Abstract
Objectives
Vancomycin-intermediate Staphylococcus aureus (VISA) is increasingly being reported. Previous studies have shown that vraC and vraP may be involved in vancomycin resistance, although the molecular mechanism remains elusive.
Methods
The vraC (SAV0577), vraP (SAV0578) and vraCP mutants were constructed in Mu50 by allelic replacement. Some common VISA phenotypes were assessed in mutants, such as, susceptibility to the cell wall-associated antibiotics, cell wall thickness, autolysis activity and growth rate. RT-qPCR was performed to reveal the differential genes associated with these phenotypes. The binding abilities of VraC and VraCP to the promoters of target genes were determined by electrophoretic mobility shift assay (EMSA).
Results
VraP forms a stable complex with VraC to preserve their own stability. The vraC, vraP and vraCP mutants exhibited increased susceptibility to the cell wall-associated antibiotics and thinner cell walls compared with the WT strain. Consistent with these phenotypes, RT-qPCR revealed downregulated transcription of glyS, sgtB, ddl and alr2, which are involved in cell wall biosynthesis. Moreover, the transcription of cell wall hydrolysis genes, including sceD, lytM and isaA, was significantly downregulated, supporting the finding that mutants exhibited reduced autolysis rates. EMSA confirmed that both VraC and VraCP can directly bind to the sceD, lytM and isaA promoter regions containing the consensus sequence (5′-TTGTAAN2AN3TGTAA-3′), which is crucial for the binding of VraCP with target genes. GFP-reporter assays further revealed VraC and VraCP can enhance promoter activity of sceD to positively regulate its expression.
Conclusions
vraCP plays a significant role in cell wall metabolism and antibiotic resistance in Mu50.
Introduction
Staphylococcus aureus is an important human pathogen that can cause a variety of infections ranging from mild skin and soft-tissue infections to life-threatening infections, such as endocarditis, bacteraemia, pneumonia and chronic osteomyelitis.1–4 The advent and use of antibiotics initially proved effective against S. aureus. However, the rise and widespread prevalence of MRSA poses a serious threat to human health.5,6 Vancomycin has been introduced to treat severe MRSA infections.7 However, S. aureus strains with reduced susceptibility to vancomycin have been increasingly reported worldwide.8–11
Based on levels of vancomycin resistance, vancomycin-non-susceptible S. aureus has been classified into vancomycin-intermediate S. aureus (VISA, MIC = 4–8 mg/L) and vancomycin-resistant S. aureus (VRSA, MIC ≥16 mg/L).12 Because of high prevalence of VISA, VISA is a much greater problem in the clinic than VRSA.13
The cell wall is crucial to bacterial survival.14 Any compromise of the cell wall plays an integral part in antibiotic resistance, because it is targeted by many antibiotics, including β-lactams (oxacillin), glycopeptides (vancomycin and teicoplanin) and other cell wall-associated antibiotics (daptomycin).15,16 Vancomycin interferes with late-stage peptidoglycan synthesis by forming non-covalent hydrogen bonds with the penultimate d-Ala-d-Ala residues of newly synthesized UDP-MurNAc-pentapeptides, thereby disrupting downstream peptidoglycan assembly, cell wall synthesis is ultimately inhibited.17 VISA strains share some common characteristics, including thickened cell wall, decreased autolytic activity, reduced cross-linking of peptidoglycan, slower growth rate and attenuated virulence.18–22
Since VISA strain was first reported in 1997,8 multiple approaches had been used to investigate the molecular genetic basis of the VISA phenotype.18,23–27 These studies revealed several genes and/or mutations in genes that contribute to the development of VISA, such as graRS,28,29vraSR,30walKR,31,32sigB,33,34rpoB,35,36lytM,37sceD,38 and mprF.39 So far, however, the molecular mechanisms underlying vancomycin resistance in VISA have been incompletely defined. It is clear that resistance to vancomycin is complex and involves more genes than those that have been identified to date.
Previously, Kuroda et al.23 reported that the transcription of vraC (measured by cDNA differential hybridization) was remarkably up-regulated in Mu3 and Mu50 as compared with VSSA strain Mu50ω. Many differently expressed genes observed in VISA/VSSA pairs could be induced by treating the corresponding parental strain with vancomycin.33 The transcription profiles of N315 strain exposed to vancomycin revealed that 139 genes are induced, including SA0536,30 whose sequence is identical to that of SAV0578 (vraP) in Mu50. SAV0578 is located 2 nt downstream of vraC (SAV0577). These findings indicated that vraC and vraP may collaborate and contribute to vancomycin resistance. In this study, we investigated the mechanism behind increased susceptibility to the cell wall-associated antibiotics, reduced cell wall thickness and decreased autolysis in vraC, vraP and vraCP mutants with the Mu50 strain background.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1 (available as Supplementary data at JAC Online). Unless otherwise indicated, S. aureus strains were grown with shaking (220 rpm) in tryptic soy broth (TSB; Difco) or on tryptic soy agar (TSA) at 37°C. Escherichia coli strains were grown in lysogeny broth (LB; Oxoid) medium or on lysogeny broth agar (LA) at 37°C. For plasmid maintenance, media were supplemented with ampicillin (150 mg/L) or kanamycin (50 mg/L) for E. coli and chloramphenicol (15 mg/L) for S. aureus, as needed.
Construction of mutant strains
To construct the vraC, vraP and vraCP mutants, the temperature-sensitive shuttle vector pBTs was used as previously described.25 Brifely, the upstream and downstream fragments of individual genes were amplified and ligated by overlap to form up-down fragments. The product fragments were digested with KpnI/SalI and then ligated into the plasmid pBTs with T4 ligase. The resulting plasmids were first transformed into S. aureus RN4220 and then transformed into S. aureus Mu50. Allelic replacement mutants were screened using a previously described method and were further confirmed by PCR and sequencing.40
Growth curves
Overnight cultures of S. aureus were diluted into fresh TSB and grown at 37°C with shaking. The optical density was measured at 600 nm each hour using a microplate reader (Elx800; Bio-Tek). To analyse the growth defect of the mutants in vancomycin-containing medium, Mueller-Hinton broth supplemented with vancomycin at concentrations of 1, 2 and 3 mg/L was used.
Antibiotic susceptibility assay
Antibiotic susceptibility testing was performed by the broth microdilution method, as recommended by the CLSI.41 Population analysis was performed as described previously,42 which is established by plating appropriate dilutions of direct colony suspension on TSA containing increasing concentrations of antibiotics. The numbers of cfu were determined after incubation at 37°C.
Triton X-100-induced autolysis assay
Overnight cultures were diluted to an OD600 of 0.05 in TSB. Cells were harvested at an OD600 of 0.5, washed twice with PBS, resuspended in 0.05 M Tris-HCl buffer (pH 7.5) containing 0.2% (vol/vol) Triton X-100, incubated at 37°C with shaking, and tested for lysis by measuring the absorbance (OD600) each hour using a microplate reader (Elx800; Bio-Tek). The experiment was repeated at least three times.
Expression and purification of proteins
E. coli BL21 containing protein expression plasmid was cultivated in LB at 37°C to an OD600 of 0.5 and induced with 0.5 mM IPTG at 16°C for 20 h. Cells were harvested, resuspended in protein buffer (50 mM Tris-HCl, 300 mM NaCl, pH 8.0) and lysed by sonication on ice. Proteins with C-terminal His tag were purified using a nickel-nitrilotriacetic acid agarose solution (Novagen) according to the manufacturer’s instructions. The bound protein was eluted with elution buffer (50 mM Tris-HCl, 300 mM NaCl, 200 mM imidazole, pH 8.0). The imidazole in the eluent was removed with the protein buffer. Proteins were stored at −80°C until use.
RNA extraction, reverse transcription and quantitative reverse transcription-PCR
Overnight cultures of S. aureus were diluted to an OD600 of 0.05 in TSB, grown to the indicated cell density and collected by centrifugation. The pellet was treated with RNAiso plus (TaKaRa) and lysed with 0.1 mm diameter silica beads in the FastPrep-24 system (MP Biomedicals). The total RNA was extracted. cDNA was synthesized using a PrimeScript first-strand cDNA synthesis kit (TaKaRa), RT-qPCR was performed with SYBR Ex Taq premix (TaKaRa) using the StepOne real-time PCR system (Applied Biosystems) to quantify relative gene expression. pta served as an internal reference gene to normalize the gene expression abundance.43 All RT-qPCR assays were repeated at least three times. The primers used in this study are listed in Table S2.
Transmission electron microscopy
Strains were cultured until exponential growth phase and harvested. Samples were prepared as described previously,25 and sent to the Core Facility Center for Life Science (USTC, China). Specimens were examined with a transmission electron microscope (TEM) operated at an accelerating voltage of 120 kV. Fifteen cells of each strain were measured by Image J.
Electrophoretic mobility shift assay (EMSA)
The biotin-labelled DNA fragments containing promoter regions of target genes were amplified from Mu50 genomic DNA. Various amounts of proteins were incubated with the biotin-labelled probes at 25°C for 30 min. Samples were mixed with gel loading buffer and electrophoresed in a native polyacrylamide gel in 1× Tris-borate-EDTA (TBE) buffer, and then transferred to a nylon membrane in 0.5× TBE buffer. The band shifts were detected according to the manufacturer’s instructions. The images were obtained using ImageQuant LAS 4000 mini (GE, Piscataway, NJ). The unlabelled fragment of each promoter was added as specific competitors. The unlabelled DNA fragment of hu ORF was added as a non-specific competitor.
Fluorescence-based promoter activity assay
Bacteria were cultivated, harvested and washed twice with PBS buffer. Promoter activities were analysed by measuring OD600 and GFP fluorescence (excitation, 488 ± 9 nm; emission, 518 ± 20 nm) using CLARIOstar plate reader (BGM labtech).
Zymographic analysis
Zymographic analyses were performed as described previously.44 Cells were grown until exponential growth phase and harvested. Substrate cells were autoclaved for 15 min at 121°C, lyophilized overnight in a speed-vac, then resuspended thoroughly in water. Proteins were loaded onto 15% SDS-PAGE gels containing 2 mg/L substrate from S. aureus NCTC8325-4. The gels were washed and incubated in renaturation buffer (0.1% Triton X-100, 10 mM CaCl2, 10 mM MgCl2, 50 mM Tris-HCl, pH 7.5) at 37°C with gentle agitation.
Statistical analysis
All experiments were performed in biological triplicates. Graphing and analysis were performed using Origin 2019. Statistically significant differences calculated by the unpaired two-tailed Student’s t test are indicated: NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Results
vraC and vraP are co-transcribed and induced by vancomycin treatment
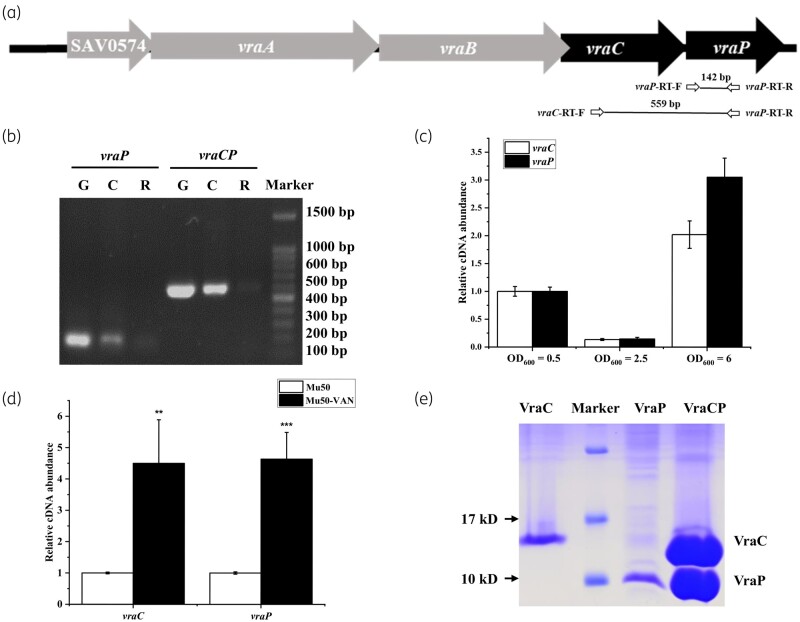
vraCP encodes two proteins of uncharacterized function since no gene homologue was found in extant genes.23vraP (SAV0578) is located 2 nt downstream of vraC (SAV0577) (Figure 1a). BioCyc software predicted that vraC and vraP are co-transcribed.45 To verify this, PCR analysis was performed with primer pairs spanning ORFs of vraC and vraP. Genomic DNA, reverse-transcription cDNA and the same amount of total RNA from Mu50 were used as templates. This result suggested that vraC and vraP could be co-transcribed (Figure 1b).
Figure 1.
vraC and vraP are co-transcribed and induced by vancomycin treatment. (a) Schematic of the vraCP locus in S. aureus Mu50 and the positions of primers used in the PCR assay. The black arrows indicate the genes we studied, and the grey arrows indicate neighbouring genes. (b) Gel electrophoresis analysis of PCR products amplified with primers spanning fragments of vraP and vraCP. Genomic DNA, reverse-transcription cDNA and the same amount of total RNA from strain Mu50 were used as templates. G, Genomic DNA; C, reverse-transcription cDNA; R, RNA. (c) Transcription of vraC and vraP is growth-phase dependent. (d) Transcription of vraC and vraP was induced by vancomycin treatment. (e) Protein electrophoresis of VraC, VraC and VraCP by SDS-PAGE.
Besides, the transcription profiles of vraC and vraP in Mu50 were determined by RT-qPCR. As shown in Figure 1c, the transcriptional levels of vraC and vraP are similar in the same growth phase, and their expression is growth-phase dependent. Meanwhile, vancomycin treatment induced similar transcriptional levels of vraC and vraP (Figure 1d), which further confirmed that vraC and vraP are co-transcribed.
To study the function of these genes, the expression plasmids pEvraC, pEvraP and pEvraCP were constructed. pEvraC and pEvraP contained a C-terminal His6 fusion tag, respectively, pEvraCP only contained a C-terminal His6 fusion tag at vraP gene. We found that VraC and VraP are unstable during the protein purification. However, the expression product of pEvraCP displayed two clear bands corresponding to VraC and VraP (Figure 1e), which indicated that VraC and VraP could form a stable complex by protein–protein interaction. Thus, we named the SAV0578 protein as a partner of VraC (VraP).
The vraC, vraP and vraCP mutants exhibit increased susceptibility to cell wall-associated antibiotics
To understand the effects of vraC, vraP and vraCP on antibiotic resistance in VISA, we constructed the vraC, vraP and vraCP mutants in hospital-associated VISA strain Mu50. Antibiotic susceptibilities of these mutants were evaluated by measuring the MICs of antibiotics according to CLSI.41 Significant decreases in the vancomycin, teicoplanin, oxacillin and daptomycin MICs were detected in the vraC, vraP and vraCP mutants compared with that of WT strain, however, the susceptibilities to other classes of antibiotics exhibited no obvious change (Table S3). Specifically, the MICs of all three mutants decreased from a vancomycin intermediate resistant level of 8 mg/L to a vancomycin susceptible level of 2 mg/L, and the vraCP chromosomal complemented strain exhibited increased MIC, phenocopying the parent strain, which confirmed that the MIC decrease was due to deletion of the vraCP (Table 1).
Table 1.
MICs for different cell wall-targeting antimicrobials in the vraC, vraP and vraCP mutants and Mu50 WT strain
| Strain | MIC (mg/L) of indicated antibiotic |
|||
|---|---|---|---|---|
| Vancomycin | Daptomycin | Teicoplanin | Oxacillin | |
| WT | 8 | 8 | 4 | 512 |
| ΔvraCP | 2 | 4 | 1 | 256 |
| ΔvraC | 2 | 4 | 1 | 256 |
| ΔvraP | 2 | 4 | 2 | 256 |
| ΔvraCP-compl. | 8 | 8 | 4 | 512 |
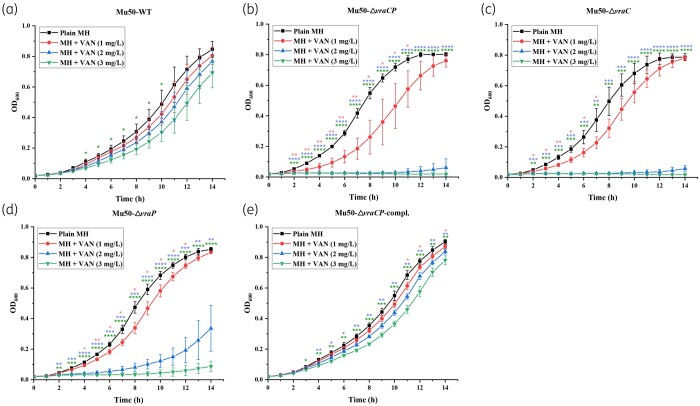
One of the characteristics of VISA is a significant slow-down in growth rates relative to their susceptible counterparts.13,46 Thus, we measured the growth rates of the mutants and WT strain in TSB, and found that the mutants exhibited faster growth rates compared with the WT strain (Figure S1). In addition, we also tested cell growth in MH broth without antibiotic or exposed to different vancomycin concentrations (1, 2 and 3 mg/L). This result showed that the WT and complemented strains displayed slow and steady growth rates at all three concentrations of vancomycin, whereas the three mutants did not display any growth at 2 mg/L or higher vancomycin concentrations (Figure 2). These data confirmed the increased susceptibility of the vraC, vraP and vraCP mutants to vancomycin.
Figure 2.
Growth curves of S. aureus Mu50 and its derivative strains in different concentrations of vancomycin. Growth of the WT strain (a), the vraCP mutant (b), the vraC mutant (c), the vraP mutant (d) and the vraCP complemented strain (e) in MH broth at 37°C containing 0, 1, 2 and 3 mg/L of vancomycin.
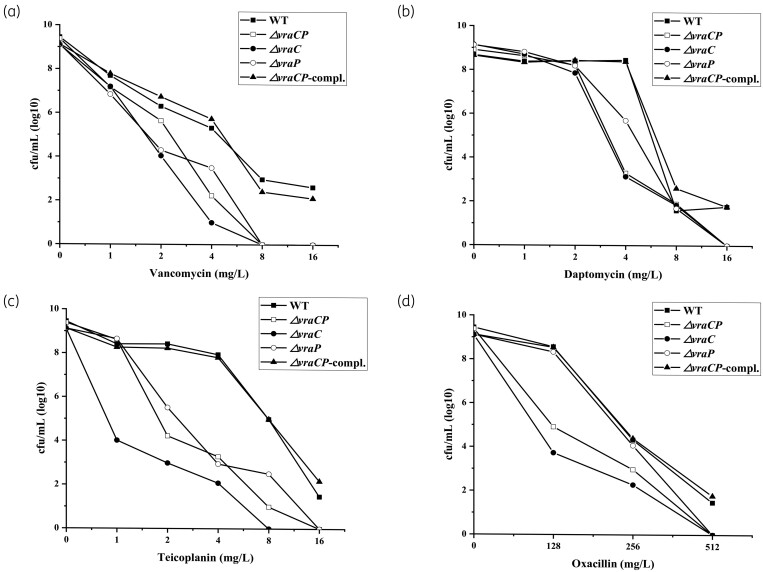
To further characterize the population of the mutants with regard to the cell wall-associated antibiotics resistance, population analysis was performed as described previously,25,47 and the results showed that resistance to the cell wall-associated antibiotics (vancomycin, teicoplanin, daptomycin and oxacillin) in these mutants was significantly reduced (Figure 3), which is consistent with the MIC assay results. The increased susceptibilities of the mutants to cell wall-associated antibiotics suggested that the cell walls of the mutants were compromised.
Figure 3.
Population analysis profiles of S. aureus Mu50 and its derivative strains. The vancomycin (a), daptomycin (b), teicoplanin (c) and oxacillin (d) resistance levels were evaluated by population analysis profiles. The colonies were counted after incubation at 37°C for 48 h.
Deletion of the vraC, vraP and vraCP decreases cell wall thickness
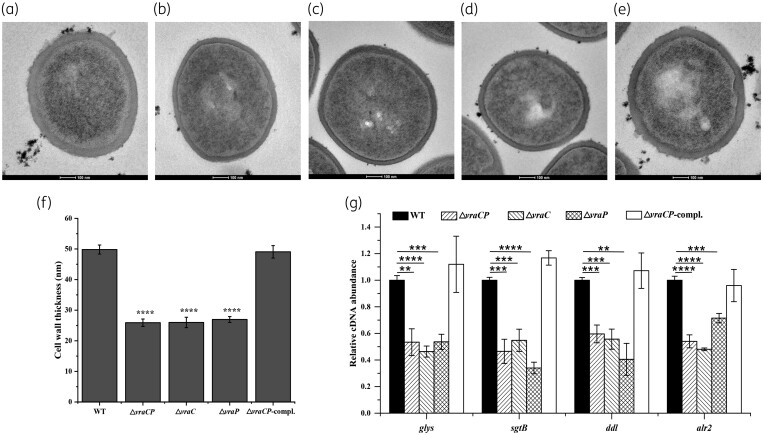
To confirm whether cell wall defects exist in the vraC, vraP and vraCP mutants, cell wall morphology was evaluated using TEM. These data revealed that the mutants showed significant reduction (approximately 46%) in cell wall thickness compared with the WT strain (Figure 4a–f and Table 2). These results suggested that vraC, vraP and vraCP play important roles in cell wall synthesis and maintenance in Mu50.
Figure 4.
Transmission electron micrographs of cell walls of S. aureus Mu50 and its derivative strains. TEM of the WT strain (a), the vraCP mutant (b), the vraC mutant (c), the vraP mutant (d) and the vraCP complemented strain (e) at the early exponential phase. (f) Comparison of cell wall thickness between Mu50 and its derivative strains. Data are expressed as the mean ± SD for 15 cells of each strain. (g) Transcriptional analysis of cell wall biosynthesis-related genes. The transcriptional levels of glys, sgtB, ddl and alr2 significantly decreased in the vraC, vraP and vraCP mutants compared with that of Mu50.
Table 2.
Cell wall thicknesses of the vraC, vraP and vraCP mutants and Mu50 WT strain
| Strain | Cell wall thickness (mean±SD) (nm) |
|---|---|
| WT | 49.81 ± 1.48 |
| ΔvraCP | 25.91 ± 1.21 |
| ΔvraC | 26.00 ± 1.70 |
| ΔvraP | 26.00 ± 1.70 |
| ΔvraCP-compl. | 49.07 ± 2.03 |
Deletion of the vraC, vraP and vraCP influences the expression of cell wall biosynthesis genes
The above data clearly indicated that vraC, vraP and vraCP participate in the cell wall-associated antibiotic resistance and cell wall synthesis. Many genes involved in cell wall synthesis and antibiotic resistance have been reported. To further explore the mechanism behind these phenotypes, RT-qPCR was performed and revealed that the expression of glyS (glycine tRNA synthetase), sgtB (monofunctional peptidoglycan glycosyltransferase), ddl (d-alanine-d-alanine ligase) and alr2 (alanine racemase) significantly decreased in the vraC, vraP and vraCP mutants (Figure 4g), which is consistent with a thinner cell wall and decreased cell wall-associated antibiotic resistance. These data supported this opinion that vraC, vraP and vraCP play an integral part in cell wall synthesis and maintenance in Mu50.
The vraC, vraP and vraCP mutants exhibit decreased autolysis rate and downregulated transcription of cell wall hydrolysis genes
Beyond the thickened cell wall, reduced antibiotic susceptibility and slower growth rate, the change in autolytic activity is also a common feature of VISA.13,21 To determine whether vraC, vraP and vraCP also confer this phenotype, we examined Triton X-100-induced autolytic activity of Mu50 strain as well as its derivatives. These mutants showed reduced autolytic activity compared with the WT strain, And the phenotype was restored in vraCP complemented strain (Figure 5a). Meanwhile, we observed significantly downregulated transcription of sceD, lytM and isaA (Figure 5b). Zymographic assays confirmed cell wall hydrolytic activity of SceD, LytM and IsaA proteins (Figure 5c,d). We therefore hypothesized that vraC, vraP and vraCP influenced in cell wall hydrolysis by regulating the expression of sceD, lytM and isaA.
Figure 5.
The vraC, vraP and vraCP mutants displayed decreased autolysis rate. (a) 0.2% Triton X-100-induced autolysis assay showed that the vraC, vraP and vraCP mutants exhibited reduced autolysis rates than the WT strain. (b) Genes involved in cell wall degradation, such as sceD, lytM and isaA, have decreased expression in the vraC, vraP and vraCP mutants compared with that of Mu50. (c) SDS-PAGE analysis of purified SceD, LytM and IsaA proteins. (d) Zymographic analyses were performed to confirm cell wall hydrolytic activity of SceD, LytM and IsaA proteins. Purified SceD, LytM and IsaA proteins were separated by 15% SDS-PAGE with 2 mg/L S. aureus purified peptidoglycan. (e) Promoter PsceD shows dramatically weaker activity in the vraC, vraP and vraCP mutants compared with that of WT strain. Promoter activities of strains harbouring a transcriptional fusion of PsceD with GFP (pPsceD-GFP) were assessed at different growth phases by measuring fluorescence as well as the OD600. Depicted is GFP emission normalized to the OD600 (GFP/OD600).
VraC and VraCP can enhance promoter activity of sceD
To further verify the regulatory role of vraCP in the expression of sceD, we constructed the sceD promoter-GFP fusion reporter plasmid to detect the promoter activity in Mu50 as well as its derivatives by measuring GFP fluorescence intensity. As predicted, the sceD promoter activity was dramatically weaker in the vraC, vraP and vraCP mutants than WT strain (Figure 5e). These data suggested that VraCP can enhance the promoter activity of sceD, and then promote its expression.
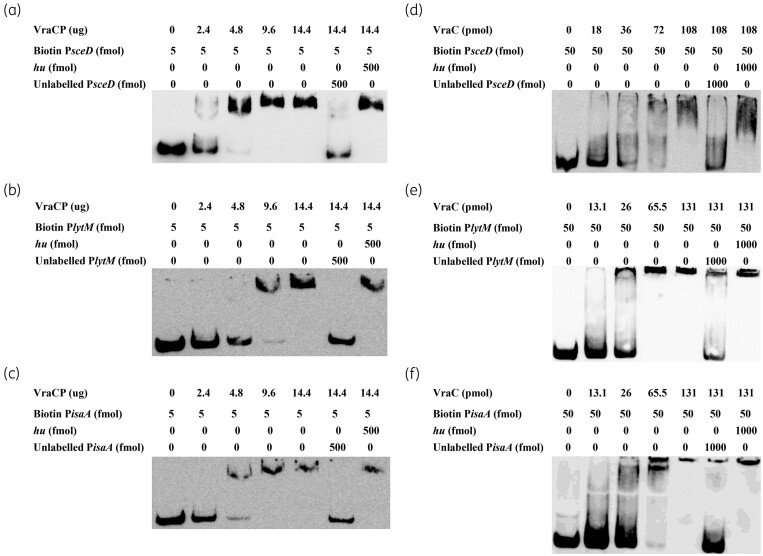
Both VraC and VraCP can directly bind to the promoter regions of sceD, lytM and isaA
To investigate the mechanism underlying altered expression of sceD, lytM and isaA, EMSA was performed with biotin-labelled putative promoter regions and the recombinant VraC and VraCP. The results showed that VraCP can retard the mobility of the sceD, lytM and isaA promoters in a dose-dependent manner (Figure 6a–c). These shifted bands disappeared in the presence of an approximately 100-fold excess of individual unlabelled promoter regions, but not in the presence of a 100-fold excess of an unlabelled coding sequence DNA of hu. As for VraC, similar band shift patterns were observed (Figure 6d–f). These data suggested that both VraC and VraCP can directly bind to sceD, lytM and isaA promoter regions.
Figure 6.
EMSA of VraCP or VraC with the biotin-labelled promoters PsceD, PlytM and PisaA. The promoter regions of sceD, lytM and isaA were amplified by PCR, and incubated with purified VraCP (a–c) and VraC (d–f), respectively. The unlabelled probes were used as the specific competitors, and the unlabelled partial fragment of hu ORF region was used as the non-specific competitor.
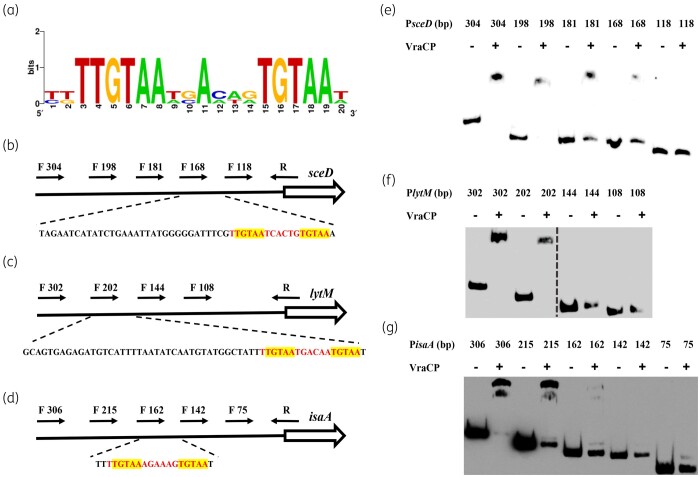
The consensus sequence (5′-TTGTAAN2AN3TGTAA-3′) plays the important role in the binding of VraCP with target genes
Sequence alignment revealed that the potential consensus sequence (5′-TTGTAAN2AN3TGTAA-3′) is present in the promoters of sceD, lytM and isaA, this sequence contains two pentanucleotide direct repeats (TGTAA) (Figure 7a).48 To confirm the role of the consensus sequence in the binding of VraCP, truncated probes with different lengths were designed to perform EMSA (Figure 7b–d). The binding ability of VraCP to PsceD was abolished when the probe length was truncated from 168 bp to 118 bp (Figure 7e). The binding ability of VraCP to PlytM was abolished when the probe length was truncated from 202 bp to 144 bp (Figure 7f). The binding ability of VraCP to PisaA was abolished when the probe length was truncated from 162 bp to 142 bp (Figure 7g). These data demonstrated that the consensus sequence (5′-TTGTAAN2AN3TGTAA-3′) is crucial for the binding of VraCP with target genes.
Figure 7.
The consensus sequence (5′-TTGTAAN2AN3TGTAA-3′) plays an important role in the binding of VraCP with target genes. (a)The consensus sequence (5′-TTGTAAN2AN3TGTAA-3′) in promoter regions of sceD, lytM and isaA was identified by sequences alignment. The locations of PsceD, PlytM and PisaA truncated probes with different lengths are marked by black arrows. The partial relative dispositions of sequence are illustrated in the lower panel. The consensus sequences (5′-TTGTAAN2AN3TGTAA-3′) are marked in red, the two pentanucleotide direct repeats (TGTAA) are highlighted in yellow (b–d). EMSA analysis of VraCP with PsceD, PlytM and PisaA truncated probes (e–g).
Discussion
VISA has attracted considerable attention because of widespread prevalence and treatment failure.9,11 Comparative genome, transcriptome and proteome have been employed to explore the mechanism behind vancomycin resistance.23,25,26,38 Hundreds of mutations were discovered by comparing the genomes of VSSA/VISA pairs. Allelic exchange experiments replacing the normal allele in VSSA with the mutated allele from VISA were performed to evaluate whether allele swapping is responsible for vancomycin resistance. However, reverse-direction studies are rare, because genetic manipulation is challenging in clinical VISA strains, such as Mu50. Meanwhile, several differential expression genes were also detected by transcriptome and proteome analysis.23,26,49 It is obvious that the mechanism of the generation of VISA is complex and varied.
Herein, we explored the mechanism through which vraCP contributes to antibiotic resistance and cell wall metabolism in VISA strain Mu50. The vraC, vraP and vraCP mutants exhibited increased susceptibility to the cell wall-associated antibiotics and reduced cell wall thickness, which are consistent with the VSSA phenotypes. Previous studies revealed that the cell wall thickness and antibiotic resistance are strongly correlated.46,50 VISA strains display thickened cell wall and increased binding of vancomycin to ‘false targets’, thereby contributing to reduced vancomycin susceptibility.22,51–53 Daptomycin needs to penetrate through the cell wall before reaching lethal targets.54 Therefore, one possible reason leading to increased vancomycin and daptomycin susceptibility in mutants is the thinner cell-wall. RT-qPCR was performed to examine the expression of genes involved in cell wall biosynthesis (Figure 4g, Figure S2a), and revealed that the transcriptional levels of glyS, sgtB, ddl and alr2 were decreased in mutants, which may result in thinner cell wall. Ddl and Alr are responsible for terminal stem peptide d-Ala-d-Ala.55–57 The decreased transcription of ddl and alr2 results in reduced free d-Ala-d-Ala residues, which are ‘false targets’ for vancomycin.52 The reduced cell wall thickness and the decreased proportion of ‘false targets’ accelerate penetration of vancomycin molecules to lethal targets, which may be the reason for increased susceptibility to vancomycin and teicoplanin in mutants.
In addition, the mutants exhibited reduced autolytic activity, which seems contradictory to their VSSA-like phonotype. Previous studies revealed that reduced autolytic activity of VISA resulted from decreased expression of cell wall hydrolytic genes, such as atlA, sle1, lytM and lytN.24,25,58 However, in this study, the transcriptional levels of atlA, sle1 and lytN showed no difference between mutants and WT strain (Figure S2b). In contrast, the transcriptional levels of sceD, lytM and isaA were significantly downregulated, which may play the leading roles in reduced autolytic activity of mutants. Among the differentially regulated genes, the change of sceD was the most pronounced. Studies have reported that high expression of sceD is related to vancomycin resistance in VISA,38,49 which is consistent with our study. Both VraC and VraCP can directly bind to sceD, lytM and isaA promoter regions, while VraP is too unstable to be employed for EMSA. The consensus sequence (5′-TTGTAAN2AN3TGTAA-3′) in sceD, lytM and isaA promoter regions is crucial for the binding of VraCP to target genes, which is similar to the WalR binding site (5′-TGTWAH-N5-TGTWAH-3′).59 However, whether an association between VraCP and WalKR exists in the Staphylococcus regulatory network needs further exploration. Meanwhile, GFP-reporter assays revealed that VraC, VraP and VraCP can enhance the sceD promoter activity to promote its expression. However, GFP fluorescence intensity has not been detected in all strains carrying the pPlytM-GFP plasmid. This might be due to the lytM promoter region containing 15 ATG codons, which affects the expression of GFP. Besides, only 4-fold difference of GFP fluorescence intensity in mutants carrying the pPsceD-GFP plasmid was detected compared with the WT strain, but a 20-fold difference was revealed by RT-qPCR. Thus, it is possible that the change of isaA promoter activity is not sufficient to be detected.
Moreover, vraCP was induced by vancomycin and regulated by GraSR and VraSR (Figure S3a, b). Meanwhile, vraCP was expressed at slightly higher levels in Mu50 than its isogenic strain Mu3, overexpression of vraCP in Mu3 could increase vancomycin resistance (Figure S3c, d, Table S4), indicating that the expression of vraCP plays important roles in vancomycin resistance with the Mu50 strain background. However, we constructed vraCP knockdown strains in clinical VISA strain XN108 by CRISPRi,11,60 and no vancomycin resistance change was observed in knockdown strains (Figure S4, Table S5), which is due to the difference in vancomycin resistance mechanism between Mu50 and XN108.
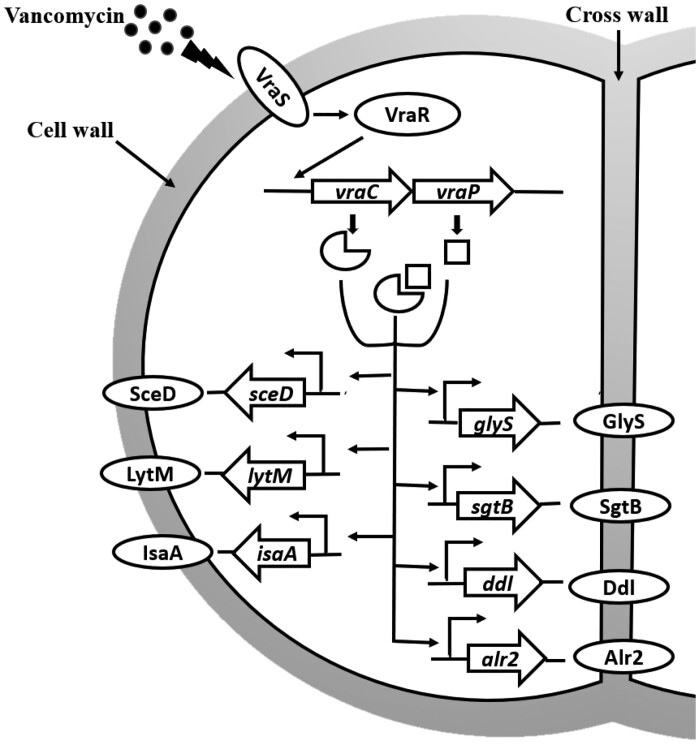
In conclusion, our data revealed that vraCP plays a key role in cell wall metabolism and antibiotic resistance in VISA strain Mu50. Deletion of vraC, vraP and vraCP leads to a series of phenotypic alterations, including increased susceptibility to the cell wall-associated antibiotics, reduced cell wall thickness and decreased autolysis. Interestingly, vraCP can simultaneously promote the expression of cell wall synthesis and hydrolytic genes, which is rare, supporting phenotypic differences as described herein (Figure 8). These results deepen our knowledge of the promotion of vancomycin resistance in Mu50 and provide new insights into the generation and development of VISA strains.
Figure 8.
VraCP is involved in cell wall metabolism and antibiotic resistance in vancomycin-intermediate S. aureus strain Mu50. VraSR can induce the expression of vraCP to response to vancomycin treatment. VraCP promotes the expression of both cell wall synthesis genes (glyS, sgtB, ddl and alr2) and hydrolysis genes (sceD, lytM and isaA), and thus influences cell wall thickness, antibiotic resistance and autolysis rate.
Supplementary Material
Acknowledgements
We thank Xiancai Rao and Xueer Liu for supplying strains.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29020000).
Transparency declarations
None to declare.
Supplementary data
Figures S1 to S4 and Tables S1 to S5 are available as Supplementary data at JAC Online.
References
- 1. Lowy FD. Medical progress - Staphylococcus aureus infections. N Engl J Med 1998; 339: 520–32. [DOI] [PubMed] [Google Scholar]
- 2. Roberts S, Chambers S.. Diagnosis and management of Staphylococcus aureus infections of the skin and soft tissue. Intern Med J 2005; 35: S97–105. [DOI] [PubMed] [Google Scholar]
- 3. Murray RJ. Staphylococcus aureus infective endocarditis: diagnosis and management guidelines. Intern Med J 2005; 35: S25–44. [DOI] [PubMed] [Google Scholar]
- 4. Murdoch DR, Corey GR, Hoen B. et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century. Arch Intern Med 2009; 169: 463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Malani PN. National burden of invasive methicillin-resistant Staphylococcus aureus infection. JAMA 2014; 311: 1438–9. [DOI] [PubMed] [Google Scholar]
- 6. de Kraker MEA, Jarlier V, Monen JCM. et al. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect 2013; 19: 860–8. [DOI] [PubMed] [Google Scholar]
- 7. Sorrell TC, Packham DR, Shanker S. et al. Vancomycin therapy for methicillin-resistant Staphylococcus-Aureus. Ann Intern Med 1982; 97: 344–50. [DOI] [PubMed] [Google Scholar]
- 8. Hiramatsu K, Aritaka N, Hanaki H. et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 1997; 350: 1670–3. [DOI] [PubMed] [Google Scholar]
- 9. Ward PB, Johnson PDR, Grabsch EA. et al. Treatment failure due to methicillin-resistant Staphylococcus aureus (MRSA) with reduced susceptibility to vancomycin. Med J Aust 2001; 175: 480. [DOI] [PubMed] [Google Scholar]
- 10. Lessing MPA, Raftery MJ.. Vancomycin-resistant Staphylococcus aureus. Lancet 1998; 351: 601–2. [DOI] [PubMed] [Google Scholar]
- 11. Zhang X, Hu QW, Yuan WC. et al. First report of a sequence type 239 vancomycin-intermediate Staphylococcus aureus isolate in Mainland China. Diagn Microbiol Infect Dis 2013; 77: 64–8. [DOI] [PubMed] [Google Scholar]
- 12. Tenover FC, Moellering RC.. The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis 2007; 44: 1208–15. [DOI] [PubMed] [Google Scholar]
- 13. McGuinness WA, Malachowa N, DeLeo FR.. Vancomycin Resistance in Staphylococcus aureus. Yale J Biol Med 2017; 90: 269–81. [PMC free article] [PubMed] [Google Scholar]
- 14. Rajagopal M, Walker S.. Envelope structures of Gram-Positive bacteria. Curr Top Microbiol Immunol 2017; 404: 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sewell EWC, Brown ED.. Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot (Tokyo) 2014; 67: 43–51. [DOI] [PubMed] [Google Scholar]
- 16. Bibek GC, Sahukhal GS, Elasria MO.. Role of the msaABCR operon in cell wall biosynthesis, autolysis, integrity, and antibiotic resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2019; 63: e00680-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barna JCJ, Williams DH.. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol 1984; 38: 339–57. [DOI] [PubMed] [Google Scholar]
- 18. Hanaki H, Kuwahara-Arai K, Boyle-Vavra S. et al. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother 1998; 42: 199–209. [DOI] [PubMed] [Google Scholar]
- 19. Daum RS, Gupta S, Sabbagh R. et al. Characterization of Staphylococcus-Aureus isolates with decreased susceptibility to vancomycin and teicoplanin - isolation and purification of a constitutively produced protein associated with decreased susceptibility. J Infect Dis 1992; 166: 1066–72. [DOI] [PubMed] [Google Scholar]
- 20. Boyle-Vavra S, Challapalli M, Daum RS.. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob Agents Chemother 2003; 47: 2036–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boyle-Vavra S, Labischinski H, Ebert CC. et al. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob Agents Chemother 2001; 45: 280–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui L, Ma XX, Sato K. et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 2003; 41: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuroda M, Kuwahara-Arai K, Hiramatsu K.. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun 2000; 269: 485–90. [DOI] [PubMed] [Google Scholar]
- 24. Peng HG, Hu QW, Shang WL. et al. WalK(S221P), a naturally occurring mutation, confers vancomycin resistance in VISA strain XN108. J Antimicrob Chemoth 2017; 72: 1006–13. [DOI] [PubMed] [Google Scholar]
- 25. Hu JF, Zhang X, Liu XY. et al. Mechanism of reduced vancomycin susceptibility conferred by walK mutation in community-acquired methicillin-resistant Staphylococcus aureus strain MW2. Antimicrob Agents Chemother 2015; 59: 1352–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scherl A, Francois P, Charbonnier Y. et al. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics 2006; 7: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfeltz RF, Singh VK, Schmidt JL. et al. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob Agents Chemother 2000; 44: 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neoh HM, Cui L, Yuzawa H. et al. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob Agents Chemother 2008; 52: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howden BP, Stinear TP, Allen DL. et al. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob Agents Chemother 2008; 52: 3755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuroda M, Kuroda H, Oshima T. et al. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 2003; 49: 807–21. [DOI] [PubMed] [Google Scholar]
- 31. Howden BP, McEvoy CRE, Allen DL. et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. Plos Pathog 2011; 7: e1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shoji M, Cui LZ, Iizuka R. et al. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob Agents Chemother 2011; 55: 3870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McAleese F, Wu SW, Sieradzki K. et al. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S-aureus-type resistance to vancomycin. J Bacteriol 2006; 188: 1120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui LZ, Lian JQ, Neoh HM. et al. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2005; 49: 3404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cui LZ, Isii T, Fukuda M. et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother 2010; 54: 5222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsuo M, Hishinuma T, Katayama Y. et al. Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA-to-VISA phenotypic conversion of strain Mu3. Antimicrob Agents Chemother 2011; 55: 4188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Utaida S, Pfeltz RF, Jayaswal RK. et al. Autolytic properties of glycopeptide-intermediate Staphylococcus aureus Mu50. Antimicrob Agents Chemother 2006; 50: 1541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Drummelsmith J, Winstall E, Bergeron MG. et al. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J Proteome Res 2007; 6: 4690–702. [DOI] [PubMed] [Google Scholar]
- 39. Ruzin A, Severin A, Moghazeh SL. et al. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Bba-Gen Subjects 2003; 1621: 117–21. [DOI] [PubMed] [Google Scholar]
- 40. Bae T, Schneewind O.. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 2006; 55: 58–63. [DOI] [PubMed] [Google Scholar]
- 41.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Sixth Edition: M100.2016.
- 42. Wootton M, Howe RA, Hillman R. et al. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 2001; 47: 399–403. [DOI] [PubMed] [Google Scholar]
- 43. Valihrach L, Demnerova K.. Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J Microbiol Methods 2012; 90: 214–6. [DOI] [PubMed] [Google Scholar]
- 44. Vaz F, Filipe S.. Preparation and Analysis of Crude Autolytic Enzyme Extracts from Staphylococcus aureus. Bio-Protocol 2015; 5: e1687. [Google Scholar]
- 45. Romero PR, , Karp PD. Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics 2004; 20: 709–17. [DOI] [PubMed] [Google Scholar]
- 46. Cui LZ, Murakami H, Kuwahara-Arai K. et al. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob Agents Chemother 2000; 44: 2276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu XY, Zhang SJ, Sun BL.. SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-resistant Staphylococcus aureus strain N315. Antimicrob Agents Chemother 2016; 60: 3455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crooks GE, Hon G, Chandonia JM. et al. WebLogo: a sequence logo generator. Genome Res 2004; 14: 1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pieper R, Gatlin-Bunai CL, Mongodin EF. et al. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 2006; 6: 4246–58. [DOI] [PubMed] [Google Scholar]
- 50. Thitiananpakorn K, Aiba Y, Tan XE. et al. Association of mprF mutations with cross-resistance to daptomycin and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA). Sci Rep 2020; 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sieradzki R, Pinho MG, Tomasz A.. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J Biol Chem 1999; 274: 18942–6. [DOI] [PubMed] [Google Scholar]
- 52. Pereira PM, Filipe SR, Tomasz A. et al. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant staphylococcus aureus. Antimicrob Agents Chemother 2007; 51: 3627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cui LZ, Iwamoto A, Lian JQ. et al. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2006; 50: 428–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schriever CA, Fernandez C, Rodvold KA. et al. Daptomycin: a novel cyclic lipopeptide antimicrobial. Am J Health Syst Pharm 2005; 62: 1145–58. [DOI] [PubMed] [Google Scholar]
- 55. Sobral RG, Ludovice AM, Gardete S. et al. Normally functioning murF is essential for the optimal expression of methicillin resistance in Staphylococcus aureus. Microb Drug Resist 2003; 9: 231–41. [DOI] [PubMed] [Google Scholar]
- 56. Sobral RG, Ludovice AM, de Lencastre H. et al. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J Bacteriol 2006; 188: 2543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gallagher LA, Shears RK, Fingleton C. et al. Impaired alanine transport or exposure to D-cycloserine increases the susceptibility of MRSA to beta-lactam antibiotics. J Infect Dis 2020; 221: 1006–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Katayama Y, Sekine M, Hishinuma T. et al. Complete reconstitution of the vancomycin-intermediate Staphylococcus aureus phenotype of strain Mu50 in vancomycin-susceptible S-aureus. Antimicrob Agents Chemother 2016; 60: 3730–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dubrac S, Msadek T.. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 2004; 186: 1175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao CL, Shu XQ, Sun BL.. Construction of a gene knockdown system based on catalytically inactive ("Dead") Cas9 (dCas9) in Staphylococcus aureus. Appl Environ Microb 2017; 83: e00291-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bao Y, Li YJ, Jiang Q. et al. Methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs) of Staphylococcus aureus is essential for the virulence independent of LuxS/AI-2 system. Int J Med Microbiol 2013; 303: 190–200. [DOI] [PubMed] [Google Scholar]
- 62. Corrigan RM, Foster TJ.. An improved tetracycline inducible expression vector for Staphylococcus aureus. Plasmid 2009; 61: 126–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.