., Watcham S., Wilson N.K., Eilers K., Lipien A., Ang H., Birch F., Anton S.G., Pirillo C., Ruivo N., et al. 2020. Manipulating niche composition limits damage to haematopoietic stem cells during Plasmodium infection. Nat. Cell Biol. 22:1399–1410. 10.1038/s41556-020-00601-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Ando K., Tsurumi H., and Moriwaki H.. 2004. Excessive production of tumor necrosis factor-alpha by bone marrow T lymphocytes is essential in causing bone marrow failure in patients with aplastic anemia. Eur. J. Haematol. 73:10–16. 10.1111/j.1600-0609.2004.00259.x [DOI] [PubMed] [Google Scholar]
- Hasan, S., Lacout C., Marty C., Cuingnet M., Solary E., Vainchenker W., and Villeval J.L.. 2013. JAK2V617F expression in mice amplifies early hematopoietic cells and gives them a competitive advantage that is hampered by IFNα. Blood. 122:1464–1477. 10.1182/blood-2013-04-498956 [DOI] [PubMed] [Google Scholar]
- Heaton, W.L., Senina A.V., Pomicter A.D., Salama M.E., Clair P.M., Yan D., Bell R.N., Gililland J.M., Prchal J.T., O’Hare T., and Deininger M.W.. 2018. Autocrine Tnf signaling favors malignant cells in myelofibrosis in a Tnfr2-dependent fashion. Leukemia. 32:2399–2411. 10.1038/s41375-018-0131-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbling, P.M., Piñeiro-Yáñez E., Gerosa R., Boettcher S., Al-Shahrour F., Manz M.G., and Nombela-Arrieta C.. 2019. Global transcriptomic profiling of the bone marrow stromal microenvironment during postnatal development, aging, and inflammation. Cell Rep. 29:3313–3330.e4. 10.1016/j.celrep.2019.11.004 [DOI] [PubMed] [Google Scholar]
- Henry, C.J., Casás-Selves M., Kim J., Zaberezhnyy V., Aghili L., Daniel A.E., Jimenez L., Azam T., McNamee E.N., Clambey E.T., et al. 2015. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J. Clin. Invest. 125:4666–4680. 10.1172/JCI83024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, G., Mills T.S., Rabe J.L., Chavez J.S., Kuldanek S., Kirkpatrick G., Noetzli L., Jubair W.K., Zanche M., Myers J.R., et al. 2020. Pro-inflammatory cytokine blockade attenuates myeloid expansion in a murine model of rheumatoid arthritis. Haematologica. 105:585–597. 10.3324/haematol.2018.197210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa, K.C., Goodspeed A., Chavez J.S., De Dominici M., Danis E., Zaberezhnyy V., Rabe J.L., Tenen D.G., Pietras E.M., and DeGregori J.. 2021. Chronic interleukin-1 exposure triggers selection for Cebpa-knockout multipotent hematopoietic progenitors. J. Exp. Med. 218:e20200560. 10.1084/jem.20200560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirche, C., Frenz T., Haas S.F., Döring M., Borst K., Tegtmeyer P.K., Brizic I., Jordan S., Keyser K., Chhatbar C., et al. 2017. Systemic virus infections differentially modulate cell cycle state and functionality of long-term hematopoietic stem cells in vivo. Cell Rep. 19:2345–2356. 10.1016/j.celrep.2017.05.063 [DOI] [PubMed] [Google Scholar]
- Hoermann, G., Greiner G., and Valent P.. 2015. Cytokine regulation of microenvironmental cells in myeloproliferative neoplasms. Mediators Inflamm. 2015:869242. 10.1155/2015/869242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaechea-Agulla, D., Matatall K.A., Le D.T., Kain B., Long X., Kus P., Jaksik R., Challen G.A., Kimmel M., and King K.Y.. 2021. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell.:S1934-5909(21)00108-9. 10.1016/j.stem.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgrò, A., Mezzaroma I., Aiuti A., Fantauzzi A., Pinti M., Cossarizza A., and Aiuti F.. 2004. Decreased apoptosis of bone marrow progenitor cells in HIV-1-infected patients during highly active antiretroviral therapy. AIDS. 18:1335–1337. 10.1097/00002030-200406180-00013 [DOI] [PubMed] [Google Scholar]
- Isringhausen, S., Kovtonyuk L., Suessbier U., Kraeutler N.J., Gomariz A., Helbling P.M., Wong H.C., Nagasawa T., Manz M.G., Oxenius A., et al. 2020. CD8 T cells induce destruction of bone marrow stromal niches and hematopoietic stem cell dysfunction in chronic viral infections. bioRxiv. (Preprint posted May 8, 2020) 10.1101/2020.05.07.076083 [DOI]
- Ito, S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., and Zhang Y.. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 466:1129–1133. 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamura, C., Bouladoux N., Belkaid Y., Sher A., and Jankovic D.. 2017. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood. 129:171–176. 10.1182/blood-2016-06-723742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371:2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, M., Sun D., Luo M., Huang Y., Challen G.A., Rodriguez B., Zhang X., Chavez L., Wang H., Hannah R., et al. 2014. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 46:17–23. 10.1038/ng.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, M., Park H.J., Celik H., Ostrander E.L., Reyes J.M., Guzman A., Rodriguez B., Lei Y., Lee Y., Ding L., et al. 2018. Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 23:1–10. 10.1016/j.celrep.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsdottir, K.S., Baldridge M.T., Kadmon C.S., and King K.Y.. 2017. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 129:729–739. 10.1182/blood-2016-03-708594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasinen, E., Kuismin O., Rajamäki K., Ristolainen H., Aavikko M., Kondelin J., Saarinen S., Berta D.G., Katainen R., Hirvonen E.A.M., et al. 2019. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 10:1252. 10.1038/s41467-019-09198-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama, M., Izumi Y., Yamauchi Y., Kuroda S., Shin T., Ishikawa S., Sato T., Kajita M., and Ohteki T.. 2020. CD86-based analysis enables observation of bona fide hematopoietic responses. Blood. 136:1144–1154. 10.1182/blood.2020004923 [DOI] [PubMed] [Google Scholar]
- Kent, D.G., Li J., Tanna H., Fink J., Kirschner K., Pask D.C., Silber Y., Hamilton T.L., Sneade R., Simons B.D., and Green A.R.. 2013. Self-renewal of single mouse hematopoietic stem cells is reduced by JAK2V617F without compromising progenitor cell expansion. PLoS Biol. 11:e1001576. 10.1371/journal.pbio.1001576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldoyanidi, S., Sikora L., Orlovskaya I., Matrosova V., Kozlov V., and Sriramarao P.. 2001. Correlation between nicotine-induced inhibition of hematopoiesis and decreased CD44 expression on bone marrow stromal cells. Blood. 98:303–312. 10.1182/blood.V98.2.303 [DOI] [PubMed] [Google Scholar]
- Khan, N., Downey J., Sanz J., Kaufmann E., Blankenhaus B., Pacis A., Pernet E., Ahmed E., Cardoso S., Nijnik A., et al. 2020. M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell. 183:752–770.e22. 10.1016/j.cell.2020.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi, A., Yáñez A., Price J.G., Chow A., Merad M., Goodridge H.S., and Mazmanian S.K.. 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 15:374–381. 10.1016/j.chom.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.Y., and Goodell M.A.. 2011. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11:685–692. 10.1038/nri3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.Y., Baldridge M.T., Weksberg D.C., Chambers S.M., Lukov G.L., Wu S., Boles N.C., Jung S.Y., Qin J., Liu D., et al. 2011. Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling. Blood. 118:1525–1533. 10.1182/blood-2011-01-328682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, M., Bandukwala H.S., An J., Lamperti E.D., Thompson E.C., Hastie R., Tsangaratou A., Rajewsky K., Koralov S.B., and Rao A.. 2011. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA. 108:14566–14571. 10.1073/pnas.1112317108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtonyuk, L.V., Fritsch K., Feng X., Manz M.G., and Takizawa H.. 2016a. Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front. Immunol. 7:502. 10.3389/fimmu.2016.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtonyuk, L.V., Manz M.G., and Takizawa H.. 2016b. Enhanced thrombopoietin but not G-CSF receptor stimulation induces self-renewing hematopoietic stem cell divisions in vivo. Blood. 127:3175–3179. 10.1182/blood-2015-09-669929 [DOI] [PubMed] [Google Scholar]
- Kueh, H.Y., Champhekar A., Nutt S.L., Elowitz M.B., and Rothenberg E.V.. 2013. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science. 341:670–673. 10.1126/science.1240831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranda, K., Vargaftig J., de la Rochere P., Dosquet C., Charron D., Bardin F., Tonnelle C., Bonnet D., and Goodhardt M.. 2011. Age-related changes in human hematopoietic stem/progenitor cells. Aging Cell. 10:542–546. 10.1111/j.1474-9726.2011.00675.x [DOI] [PubMed] [Google Scholar]
- Laconi, E., Marongiu F., and DeGregori J.. 2020. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br. J. Cancer. 122:943–952. 10.1038/s41416-019-0721-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti, E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.F., MacDonald H.R., Eisenman R.N., et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 3:611–624. 10.1016/j.stem.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.K., Menezes J.S., Umesaki Y., and Mazmanian S.K.. 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA. 108(Suppl 1):4615–4622. 10.1073/pnas.1000082107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Kim H., You G., Kim Y.M., Lee S., Le V.H., Kwon O., Im S.H., Kim Y.M., Kim K.S., et al. 2019. Bone marrow CX3CR1+ mononuclear cells relay a systemic microbiota signal to control hematopoietic progenitors in mice. Blood. 134:1312–1322. 10.1182/blood.2019000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni, C., Montagner S., Rinaldi A., Bertoni F., Polletti S., Balestrieri C., and Monticelli S.. 2017. Dnmt3a restrains mast cell inflammatory responses. Proc. Natl. Acad. Sci. USA. 114:E1490–E1499. 10.1073/pnas.1616420114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Zhao J., Cheng L., Jiang Q., Kan S., Qin E., Tu B., Zhang X., Zhang L., Su L., and Zhang Z.. 2017. HIV-1 infection depletes human CD34+CD38- hematopoietic progenitor cells via pDC-dependent mechanisms. PLoS Pathog. 13:e1006505. 10.1371/journal.ppat.1006505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loberg, M.A., Bell R.K., Goodwin L.O., Eudy E., Miles L.A., SanMiguel J.M., Young K., Bergstrom D.E., Levine R.L., Schneider R.K., and Trowbridge J.J.. 2019. Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis. Leukemia. 33:1635–1649. 10.1038/s41375-018-0368-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussana, F., and Rambaldi A.. 2017. Inflammation and myeloproliferative neoplasms. J. Autoimmun. 85:58–63. 10.1016/j.jaut.2017.06.010 [DOI] [PubMed] [Google Scholar]
- MacNamara, K.C., Jones M., Martin O., and Winslow G.M.. 2011. Transient activation of hematopoietic stem and progenitor cells by IFNγ during acute bacterial infection. PLoS One. 6:e28669. 10.1371/journal.pone.0028669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz, M.G., and Boettcher S.. 2014. Emergency granulopoiesis. Nat. Rev. Immunol. 14:302–314. 10.1038/nri3660 [DOI] [PubMed] [Google Scholar]
- Marandin, A., Katz A., Oksenhendler E., Tulliez M., Picard F., Vainchenker W., and Louache F.. 1996. Loss of primitive hematopoietic progenitors in patients with human immunodeficiency virus infection. Blood. 88:4568–4578. 10.1182/blood.V88.12.4568.bloodjournal88124568 [DOI] [PubMed] [Google Scholar]
- Martincorena, I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D.C., Fullam A., Alexandrov L.B., Tubio J.M., et al. 2015. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 348:880–886. 10.1126/science.aaa6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, A., Bono C., Megías J., Yáñez A., Gozalbo D., and Gil M.L.. 2018. Systemic candidiasis and TLR2 agonist exposure impact the antifungal response of hematopoietic stem and progenitor cells. Front. Cell. Infect. Microbiol. 8:309. 10.3389/fcimb.2018.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Shen C.-C., Challen G.A., and King K.Y.. 2014. Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells. 32:3023–3030. 10.1002/stem.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., and King K.Y.. 2016. Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep. 17:2584–2595. 10.1016/j.celrep.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKelvey, K.J., Hudson A.L., Back M., Eade T., and Diakos C.I.. 2018. Radiation, inflammation and the immune response in cancer. Mamm. Genome. 29:843–865. 10.1007/s00335-018-9777-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, M.D., Ghisi M., Oxley E.P., Ngo S., Cimmino L., Esnault C., Liu R., Salmon J.M., Bell C.C., Ahmed N., et al. 2019. Interconversion between tumorigenic and differentiated states in acute myeloid leukemia. Cell Stem Cell. 25:258–272.e9. 10.1016/j.stem.2019.07.001 [DOI] [PubMed] [Google Scholar]
- Medzhitov, R. 2008. Origin and physiological roles of inflammation. Nature. 454:428–435. 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- Meisel, M., Hinterleitner R., Pacis A., Chen L., Earley Z.M., Mayassi T., Pierre J.F., Ernest J.D., Galipeau H.J., Thuille N., et al. 2018. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 557:580–584. 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitroulis, I., Ruppova K., Wang B., Chen L.S., Grzybek M., Grinenko T., Eugster A., Troullinaki M., Palladini A., Kourtzelis I., et al. 2018. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell. 172:147–161.e12. 10.1016/j.cell.2017.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin, M., Bourke E., Alexander D., Warr M.R., Barry-Holson K., Le Beau M.M., Morrison C.G., and Passegué E.. 2010. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 7:174–185. 10.1016/j.stem.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio, K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M.E., Vasanthakumar A., Patel J., Zhao X., et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 20:11–24. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, B.U., Pabst T., Fos J., Petkovic V., Fey M.F., Asou N., Buergi U., and Tenen D.G.. 2006. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 107:3330–3338. 10.1182/blood-2005-07-3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally, A., Lane S.W., Ball B., Megerdichian C., Okabe R., Al-Shahrour F., Paktinat M., Haydu J.E., Housman E., Lord A.M., et al. 2010. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 17:584–596. 10.1016/j.ccr.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally, A., Bruedigam C., Poveromo L., Heidel F.H., Purdon A., Vu T., Austin R., Heckl D., Breyfogle L.J., Kuhn C.P., et al. 2013. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-α in a murine model of polycythemia vera. Blood. 121:3692–3702. 10.1182/blood-2012-05-432989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, T., Walker C.S., Choi K., Hueneman K., Smith M.A., Gul Z., Garcia-Manero G., Ma A., Zheng Y., and Starczynowski D.T.. 2020. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol. 21:535–545. 10.1038/s41590-020-0663-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, H., Fang G., Tang Y., Xie L., Yang H., Morel L., Diamond B., and Zou Y.R.. 2013. The function of hematopoietic stem cells is altered by both genetic and inflammatory factors in lupus mice. Blood. 121:1986–1994. 10.1182/blood-2012-05-433755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera, N.I., Piredda M.L., Taulli R., Catalano G., Angelini G., Gaur G., Nervi C., Voso M.T., Lunardi A., Pandolfi P.P., and Lo-Coco F.. 2016. PML/RARa inhibits PTEN expression in hematopoietic cells by competing with PU.1 transcriptional activity. Oncotarget. 7:66386–66397. 10.18632/oncotarget.11964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela-Arrieta, C., and Manz M.G.. 2017. Quantification and three-dimensional microanatomical organization of the bone marrow. Blood Adv. 1:407–416. 10.1182/bloodadvances.2016003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oduro, K.A. Jr., Liu F., Tan Q., Kim C.K., Lubman O., Fremont D., Mills J.C., and Choi K.. 2012. Myeloid skewing in murine autoimmune arthritis occurs in hematopoietic stem and primitive progenitor cells. Blood. 120:2203–2213. 10.1182/blood-2011-11-391342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano, M., Bell D.W., Haber D.A., and Li E.. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Pandit, T.S., Sikora L., Muralidhar G., Rao S.P., and Sriramarao P.. 2006. Sustained exposure to nicotine leads to extramedullary hematopoiesis in the spleen. Stem Cells. 24:2373–2381. 10.1634/stemcells.2005-0447 [DOI] [PubMed] [Google Scholar]
- Pang, W.W., Price E.A., Sahoo D., Beerman I., Maloney W.J., Rossi D.J., Schrier S.L., and Weissman I.L.. 2011. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 108:20012–20017. 10.1073/pnas.1116110108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki, H.A., Boumpas D.T., Gibson F.M., Jayne D.R., Axford J.S., Gordon-Smith E.C., Marsh J.C.W., and Eliopoulos G.D.. 2001. Increased apoptosis of bone marrow CD34(+) cells and impaired function of bone marrow stromal cells in patients with systemic lupus erythematosus. Br. J. Haematol. 115:167–174. 10.1046/j.1365-2141.2001.03076.x [DOI] [PubMed] [Google Scholar]
- Papadaki, H.A., Kritikos H.D., Valatas V., Boumpas D.T., and Eliopoulos G.D.. 2002. Anemia of chronic disease in rheumatoid arthritis is associated with increased apoptosis of bone marrow erythroid cells: improvement following anti-tumor necrosis factor-α antibody therapy. Blood. 100:474–482. 10.1182/blood-2002-01-0136 [DOI] [PubMed] [Google Scholar]
- Pascual, V., Allantaz F., Arce E., Punaro M., and Banchereau J.. 2005. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J. Exp. Med. 201:1479–1486. 10.1084/jem.20050473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Lakshminarasimhan R., Techner J.M., Fong S., Flach J., Binnewies M., and Passegué E.. 2014. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211:245–262. 10.1084/jem.20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli, P.D., Casero D., Montecino-Rodriguez E., Morrison S.L., and Dorshkind K.. 2019. Plasma cells are obligate effectors of enhanced myelopoiesis in aging bone marrow. Immunity. 51:351–366.e6. 10.1016/j.immuni.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta, C., Caporali R., Epis O., Ramaioli I., Invernizzi R., Rovati B., Comolli G., Danova M., and Montecucco C.. 2004. Impaired bone marrow hematopoietic progenitor cell function in rheumatoid arthritis patients candidated to autologous hematopoietic stem cell transplantation. Bone Marrow Transplant. 33:721–728. 10.1038/sj.bmt.1704407 [DOI] [PubMed] [Google Scholar]
- Pronk, C.J.H., Veiby O.P., Bryder D., and Jacobsen S.E.W.. 2011. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J. Exp. Med. 208:1563–1570. 10.1084/jem.20110752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrovolaki, K., Mavroudi I., Sidiropoulos P., Eliopoulos A.G., Boumpas D.T., and Papadaki H.A.. 2009. Increased expression of CD40 on bone marrow CD34+ hematopoietic progenitor cells in patients with systemic lupus erythematosus: contribution to Fas-mediated apoptosis. Arthritis Rheum. 60:543–552. 10.1002/art.24257 [DOI] [PubMed] [Google Scholar]
- Rabe, J.L., Hernandez G., Chavez J.S., Mills T.S., Nerlov C., and Pietras E.M.. 2020. CD34 and EPCR coordinately enrich functional murine hematopoietic stem cells under normal and inflammatory conditions. Exp. Hematol. 81:1–15.e6. 10.1016/j.exphem.2019.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, T.N., Hansen N., Stetka J., Luque Paz D., Kalmer M., Hilfiker J., Endele M., Ahmed N., Kubovcakova L., Rybarikova M., et al. 2021. JAK2-V617F and interferon-α induce megakaryocyte-biased stem cells characterized by decreased long-term functionality. Blood. 137:2139–2151. 10.1182/blood.2020005563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan-Komito, D., Swann J.W., Demetriou P., Cohen E.S., Horwood N.J., Sansom S.N., and Griseri T.. 2020. GM-CSF drives dysregulated hematopoietic stem cell activity and pathogenic extramedullary myelopoiesis in experimental spondyloarthritis. Nat. Commun. 11:155. 10.1038/s41467-019-13853-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Moreira, S., Moreno S.G., Ghinatti G., Lewandowski D., Hoffschir F., Ferri F., Gallouet A.S., Gay D., Motohashi H., Yamamoto M., et al. 2017. Low-dose irradiation promotes persistent oxidative stress and decreases self-renewal in hematopoietic stem cells. Cell Rep. 20:3199–3211. 10.1016/j.celrep.2017.09.013 [DOI] [PubMed] [Google Scholar]
- Rozhok, A.I., Salstrom J.L., and DeGregori J.. 2014. Stochastic modeling indicates that aging and somatic evolution in the hematopoetic system are driven by non-cell-autonomous processes. Aging (Albany NY). 6:1033–1048. 10.18632/aging.100707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhok, A.I., Salstrom J.L., and DeGregori J.. 2016. Stochastic modeling reveals an evolutionary mechanism underlying elevated rates of childhood leukemia. Proc. Natl. Acad. Sci. USA. 113:1050–1055. 10.1073/pnas.1509333113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundberg Nilsson, A., Soneji S., Adolfsson S., Bryder D., and Pronk C.J.. 2016. Human and murine hematopoietic stem cell aging is associated with functional impairments and intrinsic megakaryocytic/erythroid bias. PLoS One. 11:e0158369. 10.1371/journal.pone.0158369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallman, D.A., and List A.. 2019. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 133:1039–1048. 10.1182/blood-2018-10-844654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, S., Oshima K., Wang Y., Katanasaka Y., Sano M., and Walsh K.. 2018. a. CRISPR-mediated gene editing to assess the roles of TET2 and DNMT3A in clonal hematopoiesis and cardiovascular disease. Circ. Res. 123:335–341. 10.1161/CIRCRESAHA.118.313225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, S., Oshima K., Wang Y., MacLauchlan S., Katanasaka Y., Sano M., Zuriaga M.A., Yoshiyama M., Goukassian D., Cooper M.A., et al. 2018b. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J. Am. Coll. Cardiol. 71:875–886. 10.1016/j.jacc.2017.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Onai N., Yoshihara H., Arai F., Suda T., and Ohteki T.. 2009. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 15:696–700. 10.1038/nm.1973 [DOI] [PubMed] [Google Scholar]
- Saxton, R.A., and Sabatini D.M.. 2017. mTOR signaling in growth, metabolism, and disease. Cell. 168:960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettpelz, L.G., Borgerding J.N., Christopher M.J., Gopalan P.K., Romine M.P., Herman A.C., Woloszynek J.R., Greenbaum A.M., and Link D.C.. 2014. G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia. 28:1851–1860. 10.1038/leu.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch, C.M., Riether C., and Ochsenbein A.F.. 2014. Cytotoxic CD8+ T cells stimulate hematopoietic progenitors by promoting cytokine release from bone marrow mesenchymal stromal cells. Cell Stem Cell. 14:460–472. 10.1016/j.stem.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Scognamiglio, R., Cabezas-Wallscheid N., Thier M.C., Altamura S., Reyes A., Prendergast Á.M., Baumgärtner D., Carnevalli L.S., Atzberger A., Haas S., et al. 2016. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell. 164:668–680. 10.1016/j.cell.2015.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scumpia, P.O., Kelly-Scumpia K.M., Delano M.J., Weinstein J.S., Cuenca A.G., Al-Quran S., Bovio I., Akira S., Kumagai Y., and Moldawer L.L.. 2010. Cutting edge: bacterial infection induces hematopoietic stem and progenitor cell expansion in the absence of TLR signaling. J. Immunol. 184:2247–2251. 10.4049/jimmunol.0903652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender, R., and Milo R.. 2021. The distribution of cellular turnover in the human body. Nat. Med. 27:45–48. 10.1038/s41591-020-01182-9 [DOI] [PubMed] [Google Scholar]
- Shao, L., Wang Y., Chang J., Luo Y., Meng A., and Zhou D.. 2013. Hematopoietic stem cell senescence and cancer therapy-induced long-term bone marrow injury. Transl. Cancer Res. 2:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W., Vu T., Boucher D., Biernacka A., Nde J., Pandita R.K., Straube J., Boyle G.M., Al-Ejeh F., Nag P., et al. 2017. Ssb1 and Ssb2 cooperate to regulate mouse hematopoietic stem and progenitor cells by resolving replicative stress. Blood. 129:2479–2492. 10.1182/blood-2016-06-725093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., Wei S., Simms K.J., Cumpston D.N., Ewing T.J., and Zhang P.. 2018. Sonic hedgehog signaling regulates hematopoietic stem/progenitor cell activation during the granulopoietic response to systemic bacterial infection. Front. Immunol. 9:349. 10.3389/fimmu.2018.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush, L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. HALT Pan-Leukemia Gene Panel Consortium . 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 506:328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggins, R.W., Hossain F., Rehman T., Melvan J.N., Zhang P., and Welsh D.A.. 2014. Cigarette smoke alters the hematopoietic stem cell niche. Med. Sci. (Basel). 2:37–50. 10.3390/medsci2010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirecki, T., Kawiak J., Machaj E., Pojda Z., Wasilewska D., Czubak J., and Hoser G.. 2015. Early severe impairment of hematopoietic stem and progenitor cells from the bone marrow caused by CLP sepsis and endotoxemia in a humanized mice model. Stem Cell Res. Ther. 6:142. 10.1186/s13287-015-0135-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirecki, T., Mikaszewska-Sokolewicz M., Godlewska M., Dołęgowska B., Czubak J., Hoser G., Kawiak J., and Zielińska-Borkowska U.. 2019. Mobilization of stem and progenitor cells in septic shock patients. Sci. Rep. 9:3289. 10.1038/s41598-019-39772-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.N.P., Zhang Y., Li J.J., McCabe A., Jo H.J., Maloney J., and MacNamara K.C.. 2018. Type I IFNs drive hematopoietic stem and progenitor cell collapse via impaired proliferation and increased RIPK1-dependent cell death during shock-like ehrlichial infection. PLoS Pathog. 14:e1007234. 10.1371/journal.ppat.1007234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber, P.B., Zhang P., Ye M., Welner R.S., Nombela-Arrieta C., Bach C., Kerenyi M., Bartholdy B.A., Zhang H., Alberich-Jordà M., et al. 2013. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell. 49:934–946. 10.1016/j.molcel.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffas, A., Burgos da Silva M., Slingerland A.E., Lazrak A., Bare C.J., Holman C.D., Docampo M.D., Shono Y., Durham B., Pickard A.J., et al. 2018. Nutritional support from the intestinal microbiota improves hematopoietic reconstitution after bone marrow transplantation in mice. Cell Host Microbe. 23:447–457.e4. 10.1016/j.chom.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetka, J., Vyhlidalova P., Lanikova L., Koralkova P., Gursky J., Hlusi A., Flodr P., Hubackova S., Bartek J., Hodny Z., and Divoky V.. 2019. Addiction to DUSP1 protects JAK2V617F-driven polycythemia vera progenitors against inflammatory stress and DNA damage, allowing chronic proliferation. Oncogene. 38:5627–5642. 10.1038/s41388-019-0813-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., and Camargo F.D.. 2014. Clonal dynamics of native haematopoiesis. Nature. 514:322–327. 10.1038/nature13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, T., Yamamura S., Kuwano Y., and Abo T.. 1996. Level of myelopoiesis in the bone marrow is influenced by intestinal flora. Cell. Immunol. 173:155–161. 10.1006/cimm.1996.0261 [DOI] [PubMed] [Google Scholar]
- Takizawa, H., Regoes R.R., Boddupalli C.S., Bonhoeffer S., and Manz M.G.. 2011. Dynamic variation in cycling of hematopoietic stem cells in steady state and inflammation. J. Exp. Med. 208:273–284. 10.1084/jem.20101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, H., Boettcher S., and Manz M.G.. 2012. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 119:2991–3002. 10.1182/blood-2011-12-380113 [DOI] [PubMed] [Google Scholar]
- Takizawa, H., Fritsch K., Kovtonyuk L.V., Saito Y., Yakkala C., Jacobs K., Ahuja A.K., Lopes M., Hausmann A., Hardt W.D., et al. 2017. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell. 21:225–240.e5. 10.1016/j.stem.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Talhout, R., Schulz T., Florek E., van Benthem J., Wester P., and Opperhuizen A.. 2011. Hazardous compounds in tobacco smoke. Int. J. Environ. Res. Public Health. 8:613–628. 10.3390/ijerph8020613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraldsrud, E., Grøgaard H.K., Solheim S., Lunde K., Fløisand Y., Arnesen H., Seljeflot I., and Egeland T.. 2009. Age and stress related phenotypical changes in bone marrow CD34+ cells. Scand. J. Clin. Lab. Invest. 69:79–84. 10.1080/00365510802419447 [DOI] [PubMed] [Google Scholar]
- Trumpp, A., Essers M., and Wilson A.. 2010. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 10:201–209. 10.1038/nri2726 [DOI] [PubMed] [Google Scholar]
- Ueda, Y., Cain D.W., Kuraoka M., Kondo M., and Kelsoe G.. 2009. IL-1R type I-dependent hemopoietic stem cell proliferation is necessary for inflammatory granulopoiesis and reactive neutrophilia. J. Immunol. 182:6477–6484. 10.4049/jimmunol.0803961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainieri, M.L., Blagborough A.M., MacLean A.L., Haltalli M.L.R., Ruivo N., Fletcher H.A., Stumpf M.P.H., Sinden R.E., and Celso C.L.. 2016. Systematic tracking of altered haematopoiesis during sporozoite-mediated malaria development reveals multiple response points. Open Biol. 6:160038. 10.1098/rsob.160038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valletta, S., Thomas A., Meng Y., Ren X., Drissen R., Sengül H., Di Genua C., and Nerlov C.. 2020. Micro-environmental sensing by bone marrow stroma identifies IL-6 and TGFβ1 as regulators of hematopoietic ageing. Nat. Commun. 11:4075. 10.1038/s41467-020-17942-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer, J.W.M., Barza M., Wolff S.M., and Dinarello C.A.. 1988. A low dose of recombinant interleukin 1 protects granulocytopenic mice from lethal gram-negative infection. Proc. Natl. Acad. Sci. USA. 85:1620–1623. 10.1073/pnas.85.5.1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangala, R.K., Heiss-Neumann M.S., Rangatia J.S., Singh S.M., Schoch C., Tenen D.G., Hiddemann W., and Behre G.. 2003. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 101:270–277. 10.1182/blood-2002-04-1288 [DOI] [PubMed] [Google Scholar]
- Vasto, S., Candore G., Balistreri C.R., Caruso M., Colonna-Romano G., Grimaldi M.P., Listi F., Nuzzo D., Lio D., and Caruso C.. 2007. Inflammatory networks in ageing, age-related diseases and longevity. Mech. Ageing Dev. 128:83–91. 10.1016/j.mad.2006.11.015 [DOI] [PubMed] [Google Scholar]
- Vyas, D., Laput G., and Vyas A.K.. 2014. Chemotherapy-enhanced inflammation may lead to the failure of therapy and metastasis. OncoTargets Ther. 7:1015–1023. 10.2147/OTT.S60114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., et al. 2015. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 520:549–552. 10.1038/nature14131 [DOI] [PubMed] [Google Scholar]
- Wang, W., Liu W., Fidler T., Wang Y., Tang Y., Woods B., Welch C., Cai B., Silvestre-Roig C., Ai D., et al. 2018. Macrophage inflammation, erythrophagocytosis, and accelerated atherosclerosis in JAK2V617F mice. Circ. Res. 123:e35–e47. 10.1161/CIRCRESAHA.118.313283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Sano S., Yura Y., Ke Z., Sano M., Oshima K., Ogawa H., Horitani K., Min K.D., Miura-Yura E., et al. 2020. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight. 5:e135204. 10.1172/jci.insight.135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Wang J., Li Y.H., Wang L., Shang H.C., and Wang J.X.. 2021. Phenotypical changes of hematopoietic stem and progenitor cells in sepsis patients: correlation with immune status? Front. Pharmacol. 11:640203. 10.3389/fphar.2020.640203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser, M., Demel U.M., Stein S., Chen-Wichmann L., Touzot F., Santilli G., Sujer S., Brendel C., Siler U., Cavazzana M., et al. 2016. Hyperinflammation in patients with chronic granulomatous disease leads to impairment of hematopoietic stem cell functions. J. Allergy Clin. Immunol. 138:219–228.e9. 10.1016/j.jaci.2015.11.028 [DOI] [PubMed] [Google Scholar]
- Wilson, A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., and Trumpp A.. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763. 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129. 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Woolthuis, C.M., Mariani N., Verkaik-Schakel R.N., Brouwers-Vos A.Z., Schuringa J.J., Vellenga E., de Wolf J.T.M., and Huls G.. 2014. Aging impairs long-term hematopoietic regeneration after autologous stem cell transplantation. Biol. Blood Marrow Transplant. 20:865–871. 10.1016/j.bbmt.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Yamashita, M., and Passegué E.. 2019. TNF-α Coordinates Hematopoietic Stem Cell Survival and Myeloid Regeneration. Cell Stem Cell. 25:357–372.e7. 10.1016/j.stem.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Liang H., Yan J.S., Tao R., Hao S.G., and Ma L.Y.. 2012. Down-regulation of hematopoiesis master regulator PU.1 via aberrant methylation in chronic myeloid leukemia. Int. J. Hematol. 96:65–73. 10.1007/s12185-012-1106-x [DOI] [PubMed] [Google Scholar]
- Zhang, X., Sejas D.P., Qiu Y., Williams D.A., and Pang Q.. 2007. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J. Cell Sci. 120:1572–1583. 10.1242/jcs.003152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Yin L., Zhang K., Sun W., Yang S., Zhang B., Salzman P., Wang W., Liu C., Vidyasagar S., et al. 2012. Response patterns of cytokines/chemokines in two murine strains after irradiation. Cytokine. 58:169–177. 10.1016/j.cyto.2011.12.023 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Zhao K., Shen Q., Han Y., Gu Y., Li X., Zhao D., Liu Y., Wang C., Zhang X., et al. 2015. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 525:389–393. 10.1038/nature15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.R.C., Nix D., Gregory M., Ciorba M.A., Ostrander E.L., Newberry R.D., Spencer D.H., and Challen G.A.. 2019. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp. Hematol. 80:36–41.e3. 10.1016/j.exphem.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Tao F., Venkatraman A., Li Z., Smith S.E., Unruh J., Chen S., Ward C., Qian P., Perry J.M., et al. 2019. N-cadherin-expressing bone and marrow stromal progenitor cells maintain reserve hematopoietic stem cells. Cell Rep. 26:652–669.e6. 10.1016/j.celrep.2018.12.093 [DOI] [PMC free article] [PubMed] [Google Scholar]