Abstract
Plant association with arbuscular mycorrhizal fungi (AMF) can increase their ability to overcome multiple stresses, but their impact on plant interactions with herbivorous insects is controversial. Here we show higher mortality of the leaf-chewer Spodoptera exigua when fed on tomato plants colonized by the AMF Funneliformis mosseae, evidencing mycorrhiza-induced resistance. In search of the underlying mechanisms, an untargeted metabolomic analysis through ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS) was performed. The results showed that mycorrhizal symbiosis had a very limited impact on the leaf metabolome in the absence of stress, but significantly modulated the response to herbivory in the damaged area. A cluster of over accumulated metabolites was identified in those leaflets damaged by S. exigua feeding in mycorrhizal plants, while unwounded distal leaflets responded similar to those from non-mycorrhizal plants. These primed-compounds were mostly related to alkaloids, fatty acid derivatives and phenylpropanoid-polyamine conjugates. The deleterious effect on larval survival of some of these compounds, including the alkaloid physostigmine, the fatty acid derivatives 4-oxododecanedioic acid and azelaic acid, was confirmed. Thus, our results evidence the impact of AMF on metabolic reprograming upon herbivory that leads to a primed accumulation of defensive compounds.
Keywords: Arbuscular mycorrhiza, defence priming, herbivory, metabolomics, mycorrhiza induced resistance, Spodoptera exigua
Mycorrhizal symbiosis primes the accumulation of several antiherbivore compounds in response to Spodoptera exigua attack, increasing larval mortality.
Introduction
Plants are highly dynamic systems that are able to react to, and interact with a broad range of organisms that may have an impact on plant health (Mendes et al., 2013; Gruden et al., 2020; Pozo et al., 2020). Among the many positive interactions with soil-borne organisms, the mutualistic association with arbuscular mycorrhizal fungi (AMF), known as arbuscular mycorrhizal (AM) symbiosis, deserves special attention. Due to the obligate biotrophy of these fungi, host plants are required to provide them with photosynthates and lipids for the development, maintenance and function of mycorrhizal structures (Bago et al., 2000; Keymer et al., 2017). In return, the AMF assist the host plant in the acquisition of water and mineral nutrients, becoming extremely important for the uptake of inorganic phosphate, nitrogen and various micronutrients (Smith and Smith, 2011; Hodge and Storer, 2015; Ezawa and Saito, 2018). In addition, AM symbiosis also result in an enhanced ability of the host plant to overcome adverse conditions, including abiotic and biotic stress factors (Sánchez-Bel et al., 2016; He et al., 2017; Rivero et al., 2018, Fracasso et al., 2020; Miozzi et al., 2020; Sanmartín et al., 2020a; b).
The association of plants with AMF and other beneficial microbes can stimulate the plant’s immune system, rendering the plant more resistant to the attack by different aggressors. This ‘alert’ state is known as induced systemic resistance (ISR; Choudhary et al., 2007; Pieterse et al., 2014). Usually, ISR implies a faster and more efficient activation of the plant defence responses upon pathogen or pest attacks. Thus, defence priming is an intrinsic part of ISR, conditioning the plants for the super activation of defences against environmental challenges (Martínez-Medina et al., 2016; Mauch-Mani et al., 2017). Priming is a low-cost defensive strategy since plant defences are not (or only slightly) activated in the absence of stress, but they are strongly triggered in response to a challenge (Mauch-Mani et al., 2017).
The ISR achieved in mycorrhizal plants is known as mycorrhiza-induced resistance (MIR; Pozo and Azcón-Aguilar, 2007). Several reports have demonstrated the functionality of MIR in protecting plants against a wide range of belowground aggressors such as soil-borne pathogens, nematodes or root-chewing insects (Currie et al., 2011; Jung et al., 2012; Schouteden et al., 2015; Olowe et al., 2018; Pozo et al., 2020). However, contrasting results have been reported regarding the effect of mycorrhiza on aboveground attackers, and the efficiency of MIR seems to be related to the lifestyle and feeding guild of the aggressors (Pozo and Azcón-Aguilar, 2007). Indeed, MIR is generally accepted to be effective mainly against necrotrophic pathogens and generalist leaf-chewing insects (Hartley and Gange, 2009; Jung et al., 2012; Roger et al., 2013; Song et al., 2013; Nair et al., 2015; Sánchez-Bel et al., 2016; He et al., 2017). Remarkably, those organisms are usually sensitive to the plant defences regulated by jasmonic acid (JA) signalling. Therefore, the priming of JA-dependent responses in AM plants has been proposed as the main molecular mechanism underlying MIR (Jung et al., 2012; Song et al., 2013; Minton et al., 2016; He et al., 2017).
JA signalling is a key regulator of plant defences against chewing herbivores (Erb and Reymond, 2019; Gruden et al., 2020), often in synergy with abscisic acid (ABA)-related signalling (Vos et al., 2013). For example, proteinase inhibitors (PI), antifeedant proteins that negatively impact the development of herbivores, as well as several toxic specialized metabolites like alkaloids or glucosinolates, are known to be induced after herbivory in a JA-dependent manner (Orozco-Cardenas et al., 1993; Wasternack and Hause, 2013; Erb and Reymond, 2019). The oxylipin pathway, which is responsible of the biosynthesis of JA and its derivatives, is positively regulated by mycorrhization in tomato roots (López-Ráez et al., 2010; Fernández et al., 2014; Rivero et al., 2015). Tomato plants colonized by the AMF Funneliformis mosseae negatively affected the performance of larvae from the generalist herbivore Helicoverpa armigera (Song et al., 2013); the mycorrhizal plants displayed JA-primed accumulation and a stronger induction in the expression of several genes of the oxylipin pathway, after the herbivore attack.
Besides the study of JA and its marker genes, the potential effect of AM establishment in the rearrangement of plant secondary metabolism after herbivory, has not been explored. Most studies dealing with herbivory have focused on targeted analysis towards secondary metabolites with known toxicity against herbivores, such as some alkaloids or iridoid glycosides (Andrade et al., 2013; Tomczak et al., 2016). However, a global overview of the metabolic reprogramming in AM plants in response to herbivory is still missing.
Untargeted metabolomics approaches have significantly contributed to understanding the impact of AM symbiosis on the host plant. Remarkable changes on the metabolomic profile have been evidenced in mycorrhizal roots (Schliemann et al., 2008; Laparre et al., 2014, Rivero et al., 2015; 2018, Saia et al., 2015). In tomato, untargeted metabolomics revealed up-regulation of the oxylipin and phenylpropanoid biosynthetic pathways in mycorrhizal roots (Rivero et al., 2015; 2018). This methodology has also been applied to identify potential primed compounds that can mediate the enhanced stress tolerance achieved in mycorrhizal plants (Rivero et al., 2018, Tugizimana et al., 2018). We previously identified several primed metabolites involved in the increased tolerance of mycorrhizal tomato against salinity stress, such as B6 vitamers or the flavonoid catechin (Rivero et al., 2018). Similarly, phenolic compounds, lipids, and sugars have been shown to be modulated in response to drought in mycorrhizal roots (Bernardo et al., 2019).
The modulation of the metabolome by AM in aboveground tissues has been less explored, and its impact seems to be highly dependent on the plant species (Schweiger et al., 2014). The impact of AM on the metabolic profiles in shoots seems to be much lower than in roots. In fact, Senecio jacobea plants colonized by Rhizoglomus irregulare showed significant changes in the root metabolome while no alteration was described in the shoots (Hill et al., 2018). Remarkably, a combination of untargeted and targeted metabolomics has recently allowed the identification of a group of blumenol derivates that seem to exclusively accumulate in shoots of mycorrhizal plants, independent of the plant species or any stresses (Wang et al., 2018), confirming that a very limited set of compounds is consistently altered in shoots of mycorrhizal plants in the absence of stress.
Less information is available about the metabolic rearrangement in shoots of AM plants following biotic challenges. Overaccumulation of phenolic acids, amino acids, indoles and several intermediates in the oxylipin pathway were observed in leaves of tomato AM plants upon B. cinerea infection, suggesting that metabolic changes in the leaves also contribute to the final output of MIR (Sánchez-Bel et al., 2016). Regarding interactions with herbivores, modulation of metabolic reprogramming by ectomycorrhizas has been shown in poplar, increasing its resistance to the leaf beetle Chrysomela populi (Kaling et al., 2018). However, no information is available about the possible regulation of host plant metabolism by AM symbiosis in response to herbivory.
In the present study, we investigated whether Funneliformis mosseae colonization in tomato plants has a negative impact on the common pest Spodoptera exigua. To explore the potential molecular mechanisms regulating such effects, it should be considered that the plant defences triggered differ according to the proximity to the damaged area (Koo et al., 2009). In fact, the defensive response (such as phytohormone signalling activation or accumulation of toxic metabolites) differs between damaged tissues (local response) and those still non-attacked (systemic response) that may prepare the plant for a potential subsequent aggression (Dugé de Bernonville et al., 2017). Thus, an in-depth study of the metabolic response to herbivory requires a separate analysis of the local and systemic responses. In this study we compared the local and systemic responses to herbivory by S. exigua in mycorrhizal and non-mycorrhizal plants following an untargeted metabolomic approach. We identified primed accumulation of defensive compounds locally in the damaged leaf tissues, which may be responsible for the enhanced ability of AM plants to cope with this aggressor.
Materials and methods
Plant and fungal materials and growing conditions
The AMF Funneliformis mosseae BEG12 (F. mosseae, formerly Glomus mosseae, International Bank of Glomeromycota, https://www.i-beg.eu/cultures/BEG12.htm), was maintained in an open-pot culture of Trifolium repens L. mixed with Sorghum vulgare Pers. plants in a greenhouse. The inoculum consisted of substrate (vermiculite/sepiolite, 1:1), spores, mycelia and infected root fragments from those cultures. Tomato seeds (Solanum lycopersicum L. ‘Moneymaker’) were surface sterilized by immersion in 4% NaHClO (10 min) containing 0.02% (v/v) Tween20 ®, rinsed thoroughly with sterile water and incubated for seven days in an open container with sterile vermiculite at 25 ºC. Tomato plantlets were transferred to 250 ml pots containing a sterile sand: vermiculite (1:1) mixture. Pots for mycorrhizal treatments were inoculated by adding 10% (v/v) F. mosseae inoculum. Uninoculated plants received an aliquot of a filtrate (<20 µm) from F. mosseae inoculum in order to provide the general microbial population, but free of AMF propagules.
A total of ten plants were used for each treatment, randomly distributed and grown in a greenhouse at 24 °C/16 ºC with a 16 h/8 h diurnal photoperiod and 70% humidity. Plants were watered three times a week with Long Ashton nutrient solution (Hewitt, 1966) containing 25% of standard phosphorus (P). Plants were harvested after eight weeks, the fresh weight of shoots and roots was determined, and the leaf material immediately frozen in liquid nitrogen and stored at −80 ºC. An aliquot of each individual root system was reserved for mycorrhizal quantification.
Determination of mycorrhizal colonization
Mycorrhizal colonization was estimated after clearing washed roots in KOH (10%) and subsequent staining of fungal structures with 5% ink (Lamy, Germany) in 2% acetic acid (Vierheilig et al., 2005). The extent of mycorrhizal colonization (expressed as percentage of total root length colonized by the AMF) was calculated according to the gridline intersection method (Giovannetti and Mosse, 1980) using an Eclipse 50i microscope (Nikon, Japan) and brightfield conditions.
Insect rearing and herbivore performance set-up in tomato plants
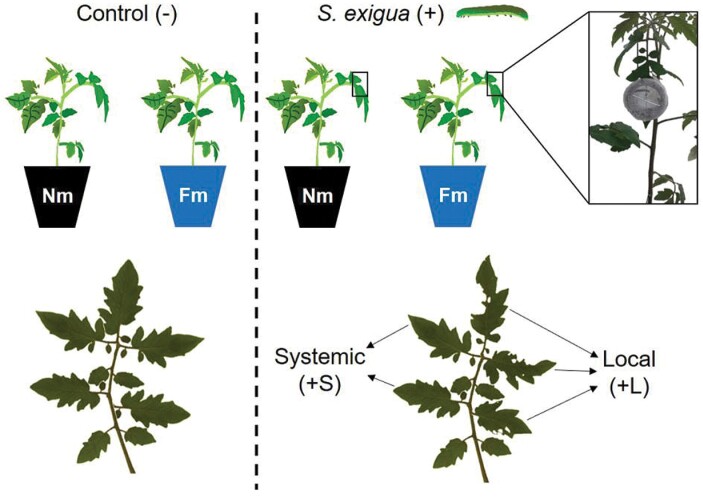
Batches from the same S. exigua (Lepidoptera: Noctuidae) laboratory colony were used for the different experiments. The colony was established with eggs provided by Andermatt Biocontrol AG (Grossdietwil, Switzerland) and reared in the laboratory for more than 50 generations. Larvae were reared on artificial diet (Greene et al., 1976) at 25±3 ºC with 70±5% relative humidity and a 16 h/8 h diurnal photoperiod. For the in planta experiments, once they reached second instar stage of development, two larvae were placed on a leaflet from the third true leaf of each six week-old tomato plant, using 30 mm Ø clip-cages to limit the feeding area and avoid their escape. Clip-cages were moved into new leaflets every two days to make sure they always had food available. Infestation was maintained for two weeks. To monitor insect development, larvae biomass and mortality were measured every day. Dead larvae were replaced by new ones in order to keep the same stimulus in the plant. Finally, eight week-old plants with two weeks of herbivory infestation were harvested. Leaflets from non-infested plants (control, –), leaflets from infested plants where S. exigua directly fed (local, +L) and leaflets contiguous to those with direct damage (systemic, +S) were harvested separately and immediately frozen in liquid nitrogen to be stored at –80 °C until use for molecular analysis (Fig. 1).
Fig. 1.
Experimental set-up and samples used. Harvested shoot material were from Funneliformis mosseae-colonized (Fm) or non-mycorrhizal (Nm) tomato plants, infested (+) or not (-) with Spodoptera exigua. Two second instar larvae were placed into a clip-cage per plant. Clip-cages were moved every day into new leaflets and larval mortality was recorded daily, for two weeks. At the end of the assay, shoot material was collected from each treatment, differentiated into leaflets from non-stressed plants (control; -), leaflets with damage from S. exigua feeding (local, +L) and those leaflets without damage but contiguous to where S. exigua fed (systemic, +S).
Liquid chromatography and electro-spray ionization (LC-ESI) mass spectrometry
LC–ESI full scan mass spectrometry
Freeze-dried leaves (50 mg) were homogenized on ice in 1 ml of MeOH:H2O (10:90) containing 0.01% of HCOOH. The homogenate was centrifuged at 15 000 × g for 15 min at 4 ºC, and the supernatant was recovered and filtered through 0.2 μm cellulose filters (regenerated cellulose filter, 0.20 μm, 13 mmØ. pk/100; Teknokroma, St Cugat, Spain). Subsequently, 20 μl of the filtered supernatant was injected into an Acquity ultra-performance liquid chromatography system (UPLC; Waters, Mildford, MA, USA), which was interfaced with a hybrid quadrupole time-of-flight equipment (Q-TOF-MS Premier, Waters, Mildford, MA, USA). Analytes were eluted with an aqueous methanol gradient containing 0.01% HCOOH. Solvent gradients and further chromatographic conditions were performed as previously described (Agut et al., 2014). Five biological replicates were randomly injected for every treatment. LC separation was performed using an ultra-performance liquid chromatography (UPLC) Kinetex C18 analytical column with a 5 μm particle size, 2.1 × 100 mm (Phenomenex, Madrid, Spain). To accurately identify the signals detected, a second fragmentation function was introduced into the TOF analyser. This function was programmed in a t-wave ranging from 5 to 45 eV to obtain a fragmentation spectrum of each analyte (Agut et al., 2014; Gamir et al., 2014).
To precisely identify metabolites, a library of plant metabolites was generated using chemical standards. The compounds from this library were characterized at the level of retention time, exact mass, and spectrum fragmentation (Schymanski et al., 2014). Up to 84 compounds were prepared at a final concentration of 100 ppb in a composite solution (Rivero et al., 2015) and injected through the UPLC in both positive and negative electro-spray ionization (ESI+; ESI−) modes. For those compounds that were not represented in the internal library, the signals obtained in the untargeted metabolomic analysis were confirmed by comparing the fragmentation spectrum in the Massbank, Metlin or Human Metabolome databases (www.massbank.jp; www.masspec.scripps.edu; www.hmdb.ca).
Full scan data analysis
Data were acquired in centroid mode and subsequently transformed into .cdf files using the Databridge from MassLynx 4.1 software (MassLynx 4.1, Waters, USA). Chromatographic signals were processed using the software R for statistical purposes. Signals from positive and negative electrospray ionization (ESI+; ESI-) were processed separately. Peak peaking, grouping and signal corrections were performed using the XCMS algorithm (Smith et al., 2006). A total of 1132 signals were obtained, 793 in ESI+ mode and 339 ESI- mode (Supplementary Table S1). Metabolite amounts were analysed based on normalized peak area units relative to the dry weight. Adduct, isotope correction, and Kruskal–Wallis test (P<0.05) were performed by using the MarVis Suit 2.0 software tool (Kaever et al., 2015). To determine a global behaviour of the signals, data obtained from positive and negative ESI were combined using MarVis Suit 2.0 software. Subsequently, data were normalized by sum, transformed by cube root, and scaled by Pareto method in order to obtain the sparse partial least squares discriminant analysis (sPLSDA) and heatmap plots, which were generated using MetaboAnalyst 4.0 software (www.metaboanalyst.ca), a comprehensive web-based package for a range of metabolomics applications (Chong et al., 2019).
Antiherbivore properties of selected compounds
Metabolites with primed accumulation in mycorrhizal plants upon herbivory were selected from the metabolome analyses, and those commercially available were purchased to test their effect on S. exigua larvae mortality in different experimental set ups. Treatments were: (i) physostigmine (PHYS; AnalytiCon Discovery GmbH, Potsdam, Germany); (ii) azelaic acid (AZA; Sigma-Aldrich, Germany); (iii) 4-oxododecanedioic acid (4-OXO; Biocrick, Chengdu, China); and (iv) feruloylputrescine (FP; AKos Consulting & Solutions Deutschland GmbH, Steinen, Germany).
In planta bioassay
Non-mycorrhizal tomato plants (Solanum lycopersicum L. ‘Moneymaker’) were grown in 500 ml pots containing a sterile sand: vermiculite (1:1) mixture under greenhouse conditions, as described above. Chemical treatments were applied five weeks after transplanting by spraying the third true leaf of each plant (n=12) with an aqueous solution containing 100 ppm of the compounds, 2% MeOH and 0.02% Tween 20 ®. Control plants were mock-treated with the same solution. For each plant and treatment, two S. exigua larvae at the third instar stage (previously reared on artificial diet) were placed on two different leaflets from the treated leaf using 30 mm Ø clip-cages. Larvae that did not feed in the first 24 h were discarded. Larval mortality was monitored every 72 h during the following six days.
Bioassay on artificial diet
S. exigua larvae were reared on artificial diet (Hoffman and Lawson, 1964) until they became second instar. Sixteen newly molted second instar larvae were transferred to diet supplemented with the different selected compounds at a final concentration of 30 ppm or 3 ppm (from stock solutions in methanol or water). Control larvae were transferred to diet containing the same concentration of the solvent used for the preparation of the supplemented diets (0.6% methanol). For those compounds dissolved in water (AZA and FP) methanol was added to the diet to a final concentration of 0.6% to get an equivalent concentration of methanol in all the treatments. In addition, a group of larvae were also treated with a mixture of the four tested compounds (final concentration of 3 ppm for each compound). Larval mortality was recorded daily during the following 6 d. Sixteen larvae were used per dose and treatment, and the bioassay was repeated twice.
Statistical analyses
Besides the methods and software for metabolomic analysis described above, all statistical analyses (two-way ANOVAs and post hoc tests applied when appropriate, as indicated in the corresponding figure legends) were conducted using Statgraphics Plus 3.1 (Rockville, MD, USA), R software version 2.9.2 (R Development Core Team) and the XCMS package. Larval survival distribution comparisons between treatments were performed using the Logrank (Mantel-Cox) non-parametric test. The reported error bars represent the SE of the survival fractions. Statistical analysis and graphs were generated with GraphPad Prism v.7.0.1.
Results
Arbuscular mycorrhizal symbiosis increases mortality of S. exigua larvae
In order to determine whether root colonization by F. mosseae affects S. exigua performance, tomato plants displaying a well-established mycorrhizal symbiosis were challenged with S. exigua larvae. The percentage of root colonization by the mycorrhizal fungus was ~13% by the end of the experiment (Supplementary Fig. S1A). The symbiosis had no effect on plant growth (Supplementary Fig. S1B). This is of interest for studies on defence mechanisms, by avoiding the potential effects of improved plant growth on insect performance. AM establishment was clearly detrimental to the development of S. exigua. Larvae fed on F. mosseae-colonized tomato plants (Fm) showed a higher mortality than those feeding in non-mycorrhizal plants (Nm; Fig. 2). In fact, 50% of the larvae in Fm plants were dead by day 10, while the mortality was only 15% for larvae on Nm plants.
Fig. 2.
Percentage survival of Spodoptera exigua larvae fed on non-mycorrhizal plants (Nm, black line) or on plants colonized by AMF Funneliformis mosseae (Fm, blue line). Tomato plants colonized or not colonized by F. mosseae (n=20) were infested with second instar S. exigua larvae (two per plant) using clip cages, and mortality was recorded daily. Asterisks indicate statistically significant differences between treatments [***P <0.0005, Logrank (Mantel-Cox)].
Despite the differences in the gain of weight of the survivors between Fm and Nm plants not being statistically significant (P>0.05), there was a clear trend showing reduced larval weight gain in Fm plants (Supplementary Fig. S1C). In addition, 11 days following infestation, 17% of the larvae fed on Nm plants started to pupate, while in Fm plants none of the larvae reached this developmental stage.
S. exigua herbivory had an important impact on the leaf metabolome
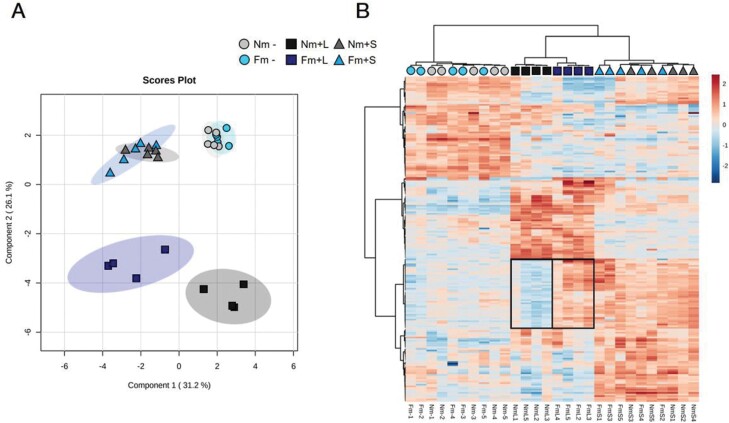
We hypothesized that the reduced larval survival rate and performance in Fm plants compared with non-colonized ones was a result of changes in the defence response in the mycorrhizal plants. To test this possibility, we compared the plant responses of Nm- and Fm-colonized plants subjected to (+) or not subjected to (–) S. exigua feeding during 14 days, through an untargeted metabolomic analysis. To have a more precise overview of the metabolic regulation upon herbivory, we harvested the leaflets where the larvae fed separately from the other unwounded leaflets of the same leaves. Thus, herbivory-related changes were explored in local wounded leaflets (+L) or in unwounded leaflets (systemic response, +S), as shown in Fig. 1. A total of 1132 signals (potential compounds) were registered (Supplementary Table S1), and subsequent statistical analysis showed 200 signals with significantly different accumulation in at least one of the different treatments (P<0.05, Supplementary Table S2). A representation of a supervised analysis (sPLSDA) of these 200 signals revealed that, as expected, herbivory had a strong impact on plant metabolism (Fig. 3A). According to the two main components contributing to data variation, the differences were more pronounced locally at the feeding sites than systemically in leaflets that were not exposed to direct herbivory, since the group of these signals were more distant compared with undamaged leaves (Fig. 3A). These observations were further confirmed by a heatmap analysis (Fig. 3B). This analysis clearly showed the rearrangement of the leaf metabolomic profile following herbivory. Remarkably, in the absence of herbivory (Nm– versus Fm–), AM symbiosis had very low impact on the foliar metabolic profile, since only one hit out of the 1132 detected displayed significantly altered accumulation (P<0.05) in mycorrhizal plants (Supplementary Fig. S2A, B). Although still not fully characterized, exact mass identification of this metabolite matched with 11-carboxyblumenol C-9-O-Glc, a metabolite recently reported as a shoot marker of AM symbiosis (Wang et al., 2018).
Fig. 3.
Overview of the metabolomic reprogramming in tomato leaves subjected to Spodoptera exigua feeding for 15 days. Six weeks old tomato plants non-mycorrhizal (Nm) or colonized by the arbuscular mycorrhizal fungi Funneliformis mosseae (Fm) were infested with S. exigua second instar larvae, and leaves were harvested after two weeks of systemic herbivory. Lyophilized leaf material from control non-infested plants (-), infested leaflets (local response to herbivory, +L) and non-damaged leaflets from infested plants (systemic response to herbivory, +S) were analysed through ultra-high performance liquid chromatography interfaced with a quadrupole time-of-flight mass spectrometer (UPLC-Q-TOF-MS), in order to monitor metabolomic changes. Signal intensity was determined in all samples after normalizing the chromatographic area for each compound to the dry weight of the sample. All 1132 registered signals (Supplementary Table S1), from combination of both positive and negative electrospray ionization (ESI), were compared using non-parametric Kruskal–Wallis test, and only data with significant differences between groups at P<0.05 were used for the supervised analysis (n=5 for - and +S, n=4 for +L), obtaining 200 signals (Supplementary Table S2). Grey and blue colour scales represent Nm and Fm treatments, respectively, and for each case, circles represent non-stress (-), dark coloured squares represent changes in infested, damaged tissue (L), and triangles represent changes in systemic responses to herbivory (S). (A) Supervised three-dimensional sparse partial least squares discriminant analysis (sPLSDA) representation of the major sources of variability. (B) Heatmap and clustering representation of the 200 signals showing differences among treatments. Cluster of metabolites showing an over-accumulation pattern upon herbivory in L tissue only in Fm plants (priming pattern) is highlighted.
Funneliformis mosseae colonization primes the accumulation of specific metabolites in leaves damaged by S. exigua
To explore potential changes in the chemistry of the leaves leading to enhanced larval mortality in mycorrhizal plants, we compared the metabolic profile of mycorrhizal and non-mycorrhizal plants upon herbivory. Few changes in the metabolite profiles were observed between Nm and Fm in the systemic response to herbivory (Nm+S versus Fm+S), as confirmed by the overlap in both sPLSDA and heatmap plots (Fig. 3A, B). In contrast, the local responses to herbivory were modulated as illustrated by the clear separation between the Nm+L and Fm+L samples in the sPLSDA (Fig. 3A). The heatmap analysis confirmed these clearly separated profiles, while uninfested Nm and Fm, and systemic Nm+S and Fm+S clustered together. The distinct pattern of metabolite accumulation in Fm+L revealed a set of compounds with a stronger accumulation, resembling a priming profile (highlighted by a black square, Fig. 3B). In contrast, these metabolites were not modulated by the presence of AM symbiosis in the absence of herbivory. Intriguingly, as shown by their similar patterns in the heatmap, most of those compounds seemed to accumulate to similar levels in systemic tissues of both Nm and Fm plants (Nm+S, Fm+S).
Characterization of the metabolic pathways differentially regulated in response to herbivory
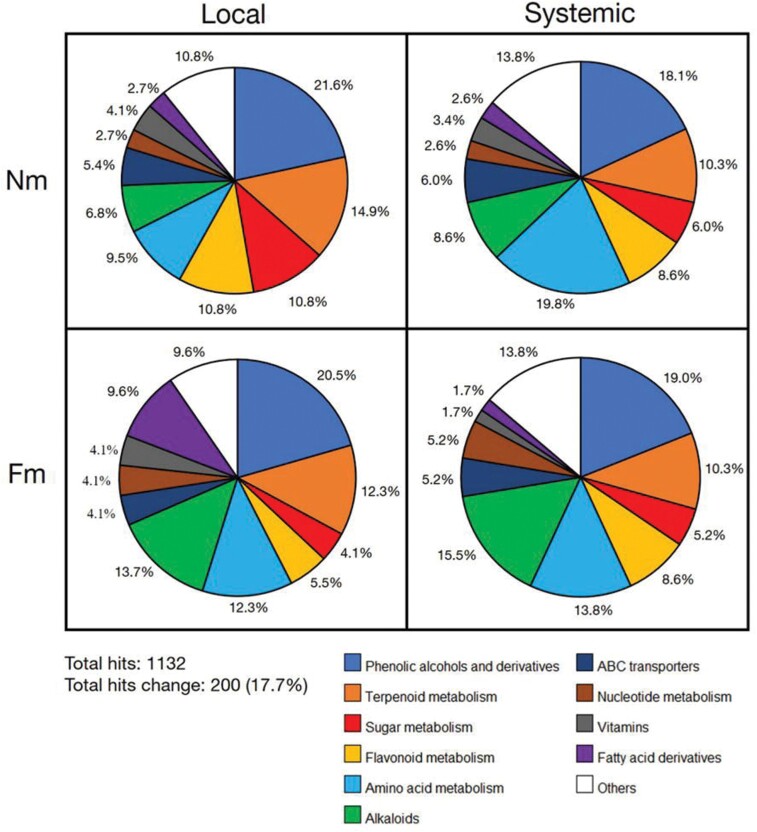
Following untargeted metabolomic analysis, we aimed to identify the 200 signals (Supplementary Table S2) whose accumulation differed in response to herbivory by exact m/z and/or individual spectrum match with databases. For such classification, we used the S. lycopersicum KEGG and internal databases, putatively identifying 107 signals by m/z or/and fragmentation spectrum (Supplementary Table S3). Subsequently, we classified them into the major metabolic pathways (Supplementary Fig. S3), representing the percentage of differentially regulated metabolites in each of these categories for each treatment condition (Fig. 4). This classification illustrates that the local response to S. exigua herbivory resulted in altered accumulation of compounds in multiple pathways, but this consisted mostly of phenolic compounds (21%), terpenoids (14.9%), carbohydrates (10.8%), and flavonoids (10.8%). In distal leaflets, the systemic response resulted in activation of those pathways too, but with major changes in metabolites related to amino acid metabolism. In mycorrhizal plants, these response patterns were different. In local damaged leaflets Fm plants showed a higher proportion of metabolites within the fatty acid metabolism (9.6%) and alkaloids (13.7%) than Nm plants, where these categories only reached 2.7% and 6.8%, respectively (Fm+L versus Nm+L). In contrast, the proportion of herbivore-modulated compounds related to sugar and flavonoid metabolism was lower in Fm+L than in Nm+L samples. Regarding the systemic responses, the proportion of alkaloids and metabolites related to nucleotide metabolism was almost doubled in Fm (Fm+S) compared with Nm plants (Mm+S).
Fig. 4.
Composition of altered metabolic pathways from tomato plants leaves under S. exigua herbivory. Six weeks old tomato plants non-mycorrhizal (Nm) or colonized by the arbuscular mycorrhizal fungi Funneliformis mosseae (Fm) were infested with S. exigua second instar larvae, and leaves were harvested after two weeks of systemic herbivory. After untargeted metabolomic analysis through ultra-high UPLC-Q-TOF-MS (described in Fig. 3 legend), obtained signals were compared using non-parametric Kruskal Wallis test (P<0.05), and only data with differences between herbivory treatments (local or systemic) and non-stress conditions were used for the supervised analysis (n=5 for - and +S, n=4 for +L). The resulting hits were identified by exact m/z and/or spectra coincidence and subsequently grouped into the different S. lycopersicum metabolic pathways from KEGG databases included in software MarVis 2.0. Finally, the pie charts with the percentage of main metabolic pathways altered in each treatment were represented.
Thus, these results illustrate that mycorrhization implies mostly a stronger activation of compounds related to alkaloid and fatty acid metabolism upon herbivory.
Mycorrhiza-primed metabolites in local response to S. exigua herbivory
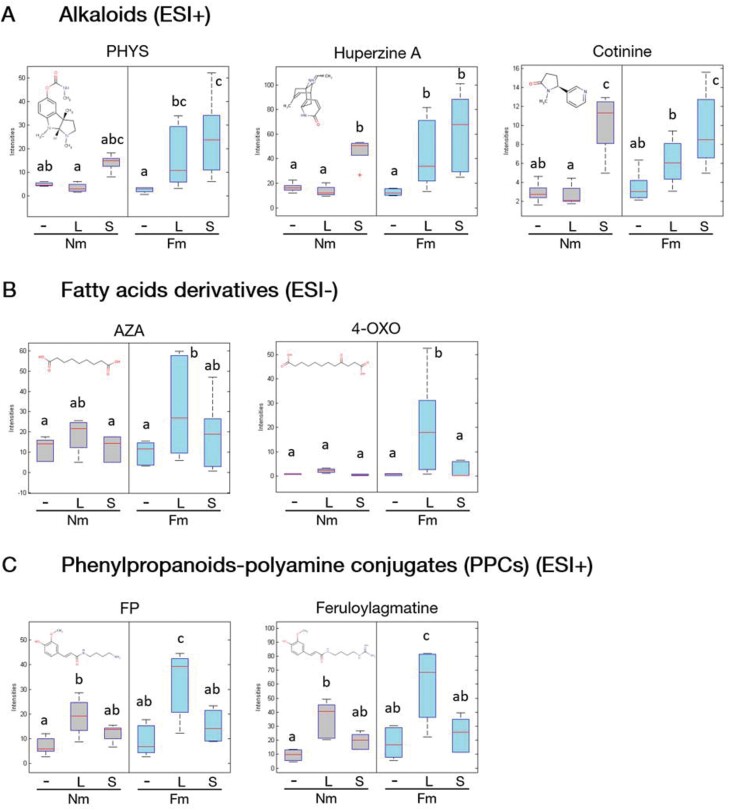
As described above, the most remarkable difference between the response to herbivory in Nm and Fm plants is the cluster of compounds displaying a primed accumulation pattern in the damaged tissues (Fig. 3B, highlighted cluster). Identification through exact mass and/or fragmentation revealed alkaloids, fatty acid derivatives and phenylpropanoid-polyamine conjugates (PPCs) among the metabolites within this primed cluster (Table S3, Fig. 5A-C). The alkaloids, including physostigmine (PHYS, also known as eserine), huperzine A and cotinine, appeared to be over accumulated locally after larvae feeding only in Fm plants, while remaining unaltered locally in Nm plants. Interestingly, the priming profile of these compounds was only observed in local tissues, since they accumulated in systemic leaves upon herbivory in both Nm and Fm plants (Fig. 5A).
Fig. 5.
Boxplots of selected metabolites with a primed accumulation pattern in response to local herbivory. Six week-old tomato plants, previously colonized by arbuscular mycorrhizal fungi (AMF) Funneliformis mosseae (Fm) or not (non-mycorrhizal, Nm), were infested with second instar larvae of S. exigua, and infestation maintained for two weeks more. Leaves from plants in the absence of stress, or with local (L) or systemic (S) damage by S. exigua feeding were analysed through an untargeted metabolic assay in order to monitor metabolic changes (data acquisition as described in Fig. 3 legend). Grey colour represents accumulation in Nm plants and blue colour represents accumulation in Fm-colonized plants. Boxes represent the interquartile range, thick red-lines represent the median, whiskers represent maxima and minima within 1.5 times the interquartile range, and red-crosses show outliers. Putative metabolites were classified into three main metabolic groups: (A) alkaloids family; (B) fatty acids derivatives family; and (C) phenylpropanoid-polyamine conjugates (PPCs) family. Two-way factorial ANOVA (using Fm colonization and herbivory treatments as factors) were performed. Data not sharing a letter in common differ significantly according to the Fisher’s LSD test (P<0.05, n=6).
Among the fatty acid derivatives showing a primed accumulation profile in Fm leaves, we found azelaic acid (AZA) and 4-oxododecanedioic acid (4-OXO). These compounds, unlike alkaloids, were not altered at all in non-mycorrhizal plants, and only marginally in systemic leaves of mycorrhizal plants (Fig. 5B).
Finally, a group of phenylpropanoid-polyamine conjugates (PPCs), also known as hydroxycinnamic amides or phenolamides, formed by condensation of phenolic acids with amines such as polyamines and arylamines, were found to be primed in local leaves of Fm plants (Fig. 5C). This was the case for feruloylputrescine (FP) and feruloylagmatine that showed significantly higher (P<0.05) accumulation only in Fm leaves in response to herbivory (Fm+L). In contrast, other compounds in this group, tricoumaroylspermidine and feruloyl-2-hydroxyputrescine, besides being locally accumulated in response to herbivory in Fm plants, were also accumulated in systemic tissues, regardless of the mycorrhizal status of the plant (Supplementary Fig. S4).
Effect of selected primed compounds on S. exigua mortality
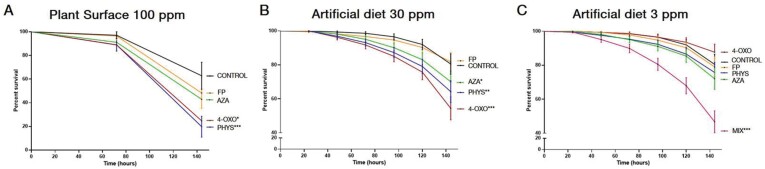
Four of the main primed compounds were tested for their effects on insect survival by applying the synthetic compounds (PHYS, AZA, 4-OXO, and FP) to the plant surface as well as in combination with an artificial diet. Both experimental set ups revealed a significant detrimental effect (P<0.05) on larvae for the PHYS and the 4-OXO treatments (Fig. 6A, B). In addition, AZA also led to a reduction in larvae survival, but only when combined with artificial diet. No effect of FP was observed for any of the treatments. These results confirm the antiherbivore properties of most of the primed compounds tested, and suggest that their action occurs by direct negative effects on the herbivore and not through the activation of antiherbivore defences in the plant, as the addition of the compound in artificial diet, in the absence of plant tissues, was sufficient for increasing larval mortality.
Fig. 6.
Antiherbivore effect of selected compounds. (A) Survival of Spodoptera exigua larvae when fed on plants sprayed with 100 ppm each of physostigmine (PHYS), azelaic acid (AZA), 4-oxododecanedioic acid (4-OXO), feruloylputrescine (FP), or a mixture (MIX) of the four compounds. (B) Survival of Spodoptera exigua larvae when fed on artificial diet supplemented with 30 ppm each the different compounds listed in (A). (C) Survival of Spodoptera exigua larvae when fed on artificial diet supplemented with 3 ppm each of the different compounds listed in (A). For each curve, 12 plants, with two larvae each were used with the different compounds (n=16). Larval mortality was recorded every 72 h (A) or daily (B, C) during the following 6 d. For (B) and (C), bioassays were repeated twice. Asterisks indicate statistically significant differences between control and compound-fed samples (Logrank (Mantel-Cox). *P<0.05, **P<0.005, ***P <0.0005, ****P<0.00005.
The assay on artificial diet was carried out at two different doses of the primed compounds (30 ppm and 3 ppm). At the lower dose, none of the compounds applied individually had an impact on larval survival (Fig. 6C). However, the simultaneous addition of the four compounds into the diet resulted in a significant increase (P<0.0005) in larval mortality compared with the non-supplemented diet, suggesting a potential synergistic effect of the active compounds.
Discussion
In the present study, we demonstrated that tomato root colonization by the AMF F. mosseae has a significant impact on the host interaction with the generalist pest S. exigua. The larvae feeding in these mycorrhizal plants showed an increased mortality and delayed development (Fig. 2), thus these plants are more efficient in containing the herbivore. Although the protective role of AM against pathogens and the underlying molecular mechanisms have been studied previously (Nair et al., 2015; Sánchez-Bel et al., 2016; Jung et al., 2012; Sanmartín et al., 2020a; b), the influence of symbiosis against insects at the mechanistic level are less studied (Song et al., 2013; Minton et al., 2016). Contrasting effects have been reported against leaf chewers, explained as the result of the potential positive effects of symbiosis on the larvae by increasing the plant nutritional value, or the negative effects in the case of improved plant defences (Pozo et al., 2020). Indeed, it has been proposed that AM plants display an optimal tuning of their defences, leading to improved resistance.
Mycorrhization has been reported to decrease the performance of the generalist chewing insects Helicoverpa armigera (Song et al., 2013) and several Spodoptera spp. (Roger et al., 2013; He et al., 2017). Mycorrhizal protection against herbivores (Spodoptera litura) was shown even to be transferred to neighbouring plants by warning them through common mycorrhizal networks (Song et al., 2014). However, benefits in larval fitness from the generalist Mamestra brassicae were observed on mycorrhizal Plantago, despite the strongest induction of aucubin (toxic metabolite against herbivores) in AM-plants (Tomczak et al., 2016). The authors hypothesized that this effect could be due to the better plant quality found in mycorrhizal plants. Mycorrhizal rice also exhibited higher susceptibility to the generalist Spodoptera frugiperda, where mycorrhization resulted in plant-growth promotion (Bernaola et al., 2018). In our experimental system there were no differences in plant growth in response to the mycorrhizal symbiosis, so that we could evaluate, without the interference of significant plant growth effects, the contribution of the plant defence responses to the impact of mycorrhiza on the larvae.
A strongest defensive capacity in AM plants has been shown in previous studies, usually through phytohormone measurements or quantification of metabolites with known toxic effects against insects (Andrade et al., 2013; Song et al., 2013; Minton et al., 2016; Tomczak et al., 2016; Sanmartín et al., 2020b). However, a more holistic overview of the possible modulation of metabolism in the host plant is missing. Here, by using untargeted metabolomics analysis we attempted to profile the metabolic tuning in Nm and Fm plants when interacting with S. exigua, aiming to uncover the mechanisms underlying the enhanced mortality of the larvae in Fm plants.
In previous studies, we showed that AMF colonization resulted in a strong metabolic rearrangement in tomato plant roots in absence of stress (Rivero et al., 2015; 2018, Sanmartín et al., 2020b). Here, we found a minimal alteration of the metabolome in aboveground tissues. Indeed, only one out of 1132 signals registered during the analysis showed differential accumulation in AM plants. The levels of the compound were not altered by herbivory. Its tentative exact mass identification matches with 11-carboxyblumenol C-9-O-Glc, recently reported as a mycorrhization marker in shoots (Supplementary Fig. S2; Wang et al., 2018).
When examining the effect of S. exigua feeding on the metabolome, the bidimensional sPLSDA analysis showed that plant responses upon herbivory clearly resulted in a rearrangement of the metabolic profiles in local and systemic leaves, compared with those from uninfested plants (Fig. 3A). These modifications were different, depending on the distance to the damaged area (local or systemic responses). These distinct responses are in agreement with previous reports, where metabolic differences between damaged and non-attacked tissues were also found (Dugé de Bernonville et al., 2017).
When comparing the profiles in response to herbivory in Fm and Nm plants, differences were only observed in damaged leaflets, while the systemic responses were similar in Nm and Fm plants. The heatmap exhibited a cluster of compounds with a clear primed accumulation profile in the local response to herbivory (Fig. 3B), including several metabolites accumulated to a higher level in Fm plants than in non-colonized ones. Strikingly, this cluster showing a primed profile locally upon herbivory in Fm plants resembled the systemic response. The stronger accumulation of compounds in the damaged area, together with the absence of significant modulation under non-stress conditions, reinforced the hypothesis of defence priming as being responsible for the enhanced mortality observed: the modulation of plant metabolism only takes place in response to herbivory challenge.
A deeper characterization of the response to herbivory revealed phenylpropanoids, terpenoids, flavonoids, sugars and amino acid metabolic pathways as the major groups altered in the local responses to damage by S. exigua attack in both Nm and Fm plants (Fig. 4). In systemic tissues, the same metabolic pathways were targeted, but the proportion of the amino acids and alkaloids was higher. The increase in alkaloids as a local and systemic response to damage was more pronounced in mycorrhizal plants. Considering the insecticidal properties of these compounds, it is tempting to speculate their potential role in the observed protection. In addition, there was a marked 3-fold higher accumulation of fatty acid-related compounds in the local response of Fm plants. This group could be also of great importance for plant defences since it comprises JA precursors from oxylipin biosynthetic pathway, among others.
We focused on those compounds showing a priming profile in mycorrhizal plants, as they could be related to the higher mortality of the larvae feeding on these plants. The three over-represented families of compounds within this cluster were alkaloids (Fig 5A), fatty acid derivatives (Fig 5B), and PPCs (Fig. 5C). Alkaloids are a well characterized group of compounds with toxic effects against herbivores (Mithöfer and Maffei, 2016). Besides their strong clear accumulation induction in local damaged leaves of Fm plants, alkaloids also showed high accumulation in systemic leaves, suggesting a higher mobility of these compounds that may contribute to improved defensive responses against subsequent infestations in distal tissues. Amongst them, cotinine, a common metabolite from nicotine catabolism (Saremba et al., 2018), was found. In addition, two acetylcholinesterase inhibitors were found: PHYS and huperzine A (Luo et al., 2010; Rattan, 2010).
Different studies have addressed the involvement of fatty acids as chemical signals triggering defences during negative interactions (Lim et al., 2017). In our experiment we found a higher local accumulation of AZA in Fm plants upon herbivory, and 4-OXO was exclusively found as a local response to herbivory in those plants. AZA was previously reported to accumulate upon Spodoptera littoralis herbivory in maize (Marti et al., 2013. While no antiherbivore properties have been described for AZA, it has been proposed to be a component of plant systemic immunity regulating defence priming (Jung et al., 2009). Interestingly, exogenous application of AZA in tobacco cells resulted in primed expression of genes involved in systemic acquired resistance, and enhanced synthesis of PPCs (Djami-Tchatchou et al., 2017), supporting the role of AZA in the priming of this family of secondary metabolites. As far as we know, no direct effect of AZA nor 4-OXO acid on plant pathogens or pests has been reported.
Lastly, PPCs (also known as phenolamides) are a diverse class of secondary metabolites present ubiquitously in plants. In our conditions, both tricoumaroylspermidine and feruloyl-2-hydroxyputrescine metabolites showed a similar pattern than alkaloids, with primed-behaviour in Fm plants locally, but high accumulation in the systemic tissues in both Nm and Fm plants. Conversely, feruloylagmatine and FP only over accumulated locally in Fm plants, with their levels remaining unaltered in distal tissues. These compounds were suggested to play an important role in plant development and defence (Edreva et al., 2007; Macoy et al., 2015), especially after challenge with pathogens (López-Gresa et al., 2011; Yogendra et al., 2015) or virus (López-Gresa et al., 2016). Regarding the role of PPCs in anti-herbivore defence, an enhanced growth of S. littoralis reared on Nicotiana mutant plants lacking PPCs was reported (Kaur et al., 2010). In the same study, Nicotiana plants sprayed with caffeoylputrescine reduced Manduca sexta larval growth, suggesting that PPCs are important players in plant defence against leaf chewers.
To test the potential role of the identified primed compounds in the defence response, we selected commercially available compounds representative of these groups and performed different bioassays (Fig. 6). Plant treatments with the alkaloid physostigmine or the fatty acid derivative 4-OXO acid significantly enhanced larval mortality. To test whether this was due to a direct effect on the larvae, rather than to a defence elicitation role, we tested the effect of the compounds on artificial diet (Fig 6B,C). Both PHYS and 4-OXO acid also had a negative effect when provided an artificial diet. In addition, AZA showed a negative effect on the larvae too, but only in artificial diet. Accordingly, the data confirm a direct negative effect of the compounds on larval performance, although we cannot rule out some eliciting activity of plant defences. No effect on mortality was found for FP treatments in any of the bioassays. However, we cannot rule out indirect effects through interaction with other toxic compounds, or by altering larvae immunity as it has been reported in other systems (Bandoly et al., 2016). It is remarkable that treatments with the individual compounds at lower doses (3 ppm) did not have any effect on larval survival, but their combination had a significant deleterious effect, pointing to potential synergistic effects that would support a stronger effect in planta, as several defensive metabolites co-occur. In fact, recent meta-analyses pinpoint to the relevance of synergistic effects of phytochemical mixtures on generalist herbivores (Richards et al., 2016).
In conclusion, our results indicate that F. mosseae colonization of tomato roots increase plant resistance against S. exigua. Our study shows evidence that AM symbiosis modulates host metabolism in response to herbivory, leading to a primed accumulation of defensive compounds including fatty acid derivatives, alkaloids and PPCs. The functional analysis of some of these compounds as PHYS and 4-OXO demonstrate their negative effect on larval survival. Thus, the higher accumulation upon herbivory of these defensive compounds in mycorrhizal plants likely contributes to the enhanced larval mortality. Future studies will aim to uncover the transcriptional and post-transcriptional regulation and key regulatory elements underlying this primed accumulation of defences. Our study contributes to the identification of new functional compounds with anti-herbivore properties and illustrates that mycorrhizal colonization can prime plant metabolic responses to herbivore attack.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Physiological parameters measured.
Fig. S2. Accumulation pattern and fragmentation profile of the only signal over accumulated in F. mosseae-colonized tomato plants in the absence of herbivory.
Fig. S3. Classification of the major metabolic pathways according to S. lycopersicum KEGG and internal databases.
Fig. S4. Boxplots of selected metabolites with a primed accumulation pattern in response to local herbivory.
Table S1. Dataset containing metabolic profiles from tomato leaves.
Table S2. Dataset containing 200 signals with significantly different accumulation in at least one of the different treatments.
Table S3. Dataset containing 107 signals putatively identified by m/z and/ or fragmentation spectrum.
Acknowledgements
We acknowledge Serveis Centrals d’Instrumentació Científica (SCIC) at the Universitat Jaume I, J. Palenzuela (responsible and curator of the EEZ germplasm collection), E. Berrio and J. M. García for excellent technical support, and Dr C. Azcón J. A. López-Ráez and Pierre Petriarq for helpful discussions. This research has been funded by project AGL2015-64990-C2-1-R, RTI2018-094350-B-C31/32/33 and the research network Ref. RED2018-102407-T from the Spanish Ministry of Science, Innovation and Universities. J. Rivero and J. Lidoy were supported by Ph.D. fellowship BES-2013–062638 and BES-2016–077850. We acknowledge ideas exchange under COST Action FA1405.
Author contributions
VF, JR and MJP conceptualized the study; JR and JL performed plant experiments; ALG and SH performed insect bioassays; JR and VF performed metabolomics and statistical analyses; JR and MJP wrote the manuscript.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
References
- Agut B, Gamir J, Jacas JA, Hurtado M, Flors V. 2014. Different metabolic and genetic responses in citrus may explain relative susceptibility to Tetranychus urticae. Pest Management Science 70, 1728–1741. [DOI] [PubMed] [Google Scholar]
- Andrade SAL, Malik S, Sawaya ACHF, Bottcher A, Mazzafera P. 2013. Association with arbuscular mycorrhizal fungi influences alkaloid synthesis and accumulation in Catharanthus roseus and Nicotiana tabacum plants. Acta Physiologiae Plantarum 35, 867–880. [Google Scholar]
- Bago B, Pfeffer PE, Shachar-Hill Y. 2000. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiology 124, 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandoly M, Grichnik R, Hilker M, Steppuhn A. 2016. Priming of anti-herbivore defence in Nicotiana attenuata by insect oviposition: herbivore-specific effects. Plant, Cell & Environment 39, 848–859. [DOI] [PubMed] [Google Scholar]
- Bernaola L, Cosme M, Schneider RW, Stout M. 2018. Belowground inoculation with arbuscular mycorrhizal fungi increases local and systemic susceptibility of Rice plants to different pest organisms. Frontiers in Plant Science 9, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo L, Carletti P, Badeck FW, Rizza F, Morcia C, Ghizzoni R, Rouphael Y, Colla G, Terzi V, Lucini L. 2019. Metabolomic responses triggered by arbuscular mycorrhiza enhance tolerance to water stress in wheat cultivars. Plant Physiology and Biochemistry 137, 203–212. [DOI] [PubMed] [Google Scholar]
- Chong J, Wishart DS, Xia J. 2019. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Current Protocols in Bioinformatics 68, e86. [DOI] [PubMed] [Google Scholar]
- Choudhary DK, Prakash A, Johri BN. 2007. Induced systemic resistance (ISR) in plants: mechanism of action. Indian Journal of Microbiology 47, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie AF, Murray PJ, Gange AC. 2011. Is a specialist root-feeding insect affected by arbuscular mycorrhizal fungi? Applied Soil Ecology 47, 77–83. [Google Scholar]
- Djami-Tchatchou AT, Ncube EN, Steenkamp PA, Dubery IA. 2017. Similar, but different: structurally related azelaic acid and hexanoic acid trigger differential metabolomic and transcriptomic responses in tobacco cells. BMC Plant Biology 17, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugé de Bernonville T, Carqueijeiro I, Lanoue A, et al. 2017. Folivory elicits a strong defense reaction in Catharanthus roseus: metabolomic and transcriptomic analyses reveal distinct local and systemic responses. Scientific Reports 7, 40453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edreva AM, Velikova VB, Tsonev TD. 2007. Phenylamides in plants. Russian Journal of Plant Physiology 54, 287–301. [Google Scholar]
- Erb M, Reymond P. 2019. Molecular interactions between Plants and Insect Herbivores. Annual Review of Plant Biology 70, 527–557. [DOI] [PubMed] [Google Scholar]
- Ezawa T, Saito K. 2018. How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New phytologist 220, 1116–1121. [DOI] [PubMed] [Google Scholar]
- Fernández I, Merlos M, López-Ráez JA, Martínez-Medina A, Ferrol N, Azcón C, Bonfante P, Flors V, Pozo MJ. 2014. Defense related phytohormones regulation in arbuscular mycorrhizal symbioses depends on the partner genotypes. Journal of Chemical Ecology 40, 791–803. [DOI] [PubMed] [Google Scholar]
- Fracasso A, Telò L, Lanfranco L, Bonfante P, Amaducci S. 2020. Physiological beneficial effect of Rhizophagus intraradices inoculation on tomato plant yield under water deficit conditions. Agronomy, 10, 71. [Google Scholar]
- Gamir J, Pastor V, Kaever A, Cerezo M, Flors V. 2014. Targeting novel chemical and constitutive primed metabolites against Plectosphaerella cucumerina. The Plant Journal 78, 227–240. [DOI] [PubMed] [Google Scholar]
- Giovannetti M, Mosse B. 1980. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist 84, 489–500. [Google Scholar]
- Greene GL, Leppla NC, Dickerson WA. 1976. Velvetbean caterpillar: A rearing procedure and artificial medium. Journal of Economic Entomology 69, 487–488. [Google Scholar]
- Gruden K, Lidoy J, Petek M, et al. 2020. Ménage à Trois: Unraveling the mechanisms regulating plant-microbe-arthropod interactions. Trends in Plant Science 25, 1215–1226. [DOI] [PubMed] [Google Scholar]
- Hartley SE, Gange AC. 2009. Impacts of plant symbiotic fungi on insect herbivores: mutualism in a multitrophic context. Annual Review of Entomology 54, 323–342. [DOI] [PubMed] [Google Scholar]
- He L, Li C, Liu R. 2017. Indirect interactions between arbuscular mycorrhizal fungi and Spodoptera exigua alter photosynthesis and plant endogenous hormones. Mycorrhiza 27, 525–535. [DOI] [PubMed] [Google Scholar]
- Hewitt EJ. 1966. Sand and water culture methods used in the study of plant nutrition. Technical Communication No. 22. East Malling, UK: Commonwealth Bureau of Horticulture and Plantation Crops [Google Scholar]
- Hill EM, Robinson LA, Abdul-Sada A, Vanbergen AJ, Hodge A, Hartley SE. 2018. Arbuscular mycorrhizal fungi and plant chemical defence: effects of colonisation on aboveground and belowground metabolomes. Journal of Chemical Ecology 44, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge A, Storer K. 2015. Arbuscular mycorrhiza and nitrogen: implications for individual plants through to ecosystems. Plant and Soil 386, 1–19. [Google Scholar]
- Hoffman DJ, Lawson FR. 1964. Preliminary Studies on Mass Rearing of the Tobacco Hornworm. Journal of Economic Entomology, 57(3), 354-355. [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. 2009. Priming in systemic plant immunity. Science 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ. 2012. Mycorrhiza-induced resistance and priming of plant defenses. Journal of Chemical Ecology 38, 651–664. [DOI] [PubMed] [Google Scholar]
- Kaever A, Landesfeind M, Feussner K, Mosblech A, Heilmann I, Morgenstern B, Feussner I, Meinicke P. 2015. MarVis-Pathway: integrative and exploratory pathway analysis of non-targeted metabolomics data. Metabolomics 11, 764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaling M, Schmidt A, Moritz F, Rosenkranz M, Witting M, Kasper K, Janz D, Schmitt-Kopplin P, Schnitzler JP, Polle A. 2018. Mycorrhiza-triggered transcriptomic and metabolomic networks impinge on herbivore fitness. Plant Physiology 176, 2639–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schöttner M, Baldwin IT, Gális I. 2010. R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiology 152, 1731–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer A, Pimprikar P, Wewer V, et al. 2017. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6, e29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Gao X, Jones AD, Howe GA. 2009. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. The Plant Journal 59, 974–986. [DOI] [PubMed] [Google Scholar]
- Laparre J, Malbreil M, Letisse F, Portais JC, Roux C, Bécard G, Puech-Pagès V. 2014. Combining metabolomics and gene expression analysis reveals that propionyl- and butyryl-carnitines are involved in late stages of arbuscular mycorrhizal symbiosis. Molecular Plant 7, 554–566. [DOI] [PubMed] [Google Scholar]
- Lim GH, Singhal R, Kachroo A, Kachroo P. 2017. Fatty acid- and lipid-mediated signaling in plant defense. Annual Review of Phytopathology 55, 505–536. [DOI] [PubMed] [Google Scholar]
- López-Gresa MP, Torres C, Campos L, Lisón P, Rodrigo I, Bellés JM, Conejero V. 2011. Identification of defence metabolites in tomato plants infected by the bacterial pathogen Pseudomonas syringae. Environmental and Experimental Botany 74, 216–228. [Google Scholar]
- López-Gresa MP, Lisón P, Yenush L, Conejero V, Rodrigo I, Bellés JM. 2016. Salicylic acid is involved in the basal resistance of tomato plants to citrus exocortis viroid and tomato spotted wilt virus. PLoS One 11, e0166938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Verhage A, Fernández I, García JM, Azcón-Aguilar C, Flors V, Pozo MJ. 2010. Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. Journal of Experimental Botany 61, 2589–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Sun C, Li Y, Wu Q, Song J, Wang D, Jia X, Li R, Chen S. 2010. Analysis of expressed sequence tags from the Huperzia serrata leaf for gene discovery in the areas of secondary metabolite biosynthesis and development regulation. Physiologia Plantarum 139, 1–12. [DOI] [PubMed] [Google Scholar]
- Macoy DM, Kim W-Y, Lee SY, Kim MG. 2015. Biotic stress related functions of hydroxycinnamic acid amide in plants. Journal of Plant Biology 58, 156–163. [Google Scholar]
- Marti G, Erb M, Boccard J, Glauser G, Doyen GR, Villard N, Robert CA, Turlings TC, Rudaz S, Wolfender JL. 2013. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant, Cell & Environment 36, 621–639. [DOI] [PubMed] [Google Scholar]
- Martínez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CMJ, Pozo MJ, Ton J, van Dam NM, Conrath U. 2016. Recognizing plant defense priming. Trends in Plant Science 21, 818–822. [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Baccelli I, Luna E, Flors V. 2017. Defense priming: An adaptive part of induced resistance. Annual Review of Plant Biology 68, 485–512. [DOI] [PubMed] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM. 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiology Reviews 37, 634–663. [DOI] [PubMed] [Google Scholar]
- Minton MM, Barber NA, Gordon LL. 2016. Effects of arbuscular mycorrhizal fungi on herbivory defense in two Solanum (Solanaceae) species. Plant Ecology and Evolution 149, 157–164. [Google Scholar]
- Miozzi L, Vaira AM, Brilli F, Casarin V, Berti M, Ferrandino A, Nerva L, Accotto GP, Lanfranco L. 2020. Arbuscular mycorrhizal symbiosis primes tolerance to cucumber mosaic virus in tomato. Viruses 12, 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Maffei ME. 2016. General mechanisms of plant defense and plant toxins. In: Gopalakrishnakone P, Carlini CR, Ligabue-Braun R, eds. Plant Toxins. Dordrecht: Springer. https://doi.org/10.1007/978-94-007-6728-7_21-1 [Google Scholar]
- Nair A, Kolet SP, Thulasiram HV, Bhargava S. 2015. Systemic jasmonic acid modulation in mycorrhizal tomato plants and its role in induced resistance against Alternaria alternata. Plant Biology 17, 625–631. [DOI] [PubMed] [Google Scholar]
- Olowe OM, Olawuyi OJ, Sobowale AA, Odebode AC. 2018.Role of arbuscular mycorrhizal fungi as biocontrol agents against Fusarium verticillioides causing ear rot of Zea mays L. (maize). Current Plant Biology 15, 30-37 [Google Scholar]
- Orozco-Cardenas M, McGurl B, Ryan CA. 1993. Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proceedings of the National Academy of Sciences, USA 90, 8273–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA. 2014. Induced systemic resistance by beneficial microbes. Annual Review of Phytopathology 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Azcón-Aguilar C. 2007. Unraveling mycorrhiza-induced resistance. Current Opinion in Plant Biology 10, 393–398. [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Albrectsen BR, Bejarano ER, de la Peña E, Herrero S, Martinez-Medina A, Pastor V, Ravnskov S, Williams M, Biere A (2020). Three-way interactions between plants, microbes, and arthropods (PMA): Impacts, mechanisms, and prospects for sustainable plant protection. Teaching tools in plant biology: Lecture Notes. The Plant Cell 32, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan RS. 2010. Mechanism of action of insecticidal secondary metabolites of plant origin. Crop Protection 29, 913–920. [Google Scholar]
- Richards LA, Glassmire AE, Ochsenrider KM, Smilanich AM, Dodson CD, Jeffrey CS, Dyer LA. 2016. Phytochemical diversity and synergistic effects on herbivores. Phytochemistry Reviews 15, 1153–1166. [Google Scholar]
- Rivero J, Gamir J, Aroca R, Pozo MJ, Flors V. 2015. Metabolic transition in mycorrhizal tomato roots. Frontiers in Microbiology 6, 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero J, Álvarez D, Flors V, Azcón-Aguilar C, Pozo MJ. 2018. Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New phytologist 220, 1322–1336. [DOI] [PubMed] [Google Scholar]
- Roger A, Gétaz M, Rasmann S, Sanders IR. 2013. Identity and combinations of arbuscular mycorrhizal fungal isolates influence plant resistance and insect preference. Ecological Entomology 38, 330–338. [Google Scholar]
- Saia S, Ruisi P, Fileccia V, Di Miceli G, Amato G, Martinelli F. 2015. Metabolomics suggests that soil inoculation with arbuscular mycorrhizal fungi decreased free amino acid content in roots of durum wheat grown under N-jimited, P-rich field conditions. PLoS One 10, e0129591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Bel P, Troncho P, Gamir J, Pozo MJ, Camañes G, Cerezo M, Flors V. 2016. The nitrogen availability interferes with mycorrhiza-induced resistance against Botrytis cinerea in tomato. Frontiers in Microbiology 7, 1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín N, Pastor V, Pastor-Fernández J, Flors V, Pozo MJ, Sánchez-Bel P. 2020a. Role and mechanisms of callose priming in mycorrhiza-induced resistance. Journal of Experimental Botany 71, 2769–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín N, Sánchez-Bel P, Pastor V, Pastor-Fernández J, Mateu D, Pozo MJ, Cerezo M, Flors V. 2020b. Root-to-shoot signalling in mycorrhizal tomato plants upon Botrytis cinerea infection. Plant Science 298, 110595. [DOI] [PubMed] [Google Scholar]
- Saremba BM, Murch SJ, Tymm FJM, Rheault MR. 2018. The metabolic fate of dietary nicotine in the cabbage looper, Trichoplusia ni (Hübner). Journal of Insect Physiology 109, 1–10. [DOI] [PubMed] [Google Scholar]
- Schliemann W, Ammer C, Strack D. 2008. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 69, 112–146. [DOI] [PubMed] [Google Scholar]
- Schouteden N, De Waele D, Panis B, Vos CM. 2015. Arbuscular mycorrhizal fungi for the biocontrol of plant-parasitic nematodes: a review of the mechanisms involved. Frontiers in Microbiology 6, 1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger R, Baier MC, Persicke M, Müller C. 2014. High specificity in plant leaf metabolic responses to arbuscular mycorrhiza. Nature Communications 5, 3886. [DOI] [PubMed] [Google Scholar]
- Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, Hollender J. 2014. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environmental Science & Technology 48, 2097–2098. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry 78, 779–787. [DOI] [PubMed] [Google Scholar]
- Smith SE, Smith FA. 2011. Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annual Review of Plant Biology 62, 227–250. [DOI] [PubMed] [Google Scholar]
- Song YY, Ye M, Li CY, Wang RL, Wei XC, Luo SM, Zeng RS. 2013. Priming of anti-herbivore defense in tomato by arbuscular mycorrhizal fungus and involvement of the jasmonate pathway. Journal of Chemical Ecology 39, 1036–1044. [DOI] [PubMed] [Google Scholar]
- Song YY, Ye M, Li C, He X, Zhu-Salzman K, Wang RL, Su YJ, Luo SM, Zeng RS. 2014. Hijacking common mycorrhizal networks for herbivore-induced defence signal transfer between tomato plants. Scientific Reports 4, 3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak VV, Schweiger R, Müller C. 2016. Effects of arbuscular mycorrhiza on plant chemistry and the development and behavior of a generalist herbivore. Journal of Chemical Ecology 42, 1247–1258. [DOI] [PubMed] [Google Scholar]
- Tugizimana F, Mhlongo M, Piater L, Dubery I. 2018. Metabolomics in plant priming research: The way forward? International Journal of Molecular Sciences 19, 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H, Schweiger P, Brundrett M. 2005. An overview of methods for the detection and observation of arbuscular mycorrhizal fungi in roots. Physiologia Plantarum 125, 393–404. [Google Scholar]
- Vos IA, Verhage A, Schuurink RC, Watt LG, Pieterse CM, Van Wees SC. 2013. Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Frontiers in Plant Science 4, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Schäfer M, Li D, et al. 2018. Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi. eLife 7, e37093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogendra KN, Kumar A, Sarkar K, Li Y, Pushpa D, Mosa KA, Duggavathi R, Kushalappa AC. 2015. Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. Journal of Experimental Botany 66, 7377–7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.