Abstract
Normal contractile function of the heart depends on a constant and reliable production of ATP by cardiomyocytes. Dysregulation of cardiac energy metabolism can result in immature heart development and disrupt the ability of the adult myocardium to adapt to stress, potentially leading to heart failure. Further, restoration of abnormal mitochondrial function can have beneficial effects on cardiac dysfunction. Previously, we identified a novel protein termed Perm1 (PGC-1 and estrogen-related receptor (ERR)-induced regulator, muscle 1) that is enriched in skeletal and cardiac-muscle mitochondria and transcriptionally regulated by PGC-1 (peroxisome proliferator-activated receptor gamma coactivator 1) and ERR. The role of Perm1 in the heart is poorly understood and is studied here. We utilized cell culture, mouse models, and human tissue, to study its expression and transcriptional control, as well as its role in transcription of other factors. Critically, we tested Perm1's role in cardiomyocyte mitochondrial function and its ability to protect myocytes from stress-induced damage. Our studies show that Perm1 expression increases throughout mouse cardiogenesis, demonstrate that Perm1 interacts with PGC-1α and enhances activation of PGC-1 and ERR, increases mitochondrial DNA copy number, and augments oxidative capacity in cultured neonatal mouse cardiomyocytes. Moreover, we found that Perm1 reduced cellular damage produced as a result of hypoxia and reoxygenation-induced stress and mitigated cell death of cardiomyocytes. Taken together, our results show that Perm1 promotes mitochondrial biogenesis in mouse cardiomyocytes. Future studies can assess the potential of Perm1 to be used as a novel therapeutic to restore cardiac dysfunction induced by ischemic injury.
Keywords: cardiomyocytes, mitochondrial biogenesis, Perm1, oxidative metabolism
Abbreviations: ATP5b, ATP synthase F1 subunit beta; Cox, cytochrome c oxidase; COXIV, cytochrome c oxidase subunit IV; ERR, estrogen-related receptor; H/R, hypoxia-reoxygenation; LDH, lactate dehydrogenase; Mito, mitochondrial; NCM, neonatal cardiomyocytes; NDUFS3, NADH dehydrogenase [ubiquinone] iron–sulfur protein 3; OCR, oxygen consumption rate; OxPhos, oxidative phosphorylation; Perm1, peroxisome proliferator-activated receptor coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator, muscle 1; PGC-1, PPARgamma coactivator 1; SDHb, succinate dehydrogenase complex iron–sulfur subunit B; SERCA, Sarco/endoplasmic reticulum Ca2+-ATPase; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; UQCRC2, ubiquinol-cytochrome C reductase core protein 2
Proper homeostasis of cellular energy is central to maintain normal heart development and cardiac muscle function (1, 2). Impaired capacity for energy production, as seen in patients with mutations in mitochondrial DNA or nuclear genes encoding lipid oxidation enzymes or respiratory chain components, can lead to cardiac dysfunction (3, 4). An inability to match energy supply to energy needs is also seen in animal models of heart failure (HF) as well as cardiomyopathies in humans (5, 6). For instance, cardiac mitochondrial number and activity are downregulated in myocardium in both animal models of HF and HF patients (7, 8, 9). A similar phenomenon is seen in animals challenged by myocardial infarction (MI) or ischemia-reperfusion (I/R) injury (10). By contrast, restoring mitochondrial function has beneficial effects on HF caused by I/R injury and MI (11, 12). Thus, understanding the molecular networks that regulate mitochondrial biogenesis, fueling, and function may lead to future novel therapeutic targets that can be manipulated for the treatment of diseases associated with cardiac bioenergetic defects.
Peroxisome proliferator-activated receptor (PPAR) gamma coactivator 1 (PGC-1) and the nuclear receptor, estrogen-related receptor (ERR), play central roles in cardiac metabolism and bioenergetics (13). The expression of these factors in cardiac myocytes (CM) parallels the increase in mitochondrial biogenesis and becomes decreased in HF, concurrent with the energetic deficiencies found in this pathological state (14). Mice lacking PGC-1α or PGC-1β show energetic defects in the heart, and PGC-1α null mice develop HF when subjected to hemodynamic stress (15). Further, combined global knockout (KO) of both PGC-1α and PGC-1β causes perinatal lethality, with mice showing bradycardia, heart block, reduced heart size, and ultimately evidence of HF (16). ERRα-null mice also develop signs of HF when challenged with cardiac pressure overload, while ERRγ-null mice show metabolic defects in the perinatal switch from glucose to fatty acid oxidation (17, 18). Thus, identifying and defining the role of novel regulators that work along with PGC-1s and ERRs is necessary to fully understand the regulation of the cardiac energetic state, both at rest and with disease.
Previously, we isolated a novel muscle enriched PGC-1/ERR target termed Perm1 (PGC-1 and ERR-induced regulator, muscle 1) (19). Perm1 is highly expressed in the heart and skeletal muscle, and in skeletal muscle, it is induced directly by PGC-1/ERR as well as by increased energetic demand, such as endurance exercise (19). In cultured skeletal myotubes, Perm1 is required for PGC-1α-induced mitochondrial biogenesis and selected PGC-1α and ERRγ target gene expression. Overexpression of Perm1 in skeletal muscle also causes increased mitochondrial biogenesis and maximal oxidative capacity (20). By contrast, reduction of skeletal muscle Perm1 attenuates endurance exercise-induced Mito biogenesis (21). Together this work shows that Perm1 plays an important role in skeletal muscle oxidative metabolism.
We pursued the current work since Perm1 is highly expressed in the heart and its CM, as well as skeletal muscle, but its role in CM and the intact heart is largely unknown (22). Here we assess its expression in the developing embryo and postnatal heart. Then we employ a primary mouse neonatal cardiomyocyte (NCM) culture model. Our findings show that Perm1 is a positive regulator of mitochondrial biogenesis and oxidative capacity in CM and protects CM from cellular damage induced by hypoxia reoxygenation.
Results
Perm1 protein increases during cardiac development and maturation
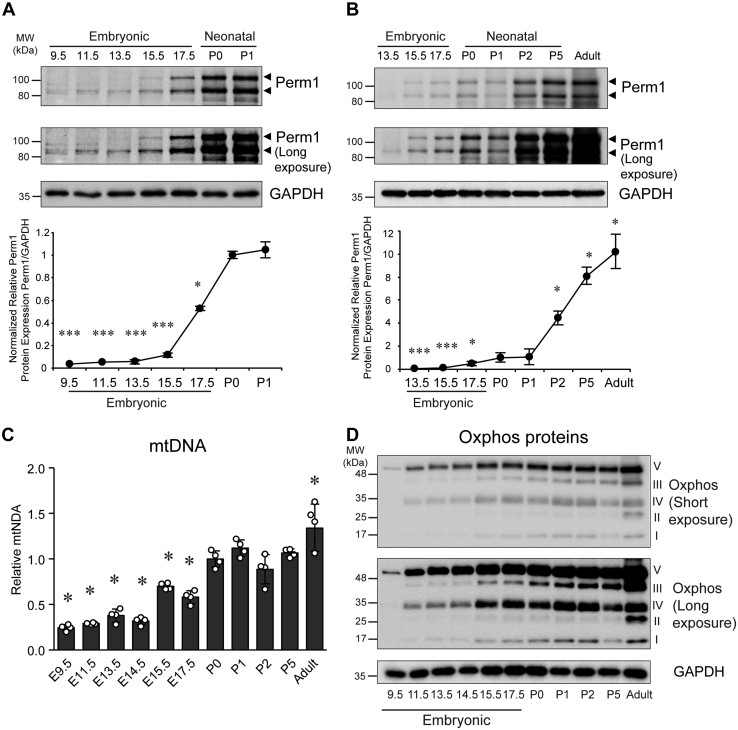
To begin to understand the role of Perm1 in the heart, we first studied if Perm1 is involved in the physiologic regulation of mitochondrial (Mito) function and biogenesis in the developing heart. Perm1 protein expression was analyzed in embryonic and postnatal hearts. Evaluating mitochondrial protein expression during development is important since mitochondrial number and activity rise to meet the increased energy demand and contractile activity of the heart as it transitions to its postnatal state (23). Using mouse tissue from defined stages of cardiogenesis, we evaluated cardiac tissues at embryonic days (E) 9.5–17.5, postnatal days (P) 0–5, and in the adult heart (10-week-old). Whole heart tubes were assessed at E9.5, with ventricular tissue sampled at later time points. Of note, there are two major isoforms of Perm1 protein, the short isoform is about 85 kDa, and the long isoform is 105 kDa. While we have shown that these isoforms arise given varied transcriptional start sites, their functional differences are currently not understood (see below). At early embryonic stages, E9.5 and E11.5, only the short Perm1 form was detectable, albeit weakly. In addition, we also observed an additional faint band (95 kDa) at E9.5 and E11.5. By E13.5 both major isoforms were expressed, and expression was seen to increase significantly through to birth (P0). (Fig. 1, A and B). With additional maturation (P0 to adult), Perm1 protein expression continued to increase. These results show that Perm1 protein expression is developmentally upregulated from embryonic stages to adult, suggesting its increased functional importance as the heart matures.
Figure 1.
Expression level of Perm1 protein, mitochondrial DNA copy number, and Oxphos proteins increase during cardiac development. Whole tissue protein and DNA were extracted from cardiac tissues at various embryonic stages or from neonatal pups and adult mice. A and B, Upper panel, western blot analyses with antibodies indicated. Long-exposure images are shown to highlight the differences of Perm1 between embryonic, postnatal, and adult hearts. Lower panel, quantification of Perm1 protein level. (Expression of Perm1 protein was normalized first to GAPDH in each sample and then relative amounts were expressed versus that in P0 heart samples, which was set = 1.) (n = 4). C, the relative mtDNA content was determined as the ratio of mitochondrial (CoxII) to genomic (Dio3) DNA copy numbers and expressed relative to the ratio seen in P0 hearts. Data are the mean ± SD, expressed relative to P0 (n = 4). ∗p < 0.05; ∗∗∗p < 0.001 versus P0. D, representative images of western blots of OxPhos proteins are shown. Quantification is shown in Figure S1. (For all studies, whole heart tubes were sampled at E9.5, with ventricular tissue used at later stages.)
To begin to evaluate the functional role that Perm1 might have in the maturing heart, we evaluated Mito biogenesis during cardiac development. As shown in Figure 1C, Mito DNA (mtDNA) levels were seen to significantly increase in heart tissue as it matured from embryonic stages to adult hearts, in parallel with the Perm1 changes.
It is well known that Mito oxidative phosphorylation (OxPhos) is a crucial metabolic pathway during which 5 MDa enzyme complexes transfer electrons to oxygen, resulting in ATP generation. Studies show that an increase in OxPhos protein is accompanied by Mito biogenesis (23, 24). Thus, we next analyzed OxPhos protein expression during cardiac development. As shown in Figures 1D and S1, similar to Perm1 protein levels and mtDNA copy number, OxPhos proteins also became upregulated during cardiac development and postnatally. OxPhos protein levels significantly increased through embryonic stages and were generally dramatically increased as the heart fully matured in the adult form.
Next, given that our prior work showed that Perm1 is a target of PGC-1 and ERR in skeletal muscle, we analyzed mRNA levels of Perm1, PGC-1s, and ERRs using hearts from the different embryonic and postnatal stages. As shown in Figure 2A, consistent with the protein expression of Perm1, the level of Perm1 mRNA in cardiac tissue significantly increased as heart maturation progressed from embryonic stages to newborn (P0). Adult hearts showed a 2-fold higher Perm1 mRNA level as compared with P0 hearts, indicating that Perm1 mRNA increases even postnatally. As displayed in Figure 2B, the level of PGC-1α mRNA gradually increased through cardiac development till P1 and then decreased at P2 and P5. Adult hearts showed similar PGC-1α levels as P0, whereas PGC-1β, ERRα, and ERRγ had similar expression patterns as PGC-1α (Fig. 2, C–E). Interestingly, although ERRβ mRNA remained at a low level from P0 to P5, in the adult heart its levels increased 6-fold compared with P0 (Fig. 2F). These results indicate that Perm1, PGC-1s, and ERRs gradually increase throughout cardiac development toward birth, but also show that these factors exhibit unique expression patterns in the postnatal heart.
Figure 2.
Transcript level of Perm1, PGC-1s, and ERRs increased during cardiac development and maturation. Protein and RNA were extracted from cardiac tissues at a different time points of embryogenesis, from neonatal pups and adult mice. mRNA levels for: (A) Perm1, (B) PGC-1α, (C) PGC-1β, (D) ERRα, (E) ERRγ, and (F) ERRβ were determined by RT-qPCR, normalized by 36B4 levels, and expressed relative to levels of each gene in P0 heart, which was set = 1. Data are the mean ± SD, expressed relative to P0 (n = 4). ∗p < 0.05 versus P0. (Whole heart tubes were sampled at E9.5, with ventricular tissue used at later stages.)
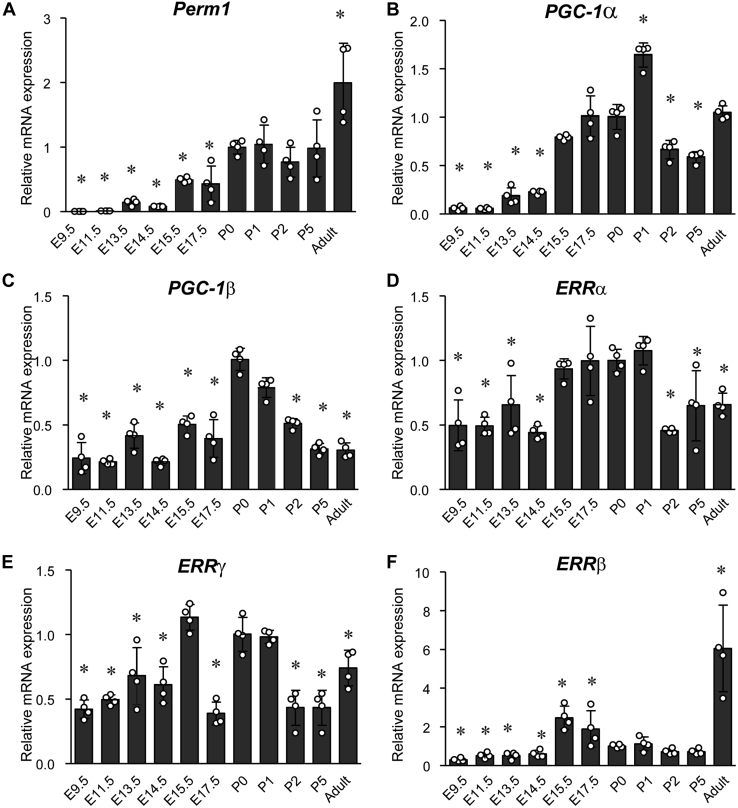
Perm1 is enriched in the ventricle versus atria and downregulated in failing human heart
The expression pattern of Perm1 in specific cardiac chambers, as well as its subcellular localization in CM, has not been previously explored. Also, it is not clear if Perm1 expression is altered in myocardial disease in animals and humans. Perm1 protein levels were first analyzed in mouse atria and ventricles and then compared with samples from gastrocnemius (GAST) skeletal muscle. GAST was used as a control sample since it is known to highly express Perm1 based on our prior work (19, 21). As shown in Figure 3A, ventricular tissue showed 2-fold higher expression of Perm1 protein as compared with atria or GAST. Next, we performed biochemical fractionation of mouse ventricular tissue to study the subcellular localization of Perm1 in that cardiac chamber. Ventricular homogenates were fractionated into cytosolic, Mito, sarcoplasmic reticulum (SR), and nuclear fractions by differential centrifugation. Antibodies against GAPDH (cytosolic), Cox (Mito), SERCA2a (SR), and Lamin A/C (Nuclear) were used to demonstrate the purity of each fraction. As shown in Figure 3B, we found Perm1 present in all fractions, though with relatively higher enrichment in Mito and SR fractions. A recent study also showed that Perm1 protein is localized in part to Mito in CM (25). As Perm1 does not have a Mito localization signal, it is possible that Perm1 may interact with other Mito proteins, leading to its localization in Mito. These findings suggest that Perm1 protein expression is not absolutely restricted to Mito, indicating it may have a unique function in different subcellular components of the ventricular myocyte.
Figure 3.
Cardiac Perm1 is enriched in the ventricle and becomes decreased in failing human heart. A, the protein levels of Perm1 in the atrium, ventricle, and GAST muscles were determined by western blot analysis and quantified (right panel). Arrows indicate the two major protein isoforms encoded for by Perm1. (Expression of Perm1 protein was first normalized to GAPDH in each sample and then relative amounts were determined relative to atrial expression, which was set = 1.) Data are the mean ± SD, expressed relative to the atrium (n = 3). ∗p < 0.05 versus ventricle. B, adult mice heart tissue was subjected to subcellular fractionation, followed by western blotting. Antibodies against Perm1 (top panel), cytoplasmic GAPDH, mitochondrial Cox, sarcoplasmic reticulum SERCA2a, and nuclear lamins (shown as four western blots beneath Perm1) were used to assess the purity of the cytoplasmic, mitochondrial, SR (sarcoplasmic reticulum), and nuclear fractions. Representative images from three independent experiments are shown. C, total RNA was extracted from the ventricles of adult mice subjected to Sham or TAC surgery. mRNA levels for Perm1 were determined by RT-qPCR, normalized by 36B4 levels, and expressed relative to normalized expression in Sham-treated mice that were set = 1. (n = 6–8). D, whole tissue protein extracted from the ventricles of Sham or TAC mice was subjected to western blot with antibodies indicated (Perm1 and GAPDH). (n = 4). E, whole tissue protein extracted from the left ventricle of DCM (dilated cardiomyopathy) patients and normal subjects (Control) was evaluated with western blot using the antibodies indicated. (n = 6) Data are the mean ± SD, expressed relative to sham or control. ∗p < 0.05; ∗∗p < 0.01.
Cardiomyopathic hearts in humans and animals show a reduction in oxidative metabolism and Mito dysfunction (7, 8, 9, 26). Also, the expression and activity of cardiac PGC-1α and ERRs are downregulated in cardiac tissue from HF patients and animal models of HF (14), indicating that the downregulation of PGC-1/ERR signaling may contribute to the pathogenesis of HF. Given this, we next analyzed Perm1 expression using C57Bl6 mouse heart tissue from sham-operated mice (control) versus mice with HF induced by pressure overload produced by transverse aortic constriction (TAC) (27, 28). Mice showed significantly decreased cardiac function 8 weeks post-TAC (fractional shortening—TAC = 15%, versus Sham = 40%) with associated cardiac hypertrophy (Table S1). As shown in Figure 3, C and D, Perm1 mRNA and protein were significantly decreased in TAC compared with Sham hearts, showing that decreased Perm1 levels are associated with cardiac dysfunction. Furthermore, we also analyzed PERM1 protein levels in ventricular tissue from normal human heart as compared with samples from explanted tissue obtained from patients with dilated cardiomyopathy (DCM) (28, 29). As shown in Figure 3E, DCM tissue exhibited significantly decreased PERM1 protein levels compared with normal control myocardium. These data clearly show that Perm1 is downregulated in cardiac tissue from failing as compared with normally functioning heart, in both mice and humans.
An internal translational initiation site leads to production of the short isoform of Perm1
As discussed above, there are two major isoforms of Perm1 protein (Fig. 1A and (19)). The larger-size protein isoform (termed Perm1-L, in contrast to the shorter isoform, Perm1-S) was found to be the predominant isoform expressed when our prior work forced overexpression of full-length Perm1 cDNA in skeletal muscle and heart (19). Bioinformatic analysis of human and mouse Perm1 sequences showed two conserved AUGs (methionine/putative translational start sites) that were both in-frame (Fig. S2). Therefore, we studied if the two Perm1 forms detected by western blotting might originate from these two AUG sites, with the longer form starting at the upstream site and the shorter isoform, beginning from the downstream, internal translational initiation at the second in-frame AUG.
First, we employed cDNA cloning and mutagenesis (Fig. S3, A–C). A Perm1 cDNA coding from Exon1 through the stop codon (plasmid A) was cloned into plasmid vector pcDNA3. This sequence included an untranslated 5′ region of Perm1, as well as its full-length coding sequence. Next, the second AUG, which still remained in the Plasmid A clone, was mutated to CGA (Alanine), forming Plasmid B. CGA cannot function as a translational initiation site. We also cloned Perm1 cDNA without the 5′ untranslated region into pcDNA3, beginning with the first AUG (termed plasmid C) or starting at the second AUG (termed plasmid D) (Fig. S3A). The various plasmids were then transfected into HEK293 cells. Protein lysates from the transfected cells were obtained and subjected to western blotting using an anti-PERM1 antibody, which recognizes the C-terminal region of Perm1. As shown in (Fig. S3B), plasmid A samples were shown to express two isoforms of Perm1, of identical sizes as endogenous Perm1, suggesting that a single Perm1 cDNA generates two isoforms of Perm1. Mutation of the second AUG to CGA (plasmid B) resulted in detection of the 105 kDa long isoform, without the shorter 85 kDa short isoform, suggesting that the “short” Perm1 form results from start-site usage of the downstream, in-frame AUG. Like plasmid A, plasmid C expressed two major isoforms of Perm1. Plasmid D, which only contained sequence beginning at the second AUG, expressed only the short isoform. These results show that translational initiation from the second AUG (30, 31, 32) generates the short isoform of Perm1.
Next, we tested if a Kozak sequence affects the translation of the Perm1 isoforms. A Kozak sequence (ACC) (33) was introduced into Perm1 cDNA clone beginning with the first AUG (Plasmid E) or the second AUG (Plasmid F). Following similar HEK293 transfection and western blot analysis, Plasmid E expressed both isoforms with higher expression of the long isoform of Perm1, and Plasmid F expressed only the short isoform of Perm1 (Fig. S3C). These results show that the Kozak sequence can determine translational initiation of Perm1. When a Kozak is adjacent to the first AUG, the long isoform will be predominantly expressed.
In addition to Perm1-L and Perm1-S, there are two lower-molecular-weight bands detected below Perm1-S, at approximately 70 and 75 kDa (Perm1-70 and Perm1-75), respectively. Endogenous levels of Perm1-70 and 75 are approximately 10-fold lower than Perm1-L and Perm1-S. However, when the second AUG is mutated (Plasmid B), the protein level of Perm1-70 and 75 increased in the transfected cells (Fig. S3B). These results raise the possibility that Perm1-70 and 75 could be derived from other internal AUGs.
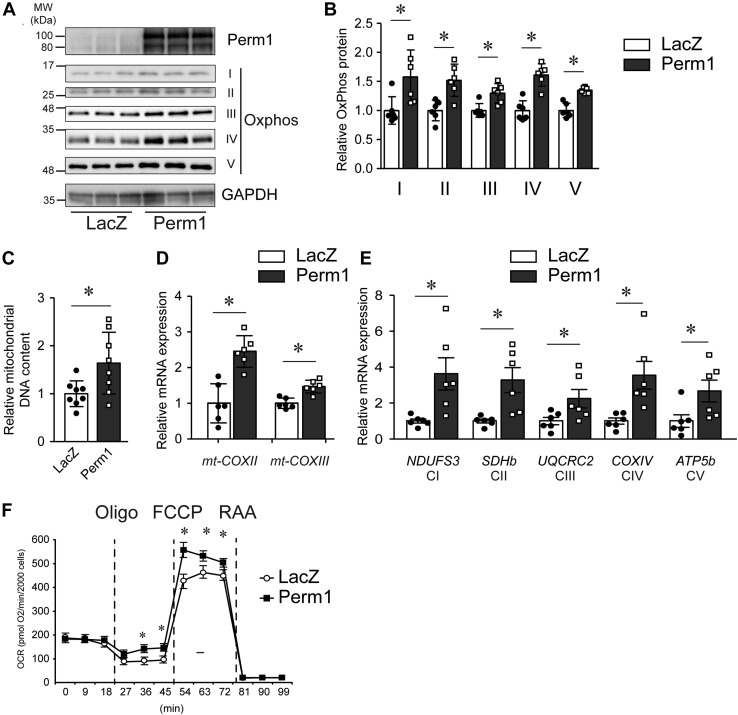
Perm1 overexpression increases cardiomyocyte mitochondrial DNA content and enhances cellular oxidative capacity
Given the potential role of Perm1 in CM Mito biology and function, next we tested if Perm1 regulates Mito biogenesis and oxidative metabolism in CM. Mouse neonatal CM (NCM) were infected with recombinant adenoviruses expressing Perm1 or β-galactosidase (LacZ) as control. Forty-eight hours postinfection, cells were harvested. First, we evaluated OxPhos protein expression and Mito DNA (mtDNA) copy number. As shown in Figure 4, A and B, Perm1 overexpression significantly increased OxPhos protein expression (complexes I–IV). In addition, Perm1 increased mtDNA copy number, which was accompanied by higher expression levels of genes encoded for by the Mito genome, mt-CoxII (cytochrome c oxidase subunit 2), and mt-CoxIII (cytochrome c oxidase subunit 3) (Fig. 4, C and D). Furthermore, Perm1 increased nuclear-encoded OxPhos genes (NDUFS3, SDHb, UQCRC2, COXIV, and ATP5b, CI–CV) (Fig. 4E).
Figure 4.
Perm1 enhances mitochondrial biogenesis in neonatal mouse cardiomyocytes. Primary cultured neonatal mouse cardiomyocytes (NCM) were infected with adenoviruses expressing LacZ or Perm1. Forty-eight hours later, the cells were harvested. A and B, whole cell protein lysates were subjected to western blot using antibodies as indicated. OxPhos protein levels were quantified with expression level of LacZ (control) infected cells = 1. C, the relative mtDNA content was determined in Perm1-infected cells, relative to that detected in control (LacZ)-infected NCM. (LacZ expression was set =1.) D and E, mRNA levels for the indicated mitochondrial genome-encoded OxPhos genes mt-CoxII and mt-CoxIII and other OxPhos component genes, NDUFS3, SDHb, UQCRC2, COXIV, and ATP5b were determined by RT-qPCR, normalized to 36B4 levels, and expressed relative to levels of each gene in control (LacZ) NCM, set = 1. B–E, data are the mean ± SD, expressed relative to LacZ. ∗p < 0.05 versus LacZ. Data are the mean of six experimental replicates from two representative experiments. F, oxygen consumption rates of NCM were measured in the absence and presence of 1 μM oligomycin (Oligo), 800 nM FCCP, and 1 μM rotenone/antimycin (RAA). Rates are normalized by 20 × 103 cells. Data are the mean ± SD. ∗p < 0.05 versus LacZ. Data are the mean of nine experimental replicates from two representative experiments. Data are the mean ± SD, expressed relative to LacZ. ∗p < 0.05 versus LacZ.
Importantly, to show the functional effects of the increased Mito DNA content and OxPhos protein produced by Perm1, oxygen consumption was next evaluated. As shown in Figure 4F, while Perm1 overexpression had no effect on the basal respiration rate of the NCM, it significantly enhanced maximal oxygen consumption when NCM were treated with the uncoupling agent trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP). Uncoupled respiration produced by oligomycin (Oligo) was also increased by Perm1 overexpression. These results suggest that Perm1 enhances functional Mito biogenesis in the CM and can result in increased maximal oxygen consumption.
Perm1 modulates selective genes involved in oxidative metabolism in cardiomyocytes
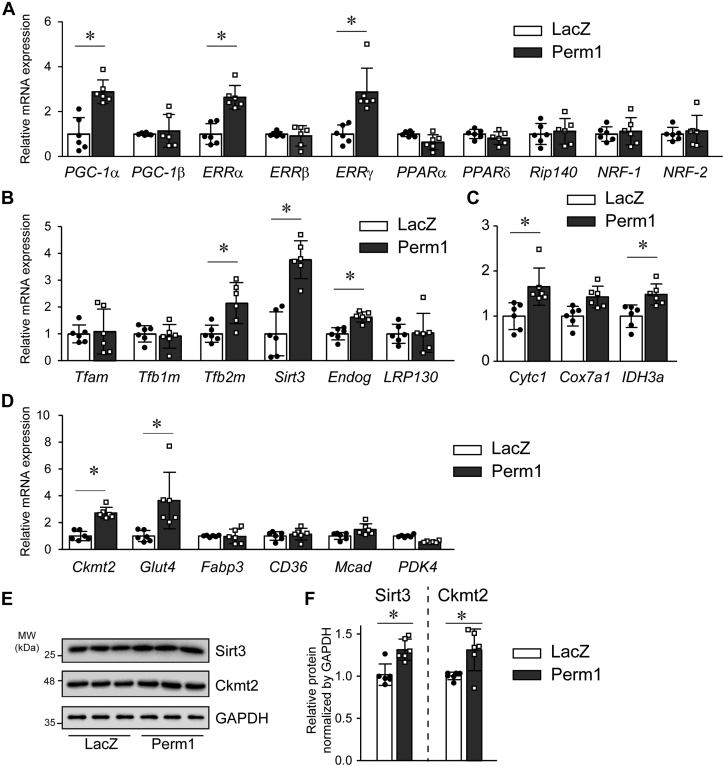
Next, we sought to gain insights into the pathways by which Perm1 modulates oxidative metabolism in NCM. We assessed multiple genes that are critical for Mito biogenesis and oxidative function in the CM. First, as shown in Figure 5A, Perm1 overexpression led to increases in PGC-1α, ERRα, and ERRγ mRNA levels. Perm1 also increased expression of the mtDNA replication/transcription factors Tfb2m (transcription factor B2, mitochondrial), deacetylase Sirt3 (Sirtuin 3), and mitochondrial endonuclease Endog (Endonuclease G), which are PGC-1/ERR targets (34, 35, 36, 37) (Fig. 5B). Further, Perm1 increased expression of other genes known to be essential for electron transfer, such as Cytc1 (cytochrome c1) and IDH3a (isocitrate dehydrogenase [NAD] subunit alpha) (Fig. 5C), nutrient utilization, and energy transduction such as Glut4 (glucose transporter type 4) and Ckmt2 (creatine kinase mitochondria) (17, 38) (Fig. 5D). Interestingly, we did not observe significant changes in genes essential for fatty acid transport and metabolism, such as Fabp3 (fatty acid binding protein 3), CD36 (cluster of differentiation 36), Mcad (medium-chain acyl-coA dehydrogenase), and Pdk4 (pyruvate dehydrogenase kinase 4), suggesting that Perm1 may not be essential for modifying fatty acid metabolism in the CM.
Figure 5.
Perm1 selectively regulates genes essential for oxidative metabolism. Primary cultured NCM were infected with adenoviruses expressing LacZ or Perm1. Forty-eight hours after infection later, the cells were harvested, followed by RNA extraction. A–D, mRNA levels for indicated genes were determined by qRT-PCR, normalized by 36B4 levels, and expressed relative to levels of each gene in LacZ infected cells = 1. Data are the mean ± SD. ∗p < 0.05 versus LacZ. Data are the mean of six experimental replicates from two representative experiments. E and F, whole cell protein lysates were subjected to western blot using antibodies as indicated (E). Sirt3 and Ckmt2 protein levels were quantified relative to expression level of LacZ (control) infected cells = 1 (F). Data are the mean of six experimental replicates from two representative experiments. ∗p < 0.05 versus LacZ.
In addition to gene expression, we also analyzed the effect of Perm1 on protein expression of Sirt3 and Ckmt2, whose gene expression was upregulated by Perm1. As shown in Figure 5, E and F, Perm1 increased Sirt3 and Ckmt2 protein levels in NCM. These results indicate that Perm1 may act by changing PGC-1 and ERR levels, subsequently modifying specific PGC-1 and ERR targets to enhance mitochondrial biogenesis and function.
Perm1 binds PGC-1α, increases ERR-dependent transcriptional activity, and enhances recruitment of PGC-1α to target gene promoters
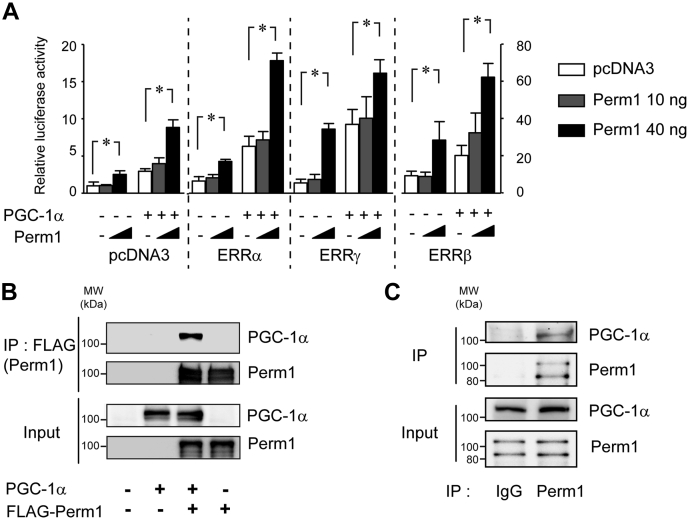
To assess potential mechanisms of how Perm1 might modify cellular function, we next performed a series of biochemical and molecular studies, particularly focused on its role as a nuclear factor. Both human PERM1 and mouse PERM1 contain a putative nuclear localization signal (NLS) and a nuclear export signal (NES) that are conserved between species (19). No other prominent protein motifs are evident that provide insights into the Perm1 function. To test whether Perm1 might have a nuclear function in PGC-1/ERR-induced transcription, we transfected U2OS sarcoma cells with an ERR-responsive luciferase reporter (ERRE-LUC) (39), in the presence or absence of expression vectors for Perm1, PGC-1α, ERRs, and combinations thereof. U2OS cells are a useful model to evaluate the effect of Perm1 on ERR activity as they have detectable endogenous ERR activity (39). As shown in Figure 6A, first without exogenous ERRs, where the luciferase reporter depends only on endogenous ERR activity, PGC-1α increased the luciferase activity 3.5-fold compared with pcDNA3. Perm1 dose-dependently increased the luciferase activity. High-dose Perm1 increased the activity 3.0-fold both in the absence and in the presence of PGC-1α, compared with control (pcDNA3) vectors. Second, in the presence of exogenous ERRs, Perm1 showed a similar trend as seen in samples without exogenous ERRα and ERRβ, where high-dose Perm1 increased luciferase activity 3.0–3.5-fold compared with control. Interestingly, high-dose Perm1 increased ERRγ-dependent transcriptional activity 6.5-fold in the absence of PGC-1α, indicating Perm1 may have preference for specific ERR. These effects of Perm1 were found in cells that only had endogenous ERR activity (i.e., when transfected with control plasmids instead of ERR containing vectors) or when exogenous ERRs were expressed. To further confirm this effect, we documented that no reporter induction was present when the reporter construct lacked the ERR-responsive element (ERRE) (Fig. S4).
Figure 6.
Perm1 binds PGC-1α and increases ERR-dependent transcriptional activity. A, U2OS cells were transfected with the pERRE-LUC reporter (40 ng) and pcDNA3 (control) or ERR expression plasmids (10 ng), together with pcDNA3 control (−), PGC-1α (5 ng) (+), in the presence of pcDNA3 control (white bars), Perm1 (10 ng) (gray bars), or Perm1 (40 ng) (black bars), plasmids. ∗p < 0.05. Data are the mean ± SD, of eight experimental replicates from two representative experiments. B, interaction of Perm1 and PGC-1α in vitro. HEK293 cells were transfected with pcDNA3-PGC-1α in combination with control or FLAG-Perm1 plasmid (A Flag-tag with a Kozak sequence was inserted into the N-terminus of full-length Perm1). Protein complexes were immunoprecipitated with an anti-FLAG antibody and then immunoblotted with an anti-PGC-1α antibody or anti-Perm1 antibody. C, interaction of Perm1 and PGC-1α in vivo. Whole ventricular tissue protein lysates from adult C57Bl6 mice were subjected to immunoprecipitation with anti-PERM1 antibody and then then immunoblotted with an anti-PGC-1α antibody or anti-Perm1 antibody. Representative blots are from two independent experiments. IP, immunoprecipitation.
Based on Perm1's ability to enhance ERR-dependent transcription, we hypothesized that Perm1 might interact with ERR and/or PGC-1. HEK293 cells were transfected with PGC-1α and/or FLAG-Perm1 plasmids. Cell lysates were subjected to immunoprecipitation (IP) by using a FLAG antibody, followed by western blotting. As shown in Figure 6B, Flag-tagged Perm1 co-immunoprecipitated with PGC-1α, suggesting that Perm1 may directly interact with PGC-1α.
To further illustrate the physiological importance of this interaction, we next assessed the interaction of Perm1 and PGC-1α in the heart tissue, in vivo. Ventricular tissue protein lysates were subjected to IP by using an anti-PERM1 antibody, followed by western blotting. As shown in Figure 6C, endogenous Perm1 co-immunoprecipitated with PGC-1α, suggesting that Perm1 interacts with PGC-1α in the intact heart.
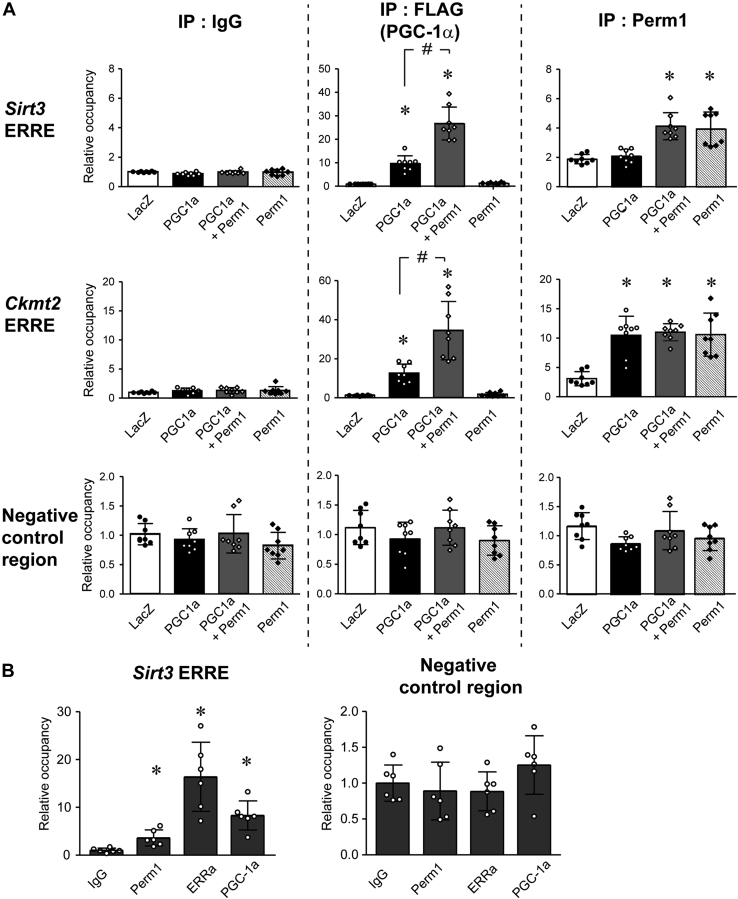
Given that Perm1 increases PGC-1 and ERR targets, interacts with PGC-1α, and enhances ERR-dependent transcriptional activity, we next used chromatin immunoprecipitation assays (ChIP) to test if Perm1 is present on ERRE and modulates recruitment of PGC-1α to target gene promoters. Since there is no ideal continuous cardiac myocyte cell line, we used cultured skeletal muscle C2C12 cells, differentiated into myotubes, where Perm1 regulates PGC-1/ERR target genes and Mito biogenesis is similar to cardiomyocytes (19). First, C2C12 myotubes were infected with adenoviruses expressing LacZ (control), Flag-tagged PGC-1α, or HA-tagged Perm1 singularly or with both PGC-1α and Perm1 together. Forty-eight hours following infection, cell lysates were harvested and subjected to ChIP assay using normal IgG, anti-FLAG, or anti-PERM1 antibodies. Purified DNA was subjected to real-time qRT-PCR to quantify the occupancy of Flag-tagged PGC-1α and Perm1, on gene promoters. We studied ERREs present in the Sirt3 and Ckmt2 genes. We chose to study these genes since as shown above (Fig. 5D), they are upregulated by Perm1 overexpression. ERREs of Sirt3 and Ckmt2 were identified based upon prior published work (17, 35). As shown in Figure 7A, upon the introduction of exogenous PGC-1α, the occupancy of PGC-1α on ERREs was significantly increased, compared with LacZ, showing that PGC-1α is recruited onto the promoter of Sirt3 and Ckmt2. Coexpression of Perm1 along with PGC-1α significantly enhanced the occupancy of PGC-1α on the ERREs compared with PGC-1α alone. Interestingly, we also found that occupancy of Perm1 on the ERREs is increased by overexpression of Perm1 compared with LacZ, indicating that Perm1 is also recruited onto the Sirt3 and Ckmt2 promoters. There were no changes in occupancy of PGC-1α or Perm1 detected in a negative control region that did not have ERREs.
Figure 7.
Perm1 enhances recruitment of PGC-1α to target gene promoter. A, C2C12 myotubes were infected with LacZ (white bars), PGC-1α (Flag-tagged) (black bars), Perm1 (striped bars), or both PGC-1α (Flag-tagged) and Perm1 (gray bars). ChIP assays were performed using anti-FLAG antibody, anti-PERM1 antibodies, or IgG (control). B, ChIPs were performed with adult mouse ventricular tissues using anti-PERM1, anti-ERRα, or anti-PGC-1α antibodies. The abundance of the Sirt3 ERRE, the Ckmt2 ERRE, and a negative control genomic region in the ChIPs were quantified by qPCR, normalized to the input signal, and expressed relative to the levels of each region in the control IgG sample. In A and B, data are the mean ± SD. A, ∗p < 0.05 versus LacZ. #p < 0.05 versus FLAG-PGC-1α. The data are the mean of eight experimental replicates from two representative experiments. B, ∗p < 0.05 versus IgG. The data are the mean of six different heart samples.
To extend the findings determined in cell culture to an in vivo model, we performed ChIP assays using adult heart tissue with an anti-PERM1 antibody, to assess endogenous occupancy. We evaluated the recruitment of Perm1 onto ERRE in the Sirt3 promoter, again using a ChIP assay, but now with adult heart tissue. Anti-ERRα and anti-PGC-1α antibodies were used as positive controls. As shown in Figure 7B, The Sirt3 promoter occupancy was significantly increased with anti-PERM1, anti-ERRα, and anti-PGC-1α antibodies, compared with control IgG. When we evaluated a negative control region, we did not observe increased occupancy of Perm1, ERRα, or PGC-1α, compared with control IgG. These results suggest that Perm1 is recruited to the Sirt3 promoter in the intact heart.
Perm1 protects cardiomyocytes from cellular damage induced by hypoxia reoxygenation
In cardiac myocytes, hypoxia/reoxygenation (H/R) induces irreversible Mito dysfunction, leading to CM death. Likewise, in vivo, similar events occur during myocardial I/R injury (40). Studies have shown that restoring Mito function and increasing Mito number protects CM from cell death and cardiac dysfunction induced by I/R (11, 41). Given that Perm1 increases Mito biogenesis, we hypothesized that Perm1 could exert protective effects against cellular damage caused by I/R. To test this hypothesis, we employed an in vitro model of I/R, using H/R.
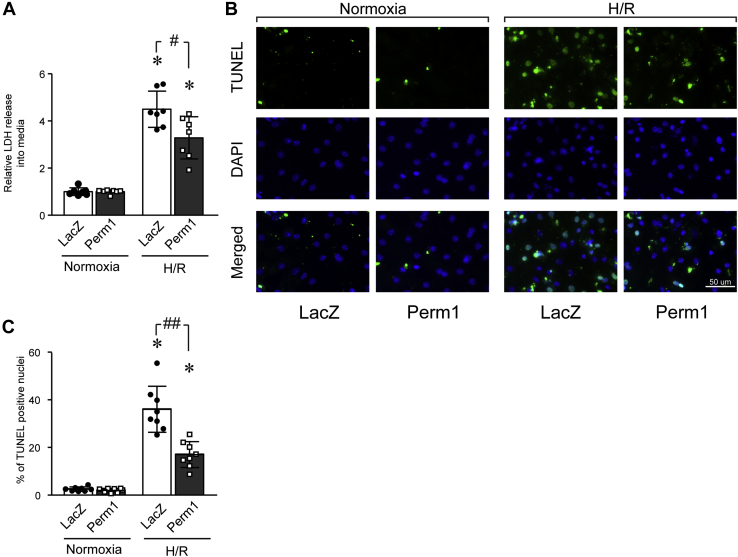
NCM were infected with recombinant adenoviruses expressing Perm1 or LacZ (control). Forty-eight hours postinfection, the NCM were subjected to H/R or maintained in normoxic control culture conditions. Cellular damage was analyzed first by evaluating release of lactate dehydrogenase (LDH) into the culture media. As shown in Figure 8A, while H/R significantly increased LDH released into the cell culture media in both Perm1 and LacZ-infected cells, Perm1 overexpression significantly decreased the amount of H/R-induced cellular damage as monitored by LDH levels, versus control (LacZ). To extend this work, we next examined if Perm1 would affect H/R-induced apoptosis. For this, we performed TUNEL staining of NCM in this H/R model system. As shown in Figure 8, B and C, when cells were subjected to H/R, the number of TUNEL-positive nuclei were significantly diminished in Perm1-induced NCM compared with LacZ cells (Fig. 8, B and C). Together, these results suggest that Perm1 protects CM from H/R.
Figure 8.
Perm1 protects cardiomyocytes from cellular damage induced by hypoxia-reoxygenation. Primary cultured NCM were infected adenoviruses (Ad) expressing LacZ or Perm1. Forty-eight hours later, the cells were subjected to hypoxia and reoxygenation (H/R). A, LDH released into the media was analyzed and Perm1-infected cell levels were detected, were quantified relative to that detected in Ad-LacZ infected cells. Data are the mean ± SD. ∗p < 0.05 versus Normoxia. #p < 0.05 versus LacZ. Data are the mean of eight experimental replicates from two representative experiments. B, cardiomyocyte apoptosis was determined by staining with TUNEL and DAPI, and representative images of TUENL-positive (green) and DAPI (blue) are shown. Scale bar: 50 μm. C, quantitative analysis of TUNEL-positive nuclei is shown. Data are the mean ± SD. ∗p < 0.05 versus Normoxia. #p < 0.05 and ##p < 0.01 versus LacZ. Data are the mean of eight experimental replicates from two representative experiments.
Discussion
In the present work, we have advanced our understanding of Perm1 expression and function in the heart. For this, we employed tissue from the embryonic, perinatal, and adult mouse heart. In addition, we used a neonatal mouse CM culture model to study Perm1 in normal and stressed states of the myocardium. Human tissue was also interrogated.
First, we found that Perm1 increased along with cardiac development in mouse heart. This expression pattern is similar to mtDNA, Oxphos proteins, and gene expression of PGC-1s and ERRs. We also found that Perm1 was downregulated in a mouse model of heart failure and also in human heart samples from DCM patients, in contrast to the ones from normally functioning hearts. Second, we found that Perm1 increased mitochondrial DNA copy number, OxPhos proteins, and oxygen consumption rate in NCM, showing that Perm1 promotes Mito biogenesis in NCM. Moreover, our results showed that Perm1 increased select PGC-1/ERR targets important for Mito biogenesis and oxidative metabolism. We also found that Perm1 bound PGC-1α both in vitro and in intact heart samples in vivo, increased ERR-dependent transcriptional activity, and enhanced recruitment of PGC-1α to target gene promoters. This established Perm1 as a new component of Mito biogenesis involved in PGC-1 signaling in the heart. Importantly, we found that Perm1 had a protective effect against cellular damage induced by H/R in NCM.
An increase in Mito oxidative capacity is a crucial feature of cardiac development, and Mito biogenesis plays a pivotal role in the metabolic maturation process of the heart (23). In the early embryonic stages, the Mito oxidative metabolism is poorly developed in CM, and the CM mainly depend on glycolysis as a source of energy (23, 24). During cardiac development and particularly postnatally, the Mito oxidative capacity increases and the heart shifts from glycolysis to Mito-mediated oxidative phosphorylation as a source of energy, in order to meet energy demands of the maturing heart (23). Evidence from a number of studies shows that dysregulation of Mito metabolism not only causes defects in metabolic maturation of CM, but also causes defects in cardiac growth (42, 43, 44). Supporting this notion are studies where PGC-1α/β and ERRγ are ablated from expression in the embryonic heart in mice, resulting in metabolic abnormalities and development of postnatal cardiomyopathy (16).
In this study, we found that the level of Perm1 protein parallels increases in Oxphos proteins and mtDNA, throughout cardiac development. This expression pattern was correlated with enhanced gene expression of PGC-1s and ERRs (Figs. 1D and S1). Together, these results raise the possibility that Perm1 may contribute to Mito biogenesis and metabolic maturation, in concert with PGC-1 and ERR, during cardiac development.
In contrast to upregulation of oxidative metabolism in cardiac development, downregulation of Mito function is well documented in the failing heart in animals and humans (45, 46). In rodents, loss of the mitochondrial DNA replication factor Tfam causes HF (46). In humans, Mito respiration is downregulated in heart tissue samples from both dilated and ischemic cardiomyopathy patients, compared with that obtained from normal subjects (9, 47). Demonstration of the critical importance of maintaining normal mitochondrial respiration is illustrated in Barth Syndrome, an inherited disorder, which produces left ventricular noncompaction, a rare congenital cardiomyopathy characterized by extensive endomyocardial trabeculation (48). Aligned with this, we found that Perm1 expression was downregulated in both a mouse heart failure model and human heart samples from patients with DCM, as opposed to control samples from normal functioning hearts (Fig. 3D). It is possible that downregulation of Perm1 in the failing heart is the consequence of decreased PGC-1 signaling. However, since Perm1-regulated genes, such as Ckmt2 and Glut4, are also downregulated in animal models with HF (17), these results indicate that the downregulation of PERM1 may also contribute to metabolic dysfunction as seen in HF in humans and animals.
To extend the above results, we also showed that Perm1 increased the recruitment of PGC-1α to target gene promoters. PGC-1α enhances target gene expression through binding with histone acetyltransferases, including CREB-binding protein/p300 and steroid receptor coactivator-1 (SRC-1) (49), chromatin remodeling, and mediator complexes, such as the thyroid hormone receptor–associated protein/vitamin D receptor–interacting protein (TRAP/DRIP) coactivator complex (50). Studies showed that posttranslational modifications regulate PGC-1α activity, such as phosphorylation and acetylation (51, 52). In our previous report, we showed that Perm1 was associated with Ca2+ and calmodulin-dependent protein kinase II (CaMKII) and modulated CaMKII activity in skeletal muscle (21). We are aware that CaMKII isoforms are differentially expressed and have distinct functions in skeletal muscle as compared with the heart (53). While CaMKIIβ, γ, and δ isoforms are expressed in skeletal muscle and are required for its contractile and metabolic function (54), CaMKIIδ is dominantly expressed in CM (55). Thus, future studies are needed to study if cardiac CaMKIIδ activity is affected by Perm1, modulates PGC-1α activity, and affects the association of PGC-1α/ERRs.
In addition to supporting the continuous requirements for high energy demands necessary for even the basal cardiac workload, mitochondria also play a pivotal role in the heart stressed by insults such as I/R injury, as can occur during cardiac bypass surgeries, or in the face of coronary occlusion that can produce myocardial infarction. As the heart is subjected to ischemia, damage to the mitochondrial electron transport chain occurs, and production of reactive oxygen species (ROS) increases. Then, during the reperfusion phase (e.g., as a cardiac surgery patient is taken off artificial bypass or when coronary artery stenting is performed) mitochondrial-driven injury proceeds and opening of the mitochondrial permeability transition pore (MPTP) occurs. This leads to release of mitochondrial enzymes and cytochrome c from the damaged mitochondria, resulting in CM cell death (40). Interestingly, studies showed that preserving Oxphos function can provide protective effects against I/R injury associated with reduced ROS production upon I/R (56, 57). Since Perm1 increased Oxphos proteins in our model using neonatal CM H/R, we suggest that Perm1 may protect the intact heart subjected to I/R, by preservation of Oxphos proteins during these stresses.
To date, there are no specific motifs or domains detected in Perm1 that allow for clearly predictable modulation of its function. Yet, it is important to note that as we submitted this paper, Aravamudhan et al. (25) published work studying the phospho-proteome in the postnatal heart. From this survey work, the authors elected to pursue studies on Perm1. They identified that Perm1 is positioned in the outer Mito membrane and undergoes rapid changes via the ubiquitin-proteasome system by phosphorylation of its PEST motif. Loss of Perm1 expression alters lipid and amino acid metabolites in the mouse heart.
Since we could not find a canonical mitochondrial localization signal in the Perm1 sequence, it is possible that Perm1 may interact with other Mito protein(s), leading to its localization in Mito. Our study found that multiple isoforms of Perm1 are present, with two major forms identified. The functional differences of even these two major forms are not currently known, but it may be possible that the shorter isoform of Perm1 has a distinct function compared with the longer isoform. To date, we have not identified a functional Kozak sequence in either AUG. Previous studies showed that the sequences upstream of the Kozak and downstream of the AUG also affect translational initiation (33). Thus, future mutagenesis studies modifying nucleotides surrounding the AUGs in Perm1 will be necessary to fully address these questions.
It is important to consider that Perm1 may increase the protein stability of PGC-1α. Still, we would suggest that Perm1 functions as a chaperon protein to stabilize the PGC-1/ERR complex, such as p300 (50). Further, it is also interesting to note that Perm1 preferentially modulates genes important for Mito biogenesis and OxPhos, but not for fatty acid oxidation (Pdk4 and CD36). These data suggest that Perm1 may define specific PGC-1 and ERR-regulated genes and pathways. Future studies using ChIP and other techniques/systems, will be required to identify the gene promoters that are regulated by Perm1 in CM.
In conclusion, our results are the most extensive to date evaluating the role of the striated muscle-specific protein, Perm1, in cardiac myocytes and heart. We showed how Perm1 expression is increased throughout development and postnatally in adult CM and the heart. Most importantly, we demonstrated that Perm1 promotes mitochondrial biogenesis and reduces cellular damage caused by H/R in isolated cardiac myocytes. The full role of Perm1 in the intact mammalian heart remains to be elucidated. Studies that employ models such as cardiomyocyte-specific Perm1 knockout and transgenic animals that increase Perm1 expression will be needed to more fully determine the function of Perm1 in the heart under physiological and pathological states, such as pressure overload (17), HF induced by aging (58), and ischemic heart disease (59).
Experimental procedures
Animals and approvals
C57BL/6 mice were housed under a 12-h light/dark cycle at constant temperature and given food and water ad libitum. Mouse embryos were harvested at defined stages from timed-mating pairs. Morphology and somite count were used as inclusion criteria for each developmental time point. Cardiac tissues were micro-dissected from embryos at embryonic days (E) 9.5–E17.5, postnatal day (P) 0–day 5 (P0–P5), and from adult mice (10-week-old male) (60). Entire heart tubes from E9.5 embryos were sampled, while ventricular tissue was used from later stage embryos and all postnatal and adult mouse hearts. The Institutional Animal Care and Use Committee (IACUC) of the University of California, San Diego, approved all animal experiments.
Human ventricular tissue
Left ventricle tissue samples were from the Loyola University Medical Center's (LUMC's) Cardiovascular Institute Tissue Repository and from the Gift of Hope Organ and Tissue Donor Network, as outlined previously (28, 29). All collections were approved to conform with the Declaration of Helsinki and by a protocol and informed consent document reviewed by LUMC's Institutional Review Board. Explanted LV tissue was obtained from patients with nonischemic cardiomyopathy who were undergoing heart transplantation. Donor hearts judged unsuitable for cardiac transplantation were used as control samples and were also obtained through the Gift of Hope Organ and Tissue Donor Network. All samples were frozen in liquid N2 and stored at −80 °C for future use.
Primary culture of mouse neonatal cardiomyocytes and adenovirus infection
Primary mouse neonatal ventricular cardiomyocytes (NCM) were isolated from postnatal day 0 to 2 (P0–P2) C57BL/6 mice (61). Briefly, newborn pups were decapitated, and ventricles were excised, cut into eight pieces, and washed in ice-cold HBSS without Ca2+ and Mg2+. After washing, hearts were placed in 20 ml HBSS containing trypsin and incubated for 4 h at 4 °C. Hearts were digested with HBSS containing collagenase at 37 °C. Isolated cells were plated in noncoated plastic dishes with NCM culture media [30% DMEM, 30% M199, 25% FBS, and 15% horse serum] and cultured for 2 h at 37 °C (termed preplating) to remove fibroblasts and endothelial cells. After preplating, nonadherent cardiomyocytes were seeded at a density of 5 × 105 cells/well in 12-well plates. After 24 h incubation, cells were washed with PBS to remove dead cells and debris. For experiments as described, NCM were infected with adenoviruses expressing LacZ or Perm1 at a multiplicity of infection (M.O.I.) of 50. After 48-h of infection, cells were subjected to further assays as discussed.
Oxygen consumption assay using Seahorse XFe96
Oxygen consumption assays were performed by using a Seahorse XFe96 device (Agilent) (61). Briefly, isolated NCM were seeded into the Seahorse 96-well culture plates at a cell density of 20 × 103/well. Twenty-four hours later, NCM were infected with Ad-LacZ or Ad-Perm1 at MOIs of 50. Forty-eight hours postinfection, cell culture media was switched to NCM culture media without serum, and cells were cultured for an additional 3 h at 37 °C. NCM media was then changed to Seahorse Bioscience assay buffer [phenol red-free DMEM with 10 mM HEPES, 5 mM glucose, 1 mM sodium pyruvate, and 2 mM glutamine] and incubated for 1 h at 37 ° C, followed by measurement of oxygen consumption rates (OCRs), using an XFe96 Extracellular Flux Analyzer. 1 μM Oligomycin (Oligo), 800 nM FCCP (carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone), and 1 μM each of rotenone and antimycin A (RAA) were injected during the OCR assay. OCRs were normalized to the cell number and expressed as OCR per 20 × 103 cells. Data shown are the mean ± SD of two independent experiments; n = 9.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed with adenoviral-infected C2C12 myotubes and adult mouse heart (10-week-old) as described previously (19). Briefly, C2C12 myotubes were infected on day 4 after differentiation with adenoviruses at a total MOI of 50, such that conditions were (1) LacZ (control) at an MOI = 50, (2) FLAG-PGC-1α (MOI = 25) + LacZ (MOI = 25) for a total MOI = 50, (3) FLAG-PGC-1α + Perm1 (MOI = 25), or (4) Perm1 (MOI = 25) + LacZ (MOI = 25) for a total MOI = 50. Forty-eight hours after infection, cells were cross-linked for 10 min at 37 °C in 1% formaldehyde in PBS.
For adult heart ChIP assays, C57Bl6 WT mouse ventricular tissues were cut into pieces and cross-linked for 10 min at 37 °C in 1% formaldehyde in PBS. After quenching, sonication was performed to generate approximate 500-bp fragments, followed by preclearing with protein A/G-Sepharose. For C2C12 myotubes, soluble chromatin was immunoprecipitated with the following antibodies: normal IgG, anti-FLAG (Clone M2, Sigma) for the immunoprecipitation of FLAG-tagged PGC-1α, and anti-PERM1. For adult heart tissues, soluble chromatin was immunoprecipitated with normal IgG, anti-PERM1, anti-ERRα, and anti-PGC-1α antibodies. Genomic DNA from each immunoprecipitation was quantified by quantitative PCR using primers flanking an ERRE in the promoter region of Sirt3 (35), Ckmt2 (17), and also negative control regions that lack ERREs and are distal to the Esrra promoter (19). Primers are described in Table S2. Data were first normalized against total genomic input and then expressed relative to levels of immunoprecipitated DNA in IgG control samples (19).
Hypoxia/reoxygenation, LDH assay, and TUNEL staining
NCM were infected with adenoviruses expressing LacZ or Perm1 at an MOI of 50. Forty-eight hours after infection, the media were changed to DMEM without glucose, glutamine, and pyruvate. NCM were cultured in normoxic (5% CO2-95% air) or hypoxic (5% CO2-95% N2) conditions for an additional 4 h, after which media was replaced with DMEM containing 5 mM glucose. Cells were then cultured in normoxic conditions for 2 h to subject them to reoxygenation. Finally, at the termination of the culture period, media was collected and analyzed for LDH, using a commercial assay kit (Roche, 04744926001).
Cells incubated in the above manner were also subjected to TUNEL staining (Thermo Fisher, A23210). Briefly, NCM were plated on Lab-Tek chamber glass slides (Thermo Fisher), treated in the above manner and washed with PBS, followed by fixation in 4% paraformaldehyde for 10 min at room temperature. After being washed with PBS, the cells were incubated with a TUNEL reaction buffer for 1 h at 37 °C in a humidified chamber. The percentage of CM with DNA nick-end labeling was determined by counting cells exhibiting yellow-green nuclei among 500 nuclei in triplicate plates in two independent experiments.
Other experimental procedures are described in the supporting information.
Statistics
Data shown are mean ± SD and were analyzed with a two-tailed Student's t-test for single variables. For comparisons between multiple variables, p-values were determined using a two-way ANOVA test, followed by a Bonferroni analysis.
Data availability
All data are included within the article and associated supporting information.
Supporting information
This article contains supporting information (19, 20, 39, 62).
Conflict of interest
The authors declare that they have no conflict of interest with the contents of the article.
Acknowledgments
We thank Ross laboratory members for technical help and discussions, as well as Sarah Schurr and Cameron Hill for technical help. We also thank Dr Anastasia Kralli (Johns Hopkins University, Baltimore, MD) for providing ERR expressing plasmid DNA constructs and Dr Alan Samarel (Loyola University, Chicago, IL) for providing human tissue samples.
Author contributions
Y. C. and R. S. R. conceptualization; Y. C., S. T., A. A., and A. N. M. resources; Y. C., S. T., K. L., Y. A., S. K., O. Z., A. L., R. L., and A. A. data curation; Y. C. and A. N. M. software; Y. C. and S. T. formal analysis; Y. C. and R. S. R. supervision; Y. C. and R. S. R. funding acquisition; Y. C. and S. T. validation; Y. C., S. T., K. L., Y. A., S. K., O. Z., A. L., R. L., and R. S. R. investigation; Y. C. and S. T. visualization; Y. C., S. T., K. L., and Y. A. methodology; Y. C. and S. T. writing-original draft; Y. C. and R. S. R. project administration; Y. C. and R. S. R. writing-review and editing.
Funding and additional information
This work is supported by NHLBI, National Institute of Health (NIH) Grant R01HL151239 (to Y. C. and R. S. R.). R. S. R was also funded by the Veterans Administration (I01 BX004897), and Y. C. was also funded by Tobacco-Related Disease Research Program (TRDRP) T31IP1606. The content is solely the responsibility of the authors and does not represent official views of the National Institute of Health.
Edited by John Denu
Footnotes
Present address for Anne N. Murphy: Cytokinetics, Inc, South San Francisco, California, USA.
Supporting information
References
- 1.Stanley W.C., Recchia F.A., Lopaschuk G.D. Myocardial substrate metabolism in the normal and failing heart. Physiol. Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 2.Taegtmeyer H., Young M.E., Lopaschuk G.D., Abel E.D., Brunengraber H., Darley-Usmar V., Des Rosiers C., Gerszten R., Glatz J.F., Griffin J.L., Gropler R.J., Holzhuetter H.G., Kizer J.R., Lewandowski E.D., Malloy C.R. Assessing cardiac metabolism: A scientific statement from the American Heart Association. Circ. Res. 2016;118:1659–1701. doi: 10.1161/RES.0000000000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Wilhelmsson H., Graff C., Li H., Oldfors A., Rustin P., Bruning J.C., Kahn C.R., Clayton D.A., Barsh G.S., Thoren P., Larsson N.G. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 4.Park C.B., Larsson N.G. Mitochondrial DNA mutations in disease and aging. J. Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasquez-Trincado C., Garcia-Carvajal I., Pennanen C., Parra V., Hill J.A., Rothermel B.A., Lavandero S. Mitochondrial dynamics, mitophagy and cardiovascular disease. J. Physiol. 2016;594:509–525. doi: 10.1113/JP271301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventura-Clapier R., Garnier A., Veksler V. Energy metabolism in heart failure. J. Physiol. 2004;555:1–13. doi: 10.1113/jphysiol.2003.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnier A., Fortin D., Delomenie C., Momken I., Veksler V., Ventura-Clapier R. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. J. Physiol. 2003;551:491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mettauer B., Zoll J., Garnier A., Ventura-Clapier R. Heart failure: A model of cardiac and skeletal muscle energetic failure. Pflugers Arch. 2006;452:653–666. doi: 10.1007/s00424-006-0072-7. [DOI] [PubMed] [Google Scholar]
- 9.Sharov V.G., Todor A.V., Silverman N., Goldstein S., Sabbah H.N. Abnormal mitochondrial respiration in failed human myocardium. J. Mol. Cell. Cardiol. 2000;32:2361–2367. doi: 10.1006/jmcc.2000.1266. [DOI] [PubMed] [Google Scholar]
- 10.Paradies G., Paradies V., Ruggiero F.M., Petrosillo G. Mitochondrial bioenergetics and cardiolipin alterations in myocardial ischemia-reperfusion injury: Implications for pharmacological cardioprotection. Am. J. Physiol. Heart Circ. Physiol. 2018;315:H1341–H1352. doi: 10.1152/ajpheart.00028.2018. [DOI] [PubMed] [Google Scholar]
- 11.Ikeuchi M., Matsusaka H., Kang D., Matsushima S., Ide T., Kubota T., Fujiwara T., Hamasaki N., Takeshita A., Sunagawa K., Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 12.Camara A.K., Bienengraeber M., Stowe D.F. Mitochondrial approaches to protect against cardiac ischemia and reperfusion injury. Front. Physiol. 2011;2:13. doi: 10.3389/fphys.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leone T.C., Kelly D.P. Transcriptional control of cardiac fuel metabolism and mitochondrial function. Cold Spring Harb. Symp. Quant. Biol. 2011;76:175–182. doi: 10.1101/sqb.2011.76.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sihag S., Cresci S., Li A.Y., Sucharov C.C., Lehman J.J. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J. Mol. Cell. Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arany Z., Novikov M., Chin S., Ma Y., Rosenzweig A., Spiegelman B.M. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai L., Leone T.C., Zechner C., Schaeffer P.J., Kelly S.M., Flanagan D.P., Medeiros D.M., Kovacs A., Kelly D.P. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huss J.M., Imahashi K., Dufour C.R., Weinheimer C.J., Courtois M., Kovacs A., Giguere V., Murphy E., Kelly D.P. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Alaynick W.A., Kondo R.P., Xie W., He W., Dufour C.R., Downes M., Jonker J.W., Giles W., Naviaux R.K., Giguere V., Evans R.M. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Cho Y., Hazen B.C., Russell A.P., Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J. Biol. Chem. 2013;288:25207–25218. doi: 10.1074/jbc.M113.489674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho Y., Hazen B.C., Gandra P.G., Ward S.R., Schenk S., Russell A.P., Kralli A. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. FASEB J. 2016;30:674–687. doi: 10.1096/fj.15-276360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho Y., Tachibana S., Hazen B.C., Moresco J.J., Yates J.R., 3rd, Kok B., Saez E., Ross R.S., Russell A.P., Kralli A. Perm1 regulates CaMKII activation and shapes skeletal muscle responses to endurance exercise training. Mol. Metab. 2019;23:88–97. doi: 10.1016/j.molmet.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oka S.I., Sabry A.D., Horiuchi A.K., Cawley K.M., O'Very S.A., Zaitsev M.A., Shankar T.S., Byun J., Mukai R., Xu X., Torres N.S., Kumar A., Yazawa M., Ling J., Taleb I. Perm1 regulates cardiac energetics as a downstream target of the histone methyltransferase Smyd1. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piquereau J., Ventura-Clapier R. Maturation of cardiac energy metabolism during perinatal development. Front. Physiol. 2018;9:959. doi: 10.3389/fphys.2018.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopaschuk G.D., Jaswal J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 25.Aravamudhan S., Turk C., Bock T., Keufgens L., Nolte H., Lang F., Krishnan R.K., Konig T., Hammerschmidt P., Schindler N., Brodesser S., Rozsivalova D.H., Rugarli E., Trifunovic A., Bruning J. Phosphoproteomics of the developing heart identifies PERM1 - an outer mitochondrial membrane protein. J. Mol. Cell. Cardiol. 2021;154:41–59. doi: 10.1016/j.yjmcc.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Sharov V.G., Goussev A., Lesch M., Goldstein S., Sabbah H.N. Abnormal mitochondrial function in myocardium of dogs with chronic heart failure. J. Mol. Cell. Cardiol. 1998;30:1757–1762. doi: 10.1006/jmcc.1998.0739. [DOI] [PubMed] [Google Scholar]
- 27.Rockman H.A., Ross R.S., Harris A.N., Knowlton K.U., Steinhelper M.E., Field L.J., Ross J., Jr., Chien K.R. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manso A.M., Li R., Monkley S.J., Cruz N.M., Ong S., Lao D.H., Koshman Y.E., Gu Y., Peterson K.L., Chen J., Abel E.D., Samarel A.M., Critchley D.R., Ross R.S. Talin1 has unique expression versus talin 2 in the heart and modifies the hypertrophic response to pressure overload. J. Biol. Chem. 2013;288:4252–4264. doi: 10.1074/jbc.M112.427484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deacon D.C., Happe C.L., Chen C., Tedeschi N., Manso A.M., Li T., Dalton N.D., Peng Q., Farah E.N., Gu Y., Tenerelli K.P., Tran V.D., Chen J., Peterson K.L., Schork N.J. Combinatorial interactions of genetic variants in human cardiomyopathy. Nat. Biomed. Eng. 2019;3:147–157. doi: 10.1038/s41551-019-0348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C.C., Simonsen C.C., Levinson A.D. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984;309:82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- 31.Lin F.T., MacDougald O.A., Diehl A.M., Lane M.D. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: Transcriptional activator lacking antimitotic activity. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong W., Hsieh C.C., Kurtz A.J., Rabek J.P., Papaconstantinou J. Regulation of CCAAT/enhancer-binding protein-beta isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res. 2001;29:3087–3098. doi: 10.1093/nar/29.14.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984;308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- 34.Gleyzer N., Vercauteren K., Scarpulla R.C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giralt A., Hondares E., Villena J.A., Ribas F., Diaz-Delfin J., Giralt M., Iglesias R., Villarroya F. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J. Biol. Chem. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDermott-Roe C., Ye J., Ahmed R., Sun X.M., Serafin A., Ware J., Bottolo L., Muckett P., Canas X., Zhang J., Rowe G.C., Buchan R., Lu H., Braithwaite A., Mancini M. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wende A.R., Kim J., Holland W.L., Wayment B.E., O'Neill B.T., Tuinei J., Brahma M.K., Pepin M.E., McCrory M.A., Luptak I., Halade G.V., Litwin S.E., Abel E.D. Glucose transporter 4-deficient hearts develop maladaptive hypertrophy in response to physiological or pathological stresses. Am. J. Physiol. Heart Circ. Physiol. 2017;313:H1098–H1108. doi: 10.1152/ajpheart.00101.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gantner M.L., Hazen B.C., Conkright J., Kralli A. GADD45gamma regulates the thermogenic capacity of brown adipose tissue. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11870–11875. doi: 10.1073/pnas.1406638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lesnefsky E.J., Chen Q., Tandler B., Hoppel C.L. Mitochondrial dysfunction and myocardial ischemia-reperfusion: Implications for novel therapies. Annu. Rev. Pharmacol. Toxicol. 2017;57:535–565. doi: 10.1146/annurev-pharmtox-010715-103335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikeda M., Ide T., Fujino T., Arai S., Saku K., Kakino T., Tyynismaa H., Yamasaki T., Yamada K., Kang D., Suomalainen A., Sunagawa K. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Carvalho A., Bassaneze V., Forni M.F., Keusseyan A.A., Kowaltowski A.J., Krieger J.E. Early postnatal cardiomyocyte proliferation requires high oxidative energy metabolism. Sci. Rep. 2017;7:15434. doi: 10.1038/s41598-017-15656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papadopoulou-Legbelou K., Gogou M., Evangeliou A. Cardiac manifestations in children with inborn errors of metabolism. Indian Pediatr. 2017;54:667–673. doi: 10.1007/s13312-017-1131-1. [DOI] [PubMed] [Google Scholar]
- 44.Hom J.R., Quintanilla R.A., Hoffman D.L., de Mesy Bentley K.L., Molkentin J.D., Sheu S.S., Porter G.A., Jr. The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev. Cell. 2011;21:469–478. doi: 10.1016/j.devcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou B., Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. J. Clin. Invest. 2018;128:3716–3726. doi: 10.1172/JCI120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansson A., Hance N., Dufour E., Rantanen A., Hultenby K., Clayton D.A., Wibom R., Larsson N.G. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karamanlidis G., Nascimben L., Couper G.S., Shekar P.S., del Monte F., Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ. Res. 2010;106:1541–1548. doi: 10.1161/CIRCRESAHA.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh S., Iadarola D.M., Ball W.B., Gohil V.M. Mitochondrial dysfunctions in barth syndrome. IUBMB Life. 2019;71:791–801. doi: 10.1002/iub.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 50.Wallberg A.E., Yamamura S., Malik S., Spiegelman B.M., Roeder R.G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cel. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 51.Knutti D., Kressler D., Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lerin C., Rodgers J.T., Kalume D.E., Kim S.H., Pandey A., Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Anderson M.E., Brown J.H., Bers D.M. CaMKII in myocardial hypertrophy and heart failure. J. Mol. Cell. Cardiol. 2011;51:468–473. doi: 10.1016/j.yjmcc.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rose A.J., Hargreaves M. Exercise increases Ca2+-calmodulin-dependent protein kinase II activity in human skeletal muscle. J. Physiol. 2003;553:303–309. doi: 10.1113/jphysiol.2003.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T., Johnson E.N., Gu Y., Morissette M.R., Sah V.P., Gigena M.S., Belke D.D., Dillmann W.H., Rogers T.B., Schulman H., Ross J., Jr., Brown J.H. The cardiac-specific nuclear delta(B) isoform of Ca2+/calmodulin-dependent protein kinase II induces hypertrophy and dilated cardiomyopathy associated with increased protein phosphatase 2A activity. J. Biol. Chem. 2002;277:1261–1267. doi: 10.1074/jbc.M108525200. [DOI] [PubMed] [Google Scholar]
- 56.Lesnefsky E.J., Tandler B., Ye J., Slabe T.J., Turkaly J., Hoppel C.L. Myocardial ischemia decreases oxidative phosphorylation through cytochrome oxidase in subsarcolemmal mitochondria. Am. J. Physiol. 1997;273:H1544–1554. doi: 10.1152/ajpheart.1997.273.3.H1544. [DOI] [PubMed] [Google Scholar]
- 57.Chen Q., Moghaddas S., Hoppel C.L., Lesnefsky E.J. Reversible blockade of electron transport during ischemia protects mitochondria and decreases myocardial injury following reperfusion. J. Pharmacol. Exp. Ther. 2006;319:1405–1412. doi: 10.1124/jpet.106.110262. [DOI] [PubMed] [Google Scholar]
- 58.Yeh C.H., Shen Z.Q., Hsiung S.Y., Wu P.C., Teng Y.C., Chou Y.J., Fang S.W., Chen C.F., Yan Y.T., Kao L.S., Kao C.H., Tsai T.F. Cisd2 is essential to delaying cardiac aging and to maintaining heart functions. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imahashi K., Pott C., Goldhaber J.I., Steenbergen C., Philipson K.D., Murphy E. Cardiac-specific ablation of the Na+-Ca2+ exchanger confers protection against ischemia/reperfusion injury. Circ. Res. 2005;97:916–921. doi: 10.1161/01.RES.0000187456.06162.cb. [DOI] [PubMed] [Google Scholar]
- 60.Arita Y., Nakaoka Y., Matsunaga T., Kidoya H., Yamamizu K., Arima Y., Kataoka-Hashimoto T., Ikeoka K., Yasui T., Masaki T., Yamamoto K., Higuchi K., Park J.S., Shirai M., Nishiyama K. Myocardium-derived angiopoietin-1 is essential for coronary vein formation in the developing heart. Nat. Commun. 2014;5:4552. doi: 10.1038/ncomms5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tachibana S., Chen C., Zhang O.R., Schurr S.V., Hill C., Li R., Manso A.M., Zhang J., Andreyev A., Murphy A.N., Ross R.S., Cho Y. Analyzing oxygen consumption rate in primary cultured mouse neonatal cardiomyocytes using an extracellular Flux analyzer. J. Vis. Exp. 2019 doi: 10.3791/59052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho Y., Noshiro M., Choi M., Morita K., Kawamoto T., Fujimoto K., Kato Y., Makishima M. The basic helix-loop-helix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Mol. pharmacol. 2009;76:1360–1369. doi: 10.1124/mol.109.057000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included within the article and associated supporting information.