Abstract
Heterosis refers to the improved agronomic performance of F1 hybrids relative to their parents. Although this phenomenon is widely employed to increase biomass, yield, and stress tolerance of plants, the underlying molecular mechanisms remain unclear. To dissect the metabolic fluctuations derived from genomic and/or environmental differences contributing to the improved biomass of F1 hybrids relative to their parents, we optimized the growth condition for Arabidopsis thaliana F1 hybrids and their parents. Modest but statistically significant increase in the biomass of F1 hybrids was observed. Plant samples grown under the optimized condition were also utilized for integrated omics analysis to capture specific changes in the F1 hybrids. Metabolite profiling of F1 hybrids and parent plants was performed using gas chromatography-mass spectrometry. Among the detected 237 metabolites, 2-oxoglutarate (2-OG) and malate levels were lower and the level of aspartate was higher in the F1 hybrids than in each parent. In addition, microarray analysis revealed that there were 44 up-regulated and 12 down-regulated genes with more than 1.5-fold changes in expression levels in the F1 hybrid compared to each parent. Gene ontology (GO) analyses indicated that genes up-regulated in the F1 hybrids were largely related to organic nitrogen (N) process. Quantitative PCR verified that glutamine synthetase 2 (AtGLN2) was upregulated in the F1 hybrids, while other genes encoding enzymes in the GS-GOGAT cycle showed no significant differences between the hybrid and parent lines. These results suggested the existence of metabolic regulation that coordinates biomass and N metabolism involving AtGLN2 in F1 hybrids.
Keywords: Arabidopsis, glutamine synthetase, heterosis, nitrogen metabolism, 2-oxoglutarate
Introduction
Heterosis, or hybrid vigor, refers to the phenomenon in which F1 hybrids exhibit superior traits compared to their parental lines and occurs in many eukaryotes, including plants. Since heterosis helps improve valuable agronomic traits such as biomass size, yield, and biotic and abiotic stress tolerance in crops and vegetables, there is strong motivation to understand the mechanisms underlying heterosis. Many studies have been performed over multiple decades to attempt to uncover how heterosis occurs (Chen 2013). From a number of studies on hybrid vigor in maize (Zea mays), rice (Oryza sativa), tomato (Solanum lycopersicum), and Arabidopsis thaliana (Arabidopsis), various models have been proposed, including the dominant theory, the super-dominant theory, and the epistasis theory. However, obtaining a complete picture of heterosis is still challenging.
Arabidopsis is a model plant and has been the subject of many studies of heterosis (Fujimoto et al. 2012; Groszmann et al. 2015; Meyer et al. 2004, 2012; Vasseur et al. 2019). For example, several studies have reported that specific combinations of Arabidopsis accessions, like C24×Col or C24×Landsberg erecta (Ler), show heterosis at the F1 generation (Groszmann et al. 2014, 2015; Meyer et al. 2004). In Arabidopsis, timing of seed germination and levels of growth are different between accessions and their F1 hybrids (Groszmann et al. 2014; Meyer et al. 2012). The increased biomass of F1 hybrids seems to arise from two different sources: enhanced growth stage progress and greater amounts of biomass increase at each growth stage. Growth promotion in C24×Ler F1 hybrids was associated with early activation of photosynthesis and auxin-pathway genes compared with parental lines (Wang et al. 2019). Another study demonstrated that C24×Col F1 hybrids have higher metabolic activity than parental lines (Meyer et al. 2012). Such acceleration could be explained by the existence of links involving non-additive gene expression or changes in metabolic activities affected by the heterodimerization of enzymes (Chen 2013; Ng et al. 2017). Metabolic fluxes in F1 hybrids may also be influenced by unknown factors at multiple loci in each parental line (Fiévet et al. 2018; Vasseur et al. 2019).
Amino acids, organic acids, and carbohydrates (e.g., sugars) are important building blocks of chlorophylls, proteins, nucleic acids, starch, cellulose, lipids, and secondary metabolites in plants. The level of metabolite production is thus thought to influence biomass and yield. To capture comprehensive metabolite changes, omics analyses have been utilized (Fukushima et al. 2009; Kusano et al. 2007, 2011; Le et al. 2019; Li et al. 2020; Meyer et al. 2012). Sucrose feeding experiments to Arabidopsis seedlings demonstrated that leaf size is increased by the addition of exogenous sucrose (Van Dingenen et al. 2016). The study of different carbon and nitrogen (N) supply regimes applied to 97 Arabidopsis accessions revealed that metabolite changes can be used as traits for predicting biomass size under different growth conditions (Sulpice et al. 2013).
In this study, we aimed to obtain novel insights into the extent of metabolomic effects on Arabidopsis Col, C24 and their F1 hybrids grown under the optimized conditions that minimize effects on biomass that are not dependent on heterosis. Aerial parts of F1 and parental plants were harvested at approximately two weeks after sowing and then used for metabolite and transcript profiling. This time point allowed us to avoid the influence of specific life events occurring during early stages of growth including germination, the start of photosynthesis, and the switch between heterotrophic and autotrophic stages.
Metabolite profiling demonstrated that metabolites from the TCA cycle and N metabolism showed different accumulation patterns between F1 hybrid and parents. Transcriptome analysis revealed that N-related genes, in addition to stress response genes, are over-represented in the genes up-regulated in F1 hybrid. This trend of N-related transcript changes was well-associated with that of metabolite changes. Overall, these results can provide novel insights into effects of heterosis on N metabolism in Arabidopsis.
Materials and methods
Plant materials and growth conditions
We used Arabidopsis plants from the accessions Col-0 (Col), C24, and their reciprocal F1 hybrids. F1 hybrid seeds were obtained from hand-pollinated crosses between Col and C24, and parental seeds were obtained via self-pollination of parents with a restricted numbers of flowers (Meyer et al. 2004). Sterilized seeds were stored at 4°C in the dark for three days, then placed onto soil or agar containing half-strength Murashige and Skoog medium (pH 5.7, no sucrose) under short-day conditions at 22°C with a 12 h light/12 h dark cycle (light intensity 120 mmol m−2 s−1) in the study. For phenotyping experiments in different concentrations of Murashige and Skoog salts, sterilized seeds were sown on agar plates with different concentrations of Murashige and Skoog salts (1, 1/2, 1/4, 1/16, 1/32, 1/64). To minimize any biases of growth conditions for each plant, the position of each agar plate in the growth chamber was rotated daily. Additionally, to standardize conditions between F1 hybrids and their parents, 5 days after sowing (DAS), each seedling was transferred onto a 150-mm diameter agar plate and outliers in germination timing were removed. The selection of plants for phenotyping on soil was performed using the same criteria.
Metabolite profile analysis using gas chromatography-time-of-flight mass spectrometry (GC-TOF-MS) analysis
Whole rosettes from F1 hybrid (female C24) plants and their parents were harvested at zeitgeber time (ZT)=6 at 13, 15, and 17 DAS, with two plants bulked to generate six replicates samples for gas-chromatography-time-of-flight mass spectrometry (GC-TOF-MS) analysis. Extraction, derivatization, GC-TOF-MS measurements, and metabolite identification and quantification were performed as described previously (Kusano et al. 2007).
Microarray-based transcriptome analysis
Total RNA was isolated from whole rosettes of F1 hybrid (female C24) plants and their parents at 15 DAS using an RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s manual. GeneChip Arabidopsis ATH1 Genome Arrays (Affymetrix) were used for comprehensive transcript profiling. Hybridization and scanning were performed according to the manufacturer’s instructions. Three biological replicates were conducted for each genotype. Microarray signals were analyzed using the Bioconductor affy package and related MAS5 file (Gautier et al. 2004) in R software (http://www.R-project.org). The microarray data was deposited in Gene Expression Omnibus (GEO) Database in NCBI (http://www.ncbi.nlm.nih.gov/geo/). The accession number in the GEO database is GSE160630.
Quantitative real-time PCR (qPCR)
Total RNA was isolated from whole rosettes of 15 DAS seedlings using RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s manual. First-strand cDNA was synthesized using 1 µg of total RNA with a SuperScript III First-Strand Synthesis System (Invitrogen, US). qPCR was performed using a LightCycler 480 II (Roche diagnostics, Germany) with LightCycler 480 SYBR Green I Master Mix (Roche diagnostics, Germany). Gene-specific primer sequences used for qPCR are listed in Supplementary Table S1. Six biological replicates were analyzed for each genotype, with three technical replicates used per biological replicate. The mid-parent value (MPV) was defined as the mean parental expression value for each replicate.
Statistical analyses
Microarray data were analyzed using R software (http://www.R-project.org) and the Bioconductor package limma. Differentially-expressed genes were examined using GO analysis performed using singular enrichment analysis (SEA) in the agriGO platform (Tian et al. 2017). Transcript change overviews were visualized by MapMan software using log2-fold change values generated by comparing F1 hybrid to the MPV (Usadel et al. 2005). Changes in levels of 237 metabolites detected by GC-TOF-MS were visualized by principal component analysis (PCA) using pcaMethods and ggplot2 in R software. Two-way ANOVA and one-way ANOVA with Tukey’s multiple comparisons tests were performed using R software. The distribution of compounds in the central metabolic pathway for each line was visualized using VANTED software (Rohn et al. 2012).
Results
Optimizing growth conditions for the statistical evaluation of biomass differences between F1 hybrids and their parental lines
It is known that Arabidopsis F1 hybrids obtained from reciprocal crosses of Col and C24 accessions show significant heterosis in biomass size (Groszmann et al. 2014; Le et al. 2019; Meyer et al. 2004, 2012). However, little is known about which factors might generate such differences. We compared seed sizes of F1 hybrids, Col, and C24 plants because seeds act as sink tissue that provide energy and building blocks affecting plant biomass (Supplementary Figure S1). As a result, F1 hybrids did not show heterosis in mature seed stage, suggesting that any carryover of organic materials from seeds can be ignored, at least at the stage we assayed.
Biomass size is thought to be highly dependent on growth conditions. Major factors contributing to plant growth in a climate-controlled chamber (i.e., a closed growth condition) are (i) light quality, (ii) nutrition, and (iii) growth medium (the culture medium and soil). Changes in photoperiod and light strength, as well as the addition of sucrose to growth media, can modulate the composition of metabolites available as carbon sources (Le et al. 2019; Sulpice et al. 2013). We employed the same light conditions used in a previous metabolite profiling study (Le et al. 2019) without supplementing sucrose.
We next evaluated the effects of different N concentrations on the biomass of F1 hybrid and parental line plants (Figure 1A). The shoot fresh weight (FW) of F1 hybrid samples at grown under the three different N concentrations generated by using full-strength, half-strength, and quarter-strength MS media was higher than that of the parental lines. The rosette diameters of F1 hybrid and parental line plants grown under severely N-limited conditions were not significantly different (Supplementary Figure S2), suggesting that the amount of inorganic N may influence biomass size.
Figure 1. Phenotyping of Col, C24, and F1 hybrids in different growth conditions. (A) Fresh weight (FW) of F1 hybrids and parents grown in different levels of N. Murashige and Skoog salt concentrations were modified for each condition. FW of Col, C24, and F1 hybrids for each condition was measured at 15 DAS. Two plants were combined into a single sample for FW measurements. n=6. (B) FW of rosette leaves at 15 DAS of plants grown on soil. n≥21. (C) FW of rosette leaves from plants grown on agar plates at 15 DAS. n=12. (D) The relative FW was calculated by dividing the average of each sample weight by the MPV. The RSD was calculated by dividing the SD value by the mean FW. Error bars represent SD. Asterisks (*) indicate significant differences from MPV based on Student’s t-tests (p<0.05).
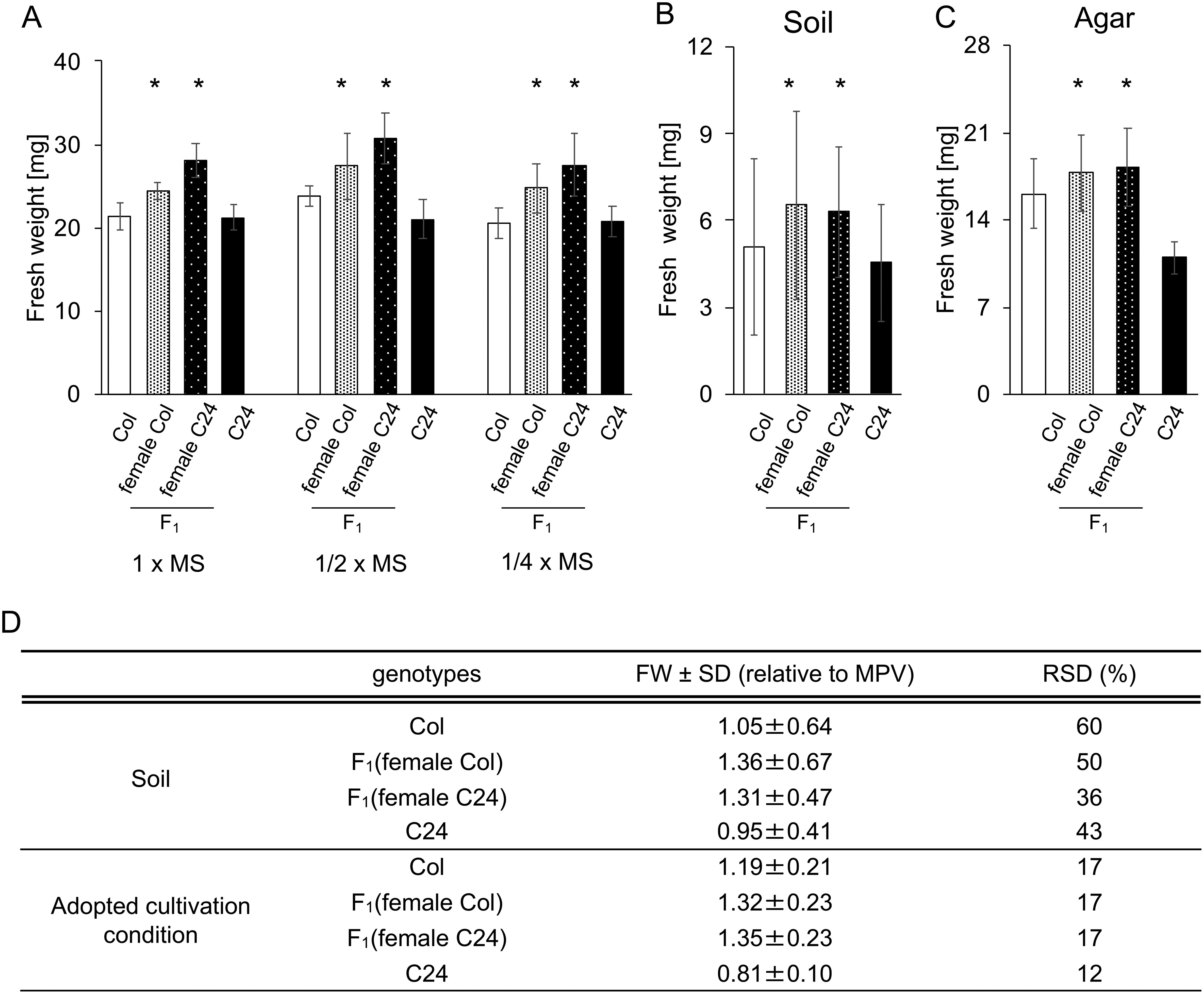
Variations in soil conditions have been often utilized for metabolome analyses in heterosis studies (Korn et al. 2010; Meyer et al. 2007, 2012). Comparing shoot FW of plants grown on culture medium and on soil indicated that the biomass of F1 hybrids were tends to be higher than that of parental lines under both conditions. In addition, the relative standard deviation (RSD) of the FW of plants grown on the culture media ranged from 12 to 17% while the RSD of the FW of plants grown on soil ranged from 36 to 60% (Figure 1B, D). Based on these results, we chose the grow plants on culture media for the remaining experiments.
Metabolomic evaluation of the F1 hybrid and the parental lines
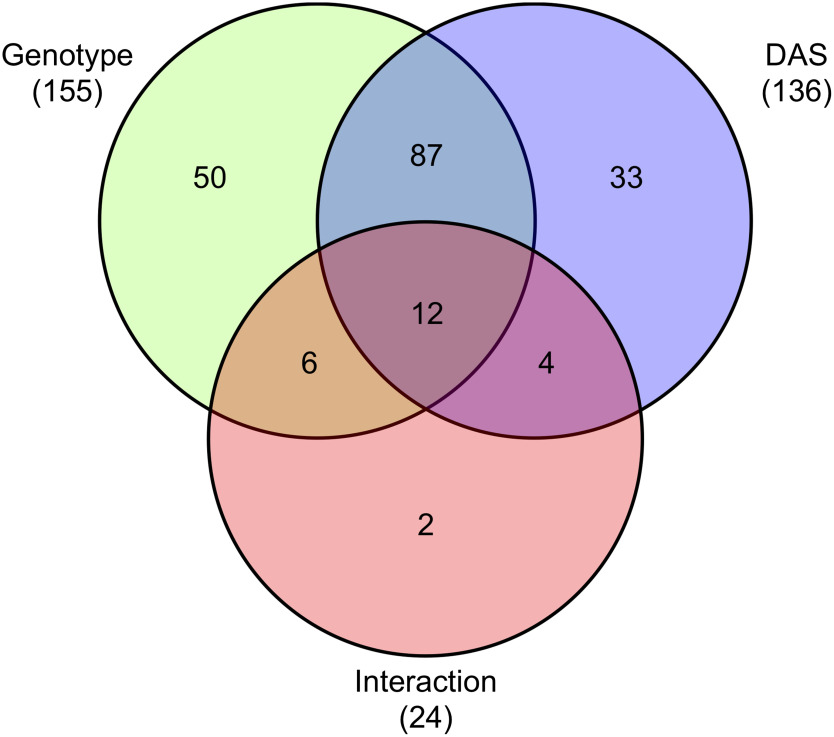
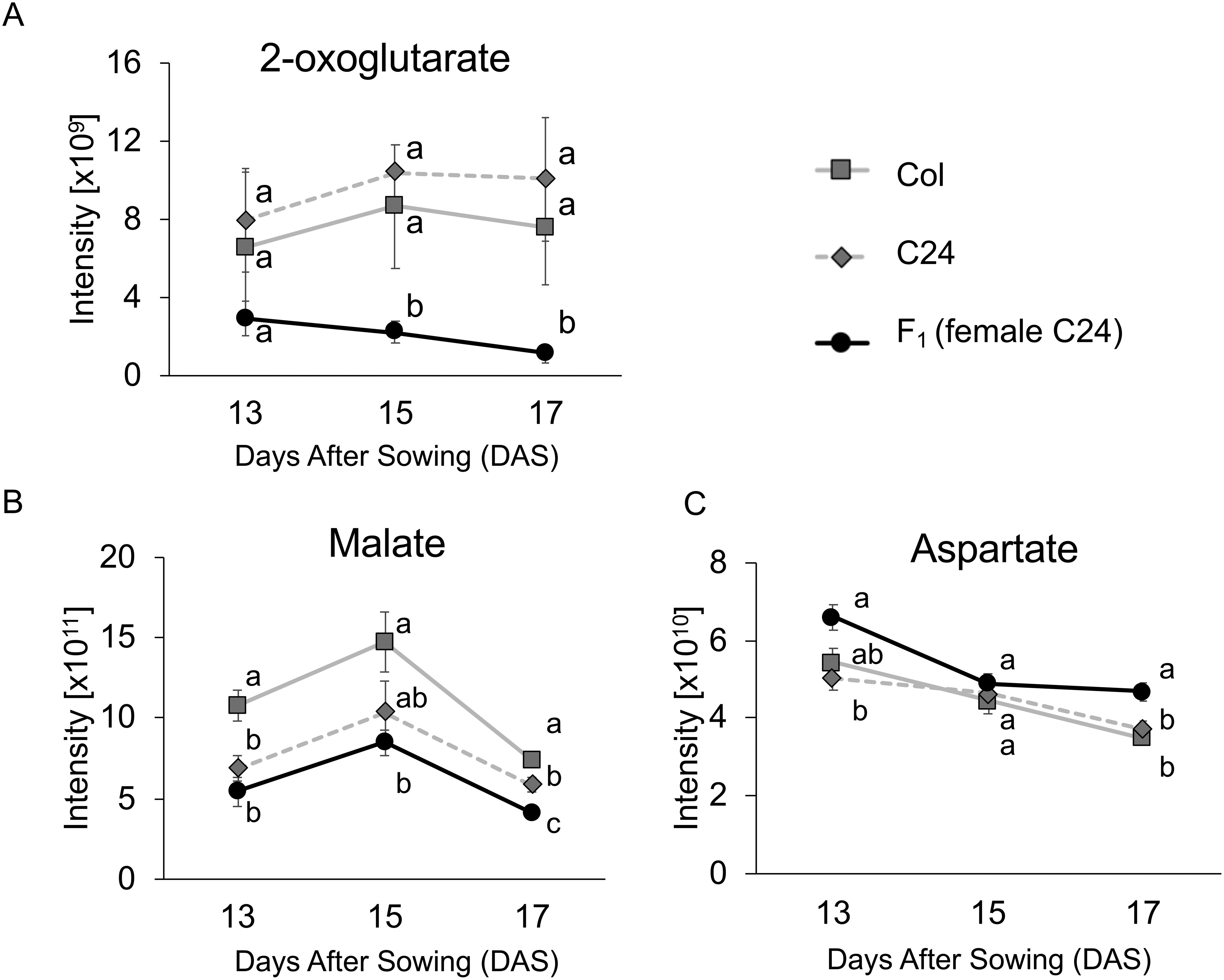
For metabolite profiling, we used a total of 54 samples from whole rosettes of three genotypes (F1, Col, and C24), with six biological replicates for each time point (13, 15, and 17 DAS) (Kusano et al. 2007, 2011). Of the 237 metabolites detected from all three genotypes, 88 were identified or annotated as metabolites (Supplementary Table S2). The scatter plot of scores for PCA was used to visualize genotype-dependent separation on PC1 (30.05% of explained variance) and sampling time (DAS) dependence on PC2 (10.47% explained variance) (Figure 2). Two-way ANOVA was conducted to determine the dependence of metabolite changes on genotype and growth-dependent (DAS). Result showed that 155 and 136 metabolites were significantly altered by each factor independently, while 24 metabolites showed significant interaction between both factors (Figure 3). Metabolomic profile changes in the F1 hybrid and parental lines during the three different time periods were overlaid onto a metabolic map (Figure 4) and F1 line metabolite levels that were above or below the corresponding levels in both parents were further validated. Levels of malate and 2-oxoglutarate (2-OG) in the F1 hybrid were lower than in the parental lines for all three time periods (Figure 5A, B) while the level of aspartate in the F1 plants was higher than in each parental line (Figure 5C).
Figure 2. The PCA score plot generated using metabolite profile data of Col, C24, and F1 hybrid at 13, 15, and 17 DAS. PCA was conducted using a 237 detected peak×54 samples matrix. Six biological replicates were used for each sample.
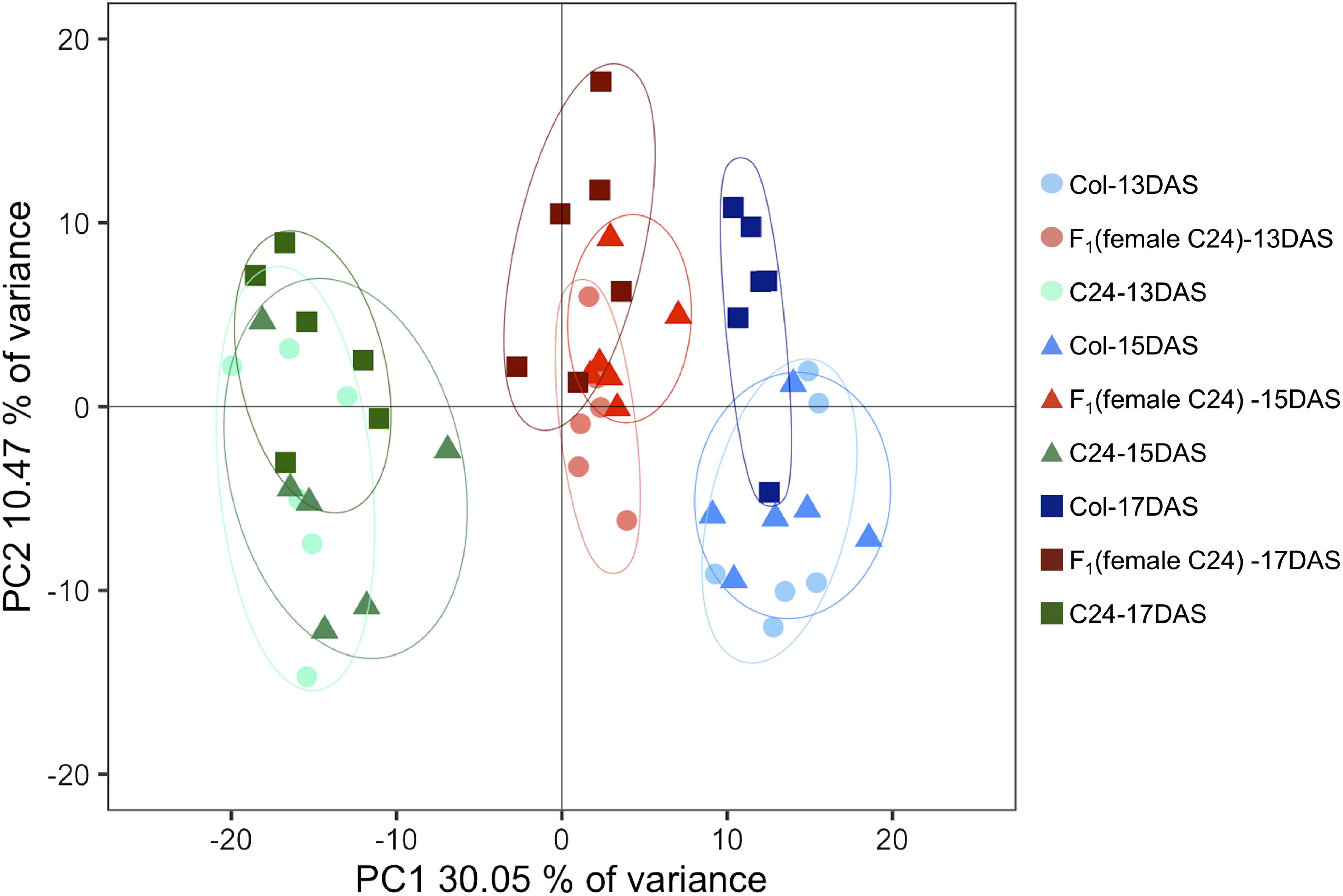
Figure 3. Significant differences in levels of metabolites detected using GC-TOF-MS analysis based on two-way ANOVA. Numbers of the intersection on the Venn diagram indicate the common metabolites, while those of each complement of genotype, DAS, or interaction shows specific metabolites associated with differences in genotype, DAS, or both (two-way ANOVA; p<0.05).
Figure 4. Metabolite distribution in central metabolic pathways of Col, C24, and F1 hybrid. Patterns for Col, C24, and F1 hybrid at 13, 15, and 17 DAS were projected onto the metabolic map. Blue, green, and red line colors correspond to Col, C24, and F1 hybrid (female C24), respectively. Error bars represent the SD of six biological replicates. Asterisks (*) indicate that the particular metabolite was significantly different in F1 hybrid samples when compared to each parent based on Tukey’s test (p<0.05). 2-OG: 2-oxoglutarate, β-Ala: β-alanine, Cit: citrate, F6P: fructose-6-phosphate, Fum: fumarate, GABA: 4-aminobutyrate, G6P: glucose-6-phosphate, Hmser: homoserine, Ino: inositol, InoP: inositol-1-phosphate, Isocit: isocitrate, Isomalt: isomaltose, Mal: malate, Tre: trehalose, T6P: trehalose-6-phosphate, Orn: ornithine and Raf: raffinose.
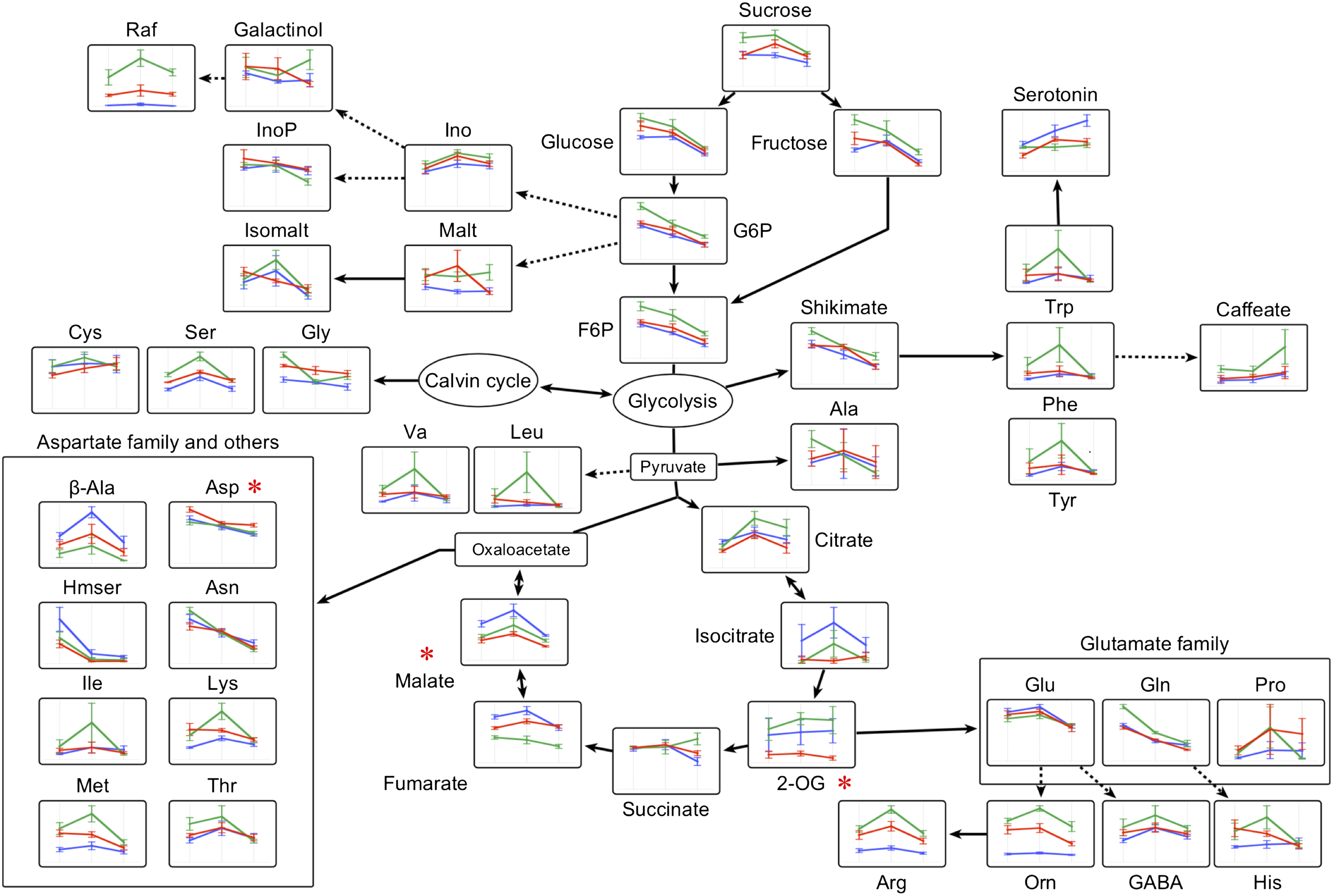
Figure 5. Unique changes in levels of metabolites involved in the TCA cycle and N metabolism in F1 hybrid. X-axis represents sampling periods, while Y-axis shows normalized intensity of each metabolite peak. The levels of 2-OG (A) and malate (B) were significantly lower in F1 hybrid compared to in parent plants. The level of aspartate (C) was significantly higher in F1 hybrid than parent plants. Different letters indicate significant differences based on Tukey’s test (p<0.05). Error bars represent the SE of six biological replicates.
Identification of over-represented pathways at the transcript level in F1 hybrid
Microarray analysis was conducted to investigate transcript changes in the F1 hybrid and parental lines. We focused on genes that were altered in F1 profiles compared to both Col and C24 lines. There were 44 genes whose levels were significantly increased and 12 genes whose levels were significantly decreased in the F1 lines compared to both parents (Supplementary Table S3). SEA was performed for the 44 up- and 12 down-regulated genes in the F1 hybrid line (Supplementary Figures S3, S4, Supplementary Table S4). One of the over-represented GO terms for the genes up-regulated in F1 hybrid plants was “response to chemicals” in the GO category “biological process”. The GO terms “response to organonitrogen compound”, “response to nitrogen compound”, and “response to organic substance” in the subclass “biological process” were significantly over-represented. Additionally, seven of the 12 down-regulated genes in the F1 hybrid were chloroplast-encoded genes. The most over-represented GO term in the down-regulated genes in F1 hybrid was “transport” in the GO category “biological process”, as well as “establishment of localization” and “localization”. In addition, the most over-represented terms in the GO category “cellar component” were “plastid” and “chloroplast”.
Evaluation of transcript changes related to N metabolism in the F1 hybrids
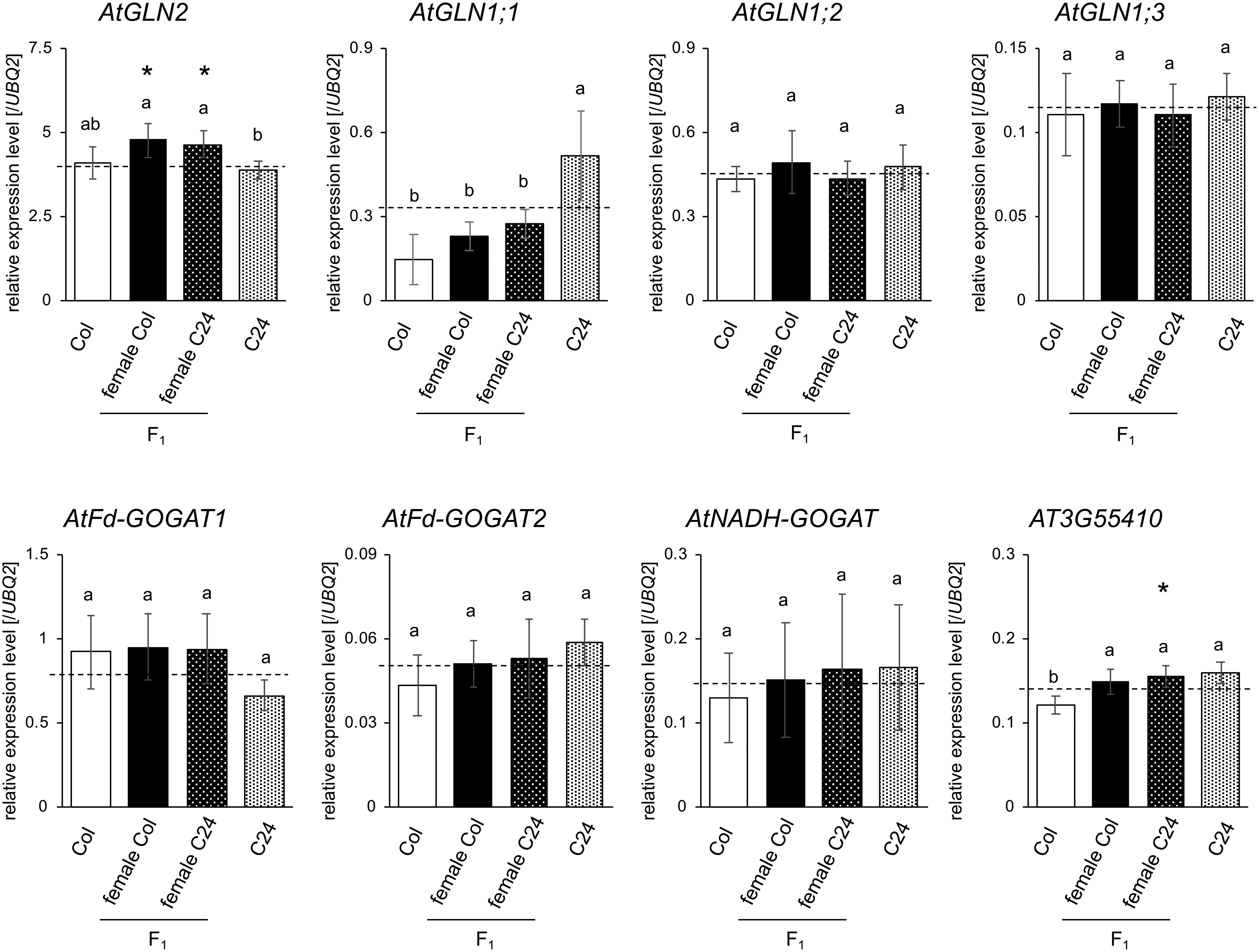
The results of the metabolomic analyses and GO analysis of transcriptome analyses of the F1 hybrid and parental lines suggest that N metabolism would be important to alterations of biomass production in Arabidopsis resulting from heterosis. However, out of the 44 up-regulated genes and the 12 down-regulated genes in the F1 hybrid from our microarray data, we could not find any enzymatic genes related to N metabolism (Supplementary Table S3). Therefore, we checked the trend of enzymatic genes related to N-metabolism using the MapMan tool to investigate the gene expression directly affecting metabolite contents. As a result, although prominent changes in specific metabolic pathways were not observed in F1 hybrid plants in our transcript profile overview, slightly increased expression in several N metabolism-related genes in the F1 hybrid was detected (Supplementary Figure S5). Additionally, in silico analysis of N metabolism-related genes in Arabidopsis using data extracted from a previously published paper (see Fujimoto et al. 2012) might suggest that AtGLN1;1, AtGLN1;2, AtGLN2, AtFd-GOGAT1, AtNADH-GOGAT, and the gene encoding the E1 subunit of the 2-OG dehydrogenase complex (AT3G55410) are differentially expressed in the F1 hybrid. Therefore, the expression levels of the genes described above were analyzed by qPCR (Figure 6). As a result, there were no significant differences in mRNA levels of AtGLN1;2, AtGLN1;3, and the three GOGAT-coding genes. Gene expression levels between Col and C24 showed significant differences and the expression value of F1 hybrids showed intermediate value between both parental values in AtGLN1;1 and AT3G55410. In contrast, AtGLN2 mRNA level was approximately 20% higher in the F1 hybrids compared with their parents. These results imply that the alterations in AtGLN2 levels could impact N metabolism and increase biomass in F1 hybrids.
Figure 6. Expression patterns of genes related to N metabolism assessed using transcript levels in Col, C24, and F1 hybrids at 15 DAS. Error bars indicate the SD of six biological replicates. Different letters indicate significant differences based on Tukey’s test (p<0.05). Dashed line represents the MPV. Asterisks (*) indicate significant differences between F1 hybrid values and MPV based on results of Student’s t-tests (p<0.05).
Discussion
Fine tuning of growth conditions to evaluate heterosis for biomass size using omics analyses
Plant growth is influenced by light intensity and quality, photoperiod, temperature, and humidity (Matsubara 2018). Metabolic fluctuations can be observed even when plant samples were grown under strictly-controlled conditions (Kusano et al. 2007; Weckwerth et al. 2004). Careful sample preparation is thus critical when evaluating metabolite and transcript changes while excluding as many extraneous and unknown factors as possible.
In this study, we investigated the metabolite and transcript changes in F1 samples showing significant increases of biomass size compared with parental lines. Many previous studies focused on the influence of heterosis on biomass-related traits in Arabidopsis (Fujimoto et al. 2012; Meyer et al. 2004; Miller et al. 2015; Wang et al. 2019), with biomass levels of C24×Col F1 hybrids plants ranging from 26 to 161% of parental line biomasses. Although the average FW of our F1 samples grown in culture medium under optimized conditions was about 1.3 times greater than MPV, the RSD of FW was much lower than that of plants grown on soil (Figure 1). These results show that optimized growth condition can produce F1 samples with statistically greater biomasses than their parental lines.
The dissection of genotype- and growth-dependent metabolite changes highlights specific metabolites in F1 hybrid
Metabolite composition is greatly altered during different transitional periods in the lifetime of a plant (Allen et al. 2010; Meyer et al. 2012; Wahl et al. 2013). During transitions between heterotrophic and autotrophic stages as well as vegetative and reproductive stages, plant biomass and morphology change dramatically. Since visible phenotypic changes have large impacts on metabolism in plants, sampling points were carefully determined to maximize extraction of candidate metabolites with heterosis-dependent changes in the F1 samples compared with parental lines. In Arabidopsis, the speed of germination and growth are different between accessions and their F1 hybrids (Groszmann et al. 2014; Meyer et al. 2012). For example, heterosis in C24×Col F1 hybrids has been observed at the photosynthetic initiation stage at around 3 DAS (Fujimoto et al. 2012; Meyer et al. 2012). In addition, various patterns of metabolite changes have been observed during the young seedling stage (Allen et al. 2010). Moreover, flowering time in Arabidopsis is affected by photoperiod, and Arabidopsis accessions and their F1 hybrids tend to show different flowering patterns when grown in long-day conditions (Blázquez et al. 2001; Fujimoto et al. 2012; Groszmann et al. 2014). In this study, we used short-day conditions to inhibit flowering in all genotypes and as a result, flower formation was not detected until at least 17 DAS (data not shown). Since samples harvested at approximately 15 DAS were used in previous transcriptome studies (Groszmann et al. 2014, 2015; Zhang et al. 2016), we examined plants at 15±2 DAS for metabolite profiling.
We conducted two-way ANOVA analyses in order to generate information about to what extent the metabolite changes we detected are likely to arise from genotype, growth, and interactions between the two (Figures 2, 3). Our results indicated that genotype-dependent factors had the greatest impact on the metabolite composition of F1 hybrid and parental lines (Figures 3, 4). Levels of metabolites including sugars, sugar phosphates, and several amino acids (Figure 4) were similar between F1 hybrid and parental lines. In contrast, levels of 2-OG, malate, and aspartate were different between F1 hybrid and parental lines. 2-OG is both a TCA cycle intermediate and can be synthesized via the photorespiratory pathway (Dellero et al. 2016; Huergo and Dixon 2015). Photorespiration starts when Rubisco fixes O2 instead of CO2. C4 plants have developed a CO2 pump and inhibit this oxygenation reaction by increasing the relative intracellular concentration of CO2 compared O2 around Rubisco (Bräutigam and Gowik 2016). A recent investigation demonstrated that the photorespiratory pathway is altered in maize F1 hybrids compared with parental lines (Li et al. 2020). In our study, levels of photorespiration-related metabolites such as serine, glycine, glycolate, and glycerate, as well as metabolites belonging the GC-GOGAT cycle (glutamine and glutamate), were not significantly different between the F1 hybrid and the parental lines, while clear changes were observed for 2-OG between F1 hybrid and parents (Figures 4, 5).
Higher fumarate/malate ratios have been previously associated with increased biomass in Arabidopsis (Riewe et al. 2016). In our study, F1 hybrid had decreased malate but unchanged fumarate levels, resulting in higher fumarate/malate ratios compared with parental lines. Similarly, malate content and biomass showed negative correlation in the 429 Recombinant Inbred Line (RIL) population derived from a cross between Col and C24 (Meyer et al. 2007). Overall, the decrease in the malate level is likely associated with increased biomass of F1 hybrid, although more evidence needs to be obtained.
Aspartate aminotransferase catalyzes a reversible reaction between aspartate and 2-OG that yields glutamate and oxaloacetate (Gaufichon et al. 2016). As a result, aspartate serves as a carbon source for the TCA cycle. Aspartate contributes to plant growth by transporting reducing equivalents from mitochondria and chloroplasts into the cytoplasm via the malate-aspartate shuttle (Coruzzi 2003). In addition, decreased biomass, aspartate, glutamine, and proline levels were reported in glutamate dehydrogenase β-subunit-overexpressing lines (Tercé-Laforgue et al. 2013), suggesting that aspartate may be an important metabolite for N metabolism and biomass production. The increased aspartate level in F1 hybrids may thus be associated with increased biomass.
AtGLN2 may contribute to increased biomass in F1 hybrids
Transcript profiling of F1 hybrid and their parental lines emphasized that organic N metabolism is altered in the F1 samples (Supplementary Figure S3). In particular, AtGLN2 mRNA level was higher in the F1 samples compared to in the parental lines (Figure 6). AtGLN2 helps assimilate ammonium ions during photorespiration. Ammonium released during photorespiration may be more than ten times higher than ammonium released due to primary N assimilation (Keys et al. 1978; Oliveira et al. 2002). In addition, 2-OG is utilized as a substrate in the GS-GOGAT cycle. Several reports have measured increased biomass in GS-overexpressing lines, suggesting that GS may be a rate-limiting enzyme in the GS-GOGAT cycle (Kaachra et al. 2018; Lu et al. 2018; Man et al. 2011; Migge et al. 2000). Photorespiration is initiated by the reaction of Rubisco with oxygen and is thought to occur simultaneously with photosynthesis. Combined with the fact that hybrid vigor onset is thought to be around 3 DAS, which is the same time at which photosynthesis is initiated, acceleration of photorespiration-derived ammonium re-assimilation via upregulation of chloroplast-localized AtGLN2 may contribute to increased biomass in F1 hybrids. Li et al. proposed that the photosynthetic and photorespiratory pathways of maize are temporally regulated in F1 hybrids compared with parents (Li et al. 2020). This finding has also been supported by proteome, metabolome, and enzyme activity data. One unresolved mechanism highlighted in our study is the regulatory mechanism controlling AtGLN2 expression in the F1 hybrids. Additionally, post-transcriptional and post-translational regulation can also affect metabolite levels. We thus need to proceed with further multi-approach analyses in order to understand the molecular mechanism underlying heterosis in Arabidopsis.
In conclusion, our data suggest that N metabolism and the TCA cycle in F1 hybrid plants behaves differently compared to in parental lines. N metabolism is one of the most important factors contributing to plant growth and is known that affect biomass and stress tolerance. This could allow us to generate novel insights regarding the influence of N metabolism in heterosis in F1 hybrid plants.
Acknowledgments
We thank Prof. Shinobu Satoh (Faculty of Life and Environmental Sciences, University of Tsukuba), Prof. Kazuki Saito (RIKEN center for Sustainable Resource Science), Prof. Yutaka Suzuki (Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, University of Tokyo), Dr. Nobutaka Mitsuda and Dr. Sumire Fujiwara (Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST)) for helpful discussions regarding the experiments. We are grateful to Ms. Kazuko Ito and Ms. Masumi Nakagawara for their excellent technical assistance. We would also thank to Ms. Tomoko Nishizawa at RIKEN center for Sustainable Resource Science for her technical support in microarray analysis. This work was supported in part by the Tsukuba Innovation Arena (TIA) collaborative research program “Kakehashi” to H.S. from TIA, and by the NEXT Program (GS018) to H.S. from JSPS. This research was also supported by the “Sustainable Food Security Research Project” in the form of an operational grant from the National University Corporation, Japan.
Abbreviations
- 2-OG
2-oxoglutarate
- DAS
day after sowing
- TCA
tricarboxylic acid
- GC-TOF-MS
gas chromatography-time-of-flight-mass spectrometry
- PCA
principal component analysis
- ANOVA
analysis of variance
- GS
Glutamine Synthetase
- MPV
mid-parent value
Conflicts of interest
No potential conflicts of interest are declared in our study.
Authors’ contribution
N. Sugi, and H. Shiba conceived the research plans. N. Sugi, Q.T.N. Le, M. Kobayashi, and M. Kusano performed experiments and conducted data analysis. N. Sugi, M.kusano, Q.T.N. Le and H. Shiba wrote the manuscript. All of authors critical read and approved the final manuscript.
Supplementary Data
References
- Allen E, Moing A, Ebbels TMD, Maucourt M, Tomos AD, Rolin D, Hooks MA (2010) Correlation Network Analysis reveals a sequential reorganization of metabolic and transcriptional states during germination and gene-metabolite relationships in developing seedlings of Arabidopsis. BMC Syst Biol 4: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez M, Koornneef M, Putterill J (2001) Flowering on time: Genes that regulate the floral transition. Workshop on the molecular basis of flowering time control. EMBO Rep 2: 1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Gowik U (2016) Photorespiration connects C3 and C4 photosynthesis. J Exp Bot 67: 2953–2962 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14: 471–482 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM (2003) Primary N-assimilation into amino acids in Arabidopsis. Arabidopsis Book 2: e0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellero Y, Jossier M, Schmitz J, Maurino VG, Hodges M (2016) Photorespiratory glycolate-glyoxylate metabolism. J Exp Bot 67: 3041–3052 [DOI] [PubMed] [Google Scholar]
- Fiévet JB, Nidelet T, Dillmann C, de Vienne D (2018) Heterosis is a systemic property emerging from non-linear genotype-phenotype relationships: Evidence from in vitro genetics and computer simulations. Front Genet 9: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto R, Taylor JM, Shirasawa S, Peacock WJ, Dennis ES (2012) Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc Natl Acad Sci USA 109: 7109–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K (2009) Impact of clock-associated Arabidopsis pseudoresponse regulators in metabolic coordination. Proc Natl Acad Sci USA 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaufichon L, Rothstein SJ, Suzuki A (2016) Asparagine metabolic pathways in arabidopsis. Plant Cell Physiol 57: 675–689 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy-Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Groszmann M, Gonzalez-Bayon R, Greaves IK, Wang L, Huen AK, Peacock WJ, Dennis ES (2014) Intraspecific arabidopsis hybrids show different patterns of heterosis despite the close relatedness of the parental genomes. Plant Physiol 166: 265–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Gonzalez-Bayon R, Lyons RL, Greaves IK, Kazan K, Peacock WJ, Dennis ES (2015) Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 112: E6397–E6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huergo LF, Dixon R (2015) The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol Mol Biol Rev 79: 419–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaachra A, Vats SK, Kumar S (2018) Heterologous expression of key C and N metabolic enzymes improves re-assimilation of photorespired CO 2 and NH 3, and growth. Plant Physiol 177: 1396–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ (1978) Photorespiratory nitrogen cycle. Nature 275: 741–743 [Google Scholar]
- Korn M, Gärtner T, Erban A, Kopka J, Selbig J, Hincha DK (2010) Predicting arabidopsis freezing tolerance and heterosis in freezing tolerance from metabolite composition. Mol Plant 3: 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Fukushima A, Arita M, Jonsson P, Moritz T, Kobayashi M, Hayashi N, Tohge T, Saito K (2007) Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst Biol 1: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Tohge T, Fukushima A, Kobayashi M, Hayashi N, Otsuki H, Kondou Y, Goto H, Kawashima M, Matsuda F, et al. (2011) Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J 67: 354–369 [DOI] [PubMed] [Google Scholar]
- Le QTN, Sugi N, Furukawa J, Kobayashi M, Saito K, Kusano M, Shiba H (2019) Association analysis of phenotypic and metabolomic changes in arabidopsis accessions and their F1 hybrids affected by different photoperiod and sucrose supply. Plant Biotechnol (Tokyo) 36: 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhu A, Song Q, Chen HY, Harmon FG, Chen ZJ (2020) Temporal regulation of the metabolome and proteome in photosynthetic and photorespiratory pathways contributes to maize heterosis. Plant Cell 32: 3706–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Liu L, Wei M, Liu Y, Qu Z, Yang C, Wei H, Wei Z (2018) The effect of poplar PsnGS1.2 overexpression on growth, secondary cell wall, and fiber characteristics in tobacco. Front Plant Sci 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man H, Pollmann S, Weiler EW, Kirby EG (2011) Increased glutamine in leaves of poplar transgenic with pine GS1a caused greater anthranilate synthetase α-subunit (ASA1) transcript and protein abundances: An auxin-related mechanism for enhanced growth in GS transgenics? J Exp Bot 62: 4423–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara S (2018) Growing plants in fluctuating environments: Why bother? J Exp Bot 69: 4651–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt Ä, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Törjék O, Becher M, Altmann T (2004) Heterosis of biomass production in arabidopsis. Establishment during early development. Plant Physiol 134: 1813–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Witucka-Wall H, Becher M, Blacha A, Boudichevskaia A, Dörmann P, Fiehn O, Friedel S, Von Korff M, Lisec J, et al. (2012) Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J 71: 669–683 [DOI] [PubMed] [Google Scholar]
- Migge A, Carrayol E, Hirel B, Becker TW (2000) Leaf-specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210: 252–260 [DOI] [PubMed] [Google Scholar]
- Miller M, Song Q, Shi X, Juenger TE, Chen ZJ (2015) Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat Commun 6: 7453. [DOI] [PubMed] [Google Scholar]
- Ng DWK, Chen HHY, Chen ZJ (2017) Heterologous protein-DNA interactions lead to biased allelic expression of circadian clock genes in interspecific hybrids. Sci Rep 7: 45087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM (2002) Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol 129: 1170–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D, Jeon HJ, Lisec J, Heuermann MC, Schmeichel J, Seyfarth M, Meyer RC, Willmitzer L, Altmann T (2016) A naturally occurring promoter polymorphism of the Arabidopsis FUM2 gene causes expression variation, and is associated with metabolic and growth traits. Plant J 88: 826–838 [DOI] [PubMed] [Google Scholar]
- Rohn H, Junker A, Hartmann A, Grafahrend-Belau E, Treutler H, Klapperstück M, Czauderna T, Klukas C, Schreiber F (2012) VANTED v2: A framework for systems biology applications. BMC Syst Biol 6: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Nikoloski Z, Tschoep H, Antonio C, Kleessen S, Larhlimi A, Selbig J, Ishihara H, Gibon Y, Fernie AR, et al. (2013) Impact of the carbon and nitrogen supply on relationships and connectivity between metabolism and biomass in a broad panel of Arabidopsis accessions. Plant Physiol 162: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercé-Laforgue T, Bedu M, Dargel-Grafin C, Dubois F, Gibon Y, Restivo FM, Hirel B (2013) Resolving the role of plant glutamate dehydrogenase: II. Physiological characterization of plants overexpressing the two enzyme subunits individually or simultaneously. Plant Cell Physiol 54: 1635–1647 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1): W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dingenen J, de Milde L, Vermeersch M, Maleux K, de Rycke R, de Bruyne M, Storme V, Gonzalez N, Dhondt S, Inzé D (2016) Chloroplasts are central players in sugar-induced leaf growth. Plant Physiol 171: 590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur F, Fouqueau L, De Vienne D, Nidelet T, Violle C, Weigel D (2019) Nonlinear phenotypic variation uncovers the emergence of heterosis in Arabidopsis thaliana. PLoS Biol 17: e3000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707 [DOI] [PubMed] [Google Scholar]
- Wang L, Liu PC, Wu LM, Tan J, Peacock WJ, Dennis ES (2019) Cotyledons contribute to plant growth and hybrid vigor in Arabidopsis. Planta 249: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Xu T, Srivastava AK, Wang D, Zeng L, Yang L, He L, Zhang H, Zheng Z, et al. (2016) The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov 2: 16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.