Abstract
Objective:
C-section interrupts mother-to-newborn microbial transfer at birth. Beyond the neonatal period, the impact of C-section on offspring gut microbiota and their short chain fatty acids (SCFAs) remains unclear. Here we examine delivery mode (C-section vs. vaginal) with the infant gut microbiota and fecal SCFAs measured 3 and 12 months after birth.
Design:
Longitudinal study
Setting:
North Carolina
Population:
In 2013–2015 we enrolled pregnant women and followed up their offspring for 12 months. We asked a subset of participants, enrolled over a 3-month period, to provide fecal samples at the 3-month and 12-month follow-up visits.
Methods/Main Outcomes:
We sequenced the 16S rRNA V4 region with Illumina MiSeq and quantified SCFA concentrations using gas chromatography. We examined delivery mode with differential abundance of microbiota amplicon sequence variants (ASVs) using beta-binomial regression and fecal SCFAs using linear regression. We adjusted models for confounders.
Results:
Of the 70 infants in our analytic sample, 25 (36%) were C-section delivered. C-section (vs. vaginal) delivery was associated with differential abundance of 14 infant bacterial ASVs at 3 months and 13 ASVs at 12 months. C-section infants had a higher abundance of the potential pathobionts C. neonatale (p = 0.04) and C. perfringens (p = 0.04), and a lower abundance of potentially beneficial Bifidobacterium and Bacteroides spp. C-section infants also had higher fecal butyrate at 3 months (p < 0.005).
Conclusions:
C-section infants were more likely to be colonized by pathobionts in lieu of beneficial microbes, and they excreted more butyrate at 3 months of life.
Tweetable abstract:
C-section delivery was associated with increased butyrate excretion, decreased Bifidobacterium, & colonization of the infant gut by pathobionts at 3mo. of age.
Keywords: delivery mode, infant gut microbiome, short-chain fatty acids
Introduction
C-section delivery (vs. vaginal delivery) has been associated with higher risk of chronic immune disorders (e.g. asthma and allergies),1 neurodevelopmental outcomes (i.e. attention deficit conditions),2 as well as infectious morbidity.3 We have also found that C-section delivered infants have accelerated adiposity gain over the first year of life4 and are more likely to develop overweight or obesity later in life.5 It is postulated that these myriad health associations are explained by C-section interrupting the natural mother-to-newborn sharing of microbiota at birth. Yet uncertainty remains about how long delivery-mode differences in the microbiota persist and if they are associated with changes in production of microbiota-derived short chain fatty acids (SCFAs) such as butyrate, which are involved in the education of the immune system and metabolism.6
In this study we sought to address these gaps in the literature by prospectively examining the association between delivery mode and (a) the composition and diversity of the infant gut microbiota and (b) fecal SCFAs at 3 and 12 months of life. We hypothesized that C-section delivered infants have lower relative abundances of Bifidobacterium and Bacteroides, the bacterial genera most commonly depleted in C-section infants in previous studies,7 and higher relative abundances of potential pathobiontic Clostridia species found in previous studies.7 Moreover, given that prior literature has linked fecal butyrate with excess weight gain8 and obesity,9, 10 we explored whether C-section delivery is associated with the concentration of butyrate and other SCFAs in the stool.
Methods
Study population
This study was conducted in the prospective Nurture study, which has been previously described.11 Briefly, Nurture is prospective birth cohort study designed to assess longitudinal associations of early-life factors with infant adiposity and weight in the first year of life. Grants from the National Institutes of Health (R01DK094841) and the Mid-Atlantic Nutrition Obesity Research Center, under NIH award number P30DK0372488, supported the research reported in this publication. Although it is an observational study and a core outcome set was not used, Nurture was registered at clinicaltrials.gov (NCT01788644).
Nurture was conducted according to guidelines in the Declaration of Helsinki. Duke University Medical Center IRB (human subjects committee) (Pro0036242) approved all procedures involving human subjects. The most recent IRB approval for Nurture was April 18th, 2020. At study recruitment, while women were pregnant, we obtained written informed consent. Women confirmed their willingness to participate again after delivery.
Between 2013 and 2015, Nurture investigators and staff recruited women between 20 and 36 weeks of pregnancy from a private prenatal clinic and a county health department in Durham, North Carolina. To be considered for inclusion in the study women had to be > 18 years of age, be able to speak and read English, and be having a singleton birth. They also had to confirm their intention to remain local for home visits for at least one year after delivery. Infants were excluded if they were born < 28 weeks into gestation, were born with a congenital abnormality, were not able to take food by mouth at discharge, or if they required ≥ 3 weeks of hospitalization after birth. Home visits were conducted when infants were 3, 6, 9, and 12 months of age. In addition, there were monthly telephone calls in between visits to collect data on breastfeeding, formula feeding, and timing of solid food introduction.
In total, 666 mother-infant pairs were followed from pregnancy to the first year of life. We conducted a substudy in which we invited all mothers that were enrolled over a 3 month time period to collect stool samples from their infants at the 3-month and 12-month follow-up visits. In total, 70 mother-infant pairs provided at least one stool sample that was used for microbiota and SCFA analyses. The mother-infant pairs included in the microbiome substudy had a distribution of sociodemographic and clinical characteristics similar to the entire cohort.
Exposure variables
We obtained data on mode of delivery (C-section delivery and vaginal delivery), infant sex, birth weight (kg), gestational age (weeks), and antibiotic use from medical records. Mothers’ reported (via questionnaire) their age (years), race and ethnicity, pre-pregnancy weight (kg) and height (m), level of educational achievement, household income, and cigarette smoking status in pregnancy. We used pre-pregnancy weight and height to calculate mothers’ pre-pregnancy body mass index (BMI; kg/m2) and we defined overweight or obese as BMI ≥ 25. During monthly phone calls and the home visits we had mothers prospectively report if they were still breastfeeding, if had introduced formula, and if they had introduced solid foods and if so the timing of solid food introduction.
Stool Collection and Storage
Stool samples were taken from diapers at the 3-month and the 12-month home visits. We promptly transferred collected stool to 2 ml cryogenic vials (ThermoFisher) and froze them at −80°C.
16S rRNA-based bacterial community sequencing using Illumina MiSeq platform
We sent frozen fecal samples on dry ice to Microbiome Insights. Their lab technicians used standardized methods to extract microbial DNA, to amplify the V4 region of the 16S ribosomal RNA (rRNA) gene using PCR, and to sequence the prepared libraries using the Illumina MiSeq platform. The full methologic details for the 16S rRNA gene sequencing can be found in our prior publication using this cohort.12
16S rRNA sequence pre-processing
We controlled the quality of reads and resolved amplicon sequence variants (ASVs) in the statistical programming environment R, using version 1.8 of the DADA2 package, and following author recommendations.13 First we examined sequence quality scores for forward reads and reverse reads (Figure S1). Then we trimmed all forward reads before nucleotide position 10 and after position 240, and all reverse reads before position 10 and after position 150. We required each read to have less than two expected errors given based on their quality scores, and we filtered out reads mapping to the phiX genome and reads with ambiguous base assignments (using the overlapping paired reads “filterandtrim” function). We dereplicated the reads (using the “derep” function) and denoised the reads (using the “dada” function for pooled reads) with estimated error models. To estimate our error models we used the “learnErrors” function, randomly selecting 106,431,120 bases from 462,744 reads to estimate the forward-read error model, and 102,199,160 bases from 729,994 reads to estimate the reverse-read error model (Figure S2). We merged overlapping paired reads (using the “mergePairs” function) and removed chimeric ASVs (using the “removeBimeraDenovo” function with the pooled sample setting). Our final sequence pool retained 74% of the original raw reads (Figure S3). We assigned taxonomy using the HITdb v.1.00 16S rRNA sequence database of human intestinal microbiome taxa and the DADA2 function “assignTaxonomy”.14
Phylogenetic tree generation
Next, we followed the recommendations of a recent Bioconductor microbiome data analysis workflow to generate a phylogenetic tree from the denoised ASVs.15 In brief, we first used the DECIPHER Bioconductor package to align ASVs and construct a neighbor-joining tree.16 Then we used the phangorn package to generate a Generalized time-reversible (with Gamma rate variation) maximum likelihood phylogenetic tree, which we rooted at the midpoint.17 And finally we used the phyloseq package to merge the phylogenetic tree and the microbial ASVs with the other study variables.18
Short chain fatty acid quantification
Short chain fatty acid (SCFA) concentrations were quantified using gas chromatography (Thermo Trace 1310) coupled to a flame ionization detector as previously described by Zhao et al.19 in our prior publication in this cohort.12
Statistical Analysis
Microbial community composition analysis
Our primary hypothesis was that vaginal-born infants have fecal microbiota with higher relative abundances of species in Bifidobacterium and Bacteroides genera and lower relative abundances of opportunistic pathobionts within the genus Clostridium at 3 months of age.
To test for differential relative abundance of ASVs in C-section-born vs. vaginally-born infants we used the R package corncob to employ beta-binomial regression models that account for within-sample taxa correlation and variable sequencing depth from.20 One of the strengths of these models is that they allow for multivariable adjustment; the variables included in our multivariable model are described below. To reduce the possibility of false discovery of rare ASVs, given our relatively small sample size, we removed microbial ASVs that did not have a mean count at or above the 25th percentile in at least 10% of samples. A list of all ASVs included in the study with their assigned taxonomy can be found in Appendix S1.
Beta diversity analysis
We used the phyloseq package18 to estimate weighted UniFrac distances, a measure of pairwise community composition. We then used permutational multivariate analysis of variance, using the function “adonis” in the vegan package21 with 9,999 permutations, to test for differences in weighted UniFrac distances before and after multivariable adjustment.
Alpha diversity analysis
We also used the phyloseq package18 to estimate the Shannon diversity index, a measure that combines bacterial richness (the number of unique ASVs observed within each sample) with the evenness of the relative abundance within each sample. We then tested for differences in Shannon diversity index using unadjusted and multivariable-adjusted generalized linear regression models.
SCFA analysis
Our secondary outcome was fecal concentration of the SCFA butyrate at 3 and 12 months of age. We performed a log10 + 1 transformation on butyrate, propionate, and total SCFA concentrations to normalize the distributions at both timepoints. We fitted multivariable generalized linear regression models to examine the association of delivery mode (C-section vs. vaginal delivery) with SCFA concentrations in infant fecal samples collected at 3 and 12 months of age. We then selected microbial ASVs and genera that were statistically significantly associated with delivery mode and estimated their (Spearman) correlation with SCFA concentrations.
Multivariable adjustment and statistical significance
We adjusted multivariable regression models for maternal pre-pregnancy BMI and breastfeeding (never vs. ever). We adjusted for these factors based on prior literature that indicated they were potentially associated with both our exposure and outcomes.
We considered p < 0.05 significant for models with Shannon diversity index and Weighted Unifrac as the outcomes. For the microbial ASV differential abundance analyses, we used a two-sided false discovery rate (FDR) adjusted p < 0.05 to denote statistical significance.
Results
Participant characteristics
Among the 70 mother-infant dyads included in our analysis sample, 63% of mothers identified their infants as Black, 46% reported education below the high school level, and 63% had an annual household income of ≤ $20,000. The range of gestational age was 34.7 weeks to 41.3 weeks. A total of 25 (36%) infants were C-section delivered, similar to the prevalence of this exposure in the United States.22 C-section mother-infant pairs were similar to vaginal-delivery mother-infant pairs with respect to socio-demographic and clinical factors, except for maternal pre-pregnancy BMI, which was higher among C-section pairs (32.9 vs. 27.6 kg/m2), as shown in Table 1. Of the 25 children born via C-section, 18 had data on type of C-section. Of these, 6 were scheduled C-sections (31.6%), 8 were unscheduled C-sections (42.1%), and 4 progressed from labor to C-section (21.1%). Of the 6 scheduled C-section deliveries, 2 had membrane rupture (1 with labor). Of the 8 unscheduled C-section deliveries, 6 had membrane rupture (4 with labor). Of the 4 labor to C-section deliveries, 4 had membrane rupture.
Table 1.
Participant characteristics by delivery mode.
| Delivery mode | p-value | ||
|---|---|---|---|
| Vaginal (N=45) | C-Section (N=25) | ||
| Pre-pregnancy BMI ≥ 25 kg/m2 (%) | 30 (66.7%) | 18 (72.0%) | 0.65 |
| Pre-pregnancy BMI, kg/m2 (median [IQR]) | 27.58 [22.96, 35.18] | 32.92 [24.96, 40.77] | 0.04 |
| Maternal age (median [IQR]) | 25.94 [22.75, 29.41] | 26.26 [24.79, 30.93] | 0.24 |
| Female sex | 18 (40.0%) | 14 (56.0%) | 0.20 |
| Black or African-American infant race | 30 (66.7%) | 14 (56.0%) | 0.46 |
| Breastfed ever (%) | 37 (82.2%) | 19 (76.0%) | 0.53 |
| Breastfeeding duration, weeks (median[IQR]) | 4.50 [1.00, 13.01] | 4.00 [0.50, 7.00] | 0.69 |
| Introduction to solids before 3 months of age | 12 (25.0%) | 9 (30.4%) | 0.63 |
| Maternal education below high school level (%) | 23 (51.1%) | 9 (36.0%) | 0.22 |
| Household income (%) | 0.53 | ||
| ≤ $20,000 | 26 (60.5%) | 17 (68.0%) | |
| > $20,000 | 17 (39.5%) | 8 (32.0%) | |
| No infant antibiotics at/before 3 months (%) | 37 (82.2%) | 20 (80.0%) | 0.82 |
| No current maternal smoking (%) | 33 (82.5%) | 15 (75.0%) | 0.49 |
| Birth weight, kg (median [IQR]) | 3.19 [2.89, 3.49] | 3.19 [2.82, 3.64] | 0.79 |
| Gestational age, weeks (median [IQR]) | 39.57 [38.71, 40.43] | 39.00 [38.29, 39.29] | 0.07 |
Delivery mode and gut microbiome diversity
C-section delivery explained 3.2% variation in Weighted Unifrac at 3 months, the most among the covariates included in the model, but only 0.5% of the variation at 12 months (Figure S4). Association of delivery mode with measures of gut microbiota alpha diversity (Shannon diversity index) at 3 and 12 months of age are summarized in Figure S6. C-section delivered infants trended toward lower Shannon diversity index at 3 months of age (beta = −0.18, 95% CI: −0.41, 0.05), but not at 12 months (beta = −0.03, 95% CI: −0.36, 0.30).
Delivery mode and Gut microbiome composition
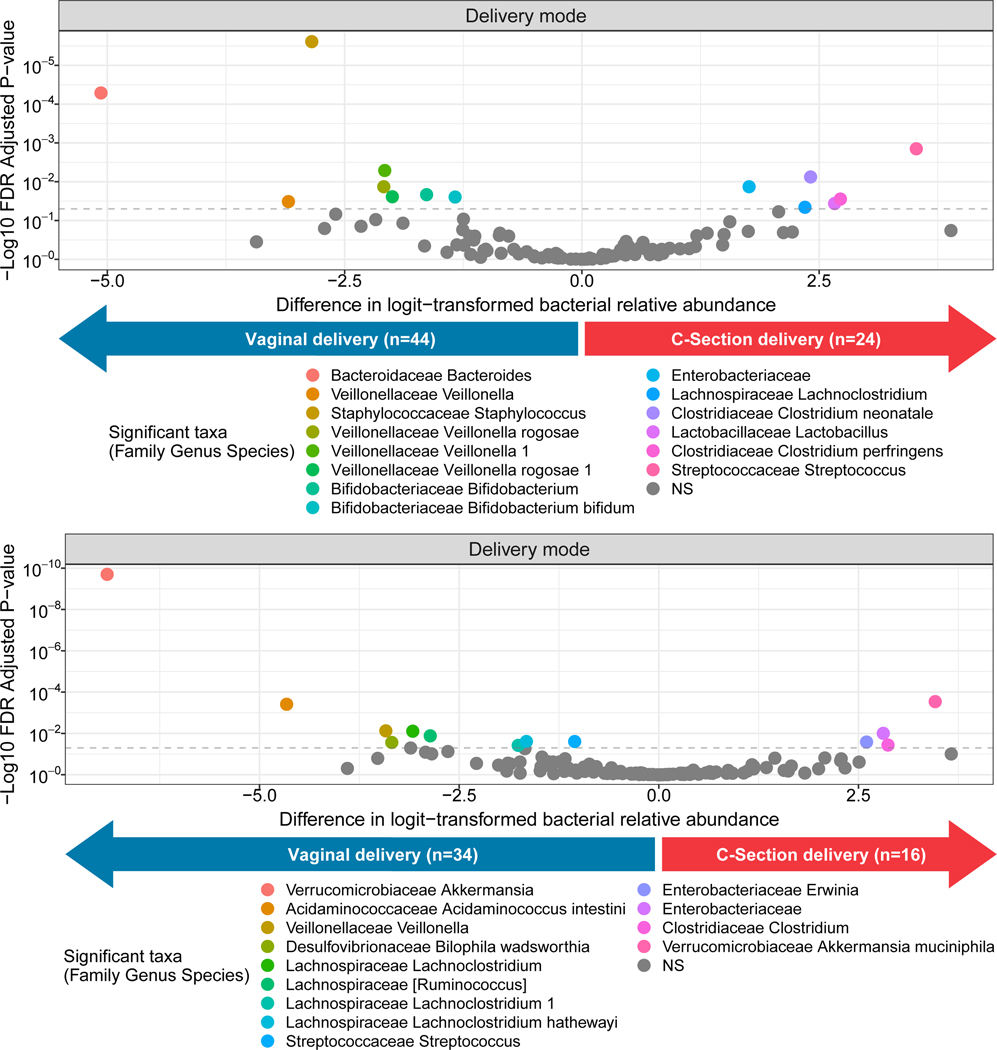
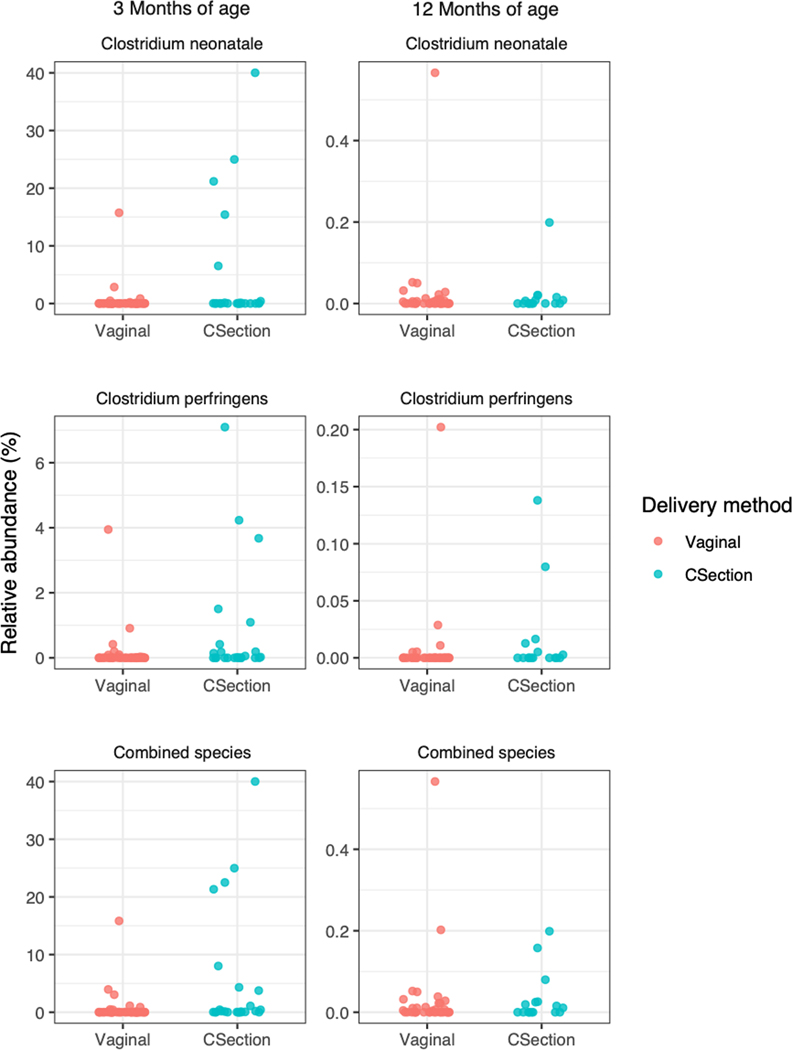
Figure S5 shows the unadjusted relative abundances of the top bacterial families and genera in our sample of infants according to delivery mode and time of stool collection. After multivariable adjustment, delivery mode was associated with 14 bacterial ASVs at 3 months (Figure 1; Table S1) and 13 ASV at 12 months (Figure 1; Table S2). Of note, at 3 months of age, C-section delivery (vs. vaginal delivery) was significantly associated with lower relative abundances of Bacteroides (sp. unknown), Bifidobacterium (sp. unknown), and Bifidobacterium bifidum, and higher relvative abundances of several opportunistic pathobionts including Clostridium perfingens and Clostridium neonatale. At 12 months, Akkermansia muciniphila and Ruminococcus (sp. unknown) were significantly lower among C-section-born infants. In Figure 2, we further show the relative abundances of the opportunistic pathobionts, C. neonatale and C. perfringens.
Figure 1.
Adjusted* associations of delivery mode (C-section and vaginal delivery) with the logit-transformed relative abundances of infant gut microbial taxa at a) 3 months of age and b) 12 months of age. *Beta-binomial regression models were adjusted for maternal pre-pregnancy body mass index and breastfeeding. NS = Not significant.
Figure 2.
Relative abundance of potential pathobionts in the infant gut microbiota according to delivery mode (C-section and vaginal delivery) and time of follow up. According to Mann-Whitney U test: p = 0.04 for difference in C. neonatale at 3 months and p = 0.94 at 12 months; p = 0.04 for difference in C. perfringens at 3 months and p = 0.08 at 12 months; and p = 0.002 for difference in combined species abundance of both C. neonatale and C. perfringens at 3 months and p = 0.59 at 12 months.
SCFA concentrations
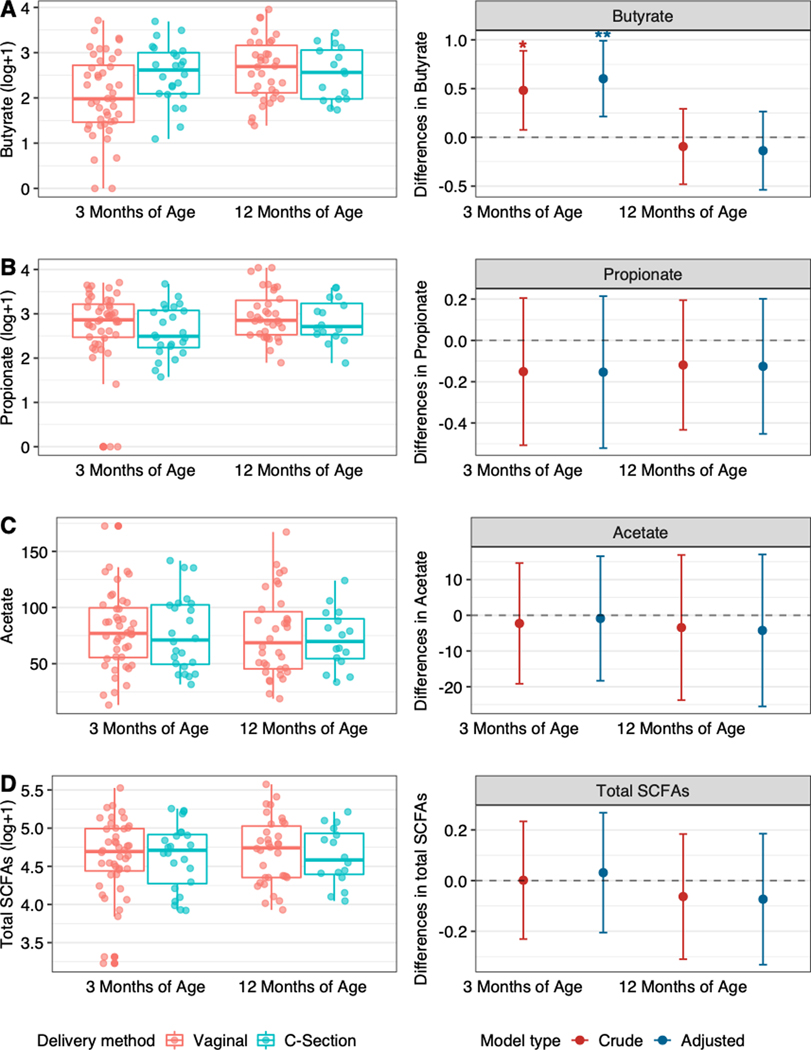
Association of delivery mode with SCFAs at 3 and 12 months is shown in Figure 3. C-section delivery was associated with higher concentration of fecal butyrate at 3 months of age (beta = 0.60, 95% CI: 0.21, 0.99), but this association was no longer significant at 12 months of age. There were no differences in concentrations of fecal propionate, fecal acetic acid, or total SCFAs measured (Figure 3; all p > 0.05).
Figure 3.
Distributions (left side) and differences (right side) in the fecal concentrations of A) butyrate, B) propionate, C) acetate, and D) total SCFAs according to delivery mode (C-section infants vs. vaginal infants [reference]). Differences on the right side of figure are presented before adjustment (crude) and after adjustment for maternal pre-pregnancy body mass index and breastfeeding. * = p <0.05, ** =p<0.01.
After FDR-correction, the only significant Spearman correlation between ASVs, that were significantly associated with delivery mode, and SCFAs was a Veillonella ASV and isovaleric acid measured at 3 months of age. Furthermore, at 3 months the relative abundance of the genera Lachnoclostridium was positively associated with butyric acid (rho=0.38, FDR p=0.05), Bacteroides was positively correlated with propionic acid (rho=0.39, FDR p=0.05), and Bifidobacterium was positively correlated with acetic acid, although this did not reach statistical significant after FDR correction (rho=0.21, FDR p=0.28). The full correlation matrices between gut microbiota and SCFAs at each time point can be found in Figures S7-S10.
Discussion
Main findings
In our racially diverse, predominantly low-income cohort of mother-child dyads from North Carolina, C-section delivery was associated with lower relative abundance of several beneficial bacteria, e.g. Bacteroides and Bifidobacterium, and higher relative abundances of the pathobiontic C. neonatale and C. perfringens bacteria. C-section was also associated with higher concentration of fecal butyrate at 3 months of age. Associations with butyrate were no longer significant at 12 months, however delivery mode was still associated with the differential abundance of 13 bacterial taxa at 12 months.
Strengths and limitations
Despite a number of strengths, including the longitudinal design, use of multivariable regression models to adjust for confounders, and measurement of both microbial communities and short chain fatty acids, there are limitations to our study. First, although we controlled for pre-pregnancy BMI and breastfeeding duration, and other participant characteristics were similar by delivery mode (Table 1), it is still possible that residual confounding by factors for which we did not have complete data (e.g. intrapartum antibiotics, chorioamnionitis, admission to NICU) could have influenced our findings. Also, future studies are needed to determine whether type of C-section has an effect on the infant microbiome, as our study was too small to rigorously conduct such an analysis. Second, while our study sample was larger than many previous studies on this topic, we cannot rule out the possibility that non-significant results could be due to not having power to detect differences. Our sample size also precluded us from stratifying on potential effect measure modifiers such as sex, prenatal antibiotic use, presence of labor, or membrane status at the time of delivery. Third, we cannot rule out the chance of false discovery; however, our results are corroborated by evidence from many other cohort studies. Finally, a limitation of the 16S rRNA sequencing is that we are only able to examine relative abundances of prokaryotic taxa. As such, culture-based studies are still needed to quantify the absolute differences in the bacteria that were found to have differential relative abundance by delivery mode in our study.
Interpretation in light of other evidence
Our results extend previous findings that delivery-mode differences in the infant microbiome emerge after meconium, when the subsequent transitional stool is passed,23 and a differential abundance of key bacterial taxa persist until 3 and 12 months of age. Our findings are largely consistent with at least 26 independent birth cohorts that have now shown that C-section delivered infants have differential abundances of gut microbiota, including delayed colonization by Bacteroides.7, 23–48 Although our study only followed infants to 12 months, a recent longitudinal study indicates that delivery-mode associated gut microbiota differences persist out to 4 years of age.40 While our study shows evidence that delivery mode affects the microbiome composition at 12 months, even if impacts to the gut microbiome converged after the first several months of life, altering the microbiome during this critical early-life window when the immune system is being formed has been shown experimentally to have long-term health consequences.49
An important finding of our analysis is the confirmation that C-section delivered infants had higher relative abundances of the potential pathobionts C. neonatale and C. perfringens. A recent study7 found that the intestinal microbiota of C-section infants (vs. vaginally delivered infants) was enriched with the pathobionts Clostridium perfringens in addition to Enterococcus faecalis, Enterococcus faecium, Staphylococcus epidermis, Streptococcus parasanguinis, Klebsiella oxytoca, Klebsiella pneumoniae, and Enterobacter cloacae. We were able to replicate the C. perfringens result, but we did not identify the other taxa at the species level (we did, however, observe associations for unclassified higher order taxa in the same direction). Future studies are needed to examine whether these bacteria mediate or modify an association between delivery mode and infant infection and diarrhea.
We also found that C-section delivered infants had a lower abundance of the potentially beneficial Bifidobacterium and a lower abundance of Akkermansia. Bifidobacterium spp. are considered to be important constituents of a healthy infant gut microbiota until the age of weaning.50, 51 They play a critical role in fermentation of non-digestible carbohydrates, such as human milk oligosaccharides found in breastmilk, and producing acetic acid and B vitamins.52–55 A lower abundance of the genera Bifidobacterium in the gut microbiota has been associated with higher levels of inflammation.53 Furthermore, Akkermansia is a mucolytic bacteria56, 57 associated with lower adiposity and improved insulin sensitivity in adults.58–61
The observation that C-section delivery is associated with higher fecal butyrate at 3 months of age is novel and provides insight into the association of delivery mode with infant health. Butyrate is an important SCFA that serves as the main energy source for the colonocytes62 and that has a complex relationships with immunometabolism.6 A higher concentration of butyrate in the feces may indicate that bacteria enriched in the gut of C-section delivered infants uniquely produce excess butyrate, not other SCFAs. This postulate is supported by the finding that several butyrate-producing bacteria, including notably the genera Lachnoclostridium,63 were not only higher in C-section infants but also positively correlated with fecal butyrate at 3 months. Alternatively, C-section delivery may decrease absorption of butyrate across the apical membrane of colonocytes, resulting in excess excretion of butyrate in the stool. Regardless, higher fecal butyrate has been associated with excess weight gain in mice8 and obesity10 and poor cardiometabolic health in humans.9 Longitudinal studies with longer follow-up are needed to shed light on higher infant fecal butyrate, among other short chain fatty acids, and its association with future health outcomes in infants and children.
Conclusions
In conclusion, after careful control for maternal pre-pregnancy BMI and breastfeeding, C-section delivery is associated with differences in gut microbiota composition and butyrate production at 3 months. Although it appears that the association of delivery mode on microbiome structure and SCFAs diminishes after 3 months of age, we still found delivery mode was significantly associated with differences in the relative abundance of 13 bacterial taxa at 12 months. Our results are complemented by the finding that C-section delivery accelerates gains in body weight until 12 months of life in this same cohort of infants.4 Future clinical trials are needed to test whether introduction of maternal microbiota to C-section delivered neonates can restore the microbiome of vaginal delivery and mitigate C-section-disease associations.
Supplementary Material
Quality scores of the aggregated forward (top panel) and reverse (bottom panel) 16S rRNA amplicon sequence reads.
Dada2 error models used for the forward (top panel) and reverse (bottom panel) 16S rRNA amplicon sequence reads.
The total number of sequences maintained in each preprocessing step for each sample.
Proportion of the variance (R2) in the infant gut microbial community structure (Weighted UniFrac) that is explained by delivery mode, breastfeeding and pre-pregnancy BMI at 3 months (top) and 12 months (bottom).
Mean relative abundance (%) of A) top 14 bacterial families and B) top 14 bacterial genera, stratified by delivery mode and time of follow up. Remaining taxa were grouped with other.
Differences between C-section-born infants and vaginal-born infants (reference) in the gut microbiome Shannon diversity index at 3 months of age and 12 months of age, before (crude) and after adjustment for maternal pre-pregnancy body mass index and breastfeeding.
Spearman correlations of delivery-mode associated ASVs with fecal SCFA concentrations at 3 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Spearman correlations of delivery-mode associated genera with fecal SCFA concentrations at 3 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Spearman correlations of delivery-mode associated ASVs with fecal SCFA concentrations at 12 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Spearman correlations of delivery-mode associated genera with fecal SCFA concentrations at 12 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Differences in the logit-transformed relative abundances of bacterial ASVs associated with delivery mode at 3 months of age, after adjustment for maternal pre-pregnancy BMI and breastfeeding in beta-binomial models. Unadjusted Mann-Whitney U test p-values provided in rightmost column.
Differences in the logit-transformed relative abundances of bacterial ASVs associated with delivery mode at 12 months of age, after adjustment for maternal pre-pregnancy BMI and breastfeeding in beta-binomial models. Unadjusted Mann-Whitney U test p-values provided in rightmost column.
A list of all ASVs included in the study with their assigned taxonomy.
Acknowledgements:
We would like to acknowledge Microbiome Insights, who collaborated with us on the extraction of microbial DNA, amplicon sequencing, and quantification of SCFAs.
Funding: This research was supported by grants from the National Institutes of Health (R01DK094841) and the Mid-Atlantic Nutrition Obesity Research Center (NORC) under NIH award number P30DK0372488. N.T.M. was also supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number K01HL141589 (PI: Mueller). M.K.D. was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health Grant Number T32 HL007024.
Footnotes
Disclosure of interest: The authors of this manuscript (NTM, MKD, TO, CH and SBN) have no conflicts of interest to disclose. Completed disclosure of interest forms are available to view online as supporting information.
Details of ethics approval: Nurture was conducted according to guidelines in the Declaration of Helsinki and all procedures involving human subjects were approved by Duke University Medical Center IRB (human subjects committee) (Pro0036242). The most recent IRB approval for Nurture was April 18th, 2020. Although an observational study, Nurture is registered at clinicaltrials.gov (NCT01788644). We obtained written informed consent at recruitment in pregnancy and women confirmed their willingness to participate again after delivery.
References
- 1.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015. January;135(1):e92–8. [DOI] [PubMed] [Google Scholar]
- 2.Zhang T, Sidorchuk A, Sevilla-Cermeno L, Vilaplana-Perez A, Chang Z, Larsson H, et al. Association of Cesarean Delivery With Risk of Neurodevelopmental and Psychiatric Disorders in the Offspring: A Systematic Review and Meta-analysis. JAMA Netw Open. 2019. August 2;2(8):e1910236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wainstock T, Walfisch A, Shoham-Vardi I, Segal I, Sergienko R, Landau D, et al. Term Elective Cesarean Delivery and Offspring Infectious Morbidity: A Population-Based Cohort Study. Pediatr Infect Dis J. 2019. February;38(2):176–80. [DOI] [PubMed] [Google Scholar]
- 4.Mueller NT, Zhang M, Hoyo C, Ostbye T, Benjamin-Neelon SE. Does cesarean delivery impact infant weight gain and adiposity over the first year of life? Int J Obes (Lond). 2019. August;43(8):1549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller NT, Whyatt R, Hoepner L, Oberfield S, Dominguez-Bello MG, Widen EM, et al. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity . Int J Obes (Lond). 2015. April;39(4):665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Wang J, He T, Becker S, Zhang G, Li D, et al. Butyrate: A Double-Edged Sword for Health? Adv Nutr. 2018. January 1;9(1):21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019. September 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005. August 2;102(31):11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Cuesta-Zuluaga J, Mueller NT, Alvarez-Quintero R, Velasquez-Mejia EP, Sierra JA, Corrales-Agudelo V, et al. Higher Fecal Short-Chain Fatty Acid Levels Are Associated with Gut Microbiome Dysbiosis, Obesity, Hypertension and Cardiometabolic Disease Risk Factors. Nutrients. 2018. December 27;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KN, Yao Y, Ju SY. Short Chain Fatty Acids and Fecal Microbiota Abundance in Humans with Obesity: A Systematic Review and Meta-Analysis. Nutrients. 2019. October 18;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamin Neelon SE, Ostbye T, Bennett GG, Kravitz RM, Clancy SM, Stroo M, et al. Cohort profile for the Nurture Observational Study examining associations of multiple caregivers on infant growth in the Southeastern USA. BMJ Open. 2017. February 8;7(2):e013939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Differding MK, Benjamin-Neelon SE, Hoyo C, Ostbye T, Mueller NT. Timing of complementary feeding is associated with gut microbiota diversity and composition and short chain fatty acid concentrations over the first year of life. BMC Microbiol. 2020. March 11;20(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016. July;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritari J, Salojarvi J, Lahti L, de Vos WM. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genomics. 2015. December 12;16(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Res. 2016;5(1492):1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright ES, Yilmaz LS, Noguera DR. DECIPHER, a Search-Based Approach to Chimera Identification for 16S rRNA Sequences. Appl Environ Microb. 2012. February;78(3):717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011. February 15;27(4):592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao G, Nyman M, Jonsson JA. Rapid determination of short-chain fatty acids in colonic contents and faeces of humans and rats by acidified water-extraction and direct-injection gas chromatography. Biomed Chromatogr. 2006. August;20(8):674–82. [DOI] [PubMed] [Google Scholar]
- 20.Martin BD, Witten D, Willis AD. Modeling microbial abundances and dysbiosis with beta-binomial regression. arXiv e-prints; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara R, et al. Package ‘vegan’. Version 2.9 ed; 2015. [Google Scholar]
- 22.Boerma T, Ronsmans C, Melesse DY, Barros AJD, Barros FC, Juan L, et al. Global epidemiology of use of and disparities in caesarean sections. Lancet. 2018. October 13;392(10155):1341–8. [DOI] [PubMed] [Google Scholar]
- 23.Mueller NT, Shin H, Pizoni A, Werlang IC, Matte U, Goldani MZ, et al. Delivery Mode and the Transition of Pioneering Gut-Microbiota Structure, Composition and Predicted Metabolic Function. Genes (Basel). 2017. December 4;8(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013. September;132(3):601–7 e8. [DOI] [PubMed] [Google Scholar]
- 25.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, et al. Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS One. 2016;11(6):e0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014. April;63(4):559–66. [DOI] [PubMed] [Google Scholar]
- 27.Hesla HM, Stenius F, Jaderlund L, Nelson R, Engstrand L, Alm J, et al. Impact of lifestyle on the gut microbiota of healthy infants and their mothers-the ALADDIN birth cohort. FEMS Microbiol Ecol. 2014. December;90(3):791–801. [DOI] [PubMed] [Google Scholar]
- 28.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015. May 13;17(5):690–703. [DOI] [PubMed] [Google Scholar]
- 29.Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015. February 3;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2016. May;123(6):983–93. [DOI] [PubMed] [Google Scholar]
- 31.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016. June 15;8(343):343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016. June 15;8(343):343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA Pediatr. 2016. March;170(3):212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O’Shea CA, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017. January 17;5(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sordillo JE, Zhou Y, McGeachie MJ, Ziniti J, Lange N, Laranjo N, et al. Factors influencing the infant gut microbiome at age 3–6 months: Findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART). J Allergy Clin Immunol. 2017. February;139(2):482–91 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravi A, Avershina E, Angell IL, Ludvigsen J, Manohar P, Padmanaban S, et al. Comparison of reduced metagenome and 16S rRNA gene sequencing for determination of genetic diversity and mother-child overlap of the gut associated microbiota. J Microbiol Methods. 2018. June;149:44–52. [DOI] [PubMed] [Google Scholar]
- 37.Wampach L, Heintz-Buschart A, Fritz JV, Ramiro-Garcia J, Habier J, Herold M, et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat Commun. 2018. November 30;9(1):5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018. October;562(7728):583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018. April;28(4):561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fouhy F, Watkins C, Hill CJ, O’Shea CA, Nagle B, Dempsey EM, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun. 2019. April 3;10(1):1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson AL, Houck KM, Jahnke JR. Pathways linking caesarean delivery to early health in a dual burden context: Immune development and the gut microbiome in infants and children from Galapagos, Ecuador. Am J Hum Biol. 2019. January 28:e23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugino KY, Paneth N, Comstock SS. Michigan cohorts to determine associations of maternal pre-pregnancy body mass index with pregnancy and infant gastrointestinal microbial communities: Late pregnancy and early infancy. PLoS One. 2019;14(3):e0213733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019. October;574(7776):117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, et al. Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life. Front Microbiol. 2017;8:738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory KE, LaPlante RD, Shan G, Kumar DV, Gregas M. Mode of Birth Influences Preterm Infant Intestinal Colonization With Bacteroides Over the Early Neonatal Period. Adv Neonatal Care. 2015. December;15(6):386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin AM, Sitarik AR, Havstad SL, Fujimura KE, Wegienka G, Cassidy-Bushrow AE, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep. 2016. August 25;6:31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee E, Kim BJ, Kang MJ, Choi KY, Cho HJ, Kim Y, et al. Dynamics of Gut Microbiota According to the Delivery Mode in Healthy Korean Infants. Allergy Asthma Immunol Res. 2016. September;8(5):471–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabeerdoss J, Ferdous S, Balamurugan R, Mechenro J, Vidya R, Santhanam S, et al. Development of the gut microbiota in southern Indian infants from birth to 6 months: a molecular analysis. J Nutr Sci. 2013;2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014. August 14;158(4):705–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014. May;80(9):2889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012. May 9;486(7402):222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008. December 2;105(48):18964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. Isme J. 2016. July;10(7):1656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, et al. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol. 2014. October;80(20):6290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011. January;3(1):118–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A. 2013. March 19;110(12):4720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017. May;106:171–81. [DOI] [PubMed] [Google Scholar]
- 58.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016. March;65(3):426–36. [DOI] [PubMed] [Google Scholar]
- 59.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med. 2019. July;25(7):1096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013. May 28;110(22):9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best Pract Res Clin Gastroenterol. 2017. December;31(6):637–42. [DOI] [PubMed] [Google Scholar]
- 62.Hague A, Butt AJ, Paraskeva C. The role of butyrate in human colonic epithelial cells: an energy source or inducer of differentiation and apoptosis? Proc Nutr Soc. 1996. November;55(3):937–43. [DOI] [PubMed] [Google Scholar]
- 63.Gutierrez N, Garrido D. Species Deletions from Microbiome Consortia Reveal Key Metabolic Interactions between Gut Microbes. mSystems. 2019. July 16;4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality scores of the aggregated forward (top panel) and reverse (bottom panel) 16S rRNA amplicon sequence reads.
Dada2 error models used for the forward (top panel) and reverse (bottom panel) 16S rRNA amplicon sequence reads.
The total number of sequences maintained in each preprocessing step for each sample.
Proportion of the variance (R2) in the infant gut microbial community structure (Weighted UniFrac) that is explained by delivery mode, breastfeeding and pre-pregnancy BMI at 3 months (top) and 12 months (bottom).
Mean relative abundance (%) of A) top 14 bacterial families and B) top 14 bacterial genera, stratified by delivery mode and time of follow up. Remaining taxa were grouped with other.
Differences between C-section-born infants and vaginal-born infants (reference) in the gut microbiome Shannon diversity index at 3 months of age and 12 months of age, before (crude) and after adjustment for maternal pre-pregnancy body mass index and breastfeeding.
Spearman correlations of delivery-mode associated ASVs with fecal SCFA concentrations at 3 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Spearman correlations of delivery-mode associated genera with fecal SCFA concentrations at 3 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Spearman correlations of delivery-mode associated ASVs with fecal SCFA concentrations at 12 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Spearman correlations of delivery-mode associated genera with fecal SCFA concentrations at 12 months of age. A white asterisk indicates an FDR-corrected p-value < 0.05.
Differences in the logit-transformed relative abundances of bacterial ASVs associated with delivery mode at 3 months of age, after adjustment for maternal pre-pregnancy BMI and breastfeeding in beta-binomial models. Unadjusted Mann-Whitney U test p-values provided in rightmost column.
Differences in the logit-transformed relative abundances of bacterial ASVs associated with delivery mode at 12 months of age, after adjustment for maternal pre-pregnancy BMI and breastfeeding in beta-binomial models. Unadjusted Mann-Whitney U test p-values provided in rightmost column.
A list of all ASVs included in the study with their assigned taxonomy.