ABSTRACT
Malaria remains one of the deadliest diseases in Africa, particularly for children. While successful in reducing morbidity and mortality, antimalarial treatments are also a major cause of adverse drug reactions (ADRs). Host genetic variation in genes involved in drug disposition or toxicity constitutes an important determinant of ADR risk and can prime for parasite drug resistance. Importantly, however, the genetic diversity in Africa is substantial, and thus, genetic profiles in one population cannot be reliably extrapolated to other ethnogeographic groups. Gabon is considered a high-transmission country, with more than 460,000 malaria cases per year. Yet the pharmacogenetic landscape of the Gabonese population or its neighboring countries has not been analyzed. Using targeted sequencing, here, we profiled 21 pharmacogenes with importance for antimalarial treatment in 48 Gabonese pediatric patients with severe Plasmodium falciparum malaria. Overall, we identified 347 genetic variants, of which 18 were novel, and each individual was found to carry 87.3 ± 9.2 (standard deviation [SD]) variants across all analyzed genes. Importantly, 16.7% of these variants were population specific, highlighting the need for high-resolution pharmacogenomic profiling. Between one in three and one in six individuals harbored reduced-activity alleles of CYP2A6, CYP2B6, CYP2D6, and CYP2C8 with important implications for artemisinin, chloroquine, and amodiaquine therapy. Furthermore, one in three patients harbored at least one G6PD-deficient allele, suggesting a considerably increased risk of hemolytic anemia upon exposure to aminoquinolines. Combined, our results reveal the unique genetic landscape of the Gabonese population and pinpoint the genetic basis for interindividual differences in antimalarial drug responses and toxicity.
KEYWORDS: Gabon, population pharmacogenetics, malaria, public health, precision medicine
TEXT
Malaria is a vector-borne tropical and subtropical disease caused by different species of Plasmodium that affects 228 million individuals, causing more than 400,000 deaths annually in Africa alone (1). Global strategies for malaria control and prevention are based on different measures, with one of the most relevant being treatment with artemisinin-based combination therapies (ACTs), recommended by the World Health Organization (WHO) as the first- and second-line treatments for uncomplicated Plasmodium falciparum malaria. ACTs combine an artemisinin derivative with a partner drug such as piperaquine, amodiaquine, lumefantrine, and mefloquine. The role of artemisinin is to reduce parasite numbers during the first 3 days of treatment (shorter half-life), while the partner drugs mainly aim at eliminating the remaining parasites due to their longer half-life in the body, thus curing the patient and providing posttreatment protection (2, 3).
Parasite resistance to antimalarial drugs remains a major challenge (4). Several factors influence the emergence and spread of drug-resistant malaria parasites, including the pharmacological selection pressure that the parasite populations are submitted to. Host genetic variability is a key factor that modulates drug disposition and clearance, resulting in differences in parasite drug exposure with regard to timing and concentration, potentially priming for drug resistance (5). Of the individual differences in drug responses, 20 to 30% have been attributed to genetic factors (6). Notably, due to limited endogenous functionality and low evolutionary constraints, there is considerable genetic ethnogeographic variation in genes involved in drug absorption, distribution, metabolism, and excretion (ADME), which can influence the pharmacokinetic profile of a medicine, thereby resulting in suboptimal efficiency of malaria treatment (7).
Multiple enzymes and transporters are involved in the disposition of the presently used ACTs, including the CYP enzymes CYP3A4, CYP2A6, CYP2C8, CYP2B6, CYP2C19, and CYP2D6; the UDP-glucuronosyltransferases UGT2B7, UGT1A8, and UGT1A9; as well as OATP1B1 (encoded by SLCO1B1), OCT1 (encoded by SLC22A1), MDR1 (P-glycoprotein, encoded by ABCB1), and BCRP (encoded by ABCG2). Importantly, most of these genes are highly variable, and a plethora of genetic polymorphisms that can impact the pharmacokinetics of antimalarials have been identified (8–10). The genetic landscape of CYPs is particularly complex in African populations, who harbor a multitude of clinically relevant high-frequency variants not found in other populations (11). Besides pharmacokinetic genes, genetic variants in KCNH2 have been associated with drug-induced prolonged QTc, possibly due to its structural characteristics that allow a broad range of drugs to bind, including the antimalarials chloroquine and piperaquine (12, 13). Similarly, genetic variations in glucose-6-phosphate dehydrogenase (G6PD) can affect the pentose phosphate shunt, thereby predisposing G6PD-deficient individuals to drug-induced hemolysis (14). Notably, while G6PD deficiency increases the risk of drug hemotoxicity, some studies suggest that it also provides partial protection from Plasmodium infection by increasing the clearance of infected erythrocytes (15, 16).
Efforts to profile the pharmacogenomic landscape have so far been conducted in populations across Europe (17, 18) and Asia (19) as well as in Native American populations (20, 21). However, to the best of our knowledge, the pharmacogenetic diversity in Gabon has not been evaluated, and even available studies in other populations of sub-Saharan Africa have interrogated only a few candidate polymorphisms in heterogeneous cohorts, focused mostly on drugs related to HIV and tuberculosis (22–25). To this end, here, we comprehensively profiled the pharmacogenomic complexity of 21 genes with relevance for antimalarial treatment using targeted sequencing of 48 pediatric patients with severe Plasmodium falciparum malaria in Gabon. In total, we identified 347 single-nucleotide variants (SNVs), the majority of which were exonic and nonsynonymous. Interestingly, 58 variants were Gabonese specific compared to other African populations, and 18 variants were novel. Translation of this pharmacogenetic variability into spectra of predicted functional variability revealed that approximately one in three individuals featured reduced activity of CYP2A6 and CYP2D6 and that one in six featured reduced activity of CYP2B6 and CYP2C8, with important implications for artemisinin, chloroquine, and amodiaquine therapy. Furthermore, one in three patients harbored at least one G6PD-deficient allele, suggesting a considerably increased risk of hemolytic anemia upon exposure to aminoquinolines.
RESULTS
The landscape of pharmacogenetic variability in Gabon.
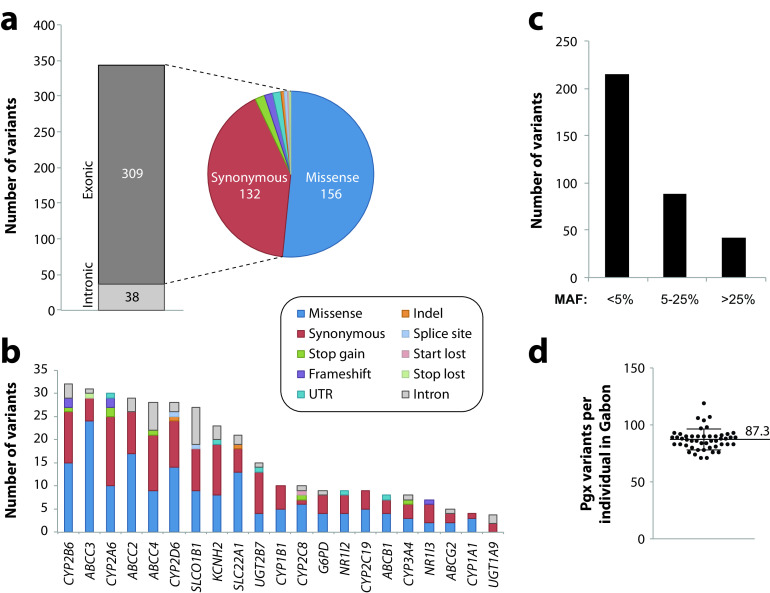
Based on targeted sequencing of the Gabonese population, we identified 347 SNVs distributed across 21 genes with relevance for antimalarial pharmacokinetics or toxicity. Due to the sequencing strategy that targeted open reading frames (ORFs), the majority of variants localized to exons (309 out of 347 variants; 89%) (Fig. 1a). Overall, 45% of variants resulted in amino acid exchanges, while 38% were synonymous and 4% resulted in gene product truncations (frameshifts or start-loss, stop-gain, or splice variations). Notably, genetic variability differed substantially across the studied genes, with CYP2B6 (32 variants), ABCC3 (31 variants), and CYP2A6 (30 variants) harboring the most variants, whereas only a few variants were detected in UGT1A9 (4 variants), CYP1A1 (4 variants), and ABCG2 (5 variants) (Fig. 1b). Across pharmacogene classes, 131 variants (81 nonsynonymous) were found in CYP genes, 101 variants (70 nonsynonymous) were found in ABC transporters, and 48 variants (34 nonsynonymous) were found in the SLC/SLCO transporters SLCO1B1 and SLC22A1 (OCT1). Across all pharmacogenes analyzed, the majority of variants were rare, with minor allele frequencies (MAFs) of <5% (Fig. 1c). When aggregating all variants in the diploid pharmacogenome, each analyzed individual harbored on average 87.3 variants in the 21 genes analyzed (Fig. 1d), demonstrating the considerable extent of pharmacogenetic variability in the Gabonese population.
FIG 1.
Overview of the pharmacogenetic landscape in the Gabonese population. (a) Across the 21 pharmacogenes analyzed, a total of 347 different variants were identified, of which 309 were exonic. The majority of exonic variants resulted in amino acid exchanges (missense) or were synonymous. (b) Stacked column plot showing the number of variants per variant class per gene. Note that variant numbers differed >10-fold between genes. (c) The majority of variants were rare, with minor allele frequencies (MAFs) of <5%. (d) On average, each individual in the Gabonese cohort harbored between 71 and 119 variants in the analyzed pharmacogenes combined (mean ± standard deviation [SD], 87.3 ± 9.2). Pgx, pharmacogenetic.
Frequencies of CYP alleles with known functional relevance.
The highly polymorphic CYP2D6 gene is the key enzyme for the metabolism of primaquine, and reduced CYP2D6 metabolism is associated with primaquine treatment failure (26, 27). CYP2D6 constitutes the most polymorphic CYP gene in the Gabonese population, with 76% of alleles harboring one or more genetic variants (Table 1). The most common variants were CYP2D6*122 (MAF = 13.5%), CYP2D6*2 (MAF = 11.5%), and the reduced-function alleles CYP2D6*17 (MAF = 10.4%) and CYP2D6*29 (MAF = 9.4%). Furthermore, we identified multiple additional variants with a reduction or loss of function, including CYP2D6*4 (MAF = 7.3%), CYP2D6*10 (MAF = 2.1%), and CYP2D6*40 (MAF = 1%).
TABLE 1.
Frequencies and putative functional consequences of CYP2D6 alleles identified in the Gabonese populationa
| Allele | Defining variant(s) | Variant type(s) (mutation[s]) | Allele frequency (%) |
Functional consequence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gabon | Africa | ESN | GWD | LWK | MSL | YRI | Global | ||||
| *1 | None | None | 24 | Neutral | |||||||
| *2 | rs16947, rs1135840 | Missense (R296C, S486T) | 11.5 | Neutral | |||||||
| *4 | rs3892097 | Splicing defect | 7.3 | 6.1 | 7.6 | 4 | 2.5 | 4.1 | 5.6 | 9.3 | Inactive |
| *10 | rs1065852, rs1135840 | Missense (P34S, S486T) | 2.1 | 11.1 | 9.1 | 11.5 | 3.5 | 15.9 | 10.7 | 23.6 | Decreased |
| *17 | rs16947, rs28371706 | Missense (R296C, T107I) | 10.4 | 21.8 | 26.3 | 16.4 | 19.2 | 28.2 | 25.6 | 5.9 | Decreased |
| *29 | rs16947, rs1135840, rs61736512, rs59421388 | Missense (R296C, S486T, V136I, V338M) | 9.4 | 9.5 | 7.6 | 13.3 | 10.6 | 9.4 | 9.7 | 2.5 | Decreased |
| *40 | rs72549356, rs28371706, rs16947, rs1135840 | In-frame insertion (FRPx2), missense (T107I, R296C, S486T) | 1 | <0.1 | 2.5 | 0 | 0 | 1.8 | 0 | <0.1 | Inactive |
| *43 | rs28371696 | Missense (R26H) | 1 | 2.3 | 2 | 4 | 4 | 0 | 0.9 | 1 | Unknown |
| *45 | rs28371710, rs16947, rs1135840 | Missense (E155K, R296C, S486T) | 6.3 | 4.6 | 5.6 | 4.9 | 7.1 | 3.5 | 3.7 | 1.3 | Neutral |
| *84 | rs148769737, rs16947, rs1135840 | Missense (P267H, R296C, S486T) | 1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | Unknown |
| *106 | rs149157808 | Missense (E418K) | 2.1 | 1.2 | 1 | 2.2 | 0 | 1.2 | 3.2 | 0.4 | Unknown |
| *122 | rs61745683 | Missense (V370I) | 13.5 | 7.1 | NA | NA | NA | NA | NA | 2.2 | Unknown |
| Novel alleles | Various combinations | 10.4 | NA | Unknown | |||||||
All star alleles identified in the Gabonese population are shown. YRI, Yoruba in Ibadan in Nigeria; LWK, Luhya in Webuye in Kenya; GWD, Gambians in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria; NA, no information available in the 1000 Genomes Project data set.
Artesunate, artemether, and arteether are prodrugs that are rapidly metabolized by CYP2A6 and, to a lesser extent, CYP2B6 and CYP3A4 into the active metabolite dihydroartemisinin (DHA). In CYP2A6, 29.2% of all alleles in the analyzed cohort harbored variants, with *17 and *31 being the most prevalent (MAF = 7.3% each). Particularly, *31 was more than 3-fold more common in the Gabonese population than in other populations in sub-Saharan Africa (Table 2). In addition, multiple other CYP2A6 variants that result in decreased CYP2A6 enzyme activity were identified, including *20 (MAF = 4.2%), *35 (MAF = 4.2%), *18 (MAF = 1%), and *23 (MAF = 1%). As a result, overall, 17.7% of CYP2A6 alleles in the Gabonese population carry variants associated with decreased activity.
TABLE 2.
Frequencies and putative functional consequences of other important CYP allelesa
| Allele | Defining variant(s) | Variant type (mutation[s]) | Allele frequency (%) |
Functional consequence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gabon | Africa | ESN | GWD | LWK | MSL | YRI | Global | ||||
| CYP2A6 | |||||||||||
| *1 | None | None | 55.2 | Neutral | |||||||
| *17 | rs28399454 | Missense (V365M) | 7.3 | 10.5 | 12.6 | 14.6 | 5.6 | 11.2 | 13.4 | 3.2 | Decreased |
| *18 | rs1809810 | Missense (Y392F) | 1 | 0.5 | 0 | 0.4 | 1 | 1.8 | 5 | 1.4 | Decreased |
| *20 | rs568811809 | Frameshift | 4.2 | 1.5 | 2.0 | 2.7 | 0 | 0.6 | 1.4 | 0.3 | Loss of function |
| *23 | rs56256500 | Missense (R203C) | 1 | 1.2 | 4.5 | 0.4 | 1 | 0.6 | 1.4 | 0.5 | Decreased |
| *25 | rs28399440 | Missense (F118L) | 1 | 1.3 | 2.5 | 0.9 | 0 | 1.2 | 3.7 | 0.5 | Unknown |
| *28 | rs8192730, rs28399463 | Missense (N418D, E419D) | 3.1 | 2.4 | NA | NA | NA | NA | NA | 0.7 | Unknown |
| *31 | rs72549432 | Missense (M6L) | 7.3 | 1.4 | 1 | 0.4 | 2 | 0.6 | 0.5 | 0.3 | Unknown |
| *35 | rs143731390 | Missense (N438Y) | 4.2 | 1.8 | NA | NA | NA | NA | NA | 5.5 | Decreased |
| Novel alleles | Various combinations | 15.6 | NA | Unknown | |||||||
| CYP2B6 | |||||||||||
| *1 | None | None | 54 | Neutral | |||||||
| *2 | rs8192709 | Missense (R22C) | 6.3 | 3.8 | 1 | 4.9 | 8.1 | 3.5 | 4.6 | 4.8 | Neutral |
| *4 | rs2279343 | Missense (K262R) | 0 | 2.4 | NA | NA | NA | NA | NA | 1.5 | Increased |
| *5 | rs3211371 | Missense (R487C) | 1 | 2.6 | 0 | 1.3 | 0 | 2.4 | 0.9 | 5.4 | Neutral |
| *6 | rs3745274, rs2279343 | Missense (Q172H, K262R) | 23 | 21.7 | NA | NA | NA | NA | NA | 13.2 | Decreased |
| *9 | rs3745274 | Missense (Q172H) | 0 | 15.4 | NA | NA | NA | NA | NA | 13.9 | Decreased |
| *17 | rs33973337, rs33980385, rs33926104, rs34284776 | Missense (T26S, D28G, R29T) | 1 | 6.5 | NA | NA | NA | NA | NA | NA | Decreased |
| *18 | rs2279343, rs28399499 | Missense (K262R, I328T) | 6.3 | 6.5 | NA | NA | NA | NA | NA | NA | Decreased |
| Novel alleles | Various combinations | 5.2 | NA | Unknown | |||||||
| CYP2C8 | |||||||||||
| *1 | None | None | 82.3 | Neutral | |||||||
| *2 | rs11572103 | Missense (I269F) | 16.7 | 14.6 | 19.7 | 23.5 | 13.6 | 17.1 | 19.9 | 5.5 | Decreased |
| *7 | rs72558195 | Stop-gain (R186X) | 1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | Loss of function |
| CYP2C19 | |||||||||||
| *1 | None | None | 78b | Neutral | |||||||
| *2 | rs4244285 | Splicing defect | 14.6 | 17 | 20.7 | 13.3 | 21.2 | 17.7 | 16.7 | 22.2 | Loss of function |
| *9 | rs17884712 | Missense (R144H) | 4.2 | 1.4 | 1 | 1.3 | 1 | 1.8 | 0.5 | 0.3 | Decreased |
| *13 | rs17879685 | Missense (R410C) | 3.1 | 1.7 | 0 | 2.7 | 4.0 | 0.6 | 1.9 | 0.6 | Neutral |
| *30 | rs145328984 | Missense (R73C) | 1 | 0.1 | 0.5 | 0 | 0 | 0 | 0 | 0 | Unknown |
| CYP3A4 | |||||||||||
| *1 | None | None | 95.9 | Neutral | |||||||
| *10 | rs4986908 | Missense (D174H) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | Unknown |
| *12 | rs12721629 | Missense (L373F) | 1 | 0.4 | 0.5 | 0 | 2 | 0 | 0 | 0.1 | Unknown |
| *15 | rs4986907 | Missense (R162Q) | 2.1 | 1.7 | 0.5 | 2.2 | 0.5 | 1.8 | 4.2 | 0.5 | Unknown |
All star alleles identified in the Gabonese population are shown. YRI, Yoruba in Ibadan in Nigeria; LWK, Luhya in Webuye in Kenya; GWD, Gambians in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria; NA, no information available in the 1000 Genomes Project data set.
CYP2C19*1 frequencies are likely overestimated due to the lack of CYP2C19*17 data.
CYP2B6 participates in the metabolism of artesunate, artemether, and arteether, and CYP2B6 variants have been shown to influence artemisinin pharmacokinetics (28). In addition to their relevance for malaria therapy, CYP2B6 polymorphisms are of high clinical relevance due to their association with elevated plasma concentrations of efavirenz and nevirapine as well as efavirenz-related neurotoxicity in individuals infected with HIV (29). The most prevalent CYP2B6 alleles in Gabon were the reduced-function alleles CYP2B6*6 (MAF = 23%) and CYP2B6*18 (MAF = 6.3%) as well as the functionally neutral allele CYP2B6*2 (MAF = 6.3%) (Table 2). Notably, 5.2% of alleles carried variant combinations without star allele designations. Of those, two patients carried the stop-gain variant p.Q286X (rs190463703), one with the putatively neutral variant p.R443C and one with a derivative of the loss-of-function allele CYP2D6*35, carrying p.I114T, p.E148D, and p.A279P but lacking p.G110V and p.M198T. In addition, we identified a novel variant constellation comprising p.I209V (rs144518874), p.S173C (rs148377536), p.E339A (rs565104467), p.H341D (rs138030127), and p.V367L (rs143979776).
Only two variants were identified in CYP2C8, encoding the main metabolizing enzyme of amodiaquine. CYP2C8*2 (MAF = 16.7%) was the most common, with frequencies very similar to those in other African populations. In addition, one patient carried the loss-of-function allele CYP2C8*7, which is very rare in all populations, including Africans, with a MAF of 0.03%. Reduced metabolism of amodiaquine to its major active metabolite N-desethylamodiaquine results in extended exposure that may lead to a considerably increased risk of amodiaquine-related adverse events, particularly in populations with high frequencies of reduced-function CYP2C8 alleles (30–32).
Four variants were found for CYP2C19, whose gene product contributes to the metabolism of chlorproguanil and proguanil, which are used mostly in combination with other antimalarials for prophylaxis and some cases of resistant parasites. The splice variant CYP2C19*2 as well as the reduced-function variant CYP2C19*9 (MAF = 4.2%) were common in Gabon (MAF = 14.6%). While CYP2C19*2 frequencies were similar across Africa (MAF = 13.3 to 21.2%), CYP2C19*9 was more common in Gabon than in other African populations (MAF = 1 to 1.8%). Notably, the increased-function variant CYP2C19*17 was not covered by the capture probes, and the fraction of rapid-metabolizer alleles in the Gabonese population thus remains unknown.
CYP3A4 constitutes the most abundant human CYP enzyme and is involved in the metabolism of approximately 30% of all clinically used drugs (8). Unlike most other drug-metabolizing CYPs, CYP3A4 also has important endogenous functions, such as the catabolism of testosterone, cortisol, and various bile acids, and CYP3A4 is thus considerably less polymorphic than most other CYP genes. In the Gabonese cohort, we identified CYP3A4*10 (MAF = 1%), CYP3A4*12 (MAF = 1%), and CYP3A4*15 (MAF = 2.1%), none of which have known functional relevance, while the remaining 95.9% of CYP3A4 alleles carried no variants.
Overall, we identified a multitude of pharmacogenetic variants in the Gabonese population with putative functional relevance for a plethora of antimalarial therapeutics. Particularly, multiple frequent reduced-function polymorphisms in CYP2A6, CYP2B6, and CYP2D6 might play important roles in the interindividual variability in treatment outcomes of patients treated with artemisinin- and primaquine-based regimens.
Genetic variability in other pharmacogenes.
Allele frequencies and functional consequences were determined for ABCB1 (encoding MDR1) and ABCG2 (encoding BCRP), whose gene products transport some antimalarial drugs such as mefloquine and its metabolites. The triallelic missense variant rs2032582 (p.A893S/T) and the synonymous variant rs1045642 (p.I1145I) are in strong linkage disequilibrium (33), and the resulting haplotype causes altered transcription and folding of the nascent polypeptide in vitro and has been shown to be associated with a risk of toxicity upon treatment with chemotherapeutic MDR1 substrates such as fluoropyrimidines or taxanes (34, 35). Albeit highly prevalent both in Africa and globally, rs2032582 variants were absent in our cohort (MAFglobal = 33.4%; MAFAfrica = 8.5%; MAFGabon = 0%) (Table 3). In contrast, rs1045642 was detected at frequencies similar to those in other African cohorts but considerably lower than those in other superpopulations (MAFGabon = 13.5%; MAFAfrica = 11.6 to 19%; MAFglobal = 39.5%). Similar observations were made for the synonymous MDR1 variant rs1128503 (p.G412G) (MAFGabon = 10.4%; MAFAfrica = 10.1 to 16.4%; MAFglobal = 41.6%). For ABCG2, we detected only rs2231137 (p.V12M) (MAF = 7.3%), a variant that is functionally neutral in vitro (36).
TABLE 3.
Allele frequencies of functionally relevant drug transporter allelesa
| Allele | Defining variant(s) | Variant type (mutation[s]) | Allele frequency (%) |
Functional consequence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gabon | Africa | ESN | GWD | LWK | MSL | YRI | Global | ||||
| ABCB1 (MDR1, Pgp) | rs2032582 | Missense (A893S) | 0 | 8.5 | 0 | 0.4 | 1.5 | 1.8 | 0 | 33.4 | Controversial |
| rs1128503 | Synonymous (G412) | 10.4 | 13.6 | 10.1 | 16.4 | 11.1 | 13.5 | 14.4 | 41.6 | Controversial | |
| rs1045642 | Synonymous (I1145) | 13.5 | 15 | 11.6 | 19 | 14.1 | 14.7 | 12.5 | 39.5 | Controversial | |
| ABCG2 (BCRP) | rs2231137 | Missense (V12M) | 7.3 | 6.3 | 5.1 | 4 | 11.6 | 8.8 | 5.6 | 15.8 | Neutral |
| SLCO1B1 (OATP1B1) | |||||||||||
| *1A | None | None | 13.5 | Neutral | |||||||
| *1B | rs2306283 | Missense (N130D) | 53.1 | 21.9 | 22.7 | 27 | 26.8 | 25.3 | 16.7 | 19.9 | Substrate specific |
| *14 | rs11045819, rs2306283 | Missense (P155T, N130D) | 3.1 | 1.7 | 1.5 | 0.5 | 1 | 1.2 | 3.7 | <0.1 | Unknown |
| *15 | rs4149056, rs2306283 | Missense (V174A, N130D) | 2.1 | 1.4 | 0 | 0 | 2 | 0 | <0.1 | 7.8 | Decreased |
| *20 | rs34671512, rs2306283, rs2291075 | Missense (L643F, N130D) | 8.3 | 5.8 | 7.6 | 4.9 | 4 | 5.3 | 5.6 | 3.3 | Unknown |
| *27 | rs59113707, rs2306283 | Missense (F400L, N130D) | 8.3 | 2.9 | 5.1 | 2.7 | 0 | 2.4 | 4.2 | 0.8 | Unknown |
| *31 | rs59502379, rs2306283 | Missense (G488A, N130D) | 4.2 | 1.7 | 1.5 | 0.5 | 1 | 1.2 | 3.7 | 5.6 | Unknown |
| *32 | rs11045852, rs2306283, rs11045819 | Missense (I245V, N130D, P155T) | 1 | 2.2 | 3.5 | <0.1 | 0 | <0.1 | 4.2 | <0.1 | Unknown |
| Novel alleles | Various combinations | 6.3 | NA | Unknown | |||||||
| SLC22A1 (OCT1) | |||||||||||
| *1 | Noneb | 58.3 | Neutral | ||||||||
| *2 | rs202220802 | Indel (M420del) | 4.2 | 4.5 | 4.6 | 3.1 | 3 | 6.5 | 1.4 | 11.8 | Inactive |
| *7 | rs34447885 | Missense (S14F) | 4.2 | 1.7 | 1.5 | 1.8 | 2.6 | 1.8 | 1.4 | 0.7 | Inactive |
| *8 | rs35270274 | Missense (R488M) | 3.1 | 3.4 | 4 | 1.3 | 5.1 | 2.4 | 5.7 | 1 | Neutral |
| rs34295611 | Missense (V461I) | 2.1 | 1.1 | 0 | 0 | 2 | 0 | 3.3 | 0.3 | Unknown | |
| rs143522800 | Missense (V400L) | 1 | 0.3 | 0 | 0 | 1.5 | 0 | 0 | <0.1 | Unknown | |
| rs628031 | Missense (M408L) | 1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | Unknown | |
| rs34570655 | Missense (L23V) | 1 | 0.2 | 0 | 0 | 0.5 | 0 | 0 | <0.1 | Unknown | |
| rs35546288 | Missense (L85P) | 1 | 0.5 | 0.5 | 1.8 | 0 | 0.6 | 0 | 0.1 | Unknown | |
| rs78899680 | Missense (V519P) | 4.2 | 3 | 4.6 | 0.4 | 2.5 | 4.8 | 4.6 | 0.8 | Unknown | |
| rs41267797 | Missense (V464I) | 18.8 | 19.5 | 21.7 | 23.5 | 18.7 | 18.2 | 19.9 | 6.3 | Unknown | |
All star alleles identified in the Gabonese population are shown. YRI, Yoruba in Ibadan in Nigeria; LWK, Luhya in Webuye in Kenya; GWD, Gambians in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria.
Variants defining SLC22A1*1 suballeles (P160L, P341L, R342H, and M408V) are not included.
Nine known and three novel alleles of SLCO1B1 encoding the drug uptake transporter OATP1B1 were identified in the Gabonese population. SLCO1B1*1B, -*20, and -*27 showed the highest frequencies, at 53.1%, 8.3%, and 8.3%, respectively. SLCO1B1*1B causes substrate-specific alterations of transport activity, whereas SLCO1B1*20 and -*27 do not have strong overt functional effects. The clinically actionable variant rs4149056 that results in reduced transporter activity was identified at frequencies of 2.1% only as part of the SLCO1B1*15 allele (in linkage with rs2306283), whereas no individuals carried the other rs4149056-containing allele SLCO1B1*5 or -*17. In addition, we identified the canonical splice variant rs77271279 as well as p.I274M (rs192911820) and p.I499V (rs74064213) in six alleles, which have not yet received a star allele designation. Additionally, the Gabonese population harbored three variant alleles of SLC22A1 (OCT1), the loss-of-function alleles *2 (MAF = 4.2%) and *7 (MAF = 4.2%), as well as the functionally neutral allele *8 (MAF = 3.1%). Furthermore, seven variant alleles were identified that were not covered by the current SLC22A1 star allele nomenclature (Table 3).
Genetic variability in genes associated with tolerance to antimalarial treatments.
G6PD deficiency, relatively common in populations where malaria is endemic, is associated with acute hemolytic anemia (AHA) upon treatment with quinolines. Four missense variants were identified in G6PD, including the likely neutral variant p.N126D (rs1050829), the African-specific variant p.V68M (rs1050828), as well as the very rare variants p.H129P (rs782402416) and p.M207T (rs1433928824) with unclear clinical significance (Table 4). Notably, frequencies of p.V68M and p.N126D have been previously studied in Gabon (37) and closely align with the results obtained here. In addition, six rare variants were identified in the potassium transporter hERG (encoded by KCNH2), two of which were not previously identified in the African subpopulations of the 1000 Genomes Project.
TABLE 4.
Allele frequencies and clinical significance of variants in genes associated with adverse events upon antimalarial treatmenta
| Gene | Defining variant | Variant type (mutation) | Allele frequency (%) |
Clinical significance | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gabon | Africa | ESN | GWD | LWK | MSL | YRI | Global | ||||
| KCNH2 | rs1805123 | Missense (K897T) | 1 | 5.2 | 0 | 0.4 | 0.5 | 0 | 0 | 22.1 | Benign |
| rs41314354 | Missense (G873S) | 1 | 0.3 | 1 | 0 | 0 | 0.6 | 0 | <0.1 | Likely benign | |
| rs370393086 | Missense (R878C) | 1 | 0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | Likely benign | |
| rs41314366 | Missense (T859M) | 1 | 0.3 | 1 | 0 | 0 | 0.6 | 0 | <0.1 | Benign | |
| rs138498207 | Missense (R791W) | 1 | 0.1 | 0 | 0 | 0 | 0 | 0.5 | <0.1 | Controversial | |
| rs150817714 | Missense (A190T) | 1 | 0.3 | 0 | 0 | 0 | 0 | 0 | 0.1 | Controversial | |
| G6PD | rs782402416 | Missense (H129P) | 1 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | Uncertain |
| rs1050829 | Missense (N126D) | 33.3 | 31.9 | 35.2 | 35.7 | 34.4 | 28.1 | 37.8 | 1.1 | Controversial | |
| rs1433928824 | Missense (M207T) | 1 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0.4 | Uncertain | |
| rs1050828 | Missense (V68M) | 16.7 | 11.6 | 15.7 | 4.1 | 17.5 | 7 | 20.7 | 1 | Pathogenic | |
YRI, Yoruba in Ibadan in Nigeria; LWK, Luhya in Webuye in Kenya; GWD, Gambians in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria.
Novel and population-specific pharmacogenetic variability in the Gabonese population.
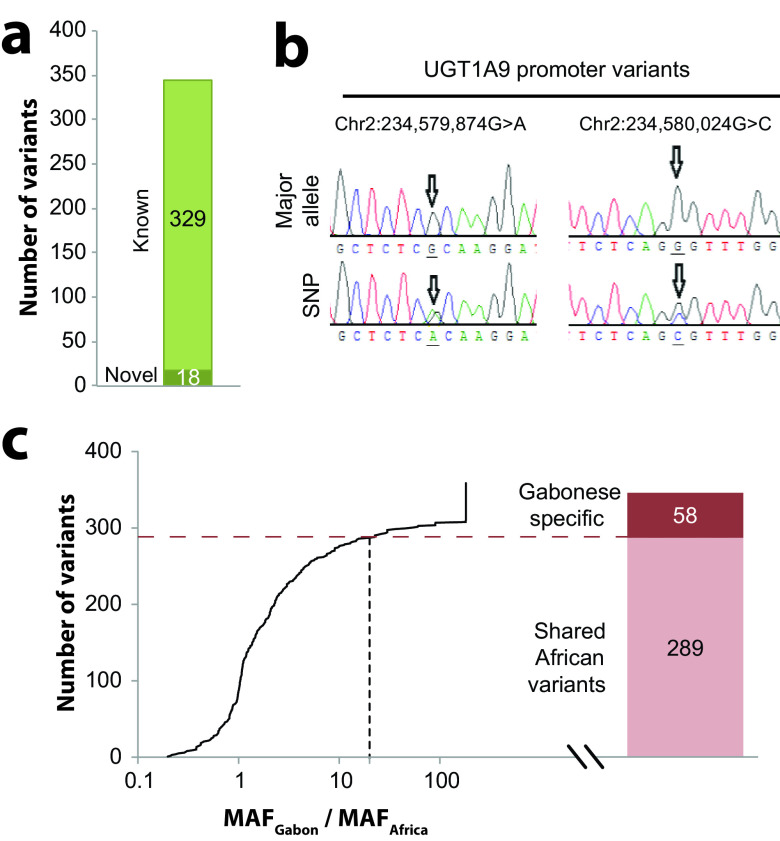
We identified a surprisingly large number of novel genetic variants in the Gabonese cohort that were not previously reported, as defined by inclusion in the latest dbSNP release. Overall, 18 novel variants were identified (5.4% of all variants) (Fig. 2a), 10 of which localized to exons (Table 5). Three novel frameshift variants were identified in CYP2A6, CYP2B6, and NR1I3 (encoding the nuclear hormone receptor CAR), and analysis using LOFTEE indicates that these variants likely abolish gene function. Furthermore, two novel missense variations were identified in CYP1A1 (p.M178I) and CYP2D6 (p.P286A). An ensemble classifier (ADME prediction framework [APF]) (see Materials and Methods) specifically developed for the functional interpretation of pharmacogenetic variants indicates that both CYP1A1M178I and CYP2D6P286A moderately affect gene function, with an estimated reduction of activity compared to the reference enzyme by 40%. Moreover, we identified two novel variants in the promoter of UGT1A9 and validated them using Sanger sequencing (Fig. 2b).
FIG 2.
The pharmacogenetic variability in Gabon is highly population specific. (a) Across all genes, 18 novel variations were identified, accounting for 4.6% of all variants. (b) Sanger sequencing data showing the nucleotide sequences (sense strands) in the promoter of UGT1A9. Major alleles are shown in the top row, while the bottom rows show the heterozygous variants. Arrows indicate the variant positions. SNP, single-nucleotide polymorphism. (c) Of all 347 detected variants, 16.7% (n = 58) were specific to the Gabonese population, as defined by a ≥20-fold increased minor allele frequency in Gabon (MAFGabon) compared to African reference populations (MAFAfrica), and each individual carried on average 2.2 population-specific pharmacogenetic variants.
TABLE 5.
Overview of novel variants and their putative functional consequencesa
| Gene | Genomic location | Reference | Alternative | Variant type (variant) | No. of alleles | Predicted consequence (estimated activity) |
|---|---|---|---|---|---|---|
| ABCC3 | Chr17:48750862 | C | T | Synonymous (P814) | 1/96 | Neutral |
| ABCC4 | Chr13:95748442 | G | GA | Intronic | 3/96 | Neutral |
| Chr13:95695981 | G | A | Synonymous (T1230) | 1/96 | Neutral | |
| CYP1A1 | Chr15:75014905 | C | G | Missense (M178I) | 1/96 | Deleterious (60% of reference enzyme) |
| Chr15:75014854 | G | C | Synonymous (T195) | 1/96 | Neutral | |
| CYP2A6 | Chr19:41354168 | TG | T | Frameshift (M204fs) | 2/96 | Deleterious (0% of reference enzyme) |
| CYP2B6 | Chr19:41497294 | CCG | C | Frameshift (R29fs) | 1/96 | Deleterious (0% of reference enzyme) |
| Chr19:41512975 | GGGAGAC | G | Intronic | 1/96 | Neutral | |
| CYP2D6 | Chr22:42523973 | G | C | Missense (P286A) | 2/96 | Deleterious (60% of reference enzyme) |
| CYP3A4 | Chr7:99364013 | A | G | Synonymous (T284) | 1/96 | Neutral |
| Chr7:99359750 | G | A | Synonymous (P389) | 1/96 | Neutral | |
| NR1I3 | Chr1:161205730 | A | AGG | Frameshift (F49fs) | 2/96 | Deleterious (0% of reference enzyme) |
| SLCO1B1 | Chr12:21375306 | GAAAAAAA | G | Intronic | 4/96 | Neutral |
| Chr12:21375306 | GAAAA | G | Intronic | 2/96 | Neutral | |
| Chr12:21375306 | GAAAAAA | G | Intronic | 1/96 | Neutral | |
| Chr12:21327652 | TA | T | Intronic | 1/96 | Neutral | |
| UGT1A9 | Chr2:234579874 | G | A | Promoter | 1/80b | Neutral |
| Chr2:234580024 | G | C | Promoter | 1/80b | Neutral | |
Residual function was estimated using the pharmacogene-specific ensemble classifier APF. Genomic locations are provided according to GRCh37.
The presence of the novel UGT1A9 variants was evaluated in only 40 individuals.
In addition to novel pharmacogenetic variants, the Gabonese population featured a large number of population-specific variants, as defined by allele frequencies >20-fold higher than those in other African populations. Overall, 58 of the 347 variants were enriched at least 20-fold in the analyzed cohort compared to other African populations, and each individual harbored approximately 2.2 pharmacogenetic variants specific for the Gabonese population (Fig. 2c).
Translation of pharmacogenetic variability into functional profiles.
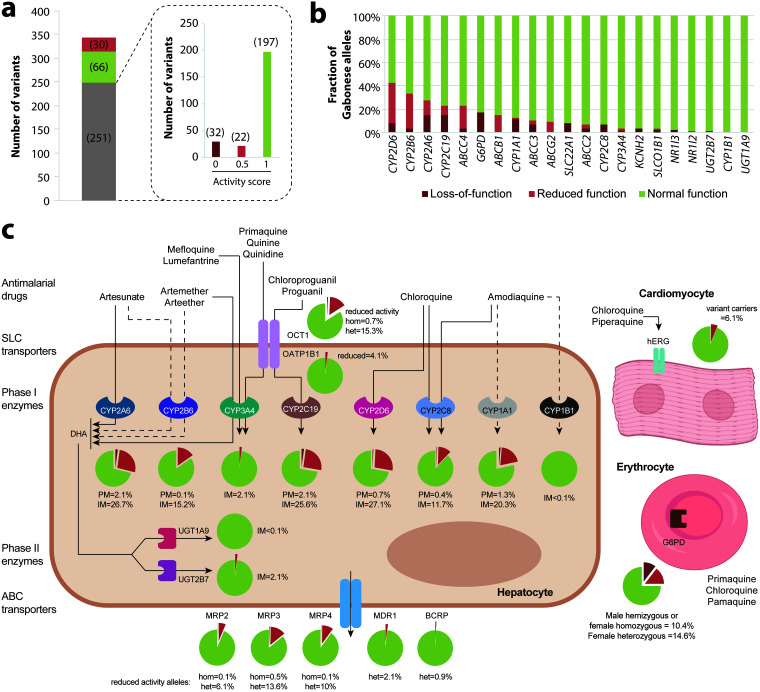
Next, we estimated the functional importance of the observed genetic variation. Of all identified variants, 30 are known to impact gene function, while 66 are functionally neutral (Fig. 3a). For the remaining 251 mostly rare or novel variants, we estimated functional consequences using a quantitative computational ensemble classifier specifically trained on pharmacogenomic data (38). Of these, 197 were predicted to be neutral, 22 were predicted to moderately decrease activity (approximately 50% of the reference allele activity; activity score = 0.5), and 32 were predicted to result in a loss of function (0% activity; activity score = 0). When aggregating the functional impact per gene, we found that altered-function alleles were most common in CYP2D6 (8.3% of loss-of-function and 34.4% of reduced-function alleles), CYP2B6 (3.1% of loss-of-function and 30.2% of reduced-function alleles), and CYP2A6 (14.6% of loss-of-function and 13.5% of reduced-function alleles) (Fig. 3b).
FIG 3.
Estimated functional consequences and clinical implications of pharmacogenetic variation in Gabon. (a) Of all 347 variants, 66 were annotated as functionally neutral (green), whereas 30 had known functional consequences on the drug response (red). The remaining 251 variants (gray) with unclear functions were analyzed using a computational prediction framework specifically developed for pharmacogenetic interpretations. Predicted outcomes of gene activity of these variants are shown in the dashed inset. Activity scores of 0, 0.5, and 1 correspond to loss-of-function variants, decreased-function variants, and neutral variants, respectively. (b) Fractions of loss-of-function alleles, reduced-function alleles, and normal-function alleles in the Gabonese population plotted for each of the 21 analyzed genes. (c) Schematic simplified depiction of the metabolic routes of antimalarials and the transporters and drug-metabolizing enzymes involved. The fractions of poor metabolizers (PM) (defined as a diplotype activity score of 0) (dark red), intermediate metabolizers (IM) (defined as a diplotype activity score of 0.5 or 1) (light red), and normal metabolizers (defined as a diplotype activity score of ≥1.5) (green) are shown as pie charts for each enzyme. For transporters, the fractions of heterozygous (het) and homozygous (hom) carriers of reduced-function alleles are shown analogously. Solid and dashed arrows indicate major and minor metabolic routes, respectively. DHA, dihydroartemisinin.
On the basis of this information, we calculated the diplotypes of the Gabonese population for all analyzed pharmacogenes involved in antimalarial disposition or toxicity risk (Fig. 3c). Overall, 2.1% and 26.7% of individuals were poor metabolizers (PMs) and intermediate metabolizers (IMs) of CYP2A6, with putatively important implications for the metabolic activation of artesunate. Similarly, 27.8% (0.7% PMs and 27.1% IMs) and 12.1% (0.4% PMs and 11.7% IMs) of patients were inferred to have reduced activity of CYP2D6 and CYP2C8, respectively, which might have pronounced impacts on the metabolism of chloroquine and amodiaquine. In contrast, only minor genetic factors were found that are predicted to impact the metabolism of the CYP3A4 substrates mefloquine and lumefantrine. Notably, however, excessive variability in CYP3A expression and activity has been reported between human livers independent of known genetic factors (39).
In addition to genetic factors of relevance for antimalarial drug disposition, carrier rates of reduced-function G6PD alleles were high in the Gabon (32.2% carried at least one G6PD-deficient allele). These effects were primarily allotted to the deleterious V68M variant (rs1050828) that has been robustly associated with erythrocyte phenotypes, including erythrocyte numbers, as well as hemoglobin and hematocrit counts (40, 41) and suggest an increased AHA risk upon primaquine, pamaquine, and chloroquine treatment in almost one-third of the Gabonese population.
DISCUSSION
Population pharmacogenomic strategies allow the comprehensive characterization of the signature of genetic and, by extension, predicted functional variability in ADME genes and drug targets in a given ethnogeographic group. The value of such methods can be assumed to be particularly high in those populations that differ considerably from commonly used reference groups, which are primarily of European ancestry, and African populations have been consistently shown to differ substantially in their pharmacogenomic repertoire from populations in Europe. Furthermore, Africa is genetically highly diverse, resulting in major differences between ethnicities. Neglecting those differences has in the past resulted in considerable public health crises, as exemplified by the change of first-line HIV treatment to efavirenz in Zimbabwe. This policy followed WHO guidance but failed to consider the local genetic predisposition, resulting in substantial numbers of efavirenz overdoses due to the particularly high numbers (20% of the general population) of Zimbabweans homozygous for the reduced-functionality allele CYP2B6*6, which caused efavirenz exposures that exceeded the recommended therapeutic levels and resulted in the population-specific failure of a globally established dosing regimen (42). It is thus of paramount importance to study ethnogeographic differences in pharmacogenes in Africa with high resolution in order to guide public health decisions. However, the pharmacogenomic diversity in Gabon or its surrounding countries had not been analyzed previously.
Antimalarials constitute some of the most widely prescribed medications in Africa. Gabon, with more than 2 million people at risk of contracting malaria, is considered a high-transmission country and accounted only last year for more than 460,000 malaria cases and 314 malaria deaths (1). While mortality rates decreased and the country recently implemented intermittent preventive treatment in pregnancy (IPTp), incidences of malaria increased over the last 5 years, and Gabon remains the central African country with the lowest malaria funding per person at risk (<0.25$) and the lowest percentage (<20%) of the population with access to either long-lasting insecticidal nets (LLINs) or indoor residual spraying (IRS), emphasizing the continued importance of improving disease control and management.
As multiple antimalarials, such as amodiaquine and artesunate, are among the most common drugs associated with adverse events across Africa (43), here, we sequenced genes involved in antimalarial disposition and toxicity in Gabon and provide the most comprehensive overview of clinically relevant pharmacogenetic diversity in central sub-Saharan Africa reported to date. We confirm the presence of multiple pharmacogenetically important haplotypes in drug-metabolizing enzymes, including CYP2A6*17, CYP2B6*6, CYP2C8*2, CYP2C19*2, CYP2D6*10, CYP2D6*17, and CYP2D6*29. Furthermore, we find important transporter polymorphisms, such as SLC22A1*2, SLCO1B1*27, and multiple ABC transporter variants (rs1128503, rs1045642, and rs2231137). Compared to African reference populations in Nigeria, Kenya, the Gambia, and Sierra Leone, our data show that the genetic makeup of the Gabonese population is distinctly different, with 16.2% of all identified variants being population specific. Overall, the Gabonese cohort was most similar to the Luhya people in Kenya and the Yoruba in Nigeria, possibly due to their relative geographic proximity. Population-specific variants are globally rare and mostly remain functionally uncharacterized. In light of the overall functional relevance of rare genetic variants (44, 45), their mapping, experimental characterization, and functional interpretation remain important frontiers for contemporary pharmacogenetics (46–48).
It has been previously shown that the human pharmacogenetic profile has the potential to influence the selection of parasite antimalarial response markers (49). These effects are likely due to a combination of pharmacokinetic differences between individuals and compliance, which can contribute to significant variations in drug exposure for the parasite, hence influencing the process of resistance development. Interestingly, a previous analysis of parasite genetics from the same cohort revealed that 48% of parasite isolates carried the Pfmdr1 N86Y mutation, which was higher than the rate in samples from Ghana or Kenya (50). This polymorphism was associated with prolonged parasite clearance in vivo (51). Notably, however, these findings contrast with other observations that associated the variant allele with higher sensitivity to lumefantrine (52). Thus, whether N86Y frequency might foreshadow the emergence of drug resistance remains to be determined.
The major limitation of this study is the relatively small size of the cohort that was sampled in a geographically confined region in and around Lambarene, Gabon. As such, the population cannot be considered representative of Gabon but rather presents only the first evidence for central African pharmacogenomic variability that should be further complemented with analyses of additional populations. The individuals analyzed were all diagnosed with P. falciparum infection severe enough that hospitalization was required and were recruited as part of a randomized controlled clinical study; while we do not expect that the frequencies of pharmacogenomic variations in this patient group would differ from those of the general population, such effects cannot be formally excluded.
Despite the small sample size, we identified 18 novel variations, 5 of which were predicted to impact gene function. These novel variants encompassed multiple frameshift and missense variants in CYP1A1, CYP2A6, CYP2B6, and CYP2D6. The novel missense variant in CYP1A1 resulted in a p.M178I amino acid exchange and was estimated to reduce enzyme activity by 40%, which might affect amodiaquine clearance. Similar reductions in enzyme activity were predicted for the novel CYP2D6 variant P286A that might impact chloroquine clearance. Novel frameshift mutations in CYP2A6 and CYP2B6 could have major impacts on the metabolism of artesunate. Similarly, the identification of novel promoter variants in UGT1A9 could be relevant for the prediction of artesunate pharmacokinetics, as previous studies demonstrated links between promoter polymorphisms and the transcriptional activity of this gene (53). Artesunate and amodiaquine are used in combination as the first-line treatment for uncomplicated malaria in Gabon, indicating the urgent need to conduct further experimental evaluations into the functional relevance of these novel variations.
The coadministration of drugs for different therapeutic uses is common in Africa due to the frequent occurrence of coinfections. For instance, CYP2C8 inhibitors (some antiretrovirals or antifungals) are in use in areas where amodiaquine (metabolized by CYP2C8) is also being administered for malaria treatment. The resulting drug-drug interactions can be amplified by genetic factors, resulting in strongly increased risks due to cotreatment, particularly in poor and intermediate metabolizers (54). Thus, future studies are needed to increase the resolution of pharmacogenetic maps across African geographical locations and ethnic groups, which might help in refining antimalarial treatment for these populations in a low-cost public health setting.
In summary, our analyses provide the first comprehensive overview of the pharmacogenetic landscape in a Gabonese population and reveal considerable population specificity. As such, these data contribute to a refined map of genetic diversity in Africa and highlight the clinical relevance of understanding the population-specific pharmacogenetic signature of African populations to optimize precision public health.
MATERIALS AND METHODS
Allele nomenclature and definitions.
CYP variant and allele designations were defined according to PharmVar (55). SLCO1B1 (encoding OATP1B1) and SLC22A1 (encoding OCT1) alleles were defined based on the available literature (56, 57). Variants with minor allele frequencies (MAFs) of <5% were defined as rare.
Sample collection and data processing.
Full open reading frames were sequenced for a set of 21 main genes in a Gabonese population with severe Plasmodium falciparum malaria from the Centre de Recherches Médicales and the Hôpital Albert Schweitzer in Lambarene, Gabon. Patient characteristics are summarized in Table 6. The analyzed patient cohort constituted a subset of the patients recruited in a randomized controlled trial evaluating the use of intramuscular artesunate for severe malaria (51). In short, the cohort comprised pediatric patients aged 6 months to 10 years with severe P. falciparum infection and clinical symptoms severe enough to require hospitalization, according to the Severe Malaria in African Children (SMAC) network definition of severe malaria. In addition, the legal guardians provided their written informed consent to participate in this study. All procedures were carried out in accordance with the Declaration of Helsinki and were reviewed and approved by the respective ethics committees. DNA from whole blood was extracted using commercial kits (Qiagen), and sequencing libraries were prepared using the Illumina SureSElect XT v.1 target enrichment kit for paired-end multiplexed sequencing, aided by an Agilent Bravo pipetting system. Sequencing was performed using MiSeq reagent kit v2 chemistry for 2- by 150-bp paired-end reads per lane on Illumina MiSeq sequencers at a sequencing depth of 90× (SciLifeLab, National Genomics Infrastructure, Stockholm, Sweden). Sequencing quality control was performed using MultiQC, and 95.8 to 97.5% of bases had Phred scores of ≥30. Bioinformatic analysis was performed by Genevia Technologies. Variants were annotated with pathogenicity data using SnpEff and SnpSift 4.3 (58).
TABLE 6.
Characteristics of the Gabonese patient cohort with severe P. falciparum malaria
| Characteristica | Value for cohort |
|---|---|
| Cohort size (no. of patients/no. of alleles) | 48/96 |
| Median age (yrs) (range) | 4.5 (0.4–10) |
| No. of female patients (%) | 23 (47.9) |
| Median asexual Plasmodium falciparum parasitemia (no. of parasites/μl) (range) | 118,459 (5,377–745,100) |
| Median parasite clearance (h) (range) | |
| PC50 | 10.7 (6–24) |
| PC90 | 15.5 (6–24) |
| Mean hemoglobin concn (g/dl) (range) | 9.9 (3.2–12.3) |
| Median body temp (°C) | 39.4 |
PC50 and PC90, 50% and 90% parasite clearance, respectively.
Sanger sequencing of UGT1A9 promoter variants.
The promoter of UGT1A9 (positions −930 to +345 relative to the translation start site) was amplified as previously described (59) and sequenced for both strands using the ABI BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). PCRs and sequencing were repeated to rule out PCR artifacts and confirmed the novel variations.
Calculation of allele frequencies.
Single-nucleotide variants (SNVs) were identified and analyzed regarding frequency and functional consequences. Allele frequencies in the Gabonese population based on our sequencing data were compared to those of other African populations from the 1000 Genomes Project, Yoruba in Ibadan, Nigeria (YRI); Luhya in Webuye, Kenya (LWK); Gambians in Western Divisions in the Gambia (GWD); Mende in Sierra Leone (MSL); and Esan in Nigeria (ESN), as well as overall African frequencies from gnomAD (n = 8,128) (60). Haplotypes defined by multiple variants were calculated based on linkage information of the African populations of the 1000 Genomes Project. Linkage disequilibria were calculated using LDLink (61). CYP2A6*28, CYP2C9*9, and CYP2C9*12 were very rare, and thus, linkage disequilibria for these variants could not be calculated. Frequencies of the wild-type allele (FWT) corresponding to *1 are calculated as FWT = 1 − ΣiFi, with Fi being the frequency of each considered variant allele i. Novel variants were defined based on dbSNP 2.0 build 153.
Computational functionality predictions.
The putative functional impacts of variants were calculated using an ensemble classifier specifically trained on pharmacogenetic variants (38). Furthermore, nonsense variants (frameshifts, splice variants, stop-gain variants, and start-loss variants) were evaluated using the LOFTEE plug-in of the Variant Effect Predictor (62). ClinVar was used for the clinical annotations of previously reported variants (63).
ACKNOWLEDGMENTS
We thank all participating patients and health care staff for their dedicated study participation.
Y.Z. and V.M.L. are cofounders and shareholders of PersoMedix AB. In addition, V.M.L. is CEO and shareholder of HepaPredict AB and discloses consultancy work for Enginzyme AB.
S.K., J.P.G., P.G.K., V.M.L., and T.P.V. designed the study and contributed to the study materials. A.A.A. is the principal investigator of the SMAC clinical trial site, Lambarene, Gabon. A.A.A. and J.F.Z. recruited children with severe malaria. L.P.-L. and Y.Z. analyzed the data. L.P.-L. and V.M.L. wrote the manuscript. J.P.G. and V.M.L. supervised the analysis. All authors reviewed and approved the final version of the manuscript.
T.P.V., P.G.K., and A.A.A. acknowledge financial support from the German Center for Infection Research (DZIF), Clinical Research Group, Lambarene, and a German Science Foundation grant (DFG Ku 775/17-1) for the German African Cooperation Project in Infectiology. Work was supported by UIDB/04046/2020 and UIDP/04046/2020 Centre grants from FCT, Portugal (to BioISI). L.P.-L. is the recipient of a fellowship from BioSys Ph.D. program PD65-2012 (reference number SFRH/BD/142860/2018) from FCT (Portugal). V.M.L. receives support from the Swedish Research Council (grant agreement numbers 2016-01153, 2016-01154, and 2019-01837), the EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant number 875510), the Swedish Strategic Research Programmes in Diabetes (SFO Diabetes), Stem Cells and Regenerative Medicine (SFO StratRegen), as well as the European Union’s Horizon 2020 research and innovation program U-PGx (grant agreement number 668353).
Contributor Information
Volker M. Lauschke, Email: volker.lauschke@ki.se.
Thirumalaisamy P. Velavan, Email: velavan@medizin.uni-tuebingen.de.
REFERENCES
- 1.World Health Organization. 2019. WHO malaria report 2019. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Cowman AF, Healer J, Marapana D, Marsh K. 2016. Malaria: biology and disease. Cell 167:610–624. 10.1016/j.cell.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 3.Ashley EA, Pyae Phyo A, Woodrow CJ. 2018. Malaria. Lancet 391:1608–1621. 10.1016/S0140-6736(18)30324-6. [DOI] [PubMed] [Google Scholar]
- 4.Wicht KJ, Mok S, Fidock DA. 2020. Molecular mechanisms of drug resistance in Plasmodium falciparum malaria. Annu Rev Microbiol 74:431–454. 10.1146/annurev-micro-020518-115546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paganotti GM, Gallo BC, Verra F, Sirima BS, Nebié I, Diarra A, Coluzzi M, Modiano D. 2011. Human genetic variation is associated with Plasmodium falciparum drug resistance. J Infect Dis 204:1772–1778. 10.1093/infdis/jir629. [DOI] [PubMed] [Google Scholar]
- 6.Lauschke VM, Zhou Y, Ingelman-Sundberg M. 2019. Novel genetic and epigenetic factors of importance for inter-individual differences in drug disposition, response and toxicity. Pharmacol Ther 197:122–152. 10.1016/j.pharmthera.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerb R, Fux R, Mörike K, Kremsner PG, Gil JP, Gleiter CH, Schwab M. 2009. Pharmacogenetics of antimalarial drugs: effect on metabolism and transport. Lancet Infect Dis 9:760–774. 10.1016/S1473-3099(09)70320-2. [DOI] [PubMed] [Google Scholar]
- 8.Zanger UM, Schwab M. 2013. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Gordon AS, Tabor HK, Johnson AD, Snively BM, Assimes TL, Auer PL, Ioannidis JPA, Peters U, Robinson JG, Sucheston LE, Wang D, Sotoodehnia N, Rotter JI, Psaty BM, Jackson RD, Herrington DM, O’Donnell CJ, Reiner AP, Rich SS, Rieder MJ, Bamshad MJ, Nickerson DA, NHLBI GO Exome Sequencing Project . 2014. Quantifying rare, deleterious variation in 12 human cytochrome P450 drug-metabolism genes in a large-scale exome dataset. Hum Mol Genet 23:1957–1963. 10.1093/hmg/ddt588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujikura K, Ingelman-Sundberg M, Lauschke VM. 2015. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet Genomics 25:584–594. 10.1097/FPC.0000000000000172. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Ingelman-Sundberg M, Lauschke VM. 2017. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102:688–700. 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez D, Ghanta A, Kauffman GW, Sanguinetti MC. 2004. Physicochemical features of the HERG channel drug binding site. J Biol Chem 279:10120–10127. 10.1074/jbc.M310683200. [DOI] [PubMed] [Google Scholar]
- 13.He F-Z, McLeod HL, Zhang W. 2013. Current pharmacogenomic studies on hERG potassium channels. Trends Mol Med 19:227–238. 10.1016/j.molmed.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Ryan K, Tekwani BL. 2020. Current investigations on clinical pharmacology and therapeutics of glucose-6-phosphate dehydrogenase deficiency. Pharmacol Ther 222:107788. 10.1016/j.pharmthera.2020.107788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopera-Mesa TM, Doumbia S, Konaté D, Anderson JM, Doumbouya M, Keita AS, Diakité SAS, Traoré K, Krause MA, Diouf A, Moretz SE, Tullo GS, Miura K, Gu W, Fay MP, Taylor SM, Long CA, Diakité M, Fairhurst RM. 2015. Effect of red blood cell variants on childhood malaria in Mali: a prospective cohort study. Lancet Haematol 2:e140–e149. 10.1016/S2352-3026(15)00043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X-Y, Chen J-T, Ma Y-B, Huang H-Y, Xie D-D, Monte-Nguba S-M, Ehapo CS, Eyi UM, Zheng Y-Z, Liu X-Z, Zha G-C, Lin L-Y, Chen W-Z, Zhou X, Lin M. 2020. Evidence of positively selected G6PD A− allele reduces risk of Plasmodium falciparum infection in African population on Bioko Island. Mol Genet Genomic Med 8:e1061. 10.1002/mgg3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Başak N, Böhm R, Borg J, Borgiani P, Bozina N, Bruckmueller H, Burzynska B, Carracedo A, Cascorbi I, Deltas C, Dolzan V, Fenech A, Grech G, Kasiulevicius V, Kádaši Ľ, Kučinskas V, Khusnutdinova E, Loukas YL, Macek M, Makukh H, Mathijssen R, Mitropoulos K, Mitropoulou C, Novelli G, Papantoni I, Pavlovic S, Saglio G, Setric J, Stojiljkovic M, Stubbs AP, Squassina A, Torres M, Turnovec M, van Schaik RH, Voskarides K, Wakil SM, Werk A, del Zompo M, Zukic B, Katsila T, Lee MTM, Motsinger-Rief A, Mc Leod HL, van der Spek PJ, Patrinos GP. 2016. A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS One 11:e0162866. 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrović J, Pešić V, Lauschke VM. 2020. Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur J Hum Genet 28:88–94. 10.1038/s41431-019-0480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Runcharoen C, Fukunaga K, Sensorn I, Iemwimangsa N, Klumsathian S, Tong H, Vo NS, Le L, Hlaing TM, Thant M, Zain SM, Mohamed Z, Pung Y-F, Capule F, Nevado J, Silao CL, Al-Mahayri ZN, Ali BR, Yuliwulandari R, Prayuni K, Zahroh H, Noor DAM, Xangsayarath P, Xayavong D, Kounnavong S, Sayasone S, Kordou Z, Liopetas I, Tsikrika A, Tsermpini E-E, Koromina M, Mitropoulou C, Patrinos GP, Kesornsit A, Charoenyingwattana A, Wattanapokayakit S, Mahasirimongkol S, Mushiroda T, Chantratita W. 2021. Prevalence of pharmacogenomic variants in 100 pharmacogenes among Southeast Asian populations under the collaboration of the Southeast Asian Pharmacogenomics Research Network (SEAPharm). Hum Genome Var 8:7. 10.1038/s41439-021-00135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson LM, Claw KG, Woodahl EL, Robinson RF, Boyer BB, Burke W, Thummel KE. 2018. P450 pharmacogenetics in indigenous North American populations. J Pers Med 8:9. 10.3390/jpm8010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Covarrubias V, Morales-Franco M, Cruz-Correa OF, Martínez-Hernández A, García-Ortíz H, Barajas-Olmos F, Genis-Mendoza AD, Martínez-Magaña JJ, Nicolini H, Orozco L, Soberón X. 2019. Variation in actionable pharmacogenetic markers in natives and mestizos from Mexico. Front Pharmacol 10:1169. 10.3389/fphar.2019.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulder N. 2017. Development to enable precision medicine in Africa. Per Med 14:467–470. 10.2217/pme-2017-0055. [DOI] [PubMed] [Google Scholar]
- 23.Nordling L. 2017. How the genomics revolution could finally help Africa. Nature 544:20–22. 10.1038/544020a. [DOI] [PubMed] [Google Scholar]
- 24.Mpye KL, Matimba A, Dzobo K, Chirikure S, Wonkam A, Dandara C. 2017. Disease burden and the role of pharmacogenomics in African populations. Glob Health Epidemiol Genom 2:e1. 10.1017/gheg.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radouani F, Zass L, Hamdi Y, da Rocha J, Sallam R, Abdelhak S, Ahmed S, Azzouzi M, Benamri I, Benkahla A, Bouhaouala-Zahar B, Chaouch M, Jmel H, Kefi R, Ksouri A, Kumuthini J, Masilela P, Masimirembwa C, Othman H, Panji S, Romdhane L, Samtal C, Sibira R, Ghedira K, Fadlelmola F, Kassim SK, Mulder N. 2020. A review of clinical pharmacogenetics studies in African populations. Per Med 17:155–170. 10.2217/pme-2019-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brasil LW, Rodrigues-Soares F, Santoro AB, Almeida ACG, Kühn A, Ramasawmy R, Lacerda MVG, Monteiro WM, Suarez-Kurtz G. 2018. CYP2D6 activity and the risk of recurrence of Plasmodium vivax malaria in the Brazilian Amazon: a prospective cohort study. Malar J 17:57. 10.1186/s12936-017-2139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladeia-Andrade S, Menezes MJ, de Sousa TN, Silvino ACR, de Carvalho JF, Salla LC, Nery OA, de Melo GNP, Corder RM, Rodrigues PT, Ferreira MU. 2019. Monitoring the efficacy of chloroquine-primaquine therapy for uncomplicated Plasmodium vivax malaria in the main transmission hot spot of Brazil. Antimicrob Agents Chemother 63:e01965-18. 10.1128/AAC.01965-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svensson U, Ashton M. 1999. Identification of the human cytochrome P450 enzymes involved in the in vitro metabolism of artemisinin. Br J Clin Pharmacol 48:528–535. 10.1046/j.1365-2125.1999.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas DW, Gebretsadik T, Mayo G, Menon UN, Acosta EP, Shintani A, Floyd M, Stein CM, Wilkinson GR. 2009. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis 199:872–880. 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gil JP, Gil Berglund E. 2007. CYP2C8 and antimalaria drug efficacy. Pharmacogenomics 8:187–198. 10.2217/14622416.8.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adjei GO, Kristensen K, Goka BQ, Hoegberg LCG, Alifrangis M, Rodrigues OP, Kurtzhals JAL. 2008. Effect of concomitant artesunate administration and cytochrome P4502C8 polymorphisms on the pharmacokinetics of amodiaquine in Ghanaian children with uncomplicated malaria. Antimicrob Agents Chemother 52:4400–4406. 10.1128/AAC.00673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernaute-Lau L, Morris U, Msellem M, Mårtensson A, Björkman A, Gil JP. 2021. Influence of cytochrome P450 (CYP) 2C8 polymorphisms on the efficacy and tolerability of artesunate-amodiaquine treatment of uncomplicated Plasmodium falciparum malaria in Zanzibar. Malar J 20:90. 10.1186/s12936-021-03620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horinouchi M, Sakaeda T, Nakamura T, Morita Y, Tamura T, Aoyama N, Kasuga M, Okumura K. 2002. Significant genetic linkage of MDR1 polymorphisms at positions 3435 and 2677: functional relevance to pharmacokinetics of digoxin. Pharm Res 19:1581–1585. 10.1023/A:1020433422259. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Haba E, García MI, Cortejoso L, López-Lillo C, Barrueco N, García-Alfonso P, Alvarez S, Jiménez JL, Martín ML, Muñóz-Fernández MA, Sanjurjo M, López-Fernández LA. 2010. ABCB1 gene polymorphisms are associated with adverse reactions in fluoropyrimidine-treated colorectal cancer patients. Pharmacogenomics 11:1715–1723. 10.2217/pgs.10.159. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, Ahn J-H, Kim S-B, Jung KH, Yoon DH, Lee JS, Ahn S-H. 2012. Prospective evaluation of the drug-metabolizing enzyme polymorphisms and toxicity profile of docetaxel in Korean patients with operable lymph node-positive breast cancer receiving adjuvant chemotherapy. Cancer Chemother Pharmacol 69:1221–1227. 10.1007/s00280-011-1816-4. [DOI] [PubMed] [Google Scholar]
- 36.Kim H-S, Sunwoo YE, Ryu J-Y, Kang H-J, Jung H-E, Song I-S, Kim E-Y, Shim J-C, Shon J-H, Shin J-G. 2007. The effect of ABCG2 V12M, Q141K and Q126X, known functional variants in vitro, on the disposition of lamivudine. Br J Clin Pharmacol 64:645–654. 10.1111/j.1365-2125.2007.02944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguetse CN, Meyer CG, Adegnika AA, Agbenyega T, Ogutu BR, Kremsner PG, Velavan TP. 2016. Glucose-6-phosphate dehydrogenase deficiency and reduced haemoglobin levels in African children with severe malaria. Malar J 15:346. 10.1186/s12936-016-1396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Mkrtchian S, Kumondai M, Hiratsuka M, Lauschke VM. 2019. An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J 19:115–126. 10.1038/s41397-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, Edwards RJ, Boobis AR, Ingelman-Sundberg M. 2003. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos 31:755–761. 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 40.Lo KS, Wilson JG, Lange LA, Folsom AR, Galarneau G, Ganesh SK, Grant SFA, Keating BJ, McCarroll SA, Mohler ER, O’Donnell CJ, Palmas W, Tang W, Tracy RP, Reiner AP, Lettre G. 2011. Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum Genet 129:307–317. 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Tang H, Qayyum R, Schick UM, Nalls MA, Handsaker R, Li J, Lu Y, Yanek LR, Keating B, Meng Y, van Rooij FJA, Okada Y, Kubo M, Rasmussen-Torvik L, Keller MF, Lange L, Evans M, Bottinger EP, Linderman MD, Ruderfer DM, Hakonarson H, Papanicolaou G, Zonderman AB, Gottesman O, BioBank Japan Project, CHARGE Consortium, Thomson C, Ziv E, Singleton AB, Loos RJF, Sleiman PMA, Ganesh S, McCarroll S, Becker DM, Wilson JG, Lettre G, Reiner AP. 2013. Genome-wide association analysis of red blood cell traits in African Americans: the COGENT Network. Hum Mol Genet 22:2529–2538. 10.1093/hmg/ddt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauschke VM, Milani L, Ingelman-Sundberg M. 2017. Pharmacogenomic biomarkers for improved drug therapy—recent progress and future developments. AAPS J 20:4. 10.1208/s12248-017-0161-x. [DOI] [PubMed] [Google Scholar]
- 43.Rajman I, Knapp L, Morgan T, Masimirembwa C. 2017. African genetic diversity: implications for cytochrome P450-mediated drug metabolism and drug development. EBioMedicine 17:67–74. 10.1016/j.ebiom.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozyra M, Ingelman-Sundberg M, Lauschke VM. 2017. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med 19:20–29. 10.1038/gim.2016.33. [DOI] [PubMed] [Google Scholar]
- 45.Ingelman-Sundberg M, Mkrtchian S, Zhou Y, Lauschke VM. 2018. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genomics 12:26. 10.1186/s40246-018-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauschke VM, Ingelman-Sundberg M. 2016. Precision medicine and rare genetic variants. Trends Pharmacol Sci 37:85–86. 10.1016/j.tips.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Lauschke VM, Ingelman-Sundberg M. 2018. How to consider rare genetic variants in personalized drug therapy. Clin Pharmacol Ther 103:745–748. 10.1002/cpt.976. [DOI] [PubMed] [Google Scholar]
- 48.Lauschke VM, Ingelman-Sundberg M. 2020. Emerging strategies to bridge the gap between pharmacogenomic research and its clinical implementation. NPJ Genom Med 5:9. 10.1038/s41525-020-0119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cavaco I, Mårtensson A, Fröberg G, Msellem M, Björkman A, Gil JP. 2013. CYP2C8 status of patients with malaria influences selection of Plasmodium falciparum pfmdr1 alleles after amodiaquine-artesunate treatment. J Infect Dis 207:687–688. 10.1093/infdis/jis736. [DOI] [PubMed] [Google Scholar]
- 50.Nguetse CN, Adegnika AA, Agbenyega T, Ogutu BR, Krishna S, Kremsner PG, Velavan TP. 2017. Molecular markers of anti-malarial drug resistance in Central, West and East African children with severe malaria. Malar J 16:217. 10.1186/s12936-017-1868-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremsner PG, Adegnika AA, Hounkpatin AB, Zinsou JF, Taylor TE, Chimalizeni Y, Liomba A, Kombila M, Bouyou-Akotet MK, Mawili Mboumba DP, Agbenyega T, Ansong D, Sylverken J, Ogutu BR, Otieno GA, Wangwe A, Bojang KA, Okomo U, Sanya-Isijola F, Newton CR, Njuguna P, Kazungu M, Kerb R, Geditz M, Schwab M, Velavan TP, Nguetse C, Köhler C, Issifou S, Bolte S, Engleitner T, Mordmüller B, Krishna S. 2016. Intramuscular artesunate for severe malaria in African children: a multicenter randomized controlled trial. PLoS Med 13:e1001938. 10.1371/journal.pmed.1001938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duraisingh MT, Roper C, Walliker D, Warhurst DC. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol 36:955–961. 10.1046/j.1365-2958.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- 53.Yamanaka H, Nakajima M, Katoh M, Hara Y, Tachibana O, Yamashita J, McLeod HL, Yokoi T. 2004. A novel polymorphism in the promoter region of human UGT1A9 gene (UGT1A9*22) and its effects on the transcriptional activity. Pharmacogenetics 14:329–332. 10.1097/00008571-200405000-00008. [DOI] [PubMed] [Google Scholar]
- 54.Parikh S, Ouedraogo J-B, Goldstein JA, Rosenthal PJ, Kroetz DL. 2007. Amodiaquine metabolism is impaired by common polymorphisms in CYP2C8: implications for malaria treatment in Africa. Clin Pharmacol Ther 82:197–203. 10.1038/sj.clpt.6100122. [DOI] [PubMed] [Google Scholar]
- 55.Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE, PharmVar Steering Committee . 2018. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharmacol Ther 103:399–401. 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang B, Lauschke VM. 2019. Genetic variability and population diversity of the human SLCO (OATP) transporter family. Pharmacol Res 139:550–559. 10.1016/j.phrs.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 57.Seitz T, Stalmann R, Dalila N, Chen J, Pojar S, dos Santos Pereira JN, Krätzner R, Brockmöller J, Tzvetkov MV. 2015. Global genetic analyses reveal strong inter-ethnic variability in the loss of activity of the organic cation transporter OCT1. Genome Med 7:56. 10.1186/s13073-015-0172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korprasertthaworn P, Udomuksorn W, Yoovathaworn K. 2009. Three novel single nucleotide polymorphisms of UGT1A9 in a Thai population. Drug Metab Pharmacokinet 24:482–485. 10.2133/dmpk.24.482. [DOI] [PubMed] [Google Scholar]
- 60.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443. 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Machiela MJ, Chanock SJ. 2015. LDlink: a Web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31:3555–3557. 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl variant effect predictor. Genome Biol 17:122. 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. 2014. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980–D985. 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]