Abstract
We describe the use of high-fidelity single molecule sequencing to assemble the genome of the psychoactive Psilocybe cubensis mushroom. The genome is 46.6Mb, 46% GC, and in 32 contigs with an N50 of 3.3Mb. The BUSCO completeness scores are 97.6% with 1.2% duplicates. The Psilocybin synthesis cluster exists in a single 3.2Mb contig. The dataset is available from NCBI BioProject with accessions PRJNA687911 and PRJNA700437.
Keywords: Psilocybe cubensis, Genome, Single molecule sequencing, Psilocybin
Introduction
There are several mushrooms capable of synthesizing the psychoactive compound psilocybin. This compound has been classified as a “breakthrough therapy” for depression by the FDA ( Johnson and Griffiths 2017). The psilocybin pathway was identified by Fricke et al., but to date no public references exist in NCBI with N50s longer than 50kb ( Fricke et al. 2017; Blei et al. 2018; Fricke et al. 2019a; Fricke et al. 2019b; Blei et al. 2020; Demmler et al. 2020; Fricke et al. 2020). A more contiguous genome assembly can assist in further resolution of the genetic diversity in the fungi’s secondary metabolite production.
Methods
DNA isolation
Dried stems from Psilocybe cubensis strain P.envy (strain name is anecdotal and obtained in Somerville, MA, USA) were used for isolation of high molecular weight (HMW) DNA using a modified CTAB/Chloroform and SPRI protocol. Briefly, 300mg of stem sample were ground to a fine powder using a -80C frozen mortar and pestle. 150 mg of powder was then aliquoted into 2 mL conical tubes (USA Scientific) with 1.5 mL cetrimonium bromide. These tubes were then incubated at room temperature on a tube rotator for 10 minutes. 6 uL of RNase A (Promega 4 mg/mL) was then added and both tubes were incubated at 37°C for one hour, vortexing every 15 minutes. Following this incubation, 7.5 uL Proteinase K (New England Biolabs 20 mg/mL) was added and the tubes were incubated at 60°C for 30 minutes, vortexing every 10 minutes. At the conclusion of the Proteinase K incubation, both tubes were incubated on ice for 10 minutes. The samples were then centrifuged for 5 minutes at 14000 rpm. 600 uL of supernatant was removed from each tube and added to 600 uL chloroform. The tubes were then vortexed until opaque and spun for 5 minutes at 14000 rpm. 400 uL of the aqueous layer was removed using a wide bore tip and added to a 1.5 mL Eppendorf tube. 400 uL MIP (marijuana infused products) Solution B and 400 uL DNA Binding Beads (Medicinal Genomics PN 420004) were added to the Eppendorf tube and inverted to homogenize. The tubes were then incubated at room temperature on the tube rotator for 15 minutes. The tubes were then removed from the rotator and placed on a magnetic tube rack for 3 minutes. The supernatant was removed, the beads were washed twice with 1 mL of 70% ethanol and allowed to dry for 5 minutes. The beads were then eluted in 100 uL of 56°C Elution Buffer (Medicinal Genomics PN 420004) using a wide bore tip and incubated at 56°C for 5 minutes. Following this incubation, the tubes were returned to the magnetic rack, the supernatant of both tubes were removed using a wide bore tip and pooled in a fresh Eppendorf tube. HMW DNA length was quantified on an Agilent TapeStation and produced a DIN of 8.1. Qubit Fluorometer (Thermo Fisher Scientific) quantified 55ng/ul. Nanodrop Spectrophotometer (Thermo Fisher Scientific) quantified 104ng/ul with 260/280nm ratio of 1.85 and 260/230nm of 0.95.
Sequencing
Sequencing libraries were constructed according to the manufacturer’s instructions for Pacific Biosciences Sequel II HiFi sequencing. 773,735 CCS reads were generated. Quast ( Gurevich et al. 2013) was used to assess the quality of the input fasta sequence file (N50 = 13.9Kb) and the output assembly fasta file (3.33Mb N50).
Assembly and annotation
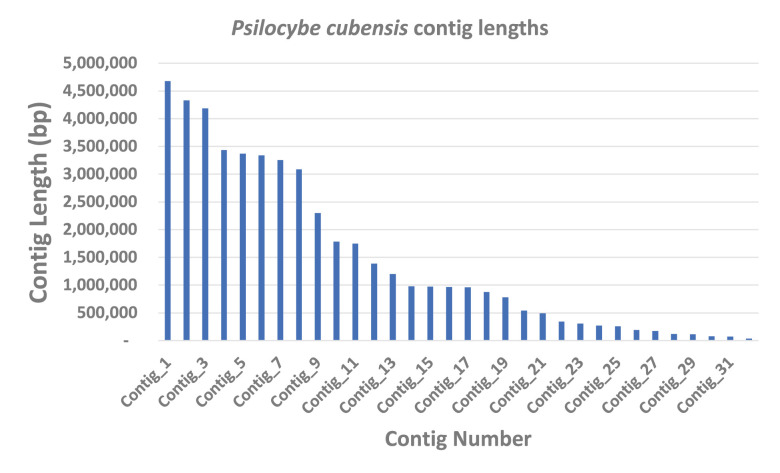
The unfiltered CCS data was assembled using the Peregrine assembler (pg_asm 0.3.5,arm_config5e69f3d+) ( Chin 2019). Reads were assembled into 32 contigs with lengths ranging from 32 kilobases to 4.6 megabases ( Figure 1). BUSCO v3.0.2 completeness scores (97.6%) were measured using agaricales_odb10.2020-08-05 BUSCO lineage database ( Table 1) ( Simao et al. 2015; Waterhouse et al. 2018). FunAnnotate v1.8.4 was used to annotate the genome ( Li and Wang 2021) resulting in 13,478 genes.
Figure 1. Psilocybe cubensis P.envy contig length distribution (n=32).
Table 1. BUSCO completeness scores using agaricales_odb10.2020-08-05.
| Total BUSCOs | Single-copy | Duplicated | Fragmented | Missing |
|---|---|---|---|---|
| 3870 | 3729 | 45 | 9 | 87 |
| 97.60% | 96.40% | 1.20% | 0.20% | 2.20% |
The final genome assembly was aligned to three other public Psilocybe cubensis datasets ( Fricke et al. 2017; Torrens-Spence et al. 2018; Reynolds et al. 2018) and one different Psilocybe species ( Psilocybe cyanescens) to verify taxonomic identification ( Table 2). In total, 96-98.75% of these Psilocybe cubensis sequences align to the new HiFi generated Psilocybe cubensis P.envy reference using minimap2 and bwa-mem ( Li and Durbin 2010; Li 2018). Mapping rates were determined using samtools flagstat on bam files ( Li et al. 2009). Alignments were visualized with MUMmer V4.0.0beta2 and Integrative Genomics Viewer v2.4.16 ( Delcher et al. 2003; Robinson et al. 2011; Thorvaldsdottir et al. 2013).
Table 2. Three Psilocybe cubensis data sets in NCBI and JGI were aligned to the P.envy HiFi reference.
A different Psilocybe species ( Psilocybe cyanescens) genome was also mapped with much lower mapping efficiency.
| Author | Accession | Data type | Mapping rate | Tool | Species |
|---|---|---|---|---|---|
| Fricke et al. 2017 | https://mycocosm.jgi.doe.gov/Psicub1_1/Psicub1_1.home.html | Illumina Assembly | 98.8% | Minimap2 | P. cubensis |
| McKernan et al. 2020 | NCBI Project: PRJNA687911 | Illumina FastQ | 96.0% | bwa-mem | P. cubensis |
| Torrens-Spence et al. 2018 | NCBI Project: PRJNA450675 | RAN-Seq Assembly | 98.5% | Minimap2 | P. cubensis |
| Reynolds et al. 2018 | NCBI Project: PRJNA387735 | Illumina Assembly | 56.8% | Minimap2 | P. cyanescens |
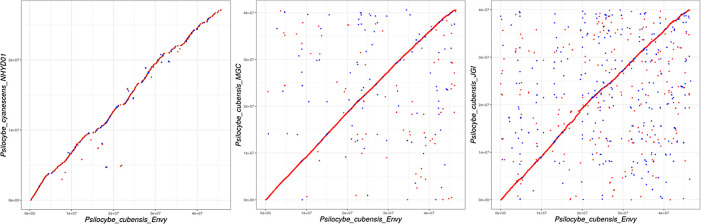
Three Illumina genome assemblies (Reynolds et al., McKernan et al., Fricke et al.) were additionally aligned using MUMmer for whole genome alignment plots ( Figure 2).
Figure 2. Whole genome alignments of short read Illumina assemblies to Psilocybe cubensis strain P. envy.
Left is Psilocybe cyanescens from Reynolds et al. Middle is McKernan et al. (MGC) Illumina assembly. Right is Fricke et al. or JGI assembly.
Polymorphisms
183,430 SNPs were identified with GATK v4.1.6.0 by comparing Illumina whole genome shotgun data (McKernan et al. NCBI Project: PRJNA687911) with the HiFi reference assembly. This equates to a SNP every 243 bases.
Structural variation
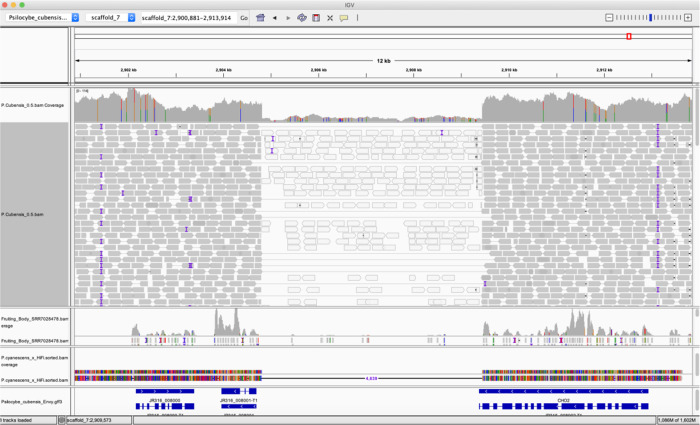
The N-methyltransferase gene responsible for Psilocybin production in P.envy contains a structural variation not seen in previous P. cubensis surveys ( Figure 3). Illumina read mapping of the McKernan et al. P. cubensis assembly in NCBI (NCBI Project: PRJNA687911) demonstrates multiple read pairs spanning a 4.6kb insertion in the HiFi P. cubensis strain P.envy. This insertion extends the 3’ end of the P.envy N-methyltransferase gene. The 4.6kb insertion is also observed as a deletion in Psilocybe cyanescens and as a deletion in RNA-Seq data from Torrens-Spence et al. (NCBI Project: PRJNA450675) ( Reynolds et al. 2018). Further work is required to understand the biological significance of this variation.
Figure 3. IGV display of Illumina reads mapped to HiFi Psilocybe cubensis P.envy assembly.
Top track is Medicinal Genomics Illumina whole genome shotgun data of a different P. cubensis (strain name unknown: NCBI Project: PRJNA687911) mapped to the HiFi P. cubensis strain P.envy. Second track contains RNA-Seq data from a third P. cubensis genome (strain name also unknown: NCBI Project: PRJNA450675) hosted at JGI. Third track is Psilocybe cyanescens genome mapped to HiFi P. cubensis P.envy reference genome. Fourth track is FunAnnotate GFF3 annotation of the HiFi P. cubensis P.envy genome.
Conclusions
A highly contiguous Psilocybe cubensis genome has been generated. The N50 contigs lengths are 75 fold more contiguous than the existing assembly available at JGI. This reference can aid in the identification of genetic variation that may impact psilocybin, psilocin, norpsilocin, baeocystin, norbaeocystin and aeruginascin production.
Data availability
GenBank: Psilocybe cubensis strain MGC-MH-2018, whole genome shotgun sequencing project, Accession number JAFIQS000000000.1: https://www.ncbi.nlm.nih.gov/nuccore/JAFIQS000000000.1/.
BioProject: Psilocybe cubensis, Accession number PRJNA687911: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA687911
BioProject: Psilocybe cubensis strain: MGC-MH-2018, Accession number PRJNA700437: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA700437
CoGe genome browser: Psilocybe cubensis (Psilocybe cubensis P.envy), https://genomevolution.org/coge/GenomeInfo.pl?gid=60487
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; peer review: 1 approved
References
- Blei F, Baldeweg F, Fricke J, et al. : Biocatalytic Production of Psilocybin and Derivatives in Tryptophan Synthase-Enhanced Reactions. Chemistry. 2018. 10.1002/chem.201801047 [DOI] [PubMed] [Google Scholar]
- Blei F, Dorner S, Fricke J, et al. : Simultaneous Production of Psilocybin and a Cocktail of beta-Carboline Monoamine Oxidase Inhibitors in “Magic” Mushrooms. Chemistry. 2020;26:729–734. 10.1002/chem.201904363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS: Human Genome Assembly in 100 Minutes. bioRxiv. 2019. 10.1101/705616 [DOI]
- Delcher AL, Salzberg SL, Phillippy AM: Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;Chapter 10:Unit 10 13. [DOI] [PubMed] [Google Scholar]
- Demmler R, Fricke J, Dorner S, et al. : S-Adenosyl-l-Methionine Salvage Impacts Psilocybin Formation in “Magic” Mushrooms. Chembiochem. 2020;21:1364–1371. 10.1002/cbic.201900649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke J, Blei F, Hoffmeister D: Enzymatic Synthesis of Psilocybin. Angewandte Chemie. 2017;56:12352–12355. 10.1002/anie.201705489 [DOI] [PubMed] [Google Scholar]
- Fricke J, Kargbo R, Regestein L, et al. : Scalable Hybrid Synthetic/Biocatalytic Route to Psilocybin. Chemistry. 2020;26:8281–8285. 10.1002/chem.202000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke J, Lenz C, Wick J, et al. : Production Options for Psilocybin: Making of the Magic. Chemistry. 2019a;25:897–903. 10.1002/chem.201802758 [DOI] [PubMed] [Google Scholar]
- Fricke J, Sherwood A, Kargbo R, et al. : Enzymatic Route toward 6-Methylated Baeocystin and Psilocybin. Chembiochem. 2019b;20:2824–2829. 10.1002/cbic.201900358 [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, et al. : QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MW, Griffiths RR: Potential Therapeutic Effects of Psilocybin. Neurotherapeutics. 2017;14:734–740. 10.1007/s13311-017-0542-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H: Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R: Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. : The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Wang TF: PacBio Long-Read Sequencing, Assembly, and Funannotate Reannotation of the Complete Genome of Trichoderma reesei QM6a. Methods Mol Biol. 2021;2234:311–329. 10.1007/978-1-0716-1048-0_21 [DOI] [PubMed] [Google Scholar]
- Reynolds HT, Vijayakumar V, Gluck-Thaler E, et al. : Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evol Lett. 2018;2:88–101. 10.1002/evl3.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. : Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP: Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens-Spence MP, Liu CT, Pluskal T, et al. : Monoamine Biosynthesis via a Noncanonical Calcium-Activatable Aromatic Amino Acid Decarboxylase in Psilocybin Mushroom. ACS Chem Biol. 2018;13:3343–3353. 10.1021/acschembio.8b00821 [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simao FA, et al. : BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol Biol Evol. 2018;35:543–548. 10.1093/molbev/msx319 [DOI] [PMC free article] [PubMed] [Google Scholar]