Significance
Renal cell carcinomas (RCCs) are heterogeneous malignancies thought to arise from kidney tubular epithelial cells, and clear cell RCC is the most common entity. This study demonstrates that cell atlases generated from benign kidney and two common RCCs using single-cell RNA sequencing can predict putative cells of origin for more than 10 RCC subtypes. A focused analysis of distinct cell-type compartments reveals the potential role of tumor epithelia in promoting immune infiltration and other molecular attributes of the tumor microenvironment. Finally, an observed association between the lack of immunotherapy response and endothelial cell fraction has important clinical implications. The current study, therefore, significantly contributes toward understanding disease ontogenies and the molecular dynamics of tumor epithelia and the microenvironment.
Keywords: renal cell carcinoma, clear cell renal cell carcinoma, single-cell RNA sequencing, cell of origin, tumor microenvironment
Abstract
Diverse subtypes of renal cell carcinomas (RCCs) display a wide spectrum of histomorphologies, proteogenomic alterations, immune cell infiltration patterns, and clinical behavior. Delineating the cells of origin for different RCC subtypes will provide mechanistic insights into their diverse pathobiology. Here, we employed single-cell RNA sequencing (scRNA-seq) to develop benign and malignant renal cell atlases. Using a random forest model trained on this cell atlas, we predicted the putative cell of origin for more than 10 RCC subtypes. scRNA-seq also revealed several attributes of the tumor microenvironment in the most common subtype of kidney cancer, clear cell RCC (ccRCC). We elucidated an active role for tumor epithelia in promoting immune cell infiltration, potentially explaining why ccRCC responds to immune checkpoint inhibitors, despite having a low neoantigen burden. In addition, we characterized an association between high endothelial cell types and lack of response to immunotherapy in ccRCC. Taken together, these single-cell analyses of benign kidney and RCC provide insight into the putative cell of origin for RCC subtypes and highlight the important role of the tumor microenvironment in influencing ccRCC biology and response to therapy.
Renal cell carcinoma (RCC) encompasses several histologically and molecularly diverse tumor groups (1). The past two decades of research have uncovered a variety of genomic drivers in diverse renal tumor subtypes including the most common subtype, clear cell RCC (ccRCC), and its rare renal tumor counterparts. For instance, the frequent biallelic loss of tumor suppressor genes on chromosome 3p, such as VHL (∼90%), PBRM1, SETD2, and BAP1, is a unique characteristic of ccRCCs (2), while recurrent allelic loss of heterozygosity of chromosomes 1, 2, 6, 10, 13, and 17 is a signature event of classic chromophobe RCC (chRCC), along with frequent TP53 mutations (3).
It has long been hypothesized that diverse RCC subtypes originate from distinct types of nephron tubular epithelial cells (4, 5). Thus, identification of cellular orthologs in the benign tissues that share transcriptional signatures with the tumor epithelia of specific RCC subtypes may indicate a putative cell of origin (P-CO). Identifying P-CO transcriptomes provides an appropriate reference to investigate gene expression patterns that are either retained or altered in the tumor epithelia. This knowledge will help refine in vivo disease models and facilitate the exploration of phenotype–genotype associations of disease subtypes. For example, the highly vascularized ccRCC subtype displays high levels of immune cell infiltration (2), and thus metastatic ccRCC often responds favorably to antiangiogenesis therapies and immunotherapy (6–9). By contrast, chRCC and nearly 50% of papillary RCCs (pRCCs) are relatively immune cell-poor/cold. However, the molecular underpinnings dictating whether certain RCC subtypes clinically present as immune cell-rich (hot tumors) or -poor (cold tumors) remain to be fully defined (10).
Transcriptional landscapes of benign kidney and RCC subtypes curated with bulk RNA sequencing (RNA-seq) provide the average gene expression of all cell types within each tissue (4, 10, 11). Single-cell sequencing methodologies have been increasingly adopted as a higher-resolution alternative to study gene expression, genomic aberrations, and epigenetic modifications in the constituent cells of various malignancies and their benign counterparts. These methods enable investigations into the significant variations of cell types observed in the tumor and microenvironment across different renal tumor entities, as well as among patients with a given renal tumor. A few studies have examined murine and human benign kidney cell types using single-cell mRNA sequencing (scRNA-seq). Park et al. generated a murine kidney cell atlas and discovered a novel nephron tubular epithelial transitional cell type (12). The data also helped them map expression of genes associated with chronic kidney diseases to specific cellular compartments. Other recent reports (13, 14) identified a P-CO for pediatric renal Wilms tumor (n = 3 specimens), ccRCC (n = 3), and pRCC (n = 1) and also an intriguing association between the zonation pattern of immune cells and the anatomical location of benign kidney tissues.
To gain a mechanistic understanding of common adult kidney tumors at single-cell resolution, we generated gene expression atlases of benign kidney and RCC tumor samples using a microfluidic droplet-based scRNA-seq platform. These atlases allowed us to address several outstanding questions in RCC pathobiology, including defining the P-CO for diverse RCC molecular subtypes, determining the pathways regulated by tumor epithelial cells, and examining the role of cells constituting the tumor microenvironment in disease pathogenesis and treatment response.
Results
Cell Atlas of Human Benign Adjacent Kidney Tissue.
Recent scRNA-seq of murine and human renal tissue has identified gene signatures of constituent cell types and furthered our understanding of genes whose expressions are linked with genetic traits of chronic kidney diseases and renal cancers (12, 14, 15). To expand scRNA-seq efforts in human renal cancers, we performed comprehensive genomic profiling of dissociated tissues by scRNA-seq on a cohort of RCCs and benign adjacent kidney tissues (14 samples from nine patients) for which we also had paired whole-exome sequencing (WES) and RNA-seq of corresponding bulk specimens. The benign samples were derived from either cortex or medullary regions, and the tumor specimens represented ccRCC and chRCC (Datasets S1 and S2). As an important analytical caveat, the tubular cells of distal nephron origin were found to display a higher mitochondrial content, reflecting their cellular biology. This observation necessitated separate mitochondrial read thresholds for benign and tumor samples (SI Appendix, Fig. S1 A–C). This difference was corroborated by bulk tissue WES data, which showed higher mitochondrial read coverage in benign compared with tumor tissues, and by previous kidney single-cell (13, 16) and bulk RNA-seq data from the Genotype-Tissue Expression (GTEx) Consortium (17). A high concordance in the data from bulk RNA-seq and averaged scRNA-seq from the corresponding samples showed that the tissue dissociation step did not significantly alter gene expression patterns. Thus, we proceeded to generate benign and tumor gene expression cell atlases with cell clusters based on data from multiple samples (SI Appendix, Fig. S1 B and C).
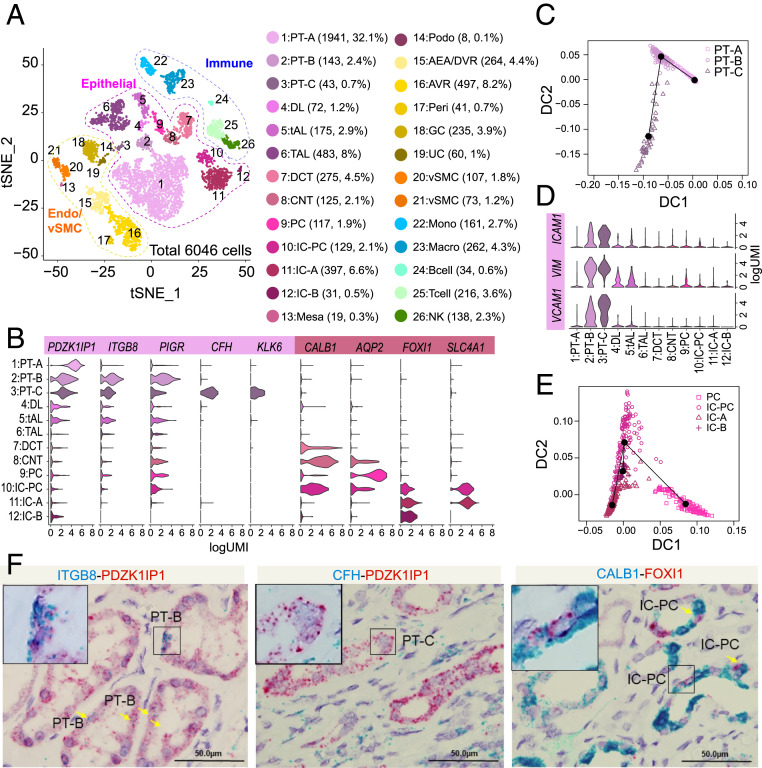
The benign renal scRNA-seq atlas demonstrated 26 cell clusters formed by 6,046 cells derived from six samples (representing five patients) (Fig. 1A). We found clusters based on previously characterized lineage-specific markers (Dataset S2), and they spanned both common and rare cell types, including tubular epithelial (clusters 1 to 14), endothelial (clusters 15 to 19), and stromal cells (clusters 20 and 21), as well as immune cells of myeloid and lymphoid branches (clusters 22 to 26). Proximal tubule (PT) cells comprising clusters 1, 2, and 3 (henceforth labeled PT-A, PT-B, and PT-C) expressed known PT markers, including PDZK1IP1 (18) (Fig. 1B). Previous studies have noted stochastic variation in the abundance of PT cells within benign renal cortical and medullary regions (4, 13). This phenotypic variation was recapitulated in our benign scRNA-seq data, where PT cells accounted for more than 50% of the cells from the cortex region but were less than 10% in medullary (SI Appendix, Fig. S1D and Dataset S1). The PT has been classically divided into three segments (S1 to S3) based on anatomical location and variation in cellular ultrastructure. PT-A cells from these three regions were captured by our data, as evidenced by the distinct expression pattern of markers previously known to be associated with S1, S2, and S3 segments (SI Appendix, Fig. S1E).
Fig. 1.
Single-cell analysis of benign human kidney reveals novel nephron tubular epithelial cell types. (A) Single-cell atlas of human kidney. t-SNE plot of scRNA-seq data from 6,046 cells obtained from six benign kidney samples. Cell clusters found therein representing 26 cell types are shown. DL, descending limb; DCT, distal convoluted tubule; CNT, connecting duct; Mesa, mesangial cells; Podo, podocytes; Peri, pericytes; vSMC, vascular smooth muscle cells; Mono, monocytes; Macro, macrophages; NK, natural killer cells. (B) Violin plots depicting gene expression patterns of select cell type markers: PDZK1IP1 (all PT cells), ITGB8 (PT-B and -C), PIGR (PT-B and -C), CFH (PT-C), KLK6 (PT-C), CALB1 (IC-PC, CNT), AQP2 (PC), FOXI1 (IC-PC, IC-A, IC-B), and SLC4A1 (IC-PC, IC-A). (C) Trajectory analysis of the three PT cell clusters identified: the common PT-A and rare/novel PT-B and PT-C. (D) As in B except showing stem/progenitor cell markers (VCAM1, VIM, ICAM1) across different cell types. (E) Trajectory analysis of distal tubule IC, PC, and IC-PC populations. (F) Validation of PT-B, PT-C, and IC-PC cells in benign adjacent kidney by RNA-ISH dual staining. PT-B marker ITGB8 (Left) and PT-C marker CFH (Middle) in blue channel and pan-PT cell marker PDZK1IP1 in red channel. IC-PC marker CALB1 in blue channel and IC marker FOXI1 in red channel (Right). (Scale bars for all images, 50 μm.)
From the benign cell atlas we identified three uncharacterized cell type clusters. Among the PT cells, while PT-A (32%) corresponded to the previously studied most common PT cell type (14), two related but distinct clusters termed PT-B (2.4%) and PT-C (0.7%) represented rarer populations (Fig. 1 A and C). Slingshot trajectory analysis (a trajectory inference method, also called pseudotime analysis from single-cell gene expression data, which orders cells along a trajectory based on similarities in their expression patterns and determines lineage structure by identifying branching events) revealed that the PT-B cluster was more closely related to PT-A cells (Fig. 1C). Notably, both PT-B and PT-C cells were characterized by distinct marker expression, such as ITGB8 and PIGR in PT-B and CFH and KLK6 in PT-C cells (Fig. 1B). Interestingly, PT-B and PT-C cells showed high expression of various renal stem cell marker genes, such as ICAM1, VIM, and VCAM1 (Fig. 1D) (19). In addition to the PT-B and PT-C clusters, the non-PT epithelial cell cluster 10 (2.1%) expressed markers of both intercalated cells (IC) and principal cells (PC), and we have thus termed this third cell type IC-PC (Fig. 1A). This cluster may represent the human equivalent of the transitional cell type between PC and IC cells, which was recently described in an scRNA-seq study of murine kidneys (12). Supporting this notion, slingshot trajectory analysis showed IC-PC cells as related to both PC and IC cells (Fig. 1E). Finally, RNA in situ hybridization (RNA-ISH) on independent benign kidney tissue samples with select markers confirmed the presence of the PT-B, PT-C, and IC-PC cells (Fig. 1F).
RCC Cell Atlases and P-CO.
Previous studies have hypothesized the P-CO for major RCCs based on their anatomical location, protein expression by immunohistochemistry, and bulk RNA-seq (4, 5). Determination of the P-CO for various RCC subtypes will help distinguish transcriptomic events in the tumor epithelia that are shared with benign cell types from those which are tumor-specific, uncovering novel tumor biomarkers and disease-specific molecular mechanisms. Toward this goal, RCC tumor atlases were constructed with scRNA-seq data from ∼20,500 cells derived from seven ccRCC samples (Dataset S1 and SI Appendix, Fig. S2 A and B) and ∼2,500 cells from one chRCC sample (SI Appendix, Fig. S2C). Tumor cell clusters (Dataset S3) were annotated based on known cell type-specific markers. The neoplastic cells representing ccRCC in our cohort overexpressed classic biomarkers of this disease like CA9, ANGPTL4, NDUFA4L2, and NNMT (20–22), while chRCC tumor epithelia overexpressed KIT, RHCG, and FOXI1 (23). We examined the gene expression patterns of both erythropoietin (EPO) and its receptor (EPOR) (24) in our scRNA-seq data, where only endothelial cells (afferent/efferent arterioles/descending vasa recta [AEA-DVR]) in the benign kidney showed EPO expression, while EPO was detected mostly in the tumor cells in ccRCC. EPOR expression was detected in most cell types at varying levels in both benign and tumor tissues (SI Appendix, Fig. S2D). Consistent with the phenotype previously deduced by histology and bulk RNA-seq (10, 11), we observed high immune cell infiltration in ccRCC samples (∼30% of total cells sequenced) compared with chRCC (∼5% of total cells; SI Appendix, Fig. S2 A–C); a smaller endothelial cell fraction (1.4%) was also observed in chRCC compared with ccRCC.
The tumor cell atlases (SI Appendix, Fig. S2 B and C) were generated from batch-corrected data in which all tumor epithelia clustered together. However, WES of the tumor and matched normal samples revealed somatic copy-number variations (CNV) specific to individual patients (SI Appendix, Fig. S3). To study the effect of CNV on tumor epithelia gene expression, we performed clustering without batch correction. While cells of the tumor microenvironment from different patients clustered according to type, tumor epithelia formed distinct patient-specific clusters (Fig. 2A). Gene expression of the tumor epithelia was specifically influenced by chromosomal aneuploidy, where genes in regions with copy gains and losses showed a corresponding increase or decrease in mRNA expression (Fig. 2B and SI Appendix, Fig. S3). This effect was restricted to the tumor epithelia and not seen among cells comprising the microenvironment (SI Appendix, Fig. S3).
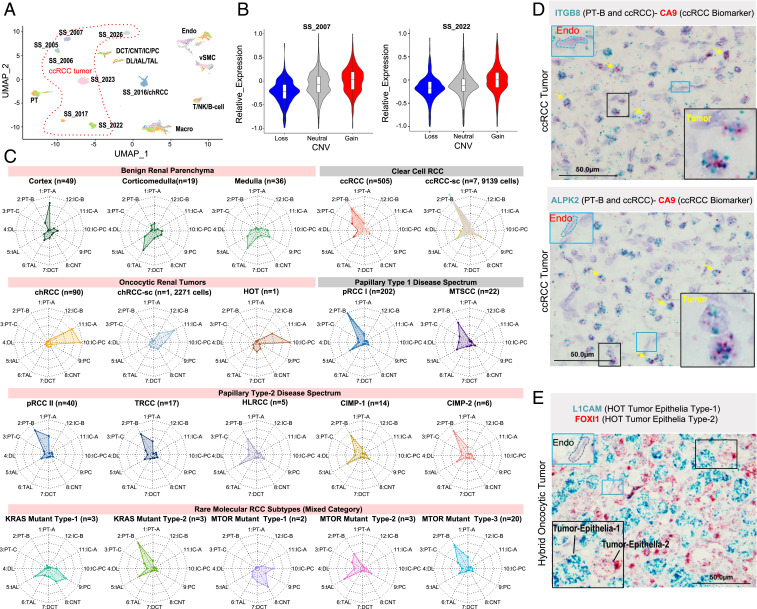
Fig. 2.
Cell of origin predictions for RCCs. (A) Impact of patient-specific CNV on tumor epithelial cell gene expression. UMAP plot of cell types captured from seven different ccRCC samples, where tumor epithelial cells clustered according to patient, while nontumor cells from different patients clustered according to cell types. (B) Individual examples reemphasize the association between genome-wide CNV gains and losses and single-cell gene expression patterns in the tumor epithelia. (C) Delineation of the P-CO for various RCCs. The “radar” plots indicate the probabilities based on a random forest classifier of a given query gene expression dataset (single-cell data from tumor epithelia of RCCs or bulk data from benign renal tissues, different anatomic locations, and tumors) to resemble a given benign epithelial cell type (periphery), as depicted by the spokes/radii. The predicted closest normal cell types for the various tumor tissues analyzed include the following. (Row 1) benign: bulk renal cortex, bulk cortico-medullary, bulk medullary; ccRCC tumors: bulk ccRCC, single-cell ccRCC; (row 2) oncocytic renal tumors: bulk chRCC, single-cell chRCC, HOT; papillary type-1 tumors: bulk pRCC type-1, bulk MTSCC; (row 3) papillary type-2 tumors: bulk pRCC type-2, TRCC, HLRCC, CIMP (-1, -2); (row 4) rare molecular subtypes: KRAS-mutant types 1 and 2 and MTOR-mutant types 1 to 3. (D) Lineage-specific marker validation by RNA-ISH dual staining. ITGB8 expression (blue) validates PT-B as P-CO for ccRCC (Top). CA9 (red) is a general biomarker of ccRCC. As in Top, except using ALPK2 as a second PT-B marker (Bottom). (E) Mutual exclusivity observed in FOXI1 and L1CAM dual stains reveals the distinct identity of two tumor epithelial cell types in a HOT. (Scale bar, 50 μm.)
To nominate P-COs for different RCC subtypes, we trained a random forest model with expression profiles of the 12 benign tubular epithelial cell types. This model was then validated using bulk RNA-seq data obtained from TCGA (The Cancer Genome Atlas) benign renal tissues that Lindgren et al. reclassified to distinct anatomical locations, including 49 cortex, 19 corticomedullary, and 36 medullary samples (4). As expected, we observed abundant PT and thick ascending limb (TAL) cells from cortex and medullary regions, respectively, in the TCGA benign renal samples (Fig. 2C); in the radar plots, spoke length depicts the probability of shared gene signatures between the samples of interest and the 12 benign cell types from our atlas.
We used this model to identify similarities between the tumor epithelia scRNA-seq data from ccRCC and chRCC samples to predict their corresponding P-COs. The ccRCC data from both the tumor epithelia scRNA-seq and bulk RNA-seq (TCGA) showed the highest probability of matching to the RNA expression profile of the rare PT-B cells (Fig. 2C). To corroborate this observation, we performed dual RNA-ISH with the classic ccRCC marker CA9 and two markers of PT-B cells (ITGB8 and ALPK2) on ccRCC tumor tissues and observed coexpression of CA9/ITGB8 and CA9/ALPK2 (Fig. 2D).
In contrast, among the oncocytic renal tumors analyzed, data from both chRCC tumor epithelia scRNA-seq and bulk chRCC RNA-seq (TCGA) were similar to the RNA expression of IC (Fig. 2C). Hybrid oncocytic tumors (HOTs) are intriguing renal tumors found predominantly in patients with Birt–Hogg–Dubé syndrome that demonstrate morphologic and immunohistochemical features overlapping with renal oncocytoma and chRCC. Our in-house RNA-seq data from a HOT was most similar to that of IC-PC cells (Fig. 2C). Intrigued by these results, we performed RNA-ISH for FOXI1, a key transcription factor expressed in IC cells and oncocytic/chromophobe tumors (Fig. 2E). We found FOXI1 staining in ∼50% of the tumor epithelia and reasoned that subtracting the IC-A gene signature from the tumor RNA-seq data may identify markers corresponding to the second (FOXI1-negative) tumor epithelial population. Using this approach, we identified L1CAM overexpression in HOTs, wherein it is relevant to note that L1CAM is expressed in PC cells in the benign kidney. Dual RNA-ISH of FOXI1 and L1CAM showed mutually exclusive expression within HOT epithelia, thereby validating markers specific for these distinct epithelial populations within the same HOT (Fig. 2E) (23).
In P-CO analysis of additional RCC subtypes, most subtypes from the pRCC types 1 and 2 disease spectrum showed the highest probability of matching to the gene signature of the PT-B cluster, while some subtypes (hereditary leiomyomatosis and renal cell carcinoma (HLRCC), CpG island methylator phenotype (CIMP), and pRCC type 1) showed additional similarity to thin ascending limb (tAL) cells to varying degrees (Fig. 2C). The presence of activating hotspot mutations in KRAS (∼0.6% recurrence) and MTOR (∼2.0%) from the TCGA pan-kidney cancer data are infrequent events mainly associated with the understudied rare RCC subtypes that lack the classic molecular and histologic features that define the more common renal tumor subtypes (e.g., chromosome 3p loss and VHL and MET mutations). Applying our model to these rare molecular subtypes revealed five different patterns with high probabilities of matching to either tAL/PC or PT-B/tAL signatures among KRAS-mutated cases, as well as to PT-B/tAL and connecting tubule (CNT)/IC-PC among MTOR-mutated cases, suggesting distinct cells of origin for these diverse subtypes (Fig. 2C).
Tumor Epithelial Cells Drive Immune Cell Infiltration in ccRCC.
Immune cell infiltration-associated phenomena in the tumor microenvironment are robust predictors of response to immunotherapy (25–27) and have been strongly linked with high somatic mutation/neoantigen burden (28–31). RCCs, however, are typically characterized by an immune-hot phenotype despite a low mutation burden. To gain insights into this paradox and better understand the milieu of RCC cells and their microenvironment, we compared differentially expressed genes (>0.5 log2FC and <FDR [false discovery rate] 0.05) and resulting pathway enrichment for benign cell types against their tumor counterparts. As the ccRCC P-CO, PT-B cells had less differentially expressed genes (6.8% up/6% down) compared with the tumor epithelia than PT-A cells (19.5% up/17.3% down; SI Appendix, Fig. S4A). Comparison between bulk RNA-seq of ccRCC tumor/normal adjacent tissue (NAT) samples identified the most differentially expressed genes (22.7% up/26.5% down; SI Appendix, Fig. S4A); the higher number of differentially expressed genes identified in the bulk tumor/NAT comparison can be attributed to differences in cell type composition (such as higher stromal cell fraction in benign tissues and infiltrating immune cells in tumor). Likewise, bulk chRCC vs. bulk NAT analysis (30.6% up/25.9% down) showed a higher percentage of differentially expressed genes compared with scRNA-seq of chRCC tumor epithelia vs. its P-CO, IC-A cells (2.8% up/2.7% down; SI Appendix, Fig. S4 A and B).
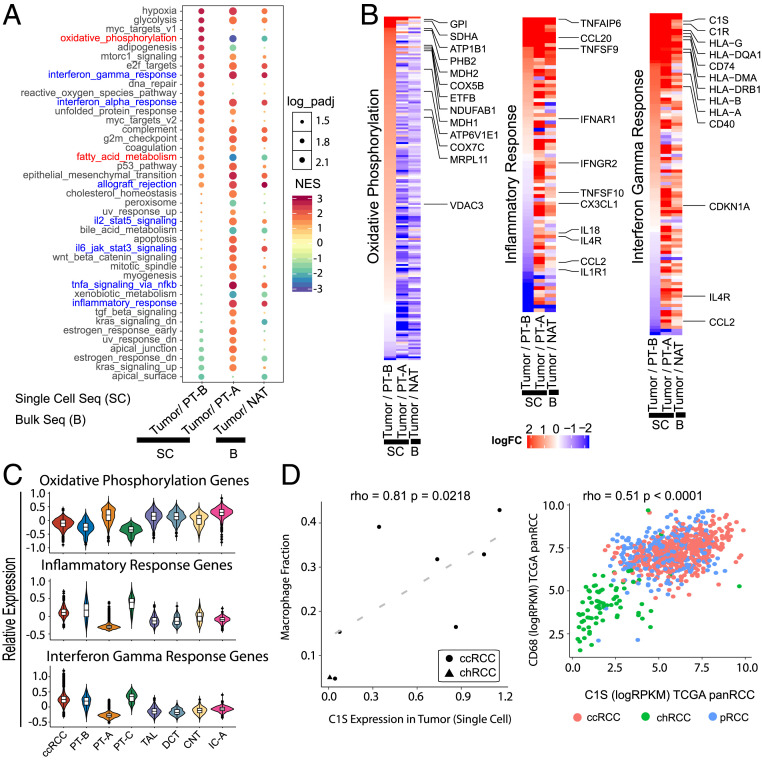
Examining the pathway enrichment analysis results from bulk RNA-seq of ccRCC tumor vs. NAT and tumor epithelia vs. PT-A and PT-B from our single-cell atlas led to several interesting observations (Fig. 3A and Dataset S4 A–C). A negative enrichment of the oxidative phosphorylation (OxPhos) gene set was noted in the ccRCC bulk tumor/NAT and tumor epithelia/PT-A comparison; in contrast, a positive enrichment of the same gene set was seen in the tumor epithelia/PT-B comparison (Fig. 3A). This enrichment was not observed in the chRCC vs. IC analysis (SI Appendix, Fig. S4C). The hallmark_oxidative_phosphorylation concept, which showed enrichment in ccRCC epithelia when compared with the PT-B population, consists of 200 genes that includes 74 nuclear oxidative phosphorylation (nuOxPhos) genes (Fig. 3 B and C and Dataset S4A) (32). While the increased glycolysis pathway enrichment we observed in ccRCC confirms the widely studied “Warburg effect” in cancer (Fig. 3A), several previous bulk tissue analyses also noted a corresponding decrease in OxPhos (2, 11). To further investigate the discrepancy in OxPhos enrichment, we examined mitochondrial oxidative phosphorylation (mtOxPhos) and nuOxPhos gene expression separately. While the nuOxPhos genes showed positive enrichment in ccRCC vs. PT-B, the mtOxPhos genes showed negative enrichment (SI Appendix, Fig. S4 D and E). This observation places ccRCC among tumors such as breast, endometrial, and lung adenocarcinomas that show increased expression of nuOxPhos and decreased levels of mtOxPhos (32). The reason for this discrepancy in OxPhos concept enrichments can now be attributed to the fact that bulk tissue analyses do not factor in the cellular heterogeneity of the samples and the relative abundance of mitochondria in various tubular epithelial cells, which comprise the major cell types found in benign kidney tissues that were used as reference samples in previous studies. The positive enrichment of hypoxia and epithelial to mesenchymal transition (EMT) concepts in analyses using all comparisons concurs with previous studies (Fig. 3A) (2, 11).
Fig. 3.
ccRCC tumor epithelial cells actively promote immune infiltration. (A) Pathway enrichments identified by GSEA of single-cell (SC) data for ccRCC tumor epithelial cells vs. the P-CO PT-B cells (first column) or the common PT-A cell population (second column). The tumor epithelia vs. P-CO showed fewer concepts, as compared with tumor epithelia vs. PT-A. Results from bulk RNA-seq (B) data for tumor vs. benign NAT is displayed alongside (third column). Concept names in red show reversal in patterns based on reference used. All immune/inflammation-related concepts are in blue. (B) Expression pattern of genes that constitute the “Hallmark Oxidative Phosphorylation” (Left), “Hallmark Inflammatory Response” (Middle), and “Hallmark Interferon Gamma Response” (Right) concepts from A. Columns within each subpanel (left to right) show the fold-change ratios in tumor vs. PT-B, tumor vs. PT-A, or bulk tumor vs. bulk NAT analysis, respectively. (C) Violin plots representing average of absolute gene expression values of all genes that constitute the three concepts across the tumor epithelia and various normal cell types. (D) C1S expression in tumor epithelia is associated with macrophage infiltration as represented by scatter plots based on both scRNA-seq (Left) and TCGA pan-RCC bulk RNA-seq (Right).
Next, we found several immune/inflammation-related concepts positively enriched in bulk tumor/NAT and tumor epithelia/PT-A analyses but not in the tumor/PT-B comparison (Fig. 3A). Again reflecting closer alignment of PT-B cells with tumor epithelia, we noted significantly elevated expression of several immunoregulatory factors in the PT-B population (Fig. 3 B and C). There were also several immune/inflammation regulators expressed by both the benign PT-B cells and tumor epithelia, while others were exclusively expressed in the tumor epithelia. Considering PT-B as the P-CO of ccRCC, these data suggest that the tumor epithelia retain the expression of a subset of immune regulators and also actively contribute additional ones to support the high immune cell infiltration noted in ccRCC. Further supporting this notion, we observed that several genes in the “inflammatory response” and “interferon gamma response” pathways were commonly expressed in PT-B and tumor epithelial populations, which was distinct from PT-A cells or other tubular epithelial cells (Fig. 3 B and C and Dataset S4 B and C). Interestingly, expression of complement serine protease C1S in tumors positively correlated with levels of macrophage infiltration both in scRNA-seq (macrophage fraction) and bulk RNA-seq (CD68 expression) (Fig. 3D). In contrast to ccRCC, the chRCC (immune cold) vs. IC-A cell comparison did not show enrichment of the numerous immune/inflammation regulatory concepts that were positively enriched in ccRCC. The only relevant concept “tnf_signaling_via_nfkb” that was significant in chRCC showed a negative enrichment, opposite of the trend observed in ccRCC (SI Appendix, Fig. S4C).
Finally, for a better understanding of tumor–microenvironment interactions, we mapped all canonical ligand–receptor pairs that were expressed across various cell types in ccRCC tumor samples. We identified receptor–ligand pairs expressed in the tumor epithelia and several cell types, including macrophages, B cells, T cells, and endothelial cells (SI Appendix, Fig. S5A). Expression of oncostatin M receptor (OSMR) in the ccRCC compartment and its ligand OSM in macrophage is of particular significance. Macrophages in the tumor microenvironment have been shown to be a major source of OSM in breast and cervical cancers (33, 34) and is thought to induce an EMT phenotype in breast and pancreatic tumor cells (34, 35). As evident from our ccRCC scRNA-seq atlas (SI Appendix, Fig. S2A), the macrophage compartment comprises more than 20% (range: 5 to 43%). EMT marker genes such as CDH2, ZEB1, and SNAI1 are highly expressed in ccRCC, and the reason behind this has not been fully characterized. Our data now show that the OSM–OSMR signaling axis between tumor-infiltrated macrophages and ccRCC epithelia may be a major factor influencing this phenomenon. Finally, as an example validation of a ligand–receptor pair, we observed expression of endothelin ligand 1 (EDN1) in tumor epithelia in sync with its cognate receptor EDNRB expressed in endothelial cells of the tumor vasculature visualized by RNA-ISH (SI Appendix, Fig. S5B).
Myeloid Cell Types in ccRCC and Association with Clinical Outcome.
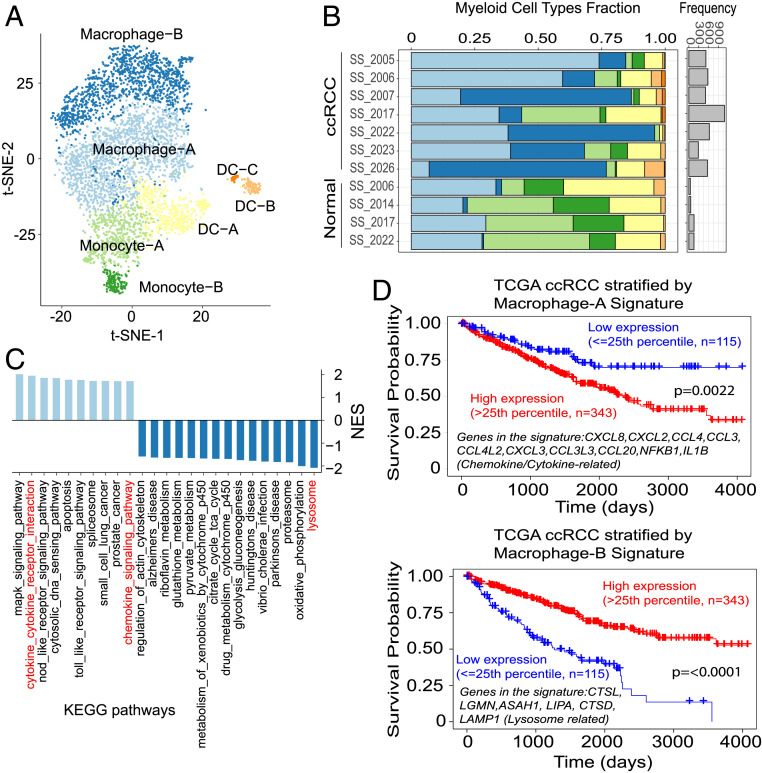
Clustering of all myeloid cells in the ccRCC scRNA-seq data revealed seven cell populations, distributed among two monocyte, three dendritic cell, and two macrophage clusters (Fig. 4 A and B). Clusters were annotated based on previously reported “xCell” signature genes, identified from 22 functionally defined human immune subsets (36) (SI Appendix, Fig. S6A). While monocytes in general accounted for smaller fractions and macrophages for larger fractions in tumor samples compared with normal, specific differences in tumor macrophage infiltration patterns were evident across cases (Fig. 4B). It is well recognized that macrophages can polarize into proinflammatory M1 and antiinflammatory M2 phenotypes (37). However, the two macrophage populations identified in this study (macrophage-A and macrophage-B) could not be clearly differentiated by conventional M1/M2 markers (SI Appendix, Fig. S6 A and B). The macrophage-A cluster showed higher expression of proinflammatory genes like IL1B, CCL3, CCL4, CCL20, and IL23A compared with the macrophage-B population. To better understand the difference between the two macrophage populations, we performed gene set enrichment analysis (GSEA) on KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. This analysis indicated that the top enriched pathways in the macrophage-A group included chemokine/cytokine pathways, while in the macrophage-B cluster lysosome and OxPhos pathway enrichments were noted (Fig. 4C). Interestingly, Kaplan–Meier analyses with the macrophage-A gene signature showed an association between high expression and worse overall survival in ccRCC (TCGA-KIRC). In contrast, high expression of the macrophage-B gene signature was a positive survival indicator (Fig. 4D). These findings suggest that scRNA-seq analysis of distinct myeloid populations of the tumor microenvironment can inform outcome in ccRCC patients.
Fig. 4.
Myeloid cell types detected through single-cell analyses of ccRCC and their associations with survival. (A) t-SNE plot depicts the seven major myeloid lineage cell types captured by scRNA-seq: macrophages-A and -B, monocytes-A and -B, and dendritic cells (DC)-A, -B, and -C. (B) Stacked bar plot shows samplewise frequency/composition of the myeloid cell types. Myeloid cell numbers in general are much higher in tumors compared with adjacent normal tissues (bar plot, Right). Macrophage-B population is enriched in tumors compared with normal. (C) Significantly enriched KEGG pathways (FDR < 0.05) in macrophage-B vs. macrophage-A population comparison. Light blue bars: positively enriched in macrophage-A; dark blue bars: positively enriched in macrophage-B. (D) Survival plots (TCGA-ccRCC bulk RNA-seq data) based on signature genes that are differentially expressed between macrophage-A and macrophage-B populations.
Endothelial Cell Diversity in ccRCC and Association with Therapy Response.
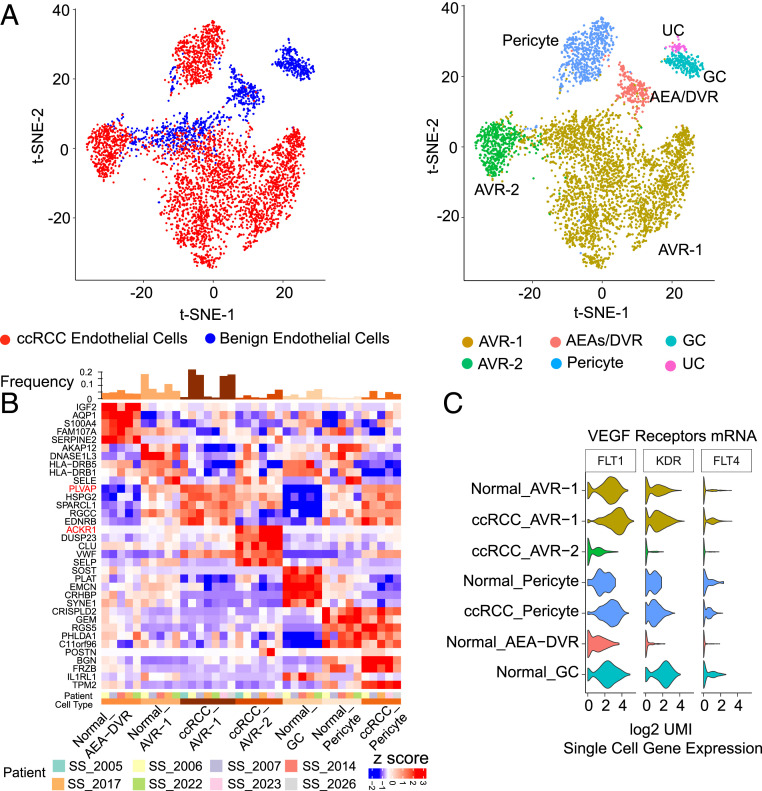
ccRCCs commonly present as highly vascularized neoplasms largely due to overexpression of vascular endothelial growth factor (VEGF), a gene target of hypoxia inducible factor (HIF) which is stabilized by genetic loss of the VHL gene (38, 39). Benign renal parenchyma also contains rich vasculature to facilitate nephron function, and ccRCC tumors and benign adjacent kidney cell atlases in our study were found to have similar fractions of endothelial cells and pericytes (Fig. 1A and SI Appendix, Fig. S2B). The benign adjacent tissue revealed five endothelial subpopulations, clusters 15 to 19 (Figs. 1A and 5 A and B), based on known marker expression. Four of the clusters were annotated as AEA/DVR (CLDN5+/AQP1+), AVR (ascending vasa recta, PLVAP+), pericyte (PDGFB+, RGS5+), and GC (glomerular capillaries, ITGA8+). The fifth endothelial subpopulation was uncharacterized (UC), as it expressed both the general endothelial marker PECAM1 and the cell adhesion gene POSTN. We also identified other markers for these subtypes, including IGF2 and SERPINE2 for AEA/DVR, DNASE1L3 for AVR, and SOST and CRHBP for GC (Fig. 5B).
Fig. 5.
Endothelial cell types in ccRCC. (A, Left) t-SNE plot of the endothelial cell types in benign adjacent kidney and ccRCC samples. (A, Right) t-SNE plot showing the diversity in endothelial cell type in benign adjacent kidney and ccRCC tumor tissues. The predominant endothelial population in tumors was AVR-1, followed by AVR-2. (B) Heat map depicting the top markers associated with the different endothelial subtypes. PLVAP, a marker of the AVR-1 population, and ACKR1, a marker of the AVR-2 population, are highlighted in red. (C) Messenger RNA expression of VEGF receptors FLT1, KDR, and FLT4 shows low expression in ACKR1-positive ccRCC AVR-2 population (violin plots).
The major cluster in ccRCC, ccRCC-AVR-1, was similar to the AVR population in benign tissue, while the minor cluster, ccRCC-AVR-2, specifically expressed ACKR1 and SELP (SI Appendix, Fig. S2B and Fig. 5B), thereby implying that the feeder vessels of tumor vasculature may be most composed of PLVAP+ AVR1-rich vessels. Curiously, ccRCC-AVR-1 cells had higher expression of VEGF receptor mRNAs FLT1, KDR, and FLT4 compared with ccRCC-AVR-2 cells (Fig. 5C), likely reflective of variations in tumor-specific VEGF signaling. Comparing the expression profiles of the major ccRCC-AVR-1 population in the tumor with AVR in the normal, we observed up-regulation of endothelial genes, such as endothelin receptor type B (EDNRB), von Willebrand factor (VWF) involved in hemostasis and possibly angiogenesis (40, 41), and heparan sulfate proteoglycan 2 (HSPG2) that can stimulate endothelial growth and regeneration (42) (Fig. 5B). GSEA indicated down-regulation of the hallmark interferon gamma response and up-regulation of EMT pathways in the ccRCC-AVR-1 population (SI Appendix, Fig. S7A). This observation is noteworthy as tumor endothelium forms the primary interface between circulating immune cells and the tumor. A previous study in lung cancer also found down-regulation of inflammatory concepts in tumor endothelium, which is thought to promote tumor immune tolerance via down-regulation of antigen presentation and immune cell homing (43). To substantiate existence of these endothelial subpopulations, we validated coexpression of endothelial lineage-specific marker PECAM1 and other endothelial subtype-specific markers, including PLVAP, HSPG2, SOST, ACKR1, and VWF, in benign and tumor tissues using two-color RNA-ISH (SI Appendix, Fig. S7B).
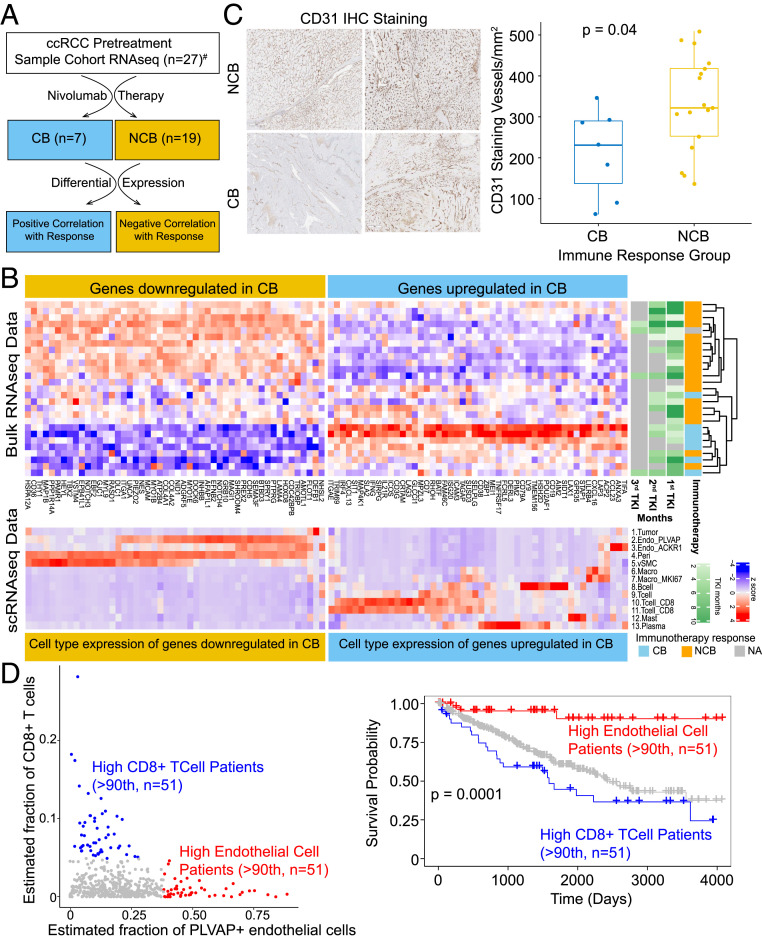
Finally, we explored the association between tumor microenvironment cellular content and clinical outcomes, beginning with an examination of immunotherapy response among ccRCC patients. We analyzed bulk RNA-seq data (n = 27) from pretreatment ccRCC primary tumor samples from patients with metastatic disease who underwent tyrosine kinase inhibitor followed by nivolumab therapy (Fig. 6A). These patients later developed progressive disease (PD, n = 15), exhibited stable disease (SD, n = 4), or showed complete or partial response to immunotherapy (n = 7, defined as clinical benefit). We then identified genes associated with either clinical benefit or no clinical benefit (SD or PD) (Fig. 6B). Next, we examined the expression pattern of these genes in the tumor microenvironment cell types from our ccRCC scRNA-seq data (Fig. 6B). As anticipated, genes positively associated with response were predominantly expressed among T cells and macrophages from the immune compartment. However, genes negatively associated with response were predominantly expressed among AVR-1 cells and pericytes of the endothelial compartment (Fig. 6B), showing an enrichment of an endothelial cell population in this group as opposed to the enrichment of immune cells in the former. We also performed CD31/PECAM1 immunohistochemistry to assess the endothelial fraction in the tumor tissues by an orthogonal method and found significantly lower CD31/PECAM1 staining in the group of patients that responded to nivolumab (Fig. 6C).
Fig. 6.
Endothelial cell content informs response to immunotherapy and survival in ccRCC patients. (A) Schematic workflow of the integrative analysis; CB, clinical benefit (patients with complete or partial response); NCB, no clinical benefit (patients with progressive disease or stable disease); #response information not available for one patient. (B, Top) Heat map of up- and down-regulated genes associated with treatment response. Tyrosine kinase inhibitor (TKI) treatment durations prior to immunotherapy are provided in the TKI annotation tracks on the right, and immunotherapy response category is shown next to that. NA, immunotherapy response information not available for one patient. (B, Bottom) Expression of response-associated genes among the various cell types identified by scRNA-seq. Genes down-regulated in CB category are mostly expressed among the endothelial cell types (yellow box), while genes up-regulated in CB (blue box) are predominantly expressed in immune cell compartments. (C) CD31/PECAM1 immunohistochemistry, representative images (Left) and quantitation (Right). (D) Scatter plot (Left) shows the mutual exclusivity of outlier samples with high estimated fraction of CD8+ T cells (blue dots) and patients with high estimated endothelial cells (red dots) in TCGA ccRCC bulk RNA-seq data. Kaplan–Meier plot shows the survival probability between the two groups. Patients with high endothelial cell fraction showed significant difference (P = 0.0001) in survival outcome compared with patients enriched with CD8+ T cells.
We next assessed whether the tumor microenvironment fraction could have prognostic utility in the setting of localized disease by examining primary ccRCC (KIRC) tumor data from TCGA. Interestingly, we noted that cases in the outlier endothelial fraction were largely exclusive of those in the outlier CD8+ T cell fraction (Fig. 6D). Between these two groups, better overall survival among patients in the higher tumor endothelial cell fraction was noted. In summary, a dichotomy in tumor microenvironment phenotype exists, whereby patients with higher endothelial cell content in primary disease tend to have better overall survival, but metastatic ccRCC patients with high endothelial cell content respond poorly to immunotherapy. Conversely, individuals with a higher tumor immune fraction had poorer overall survival in the localized setting but benefited more from immunotherapeutic intervention for metastatic ccRCC.
Discussion
Comprehensive cell atlases of benign and tumor tissues generated through scRNA-seq provide a rare glimpse into the unique composition of constituent cell types of complex organs and solid tumors that arise therein (44–46), such as histologically complex kidney tissue and diverse subtypes of RCCs (14, 47, 48). Our deep-coverage benign human renal cell atlas captured all previously characterized cell types as well as rarer epithelial cells—PT-B, PT-C, and IC-PC. Trajectory analysis of the PT-A population showed the presence of subpopulations (SI Appendix, Fig. S1E) with distinct expression patterns of genes previously ascribed to the S1, S2, and S3 PT regions (49). The two rarer populations of PT clusters, PT-B and PT-C, do not completely overlap with populations identified thus far. The recent study by Young et al. described the presence of a rare population of PT cells they termed “PT1” characterized by VCAM1+, SLC17A3+, and SLC7A13− expression (14). While there are certain similarities (VCAM1+ and SLC7A13−) between the two, our PT-B population is SLC17A3−. Only a small subset of PT-A cells express SLC17A3, which has been demonstrated as a marker of the PT S3 region (50). More recently, renal single-nuclei sequencing carried out by Lake et al. (15) showed two rare PT clusters they termed “PT-4” and “cluster-29.” In our analysis, PT-4 markers ITGB8 and ALPK2 were shared by our PT-B cluster, while their cluster-29 genes (e.g., CFH and LINC01435) were found in our PT-C cluster, suggesting these rare PT populations may represent similar cell types. However, several genes reported as renal stem cell-like markers (VCAM1, VIM, and ICAM1) are expressed in our PT-B and PT-C clusters, which together have not been previously described (Fig. 1D).
Our comprehensive P-CO analysis revealed three distinct prediction patterns among the three common RCC histologic subtypes that we examined (Fig. 2). Namely, pattern-1 showed ccRCC similarity to PT-B cells, pattern-2 revealed chRCC similarity to IC, and pattern-3 showed shared similarity of pRCC type 1 to both PT-B and tAL cells. Among the rare RCC histologic subtypes, mucinous tubular and spindle cell carcinoma (MTSCC, which is in the differential diagnosis for type 1 pRCC) and translocation RCC (TRCC, which is in the differential diagnosis for pRCC type 2) both showed high similarity to PT-B cells. Interestingly, HLRCC-associated RCC and CIMP RCC, two aggressive disease subsets characterized by DNA hypermethylation phenotypes, showed shared similarity to both tAL and PT-B cells. We further examined for differential molecular features among the samples in the two CIMP groups, and a surprising enrichment of FAT1/NF2 biallelic loss only in CIMP1 (Fisher exact test P = 0.0141) was noted (SI Appendix, Fig. S8); however, confirmation from larger sample sizes is required. Our bulk RNA-seq analysis of a rare HOT indicated IC-PC cells as a P-CO. We show that FOXI1 is expressed within one of the epithelial cell populations of a HOT, which constitutes ∼50% of the tumor epithelia. Subsequent analysis subtracting genes associated with IC cells narrowed down L1CAM as a marker specific for the second epithelial population, and dual RNA-ISH staining showed mutually exclusive checkered staining of L1CAM and FOXI1, thereby resolving a long-standing cellular molecular identity question in this disease entity (Fig. 2E). Finally, RCCs with activating mutations in KRAS and MTOR represent a much rarer group, and they pose diagnostic challenges because they lack signature genomic features of the major RCC histologic subtypes but also share some histomorphologic features with them. Within this group, we observed samples with distinct P-COs, which could render some clarification of their ontogeny. It is also noteworthy that identification of the closest benign cellular correlates for different RCC subtypes likely reveals a vulnerable benign population with a similar transcriptome profile that might be affected by therapeutics that target a lineage-related cancer. Overall, the lineage-specific biomarkers (e.g., ALPK2 and ITGB8 expressed in PT-B cells and retained in ccRCC) identified in our study form a valuable resource for future lineage tracing studies and for creating appropriate tumor models.
Our study demonstrates that patient-specific CNVs impact gene expression changes specifically in the tumor epithelia, a phenomenon which is often not factored into cohort-wide bulk RNA-seq analysis. Understanding the broader genomic context in which a given patient’s oncogenic aberration is placed will help in interpreting the differences observed in disease and treatment course. For example, ccRCC patients whose tumors contain signature biallelic loss of chromosome 3p tumor suppressor genes with or without the losses in chromosomes 9 or 14 show different disease outcomes (51). Most CNVs in ccRCC are arm-level and/or whole-chromosomal events and rarely focal, and our scRNA-seq data now show that expression of most genes harbored in the CNV regions are affected in most tumor cells. Understanding how this dosage effect impacts disease course will be a subject of future study.
The tumor cell atlas effectively captured features of high immune cell infiltration in primary ccRCC, with potential immunotherapeutic utility for metastatic ccRCCs. In contrast, the chRCC tumor atlas showed much less immune infiltration, as previously determined (10, 11). As RCCs harbor a low somatic mutation burden, the mechanisms behind ccRCC being an immune-hot tumor while chRCC is immune-cold have not been defined. Direct comparison of tumor epithelia against benign PT-A and PT-B populations along with bulk tumor/normal analysis presented here suggests that the tumor epithelial gene expression itself partly contributes to this phenomenon in addition to the tumor neoantigens derived from somatic aberrations (52). We uncovered that ccRCC tumor epithelia share the highest gene expression similarity with PT-B cells, a stem-like population which expresses several genes that regulate immune and inflammation processes. Our discovery that complement factor C1S (Fig. 3D) is expressed in the ccRCC epithelia and associates with tumor macrophage infiltration lends strong support to this notion. Interestingly, our ligand receptor analysis showed the presence of the OSM-OSMR signaling axis between the infiltrating macrophages and the tumor epithelia, respectively. We believe this might be a major regulator of the high EMT gene expression noted particularly in ccRCCs. OSM signaling has been explored as a therapeutic avenue in some cancers (33, 53, 54), and it may be warranted to investigate a combinatorial approach along with antiangiogenesis drugs and immunotherapy in ccRCC. Concept analysis of differentially expressed genes (Fig. 3 A–C and Dataset S4 A–C) also showed differences in enrichment of immune and inflammation processes based on the reference used, again highlighting the importance of scRNA-seq in enabling these findings. Interestingly, this also rectified previous conclusions based on bulk RNA-seq data (2, 10, 11), where several groups showed increased glycolysis and decreased OxPhos, indirectly suggesting defective OxPhos in tumors. As recently speculated (55), inclusion of cell type-specific data revised our current understanding of nuOxPhos which shows a corresponding increase in tumors. Finally, a recent study reported an association between endogenous retrovirus (ERV) expression in ccRCC and response to immunotherapy from bulk RNA-seq data (56). We examined our scRNA-seq data to see if ERV expression patterns could be detected among PT-A, PT-B, and tumor epithelial cells and noted a higher diversity of ERV expression in tumor cells (SI Appendix, Fig. S9).
Among the cell types comprising the tumor immune microenvironment we did not find a large representation of lymphoid cells in the ccRCC cohort; instead, a large number of myeloid populations were detected. Consistent with recent studies that have questioned the dichotomic M1/M2 model of macrophage polarization (37, 57), although we saw some expression of M1 and M2 genes in a small subset of macrophage populations we did not observe clusters with the expected coexpression of genes that constitute the M1 or M2 signatures that would be needed to support annotation as M1 or M2 macrophages, thus emphasizing the greater heterogeneity of macrophages in vivo. Similar conclusions have been independently arrived at through several scRNA-seq studies that characterized enriched immune populations from ccRCC patient samples (58–60). Our study showed marked enrichment specifically in the average macrophage-B population (normalized to total myeloid cells) in tumor (37%) compared with benign kidney (1%), while monocytes exhibited an opposite trend, with enrichment in benign tissues (Fig. 4B). These findings concur with recent studies (58–60) which provide important insights by collectively profiling tissues representing the ccRCC disease spectrum and treatment response. Integrative analysis of these public datasets with ccRCC cellular indexing of transcriptomes and epitopes by sequencing data (61) (which provides superior cell-type annotation based on simultaneous assessments of surface marker protein expression and scRNA-seq) is currently unexplored but one of our future goals. Unsupervised clustering of macrophages revealed two distinct populations, one with relatively higher chemokine/cytokine signaling (macrophage-A) and the other with higher lysosome gene expression (macrophage-B). Top markers in our macrophage-A and -B populations highly overlapped with the recently reported ccRCC tumor-associated macrophages (TAM) 1 and 2, respectively (58), and this prompts a more detailed follow-up study that is required to integrate all these findings. Interestingly, expression of the IL1B-containing macrophage-A signature gene set was associated with reduced overall patient survival. An independent study also confirmed that expression of IL1B by macrophages played a tumor promoting role in RCC (62).
Among the other notable tumor microenvironment cell types, such as endothelial, stromal, and various immune cells, we noted differential expression in the tumor-infiltrated cells compared with their matched benign counterparts. For example, while benign tissues had five different endothelial cell types, the tumor vasculature was predominantly composed of AVR-1–type endothelial cells (Fig. 5A). The tumor endothelium contains two distinct subpopulations which include the major AVR-1 (EDNRB+) and minor AVR-2 (ACKR1+) groups. A similar observation was made by Lambrechts et al., who noted enrichment of tumor-specific ACKR1+ endothelial cells in lung cancers as compared with benign lung tissues by single cell sequencing (43). Intriguingly, the ACKR1+ endothelial cells showed relatively less expression of VEGF receptors KDR and FLT1 compared with the EDNRB+ AVR-1 cells, suggesting these cells may evade angiogenesis inhibitors. Notably, the endothelial component of the tumor microenvironment may have utility in prognostication of survival and immunotherapy response in ccRCC patients, as previously observed (8, 9). Our analysis suggests that ccRCC patients with primary disease that have a high fraction of endothelial cells have better overall survival, an intriguing concept given that these patients also mostly display low CD8+ T cell expression (Fig. 6E). Conversely, in the setting of immunotherapy with nivolumab, higher endothelial cell fractions were found in metastatic ccRCC patients who did not respond to treatment (Fig. 6D), which we could posit may result from decreased interferon gamma signaling in the tumor endothelium (SI Appendix, Fig. S7A) and, thus, lowered immune cell homing.
The genomic landscape of kidney cancer has considerably evolved over the last decade, and this scRNA-seq–based study expands our understanding of the clinicopathologic and molecular features of RCC. Despite significant discovery of new biomarkers in the literature over recent years, the current National Comprehensive Cancer Network guidelines do not discuss the routine use of biomarkers for RCC and are primarily driven by clinical criterion (63). It will be important to incorporate new expression-based diagnostic or prognostic biomarker tools into the RCC clinical management algorithms as distinct genomic alterations and associated therapeutic sensitivities continue to be elucidated.
Materials and Methods
Sample Cohort Description.
Kidney samples were collected from patients who underwent partial nephrectomy at the University of Michigan Health System. The study was approved by the University of Michigan Institutional Review Board, and all patients provided informed consent. Tumor specimens were subjected to imprint/squash on glass slides, air-dried, and stained by the “Quickdiff” method for initial assessment before single-cell dissociation. All benign and tumor specimens were subsequently assessed with hematoxylin and eosin (H&E)-stained sections by pathologists. Where clinically possible, separate cortical and medullary specimens were obtained. Fresh specimens were collected in RPMI (serum and phenol red-free) media and processed for single-cell preparation within 1 to 2 h of the clinical procedure. Major portions of tissues (∼100 mg) were used for single-cell preparation. Remaining tissues were used for H&E staining and bulk tissue DNA/RNA isolation. Seven ccRCC, one chRCC, and six benign samples were collected from both cortex and medulla regions (Dataset S1).
Preparation of Single-Cell Suspension.
Freshly collected benign and tumor kidney samples were used for single-cell isolation. Up to 100 mg of collected normal and tumor kidney tissues were digested with 2 mg/mL final concentration collagenase type II in 10 mL of RPMI (serum and phenol red-free) media and incubated for 20 to 30 min at 37 °C on a rotator. After incubation, the solution was passed through a 40-µm cell strainer. The flow-through was centrifuged at 300 × rcf for 5 min. The cell pellet was incubated with 5 mL of RBC lysis buffer at room temperature for 2 min. Cells were washed with an ice-cold resuspension buffer (0.5% bovine serum albumin in 1× phosphate-buffered saline) for two times at 300 × rcf for 5 min. Cell numbers and viability were analyzed using a Bio-Rad cell counter. This method yielded a single cell suspension with greater than 70 to 80% viability.
Single-Cell 3′ mRNA Sequencing Library Preparation.
The 10x barcoding and complementary DNA (cDNA) synthesis were performed using 10x chromium 3′ scRNA-seq V2 chemistry according to the manufacturer’s instructions. Briefly, an appropriate volume of single-cell suspension/enzyme mix, barcoded beads, and partitioning oil were loaded in distinct lanes on the chip and processed in a 10x chromium controller instrument. The 10x GemCode Technology uses a droplet-based method to partition thousands of cells to generate nanoliter-scale Gel Bead-In-Emulsions (GEMs). Inside each GEM, cDNA generated from an individual cell shares a common 10x barcode. The prepared GEMs were incubated in a thermomixer to produce cDNA. Recovered cDNA was cleaned up and amplified by polymerase chain reaction to generate enough quantity for library construction. The size and yield were evaluated using a high-sensitivity bioanalyzer assay chip (Agilent). A set of recommended primers (P5, P7, and R2) in the manufacturer’s standard protocol were added to prepare the Illumina-ready sequencing library. The final library was evaluated for size and yield using a high-sensitivity bioanalyzer assay chip (Agilent). The final libraries were sequenced with the Illumina Hiseq2500 according to recommended specifications.
Whole-Exome Capture Genome Sequencing (WES) and Whole-Transcriptome RNA Sequencing (WXS).
WES and WXS data from the samples in this study are a part of a larger pan-RCC study that is currently being prepared for an independent submission. Processed data for select samples that were profiled by single-cell sequencing were used here for integrative analysis. See SI Appendix, Supplementary Methods for detailed descriptions of data analyses and additional assays performed in this study.
Supplementary Material
Acknowledgments
We thank Jin Chen and Jamie Estill for assistance with computational resources. We thank Deborah Postiff and Jaikissoon Jacky for assistance with sample procurement. We also thank all members of the A.M.C. laboratory and the Michigan Center for Translational Pathology. S.M.D. and A.M.C. are supported by funding from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium (grant number U24CA210967). A.M.C. is an HHMI Investigator, A. Alfred Taubman Scholar, American Cancer Society Professor, and NCI Outstanding Investigator.
Footnotes
Competing interest statement: A.A. and P.M. are coauthors on an article [S. M. Esagian et al. BJU International, 10.1111/bju.15324 (2021)].
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103240118/-/DCSupplemental.
Data Availability
Anonymized single-cell gene expression count matrix data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE159115). All other study data are included in the article and/or supporting information.
References
- 1.Udager A. M., Mehra R., Morphologic, molecular, and taxonomic evolution of renal cell carcinoma: A conceptual perspective with emphasis on updates to the 2016 World Health Organization classification. Arch. Pathol. Lab. Med. 140, 1026–1037 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Clark D. J.et al.; Clinical Proteomic Tumor Analysis Consortium , Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179, 964–983.e31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis C. F.et al.; The Cancer Genome Atlas Research Network , The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26, 319–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindgren D., et al., Cell-type-specific gene programs of the normal human nephron define kidney cancer subtypes. Cell Rep. 20, 1476–1489 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Prasad S. R., et al., Segmental disorders of the nephron: Histopathological and imaging perspective. Br. J. Radiol. 80, 593–602 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Li L., Kaelin W. G. Jr, New insights into the biology of renal cell carcinoma. Hematol. Oncol. Clin. North Am. 25, 667–686 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer R. J., et al., Nivolumab versus everolimus in patients with advanced renal cell carcinoma: Updated results with long-term follow-up of the randomized, open-label, phase 3 checkMate 025 trial. Cancer 126, 4156–4167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott D. F., et al., Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 24, 749–757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motzer R. J., et al., Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: Biomarker analysis of the phase 3 JAVELIN renal 101 trial. Nat. Med. 26, 1733–1741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricketts C. J.et al.; Cancer Genome Atlas Research Network , The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 23, 3698 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Chen F., et al., Multilevel genomics-based taxonomy of renal cell carcinoma. Cell Rep. 14, 2476–2489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J., et al., Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart B. J., et al., Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young M. D., et al., Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake B. B., et al., A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 10, 2832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coorens T. H. H., et al., Embryonal precursors of Wilms tumor. Science 366, 1247–1251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melé M.et al.; GTEx Consortium , Human genomics. The human transcriptome across tissues and individuals. Science 348, 660–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calado J., et al., The Na+-coupled glucose transporter SGLT2 interacts with its accessory unit MAP17 in vitro and their expressions overlap in the renal proximal tubule. FEBS Lett. 592, 3317–3326 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Huling J., Yoo J. J., Comparing adult renal stem cell identification, characterization and applications. J. Biomed. Sci. 24, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., et al., NDUFA4L2 is associated with clear cell renal cell carcinoma malignancy and is regulated by ELK1. PeerJ 5, e4065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campagna R., et al., Involvement of transforming growth factor beta 1 in the transcriptional regulation of nicotinamide N-methyltransferase in clear cell renal cell carcinoma. Cell. Mol. Biol. 64, 51–55 (2018). [PubMed] [Google Scholar]
- 22.Verine J., et al., Determination of angptl4 mRNA as a diagnostic marker of primary and metastatic clear cell renal-cell carcinoma. PLoS One 5, e10421 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skala S. L., et al., Next-generation RNA sequencing-based biomarker characterization of chromophobe renal cell carcinoma and related oncocytic neoplasms. Eur. Urol. 78, 63–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westenfelder C., Baranowski R. L., Erythropoietin stimulates proliferation of human renal carcinoma cells. Kidney Int. 58, 647–657 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Chen P. L., et al., Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galon J., et al., Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Grivennikov S. I., Greten F. R., Karin M., Immunity, inflammation, and cancer. Cell 140, 883–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samstein R. M., et al., Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Q., et al., Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat. Commun. 9, 5361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cristescu R., et al., Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F., et al., Quantifying immune-based counterselection of somatic mutations. PLoS Genet. 15, e1008227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reznik E., Wang Q., La K., Schultz N., Sander C., Mitochondrial respiratory gene expression is suppressed in many cancers. eLife 6, e21592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caffarel M. M., Coleman N., Oncostatin M receptor is a novel therapeutic target in cervical squamous cell carcinoma. J. Pathol. 232, 386–390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo L., et al., Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene 32, 5272–5282 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Smigiel J. M., Parameswaran N., Jackson M. W., Potent EMT and CSC phenotypes are induced by oncostatin-M in pancreatic cancer. Mol. Cancer Res. 15, 478–488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman A. M., et al., Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez F. O., Gordon S., The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor A. S., Spratt D. E., Dhanasekaran S. M., Mehra R., Contemporary renal tumor categorization with biomarker and translational updates: A practical review. Arch. Pathol. Lab. Med. 143, 1477–1491 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Choueiri T. K., Kaelin W. G. Jr, Targeting the HIF2-VEGF axis in renal cell carcinoma. Nat. Med. 26, 1519–1530 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T., et al., Endothelin B receptor expression correlates with tumour angiogenesis and prognosis in oesophageal squamous cell carcinoma. Br. J. Cancer 110, 1027–1033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanetta L., et al., Expression of Von Willebrand factor, an endothelial cell marker, is up-regulated by angiogenesis factors: A potential method for objective assessment of tumor angiogenesis. Int. J. Cancer 85, 281–288 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Lord M. S., Jung M., Cheng B., Whitelock J. M., Transcriptional complexity of the HSPG2 gene in the human mast cell line, HMC-1. Matrix Biol. 35, 123–131 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Lambrechts D., et al., Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Tirosh I., et al., Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puram S. V., et al., Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halpern K. B., et al., Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K. T., et al., Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 17, 80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao J., et al., Single-cell RNA sequencing of human kidney. Sci. Data 7, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L., et al., Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc. Natl. Acad. Sci. U.S.A. 114, E9989–E9998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jutabha P., et al., Human sodium phosphate transporter 4 (hNPT4/SLC17A3) as a common renal secretory pathway for drugs and urate. J. Biol. Chem. 285, 35123–35132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turajlic S.et al.; PEACE; TRACERx Renal Consortium , Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 173, 581–594.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turajlic S., et al., Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Stroeder R., et al., Oncostatin M treatment increases the responsiveness toward cisplatin-based chemoradiotherapy in cervical cancer cells in a STAT3-dependent manner. Oncol. Lett. 16, 3351–3358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smigiel J., Parvani J. G., Tamagno I., Polak K., Jackson M. W., Breaking the oncostatin M feed-forward loop to suppress metastasis and therapy failure. J. Pathol. 245, 6–8 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Creighton C. J., Proteomic signatures of clear cell renal cell carcinoma. Nat. Rev. Nephrol. 16, 133–134 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Panda A., et al., Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI Insight 3, 121522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azizi E., et al., Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borcherding N., et al., Mapping the immune environment in clear cell renal carcinoma by single-cell genomics. Commun. Biol. 4, 122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bi K., et al., Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 39, 649–661.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun D. A., et al., Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 39, 632–648.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoeckius M., et al., Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chittezhath M., et al., Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity 41, 815–829 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Motzer R. J.et al.; CGC , NCCN guidelines insights: Kidney cancer, version 2.2020. J. Natl. Compr. Canc. Netw. 17, 1278–1285 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized single-cell gene expression count matrix data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE159115). All other study data are included in the article and/or supporting information.