Abstract
The development of the TMTpro-16plex series expanded the breadth of commercial isobaric tagging reagents by nearly 50% over classic TMT-11plex. In addition to the described 16plex reagents, the proline-based TMTpro molecule can accommodate two additional combinations of heavy carbon and nitrogen isotopes. Here, we introduce the final two labeling reagents, TMTpro-134C and TMTpro-135N, which permit the simultaneous global protein profiling of 18 samples with essentially no missing values. For example, six conditions with three biological replicates can now be perfectly accommodated. We showcase the 18plex reagent set by profiling the proteome and phosphoproteome of a pair of isogenic mammary epithelial cell lines under three conditions in triplicate. We compare the depth and quantitative performance of this data set with a TMTpro-16plex experiment in which two samples were omitted. Our analysis revealed similar numbers of quantified peptides and proteins, with high quantitative correlation. We interrogated further the TMTpro-18plex data set by highlighting changes in protein abundance profiles under different conditions in the isogenic cell lines. We conclude that TMTpro-18plex further expands the sample multiplexing landscape, allowing for complex and innovative experimental designs.
Keywords: TMTpro, 18plex, real-time search, eclipse, PTEN, MCF10A, KIN-193, BYL-719
Graphical Abstract

INTRODUCTION
Isobaric tagging reagents, such as tandem mass tags (TMT), permit the multiplexed sample profiling of protein abundance using mass spectrometry-based proteomic techniques.1 These tags consist of an amine reactive group, as well as a variable mass, isotope-encoded reporter ion group and a corresponding mass normalization group. The structure and nominal mass of each amine-reactive TMT reagent are identical. So, labeled peptides co-elute chromatographically and appear as one peak at the MS1 stage, while their fragmentation during an MS2 or MS3 event generates reporter ion peaks of different masses, which are used for quantification across samples.1 Sample multiplexing reduces data acquisition time and decreases missing values across experiments. As such, a key advancement in the technology is the increase in multiplexing capacity.
The feasibility to distribute heavy isotopes across the reporter ion and mass normalization groups sets the limit for the final multiplexing capability of these isobaric labels. The TMTpro-16plex isobaric reagent set has been released.2,3 However, it was noted that two additional reagents (TMTpro-134C and TMTpro-135N) could be synthesized.2 These reagents would not technically be isobaric with the other 16 and would require further investigation into their potential use alongside the 16plex reagents. While their molecular structure would be identical to that of the TMTpro-16plex set (and labeled peptides co-elute by reversed-phase chromatography), the two additional reagents would have one less 15N atom and one more 13C atom per tag, resulting in a tiny net mass shift of +6 mDa (0.0063 Da). This shift is the same as the mass difference between TMT-131C and TMT-10plex reagents.
PTEN (phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase) is a dual specificity phosphatase that dephosphorylates protein and phosphoinositide substrates and functions as a tumor suppressor by negatively regulating the PI3K signaling pathway.4 Specifically, PTEN dephosphorylates phosphatidylinositol(3,4,5)-trisphosphate (PIP3) which results in the biphosphate product PIP2. This mechanism inhibits multiple signaling outputs, including most notably the AKT signaling pathway that regulates cellular processes including cell growth, apoptosis, and migration.5 Moreover, PTEN is altered in a wide range of human cancers, including, prostate cancer, glioblastoma, endometrial cancer, and breast cancer.6,7
Here, we have evaluated the feasibility of including the 17th and 18th TMTpro reagents in sample multiplexing studies. Six conditions with three replicates are perfectly accommodated. To test this, we compared the mammary epithelial cell line MCF10A with and without a biallelic PTEN gene deletion for protein expression and phosphorylation changes after three treatments [KIN-193, BYL-719, or dimethyl sulfoxide (DMSO)]. We show that the 6 mDa mass difference is negligible for routine global proteome and phosphoproteome profiling and fully compatible with existing streamlined (SL)-TMT8 workflows. We conclude that TMTpro-134C and TMTpro-135N can be seamlessly integrated into the current TMTpro isobaric series to expand further the breadth of isobaric tagging experiments.
METHODS
Cell Culture and Drug Treatment
MCF10A-sgAAVS1 (control MCF10A) and MCF10A-sgPTEN (MCF10A PTEN−/−) cell lines were gifts from Yubao Wang (Dana Farber Cancer Institute, Boston, MA). The parental MCF10A used to create the CRISPR-Cas9 knock out cell lines was purchased from ATCC (CRL-10317). The sgRNA sequences targeting adeno-associated virus integration site 1 (AAVS1) and human PTEN are sgAAVS1: 5′-GTCCCCTCCACCCCACAGTG-3′ and sgPTEN: 5′-TCATCTGGATTATAGACCAG-3′. We used the sgRNA targeting AAVS1 as negative control because it causes no phenotypic changes.9,10
Cells were counted with the Countess Automated Cell Counter (C10281, Invitrogen) and an equal number of control MCF10A and MCF10A PTEN−/− cells were seeded into 10 cm dishes to approximately 70% confluence in complete medium [DMEM/F12+GlutaMAX-1 (10565-018, GIBCO) supplemented with 5% horse serum (16050-114, GIBCO), 20 ng/mL epidermal growth factor (E9644, Sigma-Aldrich), 10 μg/mL insulin (12585-014, GIBCO), 0.5 μg/mL hydrocortisone (H0888, Sigma-Aldrich), 0.1 μg/mL cholera toxin (C8052, Sigma-Aldrich), and 1% penicillin–streptomycin (15140, GIBCO)]. After 16 h, complete medium was removed and cells were washed with phosphate-buffered saline (PBS) once, then starvation medium [DMEM/F12+GlutaMAX-1 (10565-018, GIBCO)] supplemented with 0.5 μg/mL hydrocortisone (H0888, Sigma-Aldrich), 0.1 μg/mL cholera toxin (C8052, Sigma-Aldrich) and 1% penicillin–streptomycin (15140, GIBCO) was added. After 24 h starvation, BYL-719 (CAS 1217486-61-7, MedChemexpress) or KIN-193 (CAS 1173900-33-8, MedChemexpress) was added at a final concentration of 1 μM for 24 h. An equal volume of DMSO was added to control groups for 24 h. After treatment, cells were washed thrice with PBS, then harvested and lysed by scraping into 8 M urea lysis buffer (8 M urea, 200 mM EPPS, pH 8.5) with protease (11836170001, Roche) and phosphatase inhibitors (04906837001, Roche). Cell extracts were syringe-lysed and sonicated. Protein concentration was determined with BCA assays.
Western Blotting
Proteins were loaded onto a 10% SDS-PAGE gel and separated using Tris-glycine SDS-PAGE running buffer. PageRuler Plus pre-stained protein ladder (26619, ThermoFisher Scientific) was used to indicate the molecular weight of target proteins. After electrophoresis, proteins were transferred to 0.45 μM nitrocellulose membranes (1620115, BioRad) with Towbin transfer buffer. Membranes were blocked with 5% bovine serum albumin and then incubated with corresponding primary antibodies. Membranes were then washed with Tris-buffered saline with 0.5% Tween 20 and incubated with IRDye secondary antibodies. Membranes were scanned and images were collected on an Odyssey CLX (LICOR) imaging system. Membranes were stripped with ReBlot Plus strong antibody stripping solution (2504, Millipore) for the detection of other targets, if necessary. Anti-IRS1 (2390), anti-IRS2 (3089), and anti-PTEN (9559) antibodies were purchased from Cell Signaling Technology. Anti-β-actin (A5441) and anti-Vinculin (V9131) antibodies were purchased from Sigma-Aldrich.
Mass Spectrometry Sample Preparation
Samples were reduced with 5 mM TCEP for 30 min, alkylated with 10 mM iodoacetamide for 30 min in the dark and then quenched with 10 mM DTT for 15 min at room temperature. The streamlined (SL)-TMT protocol was used for sample preparation.8 The SL-TMT protocol requires minimal individual sample processing and permits the seamless addition of a phosphopeptide enrichment step (“mini-phos”) with little deviation from the standard TMT sample preparation or the deep proteome analysis.8 Briefly, 100 μg of protein lysate were chloroform–methanol-precipitated and reconstituted in 100 μL of 200 mM EPPS (pH 8.5). The samples were digested by Lys-C overnight at room temperature and then with trypsin for 6 h at 37 °C, both at a 1:100 protease-to-protein ratio.
Peptides corresponding to 50 μg of protein were labeled with TMTpro reagents in the presence of 29% acetonitrile for 90 min at room temperature (see Figure 2A for the experimental layout). Two microliters of each sample were pooled, desalted, and analyzed on a Q Exactive mass spectrometer (ThermoFisher Scientific) to check labeling efficiency. After labeling efficiency was verified, samples were quenched by adding 5% hydroxylamine and pooled. Pooled samples were then desalted with 100 mg Sep-Pak solid-phase extraction columns. Pierce High-Select Fe-NTA phosphopeptide enrichment kit (A32992, ThermoFisher Scientific) was used to enrich phosphopeptides from the pooled samples following manufacturer’s instruction in a “mini-phos” format.8 Unbound fractions were desalted and then fractionated with basic-pH reversed-phase chromatography. Fractions were collected in a 96-well plate and combined for a total of 24 fractions prior to desalting and subsequent LC–MS/MS analysis.
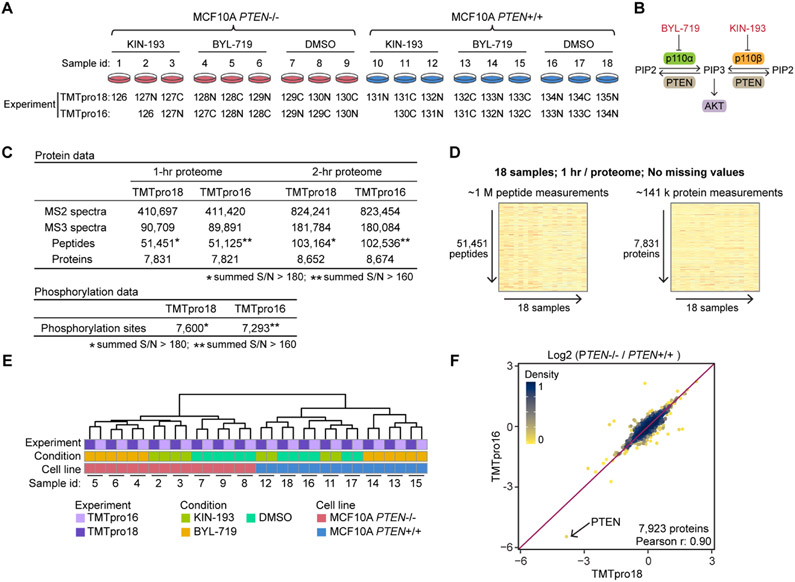
Figure 2.
TMTpro-18plex reagents facilitate proteome analysis at an effective rate of 1 h per proteome. (A) Set of 18 samples of drug-treated control MCF10A and MCF10A PTEN−/− cell lines was used to benchmark the TMTpro-18plex reagents. The cell lines were treated with a PI3K inhibitor (BYL-719 or KIN-193) or DMSO, in biological triplicate (n = 3). Samples were prepared following the SL-TMT protocol using 50 μg protein and labeling with either the full 18 reagents or only 16 reagents as indicated. Peptides were fractionated and then concatenated into 24 “super” fractions. Fractionated samples were analyzed with FAIMS and real-time search-synchronous-precursor-selection-MS3 on an Orbitrap Eclipse mass spectrometer. An equal amount of fractionated peptide was analyzed in both experiments. (B) PI3K overview. KIN-193 and BYL-719 inhibit the kinases p110α and p110β, respectively. p110α and p110β are catalytic subunits of PI3K. PI3K-mediated PIP3 production leads to the activation of the downstream AKT/mTOR pathway. (C) Data set overview. Analysis of 12 fractions (1 h per proteome) or 24 fractions (2 h per proteome) yielded ~7.8k and ~8.6k quantified proteins in both experiments, respectively. A total amount of 900 and 800 μg of peptide was used to enrich phosphorylated peptides in the 18- and 16-plex experiments, respectively. The numbers of localized and quantified phosphorylation sites are reported (AScore > 13). (D) 1 h human cell line proteome. The TMTpro-18plex reagents facilitated the collection of a 1 h per proteome analysis that included ~1 million peptide measurements and ~141,000 protein measurements across 18 samples without missing values. (E) HCA of the protein data; (F) log 2 protein fold changes between the two cell lines treated with DMSO in both experiments showed good agreement.
Mass Spectrometry Analysis
Data were collected on an Orbitrap Eclipse mass spectrometer (ThermoFisher Scientific) coupled to a Proxeon EASY-nLC 1200 LC pump (ThermoFisher Scientific). Fractionated peptides were separated using a 90 min gradient at 525 nL/min on a 35 cm column (i.d. 100 μm, Accucore, 2.6 μm, 150 Å) packed in-house. High-field asymmetric-waveform ion mobility spectrometry (FAIMS) was enabled during data acquisition with compensation voltages set as −40, −60, and −80 V.11 MS1 data were collected in the Orbitrap (60,000 resolution; maximum injection time 50 ms; AGC 4 × 105). Charge states between 2 and 5 were required for MS2 analysis, and a 120 s dynamic exclusion window was used. Cycle time was set at 1.2 s. MS2 scans were performed in the ion trap with CID fragmentation (isolation window 0.5 Da; Turbo; NCE 35%; maximum injection time 35 ms; AGC 1 × 104). An on-line real-time search algorithm (Orbiter) was used to trigger MS3 scans for quantification.12 MS3 scans were collected in the Orbitrap using a resolution of 50,000, NCE of 45%, maximum injection time of 150 ms, and AGC of 1.5 × 105. The close out was set at two peptides per protein per fraction.12
Phosphorylated peptides were separated using a 150 min gradient at 520 nL/min on a 35 cm column (i.d. 100 μm, Accucore, 2.6 μm, 150 Å) packed in-house. FAIMS was enabled during data acquisition with compensation voltages set as −40, −60, and −80 V for the first shot and −45 and −65 V for the second shot.11 MS1 data were collected in the Orbitrap (120,000 resolution; maximum injection time 50 ms; AGC 4 × 105). Charge states between 2 and 5 were required for MS2 analysis, and a 90 s dynamic exclusion window was used. Cycle time was set at 1.25 s. MS2 scans were performed in the Orbitrap with HCD fragmentation (isolation window 0.5 Da; 50,000 resolution; NCE 36%; maximum injection time 250 ms; AGC 1.5 × 105).
Mass Spectrometry Data Analysis
Raw files were converted to mzXML, and monoisotopic peaks were re-assigned using Monocle.13 The protein database included all human entries from Uniprot (downloaded on 02/15/2020). The database was concatenated with one composed of all protein sequences in the reversed order. Common contaminant proteins (e.g., trypsin and keratins) were appended as well. Searches were performed using the comet search algorithm. We used a 50 ppm precursor ion tolerance and 0.9 Da product ion tolerance for MS2 scans collected in the ion trap and 0.02 Da product ion tolerance for MS2 scans collected in the Orbitrap. TMTpro on lysine residues and peptide N-termini (+304.2071 Da) and carbamidomethylation of cysteine residues (+57.0215 Da) were set as static modifications (except when testing for labeling efficiency, in which the TMTpro modification was set to variable), while oxidation of methionine residues (+15.9949 Da) was set as a variable modification. For phosphorylated peptide analysis, +79.9663 Da was set as a variable modification on serine, threonine, and tyrosine residues.
Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR).14 PSM filtering was performed using linear discriminant analysis (LDA) as described previously,15 while considering the following parameters: comet log expect, different sequence delta comet log expect (percent difference between the first hit and the next hit with a different peptide sequence), missed cleavages, peptide length, charge state, precursor mass accuracy, and fraction of ions matched. Only phosphorylated peptides were considered in LDA for phosphorylated peptide analysis. Each run was filtered separately. Protein-level FDR was subsequently estimated. The posterior probabilities reported by the LDA model for each peptide were multiplied to give a protein-level probability estimate. Proteins were filtered to the target 1% FDR level across the entire combined data set using the picked FDR method.16
Phosphorylation site localization was determined using the AScore algorithm.17 Phosphorylation sites with an AScore higher than 13 were reported. The reporter ion masses for TMTpro-134C (134.154557) and TMTpro-135N (135.151592) were added to the list of TMTpro-16plex reporter ions. For reporter ion quantification, a 0.003 Da window around the theoretical m/z of each reporter ion was scanned, and the most intense m/z was used. Isotopic impurities of TMTpro-134C and TMTpro-135N (Table S4) were appended to the table of TMTpro-16plex isotopic impurities (ThermoFisher, product number: A44520R, lot number: VI313212). Reporter ion intensities were adjusted to correct for the isotopic impurities of the different TMTpro reagents according to manufacturer specifications. Peptides were filtered to include only those with a summed signal-to-noise (SN) ≥ 160 across 16 TMTpro channels or ≥180 across 18 TMTpro channels. The SN cutoff (set at an average of greater than 10 SN units per channel) can remove peptide measurements having high variability.18 An extra filter of an isolation specificity (“isolation purity”) of at least 0.7 in the MS1 isolation window was applied for the phosphorylated peptide analysis. For each protein or phosphorylation site, the filtered peptide TMTpro SN values were summed to generate protein or phosphorylation site quantification values. To control for different total protein loading within a TMTpro experiment, the summed protein quantities of each channel were adjusted to be equal within the experiment. Phosphorylation site quantities of each channel were also adjusted with respective normalization factors generated in the protein data normalization process. For each protein or phosphorylation site within a TMTpro experiment, the SN was scaled to sum to 100 for subsequent analyses unless stated otherwise.
Bioinformatics Analysis
Protein data from the analysis of 24 fractions (2 h proteome) were used for the comparison between TMTpro16 and TMTpro18 experiments (Figures 2 and S2), as well as the biological analysis in Figures 3 and S3. Hierarchical clustering analysis (HCA) was performed with the R package pheatmap (distance measure: Euclidean distance; clustering method: complete). Only samples in both TMTpro16 and TMTpro18 experiments were included in HCA. For each protein within the TMTpro18 experiment, the SN of the 16 samples in both experiments was scaled to sum to 100 to compare with the TMTpro16 experiment in HCA. Gene ontology enrichment analysis was performed with DAVID (6.8).19 Significance analysis of microarray (SAM) was performed with R package samr.20 Significant proteins were filtered at 1% FDR. Then, an additional fold change cutoff of 1.4 was applied.
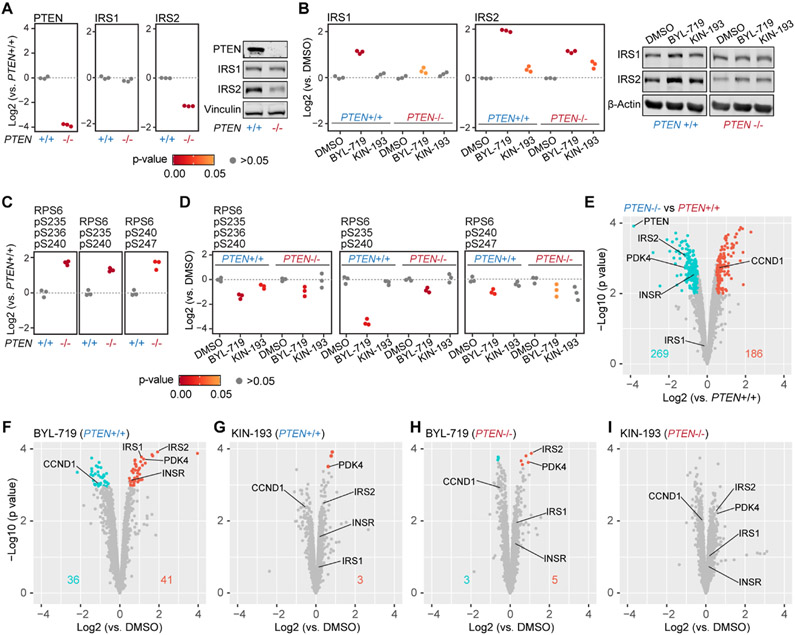
Figure 3.
Overview of alterations in protein abundance and phosphorylation levels in control and PTEN−/− cells upon treatment with two PI3K inhibitors. (A) Absence of PTEN and down-regulation of IRS2 in MCF10A PTEN−/− cells, and the equivalent expression level of IRS1 in both cell lines were recapitulated by the mass spectrometry data. Welch’s t-test p-values are shown as different colored dots to provide an estimate of the significance. (B) IRS1 and IRS2 were upregulated after BYL-719 treatment in both cell lines. IRS2 was also up-regulated under KIN-193 treatment in both cell lines. (C,D) Two doubly-phosphorylated RPS6 peptides and one triply phosphorylated RPS6 peptide were quantified. These phosphorylation events were upregulated in MCF10A PTEN−/− cells and were inhibited by BYL-719 and KIN-193 to a different extent in both cell lines. (E) Volcano plot showing protein expression level differences between control and PTEN−/− cells. SAM was used to evaluate up- or down-regulated proteins. Negative log 10-transformed estimated p-values for each protein from the SAM analysis are shown on the y-axis. Significant proteins were filtered at 1% FDR and a minimum of 40% change. Numbers on the plot indicate significantly up-regulated (red) and down-regulated (cyan) proteins in MCF10A PTEN−/− cells. (F–I) Volcano plots showing the protein level comparison between DMSO and 24 h drug treatment in both cell lines. Numbers on the plot indicate significantly up-regulated (red) and down-regulated (cyan) proteins. Inhibition of p110β by KIN-193 caused only minor protein perturbations compared with the inhibition of p110α by BYL-719 in both cell lines. PTEN knockout conferred p110α inhibitor BYL-719 resistance, as protein level changes by BYL-719 were not changed in the PTEN knockout.
Data Availability
The mass spectrometry data have been deposited in the ProteomeXchange Consortium with the data set identifier PXD024275.
RESULTS AND DISCUSSION
TMTpro-134C and TMTpro-135N Complete the Set of 18 Heavy Isotope-Labeled TMTpro Tags
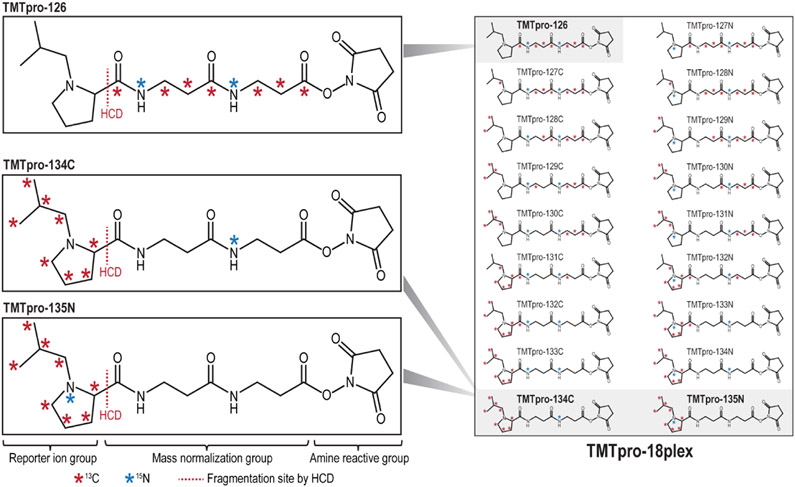
The commercial release of TMTpro-16plex offered unprecedented sample multiplexing capacity integrated into the SL-TMT workflow. The expansion to 16 samples represented a nearly 50% increase in multiplexing capability over the original TMT-6/10/11plex reagents. Higher multiplexing capability reduces data acquisition time and results in fewer missing values across experiments, thereby increasing efficiency without loss in quantitative performance. The unreleased TMTpro reagents (TMTpro-134C and TMTpro-135N) have a molecular structure identical to the TMTpro-16plex set but differ in their number of 13C and 15N heavy isotopes (Figure 1). Specifically, the isobaric TMTpro-16plex molecular composition included seven 13C and two 15N heavy isotopes distributed across the reporter ion and mass normalization region of each reagent, while the two additional reagents had eight 13C and only one 15N heavy isotopes. As such, the masses of the additional two tags were increased by 6 mDa.
Figure 1.
Chemical structures and heavy isotope positions of the TMTpro-134C and TMTpro-135N reagents. The full set of TMTpro-18plex reagents is shown on the right. The TMTpro-134C and TMTpro-135N reagents have the same chemical structure as isobaric TMTpro-16plex reagents (TMTpro-126 is shown as an example). However, the two new reagents differ from the TMTpro-16plex reagents in the number of 13C and 15N atoms. The TMTpro-16plex reagents incorporate seven 13C and two 15N atoms, while the two additional reagents incorporate eight 13C and only one 15N atoms that are distributed across the reporter ion and mass normalization region of each reagent. This arrangement enables the complete utilization of the positions in the reporter ion group, creating the 17th and 18th channels in the TMTpro-18plex reagent set.
We posited that the 6 mDa deviation of the TMTpro tag would not hinder peptide identification or quantification. With respect to identification, the resolution necessary to identify a 6 mDa shift is >133,000 at 400 m/z for a doubly-charged ion. In a routine multiplexed isobaric tag-based whole proteome analysis, the typical resolution setting for the MS1 scan is generally 60,000 or 120,000. As tryptic peptides have charge states of generally 2+ or higher, it is conceivable that the 6 mDa difference between tag isotopologues was beyond the actual resolution value at a given peptide’s m/z and thus negligible. To support this claim, we created a multiple experiment acquisition method in which we analyzed a sample labeled with all 18 TMTpro reagents. The unfractionated sample was arranged as the TMTpro18 experiment (as in Figure 2A). We used MS1 resolution settings of 30,000, 60,000, 120,000, and 500,000 and collected the data in profile mode (i.e., without centroiding). We selected one doubly-charged peptide (ALVILAK) from the whole proteome mixture and extracted the ion peaks for all resolution settings used. We noted that a single peak was observed at resolution setting of 30,000 (Figure S1A), 60,000 (Figure S1B), and 120,000 (Figure S1C). On certain peptides, such as a doubly-charged lysine-terminating peptide (which binds up two TMTpro molecules, as in the ALVILAK peptide), two peaks could be observed (depending on coalescence) using the resolution setting of 500,000 (Figure S1D). However, the resulting mass shift is small and should not affect protein identification for typical operating conditions and data analysis workflows.
Similarly, we anticipated that the two additional labels would not adversely impact quantitative performance. TMTpro reporter ions are quantified typically using either high-resolution MS2 or SPS-MS3 methods. The minimum isolation window setting for either scan type is 0.4 Th, which is well above the 6 mDa difference regardless of peptide mass. Moreover, the 6 mDa mass difference is located on the mass normalizer region and not on the reporter ion, so no deviation of reporter ion measurement is expected. As such, we anticipate no distortion of relative protein abundance ratios with the addition of the TMTpro-134C and TMTpro-135N reagents to the isobaric TMTpro-16plex reagent set. We next set out to examine the performance using full data sets which included the new 17th and 18th labels of the TMTpro reagent set.
Data set Depth and Quantitative Precision of TMTpro-18plex Mirrors that of TMTpro-16plex
We showcase the TMTpro-18plex reagents by investigating the effects of two PI3K inhibitor treatments (KIN-193 and BYL-719) and DMSO on two mammary epithelial cell lines (control MCF10A and MCF10A PTEN−/−) (Figure 2A). BYL-719 and KIN-193 preferentially inhibit the kinases p110α and p110β, respectively. p110α and p110β are catalytic subunits of PI3K that convert PIP2 to PIP3. PI3K-mediated PIP3 production promotes the activation of the downstream AKT/mTOR pathway.6,7 These kinases act in opposition to the action of PTEN that dephosphorylates PIP3 and converts it to PIP2 (Figure 2B). We assembled an 18plex experiment to compare the global proteomic and phosphoproteomic alterations resulting from the treatment of control MCF10A and isogenic PTEN−/− cell lines. Six total conditions were investigated in biological triplicate, which included each cell line treated with each drug and DMSO, for a total of 18 samples. We also assembled a 16plex experiment for comparison of depth and quantitative performance. For the 16plex experiment, we omitted one treatment of KIN-193 from each cell line (Figure 2A). We staggered the isobaric labels such that no triplicates across the two multiplexed experiments had identical labels. Pooled samples were fractionated, and each concatenated super-fraction was analyzed during a 90 min gradient on a FAIMS-equipped11,21 Orbitrap Eclipse mass spectrometer22 using real-time database searching.12,23 Each super-fraction was concatenated from the fractionation of the pooled samples into a 96-well plate (collecting every 12th sample).24 We assembled the data as both 1- and 2 h proteome data sets for the TMTpro18 experiment. The 1 h proteome consisted of 12 non-adjacent super-fractions (1.5 h analyses × 12 fractions/18 samples = 1 h per proteome), while the 2 h proteome consisted of those 12 super-fractions plus the remaining 12 super-fractions (1.5 h × 24 fractions/18 samples = 2 h per proteome).
We first evaluated overall proteome depth between the 16plex and 18plex experiments. Proteome depth was remarkably similar between the two data sets (Figure 2C). For the 1 h proteome analysis, we collected ~410,000 MS2 and triggered ~90,000 MS3 spectra for quantification. In total, ~7,800 proteins were quantified from ~50,000 quantified peptides. Note that across the 18 samples for the 1 h per proteome, essentially no missing values were present. The peptide quantitative matrix was made up of nearly 1 million abundance measurements which were combined to ~141,000 protein measurements (Figure 2D). As expected, proteome depth increased when all 24 fractions were analyzed for the 2 h per proteome data set. More than 8,600 proteins were present in the final quantitative matrix. To emphasize further the similarities in depth between 16 and 18plex workflows, we compared the number of MS2 spectra, peptide-spectrum-matches, MS3 spectra, peptides, and proteins, as well as success rate (MS2 spectra matched/acquired), for each fraction (Figure S2A). The median values for each of these parameters were similar.
To evaluate if the new reagents caused any bias in quantification, we performed unsupervised hierarchical clustering on the TMTpro reporter ion relative abundance values (Figure 2E). We observed that pairs of samples originating from the same cell line and receiving the same treatment clustered despite using different TMTpro labels (the labels were staggered, as illustrated in Figure 2A). The samples were further grouped by treatment and cell line. This clustering analysis indicated that the quantitative performance of the TMTpro-134C and TMTpro-135N was consistent with the other 16 reagents. Finally, we noted that the some of the KIN-193-treated samples clustered closely with the DMSO treatment. This was likely a result of KIN-193 treatment not triggering marked protein abundance alterations (discussed later in Figure 3).
Next, we examined the precision of replicate measurements using the coefficient of variation (CV) as a measure of the reproducibility. We found that overall, the median of measured %CV for all conditions was ~6% in the protein data, showing very low variance across replicates regardless of experiment (Figure S2B). Of particular interest were the DMSO channels of the control MCF10A cells. As we staggered the labels (Figure 2A), the last three channels from the TMTpro16 experiment were labeled with the TMTpro-133N, TMTpro-133C, and TMTpro-134N reagents, whereas those for the TMTpro18 experiment were labeled with the TMTpro-134N, TMTpro-134C, and TMTpro-135N reagents. As such, the last triplicate condition of the TMTpro18 experiment incorporated the two new labels. We noted no difference in the %CV for this condition (Figure S2C). This finding supported further our claim that these two additional labels were virtually indistinguishable from the 16 isobaric TMTpro labels when profiling protein abundance with the SL-TMT protocol. Similarly low variance across replicates was also observed in the phosphorylation data (Figure S2D,E).
Finally, we examined the correlation of the log2 fold changes in protein relative abundance values of the MCF10A PTEN−/− versus control cells for the two data sets (n = 7,923). We focused on the differences between the two cell lines treated with DMSO, as the TMTpro channels for the control MCF10A cells treated with DMSO were labeled with the TMTpro-134N reagent along with the two new tags. We plotted the log 2 fold change of the average reporter ion relative abundance of the three replicates from each data set (Figure 2F). The data were strongly correlated between the TMTpro18 and TMTpro16 data sets with a Pearson coefficient of 0.9. In all, our data revealed consistency in both the proteome depth and the quantitative performance with the addition of the TMTpro-134C and TMTpro-135N reagents as the 17th and 18th channels to the isobaric TMTpro16-plex reagent set.
Along with the whole proteome analysis, we performed a “mini-phos” analysis, as outlined in the SL-TMT protocol.8 Here, data were collected with two consecutive analyses each for 2.5 h. The analyses differed from each other with respect to the FAIMS compensation voltages (CV) (one method with CV = −45 and −65 V and the second with CV = −40, −60 and −80 V) in effort to select for complementary peptide populations via gas-phase separation. In total, the 5 h analysis yielded quantitative measurements for 7,600 and 7,293 localized phosphorylation events for the TMTpro18 and TMTpro16 experiments, respectively (Figure 2B). These values represent a modest number of phosphorylation sites, in light of recent large-scale studies.25-28 However, we began with only 900 μg of peptide and acquired 136,800 phosphorylation site measurements (18 samples × 7,600 sites) with only 5 h of analysis time. Likewise, additional sample preparation time for the “mini-phos” analysis was minimal as the protocol was performed in under 1 h and was integrated into the SL-TMT protocol. As in the whole proteome workflow, the TMTpro-134C and TMTpro-135N reagents can be seamlessly incorporated into phosphorylation profiling or the analysis of virtually any other post-translational modification.
TMTpro-18plex Experiment Recapitulated Known Alterations in Protein Abundance and Phosphorylation Events in Control and PTEN−/− Cells upon Treatment with Two Inhibitors
We next sought to interrogate the data set further to examine how a pair of isogenic mammary epithelial cell lines responded to treatment with two drugs. We investigated how well these alterations in protein abundance recapitulated known effects on the abundance of certain proteins and associated phosphorylation events related to the PTEN/PI3K/AKT signaling pathway (Figure 3). We first focused on the proteome abundance alterations related to insulin receptor substrate, as it was well known that the activation of the AKT signaling triggers a feedback mechanism that decreases IRS2 (insulin receptor substrate 2) expression levels.29,30 In agreement with previous studies, our data showed a decrease of IRS2 (Figure 3A) in MCF10A PTEN−/− cells, and subsequently, the expected induction of IRS1 (insulin receptor substrate 1) and IRS2 (Figure 3B) upon inhibition of p110α by BYL-719. We validated these mass spectrometry results by Western blotting in an independent experiment (Figures 3A,B, S4). Likewise, the loss of PTEN function results in the accumulation of activated AKT, which induces the phosphorylation of several sites within RPS6 (S235, S236, S240, S244 and S247).6,7,31 Indeed, RPS6 phosphorylation increased in MCF10A PTEN−/− cells (Figures 3C, S3A) and was inhibited by BYL-719 (Figures 3D, S3A).
In addition, we used our proteome-wide data set to determine the overall effect of each drug on a given cell line, which we illustrated by a series of volcano plots [showing −log 10 (p-value) versus the log 2 fold change] (Figure 3E-I). When we compared MCF10A PTEN−/− versus control MCF10A cells, we noted 269 proteins were significantly down-regulated and 186 proteins were significantly up-regulated (1% FDR and a minimum of 40% change). Among these proteins, PDK4 (pyruvate dehydrogenase lipoamide kinase isozyme 4) and INSR (insulin receptor) were down-regulated, while CCND1 (Cyclin D1) was up-regulated in MCF10A PTEN−/− cells, as expected30,32,33 (Figure 3E). Using gene ontology analysis, we determined that regulated proteins in MCF10APTEN−/− cells were mostly enriched in extracellular matrix-related categories (e.g., cell adhesion and extracellular region), reflecting the known roles of PTEN in cell migration and epithelial–mesenchymal transition7 (Figure S3B).
We also examined the effect of PI3K inhibition (24 h) on protein expression changes in the two cell lines (Figure 3F-I). Inhibition of p110α by BYL-719 (Figure 3F) displayed the largest effect on the proteome, but only in control MCF10A cells, as reflected by a higher number of significantly regulated proteins and overall greater fold changes in control MCF10A cells. These large protein abundance alterations demonstrated that p110α likely played a prominent role in the PI3K signaling pathway in control MCF10A cells.6 We also observed that the inhibition of p110α by BYL-719 exerted a more profound impact on control MCF10A cells than on MCF10A PTEN−/− cells (Figure 3H), which agreed well with the accepted notion that the loss of PTEN confers resistance to inhibition by p110α.34 Additionally, KIN-193 (p110β inhibitor) had little effect on the protein profiles of either cell line (Figure 3G,I). These data were consistent with previous data that emphasized that the loss of PTEN alone in vitro was not sufficient to render cells p110β-dependent.35
CONCLUSIONS
The (phospho)proteome-wide protein expression responses to BYL-719 and KIN-193 in control and isogenic MCF10A PTEN−/− cells have not been reported previously. This data set could provide valuable information for developing a better understanding of PTEN/PI3K/AKT signaling, the mechanisms related to BYL-719 resistance, and the effect of PTEN loss.
With this experiment, we have shown that the TMTpro-134C and TMTpro-135N reagents can be added to the current set of TMTpro isobaric tags to generate a TMTpro-18plex reagent set without sacrificing proteome depth or quantitative performance. The true power of sample multiplexing was the ability to analyze data within a closed experimental system with essentially no missing values. Here, we examined six conditions in triplicate. However, many other experimental arrangements can be devised. For instance, one cell line can be treated with 18 different perturbations or drug concentrations, or an 18-point time course can be acquired. Flexibility in replicates was also possible with the 18-plex with sample layouts of 9 × 2, 3 × 3 × 2, and 6 × 3, among other potential experimental arrangements. We also noted that the TMTpro18-plex reagent set incorporated ten reagents that are 1 Da apart and thus can be used for isobaric tag-based analyses in mass spectrometers not capable of resolving the full 18 reagents, such as an ion trap36,37 or a TIMS-TOF38,39 instrument. Advantages of the TMTpro-18plex reagents also include its commercial availability and seamless integration into current TMTpro workflows. In all, our data demonstrated that the TMTpro-18plex has expanded further the sample multiplexing landscape while preserving high-quality protein quantification, thereby facilitating innovative experimental designs.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the members of the Gygi Lab at Harvard Medical School. We thank Yubao Wang for sharing the control MCF10A and MCF10A PTEN−/− cells. This work was funded in part by NIH/NIGMS grant R01 GM132129 (J.A.P.), GM67945 (S.P.G.), and NIH/NCI grant R35 CA231945 (T.M.R.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.1c00168.
MS1 ion peaks for a TMTpro-18plex reagent-labeled precursor scanned with different MS1 resolutions in profile mode; evaluation of figures-of-merit in TMTpro16 and TMTpro18 experiments; RPS6 protein abundance and gene ontology enrichment; raw Western blot images; protein quantifications in the TMTpro18 experiment; protein quantifications in the TMTpro16 experiment; phosphorylation quantifications in TMTpro18 and TMTpro16 experiments; and isotopic impurities of TMTpro-134C and TMTpro-135N (PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
The authors declare the following competing financial interest(s): The TMTpro reagents were commercialized by Thermo Fisher Scientific in September 2019.
Contributor Information
Jiaming Li, Department of Cell Biology, Harvard Medical School, Boston 02115, Massachusetts, United States.
Zhenying Cai, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston 02215, Massachusetts, United States; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston 02115, Massachusetts, United States.
Ryan D. Bomgarden, Thermo Fisher Scientific, Rockford 61101-9316, Illinois, United States
Ian Pike, Proteome Sciences, London WC1H 9BB, U.K..
Karsten Kuhn, Proteome Sciences, London WC1H 9BB, U.K..
John C. Rogers, Thermo Fisher Scientific, Rockford 61101-9316, Illinois, United States
Thomas M. Roberts, Department of Cancer Biology, Dana-Farber Cancer Institute, Boston 02215, Massachusetts, United States; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston 02115, Massachusetts, United States
Steven P. Gygi, Department of Cell Biology, Harvard Medical School, Boston 02115, Massachusetts, United States.
Joao A. Paulo, Department of Cell Biology, Harvard Medical School, Boston 02115, Massachusetts, United States
REFERENCES
- (1).Rauniyar N; Yates JR 3rd Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res 2014, 13, 5293–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Li J; Van Vranken JG; Pontano Vaites L; Schweppe DK; Huttlin EL; Etienne C; Nandhikonda P; Viner R; Robitaille AM; Thompson AH; Kuhn K; Pike I; Bomgarden RD; Rogers JC; Gygi SP; Paulo JA TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 2020, 17, 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Thompson A; Wölmer N; Koncarevic S; Selzer S; Böhm G; Legner H; Schmid P; Kienle S; Penning P; Höhle C; Berfelde A; Martinez-Pinna R; Farztdinov V; Jung S; Kuhn K; Pike I TMTpro: Design, Synthesis, and Initial Evaluation of a Proline-Based Isobaric 16-Plex Tandem Mass Tag Reagent Set. Anal. Chem 2019, 91, 15941–15950. [DOI] [PubMed] [Google Scholar]
- (4).Chu EC; Tarnawski AS PTEN regulatory functions in tumor suppression and cell biology. Med. Sci. Monit 2004, 10, Ra235–41. [PubMed] [Google Scholar]
- (5).Simpson L; Parsons R PTEN: life as a tumor suppressor. Exp. Cell Res 2001, 264, 29–41. [DOI] [PubMed] [Google Scholar]
- (6).Thorpe LM; Yuzugullu H; Zhao JJ PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chang H; Cai Z; Roberts TM The Mechanisms Underlying PTEN Loss in Human Tumors Suggest Potential Therapeutic Opportunities. Biomolecules 2019, 9, 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Navarrete-Perea J; Yu Q; Gygi SP; Paulo JA Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. J. Proteome Res 2018, 17, 2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ogata T; Kozuka T; Kanda T Identification of an insulator in AAVS1, a preferred region for integration of adeno-associated virus DNA. J. Virol 2003, 77, 9000–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wang T; Wei JJ; Sabatini DM; Lander ES Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 2014, 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Schweppe DK; Prasad S; Belford MW; Navarrete-Perea J; Bailey DJ; Huguet R; Jedrychowski MP; Rad R; McAlister G; Abbatiello SE; Woulters ER; Zabrouskov V; Dunyach J-J; Paulo JA; Gygi SP Characterization and Optimization of Multiplexed Quantitative Analyses Using High-Field Asymmetric-Waveform Ion Mobility Mass Spectrometry. Anal. Chem 2019, 91, 4010–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Schweppe DK; Eng JK; Yu Q; Bailey D; Rad R; Navarrete-Perea J; Huttlin EL; Erickson BK; Paulo JA; Gygi SP Full-Featured, Real-Time Database Searching Platform Enables Fast and Accurate Multiplexed Quantitative Proteomics. J. Proteome Res 2020, 19, 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rad R; Li J; Mintseris J; O’Connell J; Gygi SP; Schweppe DK Improved Monoisotopic Mass Estimation for Deeper Proteome Coverage. J. Proteome Res 2021, 20, 591–598. [DOI] [PubMed] [Google Scholar]
- (14).Elias JE; Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [DOI] [PubMed] [Google Scholar]
- (15).Huttlin EL; Jedrychowski MP; Elias JE; Goswami T; Rad R; Beausoleil SA; Villén J; Haas W; Sowa ME; Gygi SP A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Savitski MM; Wilhelm M; Hahne H; Kuster B; Bantscheff M A Scalable Approach for Protein False Discovery Rate Estimation in Large Proteomic Data Sets. Mol. Cell. Proteomics 2015, 14, 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Beausoleil SA; Villén J; Gerber SA; Rush J; Gygi SP A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol 2006, 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- (18).McAlister GC; Nusinow DP; Jedrychowski MP; Wühr M; Huttlin EL; Erickson BK; Rad R; Haas W; Gygi SP MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 2014, 86, 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Huang DW; Sherman BT; Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 2009, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- (20).Tusher VG; Tibshirani R; Chu G Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Schweppe DK; Rusin SF; Gygi SP; Paulo JA Optimized Workflow for Multiplexed Phosphorylation Analysis of TMT-Labeled Peptides Using High-Field Asymmetric Waveform Ion Mobility Spectrometry. J. Proteome Res 2020, 19, 554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yu Q; Paulo JA; Naverrete-Perea J; McAlister GC; Canterbury JD; Bailey DJ; Robitaille AM; Huguet R; Zabrouskov V; Gygi SP; Schweppe DK Benchmarking the Orbitrap Tribrid Eclipse for Next Generation Multiplexed Proteomics. Anal. Chem 2020, 92, 6478–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Erickson BK; Mintseris J; Schweppe DK; Navarrete-Perea J; Erickson AR; Nusinow DP; Paulo JA; Gygi SP Active Instrument Engagement Combined with a Real-Time Database Search for Improved Performance of Sample Multiplexing Workflows. J. Proteome Res 2019, 18, 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Paulo JA; O’Connell JD; Everley RA; O’Brien J; Gygi MA; Gygi SP Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. J. Proteomics 2016, 148, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bekker-Jensen DB; Bernhardt OM; Hogrebe A; Martinez-Val A; Verbeke L; Gandhi T; Kelstrup CD; Reiter L; Olsen JV Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun 2020, 11, 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Humphrey SJ; Karayel O; James DE; Mann M High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat. Protoc 2018, 13, 1897–1916. [DOI] [PubMed] [Google Scholar]
- (27).Sharma K; D’Souza RCJ; Tyanova S; Schaab C; Wiśniewski JR; Cox J; Mann M Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [DOI] [PubMed] [Google Scholar]
- (28).Zhou H; Di Palma S; Preisinger C; Peng M; Polat AN; Heck AJR; Mohammed S Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res 2013, 12, 260–271. [DOI] [PubMed] [Google Scholar]
- (29).Simpson L; Li J; Liaw D; Hennessy I; Oliner J; Christians F; Parsons R PTEN expression causes feedback upregulation of insulin receptor substrate 2. Mol. Cell. Biol 2001, 21, 3947–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lackey J; Barnett J; Davidson L; Batty IH; Leslie NR; Downes CP Loss of PTEN selectively desensitizes upstream IGF1 and insulin signaling. Oncogene 2007, 26, 7132–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Meyuhas O Ribosomal Protein S6 Phosphorylation. Int. Rev. Cell Mol. Biol 2015, 320, 41–73. [DOI] [PubMed] [Google Scholar]
- (32).Puthanveetil P; Wang Y; Wang F; Kim MS; Abrahani A; Rodrigues B The increase in cardiac pyruvate dehydrogenase kinase-4 after short-term dexamethasone is controlled by an Akt-p38-forkhead box other factor-1 signaling axis. Endocrinology 2010, 151, 2306–2318. [DOI] [PubMed] [Google Scholar]
- (33).Weng L-P; Brown JL; Eng C PTEN coordinates G1 arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Hum. Mol. Genet 2001, 10, 599–604. [DOI] [PubMed] [Google Scholar]
- (34).Juric D; Castel P; Griffith M; Griffith OL; Won HH; Ellis H; Ebbesen SH; Ainscough BJ; Ramu A; Iyer G; Shah RH; Huynh T; Mino-Kenudson M; Sgroi D; Isakoff S; Thabet A; Elamine L; Solit DB; Lowe SW; Quadt C; Peters M; Derti A; Schegel R; Huang A; Mardis ER; Berger MF; Baselga J; Scaltriti M Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 2015, 518, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sethi I; Cai Z; Roberts TM; Yuan G-C Molecular Profiling Establishes Genetic Features Predictive of the Efficacy of the p110β Inhibitor KIN-193. Cancer Res. 2019, 79, 4524–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Liu JM; Sweredoski MJ; Hess S Improved 6-Plex Tandem Mass Tags Quantification Throughput Using a Linear Ion Trap-High-Energy Collision Induced Dissociation MS3 Scan. Anal. Chem 2016, 88, 7471–7475. [DOI] [PubMed] [Google Scholar]
- (37).Paulo JA; Navarrete-Perea J; Guha Thakurta S; Gygi SP TKO6: A Peptide Standard To Assess Interference for Unit-Resolved Isobaric Labeling Platforms. J. Proteome Res 2019, 18, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Meier F; Brunner A-D; Koch S; Koch H; Lubeck M; Krause M; Goedecke N; Decker J; Kosinski T; Park MA; Bache N; Hoerning O; Cox J; Räther O; Mann M Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteomics 2018, 17, 2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Ogata K; Ishihama Y Extending the Separation Space with Trapped Ion Mobility Spectrometry Improves the Accuracy of Isobaric Tag-Based Quantitation in Proteomic LC/MS/MS. Anal. Chem 2020, 92, 8037–8040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited in the ProteomeXchange Consortium with the data set identifier PXD024275.