ABSTRACT
Staphylococcus saprophyticus is a common pathogen of the urinary tract, a heavy metal-rich environment, but information regarding its heavy metal resistance is unknown. We investigated 422 S. saprophyticus isolates from human infection and colonization/contamination, animals, and environmental sources for resistance to copper, zinc, arsenic, and cadmium using the agar dilution method. To identify the genes associated with metal resistance and assess possible links to pathogenicity, we accessed the whole-genome sequence of all isolates and used in silico and pangenome-wide association approaches. The MIC values for copper and zinc were uniformly high (1,600 mg/liter). Genes encoding copper efflux pumps (copA, copB, copZ, mco, and csoR) and zinc transporters (zinT, czrAB, znuBC, and zur) were abundant in the population (20 to 100%). Arsenic and cadmium showed various susceptibility levels. Genes encoding the ars operon (arsRDABC), an ABC transporter and a two-component permease, were linked to resistance to arsenic (MICs ≥ 1,600 mg/liter; 14% [58/422]; P < 0.05). At least three cad genes (cadA or cadC and cadD-cadX or czrC) and genes encoding multidrug efflux pumps and hyperosmoregulation in acidified conditions were associated with resistance to cadmium (MICs ≥ 200 mg/liter; 20% [85/422]; P < 0.05). These resistance genes were frequently carried by mobile genetic elements. Resistance to arsenic and cadmium were linked to human infection and a clonal lineage originating in animals (P < 0.05). Altogether, S. saprophyticus was highly resistant to heavy metals and accumulated multiple metal resistance determinants. The highest arsenic and cadmium resistance levels were associated with infection, suggesting resistance to these metals is relevant for S. saprophyticus pathogenicity.
KEYWORDS: Staphylococcus saprophyticus, urinary tract infection, environment, heavy metals, whole-genome sequencing, pan-GWAS, metal resistance determinants, copper, zinc, arsenic, cadmium, metal resistance
INTRODUCTION
Antimicrobial resistance is of global concern, with the constant use of heavy metals recognized as a player contributing to this public health challenge of multiresistant bacteria (1, 2). Heavy metals are naturally found in soil but are also common environmental pollutants derived from human activities like industry, sewage, war (defoliants), and agriculture (3). The majority of metals are toxic to bacteria at low concentrations (4–7), and for this reason, they have been massively used in animal production as prophylactics or in agriculture as pesticides, herbicides, and insecticides (8). Moreover, some of them are claimed to promote growth of production animals (4, 5), which has further increased their use in the veterinary setting. Copper and zinc are examples of metals that have been continuously used as feed supplements in the animal production chain to promote growth and as prophylactics (5, 9). Cadmium and arsenic have been mainly used in agriculture, leading to environmental contamination (10).
Heavy metals are also used by the human immune system as a means of defense against bacteria. In particular, copper and zinc play an essential role in optimal immune defense mechanism against pathogen colonization and infection. The mobilization of these metals (copper and zinc) to intracellular locations is described to control inflammation, and their accumulation in the phagosomes, in combination with reactive oxygen species, promotes innate immune function by enhancing the killing of pathogens engulfed by macrophages (9, 11, 12).
Either as a consequence of human activities or by facing human immunity, human and animal microbiota, as well as environmental bacteria, are constantly exposed to selective pressure of subinhibitory concentrations of these metals (2, 9). To survive metal toxicity and overcome human immune barriers, bacteria developed different mechanisms such as enzyme detoxification, biotransformation, compartmentalization to membranes, and extrusion using efflux pumps (8). These efflux pump systems may also result in a higher potential to select for multiple resistance to two or more metals (13). Resistance to cadmium and arsenic are mediated by cad-system genes, namely, cadA, cadD, cadX, cadC and czrC, and arsRDABC operon, respectively, that are either chromosomal (14) or often plasmid-borne (9, 15, 16). In particular, uropathogens such as Escherichia coli are able to overcome the human immune barrier with the extrusion of the toxic metal using copper efflux pump systems (17) and specific zinc exporters (18).
The exposure of bacteria to heavy metals has been hypothesized to coselect for antibiotic and biocide resistance (13) because resistance determinants for these antimicrobials usually co-occur in the same mobile genetic elements, namely, plasmids (1, 10, 13). In staphylococci, metal resistance determinants are often associated with staphylococcal cassette chromosome mec (SCCmec) types such as type VIII (4A), IX (1C2), X (7C1), and XI (8E) (19, 20). Determinants encoding copper, arsenic, and cadmium resistance have also been detected in composite SCC elements in methicillin-resistant Staphylococcus aureus (MRSA; especially livestock, MRSA ST398) and also in mecA-positive coagulase-negative staphylococci (CoNS), namely, Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus capitis, among others (5, 19).
Evidence has been gathered supporting the role of metal resistance in both environmental bacterial persistence and evasion from the human immune system. What remains to be determined is whether environmental exposure of bacteria to metals and the consequent increase in resistance can also contribute to an increase in bacterial pathogenesis. To address this hypothesis, we selected as a model Staphylococcus saprophyticus, a bacterium that has a widespread distribution in the environment (21–23), is a commensal of humans and production animals (24), is a frequent contaminant of food (25), and is, additionally, a common human uropathogen associated with 10 to 20% of all urinary tract infections (UTIs) in sexually active women worldwide (23).
We have previously defined the population structure of S. saprophyticus and identified two clonal lineages, lineages G and S, that were disseminated worldwide and had distinctive genetic features. We also provided evidence for a possible animal origin for lineage G and human origin for lineage S (26). In this study, the analysis of an S. saprophyticus collection of diverse origins by phenotypic and genomic tools allowed us to determine the MIC of S. saprophyticus to four heavy metals, assess the distribution of metal resistance determinants among the S. saprophyticus population, and identify previously unknown associations between specific genetic determinants, metal resistance, and pathogenicity in this bacterial species.
RESULTS
Assessment of heavy metal MIC and resistance determinants in S. saprophyticus population.
The MIC for copper for the great majority of strains (99%, 418/422) was 1,600 mg/liter. The only exceptions were four strains that had MICs of 400 mg/liter (n = 1) or 800 mg/liter (n = 3). We found copA, copB, and/or copZ genes that encode resistance to copper (17) in all isolates. Other copper resistance determinants found include csoR (21%, 88/422), a copper-sensing transcriptional repressor, and mco (20%, 83/422), a multicopper oxidase.
For zinc, the MICs ranged from 200 to 3,200 mg/liter, but 78% of strains (329/422) had MICs of 1,600 mg/liter (Fig. 1a and b). Zinc resistance determinants, including the czrA-czrB operon that encodes zinc transporter (27) and other genes such as zinT, znuB-znuC, and zur that have been described to be essential for zinc homeostasis in bacteria (28), were present in all isolates.
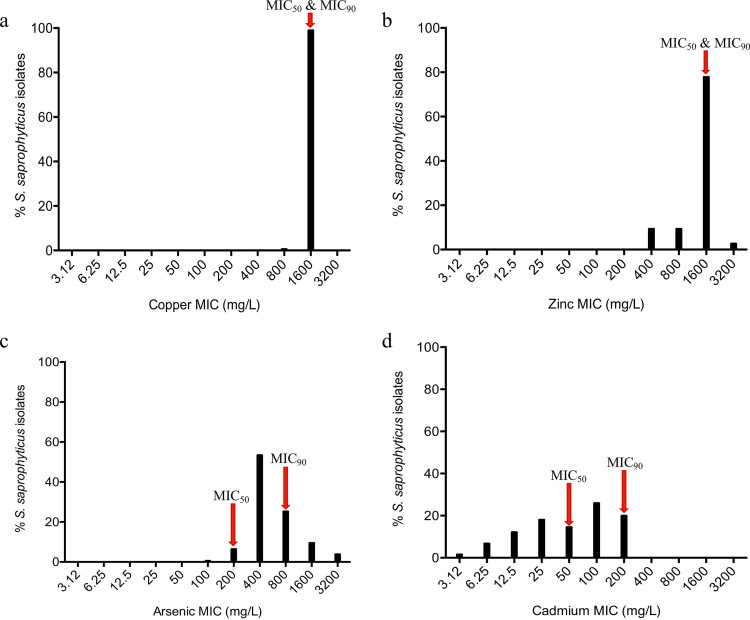
FIG 1.
MIC distribution of 422 Staphylococcus saprophyticus isolates recovered from human infection and colonization/contamination and environmental sources using the agar dilution method. Mueller-Hilton agar was supplemented with different concentrations (3.12 to 3,200 mg/liter) of copper (copper sulfate; CuSO4), zinc (zinc sulfate; ZnSO4), arsenic (sodium arsenite; AsNaO2), and cadmium (cadmium chloride; CdCl2). S. saprophyticus showed high resistance to copper (a) and zinc (b) (1,600 mg/liter). For arsenic (c), unimodal distribution was observed, which suggested epidemiological cutoff value (ECV) of ≥1,600 mg/liter. (d) A bimodal distribution was apparent for cadmium with a suggested ECV of ≥200 mg/liter.
Of note, copper and zinc had the same MIC50 and MIC90 (1,600 mg/liter) (Table S1 in the supplemental material). To understand if differences in MIC could be related to alterations in the sequences of genes or proteins, nucleotide and amino acid sequences of strains with low and high MICs were aligned. We did not find any truncation, insertion, or deletion or any mutation that could explain the low MICs. Differences in MICs in S. saprophyticus for these metals should most probably be related to the level of expression of any of the resistance genes or to posttranslational modifications. Additionally, it is possible that differences in MICs might be due to the differential distribution of yet-unknown resistance genes.
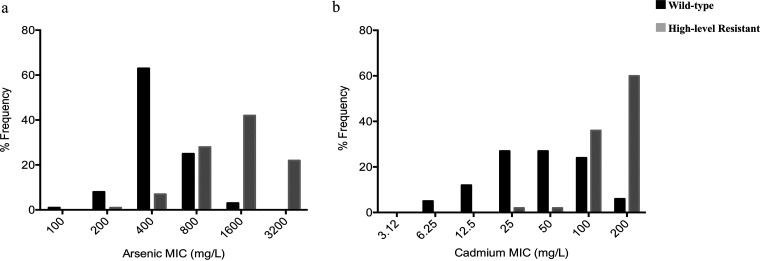
Regarding arsenic, we found that the MICs ranged from 100 to 3,200 mg/liter, with more than half (53%, 226/422) of the isolates having MICs of 400 mg/liter (MIC50), while the MIC90 was 1,600 mg/liter (Fig. 1a and b). Arsenic resistance determinants, namely, arsR, arsB, and arsC (100%, 422/422), as well as arsM (9%, 40/422), were found in the isolates studied. Additionally, 17% (72/422) of the isolates carried a complete ars operon (arsRDABC), the great majority (92%; 66/72) of which had a high MIC value of ≥1,600 mg/liter (46/72). Hence, an epidemiological cutoff value (ECV) of ≥1,600 mg/liter was suggested for arsenic (Table S1; Fig. 2a).
FIG 2.
(a) Distribution of arsenic MICs in 422 S. saprophyticus of diverse origins as determined by agar dilution. Wild type, proportion of isolates carrying arsRBC only; high-level resistant, proportion of S. saprophyticus strains carrying a complete ars operon (arsRDABC) for arsenic. (b) Distribution of cadmium MICs in 422 S. saprophyticus isolates of diverse origins, as determined by agar dilution. Wild type, proportion of isolates with less than three cad genes; high-level resistant, proportion of S. saprophyticus strains carrying at least three cad genes, including cadA and/or cadC.
For cadmium, the MICs varied between 3.12 and 200 mg/liter, with MIC50 and MIC90 of 50 and 200 mg/liter, respectively (Fig. 1c and d). Genes that encode resistance to cadmium were present in the following frequencies: cadD (58%, 243/422), cadX (56%, 237/422), cadA (24%, 100/422), cadC (23%, 96/422), and czrC (6%, 26/422). Of note, only 64% (272/422) of all isolates carried at least one known cadmium resistance gene (Table S1).
Although unimodal distributions of MICs were detected for copper, zinc, and arsenic, a bimodal distribution was observed for cadmium. Hence, the isolates were categorized into cadmium susceptible (MIC ≤ 50 mg/liter; 54%, 227/422), intermediate (MIC = 100 mg/liter; 26%, 110/422), and resistant (MIC ≥ 200 mg/liter; 20%, 85/422) groups. Notably, the great majority of the isolates in the resistant group (MIC ≥ 200 mg/liter; 88%, 75/85) carried at least three cad genes that included a combination of cadA or cadC and cadD and others (Table S1; Fig. 2b). Thus, an ECV of ≥200 mg/liter was suggested for cadmium.
Interestingly, the mco gene linked to copper resistance and oxidative stress response (29) was strongly associated with isolates with MICs of ≥800 mg/liter and ≥100 mg/liter for arsenic and cadmium, respectively (P < 0.05). This might be due to the fact that mco, cad, and/or ars genes were found to be colocated in the same plasmids (Table 1). However, we cannot disregard the hypothesis that mco in S. saprophyticus might provide cross-resistance to Cd, As, and Cu, as it was observed for some efflux pumps (30, 31). We did not test S. saprophyticus isolates for susceptibility to mercury, but we did find its resistance determinant (merB) in <10% (n = 35/422) in the collection.
TABLE 1.
Putative plasmids associated with metal resistance determinants in Staphylococcus saprophyticus in this study
| Rep gene | Plasmid | Frequency (% [no. of isolates carrying the plasmid]) | Plasmid size (bp) | Gene content (total no. of other coding sequences present in each of the plasmids) | Closest homology (BLASTn) | Size (bp) | Coverage (%) | Identity (%) | Reference no. or source |
|---|---|---|---|---|---|---|---|---|---|
| rep20_14 (pSSAP2) | KS40p1a | 57 (235) | 36,709 | rep, repA, arsC (30) | S. saprophyticus subsp.saprophyticus MS1146 plasmid pSSAP2 | 36,907 | 100 | 100 | 26, 41 |
| rep19c_3 (pETB) | KS345p_9 | 26 (109) | 30,309 | rep, repA, qacC, cadD, cadX, bin, tnpC, IS6-like (22) | S. saprophyticus strain UTI-050 plasmid pUTI-050-1 | 41,251 | 55 | 98 | This study |
| repUS10_1 (pSSP1) | KS275p_14 | 17 (71) | 34,497 | rep, repA, tnpC, cadA, cadD, cadX, IS6-like (31) | S. saprophyticus strain UTI-050 plasmid pUTI-050-1 | 41,251 | 55 | 99 | This study |
| rep21_26 (pSK108) | KS244p_29 | 15 (64) | 28,635 | rep, repA, blaZ, bin, copA, csoR, czrB, IS6-like, qacC (22) | S. warneri strain 22.1 plasmid pSW22.1 | 25,886 | 90 | 99.68 | NAb |
| rep19c_2_rep (pvSw4) | KS316p_8 | 8 (35) | 35,941 | rep, repA, cadA, arsR, arsM, bin (31) | S. equorum strain C2014 plasmid pC2014-1 | 80,302 | 38 | 96.76 | This study |
| rep19c_5 (SAP108A) | KS40p3a | 6 (25) | 29,692 | rep, repA, czcD, csoR, copA, cadC, cadD, cadX, bin, mph(C), msr(A), IS6 (13) | S. saprophyticus strain FDAARGOS_168 plasmid unnamed2 | 29,612 | 100 | 100 | 26 |
| rep13_2 (pSSP1) | KS170p_18 | 3 (13) | 3,989 | rep, cadC, cadX, cadD (0) | S. aureus subsp.aureus ST398 plasmid pUR3912 | 6,176 | 76 | 94.28 | This study |
| Rep | KS40p2a | 2 (7) | 33,002 | rep, repA, merA, merB, merT, IS200, tnpB, IS6, bin, cadC, cadD, IS6-like (23) | S. cohnii strain FDAARGOS_334 plasmid unnamed2 | 32,058 | 62 | 99.68 | 26 |
| repUS70_1 (SAP047A) | KS103p_20 | 1 (6) | 16,772 | rep, repA, cadD, cadX, cadA, arsC, arsB, arsA, arsD, arsR (10) | S. saprophyticus subsp.saprophyticus MS1146 plasmid pSSAP1 | 66,104 | 85 | 98 | This study |
| rep19c_4 (pSSP2) | KS217p_11 | 1 (4) | 34,121 | rep, repA, mco, copA, merB, cadD, cadA, cadX, arsC, arsB, arsR, arsA, arsD, arsR (19) | S. equorum strain KM1031 plasmid unnamed1 | 45,912 | 26 | 98 | This study |
| rep13_8 (pUR3912) | KS319p_14 | <1 (3) | 3,747 | rep, repA, cadD, cadX (1) | S. aureus subsp. aureus ST398 plasmid pUR3912 | 6,176 | 73 | 96.13 | This study |
| rep5b_1 (pUR2355) | KS203p_15 | <1 (2) | 32,211 | rep, repA, copA, czrB, copY, IS6-like (34) | S. warneri SG1 plasmid clone pvSw4 | 19,866 | 44 | 99.98 | This study |
| rep16_7 (pBORa53) | KS155p_8 | <1 (1) | 27,852 | rep, repA, cadD, cadX, bin (24) | S. aureus plasmid pMS97 merB gene | 33,347 | 33 | 98.48 | This study |
| repUS23_1 (SAP099B) | KS262p_11 | <1 (1) | 28,564 | rep, repA, merA, merR, merT, czrB, csoR, copA, copZ, qacA, qacR, mph(C), msr(A), bin, IS4 (17) | S. epidermidis strain 107.2 plasmid pAQZ1 | 29,431 | 68 | 99.94 | This study |
| rep19b_1 (SAP105A) | KS45p_30 | <1 (1) | 26,220 | rep, repA, cadD, cadX, bin (20) | S. hominis strain FDAARGOS_762 plasmid unnamed1 | 25,845 | 42 | 99.75 | This study |
| rep19c_1 (pLEW6932) | KS161p_10 | <1 (1) | 55,367 | rep, repA, mco, copA, bin, merB, ricR, cadX, cadD, IS6-like (49) | S. saprophyticus strain UTI-035 plasmid pUTI-035-2 | 35,615 | 27 | 92.25 | This study |
Sequenced with Nanopore.
NA, not applicable.
Overall, S. saprophyticus had higher MIC values for copper and zinc, while there was an increased diversity in the MICs for arsenic and cadmium in this bacterial population.
Different susceptibility levels to arsenic and cadmium in S. saprophyticus.
Based on the diversity in MICs observed for arsenic and cadmium, we further categorized the isolates’ MICs into three levels of susceptibility to these heavy metals. MICs of isolates without known or combination of known acquired resistance determinants were classified as susceptible, those with acquired resistance determinants known to confer high-level resistance, and those with MICs value greater than or equal to the ECV were grouped as resistant. Intermediate was defined as the MIC value that separates the other two susceptibility groups. Therefore, isolates with MIC values of ≤400 mg/liter, 800 mg/liter, and ≥1,600 mg/liter were classified as susceptible, intermediate, and resistant to arsenic, respectively. Likewise, for cadmium, isolates with MIC values of ≤50 mg/liter, 100 mg/liter, and ≥200 mg/liter were categorized as being susceptible, intermediate susceptible, and resistant to this metal (Fig. 2a and b; Table S1; Fig. S1). According to this classification, 14% (58/422) showed a resistant phenotype to arsenic, while 25% (800 mg/liter; 107/422) of the isolates in the collection showed an intermediate phenotype. In the case of cadmium, 20% (85/422) showed a resistant phenotype, whereas 26% (110/422) and 54% (227/422) showed intermediate and susceptible phenotypes, respectively (Table S1).
S. saprophyticus belonging to clonal lineage G showed a higher resistant phenotype to cadmium and arsenic.
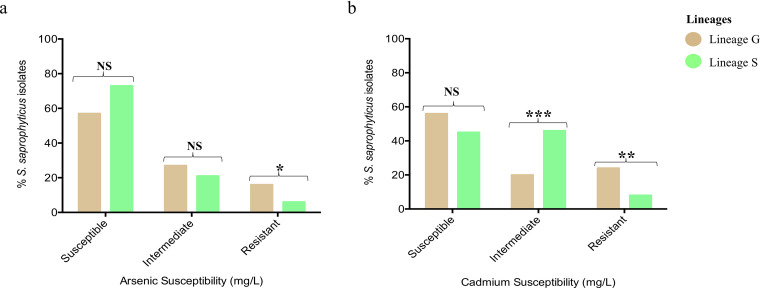
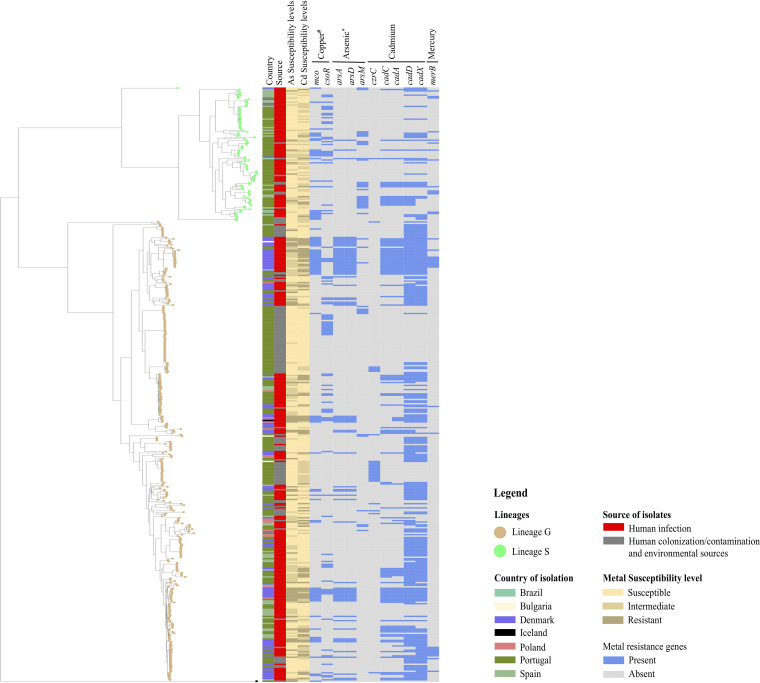
Given that the S. saprophyticus population is mainly composed of two distinct lineages of different origins (26), we looked for a possible link between the level of arsenic and cadmium resistance and S. saprophyticus genetic background. We found that resistant phenotypes to both metals were associated with isolates belonging to lineage G (arsenic: 16%; 52/327, lineage S: 6%; 6/95, P < 0.0330; cadmium: 24%; 77/327, lineage S: 8%; 8/95, P < 0.0047, respectively). Conversely, an intermediate phenotype (MIC, 100 mg/liter) to cadmium was associated with isolates belonging to lineage S (46% [44/95]; 20% [66/327] associated with lineage G; P < 0.0014) (Table S1; Fig. 3a and b; Fig. 4). The association of lineage G and cadmium resistance was independent of the geographic location. Although isolates from Denmark had a high resistance rate to these two metals (Cd, 48/85; As, 25/58), when only isolates from Portugal and Spain were considered, still over 70% (Cd, 23/30; As, 21/28) of the cadmium/arsenic resistant isolates belonged to lineage G.
FIG 3.
Lineage-based distribution of heavy metal susceptibility levels of 422 Staphylococcus saprophyticus isolates recovered from human infection and colonization/contamination and environmental sources. Resistance to arsenic (sodium arsenite) (a) and cadmium (cadmium chloride) (b) were associated with isolates belonging to lineage G (P < 0.0330 and P < 0.0047, respectively), whereas intermediate phenotype to cadmium was associated with isolates belonging to lineage S (P < 0.0014). Susceptible, ≤50 mg/liter; intermediate, 100 mg/liter; resistant, ≥200 mg/liter.
FIG 4.
Distribution of arsenic and cadmium susceptibility levels and associated resistance determinants in 422 Staphylococcus saprophyticus isolates as determined by in silico analysis. Phylogenetic tree was constructed based on core-genome single-nucleotide polymorphisms without recombination. Phylogenetic tree was visualized using a Microreact web tool. Susceptibility levels to arsenic were classified as follows: susceptible, ≤400 mg/liter; intermediate, 800 mg/liter; resistant, ≥1,600 mg/liter. Susceptibility levels to cadmium were classified as follows: susceptible, ≤50 mg/liter; intermediate, 100 mg/liter; resistant, ≥200 mg/liter. *, all isolates in the collection carried arsRBC; #, all isolates in the collection carried copABZ. Data in interactive format are accessible at https://microreact.org/project/uDDq7sLSj3n1Fax6iw8NrY.
S. saprophyticus isolates of human origin are resistant to high concentrations of arsenic and cadmium.
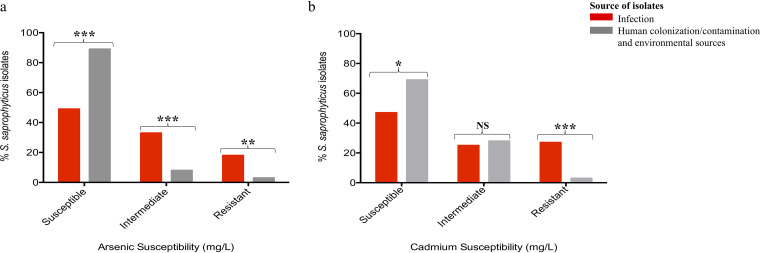
Bacterial lifestyle has been reported to play a significant role in niche adaptation (32, 33). To understand if isolates from different sources differ in their susceptibility to heavy metals, we categorized the isolates into either infection or colonization/contamination/environmental sources. Isolates from human colonization were included in the group of environmental isolates because the great majority of them (32/33) were from the hands of slaughterhouse workers, and in this case, it is not possible to distinguish a situation of colonization from that of contamination with slaughterhouse flora. Indeed, we found different resistant phenotypes to heavy metals to be significantly associated with the source of the isolates. For example, resistance to arsenic (infection, 18% [54/297]; colonization/contamination/environmental sources, 3% [4/125]; P < 0.0011) and cadmium (infection, 27% [81/297]; colonization/contamination/environmental sources, 3% [4/125]; P < 0.0001) were associated with human urinary tract infection isolates (Fig. 4; Fig. 5a and b). A similar observation was noted for an intermediate phenotype to arsenic that was strongly associated with human infection isolates (infection, 33% [97/297]; colonization/contamination/environmental sources, 8% [10/125]; P < 0.0001). Conversely, isolates that were susceptible to arsenic and cadmium were associated with colonization/contamination/environmental sources (P < 0.0007 and P < 0.0411, respectively) (Fig. 4; Fig. 5a and b).
FIG 5.
Heavy metal susceptibility levels in 422 Staphylococcus saprophyticus isolates based on origin. (a) Resistant and intermediate phenotypes to arsenic (sodium arsenite) were associated with human infection isolates (P < 0.0011 and P < 0.0001, respectively). Susceptible, ≤400 mg/liter; intermediate, 800 mg/liter; resistant, ≥1,600 mg/liter. (b). Human infection isolates were strongly associated with high-level tolerance to cadmium (cadmium chloride) (P < 0.0001). Overall, susceptible phenotypes to arsenic and cadmium were associated with isolates from colonization/contamination/environmental sources (P < 0.0007 and P < 0.0411, respectively). Susceptible, ≤50 mg/liter; intermediate, 100 mg/liter; resistant, ≥200 mg/liter.
Of note, a similar observation was found when we grouped isolates into human origin (human infection and colonization/contamination) and other environmental sources. Also, when isolates from human infection and colonization/contamination were compared, we noted that resistance to arsenic (MIC, 3,200 mg/liter) and cadmium (MIC, 200 mg/liter) was exclusively from human infection isolates (P < 0.05). This result further suggests that resistance to these metals (arsenic and cadmium) was not only linked with isolates from human origin but associated specifically with those causing infections. The two isolates recovered from small monkeys had similar phenotypes for copper and zinc (1,600 mg/liter each) and an intermediate phenotype to arsenic (200 mg/liter) but had different susceptibility levels for cadmium (100 mg/liter and 200 mg/liter, respectively). The only isolate from live pig also had an MIC of 1,600 mg/liter for copper, but low MICs for zinc and arsenic (400 mg/liter) relative to other isolates in the collection, and an intermediate phenotype (100 mg/liter) for cadmium (Table S1). Overall, S. saprophyticus strains causing infection in human are highly resistant to heavy metals.
S. saprophyticus isolates from different countries differed in their susceptibility to arsenic and cadmium.
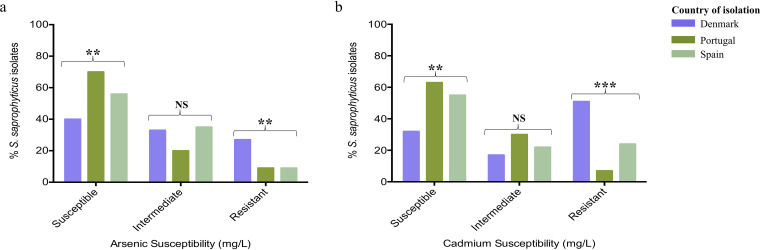
To understand if the level of resistance to metals clustered geographically, we assessed the distribution of heavy metal MICs in three countries for which ≥55 S. saprophyticus isolates were obtained, (Denmark (n = 94), Portugal (n = 250), and Spain (n = 55). We noted that the resistant phenotype to arsenic was linked to isolates recovered from a northern European country (Denmark; P < 0.05), while susceptible isolates were associated with southern European countries (Portugal and Spain; P < 0.05). Similarly, we did find that more than 50% (51%; 48/94) of isolates recovered from Denmark showed a resistant phenotype to cadmium (200 mg/liter; P < 0.05) (Fig. 4; Fig. 6a and b) and also belonged to lineage G, a lineage reported to be globally disseminated and also prevalent in northern European countries (26).
FIG 6.
Heavy metal susceptibility level in Staphylococcus saprophyticus recovered from three European countries. (a) Resistance to arsenic (sodium arsenite) was significantly associated with isolates recovered from a northern European country (Denmark), while isolates showing susceptible phenotypes were associated with southern European countries (Portugal and Spain) (P < 0.05). Susceptible, ≤400 mg/liter; intermediate, 800 mg/liter; resistant, ≥1,600 mg/liter. (b) Isolates from Denmark were significantly associated with resistance to cadmium (P < 0.05). Susceptible, ≤50 mg/liter; intermediate, 100 mg/liter; resistant, ≥200 mg/liter.
Identification of new genes putatively linked to arsenic and cadmium resistance in S. saprophyticus.
Bacterial susceptibility to different concentrations of heavy metals could be associated with different mechanisms. To identify other candidate determinants that could be associated with resistance to arsenic and cadmium to complement the in silico analysis described above, we employed a pangenome-wide association study (pan-GWAS) approach. The association between different metal susceptibility levels described above was tested against 9,182 genes that constituted the accessory genome defined as pan less core (n = 1,885/11,067) using Bonferroni and pairwise comparison P values of <0.05 and odds ratio (OR) of <1 (see Materials and Methods). We excluded the MICs of copper and zinc from this analysis since resistance to these metals was intrinsic in the collection analyzed. In addition to the ars operon (arsRDABC) that was strongly linked to resistance to arsenic (≥1,600 mg/liter) in S. saprophyticus, we found genes that encode a putative ABC-F family ATP-binding cassette (group_3565) and five hypothetical proteins also associated with high MICs. Notably, another gene that was enriched in isolates showing a resistant phenotype to arsenic was group_1381 that encoded two-component membrane permeases that have been described in Streptococcus mutans to be involved in the survival of this bacterium in general stress conditions, particularly in acidified environments (34) (Table 2; Fig. S2; Table S2).
TABLE 2.
Genes positively associated with resistance (MIC ≥ 1,600 mg/liter) to arsenic in Staphylococcus saprophyticus in this studya
| Gene annotation | Functional annotation | Frequency (%) | Other (%)b | Reference no. or source |
|---|---|---|---|---|
| arsRDABC | Arsenical resistance operon | 79 | 7 | 8 |
| group_2340 | Hypothetical protein | 79 | 7 | NAc |
| group_6095 | Hypothetical protein | 79 | 7 | NA |
| group_1381 | Putative two-component membrane permease complex subunit SMU_747c | 78 | 7 | 33 |
| group_3121 | Hypothetical protein | 50 | 15 | NA |
| group_3565 | Putative ABC transporter ATP-binding protein | 34 | 3 | NA |
| group_6309 | Hypothetical protein | 22 | 0 | NA |
| group_6308 | Hypothetical protein | 22 | 0 | NA |
Bonferroni P < 0.05 and OR > 1; n = 58.
% isolates in the population that carried the genetic determinant but susceptible to the heavy metals.
NA, not applicable.
In addition to the cad genes that were identified by in silico analysis to be associated with a resistant phenotype to cadmium, we found genes (group_6201 and group_7208 and group_7210) encoding multidrug efflux pumps, three transposases, and four genes with uncharacterized functions that were strongly associated with resistance to this metal. Also included was a gene (opuAC) that encodes glycine betaine-binding protein that has been described previously in E. coli to be involved in adaptation and resistance in high-osmolarity environments (35) (Table 3; Fig. S3; Table S2). Likewise, a gene (group_7205) that encodes a transporter belonging to a broad LysE transporter superfamily that is related to CadD family (36) and was previously described to be involved in the amino acids and heavy metal efflux in Corynebacterium glutamicum (36, 37) was included (Table 3; Fig. S3; Table S2).
TABLE 3.
Genes positively associated with resistance to cadmium (MIC ≥ 200 mg/liter) in Staphylococcus saprophyticus in this studya
| Gene annotation | Nonunique gene name | Functional annotation | Frequency (%) | Others (%)b | Reference no. or source |
|---|---|---|---|---|---|
| cadX_1 | Putative cadmium efflux system accessory protein | 72 | 7 | 5 | |
| group_3121 | Hypothetical protein | 49 | 13 | NAc | |
| group_2412 | Hypothetical protein/transposase | 45 | 11 | NA | |
| group_2065 | Hypothetical protein/resolvase | 45 | 4 | NA | |
| group_3485 | cadD | Cadmium resistance permease (CadD) | 44 | 4 | 5 |
| group_1830 | cadA_2 | Cadmium-transporting ATPase (CadA) | 44 | 4 | 5 |
| cadD_1 | Cadmium resistance permease (CadD) | 44 | 4 | 5 | |
| tnpC_1 | Transposase for transposon Tn554 | 35 | 2 | NA | |
| group_6073 | MerR family transcriptional regulator | 35 | 2 | NA | |
| group_6075 | Hypothetical protein | 34 | 2 | NA | |
| group_7205 | Hypothetical protein/lysE family transporter | 18 | 1 | 35, 36 | |
| opuAC | Glycine betaine-binding protein OpuAC | 18 | 1 | 34 | |
| group_7208 | RND superfamily resistance-nodulation-cell division, proton (H+) antiporter | 18 | 1 | NA | |
| group_7209 | Hypothetical protein | 18 | 1 | NA | |
| group_7210 | Hypothetical protein/ABC transporter permease | 18 | 1 | NA | |
| group_2701 | Hypothetical protein | 16 | 1 | NA | |
| group_6201 | Hypothetical protein/DHA2 family multidrug efflux MFS transporter permease subunit | 15 | 1 | NA |
Bonferroni P < 0.05 and OR > 1; n = 85.
% isolates in the population that carried the genetic determinant but susceptible to the heavy metals.
NA, not applicable.
Besides the metal resistance genes, the putative new resistance determinants identified by GWAS were not part of the representative or putative plasmids identified in this study, with the exception of group_1381, suggesting that their association with high-level resistance should be functional and not due to colocation in the same mobile genetic element.
The fact that we found the genes identified by the in silico analysis to be associated with resistant phenotype using the pan-GWAS approach further supports the ECVs defined for arsenic and cadmium (Fig. S2 and S3; Table S2). We noted that the great majority of the new genes identified were present together with other known resistance determinants to arsenic and cadmium in isolates that showed resistant phenotypes to these metals (Fig. S2 and S3).
Metal resistance determinants in S. saprophyticus are either chromosomal and/or plasmid-borne.
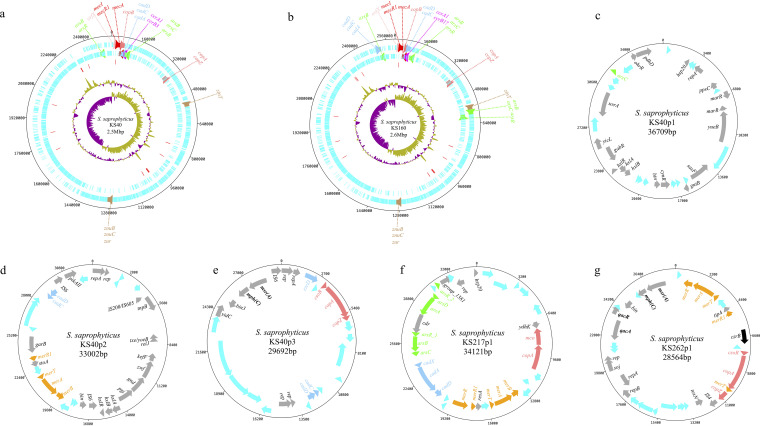
To determine the location of metal resistance genes in S. saprophyticus, first, we accessed the Nanopore closed genomes of KS40, representative of lineage G, and KS160, representative of lineage S (26). The chromosomes have an average size of 2.6 Mb and 2,481 coding sequences (CDS). Noteworthy, the two strains were methicillin resistant and carried mecA. While one (KS160) of the SCCmec types was nontypeable by SCCmecFinder (38), the other (KS40) carried SCCmec type III, a type commonly found in S. saprophyticus (39). These two strains were also different in terms of metal susceptibility phenotypes. KS160 showed an intermediate phenotype to both arsenic and cadmium, while KS40 showed susceptible and resistant phenotypes to these metals, respectively (Table S1).
The visualization of the two closed genomes using Artemis v17.0.1 (40) showed that both strains had resistance determinants for copper (copB) and cadmium (cadD, cadC, and/or cadA) located within the SCCmec region. Other resistance determinants to copper (copA, copZ), zinc (zinT, znuB-znuC, and zur), arsenic (arsB, arsR, and/or arsC), and cadmium (cadC, cadD, and/or cadA) were found in the same chromosomal location in the two strains. There were, however, some differences in the location of extra copies of arsenic resistance determinants (arsB, arsC, and/or arsR) in the chromosomes of the two representative strains (Fig. 7a and b). Additionally, KS160 strain carried extra copies of cadmium resistance determinants (cadA, cadC, and cadD) in its chromosome (Fig. 7b).
FIG 7.
Staphylococcus saprophyticus genomes showing the chromosome and plasmid-borne genetic determinants that encode resistance to heavy metals. (a to b) S. saprophyticus KS40 (a) and KS160 (b) chromosomes showing the chromosome-borne metal resistance determinants of representative strains of lineages G and S, respectively. The tracks from the outside represent (i) chromosome tick marks, (ii) forward CDS, (iii) reverse CDS, (iv) tRNA (red), (v) percent GC plot, and (vi) GC skew (GC)/(G+C). orfX-mecA complex and cassette recombinase genes within the SCCmec are depicted with pink-, red-, and magenta-colored arrows, respectively. Other genes encoding resistance to copper (copA, copB, copZ), zinc (zinT, znuB, znuC, zur), arsenic (arsB, arsC, arsR), and cadmium (cadA, cadC, cadD) are highlighted with different-colored arrows. Arrows with the same color encode resistance to the same heavy metal. (c to g) Representative S. saprophyticus plasmids that carried a single (c) or multiple (d and f) heavy metal resistance determinants in addition to determinants that encode resistance to antibiotics and/or biocides (e and g). Colored arrows represent forward and reverse CDS. Cyan arrow, hypothetical proteins; light red arrows, copper resistance determinants; green arrows, arsenic resistance determinants; light sky-blue arrow, cadmium resistance genes; orange arrows, mercury resistance genes; black arrow, cobalt, zinc, and cadmium resistance gene; dark gray arrow, other CDS. Gene names in bold form represent biocides and antibiotics resistance genes.
The KS160 strain carried no plasmids and more chromosomally encoded heavy metal resistance determinants, but KS40 strain carried three metal-containing plasmids (KS40p1, KS40p2, and KS40p3) (Fig. 7c to e) with sizes ranging from 29,628 bp to 36,856 bp with 27 to 36 CDS. The best hits in a BLAST search against the NCBI (BLASTn) database for each plasmid identified KS40p1 as pSSAP2 (100% nucleotide identity) (GenBank accession number HE616681.1) (41) and KS40p3 as FDAARGOS_168 plasmid unnamed2 (GenBank accession number CP014115.1)—two plasmids previously found in S. saprophyticus. The KS40p1 was present in 57% (235/422) of the isolates in the collection. It contained the gene encoding plasmid replication protein 20 (rep20), genes encoding arsenic reductase (arsC), and surface protein F (sssF), among other metabolic genes (Table 1; Fig. 7c). On the other hand, KS40p3 was found in 6% (n = 25/422) of the collection and carried rep19c genes, genes encoding transposases for insertion sequences, and several resistance determinants to copper (copA, copZ, csoR), cadmium and zinc (cadC, cadD, and czcD), and macrolides [mph(C), msr(A)], among other metabolic genes as well (Table 1; Fig. 7e). The third KS40 plasmid (KS40p2) was present in only seven isolates and had a low homology (coverage, 62%) to those in the NCBI database and hence could be a novel S. saprophyticus plasmid. This novel plasmid contained multiple resistance determinants to heavy metals (cadmium; cadC and cadD; mercury, merR, merA, and merB), IS6-like elements, rep genes, and other metabolic genes (Table 1; Fig. 7d).
Other putative plasmids carrying metal resistance determinants in the entire S. saprophyticus collection were identified using PlasmidFinder (see Materials and Methods). We identified 13 rep gene families (Table S3). Next, we selected one representative isolate from each rep family and blasted the contigs containing these rep genes using BLASTn nonredundant/nucleotide (nr/nt) against the NCBI database and the Nanopore sequences obtained for the KS40 and KS160 strains. We found 13 putative plasmids that were associated with at least one metal resistance determinant in addition to the three plasmids described above (Table 1). One of these putative plasmids, KS244p, rep21_26 (pSK108), 28.6 Kbp, present in 15% (n = 64/422) of the isolates in this collection, was similar (>90% nucleotide identity and coverage) to that in Staphylococcus warneri (pSW22.1) (NBCI nucleotide accession no. NZ_CP032158.1) and carried copper (copA, csoR) and zinc (czrB) resistance determinants in addition to biocide (qacC) and penicillin (blaZ) resistance genes. The remaining putative plasmids (n = 12) appear to be new, as they had low homology compared to those in NCBI database and present in <10% of the isolates in the collection. Five of these putative plasmids carried only cad-system genes (cadA, cadD, cadX, and/or cadC), and two carried both ars operon and cad-system genes, while others carried genes that encoded resistance to macrolides [mph(C), msr(A)], biocides (qacA, qacR), and multiple heavy metals, including copper (copA, csoR, mco), arsenic (arsRDABC), cadmium (cad-system), and mercury (merB) (Table 1; Fig. 7f and g). However, we did not find a direct correlation between the carriage of rep genes or the putative plasmids identified and the metal resistance genes in our study. Additionally, we could not ascertain by the methodology used if the plasmids in the collection analyzed were located in the chromosome or autonomously replicating in the cytoplasm.
DISCUSSION
S. saprophyticus is a common cause of uncomplicated community-acquired UTIs worldwide. An association between metal resistance and antimicrobial resistance has been previously established (1), and metal resistance has been described to be a means of evading human immune response (12), potentially contributing to pathogenicity. To better evaluate the clinical significance of resistance to metals, we determined the susceptibility to metals of a diverse collection of S. saprophyticus, comprising isolates from human infection and colonization/contamination and food and environmental contamination. Moreover, we explored the heavy metal resistome and identified genes that were never before linked to resistance using a pan-GWAS approach.
Our study showed that S. saprophyticus was highly resistant to copper and zinc and carried, in parallel, resistance determinants for these metals. Copper is acknowledged as a host effector mobilized to urine during UTI to impair bacterial colonization (42, 43). All isolates in this study carried copA and copB that were previously described to mediate efflux and maintain intracellular concentrations of copper necessary for growth in staphylococci (28). These same genes in E. coli were additionally described to be involved in detoxification, colonization, and adhesion of bacteria to the epithelial cells (18). Zinc, on the other hand, is an essential element that plays an important catalytic and structural role in a number of proteins (44). In humans, in particular, the trafficking of zinc to intracellular locations may enhance the bactericidal capability of macrophages and neutrophils (9, 11, 12). All isolates in this study carried genes (zinT, czrA, czrB, znuB, znuC, and zur) which are all part of already described zinc extrusion mechanisms in S. aureus (27). The fact that carriage of these copper and zinc detoxification systems was associated with a high MIC in our study suggests that these systems are probably responsible for the high resistance of S. saprophyticus to copper and zinc. On the other hand, they could additionally contribute to mediating resistance to phagocytosis in infection (11, 43). However, the establishment of a definitive link between these genetic determinants and antibiotic resistance or pathogenicity in S. saprophyticus requires further genetic studies.
In contrast to copper and zinc, for which all S. saprophyticus isolates had high MICs, susceptibility to arsenic and cadmium had diverse levels in the population. In particular, 14% of the isolates in this study showed resistance to arsenic (≥1,600 mg/liter), which was correlated with the corresponding carriage of a complete ars operon (arsRDABC). This system had been described in S. aureus and E. coli, among others, to mediate resistance to arsenic (8). Like for arsenic, for cadmium, there was a strong association between resistance present in 20% of the population and the presence of a specific cad gene system (cadA or cadC and cadD-cadX) that encodes resistance to cadmium in S. aureus (27, 45, 46). For these reasons, we proposed that the ECV of ≥1,600 mg/liter for arsenic and ≥200 mg/liter for cadmium should be considered in S. saprophyticus. Another phenomenon that we observed was that the accumulation of previously described arsenic and cadmium resistance genes were associated with a steady increase in MIC for these two metals, which could be an alternative strategy to achieve high-level resistance.
Using pan-GWAS, we additionally identified other putative determinants that were strongly linked to resistance to arsenic and cadmium. This included a putative ABC-F family ATP-binding cassette and a gene encoding a two-component membrane permease associated with high-level resistance to arsenic and a multidrug efflux pump and an ABC transporter linked to high-level resistance to cadmium. These determinants were previously described to contribute to survival in acidified environments (34) and stress conditions in other bacterial species (34, 36, 37) and can also be important for arsenic and cadmium resistance in S. saprophyticus, contributing to its persistence in low-pH environments such as urine. Most of these new genes were carried together with other known resistance determinants, suggesting that the accumulation of different resistance mechanisms could probably be responsible for the high-level resistance to these metals (arsenic and cadmium) in this bacterium. Indeed, a similar phenomenon has been previously described for high-level resistance to linezolid in S. epidermidis (47). Further studies on the functional characterization of these candidate protein-coding genes and the hypothetical proteins identified are required to give insight into their roles in heavy metal resistance in S. saprophyticus. Surprisingly, for the few cadmium-intermediate-resistant isolates that do not carry any known resistance genes, no genes were found to be associated with this phenotype. The phenotypes observed in these isolates could probably be due to cross-resistance from resistance mechanisms from other heavy metals or unknown mechanisms yet unexplored.
Resistance to arsenic and cadmium was statistically associated with isolates belonging to clonal lineage G, a lineage previously reported to have originated in food production animals (26). Likewise, livestock-associated MRSA (LA-MRSA) isolates belonging to clonal complex 398 (CC398) have been previously reported to be associated with resistance to cadmium (48). Arsenic and cadmium are ubiquitous in nature, and the continuous use of these metals in human industrial activities could have contributed to their abundance (8). This could have been the driving force for the emergence of arsenic and cadmium resistance of bacteria in the farm environment (8). In addition, resistance to arsenic and cadmium was additionally strongly linked to isolates from northern Europe (Denmark). The causal link between arsenic resistance and Denmark is not easy to explain and could derive from either the high prevalence of lineage G in this country (26) or selection by exposure to arsenic. Prevalence of lineage G, a lineage associated with the pig meat processing chain, can be a result of the fact that Denmark is one of the major producers of pigs and pig meat (49). On the other hand, studies assessing the concentration of metals in food products showed that contamination of these products (fish, meat, and milk) with arsenic is apparently higher in Denmark than in other countries such as Portugal (50). Since S. saprophyticus can be a colonizer or contaminant of these food products (7), resistance to arsenic could have resulted from prolonged exposition to arsenic and cadmium in these food products. Nevertheless, we could not establish if there were differences in the use of heavy metals between the southern and northern European countries sampled in this study that could explain these results, as there are scarce reports addressing this.
Interestingly, resistance to arsenic and cadmium was associated with human infection, an observation that was still maintained when only isolates outside Portugal (infection and environment) were considered. This finding could be the consequence of continuous interaction of human microbiota with these heavy metals in the environment where they are found as common pollutants (51) or from their use as topical antiseptics, disinfectants, and preservatives in consumer products (52, 53). On the other hand, the link between arsenic resistance and infection might derive from previous colonization with S. saprophyticus strains belonging to lineage G. This lineage was found in this study to be associated with arsenic and cadmium resistance. Furthermore, lineage G was previously shown to have originated in the food production chain and to be transmitted to humans, probably as a result of poor hygienic procedures during raw meat handling or contact with the food processing chain setting (26). Since urine is the main route of excretion of most metals (54–56), resistance of uropathogenic S. saprophyticus to these metals could also contribute to its survival in the bladder. On the other hand, genetic determinants associated with resistance to these metals can provide a better adaptation of S. saprophyticus to the acidic environment of the urine as mentioned above, functioning as virulence determinants under these conditions.
Some of the resistance determinants for copper, arsenic, and cadmium were found in our collection to be part of the core nonmobile genome, and some have been found in mobile genetic elements like putative plasmids and SCCmec, as previously described in S. aureus (46, 57). In particular, a high prevalence of putative plasmids harboring metal resistance genes was estimated among the S. saprophyticus collections analyzed, reaching 57% for some plasmids. In some of these putative plasmids, there was co-occurrence of multiple metal resistance determinants (ars operon, cad gene system, copA, mco, and merB) in addition to biocides (qacC, qacA) and antibiotic resistance genes [mph(C), msr(A), and blaZ]. This suggests that these determinants could probably be transferred between strains or to other related staphylococcal species, providing resistance not only to multiple heavy metals but also to biocides and antibiotics in a single genetic event. Indeed, the exposure of bacteria to heavy metals has been suggested to coselect for antibiotic and biocide resistance owing to the co-occurrence of their resistance determinants that could be transported in the same plasmid (10, 13).
In this study, we found high levels of resistance to heavy metals in S. saprophyticus, identified putative resistance determinants, found geographic and clonal lineage associations, and established a link between high heavy metal resistance and pathogenicity.
MATERIALS AND METHODS
Ethical considerations.
The human isolates were recovered as part of the routine clinical diagnostic testing; ethical approval and informed consent were not required. All data were handled anonymously. Isolates from slaughterhouse samples were recovered as part of the routine control practices for evaluation of good hygiene practices and programs to ensure meat safety (CE no. 853/2004).
Bacterial collection.
A diverse collection of 422 S. saprophyticus isolates recovered from different sources between 1997 and 2017 in seven countries was assembled. UTI-causing isolates (n = 285) were recovered from Denmark (n = 91), Poland (n = 13), Portugal (n = 128), and Spain (n = 54). Twelve isolates recovered from human blood in Brazil (n = 2), Bulgaria (n = 3), Denmark (n = 4), Iceland (n = 1), Poland (n = 1), and Spain (n = 1), as well as one isolate from human nasal swab in Poland, were included in the collection. Additionally, we included 104 isolates recovered from human hands (n = 32), slaughterhouse equipment (n = 32), pork meat samples (n = 39), and a live pig (n = 1). We also included 18 isolates recovered from household kitchen equipment (n = 13) and staple food products (n = 5). Lastly, two isolates recovered from small monkeys in Brazil were also included (Table S1 in the supplemental material).
Metal susceptibility testing.
MICs to four heavy metals were determined for all isolates using agar dilution method with Mueller-Hinton agar (58) containing copper (copper sulfate; Merck, Darmstadt, Germany), zinc (zinc sulfate; Merck, Darmstadt, Germany), arsenic (sodium arsenite; Sigma-Aldrich, St. Louis, MO), and cadmium (cadmium chloride; Sigma-Aldrich, St. Louis, MO) and at concentrations ranging from 3.12 to 3,200 mg/liter (58). Bacteria were grown overnight, and cultures were emulsified in 0.85% NaCl to a turbidity equivalent to a 0.5 McFarland standard suspension, inoculated with 2-μl spots on the top of the agar plates supplemented with metals, and incubated at 37°C for 24 h. Assays were carried out in duplicate. The MIC was determined as the lowest concentration of metal that completely inhibited growth (58).
Determination of ECV.
The ECV was defined as the MIC value that separates the isolates into a wild-type population and those with acquired or unknown resistance mechanisms to the heavy metals, as previously suggested by EUCAST (59).
Whole-genome sequencing and assembly.
Pair-end sequence reads produced on an Illumina MiSeq with an average coverage of 103x per genome reported in Lawal et al. (26) with the accession number PRJNA604222 were retrieved from Sequence Read Archive. Low Q score ends (Q score < 20) were trimmed of the Illumina reads using Trimmomatic v0.36 (60). Reads were assembled using SPAdes v3.11.1 (61). The quality of the assemblies was evaluated using QUAST v5.1 (62). Contig sizes of less than 200 bp were discarded.
Detection of metal resistance genes and putative plasmids.
The draft genomes were screened for known metal resistance determinants using ABRicate v0.5 pipeline (https://github.com/tseemann/abricate). Genes that were reported to be putatively associated with resistance to metals in staphylococci were searched for in literature, and respective nucleotide sequences were extracted from NCBI to create a custom database that was embedded in ABRicate (Appendix I in the supplemental material). Genes with minimum coverage length of ≥70% and identity of ≥90% were considered present. Moreover, to determine whether metal resistance genes were carried within plasmids, genomes were first screened for genes encoding plasmid replication proteins (rep) using the PlasmidFinder database (https://cge.cbs.dtu.dk/services/PlasmidFinder/). Furthermore, contigs containing the rep genes were blasted against the NCBI database and against closed genomes of representative strains that had previously been sequenced with Nanopore (26) (accession number PRJNA604222). The chromosomes and plasmids were visualized using Artemis v17.0.1 (40) and DNAPlotter v1 (63).
Phylogenetic construction.
Sequence reads were mapped to the S. saprophyticus strain (ATCC 15305) and generated core genome alignment using Snippy v4.3.6 (https://github.com/tseemann/snippy). Unknown characters from the sequence alignment were removed, and the cleaned alignment was used as input for the recombination detection pipeline Gubbins v2.3.4 (64) with default parameters to remove the polymorphic sites. The recombination-free alignment was used to construct a maximum-likelihood tree using RAxML v8.2.18 (65). The generalized time-reversible nucleotide substitution with gamma correction for rate heterogeneity was performed with 100 bootstrap replicates random resampling for node support. Phylogenetic tree was visualized using Microreact web tool (66).
Genome annotation and pan-GWAS analysis.
To further identify putative genetic determinants associated with different metal resistance levels (high, medium, and low), we annotated the draft genomes using the gene function prediction pipeline, Prokka v1.13 (67). Homologues were clustered at 85% amino acid identity to generate the pangenome using Roary (68). We accessed the accessory genome, which was defined as the pan less the core. To determine genetic determinants for heavy metal resistance, the three different phenotypes (susceptible, intermediate, and resistant) were used as predefined traits for pan-GWAS using Scoary (69). Only genes with a Bonferroni and best pairwise comparison P values of <0.05 and odds ratio (OR) >1, with no duplicated function in the pangenome, were considered.
Statistical analysis.
The mean and standard error values of replicates of each strain were calculated. The statistical significance was assessed using unpaired Student's t test or chi-square test. Statistical analyses were performed with GraphPad Prism v6.0 (GraphPad Software, Inc., San Diego, CA).
ACKNOWLEDGMENTS
O.U.L. and O.B. performed the isolation. O.U.L. performed the phenotypic experiments and bioinformatics analysis. O.U.L., H.W., P.W., and M.D.B. performed the sequencing of the isolates. O.U.L. and M.M. carried out the data analysis and interpretation and wrote the first draft of the manuscript. O.U.L. and M.M. revised the manuscript. M.J.F., L.G., P.P., E.G., C.T., J.E., M.U., H.d.L., H.W., and M.D.B. provided the isolates. M.J.F., L.G., P.P., E.G., C.T., J.E., M.U., H.d.L., H.W., P.W., and M.D.B. were involved in manuscript revision. All authors read and approved the final manuscript.
O.U.L. was supported by PhD grants from the Fundação para a Ciência e Tecnologia (FCT) PD/BD/113992/2015. This work was partially supported by project PTDC/CVT-CVT/29510/2017 from Fundação para a Ciência e Tecnologia, projects LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) and UID/Multi/04378/2019) funded by FEDER funds through COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI), by ONEIDA project (LISBOA-01-0145-FEDER- 016417) cofunded by FEEI (Fundos Europeus Estruturais e de Investimento) from Programa Operacional Regional Lisboa2020, and by national funds through FCT, Operacional Competitividade e Internacionalização, Programa Operacional Regional de Lisboa (FEDER), and Fundação para a Ciência e a Tecnologia.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol 14:176–182. 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Lemire JA, Harrison JJ, Turner RJ. 2013. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol 11:371–384. 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- 3.Bencko V, Foong FYL. 2017. The history of arsenical pesticides and health risks related to the use of Agent Blue. Ann Agric Environ Med 24:312–316. 10.26444/aaem/74715. [DOI] [PubMed] [Google Scholar]
- 4.Adamse P, Van der Fels-Klerx HJI, de Jong J. 2017. Cadmium, lead, mercury and arsenic in animal feed and feed materials-trend analysis of monitoring results. Food Addit Contam Part A 34:1298–1311. 10.1080/19440049.2017.1300686. [DOI] [PubMed] [Google Scholar]
- 5.Argudín MA, Butaye P. 2016. Dissemination of metal resistance genes among animal methicillin-resistant coagulase negative staphylococci. Res Vet Sci 105:192–194. 10.1016/j.rvsc.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Adekanmbi AO, Falodun OI. 2015. Heavy metals and antibiotics susceptibility profiles of Staphylococcus aureus isolated from several points receiving daily input from the Bodija abattoir in Ibadan, Oyo State, Nigeria. Adv Microbiol 5:871–880. 10.4236/aim.2015.513091. [DOI] [Google Scholar]
- 7.Turner RJ. 2017. Metal-based antimicrobial strategies. Microb Biotechnol 10:1062–1065. 10.1111/1751-7915.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalia K, Joshi DN. 2009. Detoxification of arsenic, p 1083–1100. In Gupta RC (ed), Handbook of toxicology of chemical warfare agents. Elsevier, London, UK. [Google Scholar]
- 9.Hobman JL, Crossman LC. 2014. Bacterial antimicrobial metal ion resistance. J Med Microbiol 64:471–497. 10.1099/jmm.0.023036-0. [DOI] [PubMed] [Google Scholar]
- 10.Seiler C, Berendonk TU. 2012. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol 3:399. 10.3389/fmicb.2012.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djoko KY, Ong CY, Walker MJ, McEwan AG. 2015. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem 290:18954–18961. 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.German N, Doyscher D, Rensing C. 2013. Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol 8:1257–1264. 10.2217/fmb.13.100. [DOI] [PubMed] [Google Scholar]
- 13.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. 2015. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16:1–14. 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, Kuhara S, Hattori M, Ohta T. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci U S A 102:13272–13277. 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons C, Lee S, Kathariou S. 2020. Dissemination and conservation of cadmium and arsenic resistance determinants in Listeria and other Gram-positive bacteria. Mol Microbiol 113:560–569. 10.1111/mmi.14470. [DOI] [PubMed] [Google Scholar]
- 16.Argudín MA, Hoefer A, Butaye P. 2019. Heavy metal resistance in bacteria from animals. Res Vet Sci 122:132–147. 10.1016/j.rvsc.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Sitthisak S, Knutsson L, Webb JW, Jayaswal RK. 2007. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology 153:4274–4283. 10.1099/mic.0.2007/009860-0. [DOI] [PubMed] [Google Scholar]
- 18.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci U S A 97:652–656. 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simões PM, Rasigade JP, Lemriss H, Butin M, Ginevra C, Lemriss S, Goering RV, Ibrahimi A, Picaud JC, El Kabbaj S, Vandenesch F, Laurent F. 2013. Characterization of a novel composite staphylococcal cassette chromosome mec (SCCmec-SCCcad/ars/cop) in the neonatal sepsis-associated Staphylococcus capitis pulsotype NRCS-A. Antimicrob Agents Chemother 57:6354–6357. 10.1128/AAC.01576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yagaguchi T, Ono D, Sato A. 2020. Staphylococcal cassette chromosome mec (SCCmec) analysis of MRSA. Methods Mol Biol 2069:59–78. 10.1007/978-1-4939-9849-4_4. [DOI] [PubMed] [Google Scholar]
- 21.Widerström M, Wiström J, Ferry S, Karlsson C, Monsen T. 2007. Molecular epidemiology of Staphylococcus saprophyticus isolated from women with uncomplicated community-acquired urinary tract infection. J Clin Microbiol 45:1561–1564. 10.1128/JCM.02071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Sousa VS, Da-Silva APDS, Sorenson L, Paschoal RP, Rabello RF, Campana EH, Pinheiro MS, Dos Santos LOF, Martins N, Botelho ACN, Picão RC, Fracalanzza SEL, Riley LW, Sensabaugh G, Moreira BM. 2017. Staphylococcus saprophyticus recovered from humans, food, and recreational waters in Rio de Janeiro, Brazil. Int J Microbiol 2017:1–11. 10.1155/2017/4287547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker K, Heilmann C, Peters G. 2014. Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedman P, Ringertz O, Lindström M, Olsson K. 1993. The origin of Staphylococcus saprophyticus from cattle and pigs. Scand J Infect Dis 25:57–60. 10.1080/00365549309169670. [DOI] [PubMed] [Google Scholar]
- 25.Lee B, Jeong D-W, Lee J-H. 2015. Genetic diversity and antibiotic resistance of Staphylococcus saprophyticus isolates from fermented foods and clinical samples. J Korean Soc Appl Biol Chem 58:659–668. 10.1007/s13765-015-0091-1. [DOI] [Google Scholar]
- 26.Lawal OU, Fraqueza MJ, Bouchami O, Worning P, Bartels MD, Gonçalves ML, Paixão P, Gonçalves E, Toscano C, Empel J, Urbaś M, Domínguez MA, Westh H, de Lencastre H, Miragaia M. 2021. Foodborne origin and local and global spread of Staphylococcus saprophyticus causing human urinary tract infections. Emerg Infect Dis 27:880–893. 10.3201/eid2703.200852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda M, Hayashi H, Ohta T. 1999. Chromosome-determined zinc-responsible operon czr in Staphylococcus aureus strain 912. Microbiol Immunol 43:115–125. 10.1111/j.1348-0421.1999.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 28.Gaupp R, Ledala N, Somerville GA. 2012. Staphylococcal response to oxidative stress. Front Cell Infect Microbiol 2:33. 10.3389/fcimb.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sitthisak S, Howieson K, Amezola C, Jayaswal RK. 2005. Characterization of a multicopper oxidase gene from Staphylococcus aureus. Appl Environ Microbiol 71:5650–5653. 10.1128/AEM.71.9.5650-5653.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruins MR, Kapil S, Oehme FW. 2000. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf 45:198–207. 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 31.Kosmidis C, Schindler BD, Jacinto PL, Patel D, Bains K, Seo SM, Kaatz GW. 2012. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int J Antimicrob Agents 40:204–209. 10.1016/j.ijantimicag.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Espadinha D, Sobral RG, Mendes CI, Méric G, Sheppard SK, Carriço JA, de Lencastre H, Miragaia M. 2019. Distinct phenotypic and genomic signatures underlie contrasting pathogenic potential of Staphylococcus epidermidis clonal lineages. Front Microbiol 10:1971. 10.3389/fmicb.2019.01971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wendlandt S, Shen J, Kadlec K, Wang Y, Li B, Zhang WJ, Feßler AT, Wu C, Schwarz S. 2015. Multidrug resistance genes in staphylococci from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol 23:44–54. 10.1016/j.tim.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Król JE, Biswas S, King C, Biswas I. 2014. SMU.746-SMU.747, a putative membrane permease complex, is involved in aciduricity, acidogenesis, and biofilm formation in Streptococcus mutans. J Bacteriol 196:129–139. 10.1128/JB.00960-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perroud B, Le Rudulier D. 1985. Glycine betaine transport in Escherichia coli: osmotic modulation. J Bacteriol 161:393–401. 10.1128/JB.161.1.393-401.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsu BV, Saier MH. 2015. The LysE superfamily of transport proteins involved in cell physiology and pathogenesis. PLoS One 10:e0137184-22. 10.1371/journal.pone.0137184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vrljic M, Sahm H, Eggeling L. 1996. A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826. 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- 38.Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. 2018. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soderquist C, Berglund B. 2009. Methicillin-resistant Staphylococcus saprophyticus in Sweden carries various types of staphylococcal cassette chromosome mec (SCCmec). Clin Microbiol Infect 15:1176–1178. 10.1111/j.1469-0691.2009.02771.x. [DOI] [PubMed] [Google Scholar]
- 40.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King NP, Sakinç T, Ben Zakour NL, Totsika M, Heras B, Simerska P, Shepherd M, Gatermann SG, Beatson SA, Schembri MA. 2012. Characterisation of a cell wall-anchored protein of Staphylococcus saprophyticus associated with linoleic acid resistance. BMC Microbiol 12:1–12. 10.1186/1471-2180-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subashchandrabose S, Mobley HLT. 2015. Back to the metal age: battle for metals at the host–pathogen interface during urinary tract infection. Metallomics 7:935–942. 10.1039/c4mt00329b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyre AN, Kavanagh K, Kock ND, Donati GL, Subashchandrabose S. 2017. Copper is a host effector mobilized to urine during urinary tract infection to impair bacterial colonization. Infect Immun 85:e01041-16. 10.1128/IAI.01041-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blencowe DK, Morby AP. 2003. Zn(II) metabolism in prokaryotes. FEMS Microbiol Rev 27:291–311. 10.1016/S0168-6445(03)00041-X. [DOI] [PubMed] [Google Scholar]
- 45.Oger C, Mahillon J, Petit F. 2003. Distribution and diversity of a cadmium resistance (cadA) determinant and occurrence of IS257 insertion sequences in staphylococcal bacteria isolated from a contaminated estuary (Seine, France). FEMS Microbiol Ecol 43:173–183. 10.1111/j.1574-6941.2003.tb01056.x. [DOI] [PubMed] [Google Scholar]
- 46.Feßler AT, Zhao Q, Schoenfelder S, Kadlec K, Brenner Michael G, Wang Y, Ziebuhr W, Shen J, Schwarz S. 2017. Complete sequence of a plasmid from a bovine methicillin-resistant Staphylococcus aureus harbouring a novel ica-like gene cluster in addition to antimicrobial and heavy metal resistance genes. Vet Microbiol 200:95–100. 10.1016/j.vetmic.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 47.Ikonomidis A, Grapsa A, Pavlioglou C, Demiri A, Batarli A, Panopoulou M. 2016. Accumulation of multiple mutations in linezolid-resistant Staphylococcus epidermidis causing bloodstream infections; in silico analysis of L3 amino acid substitutions that might confer high-level linezolid resistance. J Chemother 28:465–468. 10.1080/1120009X.2015.1119373. [DOI] [PubMed] [Google Scholar]
- 48.Dweba CC, Zishiri OT, El Zowalaty ME. 2019. Isolation and molecular identification of virulence, antimicrobial and heavy metal resistance genes in Staphylococcus aureus. Pathogens 8:79. 10.3390/pathogens8020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamann K. 2006. An overview of Danish pork industry integration and structure. Adv Pork Prod 17:93–97. [Google Scholar]
- 50.Directorate-General Health and Consumer Protection. 2004. Assessment of the dietary exposure to arsenic, cadmium, lead and mercury of the population of the EU Member States. Commission of the European Communities, Brussels, Belgium. [Google Scholar]
- 51.Nguyen CC, Hugie CN, Kile ML, Navab-Daneshmand T. 2019. Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: a review. Front Environ Sci Eng 13:46. 10.1007/s11783-019-1129-0. [DOI] [Google Scholar]
- 52.Sutterlin S, Tellez-Castillo CJ, Anselem L, Yin H, Bray JE, Maiden MCJ. 2018. Heavy metal susceptibility of Escherichia coli isolated from urine samples from Sweden, Germany, and Spain. Antimicrob Agents Chemother 62:e00209-18. 10.1128/AAC.00209-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cambiaso-Daniel J, Boukovalas S, Bitz G, Branski L, Herndon D, Culnan D. 2018. Topical antimicrobials in burn care: part I – topical antiseptics. Ann Plast Surg 1:1–26. 10.1097/SAP.0000000000001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams SV, Newcomb PA. 2014. Cadmium blood and urine concentrations as measures of exposure: NHANES 1999–2010. J Expo Sci Env Epidemiol 24:163–170. 10.1038/jes.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kales SN, Huyck KL, Goldman RH. 2006. Elevated urine arsenic: un-speciated results lead to unnecessary concern and further evaluations. J Anal Toxicol 30:80–85. 10.1093/jat/30.2.80. [DOI] [PubMed] [Google Scholar]
- 56.Vacchi-Suzzi C, Kruse D, Harrington J, Levine K, Meliker JR. 2016. Is urinary cadmium a biomarker of long-term exposure in humans? A review. Curr Environ Heal Rep 3:450–458. 10.1007/s40572-016-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito T, Hiramatsu K, Oliveira DC, De Lencastre H, Zhang K, Westh H, O’Brien F, Giffard PM, Coleman D, Tenover FC, Boyle-Vavra S, Skov RL, Enright MC, Kreiswirth B, Kwan SK, Grundmann H, Laurent F, Sollid JE, Kearns AM, Goering R, John JF, Daum R, Soderquist B. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akinbowale OL, Peng H, Grant P, Barton MD. 2007. Antibiotic and heavy metal resistance in motile aeromonads and pseudomonads from rainbow trout (Oncorhynchus mykiss) farms in Australia. Int J Antimicrob Agents 30:177–182. 10.1016/j.ijantimicag.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 59.EUCAST. 2017. MIC distributions and the setting of epidemiological cut-off value (ECOFF). https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/EUCAST_SOP_10.1_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20191130.pdf.
- 60.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt G, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 68.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1, Fig S2, Fig.S3, Appendix 1. Download AAC.02685-20-s0001.pdf, PDF file, 2.2 MB (2.2MB, pdf)
Table S1. Download AAC.02685-20-s0002.xlsx, XLSX file, 0.04 MB (44.3KB, xlsx)
Table S2. Download AAC.02685-20-s0003.xlsx, XLSX file, 0.06 MB (66.1KB, xlsx)
Table S3. Download AAC.02685-20-s0004.xlsx, XLSX file, 0.04 MB (39.7KB, xlsx)