Abstract
Understanding how organisms adapt to new environments is a key problem in evolution, yet it remains unclear whether phenotypic plasticity generally facilitates or hinders this process. Here we studied evolved and plastic responses to water-stress in lab-born descendants of wild house mice (Mus musculus domesticus) collected from desert and non-desert environments and measured gene expression and organismal phenotypes under control and water-stressed conditions. After many generations in the lab, desert mice consumed significantly less water than mice from other localities, indicating that this difference has a genetic basis. Under water-stress, desert mice maintained more weight than non-desert mice, and exhibited differences in blood chemistry related to osmoregulatory function. Gene expression in the kidney revealed evolved differences between mice from different environments as well as plastic responses between hydrated and dehydrated mice. Desert mice showed reduced expression plasticity under water-stress compared to non-desert mice. Importantly, non-desert mice under water-stress generally showed shifts towards desert-like expression, consistent with adaptive plasticity. Finally, we identify several co-expression modules linked to phenotypes of interest. These findings provide evidence for local adaptation after a recent invasion and suggest that adaptive plasticity may have facilitated colonization of the desert environment.
Keywords: desert, plasticity, adaptation, Mus, gene expression
Introduction
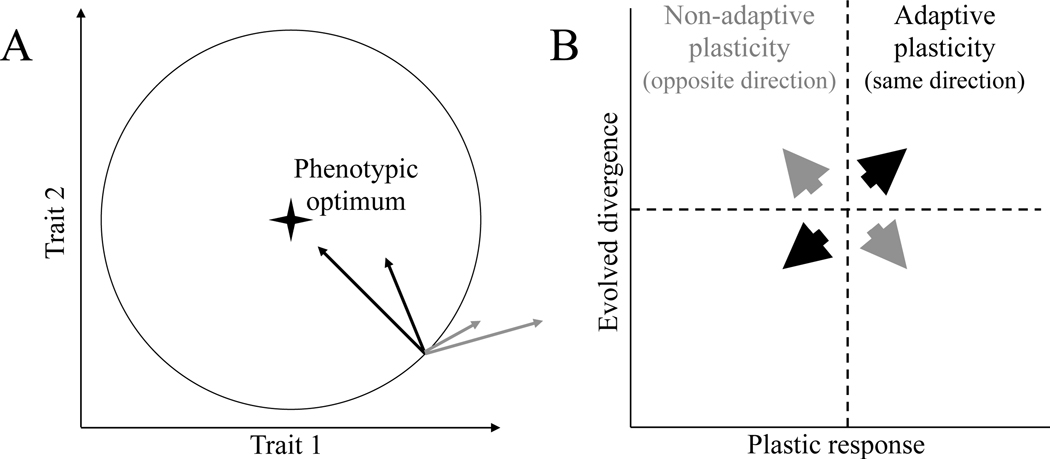
Understanding the origin and genetic architecture of complex traits associated with local adaptation is a central goal of evolutionary biology. One ongoing debate concerns the extent to which phenotypic plasticity may facilitate or constrain adaptation to new environments (Baldwin 1896; Price et al. 2003; Ghalambor et al. 2007; Levis and Pfennig 2016). Plasticity, defined as an environmentally induced phenotypic change, may be adaptive or non-adaptive when compared with a local phenotypic optimum. Adaptive plasticity, change that brings the phenotype closer to the local optimum, may enable organisms to invade new environments (Figure 1A). Subsequent genetically encoded changes in the same direction as the plastic changes may then accrue, bringing individuals even closer to the optimum, as seen for coloration in lizards living on dark substrates (Corl et al. 2018). Conversely, plastic changes may be non-adaptive if they move phenotypes farther from the local optimum. In such cases, selection is expected to favor genetic changes underlying phenotypes that go in the opposite direction of the plastic change and thereby bring the individual closer to the optimum. This pattern of non-adaptive plasticity is seen for gene expression changes in guppies reared in the absence of predators (Ghalambor et al. 2015). Which of these two outcomes is most likely remains unresolved and may depend both on the phenotype in question and the environmental heterogeneity to which populations have been exposed (e.g. Huang and Agrawal 2016).
Figure 1. Examples of adaptive and non-adaptive plasticity.
Illustration of adaptive and non-adaptive phenotypic plasticity in general (A) and for gene expression (B). (A) Plastic responses that bring individuals closer to the local optimum (black) are examples of adaptive plasticity. Conversely, responses that bring individuals farther from the local optimum (grey) reflect non-adaptive plasticity (modified from Gibert et al. 2019). The diagram depicts two dimensions of phenotype space, and the optimal combination of phenotypic values for a particular environment is shown. (B) Plastic changes in gene expression (X axis) represent changes within an individual (or line) when measured in an ancestral environment and a novel environment, while evolved changes in gene expression (Y axis) represent differences between individuals (or lines) when measured in the same environment. In this case, the evolved changes are assumed to bring the derived population closer to its optimum. Under this assumption, plastic and evolved responses will be positively correlated when plasticity is adaptive, and negatively correlated plasticity is non-adaptive (Ghalambor et al. 2015). Modified from Fischer et al. (2016). See methods for further details.
House mice (Mus musculus domesticus) provide an opportunity to study plastic and evolved changes in the context of adaptation to novel environments. House mice are native to western Europe but were recently introduced to the Americas with European colonization, approximately 400–600 generations ago (Phifer-Rixey and Nachman 2015). In this short time, they have colonized a wide variety of different environments. In eastern North America, house mice show strong evidence of local adaptation for several complex phenotypes such as body size, activity, and nest-building behavior (e.g. Lynch 1992; Mack et al. 2018; Phifer-Rixey et al. 2018). Recently, house mice have also invaded the Sonoran Desert of North America where they must contend with low to seasonally-absent water as well as extreme heat and aridity. Although house mice are human commensals, they frequently live in sheds, grain storage areas, barns, and other habitats where they are not well shielded from the environment. They can also live in situations where they are not associated with humans (Sage 1981). Consequently, colonization of the Sonoran Desert may have imposed strong selective pressure on house mice, potentially resulting in rapid adaptation to mitigate water stress.
Many previous studies have examined the behavioral, morphological, and physiological adaptations that allow desert mammals to persist under xeric conditions (reviewed in Schmidt-Nielsen 1964; Degen 1997; Donald and Pannabecker 2015). Recent work has also begun to identify some of the genes associated with these phenotypes (Marra et al. 2012, 2014; Wu et al. 2014; MacManes 2017; Giorello et al. 2018; Tigano et al. 2020). However, far less is known about the role of phenotypic plasticity in facilitating or impeding desert adaptation. Since all mice, including those living in more mesic environments, occasionally go through periods of water stress, selection may have favored plastic responses that enable mice to survive periods of water shortage (i.e. adaptive plasticity). Here we use house mice collected from the Sonoran Desert and mice from non-xeric environments to ask (1) whether house mice from the desert show genetically based differences in kidney gene expression and organismal phenotypes when compared to mice from other environments in a common lab setting, and (2) whether plastic responses to water stress in house mice are adaptive.
To understand the contribution of plastic and evolved changes to the colonization of xeric environments, we studied lab-born descendants of wild mice from two populations: Edmonton, Canada and Tucson, Arizona, USA. The annual precipitation in Edmonton is 52% more than in Tucson, the average precipitation of the driest month in Edmonton is 300% more than in Tucson, and these two locations differ dramatically in average temperature (Figure S1). Using Edmonton mice as a proxy for the non-desert-adapted ancestral state of North American house mice, we compared these to the putatively desert-adapted Tucson population to test three hypotheses. First, we reasoned that selection in the desert favored mice that were less dependent on free water. This hypothesis predicts that the progeny of mice from the Tucson population would show decreased water consumption in a common lab environment when compared with mice from other populations. Second, we hypothesized that mice from xeric and non-xeric environments would have differing plastic responses to water stress in a lab setting. In particular, we expected that mice from Tucson would have an attenuated physiological and transcriptional response to water deprivation as they must contend with a water-limited environment in nature, and have likely been exposed to much more regular periods of water deprivation than mice from non-desert populations. Third, we hypothesized that plasticity under conditions of water stress in the ancestral population may have been adaptive and thereby facilitated the colonization of the desert environment. This hypothesis predicts that the direction of plastic gene expression change (i.e. increased expression or decreased expression) in the ancestral population should correlate with the direction of evolved gene expression differences between the ancestral population and the desert-adapted population (Figure 1B). Finally, we used weighted gene co-expression network analysis (Langfelder and Horvath 2008) to associate phenotypic variation with transcriptional variation. We show that in a common environment, Tucson mice drank less water than Edmonton mice and differed in blood chemistry and gene expression in the kidney. These same traits exhibited significant plasticity when mice were hydrated compared to mice under water stress, although Tucson mice exhibited less plasticity than Edmonton mice. Evolved differences were generally in the same direction as plastic differences, both for gene expression and for organism-level phenotypes. Finally, co-expression networks identify groups of genes whose patterns of expression are associated with phenotypic variation measured in this study and differences between xeric and non-xeric individuals. These findings suggest an important role for adaptive plasticity in the colonization of the desert environment.
Materials and Methods
Mice
To assess whether mice from the Sonoran Desert differ in water consumption compared to mice from other habitats, we used wild-derived mouse lines developed in our lab from a range of localities in the Americas. Wild house mice were caught for these lines from five populations in different habitats and inbred through sib-sib mating over multiple generations as part of a larger project on the genetic basis of environmental adaptation in house mice (e.g. Phifer-Rixey et al. 2018; Suzuki et al. 2020). The five localities were Tucson, AZ, USA (TUC); Edmonton, Alberta, Canada (EDM); Gainesville, FL, USA (GAI); Saratoga Springs, NY, USA (SAR); and Manaus, Amazonas, Brazil (MAN). These locations represent a range of habitats and differ in a variety of environmental metrics (Figures S1A–E). In nearly all cases, lines were established from unrelated individuals. Lines were maintained in the laboratory for 6–19 generations. All mice were handled in accordance with a UC Berkeley Animal Care and Use protocol (protocol AUP-2016-03-8548-1).
Measuring water consumption
To determine whether desert Tucson mice consume less water than mice from more mesic environments, we measured water consumption over 72 hours in 163 adult males representing 45 different inbred lines (Table S1). In total, 40 individuals were measured from Tucson, 24 from Gainesville, 48 from Edmonton, 23 from Saratoga Springs, and 28 from Manaus, all between 90 and 200 days of age. Mice were housed at 23°C with a 10 hour dark and 14 hour light cycle on standard Teklad Global rodent chow (18% protein, 6% fat) with water ad libitum. During the assay, mice were house singly to record individual measurements. The amount of water consumed was measured by weighing the water apparatus (Hydropac® holder and Hydropac®) on a digital scale before and after a 72 hour period and calculating the difference. This was the only water the mouse had access to over the course of the experiment. Body weight was also measured on a digital scale and recorded. To account for population-level differences in body weight, we calculated relative water consumption (RWC, grams of water consumed per gram of mouse). We tested the relationship of body weight to water consumption using Pearson’s correlation, and we compared RWC among populations using a Kruskal-Wallis test followed by post hoc pairwise comparisons with a Mann-Whitney U test implemented in R.
Quantifying response to dehydration
To study evolved and plastic responses to xeric conditions, we chose one wild-derived inbred line each from Tucson and Edmonton [Tucson: TUSA4xA8 (TUCC/Nach), Edmonton: TAS111×165 (EDMA/Nach)]. These lines were chosen because they showed large differences in water consumption between lines as well as little variance between individuals within lines. Males from the same litter were assigned at random to either control or water restriction treatments and housed individually post-weaning according to standard husbandry procedures. This was done to remove the effects of dominance between male siblings, which often results in differing body weights between individuals. After 90 days of age, mice were weighed and phenotyped for relative water consumption as above. Following phenotyping (average age = 99 days), mice assigned to the water restriction treatment (Edmonton n=11, Tucson n=7) were restricted from all water consumption for 72 hours. Mice assigned to the control treatment (Edmonton n=10, Tucson n=6) were maintained with water ad libitum. All mice were weighed every 24 hours and monitored for declining health. Weight loss is a classic marker of condition used to monitor health in husbandry settings and has been used to compare the overall effects of dehydration (Haines and Schmidt-Nielsen 1967; MacMillen and Lee 1967; Foltz and Ullman-Cullere 1998; Bekkevold et al. 2013). All experiments with animals were conducted in accordance with a protocol approved by the UC Berkeley Animal Care and Use Committee (protocol AUP-2017–05-9940). After 72 hours, mice were sacrificed with isofluorane, and the left kidney, liver, and caecum were immediately removed and stored in RNAlater according to manufacturer’s instructions.
mRNA library preparation and sequencing
RNA was extracted from half of a kidney preserved in RNAlater from twenty mice total (five mice per population per treatment). The kidney was dissected longitudinally in an attempt to minimize differences in cellular composition between samples of this heterogenous tissue. A MoBio Laboratories Powerlyzer Ultraclean Tissue & Cells RNA Isolation Kit was used to extract RNA. RNA libraries were prepared using the KAPA Hyper Prep Kit and then pooled and sequenced across two lanes of 100bp PE Illumina HiSeq4000 at the Vincent J. Coates Genomics Sequencing Center at UC Berkeley.
mRNA read mapping and quantification of gene expression differences after water stress
Reads were trimmed for quality and adaptor contamination with Trimmomatic v0.36 (Bolger et al. 2014) and mapped to the Mus musculus reference genome (GRCm38/mm10) using STAR v2.6.0c (Dobin et al. 2013). Reads overlapping exons were counted using the program HTSeq 0.6.1 (Anders et al. 2015) to estimate per-gene mRNA abundance. We removed genes with a mean fewer than ten reads across samples and normalized read counts with DESeq2 for future analyses.
To quantify expression changes, the R package DESeq2 (Love et al. 2014) was used to test for differential expression between control and water-stressed individuals from each population. Genes were retained as significant at a false-discovery rate of 5%. Independent contrasts between control and water-stressed individuals in each population (i.e., hydrated-Tucson vs. dehydrated-Tucson and hydrated-Edmonton vs. dehydrated-Edmonton) allowed us to compare the magnitude of the transcriptional responses between populations. To compare the proportion of genes that were differentially expressed in response to water restriction in each population, we used a Chi-square test with Yates Correction.
Identifying adaptive vs. non-adaptive plasticity in gene expression
To understand whether plastic changes in gene expression under conditions of water stress are adaptive or non-adaptive, we compared plastic changes in Edmonton mice to evolved expression differences between Edmonton and Tucson mice. The logic underlying this comparison assumes that the Edmonton mice represent a population closer to the ancestral (non-desert) condition with more frequent exposure to free water, whereas the evolved differences seen in the Tucson mice represent changes that have brought the population closer to its optimum following colonization of the desert. If gene expression plasticity is adaptive, then plastic changes in gene expression in the ancestral population should be in the same direction as the evolved differences between populations. In contrast, if gene expression plasticity is non-adaptive, then plastic changes in gene expression in the ancestral population should be in the opposite direction as the evolved differences between populations (Ghalambor et al. 2015). These alternative scenarios are depicted in Figure 1B. In these comparisons, direction refers to whether a gene shows increased expression or decreased expression compared to hydrated Edmonton mice. First, we identified genes that were differentially expressed between hydrated Tucson mice and hydrated Edmonton mice. For these genes, we compared the log2 fold change in expression between dehydrated and hydrated Edmonton mice (EDM dehydrated/ EDM hydrated) to the log2 fold change in expression between hydrated Tucson mice and hydrated Edmonton mice (TUC hydrated/ EDM hydrated). Since this comparison involves two ratios that share a common denominator, correlations may arise by chance. To account for this, we randomly chose two different sets of two replicates of the hydrated Edmonton mice for each comparison. Consequently, the numerator of each axis was divided by averages of different replicates ensuring that the denominators were different between the two axes. We also compared the observed correlation to a permuted distribution to assess whether the correlation between plastic and evolved changes exceeded that of a random set of genes. We randomly sampled 490 genes with expression in the kidney 1,000 times and tested for the correlation between plastic and evolved divergence (log2(TUC hydrated/ EDM hydrated) vs. log2(TUC hydrated/ EDM hydrated)) to create a distribution for comparison to the observed correlation (Pearson’s correlation). As in the above test, we randomly chose two different sets of two replicates of the hydrated Edmonton mice for each comparison to avoid spurious correlations.
Measuring morphological and physiological phenotypes in response to water stress
To better understand the physiological and morphological consequences of both evolved and plastic changes in these lines, we measured circulating serum solute levels and aspects of kidney morphology. Following euthanasia, blood was extracted from the heart and body cavity using a syringe and centrifuged in BD SST Microtainer tubes. To separate serum, blood was left undisturbed for 30 minutes and then spun for 15 minutes in a centrifuge at >3500RPM. Serum was then frozen. Levels of blood urea nitrogen (BUN), total protein, creatinine, chloride, potassium, and sodium levels were analyzed at the UC Davis Comparative Pathology Laboratory for 20 mice (five individuals per population per treatment) using a Roche Diagnostics Cobas Integra 400 Plus. For each solute, we compared levels among populations and treatments with a Kruskal-Wallis test. We followed these with post hoc pairwise comparisons using Mann-Whitney U tests. These serum solutes were chosen to quantify kidney health, glomerular function, and general effects of dehydration in treated and control samples.
To measure morphological differences in the kidney, the right kidney was extracted following euthanasia, weighed, and stored in 10% neutral buffered formalin for 24 hours, then transferred to a 70% ethanol solution, and sent for morphological analysis to the UC Davis Comparative Pathology Laboratory. Kidneys from five hydrated mice per line (ten mice total) were processed for histopathology. These were embedded in paraffin blocks, sectioned 5 uM thick on glass slides and stained with Hematoxylin and Eosin. Renal cortical and medullary measurements were achieved by light microscopy (Olympus BX43) using Olympus cellSens Count and Measure software and the ratio of these was compared. Replicates of these measurements between populations were compared with a Mann-Whitney U test. We were interested in determining if there were gross morphological differences in the adult kidneys of healthy animals and thus used hydrated mice for this analysis to give a clear picture of evolved differences in kidney morphology between the two lines when not under water stress.
Gene co-expression analyses
To identify sets of genes with similar expression patterns and relate these to phenotypic differences across individuals, we used the program Weighted Gene Co-expression Network Analysis (WGCNA) to construct a co-expression network (Langfelder and Horvath 2008). Expression was normalized across individuals with DESeq2 and genes with fewer than 20 reads across all individuals were excluded. We constructed a gene co-expression network, represented by an adjacency matrix, which denotes co-expression similarity between pairs of genes among different individuals. Modules were identified using unsupervised clustering. Dissimilarly between clusters is measured based on topological overlap and defined by cutting branches off the dendrogram (Zhang and Horvath 2005; Langfelder et al. 2008). A soft-thresholding power of 8 was chosen based on the pickSoftThreshold function in WGCNA (Figure S2). We chose a minimum module size of 30 genes. For clarity, we arbitrarily assigned each module a number for reference throughout the text.
To associate modules with traits of interest, we then tested for correlations between eigengenes (the first principle component of a module) and each of the nine phenotypes described (RWC, kidney weight, proportion of weight maintained, serum BUN, serum creatinine, serum total protein, serum chloride, serum potassium, and serum sodium) as well as population of origin and treatment group using Pearson’s correlations. Hub genes were identified by estimating each gene’s module membership. Module membership was estimated based on the correlation between that gene’s expression and the expression of the module eigengene (Langfelder and Horvath 2008). Genes where module membership was greater than 0.8 were considered “hub genes” for subsequent analyses (as in Mack et al. 2019).
To identify genes that show differential co-expression between Tucson and Edmonton we used the program DGCA (Differential Gene Correlation Analysis) (McKenzie et al. 2016), which calculates the average change in correlation between the two lines across all gene pairs
Enrichment analyses
GO category enrichment on gene sets of interests was performed with GOrilla (Eden et al. 2009) by testing the foreground set against the background set of all genes expressed in the kidney. Phenotype enrichment tests were performed with modPhea (Weng and Liao 2017) by comparing the foreground set against the background set of all genes expressed in the kidney.
Results
Relative water consumption is lowest in mice from the Sonoran Desert
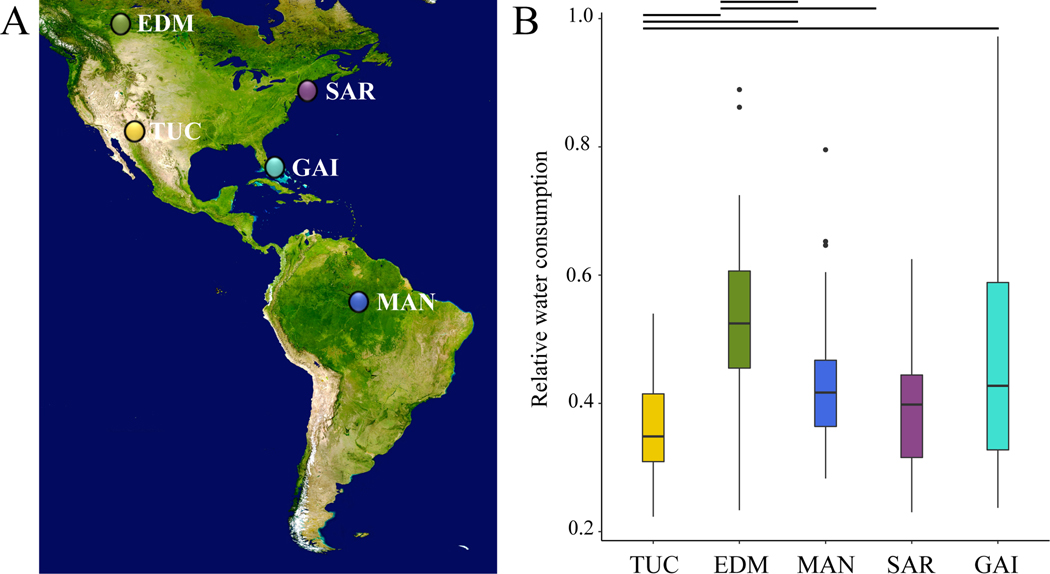
To determine whether mice from Tucson, Arizona consume less water compared to mice from other populations, we took advantage of a set of wild-derived inbred lines of mice from five localities across the Americas (Figure 2A). We assayed body weight and amount of water consumed over 72 hours to determine relative water consumption (RWC) for 163 male mice representing 45 wild derived inbred lines from five founder populations: (Tucson (TUC), Edmonton (EDM), Saratoga Springs (SAR), Gainesville (GAI), and Manaus (MAN). Some lines from different populations differed in body weight, with significant differences between Tucson and both Manaus and Gainesville (Median (g): TUC: 22.34, SAR: 21.97, MAN: 17.84, GAI: 18.65, EDM: 21.79)(Figure S3). Water consumption was significantly correlated with body weight (R2 = 0.169, Pearson’s correlation, p = 0.03)(Figure S4), consistent with previous findings in house mice (Bachmanov et al. 2002). Mice from Tucson drank significantly less water than mice from any other population except Saratoga Springs (Median RWC: TUC: 0.34, SAR: 0.40, MAN: 0.42, GAI: 0.43, EDM: 0.52, Kruskal-Wallis test, p = 5×10−8)(Figure 2B). The greatest difference was seen between mice from Tucson and mice from Edmonton (Mann-Whitney U, p < 0.00001)(Figure 2B). For this reason, and because these lines are similar in body weight, we chose to focus on comparisons between lines from these two populations in all subsequent analyses.
Figure 2. Relative water consumption in lab-born descendants of wild mice from different environments.
(A) Sampling localities of wild-caught mice used to establish inbred lines in this study (map obtained from Google satelites.pro): Edmonton, Canada (EDM), Tucson, AZ (TUC), Gainesville, FL (GAI), Saratoga Springs, NY (SAR), and Manaus, Brazil (MAN). (B) Relative water consumption (g water consumed/ g mouse) in descendants of mice from different localities. Lines indicate comparisons that are significant (p < 0.05; Mann-Whitney U tests). Vertical lines denote 1.5x the interquartile range.
Desert house mice maintain more weight under water stress
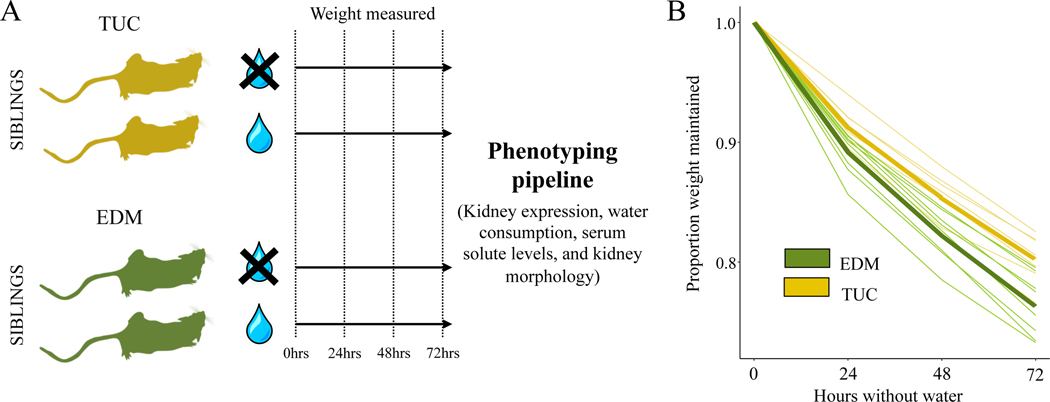
Since mice from Tucson drank less water on average than mice from other populations, we next asked whether these mice also showed attenuated responses to short-term water stress. Measuring weight loss in response to water stress is a classic method for assessing the response to dehydration, and desert animals typically lose less weight under water restriction (Shkolnik and Borut 1969; MacMillen and Hinds 1983).To compare the response to water stress in mice from a desert and non-desert environment, we compared the inbred line from Tucson with the lowest average water consumption (TUCC/Nach) to the inbred line from Edmonton with the greatest average water consumption (EDMA/Nach). This comparison between two lines revealed the same general pattern as seen above for population-level comparisons (i.e. Figure 2B), except that we observed lower variance among these age matched individuals from single inbred lines (Figure S5). We took male full-siblings from the same litter as mice from our hydrated comparison and withheld water from these mice for 72-hours, weighing individuals every 24-hours (Figure 3A). Hereafter, we refer to the water-restricted group as “dehydrated.”
Figure 3. Experimental design and amount of weight maintained after water stress in two populations.
(A) Schematic of the experimental design utilized in this study. Siblings from the same litter were split between control (hydrated) and treatment (dehydrated) trials. They were weighed every 24 hours for 72 hours and then put through a phenotyping pipeline. (B) Tucson mice maintain significantly more weight after 72 hours of water restriction (p=0.027) exhibiting an attenuated response to water stress.
While mice showed some signs of dehydration, they were alert and remained active throughout the experiment. We found that Tucson mice lost significantly less weight than Edmonton mice over the course of 72 hours without access to water (median proportion weight maintained: Tucson: 0.82, Edmonton: 0.78, Mann-Whitney U, p=0.027, Figure 3B). These results suggest that Tucson mice are less affected by dehydration and are more buffered against water stress.
Evolved and plastic transcriptional responses to xeric conditions
Changes in gene expression provide a flexible mechanism for rapidly responding to changes in the local environment, and can also underlie evolutionary divergence. Kidneys are the primary osmoregulatory organ and are essential for water and salt homeostasis and solute excretion. To identify expression differences that have arisen over short evolutionary timescales as well as plastic responses to water restriction, we sequenced mRNA from kidneys of ten Tucson and ten Edmonton mice, five from the dehydrated and five from the hydrated treatment. Differences in expression between Tucson and Edmonton mice in a common environment represent evolved differences, while differences between dehydrated and hydrated treatments represent a plastic response to water restriction.
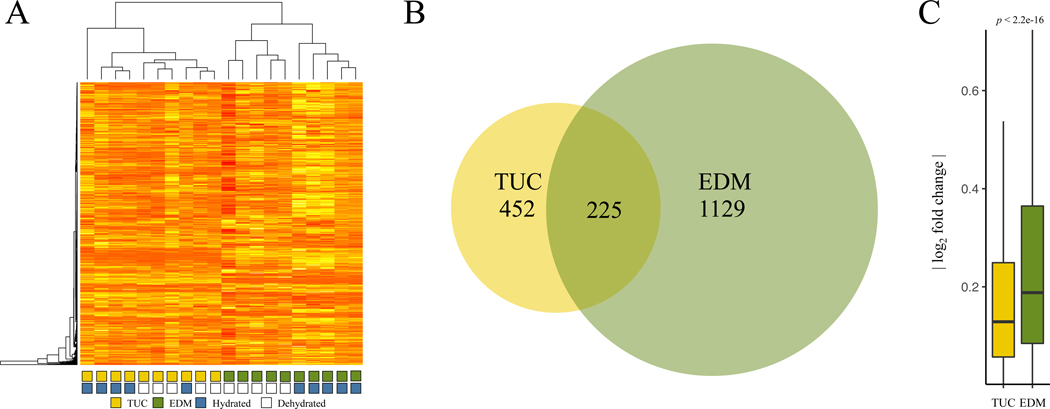
We sequenced a total of ~1.3 billion reads for an average of 26,406,068 uniquely mapped reads per sample which were used to quantify mRNA expression levels and differential expression between samples. 19,105 genes were expressed over a mean of ten reads per sample. Sampling the 1,000 genes with the greatest variance, we found that Tucson and Edmonton individuals clustered separately, indicating that more of the gene expression variation was partitioned between line-of-origin than between treatment groups (Figure 4A). Edmonton samples clustered into two distinct groups based on treatment (dehydrated vs. hydrated individuals), but dehydrated and hydrated Tucson samples did not form distinct clusters (Figure 4A). These patterns were also seen in a principal component analysis (PCA) based on expression co-variance (Figure S6) where PC1 (which accounts for 69% of the variance) reflects the difference between the two lines and PC2 (which accounts for 9% of the variance) largely accounts for differences associated with treatment type. Variance associated with treatment type was greater in the Edmonton line than in the Tucson line.
Figure 4. Evolved and plastic gene expression variation among hydrated and dehydrated mice from desert and non-desert environments.
(A) Heat map depicting relationships among samples for the top 1000 genes with greatest variance in expression. Expression patterns form two major groups, corresponding to line of origin (Tucson versus Edmonton). Edmonton samples also form clusters based on treatment (hydrated versus dehydrated) while Tucson samples do not. (B) Numbers of differentially expressed genes between dehydrated and hydrated samples in Tucson and Edmonton. Edmonton mice exhibit twice as many genes with differential expression between dehydrated and hydrated conditions compared to Tucson mice. The 225 genes at the intersection represent the shared transcriptional response to water stress. (C) Magnitude of fold changes in each population between dehydrated and hydrated samples. Vertical lines denote 1.5x the interquartile range.
To further characterize the transcriptional response of the Tucson and Edmonton lines to water stress, we used DESeq2 (Love et al. 2014) to perform independent pairwise contrasts between treatments (dehydrated vs. hydrated) within each line. Comparing the hydrated and dehydrated groups within each line (Tucson dehydrated vs. Tucson hydrated, and Edmonton dehydrated vs. Edmonton hydrated), we found that twice as many genes were differentially expressed in the Edmonton (1,354 genes) than in the Tucson comparisons (677 genes) (Chi-square test with Yates Correction, p < 0.0001, Figure 4B), with a 225 gene overlap. This 225 gene overlap represents the shared transcriptional response to dehydration with respect to these two lines. This group of genes is enriched for phenotypes including dehydration (q = 9×10−3) and decreased vasodilation (q =3.8×10−2) and GO terms involved in regulation of blood pressure (q = 4.99×10−2). Genes differentially expressed between hydrated and dehydrated Edmonton mice were also enriched for GO terms relevant to water stress, such as renal system processes (q = 5.08 × 10−2) and regulation of body fluids (q = 1.30×10−2). Within genes solely differentially expressed between the Tucson groups, we saw enrichment for homeostasis related GO terms (see File S1), but not for any kidney-specific categories. In addition to having a greater number of differentially expressed genes in the Edmonton comparison, we also found that the average magnitude of expression differences between hydrated and dehydrated treatments (|log2 fold change|) was significantly greater for Edmonton samples than for Tucson samples (Mann-Whitney U, p < 2.2 × 10−16) (Figure 4C). This result is consistent with an attenuated transcriptional response to short term water stress in desert Tucson mice.
Dehydrated Edmonton mice show shifts towards Tucson-like expression
The results presented above identified phenotypic divergence in Tucson mice consistent with adaptation to water-restricted environments, with Tucson mice drinking less water on average, maintaining more weight under water stress, and showing attenuated transcriptional responses to dehydration. Consequently, we asked whether plastic responses to water stress in non-xeric populations of house mice are adaptive or non-adaptive. Since plasticity is considered adaptive when a phenotype is altered in the same direction as under natural selection, we asked whether the transcriptional response of Edmonton mice under water stress is in the same or opposite direction as the expression divergence between Tucson and Edmonton mice (Figure 1B).
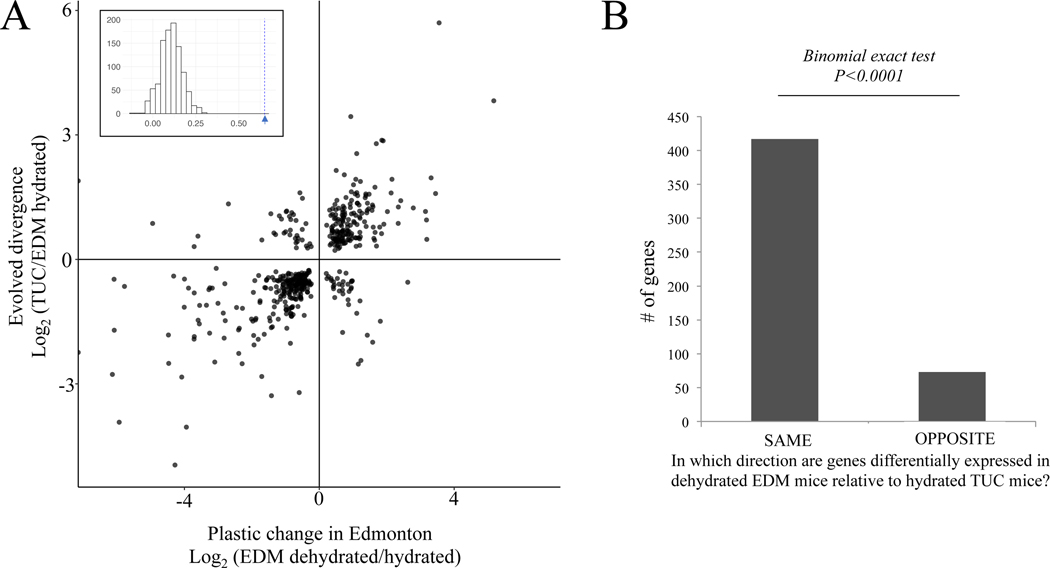
First, we used a pairwise contrast to identify genes that were differentially expressed between the Edmonton and Tucson lines (hydrated-Tucson vs. hydrated-Edmonton). This set of 3,935 genes represents evolved divergence between the Tucson and Edmonton lines. Of these genes, we asked how many showed evidence of a plastic response in Edmonton (i.e., were differentially expressed between hydrated and dehydrated Edmonton mice) in the same vs. opposite directions. For example, if a gene showed significantly higher expression in response to water stress in Edmonton mice and showed significantly higher expression in Tucson compared to Edmonton mice, the plastic and evolved response were said to be in the same direction. We found that the majority of these genes (85%, 416 genes of 490 genes) showed changes in the same direction [log2(hydrated-Tucson/hydrated-Edmonton) vs. log2(dehydrated-Edmonton /hydrated-Edmonton) Pearson’s correlation = 0.65, p < 2.2e-16]. Only 15% (74 genes) showed changes in opposite directions (Figure 5). We compared this result to a permutation test where we sampled a random set of 490 genes out of all genes 1,000 times. The observed correlation of genes showing evolved and plastic differences (Pearson’s correlation = 0.65) fell far above the distribution of permuted correlations of genes showing evolved and plastic differences (Permutation test, p < 0.001) (Figure 5A).
Figure 5. Plastic responses to dehydration in non-desert (Edmonton) mice are mostly in the same direction as evolved differences between non-desert (Edmonton) and desert (Tucson) hydrated mice.
(A) Scatterplot comparing the evolved and plastic changes in gene expression. Each point represents a gene and the log2 fold change between Edmonton dehydrated vs. hydrated on the x-axis (plastic response) and Tucson hydrated vs. Edmonton hydrated on the y-axis (evolved divergence). Inset shows the observed correlation coefficient (blue arrow) and the distribution of correlation coefficients from randomly sampled genes. See methods for further details. (B) Number of genes in which the evolved and plastic transcriptional responses go in the same direction, and number of genes in which the evolved and plastic transcriptional responses go in the opposite direction.
The group of 416 genes with evolved and plastic changes in the same direction was enriched for GO terms involved in ion transport (q = 0.03) and for mutant renal/urinary system phenotypes (q = 0.035). For example, one of these genes, Aquaporin 1 (Aqp1), showed differences in expression between hydrated and dehydrated Edmonton mice (q = 0.0016) and between the hydrated conditions of both lines (q = 0.00019)(Figure S7A). In Tucson mice, there was no significant effect of treatment on expression level, but in Edmonton mice, expression increased in response to water stress recapitulating the constitutive expression level of the mice from Tucson. Aquaporins are a family of membrane proteins which form channels used to transport water and small solutes across cell membranes. Aqp1 is expressed in the descending loop of Henle, and channels formed from this protein are the main route through which water is reabsorbed in this region (Chou et al. 1999). This gene is known to affect urine concentrating ability, response to dehydration, and renal water transport in lab lines of house mice (Ma et al. 1998; Sohara et al. 2005) and this protein family has been identified in a several studies related to desert adaptation in rodents (reviewed in Pannabecker, 2015).
Of the 416 genes where dehydrated Edmonton mice showed shifts towards Tucson-like expression, the majority (87%) were not differentially expressed between hydrated and dehydrated Tucson mice. For the 54 of these genes that were differentially expressed in the Tucson comparison, in all cases the plastic response was observed to change in the same direction as in Edmonton mice (i.e., increased or decreased in both lines in response to water stress).
Evolved and plastic phenotypic differences in blood chemistry associated with xeric conditions
Water consumption is a complex trait with many factors contributing to the ultimate phenotype. To understand plastic and evolved responses to water stress beyond gene expression changes, we next compared several blood chemistry measures under hydrated and dehydrated conditions. Most blood serum measures varied by population, treatment, or both (Kruskall-Wallis, BUN: p = 0.012, Creatinine: p = 0.07, Total Protein: p = 0.001, Chloride: p = 0.02, Potassium: p = 0.02, Sodium: p = 0.049; Figures 6 and S8). Hydrated mice from Tucson and Edmonton showed several differences in blood chemistry, some of which were correlated (Table S2). Tucson mice had higher chloride levels (median mmol/L: Tucson: 114.9, Edmonton: 112.5, p=0.03, Figure 6A). Chloride, an electrolyte, is a marker of dehydration and thus expected to be at higher concentrations in the blood of dehydrated animals (e.g., MacManes 2017). The increased levels of serum chloride in healthy Tucson mice suggests that under hydrated conditions their baseline kidney function differs from that of Edmonton mice and they may be able to function normally despite higher blood osmolarity.
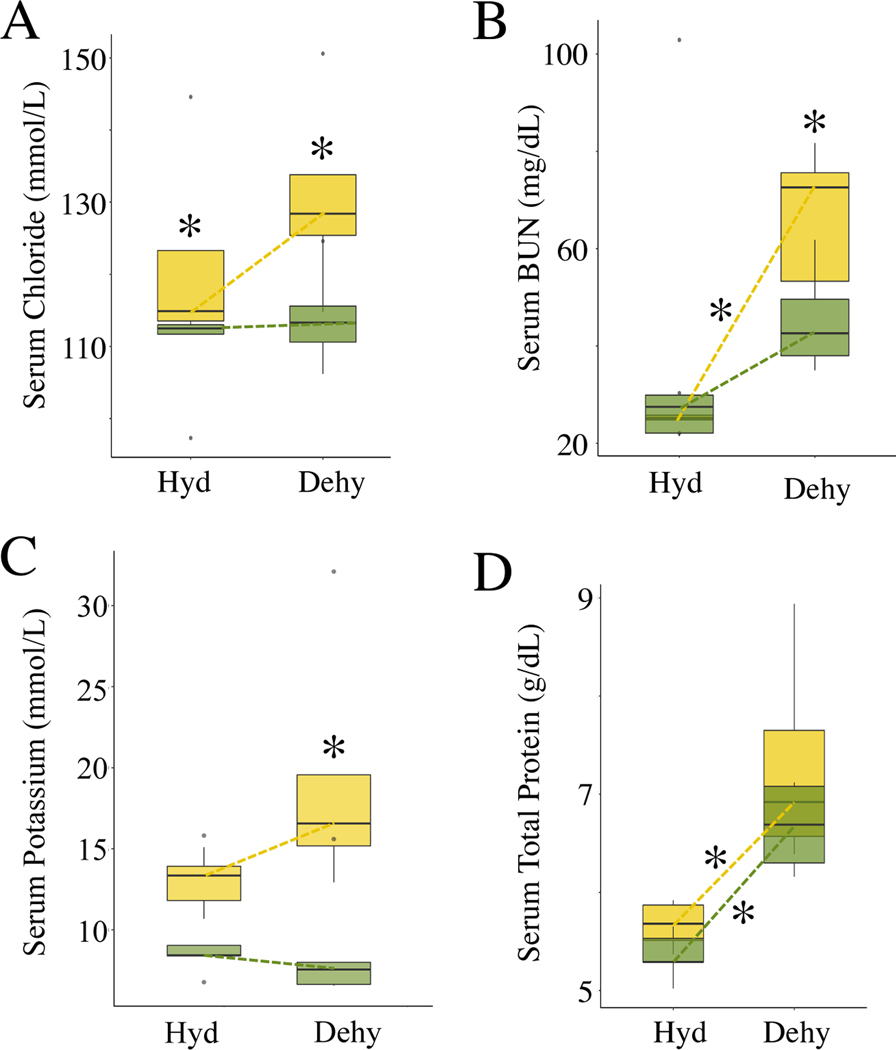
Figure 6. Evolved and plastic phenotypic variation among hydrated and dehydrated mice from desert and non-desert environments.
Reaction norms between hydrated and dehydrated states of both lines (Tucson, Edmonton) are shown for each phenotype. Dotted lines connecting the medians represent the pattern of phenotypic expression as a response to the treatment (hydrated, dehydrated). Asterisks denote significant comparisons (p < 0.05) either between lines (above boxes) or between treatments (next to dotted lines). (A) Tucson mice show higher levels of serum chloride than Edmonton mice, both when hydrated and when dehydrated (p=0.03 for both). (B) Significant differences in serum BUN between hydrated and dehydrated Tucson mice (p = 0.008) and dehydrated Tucson and Edmonton mice (p = 0.03). (C) Significant differences in serum potassium between dehydrated Tucson Edmonton mice (p = 0.03). (D) Significant differences in total protein between hydrated and dehydrated Tucson mice (p=0.008) and between hydrated and dehydrated Edmonton mice (p=0.01). Vertical lines denote 1.5x the interquartile range.
Comparing blood chemistry measures in dehydrated mice we found that Tucson mice measured higher in chloride (median mmol/L: Tucson: 129.60, Edmonton: 111.95, p=0.03, Figure 6A), BUN (median mg/dL: Tucson: 64.45, Edmonton: 43.80, p=0.03, Figure 6B), and potassium (median mmol/L: Tucson: 18.06, Edmonton: 7.78, p=0.03, Figure 6C) than Edmonton mice. BUN is a waste product from the liver during the metabolism of protein and increases as glomerular filtration rate and blood volume decreases (Baum et al. 1975). Similarly, high levels of serum potassium often reflect a decrease in filtration of the solute from the blood and thus decreased kidney function (Schwartz 1955) although this measure is particularly sensitive to lysed blood cells during collection and could reflect the challenge of collecting blood from dehydrated animals. Regardless of treatment, Tucson mice maintained higher serum chloride levels than Edmonton mice. In contrast, serum potassium levels were higher in Tucson mice only in the dehydrated treatment. High levels of both of these electrolytes are consistent with dehydration. Dehydrated mice from Tucson showed greater indicators for dehydration and kidney dysfunction than Edmonton mice but lost less weight when water stressed. This may reflect a greater evolved capacity to respond to the stress of dehydration through greater tolerance or lessened water requirements. That phenotypic differences in both hydrated and dehydrated animals persist in a common environment indicates that they have a genetic basis.
While differences between the Tucson and Edmonton lines represent evolved differences, differences between hydrated and dehydrated mice reflect plastic responses to water stress. Many of the traits that differed between lines also exhibited phenotypic plasticity in comparisons between hydrated and dehydrated mice within lines. Dehydrated Tucson mice had higher levels of serum BUN compared with hydrated Tucson mice (median mg/dL: Hydrated: 25.20, Dehydrated: 64.45, p=0.008, Figure 6B). BUN differed both between water stressed and control mice from Tucson and was higher than in Edmonton mice when water stressed. Dehydrated Edmonton mice had higher levels of serum sodium (mean mmol/L: Hydrated: 153, Dehydrated: 161, p=0.01, Figure S8A) than hydrated Edmonton mice, reaching the levels seen in Tucson hydrated and dehydrated animals. The only solute that responded to water stress in both lines was total protein (median g/dL: Edmonton: Hydrated: 5.29, Dehydrated: 6.89, p=0.01; Tucson: Hydrated: 5.68, Dehydrated: 7.29, p=0.008, Figure 6D). Total protein measures the concentration of both albumin and gobulin in the blood which increases with dehydration (Senay and Christensen 1965). These results indicate that while both lines react physiologically to the stress of dehydration, they may do this through different mechanisms.
Differences in kidney morphology
Edmonton mice had heavier kidneys relative to their body weight than Tucson mice (p = 0.00015) (Figure S9). There were no significant differences in the ratio of papilla to cortex thickness from the samples analyzed, however, the pattern follows that of many desert specialists with a higher ratio in Tucson individuals (Figure S10). The relative thickness of the papillae in the medulla to the cortex is correlated with urine concentrating ability; thicker medullas are often associated with animals inhabiting more arid climates (Sperber 1944; Beuchat 1990; Al‐kahtani et al. 2004). Anecdotally, mice from Tucson appeared to produce far less urine than mice from Edmonton, consistent with many desert rodents, but this was not measured in this study due to difficulty in obtaining urine from Tucson mice.
Co-expressed sets of genes are associated with phenotypic variation in Tucson and Edmonton mice
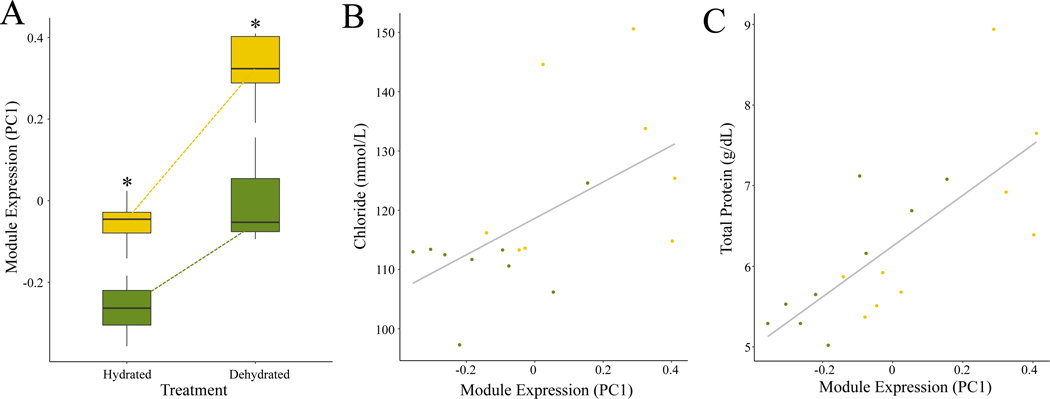
Finally, we sought to identify gene co-expression networks associated with the phenotypes measured in this study. To identify transcriptional networks associated with phenotypic variation in Tucson and Edmonton mice, we performed a weighted gene co-expression network analysis (WGCNA). This analysis uncovers genes with highly correlated expression profiles and groups them into modules. We identified 51 co-expression modules, 15 of which were associated with population, 12 of which were associated with treatment, and 30 of which were associated with blood chemistry phenotypes (Figure S11). Of these, module 7 (Figure S12) was of particular interest because it was significantly associated with eight different phenotypes (BUN, p=0.04; Creatinine, p=0.001; total protein, p = 2 × 10−4; Chloride, p = 0.01; Potassium, p = 0.006; weight loss, p = 0.003; RWC, p= 0.01; water consumption p=0.01) as well as population of origin (p = 0.005) and treatment (p = 4 × 10−4)(Figure 7). Genes in this module were enriched for several metabolic processes, including glyceraldehyde-3-phosphate metabolic process (q = 6.85 × 10−9), ribose phosphate metabolic process (q = 1.45× 10−8), and carbohydrate derivative metabolic process (q = 1.3 × 10−7). Visualizing the genes in module 7, we identified Tmtc1, Apoe, and Sult1a1 as the hub genes with the greatest number of connections (Figure S12). All three of these genes were identified in our previous analysis as genes for which dehydrated Edmonton mice showed shifts towards Tucson-like expression (Figure 5). Apoe is of particular interest because of its documented role in kidney function. It is thought to play an important role in renal damage protection (Wen et al. 2002; Bonomini et al. 2011), and laboratory mutants show a number physiological and morphological changes similar to kidney disease phenotypes (Bonomini et al. 2011). Apoe expression responded in the same direction and in a similar magnitude in both lines under water stress. Interestingly, under water stress, the expression level in Edmonton recapitulated the constitutive expression level of hydrated Tucson mice (Figure S7B).
Figure 7. Module 7 expression and associations with phenotypes of interest.
Expression of module 7 is associated with plastic and evolved responses to water stress as well as phenotypes of interest. (A) Expression levels comparing the evolved and plastic changes in response to water stress in TUC (green) and EDM (gold). Vertical lines denote 1.5x the interquartile range. (B) Chloride levels (mmol/L) are positively correlated with PC1 of module 7 expression (p = 0.01). (C) Total protein (g/dL) levels are positively correlated with PC1 of module 7 expression (p = 2 × 10−4).
In addition to shifts in the expression of entire co-expression modules, populations may differ as a consequence of altered co-expression between pairs of genes, called differential co-expression. In order to identify genes that show differential co-expression between Tucson and Edmonton, we calculated the average change in correlation between the two lines across all gene pairs using the program DGCA (Differential Gene Correlation Analysis) (McKenzie et al. 2016). We identified 182 genes that showed significantly different co-expression between Tucson and Edmonton individuals (q <0.10, see methods). These genes were enriched for several GO categories including glycolysis: generation of precursor metabolites and energy (q = 0.0013), cellular amino acid metabolic process (q = 0.00046), and lipid metabolic process (q = 0.0014). This gene set was also enriched for mutant phenotypes related to renal/urinary system (q = 0.00001), abnormal urine homeostasis (q = 0.005), and abnormal ion homeostasis (q = 0.01). One of the genes with significant changes in co-expression was Aqp1, which is involved in renal water transport. We also found that several solute carriers (Slc22a19, Slc11a2, Slc36a1, Slc47a1, Slc12a1, Slco3a1) showed evidence of differential co-expression, including one (Slc47a1) that has been shown to be under positive selection in the desert-adapted cactus mouse, Peromyscus eremicus (Kordonowy et al. 2017). Slc47a1 mouse mutants are also associated with increased BUN, increased circulating creatinine level, and kidney degeneration (Tsuda et al. 2009).
Discussion
Here we have described phenotypic and transcriptional divergence between descendants of mice from a desert environment and descendants of mice from a more mesic environment when reared under identical laboratory conditions. We also described plastic responses in these mice under conditions of water stress. First, we showed that inbred lines derived from the Sonoran Desert consume less water than do mice from other populations in the Americas. Next, comparisons between a single line from Tucson and a single line from Edmonton revealed many phenotypic differences in a common environment, both at the organismal level and at the level of gene expression in the kidney. The fact that these differences were present after multiple generations in the lab indicates that they are genetically based and represent divergence between these two populations. Nonetheless, these same traits reveal considerable plasticity in comparisons between control mice and mice under conditions of water stress. Notably, we found that Tucson mice showed attenuated responses to water stress compared to mice from Edmonton. After a 72 hour period without water, Tucson mice maintained more weight, a key indicator of condition, and showed less expression divergence in the kidney. Surprisingly, the blood chemistry of Tucson mice was consistent with higher levels of dehydration and reduced kidney function, both under standard and water-restricted conditions. This result may indicate that Tucson mice have developed a greater tolerance for water stress and thus have a reduced response to dehydration. Phenotypes that appear non-adaptive in one genomic or environmental context may be adaptive in another (e.g., Riddle et al., 2018). Thus the literature from humans and laboratory mouse models may provide an incomplete context for understanding similar phenotypes in wild mice. Altogether, the phenotypic divergence in desert and non-desert mice in a common lab environment are consistent with genetic changes to Tucson mice following their colonization of a desert environment.
Water consumption is a complex trait that is influenced by multiple factors and is known to vary among lab lines of house mice (Bachmanov et al. 2002). We observed the lowest water consumption in mice from Tucson, the locality with lowest annual rainfall, however, we did not see a general association between water consumption and average precipitation in other populations. This observation may reflect a threshold effect of water consumption or an interaction between free drinking water, aridity and temperature. Tucson mice and Edmonton mice differ in a variety of phenotypes (Figure 6), and the functional and ecological significance of these differences remains unclear. Importantly, while we compared two lines that show marked differences in water consumption, we did not measure additional non-desert lines for transcriptional divergence or plasticity of gene expression. Future work aimed at characterizing mice from a variety of habitats, including those from the ancestral range in Western Europe, would be useful for establishing links between gene expression plasticity and environmental variation. We did identify gene networks associated with phenotypes of interest in this study, providing testable hypotheses about the genes underlying the transcriptional response to both short term water stress and evolutionary responses to xeric environments. We note that causative mutations may lie in cis-regulatory regions of these genes or in upstream (trans- acting) regulators of these genes.
Phenotypic plasticity may allow animals to persist in new environments if the plastic responses bring individuals closer to the local optimum (reviewed by Ghalambor et al., 2007). Adaptive plasticity can be followed by genetic changes as populations become established, in a process called “genetic assimilation” or the “Baldwin Effect” (Waddington 1942, 1952, 1953; Simpson 1953; Robinson and Dukas 1999; Price et al. 2003). While we have no evidence of genetic assimilation in this study, we do observe that the plastic response to water stress in Edmonton mice appears to be adaptive. If this plasticity reflects the ancestral state, we hypothesize that plastic responses to water stress may have helped facilitate the initial colonization of the desert environment by house mice.
Our finding that plastic responses generally go in the same direction as evolved responses is within the context of other recent studies providing examples of adaptive and non-adaptive plasticity facilitating colonization. For example, an allele of the Epas1 gene confers adaptation to high altitude in Tibetans by attenuating the maladaptive plastic response of increased hemoglobin concentration (Beall et al. 2010; Simonson et al. 2010; Yi et al. 2010; Jeong et al. 2018), and maladaptive cardiac phenotypes are lessened in deer mice at high altitude (Velotta et al. 2018). Similarly, most gene expression changes in the brains of guppies reared in the absence of predators go in the opposite direction of those seen in populations that have evolved without predators (Ghalambor et al. 2015). In both cases, the selective agent (hypoxia in mammals or absence of predators in fish) may have not been present or present in the same way in the recent history of the populations exhibiting non-adaptive plasticity in these phenotypes. However, we speculate that occasional periods of water stress are probably common in many populations of mice, including in places, like Edmonton, that are not in deserts. Under such situations, selection is expected to favor an adaptive plastic response.
While adaptive plasticity may facilitate the colonization of new environments, it can also slow or impede adaptive evolution if, by moving individuals closer to the optimum, genetic variation is shielded from natural selection (Ghalambor et al. 2007; Price et al. 2003). However, when plastic responses to a new environment are incomplete, directional selection may favor a more extreme phenotype and lead to subsequent genetic changes (Price et al. 2003). While all house mice, even those in mesic environments, likely undergo short periods of water stress, water stress is likely to be more severe in desert environments. Phenotypic comparisons between Edmonton and Tucson mice suggest that plastic responses to water stress in Edmonton mice may be suboptimal. Tucson mice drink less water on average and lose less weight in response to dehydration, indicating that these animals are buffered against water stress in a way that Edmonton mice are not. The blood chemistry comparisons reported here also suggest there are differences between Tucson and Edmonton mice in kidney function and homeostasis. Consequently, we hypothesize that while phenotypic plasticity may have helped house mice colonize the Sonoran Desert, the xeric environment still imposed sufficient selective pressure for subsequent genetic changes leading to divergence from non-desert populations in the Americas.
Supplementary Material
Acknowledgements
We thank members of the Nachman Lab for valuable comments and discussions. We thank Taichi Suzuki, Megan Phifer-Rixey, Felipe Martins, Dana Lin, Michael Sheehan, and Mallory Ballinger for help with animal husbandry or for collecting the wild mice used to establish the lines for this study. We thank Lydia Smith of UC Berkeley and Eugene Dunn of UC Davis for their technical expertise. This work was facilitated by an Extreme Science and Engineering Discovery Environment (XSEDE) allocation to M.W.N. XSEDE is supported by National Science Foundation (NSF) grant number ACI-1548562. This work was supported by an NIH grant to MWN (R01 GM127468) and a NSF Doctoral Dissertation Improvement Grant to NKJB (1601827).
Footnotes
Data Accessibility Statement
Illumina sequencing data from this study is available is available through the NCBI Sequence Read Archive under accession PRJNA614581. Phenotype data has is available through Dryad (https://doi.org/10.6078/D14T49).
Conflict of Interest Statement
The authors declare that there are no conflicts of interest regarding publication of this research.
References
- Al‐kahtani MA, Zuleta C, Caviedes‐V idal E, and Garland Theodore Jr.. 2004. Kidney Mass and Relative Medullary Thickness of Rodents in Relation to Habitat, Body Size, and Phylogeny. Physiological and Biochemical Zoology 77:346–365. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov AA, Reed DR, Beauchamp GK, and Tordoff MG. 2002. Food Intake, Water Intake, and Drinking Spout Side Preference of 28 Mouse Strains. Behav Genet 32:435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM 1896. A New Factor in Evolution. The American Naturalist 30:441–451. [Google Scholar]
- Baum N, Dichoso CC, and Carlton CE. 1975. Blood urea nitrogen and serum creatinine: Physiology and interpretations. Urology 5:583–588. [DOI] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, Montgomery HE, Pan H, Robbins PA, Shianna KV, Tam SC, Tsering N, Veeramah KR, Wang W, Wangdui P, Weale ME, Xu Y, Xu Z, Yang L, Zaman MJ, Zeng C, Zhang L, Zhang X, Zhaxi P, and Zheng YT. 2010. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. PNAS 107:11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkevold CM, Robertson KL, Reinhard MK, Battles AH, and Rowland NE. 2013. Dehydration Parameters and Standards for Laboratory Mice. J Am Assoc Lab Anim Sci 52:233–239. [PMC free article] [PubMed] [Google Scholar]
- Beuchat CA 1990. Body size, medullary thickness, and urine concentrating ability in mammals. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology 258:R298–R308. [DOI] [PubMed] [Google Scholar]
- Bittner N, Mack K, and Nachman M. 2021. Data from: Gene expression plasticity and desert adaptation in house mice. Dryad Dataset. 10.6078/D14T49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomini F, Rodella LF, Moghadasian M, Lonati C, Coleman R, and Rezzani R. 2011. Role of apolipoprotein E in renal damage protection. Histochem. Cell Biol. 135:571–579. [DOI] [PubMed] [Google Scholar]
- Chou C-L, Knepper MA, van Hoek AN, Brown D, Yang B, Ma T, and Verkman AS. 1999. Reduced water permeability and altered ultrastructure in thin descending limb of Henle in aquaporin-1 null mice. J Clin Invest 103:491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corl A, Bi K, Luke C, Challa AS, Stern AJ, Sinervo B, and Nielsen R. 2018. The Genetic Basis of Adaptation following Plastic Changes in Coloration in a Novel Environment. Current Biology 28:2970–2977.e7. [DOI] [PubMed] [Google Scholar]
- Degen AA 1997. Water Requirements and Water Balance. Pp. 93–162 in Ecophysiology of Small Desert Mammals. Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald J, and Pannabecker TL. 2015. Osmoregulation in Desert-Adapted Mammals. Pp. 191–211 in Hyndman KA and Pannabecker TL, eds. Sodium and Water Homeostasis: Comparative, Evolutionary and Genetic Models. Springer, New York, NY. [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer EK, Ghalambor CK, and Hoke KL. 2016. Can a Network Approach Resolve How Adaptive vs Nonadaptive Plasticity Impacts Evolutionary Trajectories? Integr Comp Biol 56:877–888. [DOI] [PubMed] [Google Scholar]
- Foltz C, and Ullman-Cullere M. 1998. Guidelines for Assessing the Health and Condition of Mice. Lab Animal 28. [PubMed] [Google Scholar]
- Ghalambor CK, Hoke KL, Ruell EW, Fischer EK, Reznick DN, and Hughes KA. 2015. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525:372–375. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, McKAY JK, Carroll SP, and Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology 21:394–407. [Google Scholar]
- Gibert P, Debat V, and Ghalambor CK. 2019. Phenotypic plasticity, global change, and the speed of adaptive evolution. Current Opinion in Insect Science 35:34–40. [DOI] [PubMed] [Google Scholar]
- Giorello FM, Feijoo M, D’Elía G, Naya DE, Valdez L, Opazo JC, and Lessa EP. 2018. An association between differential expression and genetic divergence in the Patagonian olive mouse (Abrothrix olivacea). Molecular Ecology 27:3274–3286. [DOI] [PubMed] [Google Scholar]
- Haines H, and Schmidt-Nielsen K. 1967. Water Deprivation in Wild House Mice. Physiological Zoology 40:424–431. [Google Scholar]
- Huang Y, and Agrawal AF. 2016. Experimental Evolution of Gene Expression and Plasticity in Alternative Selective Regimes. PLOS Genetics 12:e1006336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Witonsky DB, Basnyat B, Neupane M, Beall CM, Childs G, Craig SR, Novembre J, and Rienzo AD. 2018. Detecting past and ongoing natural selection among ethnically Tibetan women at high altitude in Nepal. PLOS Genetics 14:e1007650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordonowy L, Lombardo KD, Green HL, Dawson MD, Bolton EA, LaCourse S, and MacManes MD. 2017. Physiological and biochemical changes associated with acute experimental dehydration in the desert adapted mouse, Peromyscus eremicus. Physiol Rep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, and Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, and Horvath S. 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24:719–720. [DOI] [PubMed] [Google Scholar]
- Levis NA, and Pfennig DW. 2016. Evaluating ‘Plasticity-First’ Evolution in Nature: Key Criteria and Empirical Approaches. Trends in Ecology & Evolution 31:563–574. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CB 1992. Clinal Variation in Cold Adaptation in Mus domesticus: Verification of Predictions from Laboratory Populations. The American Naturalist 139:1219–1236. [Google Scholar]
- Ma T, Yang B, Gillespie A, Carlson EJ, Epstein CJ, and Verkman AS. 1998. Severely Impaired Urinary Concentrating Ability in Transgenic Mice Lacking Aquaporin-1 Water Channels. J. Biol. Chem. 273:4296–4299. [DOI] [PubMed] [Google Scholar]
- Mack KL, Ballinger MA, Phifer-Rixey M, and Nachman MW. 2018. Gene regulation underlies environmental adaptation in house mice. Genome Res. 28:1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack KL, Phifer-Rixey M, Harr B, and Nachman MW. 2019. Gene Expression Networks Across Multiple Tissues Are Associated with Rates of Molecular Evolution in Wild House Mice. Genes 10:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManes MD 2017. Severe acute dehydration in a desert rodent elicits a transcriptional response that effectively prevents kidney injury. American Journal of Physiology-Renal Physiology 313:F262–F272. [DOI] [PubMed] [Google Scholar]
- MacMillen RE, and Hinds DS. 1983. Water Regulatory Efficiency in Heteromyid Rodents: A Model and Its Application. Ecology 64:152–164. [Google Scholar]
- MacMillen RE, and Lee AK. 1967. Australian desert mice: independence of exogenous water. Science 158:383–385. [DOI] [PubMed] [Google Scholar]
- Marra NJ, Eo SH, Hale MC, Waser PM, and DeWoody JA. 2012. A priori and a posteriori approaches for finding genes of evolutionary interest in non-model species: osmoregulatory genes in the kidney transcriptome of the desert rodent Dipodomys spectabilis (banner-tailed kangaroo rat). Comp. Biochem. Physiol. Part D Genomics Proteomics 7:328–339. [DOI] [PubMed] [Google Scholar]
- Marra NJ, Romero A, and DeWoody JA. 2014. Natural selection and the genetic basis of osmoregulation in heteromyid rodents as revealed by RNA-seq. Molecular Ecology 23:2699–2711. [DOI] [PubMed] [Google Scholar]
- McKenzie AT, Katsyv I, Song W-M, Wang M, and Zhang B. 2016. DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Systems Biology 10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannabecker TL 2015. Aquaporins in Desert Rodent Physiology. The Biological Bulletin 229:120–128. [DOI] [PubMed] [Google Scholar]
- Phifer-Rixey M, Bi K, Ferris KG, Sheehan MJ, Lin D, Mack KL, Keeble SM, Suzuki TA, Good JM, and Nachman MW. 2018. The genomic basis of environmental adaptation in house mice. PLOS Genetics 14:e1007672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, and Nachman MW. 2015. Insights into mammalian biology from the wild house mouse Mus musculus. eLife 4:e05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TD, Qvarnström A, and Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proceedings of the Royal Society of London. Series B: Biological Sciences 270:1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle MR, Aspiras AC, Gaudenz K, Peuß R, Sung JY, Martineau B, Peavey M, Box AC, Tabin JA, McGaugh S, Borowsky R, Tabin CJ, and Rohner N. 2018. Insulin resistance in cavefish as an adaptation to a nutrient-limited environment. Nature 555:647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BW, and Dukas R. 1999. The Influence of Phenotypic Modifications on Evolution: The Baldwin Effect and Modern Perspectives. Oikos 85:582–589. [Google Scholar]
- Sage RD 1981. Wild Mice. Pp. 39–90 in Foster HL, Small J D., and Fox JG, eds. The Mouse in Biomedical Research: History, Genetics, and Wild Mice. Academic Press: New York. [Google Scholar]
- Schmidt-Nielsen K 1964. Desert Animals: Physiological Problems of Heat and Water. Dover Publications, New York. [Google Scholar]
- Schwartz WB 1955. Potassium and the Kidney. N Engl J Med 253:601–608. [DOI] [PubMed] [Google Scholar]
- Senay LC, and Christensen ML. 1965. Changes in blood plasma during progressive dehydration. Journal of Applied Physiology 20:1136–1140. [Google Scholar]
- Shkolnik A, and Borut A. 1969. Temperature and Water Relations in Two Species of Spiny Mice (Acomys). J Mammal 50:245–255. [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, Prchal JT, and Ge R. 2010. Genetic Evidence for High-Altitude Adaptation in Tibet. Science 329:72–75. [DOI] [PubMed] [Google Scholar]
- Simpson GG 1953. The Baldwin Effect. Evolution 7:110–117. [Google Scholar]
- Sohara E, Rai T, Miyazaki J, Verkman AS, Sasaki S, and Uchida S. 2005. Defective water and glycerol transport in the proximal tubules of AQP7 knockout mice. American Journal of Physiology-Renal Physiology 289:F1195–F1200. [DOI] [PubMed] [Google Scholar]
- Sperber I 1944. Studies on the mammalian kidney. Zool Bild Uppsala 22:249–431. [Google Scholar]
- Suzuki TA, Martins FM, Phifer‐Rixey M, and Nachman MW. 2020. The gut microbiota and Bergmann’s rule in wild house mice. Molecular Ecology 29:2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigano A, Colella JP, and MacManes MD. 2020. Comparative and population genomics approaches reveal the basis of adaptation to deserts in a small rodent. Molecular Ecology 29:1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Isaka Y, Takahara S, and Horio M. 2009. Discrepancy between serum levels of low molecular weight proteins in acute kidney injury model rats with bilateral ureteral obstruction and bilateral nephrectomy. Clin Exp Nephrol 13:567–570. [DOI] [PubMed] [Google Scholar]
- Velotta JP, Ivy CM, Wolf CJ, Scott GR, and Cheviron ZA. 2018. Maladaptive phenotypic plasticity in cardiac muscle growth is suppressed in high‐altitude deer mice. Evolution 72:2712–2727. [DOI] [PubMed] [Google Scholar]
- Waddington CH 1942. Canalization of Development and the Inheritance of Acquired Characters. Nature 150:563–565. [DOI] [PubMed] [Google Scholar]
- Waddington CH 1953. Genetic Assimilation of an Acquired Character. Evolution 7:118–126. [Google Scholar]
- Waddington CH 1952. Selection of the Genetic Basis for an Acquired Character. Nature 170:71–71. [DOI] [PubMed] [Google Scholar]
- Wen M, Segerer S, Dantas M, Brown PA, Hudkins KL, Goodpaster T, Kirk E, LeBoeuf RC, and Alpers CE. 2002. Renal Injury in Apolipoprotein E–Deficient Mice. Laboratory Investigation 82:999–1006. [DOI] [PubMed] [Google Scholar]
- Weng M-P, and Liao B-Y. 2017. modPhEA: model organism Phenotype Enrichment Analysis of eukaryotic gene sets. Bioinformatics 33:3505–3507. [DOI] [PubMed] [Google Scholar]
- Wu H, Guang X, Al-Fageeh MB, Cao J, Pan S, Zhou H, Zhang L, Abutarboush MH, Xing Y, Xie Z, Alshanqeeti AS, Zhang Y, Yao Q, Al-Shomrani BM, Zhang D, Li J, Manee MM, Yang Z, Yang L, Liu Y, Zhang J, Altammami MA, Wang S, Yu L, Zhang W, Liu S, Ba L, Liu C, Yang X, Meng F, Wang S, Li L, Li E, Li X, Wu K, Zhang S, Wang J, Yin Y, Yang H, Al-Swailem AM, and Wang J. 2014. Camelid genomes reveal evolution and adaptation to desert environments. Nature Communications 5:1–10. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Asan, Ni P, Tian G, Xu J, Liu X, Jiang T, Wu R, Zhou G, Tang M, Qin J, Wang T, Feng S, Li G, Huasang, Luosang J, Wang W, Chen F, Wang Y, Zheng X, Li Z, Bianba Z, Yang G, Wang X, Tang S, Gao G, Chen Y, Luo Z, Gusang L, Cao Z, Zhang Q, Ouyang W, Ren X, Liang H, Zheng H, Huang Y, Li J, Bolund L, Kristiansen K, Li Y Zhang Y, Zhang X, Li R, Li S, Yang H, Nielsen R, Wang J, and Wang J 2010. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, and Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article 17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.