Abstract
Biogenesis of ribosomal subunits involves enzymatic modifications of rRNA that fine-tune functionally important regions. The universally conserved prokaryotic dimethyltransferase KsgA sequentially modifies two universally conserved adenosine residues in helix 45 of the small ribosomal subunit rRNA, which is in proximity of the decoding site. Here we present the cryo-EM structure of Escherichia coli KsgA bound to an E. coli 30S at a resolution of 3.1 Å. The high-resolution structure reveals how KsgA recognizes immature rRNA and binds helix 45 in a conformation where one of the substrate nucleotides is flipped-out into the active site. We suggest that successive processing of two adjacent nucleotides involves base-flipping of the rRNA, which allows modification of the second substrate nucleotide without dissociation of the enzyme. Since KsgA is homologous to the essential eukaryotic methyltransferase Dim1 involved in 40S maturation, these results have also implications for understanding eukaryotic ribosome maturation.
Graphical Abstract
Graphical Abstract.
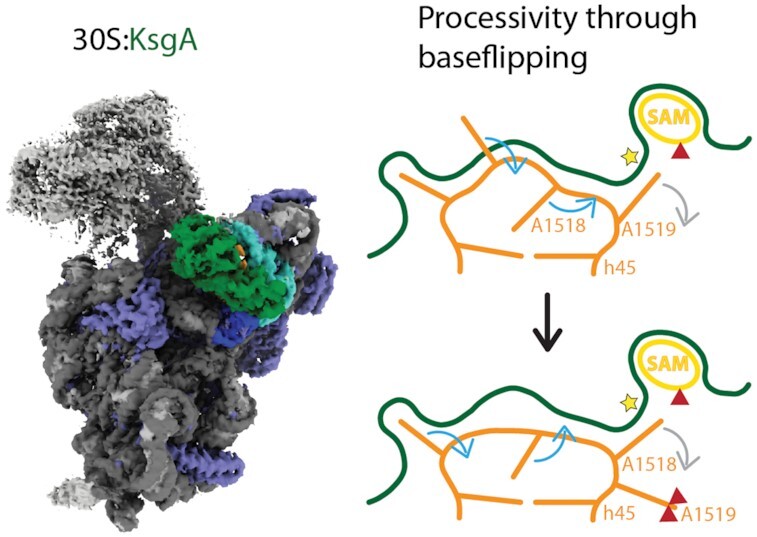
The high resolution structure of KsgA bound to the 30S ribosome reveals how KsgA recognizes immature ribosomal RNA and allows deduction of a mechanism for processive dimethylation of its two target rRNA adenosines. We suggest that the mechanism involves base-flipping of the rRNA, which allows modification of the second substrate nucleotide without dissociation of the enzyme.
INTRODUCTION
During biogenesis of the bacterial small ribosomal subunit (30S) the ribosomal RNA is modified in functionally important areas (1). KsgA, also known as RsmA, is a universally conserved SAM-dependent dimethyltransferase, which specifically modifies nucleotides A1518 and A1519 in the Escherichia coli 16S rRNA at position N6 (2). KsgA is the only methyltransferase involved in 30S biogenesis of E. coli that targets two adjacent nucleotides and modifies each target nucleotide twice (3). These nucleotides are located in the apical loop of helix 45, one of the most highly conserved areas of the rRNA (4), which plays a role in shaping the geometry of the nearby decoding center (5). The dimethylation of the small ribosomal subunit in this location is conserved throughout evolution (4) with very few exceptions known in plastid (6,7) and mitochondrial ribosomes (8–10).
KsgA deficient E. coli strains display a higher error rate during translation (11), increased resistance towards the antibiotic kasugamycin (12), and a cold-sensitive phenotype during growth (13). Deletion of KsgA impedes processing of the rRNA during ribosome maturation and leads to accumulation of the precursor 17S rRNA (13). This phenotype can be rescued by the overexpression of the homologous essential yeast enzyme Dim1 (14), which emphasizes a high degree of conservation not only for the modifying enzymes but also for the region of rRNA that the enzyme recognizes. Inactive mutants of KsgA bind to their substrate, do not dissociate and thereby inhibit the completion of 30S maturation (13), which suggests that KsgA has an additional function as a checkpoint factor during 30S biogenesis.
KsgA has homologs in all kingdoms of life, which share a similar structure: Dim1 in archaea (15) and eukaryotes (14), Pfc1 in chloroplasts (16) and mtTFB1 in mitochondria (17,18). A crystal structure of the E. coli wild type KsgA (19) reveals a two domain architecture, which can also be seen in the homologous structure of Thermus thermophilus KsgA (20). The enzymatic N-terminal domain (NTD) has a Rossmann like fold (21) and harbors the active site for methyl transfer, and the SAM binding pocket, whereas the smaller C-terminal domain (CTD) is involved in substrate rRNA recognition.
The interaction of KsgA with its substrate has been studied by determining the cryo-EM structure of KsgA bound to the submethylated 30S at 13.5 Å resolution (22). The structure revealed that KsgA binds to the intersubunit side at the edge of the platform close to the decoding center. The binding site of KsgA on the 30S overlaps with the binding site of initiation factor IF3 (23) and helix 69 of the large ribosomal subunit (22), which prevents the KsgA bound 30S from entering the translation cycle before modification of rRNA takes place. In addition, the position of KsgA excludes helix 44 from adopting its mature conformation and thereby blocks the formation of the decoding center and subunit joining for translation initiation. The enzymatic NTD of KsgA binds to the apical loop of helix 45 harboring the substrate adenosines, whereas the CTD binds to rRNA helices 24 and 27. KsgA likely recognizes the interacting helices 24 and 27, which is consistent with biochemical data suggesting KsgA acts only on the near-mature 30S where these helices are in close proximity (24). Structures of Saccharomyces cerevisiae small ribosomal subunit biogenesis intermediates (25,26) show that Dim1 is localized on the eukaryotic small ribosomal subunit at the platform, similar to KsgA on the bacterial 30S. However, the mechanistic details of how KsgA methylates two adjacent adenine bases successively, have so far not been described. Crystal structures of KsgA homologs in complex with rRNA fragments of helix 45 were solved (27,28), however, the observed interactions are unlikely to be specific since rRNA fragments are not a physiological substrate for KsgA. In these structures the substrate adenosine is located too far from the active site to be methylated and the helix 45 stem loop is seen in a different orientation compared to the ribosome bound structure. More recently, a structure of the human mitochondrial KsgA homolog mtTFB1 in complex with a helix 45 fragment has been reported (29), which shows the rRNA bound in a similar position as in the cryo-EM reconstruction at intermediate resolution (22). In this structure the substrate adenosine homologous to bacterial A1519 is flipped out of the substrate rRNA loop into the active site of the methyltransferase.
To investigate the nature of interactions of KsgA with its substrate near-mature 30S ribosomal subunit we reconstructed the structure of the KsgA:30S complex at 3.1 Å resolution using cryo-EM. The structure reveals the key regions of the enzyme responsible for binding to the rRNA on the platform of the small ribosomal subunit. Furthermore, the structure reveals a unique conformation of the substrate rRNA loop of helix 45 bound to the active site of KsgA with implications for understanding the mechanism of successive modification of consecutive adenines.
MATERIALS AND METHODS
| Material | Source | Identifier |
|---|---|---|
| Reagents | ||
| DNase I | Roche | 04716728001 |
| Grapheneoxide Suspension | Sigma | 763705 |
| HisTrap HP 5 ml | GE Healthcare | 17524801 |
| HiTrap SP 5 ml | GE Healthcare | 17115101 |
| HiLoadTM 16/60 SuperdexTM 75 prepgrade | GE Healthcare | 28989333 |
| Quantifoil R2/2 Cu holey carbon grids | Agar Scientific | AGS173-3 |
| Biological resources | ||
| E. coli BL21 (DE3) | Novagen | 69450–3 |
| E. coli ksgA− | Keio Collection | JW0050.3 |
| pET15b-His-KsgA | (30) | N/A |
| Computational resources | ||
| Relion | (31) | RRRID:SCR_016274 |
| MotionCorr 2.1 | (32) | N/A |
| GCTF | (33) | RRRID:SCR_016500 |
| Batchboxer | (34) | N/A |
| Coot | (35) | RRRID:SCR_014222 |
| PHENIX | (36) | RRRID:SCR_014224 |
| ResMAP | (37) | N/A |
| MolProbity | (38) | RRRID:SCR_014226 |
| UCSF Chimera | (39) | RRRID:SCR_004097 |
| UCSF ChimeraX | (40) | RRRID:SCR_015872 |
| PyMol | Molecular Graphics System, Version 1.8 Schrodinger, LLC | RRRID:SCR_000305 |
| Illustrator CC | Adobe | RRRID:SCR_010279 |
Preparation of KsgA
The purification of His-tagged KsgA from E. coli was inspired from a previously described purification (30). E. coli BL21 (DE3) cells were transformed with 10 ng of plasmid DNA and after recovery in antibiotic free LB medium (5 g/l yeast extract, 10 g/l tryptone, 10 g/l NaCl, autoclaved) for 30 min at 37°C an over-night pre-culture of 20 mL MDG-medium (2.5 g/l aspartic acid, 5 g/l glucose, 2 mM MgSO4, 25 mM Na2HPO4, 25 mM NaH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, filtered sterile) with 100 μg/ml ampicillin was inoculated and incubated at 37°C and 150 rpm over-night in a rotary shaker. Six liters of LB medium containing 100 μg/ml Ampicillin were inoculated with dense over-night pre-culture and grown at 37°C until absorbance at 600 nm reached 0.6. Expression was induced with 1 mM IPTG for 3 h. The following steps were performed at 4°C. The culture was harvested at 6000 rcf in an SLC-6000 rotor for 10 min. The supernatant was discarded and the cell pellet was resuspended in 50 mL of resuspension buffer (50 mM HEPES/KOH pH 7.6, 300 mM NaCl, 20 mM imidazole/HCl pH 8.0, 6 mM β-mercaptoethanol) supplemented with DNase I. Cells were lysed by sonicating 8 times 20 s at 50–70 W. The lysate was cleared by centrifuging 30 min at 48 000 rcf using an SS-34 rotor. The cleared supernatant was loaded onto a HisTrap HP 5 ml column (GE Healthcare) equilibrated in resuspension buffer using an Aekta Pure system (GE Healthcare) and washed with 5 CV (column volumes) resuspension buffer. Bound proteins were eluted with 5 CV elution buffer (50 mM HEPES/KOH pH 7.6, 50 mM NaCl, 250 mM imidazole/HCl pH 8.0, 6 mM β-Mercaptoethanol) and directly loaded onto a HiTrap SP 5 ml column (GE Healthcare) equilibrated in low salt buffer (50 mM HEPES/KOH pH 7.6, 50 mM NaCl, 6 mM β-mercaptoethanol). The HiTrap SP column was washed with 5 CV of low salt buffer and the protein was eluted in a linear gradient to high salt buffer (50 mM HEPES/KOH pH 7.6, 1 M NaCl, 6 mM β-mercaptoethanol) within 20 CV. Fractions of different sizes were collected throughout the purification. Ion exchange chromatography elution fractions containing KsgA were pooled and concentrated using a centrifugation filter with 10 kDa molecular weight cut off (Amicon) to a volume smaller than 2 ml. This sample was loaded onto a HiLoadTM 16/60 SuperdexTM 75 prepgrade column (GE Healthcare) equilibrated in SEC buffer (50 mM HEPES/KOH pH 7.6, 150 mM NH4Cl, 10 % (v/v) glycerol, 6 mM β-mercaptoethanol) and eluted at 0.2 ml/min SEC buffer while fractions of 1.2 ml were collected. The fractions containing KsgA were combined and exchanged into storage buffer using a centrifugation filter with a molecular weight cut off of 10 kDa.
Isolation of ribosomal subunits
Submethylated E. coli ribosomes were purified using similar methods described previously (41). KsgA deficient E. coli strain JW0050.3 from the Keio collection (42) was grown in 6 l of LB medium containing 50 μg/ml kanamycin at 37°C until absorbance at 600 nm reached 0.6. The following steps were performed at 4°C. The culture was harvested at 6000 rcf in a SLC-6000 rotor for 10 min. The supernatant was discarded and the cell pellet was resuspended in 20 ml of lysis buffer (50 mM HEPES/KOH pH 7.6, 10 mM MgCl2, 100 mM NH4Cl, 6 mM β-mercaptoethanol, 0.5 mM EDTA/NaOH pH 8.0) supplemented with DNase I. Cells were lysed by passing through a French Press (Thermo Fisher Scientific) twice at 1000 psi. The lysate was cleared by centrifuging twice 15 min at 20 000 rpm using a SS-34 rotor. The supernatant was layered on top of cushion buffer (50 mM HEPES/KOH pH 7.6, 10 mM MgCl2, 500 mM NH4Cl, 6 mM β-mercaptoethanol, 0.5 mM EDTA/NaOH pH 8.0, 40 % (w/v) sucrose) and ribosomes were pelleted by centrifuging 22 h at 29 000 rpm in a Type 70 Ti rotor (Beckmann Coulter). The pellets were dissolved in dissociating buffer (50 mM HEPES/KOH pH 7.6, 1 mM MgCl2, 100 mM NH4Cl, 6 mM β-mercaptoethanol) by incubation for 6 h on a rotary shaker. The subunits were separated by centrifuging 16 h at 26 000 rpm through a density gradient of 10 % (w/v) to 40 % (w/v) sucrose in dissociating buffer in a SW 32 Ti rotor (Beckmann Coulter). Sucrose gradients were prepared using a Gradient Master instrument (BioComp Instruments). The subunits were visualized by light scattering and extracted separately from the gradients.
Inactivation of 30S
For inactivation, small ribosomal subunits were exchanged into inactivation buffer (50 mM HEPES/KOH pH 7.6, 40 mM KCl, 0.5 mM MgCl2, 6 mM β-mercaptoethanol) using a centrifugation filter with a molecular weight cut off of 100 kDa (Amicon) and incubated 20 h at 4°C. Submethylated E. coli 30S were exchanged into storage buffer (50 mM HEPES/KOH pH 7.6, 40 mM KCl, 4 mM MgCl2, 6 mM β-Mercaptoethanol) using a centrifugation filter with 100 kDa molecular weight cut off (Amicon).
Cryo-EM
70 nM submethylated E. coli 30S were mixed with 700 nM KsgA and incubated 10 min at 37°C. The sample was spun 5 min at 16 000 rcf and 4°C to remove aggregates. R2/2 holey carbon grids (Quantifoil) washed with ethylacetate were glow discharged for 30 s at negative 15 mA. For each grid 3 μl of 0.2 mg/ml grapheneoxide suspension was applied on the carbon side of the grid and excess liquid was blotted after 4 min incubation at room temperature. Grids were washed once with 20 μl of MilliQ, excess liquid was blot and grids were dried. The grid was transferred to a Vitrobot (FEI Company) at 4°C and 100% relative humidity and 5 μl sample were incubated for 30 s on the graphene oxide coated grids. Grids were blotted for 6 and 9 s and frozen in a 1:2 mixture of liquid ethane and propane. The grids were transferred to a Titan Krios cryo-electron microscope (FEI Company) operated at 300 kV and a magnification of 100720 × corresponding to a physical pixel size of 1.39 Å. Micrographs were recorded from two grids on a Falcon III direct electron detector in integration mode using dose fractionation with 25 frames and a total average dose of 35 e−/Ų. Defocus values were in the range from −0.7 μm to −3 μm (Supplementary Table S1).
Image processing
The dose fractionated image stacks were aligned, dose weighted and integrated using MotionCorr 2.1 (32). CTF estimation was performed using GCTF v 1.06 (33) and power spectra showing signs of astigmatism, ice contamination or drift were discarded. Particles were semi-automatically identified and picked using Batchboxer (34), providing a cytoplasmic, yeast pre-40S ribosome reconstruction (43) as a reference. Particles were extracted from all images and subjected to 2D classification using Relion 2.1 (31). Projections showing small subunits were selected for 3D classifications. The first classification was focused on the body and KsgA. A second classification was focused on KsgA and its surrounding. All 3D classifications were carried out without image alignment following a 3D refinement of the 30S body. After sorting out micrographs with too low defocus, the remaining 165073 particles were subjected to a final 3D refinement with a mask and initial model prepared from a 3D class from earlier in the processing (data processing workflow: Supplementary Figure S1; refinement parameters: Supplementary Table S1).
Model building and validation
Model building was performed in Coot 0.9.3 (35). The structure was built using an empty 30S from an E. coli ribosome (44) (PDBID: 4YBB) and an E. coli KsgA crystal structure (19) (PDBID:1QYR) as a starting base. The rRNA domains and the individual proteins of the body—including KsgA—were rigid body fitted into the EM-map in UCSF Chimera (39). The modified nucleotides and amino acids in the ribosome were replaced by their unmodified counterpart. All residues were real space refined into the density and removed if not resolved in the EM-map. The resulting model was refined, running real space refinement with PHENIX 1.19-4080 (45). In order to preserve the secondary structure also in regions with lower local resolution, Ramachandran restraints were enabled. Structure validation was carried out with MolProbity (46) (Supplementary Table S1) and by comparing the half map vs. half map FSC curve at a threshold of FSC = 0.143 with the map vs. model FSC curve at a threshold of FSC = 0.5 (Supplementary Figure S3).
The alternative conformation of the helix 45 tetraloop with A1518 in the active site was modelled in Coot 0.9.3 (35), using the original structure as a starting point. Base flipping and building of the alternative conformation could be performed within the geometric restraints of coot.
Figures were prepared with PyMol and ChimeraX (40).
RESULTS
Structure of the KsgA:30S complex
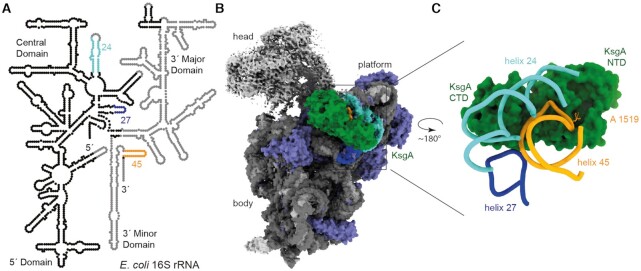
Ribosomes lacking the dimethylations on 30S helix 45 were isolated from a KsgA deficient E. coli strain and subunits were separated by magnesium depletion and subsequent sucrose gradient centrifugation. The isolated small subunits were brought into a translationally inactive conformation mimicking an assembly intermediate state by further magnesium depletion (47), which allows binding of KsgA and has been shown to function as a substrate in methylation assays (22,24,48,49). The inactivation step was described to cause destabilization of helix 44, which possibly mimics a 30S biogenesis intermediate and provides required space for KsgA to bind to the 30S platform (22,50). The destabilized helices 44 and 28 at the head to body junction furthermore cause extraordinary flexibility of the 30S head (22,50). KsgA was produced and isolated from E. coli and assembled with the 30S by mixing 10-fold excess of the enzyme with the ribosomal subunit. The structure of the KsgA:30S complex was determined to a resolution of 3.1 Å using single particle cryo-EM. The high resolution of the reconstruction for most parts of the 30S body allowed rebuilding and refinement of a molecular model for the ribosomal proteins and RNA, and KsgA (Figure 1B, Supplementary Figures S2 and S5C). The 30S head displays high flexibility and the density for helix 44 is missing, as observed before for the magnesium depleted 30S subunit (22,50), therefore no molecular model was built for these parts (Figure 1A and B). KsgA is bound on the 30S ribosomal subunit platform (Figure 1B). The high-resolution reconstruction reveals that KsgA binds exclusively rRNA helices 24, 27 and 45. The catalytic NTD of KsgA binds to the substrate helix 45, whereas the CTD binds to an rRNA helices 24 and 27 (Figure 1C).
Figure 1.
Structure of the KsgA:30S complex. (A) Secondary structure diagram of the E. coli 16S rRNA highlights helices 24 (turquoise), 27 (dark blue) and 45 (orange) which form the interaction patch for KsgA in the context of a near mature 30S. Nucleotides that are not included in the molecular structure are shown in light grey. (B) Map of KsgA (green) in complex with the hypomethylated 30S. The rRNA is colored in grey, helices that interact with KsgA are highlighted (helix 24 turquoise, helix 27 dark blue, helix 45 orange) and ribosomal proteins are colored in purple. Head and toe of the 30S could not be built, due to high flexibility in these regions, in these areas the map is colored in light grey. Platform, body and head of the 30S and KsgA are indicated. (C) Molecular model of KsgA interaction with the 30S seen from the direction of the small ribosomal subunit shows rRNA helices 24, 27 and 45 bound to KsgA.
KsgA-CTD:h24:h27 interaction confers substrate specificity
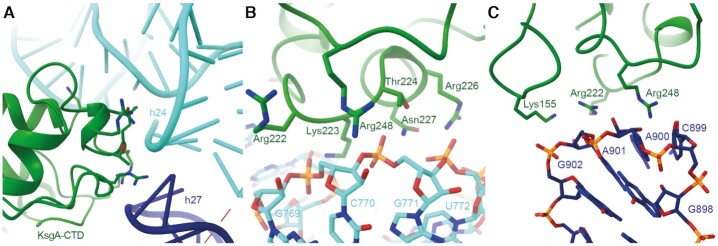
The KsgA-CTD confers substrate specificity by recognizing helices 24 and 27 that are positioned next to each other in a conformation defined by the near mature fold of the 30S subunit (Figure 2A). This is consistent with biochemical results indicating that KsgA does not act on isolated 16S rRNA (51). The structure reveals that positively charged amino acids in the CTD of KsgA (Arg222, Lys223, Arg226, Arg248) interact with the negatively charged backbone of helix 24 (nucleotides 769–771) (Figure 2B). Furthermore, amino acids Thr224 and Asn227 are in hydrogen bonding distance to helix 24. The negatively charged backbone of helix 27 (nucleotides 899–902) interacts with positively charged amino acids (Lys155, Arg222, Arg248) of KsgA (Figure 2C).
Figure 2.
Substrate specificity through tripartite interaction between KsgA-CTD:h24:h27 (A) KsgA binds at the intersection of helices 24 (turquoise) and 27 (dark blue) to the backbones of both. (B) Interactions of KsgA (green) and helix 24 (turquoise). Residues involved in interaction are indicated. (C) Interactions of KsgA (green) and helix 27 (dark blue). Residues involved in interaction are indicated.
KsgA binds to substrate adenosine A1519 in flipped out conformation
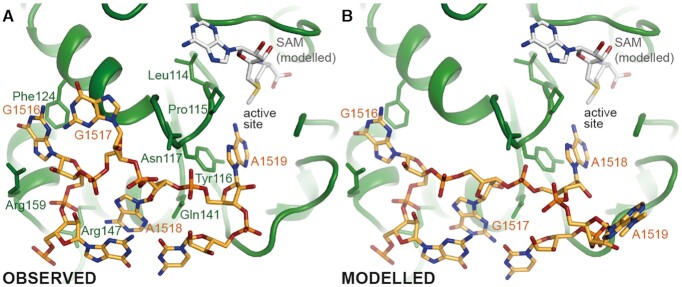
A positively charged cleft on the surface of KsgA stretches out from the CTD across the NTD where the active-site is located. This cleft is lined with positively charged amino acids of KsgA (Arg147, Lys155, Arg159, Arg221, Arg222) that interact with the negatively charged phosphate backbone of helix 45. The helix 45 loop, including the two substrate adenosines, is bound by the catalytic NTD of KsgA in the area of the active site (Figure 3A and Supplementary Figure S5A). The nucleobase of G1516 is bound in a pocket where it π-stacks onto amino acid Phe124 on the one side and is involved in π-cation interactions with Arg159 on the other side, whereas its phosphate interacts with Arg147. The ribose of G1517 is in hydrogen bonding distance with Asn117. The nucleobase of A1518 stacks onto G1515 of helix 45, whereas the ribose-phosphate backbone is in hydrogen bonding distance with Asn117 and Gln141. One of the substrate residues, A1519, is positioned in the active site by π-stacking of the adenine onto Tyr116 and through hydrogen bonding of the N6 and N7 to the peptide backbone of amino acids Leu114 and Tyr116, respectively. A1519 is positioned in the active site of KsgA such that it could be methylated by a bound SAM. Although SAM was not present in the sample, its position could be modelled with confidence into the structure by using the crystal structure of the archaeal homologue Dim1 in complex with SAM as a guide (52). The position is in line with the structure of the eukaryotic mitochondrial homolog mtTFB1 in complex with SAM and helix 45 (29) (Supplementary Figure S4). The fitted SAM is in close proximity to the active site pocket (Figure 3A) and the distance between donor methyl group of SAM and acceptor N6 of the adenosine is 2.7 Å, which would be optimal for enzymatic transfer of the methyl group. The structure of the complex reveals that SAM could dissociate and associate from KsgA while bound to the 30S, allowing the sequential dimethylation of both target adenine bases without dissociation of KsgA, as the access to the binding pocket is not obstructed by the small subunit. This is in agreement with biochemical data indicating that KsgA is a processive enzyme which produces primarily the dimethylated adenosines without releasing the monomethylated intermediate (49,53).
Figure 3.
Binding of Substrate helix 45 by KsgA-NTD and its possible alternative conformation (A) Position of the helix 45 apical loop (orange) at the KsgA (green) interface. The residues involved in interaction of KsgA and helix 45 apical loop are indicated. The modelled binding pocket of SAM (grey) is in proximity to the nucleotide A1519, which is proposed to be modified first. The active site pocket is indicated. (B) Alternative conformation of helix 45 with A1518 in the active site pocket, A1519 moved out of the pocket and G1517 replacing the former position of A1518.
Model for modification of the second substrate adenosine A1518
In our structure adenosine A1519 is bound in the active site of KsgA (Figure 3A and Supplementary Figure S5A, B), which indicates that KsgA dimethylates A1519 first. This is in agreement with biochemical studies, where under limiting SAM levels KsgA was observed to methylate only A1519 (6) and where rRNA mutation studies indicated that A1519 is the kinetically preferred substrate (54). Furthermore, the crystal structure of human mitochondrial KsgA homolog mtTFB1 in complex with SAM and helix 45 displays the same conformation of flipped out A1519 (29) (Supplementary Figure S4). Based on mutational studies it has been suggested that the first methylation weakens the hydrogen bonding with the conserved KsgA motif IV, while the second methylation abolishes all hydrogen bonding interactions of the substrate adenosine and would expel the modified nucleotide from the active site pocket (3). Successive modification of A1518 requires it to be positioned into the active site. The conformation of the helix 45 - KsgA interface suggests a plausible alternative conformation with A1518 placed in the active site (Figure 3B). It requires re-arrangement of the positions of nucleotides G1517–A1519, which is geometrically reasonable and shows no clashes after regularization. The geometry in the active site occupied by A1518 in this proposed model is comparable to that of A1519 in our reconstruction, which would allow SAM-dependent dimethylation of A1518 by a similar mechanism.
DISCUSSION
Base flipping mechanism of successive modification
Our structure reveals a partly flipped out conformation of the substrate loop bound to KsgA suggesting that the methylations occur by sequential base flipping rearrangements in helix 45. Base flipping has first been described to make a single base from a DNA double helix accessible for modification in the bacterial HhaI DNA cytosine-5-methyltransferase (55). It has since been discovered to be a well-established mechanism among DNA methyltransferases and other single base targeted enzymes (56,57). Among RNA methyltransferases base flipping has also been discovered for example in RumA (58), TrmA (59) and METTL16 (60). The more versatile secondary structures of RNA allow a wider variety of base flipping conformational changes than discovered in DNA. The mechanism we suggest for KsgA:h45 uses two coupled base flippings upon KsgA binding, substrate relocation and reorganization into the mature conformation:
KsgA binding
Comparison of the conformation in the hypo-methylated 30S of T. thermophilus (61) with the conformation of helix 45 bound to KsgA reveals two base flipping rearrangements (Figure 4A and B). The hypomethylated helix 45 tetraloop, folds into a classical GNRA tetraloop conformation (62), where G1516 stacks onto G1515 in the loop and nucleobases 1517–1519 form a three base stack in flipped out conformation (61) (Figure 4A). In our structure, G1516 is in a flipped out conformation tightly anchored in a small KsgA binding pocket (Figure 4B) as well as the substrate base A1519 is in the active site, which is in line with the structural findings in the human mitochondrial KsgA homolog mtTFB1 with helix 45 (29) (Supplementary Figure S4). It allows the hypothesis that G1516 anchoring is likely required for correct positioning of the substrate bases. Its flipping is probably energetically compensated by (a) the stronger interaction of G1516 with its KsgA binding pocket than with G1515 and (b) the second base flip of A1518 to replace G1516 on top of the last base pair of h45 stem. While replacement of original base stacking interactions with amino acid side chains has been observed in several nucleic acid modifying enzymes to compensate the energetically expensive base flipping process (55,63,64), the RNA methyltransferases RumA and TrmA also utilize replacement with other bases as seen here for KsgA (58,59). A1519 inserts into the active site of KsgA, and the third member of the orginal base stack G1517 interacts with the surface of the enzyme (Figure 4B).
Figure 4.
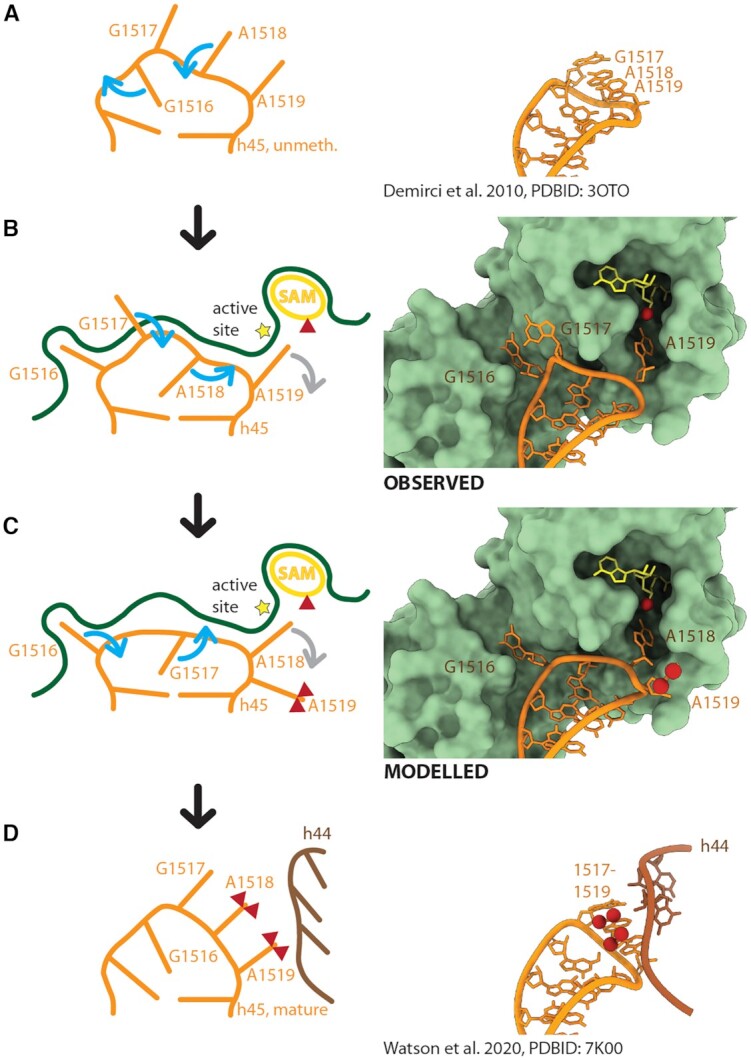
Proposed Base Flipping Mechanism for Successive Dimethylation Left panels: Mechanistic representation indicating helix 45 (orange) conformational changes upon KsgA (green) binding (A, B), substrate relocation (B, C) and adoption of the mature conformation (C, D) with helix 44 (brown) interaction. Base-flipping is indicated by blue arrows and base expulsion from the active site by grey arrows. Co-factor SAM is depicted in yellow and methyl groups are represented by red triangles. Right panels: (A) Structure of the hypomethylated helix 45 (61), (B) observed structure of KsgA bound helix 45 and (C) modelled structure of relocated RNA, (D) structure of mature helix 45 (67). The conformation of mature helix 45 allows—in contrast to the hypomethylated conformation—hydrogen bond interactions with helix 44 (brown). Helix 45 is depicted in orange, helix 44 in brown, KsgA in pale green, SAM in yellow, methyl groups are indicated by red spheres.
Substrate relocation
After A1519 is dimethylated, consuming two SAM molecules, resulting changes in polarity and dimension of the nucleobase likely cause its expulsion from the active site (Figure 4B and C) (58,65). A1519 movement then might be able to build up a tension on the helix 45 backbone, which causes A1518 to flip out of the tetraloop into the active site and G1517 into the tetraloop, replacing A1518 (Figure 4B). According to this scenario G1516 still remains in its flipped-out conformation, anchoring KsgA to helix 45 (Figure 4C). A relatively higher conformational flexibility of bases G1517 and A1518 compared to G1516 bound to KsgA was also observed in crystal structures of the homologous mitochondrial system (29), supporting our mechanistic model.
Reorganization into mature conformation
After A1518 has been dimethylated consuming further two SAM molecules, it would be expected to be expelled from the active site similar to the first-modified A1519. At this stage the extent of interactions with KsgA would be sufficiently reduced, to allow dissociation of the factor and formation of a three-base stack between G1517, A1518 and A1519, closing of the tetraloop through insertion of G1516, where it would replace G1517 (Figure 4C and D). Helix 45 in its fully folded conformation will then form interactions with the backbone of folded helix 44 as seen in the mature 30S subunit (Figure 4D) (66,67).
The ability of the helix 45 RNA loop to adopt different conformations by base flipping appears to be a pre-requisite for the successive dimethylation to occur, indicating that the flexibility of the RNA substrate is critical for the modification mechanism. A related RNA-structure specific recognition mechanism has been reported for the RumA catalyzed 23S U1939 methylation (58). The concept that conformational changes mediated by the flexibly disposed loops or single stranded ends of RNA/DNA substrates are critical for a number of important cellular reactions have been described previously, including the processes catalyzed by the CCA-adding enzyme that modifies immature tRNAs (68), and proofreading mechanisms of tRNA synthetases (69) and DNA polymerases (70,71).
KsgA CTD provides substrate specificity and stability in binding
The organization of KsgA in two separate but closely connected domains and the proposed reaction mechanism that involves exchange of the modified nucleobases in the active site of the enzyme, suggest that the CTD has some level of affinity for the rRNA on its own. This is in accordance with mutation studies, where positively charged amino acids of the CTD were mutated to alanine (R248A and 221RRK223 to 221AAA223) and resulted in reduced substrate binding affinity (22). Generally, a two-domain architecture is common not only among SAM dependent RNA methyltransferases, but also in SAM dependent DNA, protein and small molecule methyltransferases, where also one domain has a conserved methyltransferase fold and the other is a recognition domain that confers additional substrate specificity (56).
A structurally homologous two domain architecture has been discovered for ErmC’, an N6-methyltransferase, which targets the large bacterial subunit 23S rRNA at helix 73 (72,73). The KsgA and Erm families carry the same conserved methyltransferase motifs in the N-terminal domain, while the C-terminal domains differ. The conserved positively charged amino acids of KsgA CTD involved in helix 24 and 27 binding (R248 and 221RRK223) are not conserved in the Erm family, consistent with the observation that the two CTD are responsible for recognition of different substrates (19,22). Furthermore, the M. tuberculosis variant Erm37 is lacking the CTD and initially targets the conserved Erm substrate A2058, but then proceeds to less specifically methylate neighboring adenosines (Madsen et al. 2005). Studies with KsgA-ErmC’ chimeras show that switching the KsgA CTD and two additional loop regions to the respective ErmC’ sequences in vivo results in a methylation of the ErmC’ target nucleoside by the KsgA methyltransferase domain (74).
The KsgA CTD binds via stretches of positively charged amino acids to the adjacent backbones of helix 24 and 27, which suggests that the interaction is specific for the RNA fold rather than sequence. This explains the observations that KsgA can only use 16S rRNA as a substrate if it is folded in the context of the close-to-mature 30S (24,51). Additionally, our data suggest that G1516 acts as an anchor for the precise positioning of A1519 and A1518 in the catalytic pocket of KsgA, both during initial enzyme attachment and the switching of A1519 with A1518. Together, these results suggest that binding specificity is conferred by three 16S RNA helices in close proximity, which probably only happens at a specific point during SSU maturation. A recognition site built from sequentially distant modules but located in spatial proximity in the context of the 30S has been termed ‘proximity model’ (75).
KsgA/Dim1 might have a checkpoint and timer function during small subunit biogenesis
KsgA introduces a unique rRNA modification by dimethylating two adjacent nucleobases, the reason for which is not clear. Our data provide some indications for why this might be the case. First, KsgA is capable of detecting a particular stage of SSU maturation by sensing the spatial proximity of helices 24, 27 and 45, which then allows binding of G1516 into its KsgA binding pocket triggering a cascade of methylation reactions. Second, since the enzyme needs to dimethylate two adjacent bases with a putative substrate re-arrangement in between, this process likely requires more time than a more common rRNA monomethylation. Consequently, as long as KsgA is bound, subunit joining is impaired (13) and helix 44 cannot adopt its position as in the mature 30S ribosomal subunit (22), potentially providing time for other 30S modifications, re-arrangements or folding processes to occur. Once KsgA finished the dimethylation reactions and dissociates from the ribosome, helix 45 adopted its mature conformation and provides an interaction site for helix 44. Therefore, KsgA binding may serve as a checkpoint for the integrity of the platform fold, whereas successive methylation reactions may function as a timer to provide a time window for other maturation processes to occur before KsgA dissociates.
The high conservation between KsgA and its eukaryotic cytosolic homologue Dim1 suggests that the eukaryotic enzymes modify rRNA by a similar mechanism, which is in line with a complementation analysis that showed KsgA deletion phenotypes in E. coli can be rescued by overexpression of the homologous S. cerevisiae enzyme Dim1 (14). Consequently, the mode of interaction between KsgA and the rRNA that we observe is consistent with previous low resolution studies on eukaryotic ribosomal complexes that include Dim1 (25,43). Therefore, our structural data and the proposed mechanism have implications for understanding ribosome maturation in general.
DATA AVAILABILITY
The 3.1 Å cryo-EM map of the complex between submethylated E. coli 30S and E. coli KsgA generated during this study is available at EMDB EMD-12736. The refined coordinates generated during this study are available at PDB 7O5H. Materials are available from the corresponding author upon request.
Supplementary Material
ACKNOWLEDGEMENTS
Cryo-EM data were collected at the electron microscopy facility of ETH Zurich (ScopeM). We thank Peter Tittmann for technical support. We thank Dr David Ramrath for support with cryo-EM data processing and model building, Dr Marc Leibundgut for support with model building and validation and Dr Ahmad Jomaa for valuable input on the manuscript.
Author contributions: Conceptualization, D.B. and N.B.; Data Curation, N.C.S. and A.B.R.; Methodology, D.B.; Formal Analysis, N.C.S. and A.B.R.; Investigation, N.C.S. and D.B.; Validation, N.C.S., A.B.R. and D.B.; Resources, D.B. and N.B.; Writing - Original Draft, N.C.S., A.B.R. and D.B.; Writing - Review & Editing, N.C.S, A.B.R., D.B. and N.B.; Visualization, N.C.S and A.B.R.; Supervision, D.B. and N.B.; Project Administration, N.C.S., A.B.R., D.B. and N.B.; Funding Acquisition, N.B.
Notes
Present address: Daniel Boehringer, Cryo-EM Knowledge Hub, ETH Zurich (Swiss Federal Institute of Technology) Zürich, Otto-Stern-Weg 3, Zürich 8093, Switzerland.
Contributor Information
Niklas C Stephan, Institute of Molecular Biology and Biophysics, ETH Zurich (Swiss Federal Institute of Technology), Zürich, Otto-Stern-Weg 5, Zürich 8093, Switzerland.
Anne B Ries, Institute of Molecular Biology and Biophysics, ETH Zurich (Swiss Federal Institute of Technology), Zürich, Otto-Stern-Weg 5, Zürich 8093, Switzerland.
Daniel Boehringer, Institute of Molecular Biology and Biophysics, ETH Zurich (Swiss Federal Institute of Technology), Zürich, Otto-Stern-Weg 5, Zürich 8093, Switzerland.
Nenad Ban, Institute of Molecular Biology and Biophysics, ETH Zurich (Swiss Federal Institute of Technology), Zürich, Otto-Stern-Weg 5, Zürich 8093, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [31003A_182341]; National Centre of Competence in Research RNA and Disease [182880]; Roessler Prize, Ernst Jung Prize and Otto Naegeli Prize for Medical Research (to N.B., in part). Funding for open access charge: SNSF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kimura S., Suzuki T.. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010; 38:1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Poldermans B., Goosen N., Van Knippenberg P.H.. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. I. The effect of kasugamycin on initiation of protein synthesis. J. Biol. Chem. 1979; 254:9085–9089.383710 [Google Scholar]
- 3. O’Farrell H.C., Musayev F.N., Scarsdale J.N., Rife J.P.. Control of substrate specificity by a single active site residue of the KsgA methyltransferase. Biochemistry. 2012; 51:466–474. [DOI] [PubMed] [Google Scholar]
- 4. Van Knippenberg P.H., Van Kimmenade J.M., Heus H.A.. Phylogeny of the conserved 3' terminal structure of the RNA of small ribosomal subunits. Nucleic Acids Res. 1984; 12:2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wimberly B.T., Brodersen D.E., Clemons W.M. Jr, Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V.. Structure of the 30S ribosomal subunit. Nature. 2000; 407:327–339. [DOI] [PubMed] [Google Scholar]
- 6. Van Buul C.P., Hamersma M., Visser W., Van Knippenberg P.H.. Partial methylation of two adjacent adenosines in ribosomes from Euglena gracilis chloroplasts suggests evolutionary loss of an intermediate stage in the methyl-transfer reaction. Nucleic Acids Res. 1984; 12:9205–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noon K.R., Bruenger E., McCloskey J.A.. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 1998; 180:2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klootwijk J., Klein I., Grivell L.A.. Minimal post-transcriptional modification of yeast mitochondrial ribosomal RNA. J. Mol. Biol. 1975; 97:337–350. [DOI] [PubMed] [Google Scholar]
- 9. Schnare M.N., Heinonen T.Y., Young P.G., Gray M.W.. A discontinuous small subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J. Biol. Chem. 1986; 261:5187–5193. [PubMed] [Google Scholar]
- 10. Fan J., Schnare M.N., Lee R.W.. Characterization of fragmented mitochondrial ribosomal RNAs of the colorless green alga Polytomella parva. Nucleic Acids Res. 2003; 31:769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Buul C.P., Visser W., van Knippenberg P.H.. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett. 1984; 177:119–124. [DOI] [PubMed] [Google Scholar]
- 12. Poldermans B., Roza L., Van Knippenberg P.H.. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. III. Purification and properties of the methylating enzyme and methylase-30 S interactions. J. Biol. Chem. 1979; 254:9094–9100.383712 [Google Scholar]
- 13. Connolly K., Rife J.P., Culver G.. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008; 70:1062–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lafontaine D., Delcour J., Glasser A.L., Desgres J., Vandenhaute J.. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3'-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994; 241:492–497. [DOI] [PubMed] [Google Scholar]
- 15. Pulicherla N., Pogorzala L.A., Xu Z., HC O.F., Musayev F.N., Scarsdale J.N., Sia E.A., Culver G.M., Rife J.P.. Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases. J. Mol. Biol. 2009; 391:884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tokuhisa J.G., Vijayan P., Feldmann K.A., Browse J.A.. Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA dimethylase. Plant Cell. 1998; 10:699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schubot F.D., Chen C.J., Rose J.P., Dailey T.A., Dailey H.A., Wang B.C.. Crystal structure of the transcription factor sc-mtTFB offers insights into mitochondrial transcription. Protein Sci. 2001; 10:1980–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metodiev M.D., Lesko N., Park C.B., Camara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.G.. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009; 9:386–397. [DOI] [PubMed] [Google Scholar]
- 19. O’Farrell H.C., Scarsdale J.N., Rife J.P.. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J. Mol. Biol. 2004; 339:337–353. [DOI] [PubMed] [Google Scholar]
- 20. Demirci H., Belardinelli R., Seri E., Gregory S.T., Gualerzi C., Dahlberg A.E., Jogl G.. Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine. J. Mol. Biol. 2009; 388:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao S.T., Rossmann M.G.. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973; 76:241–256. [DOI] [PubMed] [Google Scholar]
- 22. Boehringer D., O’Farrell H.C., Rife J.P., Ban N.. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J. Biol. Chem. 2012; 287:10453–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hussain T., Llacer J.L., Fernandez I.S., Munoz A., Martin-Marcos P., Savva C.G., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014; 159:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thammana P., Held W.A.. Methylation of 16S RNA during ribosome assembly in vitro. Nature. 1974; 251:682–686. [DOI] [PubMed] [Google Scholar]
- 25. Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L. 3rd, Karbstein K., Skiniotis G.. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011; 333:1449–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitterer V., Shayan R., Ferreira-Cerca S., Murat G., Enne T., Rinaldi D., Weigl S., Omanic H., Gleizes P.E., Kressler D.et al.. Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism. Nat. Commun. 2019; 10:2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tu C., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.. Structural basis for binding of RNA and cofactor by a KsgA methyltransferase. Structure. 2009; 17:374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tu C., Zhou X., Tarasov S.G., Tropea J.E., Austin B.P., Waugh D.S., Court D.L., Ji X.. The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10156–10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X., Shen S., Wu P., Li F., Liu X., Wang C., Gong Q., Wu J., Yao X., Zhang H.et al.. Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function. Nucleic. Acids. Res. 2019; 47:7648–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O’Farrell H.C., Musayev F.N., Scarsdale J.N., Wright H.T., Rife J.P.. Crystallization and preliminary X-ray diffraction analysis of KsgA, a universally conserved RNA adenine dimethyltransferase in Escherichia coli. Acta Crystallogr. D. Biol. Crystallogr. 2003; 59:1490–1492. [DOI] [PubMed] [Google Scholar]
- 31. Kimanius D., Forsberg B.O., Scheres S.H., Lindahl E.. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016; 5:e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016; 193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ludtke S.J., Baldwin P.R., Chiu W.. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999; 128:82–97. [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kucukelbir A., Sigworth F.J., Tagare H.D.. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014; 11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. 3rd, Snoeyink J., Richardson J.S.et al.. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic. Acids. Res. 2007; 35:W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 40. Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu Z., O’Farrell H.C., Rife J.P., Culver G.M.. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008; 15:534–536. [DOI] [PubMed] [Google Scholar]
- 42. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:doi:10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scaiola A., Pena C., Weisser M., Bohringer D., Leibundgut M., Klingauf-Nerurkar P., Gerhardy S., Panse V.G., Ban N.. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 2018; 37:e98499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Noeske J., Wasserman M.R., Terry D.S., Altman R.B., Blanchard S.C., Cate J.H.. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 2015; 22:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J.et al.. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019; 75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prisant M.G., Williams C.J., Chen V.B., Richardson J.S., Richardson D.C.. New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink “waters,” and NGL Viewer to recapture online 3D graphics. Protein Sci. 2020; 29:315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moazed D., Van Stolk B.J., Douthwaite S., Noller H.F.. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J. Mol. Biol. 1986; 191:483–493. [DOI] [PubMed] [Google Scholar]
- 48. Desai P.M., Rife J.P.. The adenosine dimethyltransferase KsgA recognizes a specific conformational state of the 30S ribosomal subunit. Arch. Biochem. Biophys. 2006; 449:57–63. [DOI] [PubMed] [Google Scholar]
- 49. O’Farrell H.C., Pulicherla N., Desai P.M., Rife J.P.. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA. 2006; 12:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jahagirdar D., Jha V., Basu K., Gomez-Blanco J., Vargas J., Ortega J.. Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy. RNA. 2020; 26:2017–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Helser T.L., Davies J.E., Dahlberg J.E.. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 1972; 235:6–9. [DOI] [PubMed] [Google Scholar]
- 52. O’Farrell H.C., Musayev F.N., Scarsdale J.N., Rife J.P.. Binding of adenosine-based ligands to the MjDim1 rRNA methyltransferase: implications for reaction mechanism and drug design. Biochemistry. 2010; 49:2697–2704. [DOI] [PubMed] [Google Scholar]
- 53. Desai P.M., Culver G.M., Rife J.P.. Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly. Biochemistry. 2011; 50:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cunningham P.R., Weitzmann C.J., Nurse K., Masurel R., Van Knippenberg P.H., Ofengand J.. Site-specific mutation of the conserved m6(2)A m6(2)A residues of E. coli 16S ribosomal RNA. Effects on ribosome function and activity of the ksgA methyltransferase. Biochim. Biophys. Acta. 1990; 1050:18–26. [DOI] [PubMed] [Google Scholar]
- 55. Klimasauskas S., Kumar S., Roberts R.J., Cheng X.. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994; 76:357–369. [DOI] [PubMed] [Google Scholar]
- 56. Cheng X., Roberts R.J.. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic. Acids. Res. 2001; 29:3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hong S., Cheng X.. Jeltsch A., Jurkowska R.Z.. DNA Methyltransferases - Role and Function. 2016; Cham: Springer International Publishing; 321–341. [Google Scholar]
- 58. Lee T.T., Agarwalla S., Stroud R.M.. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005; 120:599–611. [DOI] [PubMed] [Google Scholar]
- 59. Alian A., Lee T.T., Griner S.L., Stroud R.M., Finer-Moore J.. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y.. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell. 2018; 71:1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Demirci H., Murphy F.t., Belardinelli R., Kelley A.C., Ramakrishnan V., Gregory S.T., Dahlberg A.E., Jogl G.. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010; 16:2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pley H.W., Flaherty K.M., McKay D.B.. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994; 372:68–74. [DOI] [PubMed] [Google Scholar]
- 63. Hoang C., Ferre-D’Amare A.R.. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001; 107:929–939. [DOI] [PubMed] [Google Scholar]
- 64. Slupphaug G., Mol C.D., Kavli B., Arvai A.S., Krokan H.E., Tainer J.A.. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996; 384:87–92. [DOI] [PubMed] [Google Scholar]
- 65. Lee T.T., Agarwalla S., Stroud R.M.. Crystal structure of RumA, an iron-sulfur cluster containing E. coli ribosomal RNA 5-methyluridine methyltransferase. Structure. 2004; 12:397–407. [DOI] [PubMed] [Google Scholar]
- 66. Schluenzen F., Takemoto C., Wilson D.N., Kaminishi T., Harms J.M., Hanawa-Suetsugu K., Szaflarski W., Kawazoe M., Shirouzu M., Nierhaus K.H.et al.. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat. Struct. Mol. Biol. 2006; 13:871–878. [DOI] [PubMed] [Google Scholar]
- 67. Watson Z.L., Ward F.R., Meheust R., Ad O., Schepartz A., Banfield J.F., Cate J.H.. Structure of the bacterial ribosome at 2 A resolution. Elife. 2020; 9:e60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xiong Y., Steitz T.A.. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004; 430:640–645. [DOI] [PubMed] [Google Scholar]
- 69. Silvian L.F., Wang J., Steitz T.A.. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999; 285:1074–1077. [PubMed] [Google Scholar]
- 70. Joyce C.M. How DNA travels between the separate polymerase and 3'-5'-exonuclease sites of DNA polymerase I (Klenow fragment). J. Biol. Chem. 1989; 264:10858–10866. [PubMed] [Google Scholar]
- 71. Joyce C.M., Steitz T.A.. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994; 63:777–822. [DOI] [PubMed] [Google Scholar]
- 72. Bussiere D.E., Muchmore S.W., Dealwis C.G., Schluckebier G., Nienaber V.L., Edalji R.P., Walter K.A., Ladror U.S., Holzman T.F., Abad-Zapatero C.. Crystal structure of ErmC', an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry. 1998; 37:7103–7112. [DOI] [PubMed] [Google Scholar]
- 73. Schluckebier G., Zhong P., Stewart K.D., Kavanaugh T.J., Abad-Zapatero C.. The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol. 1999; 289:277–291. [DOI] [PubMed] [Google Scholar]
- 74. Bhujbalrao R., Anand R.. Deciphering determinants in ribosomal methyltransferases that confer antimicrobial resistance. J. Am. Chem. Soc. 2019; 141:1425–1429. [DOI] [PubMed] [Google Scholar]
- 75. Rife J.P. Grosjean H. DNA and RNA Modification Enzymes. 2009; 1st edn.Boca Raton: CRC Press; 509–523. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3.1 Å cryo-EM map of the complex between submethylated E. coli 30S and E. coli KsgA generated during this study is available at EMDB EMD-12736. The refined coordinates generated during this study are available at PDB 7O5H. Materials are available from the corresponding author upon request.