Abstract
Transcription is a vital process activated by transcription factor (TF) binding. The active gene releases a burst of transcripts before turning inactive again. While the basic course of transcription is well understood, it is unclear how binding of a TF affects the frequency, duration and size of a transcriptional burst. We systematically varied the residence time and concentration of a synthetic TF and characterized the transcription of a synthetic reporter gene by combining single molecule imaging, single molecule RNA-FISH, live transcript visualisation and analysis with a novel algorithm, Burst Inference from mRNA Distributions (BIRD). For this well-defined system, we found that TF binding solely affected burst frequency and variations in TF residence time had a stronger influence than variations in concentration. This enabled us to device a model of gene transcription, in which TF binding triggers multiple successive steps before the gene transits to the active state and actual mRNA synthesis is decoupled from TF presence. We quantified all transition times of the TF and the gene, including the TF search time and the delay between TF binding and the onset of transcription. Our quantitative measurements and analysis revealed detailed kinetic insight, which may serve as basis for a bottom-up understanding of gene regulation.
INTRODUCTION
Transcription of mRNA is subject to large fluctuations. Periods of transcriptional activity of a gene, during which several mRNA molecules are produced, are followed by periods of quiescence without transcription (1). This bursting behaviour has been observed for most investigated genes (2,3), and in various species ranging from bacteria (4–6) to yeast (7,8) and mammals (9–12). Although biological processes are intrinsically stochastic due to low molecule counts and thermal forces, the large fluctuations of transcriptional bursts point to a high degree of cooperativeness within the underlying molecular mechanisms. Indeed, numerous regulatory factors of transcriptional bursting have been observed, including enhancer-promoter interactions (13,14), DNA topology (4,15), chromatin modifications and chromatin remodelers (16–18), co-activators such as p300, CBP and mediator (15), assembly of general transcription factors and the preinitiation complex, polymerase pausing and reinitiation (17,19–21). Not least, binding of specific transcription factors (TFs) to cis regulatory sequences, usually representing the first step in the cascade of transcription activation is associated with bursting (2,7,11,16,17,22–31).
A bursting gene can usually be described by the two-state or random-telegraph model (32). There, switching of the gene between the quiescent and the active state is modelled as stochastic process with on- and off-rates, and a transcription rate describes mRNA production from the active state. First insight into the mechanism of a regulatory trait, such as enhancer action or TF binding, can be obtained by revealing which of the rates of the two-state model it influences. For TFs, it is well known that TF concentration is coupled to transcriptional activity (6,26,27,33–35). Mostly, increasing concentration was associated with an increase in burst frequency (2,16,26,27,30). An effect on burst duration (29) or burst size (2,10) was also reported, although care has to be taken to avoid concatenating individual bursts (36).
Recently, in addition to concentration, TF residence time has been observed to affect bursting (16,22,23,28,30,31). As with concentration, longer residence time was associated with an increase in burst frequency (16,17,28,30,31). However, in many studies this effect was not exclusive, and an increase in burst duration (16,17,30,31) and sometimes in burst size (30,31) was also coupled to longer residence times. This mechanistic variation might trace back to the dissimilar methods of varying TF residence time. Different residence times were either achieved indirectly by studying modified TFs prevalent in certain cellular conditions (28,30,31), TF mutants with potentially different DNA binding mechanisms (22), by studying different target sequences (16) or by knocking out co-factors (17). Overall, a clear understanding of how TF binding, in particular TF residence time and concentration, affect transcriptional bursting, and which of the rates of the two-state model are altered by a TF, is still missing.
Here, we decipher the role of TF residence time and concentration on bursting of a minimal reporter gene with well-defined promoter structure. We used the Auxin-Inducible Degron (AID) system (37) to vary TF concentration without further genomic modification. To vary the residence time with minimal disturbances to the TF, we utilized the modular DNA binding domain of transcription-activator like effectors (TALEs) (23). In contrast to our previous study, where we revealed the effect of residence time of a competitive repressor (23), we here determined the effect of residence time of a transcription activator. We quantified mRNA production and bursting parameters with single molecule Fluorescence In Situ Hybridisation (smFISH) in fixed cells, live measurements of bursting with the MS2 system, and a novel analysis tool, Bursting Inference from mRNA Distributions (BIRD). We found that both TF residence time and concentration solely affected the on-rate of the gene, leaving burst size and burst duration unaffected. Interestingly, variation of TF residence time had a stronger influence than an equal variation of concentration. We could explain our observations in terms of an extended three-state model of gene transcription, in which binding of the TF switches the gene from a quiescent to a primed state, from which multiple successive transitions have to be traversed before transcription from the active state can be initiated. Overall, our measurements and analysis reveal detailed mechanistic and kinetic information on transcription of a minimal mammalian gene, which may serve as basis for a bottom-up approach in understanding regulatory traits of gene transcription.
MATERIALS AND METHODS
Cloning
Gene construct
The gene construct was synthesized by GeneArt (ThermoFisher). Afterwards, we integrated 24xMS2 stem loops from phage-CMV-CFP-24xMS2 (Wu et al., 2012) by EagI-HF and BglII restriction and ligation. To allow integration of the gene construct via the FlipIn system, we thereafter exchanged the plasmid backbone of the gene construct to pcDNA5/FRT taken from pcDNA5/FRT/TO V5 (38), using MluI-HF and SphI-HF digestion. The sequence of the gene construct with inserted 24xMS2 can be found in the Supplementary Information.
For lentivirial gene transfer, we transferred the gene construct containing 24xMS2 repeats into the pLenti backbone of pLenti-CMV-OsTIR1 (Section CMV-OsTIR1) with restriction by MreI-Fast and XhoI. To allow this, we first needed to insert a MreI and a XhoI restriction site into pLenti-CMV-OsTIR1. We achieved this by annealing the two primers Cloning_pLenti_fw and Cloning_pLenti_rev as insert and digesting both insert and backbone with BamHI-HF and ClaI followed by ligation.
tdMCP-tdGFP
To generate a tandem-GFP version of phage-ubc-nls-ha-tdMCP-gfp (39), we amplified the GFP encoding sequence by PCR with the primers Cloning_MCP-GFP_fw and Cloning_MCP-GFP_rev and inserted it into the phage-ubc-nls-ha-tdMCP-gfp plasmid via XbaI restriction and ligation to generate the tdMCP-tdGFP construct. We checked for correct orientation with sequencing.
CMV-OsTIR1
For lentiviral gene transfer of OsTIR1, we amplified the OsTIR1 coding region from pMK232 (40) via PCR with the primers Cloning_OsTIR1_fw and Cloning_OsTIR1_rev and inserted it into the pLenti backbone from pLenti-CMV-rtTA3 (Addgene plasmid #26429) by digestion with BstXI. We checked for correct orientation with sequencing.
TALE-TF backbone
We modified the pICE-Halo-VP64 plasmid from (23) for this study. To hinder regulation of TALE-TF expression due to binding of the TALE-TF to their own promoter region, we removed the Tet-operators on the plasmid by SacI-HF digestion and ligation. To apply the AID system (37) for our TALE-TFs, we inserted two copies of the mAID tag, which we amplified via PCR on pMK292 (40). We therefore used for the first copy the primers Cloning_mAID1_fw and Cloning_mAID1_rev together with HindIII-HF and MluI-HF digestion, and for the second copy Cloning_mAID2_fw and Cloning_mAID1_rev with only MluI-HF digestion. We checked for correct orientation with sequencing. To ensure nuclear localization with those modifications, we inserted another nuclear localization signal C-terminally to the TALE-DBDs by annealing the two primers Cloning_NLS_fw and Cloning_NLS_rev and digested both insert and backbone by restriction with EcoRI-HF and PacI, followed by ligation.
TALE-TF Golden Gate Reaction
We assembled TALE-TF with the Golden Gate TALEN and TAL Effector Kit2.0 (Addgene kit #1000000024) (41) as previously described (23). First, the repeat modules 1–10 were assembled into the pFUS_A plasmid and the repeat modules 11-(n-1) were assembled into the pFUS_B(n-1) plasmid, both via 10 cycles of BsaI restriction and ligation. There, n stands for the total number of repeats of the TALE-TF construct. In a second step, the arrays assembled in pFUS_A and pFUS_B(n-1) were inserted together with the last repeat into the TALE-TF backbone via 5 cycles of Esp3I restriction and ligation. For T9R, which has not enough repeats for this standard assembly strategy, we first assembled all repeats except of the last repeat into pFUS_B(n-1). In the second step of T9R assembly, we inserted those repeats together with the last repeat into the TALE-TF backbone. To achieve complete ligation of this construct, we additionally added a self-ligated version of pFUS_A (23) to the reaction. We validated all steps of the assembly for each construct via sequencing. The designed TALE-TFs are listed with their target sequences in Supplementary Table S1.
Cell culture
We performed all experiments in U2-OS based cell lines, which we cultivated at 37°C and 5% CO2 in DMEM supplemented with 10% FBS, 1% glutamax, 1% nonessential amino acids, and 1% sodium pyruvate. We supplemented all antibiotics used for selection also during normal cultivation to hinder loss of the integrated sequences.
Generation of cell lines
FlipIn U2-OS
To generate a FlipIn U2-OS cell line, we transfected U2-OS cells with linearized pFRT/lacZeo (ThermoFisher). After selection with Zeocin, we isolated colonies resulting from single cells with cloning cylinders. We screened all clones for single integration of pFRT/lacZeo by Southern Blot (performed by Lofstrand Labs Limited) using a FRT site-specific probe (18). We generated this probe by PCR with primers Probe_pFRTneo_fw and Probe_pFRTneo_rev on pFRT/lacZeo.
FlipIn reaction
For the FlipIn reaction of the gene construct, we seeded 1.5 million FlipIn U2-OS cells on a 10 cm dish without antibiotics. After 24 h, we transfected the cells with 9 μg pOG44 and 1 μg pcDNA5/FRT-gene construct using lipofectamine 2000. After 72 h, we started the selection with hygromycin. After selection, we isolated single colonies with cloning cylinders and screened them for positive FlipIn via zeocin sensitivity, lack of β-galactosidase activity and PCR (Supplementary Figure S1). To test for β-galactosidase activity, we lysed the cells with 0.5% Triton X-100 and incubated the lysates 24 h with a X-Gal containing buffer. A positive FlipIn reaction prohibited formation of a blue staining. For PCR, we isolated genomic DNA as described in (42). In brief, we resuspended 2.000 cells in 50 μl Alkaline Lysis reagent (25 mM NaOH, 0.2 mM Disodium EDTA), boiled it for 10 min and thereafter we neutralized it by the addition of 50 μl Neutralizing reagent (40 mM Tris–HCl). After that, we validated the site specific integration via FlipIn reaction by PCR with specific primers (FlipIn_Test1_fw, FlipIn_Test1_rev, FlipIn_Test2_fw and FlipIn_Test2_rev)) (Supplementary Figure S1).
tdMCP-tdGFP and OsTIR1
After FlipIn of the gene construct, we stably integrated tdMCP-tdGFP and OsTIR1 into cells using a standard lentiviral production protocol (Addgene). For lentivirus production, we seeded LentiX cells on a 10 cm dish (Sarstedt) and transfected them 48 hours before transfection with 7.5 μg psPAX2 (Addgene #12260), 2.5 μg pMD2.G (Addgene #12259), 10 μg pLenti-CMV-OsTIR1 or 10 μg phage-tdMCP-tdGFP. We harvested the lentiviruses 48 h after transfection of the LentiX cells. Thereafter, we exposed the cells for transfection to the harvested lentivirus. For this, we seeded 30 000 cells on a six-well dish (Sarstedt) and exposed them 24 h after seeding to 2 ml pLenti-CMV-OsTIR1 lentivirus and 0.5 ml phage-tdMCP-tdGFP lentivirus. We selected transfected cells with blasticidin for OsTIR1 integration and sorted them via the GFP signal for tdMCP-tdGFP expression using FACS (BD FACSAria II).
TALE-TF
We stably transfected the TALE-TF with linearized plasmids via puromycin selection after the integration of OsTIR1 and tdMCP-tdGFP. We screened and sorted the colonies via FACS for equal expression (BD FACSAria II) of the different TALE-TF using staining with 1.25 μM HaloTag-TMR ligand following the protocol of the manufacturer (Promega). Thereafter, we used those cell lines for the transcription quantification measurements (smFISH and MS2).
Stable transfection of additional copies of gene construct
For the single molecule tracking experiments, we generated cells with multiple copies of the gene construct. Therefore, we stable transfected the cell lines containing a single copy of the reporter gene, OsTIR1, tdMCP-tdGFP and one of the TALE-TF with additional copies of the gene construct via lentiviral gene transfer using a standard lentiviral production protocol (Addgene). We quantified the number of integrations by comparing the levels of transcription activation before and after lentiviral transfection of additional copies of the gene construct. We used the cells with multiple integrations of the gene construct solely for the single molecule tracking experiments, but not for the measurements concerning transcription quantification.
AID for TALE-TF degradation
To test TALE degradation with AID, we stained the TALE-TF with 1.25 μM Halo-TMR-ligand after manufacturer protocol (Promega) in cells grown with Auxinole (Aobious) for 24 h. Directly afterwards, we exchanged the medium to DMEM supplemented with 500 μM Auxin (I2886, Sigma) to degrade TALE-TF (40). We than fixed the cells at different time points after addition of Auxin. For each time point, we determined TALE-TF expression with the Spinning-Disk microscope explained in the smFISH methods section. For this, we chose the cells randomly during imaging and acquired images of the Halo-TMR stained TALE-TF via 8 averages of 500 ms exposure time with a 532 nm laser. The fluorescence signal was filtered using a 630dcxr dichroic (Chroma Technology) and a HQ585/80 filter (Chroma Technology). We then determined the TALE-TF expression level for each individual cells as the average nuclear intensity in those images. We determined the background fluorescence in cells without TALE-TF after staining with Halo-TMR-ligand.
Single molecule imaging and residence time analysis
Preparations
For single molecule imaging, we grew cells on glass bottom dishes (Delta T, Bioptechs). We stained the cells with 3–6 pM Halo-SiR ligand (43) directly before imaging according to the protocol of the manufacturer (Promega) to obtain equal molecular densities during imaging and therefore minimize differences in tracking losses.
Single molecule time-lapse imaging
We performed single molecule imaging with highly inclined and laminated optical sheet (44) illumination as described in (23) at a custom built fluorescence microscope. The microscope is built around a commercial microscope body (Nikon, TIE) and equipped with a 638 nm laser (Toptica, IBEAM-SMART-640-S, 150 mW), an AOTF (AAOptoelectronics, AOTFnC-400.650-TN), a high-NA objective (100× 1.45 Nikon Plan Apo) and an EMCCD camera (Andor, iXon Ultra DU 897U). We filtered the fluorescence signal with a dichroic mirror (AHF, F73-6866), an emission filter (AHF, F72-866) and a notch filter (AHF, F40-072). To distinguish dissociation rates of TALE-TFs independently from the photobleaching rate of the SiR-dye, we performed time-lapse microscopy (45). For each time-lapse condition, we introduced a different dark time between frame acquisitions of 50 ms exposure, resulting in frame acquisitions each 50 ms (continuous), 1 s or 5 s (time-lapses). For continuous and 1 s time-lapse movie, we took 300∼frames and for 5s time lapse movie 100 frames. To minimize differences in photobleaching rate, we controlled the laser power before each measurement to 200 mW/cm². We performed imaging up to 90 min per dish at 37°C in OptiMEM. We chose the cells for imaging at random and took a maximum of two movies per cell. During analysis, we chose only cells with an average molecule detection number per frame <2 × 10–3/pixel. We supplemented the medium for labelling and imaging with 200 μM Auxinole (AOB8812, Aobious) to prevent degradation of TALE-TF during imaging.
Tracking analysis
We analyzed the single molecule microscopy data with TrackIt (46) to obtain fluorescence survival time distributions of bound TALE-TF. We adjusted the tracking settings for a nearest neighbour tracking algorithm to minimize false-positive connections due to nearby binding events and to obtain an equal probability for tracking losses due to tracking errors and photobleaching for all time-lapse conditions. The resulting tracking settings were SNR = 5, maximal displacement of 0.6 pixels (continuous), 1.4 pixels (1 s time-lapse) and 2.0 pixels (5 s time-lapse) to separate bound from diffusing molecules and 1 gap frame for all time-lapse conditions. We defined a molecule as bound, if it stayed within the tracking radius for 2 frames (minimal track length 2) in 1 s and 5 s time-lapse conditions. For continuous movies, we quantified the effect of varying the minimal track length (Supplementary Figure S2). We chose a minimal track length of 5 (corresponding to 250 ms) for continuous movies, which approx. yields <5% of falsely assigned diffusing molecules to the bound population (Supplementary Figure S2) when determining the fluorescence survival time distributions of bound TALE-TF.
Analysis of survival time distributions using GRID
We determined the dissociation rate spectrum of bound TALE-TF by analysing the fluorescence survival time distributions obtained from time-lapse imaging with GRID (47). In brief, GRID performs an inverse Laplace transformation of a fluorescence survival time distribution to reveal the underlying dissociation rate spectrum. To account for photobleaching, the survival time distributions of different time-lapse measurements for a TALE-TF were analysed globally. GRID first results in an event spectrum, which counts the number of binding events that happen within a given time period. A state spectrum can be obtained from the event spectrum via normalizing the fractions with the underlying dissociation rates (47). The state spectrum then reflects the number of molecules present in each binding state at any time snapshot.
Single molecule RNA-FISH (smFISH)
One day before sample preparations started, we seeded cells on 35 mm glass bottom dishes (Ibidi). We performed smFISH with additional staining of TALE-TF with Halo-TMR-ligand (Promega) and of membrane with Lectin-FITC (Sigma-Aldrich) after a modified Stellaris RNA-FISH protocol (Biosearch Technologies) as described in (23). In brief, we stained TALE-TFs with 1.25 μM Halo-TMR-ligand according to the manufacturer's protocol (Promega) and then directly fixed the cells in a 3.2% PFA solution for 10 min followed by washing steps and membrane-staining with 10 μg/ml Lectin-FITC for 10 min. Membrane-staining was followed by additional washing steps and permeabilisation in 70% EtOH for at least 16 h at 4°C. After permeabilisation, we prepared the cells for hybridization with an incubation in Stellaris Wash Buffer A for 5 min. Then, we hybridized fluorescently labelled probes targeting SNAP-tag and MS2 repeats to their target mRNA for 16 h at 37°C to visualize the target RNAs. This was followed by a total of five washing steps (three times with Stellaris Wash Buffer A followed by two times with Stellaris Wash Buffer B) with a final washing time of 16 h to lower the background signal. We ordered the probes as a Stellaris Custom Probe set (Biosearch Technologies) and they are listed in Supplementary Table S2.
We performed imaging of smFISH, Halo-TMR stained TALE-TFs and membrane staining at a set-up custom-built around a CSU 10 spinning disk (Yokogawa) and equipped with a UPlanSapo 60×/1.35 Oil objective (Olympus), E-665.XP piezo amplifier (Physik Instrumente) and an iXon DV-887 EMCCD camera (Andor) (23). We picked the cells for imaging randomly and grouped them by TALE-TF concentration during the analysis. For smFISH, we acquired a z-stack covering the whole cell volume with a step size of 500 nm and eight averages of individual exposure times of 500 ms with a 635 nm laser. For the Halo-TMR stained TALE-TFs, we imaged one characteristic plane of the cells with eight averages of a 500 ms exposure time with a 532 nm laser. For the membrane-staining with Lectin-FITC, we also imaged one characteristic plane of the cells with eight averages of a 500 ms exposure time with a 473 nm laser. The fluorescence signal was filtered using a 630dcxr dichroic (Chroma Technology) and a FF01-635 LP filter (BrightLine) for smFISH and a HQ585/80 filter (Chroma Technology) for TALE-TF and membrane-staining.
We analysed the smFISH data with the Matlab toolbox FISH-quant using the membrane staining to draw the cell outlines (48). Transcription sites were detected as brightest spot in the nucleus with a minimum of four transcripts to minimize false-positive detections and with a cut-off of the 5% brightest spots. With the detected transcription sites, we were able to determine burst frequencies (# of cells with a transcription site/# of all cells) and burst heights (brightness of transcription site/average brightness of single mRNA) (48). We used the Halo-TMR staining to quantify nuclear TALE-TF concentrations, with U2-OS Halo-CTCF C32 (49) as calibrated reference (Supplementary Figure S3 and Supplementary Equation S30).
To achieve a higher expression of the TALE-TF in the same cell lines, we inhibited leaky degradation via the AID system (50) by supplementing medium with 200 μM Auxinole (AOB8812, Aobious) 24 h before sample preparation.
MS2 measurements
For live cell transcription measurements, we seeded cells on two-well dishes (Ibidi). To simultaneously quantify TALE-TF expression levels, we stained the cells directly before the measurement with 1.25 μM Halo-TMR-ligand according to the protocol of the manufacturer (Promega). We supplemented the medium for labelling and imaging with 200 μM Auxinole (Aobious) to prevent leaky degradation of TALE-TF during imaging. We omitted Halo-tag labelling and incubation with Auxinole for the cell line containing no TALE-TF.
We performed imaging with the spinning disk microscope set-up explained in the smFISH method section, but equipped with an additional cultivation chamber for temperature and CO2 control (Pecon). We imaged the cells in phenolred-free DMEM at 37°C and 5% CO2 by taking z-stacks of the entire nuclear volume (typically 12.5 μm) with a step size of 500 nm and exposure times of 200 ms with a 473 nm laser. We acquired a z-stack every 2 min for up to 6 h. Before each MS2 z-stack time series, we imaged the Halo-TMR staining for the cells expressing TALE-TFs via 8 averages of a 500 ms exposure time with a 532 nm laser. The fluorescence signal was filtered using a 530dcxr dichroic (Chroma Technology) and a HQ502/40 (Chroma Technology) for MS2 imaging and a HQ585/80 (Chroma Technology) for Halo-TMR.
We analysed the durations and intensities of the transcriptional bursts with TrackIt (46). We defined the time of active transcription, which we measured by the presence of nascent transcripts, as the burst duration. We adjusted the SNR for each cell individually depending on the brightness of the burst and the background signal. For tracking, we used a maximal displacement of 2 μm, a minimal track length of 2 and 2 gap frames to minimize premature loss of tracks.
RESULTS
Reporter cell lines to study TF residence time and concentration-dependent transcription
We aimed at studying the influence of TF binding on gene transcription, with a focus on the leverage of TF residence time and concentration and minimal blurring by other traits such as heterogeneous TF sites. Thus, we designed a reporter gene with well-defined promoter structure including a TATA box and BRE and Inr sites based on a minimal CMV promoter (Figure 1A and Supplementary information) (51). This promoter exhibits minimal background expression and high activation potential. We integrated a single copy of the Tet-operator (TetO) sequence, which is not present in the human genome, to exclude cooperative effects and binding competition with endogenous TFs. The artificial gene body consisted of 630 bp, followed by 24 copies of MS2 stem loops, which enable visualizing transcription events in living cells (9,52). As terminator, we chose the SV40 polyA sequence for an increased transcript level (53). We inserted the reporter gene into the human osteosarcoma cell line U2-OS using the FlipIn-system (54) (Materials and Methods), as this line is well suited for single molecule fluorescence in situ hybridization (smFISH) (23). We validated the integration of a single FlipIn site via Southern Blot (Supplementary Figure S4) and confirmed the positive FlipIn reaction to insert the reporter gene via gain of Hygromycin resistance, loss of Zeocin resistance, loss of lacZ activity and different PCR reactions on genomic DNA (Supplementary Figure S1). We used the cell line with one copy of the reporter gene as basis for all experiments in which parameters of transcription were quantified. After FlipIn of the gene construct, we stably transfected tdMCP-tdGFP for high signal-to-noise visualization of the MS2 stem loops (39).
Figure 1.
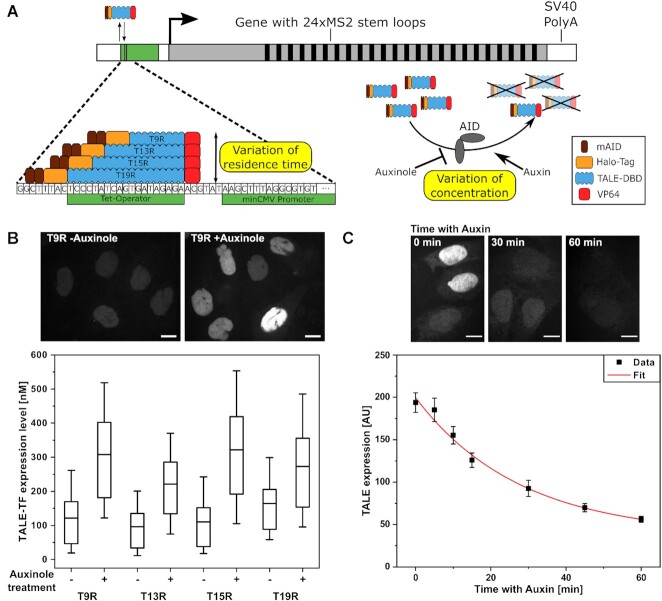
Design of reporter gene and strategy to vary TF residence time and concentration. (A) The reporter gene consists of a TetO sequence, to which TALE-TFs are targeted, upstream of a minimal CMV-derived promoter, a 630 bp gene body, 24xMS2 repeats and a SV40 poly(A) sequence. Residence time of TALE-TFs is varied by length of the TALE DBD. Concentration of TALE-TFs is varied with the AID system and addition of either Auxinole or Auxin. (B) Distribution of TALE-TF expression level with and without Auxinole treatment extracted from smFISH measurements. N: number of nuclei; N = 450 (T9R –Auxinole); N = 472 (T9R +Auxinole); N = 302 (T13R –Auxinole); N = 467 (T13R +Auxinole); N = 476 (T15R –Auxinole); N = 672 (T15R +Auxinole); N = 582 (T19R –Auxinole); N = 590 (T19R +Auxinole). Mean (line), 25th/75th percentile (box) and 10th/90th percentile (whiskers) define features of box plot. Inset: Fluorescence images of T9R expression without Auxinole (left) and after 16 h of Auxinole treatment (right). Scale bars denote 10 μm. (C) TALE-TF expression level measured by spinning disc micoscopy and corrected for background as function of time after Auxin treatment (black squares) and mono-exponential fit (red line). (N: number of nuclei; N = 207 (0 min); N = 207 (5 min); N = 215 (10 min); N = 204 (15 min); N = 205 (30 min); N = 210 (45 min); N = 213 (60 min); N = 138 (background)). Error bars denote s.e.m.. Inset: Fluorescence images of T15R expression without Auxin treatment and after 30 and 60 min of Auxin treatment. Scale bars denote 10 μm.
As TFs, we designed artificial activators based on transcription activator like effectors (TALEs) (55), which allow targeting any DNA sequence starting with thymine by varying their modular DNA binding domain (TALE-DBD) (56). By altering the length of the TALE-DBD, TFs with different residence times can be obtained (23). We therefore constructed four TALE-TFs binding to 9, 13, 15 or 19 nucleotides of the TetO sequence (called T9R, T13R, T15R and T19R; Figure 1A, target sequences in Supplementary Table S1), hypothesizing that they would also exhibit differences in their residence time. To exclude potential effects of TF position relative to the transcription start site, all TALE-DBDs ended at the same position. As activation domain, we used a C-terminal VP64 domain to achieve a high activation potential (57). The TALE-TFs further possessed an N-terminal HaloTag (58) for fluorescence imaging and a nuclear localization signal (NLS) (Figure 1B and C).
To adjust the concentration of TFs without further genetic modulation, we utilized two copies of the mAID-tag of the Auxin-inducible-Degron (AID) system (Figure 1A) (37). After stably transfecting the enzyme responsible for ubiquitination, OsTIR1, we created four cell lines, each stably transfected with one of the TALE-TFs and selected for comparable expression levels (Figure 1B) (Materials and Methods). By comparing the intensities of TMR labelled HaloTag-TALE-TF in the nucleus with those of a HaloTag-CTCF cell line calibrated for molecular numbers as standard (49), we determined the molar concentration of TALE-TFs within the nucleus of the four cell lines to be in the range of <300 nM (Figure 1B). We could increase the expression level of TALE-TFs by treating cells with 200 μM of the AID inhibitor Auxinole for 24 h to a concentration <600 nM (Figure 1B and Supplementary Figure S5) (50). A higher expression level could not be achieved with the AID system. TALE-TFs were depleted with a half time of 28 min by treatment with 500 μM Auxin (Figure 1C).
TALE-TFs show differences in DNA residence time
Before characterizing the binding kinetics of the TALE-TFs, we inserted additional reporter gene sequences in each of the TALE-TF cell lines by lentiviral gene transfer (Materials and Methods). The average number of integrated copies in each cell line was 5 as determined by quantifying the activation profiles (Supplementary Figure S6). However, we note that in particular TALE-TFs with short DBD have numerous TetO-related target sequences within the human genome, to which they are expected to bind with similar affinity (Supplementary Table S1). We used the cells with the additional copies of the reporter gene solely for the residence time measurements. For all transcription quantifications, we used the cell lines with a single copy of the reporter gene. To visualize single TALE-TFs, we labelled the HaloTag with the photostable organic SiR dye (43) at low labelling densities (59) and applied HILO microscopy (44) for single molecule tracking in nuclei of live cells (23).
To measure the dissociation rates of TALE-TFs from chromatin, we extended the time window to long observation times using time-lapse microscopy (45) with 50 ms frame acquisition time and frame cycle times of 0.05, 1 and 5 s (Materials and Methods). All constructs showed binding events ranging from less than a second up to several hundred seconds (Figure 2A, Supplementary Videos 1–3 and Supplementary Figure S7). We first confirmed that overall TALE-TF binding interactions linearly increased with increasing concentration and did not exhibit binding saturation (Supplementary Figure S8) (60). We then accumulated the binding times of each time-lapse condition in survival time distributions (Figure 2B and Supplementary Figure S7).
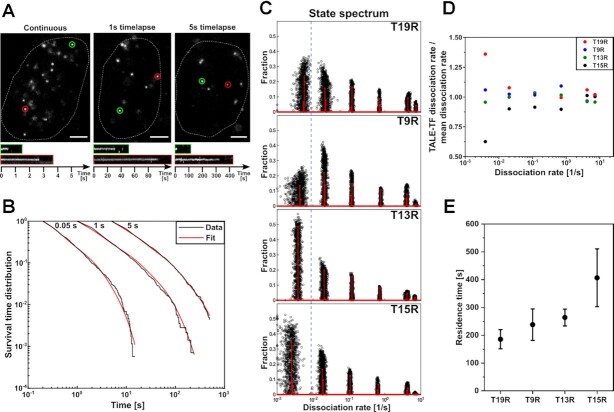
Figure 2.
Determination of residence times of TALE-TFs. (A) Example images of SiR-labelled HaloTag-T15R at time-lapse conditions of 0.05 s (continuous), 1 s and 5 s frame-cycle time. Images are extracted from exemplary movies S1, S2 and S3. Scale bars denote 4 μm. Lower panels: Kymographs of green and red circled molecules. (B) Survival time distributions of SiR-HaloTag-T15R at the time-lapse condition indicated on top (black lines) and survival time functions obtained by GRID (red lines). (Number of total/bound molecules: 32 933/1739 (continuous); 13 882/1419 (1 s time-lapse); 14 066/1608 (5 s time-lapse) in 88 cells). (C) State spectra of dissociation rates of T19R, T9R, T13R and T15R obtained by GRID using all data (red bars) and 500 resampling runs with 80% of data (black data points) as an error estimation of the spectra. The dashed blue line is given as guide to the eye. (D) Ratio of the dissociation rate of an individual TALE-TF to the mean dissociation rate of all TALE-TFs corresponding to the same dissociation rate cluster. Dissociation rates are extracted from the spectra shown in C). (E) Residence times of TALE-TFs extracted from the slowest dissociation rate cluster of the state spectra. Error bars denote standard deviation of the resampled spectrum in C).
Common analysis of the survival time distributions including two or three exponential decays (59,61–63) were not compatible with the data (Supplementary Figure S9). Thus, we used the method GRID (genuine rate identification) to extract dissociation rate spectra of TALE-TFs (Figure 2B, C and Supplementary Figure S7) by inverse Laplace transformation (47). Instead of using multi-exponential fitting, GRID optimizes amplitudes of a spectrum of 200 possible dissociation rates logarithmically spaced between 10–3 to 101 s–1. This is possible and robust due to the regularization of GRID, which includes that the 200 amplitudes are constrained to positive values and the corresponding 200 dissociation rates are not free parameters anymore. Global consideration of all three time-lapse conditions allowed correcting for photobleaching. Moreover, it enabled access to a broad range of dissociation rates, since each time-lapse condition contributed sensitivity to a certain time regime and all collectively spanned a broad temporal range.
All constructs showed six dissociation rate clusters and equal photobleaching rates (Figure 2C, Supplementary Figure S10, Supplementary Tables S3 and S4). The five dissociation rate clusters corresponding to short and intermediate binding events were similar to within ca. ±10% for all constructs (Figure 2C, D, Supplementary Figure S10 and S11). In contrast, the dissociation rate cluster corresponding to the longest binding events differed between the constructs up to ca. ± 40% (Figure 2C, D, Supplementary Figures S10 and S11). We previously demonstrated that GRID is able to correctly resolve up to 6 rate clusters in the temporal range measured here, if the underlying distribution is distinct (47). For very broad rate distributions, GRID started to insert gaps, leading to multiple broad rate clusters. However, extremal rates were still correctly obtained. Thus, the dissociation rate cluster corresponding to the longest binding time should correctly reflect dissociation events present in the system. From these dissociation rate clusters that differed among TALE-TFs we calculated the corresponding residence times and obtained an increasing series of residence times ranging from 186 s for T19R to 407 s for T15R (Figure 2E).
Since the TALE-TFs were designed to only differ in their DBD, we reasoned that the dissociation rate cluster differing strongly among the constructs reflect this structural difference and include unbinding from target sequences. Of note, TALE-TFs with shorter DBD bind only parts of the TetO sequence and have several hundreds to thousands of identical or nearly identical target sequences throughout the genome, to which they are expected to bind with similar affinity (Supplementary Table S1). This dissociation rate cluster presumably contains binding events to all such target sequences. In contrast, given the thickness of the HILO light sheet of ca. 0.6 μm and the height of the nucleus of ca. 5 μm, only ca. 0.6 of the ∼5 TetO sequences from a reporter gene construct will be in focus at any time and such sites will contribute little to this dissociation rate cluster. Since the other rate clusters differed much less among the TALE-TFs, we reasoned that they predominantly reflect binding to sequences unrelated to TetO. Consistent with this interpretation, the fraction of those clusters in the event spectrum of GRID (Supplementary Figure S10 and Supplementary Table S4) was up to several orders of magnitude larger than the fraction of the cluster corresponding to the longest binding time. This event spectrum counts the number of binding events that occur within a given time period. In contrast, the fractions in the state spectrum of GRID (Figure 2C and Supplementary Table S3) reflect the number of molecules present in each binding state at any time snapshot. Thus, the state spectrum appreciates the relative importance of long binding states in tying molecules down to chromatin. We currently cannot give further biological or mechanistic interpretation of the six dissociation rate clusters beyond separating them in similar and differing among TALE-TFs. Future studies are thus necessary to fully detail the mechanistic origin of the dissociation rate clusters.
We note that the dependence of residence time with the length of the DBD is non-linear, similar to our previous observations (Supplementary Figure S12) (23) and to the dependence reported for in vitro residence times of TALEs (64). The helical pitch of TALE DBDs wrapped around DNA differs from the helical pitch of DNA (65,66). This might lead to destabilization of TALE-TFs with long DBD (T19R), which have been shown to tolerate more mismatches (67) and whose C-terminal repeats are less important for efficient TALE-TF binding (68). Thus, the unexpectedly short binding construct T19R might only partially wrap around the TetO sequence and associate to multiple partial or nearly identical sites in the genome with lower residence time than the other TALE-TFs.
From the fractions of the GRID spectrum, number of binding events detected per frame, occupation frequency of the target site and labelling density, the number of binding sites contributing to the rate cluster including dissociation from target sequences can be estimated (Supplementary Information and Supplementary Table S1). This yielded ca. 3000 sites for T19R, many more than the 6 complete TetO sites present at reporter genes or nearly identical sites in the genome, which is consistent with only partial binding of the TetO sequence by this construct. For T15R, the estimate yielded ca. 4 times more and for T13R ca. 2 times less binding sites then expected from BLAST analysis of partial and related TetO sites (Supplementary Table S1). This indicates that T15R binds to additional sequences not accounted for in the BLAST analysis. On the other hand, the discrepancy could also reflect the uncertainty associated with this estimate. T9R was estimated to bind only ca. 1/3 of the sites accessible to T13R, perhaps again reflecting this uncertainty. In addition, since only a small fraction of TALE-TFs was labelled, we do not expect to be able to sample all potential binding sites, at least not for TALE-TFs with short DBD.
We further compared association of TALE-TFs to chromatin by calculating the number of binding events per area and time and normalized to concentration (pseudo-on-rate) (Supplementary Figure S13) (69). The pseudo-on-rates predominantly report binding to unspecific sites, which together with free diffusion dominates the target search time (70). We found the pseudo-on-rate was comparable for all TALE-TFs. In addition, we estimated the cycle time between free diffusion and unspecific binding from the overall bound fraction obtained by analysis of diffusion in continuous movies and from the fraction of unspecific dissociation events in the GRID spectra (Supplementary information, Supplementary Figure S14 and Supplementary Table S8) (28). We found a maximum relative difference of ca. 1.6 between constructs (Supplementary Figure S15). Together, the equal pseudo-on-rate and similar cycle time indicate that the search time is similar for all constructs.
TF residence time and concentration increase transcription
To quantify the number of mRNA molecules of the reporter gene in individual cells of the cell lines including a single copy of the reporter gene, we used smFISH (Figure 3A and Materials and Methods) (23,71). This approach also enables identifying sites of nascent mRNA transcription, the number of nascent mRNA molecules at those sites (burst height) and the percentage of cells exhibiting active nascent transcription (burst frequency) (Materials and Methods) (48). FISH probes were targeted to the SNAP gene and MS2 stem loop sequences, which are not present in the human genome and could be detected without false-positives (Supplementary Figure S16). Besides mRNA, we also stained the cell membrane using Lectin-FITC and the HaloTag-TALE-TF using TMR to quantify their nuclear concentration in individual cells with U2-OS Halo-CTCF C32 (49) as calibrated standard (Figure 3A and Supplementary Figure S3).
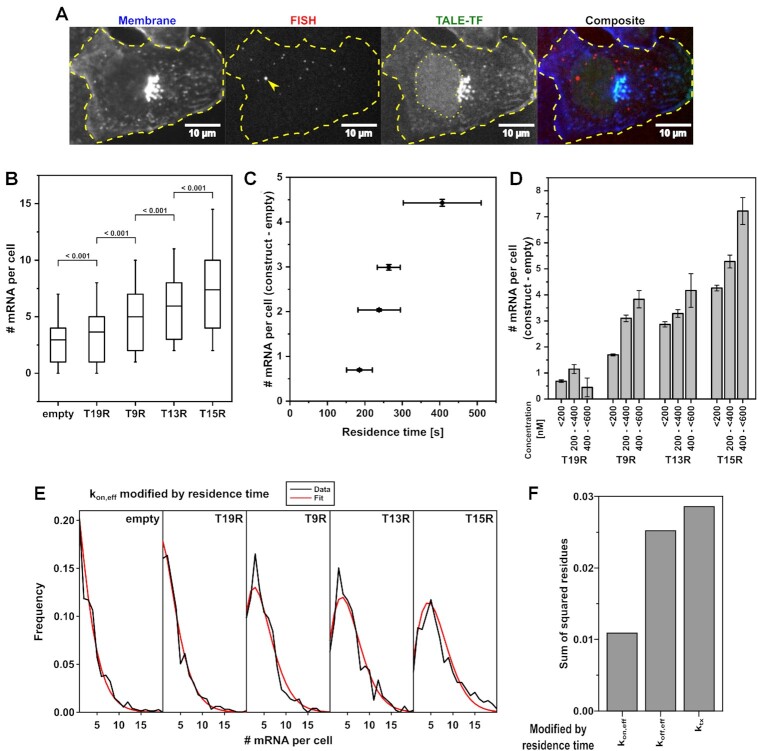
Figure 3.
TF residence time and concentration stimulate transcription activation. (A) Example images of the smFISH methodology: Lectin-FITC-stained cell membrane (blue; contrast 0 3000), smFISH of specific mRNA transcripts and nascent sites of transcription (arrow) (red; z-projection; contrast 2 40), TMR-stained HaloTag-TALE-TF (green; contrast 0 500) and composite image. Dashed line indicates the cell outline. Dotted line indicates the nucleus. The bright aggregate visualized by membrane stain and seen in the TALE-TF channel due to bleed-through probably corresponds to endoplasmic reticulum or Golgi apparatus. (B) mRNA distribution in cells expressing no TALE-TF (empty) and in cells expressing the different TALE-TF with < 300 nM. Cell numbers: N = 752 (empty); N = 888 (T19R); N = 690 (T9R); N = 664 (T13R); N = 790 (T15R). Mean (line), 25th/75th percentile (box) and 10th/90th percentile (whiskers) define features of box plot. Significance was determined with Wilcoxon–Mann–Whitney two-sample rank test. (C) Mean mRNA numbers from B) of cells expressing TALE-TF subtracted with the mean mRNA number of cells expressing no TALE-TF plotted versus TALE-TF residence time. Error bars denote s.e.m. for mean mRNA numbers and resampling error estimation for residence times. (D) Mean mRNA numbers of cells expressing TALE-TF at the indicated concentration subtracted with the mean mRNA number of cells expressing no TALE-TF. Cell numbers (<200 nM, 200 to <400 nM, 400 to <600 nM): T19R N = 655, 375, 113; T9R N = 509, 282, 102; T13R N = 478, 256, 33; T15R N = 580, 365, 155). Error bars denote s.e.m. (E) mRNA distributions of TALE-TFs together with the distribution inferred by BIRD for the case where TF residence time modifies kon,eff of the gene. (F) Comparison of mRNA distributions inferred by BIRD with the measured mRNA distributions of TALE-TFs by means of the lowest sum of squared residues in cases were TF residence time modifies kon,eff, koff,eff or ktx.
To determine the effect of TF residence time on transcription, we compared the distributions of mRNA molecules measured by smFISH in a cell line without TALE-TF and in the four TALE-TF cell lines (Figure 3B). In these experiments, we minimized the effect of varying TALE-TF concentration by only considering cells with concentrations below 300 nM. All cell lines showed different mRNA levels, with mutually significant increases from empty to T19R, T9R, T13R and T15R (Figure 3B). Overall, the activation potential of the TALE-TFs is low, consistent with the minimal promoter design including a single target site. In particular, T19R with least residence time only increased transcription by ca. 30% compared to cells without TALE-TF. The mean number of mRNA produced due to TALE-TF activation above the background of leaky transcription increased with TALE-TF residence time (Figure 3C and Supplementary Figure S17A). Similar to previous findings (16,22,23,28,30,31), this suggests that TF residence time is a regulatory factor of transcription.
Next, we determined the effect of TF concentration on transcription. Therefore, we suppressed leaky AID-based degradation (50) of TALE-TFs by adding 200 μM Auxinole for 24 h before sample preparation. We excluded an effect of Auxinole on the general transcription mechanism by comparing the mRNA distributions measured by smFISH in cells without TALE-TF in absence and presence of Auxinole (Supplementary Figure S18). We then quantified the mean number of mRNA molecules in each of the TALE-TF cell lines and assigned the results to three concentration bins (Figure 3D and Supplementary Figure S17B). As expected from previous findings (6,26,27,33,34), all but one condition exhibited a larger number of mRNAs, the higher the TALE-TF expression level was. The deviation of T19R might be due to the overall low transcription potential of this construct, which led to a large relative error for the highest concentration. Altogether our data again demonstes that TF concentration is a regulatory factor of transcription.
TF residence time and concentration solely modify the burst frequency
After having shown that both TF residence time and concentration positively correlate with transcription activation, we aimed at understanding whether TF residence time affected the on-rate kon,eff, the transcription rate ktx or the off-rate koff,gene of the gene in a two-state model of transcriptional bursting (6,11,32). We therefore developed Bursting Inference from mRNA Distributions (BIRD), an inference algorithm based on iterative fitting of RNA distributions with semi-analytical solutions to the systems of differential equations describing a two-state gene bursting model (Supplementary Information and Supplementary Table S9). Previously, gene kinetics was simulated with the Gillespie algorithm (72) or modelled using hypergeometric series (73). We globally applied BIRD to the mRNA distributions obtained in the four TALE-TF cell lines at TF concentrations <200 nM, while allowing only one of the rates of the two-state model to be varied at a time. Modulation of kon,eff by the TALE-TF residence time resulted in the best representation of our data (Figure 3E) in terms of the sum of squared residues, compared to modulating one of the other rates (Figure 3F and Supplementary Figure S19).
Using the global BIRD analysis, we obtained the effective on-rate, the off-rate and the transcription rate of a two-state model from the mRNA distributions experimentally measured with smFISH (Supplementary Table S5). These rates are still normalized with the degradation rate. We tested whether the kinetic model underlying BIRD, which assumes a well-defined TF concentration, would falsely infer rates from experimental distributions that combine a finite range of TF concentration (Supplementary Table S5). However, a finite concentration range had only small effects. Importantly, extrinsic noise, for example due to cell-to-cell variations in the abundances of RNA polymerase II and other transcription cofactors, contributes to cell-to-cell variations of mRNA distributions and was shown to dominate over intrinsic noise resulting from the stochastic nature of TFs and the transcription process (35,74). Since we do not have control over such extrinsic noise contributions, we did not account for extrinsic noise when inferring effective rates of the two-state model, which limits the accuracy of inferred values. Another source of noise, which we did not account for, comes from the fraction of cells that completed replication of the reporter gene and thus might transcribe two alleles (35,75,76).
To confirm the notion obtained from the BIRD analysis that TF residence time affects the on-rate of the reporter gene, we quantified its effect and the effect of TF concentration on several bursting parameters. First, we determined both frequency and height of a nascent transcription site with smFISH in each of the TALE-TF cell lines (Materials and Methods) (48). This approach underestimates the burst frequency and overestimates the burst height, since we only considered nascent sites with four or more transcripts to minimize false positives (Materials and Methods). Nevertheless, a relative comparison of the parameters between different conditions is possible. Consistent with the BIRD analysis, both higher TF residence time and higher concentration resulted in higher burst frequency (Figure 4A and B), while neither TF residence time nor concentration had an effect on the burst height (Figure 4A and B).
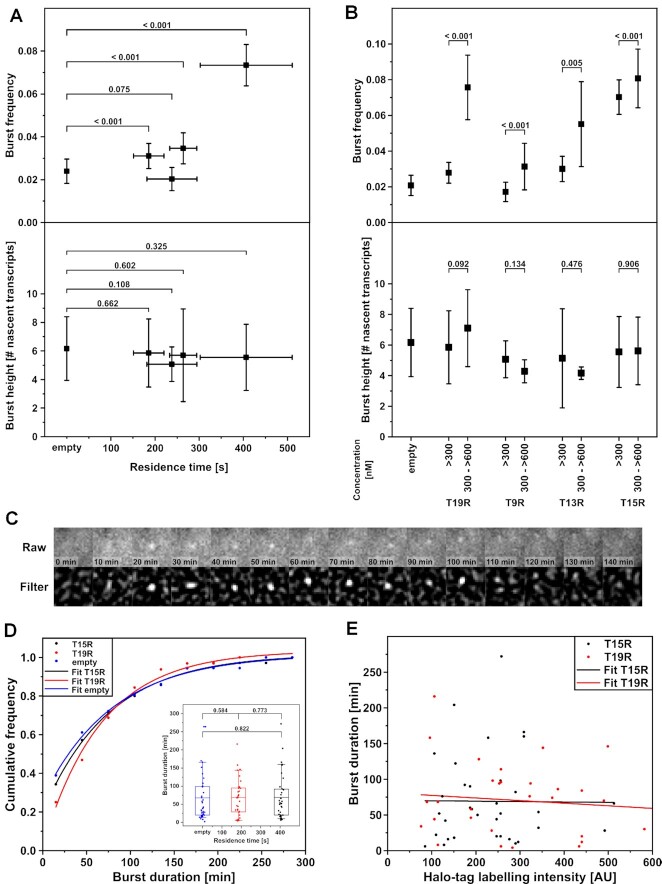
Figure 4.
Effect of TF residence time and concentration on burst parameters. (A) Effect of TF residence time on burst frequency and burst height. Both parameters were determined with smFISH in cells expressing <300 nM TALE-TF. Number of detected transcription sites: N = 19 (empty); N = 28 (T19R); N = 14 (T9R); N = 23 (T13R); N = 58 (T15R). Error bars denote sqrt(N) for burst frequencies, s.d. for burst heights and resampling error estimation for residence times. Significance was tested with unranked two sample t test. (B) Effect of TF concentration on burst frequency and burst height. Both parameters were determined with smFISH. Number of detected transcription sites (<300 nM, 300 to <600 nM): N = 19 (empty); N = 28, 19 (T19R); N = 14, 7 (T9R); N = 23, 6 (T13R); N = 58, 26 (T15R). Error bars denote sqrt(N) for burst frequencies and s.d. for burst heights. (C) Example time series of a transcription site detected with the MS2 system in a living cell expressing T15R. Raw: z-projection of complete cell height; Filter: wavelet filtered z-projection. (D) Cumulative frequency distributions of burst durations of cells expressing T15R (black symbols), T19R (red symbols), or no TALE-TF (empty, blue symbols). Lines represent mono-exponential fits. Number of transcription events: N = 35 (T15R), N = 32 (T19R), N = 36 (empty). Inset: burst duration as function of TF residence time. Mean (line), 25th/75th percentile (box) and 10th/90th percentile (whiskers) define features of box plot. Significance was tested with Wilcoxon–Mann–Whitney two-sample rank test. (E) Scatter plot of burst duration versus TF concentration of cells expressing T15R (black symbols) or T19R (red symbols). Lines represent linear fits. (Number of transcription events: N = 35 (T15R) and N = 32 (T19R).
The burst frequency changes if either the on-rate or the off-rate of the gene is altered, resulting in either more or longer bursts, respectively. We thus next applied the MS2 system to visualize transcription bursts in living cells and thereby directly assessed their duration (Figure 4C, Supplementary Video 4 and Materials and Methods) (9). Burst durations lasted for several minutes up to several hours for the TALE-TFs with longest (T15R) and shortest (T19R) residence time, with similar average burst duration of 69 min for T15R and 68 min for T19R (Figure 4D). The cumulative frequency distributions of burst durations were well described by a mono-exponential function for both T15R and T19R, indicating a single rate-determining step for the termination of bursts (Figure 4D). We also determined the effect of TF concentration on the burst duration, by quantifying the TMR-HaloTag labelling intensity of T15R and T19R (Figure 4E). As with TF residence time, burst duration was also independent of TF concentration.
We tested whether burst duration was indeed completely independent of the TF by measuring the burst duration of leaky reporter gene transcription in the cell line without TALE-TF. As predicted, the average burst duration (68 min) and the cumulative frequency distribution of burst durations were comparable to the duration and distribution in presence of TF (Figure 4D). In addition to the burst durations, we also extracted the average burst heights from our MS2 time traces. Consistently, burst height was independent of both TF residence time and concentration (Supplementary Figure S20). Overall, our experiments confirm the BIRD analysis and suggest that binding of a TF solely affects the on-rate kon,eff of the reporter gene.
Long TF residence time more efficiently activates transcription than frequent TF binding
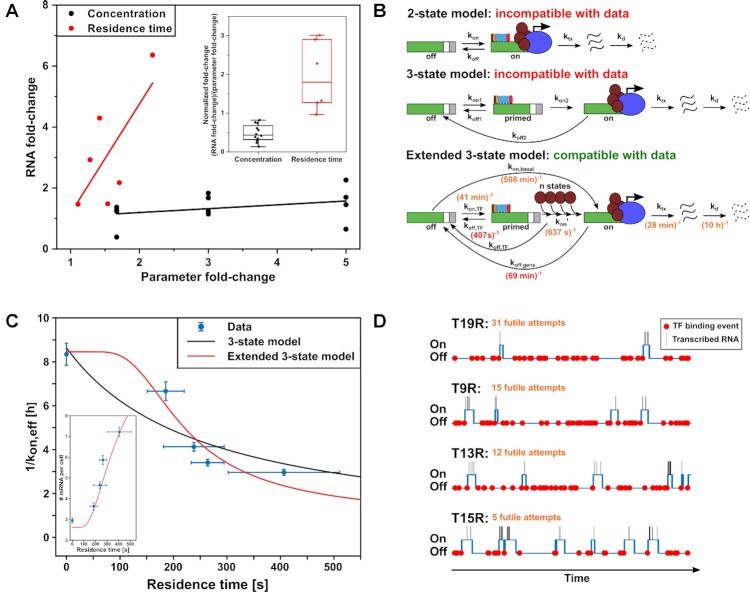
Since both high TF residence time and high TF concentration increase transcription, the question arises whether it is more important to have long residence times or frequent association events of TFs for efficient activation of transcription. To answer this question, we calculated the fold-changes in RNA production for each difference in residence time of the four TALE-TF constructs, and for each difference in concentration of the different concentration bins, at the parameter ranges comprised by our system (Figure 5A and Supplementary Figure S21). A certain fold-change in TF residence time affects a larger fold-change in RNA production compared to the same fold-change in TF concentration. To better compare the severity of this effect, we normalized each RNA fold-change by the underlying parameter fold-change (Figure 5A and Supplementary Figure S21) and found a 4 times larger effect for TF residence time.
Figure 5.
TF residence time dominates over concentration in transcription activation. (A) Effect of TF residence time and concentration on mRNA fold-change. mRNA fold-changes were calculated from mean mRNA levels (construct – empty) shown Figure 3C and D. Residence time fold-changes were calculated for each TALE-TF combination. Concentration fold-changes were calculated from the middle values of the three concentration bins shown in Figure 3D. Lines are guides to the eye. Inset: RNA fold-change normalized to corresponding parameter fold-change. (B) Sketches of the 2-state, 3-state and extended 3-state models of gene transcription. Rates indicated in the extended 3-state model were extracted from measurements (red font) or from BIRD and modeling (orange font). (C) Plot of 1/kon,eff as obtained by BIRD versus residence time (blue symbols). Lines represent fits to the 3-state model (Supplementary Equation 22, black) and to the extended 3-state model (Supplementary Equation 23, red). Error bars denote s.d. for BIRD inference and s.d. for residence time. Inset: Measured mean mRNA number per cell versus TALE-TF residence time for cells with <200 nM TALE-TF (blue data points) and predicted mRNA number by extended 3-state model (red). (D) Simulations of TF binding (red spheres), gene switching between off / primed and on-state (blue line) and transcriptional bursts (black lines) for T19R, T9R, T13R and T15R with transition rates of the extended 3-state model taken from measurements, BIRD and modelling (Supplementary Table S6). The average number of futile TF binding events before successful transcription is indicated (orange).
The two-state model consisting of an off-state and an on-state of the gene, from which transcription occurs, well describes the RNA distribution of gene transcription (Figure 3E) (6,11,32). However, it is not detailed enough to include TF dissociation, in particular as the off-rate of the gene is not equal to or determined at all by binding of the TF (Figure 5B). The next simple model, a three-state model that includes a primed state from which transcription occurs upon binding of the TF and that switches to the off-state upon TF dissociation, predicts equal importance of TF residence time and TF concentration for transcription activation. To explain a dominant effect of TF residence time over TF concentration, we thus considered a multi-state model with n successive states between the primed state and the on-state of the gene (Figure 5B and Supplementary Information). In the case of large n, this model converges to the extended three-state model that predicts a highly non-linear dependency of the time necessary to switch from the off-state to the on-state (1/kon,eff) with TF residence time (Supplementary Information). Indeed, the relation between 1/kon,eff in presence or absence of TALE-TF and the residence time (1/koff,TF) of our TALE-TFs is incompatible with the response predicted by the three-state model (Adj. R2 = 0.64), but well described by the steep response of the extended three-state model (Adj. R2 = 0.88) (Figure 5C). For TF residence time, the relation with the bursting rate in the extended 3-state model is a Hill function with coefficient 1.4 (Supplementary Equation S21 and (77)). In contrast, our model predicts a Hill function with coefficient 1 for the relation between bursting rate and concentration (Supplementary Equation S21). We could however not fully sample this concentration dependence due to the small accessible concentration range of TALE-TFs in our system. Other systems might exhibit a strong Hill behavior for concentration if they include traits such as TF dimerization or TF binding synergy of several TFs (78). We chose a system without these traits to arrive at a basic picture of transcription regulation.
By combining our values for the TF residence time and the burst duration with the values for the effective on-rate, the off-rate and the transcription rate of the gene obtained by BIRD analysis of the RNA distributions, we could assign rates and values to all the transitions and parameters of the extended three-state model (Figure 5C and Supplementary Information). To arrive at these rates, we used the values for burst frequency and burst size determined by BIRD, and not those obtained by smFISH. In smFISH an intensity threshold is used in the detection of nascent sites to avoid assigning false-positives, which overlooks small bursts. In contrast, BIRD analyses the whole mRNA distribution, not only nascent sites, and thus yields less biased values. We found an on-rate of the TF to the gene, kon,TF of (41 min)–1, burst frequencies of 0.12, 0.15, 0.22, 0.25 and 0.28 for empty, T19R, T9R, T13R and T15R, a transcription rate ktx of (28 min)–1, an average burst size of 2.5 transcripts and a degradation rate kd of (10 h)–1 (Supplementary Table S6)
Interestingly, it takes ∼637 s to pass through the n states following TF binding, which is comparable to the TF residence time. This yields mechanistic insight into the importance of the TF residence time: While the long binding T15R on average undergoes 5 futile binding event until complete transition to the on-state of the gene and successful transcription activation, the short binding T19R already undergoes 31 futile binding events (Supplementary Information). We confirmed insufficient gene activation by short TF residence time by simulating the transcription process according to the extended three-state model using all transition rates for all four TALE-TFs (Figure 5D and Supplementary Information). All TALE-TFs bind with equal frequency to the target site, but due to varying numbers of futile attempts, burst frequency increases with TALE-TF residence time.
DISCUSSION
We quantified the influence of both TF residence time and concentration on the bursting parameters of a single well-defined reporter gene. We found that both regulatory traits solely affected the rate of switching on transcription, as expected from previous reports on the role of TF binding (2,7,11,16,17,23–31). Strikingly, we found that the regulatory effect of varying TF residence time was significantly more important than the effect of varying concentration at the parameter ranges comprised in our system. This enabled us to obtain mechanistic and kinetic details about the transcription activation process beyond the common two-state or a simple three-state description of gene transcription (11,32). Rather, we found an extended three-state model was compatible with our data, in which TF binding triggers several successive transitions of the system until the gene assumes the active state and productive transcription starts. Our model is compatible with recently identified control points of transcription regulation (19) and adds to previous considerations on three-state or multi-state models (11,35,73,79,80). In our model, for which we could determine all transition rate constants, presence of the TF increases the probability of the gene to transit from the off state over a primed state to the active state. Overall, not just the quantity of TF interactions matters, but their quality: the probability for a successful transition to the active state is more sensitive to the TF residence time than to its concentration.
The burst duration of 70 min and the small burst size we observed for the artificial reporter gene agree with burst parameters reported previously for endogenous mammalian genes (Supplementary Table S7) (11,15,24,27,28,30), although shorter burst durations were also reported (31,35). At first sight, the corresponding small transcription rate with one transcript produced every 28 minutes conflicts with the fast elongation speed of RNA polymerase II of ca. 1 kb/min (12,15,81–83). However, factors such as a slow release rate of RNA transcripts from the reporter gene, or accumulation of transcripts at the gene end and common release can account for the long burst duration observed by others and us.
One of the main tasks of a TF is to recruit cofactors necessary for transcription initiation (84,85). Such recruitment processes require time and would benefit from long residence times of the TF. Indeed, successful cofactor recruitment becomes more probable the longer the TF is associated to the promoter (86,87). The activation domain of our TALE-TFs, VP64, directly interacts with several subunits of the mediator complex (88). Together, possible candidates for the transitional steps between primed and active states of the gene include recruitment of the mediator complex and subsequent assembly of general transcription factors and RNA polymerase II (89). Additional processes might include binding of histone writers and readers such as CBP, P300 and BRD4 (90–92). Of note, leaky transcription adds a parallel path to TF-mediated gene activation and leads to transcription even in the absence of the TF. This suggests that at least some steps between the off state and the active state should be able to occur independent of the TF, albeit with much lower probability. For general TFs, a self-sufficient assembly of the pre-initiation complex is possible (93). As a consequence of the additional cascade of steps in the extended three-state model, an exponential relationship between the TF residence time and the number of futile binding attempts of a TF before successful transition of the gene to the active state is predicted. While T15R only binds twice as long as T19R, it activates the gene six times more efficiently, which reflects the advantage of a long residence time for successful transcription activation.
Given the non-linear dependence of successful transcription activation on TF residence time, the question arises whether residence times of hundreds of seconds as for the TALE-TFs are generally necessary for successful transcription. Residence times comparable to the TALE-TFs have been observed for other transcription factors including GR (31,94), SRF (62), CDX2 (47), TBP (95,96), LacI (97) and TetR (98). However, considerably shorter residence times were reported for many TFs including p53 (28,59,99), p65 (22), Sox2/Oct4 (33), STAT1 (100), Zelda (101) and CREB (102). In contrast to our minimal reporter gene system with a single TF target site, endogenous genes may include additional regulatory traits that compensate for short residence times. A possibility to increase the on-rate of the gene is to increase the number of TF target sites at the promoter (11), which reduces the search time of the TF to the location (98). Enhancers additionally add a prominent regulatory level to increase the burst frequency of genes (3,13,14,35). In addition to the on-rate, the burst size or the burst duration could be increased once the gene is active. The burst size was found to be influenced by the nature of the activation domain (30) and the core promoter sequence (3). Similarly, increasing the number of TF binding sites at the promoter was shown to increase the number of nascent transcripts (11,30). Indeed, proximal promoters of many endogenous genes comprise multiple TF target sites, as do enhancer elements (103–107). With an increasing number of TF sites at promoter or enhancer, the propensity of TFs to form dynamic TF condensates via their low-complexity domains increases (108), as has been observed for various factors associated with transcription (109–113). The size of such condensates was found to correlate with transcription output (114) and GR hubs have the potential to prolong transcription bursts (31). Thus, condensate formation is an elegant way to locally increase the concentration of TFs and thereby compensate for the disadvantage of short TF residence times.
We can obtain an upper limit for the time that any of the TALE-TFs present in the nucleus needs to find the single specific target site within the promoter of the reporter gene from the on-rate of the gene and the number of futile binding events. In our experiment, this target search time is 41 min. With the concentration of TALE-TF of ca. 100 nM in a U2-OS nucleus of approximately an ellipsoidal volume of π/6 × 10 μm × 10 μm × 5 μm, there are ca. 16 000 TALE-TF in the nucleus. Thus, the time for one TALE-TF molecule to find the target sequence is ca. 10 900 h (456 days). A 1.6x shorter search time of ca. 7000 h (292 days) has been observed for TetR to find a single TetO site in U2-OS cells (98). The difference might reflect differences in the search process between both factors. For one Sox2 molecule in embryonic stem cells, a search time to find any target sequence of 377 s was reported, and estimated to be 31 days to find a single target site (33), ten times shorter than for TetR. Again, different search mechanisms might account for this difference, or differences in calculating the search time from the bound fraction, residence time and number of target sites for Sox2 versus from burst frequency and concentration for TALE-TF and the direct association measurement for TetR. Search times of ca. 100 s reported for p53 (28) and CTCF (115) to find any target sequence face similar challenges as Sox2 to convert to the search time of one specific site. The search time of TALE-TF is significantly longer than the search time of ca. 0.1 h for one Lac repressor to find a single operator in E. coli (ca. 120 s for any TF) (116), and the search time of ca. 5 h for one Mbp1p molecule to find a single target site in S. cerevisiae (ca. 50 s for any TF) (7). The difference in search time between bacterium and yeast presumably predominantly reflects differences in chromatin organization and to some extend in genome size. The difference between yeast and human corresponds within a factor of 3 to the difference in genome size. Consequently, mammalian cells need to scale up the TF numbers compared to bacteria and yeast to compensate for the very long search time of one TF to find a single target site.
Our data suggest that the TALE-TF on average should be already dissociated once the gene is in the active state and transcription starts, since TF residence time is comparable to the transition time from the primed to the active state and the transcription rate is low. Thus, while the TALE-TF helps in transiting the gene to the active state, most RNA transcription initiation events of a burst will occur without a TALE-TF bound to the promoter. Consistently, we observed that the burst duration was independent of the TF and its residence time. Our observations are also consistent with a recent finding that RNA polymerase II recruitment occurs only after burst initiation and is rather unregulated (19), and the finding of a delay of RNA synthesis compared to GR binding of ∼3 min (31). In contrast, recent reports in yeast suggest that TF residence time is directly coupled to the burst duration, while TF concentration affects the on-rate of the gene (16,17,29). Thus, transcription in yeast apparently only occurs while the TF is bound to the promoter. This is consistent with the burst duration of a few minutes in yeast (7,16,17,29), which is comparable to the elongation time of the gene and compatible with a few initiation events during the residence time of the TF. In comparison, typical burst durations in mammals are much longer than the TF residence time (11,15,24,27,31). The different effects of TF residence time in mammals and yeast point to differences in the regulatory mechanisms of transcription activation in both species. For example, it seems that co-factor recruitment and assembly of the transcription machinery is more efficient in yeast compared to mammals. This might be due to differences between components of the yeast and mammalian pre-initiation complexes, for example TFIID or mediator (117). It is further interesting to speculate that the ability to uncouple TF binding to and activation of the gene from RNA polymerase II recruitment enabled higher organisms to evolve enhancer elements, to which TFs predominantly bind. Our observations for the TALE-TFs predict that also an enhancer would only shortly, in the range of a few minutes, need to closely interact with a gene promoter for successful activation. Compatible with such a scenario, it has been observed that mediator condensates only transiently come into close proximity to a site of transcription while transcription continues to go on (110). Further studies will be necessary to appreciate to full extend the kinetic interplay of gene activation and actual gene transcription and differences in transcription regulation between yeast and mammals.
Given the similarities in burst size, burst duration and transcription rate of the artificial reporter gene to other mammalian systems, the reporter gene with one TF target site in the proximal promoter constitutes a basis for the kinetic understanding of gene transcription. In particular, the importance of the TF residence time, the delay between TF binding and the onset of mRNA transcription, and the TF-independent transcription termination will be able to inform the kinetic behaviour of endogenous mammalian genes with more complex promoter structures.
DATA AVAILABILTY
Data supporting the findings of this manuscript will be available from the corresponding author after publication upon reasonable request. All raw single particle tracking data, mRNA distributions, burst durations and AID degradation data are freely available in xlsx file format at https://doi.org/10.5061/dryad.xksn02vf4.
The BIRD algorithm is freely available. A MatLab version of BIRD is available at https://gitlab.com/GebhardtLab/BIRD.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Astrid Bellan-Koch for her help with cloning. phage-CMV-CFP-24xMS2 (Addgene plasmid #40651) and phage-ubc-nls-ha-tdMCP-gfp (Addgene plasmid #40649) were gifts from Robert Singer (Albert Einstein College of Medicine, New York, USA). pcDNA5/FRT/TO V5 (Addgene plasmid #19445) was a gift from Harm Kampinga (University of Groningen, Groningen, The Netherlands). pMK232 (CMV-OsTIR1-PURO; Addgene plasmid #72834) and pMK292 (mAID-mCherry2-NeoR; Addgene plasmid #72830) were gifts from Masato Kanemakie (National Institute of Genetics, Mishima, Japan). pLenti-CMV-rtTA3 (Addgene plasmid #26429) was a gift from Eric Campeau (University of Massachusetts Medical School, Worcester, USA). pOG44 was kindly provided by David Suter (Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). Halo-SiR ligand was kindly provided by K. Johnsson (Max Planck Institute for Medical Research, Heidelberg, Germany). We thank the Core Facility FACS of Ulm University for their help with cell sorting, with special thanks to Dr Simona Ursu and Daniela Froelich. The authors thank the Ulm University Center for Translational Imaging MoMAN for its support.
Author contributions: J.C.M.G. conceived the study; A.P.P. and J.C.M.G. designed experimental approaches; A.P.P. designed and performed cloning and cell line generation; A.P.P. performed experiments; A.P.P. and J.H. analysed data; J.H. devised the BIRD algorithm; J.H. and J.C.M.G modelled data; A.P.P. and J.C.M.G wrote the manuscript with input from all authors.
Contributor Information
Achim P Popp, Institute of Biophysics, Ulm University, Albert-Einstein-Allee 11, 89081 Ulm, Germany.
Johannes Hettich, Institute of Biophysics, Ulm University, Albert-Einstein-Allee 11, 89081 Ulm, Germany.
J Christof M Gebhardt, Institute of Biophysics, Ulm University, Albert-Einstein-Allee 11, 89081 Ulm, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Program [637987 ChromArch to J.C.M.G.]; German Research Foundation [SPP 2202 GE 2631/2-1 to J.C.M.G.]; Carl Zeiss Foundation (to A.P.P.); J.C.M.G. acknowledges support by the Collaborative Research Centre 1279 [DFG 316249678]. Funding for open access charge: European Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rodriguez J., Larson D.R.. Transcription in living cells: molecular mechanisms of bursting. Annu. Rev. Biochem. 2020; 89:189–212. [DOI] [PubMed] [Google Scholar]
- 2. Dar R.D., Razooky B.S., Singh A., Trimeloni T.V., McCollum J.M., Cox C.D., Simpson M.L., Weinberger L.S.. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:17454–17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larsson A.J.M., Johnsson P., Hagemann-Jensen M., Hartmanis L., Faridani O.R., Reinius B., Segerstolpe Å., Rivera C.M., Ren B., Sandberg R.. Genomic encoding of transcriptional burst kinetics. Nature. 2019; 565:251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chong S., Chen C., Ge H., Xie X.S.. Mechanism of transcriptional bursting in bacteria. Cell. 2014; 158:314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Golding I., Paulsson J., Zawilski S.M., Cox E.C.. Real-time kinetics of gene activity in individual bacteria. Cell. 2005; 123:1025–1036. [DOI] [PubMed] [Google Scholar]
- 6. So L., Ghosh A., Zong C., Sepúlveda L.A., Segev R., Golding I.. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011; 43:554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Larson D.R., Zenklusen D., Wu B., Chao J.A., Singer R.H.. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011; 332:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zenklusen D., Larson D.R., Singer R.H.. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 2008; 15:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chubb J.R., Trcek T., Shenoy S.M., Singer R.H.. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006; 16:1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S.. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006; 4:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suter D.M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F.. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011; 332:472–474. [DOI] [PubMed] [Google Scholar]
- 12. Yunger S., Rosenfeld L., Garini Y., Shav-Tal Y.. Single-allele analysis of transcription kinetics in living mammalian cells. Nat. Methods. 2010; 7:631–633. [DOI] [PubMed] [Google Scholar]
- 13. Bartman C.R., Hsu S.C., Hsiung C.C.-S., Raj A., Blobel G.A.. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell. 2016; 62:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fukaya T., Lim B., Levine M.. Enhancer control of transcriptional bursting. Cell. 2016; 166:358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tantale K., Mueller F., Kozulic-Pirher A., Lesne A., Victor J.-M., Robert M.-C., Capozi S., Chouaib R., Bäcker V., Mateos-Langerak J.et al.. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016; 7:12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donovan B.T., Huynh A., Ball D.A., Patel H.P., Poirier M.G., Larson D.R., Ferguson M.L., Lenstra T.L.. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019; 38:e100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mehta G.D., Ball D.A., Eriksson P.R., Chereji R.V., Clark D.J., McNally J.G., Karpova T.S.. Single-molecule analysis reveals linked cycles of RSC chromatin remodeling and Ace1p transcription factor binding in yeast. Mol. Cell. 2018; 72:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicolas D., Zoller B., Suter D.M., Naef F.. Modulation of transcriptional burst frequency by histone acetylation. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:7153–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bartman C.R., Hamagami N., Keller C.A., Giardine B., Hardison R.C., Blobel G.A., Raj A.. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell. 2019; 73:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tantale K., Garcia-Oliver E., L’Hostis A., Yang Y., Robert M.-C., Gostan T., Basu M., Kozulic-Pirher A., Andrau J.-C., Muller F.et al.. Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting. 2020; bioRxiv doi:25 August 2020, preprint: not peer reviewed 10.1101/2020.08.25.265413. [DOI] [PMC free article] [PubMed]
- 21. van den Berg A.A., Depken M.. Crowding-induced transcriptional bursts dictate polymerase and nucleosome density profiles along genes. Nucleic Acids Res. 2017; 45:7623–7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Callegari A., Sieben C., Benke A., Suter D.M., Fierz B., Mazza D., Manley S.. Single-molecule dynamics and genome-wide transcriptomics reveal that NF-kB (p65)-DNA binding times can be decoupled from transcriptional activation. PLoS Genet. 2019; 15:e1007891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clauß K., Popp A.P., Schulze L., Hettich J., Reisser M., Escoter Torres L., Uhlenhaut N.H., Gebhardt J.C.M.. DNA residence time is a regulatory factor of transcription repression. Nucleic Acids Res. 2017; 45:11121–11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fritzsch C., Baumgärtner S., Kuban M., Steinshorn D., Reid G., Legewie S.. Estrogen-dependent control and cell-to-cell variability of transcriptional bursting. Mol. Syst. Biol. 2018; 14:e7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalo A., Kanter I., Shraga A., Sheinberger J., Tzemach H., Kinor N., Singer R.H., Lionnet T., Shav-Tal Y.. Cellular levels of signaling factors are sensed by β-actin alleles to modulate transcriptional pulse intensity. Cell Rep. 2015; 11:419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ko M.S., Nakauchi H., Takahashi N.. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990; 9:2835–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larson D.R., Fritzsch C., Sun L., Meng X., Lawrence D.S., Singer R.H.. Direct observation of frequency modulated transcription in single cells using light activation. eLife. 2013; 2:e00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loffreda A., Jacchetti E., Antunes S., Rainone P., Daniele T., Morisaki T., Bianchi M.E., Tacchetti C., Mazza D.. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat. Commun. 2017; 8:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rullan M., Benzinger D., Schmidt G.W., Milias-Argeitis A., Khammash M.. An optogenetic platform for real-time, single-cell interrogation of stochastic transcriptional regulation. Mol. Cell. 2018; 70:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Senecal A., Munsky B., Proux F., Ly N., Braye F.E., Zimmer C., Mueller F., Darzacq X.. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014; 8:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stavreva D.A., Garcia D.A., Fettweis G., Gudla P.R., Zaki G.F., Soni V., McGowan A., Williams G., Huynh A., Palangat M.et al.. Transcriptional bursting and co-bursting regulation by steroid hormone release pattern and transcription factor mobility. Mol. Cell. 2019; 75:1161–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peccoud J., Ycart B.. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 1995; 48:222–234. [Google Scholar]
- 33. Chen J., Zhang Z., Li L., Chen B.-C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E.et al.. Single-molecule dynamics of Enhanceosome assembly in embryonic stem cells. Cell. 2014; 156:1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee R.E.C., Walker S.R., Savery K., Frank D.A., Gaudet S.. Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol. Cell. 2014; 53:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodriguez J., Ren G., Day C.R., Zhao K., Chow C.C., Larson D.R.. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019; 176:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y., Qi J., Shao J., Tang X.-Q.. Signaling mechanism of transcriptional bursting: a technical resolution-independent study. Biology. 2020; 9:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M.. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009; 6:917–922. [DOI] [PubMed] [Google Scholar]
- 38. Hageman J., Kampinga H.H.. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones. 2009; 14:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu B., Chao J.A., Singer R.H.. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J. 2012; 102:2936–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Natsume T., Kiyomitsu T., Saga Y., Kanemaki M.T.. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 2016; 15:210–218. [DOI] [PubMed] [Google Scholar]
- 41. Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F.. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011; 39:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A., Warman M.L.. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques. 2000; 29:52. [DOI] [PubMed] [Google Scholar]
- 43. Lukinavičius G., Umezawa K., Olivier N., Honigmann A., Yang G., Plass T., Mueller V., Reymond L., Corrêa I.R. Jr, Luo Z.-G.et al.. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 2013; 5:132–139. [DOI] [PubMed] [Google Scholar]
- 44. Tokunaga M., Imamoto N., Sakata-Sogawa K.. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008; 5:159–161. [DOI] [PubMed] [Google Scholar]
- 45. Gebhardt J.C.M., Suter D.M., Roy R., Zhao Z.W., Chapman A.R., Basu S., Maniatis T., Xie X.S.. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013; 10:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuhn T., Hettich J., Davtyan R., Gebhardt J.C.M. Single molecule tracking and analysis framework including theory-predicted parameter settings. Sci. Rep. 2021; 11:9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reisser M., Hettich J., Kuhn T., Popp A.P., Große-Berkenbusch A., Gebhardt J.C.M.. Inferring quantity and qualities of superimposed reaction rates from single molecule survival time distributions. Sci. Rep. 2020; 10:1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., Zimmer C.. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 2013; 10:277–278. [DOI] [PubMed] [Google Scholar]
- 49. Cattoglio C., Pustova I., Walther N., Ho J.J., Hantsche-Grininger M., Inouye C.J., Hossain M.J., Dailey G.M., Ellenberg J., Darzacq X.et al.. Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife. 2019; 8:e40164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yesbolatova A., Natsume T., Hayashi K., Kanemaki M.T.. Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods. 2019; 164–165:73–80. [DOI] [PubMed] [Google Scholar]
- 51. Loew R., Heinz N., Hampf M., Bujard H., Gossen M.. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 2010; 10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M.. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998; 2:437–445. [DOI] [PubMed] [Google Scholar]
- 53. Salem T.Z., Seaborn C.P., Turney C.M., Xue J., Shang H., Cheng X.-W.. The influence of SV40 polyA on gene expression of baculovirus expression vector systems. PLoS One. 2015; 10:e0145019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O’Gorman S., Fox D.T., Wahl G.M.. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991; 251:1351–1355. [DOI] [PubMed] [Google Scholar]
- 55. Boch J., Bonas U.. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010; 48:419–436. [DOI] [PubMed] [Google Scholar]
- 56. Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U.. Breaking the code of DNA binding specificity of TAL-Type III effectors. Science. 2009; 326:1509–1512. [DOI] [PubMed] [Google Scholar]
- 57. Beerli R.R., Segal D.J., Dreier B., Barbas C.F.. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Los G.V., Encell L.P., McDougall M.G., Hartzell D.D., Karassina N., Zimprich C., Wood M.G., Learish R., Ohana R.F., Urh M.et al.. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008; 3:373–382. [DOI] [PubMed] [Google Scholar]
- 59. Mazza D., Abernathy A., Golob N., Morisaki T., McNally J.G.. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012; 40:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reisser M., Palmer A., Popp A.P., Jahn C., Weidinger G., Gebhardt J.C.M.. Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development. Nat. Commun. 2018; 9:5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Agarwal H., Reisser M., Wortmann C., Gebhardt J.C.M.. Direct observation of cell-cycle-dependent interactions between CTCF and chromatin. Biophys. J. 2017; 112:2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hipp L., Beer J., Kuchler O., Reisser M., Sinske D., Michaelis J., Gebhardt J.C.M., Knöll B.. Single-molecule imaging of the transcription factor SRF reveals prolonged chromatin-binding kinetics upon cell stimulation. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Presman D.M., Ball D.A., Paakinaho V., Grimm J.B., Lavis L.D., Karpova T.S., Hager G.L.. Quantifying transcription factor binding dynamics at the single-molecule level in live cells. Methods. 2017; 123:76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Geiger-Schuller K., Mitra J., Ha T., Barrick D. Functional instability allows access to DNA in longer transcription Activator-Like effector (TALE) arrays. eLife. 2019; 8:e38298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.-K., Shi Y., Yan N.. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012; 335:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mak A.N.-S., Bradley P., Cernadas R.A., Bogdanove A.J., Stoddard B.L.. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012; 335:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guilinger J.P., Pattanayak V., Reyon D., Tsai S.Q., Sander J.D., Joung J.K., Liu D.R.. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods. 2014; 11:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meckler J.F., Bhakta M.S., Kim M.-S., Ovadia R., Habrian C.H., Zykovich A., Yu A., Lockwood S.H., Morbitzer R., Elsäesser J.et al.. Quantitative analysis of TALE–DNA interactions suggests polarity effects. Nucleic Acids Res. 2013; 41:4118–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Raccaud M., Friman E.T., Alber A.B., Agarwal H., Deluz C., Kuhn T., Gebhardt J.C.M., Suter D.M.. Mitotic chromosome binding predicts transcription factor properties in interphase. Nat. Commun. 2019; 10:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Berg O.G., Winter R.B., Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981; 20:6929–6948. [DOI] [PubMed] [Google Scholar]
- 71. Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S.. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008; 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gorin G., Wang M., Golding I., Xu H.. Stochastic simulation and statistical inference platform for visualization and estimation of transcriptional kinetics. PLoS One. 2020; 15:e0230736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ham L., Schnoerr D., Brackston R.D., Stumpf M.P.H.. Exactly solvable models of stochastic gene expression. J. Chem. Phys. 2020; 152:144106. [DOI] [PubMed] [Google Scholar]
- 74. Elowitz M.B., Levine A.J., Siggia E.D., Swain P.S.. Stochastic gene expression in a single cell. Science. 2002; 297:1183–1186. [DOI] [PubMed] [Google Scholar]
- 75. Skinner S.O., Xu H., Nagarkar-Jaiswal S., Freire P.R., Zwaka T.P., Golding I.. Single-cell analysis of transcription kinetics across the cell cycle. eLife. 2016; 5:e12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang M., Zhang J., Xu H., Golding I.. Measuring transcription at a single gene copy reveals hidden drivers of bacterial individuality. Nat. Microbiol. 2019; 4:2118–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Santillán M. On the use of the Hill functions in mathematical models of gene regulatory networks. Math. Model. Nat. Phenom. 2008; 3:85–97. [Google Scholar]
- 78. Xu H., Sepúlveda L.A., Figard L., Sokac A.M., Golding I.. Combining protein and mRNA quantification to decipher transcriptional regulation. Nat. Methods. 2015; 12:739–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Corrigan A.M., Tunnacliffe E., Cannon D., Chubb J.R.. A continuum model of transcriptional bursting. eLife. 2016; 5:e13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zoller B., Nicolas D., Molina N., Naef F.. Structure of silent transcription intervals and noise characteristics of mammalian genes. Mol. Syst. Biol. 2015; 11:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Boireau S., Maiuri P., Basyuk E., de la Mata M., Knezevich A., Pradet-Balade B., Bäcker V., Kornblihtt A., Marcello A., Bertrand E.. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007; 179:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Femino A.M., Fay F.S., Fogarty K., Singer R.H.. Visualization of single RNA transcripts in situ. Science. 1998; 280:585–590. [DOI] [PubMed] [Google Scholar]
- 83. Maiuri P., Knezevich A., De Marco A., Mazza D., Kula A., McNally J.G., Marcello A.. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011; 12:1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen K., Rajewsky N.. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007; 8:93–103. [DOI] [PubMed] [Google Scholar]
- 85. Lemon B., Tjian R.. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000; 14:2551–2569. [DOI] [PubMed] [Google Scholar]
- 86. Hettich J., Gebhardt J.C.M.. Transcription factor target site search and gene regulation in a background of unspecific binding sites. J. Theor. Biol. 2018; 454:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Biddle J.W., Nguyen M., Gunawardena J.. Negative reciprocity, not ordered assembly, underlies the interaction of Sox2 and Oct4 on DNA. eLife. 2019; 8:e41017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Poss Z.C., Ebmeier C.C., Taatjes D.J.. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013; 48:575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Soutourina J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018; 19:262–274. [DOI] [PubMed] [Google Scholar]
- 90. Bártová E., Krejčí J., Harničarová A., Galiová G., Kozubek S.. Histone modifications and nuclear architecture: a review. J. Histochem. Cytochem. 2008; 56:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li B., Carey M., Workman J.L.. The role of chromatin during transcription. Cell. 2007; 128:707–719. [DOI] [PubMed] [Google Scholar]
- 92. Wu S.-Y., Chiang C.-M.. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007; 282:13141–13145. [DOI] [PubMed] [Google Scholar]
- 93. Cramer P. Organization and regulation of gene transcription. Nature. 2019; 573:45–54. [DOI] [PubMed] [Google Scholar]
- 94. Garcia D.A., Johnson T.A., Presman D.M., Fettweis G., Wagh K., Rinaldi L., Stavreva D.A., Paakinaho V., Jensen R.A.M., Mandrup S.et al.. An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol. Cell. 2021; 81:1484–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen D., Hinkley C.S., Henry R.W., Huang S.. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell. 2002; 13:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Teves S.S., An L., Bhargava-Shah A., Xie L., Darzacq X., Tjian R.. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife. 2018; 7:e35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hammar P., Walldén M., Fange D., Persson F., Baltekin Ö., Ullman G., Leroy P., Elf J.. Direct measurement of transcription factor dissociation excludes a simple operator occupancy model for gene regulation. Nat. Genet. 2014; 46:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Normanno D., Boudarène L., Dugast-Darzacq C., Chen J., Richter C., Proux F., Bénichou O., Voituriez R., Darzacq X., Dahan M.. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat. Commun. 2015; 6:7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Morisaki T., Müller W.G., Golob N., Mazza D., McNally J.G.. Single molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014; 5:4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Speil J., Baumgart E., Siebrasse J.-P., Veith R., Vinkemeier U., Kubitscheck U.. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys. J. 2011; 101:2592–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dufourt J., Trullo A., Hunter J., Fernandez C., Lazaro J., Dejean M., Morales L., Nait-Amer S., Schulz K.N., Harrison M.M.et al.. Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat. Commun. 2018; 9:5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sugo N., Morimatsu M., Arai Y., Kousoku Y., Ohkuni A., Nomura T., Yanagida T., Yamamoto N.. Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci. Rep. 2015; 5:10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Haberle V., Stark A.. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018; 19:621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Morgunova E., Taipale J.. Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 2017; 47:1–8. [DOI] [PubMed] [Google Scholar]
- 105. Reiter F., Wienerroither S., Stark A.. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017; 43:73–81. [DOI] [PubMed] [Google Scholar]
- 106. Scruggs B.S., Gilchrist D.A., Nechaev S., Muse G.W., Burkholder A., Fargo D.C., Adelman K.. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell. 2015; 58:1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Taher L., Smith R.P., Kim M.J., Ahituv N., Ovcharenko I.. Sequence signatures extracted from proximal promoters can be used to predict distal enhancers. Genome Biol. 2013; 14:R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X.et al.. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018; 361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cho W.-K., Spille J.-H., Hecht M., Lee C., Li C., Grube V., Cisse I.I.. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018; 361:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cisse I.I., Izeddin I., Causse S.Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., Darzacq X.. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013; 341:664–667. [DOI] [PubMed] [Google Scholar]
- 112. Guo Y.E., Manteiga J.C., Henninger J.E., Sabari B.R., Dall’Agnese A., Hannett N.M., Spille J.-H., Afeyan L.K., Zamudio A.V., Shrinivas K.et al.. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019; 572:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Cho W.-K., Jayanth N., English B.P., Inoue T., Andrews J.O., Conway W., Grimm J.B., Spille J.-H., Lavis L.D., Lionnet T.et al.. RNA Polymerase II cluster dynamics predict mRNA output in living cells. eLife. 2016; 5:e13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hansen A.S., Amitai A., Cattoglio C., Tjian R., Darzacq X.. Guided nuclear exploration increases CTCF target search efficiency. Nat. Chem. Biol. 2020; 16:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Elf J., Li G.-W., Xie X.S.. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007; 316:1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Schier A.C., Taatjes D.J.. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020; 34:465–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.