Abstract
Temperature-dependent NMR experiments are often complicated by rather long magnetic-field equilibration times, for example, occurring upon a change of sample temperature. We demonstrate that the fast temporal stabilization of a magnetic field can be achieved by actively stabilizing the temperature of the magnet bore, which allows quantification of the weak temperature dependence of a proton chemical shift, which can be diagnostic for the presence of hydrogen bonds. Hydrogen bonding plays a central role in molecular recognition events from both fields, chemistry and biology. Their direct detection by standard structure-determination techniques, such as X-ray crystallography or cryo-electron microscopy, remains challenging due to the difficulties of approaching the required resolution, on the order of 1 Å. We, herein, explore a spectroscopic approach using solid-state NMR to identify protons engaged in hydrogen bonds and explore the measurement of proton chemical-shift temperature coefficients. Using the examples of a phosphorylated amino acid and the protein ubiquitin, we show that fast magic-angle spinning (MAS) experiments at 100 kHz yield sufficient resolution in proton-detected spectra to quantify the rather small chemical-shift changes upon temperature variations.
Introduction
The recording of an NMR data set, in particular for multidimensional experiments on proteins, requires hours or even days of measurement time during which the magnetic field at the sample position (B⃗0,sample) is required to stay constant within significantly better than the spectral linewidth. In this contribution, we investigate the temperature dependence of proton shifts in solid-state NMR, which is a particularly demanding application. In solution-state NMR, the field stability is achieved by a highly optimized field-frequency lock system implemented in virtually all high-resolution spectrometers.1 For magic-angle spinning (MAS) high-resolution NMR spectroscopy, the implementation of such a lock proved difficult and most spectra are recorded without a lock. Typically, the drift of the static magnetic field can be compensated by a linear (in time) field gradient. Linear-field drifts may also be applied a posteriori.2 With increasing spectral resolution, in particular also in the context of fast MAS and proton detection, nonlinear field drifts start to limit the spectral resolution. We, and others, have noticed that such effects appear, in particular, after a change of the probe, change of the sample, or even only a change of sample temperature. Typically, several hours are needed until these effects die out. We here identify the source of these instabilities and show that it can be mitigated by temperature stabilization of the bore. This allows not only recording spectra without long equilibration delays but also precisely measuring the temperature gradients of the proton resonance frequencies, which are indicators for hydrogen bonding.
Hydrogen bonds are crucial for protein folding and drive a variety of molecular recognition events, for instance, protein–drug and protein–nucleic acid binding. The determination of the exact location of hydrogen atoms engaged in hydrogen bonds in proteins by standard structure-determination techniques, such as X-ray crystallography or cryo-electron microscopy (cryo-EM), remains impossible as long as the resolution does not approach values around 1 Å (for cryo-EM, a slightly lower resolution might be sufficient3), which is often difficult or even impossible to achieve, although the O···N internuclear distance can be used as a proxy. Hydrogen bonds can be identified by spectroscopic information from infrared spectroscopy4,5 and NMR. Particularly, NMR is highly sensitive to such effects, and, for instance, the magnitude of a deshielded proton chemical-shift value encodes for hydrogen bonding.6−9 This effect alone is, however, typically not sufficient to unambiguously identify a hydrogen bond since the proton chemical-shift values might be affected by further structural parameters,7,10−14 and additional information is required. Some possibilities to directly prove the presence of the hydrogen bond include using magnetization transfers and exploiting the hydrogen-bond J-coupling,15,16 which may however be difficult to achieve in practice due to the typically small J-coupling values competing unfavorably with the short transverse coherence lifetimes in the solid state. Alternatively, also the measurement of NH distances by solid-state NMR and their elongation upon hydrogen-bond formation have been reported.17,18
The temperature dependence of proton chemical shifts has been the object of many studies in solution-state NMR, focusing on using it as an additional tool in structural characterization, for instance, in predicting hydrogen bonds in proteins19−24 and small organic molecules.25−29 It has also been observed by solid-state NMR, for instance, in disaccharides,30 in ionic liquid crystals,31 or dipeptides.32 The temperature dependence of 1H chemical-shift values is attributed to an increase in the average H···O hydrogen-bond length upon increasing the temperature, leading to a less pronounced deshielding effect induced by a hydrogen-bond acceptor.33 Note that ring current effects can also influence the temperature behavior of proton chemical shifts, even in non-hydrogen-bonded amides.19 For proteins, correlations between HN amide proton shift temperature coefficients Δδ(1H)/ΔT and involvement in hydrogen bonds have been studied.19,23,33,34 Chemical-shift temperature gradients (Δδ(1H)/ΔT) that are more positive than −4.6 ppb/K have been proposed to serve as a purely empirical criterion for hydrogen bonding.19 Additional NMR observables established by solution-state NMR as an indicator for hydrogen bonding are temperature-dependent hydrogen-bond scalar couplings.35 Despite the characterization of chemical-shift temperature gradients in solution-state NMR, no systematic study has been published, to our knowledge, using proton-detected solid-state NMR for biomolecules in the fast MAS regime. One of the reasons is the technical difficulty of the experiment as described above. We, herein, analyze the temperature dependence of proton chemical shifts in the crystalline compound ortho-phospho-l-serine as well as in the protein ubiquitin and evaluate their predictive power for hydrogen bonding, comparing the results to a similar study on the same protein carried out with solution-state NMR.34
Methods and Materials
Materials
Ortho-phospho-l-serine was purchased from Sigma Aldrich. The deuterated and 100% back-exchanged ubiquitin sample (13C and 15N labeled) was prepared using overexpression in E. coli. The protein was crystallized with MPD, as previously described.36 The sample was filled in an NMR rotor in an ultracentrifuge using home-build filling tools.37−39 DUL Cp149 was prepared as described in ref (40).
Solid-State NMR
Solid-state NMR experiments were recorded using an AVANCE III 850 MHz wide-bore spectrometer (static magnetic-field strength of 20.0 T) in a Bruker 0.7 mm probe. The MAS frequency was set to 100 kHz. The 2D spectra were processed using TOPSPIN software (version 3.5, Bruker Biospin) with a shifted (3) squared cosine apodization function. In the case of ortho-phospho-l-serine, 1D Hahn-echo spectra were recorded with a repetition time of 2 s and 4 scans using BCU temperatures ranging in between 280 and 320 K (1D spectra were acquired in regular time intervals until the magnetic-field stability was achieved). For the ubiquitin measurements, the sample temperature was set to BCU temperatures of 265–280 K, which corresponds to sample temperatures of 293–301 K determined using the water line for internal calibration.41 All spectra were analyzed using CcpNmr software42−44 and referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). The 2D hNH spectra for ubiquitin were recorded with an acquisition time of 26 ms and a spectral width of 40 ppm in an indirect dimension and 26 ms/46.7 ppm in a direct dimension, using a 130/30 kHz zero quantum CP condition for HN and NH transfers. Carriers were set to 4.8 ppm for 1H and 117.5 ppm for 15N. Overall, 5 kHz 15N WALTZ6445 and 10 kHz 1H frequency-swept TPPM decoupling46 were applied during the acquisition of the direct and indirect dimensions, respectively. Bruker BioSpin designed and installed the prototype magnet bore temperature control unit that was used for these studies.
Results and Discussion
Stabilizing the Temperature of the Magnet Bore Reduces Magnetic-Field Fluctuations
The reported temperature dependence of proton chemical-shift values for amide protons are small, in the order of a few parts per billion per Kelvin (ppb/K), and thus require well-resolved proton resonances, which can typically be achieved by performing the NMR experiments at >100 kHz MAS.47−49 An additional technical complication arises from the finding that a change of the sample temperature is accompanied by temperature changes in the body of the probe and the bore of the magnet, causing a change in the magnetic susceptibility and thus of the magnetic field at the sample position (B⃗0,sample). Inserting the sample into the probe at the beginning of the experiment, sample change, or even a sample-temperature change leads to a new temperature distribution in the entire probe and, by thermal contact, also within the bore of the magnet. As a result, B⃗0,sample is disturbed and the resonance frequency drifts before stabilizing to a new value when the entire probe–magnet system reaches again thermal equilibrium. The equilibration time is found to be in the order of hours (vide infra). If the experiment is performed before the magnetic-field stability is reached, the precise extraction of temperature coefficients is complicated. It now turns out that the major contribution to these effects comes from the magnet bore. Actively stabilizing the temperature of the bore by a Bore Temperature Control System developed by Bruker BioSpin (abbreviated as BTCS in the following) allows for a strong reduction of magnetic-field drifts and the required equilibration time upon changing the temperature. This system essentially consists of heating foils and a temperature sensor, which is attached to the outside of the room-temperature shim system and electrically connected to the Bruker Smart Variable Temperature System (BSVT). In addition to the heating foils and the temperature sensor, a thin copper foil was wrapped around the room-temperature shim system to minimize temperature gradients. Figure 1 shows 2D hNH fingerprint spectra of the deuterated and 100% back-exchanged (DUL) core protein of the Hepatitis B virus capsid, Cp149,40 recorded at 100 kHz MAS without the BTCS (a) and with the BTCS (b) (the bore temperature was adjusted to 295 K). For the measurements under the different setups, the same experimental procedure was applied: After inserting the probe into the magnet, the sample cooling was started (target sample/BCU temperature 271 K) and the first 2D spectrum was recorded after 30 min equilibration time. Afterward, a series of four subsequent spectra, each taking a measurement time of approximately 1 h, were recorded. The spectra shown in Figure 1b clearly reveal the absence of visible magnetic-field drifts over the total measurement time when the BTCS is turned on, and which are clearly present without operating the BTCS (Figure 1a). Note that without the BTCS, magnetic-field drifts are still observed even in the last 2D spectrum, which started recording 3.5 h after adjusting the probe temperature (Figure 1a). The use of the BTCS is, therefore, a great help for determining, with precision, proton temperature coefficients from solid-state NMR spectra in a reasonable measurement time.
Figure 1.
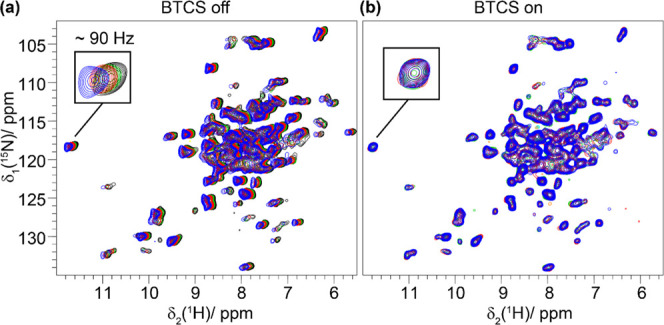
Bore temperature control system avoids magnetic-field drifts upon temperature changes. hNH correlation experiments recorded on DUL Cp149 at 100 kHz and 20 T external magnetic-field strength with (b) and without the BTCS (a). The spectra were recorded at 0.5 h (blue), 1.5 h (red), 2.5 h (green), and 3.5 h (black) after inserting the probe into the magnet bore. Each 2D experiment takes around 1 h.
Proton Chemical-Shift Temperature Coefficients Determined for an Organic Solid
We next established the extraction of a proton chemical-shift temperature coefficient on a small crystalline model system, namely, phosphorylated serine, o-phospho-l-serine, which has already been subject to solid-state NMR studies.50−52 Intermolecular hydrogen bonds exist for the carboxylic group as well as for the phosphate group,53 resulting in high-frequency-shifted proton resonances in 1H 1D spectra (see Figure 2a). The two most deshielded resonances resonate at ∼16.5 ppm (the carboxylic proton) and ∼12.5 ppm (the phosphate group proton). We recorded 1H spectra between 280 and 320 K (Figure 2a) and found, for the protons involved in an intermolecular hydrogen bond, negative proton chemical-shift temperature coefficients of −2.3 ± 0.1 ppb/K (carboxylic COOH proton) and −1.6 ± 0.1 ppb/K (phosphate PO3OH proton), whereas the further resonances do not show a significant temperature dependence, as shown in Figure 2b. Note that the BCTS dramatically reduces the equilibrium time until the stability of the magnetic field, but small shifts (in the order of a few tens of Hertz for a 10 K temperature change) in the magnetic field upon change of the probe temperature still need to be corrected. This was achieved by referencing the spectra relative to each other to the low-ppm CH2 resonance, which is not expected to participate in hydrogen bonding. The measured values are in good agreement with values extracted by solution-state NMR for small organic compounds.25
Figure 2.
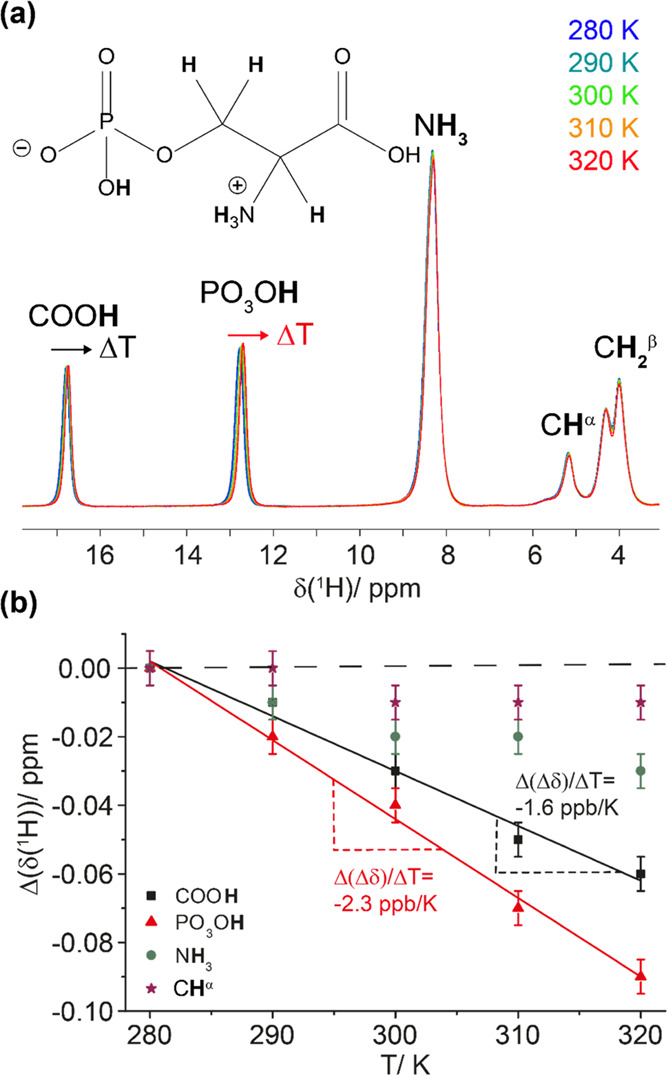
Hydrogen bonds in ortho-phospho-l-serine identified by 1H chemical-shift temperature dependences. (a) 1H MAS spectra of ortho-phospho-l-serine (the chemical structure is shown on top) recorded at 100 kHz MAS and 20 T in the temperature range from 280 to 320 K. The spectra were referenced to the low-ppm CH2 resonance (4.00 ppm). (b) Temperature dependence of proton chemical shifts extracted from the peak maxima in the spectra and the corresponding linear regression to extract the temperature coefficients for the protons involved in hydrogen bonding (solid lines). The extracted temperature coefficients correspond to the slope of the linear regression and are given in the figure. Note that the temperature-dependent chemical shifts are represented relative to the proton chemical shift at the lowest temperature (280 K). The spectra were referenced relative to each other to the low-ppm CH2 resonances (dashed line). The error bars were estimated to 0.01 ppm.
Proton Solid-State Chemical-Shift Temperature Coefficients in Ubiquitin
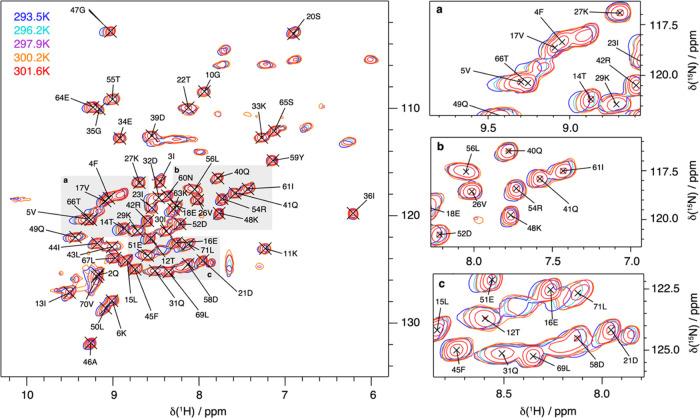
To investigate the hydrogen bonding in the protein ubiquitin, we recorded temperature-dependent hNH correlation spectra at 100 kHz MAS at different temperatures. Figure 3 shows a selection of solid-state NMR ubiquitin hNH fingerprint spectra in a temperature range of ∼20–28 °C.
Figure 3.
Temperature-dependent hNH solid-state NMR spectra of ubiquitin. Deuterated and 100% back-exchanged ubiquitin hNH fingerprint spectra recorded at 100 kHz MAS and 20 T for five different temperatures (a) and some representative extracts (b), illustrating the different temperature-dependent proton chemical-shift behaviors of the observed resonances.
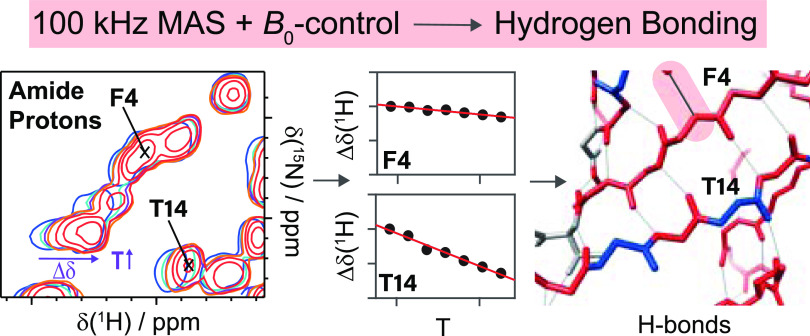
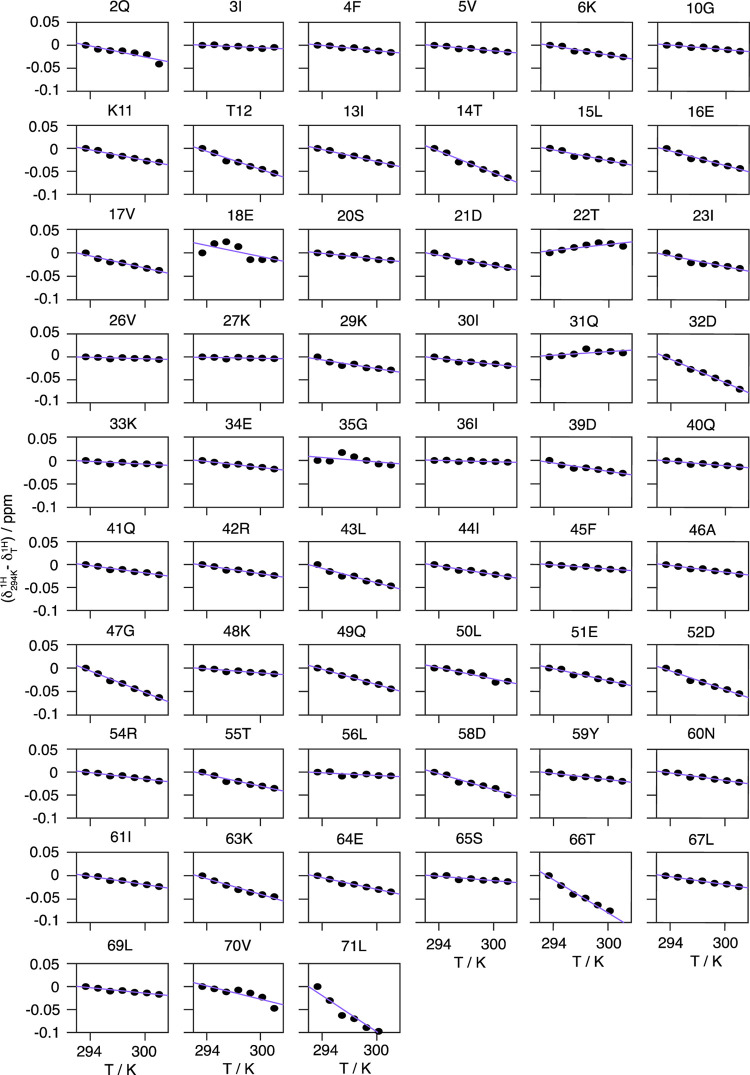
From these spectra, we evaluate the temperature dependence of the amide proton chemical shifts (see Figure 4). Some resonances, e.g., V26, K27, K33, I36, K48, and L56, show only small changes with temperature, as expected for protons in strong hydrogen bonds, while the resonances of, e.g., T14, D32, G47, T66, and L71 show strong shielding effects with increasing temperature.
Figure 4.
Site-specific temperature dependence of chemical shifts for ubiquitin. Site-specific temperature dependences of proton chemical shifts in DUL ubiquitin (black circles) measured in the sample temperature range 293–301 K. Note that the depicted chemical shifts have been referenced to the respective proton chemical shift observed at the lowest temperature of 293 K. The purple lines indicate the result of the linear regression used to extract the corresponding temperature coefficients. For each temperature, the spectra were referenced internally to DSS to correct for any additional influence of different magnetic susceptibilities of the probe head on the magnetic field experienced by the sample.
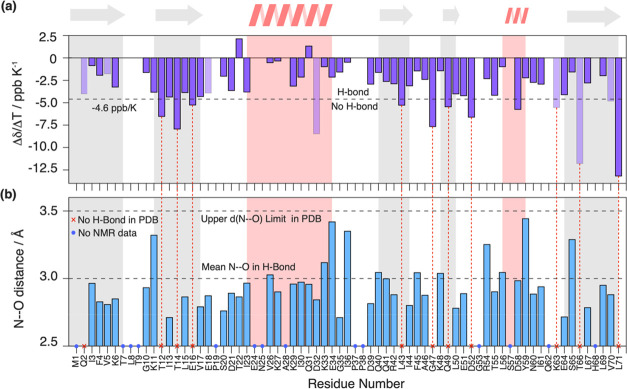
We observe no systematic nonlinearities within the experimental uncertainty, excluding the presence of low free-energy conformationally excited states and thus different, easily accessible, protein conformations, as described, for example, for the N-terminal domain of phosphoglycerate kinase, hen egg-white lysozyme, and BPTI54 in the temperature range used herein. Linear regression was used to extract the site-specific temperature coefficients Δδ(1H)/ΔT (slope) from the temperature dependence of the chemical shifts, which are plotted in Figure 5a.
Figure 5.
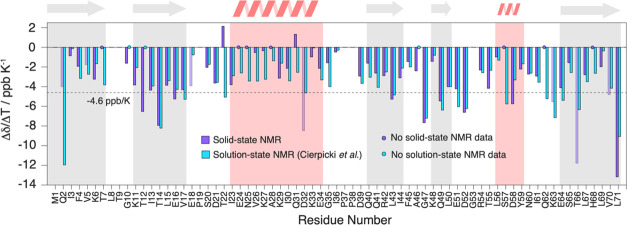
Temperature coefficients of ubiquitin and their comparison with hydrogen bonds identified from the crystal structure. (a) Site-specific Δδ(1H)/ΔT temperature coefficients for ubiquitin in the solid state. The dashed line denotes the empirical −4.6 ppb/K criterion for suspected hydrogen-bond formation found in a solution.19 Light shaded bars indicate residues, where the temperature coefficient determination may be biased by resonance peak merging at different temperatures. Gray and red background panels denote β-sheet and α-helix secondary structure elements. (b) Nearest-neighboring N–O distance determined from the crystal structure (PDB accession code 3ONS, blue bars). No distance has been determined in cases where no experimental temperature coefficient could be determined (blue circles) or where no hydrogen-bond formation is suspected from the crystal structure (d(N–O) > 3.5 Å, red crosses). Latter cases are highlighted by dashed vertical red lines, linking them to the corresponding Δδ(1H)/ΔT coefficient in (a). The mean N–O distance in H bonds and the upper distance limit (dashed lines) have been taken from refs (55) and (56).
Correlation between Temperature Coefficients and Hydrogen Bonding
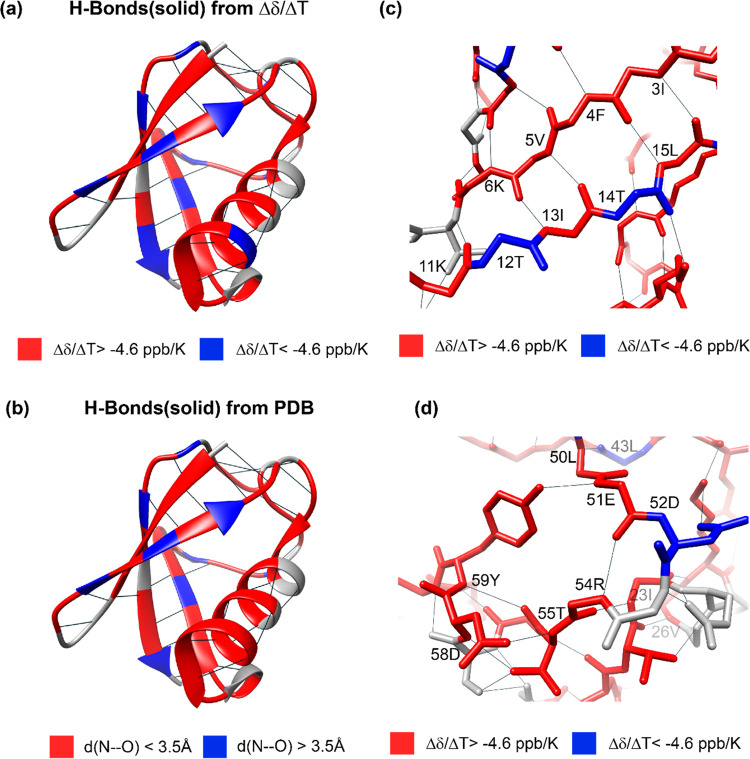
As already evident from the raw data, we observe strong differences in the temperature coefficients over the whole molecule. We next used the available X-ray crystal structure of ubiquitin (PDB accession code 3ONS(57)) to conclude on the quantitative values for which the temperature coefficients start becoming indicative for the presence of a hydrogen bond in the solid state. Since the crystal structure does not contain information on proton coordinates, we use for each residue i the presence of a short distance between the nitrogen of i to the oxygen of a close-by residue k (d(Ni–Ok) < 3.5 Å) as being indicative for the formation of a hydrogen bond.55,56 If this criterion is satisfied, the extracted distance is plotted in Figure 5b. Note that in this analysis we also included the possibility for intermolecular hydrogen bonds and bonds to HN side-chain atoms. Residue K11, for instance, forms a hydrogen bond with the side chains of T9, respectively, and the same is true for the backbone/side-chain pairs E18/D21, E51/Y59, and T55/N58. Figure 6 correlates the N–O distances extracted from the crystal structure with the proton temperature coefficients. Indeed, residues with a more positive temperature coefficient than −4.6 ppb/K are engaged in hydrogen bonds judged from N–O distances shorter than 3.5 Å, which serve as a proxy for identifying hydrogen bonds from X-ray structures.56 In contrast, for most residues that have a temperature coefficient more negative than −4.6 ppb/K, the crystal structure does not show a clear hydrogen bond to a close-by amino-acid residue. This altogether points to a similar validity of the approach as described also in solution,34 although the temperature coefficient alone is typically not sufficient to unambiguously identify hydrogen bonds.
Figure 6.
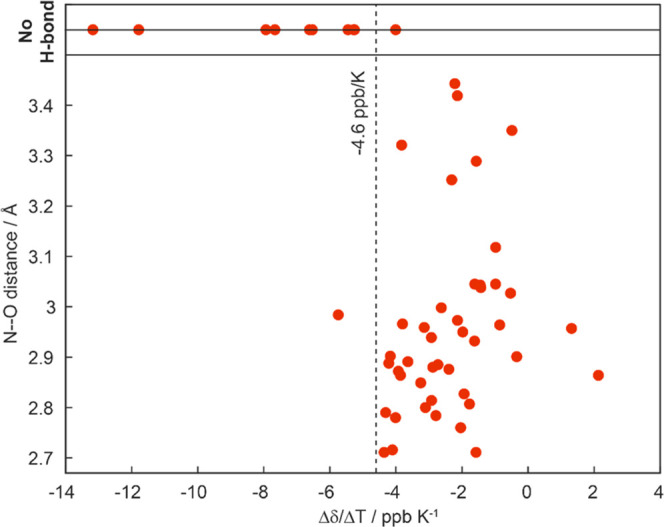
Hydrogen bonds identified from the crystal structure correlate with those identified by solid-state NMR. Correlation between N–O distances extracted from the crystal structure (PDB accession code 3ONS) and proton temperature coefficients as determined by solid-state NMR.
Comparison of Temperature Coefficients Determined in a Solution and in a Solid State
A comparison of the temperature coefficients obtained for ubiquitin with solid-state NMR in this work, with the same data from solution-state NMR,34 shows that the behavior in both aggregate states is rather similar over the whole molecule if the empirical criterion of −4.6 ppb/K is applied to identify hydrogen bonds. The numerical values of the gradient, however, show some differences that are particularly clustering in the α-helix spanning residues I23–E34, as well as residue T22 (see Figure 7 for a comparison of temperature coefficients extracted in the solid state and in solution34 and Figure S1 for the differences between the solution and the solid state). Finally, Figures 8 and S2 show the solid-state NMR temperature coefficients color-coded on the crystal structure of ubiquitin according to the −4.6 ppb/K criterion compared with the H bonds predicted from the PDB structure. This comparison highlights the high correlation of the proton chemical-shift temperature gradient with the presence of hydrogen bonds.
Figure 7.
Comparison of solution-state and solid-state proton chemical-shift temperature coefficients. Site-specific comparison of the Δδ(1H)/ΔT temperature coefficient between solid (violet) and solution-state NMR (light blue) data for ubiquitin. Light shaded bars indicate residues, where the temperature coefficient determination may be biased by resonance peak merging at different temperatures. The solution-state values have been taken from ref (34). The dashed line denotes the empirical −4.6 ppb/K criterion for suspected hydrogen-bond formation.19 Gray and red background panels denote β-sheet and α-helix secondary structure elements.
Figure 8.
Structural visualization of temperature coefficients. (a) Temperature coefficients Δδ(1H)/ΔT determined with solid-state NMR for ubiquitin and (b) determined from the crystal structure (using a criterion of 3.5 Å for the N–O distance56) color-coded on its crystal structure (PDB accession code 3ONS). In (a), values more negative than the empirical −4.6 ppb/K value are colored in blue, while values more positive than 4.6 ppb/K have been colored in red. In (b), residues with an N–O distance <3.5 Å (indicative for hydrogen bonding) are color-coded in red and residues with a distance >3.5 Å are color-coded in blue. (c) and (d) Zoomed view of the structures shown in (a). Hydrogen bonds are shown by black lines. The structures were visualized with Chimera.58
Conclusions
We have shown that temperature coefficients of a proton chemical shift are accessible by solid-state NMR due to recent technical improvements comprising fast MAS experiments and a magnet bore temperature control unit. Such a system reduces magnetic-field fluctuations and corresponding equilibration times upon temperature changes induced by magnetic susceptibility changes. The temperature coefficients have been established for the small organic molecule ortho-phospho-l-serine and the model protein ubiquitin. A criterion similar to the one used in solution has been found to predict if a hydrogen atom is engaged in a hydrogen bond or not. Such temperature coefficients determined from solid-state NMR data might complement existing strategies to directly detect hydrogen bonds by solid-state NMR (e.g., exploring the proton chemical-shift value, the chemical-shielding anisotropy, or J-coupling based polarization transfer schemes) and demonstrate the high spectral resolution that can be obtained in proton-detected solid-state NMR spectroscopy.
Acknowledgments
This work was supported by the ETH Research Grant ETH-43 17-2 (T.W.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, project number 455240421 and Heisenberg fellowship, T.W.), the ERC Advanced Grant (B.H.M., grant number 741863, Faster), the Swiss National Science Foundation (B.H.M., grant number 200020_159707 and 200020-188711), the ANRS (A.B., ECTZ100488 and ECTZ71388), and the ANR (A.B., ANR-19-CE11-0023). The authors thank Dr. Patrick Wikus (Bruker Biospin) for helpful discussion.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpcb.1c04061.
Additional figures displaying differences in temperature coefficients of ubiquitin between the solid and solution states and the temperature coefficients plotted on the PDB structure as determined from solid-state or solution-state NMR (PDF)
Author Contributions
§ A.A.M. and L.A.V. contributed equally to this work. L.L. and R.C. prepared the samples. A.A.M., L.A.V., and T.W. performed the NMR experiments. A.A.M., L.A.V., M.E., A.B., B.H.M., and T.W. analyzed the data. All authors contributed to the writing of the manuscript. B.H.M. and T.W. designed and supervised the research.
The authors declare no competing financial interest.
Supplementary Material
References
- Jiang D.; Chen H.; Chen Z.; Zheng Z. In The Digital Field-Frequency Lock System of High-Resolution NMR Spectrometer, 2010 International Conference on Electrical and Control Engineering, June 25–27, 2010; pp 2328–2331.
- Najbauer E. E.; Andreas L. B. Correcting for magnetic field drift in magic-angle spinning NMR datasets. J. Magn. Reson. 2019, 305, 1–4. 10.1016/j.jmr.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Yip K. M.; Fischer N.; Paknia E.; Chari A.; Stark H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. 10.1038/s41586-020-2833-4. [DOI] [PubMed] [Google Scholar]
- Susi H.The Strength of Hydrogen Bonding: Infrared Spectroscopy. In Methods in Enzymology; Academic Press, 1972; Vol. 26, pp 381–391. [DOI] [PubMed] [Google Scholar]
- Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta, Bioenerg. 2007, 1767, 1073–1101. 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Wagner G.; Pardi A.; Wuethrich K. Hydrogen bond length and proton NMR chemical shifts in proteins. J. Am. Chem. Soc. 1983, 105, 5948–5949. 10.1021/ja00356a056. [DOI] [Google Scholar]
- Brunner E.; Sternberg U. Solid-state NMR investigations on the nature of hydrogen bonds. Prog. Nucl. Magn. Reson. Spectrosc. 1998, 32, 21–57. 10.1016/S0079-6565(97)00018-6. [DOI] [Google Scholar]
- Asakura T.; Taoka K.; Demura M.; Williamson M. P. The relationship between amide proton chemical shifts and secondary structure in proteins. J. Biomol. NMR 1995, 6, 227–236. 10.1007/BF00197804. [DOI] [PubMed] [Google Scholar]
- Wiegand T.; Malär A. A.; Cadalbert R.; Ernst M.; Böckmann A.; Meier B. H. Asparagine and Glutamine Side-Chains and Ladders in HET-s(218–289) Amyloid Fibrils Studied by Fast Magic-Angle Spinning NMR. Front. Mol. Biosci. 2020, 7, 582033 10.3389/fmolb.2020.582033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins S. J.; Wüthrich K. Ring current effects in the conformation dependent NMR chemical shifts of aliphatic protons in the basic pancreatic trypsin inhibitor. Biochim. Biophys. Acta, Protein Struct. 1979, 576, 409–423. 10.1016/0005-2795(79)90416-1. [DOI] [PubMed] [Google Scholar]
- Case D. A. Calibration of ring-current effects in proteins and nucleic acids. J. Biomol. NMR 1995, 6, 341–346. 10.1007/BF00197633. [DOI] [PubMed] [Google Scholar]
- Sitkoff D.; Case D. A. Density Functional Calculations of Proton Chemical Shifts in Model Peptides. J. Am. Chem. Soc. 1997, 119, 12262–12273. 10.1021/ja9721430. [DOI] [Google Scholar]
- Wishart D. S.; Sykes B. D.; Richards F. M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991, 222, 311–333. 10.1016/0022-2836(91)90214-Q. [DOI] [PubMed] [Google Scholar]
- Barfield M. Structural Dependencies of Interresidue Scalar Coupling h3JNC‘ and Donor 1H Chemical Shifts in the Hydrogen Bonding Regions of Proteins. J. Am. Chem. Soc. 2002, 124, 4158–4168. 10.1021/ja012674v. [DOI] [PubMed] [Google Scholar]
- Brown S. P.; Perez-Torralba M.; Sanz D.; Claramunt R. M.; Emsley L. The Direct Detection of a Hydrogen Bond in the Solid State by NMR Through the Observation of a Hydrogen-bond Mediated 15N-15N J Coupling. J. Am. Chem. Soc. 2002, 124, 1152–1153. 10.1021/ja0172262. [DOI] [PubMed] [Google Scholar]
- Schanda P.; Huber M.; Verel R.; Ernst M.; Meier B. H. Direct Detection of 3hJNC′ Hydrogen-Bond Scalar Couplings in Proteins by Solid-State NMR Spectroscopy. Angew. Chem., Int. Ed. 2009, 48, 9322–9325. 10.1002/anie.200904411. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Sudmeier J. L.; Bachovchin W. W.; Levitt M. H. Measurement of NH Bond Lengths by Fast Magic-Angle Spinning Solid-State NMR Spectroscopy: A New Method for the Quantification of Hydrogen Bonds. J. Am. Chem. Soc. 2001, 123, 11097–11098. 10.1021/ja016328p. [DOI] [PubMed] [Google Scholar]
- Schnell I.; Saalwächter K. 15N–H Bond Length Determination in Natural Abundance by Inverse Detection in Fast-MAS Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2002, 124, 10938–10939. 10.1021/ja026657x. [DOI] [PubMed] [Google Scholar]
- Cierpicki T.; Otlewski J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR 2001, 21, 249–261. 10.1023/A:1012911329730. [DOI] [PubMed] [Google Scholar]
- Dyson H. J.; Rance M.; Houghten R. A.; Lerner R. A.; Wright P. E. Folding of immunogenic peptide fragments of proteins in water solution: I. Sequence requirements for the formation of a reverse turn. J. Mol. Biol. 1988, 201, 161–200. 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- Ohnishi M.; Urry D. W. Temperature dependence of amide proton chemical shifts: The secondary structures of gramicidin S and valinomycin. Biochem. Biophys. Res. Commun. 1969, 36, 194–202. 10.1016/0006-291X(69)90314-3. [DOI] [PubMed] [Google Scholar]
- Kopple K. D.; Ohnishi M.; Go A. Conformations of cyclic peptides. III. Cyclopentaglycyltyrosyl and related compounds. J. Am. Chem. Soc. 1969, 91, 4264–4272. 10.1021/ja01043a040. [DOI] [PubMed] [Google Scholar]
- Andersen N. H.; Neidigh J. W.; Harris S. M.; Lee G. M.; Liu Z.; Tong H. Extracting Information from the Temperature Gradients of Polypeptide NH Chemical Shifts. 1. The Importance of Conformational Averaging. J. Am. Chem. Soc. 1997, 119, 8547–8561. 10.1021/ja963250h. [DOI] [Google Scholar]
- Tomlinson J. H.; Williamson M. P. Amide temperature coefficients in the protein G B1 domain. J. Biomol. NMR 2012, 52, 57–64. 10.1007/s10858-011-9583-4. [DOI] [PubMed] [Google Scholar]
- Schaefer T.; Kotowycz G. Temperature dependence of chemical shifts of protons in hydrogen bonds. Experimental evidence on an intramolecular hydrogen bond. Can. J. Chem. 1968, 46, 2865–2868. 10.1139/v68-474. [DOI] [Google Scholar]
- Davis J. C.; Pitzer K. S. Nuclear Magnetic Resonance Studies of Hydrogen Bonding. I. Carboxylic Acids1. J. Phys. Chem. A 1960, 64, 886–892. 10.1021/j100836a016. [DOI] [Google Scholar]
- Liddel U.; Ramsey N. F. Temperature Dependent Magnetic Shielding in Ethyl Alcohol. J. Chem. Phys. 1951, 19, 1608. 10.1063/1.1748133. [DOI] [Google Scholar]
- Arnold J. T.; Packard M. E. Variations in Absolute Chemical Shift of Nuclear Induction Signals of Hydroxyl Groups of Methyl and Ethyl Alcohol. J. Chem. Phys. 1951, 19, 1608–1609. 10.1063/1.1748134. [DOI] [Google Scholar]
- Muller N.; Reiter R. C. Temperature Dependence of Chemical Shifts of Protons in Hydrogen Bonds. J. Chem. Phys. 1965, 42, 3265–3269. 10.1063/1.1696408. [DOI] [Google Scholar]
- Webber A. L.; Elena B.; Griffin J. M.; Yates J. R.; Pham T. N.; Mauri F.; Pickard C. J.; Gil A. M.; Stein R.; Lesage A.; et al. Complete 1H resonance assignment of β-maltose from 1H–1H DQ-SQ CRAMPS and 1H (DQ-DUMBO)–13C SQ refocused INEPT 2D solid-state NMR spectra and first principles GIPAW calculations. Phys. Chem. Chem. Phys. 2010, 12, 6970–6983. 10.1039/c001290d. [DOI] [PubMed] [Google Scholar]
- Mann S. K.; Devgan M. K.; Franks W. T.; Huband S.; Chan C. L.; Griffith J.; Pugh D.; Brooks N. J.; Welton T.; Pham T. N.; et al. MAS NMR Investigation of Molecular Order in an Ionic Liquid Crystal. J. Phys. Chem. B 2020, 124, 4975–4988. 10.1021/acs.jpcb.0c02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard C. J.; Salager E.; Pintacuda G.; Elena B.; Emsley L. Resolving Structures from Powders by NMR Crystallography Using Combined Proton Spin Diffusion and Plane Wave DFT Calculations. J. Am. Chem. Soc. 2007, 129, 8932–8933. 10.1021/ja071829h. [DOI] [PubMed] [Google Scholar]
- Baxter N. J.; Williamson M. P. Temperature dependence of 1H chemical shifts in proteins. J. Biomol. NMR 1997, 9, 359–369. 10.1023/A:1018334207887. [DOI] [PubMed] [Google Scholar]
- Cierpicki T.; Zhukov I.; Byrd R. A.; Otlewski J. Hydrogen Bonds in Human Ubiquitin Reflected in Temperature Coefficients of Amide Protons. J. Magn. Reson. 2002, 157, 178–180. 10.1006/jmre.2002.2597. [DOI] [PubMed] [Google Scholar]
- Cordier F.; Grzesiek S. Temperature-dependence of protein hydrogen bond properties as studied by high-resolution NMR11Edited by P. E. Wright. J. Mol. Biol. 2002, 317, 739–752. 10.1006/jmbi.2002.5446. [DOI] [PubMed] [Google Scholar]
- Schubert M.; Manolikas T.; Rogowski M.; Meier B. H. Solid-state NMR spectroscopy of 10% 13C labeled ubiquitin: spectral simplification and stereospecific assignment of isopropyl groups. J. Biomol. NMR 2006, 35, 167–173. 10.1007/s10858-006-9025-x. [DOI] [PubMed] [Google Scholar]
- Gardiennet C.; Schütz A. K.; Hunkeler A.; Kunert B.; Terradot L.; Böckmann A.; Meier B. H. A Sedimented Sample of a 59 kDa Dodecameric Helicase Yields High-Resolution Solid-State NMR Spectra. Angew. Chem., Int. Ed. 2012, 51, 7855–7858. 10.1002/anie.201200779. [DOI] [PubMed] [Google Scholar]
- Bertini I.; Luchinat C.; Parigi G.; Ravera E.; Reif B.; Turano P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 10396–10399. 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand T.; Lacabanne D.; Torosyan A.; Boudet J.; Cadalbert R.; Allain F. H.; Meier B. H.; Bockmann A. Sedimentation Yields Long-Term Stable Protein Samples as Shown by Solid-State NMR. Front. Mol. Biosci. 2020, 7, 17 10.3389/fmolb.2020.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq L.; Schledorn M.; Wang S.; Smith-Penzel S.; Malär A. A.; Callon M.; Nassal M.; Meier B. H.; Böckmann A. 100 kHz MAS Proton-Detected NMR Spectroscopy of Hepatitis B Virus Capsids. Front. Mol. Biosci. 2019, 6, 58 10.3389/fmolb.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckmann A.; Gardiennet C.; Verel R.; Hunkeler A.; Loquet A.; Pintacuda G.; Emsley L.; Meier B.; Lesage A. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR 2009, 45, 319–327. 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- Fogh R.; Ionides J.; Ulrich E.; Boucher W.; Vranken W.; Linge J. P.; Habeck M.; Rieping W.; Bhat T. N.; Westbrook J.; et al. The CCPN project: an interim report on a data model for the NMR community. Nat. Struct. Biol. 2002, 9, 416–418. 10.1038/nsb0602-416. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins: Struct., Funct., Bioinf. 2005, 59, 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Stevens T.; Fogh R.; Boucher W.; Higman V.; Eisenmenger F.; Bardiaux B.; van Rossum B.-J.; Oschkinat H.; Laue E. A software framework for analysing solid-state MAS NMR data. J. Biomol. NMR 2011, 51, 437–447. 10.1007/s10858-011-9569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.; Kümmerle R.; Qiu X.; Redwine D.; Cong R.; Taha A.; Baugh D.; Winniford B. A new decoupling method for accurate quantification of polyethylene copolymer composition and triad sequence distribution with 13C NMR. J. Magn. Reson. 2007, 187, 225–233. 10.1016/j.jmr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Vinod Chandran C.; Madhu P. K.; Kurur N. D.; Bräuniger T. Swept-frequency two-pulse phase modulation (SWf-TPPM) sequences with linear sweep profile for heteronuclear decoupling in solid-state NMR. Magn. Reson. Chem. 2008, 46, 943–947. 10.1002/mrc.2285. [DOI] [PubMed] [Google Scholar]
- Malär A. A.; Smith-Penzel S.; Camenisch G. M.; Wiegand T.; Samoson A.; Bockmann A.; Ernst M.; Meier B. H. Quantifying proton NMR coherent linewidth in proteins under fast MAS conditions: a second moment approach. Phys. Chem. Chem. Phys. 2019, 21, 18850–18865. 10.1039/C9CP03414E. [DOI] [PubMed] [Google Scholar]
- Sternberg U.; Witter R.; Kuprov I.; Lamley J. M.; Oss A.; Lewandowski J. R.; Samoson A. 1H line width dependence on MAS speed in solid state NMR – Comparison of experiment and simulation. J. Magn. Reson. 2018, 291, 32–39. 10.1016/j.jmr.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Schledorn M.; Malar A. A.; Torosyan A.; Penzel S.; Klose D.; Oss A.; Org M. L.; Wang S.; Lecoq L.; Cadalbert R.; et al. Protein NMR Spectroscopy at 150 kHz Magic-Angle Spinning Continues To Improve Resolution and Mass Sensitivity. ChemBioChem 2020, 21, 2540–2548. 10.1002/cbic.202000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuga A.; Brunner E. Phosphorylated amino acids: model compounds for solid-state 31P NMR spectroscopic studies of proteins. Magn. Reson. Chem. 2004, 42, 369–372. 10.1002/mrc.1356. [DOI] [PubMed] [Google Scholar]
- Potrzebowski M. J.; Assfeld X.; Ganicz K.; Olejniczak S.; Cartier A.; Gardiennet C.; Tekely P. An Experimental and Theoretical Study of the 13C and 31P Chemical Shielding Tensors in Solid O-Phosphorylated Amino Acids. J. Am. Chem. Soc. 2003, 125, 4223–4232. 10.1021/ja029840z. [DOI] [PubMed] [Google Scholar]
- Duma L.; Abergel D.; Tekely P.; Bodenhausen G. Proton chemical shift anisotropy measurements of hydrogen-bonded functional groups by fast magic-angle spinning solid-state NMR spectroscopy. Chem. Commun. 2008, 2361–2363. 10.1039/b801154k. [DOI] [PubMed] [Google Scholar]
- Sundaralingam M.; Putkey F. F. Molecular structures of amino acids and peptides. II. A redetermination of the crystal structure of l-O-serine phosphate. A very short phosphate–carboxyl hydrogen bond. Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem. 1970, 26, 790–800. 10.1107/S0567740870003138. [DOI] [PubMed] [Google Scholar]
- Baxter N. J.; Hosszu L. L. P.; Waltho J. P.; Williamson M. P. Characterisation of low free-energy excited states of folded proteins11Edited by P. E. Wright. J. Mol. Biol. 1998, 284, 1625–1639. 10.1006/jmbi.1998.2265. [DOI] [PubMed] [Google Scholar]
- Karle I. L. Hydrogen bonds in molecular assemblies of natural, synthetic and ‘designer’ peptides. J. Mol. Struct. 1999, 474, 103–112. 10.1016/S0022-2860(98)00564-X. [DOI] [Google Scholar]
- Jeffrey G. A.An Introduction to Hydrogen Bonding; Oxford University Press, 1997. [Google Scholar]
- Huang K. Y.; Amodeo G. A.; Tong L.; McDermott A. The structure of human ubiquitin in 2-methyl-2,4-pentanediol: A new conformational switch. Protein Sci. 2011, 20, 630–639. 10.1002/pro.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.