Abstract
Binding of antibodies to their receptors is a core component of the innate immune system. Understanding the precise interactions between antibodies and their Fc receptors has led to the engineering of novel mAb biotherapeutics with tailored biological activities. One of the most significant findings is that afucosylated monoclonal antibodies demonstrate increased affinity toward the receptor FcγRIIIa, with a commensurate increase in antibody-dependent cellular cytotoxicity. Crystal structure analysis has led to the hypothesis that afucosylation in the Fc region results in reduced steric hindrance between antibody–receptor intermolecular glycan interactions, enhancing receptor affinity; however, solution-phase data have yet to corroborate this hypothesis. In addition, recent work has shown that the fragment antigen-binding (Fab) region may directly interact with Fc receptors; however, the biological consequences of these interactions remain unclear. By probing differences in solvent accessibility between native and afucosylated immunoglobulin G1 (IgG1) using hydroxyl radical footprinting–MS, we provide the first solution-phase evidence that an IgG1 bearing an afucosylated Fc region appears to require fewer conformational changes for FcγRIIIa binding. In addition, we performed extensive molecular dynamics (MD) simulations to understand the molecular mechanism behind the effects of afucosylation. The combination of these techniques provides molecular insight into the steric hindrance from the core Fc fucose in IgG1 and corroborates previously proposed Fab–receptor interactions. Furthermore, MD-guided rational mutagenesis enabled us to demonstrate that Fab–receptor interactions directly contribute to the modulation of antibody-dependent cellular cytotoxicity activity. This work demonstrates that in addition to Fc–polypeptide and glycan-mediated interactions, the Fab provides a third component that influences IgG–Fc receptor biology.
Keywords: mAb, IgG1, FcγRIIIa, afucosylation, hydroxyl radical footprinting-mass spectrometry, fast photochemical oxidation of proteins (FPOP), antibody-dependent cellular cytotoxicity, Fab–Fc receptor interactions, molecular dynamics
Abbreviations: ADCC, antibody-dependent cellular cytotoxicity; CHO, Chinese hamster ovary; FA, formic acid; Fab, fragment antigen-binding; Fuc, fucose; HC, heavy chain; HDX, hydrogen–deuterium exchange; HRF, hydroxyl radical footprint; IgG1, immunoglobulin G1; LC, light chain; mAbs, monoclonal antibodies; N297, asparagine 297; nG0-F, normalized G0-F; SASA, solvent-accessible surface area
Monoclonal antibodies (mAbs) are the fastest growing class of biotherapeutic agents, many of which are of the human immunoglobulin G1 (IgG1) isotype (1, 2, 3). The IgG1 antibody structure consists of two homodimeric heavy chain (HC)–light chain (LC) pairs that are linked together in a parallel fashion with both noncovalent interactions and disulfide linkages (reviewed in (4)). The IgG structure consists of the fragment antigen-binding (Fab) domain and the Fc receptor–binding region (Fc), both of which contain discrete structural features.
One of the most important structural features of the Fc region for an mAb is the conserved post-translational modification of the N-glycosylation site at asparagine 297 (N297, EU numbering) (5). This N-linked glycan typically consists of a complex-type oligosaccharide, with a GlcNAc core, and varying degrees of sugar extensions made up of fucose (Fuc), mannose, galactose, and sialic acid (6). It has been shown that N-linked glycans are required for Fc receptor–facilitated antibody-dependent cellular cytotoxicity (ADCC) (7) and afucosylated antibodies display an increase in ADCC activity via an increase in affinity to FcγRIIIa (8). ADCC can be either favorable or unfavorable for biotherapeutic mAbs depending on the desired mechanism of action (9, 10); therefore, understanding the role of the N-linked glycan on effector function is critical for therapeutic design of mAbs.
Before the acquisition of structural data, there was little understanding of the clustering event for the Fcγ receptor that occurs during effector function. Woof and Burton (11) postulated the possible structural scenarios that would lead to the clustering event required for ADCC. They noted that the positioning of the receptor’s binding site at the CH2 domain would require the antibody to adopt a “bent” conformation with a dimerization of the Fc domain to enable engagement between the target and effector cell and facilitate a clustering synapse. Their pictorial representation depicts bivalent antigen binding on the cell surface, with no comparison to monovalent binding. This difference in binding may be of significance because cell-surface antigen expression and copy number viability have shown to be associated with ADCC efficiency (12).
Structural analysis has since shown that truncation of the IgG1 to the Fc-only region retains binding to FcγRIIIa. A crystal structure of this complex has been solved (13, 14), revealing that FcγRIIIa appears to interact with the CE` loop (a region containing N297) through primarily protein–protein rather than protein–glycan interactions. While intermolecular glycan interactions between both proteins were observed, these interactions did not appear to be required for binding (15, 16). More recent work by Subedi et al., has shed insight on the role of the CE` loop, hypothesizing that N297 glycosylation restricts CH2 domain conformation, thereby preorganizing the binding site in an orientation that is preferred for receptor binding (17, 18). It has since been shown that site-directed mutagenesis in the CH2 domain could modulate effector function activity regardless of the glycan (19, 20, 21), indicating that both Fc polypeptide–mediated and glycan-mediated interactions can influence Fc-receptor biology.
Through further crystal structure analysis, Ferrara et al. (15) eloquently described a hypothesis for the role of fucosylation on the molecular interactions that appear to contribute to the difference in binding affinity toward FcγRIIIa. Comparison of the fucosylated (native) versus afucosylated Fc crystal structures revealed that an N-linked glycan on FcγRIIIa (N162) is oriented toward N-linked core Fuc on the antibody, thereby requiring a large conformational change to accommodate glycan-mediated binding. Thus, the current hypothesis proposes that steric hindrance caused by the Fc-region core Fuc is responsible for weakening the glycan-mediated interactions, leading to a significant decrease in affinity.
In addition to static-phase information gained from crystallography, there has been extensive effort to understand antibody–FcγRIIIa solution-phase interactions. Both hydrogen–deuterium exchange (HDX) and the emerging technology of hydroxyl radical footprint (HRF)-MS have been used to study the IgG1–FcγRIIIa interaction. Both technologies revealed conformational changes in the Fc region outside CE` loop upon binding to the receptor (22, 23). In addition, both technologies provided structural evidence of potential interactions between the Fab and receptor; however, there was conflicting evidence between the two different HDX studies, and neither study investigated the role of afucosylation on FcγRIIIa binding directly. More recently, Yogo et al. (24) used a combination of atomic force microscopy and HDX to probe Fab–FcγRIII interactions, leading to the suggestion that the Fab may stabilize the clustering event at the cell surface. However, the extent that these interactions contribute to the binding of FcγRIII and the biological role (if any) on ADCC, remains unknown.
HDX is a well-established technology for elucidating protein dynamics and protein–protein interactions; however, HRF is an emerging technology for these applications (25, 26). Unlike HDX, which utilizes deuterium uptake to provide information on the protein amide backbone, HRF utilizes the diffusion of hydroxyl radicals to oxidize amino acid side chains in a protein. In this study, we utilize fast photochemical oxidation of proteins to generate the hydroxyl radicals (27). Hydroxyl radicals are similar in both the size and polarity to those of aqueous water, which enables diffusion to solvent-exposed regions for protein labeling. The extent of protein modification is a function of both solvent-accessible surface area (SASA) and the intrinsic reactivity of the labeled amino acid side chain. Indeed, previous data have shown that a quantitative correlation between HRF data and the protein regional SASA can be achieved (28, 29). However, because side-chain reactivity is also a major factor, HRF is best used in a direct comparative analysis in which the amino acid sequence of the region of interest remains unchanged (e.g., fucosylated versus afucosylated, receptor-bound versus unbound).
Our current solution-phase studies attempt to expand our understanding of fucosylation on the molecular mechanism of FcγRIIIa binding. We utilize HRF to probe the solvent accessibility difference in the native (fucosylated) versus afucosylated IgG1 and assess how these structural differences are reflected in the receptor-bound states. In addition, we used molecular dynamics (MD) simulations to determine the difference in molecular interactions in the absence of Fuc for the bound IgG1 structure. The corroborative data between HRF and MD enabled rational mutagenesis to probe the role for the Fab on receptor binding and ADCC activity. This work is the first to provide solution-phase data that corroborate the steric hindrance hypothesis, to show the in silico molecular detail of IgG1–receptor interactions and demonstrate that Fab–receptor interactions can modulate ADCC activity.
Results
Overview of HRF analysis of native and afucosylated IgG1 molecules
Crystal structure analysis has shown that the N-linked glycan is significantly perturbed by 2.6 Å when in its native, fucosylated state; however, the overall alignment of the native versus afucosylated Fc–receptor complex structures (3SGK and 3SGJ) results in a modest RMSD = 0.414 (PyMOL alignment) (15). This suggests that the perturbation required to accommodate the Fuc sugar has little impact on the overall Fc region conformation in the bound state. However, the effect of Fuc on IgG structure in the unbound state remains unknown.
To this end, we were first interested in determining if HRF could detect solvent accessibility differences after the removal of Fuc from the native glycan population. This required generation of a native and afucosylated IgG1 with identical glycan heterogeneity to eliminate the potential for confounding HRF data interpretation. Using a native and Fuc -transferase KO Chinese hamster ovary (CHO) cell lines, we obtained the native and afucosylated IgG molecules, respectively, with nearly identical glycan heterogeneity (Fig. S1A). We performed additional product quality assessment to ensure similar levels and/or distribution of product-related variants (e.g., aggregation, charge variants) and observed no significant differences (data not shown). These data indicate that both the primary sequence and the primary post-translational modification for these IgG1 molecules were nearly identical, and thus, our HRF data should reflect SASA differences of the IgG1 molecules that result from the Fuc glycan.
Fc fucosylation differentially affects the Fab and Fc regions of the unbound IgG1 molecules
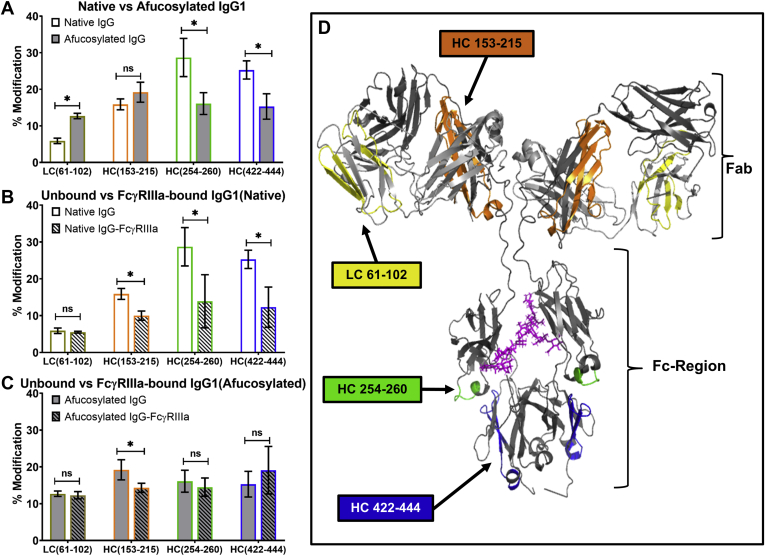
HRF analysis provides information on the SASA of peptides based on their susceptibility for oxidation. Therefore, differences observed in the oxidative footprints (the collection of peptides and their respective oxidation levels) of the unbound native and afucosylated IgG1 molecules represent changes in the protein structure because of the presence or absence of the core Fuc. While the oxidative footprints are nearly identical for both populations of molecules (Fig. S1B), indicating that the Fuc does not induce gross structural changes in the IgG1, there are distinct differences between the two molecules in the Fab and Fc regions (Fig. 1A). Specifically, the afucosylated IgG1 displays a significant increase in SASA for one tryptic peptide in the LC variable region (residue 61–102, sequential numbering for all HRF data) and a significant decrease in SASA in two tryptic peptides in the Fc region (254–260 and 422–444). Both of the tryptic peptides in the Fc region are distal from the FcγRIIIa binding site and contain residues known to interact with the neonatal Fc receptor (30). These data suggest that afucosylation results in distal changes and a more compact Fc region.
Figure 1.
Structural analysis of IgG1 by HRF-MS. The bar graph comparing the oxidative labeling footprint of the select tryptic peptides in the unbound state (A) and the bound states for the native (B) and afucosylated (C) IgG1. Full oxidative footprints are shown in Fig. S1. D, mapping of the HRF data onto the molecular model. The asterisk denotes significant changes in % modification obtained from triplicate samples (p < 0.05). HRF, hydroxyl radical footprint; IgG1, immunoglobulin G1; ns, not significant.
Conformational effect of the IgG1 structure in the bound state
HRF is an ideal tool for determining the binding interfaces between two molecules by comparing the oxidative footprints of the unbound and bound states. Peptides that demonstrate reduced SASA in the bound state relative to the unbound states provide evidence of the binding interface (25, 31, 32). Thus, we utilized HRF to analyze the solvent accessibility differences in the unbound versus FcγRIIIa-bound states for both the native and afucosylated IgG1 molecules to determine if there are differences in the IgG1–receptor interactions because of the removal of Fuc.
The receptor-bound oxidative footprints of the IgG1 show that there are only a few regions on the protein that display reduced SASA (Fig. S1, C and D). The most interesting observation is that the same 2 Fc-region tryptic peptides that showed differences in the unbound states (residues 254–260 and 422–444) also display differences upon FcγRIIIa binding, but only when the Fc region contains Fuc (Fig. 1B, peptides highlighted in green and blue). In the absence of Fuc, there are no significant changes in these Fc-region tryptic peptides upon FcγRIIIa binding (Fig. 1C). Notably, these peptides appear to have similar SASA in both the native and afucosylated Fc regions when comparing receptor-bound states. Taken together, these observations provide two significant insights into the role of Fuc on receptor binding: (1) Fc regions with glycans lacking the Fuc appear to require minimal structural changes to accommodate receptor binding; (2) the Fc regions, irrespective of core Fuc, end up in highly similar conformations once in the receptor-bound state. The former observation is consistent with structural hindrance hypothesis, and the latter is consistent with the calculated SASA for the two Fc peptides in the crystal structures (Fig. S2).
The HRF data also indicate that regardless of the Fuc, there is a Fab peptide in the CH1 region (residues 153–215) that exhibits significantly reduced solvent accessibility in the bound state (Fig. 1, B and C, peptide highlighted in orange). This peptide is in the same region of the IgG that was previously observed to have changes by Houde et al. (22) using HDX and by Shi et al. (23) using HRF. Therefore, we further investigate the role of this CH1 peptide in subsequent studies described in the MD simulations reveal insight into IgG1–FcγRIIIa molecular interactions section below.
Conformational effect of FcγRIIIa structure in the bound state
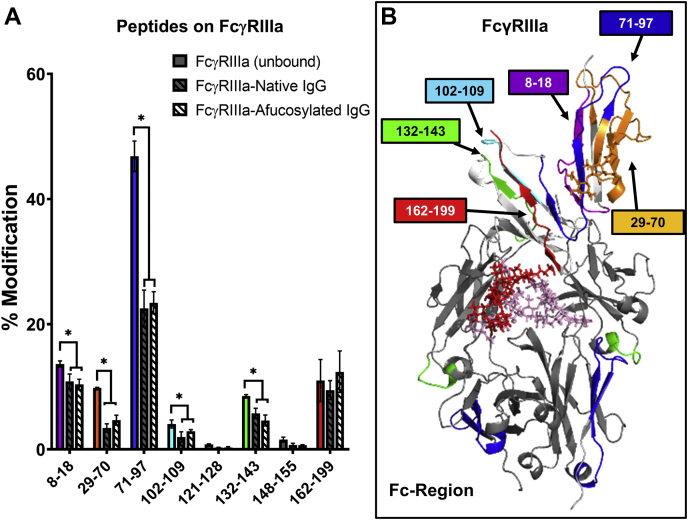
The HRF data for the receptor are nearly identical when bound to either the native or afucosylated IgG1, indicating that the decrease in the solvent accessibility appears to be independent of the IgG1 core Fc Fuc (Fig. 2). Specifically, we could distinguish eight tryptic peptides of the receptor, representing ∼80% of the receptor sequence coverage. Seven of the peptides showed reduced SASA in the bound state, albeit the modification levels were too low for two peptides to indicate statistical significance. The interactions for peptides containing residues 71 to 97 and 132 to 143 are consistent with the crystal structures (13, 15) and previously HRF data (23). The peptide containing the N-linked glycan at N162 does not appear to change upon binding (Fig. 2, red peptide). This is consistent with the idea that polypeptides on the receptor do not play a major role in binding affinity and thus reduced SASA would not be expected. However, the second N-linked glycan at N45 (orange) that is not involved in binding does display similar levels of reduced SASA in both complexes. This likely reflects distal conformation changes in the receptor that occur simply from IgG binding, independent of the Fc fucosylation.
Figure 2.
Structural analysis of FcγRIIIa receptor by HRF-MS.A, oxidative footprint of the tryptic peptides from FcγRIIIa in the unbound, native IgG-bound and afucosylated IgG-bound states. The asterisk denotes significant changes in % modification obtained from triplicate samples (p < 0.05). B, mapping of the HRF data onto the molecular model. Glycans shown as stick representation: IgG N279 (pink), receptor N45 (orange), and receptor N162 (red). HRF, hydroxyl radical footprint; IgG, immunoglobulin G.
The fact that changes in solvent accessibility for nearly all of the FcγRIIIa peptides in the bound state are independent of the IgG1 core Fc Fuc suggests that the receptor does not structurally accommodate the IgG1 fucosylation. Rather, the native IgG1 Fc region undergoes conformational structural changes to accommodate receptor binding.
MD simulations reveal insight into IgG1–FcγRIIIa molecular interactions
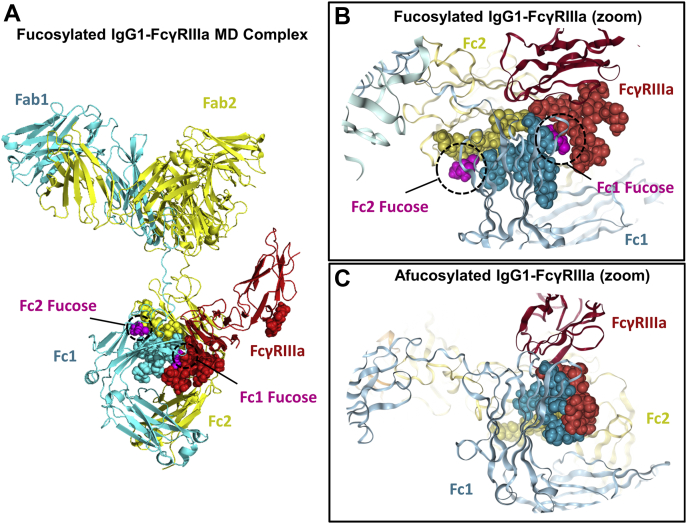
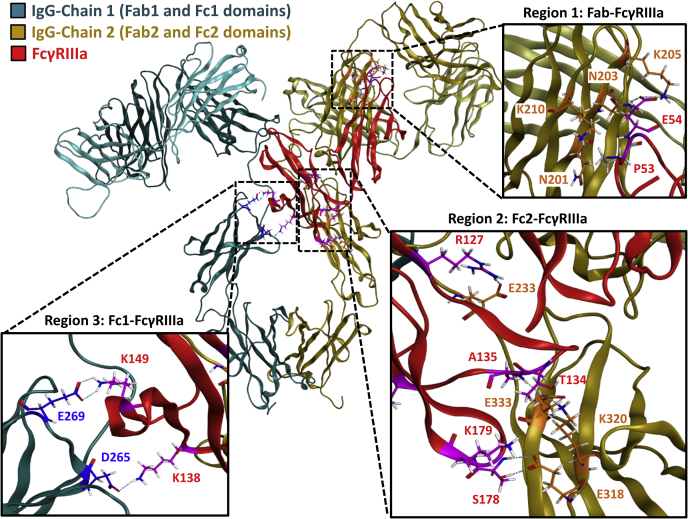
Using published crystal structures, we generated two virtual molecules of the receptor-bound IgG1 with (PDB: 3SGJ) and without (PDB: 3SGK) the core Fuc. The full-length IgG1 structures were constructed by covalently attaching the Fab homology models to the Fc domains and modifying the relative positions of the Fab and Fc domains such that the canonical Y-shape conformation was obtained (see Experimental procedures). The IgG1 molecules were constructed with an N297 G0 glycan (shown in pink in Fig. 2B), and the receptor was constructed with N45 and N162 glycans consistent with the PDB files (shown in Fig. 2B as orange and red, respectively). Extensive MD simulations were performed to generate a total of 6 μs trajectories for each structure. Figure 3A shows the full-length fucosylated IgG1–FcγRIIIa complex used for the MD simulations. Figure 3, B and C provide an expanded view of the intermolecular glycan interactions between the IgG1 (cyan and yellow spheres) and the receptor (red spheres) for the fucosylated and afucosylated complexes, respectively. Because the receptor binds the Fc region of the IgG1 asymmetrically, we have separately denoted the two Fc chains as Fc1 and Fc2 and their corresponding Fab domains as Fab1 and Fab2. For simplicity, we denote Fc1 as the HC containing the core Fuc (magenta) that is in direct proximity to the receptor glycans (Fig. 3B).
Figure 3.
Model of IgG1–FcγRIIIa complex.A, IgG1–receptor model used for MD simulations. B, expanded view of intermolecular glycan interactions with the fucosylated Fc region. C, expanded view of intermolecular glycan interactions with the afucosylated Fc region. Representative structures shown in panels B and C correspond to the most populated states obtained from the probability distribution of the intermolecular glycan distances calculated from combined MD trajectories (see Fig. S3). IgG1, immunoglobulin G1.
To understand the role of the core Fuc on the intermolecular glycan interactions, we first calculated the probability distribution of the minimum distance between the glycans in the two bound forms (Fig. S3). We observed that the presence of Fuc significantly increases the average distance within the intermolecular glycan interactions. As seen in Figure 3B, the Fc1 Fuc (magenta) is positioned directly between the rest of the Fc1 glycans (cyan spheres) and the receptor glycans (red spheres). This is the site of the hypothesized steric hindrance, which likely results in the increase in average distances between the glycans. Figure 3C visually depicts a more compact intermolecular glycan interface in the absence of Fuc, consistent with afucosylated data in Fig. S3.
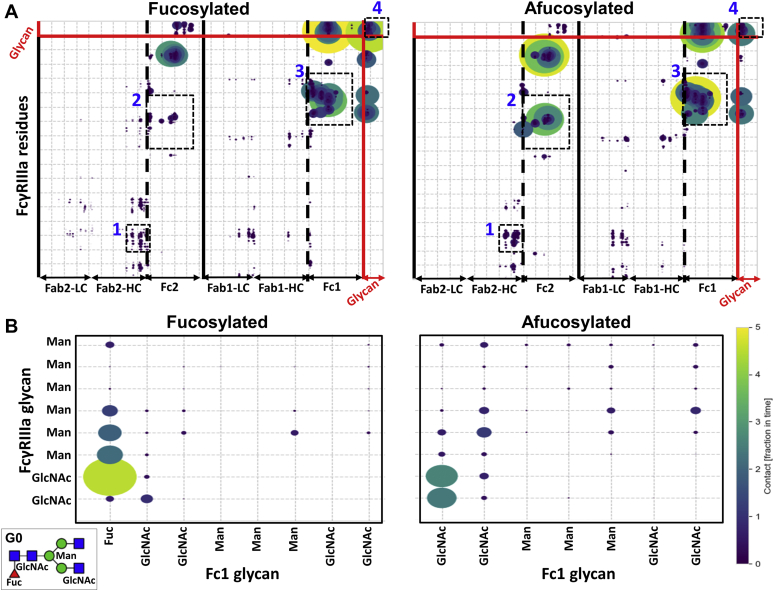
To best visualize the difference of intermolecular interactions within the two complexes, all of the polypeptide and glycan interactions between the two proteins were plotted on a 2D contour map (Fig. 4A). To distinguish which polypeptide chains are interacting with the receptor, the contour map is divided into the Fab1/Fc1 and Fab2/Fc2 for the fucosylated and afucosylated IgG–receptor complexes. Plotting the data in this fashion enabled multiple global differences to be observed.
Figure 4.
Contour representation of MD simulation data.A, full contour map of all of the IgG–receptor intermolecular polypeptide and glycan interactions. Numbering 1 to 4 depicts regions of interest. B, expanded view of region 4 showing intermolecular glycan interactions, with the inset providing pictorial representation of G0 glycan.
First, the Fab regions for both complexes appear to be nearly identical. There are minimal LC–receptor interactions, and the Fab2 domain appears to contribute to Fab–receptor interactions (Fig. 4A, region 1). This indicates that the Fab–FcγRIIIa molecular interactions are similar for both IgG1 molecules, consistent with our HRF data. Second, the interactions in the Fc1 domain are similar between the molecules (Fig. 4A, region 3), albeit with expected differences in glycan–glycan interaction between the antibody and receptor (Fig. 4A, region 4). Finally, there are apparently more polypeptide interactions between the receptor and afucosylated Fc2 domain (Fig. 4A, region 2). This suggests that the Fc2 domain may play a role in contributing to the overall interactions in the absence of Fuc. Each of these regions are discussed in detail below.
The differences in intermolecular glycan interactions due to the Fuc are shown in region 4 of the contour map (Fig. S4). Because there appears to be no significant Fc2–receptor glycan interactions, and as expected, no interactions with the N45 glycan, these regions were removed for clarity in Figure 4B. Region 4 data demonstrate a shift from Fuc-dominated glycan interactions to a more dispersed contact surface area between the proteins in the afucosylated state. The N-linked GlcNAc appears to be much more involved in interaction with the receptor glycans with the removal of Fuc.
Regions 1 to 3 of the contour map (Fig. 4A) depict the polypeptide interactions between the IgG and receptor, all of which are detailed in Figure 5 (glycans removed for clarity). Region 3 of the contour map shows that the primary Fc1–receptor interactions are conserved between the two IgG molecules. These interactions include the hinge region (residue 234–239), B/C loop (residue 264–269), and C’/E loop (residue 294–299) (Fig. S5A). However, two ionic interactions in this region become significantly stronger in the absence of Fuc: Fc1(E269):FcγRIIIa(K149) and Fc1(D265):FcγRIIIa(K138) (Fig. 5, region 3, and Fig. S5A).
Figure 5.
Structural visualization of MD simulation data. Expanded view of IgG1–receptor interactions for regions 1 to 3. Glycans removed for clarity.
Region 2 of the contour map (Fig. S5B) shows that the absence of the core Fuc appears to facilitate multiple interactions between the receptor and the Fc2 domain at the hinge region (residue 234–239), F strand (residue 315–322), and F/G loop (residue 328–338). Specifically, we observed three interactions, Fc2(K320):FcγRIIIa(T134), Fc2(K333):FcγRIIIa(T134,A135), and Fc2(E233):FcγRIIIa(R127), which are only present in the absence of Fuc (Fig. 5, region 2, and Fig. S5B).
Region 1 of the contour map (Fig. S6) not only confirms the previously identified Fab–receptor interactions (24) but also provides the molecular details into the exact nature of these interactions (Fig. 5, region 1). Regardless of the core Fuc, the major interactions appear to be conserved at the CH1 domain (residue 157–167 and residue 192–217) on the Fab2 arm. This is consistent with the HRF data that indicate this CH1 region has reduced solvent accessibility in the bound states.
It is interesting to note that the Fab–receptor interactions are primarily with the Fab2 domain, whereas the Fc–receptor interactions are primarily with the Fc1 domain. This is likely due to the 2-fold access of symmetry of the IgG1 molecule, in which the two HCs spiral around each other in a helical fashion. Because the receptor only binds in a 1:1 stoichiometry, the two HCs appear to interact with receptor through different domains (Fab1 and Fc2). This implies that heterogeneity of the HC homodimers could potentially effect IgG–receptor interactions.
Mutational analysis demonstrates Fab–FcγRIIIa binding interactions directly affect ADCC
Using rational mutagenesis of the novel Fab–receptor interface identified above, we next sought to assess the potential impact to ADCC. As shown in Table S1, 20+ different mutants were generated based on the potential Fab–FcγRIIIa interface and/or sequence alignment comparing IgG isotypes with known reduced ADCC activity (IgG1 versus IgG2/4). It is well established that each glycoform has unique affinity properties toward the receptors; however, the level of afucosylation has been shown to be nearly linear with ADCC activity (33). This means that afucosylation will likely dominate the affinity effects. Therefore, the major challenge to determine the role of site-specific mutations on Fc-effector function is ensuring that glycosylation heterogeneity for the individual mutants remains consistent.
Unfortunately, the mutants we generated exhibited significant variability in the levels of afucosylation and in the commensurate ADCC activity. These results obfuscate any potential impact the site-specific mutations have on ADCC. To circumvent this inherent challenge, we can take advantage of the knowledge that the WT protein has a direct correlation in the levels of afucosylation (normalized G0-F [nG0-F]) and the ADCC activity for our in-house ADCC assay (R2 value >0.97, data not shown). Therefore, if the nG0-F is the primary contributing factor to ADCC, the mutants should show a linear correlation. As seen in Fig. S7, the nG0-F levels of the mutants poorly correlate with ADCC (R2 value=0.57), implying the mutations themselves may be influencing the ADCC activity.
We were, however, fortunate to obtain two triple mutations that have nearly the exact same level of glycan heterogeneity as to the WT control protein (Table S2), enabling direct comparison their effect on ADCC. The two mutant proteins were generated from perceived polypeptide interactions shown in Figures 4 and 5 (region 1). Unexpectedly, both mutants (N201A/N203A/K210A and N201G/N203D/K210A) exhibit an increased ADCC activity (Table 1). The mutants displayed no distinguishable effect on the individual antigen or receptor binding, indicating that the increased ADCC activity is not directly related to the antigen or receptor binding. We anticipated the disruption of the apparent salt bridge between Fab1(K210) and FcγRIIIa(E54) would lead to reduced receptor binding affinity and a commensurate loss in ADCC, making these results puzzling. This leads us to surmise that the increase in ADCC may be due to mechanistic events occurring during ADCC (such as clustering), rather than IgG-receptor affinity. Nonetheless, these were the most convincing results we obtained that demonstrate Fab–receptor interactions can directly modulate ADCC.
Table 1.
Biological characterization of WT mAb1 and the mutants
| Mutation | aNormalized G0-F | ADCC potency | FcγRIIIa-binding affinity, KD (nM) | Antigen-binding affinity, KD (nM) |
|---|---|---|---|---|
| WT | 6.52% | 100.0 ± 6.7% | 266 ± 3 | 113 |
| HC: N201A,N203A,K210A | 6.43% | 111.5 ± 3.5% | 274 ± 8 | 119 |
| HC: N201G,N203D,K210A | 6.41% | 120.5 ± 0.5% | 273 ± 8 | 125 |
Normalized G0-F = [G0-F ÷ (G0-F + G0)] × 100%; detailed glycosylation profile is shown in Table S2.
Discussion
There has been extensive effort to elucidate the role of IgG1 N-linked glycans and their associated fucosylated glycoforms on the binding interactions with FcγRIII receptors. Early studies analyzing crystal structures of a native IgG1 have shown that the IgG1–FcγRIIIa binding site is located at the top of the CH2 domain (13, 14). The binding is primarily through polypeptide interactions that require the CH2 domain to be glycosylated (7). Ferrara et al. (15) subsequently compared the crystal structures of the fucosylated versus afucosylated Fc fragments bound to FcγRIIIa and hypothesized that the increase in affinity of afucosylated IgG1 toward FcγRIIIa is facilitated by intermolecular glycan interactions. The differences in these two crystal structures also indicated that the intermolecular glycan interactions would likely require significant conformational structural changes in the Fc to accommodate a fucosylated N-glycan. Further solution-phase data suggested there is a direct interaction between the Fab and receptor (22, 23, 24), but the biological implications of this interaction have not been shown.
In this report, we probed the IgG1–FcγRIIIa structure–function relationship with multiple solution-phase techniques to further the understanding of the role of Fuc and Fab–receptor interactions. Using HRF analysis, we observed that the core Fuc increases the solvent accessibility in a region of the Fc that is directly affected upon receptor binding. However, the overall SASA for the CH2-CH3 region for the unbound afucosylated Fc is nearly identical to that of the bound native form. This implies that in the absence of Fuc, the Fc region may be sampling conformations that require fewer conformational changes to accommodate receptor binding. In the bound states, the receptor appears to have identical interactions with the IgG regardless of Fuc, indicating that the receptor does not conform to the presence of a core Fuc, but rather that the native IgG1 must structurally accommodate receptor binding.
In addition, our HRF data support previous findings of direct Fab–receptor interactions and indicate these interactions are independent of the fucosylated state. It is important to note that HRF data reflect the average % modification for each peptide in the bound and unbound states. There are two Fab tryptic peptides (one from each HC/LC pair) that are included in the calculation of % modification. Because FcγRIIIa binds at a 1:1 M ratio with 50% probability of binding either side of the IgG1 (leaving the other side unbound), half the population of this CH1 peptide would not be interacting with FcγRIIIa. This, in effect, lowers the apparent % modification calculation if the receptor bound both sides of the IgG. Therefore, the change in the SASA from the CH1–receptor interaction is likely more pronounced than our HRF data is indicating because half of the peptide is not interacting with the receptor.
Using MD simulation, we are able to provide the molecular details of how fucosylation (or the absence of) affects IgG1–receptor interactions. In Figure 4A, region 4 of the contour plot shows that Fuc dominates the intermolecular glycan interactions, which corroborates the steric hindrance hypothesis. The absence of Fuc not only increases the number of distinct intermolecular glycan interactions but also leads to the engagement of the Fc2 domain (region 2) into the receptor polypeptide interactions. The contributions of the Fc2 domain to affinity is unclear, but these data suggest increased interactions via the F strand and F/G loop. The MD data also show that the IgG–receptor polypeptide interactions in the Fc1 domain and Fab domain are nearly identical regardless of the core Fuc. This indicates that the affinity difference between a native and afucosylated IgG appears to be primarily driven by the Fc–glycan/Fc2–domain intermolecular interactions with the receptor.
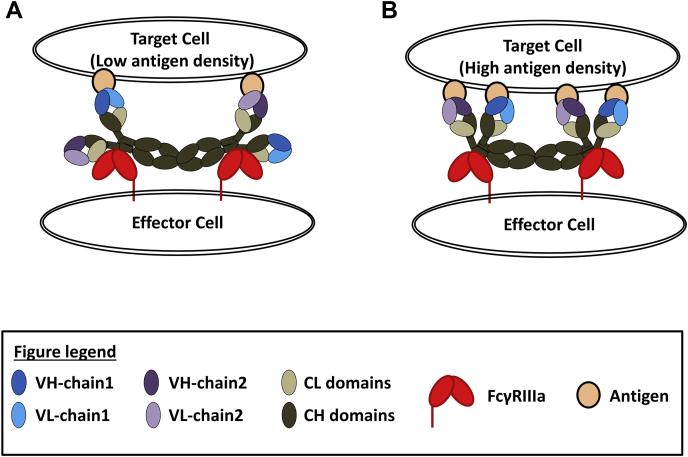
Finally, the rational design of Fab mutants has provided the first direct evidence that Fab–receptor interactions can modulate biological activity. Our triple-mutant molecules showed unexpected increase in ADCC while exhibiting virtually no effect on the individual affinities of the antigen and soluble receptor. One possible explanation is that during ADCC, multiple Fc receptors are recruited at ADCC synapses, which may amplify any Fab-induced affinity differences at cellular level. In addition, the Fab may modulate ADCC through other factors beyond affinity such as clustering of the IgG on the cell the surface as previously suggested (24). A third factor to consider is the monovalent versus bivalent antigen binding, briefly discussed in the beginning of the text. As illustrated in Figure 6, the Fab–receptor interaction may be more pronounced during monovalent binding simply because of the flexibility and degrees of freedom for the unbound Fab arm. The WIL2-S cell used in this study is known to have relatively higher level of expression of CD20 than other CD20-expressing B cells (34), but it is unknown whether monovalent or bivalent (or combination of) antigen binding is occurring during ADCC for the current work.
Figure 6.
Structural renderings of Fc–receptor clustering.A, depiction of monovalent antigen binding for the low-density surface antigen. B, depiction of bivalent antigen binding for the high-density surface antigen.
The totality of evidence across decades of research indicates that the levels of afucosylation is the primary driver for determining IgG1–FcγRIIIa affinity. Mutations in the Fc region have been shown to modulate effector function; however, to our knowledge, these are the first data showing that the CH1 domain can as well directly modulate ADCC activity. Thus, with the contributions from this work, it is becoming apparent that Fab-, Fc-, and glycan-mediated interactions likely work in concert to drive biological activity of Fc-gamma receptor biology.
Experimental procedures
All chemicals were purchased from Sigma-Aldrich unless otherwise noted. Guanidine HCl and formic acid (FA) were purchased from Pierce.
Protein expression and complex formation
The FcγRIIIa-V158 (high affinity) protein (with C-terminal histidine purification tag), the native IgG1, and afucosylated IgG1 were expressed in CHO-derived IgG1 molecules and purified at Genentech, Inc to clinical-quality drug substance. The latter protein was expressed in Fuc transferase KO CHO cell line (35). All samples were buffer exchanged into PBS for HRF-MS analysis. The IgG1–FcγRIIIa complexes were generated by mixing the IgG1 and Fc receptor in a 1:1 M ratio and incubating at 37 °C for 30 min to facilitate binding.
Glycan analysis
Glycan analysis was performed as previously described (36) with the exception that the fluorescently labeled glycans were separated using a Waters Acquity H-Class UPLC chromatography with a Waters Acquity UPLC Glycan BEH Amide Column (1.7 μm, 2.1 mm × 150 mm).
HRF labeling procedure with fast photochemical oxidation of protein
IgG1 and FcγRIIIa, unbound and bound complexes, were subject to laser-induced HRF as previously described in detail (37, 38). This work utilizes our previously described “equal-weight” strategy (39, 40) in which the total amount of protein for each sample was kept constant (0.5 mg/ml) to minimize any differences in oxidative potential between the unbound and bound antigen solutions.
LC-MS/MS analysis
Samples were diluted with 300-μl denaturing buffer (6 M guanidine HCl, 360 mM Tris, 2 mM EDTA, pH 8.6), reduced using DTT (10 mM, 45 °C for 10 min), s-carboxymethylated with sodium iodoacetate (25 mM, 45 °C for 5 min), and quenched with DTT (50 mM, room temperature). Samples were then desalted using NAP-5 columns with an elution volume of 800-μl PBS, digested with 1:20 w/w ratio of protein:trypsin (37 °C for 1 h), deglycosylated with 0.15-μg PNGaseF (37 °C for 30 min), and quenched with 100% FA. Tryptic peptides (10 μg) were separated using a Waters H-Class UPLC with Waters Acquity UPLC CSH130 C-18 column (1.7 μm, 2.1 × 150 mm). Peptide separation occurred across a gradient from 100% solvent A (water and 0.1% FA) to 35% solvent B (acetonitrile and 0.1% FA) over 60 min at a flow rate of 0.3 ml/min and column temperature of 77 °C. MS analysis was performed with a Thermo Fisher Q Exactive mass spectrometer operating in a positive mode, performing MS2 on top-10 most abundant peaks in a data-dependent mode.
Data analysis
Peak identification and quantitation of percent oxidation for each peptide were performed using Biologic Software Suite (Protein Metrics Inc). All samples were analyzed in triplicate. In addition, each sample had a corresponding “no laser” control, a sample run through the HRF system without the laser pulsing, to account for background oxidation by H2O2. The extracted-ion chromatograms for the oxidized peptide species and the parent peptide (in bound and unbound states) were used to calculate the percent oxidation with the equation as previously described (25, 37, 40). The current HRF-MS data analysis, all of the observed charge states, was included in the calculation for each peptide. Percent oxidation is presented as the average of triplicate runs after subtraction of the “no laser” background oxidation control. Error bars represent the statistical analysis by single-sample t test using a 95% confidence interval. Nonoverlapping error bars have p < 0.05 (or otherwise denoted) and are considered significant differences.
Molecular modeling and simulations
The software Molecular Operating Environment (41) from Chemical Computing Group was used to build Fab domain models for each antibody. A full-length homology model was then produced by covalently attaching the two Fab models to the hinge region of the native and afucosylated Fc–receptor complex structures (3SGK and 3SGJ), and the structures were manually adjusted to obtain a Y-shape conformation. The protonation states at pH 7.4 were assigned using the PDB2PQR tool (42).
FF14SB force field was used for proteins (43) and the Glycam06 force field for carbohydrates (44). To assign Glycam06 force fields, residues were renamed according to accepted GLYCAM (45). The bond connections were defined in the tleap script (45) to connect the residues in the glycans. The systems were then solvated using TIP3P water molecules (46) in a rectangular box extending 10 Å from protein edges. Counter ions of Na+ and Cl− with the concentration of 0.15 M were added to neutralize the system. Parameters derived by Joung and Cheatham were used for Na+ and Cl− (47).
The graphical processing unit implementation of Amber 2016 MD software package (45) with the SPFP precision model (48) was used for the MD simulation using the following protocol. First, the structure was relaxed with 2000 steps of conjugate-gradient energy minimization, using harmonic restraining potential with the force constant of 10 kcal/mol/Å2 to restrain the solute to the initial structure. Then, the solvent molecules were allowed to move using NPT ensemble with a temperature of 300 K. Another step of conjugate-gradient energy minimization was performed with 2000 steps while removing all the restrains. Next, the pressure was maintained at 1 atm and the thermostat temperature increased to 300 K over the course of 500 ps, while harmonic positional restraint of strength 10 kcal/mol/Å2 was applied to the solute. The system was then equilibrated for 1 ns with a restraint force constant of 1 kcal/mol/Å2. All restraints were removed for the production stage. The hydrogen mass repartition option of Amber was used, allowing a time step of 4 fs (49). A 10 Å cutoff radius was used for range-limited interactions, with Particle Mesh Ewald electrostatics for long-range interactions. The production simulation was carried out using NPT conditions. Langevin dynamics (50) was used to maintain the temperature at 300K with a collision frequency of 3 psˆ−1. The production stage of the MD simulation was performed for 300 ns. During dynamics, the SHAKE algorithm (51) was applied to constrain all bonds involving hydrogen atoms. Default values were used for all other simulation parameters. The protocol described above was repeated to generate 20 independent replicates of 300-ns trajectories, adding up to 6-microsecond trajectories for each structure.
The CPPTRAJ tool (52) available in AmberTools 2016 (45) was used to calculate the distance between groups of residues (i.e., Fc and receptor glycans), as well as the number of contacts between residue pairs (using “nativecontact” command in CPPTRAJ). The “savenonnative” keyword was also applied to keep track of non-native contacts (the contacts formed over the course of the MD simulations), in addition to the ones present in the starting structure. The contacts were determined by a simple distance cut-off (5 Å), that is, two residues were considered in contact if at least two atoms from the two residues were closer than 5 Å at a given frame. The circle sizes and colors in the 2D contour plots represent the total fraction of frames that the corresponding residues were in contact over the course of the simulations.
ADCC reporter assay
ADCC reporter assay was performed using NK-92 NFAT-Luc reporter cell line as effector cells, and target cells. The effector:target cell ratio was 1:2. Briefly, the effector/target cell mixture was plated in a 96-well white-bottom assay plate at 100,000 cells per well, followed by incubation with serial dilutions of antibodies. After incubation at 37 °C for 3 h, ONE-Glo Luciferase Assay Reagent (Promega) was added and luminescence was determined using SpectraMax Paradigm. The reporter signals of samples and controls were plotted against the antibody concentrations. The dose–response curves were fitted with a four-parameter model using the GraphPad Prism 8.0. For comparison, relative potency and relative efficacy of test antibodies were calculated using the following formulas: ADCC activity=EC50 of control/EC50 of sample.
Binding assay
A Biacore T200 instrument (GE Healthcare) was utilized to determine the kinetic characteristics and binding affinities of IgG1 and mutants to the FcγRIIIa receptor. The His-tagged FcγRIIIa receptor was captured on a CM5 sensor chip via immobilized anti-His antibody. Various concentrations of the test antibodies were injected during multiple cycles at a flow rate of 50 μl/min for 120 s and dissociated for 60 s. After each injection, the chip surface was regenerated by acidic condition. Analysis was performed after an in-line reference cell correction and followed by buffer sample subtraction. The dissociation equilibrium constant (KD), dissociation rate constant (kd), and association rate constant (ka) were calculated using the Biacore BIAevaluation software (version 4.1; GE Healthcare) with a 1:1 binding model.
Analysis of binding affinity to the target antigen was similar to that of binding to the FcγRIIIa receptor, except the target protein was biotinylated on the C terminus and immobilized on an SA sensor chip.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Jeongsup Shim for supplying the proteins and Christina Tsai and Tomasz Baginski for glycan profiling and acknowledge Thomas Patapoff and Jonathan Zarzar for assistance with homology modeling and molecular dynamics simulations. In addition, the authors would like to acknowledge Tilman Schlothauer and Michael Kim for helpful discussion of this article.
Author contributions
Y. S. and S. I. data curation; Y. S., M. C., and A. W. methodology; Y. S. writing–original draft; S. I. software; S. I. visualization; G. D. and A. W. conceptualization; G. D. supervision; A. W. writing–review and editing.
Biography

Yue Sun is a scientist in the bioanalytical sciences department at Genentech Inc. Her research interests focus on protein characterization, understanding protein structure and function relationships, and molecular engineering of proteins with desirable activities. She determines the higher order structure of the full-length IgG molecule, which provides molecular insight into the effect of both IgG fucosylation and Fab–receptor interactions in modulating the FcγRIIIa binding affinity and ADCC activity. She can be found at https://www.linkedin.com/in/yue-sun-6879033a/.
Edited by Peter Cresswell
Footnotes
Present address for Matthew Callahan: Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, California, USA.
Supporting information
References
- 1.Jefferis R. Isotype and glycoform selection for antibody therapeutics. Arch. Biochem. Biophys. 2012;526:159–166. doi: 10.1016/j.abb.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Satoh M., Iida S., Shitara K. Non-fucosylated therapeutic antibodies as next-generation therapeutic antibodies. Expert Opin. Biol. Ther. 2006;6:1161–1173. doi: 10.1517/14712598.6.11.1161. [DOI] [PubMed] [Google Scholar]
- 3.Ecker D.M., Jones S.D., Levine H.L. The therapeutic monoclonal antibody market. MAbs. 2015;7:9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan A.C., Carter P.J. Therapeutic antibodies for autoimmunity and inflammation. Nat. Rev. Immunol. 2010;10:301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 5.Mizuochi T., Taniguchi T., Shimizu A., Kobata A. Structural and numerical variations of the carbohydrate moiety of immunoglobulin G. J. Immunol. 1982;129:2016–2020. [PubMed] [Google Scholar]
- 6.Jefferis R. Glycosylation of natural and recombinant antibody molecules. Adv. Exp. Med. Biol. 2005;564:143–148. doi: 10.1007/0-387-25515-X_26. [DOI] [PubMed] [Google Scholar]
- 7.Raju T.S. Terminal sugars of Fc glycans influence antibody effector functions of IgGs. Curr. Opin. Immunol. 2008;20:471–478. doi: 10.1016/j.coi.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Shields R.L., Lai J., Keck R., O'Connell L.Y., Hong K., Meng Y.G., Weikert S.H., Presta L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 9.Rajasekaran N., Chester C., Yonezawa A., Zhao X., Kohrt H.E. Enhancement of antibody-dependent cell mediated cytotoxicity: A new era in cancer treatment. Immunotargets Ther. 2015;4:91–100. doi: 10.2147/ITT.S61292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung S.T., Kang T.H., Kelton W., Georgiou G. Bypassing glycosylation: Engineering aglycosylated full-length IgG antibodies for human therapy. Curr. Opin. Biotechnol. 2011;22:858–867. doi: 10.1016/j.copbio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Woof J.M., Burton D.R. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat. Rev. Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 12.Tang Y., Lou J., Alpaugh R.K., Robinson M.K., Marks J.D., Weiner L.M. Regulation of antibody-dependent cellular cytotoxicity by IgG intrinsic and apparent affinity for target antigen. J. Immunol. 2007;179:2815–2823. doi: 10.4049/jimmunol.179.5.2815. [DOI] [PubMed] [Google Scholar]
- 13.Sondermann P., Huber R., Oosthuizen V., Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 14.Radaev S., Motyka S., Fridman W.H., Sautes-Fridman C., Sun P.D. The structure of a human type III Fcgamma receptor in complex with Fc. J. Biol. Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 15.Ferrara C., Grau S., Jager C., Sondermann P., Brunker P., Waldhauer I., Hennig M., Ruf A., Rufer A.C., Stihle M., Umana P., Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima T., Yagi H., Takemoto E., Shibata-Koyama M., Isoda Y., Iida S., Masuda K., Satoh M., Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells. 2011;16:1071–1080. doi: 10.1111/j.1365-2443.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subedi G.P., Hanson Q.M., Barb A.W. Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcgammaRIIIa binding. Structure. 2014;22:1478–1488. doi: 10.1016/j.str.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subedi G.P., Barb A.W. The structural role of antibody N-glycosylation in receptor interactions. Structure. 2015;23:1573–1583. doi: 10.1016/j.str.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore G.L., Chen H., Karki S., Lazar G.A. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs. 2010;2:181–189. doi: 10.4161/mabs.2.2.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxena A., Wu D. Advances in therapeutic Fc engineering - modulation of IgG-associated effector functions and serum half-life. Front. Immunol. 2016;7:580. doi: 10.3389/fimmu.2016.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazar G.A., Dang W., Karki S., Vafa O., Peng J.S., Hyun L., Chan C., Chung H.S., Eivazi A., Yoder S.C., Vielmetter J., Carmichael D.F., Hayes R.J., Dahiyat B.I. Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houde D., Peng Y., Berkowitz S.A., Engen J.R. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol. Cell. Proteomics. 2010;9:1716–1728. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi L., Liu T., Gross M.L., Huang Y. Recognition of human IgG1 by Fcγ receptors: Structural insights from hydrogen–deuterium exchange and fast photochemical oxidation of proteins coupled with mass spectrometry. Biochemistry. 2019;58:1074–1080. doi: 10.1021/acs.biochem.8b01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yogo R., Yamaguchi Y., Watanabe H., Yagi H., Satoh T., Nakanishi M., Onitsuka M., Omasa T., Shimada M., Maruno T., Torisu T., Watanabe S., Higo D., Uchihashi T., Yanaka S. The Fab portion of immunoglobulin G contributes to its binding to Fcgamma receptor III. Sci. Rep. 2019;9:11957. doi: 10.1038/s41598-019-48323-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Wecksler A.T., Molina P., Deperalta G., Gross M.L. Mapping the binding interface of VEGF and a monoclonal antibody Fab-1 fragment with fast photochemical oxidation of proteins (FPOP) and mass spectrometry. J. Am. Soc. Mass Spectrom. 2017;28:850–858. doi: 10.1007/s13361-017-1601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Cui W., Gross M.L. Mass spectrometry for the biophysical characterization of therapeutic monoclonal antibodies. FEBS Lett. 2014;588:308–317. doi: 10.1016/j.febslet.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Rempel D.L., Gau B.C., Gross M.L. Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. J. Am. Chem. Soc. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charvatova O., Foley B.L., Bern M.W., Sharp J.S., Orlando R., Woods R.J. Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: Application to galectin-1. J. Am. Soc. Mass Spectrom. 2008;19:1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie B., Sood A., Woods R.J., Sharp J.S. Quantitative protein topography measurements by high resolution hydroxyl radical protein footprinting enable accurate molecular model selection. Sci. Rep. 2017;7:4552. doi: 10.1038/s41598-017-04689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burmeister W.P., Huber A.H., Bjorkman P.J. Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature. 1994;372:379–383. doi: 10.1038/372379a0. [DOI] [PubMed] [Google Scholar]
- 31.Yan Y., Chen G., Wei H., Huang R.Y., Mo J., Rempel D.L., Tymiak A.A., Gross M.L. Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J. Am. Soc. Mass Spectrom. 2014;25:2084–2092. doi: 10.1007/s13361-014-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiselar J.G., Chance M.R. Future directions of structural mass spectrometry using hydroxyl radical footprinting. J. Mass Spectrom. 2010;45:1373–1382. doi: 10.1002/jms.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung S., Quarmby V., Gao X., Ying Y., Lin L., Reed C., Fong C., Lau W., Qiu Z.J., Shen A., Vanderlaan M., Song A. Quantitative evaluation of fucose reducing effects in a humanized antibody on Fcgamma receptor binding and antibody-dependent cell-mediated cytotoxicity activities. MAbs. 2012;4:326–340. doi: 10.4161/mabs.19941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurjar S.A., Derrick J.P., Dearman R.J., Thorpe R., Hufton S., Kimber I., Wadhwa M. Surrogate CD16-expressing effector cell lines for determining the bioactivity of therapeutic monoclonal antibodies. J. Pharm. Biomed. Anal. 2017;143:188–198. doi: 10.1016/j.jpba.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Louie S., Haley B., Marshall B., Heidersbach A., Yim M., Brozynski M., Tang D., Lam C., Petryniak B., Shaw D., Shim J., Miller A., Lowe J.B., Snedecor B., Misaghi S. FX knockout CHO hosts can express desired ratios of fucosylated or afucosylated antibodies with high titers and comparable product quality. Biotechnol. Bioeng. 2017;114:632–644. doi: 10.1002/bit.26188. [DOI] [PubMed] [Google Scholar]
- 36.Reusch D., Haberger M., Maier B., Maier M., Kloseck R., Zimmermann B., Hook M., Szabo Z., Tep S., Wegstein J., Alt N., Bulau P., Wuhrer M. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles--part 1: Separation-based methods. MAbs. 2015;7:167–179. doi: 10.4161/19420862.2014.986000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin M., Krawitz D., Callahan M.D., Deperalta G., Wecksler A.T. Characterization of ELISA antibody-antigen interaction using footprinting-mass spectrometry and negative staining transmission electron microscopy. J. Am. Soc. Mass Spectrom. 2018;29:961–971. doi: 10.1007/s13361-017-1883-9. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y., Estevez A., Schlothauer T., Wecksler A.T. Antigen physiochemical properties allosterically effect the IgG Fc-region and Fc neonatal receptor affinity. MAbs. 2020;12:1802135. doi: 10.1080/19420862.2020.1802135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Rempel D.L., Zhang H., Gross M.L. An improved fast photochemical oxidation of proteins (FPOP) platform for protein therapeutics. J. Am. Soc. Mass Spectrom. 2015;26:526–529. doi: 10.1007/s13361-014-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia N.K., Sreedhara A., Deperalta G., Wecksler A.T. Optimizing hydroxyl radical footprinting analysis of biotherapeutics using internal standard dosimetry. J. Am. Soc. Mass Spectrom. 2020;31:1563–1571. doi: 10.1021/jasms.0c00146. [DOI] [PubMed] [Google Scholar]
- 41.Chemical Computing Group ULC . Chemical Computing Group ULC; Montreal, Canada: 2018. Molecular Operating Environment (MOE) 2013.08. [Google Scholar]
- 42.Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirschner K.N., Yongye A.B., Tschampel S.M., González-Outeiriño J., Daniels C.R., Foley B.L., Woods R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Case D.A., Betz R.M., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., Homeyer N., Izadi S., Janowski P., Kaus J., Kovalenko A., Lee T.S. University of California, San Francisco; San Francisco, CA: 2016. AMBER 2016. [Google Scholar]
- 46.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 47.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Grand S., Götz A.W., Walker R.C. SPFP: Speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 49.Hopkins C.W., Le Grand S., Walker R.C., Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theor. Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 50.Pastor R.W., Brooks B.R., Szabo A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988;65:1409–1419. [Google Scholar]
- 51.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 52.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.