Abstract
In periodontitis patients, dysbiosis of the oral microbiota is not only found at clinically diseased periodontal sites but also at clinically healthy periodontal sites, buccal mucosae, tongue, and saliva. The present study evaluated the safety and efficacy of an oral microbiota transplant (OMT) for the treatment of periodontitis in dogs. Eighteen systemically healthy beagle dogs with naturally occurring periodontitis were enrolled in the study and randomly assigned to a test or control group. A 4-y-old, periodontally healthy female beagle dog served as a universal OMT donor. To reduce periodontal inflammation, all dogs received full-mouth mechanical debridement of teeth and mucosae 2 wk before baseline. At baseline, full-mouth mechanical debridement was repeated and followed by adjunctive subgingival and oral irrigation with 0.1% NaOCl. Subsequently, test dogs were inoculated with an OMT from the healthy donor. No daily oral hygiene was performed after OMT transplantation. Adverse events were assessed throughout the observation period. Clinical examinations were performed and whole-mouth oral microbiota samples were collected at week 2, baseline, week 2, and week 12. The composition of oral microbiota samples was analyzed using high-throughput 16S ribosomal RNA gene amplicon sequencing followed by taxonomic assignment and downstream bioinformatic and statistical analyses. Results demonstrated that the intergroup difference in the primary outcome measure, probing pocket depth at week 12, was statistically insignificant. However, the single adjunctive OMT had an additional effect on the oral microbiota composition compared to the full-mouth mechanical and antimicrobial debridement alone. The OMT resulted in an “ecological shift” toward the composition of the donor microbiota, but this was transient in nature and was not observed at week 12. No local or systemic adverse events were observed throughout the study period. The results indicate that OMT may modulate the microbiota composition in dogs with naturally occurring periodontitis and can be applied safely.
Keywords: periodontal disease, biological therapy, nonsurgical therapy, host microbial interactions, microbiome, transplantation
Introduction
Humans and microorganisms have evolved together in a symbiotic relationship over thousands of years (Didelot et al. 2016). Colonizing microorganisms provide the human body with various biological functions ranging from the development of mucosal immune barriers to colonization resistance against pathogens (Avila et al. 2009; Hornef 2015). Most resident microorganisms are found within the digestive system and are estimated to approximate the number of nucleated human somatic cells (Dewhirst et al. 2010; Sender et al. 2016). While a symbiotic microbiota is beneficial to the host, the delicate balance can be disturbed by an overgrowth of accessory pathogens and pathobionts, resulting in an increased pathogenicity of the local microbial community (Hajishengallis and Lamont 2012). A dysbiosis can induce inflammation and promote a favorable environment for pathogen growth, which in turn exacerbates the host immune responses and inflammatory sequelae (Levy et al. 2017). It may cause chronic diseases, such as periodontal diseases and ulcerative colitis (Halfvarson et al. 2017).
The pathogenic potential of various microorganisms and their role in the etiology of periodontitis has been studied extensively (Haubek et al. 2009; Hajishengallis et al. 2011; Pérez-Chaparro et al. 2014). More recent reports demonstrated that there were profound changes in the relative abundance of “core” taxa and composition within the subgingival microbiota that extend beyond the species previously associated with periodontitis (Li et al. 2013; Chen et al. 2018). In that regard, a distinct partitioning of bacterial communities is commonly found within healthy and diseased periodontal sites (Griffen et al. 2012).
The ultimate goal of periodontal therapy is to reverse the oral ecological regime shift from a state of dysbiosis and to reinstall stable homeostasis with the host immune system (Moutsopoulos and Konkel 2018). Conventional periodontal therapy induces microbial community changes: nevertheless, these remain short-lasting (Sanz-Sanchez et al. 2016; Lu et al. 2019). More pronounced shifts in oral microbial ecology may result from adjunctive treatments, such as a full-mouth disinfection approach or use of systemic antibiotics, compared to mechanical debridement alone (De Soete et al. 2001).
Recently, an interesting approach for altering dysbiotic microbiota has received attention in the treatment of intestinal diseases. After cleaning the entire dysbiotic gut microbiota, fecal microbiota transplantations (FMTs) from healthy donors have been used to establish a health-compatible microbiota in recipients (Jacob et al. 2017). Initially, FMT was recommended for the treatment of persistent Clostridioides difficile (C-diff) infections (Cammarota et al. 2017), but its application expanded quickly to chronic diseases associated with gut dysbiosis, such as ulcerative colitis (Shen et al. 2018; Costello et al. 2019). Mounting evidence from pooled analyses and randomized controlled trials supports the efficacy of FMT for the remission of ulcerative colitis (Shi et al. 2016; Paramsothy et al. 2017).
Considering the demonstrated efficacy of FMT in treating some dysbiosis-associated gut diseases, the present study evaluated the safety and efficacy of an oral microbiota transplantation (OMT) as an adjunct to full-mouth debridement in the treatment of naturally occurring periodontitis in beagle dogs.
Materials and Methods
Study Animals
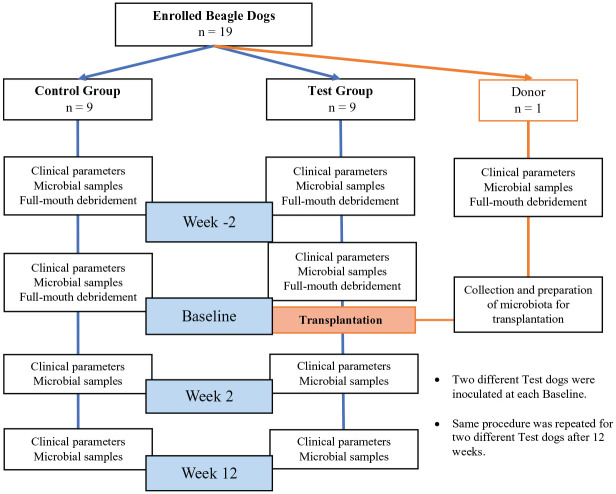
Eighteen systemically healthy 2- to 7-y-old female and male beagle dogs with naturally occurring periodontitis with at least 32 teeth were included in the study and randomly assigned to a test (n = 9) or control (n = 9) group by block randomization. One 4-y-old periodontally and systemically healthy female beagle dog with all 42 teeth was enrolled as the healthy oral microbiota transplant donor. Periodontitis severity was determined after inclusion and randomization into experimental groups to avoid allocation bias. Probing pocket depths (PPD), bleeding on probing (BOP) scores, and plaque index (PI) scores were recorded for both groups at 4 time points: 2 wk before OMT (week –2), at OMT (baseline), 2 wk after OMT (week 2), and 12 wk after OMT (week 12) by calibrated examiners under general anesthesia. Animals were housed at the Animal Research Facility of Heinrich-Heine University of Dusseldorf in Germany. To prevent any unintended transmission of oral microbiota, dogs were separated from each other during the study period.
Full-Mouth Debridement Prior to Intervention
To reduce periodontal inflammation, all dogs in both groups received full-mouth supra- and subgingival debridement using sonic scalers and hand instruments as well as full-mouth glycine powder air polishing (GPAP) 2 wk before baseline (week 2; Fig. 1). Each supra- and subgingival tooth surface was treated with GPAP for 5 s and each mucosal surface (buccal and labial mucosae, lingual and buccal alveolar mucosa, floor of the mouth, tongue, and palate) for 1 min. At baseline, this procedure was repeated except for the donor dog. To further suppress the resident oral microbiota, additional subgingival and oral irrigation with 0.1% NaOCl was performed for 5 min (Slots 2002). Then, NaOCl was inactivated by subgingival and oral rinsing with 23 µM buffered sodium ascorbate for 10 min (Pozhitkov et al. 2015). This procedure was followed by OMT in the test group as described below.
Figure 1.
Flowchart of study framework.
Oral Microbiota Transplant
The donor microbiota was collected prior to mechanical debridement from supragingival biofilms of all teeth and mucosal surfaces using a hand curette and a cotton swab under general anesthesia at week 2. At baseline, the donor dog was sedated and supragingival biofilms from all teeth and mucosal surfaces were harvested on 1 side of the mouth as described. The samples were pooled and suspended in sterile reduced transport medium. An aliquot was reserved for microbiome analysis at each visit. The collected oral microbiota was dispersed by vortexing and used immediately for transplantation to 1 dog of the test group. Test dogs were inoculated with OMT by continuous subgingival and oral irrigation over a period of 10 min. The biofilms from the other side of the mouth were analogously collected on the next day and used for transplantation to a different test dog. This general procedure was repeated on pairs of test dogs over an approximately 18-mo period, waiting a minimum of 3 mo between harvesting/transplantation procedures to allow for the reestablishment of the biofilm in the donor dog. None of the dogs received oral hygiene after OMT.
Microbial Sampling
Subgingival biofilm samples were collected from the deepest periodontal site of each quadrant using a hand curette, and mucosal biofilm samples were collected from the buccal mucosa, the dorsum of the tongue, and the palate using a cotton swab from all dogs at 4 time points: at week 2, at baseline prior to full-mouth debridement, at week 2, and at week 12. Subgingival and mucosal biofilm samples were respectively pooled for the analysis. All samples were stored at −80°C until processing for DNA extraction and 16S ribosomal RNA (rRNA) gene sequencing.
Sequencing of the Oral Microbiota
Total bacterial genomic DNA was extracted from the samples using QIAamp DNA mini kits (Qiagen) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) amplification of the hypervariable V3–V4 region of the 16S rRNA genes was performed using the universal bacterial primer pairs, 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR amplification and library construction were performed according to Illumina’s standard protocol (16S Metagenomic Sequencing Library Preparation, Part 15044223 Rev. B). The Illumina MiSeq system (300PE) was used for paired-end MiSeq sequencing of amplicon libraries, which was performed by BGI Genomics. The paired-end MiSeq sequencing reads were analyzed using the DADA2 pipeline (Callahan et al. 2016). Details of the bioinformatics analysis are reported in the appendix material. The sequences were deposited in the NCBI SRA under accession number PRJNA598540.
Clinical Measurements
Full-mouth measurements of PPD, BOP, and PI (O’Leary et al. 1972) were recorded at week 2, baseline, week 2, and week 12. PPD was measured at 6 sites (mesiobuccal, buccal, distobuccal, mesio-oral, oral, and disto-oral) at each tooth using a PCP 15 North Carolina periodontal probe. Assessment of gingival bleeding was made 30 s after PPD measurements. The measurements were rounded to the nearest integer.
Adverse Events
Examination of the intraoral tissues was carried out by 3 authors (A.K., B.W., and T.B.) before, during, and at exiting from the study. Intraoral photos were taken for a second examination by one of the abovementioned examiners who was not actively participating in oral examination at the same time point. Behavioral abnormalities of the enrolled dogs and presence of fever and diarrhea were assessed by veterinarians before, during, and at exiting from the study.
Statistical Analyses
This study evaluated the efficacy of OMT as an adjunct to mechanical and chemical debridement in the treatment of periodontitis in dogs. Outcomes following mechanical debridement in beagle dogs with naturally occurring periodontitis were used for sample size determination (Morrison et al. 1979). A total of 18 dogs were needed to detect a difference of 0.8 mm in PPD with a standard deviation of 0.4 mm at a significance level of 0.05 and a power of at least 80%. The estimated power using the exact small-sample t-distribution was 88%.
Mean PPD at week 12 was used as the primary outcome variable, as there were no preliminary data about the outcomes following OMT. All other parameters were assessed as secondary outcome variables and are presented descriptively. The dog was used as the statistical unit. All data collection and analyses were conducted in a blinded manner. PPD, BOP, and PI were analyzed using analysis of variance (ANOVA) with Fisher’s least significant difference test. Multivariate test statistics for microbiological analysis were performed using the R package GenePiper (Tong and Chan 2020) unless specified otherwise in the appendix. For microbiological data, alpha-diversity measures were calculated using the alpha-diversity module and analyzed by the Wilcoxon test. Beta-diversities were analyzed using Bray-Curtis dissimilarities. The distance matrices were visualized in principal coordinates analysis (PCoA) ordinations and analyzed with permutational multivariate analysis of variance (PERMANOVA) tests evaluated at 9,999 random permutations. Principal response curve (PRC) analysis was performed using the default parameters.
Ethics Committee Approval
The study protocol was in compliance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and approved by the appropriate local authority (Landesamt für Natur und Verbraucherschutz, #84-02.04.2014.A449). All interventions were performed under general anesthesia according to the local ethical and legal guidelines. None of the study animals were sacrificed for study purposes.
Results
Clinical Parameter Changes
One tooth was lost in a test dog (dog Q) at week 2 due to class III mobility according to Miller’s mobility index (Miller 1950). At baseline, there were a total of 366 teeth in the test group and 375 teeth in the control group. None of the enrolled dogs lost a tooth after baseline.
The mean PPD was reduced from baseline to week 12 without significant intergroup differences at week 12 (Table). BOP scores at week 2 were not significantly different, with a mean of 42% in both groups. At week 2, BOP was reduced to 33% in the test but remained at 42% in the control group (Table). However, this difference disappeared at week 12. PI scores remained high throughout the study period without any significant inter- or intragroup differences (Table). There were no significant intergroup differences at any time point for the clinical parameters.
Table.
Clinical Periodontal Measurements of the Test and Control Groups.
| PPD, mm | BOP, % | PI, % | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Time Point | Test | Control | P Value | Test | Control | P Value | Test | Control | P Value |
| Week –2 | 3.21 ± 0.65 | 2.90 ± 0.52 | 0.27 | 41.87 ± 26.73 | 42.36 ± 23.12 | 0.97 | 76.40 ± 11.37 | 77.37 ± 13.48 | 0.87 |
| Baseline | 2.98 ± 0.74 | 2.53 ± 0.46 | 0.14 | 41.24 ± 19.64 | 45.68 ± 19.64 | 0.62 | 61.32 ± 10.88 | 69.54 ± 16.31 | 0.23 |
| Week 2 | 2.69 ± 0.50 | 2.64 ± 0.40 | 0.85 | 32.90 ± 14.72 | 42.15 ± 11.34 | 0.16 | 58.57 ± 10.41 | 70.31 ± 15.42 | 0.08 |
| Week 12 | 2.69 ± 0.57 | 2.44 ± 0.28 | 0.25 | 35.99 ± 18.86 | 32.43 ± 7.78 | 0.61 | 65.66 ± 19.83 | 61.37 ± 16.17 | 0.62 |
All values are described as mean ± SD. P values describe the intergroup comparisons at each time point. P ≤ 0.05 is considered as statistically significant.
BOP, bleeding on probing; PI, plaque index; PPD, probing pocket depth.
Microbiota Profile Changes upon Transplantation
A total of 9,120,510 16S rRNA gene amplicon raw reads were retrieved from all microbiota samples collected during the study (n = 86). After stringent quality control and error modeling through the DADA2 pipeline (Callahan et al. 2016), we obtained 1,753 amplicon sequence variants (ASVs) representing a total of 1,851,136 quality-controlled, merged sequences with an average percentage output of 40.6%. Sequencing depth was equivalent among all samples at a mean of 21,525 ± 2,100 sequences per sample (Appendix Fig. 1). The levels of alpha-diversity in every microbiota sample collected at each of the 4 time points (as evaluated by Shannon entropy values) are listed in Appendix Table 2. Similarly, the levels of species richness in each sample (as evaluated by the numbers of observed ASVs) are listed in Appendix Table 3. There were no significant intergroup differences in the levels of alpha-diversity (Fig. 2) or species richness (Appendix Fig. 2) between the test and control groups at week –2. However, the levels of alpha-diversity and species richness in the periodontally healthy donor were significantly lower than those in both periodontitis groups at week –2 (P ≤ 0.01).
Figure 2.
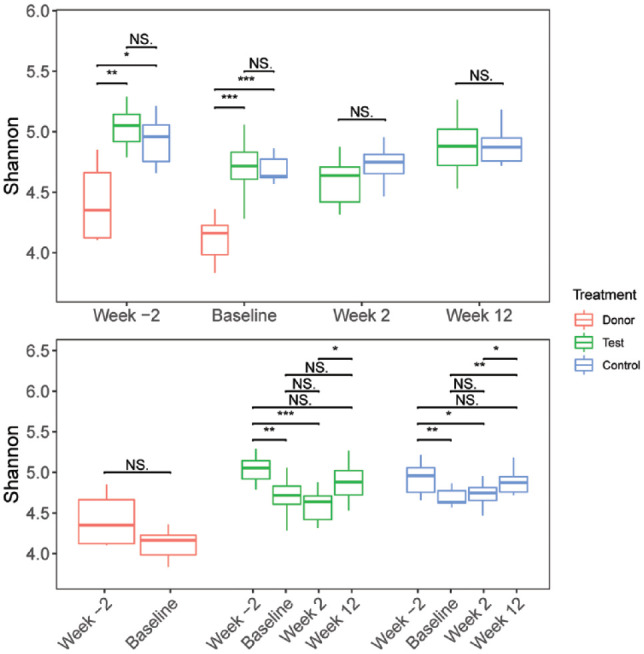
Changes in the alpha-diversities of the oral microbiota in the donor, test, and control groups throughout the study. The levels of species diversity (as determined by Shannon entropies) in the respective sets of test (green) and control (blue) oral microbiota samples were evaluated at the week –2, baseline, week 2, and week 12 time points. The median, upper and lower quartile values, and ×1.5 interquartile ranges are indicated. The corresponding levels of species diversity of the single healthy donor dog oral microbiota samples were evaluated at the week –2 and baseline time points (red). Statistically significant differences were inferred by the Wilcoxon signed-rank test at P value cutoffs: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. NS, no significant difference. The 2 panels illustrate the results of different statistical comparisons between (upper panel) and within (lower panel) the respective sets of test, control, and donor subjects at the various time points.
The median alpha-diversities of the oral microbiota in both test and control groups decreased significantly between the week –2 and baseline time points (P ≤ 0.01, Fig. 2, Appendix Table 1). Alpha-diversity levels fell further between baseline and week 2 in the test group but increased in the control group, without statistical significance. For both groups, the levels of alpha-diversity were significantly lower at week 2 compared to week –2, with considerably stronger statistical support for the test group than the control group (P ≤ 0.001 vs. P ≤ 0.05, Fig. 2). There was a statistically significant increase in the alpha-diversity levels of the test group subjects between week 2 and week 12. Overall, there were no significant changes in alpha-diversity levels between week 2 and week 12 for either group.
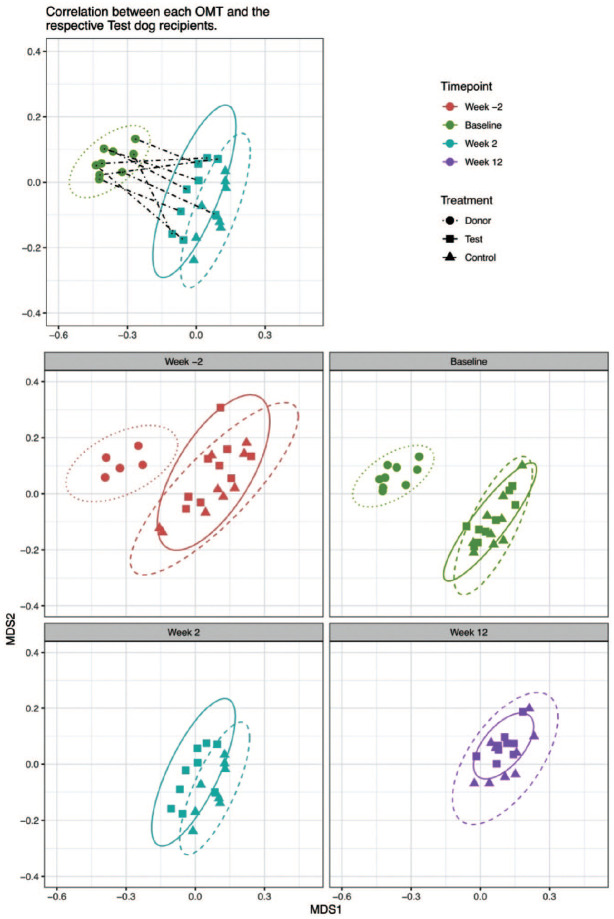
The corresponding beta-diversity levels in the oral microbiota sampled from the donor, test, and control groups throughout the experiment were analyzed using multivariate ordination analyses based on Bray-Curtis dissimilarities and visualized in PCoA plots. The oral microbiota of the donor dog was notably distinct from the test and control dogs and demonstrated stability throughout the experimental period (Fig. 3). While the composition of the oral microbiota in the control dogs displayed apparently random changes across the 4 time points, those in the test dogs shifted in a more conserved manner (Fig. 3, Appendix Figs. 3−6). Most notably, the oral microbiota of the test dogs most closely resembled that of the respective OMTs from the donor dog at the week 2 time point (Fig. 3).
Figure 3.
Principal coordinates analysis (PCoA) ordinations showing the beta-diversity of the oral microbiota in test, control, and donor groups, based on Bray-Curtis dissimilarity values. Various sets of subject data-points are shown in the 5 respective PCoA plots included in the figure panels for the sake of clarity. All PCoA plots are drawn to the same scale, with the same axes. Top left panel: PCoA plot showing oral microbiota transplant (OMT) samples collected from periodontally healthy donor dog (filled circles) immediately prior to transplantation and the oral microbiota of test dogs (filled squares) and control dogs (filled triangles) at the week 2 time point. Dashed lines link the 9 OMT samples with the 9 respective test dog recipients. Top right panel: figure key. Middle left panel: donor, test, and control dogs’ oral microbiota at the week –2 time point. Middle right panel: OMT samples collected from donor immediately prior to transplantation and oral microbiota of test and control dogs at baseline time point. Bottom left panel: oral microbiota of test and control dogs at the week 2 time point. Bottom right panel: oral microbiota of test and control dogs at the week 12 time point. Week –2 time point (red), baseline time point (green), week 2 time point (blue), and week 12 time point (purple). Ellipses were plotted at a confidence level of 0.95. The oral microbiota of the test dogs was more closely related to the respective OMT samples at the week 2 time point. Analogous PCoA plots that include different combinations of data-points are included in the appendix material.
The differences in the microbiota across the 4 time points in each group were tested by PERMANOVA tests (test group R2 = 0.163, P < 0.001; control group R2 = 0.106, P = 0.089; based on Bray-Curtis dissimilarities, Appendix Table 4). These results clearly indicated significant changes to the overall microbial community composition in the test group but not in the control group. Most significantly, the microbiota in the test group demonstrated higher resemblances to the donor microbiota at week 2 after OMT (Appendix Fig. 4). PRC analysis revealed a characteristic modulation of the oral microbiota in the test group compared to the controls. For instance, there were increases in the relative abundances of ASV40 (Chlorobi G-1 sp.), ASV150 (Neisseria sp.), and ASV44 (Bergeyella sp.) in the test group after OMT (Fig. 4, Appendix Fig. 7).
Figure 4.
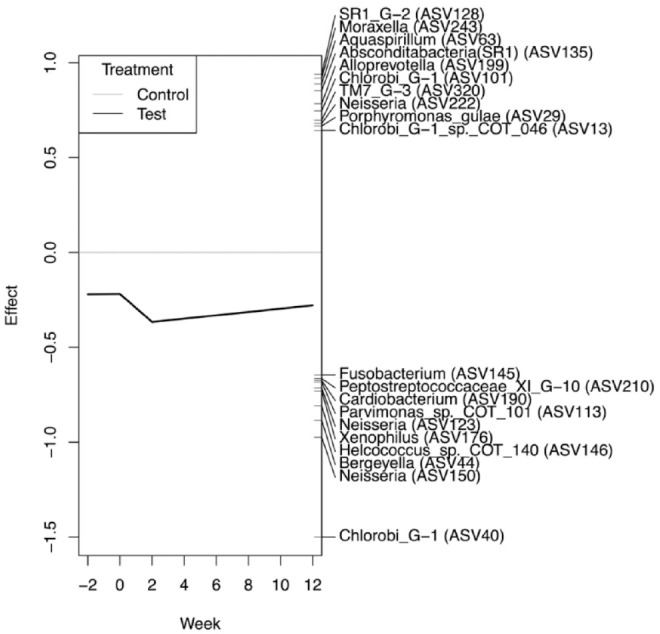
Principal response curve (PRC) analysis showing the effects of the oral microbiota transplant (OMT) in the test group when compared to the control group. Only the amplicon sequence variants (ASVs) with the top 20 scores are shown. The time point (in weeks) is shown on the x-axis. The y-axis shows the difference (effect size) between the test group and the control group. The corresponding identities of the respective 20 top-scoring ASVs are shown at the right-hand side of the plot. The effect of OMT is most apparent between the baseline and week 2 time points, with differences becoming reconciled toward the week 12 time point. Since the PRC is negative, taxa indicated with a positive score have a decrease in relative abundance in the test group compared to the controls and vice versa. ASV-40 (Chlorobi G-1) has the largest effect score at –1.5 and thus is inferred to increase significantly after OMT.
Adverse Events
No clinical signs of a local inflammatory response (i.e., redness, edema) or systemic inflammatory response (i.e., fever or diarrhea) were observed. No behavioral abnormalities that indicate animal unwellness were recorded. One control dog experienced transient cardiac arrhythmia during general anesthesia without impairment of the study conduct.
Discussion
The therapeutic alteration of a dysbiotic microbiota is a challenging undertaking. Most research has thus far focused on altering the gut microbiome, for example, using FMT to treat C-diff (Smits et al. 2013). The composition of the donor microbiota, the genetic and immunological background of an individual recipient, and the composition and inherent resilience of the recipient’s microbiota may all notably influence the ability to make long-lasting beneficial changes to the overall microbial community structure (Wilson et al. 2019). The microbiota within periodontitis niches typically has a significantly higher species diversity when compared to healthy periodontal sites, which is in contrast to the situation typically associated with gastrointestinal health and disease (Abusleme et al. 2013; Shi et al. 2018). Consistent with this relationship, the oral microbiota harvested from the donor dog had significantly lower alpha-diversity levels than test and control dogs. It should be noted that there was considerable heterogeneity in the oral microbiota in both the test and control dogs at baseline, which introduces variability to the results. This observation is consistent with the oral microbiota associated with human periodontitis, which similarly exhibits considerable compositional variability (Kirst et al. 2015).
Research on therapeutic approaches for dysbiotic oral microbiome engineering (oral microbiome transplants) has been conducted almost exclusively in murine models (Hajishengallis et al. 2011; Payne et al. 2019). The murine oral microbiota typically exhibits considerable resilience and regenerative capacity in response to (chemo)therapeutic challenges. For example, an established dysbiotic microbiota demonstrated stability to antibiotic treatment in a murine experiment of vertical and horizontal oral dysbiotic microbiota transfer (Payne et al. 2019). Therefore, we reasoned that a large reduction in the endogenous microbiota populations prior to treatment would be crucial to increase engraftment success of a transplanted microbiota. To achieve this, we conducted a full-mouth debridement and disinfection prior to transplantation to deplete the local microbial communities. However, such procedures are known to generally result in large-scale changes to the oral microbiome, which generally last several weeks, before returning to compositions similar to that originally present (Haffajee et al. 2006; Lu et al. 2019). As expected, full-mouth debridement resulted in a significant reduction in alpha-diversity levels in dogs from both groups, along with a notable change in respective microbiome compositions.
The lowering of alpha-diversity putatively corresponds to a less dysbiotic periodontal microbiome at baseline. The oral microbiota in the test group showed a closer resemblance to the microbiota of the healthy donor, evidenced by its lower alpha-diversity and changes in its ASV composition. These changes indicate that a single OMT had notably modulated the recipient microbiota composition. However, the impact of a single OMT was transient. We postulate that the increase in alpha-diversity in the test group at week 12 may be due to the resilience of the recipient’s well-established “pro-dysbiotic” microbiota counteracting the newly transplanted microbiota. In addition, the lack of oral hygiene may also have negatively influenced the colonization and establishment of the transplanted microbiota and exacerbated the high BOP and PI scores throughout the observation period. Results from FMT treatments have shown that there are notable donor-specific effects, for example, where microbiome explants from “super-donors” may be particularly effective at producing long-lasting (beneficial) changes to the recipient’s microbiome (Wilson et al. 2019). Yet, a single transplantation may be not sufficient to effect long-lasting changes in highly resilient oral microbial populations. Multiple OMTs may be required to sustain “pro-homeostatic” changes to the recipient’s dysbiotic oral microbiota (Utter et al. 2016). Moreover, the mucosae of the gut and oral cavity have highly distinct anatomical and physiological characteristics, and the resident microbiota most probably responds very differently to various OMT regimens.
The results of this dog study demonstrated that a single OMT from a periodontally healthy donor as an adjunct to mechanical and chemical full-mouth debridement has an additional modulatory effect on the composition of the oral microbiota in dogs with naturally occurring periodontitis than full-mouth mechanical and antimicrobial debridement alone. However, the effect of a single OMT was transient in nature, with the microbial composition returning to pretreatment levels after 12 wk. Future studies employing alternative transplantation strategies (e.g., with test subjects receiving multiple “healthy” OMT donations over a specific time period) are critically required to further assess the potential for OMT in the treatment of periodontal diseases.
Author Contributions
T. Beikler, contributed to conception, design, data acquisition, analysis, and interpretation, critically revised the manuscript; K. Bunte, Y. Chan, contributed to data analysis and interpretation, drafted the manuscript; B. Weiher, S. Selbach, A. Klocke, contributed to design and data acquisition, critically revised the manuscript; U. Peters, contributed to design, data acquisition, and analysis, critically revised the manuscript; R.M. Watt, contributed to design, data acquisition, analysis, and interpretation, critically revised and edited the manuscript; T.F. Flemmig, contributed to conception, design, and data interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_0022034521995423 for Oral Microbiota Transplant in Dogs with Naturally Occurring Periodontitis by T. Beikler, K. Bunte, Y. Chan, B. Weiher, S. Selbach, U. Peters, A. Klocke, R.M. Watt and T.F. Flemmig in Journal of Dental Research
Acknowledgments
The authors are indebted to Raymond Tong for technical and bioinformatic assistance and to Alexander E. Pozhitkov and Peter A. Noble for their input in the conception and design of the study.
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: T.F. Flemmig is an inventor with patents covering fine-grain, low-abrasive glycine powders such as the one used in the present study. To safeguard against the conduct and outcomes of the present study being influenced by a potential conflict of interest, T.F. Flemmig refrained from any sensitive element of the present study, including direct data scoring, statistical analysis of data, and adverse event evaluation and reporting. All other authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the German Research Council (Deutsche Forschungsgemeinschaft, grant BE5777/2-1). The data have been deposited with links to BioProject accession number PRJNA 598540 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
ORCID iDs: K. Bunte  https://orcid.org/0000-0002-9337-5279
https://orcid.org/0000-0002-9337-5279
S. Selbach  https://orcid.org/0000-0002-3031-5690
https://orcid.org/0000-0002-3031-5690
References
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila M, Ojcius DM, Yilmaz O. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28(8):405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G, Ianiro G, Tilg H, Rajilic-Stojanovic M, Kump P, Satokari R, Sokol H, Arkkila P, Pintus C, Hart A, et al. 2017. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 66(4):569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WP, Chang SH, Tang CY, Liou ML, Tsai SJ, Lin YL. 2018. Composition analysis and feature selection of the oral microbiota associated with periodontal disease. BioMed Res Int. 2018:3130607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, et al. 2019. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 321(2):156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Soete M, Mongardini C, Peuwels M, Haffajee A, Socransky S, van Steenberghe D, Quirynen M. 2001. One-stage full-mouth disinfection: long-term microbiological results analyzed by checkerboard DNA-DNA hybridization. J Periodontol. 72(3):374–382. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol. 192(19):5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. 2016. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 14(3):150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6(6):1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Teles RP, Socransky SS. 2006. The effect of periodontal therapy on the composition of the subgingival microbiota. Periodontol 2000. 42:219–258. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 27(6):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 10(5):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfvarson J, Brislawn CJ, Lamendella R, Vazquez-Baeza Y, Walters WA, Bramer LM, D’Amato M, Bonfiglio F, McDonald D, Gonzalez A, et al. 2017. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubek D, Ennibi OK, Vaeth M, Poulsen S, Poulsen K. 2009. Stability of the JP2 clone of Aggregatibacter actinomycetemcomitans. J Dent Res. 88(9):856–860. [DOI] [PubMed] [Google Scholar]
- Hornef M. 2015. Pathogens, commensal symbionts, and pathobionts: discovery and functional effects on the host. ILAR J. 56(2):159–162. [DOI] [PubMed] [Google Scholar]
- Jacob V, Crawford C, Cohen-Mekelburg S, Viladomiu M, Putzel GG, Schneider Y, Chabouni F, O’Neil S, Bosworth B, Woo V, et al. 2017. Single delivery of high-diversity fecal microbiota preparation by colonoscopy is safe and effective in increasing microbial diversity in active ulcerative colitis. Inflamm Bowel Dis. 23(6):903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst ME, Li EC, Alfant B, Chi YY, Walker C, Magnusson I, Wang GP. 2015. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol. 81(2):783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. 2017. Dysbiosis and the immune system. Nat Rev Immunol. 17(4):219–232. [DOI] [PubMed] [Google Scholar]
- Li K, Bihan M, Methe BA. 2013. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One. 8(5):e63139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhao Y, Feng X, He L, Meng H. 2019. Microbiome in maintained periodontitis and its shift over a single maintenance interval of 3 months. J Clin Periodontol. 46(11):1094–1104. [DOI] [PubMed] [Google Scholar]
- Miller SC. 1950. Textbook of periodontia: oral medicine. Philadelphia (PA): Blakiston. [Google Scholar]
- Morrison EC, Lang NP, Loe H, Ramfjord SP. 1979. Effects of repeated scaling and root planing and/or controlled oral hygiene on the periodontal attachment level and pocketdepth in beagle dogs: I. Clinical findings.J Periodontal Res. 14(5):428–437. [DOI] [PubMed] [Google Scholar]
- Moutsopoulos NM, Konkel JE. 2018. Tissue-specific immunity at the oral mucosal barrier. Trends Immunol. 39(4):276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary TJ, Drake RB, Naylor JE. 1972. The plaque control record.J Periodontal. 43(1):38. [DOI] [PubMed] [Google Scholar]
- Paramsothy S, Paramsothy R, Rubin DT, Kamm MA, Kaakoush NO, Mitchell HM, Castano-Rodriguez N. 2017. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis.J Crohns Colitis. 11(10):1180–1199. [DOI] [PubMed] [Google Scholar]
- Payne MA, Hashim A, Alsam A, Joseph S, Aduse-Opoku J, Wade WG, Curtis MA. 2019. Horizontal and vertical transfer of oral microbial dysbiosis and periodontal disease. J Dent Res. 98(13):1503–1510. [DOI] [PubMed] [Google Scholar]
- Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, Duarte P, Feres M. 2014. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 93(9):846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozhitkov AE, Leroux BG, Randolph TW, Beikler T, Flemmig TF, Noble PA. 2015. Towards microbiome transplant as a therapy for periodontitis: an exploratory study of periodontitis microbial signature contrasted by oral health, caries and edentulism. BMC Oral Health. 15:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Sanchez I, Ortiz-Vigon A, Herrera D, Sanz M. 2016. Microbiological effects and recolonization patterns after adjunctive subgingival debridement with Er: YAG laser. Clin Oral Investig. 20(6):1253–1261. [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen ZH, Zhu CX, Quan YS, Yang ZY, Wu S, Luo WW, Tan B, Wang XY. 2018. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J Gastroenterol. 24(1):5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Wei Y, Hu W, Nie Y, Wu X, Lu R. 2018. The subgingival microbiome of periodontal pockets with different probing depths in chronic and aggressive periodontitis: a pilot study. Front Cell Infect Microbiol. 8:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Dong Y, Huang W, Zhu D, Mao H, Su P. 2016. Fecal microbiota transplantation for ulcerative colitis: a systematic review and meta-analysis. PLoS One. 11(6):e0157259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J. 2002. Selection of antimicrobial agents in periodontal therapy.J Periodontal Res. 37(5):389–398. [DOI] [PubMed] [Google Scholar]
- Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. 2013. Therapeutic potential of fecal microbiota transplantation. Gastroenterology. 145(5):946–953. [DOI] [PubMed] [Google Scholar]
- Tong WM, Chan Y. 2020. Genepiper, a graphical user interface tool for microbiome sequence data mining. Microbiol Resour Announc. 9(1):e01195–e01119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utter DR, Mark Welch JL, Borisy GG. 2016. Individuality, stability, and variability of the plaque microbiome. Front Microbiol. 7:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BC, Vatanen T, Cutfield WS, O’Sullivan JM. 2019. The super-donor phenomenon in fecal microbiota transplantation. Front Cell Infect Microbiol. 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_0022034521995423 for Oral Microbiota Transplant in Dogs with Naturally Occurring Periodontitis by T. Beikler, K. Bunte, Y. Chan, B. Weiher, S. Selbach, U. Peters, A. Klocke, R.M. Watt and T.F. Flemmig in Journal of Dental Research