Abstract
Introduction
Timely matching of patients to beneficial targeted therapy is an unmet need in rheumatoid arthritis (RA). A molecular signature response classifier (MSRC) that predicts which patients with RA are unlikely to respond to tumor necrosis factor-α inhibitor (TNFi) therapy would have wide clinical utility.
Methods
The protein–protein interaction map specific to the rheumatoid arthritis pathophysiology and gene expression data in blood patient samples was used to discover a molecular signature of non-response to TNFi therapy. Inadequate response predictions were validated in blood samples from the CERTAIN cohort and a multicenter blinded prospective observational clinical study (NETWORK-004) among 391 targeted therapy-naïve and 113 TNFi-exposed patient samples. The primary endpoint evaluated the ability of the MSRC to identify patients who inadequately responded to TNFi therapy at 6 months according to ACR50. Additional endpoints evaluated the prediction of inadequate response at 3 and 6 months by ACR70, DAS28-CRP, and CDAI.
Results
The 23-feature molecular signature considers pathways upstream and downstream of TNFα involvement in RA pathophysiology. Predictive performance was consistent between the CERTAIN cohort and NETWORK-004 study. The NETWORK-004 study met primary and secondary endpoints. A molecular signature of non-response was detected in 45% of targeted therapy-naïve patients. The MSRC had an area under the curve (AUC) of 0.64 and patients were unlikely to adequately respond to TNFi therapy according to ACR50 at 6 months with an odds ratio of 4.1 (95% confidence interval 2.0–8.3, p value 0.0001). Odds ratios (3.4–8.8) were significant (p value < 0.01) for additional endpoints at 3 and 6 months, with AUC values up to 0.74. Among TNFi-exposed patients, the MSRC had an AUC of up to 0.83 and was associated with significant odds ratios of 3.3–26.6 by ACR, DAS28-CRP, and CDAI metrics.
Conclusion
The MSRC stratifies patients according to likelihood of inadequate response to TNFi therapy and provides patient-specific data to guide therapy choice in RA for targeted therapy-naïve and TNFi-exposed patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40744-021-00330-y.
Keywords: Drug response prediction, Gene expression, Precision medicine, Prospective observational study, Rheumatoid arthritis, TNF inhibitor
Plain Language Summary
A blood-based molecular signature response classifier (MSRC) integrating next-generation RNA sequencing data with clinical features predicts the likelihood that a patient with rheumatoid arthritis will have an inadequate response to TNFi therapy. Treatment selection guided by test results, with likely inadequate responders appropriately redirected to a different therapy, could improve response rates to TNFi therapies, generate healthcare cost savings, and increase rheumatologists’ confidence in prescribing decisions and altered treatment choices. The MSRC described in this study predicts the likelihood of inadequate response to TNFi therapies among targeted therapy-naïve and TNFi-exposed patients in a multicenter, 24-week blinded prospective clinical study: NETWORK-004. Patients with a molecular signature of non-response are less likely to have an adequate response to TNFi therapies than those patients lacking the signature according to ACR50, ACR70, CDAI, and DAS28-CRP with significant odds ratios of 3.4–8.8 for targeted therapy-naïve patients and 3.3–26.6 for TNFi-exposed patients. This MSRC provides a solution to the long-standing need for precision medicine tools to predict drug response in rheumatoid arthritis—a heterogeneous and progressive disease with an abundance of therapeutic options. These data validate the performance of the MSRC in a blinded prospective clinical study of targeted therapy-naïve and TNFi therapy-exposed patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40744-021-00330-y.
Key Summary Points
| Why carry out this study? |
| The many targeted therapies in rheumatoid arthritis (RA) have comparable efficacy and safety |
| Tumor necrosis factor-α inhibitor (TNFi) therapies are the most prescribed targeted therapy in RA, although only approximately one-third of patients will adequately respond to this class of treatments |
| The timely matching of patients to beneficial therapy is a critical unmet need in RA |
| What was learned from the study? |
| This study describes the performance of, and biology interrogated by a 23-feature blood-based molecular signature response classifier (MSRC) that uses RNA sequencing data and clinical features to predict which patients with rheumatoid arthritis are unlikely to adequately respond to TNFi therapies |
| Patients with a molecular signature of non-response are unlikely to respond to TNFi treatment according to ACR, CDAI, and DAS28-CRP criteria of response at 3 and 6 months |
| The MSRC stratifies patients according to their likelihood of responding to a TNFi therapy with statistically significant odds ratios of up to 8.8 among targeted therapy-naïve patients and 26.6 among TNFi-exposed patients |
| The MSRC is rooted in RA disease biology and readily generalizes to the molecular phenotypes of two independent observational cohorts |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.14718621.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by chronic inflammation that causes joint destruction. Following inadequate response to conventional synthetic disease modifying anti-rheumatic drugs (csDMARDs) such as methotrexate, clinical guidelines suggest one of many targeted therapies with comparable efficacies and safety profiles including tumor necrosis factor-α inhibitors (TNFi), interleukin-6 (IL-6) inhibitors, Janus kinase (JAK) inhibitors, and B or T cell modulators. The abundance of treatment options underscores the need for precision medicine in rheumatology [1–3]. Because clinical guidelines do not recommend one treatment over another [1, 4], therapy selection is often driven by administrative decisions and TNFi therapies remain the prevailing treatment in nearly 90% of patients with RA [5, 6]. Matching each patient with the right targeted therapy to reach treat-to-target goals of low disease activity (LDA) or remission is an unmet medical need in RA [1, 7, 8].
Only a subset of patients with RA have an adequate response to TNFi treatment: 50–70% achieve ACR20, 30–40% achieve ACR50, and 15–25% achieve ACR70 response [9–19] and only 10–25% achieve remission [20–23]. Many studies have attempted to identify biomarkers and develop models to predict response to TNFi therapy before the initiation of treatment [24–30]. Failure to validate and reproduce the performance of these predictive biomarkers in new patient populations and clinical trials was a typical outcome [31, 32]. Differing characteristics between patient populations, laboratory methods, procedures in generating molecular data, and other biases inherent to single-cohort retrospective studies have hindered precision medicine progress not only in rheumatology but in other medical specialties as well [33–35].
Mapping of disease-related proteins onto the human interactome, a network map of known pairwise protein–protein interactions that occur in human cells [36], has yielded new insights into human disease biology and response to therapy [37–40]. With the identification of molecular biomarkers involved in RA biology discovered from human interactome analyses, this study demonstrated the TNFi inadequate response prediction performance of a molecular signature response classifier (MSRC) in two prospective observational clinical studies: the CERTAIN study and NETWORK-004. Validation of the MSRC was performed in a total of 391 blood samples from targeted therapy-naïve patients and 113 blood samples from TNFi therapy-exposed patients. This precision clinical tool can help rheumatologists determine whether TNFi therapy is an appropriate treatment choice for targeted therapy-naïve patient or adjust the treatment selection for patients receiving a TNFi therapy.
Methods
Patients
See Supplementary Fig. S1 for an outline of the study design.
CERTAIN study: PAXgene blood samples and clinical measurements of 345 patients with RA were prospectively collected in the CERTAIN study, a comparative effectiveness study for patients with RA initiating a biologic [41]. Institutional review board or ethics committee approvals were obtained prior to sample collection and study participation, and patients provided informed consent. CERTAIN was a comparative effectiveness study investigating initiators of biologics. Samples were collected from patients who were naïve to targeted therapies at the time of sample collection and initiated a TNFi therapy. Consistent with the inclusion criteria of the CERTAIN study, all patients had a Clinical Disease Activity Index (CDAI) greater than ten at the time of biologic therapy initiation. The CERTAIN clinical and molecular data were used for biomarker feature selection (100 patients) and in-cohort cross-validation (245 patients).
Prospective NETWORK-004 study: Patients were determined by the treating rheumatologist to be candidates for TNFi therapy prior to enrollment. Eligible patients were at least 18 years of age, had active RA (CDAI > 10, swollen joint count ≥ 4), and were receiving a stable dose of methotrexate (at least 15 mg/week) for at least 10 weeks prior to baseline. Doses of hydroxychloroquine not exceeding 400 mg/day or leflunomide not exceeding 20 mg/day were permitted so long as the dose was stable for at least 4 weeks prior to the baseline visit. Prednisone doses of at most 10 mg/day were allowable if the dose was stable for at least 2 weeks prior to baseline. Use of intra-articular or parenteral corticosteroids less than 2 weeks prior to the first study procedure was prohibited. The study was approved by the Copernicus Group Independent Review Board (approval # 20191082) and local review boards where required. This study was conducted in accordance with the principles outlined in the Declaration of Helsinki. Written informed consent was granted by all study participants. This study was conducted at 73 rheumatology practices, distributed across the USA. Dosage and treatment of TNFi therapies were at the rheumatologist’s discretion. At the 3-month follow-up, rheumatologists were permitted to make dosing adjustments if deemed necessary for appropriate clinical care. Initiation of a second TNFi therapy resulted in subject withdrawal. The COVID-19 pandemic contributed to a higher level of attrition than initially projected. Of the 273 patients with RA enrolled, 168 completed the 24-week study; 146 patients had complete clinical and molecular data and were included in analyses. Information about patients who left the study are available in Supplementary Table S1. PAXgene blood samples collected at baseline from 146 patients were analyzed as TNFi-naïve samples and those collected at the 3-month visits from 113 patients were analyzed as TNFi-exposed samples.
Clinical Evaluation and Response to TNFi Therapies
Feature selection response definition: Clinical outcome metrics such as swollen and tender joint counts, patient and physician disease assessments have inherent variability [42–44]. To identify a subset of patients in the training cohort who have been assigned the responder and non-responder labels with high confidence, a Monte Carlo simulation approach was implemented to calculate a confidence outcome score for each patient. The clinical outcomes data for patients with at least 70% concordance between the simulations and the actual reported outcome were considered high confidence. High confidence clinical outcomes for both ACR and EULAR metrics were used for the feature selection only.
CERTAIN study: Baseline RNA sequencing data and clinical assessments to predict response to TNFi therapy at the 3- and 6-month follow-up visits according to ACR, CDAI, and DAS28-CRP criteria.
NETWORK-004: Clinical assessments were collected at baseline, 3-month, and 6-month visits: 28-joint count for tenderness and swelling, patient global assessment of pain, patient global assessment of disease activity, CDAI score, Health Assessment Questionnaire, and C-reactive protein (CRP). Rheumatoid factor (RF) and anti-cyclic citrullinated protein (anti-CCP) antibody serostatus was recorded at baseline. PAXgene RNA blood tubes were collected at all visits. The 3-month follow-up visit molecular and clinical data was used to predict response to TNFi therapy at the 6-month follow-up visit according to the ACR, CDAI, and DAS28-CRP criteria.
RNA Preparation and Sequencing Analysis
RNA was extracted from whole blood in PAXgene RNA tubes using MagMax™ for Stabilized Blood PAXgene Tubes RNA Isolation Kit (Thermo Fisher Scientific) per the manufacturer’s instructions; 100–1000 ng of RNA was processed using the KAPA RNA HyperPrep Kit with RiboErase (HMR) Globin. Samples were quantified using Agilent D1000 reagents. Libraries were sequenced to high uniform depth targeting more than 7 million protein coding reads. The CERTAIN cohort was sequenced using the Illumina NextSeq DX 500 and NovaSeq 6000 instruments. NETWORK-004 samples were sequenced using the Illumina NovaSeq 6000 instrument using a validated diagnostic assay under Clinical Laboratory Improvement Amendments (CLIA). Sequence data was processed to determine gene expression across the whole genome. To be included in analyses, samples had to have a TapeStation RIN > 4, RNA concentration ≥ 10 ng/µL, sequencing library yield ≥ 10 nM, percentage perfect basepair index > 85, percentage bases over Phred score 30 > 75, the mean quality Phred score > 30, the median Phred score > 25, and a lower quartile Phred score > 10 for all bases [45].
Human Interactome Analysis and Feature Selection
For selection of transcript biomarker features, 100 samples were randomly selected out of the cohort of 345 patients (CERTAIN study). The random forest algorithm [46] was used to rank protein coding transcripts through 96 rounds of 20% cross-validation in silico experiments. Features that were ranked in the top 100 in 70/96 iterations were further analyzed by the human interactome analysis to identify biologically relevant biomarkers. Biomarkers overlapping with the RA disease module [40] on the human interactome [37] or possessing a significant number of connections to the disease module were used in the final model. Significance of connections was assessed using the hypergeometric test.
Predictive Classification Model Training and Cross-Validation
Samples not included in biomarker feature selection were evaluated. Transcripts identified in this study were integrated with previously described gene expression biomarkers [40] using machine learning to train a response classification model. To assess model performance, tenfold cross-validation was conducted using a feedforward artificial neural network [47]. Model building was done using the MLPClassifier package available in Python’s machine learning library sklearn [48].
Statistical Analysis
Statistical analyses were performed using Python 3.7.6 and R version 3.6.1 (www.r-project.org). Continuous data were summarized with mean, standard deviation, median, minimum, maximum, and number of evaluable observations. Categorical variables were summarized with frequency counts and percentages. Confidence intervals (CI) were determined, where appropriate, using the t distribution for continuous data and an exact method for categorical variables. All tests were done in a two-sided setting. Unless otherwise specified, hypothesis testing was performed at the two-sided 0.05 significance level. All attempts were made to limit missing data. No attempt was made to impute missing data.
Results
Identification of a Molecular Signature of Non-Response to TNFi Therapies Using the Human Interactome
Transcripts predictive of inadequate response to TNFi therapies according to ACR50 and EULAR response definitions (see “Methods”) at 6 months were determined using machine learning from baseline blood sample data of 100 targeted therapy-naïve patients with RA randomly selected from the CERTAIN study (Table 2 and Supplementary Table S3). To ensure that transcripts reflected RA disease biology, the proteins encoded by selected transcripts were mapped onto the human interactome map of pairwise protein–protein interactions to identify transcripts that were significantly connected (p value < 0.05) to the RA disease module (Fig. 1) [40]. The TNFi therapy response features overlap with the same network neighborhood of the human interactome consisting of RA disease-associated proteins. These features included proteins with relevance to RA pathobiology, including JAK3 and interleukin-1 beta (IL-1β) [49, 50]. The molecular signature of non-response to TNFi therapy included 23 features: 19 RNA transcripts and 4 clinical features (Table 1).
Table 2.
Patient demographic and disease characteristics at baseline
| Characteristic | CERTAIN study | NETWORK-004 study | ||
|---|---|---|---|---|
| Feature selection (N = 100) |
Cross-validation (N = 245) |
Targeted therapy-naïve (N = 146) |
TNFi-exposed (N = 113) |
|
| Age (year), mean (SD) | 54 (12.4) | 55 (12.3) | 58 (14.1) | 57 (14.9) |
| Female, n (%) | 72 (72.0%) | 179 (73.1%) | 115 (78.8%) | 87 (77.0%) |
| Duration of disease (year), median (IQR) | 1 (1, 5) | 2 (1, 6) | 1 (0, 4.25) | 1 (0, 4) |
| Race, n (%) | ||||
| White | 83 (83.0%) | 213 (86.9%) | 113 (77.4%) | 92 (81.4%) |
| Black | 9 (9.0%) | 13 (5.3%) | 16 (11.0%) | 12 (10.6%) |
| Other | 8 (8.0%) | 19 (7.8%) | 13 (8.9%) | 9 (8.0%) |
| CCP positive, n (%) | 62 (62.0%) | 154 (62.9%) | 72 (49.3%) | 54 (47.8%) |
| RF positive, n (%) | 76 (76.0%) | 172 (70.2%) | 55 (37.7%) | 48 (54.5%) |
| Prednisone at baseline, n (%) | 30 (30.0%) | 64 (26.1%) | 33 (22.6%) | 23 (20.4%) |
| Prednisone dosage, median (IQR) | 5 (5, 10) | 5 (5, 10) | 5 (5, 9.38) | 5 (5, 5) |
| Current csDMARD, n (%) | ||||
| Methotrexate | 56 (56.0%) | 138 (56.3%) | 120 (82.2%) | 91 (80.5%) |
| ≥ 2 csDMARDs | 7 (7.0%) | 42 (17.1%) | 12 (8.2%) | 9 (8%) |
| None | 15 (15.0%) | 37 (15.1%) | 25 (17.1%) | 22 (19.5%) |
| TNFi use, n (%) | ||||
| Adalimumab | 36 (36.0%) | 98 (40.0%) | 48 (32.9%) | 40 (35.4%) |
| Etanercept | 35 (35.0%) | 76 (31.0%) | 31 (21.2%) | 25 (22.1%) |
| Infliximab | 15 (15.0%) | 48 (19.6%) | 18 (12.3%) | 16 (14.2%) |
| Certolizumab pegol | 10 (10.0%) | 17 (6.9%) | 13 (8.9%) | 7 (6.2%) |
| Golimumab | 4 (4.0%) | 6 (2.4%) | 36 (24.7%) | 25 (22.1%) |
SD standard deviation, IQR interquartile range, csDMARD conventional synthetic disease-modifying antirheumatic drug, TNFI tumor necrosis factor-α inhibitor
Fig. 1.
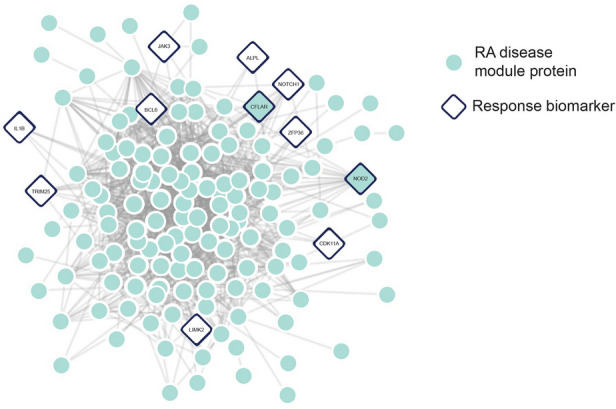
Response biomarkers are in the same network vicinity of the human interactome as the RA disease module. Proteins encoded by transcripts predictive of response were mapped onto the human interactome. Proteins are shown in circles and pairwise physical protein–protein interactions are indicated as lines. The RA disease module is composed of seed and DIAMOnD genes (teal). The proteins encoded by 11 transcript features (diamonds) were either part or significantly connected to the RA disease module (p value < 0.05)
Table 1.
Biomarkers in the MSRC
| ALPL |
| ATRAID |
| BCL6 |
| CDK11A |
| CFLAR |
| COMMD5 |
| GOLGA1 |
| IL1B |
| IMPDH2 |
| JAK3 |
| KLHDC3 |
| LIMK2 |
| NOD2 |
| NOTCH1 |
| SPINT2 |
| SPON2 |
| STOML2 |
| TRIM25 |
| ZFP36 |
| BMI |
| Sex |
| Patient global assessment |
| Anti-CCP |
In-cohort Validation of the MSRC
The MSRC was tested through in-cohort cross-validation among baseline blood samples of an independent cohort of 245 patients from the CERTAIN study who were naïve to targeted therapies (Table 2 and Supplementary Table S3). The resultant AUC for the ACR50 outcome at 6 months was 0.66 for the MSRC and 0.625 for a signature containing only the 19 RNA transcripts. To evaluate the contributions of the molecular and clinical components in response prediction, logistic regression was performed with the classifier scores from the molecular and clinical features, as two individual independent components. Both components were statistically significant using the Wald test (p value < 0.001) indicating that molecular and clinical components carry complementary information in the MSRC. In addition, the model goodness-of-fit was assessed by the Hosmer–Lemeshow test (p value = 0.27), indicating that the MSRC model is well calibrated.
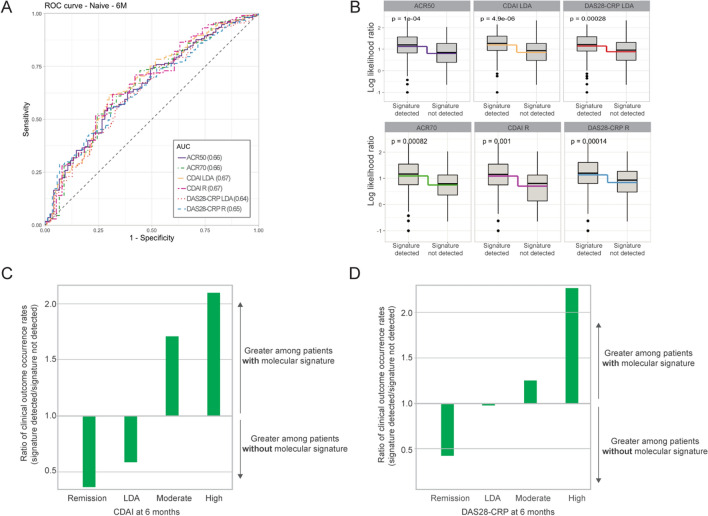
The cross-validation analysis of the 23-feature MSRC resulted in AUC values of 0.64–0.67 for ACR70, CDAI, and DAS28 responses at 6 months post treatment initiation (Fig. 2a; Supplementary Table S2). Significant differences in the log-likelihood ratio of model scores (p < 0.001) were observed between patients who did and did not have the molecular signature of non-response (Fig. 2b). Furthermore, the proportion of patients who achieved LDA or remission at 6 months per CDAI and DAS28-CRP was greater among those patients who lacked a molecular signature of non-response (Fig. 2c).
Fig. 2.
Cross-validation of the MSRC among 245 patients from the CERTAIN study. a Receiver operating characteristic curve for stratification of patients based on CDAI, DAS28-CRP, ACR70, and ACR50 clinical outcomes. b Comparison of model scores for patients with or without a detected molecular signature of non-response. Boxes and intersecting line depict interquartile range and median, respectively. Bisecting colored lines indicate change in mean. Ratio of the occurrence rates for ≤ remission, ≤ LDA, ≥ moderate or ≥ high disease activity per c CDAI and d DAS28-CRP among patients for whom a molecular signature of non-response was or was not detected. Bars indicate a greater proportion of patients with a molecular signature when above 1.0 or without a detected molecular signature when below 1.0
Validation of the MSRC in the Prospective Observational NETWORK-004 Study Among Targeted Treatment-Naïve Patient Samples
To further validate the ability of the MSRC to predict the likelihood of inadequate response to TNFi therapies, patient samples were prospectively collected in a multicenter observational clinical study. Following clinical and molecular data quality review, 146 patients completed the 24-week study and were included in analyses. These patients were predominantly female (78.8%), with a median age of 58 years (Table 2). TNFi therapy choice was at the discretion of the prescribing physician and all five therapeutic options within the class were represented (adalimumab 32.9%, certolizumab pegol 8.9%, etanercept 21.2%, infliximab 12.3%, and golimumab 24.7%). A molecular signature of non-response was detected at baseline for 44.5% (65/146) of patients.
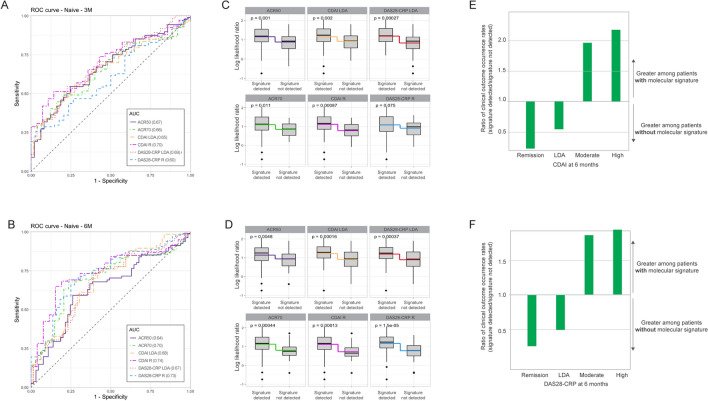
According to the primary endpoint of ACR50 response to TNFi therapy at 6 months, the MSRC stratified patients according to their likelihood of inadequate response to TNFi therapy with an AUC of 0.64 (Fig. 3a) and an odds ratio of 4.1 (95% CI 2.0–8.3; p value 0.0001) (Table 3). No significant difference in baseline demographic characteristics was observed between patients who did or did not respond to treatment (Supplementary Table S3). Additional endpoints included assessment of ACR50 response at 3 months and response to treatment at 3 and 6 months according to ACR70, DAS28-CRP remission (< 2.4) or LDA (< 2.9) and CDAI remission (≤ 2.8) or LDA (< 10). The MSRC stratified patients according to their likelihood of inadequate response at both timepoints and response criteria with AUC values ranging from 0.60 to 0.74 (Fig. 3a, b) and significant odds ratios of 3.4–8.8 (p value < 0.01) (Table 3). Odds ratios describing whether patients with a molecular signature of non-response failed to achieve DAS28-CRP remission were significant at 6 months (p value < 0.05), but not 3 months. Significant differences (p value < 0.05) in model scores between patients who did and did not have a molecular signature of non-response were observed for all response criteria except DAS28-CRP remission (Fig. 3b, c). Furthermore, the fraction of patients who achieved remission and LDA at 6 months per CDAI and DAS28-CRP definitions was greater among those patients who did not have a molecular signature of non-response (Fig. 3e, f, Supplementary Table S4).
Fig. 3.
Validation of the MSRC to identify patients naïve to targeted therapies who are unlikely to respond to TNFi therapy. Receiver operating characteristic curve for stratification of patients based on CDAI, DAS28-CRP, ACR70, and ACR50 clinical outcomes at a 3 months and b 6 months. Comparison of model scores at c 3 months and d 6 months for patients with or without a detected molecular signature of non-response. Boxes and intersecting line depict interquartile range and median, respectively. Bisecting colored lines indicate change in mean. Ratio of the occurrence rates for ≤ remission, ≤ LDA, ≥ moderate or ≥ high disease activity per e CDAI and f DAS28-CRP among patients for whom a molecular signature of non-response was or was not detected. Bars indicate a greater proportion of patients with a molecular signature when above 1.0 or without a detected molecular signature when below 1.0
Table 3.
Odds of patient with a molecular signature of non-response having an inadequate response to TNFi therapy
| AUC | Odds ratio (95% CI; p value) | |
|---|---|---|
| Prospective observational, naïve | ||
| ACR50, 3 months | 0.67 | 3.7 (1.7–8.0; 0.0007) |
| ACR50, 6 months | 0.64 | 4.1 (2.0–8.3; 0.0001) |
| ACR70, 3 months | 0.66 | 2.5 (1.0–6.2; 0.05) |
| ACR70, 6 months | 0.70 | 6.7 (2.7–16.7; < 0.0001) |
| CDAI LDA, 3 months | 0.65 | 3.4 (1.6–7.0; 0.0009) |
| CDAI LDA, 6 months | 0.68 | 3.6 (1.8–7.2; 0.0002) |
| CDAI remission, 3 months | 0.70 | 2.6 (1.1–6.1; 0.03) |
| CDAI remission, 6 months | 0.74 | 8.8 (2.9–27.3; < 0.0001) |
| DAS28-CRP LDA, 3 months | 0.69 | 4.1 (1.9–9.1; 0.0005) |
| DAS28-CRP LDA, 6 months | 0.67 | 3.6 (1.8–7.3; 0.0003) |
| DAS28-CRP remission, 3 months | 0.60 | 1.4 (0.6–3.1; 0.43) |
| DAS28-CRP remission, 6 months | 0.73 | 5.8 (2.6–13.0; < 0.0001) |
| Prospective observational, TNFi-exposed | ||
| ACR50, 6 months | 0.66 | 3.3 (1.5–7.4; 0.0038) |
| ACR70, 6 months | 0.73 | 6.0 (2.0–17.7; 0.0007) |
| CDAI LDA, 6 months | 0.72 | 5.5 (2.4–12.4; < 0.0001) |
| CDAI remission, 6 months | 0.83 | 26.6 (3.4–209.8; < 0.0001) |
| DAS28-CRP LDA, 6 months | 0.69 | 6.7 (2.8–16.1; < 0.0001) |
| DAS28-CRP remission, 6 months | 0.62 | 2.1 (0.9–5.0; 0.0878) |
AUC area under the curve, CI confidence interval, ACR American College of Rheumatology, CDAI clinical disease activity index, LDA low disease activity, DAS28-CRP disease activity score 28-joint count with C-reactive protein, TNFi tumor necrosis factor-α inhibitor
Validation of the MSRC in the Prospective Observational NETWORK-004 Study Among TNFi-Exposed Patient Samples
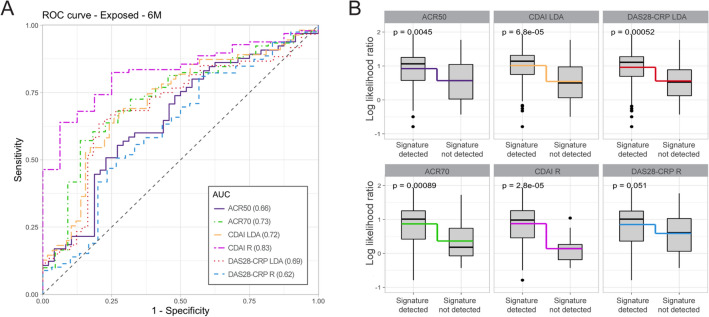
Among patients who completed the 24-week study, RNA blood samples at 3 months were available for 113 patients. The same MSRC as in the targeted therapy-naïve analyses and 3-month patient samples were used to predict inadequate response to TNFi therapy. The molecular signature in these TNFi-exposed samples stratified patients according to inadequate response to treatment with AUC values of 0.62–0.83 (Fig. 4a). A molecular signature of non-response was detected for 40.7% (46/113) of TNFi-exposed patients and a significant difference in model scores (p values < 0.05) was observed between patients who did and did not have a molecular signature of non-response (Fig. 4b). This corresponded to significant odds ratios of 3.3–26.6 among patients with a molecular signature failing to have a response to treatment according to all criteria except for DAS28-CRP remission (Table 3).
Fig. 4.
Validation of the MSRC to identify TNFi-exposed patients who are unlikely to respond to TNFi therapy. a Receiver operating characteristic curve for stratification of patients who are receiving a TNFi therapy based on achievement of CDAI remission or DAS28-CRP remission 3 months after test results. b Comparison of model scores for patients with or without a detected molecular signature of non-response. Boxes and intersecting line depict interquartile range and median, respectively. Bisecting colored lines indicate change in mean
Discussion
Many targeted therapeutic options are available in RA, but therapy selection is a challenge because these options have similar treatment outcomes. Precision medicine tools are greatly needed to identify which patients have the appropriate disease biology for each targeted therapy [1–3, 7, 8]. The blood-based MSRC analyzes gene expression data along with common clinical features to accurately identify targeted-therapy naïve and TNFi-exposed patients who were unlikely to have an adequate response to TNFi therapy. Among patients initiating their first targeted therapy, those with a molecular signature of non-response were three to nine times less likely to have an adequate response to a TNFi therapy (Table 3). When testing was performed after the patient had been receiving TNFi therapy for at least 3 months, patients with a molecular signature of non-response were as much as 26 times less likely to achieve remission. Furthermore, the molecular signature of non-response was predictive of inadequate response to TNFi therapy according to multiple clinically validated measures including ACR50, ACR70, DAS28-CRP, and CDAI. The MSRC could inform provider decision-making at multiple occasions in the care pathway, such as before initial therapy selection or when targeted TNFi therapy does not result in treatment goals and a second therapy or dose escalation is being considered. By validating multiple response target definitions, the MSRC fits within multiple practice protocols, making it easy to understand, act upon, and operationalize within clinical settings.
Precision medicine has improved patient outcomes in oncology and hematology by matching therapy selection to patients’ unique biology. Yet even in these fields, predicting drug response, particularly from blood, has remained a challenging technical problem [51–53]. Unlike in oncology, DNA analyses and biopsies of disease tissue are not readily available in the care of patients with RA because synovial biopsies outside of clinical studies are rare and DNA sequence variations provide limited actionable information in RA [54–56]. In the R4RA trial, RNA sequencing-based identification of patients with RA with a low or absent B cell lineage gene expression signature in synovial tissue significantly correlated with a superior response to tocilizumab than rituximab, thus suggesting that gene expression may be used to predict response to other targeted therapies in RA [57]. Studies of the AMPLE, AVERT, GO‐BEFORE, and GO‐FORWARD trials have used baseline disease assessments such as DAS28, RAPID3, CDAI, or SDAI to predict radiographic progression or magnetic resonance imaging-detected synovitis in response to treatment with a targeted therapy [58, 59]. Comparable AUC values were reported (0.54–0.72) but the odds ratios (1.01–1.65) were lower than those observed in this study (2.1–26.6). The odds ratios in this study were consistent between the cross-validation CERTAIN cohort and the prospective NETWORK-004 cohort indicating that the MSRC is reproducible and generalizable across studies and patient populations.
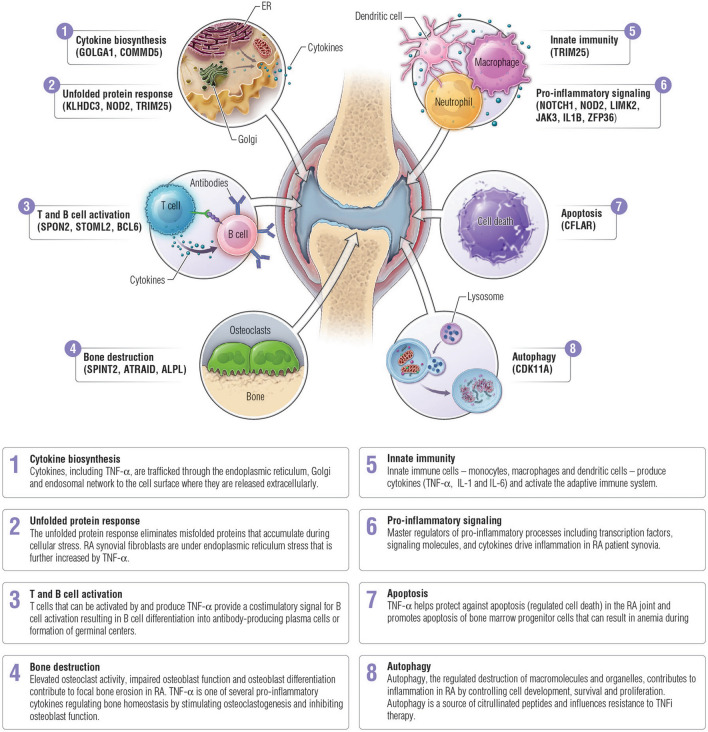
The network biology-based methods used in this study uncovered novel connections within disease biology. The RNA transcripts in the MSRC evaluate seemingly disparate aspects of the disease that are nonetheless unified in the same network neighborhood in the human interactome and capture the diverse biology of RA and response to TNFi therapy. The proteins encoded by these transcripts influence biological processes including cellular homeostasis for adaptive and innate immune cells, production of TNFα and other secreted signaling molecules, synovitis, and bone destruction (Fig. 5 and Supplementary Discussion). TNFα biology is robustly captured in the MSRC and features are involved in the production and release of TNFα (e.g., COMMD5), and upstream or downstream TNFα signaling events (e.g., NOTCH1). Identification of molecular characteristics expressed in circulating blood cells suggests that direct evaluation of joint physiology or biochemistry may not be essential to evaluating synovial phenotypes or response to treatment [60, 61].
Fig. 5.
Biology of inadequate response to TNFi therapies. The MSRC includes transcripts that encode proteins involved in many aspects of RA pathophysiology: innate immune response, cytokine biosynthesis, T and B cell homeostasis, bone homeostasis, the unfolded protein response, autophagy, apoptosis, and pro-inflammatory signaling. Detailed text and additional references can be found in the Supplemental Discussion
The MSRC has the potential to be transformative in RA. A survey of 248 USA-based rheumatologists demonstrated that they would welcome precision medicine advances in autoimmune diseases and would find value in the MSRC [62]. Selection of TNFi therapy declined by more than 80% (from 79.8% to as low as 11.3%) when rheumatologists were presented with a sample MSRC result indicating an unlikely response outcome [62]. Moreover, the majority of rheumatologists surveyed reported that test results would increase their confidence in prescribing decisions, improve medical decision-making, and alter their treatment choices [62].
A model of treatment selection guided by MSRC results was shown to improve response rates to targeted therapies and result in healthcare cost savings [63]. If MSRC test results were used to guide TNFi treatment decisions, patients lacking a molecular signature of non-response could be prescribed a TNFi therapy and would achieve ACR50 response, LDA, and remission (bars below x-axis in Figs. 2c, d, 3e, 3f). Among the 146 targeted therapy-naïve patients, 17.8% (26/146) achieved CDAI remission at 6 months. If these patients are stratified according to MSRC results, 32.4% (22/68) of patients without a molecular signature of non-response achieved remission in response to TNFi therapy, while only 5.1% (4/78) of those with a molecular signature achieved remission.
Patients with a molecular signature of non-response could be directed to one of the many alternative treatment options. This study demonstrated that the MSRC is predictive of non-response in patients already exposed to TNFi therapy. Furthermore, it would identify patients who are unlikely to benefit from a second or third TNFi therapy, thereby preventing TNFi cycling. TNFi cycling is not recommended by treatment guidelines because patients often do not reach their treatment goals within the shortest timeline [4, 64, 65]. In claims-based analyses of commercial and Medicare Advantage data, in comparison to patients who changed to an alternative targeted therapy, patients who cycled on TNFi therapies had a lower median treatment persistence, lower 1-year treatment persistence rate, lower likelihood of restarting treatment after a 60-day treatment gap, higher mean 1-year healthcare costs, and trended toward reduced improvement in treatment response per CDAI [66–68].
Given the progressive and debilitating nature of RA, identifying beneficial treatment for patients and avoiding prolonged periods of ineffective treatment is critical. Patients with RA with persistent high disease activity face an increased risk of mortality compared to those having persistent low disease activity [69]. As RA progresses, disease-induced inflammation and subsequent tissue damage can result in debilitating chronic pain and impair patients’ ability to perform activities of daily living [70]. Prescribing therapy that is appropriate for patient’s individual disease biology could more rapidly halt the progression of RA.
This study has the following limitations. This is an observational study and the MSRC test results were not used to inform treatment selection. This study did not investigate development of anti-drug antibodies; thus, the impact on response predictions could not be assessed for the TNFi-exposed patient samples. Finally, some of the clinical outcome measures used in RA rely on subjective assessments of disease activity. The variability in these assessments could have impacted the true patient outcome and the MSRC accuracy estimation. As future development, it is imperative to expand this approach to molecular signatures correlating with the response to alternative targeted therapies in RA.
Conclusions
Validation of the MSRC involved analysis of gene expression data derived from blood samples of 391 patients with RA who were treated with a TNFi therapy from two independent studies and patient populations, reproducing for the first time the predictive ability of molecular biomarkers using seemingly disparate RA biology. Among patients who are naïve to targeted therapy and those who are TNFi-exposed, patients with a molecular signature of non-response are unlikely to respond to TNFi therapy at 3 or 6 months as assessed by ACR50, ACR70, DAS28-CRP, and CDAI. When providers use MSRC test results to stratify patients to treatment, patients who have a molecular signature of non-response to TNFi therapies can be directed to an alternative therapy to avoid expenses and potential toxicities without possible benefit. Those who lack this signature can proceed with TNFi therapy and possibly achieve an increased response rate relative to the unstratified population [40]. Access to precision medicine at multiple points during care expands the clinical information available to providers, thereby providing patient-specific data to improve care and shorten the time necessary to achieve patients’ treatment goals. Future studies will further address the impact on clinical outcomes of MSRC-guided treatment selection and identify biomarkers that predict inadequate response to other targeted therapies in RA.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank all patients and providers who have participated in the NETWORK-004 and CERTAIN studies.
Funding
All work and the journal’s Rapid Service Fee was funded by Scipher Medicine Corporation.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authorship Contributions
Conceptualization: TM, VRA; Data curation: LZ, AJ, MW; Formal Analysis: LZ, TM, AJ, SDG, JW, MW; Funding acquisition: VRA, AS; Investigation: ECS, JRC, SC, RD, AFW, ZG, SR; Methodology: TM, LZ, SDG; Resources: SR, JMK, DAP; Supervision: VRA, AS; Writing—original draft: JBW, LZ, TM, SDG; Writing—review & editing: all authors.
Disclosures
Stanley Cohen: Consultant and investigator for Scipher Medicine. Theodore Mellors, Alex Jones, Mengran Wang, Susan Ghiassian, Lixia Zhang, Johanna Withers, Sarah Rapisardo, Viatcheslav Akmaev and Alif Saleh are full-time employees of and have stock ownership in Scipher Medicine. Zoran Gatalica is a consultant of Scipher Medicine. Jeffrey Curtis: research grants or contracts with Amgen, BMS, Corrona, Lilly, Pfizer, Sanofi. Consultant for Corrona LLC, AbbVie, Amgen, Bristol-Myers Squibb, Genentech, Gilead, GlaxoSmithKline, Eli Lilly and Co., Pfizer, Regeneron, Sanofi. Dimitrios Pappas: employee of Corrona and has equity interest. Board of Directors for Corrona CRF. Consulting honoraria by AbbVie, Gtech, Roche Hellas, Sanofi, Novartis. Joel Kremer: shareholder of Corrona, LLC; consultant for AbbVie, Bristol-Myers Squibb, Eli Lilly and Co., Novartis and Pfizer; grants or contracts from AbbVie, Eli Lilly and Co., Novartis and Pfizer. Corrona has been supported through contracted subscriptions in the last 2 years by AbbVie, Amgen, Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Eli Lilly and Company, Genentech, Gilead, Janssen, Momenta, Novartis, Ortho Dermatologics, Pfizer Inc., Regeneron, Sanofi, and Sun. Rajat Dhar and Alvin Wells have nothing to declare.
Compliance with Ethics Guidelines
Institutional review board or ethics committee approvals were obtained prior to sample collection and CERTAIN study participation, and patients provided written informed consent. The NETWORK-004 study was governed by both a central institutional review board (Copernicus Group Independent Review Board) as well as local review boards if required at individual sites. This study was conducted in accordance with the principles outlined in the Declaration of Helsinki.
Data Availability
The datasets generated and analyzed during the current study are not publicly available because participants did not agree for their RNA sequencing data to be shared publicly. The algorithm underlying the MSRC is proprietary to Scipher Medicine Corporation.
References
- 1.Smolen JS, Landewe RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis. 2020;79(6):685–99. [DOI] [PubMed]
- 2.Aletaha D. Precision medicine and management of rheumatoid arthritis. J Autoimmun. 2020;110:102405. doi: 10.1016/j.jaut.2020.102405. [DOI] [PubMed] [Google Scholar]
- 3.Bluett J, Barton A. Precision medicine in rheumatoid arthritis. Rheum Dis Clin North Am. 2017;43(3):377–387. doi: 10.1016/j.rdc.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Singh JA, Saag KG, Bridges SL, Jr, et al. 2015 American College of Rheumatology guideline for the treatment of rheumatoid arthritis. Arthritis Rheumatol. 2016;68(1):1–26. doi: 10.1002/art.39480. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, Desai RJ, Liu J, Choi NK, Kim SC. Factors associated with initial or subsequent choice of biologic disease-modifying antirheumatic drugs for treatment of rheumatoid arthritis. Arthritis Res Ther. 2017;19(1):159. doi: 10.1186/s13075-017-1366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis JR, Zhang J, Xie F, et al. Use of oral and subcutaneous methotrexate in rheumatoid arthritis patients in the United States. Arthritis Care Res (Hoboken) 2014;66(11):1604–1611. doi: 10.1002/acr.22383. [DOI] [PubMed] [Google Scholar]
- 7.Triaille C, Lauwerys BR. Synovial tissue: turning the page to precision medicine in arthritis. Front Med (Lausanne) 2019;6:46. doi: 10.3389/fmed.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SL, Plant D, Eyre S, Barton A. The potential use of expression profiling: implications for predicting treatment response in rheumatoid arthritis. Ann Rheum Dis. 2013;72(7):1118–1124. doi: 10.1136/annrheumdis-2012-202743. [DOI] [PubMed] [Google Scholar]
- 9.van de Putte LB, Atkins C, Malaise M, et al. Efficacy and safety of adalimumab as monotherapy in patients with rheumatoid arthritis for whom previous disease modifying antirheumatic drug treatment has failed. Ann Rheum Dis. 2004;63(5):508–516. doi: 10.1136/ard.2003.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furst DE, Schiff MH, Fleischmann RM, et al. Adalimumab, a fully human anti tumor necrosis factor-alpha monoclonal antibody, and concomitant standard antirheumatic therapy for the treatment of rheumatoid arthritis: results of STAR (Safety Trial of Adalimumab in Rheumatoid Arthritis) J Rheumatol. 2003;30(12):2563–2571. [PubMed] [Google Scholar]
- 11.Breedveld FC, Weisman MH, Kavanaugh AF, et al. The PREMIER study: a multicenter, randomized, double-blind clinical trial of combination therapy with adalimumab plus methotrexate versus methotrexate alone or adalimumab alone in patients with early, aggressive rheumatoid arthritis who had not had previous methotrexate treatment. Arthritis Rheum. 2006;54(1):26–37. doi: 10.1002/art.21519. [DOI] [PubMed] [Google Scholar]
- 12.Maini R, St Clair EW, Breedveld F, et al. Infliximab (chimeric anti-tumour necrosis factor alpha monoclonal antibody) versus placebo in rheumatoid arthritis patients receiving concomitant methotrexate: a randomised phase III trial ATTRACT Study Group. Lancet. 1999;354(9194):1932–1939. doi: 10.1016/s0140-6736(99)05246-0. [DOI] [PubMed] [Google Scholar]
- 13.St Clair EW, van der Heijde DM, Smolen JS, et al. Combination of infliximab and methotrexate therapy for early rheumatoid arthritis: a randomized, controlled trial. Arthritis Rheum. 2004;50(11):3432–3443. doi: 10.1002/art.20568. [DOI] [PubMed] [Google Scholar]
- 14.Schiff M, Keiserman M, Codding C, et al. Efficacy and safety of abatacept or infliximab vs placebo in ATTEST: a phase III, multi-centre, randomised, double-blind, placebo-controlled study in patients with rheumatoid arthritis and an inadequate response to methotrexate. Ann Rheum Dis. 2008;67(8):1096–1103. doi: 10.1136/ard.2007.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreland LW, Schiff MH, Baumgartner SW, et al. Etanercept therapy in rheumatoid arthritis. A randomized, controlled trial. Ann Intern Med. 1999;130(6):478–486. doi: 10.7326/0003-4819-130-6-199903160-00004. [DOI] [PubMed] [Google Scholar]
- 16.Bathon JM, Martin RW, Fleischmann RM, et al. A comparison of etanercept and methotrexate in patients with early rheumatoid arthritis. N Engl J Med. 2000;343(22):1586–1593. doi: 10.1056/NEJM200011303432201. [DOI] [PubMed] [Google Scholar]
- 17.Weinblatt M, Schiff M, Goldman A, et al. Selective costimulation modulation using abatacept in patients with active rheumatoid arthritis while receiving etanercept: a randomised clinical trial. Ann Rheum Dis. 2007;66(2):228–234. doi: 10.1136/ard.2006.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klareskog L, van der Heijde D, de Jager JP, et al. Therapeutic effect of the combination of etanercept and methotrexate compared with each treatment alone in patients with rheumatoid arthritis: double-blind randomised controlled trial. Lancet. 2004;363(9410):675–681. doi: 10.1016/S0140-6736(04)15640-7. [DOI] [PubMed] [Google Scholar]
- 19.Keystone EC, Kavanaugh AF, Sharp JT, et al. Radiographic, clinical, and functional outcomes of treatment with adalimumab (a human anti-tumor necrosis factor monoclonal antibody) in patients with active rheumatoid arthritis receiving concomitant methotrexate therapy: a randomized, placebo-controlled, 52-week trial. Arthritis Rheum. 2004;50(5):1400–1411. doi: 10.1002/art.20217. [DOI] [PubMed] [Google Scholar]
- 20.Flouri I, Markatseli TE, Voulgari PV, et al. Comparative effectiveness and survival of infliximab, adalimumab, and etanercept for rheumatoid arthritis patients in the Hellenic Registry of Biologics: low rates of remission and 5-year drug survival. Semin Arthritis Rheum. 2014;43(4):447–457. doi: 10.1016/j.semarthrit.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Hetland ML, Christensen IJ, Tarp U, et al. Direct comparison of treatment responses, remission rates, and drug adherence in patients with rheumatoid arthritis treated with adalimumab, etanercept, or infliximab: results from eight years of surveillance of clinical practice in the nationwide Danish DANBIO registry. Arthritis Rheum. 2010;62(1):22–32. doi: 10.1002/art.27227. [DOI] [PubMed] [Google Scholar]
- 22.Romao VC, Santos MJ, Polido-Pereira J, et al. Comparative effectiveness of tocilizumab and TNF inhibitors in rheumatoid arthritis patients: data from the Rheumatic Diseases Portuguese Register, Reuma.pt. Biomed Res Int. 2015;2015:279890. [DOI] [PMC free article] [PubMed]
- 23.Conigliaro P, Chimenti MS, Triggianese P, et al. Remission and low disease activity in a cohort of real-life patients with rheumatoid arthritis treated with first-line antitumour necrosis factor. J Int Med Res. 2016;44(1 suppl):90–94. doi: 10.1177/0300060515593262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bienkowska JR, Dalgin GS, Batliwalla F, et al. Convergent random forest predictor: methodology for predicting drug response from genome-scale data applied to anti-TNF response. Genomics. 2009;94(6):423–432. doi: 10.1016/j.ygeno.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julia A, Erra A, Palacio C, et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS ONE. 2009;4(10):e7556. doi: 10.1371/journal.pone.0007556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lequerre T, Gauthier-Jauneau AC, Bansard C, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8(4):R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiguchi N, Kawauchi S, Furuya T, et al. Messenger ribonucleic acid expression profile in peripheral blood cells from RA patients following treatment with an anti-TNF-alpha monoclonal antibody, infliximab. Rheumatology (Oxford) 2008;47(6):780–788. doi: 10.1093/rheumatology/ken083. [DOI] [PubMed] [Google Scholar]
- 28.Stuhlmuller B, Haupl T, Hernandez MM, et al. CD11c as a transcriptional biomarker to predict response to anti-TNF monotherapy with adalimumab in patients with rheumatoid arthritis. Clin Pharmacol Ther. 2010;87(3):311–321. doi: 10.1038/clpt.2009.244. [DOI] [PubMed] [Google Scholar]
- 29.Tao W, Concepcion AN, Vianen M, et al. Multiomics and machine learning accurately predict clinical response to adalimumab and etanercept therapy in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021;73(2):212–22. [DOI] [PMC free article] [PubMed]
- 30.Farutin V, Prod'homme T, McConnell K, et al. Molecular profiling of rheumatoid arthritis patients reveals an association between innate and adaptive cell populations and response to anti-tumor necrosis factor. Arthritis Res Ther. 2019;21(1):216. doi: 10.1186/s13075-019-1999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson TM, Lescarbeau RM, Drubin DA, et al. Blood-based identification of non-responders to anti-TNF therapy in rheumatoid arthritis. BMC Med Genomics. 2015;8:26. doi: 10.1186/s12920-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toonen EJ, Gilissen C, Franke B, et al. Validation study of existing gene expression signatures for anti-TNF treatment in patients with rheumatoid arthritis. PLoS ONE. 2012;7(3):e33199. doi: 10.1371/journal.pone.0033199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lightbody G, Haberland V, Browne F, et al. Review of applications of high-throughput sequencing in personalized medicine: barriers and facilitators of future progress in research and clinical application. Brief Bioinform. 2019;20(5):1795–1811. doi: 10.1093/bib/bby051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prosperi M, Min JS, Bian J, Modave F. Big data hurdles in precision medicine and precision public health. BMC Med Inform Decis Mak. 2018;18(1):139. doi: 10.1186/s12911-018-0719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulielmos GN, Zervou MI, Myrthianou E, Burska A, Niewold TB, Ponchel F. Genetic data: the new challenge of personalized medicine, insights for rheumatoid arthritis patients. Gene. 2016;583(2):90–101. doi: 10.1016/j.gene.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Rolland T, Tasan M, Charloteaux B, et al. A proteome-scale map of the human interactome network. Cell. 2014;159(5):1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghiassian SD, Menche J, Barabasi AL. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput Biol. 2015;11(4):e1004120. doi: 10.1371/journal.pcbi.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menche J, Sharma A, Kitsak M, et al. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347(6224):1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma A, Menche J, Huang CC, et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum Mol Genet. 2015;24(11):3005–3020. doi: 10.1093/hmg/ddv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellors T, Withers JB, Ameli A, et al. Clinical validation of a blood-based predictive test for stratification of response to tumor necrosis factor inhibitor therapies in rheumatoid arthritis patients. Network Syst Med. 2020;3(1):91–104. [Google Scholar]
- 41.Pappas DA, Kremer JM, Reed G, Greenberg JD, Curtis JR. Design characteristics of the CORRONA CERTAIN study: a comparative effectiveness study of biologic agents for rheumatoid arthritis patients. BMC Musculoskelet Disord. 2014;15:113. doi: 10.1186/1471-2474-15-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlig T, Kvien TK, Pincus T. Test-retest reliability of disease activity core set measures and indices in rheumatoid arthritis. Ann Rheum Dis. 2009;68(6):972–975. doi: 10.1136/ard.2008.097345. [DOI] [PubMed] [Google Scholar]
- 43.Cheung PP, Gossec L, Mak A, March L. Reliability of joint count assessment in rheumatoid arthritis: a systematic literature review. Semin Arthritis Rheum. 2014;43(6):721–729. doi: 10.1016/j.semarthrit.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Maska L, Anderson J, Michaud K. Measures of functional status and quality of life in rheumatoid arthritis: health assessment questionnaire disability index (HAQ), modified health assessment questionnaire (MHAQ), multidimensional health assessment questionnaire (MDHAQ), health assessment questionnaire II (HAQ-II), improved health assessment questionnaire (Improved HAQ), and rheumatoid arthritis quality of life (RAQoL) Arthritis Care Res (Hoboken) 2011;63(Suppl 11):S4–13. doi: 10.1002/acr.20620. [DOI] [PubMed] [Google Scholar]
- 45.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 46.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. doi: 10.1023/a:1010933404324. [DOI] [Google Scholar]
- 47.TheHastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York, NY, USA: Springer, New York Inc.; 2009. [Google Scholar]
- 48.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2012;12(2011):2825–30.
- 49.Kay J, Calabrese L. The role of interleukin-1 in the pathogenesis of rheumatoid arthritis. Rheumatology (Oxford). 2004;43(Suppl 3):iii2-iii9. [DOI] [PubMed]
- 50.Malemud CJ. The role of the JAK/STAT signal pathway in rheumatoid arthritis. Ther Adv Musculoskelet Dis. 2018;10(5–6):117–127. doi: 10.1177/1759720X18776224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan R, Chen S, Wang Y. Computational prediction of drug responses in cancer cell lines from cancer omics and detection of drug effectiveness related methylation sites. Front Genet. 2020;11:917. doi: 10.3389/fgene.2020.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adam G, Rampasek L, Safikhani Z, Smirnov P, Haibe-Kains B, Goldenberg A. Machine learning approaches to drug response prediction: challenges and recent progress. NPJ Precis Oncol. 2020;4:19. doi: 10.1038/s41698-020-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schatzle LK, Hadizadeh Esfahani A, Schuppert A. Methodological challenges in translational drug response modeling in cancer: a systematic analysis with FORESEE. PLoS Comput Biol. 2020;16(4):e1007803. doi: 10.1371/journal.pcbi.1007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yarwood A, Huizinga TW, Worthington J. The genetics of rheumatoid arthritis: risk and protection in different stages of the evolution of RA. Rheumatology (Oxford) 2016;55(2):199–209. doi: 10.1093/rheumatology/keu323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferreiro-Iglesias A, Montes A, Perez-Pampin E, et al. Evaluation of 12 GWAS-drawn SNPs as biomarkers of rheumatoid arthritis response to TNF inhibitors. A potential SNP association with response to etanercept. PLoS ONE. 2019;14(2):e0213073. doi: 10.1371/journal.pone.0213073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smits M, van de Groes S, Thurlings RM. Synovial tissue biopsy collection by rheumatologists: ready for clinical implementation? Front Med (Lausanne) 2019;6:138. doi: 10.3389/fmed.2019.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Humby F, Durez P, Buch MH, et al. Rituximab versus tocilizumab in anti-TNF inadequate responder patients with rheumatoid arthritis (R4RA): 16-week outcomes of a stratified, biopsy-driven, multicentre, open-label, phase 4 randomised controlled trial. Lancet. 2021;397(10271):305–317. doi: 10.1016/S0140-6736(20)32341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keystone EC, Ahmad HA, Yazici Y, Bergman MJ. Disease activity measures at baseline predict structural damage progression: data from the randomized, controlled AMPLE and AVERT trials. Rheumatology (Oxford) 2020;59(8):2090–2098. doi: 10.1093/rheumatology/kez455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker JF, Conaghan PG, Smolen JS, et al. Development and validation of modified disease activity scores in rheumatoid arthritis: superior correlation with magnetic resonance imaging-detected synovitis and radiographic progression. Arthritis Rheumatol. 2014;66(4):794–802. doi: 10.1002/art.38304. [DOI] [PubMed] [Google Scholar]
- 60.Coras R, Narasimhan R, Guma M. Liquid biopsies to guide therapeutic decisions in rheumatoid arthritis. Transl Res. 2018;201:1–12. doi: 10.1016/j.trsl.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis MJ, Barnes MR, Blighe K, et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 2019;28(9):2455–70.e5. [DOI] [PMC free article] [PubMed]
- 62.Pappas DA, Brittle C, Mossell JE 3rd, Withers JB, Lim-Harashima J, Kremer JM. Perceived clinical utility of a test for predicting inadequate response to TNF inhibitor therapies in rheumatoid arthritis: results from a decision impact study. Rheumatol Int. 2021;41(3):585–93. [DOI] [PMC free article] [PubMed]
- 63.Bergman MJ, Kivitz AJ, Pappas DA, et al. Clinical utility and cost savings in predicting inadequate response to anti-TNF therapies in rheumatoid arthritis. Rheumatol Ther. 2020;7(4):775–92. [DOI] [PMC free article] [PubMed]
- 64.Chastek B, Becker LK, Chen CI, Mahajan P, Curtis JR. Outcomes of tumor necrosis factor inhibitor cycling versus switching to a disease-modifying anti-rheumatic drug with a new mechanism of action among patients with rheumatoid arthritis. J Med Econ. 2017;20(5):464–473. doi: 10.1080/13696998.2016.1275653. [DOI] [PubMed] [Google Scholar]
- 65.Harrold LR, Reed GW, Kremer JM, et al. The comparative effectiveness of abatacept versus anti-tumour necrosis factor switching for rheumatoid arthritis patients previously treated with an anti-tumour necrosis factor. Ann Rheum Dis. 2015;74(2):430–436. doi: 10.1136/annrheumdis-2013-203936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chastek B, Chen CI, Proudfoot C, Shinde S, Kuznik A, Wei W. Treatment persistence and healthcare costs among patients with rheumatoid arthritis changing biologics in the USA. Adv Ther. 2017;34(11):2422–2435. doi: 10.1007/s12325-017-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu T, Shah N, Deshpande G, Tang DH, Eisenberg DF. Comparing biologic cost per treated patient across indications among adult US managed care patients: a retrospective cohort study. Drugs Real World Outcomes. 2016;3(4):369–381. doi: 10.1007/s40801-016-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei W, Knapp K, Wang L, et al. Treatment persistence and clinical outcomes of tumor necrosis factor inhibitor cycling or switching to a new mechanism of action therapy: real-world observational study of rheumatoid arthritis patients in the United States with prior tumor necrosis factor inhibitor therapy. Adv Ther. 2017;34(8):1936–1952. doi: 10.1007/s12325-017-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Listing J, Kekow J, Manger B, et al. Mortality in rheumatoid arthritis: the impact of disease activity, treatment with glucocorticoids, TNFalpha inhibitors and rituximab. Ann Rheum Dis. 2015;74(2):415–421. doi: 10.1136/annrheumdis-2013-204021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibofsky A, Galloway J, Kekow J, et al. Comparison of patient and physician perspectives in the management of rheumatoid arthritis: results from global physician- and patient-based surveys. Health Qual Life Outcomes. 2018;16(1):211. doi: 10.1186/s12955-018-1035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available because participants did not agree for their RNA sequencing data to be shared publicly. The algorithm underlying the MSRC is proprietary to Scipher Medicine Corporation.