Abstract
Skeletal development and bone formation are regulated by epigenetic mechanisms that either repress or enhance osteogenic commitment of mesenchymal stromal/stem cells and osteoblasts. The transcriptional suppressive trimethylation of histone 3 lysine 27 (H3K27me3) hinders differentiation of pre-committed osteoblasts. Osteoblast maturation can be stimulated by genetic loss of the H3K27 methyltransferase Ezh2 which can also be mimicked pharmacologically using the classical Ezh2 inhibitor GSK126. Identification of other Ezh2 inhibitors (iEzh2) that enhance osteogenic potential would increase chemical options for developing new bone stimulatory compounds. In this study, we examined a panel of iEzh2s and show that all eight inhibitors we tested are capable of accelerating osteoblast differentiation to different degrees at concentrations that are well below cytotoxic concentrations. Inhibition of Ezh2 is commensurate with loss of cellular H3K27me3 levels while forced expression of Ezh2 reverses the effect of Ezh2 suppression. Reduced Ezh2 function by siRNA depletion of Ezh2 mRNA and protein levels also stimulates osteoblastogenesis, consistent with the specificity of iEzh2 to target the active site of Ezh2. Diminished Ezh2 levels preempt the effects of iEzh2s on H3K27me3. GSK126, EPZ-6438 and siRNA depletion of Ezh2 each are effective in reducing H3K27me3 levels. However, EPZ-6438 is more potent than GSK126 in stimulating osteoblastogenesis, as reflected by increased extracellular matrix mineralization. Collectively, our data indicate that Ezh2 inhibitors properly target Ezh2 consistent with their biochemical affinities. The range of compounds capable of promoting osteogenesis presented in this study offers the opportunity to develop diverse bone anabolic strategies for distinct clinical scenarios, including spine fusion, non-union of bone and dental implant enhancement.
Keywords: Bone, Skeletal development, Osteoblast, Osteogenesis, Osteoporosis, Differentiation, Chromatin, Nucleosome, Methylation, Histone, Ezh2, Polycomb, Inhibitor, Pharmacology, Pharmacotherapy
1]. INTRODUCTION
The mature skeleton is key for structural support and movement in vertebrates. As such, deterioration in the mechanical properties of bone with aging and disease can profoundly affect a person’s quality of life. Disorders affecting bone development and maintenance, including congenital defects, physical trauma and degenerative disease affect millions of individuals worldwide. Most significantly, osteoporosis which is characterized by low bone mineral density (BMD) and a higher risk of bone fractures, affects more than 10 million people in the United States and approximately 200 million women around the world [(1), (2)]. Because of the aging population, osteoporosis represents an economic burden and it has a major impact on public health [(3)]. Despite the high occurrence of osteoporosis and other degenerative bone diseases, determining the genetic and molecular backgrounds and elucidating the complex mechanisms of bone formation and maintenance still represents a challenge.
Bone tissue is remarkably metabolically active and its homeostasis is controlled primarily by the interaction of osteoblasts, osteocytes and osteoclasts as the three main types of cells that control bone remodeling. A disruption in the balanced interaction of these cells plays an important role in the pathogenesis of osteoporosis and other bone disorders [(4)]. Bone-producing osteoblasts are derived from immature mesenchymal stem cells (MSC) during early vertebrate embryogenesis [(5), (6)]. Osteoblast lineage commitment of MSC requires major developmental signals and the activity of specific transcriptional and post-transcriptional epigenetic factors [(7), (8)]. Osteogenic differentiation is regulated by a crosstalk between multiple major signaling pathways (such as Wnt and Bmp signaling) and transcription factors (such as Runx2 and Osx/Sp7) [(9)] as well as epigenetic mechanisms, including microRNAs, DNA methylation and post translational modifications of histones [(10), (11), (12), (13), (14), (15), (16)].
Post-translational modifications (PTMs) of histones such as acetylation and methylation play important roles in regulating the expression of critical osteoblastic genes. Depending on the type of modification, PTMs can either inhibit or stimulate transcription of osteogenic genes. Trimethylation of histone 3 lysine 4 (H3K4me3), for example, is associated with gene activation while trimethylation of histone 3 lysine 27 (H3K27me3) or lysine 9 (H3K9me3) represses gene expression by inducing heterochromatin formation [(17), (18)]. Multiple epigenetic regulators and mechanisms have been characterized in mesenchymal stem cells [(19), (20)], osteoblasts and chondrocytes [(11), (12), (13), (14)] and have been shown to control distinct histone methylation marks such as H3K4me3 [(21), (22)], H3K9me3 [(23), (24)], H3K27me3 [(25), (26), (27), (28), (29), (30), (31)], histone acetylation marks [(32), (33), (34), (35), (36), (37), (38)], as well as DNA hydroxymethylation [(39), (40)].
Bone specific genes are inactivated during embryonic development in part by generation of facultative heterochromatin involving H3K27me3. The latter modification is generated by the Polycomb Repressive Complex 2 (PRC2), which comprises four core subunits: Ezh1/2, Eed, Suz12 and RbAp46/48 [(41)]. Immature osteoblasts express Ezh2 (enhancer of zeste homolog 2), which is the catalytic subunit of the PRC2 methyltransferase complex. PRC2 establishes the mono-, di-, and trimethylation states of Lysine 27 at histone H3 (H3K27) [(42), (43)], which are recognized and functionally interpreted by CBX proteins [(31)].
While Ezh2 activity is required for appropriate skeletal patterning and bone formation [(27), (28), (30), (44), (45), (46), (47)], inhibiting the activity of Ezh2 and thus reduction in H3K27 trimethylation increases osteogenic commitment and differentiation in vitro and bone formation and maintenance in vivo [(25), (29), (30), (48), (49), (50), (51), (52), (53), (54)]. Established evidence suggests that inactivation results in the up-regulation of key osteogenic genes and enhances activities of established bone-stimulatory pathways (e.g., Wnt, Pth and Bmp signaling) [(25), (28), (29), (30), (55)]. These findings collectively establish that the normal function of Ezh2 is to attenuate osteoblast differentiation. The potential biomedically relevant ramification of this conclusion is that Ezh2 inhibitors may have bone anabolic potential in vivo.
Because Ezh2 inhibitors have potential utility in a range of clinical applications that stimulate bone mass accrual, it is of considerable interest to develop Ezh2 inhibitors (iEzh2) for bone stimulatory applications. Apart from the formidable challenge of meeting pharmacokinetic criteria for use of epigenetic drugs in patients, development of clinically viable compounds for bone-related applications requires optimization of their specific inhibition of Ezh2 activity (which blocks formation of H3K27me3), while reducing any secondary effects on cell viability and metabolism. As a first step in this process, we examined a panel of eight different commercially available compounds that target Ezh2 activity and we compared their relative osteogenic efficacy. These experiments used the well-established MC3T3 cell culture model to test the hypothesis that each of these drugs exhibits intrinsic differences in effects on cell viability and osteogenic activity. We also compared the osteogenic effects of Ezh2 inhibitors to the effects modulating Ezh2 protein levels by RNA interference or forced expression. The combined results of these studies indicate that pharmacological inhibition of Ezh2 using any of these compounds is capable of stimulating osteogenic differentiation. Our findings also validate the hypothesis that each compound exhibits distinguishable biological effects, which permits selection of preferred Ezh2 inhibitors for follow up studies.
2]. RESULTS
2.1. Effects of Ezh2 inhibitors on metabolic activity and cell number of preosteoblastic cells
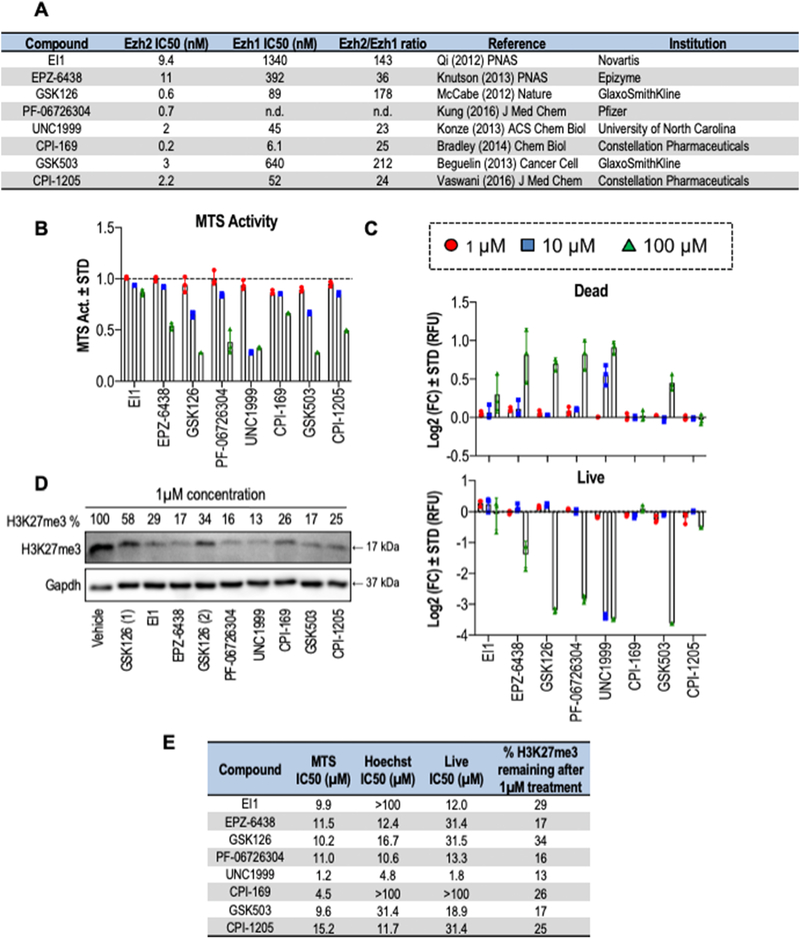
To understand the osteogenic efficacy of distinct Ezh2 inhibitors (iEzh2), we first assessed any adverse secondary effects on cell viability. These studies were guided by the known half maximum inhibitory concentrations for each inhibitor (Fig. 1A). To ensure adequate inhibition of Ezh2 while preserving cell viability, we first assessed the cytotoxic effects for a total of eight Ezh2 inhibitors on MC3T3 pre-osteoblasts. We administered a logarithmic range of concentrations (1μM, 10μM and 100μM) to subconfluent proliferating MC3T3 cells for 3 days. We then monitored potential cytotoxic effects on metabolic activity using MTS assays (which measures biochemical conversions in mitochondria) (Fig. 1B), absolute cell number by Hoechst staining (which measures DNA content) (Fig. S1) and the fraction of viable cells using live/dead staining (which measures sequestration of green and red dyes in live versus dead cells) (Figs. 1C & S1).
Figure 1: Cytotoxicity profiles of Ezh2 inhibitors on MC3T3 cells.
(A) Tabular summary of Ezh2 inhibitors with the name of each compound (first column), the concentrations that cause 50% enzymatic inhibition (IC50 in nM) for either Ezh2 (column 2) or Ezh1 (column 3), the selectivity for Ezh2 (fold change of the IC50s for Ezh2 relative to Ezh1) (column 4), key references for each inhibitor (column 5), and the entity that initially developed the compound (column 6). To obtain IC50 values, GraphPad prism was used to create a non-linear regression curve fit in which data from each concentration response curve was converted into percentage values that were log10 transformed and plotted as a curved XY non-linear graph with variable (see also Figures S1 and S2). (B & C) Subconfluent MC3T3 cells were treated with different concentrations of each one of the Ezh2 inhibitors; 1μM, 10μM and 100μM for three days. (B) Total metabolic activity measured by MTS assay of MC3T3 cells treated with Ezh2 inhibitors for three days and normalized to DMSO treated control (1:100 dilution). (C) Cell quantification by Live/Dead staining of MC3T3 cells after three days of treatment. Results were normalized to DMSO treated control (1:100 dilution). Data represents n = 3, mean ± STD. (D) Western blot analysis of histone H3K27me3 levels after treatment with the indicated inhibitors was used to determine inhibitory effects of a single dose of Ezh2 inhibitor (1μM) on histone methylation after 24 hour treatment. Inhibitory values for H3K27me3 levels (% retained after inhibitor treatment) were calculated relative to Gapdh. GSK126 treatment was performed with two different preparations (labeled 1 and 2). (E) Tabular summary of the results presented in panels B, C and Sup. Fig 1. The metrics were done using GraphPad Prism; a concentration response curve was created using a non-linear regression (curve fit). Drugs showing an IC50>100 μM were not included in the graphs in panels A-C, but are included in panel D.
The metabolic activity of MC3T3 cells exhibits a notable decrease for most of the compounds at concentrations of 10μM and higher as measured by MTS activity assays (Fig. 1B). Exceptions to this observation are EI1 and EPZ-6438 which appear to be less harmful to metabolic cell activity at a concentration of 10μM.
Similarly to the metabolic activity, there are significant reductions in cell numbers (Fig. S1) and fraction of living cells (Fig. 1C) at the 100μM concentration for five inhibitors (EPZ-6438, GSK126, PF-06726304, UNC1999 and GSK503) as measured by Hoechst and live/dead staining, respectively, but these differences are not observed for two other inhibitors (EI1 and CPI-169). Hence, Ezh2 inhibitors at a concentration of 100μM are substantially toxic, but treatments of 1μM and 10μM concentrations generally have minimal or no effects on cell viability. Taken together, our results suggest that the toxicity of Ezh2 inhibitors is clearly dose dependent and that low molar concentrations (<10μM) are generally nontoxic in MC3T3 cells.
To complement our assessment of relative toxicity, we assessed the efficacy of each inhibitor in the MC3T3 cell type by evaluating H3K27me3 levels via western blot (Fig. 1D). These data show that all inhibitors are effective at reducing cellular levels of H3K27me3, although some inhibitors (e.g., UNC1999, EPZ-6438, GSK503) are highly inhibitory (>5 fold reduction in H3K27me3) and others are slightly less effective (e.g., GSK126 and EI1; 2–3 fold inhibition). We then graphically assessed and mathematically interpolated our concentration curves (Fig. S2) to define the midpoint of the inhibitory doses in each assay where half the cells are affected. Collation of the experimental data (Fig. 1E) shows that cellular toxicity is generally proportional to the measured IC50 of the enzymatic activity of Ezh2 in vitro and the ability to reduce cellular H3K27me3 levels in culture. Hence, toxicity of these inhibitors is at least in part attributable to loss of Ezh2 function, rather than merely non-specific toxicity of these agents as xenobiotic compounds.
2.2. Ezh2 inhibition enhances osteogenic differentiation of MC3T3 cells
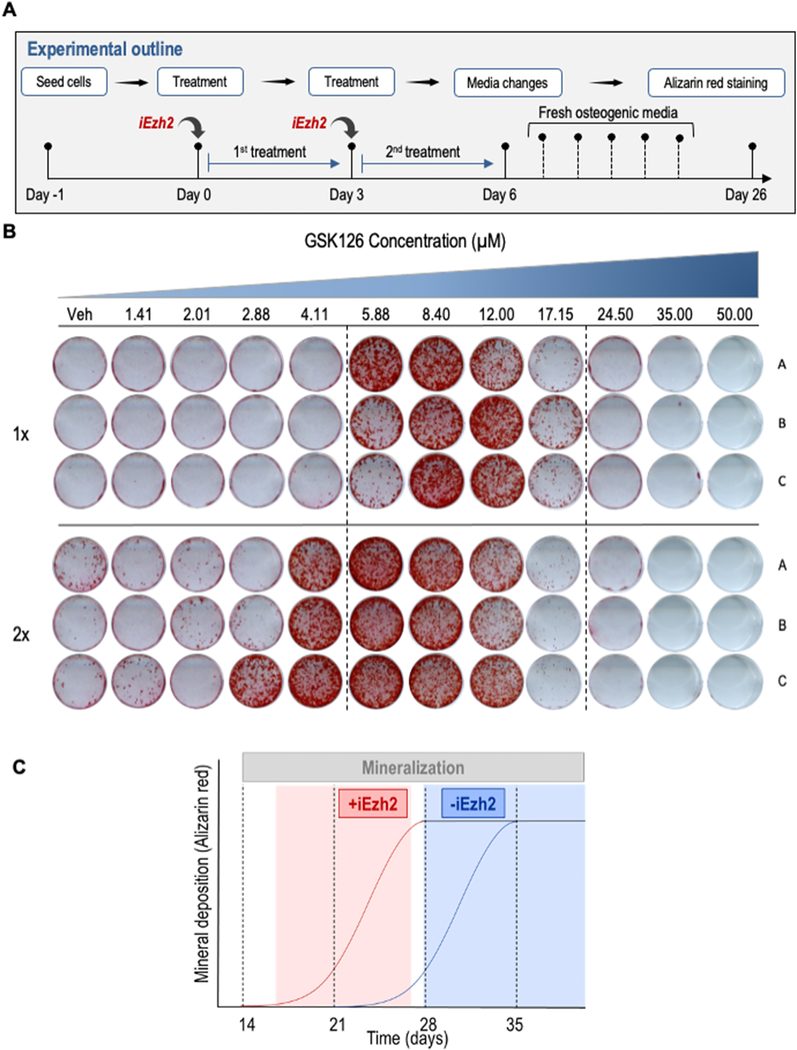
To investigate whether Ezh2 inhibition, and consequently reduction of H3K27me3, stimulates osteogenic differentiation, we treated MC3T3 pre-osteoblasts with vehicle or Ezh2 inhibitors during osteogenic differentiation for either the first three (1X dose) or six days (2X dose) (Fig. 2A). To identify the optimal concentration for each drug, we treated MC3T3 cells with vehicle (DMSO) or increasing concentrations of drugs in approximately ~0.7 fold increments and measured alizarin red staining after three weeks (i.e., between days 24 and 31)(Fig. 2B) The timing of harvest was selected to optimize differences in staining between control and iEzh2 treated samples, with control samples typically harvested just prior to the initiation of mineralization (Fig. 2C). Our results show a more robust ECM mineralization on cells treated with Ezh2 inhibitors when compared to the control group as evidenced by increased alizarin red staining reflecting increased calcium deposits (Figs. 2B, 3 & 4). These effects are observed after merely a single treatment of three days for all inhibitors. Because each compound exhibits cytotoxic effects at higher concentrations (see Fig. 1E), increasing the drug concentration enhances alizarin red staining up to a point and then tapers off at the highest doses. These results show therefore that Ezh2 inhibitors have an optimal dosing that balances the osteogenic versus cytotoxic effects. Interestingly, GSK126, which is one of the most potent and selective Ezh2 inhibitors, shows more intense alizarin red staining when compared to the other drugs as is also reflected to some degree by quantification of IC50 doses for each of the inhibitors (see Fig. 1E).
Figure 2: Mineral deposition of MC3T3 cells treated with increasing concentrations of the Ezh2 inhibitor GSK126.
(A) Experimental outline showing the treatment regimens in which MC3T3 cells are treated with vehicle (DMSO, 1:2000 dilution) or Ezh2 inhibitors for three days (1X dose) or six days (2X dose) with increasing concentrations of inhibitor (in 1.5 fold increments). (B) Alizarin red staining of MC3T3 cells treated with either 1X or 2X doses of increasing concentrations of GSK126 at day 26 of osteogenic differentiation (biological triplicates are shown). (C) Hypothetical schematic illustrating the principle of time-dependent (x-axis) variation in the detection of mineralization (y-axis) in the presence (+iEzh2, red) or absence (−iEzh2, blue) of Ezh2 inhibitors. Although all cultures eventually show maximal alizarin red staining (black horizontal slope in graph) after 35 days under our osteogenic experimental conditions, we selected Day 26 for analysis because this time point maximizes the observed differences in mineralization between absence or presence of Ezh2 inhibitors.
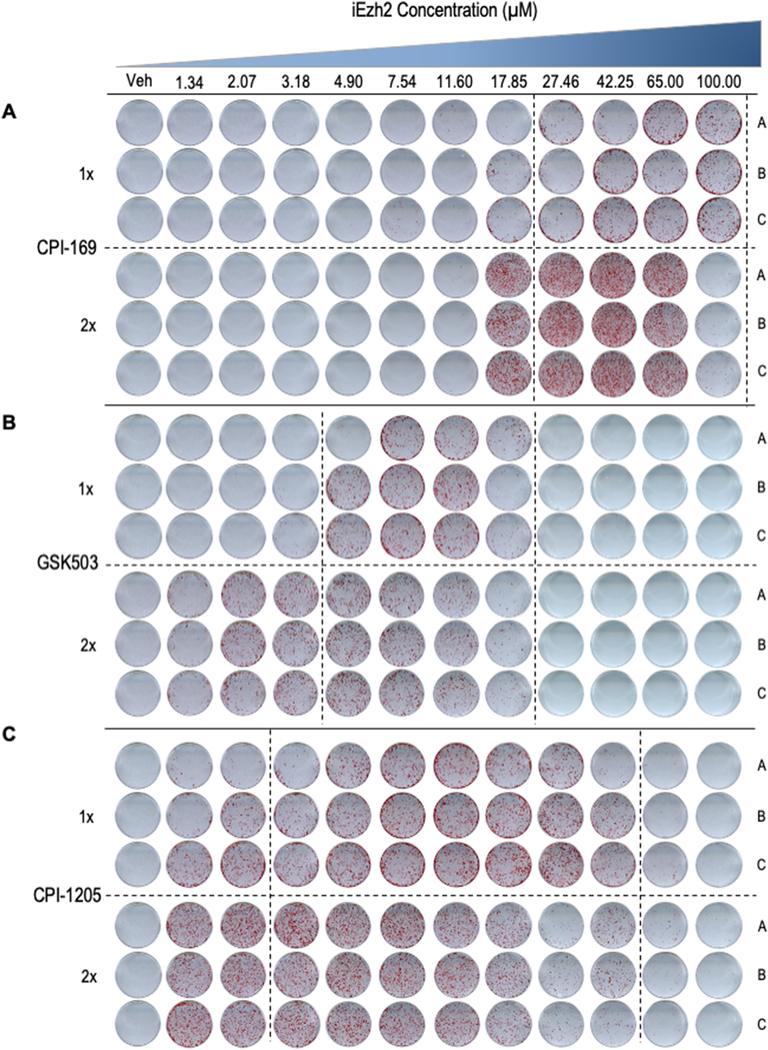
Figure 3: Mineral deposition of MC3T3 cells treated with increasing concentrations of the Ezh2 inhibitors CPI-169, GSK503 and CPI-1205.
Alizarin red staining of triplicate MC3T3 cell cultures at day 26 of osteogenic differentiation after treatment in the first week with vehicle (DMSO; 1:2,000 dilution), or a 1X or a 2X dose of increasing concentrations of CPI-169, GSK503 and CPI-1205.
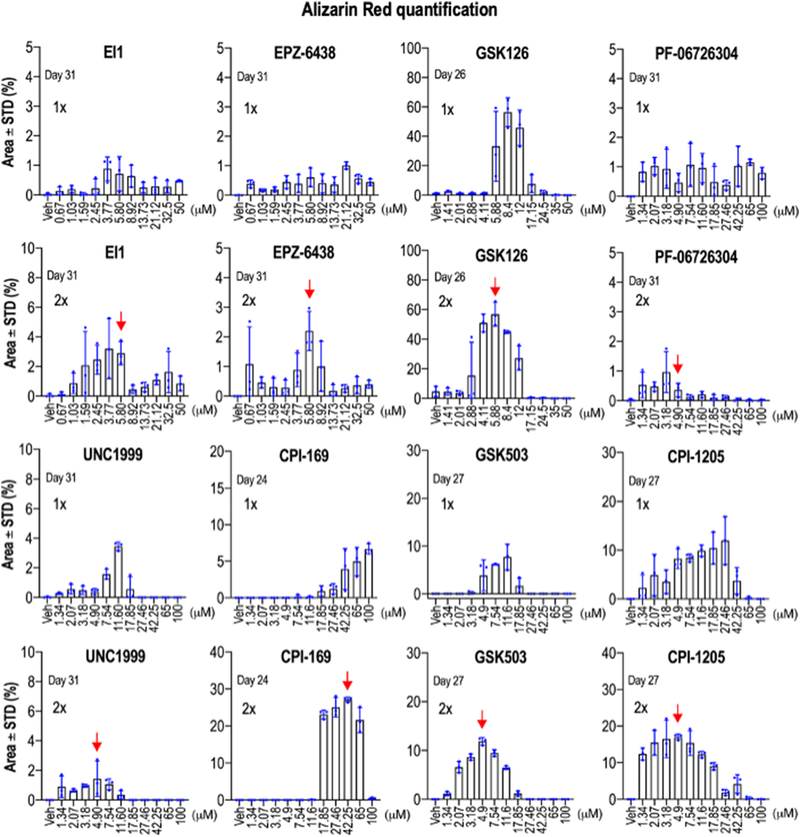
Figure 4: Quantitative analysis of mineralization.
Values for alizarin red staining in cultures of differentiated MC3T3 cells (see Figure 2) are presented as bar graphs. Quantitation is provided for experiments that received either a single dose (1X, Day 0 to 3) or two doses (2X, Day 0 to 3 and Day 3 to 6)(see Fig. 2A). Error bars represent standard deviations of triplicate measurements that are super imposed on the bar graphs. The red arrows point at optimal concentrations (in 2X dose) for compounds that were used in subsequent experiments.
Strikingly, studies in which we applied a second dose for each of these drugs demonstrate a more vigorous calcium deposition at lower drug concentrations (Figs. 2B, 3 & 4). For each inhibitor, second application results in more robust and consistent mineralization on MC3T3 cells while also favoring the usage of a lower dose concentration. Remarkably, while iEzh2 doses were only administered at the beginning of the differentiation time course, the results clearly show that each of these inhibitors still has very pronounced positive effects on osteoblast mineralization at three weeks or more after the initial treatment. The latter is consistent with the proposed mechanism of action of Ezh2 inhibitors which are thought to generate alterations in the epigenetic molecular memory of MC3T3 cells that promote osteogenesis.
2.3. Stimulation of osteoblast differentiation by Ezh2 inhibitors is facilitated by mild co-stimulatory effects of DMSO in the presence of osteogenic media.
Osteoblast differentiation is stimulated by dimethyl sulfoxide (DMSO), an aprotic solvent often used to dissolve lipophilic therapeutic agents and that was used to dissolve all of the compounds used for our experiments. DMSO is considered a relatively safe solvent that has been extensively used in the biomedical sciences. It has been shown previously that DMSO induces extracellular matrix synthesis and mineralization, while promoting osteoblast differentiation of MC3T3 cells [(39), (40), (56)]. To assess the extent to which the concentration of DMSO affects the induction of osteoblast differentiation, we exposed MC3T3 preosteoblasts to either low (0.05%) or high (0.4%) doses of DMSO for two consecutive period of three days (for a total of six days) in osteogenic media. DMSO treated cells were compared to cells receiving only osteogenic media (50 μg/ml ascorbic acid, and 10 mM β-glycerol phosphate). Gene expression studies demonstrate that DMSO has a substantial effect on MC3T3 cells by enhancing the differentiation-specific expression of several osteoblast mRNA markers (Suppl. Fig. S3). For example, mRNA levels for osteoblast differentiation markers such as Alpl, Sp7, Ibsp and Phospho1 were each significantly elevated by either 0.05% DMSO or 0.4% DMSO relative to the respective controls with no DMSO at six days after induction of differentiation in standard osteogenic medium. These co-stimulatory effects for these early differentiation markers appeared to be diminished but still trending at 10 days. Expression of the late marker Bglap which is robustly expressed on day ten did not show compelling changes with DMSO and apparently is less sensitive to the DMSO effect. We also found remarkably that low-dose DMSO induces a higher expression of osteoblast marker genes than the high-dose. The latter is consistent with DMSO having non-specific pleiotropic effects that offset its co-stimulatory effects. In sum, these results suggest that DMSO is capable of co-stimulating osteogenic differentiation of MC3T3 pre-osteoblasts and may further potentiate the effects of Ezh2 inhibitors.
2.4. H3K27me3 inhibition stimulates ECM mineralization and bone-related gene expression.
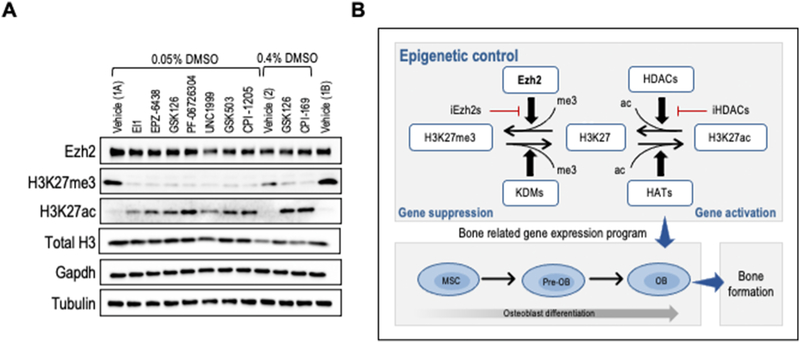
The panel of eight different Ezh2 inhibitors were analyzed simultaneously side-by-side at optimal doses determined above (see Fig. 4, red arrows; 40μM for CPI-169 and 5μM for all other compounds) to permit a direct comparison of the efficacy of enzymatic inhibition of the histone methyltransferase activity of Ezh2 and the subsequent effects on H3K27 trimethylation (Fig. 5). Results of western blot analyses revealed that H3K27me3 protein levels are significantly decreased after three days of treatment (Fig. 5A). Remarkably, the decrease in H3K27me3 levels upon Ezh2 inhibition occurs concomitant with increased H3K27ac levels (Fig. 5A). Because CPI-169 is used at a relatively high concentration (40μM) when compared to other inhibitors (5μM), a greater amount of DMSO (0.4% vs 0.05%) was used for CPI-169 studies. As an internal control for the effects of different DMSO doses, GSK126 is used at low (0.05%) and high (0.4%) DMSO concentrations. These findings suggest that global increases in either H3K27me3 or H3K27ac are mutually exclusive. Our results are consistent with a molecular concept in which loss of H3K27me3 levels is biochemically linked to H3K27ac (Fig. 5B). In this model, Ezh2 operates as an epigenetic switch that controls initial rate-limiting steps for transcriptional activation of genes required for osteogenic commitment [(50, 57)].
Figure 5: Optimal Ezh2 inhibition modulates levels of H3K27me3 and H3K27ac marks.
MC3T3 cells were treated with vehicle (Vehicle 1 = 0.05% DMSO or Vehicle 2 = 0.4% DMSO) or optimal doses of each compound (40μM for CPI-169 and 5μM for all other compounds) for the first six days of osteogenic differentiation. (A) Western blot of Ezh2 protein, H3K27me3 and H3K27ac relative to H3, Gapdh and Tubulin three days after initial treatment with each of the Ezh2 inhibitors (Vehicle 1A and 1B are both at 0.05% DMSO). (B) Diagram showing interrelationships between H3K27me3 and H3K27ac levels that are controlled as indicated by Ezh2, lysine demethylases (KDMs), histone acetyl transferases (HATs), as well as histone deacetylases (HDACs) and their cognate inhibitors (iHDACs). The western blot data in panel A are consistent with a shift from left to right to decreased total H3K27me3 levels concomitant with an increase in total H3K27ac levels that together support osteoblast differentiation.
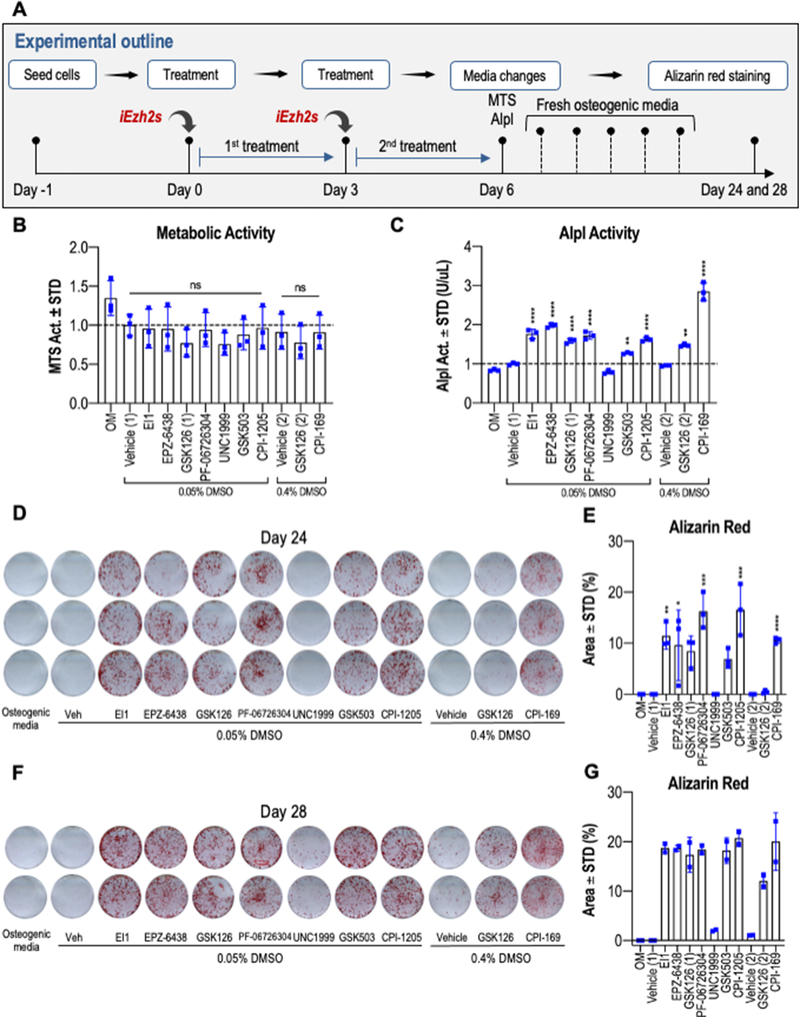
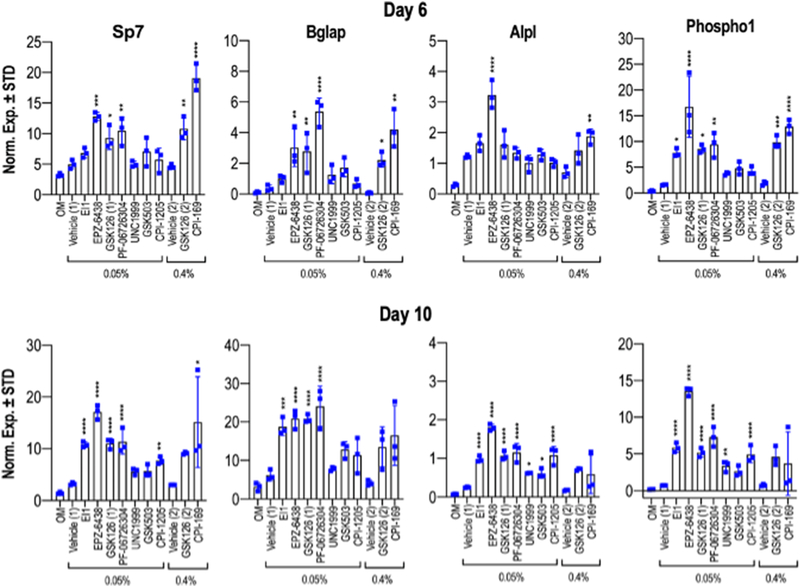
Having established that optimal dosing of each iEzh2 effectively reduces H3K27me3 levels, we validated that these concentrations do not affect cell viability significantly as measured by MTS assay at day six (Figs. 6A & B). Increased alkaline phosphatase (Alpl) enzymatic activity is observed at day six in all iEzh2-treated groups, except for UNC1999 (Fig. 6C). When compared to osteogenic medium (OM), both low and high doses of DMSO enhance Alpl enzymatic activity. Several Ezh2 inhibitors show a robust increase in Alpl activity with CPI-169 representing the most potent compound that exhibits a ~three fold stimulation in Alpl enzymatic activity at day six. We also assessed directly in parallel the stimulatory effects of iEzh2s on biomineralization of MC3T3 osteoblasts after more than three weeks in culture. Mineralization in distinct iEzh2 treated cultures differed among the various drugs we tested when compared to osteogenic media and vehicle only on day 24 (Figs. 6D & E) and day 28 (Fig. 6F & G) of osteoblast differentiation. The main finding is that ECM mineralization was significantly elevated on both days and in all Ezh2 inhibitor groups except UNC1999 and controls (Figs. 6E & G). We note that DMSO at high doses also has modest osteogenic effects (Fig. 6F), supporting Alpl enzymatic activity (Fig. 6C) and other studies presented earlier (see Fig. S3). We validated acceleration of osteoblast maturation by examining the expression of mRNAs for classical bone-related genes including Sp7, Bglap, Alpl and Phospho1 in response to each of the Ezh2 inhibitors. The results show that indeed mRNA expression of each of these osteogenic genes is enhanced on days six and ten for all inhibitors tested (Fig. 7). In sum, our findings demonstrate that reduction of H3K27me3 by multiple Ezh2 inhibitors consistently stimulates bone-specific gene expression and ECM mineralization of MC3T3 cells.
Figure 6: Ezh2 inhibition enhances ECM mineralization on MC3T3 cells.
(A) Experimental outline showing the treatment regimen in which MC3T3 cells were treated with vehicle (Vehicle 1 = 0.05% DMSO or Vehicle 2 = 0.4% DMSO) or optimal doses of each compound (40μM for CPI-169 and 5μM for all other compounds) for the first six days of osteogenic differentiation. GSK126 (1) and GSK126 (2) represent treatments in the presence of either 0.05% or 0.4% DMSO. (B) MTS activity assay of cells treated with either vehicle or Ezh2 inhibitors for six days (n = 3, mean ± STD). (C) Alkaline phosphatase (Alpl) enzymatic activity measured on day 6 (n = 3, mean ± STD). (D–G) Ezh2 inhibition enhances ECM deposition and mineralization in vitro. Alizarin red staining (D, F) and quantification (E, G) was conducted on day 24 (D, E) or day 28 (F, G) of osteogenic differentiation. Each late stage time point was sampled as five biological replicates on two different days to assess the dynamics of mineralization after three to four weeks in culture. For transparency, biological triplicates were performed for day 24 (n = 3, mean ± STD) while duplicates were performed for day 28 (n =2, mean ± range). Duplicates are provided for illustration only. The horizontal lines above the bars (see panel B) reflect statistical comparisons between distinct iEzh2 treatment groups and the respective vehicle controls (i.e., Vehicle 1 = 0.05% DMSO or Vehicle 2 = 0.4% DMSO). In panel B, these values were calculated to be non-significant (ns). Statistical significance for each iEzh2 inhibitor compared to its control (Vehicle 1 or Vehicle 2) is indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. OM, osteogenic medium.
Figure 7: Ezh2 inhibition enhances osteoblast specific gene expression.
MC3T3 cells were treated with vehicle (Vehicle 1 = 0.05% DMSO or Vehicle 2 = 0.4% DMSO) or optimal doses of each compound (40μM for CPI-169 and 5μM for all other compounds) for the first six days of osteogenic differentiation. Graphs showing mRNA expression levels of osteogenic genes determined by RT-qPCR on day six (upper) and day ten (lower) of the osteogenic differentiation timeline (n = 3, mean ± STD). Statistical significance for iEzh2 inhibitor compared to its control (Vehicle 1 or Vehicle 2) is indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. OM, osteogenic medium.
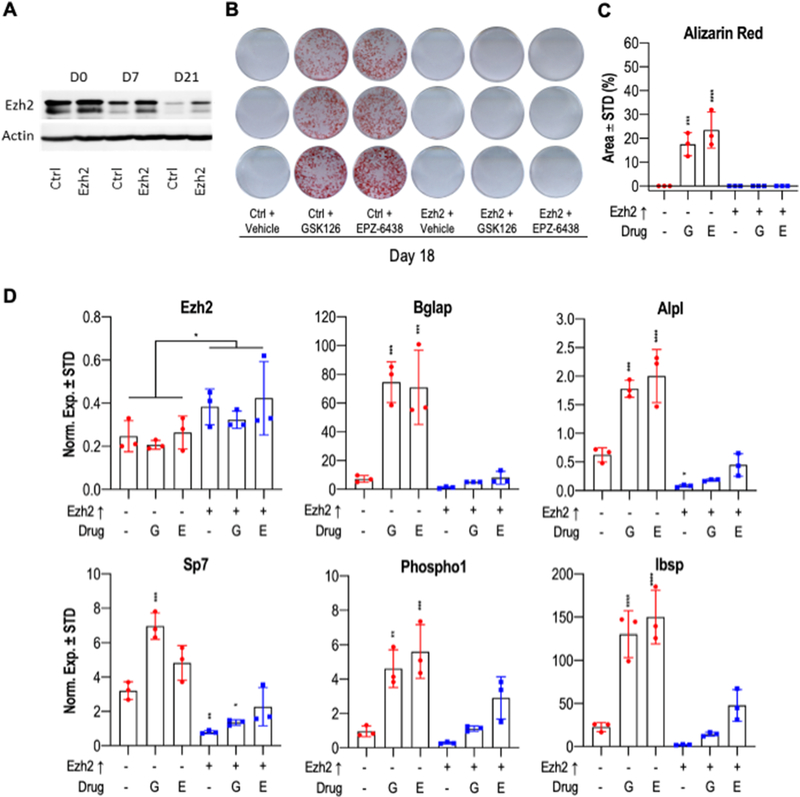
2.5. Gain-of-function analysis reveals that Ezh2 overexpression suppresses maturation of MC3T3 preosteoblasts.
Our studies using pharmacological inhibitors indicate that Ezh2 gain-of-function suppresses normal osteoblast differentiation. To test this hypothesis, we performed gain of function studies in which we forced expression of Ezh2 beyond its normal physiological levels during osteoblast maturation in MC3T3 cells using a lentiviral delivery method. We compared MC3T3 cells stably infected with a lentivirus containing the full length Ezh2 protein, versus the corresponding empty viral vector. Western blot analysis revealed that Ezh2 protein levels are initially high in control cells and then declines as differentiation progresses. However, forced expression of Ezh2 consistently elevated Ezh2 protein levels compared to the control at each time point (Fig. 8A). Subsequently, we treated Ezh2 expressing MC3T3 cells with vehicle and two Ezh2 inhibitors (5μM GSK126 and 5μM EPZ-6438) to examine whether increasing the intracellular concentration of Ezh2 can overcome the biochemical blockade of its enzymatic activity in the presence of iEzh2s. As anticipated, on day 18 of osteoblast differentiation, treatment with Ezh2 inhibitors in control cells with physiologically declining levels of Ezh2 displayed accelerated mineralization and more intense alizarin red staining as reflected by a significantly higher percent coverage area (Figs. 8B & C). In contrast, osteogenic boosting of MC3T3 cells with Ezh2 inhibitors is not observed in Ezh2 overexpressing cells which do not manifest any signs of calcium deposition (Fig. 8B & C).
Figure 8: Ezh2 overexpression impairs maturation of MC3T3 preosteoblastic cells.
MC3T3 cells overexpressing Ezh2 were treated with vehicle, 5μM GSK126 or 5μM EPZ-6438 for the first six days of osteogenic differentiation. (A) Western blot analysis showing Ezh2 protein levels over the time course. Alizarin red staining (B) and quantification (C) was performed at day 18 of osteogenic differentiation. Quantification was done using the ImageJ program (n = 3, mean ± STD). (D) Relative mRNA expression of Ezh2 and osteoblast markers in MC3T3 cells after ten days of osteoblast differentiation (n = 3, mean ± STD). The horizontal lines above the bars (see panel D, Ezh2 graph) reflect statistical comparisons between distinct iEzh2 treatment groups and the respective vehicle controls (i.e., 0.05% DMSO or 0.4% DMSO). Statistical significance to Ezh2↑ -, drug - control group indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Because samples receiving exogenous Ezh2 expression represents an independent treatment group independent of the presence of iEzh2s, we determined statistical significance of Ezh2 mRNA levels in one group of three sets of samples that was infected with empty lentiviral expression vector (n = 9 total) versus the three experimental groups that express lentiviral Ezh2 (n = 9 total). G, GSK126; E, EPZ-6438.
Expression analysis by RT-qPCR establishes that the collective levels of Ezh2 mRNA in all samples that were infected with lentiviral expression vector exhibit a modest two fold elevation in gene expression at day ten of MC3T3 osteoblast differentiation (Fig. 8D & S4). We note that these values may understate the amount of Ezh2 mRNA expression shortly after infection, and that total Ezh2 protein levels (see Fig. 8A) may be elevated by both increased mRNA levels and post-translational mechanisms (e.g., Ezh2 protein stability) [(58), (59), (60)]. The increased levels of Ezh2 mRNA and protein upon Ezh2 overexpression decelerates osteogenesis, which is not only reflected by alizarin red staining (Figs. 8B & C), but also by significant reductions in the levels of each of the osteogenic mRNA biomarkers we examined (Bglap, Alpl, Sp7, Phospho1 and Ibsp) (Fig. 8D). Optimized concentrations of 5μM GSK126 and 5μM EPZ-6438 are highly effective at increasing osteogenic markers during MC3T3 cell differentiation, but not when cells overexpress Ezh2. Collectively, our findings indicate that the negative effects of Ezh2 inhibition can be tittered by increasing the amount of expressed Ezh2 protein and that Ezh2 directly suppresses osteogenic differentiation.
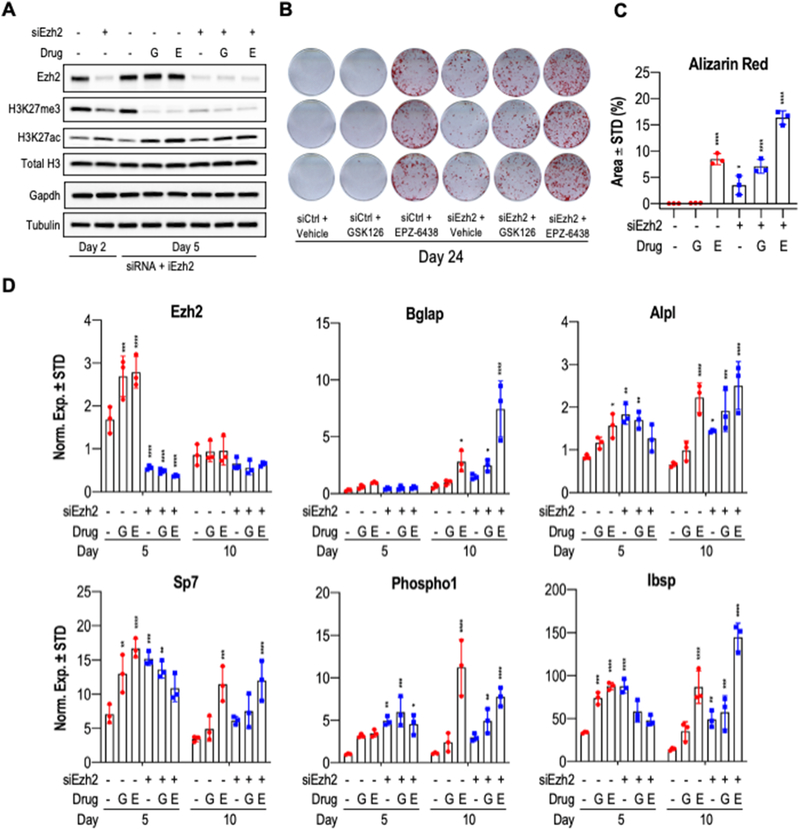
2.6. Ezh2 depletion by siRNA knockdown and Ezh2 inhibitors co-stimulate MC3T3 osteoblast differentiation.
We performed transient knockdown experiments in which we targeted Ezh2 using siRNAs in MC3T3 preosteoblasts. Additionally, we added GSK126 or EPZ-6438 two days after transfection. Osteoblast differentiation was initiated on day zero, and successful knockdown of Ezh2 was validated by Western blot analysis at both day two and day five after siRNA transfection (Fig. 9A). The reduction in Ezh2 protein levels resulted in a concomitant decrease in H3K27me3 levels at day five, and correlates with a modest increase in the levels of H3K27ac. Because siRNAs mediated knock-down nearly completely abolishes Ezh2 protein levels, we do not observe any additional effects on Ezh2 or H3K27me3 levels after treatments with Ezh2 inhibitors in cells transfected with Ezh2 siRNAs. Importantly, even though the combination of both Ezh2 siRNA and Ezh2 inhibitors were only administered in the first few days of osteoblast differentiation, we still detected major osteogenic effects on day 24 of differentiation, as reflected by enhanced mineralization as shown by alizarin red staining (Figs. 9B & C). We note that treatment with 5μM EPZ-6438 combined with Ezh2 siRNA produced a more robust staining in comparison to co-treatment with 5μM GSK126 and Ezh2 siRNA combo, suggesting that EPZ-6438 has superior osteogenic potency. RT-qPCR analysis corroborates the effectiveness of siRNA knockdown on Ezh2 mRNA levels at five days after transfection (Fig. 9D). In addition, key osteogenic markers (Bglap, Alpl, Sp7, Phospho1 and Ibsp) show elevated mRNA expression on days five and ten in response to Ezh2 loss. Taken together, the combination of Ezh2 knockdown with the inhibition of its enzymatic activity via GSK126 and EPZ-6438 co-stimulates expression of bone-related genes and enhances ECM mineralization. Collectively, our findings are consistent with the importance of Ezh2 in the regulation of osteoblast maturation in MC3T3 cells.
Figure 9: Combination of siRNA-mediated knock down of Ezh2 with GSK126 or EPZ-6438 promotes MC3T3 osteogenic differentiation.
MC3T3 cells were transfected with control and Ezh2 siRNAs on day zero and osteogenic differentiation was started on the same day. On day two, cells were treated with vehicle, GSK126 (5μM) or EPZ-6438 (5μM). Vehicle and drugs were removed on day five. (A) Western blot analysis of histone marks and reference proteins collected at day two and five of differentiation. Alizarin red staining (B) and quantification (C) was conducted on day 24 of differentiation (n = 3, mean ± STD). (D) RT-qPCR of Ezh2 and osteogenic markers at day five and ten (n = 3, mean ± STD). Statistical significance to siCtrl + Vehicle control group indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. G, GSK126; E, EPZ-6438.
3]. DISCUSSION
In this study, we evaluated the potency of distinct Ezh2 inhibitors in promoting osteoblast differentiation in vitro by comparing the biological properties of of a large panel of different Ezh2 inhibitors on the metabolic activity, cell number and viability of pre-osteoblastic cells, as well as osteoblast maturation as reflected by alkaline phosphatase activity, ECM mineralization and bone specific gene expression. Mechanistic studies using gain-of-function and loss-of-function analyses indicate that Ezh2 inhibitors function specifically by blocking the enzymatic activity of Ezh2 and reduce H3K27me3 levels. Because multiple small molecule inhibitors that target Ezh2 induce a similar phenotype (i.e., enhanced osteogenesis), our present studies demonstrate that the pro-osteogenic effects of these compounds are due to H3K27me3 suppression and less likely caused by potential off target effects of each individual inhibitor.
Cytotoxicity studies we performed with Ezh2 inhibitors indicate that osteoblasts tolerate each of these agents fairly well in the micro molar range (<10 μM). Our cell culture data corroborate previous in vivo studies in which we have shown that the potent Ezh2 inhibitor GSK126 is modestly bone anabolic in adult mice (8–12 weeks) and that there were no adverse effects in other tissues and organs (e.g., spleen, liver) [(29)]. Toxicity generally tends to correlate with the intrinsic ability of these drugs to bind the activity site (SET domain) within Ezh2. Therefore, the toxicity of these inhibitors can be attributed to the ability of Ezh2 function to methylate H3K27, rather than acting as non-specific xenobiotic compounds that trigger stress responses.
One of the most remarkable features of the osteogenic effects of Ezh2 inhibition on differentiation of MC3T3 cells is the observation that two drug doses between days one and six of differentiation accelerate osteoblastogenesis and increases the values of all histochemical and mRNA biomarkers. However, administration of a single dose between days one and three of differentiation suffices for stimulation of bone cell differentiation. Each of the inhibitors clearly has long term effects on osteoblastic markers and mineralization, which is measured as much as two to four weeks later. The observation that Ezh2 inhibitors have prolonged effects on osteoblastogenesis is entirely compatible with mechanistic models in which Ezh2 inhibitors are removing barriers that normally support the long-term suppression of genes. Hence, these compounds are in essence capable of altering the epigenetic molecular memory of MC3T3 cells through a single dose to promote osteogenesis. The latter property may be useful in translational applications that leverage the osteogenic potential of Ezh2 inhibitors.
Because all of the Ezh2 inhibitors we tested are hydrophobic compounds, we used dimethyl sulfoxide (DMSO) as the universal vehicle to dissolve them. DMSO is deemed very safe for cells and used as a common antifreeze agent in cell culture. Consistent with previous observations, we noted that DMSO itself is mildly osteogenic [(39), (56)] even at very low dilution doses (e.g., ranging from 1:2000 to 1:250). This range of concentrations is commonly used in many pharmacological studies during osteoblast differentiation and is unavoidable. Yet, the implication of the DMSO effect is that the osteogenic effects of Ezh2 inhibitors occur in the biological context of the stimulatory effects of DMSO on osteoblast differentiation. We note that our results also indicate that DMSO is not particularly toxic for osteoblasts, but may have pleiotropic non-specific effects at higher concentrations that could offset and reduce its mild osteogenic effects. Recent studies from our group indicate that DMSO has a similar biological activity as the closely related natural compound sulforaphane, which is composed of a cyanide-containing aliphatic chain with a sulfoxide moiety at one end [(40)]. Both Sulforaphane and DMSO appear to accelerate osteogenesis by affecting Tet mediated DNA hydroxylation [(40), (56)]. DNA hydroxylation in osteoblasts is also activated by Vitamin C present in osteogenic media. Because osteogenic media is used in all our experiments to drive osteogenesis, the main reason why DMSO is osteogenic may be because it enhances Vitamin C dependent hydroxylation.
It should be noted that Ezh2 inhibitors when applied to human or mouse MSCs without osteogenic stimulus will not enhance osteoblast differentiation (data not shown). One possible reason for this finding is that epigenetic inhibitors are likely to have pleiotropic effects because they may reduce stochastic kinetic barriers in chromatin. The addition of external osteogenic stimuli (e.g., differentiation cocktail with Vitamin C) may provide a more uniform biological response. For example, the cell signaling events induced by the cocktail may provide lineage direction for uncommitted MSCs and could favor which barriers in chromatin are most effected by epigenetic drugs. Hence, Ezh2 inhibitors merely may facilitate and prime the osteogenic effects of Vitamin C. The concept that Ezh2 inhibitors mediate epigenetic priming for other osteogenic stimuli is also evidenced by our recent studies showing that BMP2 induction of osteoblastogenesis is synergistically stimulated by Ezh2 inhibitors [(25)].
Our combined studies to date are consistent with a mechanistic model in which Ezh2 represents a key epigenetic regulator that tightly suppresses the transcriptional activation of bone specific genes [(50, 57),] by increasing H3K27me3 marks that generate transcriptionally silent heterochromatin. Inhibition of the methyl transferase activity of Ezh2 diminishes global H3K27me3 levels and is expected to reduce the amount of heterochromatin in maturing osteoblasts. Global loss of H3K27me3 marks obtained by any of the eight Ezh2 inhibitors we tested was accompanied by increased H3K27ac, an epigenetic mark characteristic of tissue specific super enhancers [(61)]. Hence, it is thus possible that Ezh2 inhibition may license super enhancers to support bone specific gene activation.
Our early studies on the osteogenic effects of Ezh2 inhibition heavily relied on GSK126, because human safety data were available for this compound and we had shown that it is effective for stimulation of bone accrual in normal and ovariectomized mice [(29)]. The osteogenic effects of Ezh2 loss of function were corroborated with gene knockout and siRNA studies that established the cellular specificity of the Ezh2 inhibitors [(25), (29), (30)]. We acknowledge that reliance on a single compound would not fully exclude the possibility of pharmacological side effects. Hence, the finding that eight distinct Ezh2 inhibitors have comparable osteogenic effects at non-cytotoxic concentrations increases our confidence in their utility as osteogenic reagents. Further confidence in the Ezh2 specificity and biological effects of these drugs comes from gain-of-function studies showing that Ezh2 mRNA and protein elevation impairs osteoblast differentiation and negates the effect of Ezh2 inhibition. Conversely, siRNA mediated depletion of Ezh2 promotes osteoblastogenesis and the incomplete knockdown of the protein that is observed strengthens the osteogenic effects of pharmacological inhibition.
We conclude from the combined body of data presented in the current and previous studies that Ezh2 inhibition is relatively safe and effective and can be realistically considered in skeletal regenerative strategies and bone tissue engineering. Based on our current side-by-side comparison of eight different iEzh2 compounds for cytotoxic, metabolic, epigenetic and osteogenic effects in cell culture, it appears that Ezh2 inhibitors as a class of bone stimulatory compounds have comparable merits. Further development of these compounds for bone anabolic therapies may therefore consider methods of delivery, pharmacokinetics and bioavailability in vivo for each compound.
4]. METHODS
4.1. MC3T3 cell culture
MC3T3-E1 (sc4) cells were used for all the experiments. Cells were maintained and expanded in MEM alpha ascorbic acid free media (Gibco; Waltham, MA) supplemented with 10% Fetal Bovine Serum (FBS; R&D Systems; Minneapolis, MN) and 100 units/ml penicillin (Gibco). Ezh2 inhibitors were developed by the indicated pharmaceutical companies (see Figure 1) and acquired from commercial suppliers (MilliporeSigma; St. Louis, MO). Our studies employed a panel of Ezh2 inhibitors that were obtained from Cayman Chemicals, unless indicated otherwise: CPI-1205 (Selleck Chemicals; Houston, TX; Cat.#S8353, CAS No. 1621862-70-1), CPI-169 (Cayman Chemical; Ann Arbor, MI; Cat.#18299, CAS No. 1450655-76-1), EI1 (Cat.#19146, CAS No. 1418308-27-6), EPZ-6438 (Cat.#16174, CAS No. 1403254-99-8), GSK126 (Cat.#15415, CAS No. 1346574-57-9), GSK503 (Cat.#18531, CAS No. 1346572-63-1), PF-06726304 (Sigma-Aldrich; St. Louis, MO; Cat.#PZ0346-5MG, CAS No. 1616287-82-1), UNC1999 (Cat.#1462, CAS No. 1431612-23-5). All compounds were dissolved in DMSO and stored at −20°C. At selected time points, inhibitors were diluted in pre-warmed tissue culture media and added to the cell cultures. Control groups treated with vehicle (DMSO) were supplemented within each experiment with equivalent concentrations of DMSO as appropriate.
4.2. Osteogenic differentiation
For experiments requiring osteogenic differentiation, MC3T3 cells were plated at 10,000 cells/cm2 on standard tissue culture plates (Gibco) in MEM alpha media (Gibco) supplemented with 10% FBS (R&D Systems), and 100 units/ml penicillin (Gibco). The following day, maintenance media was removed and replaced with osteogenic media which consisted of standard MEM alpha with 10% FBS, 100 units/ml penicillin, supplemented with 50 μg/ml ascorbic acid, and 10 mM β-glycerol phosphate. Treatments were performed by adding either one of the Ezh2 inhibitors or vehicle (DMSO). Media was changed every three days and fresh osteogenic media was added.
4.3. MTS Activity Assay
MTS activity assay (Promega) was performed according to manufacturer’s protocol at indicated time points. Absorbance was measured at 490 nm using a Spectra MAX Plus spectrophotometer (Molecular Devices; San Jose, CA).
4.4. Hoechst staining
Media was removed from the wells and cells were washed with Phosphate Buffered Saline (PBS). Diluted 0.1X Tris-EDTA buffer (1 mM Tris, 0.1 mM EDTA) was added to the wells to cover the cells. Subsequently, plates were stored at −80°C for a minimum of two hours and then thawed at room temperature. For each well, chromatin visualization in the nucleus was achieved by adding Hoechst staining solution (0.5 μg Hoechst 33258 [Sigma-Aldrich, Cat.# 94403] per 1ml of buffer solution [0.3 M NaCl, 5 mM Tris, pH 8.0 in H2O]). Plates were incubated in the dark at room temperature for 15 minutes. Fluorescence intensity was quantified 340/460 wavelengths using a F200 Infinite Pro plate reader (Tecan; Zürich, Switzerland). Results were fit to a standard DNA curve to establish relative DNA content.
4.5. Live/Dead Cytotoxicity Assay
Cell viability was determined using the Live/Dead assay kit with two fluorescent probes, Ethidium Homodimer-1 and Calcein-AM (Thermo Fischer Scientific). Media was removed, the probes were diluted to 2μl/ml PBS and 0.5 μl/PBS respectively, and added to the wells. Following a 20 minute incubation period, cells were observed under a fluorescent microscope (Zeiss; Oberkochen, Germany; Zeiss Axio Vert.A1) to determine viability. Proportions of live cells were measured by quantifying fluorescence intensity at 465/540 nm using a F200 Infinite Pro (Tecan) plate reader with appropriate excitation filters.
4.6. Western blotting
At indicated time points, cells were washed with PBS and lysed with radioimmunoprecipitation buffer (150 mM NaCl, 50 mM Tris (pH 7.4), 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1% Triton X-100) supplemented with protease inhibitor mixture (Sigma-Aldrich) and phenylmethylsulfonyl fluoride (Sigma-Adrich). Lysates were cleared by centrifugation and stored at −80°C. Protein concentration was quantified using the DC protein assay kit (Bio-Rad) following the manufacturer’s protocol. Western blot and membrane development were performed as previously described (31). The following primary antibodies were used: Ezh2 (Cell Signaling; Danvers, MA; Cat #5246; 1:10,000), H3K27me3 (Millipore; Cat #ABE44; 1:1000), H3K27ac (Millipore; Cat #17–683; 1:1,000), H3 (Millipore; Cat #05–928; 1:20,000), Gapdh (Cell Signaling; Cat # 5174; 1:5,000), Tubulin beta (Tubb) (Developmental Studies Hybridoma Bank; Iowa City, Iowa; Cat #E7, 1:10,000).
4.7. Alizarin red staining
At appropriate time points, media was removed and cells were washed twice with PBS. After washing, cells were fixed in 10% Neutral Buffered Formalin (NBF) for one hour. NBF was then removed, cells were washed with PBS, and stained with 2% alizarin red pH 4.2 (Thermo Fischer Scientific) for ten minutes. Stain was removed and cells were washed five times with H2O. Images of the wells were taken and staining was quantified using ImageJ program.
4.8. Alkaline phosphatase activity assay
Using the same plates from the Hoechst staining described above, 250 μl of para-nitrophenylphosphate solution (2.5 mg 4-nitrophenylphosphate disodium salt hexahydrate) (MilliporeSigma) per 1 ml of buffer (0.1 M diethanolamine, 150 mM NaCl, 2 mM MgCl2) were added to each well. Plates were incubate for 30 minutes at room temperature and the absorbance was measured at 405 nm using the Spectra MAX Plus spectrophotometer (Molecular Devices). Results were fit to a standard curved prepared using reconstituted alkaline phosphatase enzyme (Roche; Basilea, Switzerland) to determine relative enzymatic activity.
4.9. Isolation of mRNA
At indicated times, cells were lysed and RNA extracted using TRI-Reagent (Zymo Resarch; Irvine, CA). RNA was isolated using Direct-zol RNA MiniPrep (Zymo Research). Isolated RNA was quantified and quality was tested using NanoDrop 2000 spectrophotometer (Thermo Fischer Scientific).
4.10. Quantitative Real Time Reverse Transcription PCR (RT-qPCR)
RNA was isolated as described above and reverse transcription was performed to obtain cDNA using the Promega Reverse Transcription kit (Promega; Madison, WI) and protocol. Gene expression was quantified using real-time quantitative PCR; each reaction was performed with 3.3 ng of cDNA per 10 μl with QuantiTect SYBR Green PCR Kit (Qiagen; Hilden, Germany) in the CFX384 Real-Time System (Bio-Rad; Hercules, CA). Transcript levels were analyzed using the 2−ΔΔCt method and normalized to the housekeeping gene Gapdh (set at 100). Primer pairs were used at a final concentration of 0.08 μM.
4.11. Overexpression of Ezh2 in MC3T3-E1 cells
To stably overexpress murine Ezh2 in mouse pre-osteoblasts, the Ezh2 transcript was amplified from mouse cDNA with the primers GCTGCAGGTCCGATCCACCGGTGACGAAGAATAATCATG (forward) and GACTCCGGAACGAATTCTGATCACTAAGGCAGCTGTTTCAG (reverse) and cloned into the lentiviral expression vector pUltra-hot (Addgene; Watertown, MA; Cat. #24130). Thereafter, viral particles were generated following standard procedures and MC3T3-E1 cells were infected over three cycles of infection. Successful Ezh2 overexpression was tested by RT-qPCR and western blotting. As a control, an empty pUltra-hot vector was used.
4.12. siRNA-mediated Ezh2 knockdown
MC3T3 cells were seeded in maintenance media at 10,000 cells/cm2. One day later, siRNA transfections with non-targeting control (GE Healthcare; Chicago, IL; Cat # D-001810-10-20) or mouse Ezh2 (GE Healthcare; Cat # L-040882-00-005) Dharmacon ON-TARGETplus SMARTpools were performed using Lipofectamine RNAiMAX (Thermo Fischer Scientific; Waltham, MA) following manufacturer’s instructions. MC3T3 osteogenic media was added six hours after transfection. Media was changed two days after transfection and replaced with fresh osteogenic media containing vehicle (DMSO), GSK126 (5 μM) or EPZ-6438 (5 μM). Three days later, culture media for each of the treatment groups (i.e., vehicle, GSK126 and EPZ-6438) were removed and fresh osteogenic media added. Subsequently, media was changed every three days.
4.13. Statistical analysis
Experiments were performed using biological triplicates from cultures of the MC3T3 osteoblast precursor cell line. The results of the experiments are presented as mean ± STD. Statistical analysis was performed using One-way ANOVA or Two-way ANOVA and Tukey’s or Dunnett’s test for multiple comparisons. Statistical analysis was performed using GraphPad Prism Version 8.4.2 (GraphPad Software, Inc., San Diego, CA). Significance is noted in the figures, when applicable (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001).
Supplementary Material
Supplementary Figure S1. Cell counts as a measure of cytotoxicity for Ezh2 inhibitors in MC3T3 cells. Subconfluent MC3T3 cells treated with 100μM, 10μM and 1μM of each one of the Ezh2 inhibitors for three days. Cell counts were assessed by Hoechst staining (DNA content) and normalized to DMSO treated control (1:100 dilution). Data represents n = 3, mean +/−STD. See legend to Figure 1 for additional details. The data shown here were obtained in a representative preliminary experiment performed in triplicate. We note that the results for EI1 and CPI-169 show relatively large error bars (green) for very high concentrations (100 μM) of these inhibitors. The large variability in Hoechst could be due to different opposing effects of iEzh2s in cell culture. Inhibition of Ezh2 is predicted to increase Hoechst binding to chromatin (by converting heterochromatin to euchromatin). Yet, the cytotoxicity of high doses is also expected to cause cell death which reduces Hoechst staining. The balance of the two is most likely time and cell density dependent.
Supplementary Figure S2. Calculation of IC50s for cytotoxicity parameters. Quantitation values of the cytotoxicity studies (see Fig. 1 and Supplementary Fig. 1) were used for the graphical calculation of IC50s for (A) MTS metabolic activity, (B) cell number and (C) cell viability for each of the compounds. Drugs showing an IC50>100 μM were not included in the graphs in panels A-C, but are included in Fig. 1 E. GraphPad prism was used to create a non-linear regression curve fit in which data from each concentration response curve was converted into percentage values that were log10 transformed. These data were then applied to generate a curved XY plot with variable slope as a non-linear graphical representation. The estimated IC50 values from these graphs are presented in tabular form in Figure 1A.
Supplementary Figure S3: DMSO stimulates osteogenic differentiation on MC3T3 preosteoblastic cells. RT-qPCR analysis was used to measure gene expression in MC3T3 cells at day six and ten after treatment with either osteogenic media, 0.05% DMSO or 0.4% DMSO. Graphs show mRNA expression levels of osteogenic genes, with the bars representing mean values ± STD (n = 3). Normalized expression is presented as arbitrary units of values calculated using the 2−ΔΔCt method for the respective marker and Gapdh multiplied by 100. Statistical significance is indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Supplementary Figure S4: Tukey’s multiple comparisons test. The table shows statistical evaluation of results presented in Figure 8 using one-way ANOVA and Tukey’s multiple comparison test. These tests showed no significance between any of the groups. Unpaired t tests (for the averages of control (Ezh2 −) and (Ezh2 +) groups) shown in Figure 8 have a p value = 0.0127.
Highlights:
Trimethylation of histone 3 lysine 27 (H3K27me3) by Ezh2 suppresses osteogenesis.
We show that multiple Ezh2 inhibitors (iEzh2s) can accelerate osteoblast differentiation.
Any of these iEzh2s reduces H3K27me3 levels and enhances bone mRNA marker expression.
The range of iEzh2 compounds available offers opportunities for bone anabolic strategies.
ACKOWLEDGMENTS
We would like to thank all past and present members of our laboratory, as well as institutional and extramural colleagues for stimulating discussions and generously sharing ideas. We are particularly grateful to Oksana Pichurin, Catalina Galeano-Garces, and Margarita Carrasco, as well as our long term collaborators Gary Stein, Janet Stein, Jonathan Gordon, Martin Montecino, and Mario Galindo. We also thank our colleagues at Numerate Inc. (Uwe Klein, John Griffin and Brian Raimundo) for providing reagents and conceptual advice on Ezh2 inhibitors.
FUNDING
This work was supported by funding from the National Institute of Health grant R01 AR069049 (AJvW) and a Career Development Award in Orthopedics Research (AD). We also thank William and Karen Eby for their generous philanthropic support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane NE. Epidemiology, etiology, and diagnosis of osteoporosis. Am J Obstet Gynecol 2006;194(2 Suppl):S3–11. [DOI] [PubMed] [Google Scholar]
- 2.Kannus P. International osteoporosis foundation: The facts about osteoporosis and its impact. Lyon: International Osteoporosis Foundation. 2003. [Google Scholar]
- 3.Genant HK, Cooper C, Poor G, Reid I, Ehrlich G, Kanis J, et al. Interim report and recommendations of the World Health Organization Task-Force for Osteoporosis. Osteoporos Int 1999;10(4):259–64. [DOI] [PubMed] [Google Scholar]
- 4.Raisz LG. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J Clin Invest 2005;115(12):3318–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su P, Tian Y, Yang C, Ma X, Wang X, Pei J, et al. Mesenchymal Stem Cell Migration during Bone Formation and Bone Diseases Therapy. Int J Mol Sci 2018;19(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418(6893):41–9. [DOI] [PubMed] [Google Scholar]
- 7.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 2011;13(1):27–38. [DOI] [PubMed] [Google Scholar]
- 8.Yang TL, Shen H, Liu A, Dong SS, Zhang L, Deng FY, et al. A road map for understanding molecular and genetic determinants of osteoporosis. Nat Rev Endocrinol 2020;16(2):91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause C, de Gorter DJ, Karperien M, ten Dijke P. Signal transduction cascades controlling osteoblast differentiation. Primer on the metabolic bone diseases and disorders of mineral metabolism. 2008;7:10–6. [Google Scholar]
- 10.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17(8):487–500. [DOI] [PubMed] [Google Scholar]
- 11.Gordon JAR, Stein JL, Westendorf JJ, van Wijnen AJ. Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone. 2015;81:739–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Wijnen AJ, Westendorf JJ. Epigenetics as a new frontier in orthopedic regenerative medicine and oncology. Journal of Orthopaedic Research®. 2019;37(7):1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudakovic A, van Wijnen AJ. Epigenetic Control of Osteoblast Differentiation by Enhancer of Zeste Homolog 2 (EZH2). Current Molecular Biology Reports. 2017;3(2):94–106. [Google Scholar]
- 14.Pike JW, Meyer MB, John HCS, Benkusky NA. Epigenetic histone modifications and master regulators as determinants of context dependent nuclear receptor activity in bone cells. Bone. 2015;81:757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep 2013;11(2):72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 2012;8(4):212–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Wang C, Liu W, Li J, Li C, Kou X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–62. [DOI] [PubMed] [Google Scholar]
- 18.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–45. [DOI] [PubMed] [Google Scholar]
- 19.Meyer MB, Benkusky NA, Sen B, Rubin J, Pike JW. Epigenetic Plasticity Drives Adipogenic and Osteogenic Differentiation of Marrow-derived Mesenchymal Stem Cells. J Biol Chem 2016;291(34):17829–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Gordon JA, Whitfield TW, Tai PW, van Wijnen AJ, Stein JL, et al. Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochim Biophys Acta Gene Regul Mech 2017;1860(4):438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojas A, Sepulveda H, Henriquez B, Aguilar R, Opazo T, Nardocci G, et al. Mll-COMPASS complexes mediate H3K4me3 enrichment and transcription of the osteoblast master gene Runx2/p57 in osteoblasts. J Cell Physiol 2019;234(5):6244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas A, Aguilar R, Henriquez B, Lian JB, Stein JL, Stein GS, et al. Epigenetic Control of the Bone-master Runx2 Gene during Osteoblast-lineage Commitment by the Histone Demethylase JARID1B/KDM5B. J Biol Chem 2015;290(47):28329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khani F, Thaler R, Paradise CR, Deyle DR, Kruijthof-de Julio M, Galindo M, et al. Histone H4 Methyltransferase Suv420h2 Maintains Fidelity of Osteoblast Differentiation. J Cell Biochem 2017;118(5):1262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudakovic A, Gluscevic M, Paradise CR, Dudakovic H, Khani F, Thaler R, et al. Profiling of human epigenetic regulators using a semi-automated real-time qPCR platform validated by next generation sequencing. Gene. 2017;609:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudakovic A, Samsonraj RM, Paradise CR, Galeano-Garces C, Mol MO, Galeano-Garces D, et al. Inhibition of the epigenetic suppressor EZH2 primes osteogenic differentiation mediated by BMP2. J Biol Chem 2020;295(23):7877–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen B, Paradise CR, Xie Z, Sankaran J, Uzer G, Styner M, et al. β-catenin preserves the stem state of murine bone marrow stromal cells through activation of EZH2. Journal of Bone and Mineral Research. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camilleri ET, Dudakovic A, Riester SM, Galeano-Garces C, Paradise CR, Bradley EW, et al. Loss of histone methyltransferase Ezh2 stimulates an osteogenic transcriptional program in chondrocytes but does not affect cartilage development. Journal of biological chemistry. 2018;293(49):19001–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudakovic A, Camilleri ET, Paradise CR, Samsonraj RM, Gluscevic M, Paggi CA, et al. Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice. J Biol Chem 2018;293(33):12894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudakovic A, Camilleri ET, Riester SM, Paradise CR, Gluscevic M, O’Toole TM, et al. Enhancer of Zeste Homolog 2 Inhibition Stimulates Bone Formation and Mitigates Bone Loss Caused by Ovariectomy in Skeletally Mature Mice. J Biol Chem 2016;291(47):24594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, et al. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. J Biol Chem 2015;290(46):27604–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Wijnen AJ, Bagheri L, Badreldin AA, Larson AN, Dudakovic A, Thaler R, et al. Biological Functions of Chromobox (CBX) Proteins in Stem Cell Self-Renewal, Lineage-Commitment, Cancer and Development. Bone. 2020:115659. [DOI] [PubMed] [Google Scholar]
- 32.Galea GL, Paradise CR, Meakin LB, Camilleri ET, Taipaleenmaki H, Stein GS, et al. Mechanical strain-mediated reduction in RANKL expression is associated with RUNX2 and BRD2. Gene: X. 2020;5:100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paradise CR, Galvan ML, Kubrova E, Bowden S, Liu E, Carstens MF, et al. The epigenetic reader Brd4 is required for osteoblast differentiation. Journal of Cellular Physiology. 2020;235(6):5293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Najafova Z, Tirado-Magallanes R, Subramaniam M, Hossan T, Schmidt G, Nagarajan S, et al. BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire. Nucleic acids research. 2017;45(1):127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feigenson M, Shull LC, Taylor EL, Camilleri ET, Riester SM, van Wijnen AJ, et al. Histone Deacetylase 3 Deletion in Mesenchymal Progenitor Cells Hinders Long Bone Development. J Bone Miner Res 2017;32(12):2453–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carpio LR, Bradley EW, McGee-Lawrence ME, Weivoda MM, Poston DD, Dudakovic A, et al. Histone deacetylase 3 supports endochondral bone formation by controlling cytokine signaling and matrix remodeling. Sci Signal. 2016;9(440):ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudakovic A, Evans JM, Li Y, Middha S, McGee-Lawrence ME, van Wijnen AJ, et al. Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J Biol Chem 2013;288(40):28783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen J, Hovhannisyan H, Lian JB, Montecino MA, Stein GS, Stein JL, et al. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Mol Endocrinol 2003;17(4):743–56. [DOI] [PubMed] [Google Scholar]
- 39.Thaler R, Spitzer S, Karlic H, Klaushofer K, Varga F. DMSO is a strong inducer of DNA hydroxymethylation in pre-osteoblastic MC3T3-E1 cells. Epigenetics. 2012;7(6):635–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaler R, Maurizi A, Roschger P, Sturmlechner I, Khani F, Spitzer S, et al. Anabolic and Antiresorptive Modulation of Bone Homeostasis by the Epigenetic Modulator Sulforaphane, a Naturally Occurring Isothiocyanate. J Biol Chem 2016;291(13):6754–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136(21):3531–42. [DOI] [PubMed] [Google Scholar]
- 43.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyngaarden LA, Delgado-Olguin P, Su IH, Bruneau BG, Hopyan S. Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development. 2011;138(17):3759–67. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz D, Varum S, Zemke M, Schöler A, Baggiolini A, Draganova K, et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141(4):867–77. [DOI] [PubMed] [Google Scholar]
- 46.Hemming S, Cakouros D, Codrington J, Vandyke K, Arthur A, Zannettino A, et al. EZH2 deletion in early mesenchyme compromises postnatal bone microarchitecture and structural integrity and accelerates remodeling. FASEB J 2017;31(3):1011–27. [DOI] [PubMed] [Google Scholar]
- 47.Lui JC, Garrison P, Nguyen Q, Ad M, Keembiyehetty C, Chen W, et al. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat Commun 2016;7:13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol 2011;13(1):87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Meng Y, Cui Q, Qin F, Yang H, Chen Y, et al. MiR-101 Targets the EZH2/Wnt/beta-Catenin the Pathway to Promote the Osteogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. Sci Rep 2016;6:36988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem cells. 2014;32(3):802–15. [DOI] [PubMed] [Google Scholar]
- 51.Zhu XX, Yan YW, Chen D, Ai CZ, Lu X, Xu SS, et al. Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2016;7(39):63561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen YH, Chung CC, Liu YC, Yeh SP, Hsu JL, Hung MC, et al. Enhancer of Zeste Homolog 2 and Histone Deacetylase 9c Regulate Age-Dependent Mesenchymal Stem Cell Differentiation into Osteoblasts and Adipocytes. Stem Cells. 2016;34(8):2183–93. [DOI] [PubMed] [Google Scholar]
- 53.Jing H, Liao L, An Y, Su X, Liu S, Shuai Y, et al. Suppression of EZH2 Prevents the Shift of Osteoporotic MSC Fate to Adipocyte and Enhances Bone Formation During Osteoporosis. Mol Ther 2016;24(2):217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang C, Qiao Y, Mun SH, Lee MJ, Murata K, Bae S, et al. Cutting Edge: EZH2 Promotes Osteoclastogenesis by Epigenetic Silencing of the Negative Regulator IRF8. J Immunol 2016;196(11):4452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J Cell Sci 2013;126(Pt 2):565–79. [DOI] [PubMed] [Google Scholar]
- 56.Stephens AS, Stephens SR, Hobbs C, Hutmacher DW, Bacic-Welsh D, Woodruff MA, et al. Myocyte enhancer factor 2c, an osteoblast transcription factor identified by dimethyl sulfoxide (DMSO)-enhanced mineralization. J Biol Chem 2011;286(34):30071–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ren J, Huang D, Li R, Wang W, Zhou C. Control of mesenchymal stem cell biology by histone modifications. Cell Biosci 2020;10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A. 2014;111(4):1355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Long Y, Paucek RD, Gooding AR, Lee T, Burdorf RM, et al. Regulation of histone methylation by automethylation of PRC2. Genes Dev 2019;33(19–20):1416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee C-H, Yu J-R, Granat J, Saldaña-Meyer R, Andrade J, LeRoy G, et al. Automethylation of PRC2 promotes H3K27 methylation and is impaired in H3K27M pediatric glioma. Genes & development. 2019;33(19–20):1428–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346(6215):1373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Cell counts as a measure of cytotoxicity for Ezh2 inhibitors in MC3T3 cells. Subconfluent MC3T3 cells treated with 100μM, 10μM and 1μM of each one of the Ezh2 inhibitors for three days. Cell counts were assessed by Hoechst staining (DNA content) and normalized to DMSO treated control (1:100 dilution). Data represents n = 3, mean +/−STD. See legend to Figure 1 for additional details. The data shown here were obtained in a representative preliminary experiment performed in triplicate. We note that the results for EI1 and CPI-169 show relatively large error bars (green) for very high concentrations (100 μM) of these inhibitors. The large variability in Hoechst could be due to different opposing effects of iEzh2s in cell culture. Inhibition of Ezh2 is predicted to increase Hoechst binding to chromatin (by converting heterochromatin to euchromatin). Yet, the cytotoxicity of high doses is also expected to cause cell death which reduces Hoechst staining. The balance of the two is most likely time and cell density dependent.
Supplementary Figure S2. Calculation of IC50s for cytotoxicity parameters. Quantitation values of the cytotoxicity studies (see Fig. 1 and Supplementary Fig. 1) were used for the graphical calculation of IC50s for (A) MTS metabolic activity, (B) cell number and (C) cell viability for each of the compounds. Drugs showing an IC50>100 μM were not included in the graphs in panels A-C, but are included in Fig. 1 E. GraphPad prism was used to create a non-linear regression curve fit in which data from each concentration response curve was converted into percentage values that were log10 transformed. These data were then applied to generate a curved XY plot with variable slope as a non-linear graphical representation. The estimated IC50 values from these graphs are presented in tabular form in Figure 1A.
Supplementary Figure S3: DMSO stimulates osteogenic differentiation on MC3T3 preosteoblastic cells. RT-qPCR analysis was used to measure gene expression in MC3T3 cells at day six and ten after treatment with either osteogenic media, 0.05% DMSO or 0.4% DMSO. Graphs show mRNA expression levels of osteogenic genes, with the bars representing mean values ± STD (n = 3). Normalized expression is presented as arbitrary units of values calculated using the 2−ΔΔCt method for the respective marker and Gapdh multiplied by 100. Statistical significance is indicated as follows: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Supplementary Figure S4: Tukey’s multiple comparisons test. The table shows statistical evaluation of results presented in Figure 8 using one-way ANOVA and Tukey’s multiple comparison test. These tests showed no significance between any of the groups. Unpaired t tests (for the averages of control (Ezh2 −) and (Ezh2 +) groups) shown in Figure 8 have a p value = 0.0127.