Abstract
Background
Deletion of pfhrp2 and/or pfhrp3 genes cause false negatives in malaria rapid diagnostic test (RDT) and threating malaria control strategies. This systematic review aims to assess the main methodological aspects in the study of pfhrp2 and pfhrp3 gene deletions and its global epidemiological status, with special focus on their distribution in Africa; and its possible impact in RDT.
Methods
The systematic review was conducted by examining the principal issues of study design and methodological workflow of studies addressing pfhrp2 deletion. Meta-analysis was applied to represent reported prevalences of pfhrp2 and pfhrp3 single and double deletion in the World Health Organization (WHO) region. Pooled-prevalence of deletions was calculated using DerSimonnian-Laird random effect model. Then, in-deep analysis focused on Africa was performed to assess possible variables related with these deletions. Finally, the impact of these deletions in RDT results was analysed combining reported information about RDT sensitivity and deletion prevalences.
Results
49 articles were included for the systematic review and 37 for the meta-analysis, 13 of them placed in Africa. Study design differs significantly, especially in terms of population sample and information reported, resulting in high heterogeneity between studies that difficulties comparisons and merged conclusions. Reported prevalences vary widely in all the WHO regions, significantly higher deletion were reported in South-Central America, following by Africa and Asia. Pfhrp3 deletion is more prevalent (43% in South-Central America; 3% in Africa; and 1% in Asia) than pfhrp2 deletion (18% in South-Central America; 4% in Africa; and 3% in Asia) worldwide. In Africa, there were not found differences in deletion prevalence by geographical or population origin of samples. The prevalence of deletion among false negatives ranged from 0 to 100% in Africa, but in Asia and South-Central America was only up to 90% and 48%, respectively, showing substantial relation between deletions and false negatives.
Conclusion
The concerning prevalence of pfhrp2, pfhrp3 and pfhrp2/3 gene deletions, as its possible implications in malaria control, highlights the importance of regular and systematic surveillance of these deletions. This review has also outlined that a standardized methodology could play a key role to ensure comparability between studies to get global conclusions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12936-021-03812-0.
Keywords: Malaria diagnosis, Rapid diagnostic test, pfhrp2, Deletions, Malaria control, RDT
Background
Malaria is one of the major challenges for global health. Indeed, during 2019, around 229 million cases were reported worldwide and there were 409,000 deaths. Plasmodium falciparum caused the majority of cases and deaths by malaria, followed by Plasmodium vivax (2.8% of cases). Africa, where the prevalent species is P. falciparum, reported more than 94% of cases and deaths [1].
Prompt and accurate diagnosis is essential for malaria control as it allows effective and timely treatment [2]. Point-of-care testing is a good option in this regard, especially in resource-limited settings, which is typically the case for the majority of endemic malaria regions [3, 4]. The use, variety and quality of malaria rapid diagnostic tests (RDTs) have increased significantly during the last 10 years and they are currently the preferred field diagnostic test for malaria [5]. The majority of RDTs are based on detecting HRP2 (histidine-rich protein 2), a specific protein from P. falciparum encoded by the pfhrp2 gene [6]. However, in addition to detecting HRP2, there are also reports of cross-reactions with HRP3, a structural homologue of HRP2 encoded by the pfhrp3 gene [7]. As such, RDT may detect both proteins.
Previous studies have demonstrated the efficacy and advantages of RDT compared to other diagnostic techniques, including microscopy and PCR [8–10]. However, its performance has been threatened by the detection of parasites lacking the pfhrp2 and pfhrp3 genes since 2010, when these deletions were described for first time [11, 12]. Failure of RDT might also be caused by different factors, such as parasite density, although the deletion of one or both of these genes is considered to be the principal cause of false negatives [13, 14]. Some studies have also suggested that the genetic diversity in pfhrp2 and pfhrp3 genes could influence RDT results [15, 16].
It is particularly important to pay special attention to the specificity of diagnostic tests in order to identify people infected. Following a treatment based on diagnostic strategy, a wrong diagnosis means that the patient will not receive the appropriate treatment in time, and could prove fatal [17].
The first evidence of parasites carrying pfhrp2 and pfhrp3 gene deletions was reported in South America [18]. Since then, several studies have described these gene deletions in greater proportions across different countries in South and Central America [19, 20], Sub-Saharan Africa [21–23] and Asia [24, 25]. This shows the worldwide spread of parasites with such deletions, with Africa playing a particularly critical role as its high prevalence of malaria caused by P. falciparum increases the consequences of these deletions for public health.
A deletion prevalence of 5% in these genes has been defined by the World Health Organization (WHO) as the minimum prevalence for changing the RDT kit, as prevalences higher than 5% could threaten the effectiveness of the test and affect public health guidelines for malaria control [26].
The selection and spread of pfhrp2 and pfhrp3 gene deletions may be related to different factors, such as national malaria prevalence or frequencies of people seeking treatment [27], as well as to individual variables, such as age (younger age) [17] or type of symptoms [28].
Deletions are commonly detected in monoclonal infections, which are more common in low prevalence situations [29], during the low malaria transmission season and in countries with a decreasing prevalence of malaria [30, 31]. As such, the spread of these deletions will also partially depend on the epidemiology of the disease in each country. In that context, increasing the understanding of pfhrp2 and pfhrp3 gene deletions could play a key role in ensuring the efficacy of strategies for malaria control adapted to each particular region. Moreover, the use of meta-analysis may increase the accuracy of the results found in previous studies.
This paper is an original review and meta-analysis assessing the principal issues of study design and methodological workflow of studies addressing pfhrp2 deletion and the prevalence status of pfhrp2 and pfhrp3 gene deletions worldwide, with special focus on their distribution in Africa. Moreover, this study assesses the possible impact of these deletions in RDT effectiveness.
Methods
This systematic review and meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidance (http://prisma-statement.org/). The PRISMA checklist is provided in Additional file 1.
Review question
The aim of the study was to review the scientific literature characterizing the deletion of pfhrp2, pfhrp3 and double deletion pfhrp2 and 3 (pfhrp2/3) genes during the last 10 years worldwide. The review sought (1) to assess the main methodological aspects in the study of pfhrp2, pfhrp3 and pfhrp2/3 gene deletions; (2) to gather the epidemiological information reported for pfhrp2, pfhrp3 and pfhrp2/3 gene deletions worldwide; (3) to assess the epidemiological information reported for these genes in Africa; and (4) to evaluate the impact of these deletions on the results of PfHRP2-based malaria RDTs.
Search strategy and data sources
A systematic electronic search was conducted in MEDLINE (PubMed), Scopus and Cochrane, as well as a manual search, in April 2020. The search terms used included “hrp2”, “pfhrp2”, “hrp3”, “pfhrp3”, “mRDT” and “malaria rapid diagnostic test”. These terms were combined using the Boolean logical operator “OR”. The search was restricted to articles published since January 2010.
Selection of studies
Two members of the research group (IMF and PB) screened all articles using the eligibility criteria of the review independently and discussed the discrepancies. Firstly, the articles were screened by title and abstract, and finally by full text. All the inclusion and exclusion criteria used in this review are indicated in Table 1. Data analysis differed by specific objective, thus the inclusion criteria applied for each one also varied.
Table 1.
Inclusion and exclusion criteria by analysis
| Analysis where that criteria was applied | Inclusion criteria | Exclusion criteria | ||
|---|---|---|---|---|
| Meta-analysis and subgroup analysis | Meta-analysis (quantitative-synthesis) | Qualitative description |
Original articles written in English Published and peer-reviewed articles between January 2010 and April 2020 Addressed the status of pfhrp2, pfhrp3 and/or pfhrp2/3 deletions |
It had been written in language different to English Published before 2010 Field isolates were not included Major bias had been detected |
|
Primary data on pfhrp2/3 deletion Molecular methodologies to detect deletions Quality of Cross-sectional study score ≥ 5 according to The Joanna Briggs Institute (JBI) (Munn Z, 2017) Sample size of ≥ 30 |
Year of samples had not been reported Data from more than 3 years are combined |
|||
|
Placed in Sub-Saharan Africa It included malaria samples from population with different ages, not only children |
||||
Data management
The following data, summarized in an extraction table, were extracted from each study: author, title, country where samples were obtained, study design, sample characteristics, year of study collection, diagnosis of malaria and confirmation method, information about PfHRP2-RDT, methodological aspects of pfhrp2/3 gene deletion detection, prevalence of pfhrp2, pfhrp3 and flanking region deletions among all P. falciparum cases and among false negative PfHRP-based RDT results, and main conclusions of each study.
Qualitative synthesis
The descriptive summaries were structured by including all the information in an Excel spreadsheet, sorted by research topic. A qualitative narrative synthesis of findings was carried out in order to assess the main methodological aspects of the study of pfhrp2 and pfhrp3 gene deletions (first aim of this study).
Meta-analysis
The meta-analysis was performed with the studies that met the inclusion criteria and analysed the prevalence of deletion. Prevalence was first calculated for all P. falciparum samples included in each study depending on the WHO region, then for studies in Africa (origin of samples) and finally for RDT false-negatives. The prevalence of articles that analysed deletions only among disruptive samples (RDT false-negative and other diagnostic method-positive samples) was included in the general results section using the total population as denominator.
Statistical analysis
The pooled value of prevalence was established using DerSimonnian-Laird (DL) random effect model applied to proportions [32]. A random model was applied due to the differences between the studies included in terms of study design. In the analysis, the inverse variance method was used to determine the relative weight of each study and logit transformation.
Heterogeneity between studies was calculated using the I2 statistic, which describes the percentage variability due to variation between studies. I2 represents a measure of inconsistency, and I2 > 80% was considered to indicate considerable heterogeneity. To assess publication bias, contour-enhanced funnel plots were generated to study whether small studies with small effects were missing and to visualize the statistical significance of the studies included. These plots included all studies, representing their standard error and their effect. An asymmetry found in the plot could mean publication bias, as studies with more standard error reported greater effects. Additionally, the asymmetry of the funnel plot was tested using the Egger test, and possible p-hacking was tested by performing a P-curve analysis [33].
The meta-analysis was performed using the R 4.0.0 software package to analyse the deletion prevalence data extracted. The meta [34], metafor [35] and dmetar [36] R packages were used.
Meta-analysis focussed on Africa: subgroup analysis
In order to assess the epidemiological status of pfhrp2 and pfhrp3 deletions in Africa by region, only studies carried out in Africa that followed more stringent inclusion criteria were used to perform a subgroup meta-analysis. These studies were analysed independently according to the origin of samples (health facilities or from general population). For the subgroup analysis, a random-effects model was applied within and between subgroups. This model was applied because the data extracted from articles could not be considered to be representative of each African region.
In order to control the between-study heterogeneity in the subgroup analysis, a more in-depth assessment of this heterogeneity was obtained from an influential analysis and a search for outliers. The results of this influential analysis were illustrated with a Baujat Plot calculated using the leave-one-out strategy. Studies whose confidence interval did not overlap with the confidence interval of the pooled prevalence were considered outliers. The results of both analyses were then combined, and studies that overly contributed to heterogeneity and had been identified as outliers were excluded from the subgroup analysis. Once the heterogeneity had been decreased, the analysis was performed independently for each subgroup.
Results
Results of systematic review
Selection of studies
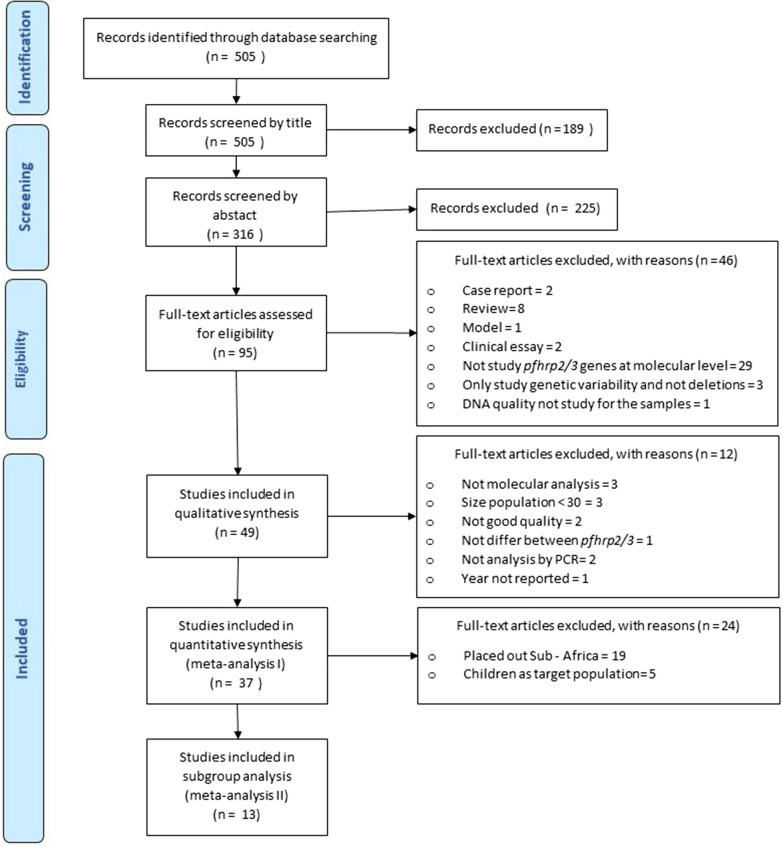
The initial search yielded 505 articles, 95 of which were found to be eligible after title and abstract screening. After full text review, 46 of these were excluded because they did not meet the inclusion criteria. Finally, 49 articles were included in the systematic review for qualitative synthesis. Out of these 49 articles, 37 were included in the quantitative synthesis and 13 in the in-depth meta-analysis with subgroup analysis (Fig. 1).
Fig. 1.
PRISMA flow diagram with the selection and inclusion process
Characteristics of studies included: population and setting
All the studies included were cross-sectional, with 22 being conducted in Africa, 12 in South America, seven in Asia, four in Central America and one in Oceania. A further study combined data from different WHO regions [15]. A total of 24 studies reported data from several regions within a country and eight studies combined and compared data from different countries [12, 15, 37–41]. The remaining studies did not specify the reporting of data from different regions.
Twenty five of the 49 studies included used only malaria-confirmed field samples. Of the studies conducted with population sampling (samples with and without malaria infection), 18 targeted the general population and 10 included only the symptomatic population suspected of malaria. The other 24 studies included samples from the general population, combining infected and non-infected samples. A total of six studies focused on children, with two of these involving symptomatic children (Table 2).
Table 2.
Characterization of articles included which study molecular deletions on pfhrp2/3
| First author | Country | Samples’ year | Collection season | Study population | Methodologies |
|---|---|---|---|---|---|
| Abdallah [19] | Honduras | 2008–09 | Both or annual | Malaria confirmed field samples | PCR, SSR/STR |
| Okoth [39] | Suriname and Guyana | 2010–11 | High transmission | Malaria confirmed field samples | PCR, SSR/STR |
| Akinyi [57] | Peru | 1998–05 | High transmission | Malaria confirmed field samples | PCR, SSR/STR |
| Amoah [74] | Ghana | 2014–15 | Low transmission | Population sampling among children, asymptomatic | PCR |
| Atroosh [48] | Yemen | 2012 | High transmission | Population sampling among all ages, symptomatic | PCR, Seq |
| Baker [15] | 18 countriesa | 2010 | Not reported | Malaria confirmed field samples | PCR, Seq |
| Baldeviano [59] | Peru | 2010–12 | Malaria outbreak | Malaria confirmed samples among symptomatic patients at health centre | PCR, SSR/STR |
| Berhane [14] | Eritrea | 2016 | High transmission | Population sampling among all ages, from health centre | PCR, SSR/STR |
| Berzosa [21] | Equatorial Guinea | 2013 | Both or annual | Population sampling among all ages | PCR, Seq., SSR/STR |
| Beshir [49] | Kenya | 2007–08 and 2014 | Not reported | Population sampling among symptomatic at health centre and among children Asymptomatic | PCR, Seq |
| Bharti [24] | India | 2014 | High transmission | Population sampling among all ages, symptomatic | PCR, Seq |
| Deme [38] | Senegal, Mali and Uganda | 2001–10 | High transmission | Malaria confirmed field samples, from health centre | Seq., SSR/STR |
| Dong [58] | China | 2013–18 | Both or annual | Population sampling among all ages, symptomatic | PCR, Seq., SSR/STR |
| Dorado [20] | Colombia | 2003–12 | Not reported | Malaria confirmed field samples | PCR, Seq., SSR/STR |
| Fontecha [41] | Honduras Guatemala and Nicaragua | 2011 and 2015 | Not reported | Malaria confirmed field samples | PCR |
| Funwei [69] | Nigeria | 2013–14 | Both or annual | Population sampling among children, Symptomatic | PCR, Seq |
| Gamboa [18] | Peru | 2003–07 and 2007 | Not reported | Malaria confirmed field samples from health facility and population sampling among symptomatic (active case detection) | PCR, Seq., ELISA |
| Gupta [22] | Mozambique | 2010–16 | Both or annual | Population sampling among all ages | PCR |
| Herman [43] | Haiti | 2012–14 | Both or annual | Population sampling among all ages, symptomatic | PCR |
| Kobayashi [52] | Zambia | 2009–11 and 2015–17 | High transmission | Population sampling among all ages | PCR |
| Koita [28] | Mali | 1996 | High transmission | Population sampling among all ages and children asymptomatic | PCR |
| Kozycki [11] | Ruanda | 2014–15 | Both or annual | Population sampling among all ages, symptomatic at health centre | PCR |
| Kreidenweiss [55] | Gabon | 2017–18 | Both or annual | Lab strains and clinical sample | PCR, Seq |
| Kumar Bharti [71] | India | 2014 | Both or annual | Malaria confirmed field samples | PCR, Seq |
| Kumar [42] | India | 2010 | Both or annual | Malaria confirmed field samples | PCR |
| Kumar [50] | India | 2009–11 | Both or annual | Malaria confirmed field samples | PCR, Seq |
| Laban [54] | Zambia | 2008–12 | Both or annual | Population sampling among all ages | PCR |
| Li [45] | China | 2011–12 | Both or annual | Malaria confirmed field samples | PCR, Seq |
| Maltha [13] | Peru | 2010–11 | Both or annual | Malaria confirmed field samples, from health centre | PCR |
| Menegon [23] | Eritrea | 2013–14 | Not reported | Population sampling among all ages | PCR |
| Murillo-Solano [63] | Colombia | 1999–09 | Both or annual | Malaria confirmed field samples | PCR, SSR/STR |
| Mussa [61] | Sudan | Not reported | Not reported | Population sampling among all ages, symptomatic | PCR, Seq |
| Nderu [9] | Kenya | 2007–16 | Not reported | Malaria confirmed field samples, from symptomatic patients | PCR, Seq |
| Nderu [60] | Kenya | 2016 | Both or annual | Malaria confirmed field samples from symptomatic patients | PCR, Seq., SSR/STR |
| Okoth [44] | Peru | 2013 | Malaria outbreak | Malaria confirmed field samples from symptomatic patients | PCR, SSR/STR |
| Owusu [47] | Ghana | 2015 | Both or annual | Population sampling among children, HIV positives and healthy | PCR |
| Parr [46] | D.R. of Congo | 2013–2014 | Both or annual | Population sampling among children, majority asymptomatic | PCR, SSR/STR |
| Pati [25] | India | 2013–16 | Both or annual | Population sampling among all ages, symptomatic | PCR, Seq |
| Plucinski [51] | Angola | 2016 | High transmission | Population sampling among all ages, at health centre | PCR |
| Rachid Viana [40] | Brazil and Bolivia | 2010–12 | Not reported | Malaria confirmed field samples, at health centre | PCR |
| Ramutton [37] | 7 countriesb | 2005–10 | Not reported | Population sampling among children with severe malaria | PCR, Seq |
| Ranadive [53] | Swaziland | 2012–15 | Both or annual | Population sampling among all ages, symptomatic | PCR |
| Sáenz [56] | Ecuador | 2012–13 | Both or annual | Malaria confirmed field samples | |
| Thomson [12] | Ghana Tanzania and Uganda | 2009–10 and 2014–15 | High transmission | Malaria confirmed field samples from symptomatic population with all ages, at health facilities and national survey | PCR |
| Trouvay [73] | French Guiana | 2009 and 2011 | Both or annual | Malaria confirmed field samples | PCR, Seq., SSR/STR |
| Willie [30] | Papua New Guinea | 2001–03 | Both or annual | Population sampling among all ages | PCR, Seq |
| Willie [62] | Madagascar | 2014–15 | Both or annual | Population sampling among all ages, mostly asymptomatic | PCR, Seq |
| Wurtz [16] | Senegal | 2009–11 | High transmission | Malaria confirmed field samples, at health centre | PCR, Seq |
Seq. sequencing, SSR/STR techniques based on microsatellite analysis
aBenin, Burkina Faso, Cameroon, Central African Republic, Gambia, Ghana, Guinea, Kenya, Liberia, Madagascar, Malawi, Nigeria, Niger, Sierra Leone, Sudan, Tanzania, Uganda, Zambia, Papua New Guinea, Solomon Is., East Timor, Vanatu, Brazil, Colombia, Ecuador, Haiti, Honduras, Peru, Santa Lucia, SUriname, Cambodia, China, Indonesia, Malaysia, Myanmar, Philippines, Thailand and Vietnam
bDemocratic Republic of Congo, Gambia, Kenya, Mozambique, Rwanda, Tanzania and Uganda
Methodological aspects of the study of pfhrp2, pfhrp3 and pfhrp2/3 deletions
All studies used molecular techniques to detect pfhrp2, pfhrp3 and pfhrp2/3 deletions (Table 3). The total sample size included in the studies ranged from 48 [42] to 9317 [43], with a median sample size of 911 [12]. The number of confirmed P. falciparum cases included ranged from 4 [44] to 3291 [11], with a median confirmed cases sample size of 169 [30].
Table 3.
Key points of the methodology to detect pfhrp2/3 deletions by molecular analysis
| First author | Method of malaria diagnosis/confirmation | N total | N cases | N pfhrp | Genes study by molecular analysis (Pfhrp2/Pfhrp3/pfhrp2/3/Flanking genes) | Method to test DNA quality | Elimination for parasitaemia ≤ 5 p/µL |
|---|---|---|---|---|---|---|---|
| Abdallah [19] | PCR | 68 | 68 | 68 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Okoth [39] | Microscopy/PCR | 203 | 175 | 175 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR, qPCR | No |
| Akinyi [57] | PCR | 188 | 188 | 188 | pfhrp2, flanking genes | PCR | NA |
| Amoah [74] | RDT, microscopy/PCR | 558 | 288 | 288 | pfhrp2, pfhrp, pfhrp2/3 | PCR | NA |
| Atroosh [48] | RDT, microscopy | 622 | 189 | 189 | pfhrp2 | PCR | NA |
| Baker [15] | RDT, microscopy | 458 | 458 | 458 | pfhrp2, pfhrp3 | NA | NA |
| Baldeviano [59] | Microscopy/PCR | 210 | 54 | 54 | pfhrp2 | PCR | NA |
| Berhane [14] | RDT, microscopy/PCR | 51 | 50 | 50 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Berzosa [21] | RDT/PCR | 1724 | 1724 | 122 | pfhrp2, pfhrp3, pfhrp2/3 | PCR | NA |
| Beshir [49] | RDT, microscopy/PCR | 274 | 131 | 131 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | qPCR | Yes |
| Bharti [24] | RDT, microscopy/PCR | 1521 | 1521 | 50 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | qPCR | No |
| Deme [38] | PCR | 74 | 74 | NA | NA | PCR | NA |
| Dong [58] | Microscopy/PCR | 306 | 306 | 306 | pfhrp2 | PCR | NA |
| Dorado [20] | RDT, microscopy/PCR | 374 | 365 | 365 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Fontecha [41] | Microscopy/PCR | 128 | 128 | 128 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Funwei [69] | RDT, microscopy/PCR | 511 | 340 | 66 | pfhrp2, pfhrp3, pfhrp2/3 | PCR | NA |
| Gupta [22] | RDT, microscopy/PCR | 9124 | 1162 | 69 | pfhrp2, pfhrp3 | PCR, qPCR | Yes |
| Gamboa [18] | RDT, microscopy/PCR | 157 | 157 | 157 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Herman [43] | RDT/PCR | 9317 | 2695 | 7 | pfhrp2 | PCR | NA |
| Kobayashi [52] | RDT, microscopy/PCR | 5167 | 1189 | 36 | pfhrp2 | qPCR | Yes |
| Koita [28] | RDT, microscopy | 723 | 480 | 37 | pfhrp2 | PCR | NA |
| Kozycki [11] | RDT, microscopy | 8757 | 3291 | 370 | pfhrp2 | PCR | Yes |
| Kreidenweiss [55] | RDT/PCR | 200 | 200 | 95 | pfhrp2 | PCR, qPCR | Yes |
| Kumar Bharti [71] | RDT/PCR | 1392 | 1392 | 1392 | pfhrp2, pfhrp3, pfhrp2/3 | PCR | NA |
| Kumar [42] | RDT, microscopy/PCR | 48 | 48 | 48 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Kumar [50] | RDT/PCR | 140 | 48 | 48 | pfhrp2, pfhrp3 | qPCR | No |
| Laban [54] | RDT, microscopy/PCR | 3292 | 61 | 61 | pfhrp2 | PCR, qPCR | Yes |
| Li [45] | RDT/PCR | 97 | 97 | 97 | pfhrp2, pfhrp2/3 | qPCR | No |
| Maltha [13] | RDT, Microscopy/PCR | 182 | 74 | 74 | pfhrp2, pfhrp3, pfhrp2/3 | qPCR | No |
| Menegon [23] | Microscopy/PCR | 144 | 144 | 144 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Murillo Solano [63] | RDT, microscopy/PCR | 115 | 100 | 100 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | NA | NA |
| Mussa [61] | RDT/PCR | 59 | 26 | 26 | pfhrp2 | PCR | NA |
| Nderu [9] | PCR | 400 | 400 | 400 | pfhrp2, pfhrp3 | PCR | NA |
| Nderu [60] | RDT, microscopy/PCR | 80 | 80 | 80 | pfhrp2 | PCR | NA |
| Owusu [47] | RDT, microscopy/PCR | 401 | 62 | 8 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Okoth [44] | Microscopy/PCR | 4 | 4 | 4 | pfhrp2, pfhrp3, pfhrp2/3 | PCR | NA |
| Parr [46] | RDT, microscopy/PCR | 7137 | 2752 | 2752 | pfhrp2, pfhrp3, pfhrp2/3 | qPCR | No |
| Pati [25] | RDT, Microscopy/PCR | 1058 | 384 | 384 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Plucinski [51] | RDT/PCR | 1267 | 458 | 5 | pfhrp2, pfhrp3, pfhrp2/3 | qPCR | No |
| Rachid Viana [40] | Two RDT, Microscopy | 223 | 223 | 223 | pfhrp2, pfhrp3, pfhrp2/3, flanking genes | PCR | NA |
| Ramutton [37] | RDT/PCR | 3826 | 77 | 77 | pfhrp2, pfhrp3 | PCR, qPCR | Yes |
| Ranadive [53] | RDT/PCR | 1353 | 162 | 9 | pfhrp2 | PCR, qPCR | Yes |
| Sáenz [56] | Microscopy/PCR | 32 | 32 | 32 | pfhrp2, pfhrp3, pfhrp2/3 | PCR | NA |
| Thomson [12] | RDT, microscopy/PCR | 911 | 718 | 411 | pfhrp2, pfhrp3, pfhrp2/3 | PCR, qPCR | Yes |
| Trouvay [73] | RDT, microscopy/PCR | 359 | 221 | 221 | pfhrp2, pfhrp3 | PCR | NA |
| Willie [30] | RDT/PCR | 169 | 169 | 137 | pfhrp2 | PCR | NA |
| Willie [62] | RDT, microscopy/PCR | 260 | 73 | 73 | pfhrp2 | PCR | NA |
| Wurtz [16] | RDT, microscopy/PCR | 136 | 125 | 125 | pfhrp2, pfhrp3 | qPCR | No |
N total total of samples included in the study, N cases total of P. falciparum confirmed cases included in the study, N pfhrp no. of samples included for molecular analysis (PCR), pfhrp2/3 double deletion pfhrp2 + pfhrp, NA not applicable
Some studies performed the molecular analysis of pfhrp2/3 deletions only on a subset of the total sample size. Ten studies only performed a molecular analysis for discordant samples with an HRP2-based RDT negative result and malaria PCR positive result [16, 21, 22, 24, 25, 42, 45–47].
Regardless of the origins of the sample size, a diagnosis of malaria was performed in all studies, and in the majority (n = 45) this diagnosis was confirmed using another diagnostic method: 27 studies used PCR and microscopy for the diagnosis, 14 PCR only, three microscopy only [11, 28, 48] and one microscopy and another malaria RDT [40]. In addition, 16 studies confirmed the density of parasite DNA by quantitative PCR to confirm that a low density was not the cause of the false negative [12, 13, 16, 22, 24, 37, 39, 45, 46, 49–55]. Moreover, samples included in 38 studies were tested with HRP2-based RDT, whereas 11 studies did not include a diagnostic by RDT [9, 19, 23, 38–41, 44, 56–59].
Deletion of the pfhrp2 gene was analysed in all studies included in the review, 14 studies only included the analysis of this gene [11, 28, 30, 31, 43, 48, 52–55, 58, 60–62]. Deletion of the pfhrp3 gene was studied in 29 articles and double deletion of the pfhrp2 and pfhrp3 genes was assessed in 26 articles. The deletion was identified by PCR in all studies except one (n = 48). Additionally, 15 articles tested the presence of deletions on pfhrp2 and pfhrp3 flanking genes [14, 18–20, 23–25, 39–42, 47, 49, 57, 63].
Results of quantitative analysis of the prevalence of deletions
The meta-analysis performed for pfhrp2, pfhrp3 and pfhrp2/3 deletions reported a high between-study heterogeneity (I2 ≥ 90%), which meant that it was difficult to obtain statistically significant results. As such, a meta-analysis was carried out as a quantitative summary of the published studies in the WHO region. To explore the potential sources of heterogeneity, a subgroup analysis was also carried out, although no significant results were obtained (Additional file 2).
Prevalence of pfhrp2, pfhrp3 and pfhrp2/3 deletions
Prevalence of pfhrp2 deletion
A total of 37 studies were included in the meta-analysis based on the eligibility criteria.
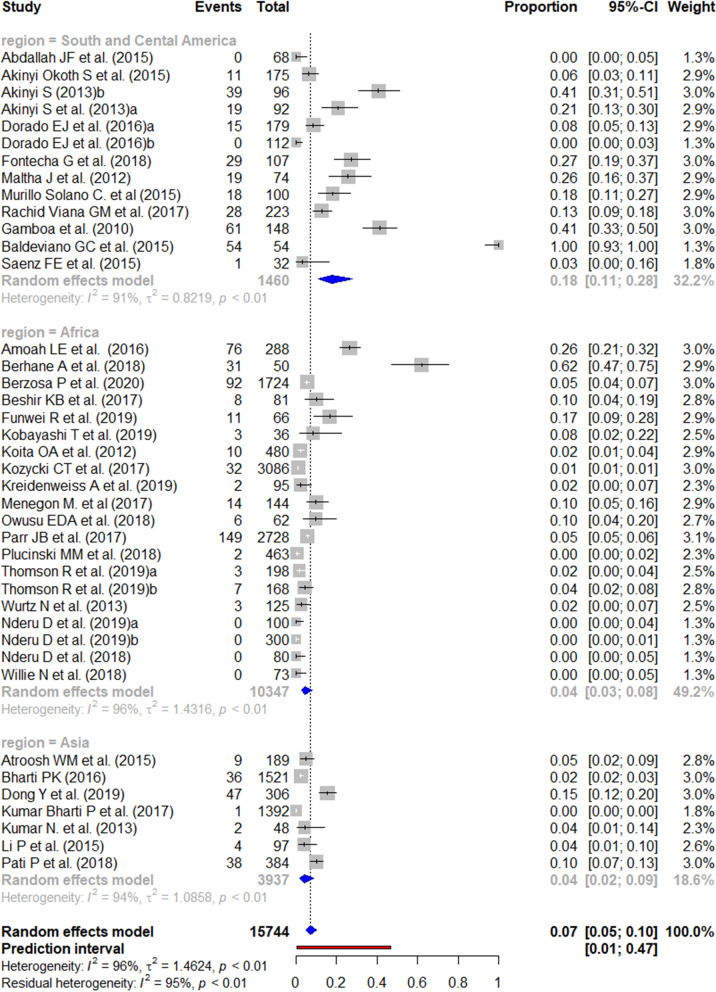
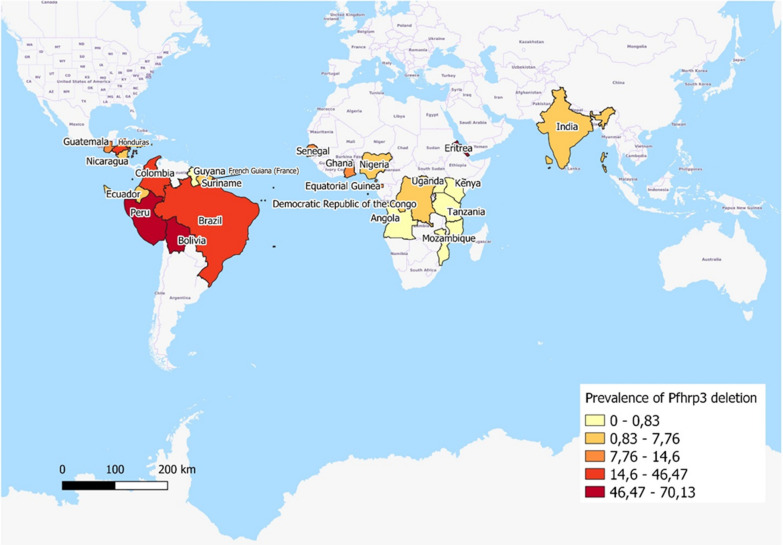
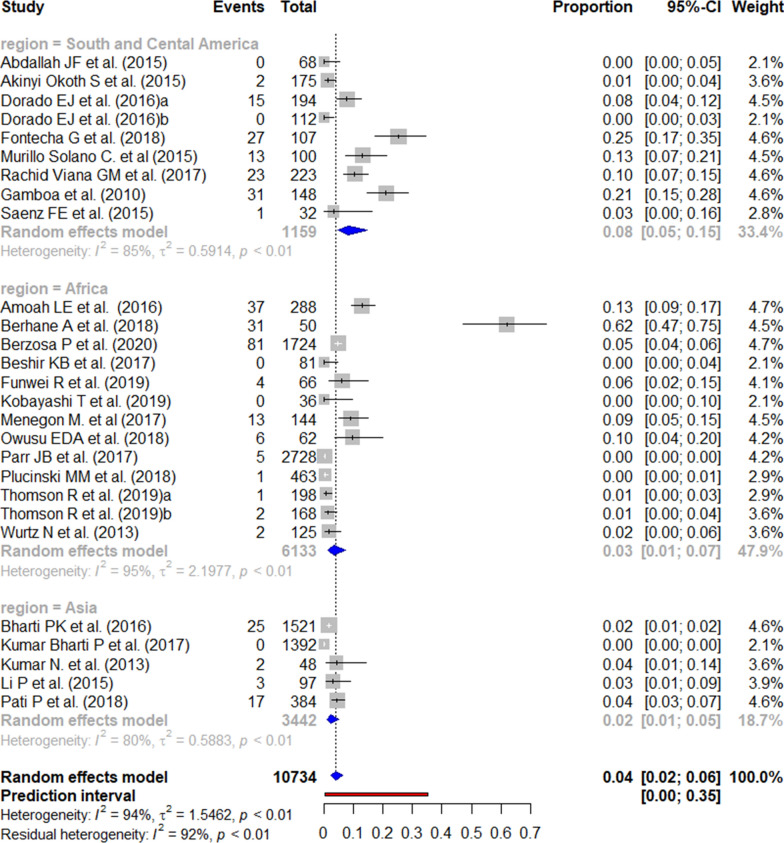
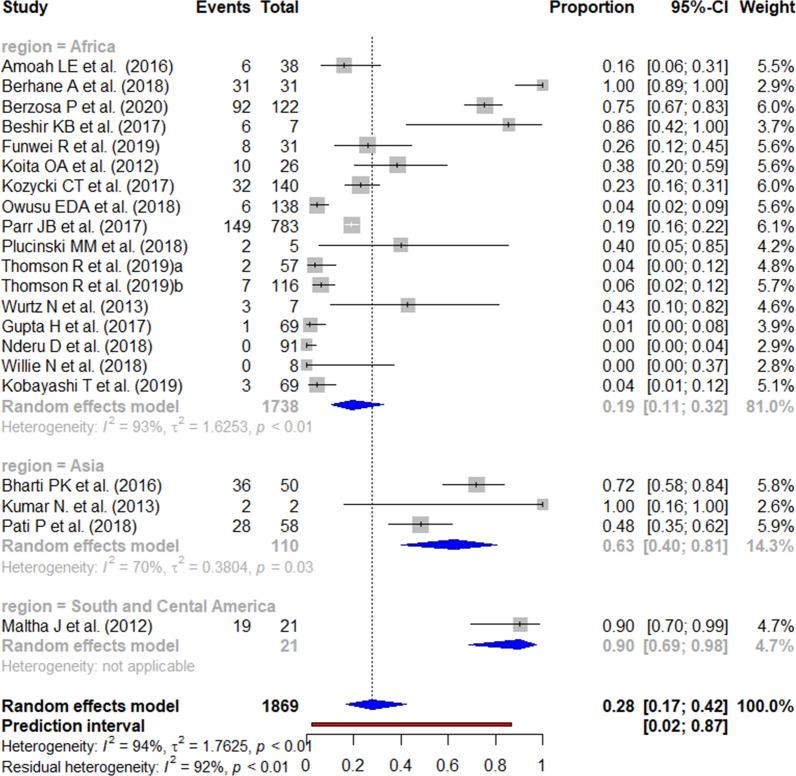
The reported prevalence of the pfhrp2 deletion varied from 0 to 100% (Figs. 2 and 3). Those studies reporting the highest prevalence (100%) were carried out in South and Central America [59], followed by Africa (62.0% in Eritrea) [14]. There were seven studies from the three WHO regions reporting an absence of deletions. The mean pooled prevalence for each WHO region was 18% in South and Central America, 4% in Africa, and 3% in Asia. No significant publication bias was found amongst the studies included (Additional file 3).
Fig. 2.
Forest plot showing the prevalence of pfhrp2 deletions worldwide and by WHO regions. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each WHO region and worldwide and the prediction interval is represented in red
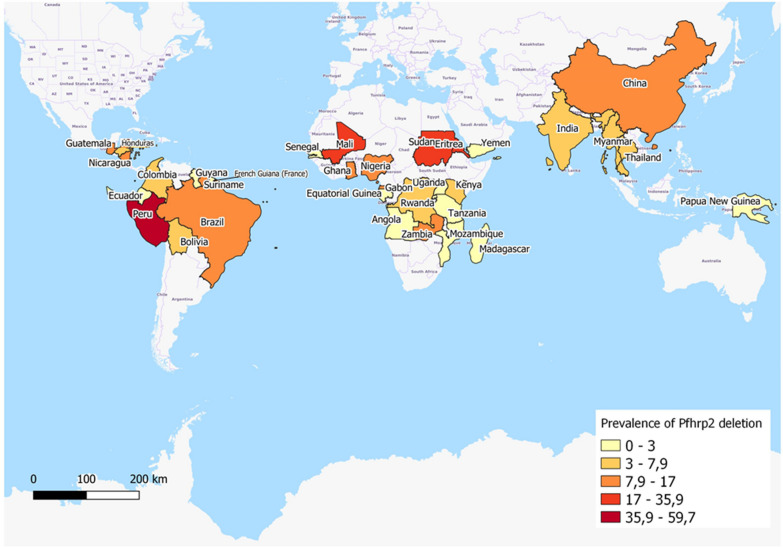
Fig. 3.
Geographical distribution for the reported prevalence of pfhrp2 deletions by country. Prevalence by country has been calculated using the mean prevalence of studies undertaken in each country. The representation was produced using jenks (natural intervals)
Prevalence of pfhrp3 deletion
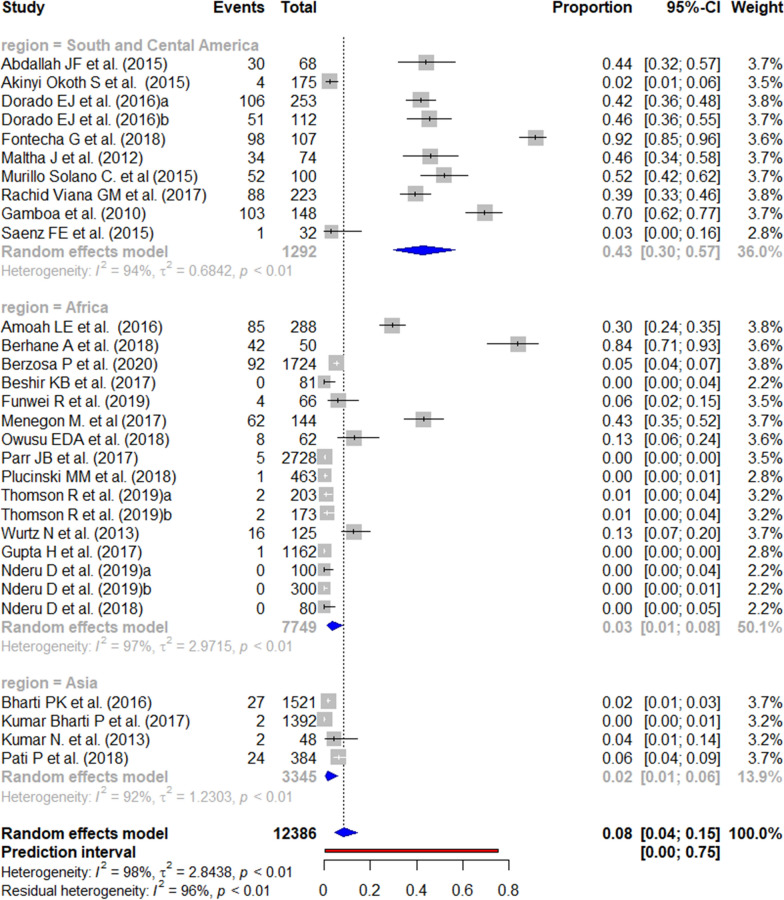
A total of 30 studies were included in the meta-analysis according to the eligibility criteria. The worldwide prevalence of pfhrp3 deletion ranged from 0 to 92%. Four studies, all from Africa, reported the absence of this deletion. In contrast, the highest prevalence of mutation was reported in South and Central America, where the prevalence ranged from 2 to 91%. The pooled prevalence of pfhrp3 deletions by WHO regions was 43% in South and Central America; 3% in Africa; and 1% in Asia (Figs. 4 and 5). The risk of publication bias was not significant (Additional file 4).
Fig. 4.
Forest plot showing the prevalence of pfhrp3 deletions worldwide and by WHO regions. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each WHO region and worldwide and the prediction interval is represented in red
Fig. 5.
Geographical distribution for the reported prevalence of pfhrp3 deletions by country. Prevalence by country has been calculated using the mean prevalence of studies undertaken in each country. The representation was produced using jenks (natural intervals)
Prevalence of pfhrp2 and pfhrp3 double deletion
A total of 23 studies were included in the meta-analysis for the pfhrp2/3 double deletion. The global pooled prevalence of reported pfhrp2/pfhrp3 double deletions was 4%, with the prevalence reported by WHO region ranging from 0 to 25% in South and Central America, from 0 to 62% in Africa, and 0% to 4% in Asia (Fig. 6).
Fig. 6.
Forest plot showing the prevalence of pfhrp2/pfhrp3 double deletion worldwide and by WHO regions. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each WHO region and worldwide and the prediction interval is represented in red
The risk of publication bias was not significant (Additional file 5).
Meta-analysis of pfhrp2, pfhrp3 and pfhrp2/3 deletions in Africa
Pooled prevalence and subgroup analysis of pfhrp2, pfhrp3 and pfhrp2/3 deletions among samples from health facilities and the general population in Africa
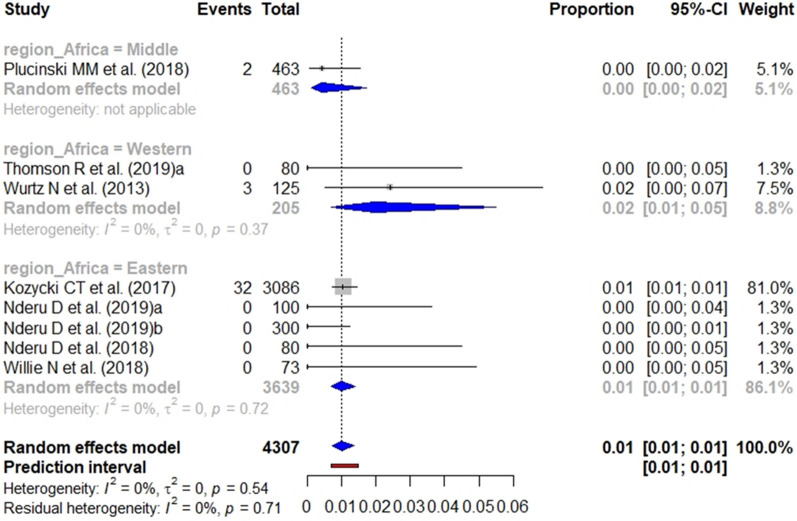
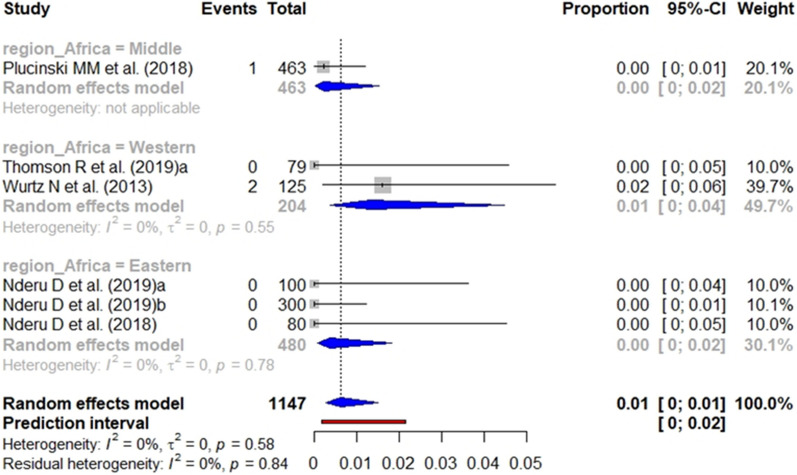
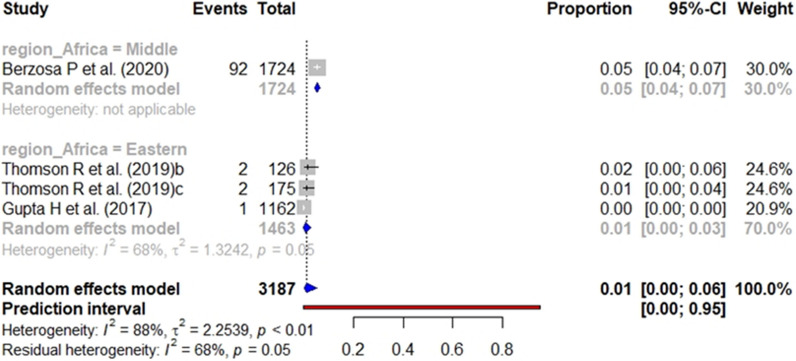
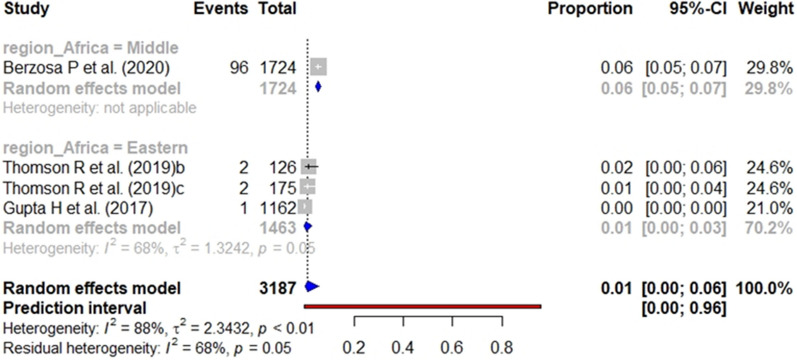
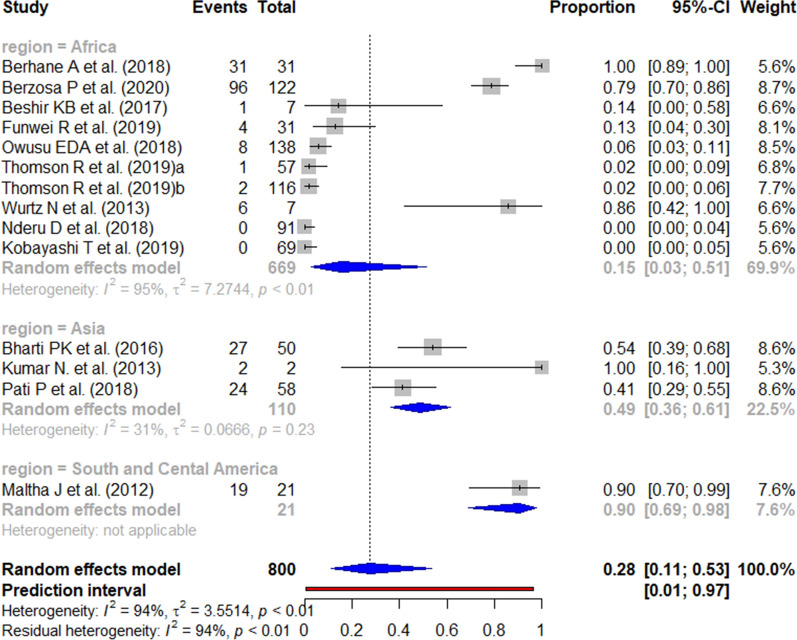
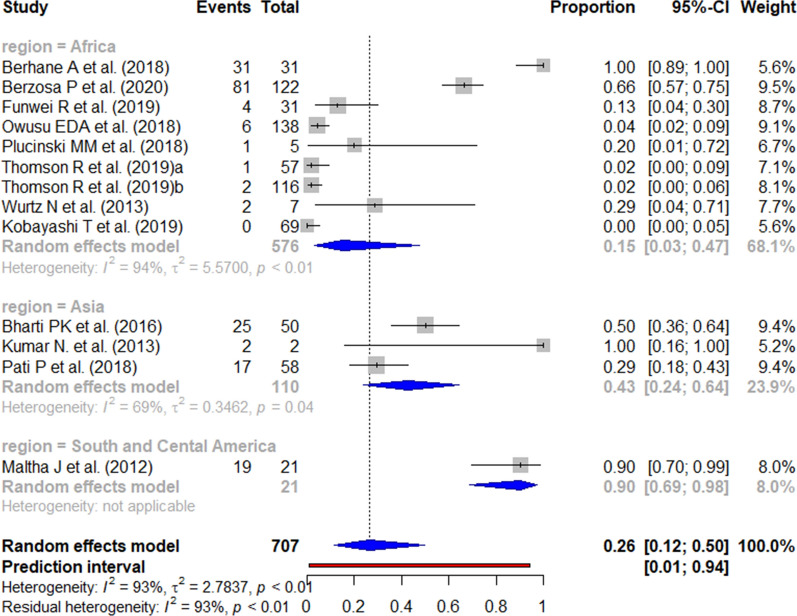
According to the heterogeneity statement for each analysis (Additional files 6, 7, 8), the subgroup analysis for pfhrp2, pfhrp3 and pfhrp2/3 deletions included 12 (8 from health facilities and 4 from general population), 10 (6 from health facilities and 4 from general population) and 12 studies (6 from health facilities and 6 from general population), respectively. The pooled prevalence of pfhrp2 (Fig. 7), pfhrp3 (Fig. 8), and pfhrp2/3 deletions (Fig. 9) from studies carried out at health facilities was 1%.
Fig. 7.
Forest plot showing the prevalence of pfhrp2 deletion among studies carried out in health facilities. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each Africa geographical region
Fig. 8.
Forest plot showing the prevalence of pfhrp3 deletion among studies carried out in health facilities. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each Africa geographical region
Fig. 9.
Forest plot showing the prevalence of pfhrp2/pfhrp3 deletion among studies carried out in health facilities. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each Africa geographical region
The subgroup analysis performed by geographical region in Africa showed a significant difference (p < 0.001) between the aggregate prevalence for the pfhrp3 single deletion by region, with the highest pfhrp3 deletion prevalence being found in Western Africa (13%). The difference for the pfhrp2 single deletion and pfhrp2/3 double deletion was not significant.
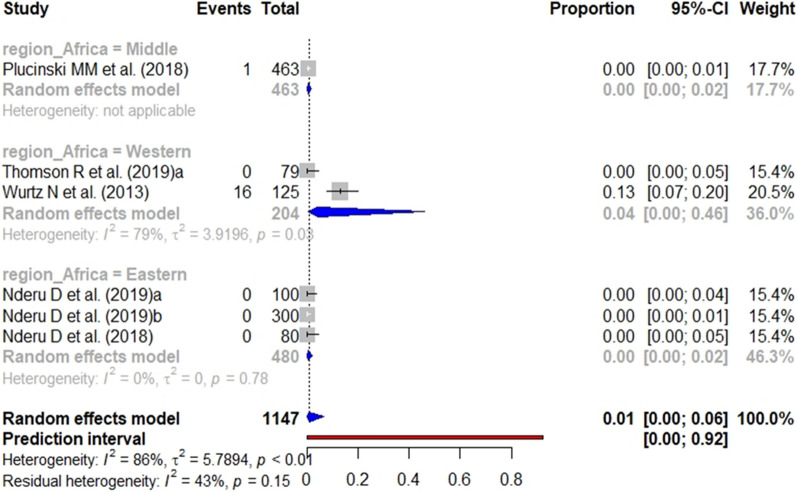
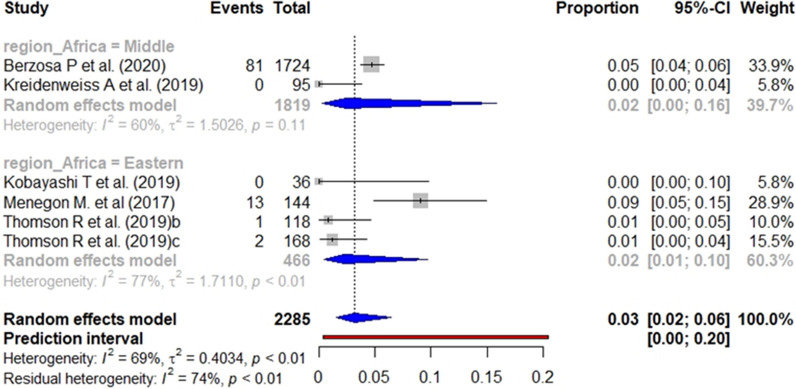
In the case of studies targeting the general population, the pooled prevalence of pfhrp2 (Fig. 10), pfhrp3 (Fig. 11), and pfhrp2/3 deletions (Fig. 12) was 4%, 1% and 3%, respectively. The subgroup analysis by geographical region found no significant differences (p value < 0.001) between the aggregate prevalence for any deletion.
Fig. 10.
Forest plot showing the prevalence of pfhrp2 deletion among studies targeting general population. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each Africa geographical region
Fig. 11.
Forest plot showing the prevalence of pfhrp3 deletion among studies targeting general population. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each Africa geographical region
Fig. 12.
Forest plot showing the prevalence of pfhrp2 & pfhrp3 double deletion among studies targeting general population. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each Africa geographical region
Impact of pfhrp2, pfhrp3 and pfhrp2/3 deletions on false-negative RDT results
Review of RDT efficacy and pfhrp2, pfhrp3 and pfhrp2/3 deletions
Published data regarding the efficacy of RDT by study is reported in Table 4. The HRP2-based RDT sensitivity was over 70% for all studies, except for one [14]. As regards the false-negative rate, the highest was 62% and the lowest 2%.
Table 4.
Reported data about pfhrp2, pfhrp3 and pfhrp2/3 deletions among RDT false
| Study | WHO region | N | HRP2-RDT sensitivity (%) | False negative rate (%) | Negative like-hood ratio | P (%) of pfhrp2 deletion (%) | P (%) of pfhrp3 deletion (%) | P (%) of pfhrp2/3 deletions (%) |
|---|---|---|---|---|---|---|---|---|
| Amoah et al. [74] | Africa | 38 | 72.66 | 27.33 | 0.49 | 15.79 | NR | NR |
| Berhane et al. [14] | Africa | 31 | 38 | 62 | NA | 100 | 100 | 100 |
| Berzosa et al. [21] | Africa | 122 | 84.38 | 15.62 | 0.18 | 75.41 | 78.69 | 66.39 |
| Beshir et al. [49] | Africa | 7 | 82.44 | 17.5 | 0.15 | 85.71 | 14.29 | NR |
| Bharti et al. [24] | Asia | 50 | 76 | NA | NA | 72.00 | 54.00 | 50.00 |
| Funwei et al. [69] | Africa | 31 | 88.53 | 9.11 | 0.001 | 25.81 | 12.90 | 12.90 |
| Gupta et al. [22] | Africa | 69 | 85.89 | 14.11 | 0.14 | 1.45 | NR | 0.0 |
| Koita et al. [28] | Africa | 26 | 95 | 5.16 | 0.063 | 38.46 | NR | 0.0 |
| Kobayashi et al. [52] | Africa | 69 | NA | 1.7 | NA | 4.35 | 0.00 | 0.0 |
| Kozycki et al. [11] | Africa | 140 | 91.8 | NA | NA | 22.86 | NR | NR |
| Kumar et al. [50] | Asia | 2 | 95.83 | 4.17 | NA | 100 | 100 | 100 |
| Maltha et al. [13] | South America | 21 | 71.60 | 28.38 | NA | 90.48 | 0.00 | 90.48 |
| Nderu et al. [9] | Africa | 91 | 93.8 | 6 | 0.08 | 0 | 0.00 | NR |
| Owusu et al. [47] | Africa | 138 | 80.78 | 20.97 | 0.002 | 4.35 | 5.80 | 4.35 |
| Parr et al. [46] | Africa | 783 | 71.55 | 28.45 | NA | 19.03 | NR | NR |
| Pati et al. [25] | Asia | 58 | 84.90 | 15.1 | 0.155 | 48.28 | 41.38 | 29.31 |
| Plucinski et al. [51] | Africa | 5 | 81 | NA | NA | 40.00 | NR | 20.00 |
| Thomson et al. [12] | Africa | 173 | 89.34 | 10.66 | 0.1368 | 5.20 | 1.73 | 1.73 |
| Willie et al. [30] | Africa | 8 | 96 | 12.9 | 0.144 | 0.00 | NR | NR |
| Wurtz et al. [16] | Africa | 7 | 94.07 | 5.90 | NA | 42.86 | 85.71 | 28.57 |
N sample size, P prevalence among RDT with negative results, NA not applicable, NR not reported
Reported prevalence of pfhrp2, pfhrp3 and pfhrp2/3 deletions for false-negative HRP2-based RDT results
The reported prevalence of pfhrp2 deletion among false-negative HRP2-based RDT results ranged from 0 to 100% in Africa (Fig. 13). In contrast, in Asia and Central and South America, the lowest published prevalence was 90% and 48%, respectively.
Fig. 13.
Forest plot showing the prevalence of pfhrp2 deletion among HRP2-based RDT false negatives worldwide. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each WHO region
The published data for the pfhrp3 deletion (Fig. 14) and pfhrp2/3 double deletion (Fig. 15) among false-negative RDT results varied widely between studies.
Fig. 14.
Forest plot showing the prevalence of pfhrp3 deletion among HRP2-based RDT false negatives worldwide. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each WHO region
Fig. 15.
Forest plot showing the prevalence of pfhrp2 & pfhrp3 double deletion among HRP2-based RDT false negatives worldwide. Each grey square represents one study (size proportional to relative weight) and black lines represent the effect and its confidence interval. Pooled prevalences are represented as blue diamonds for each WHO region
Discussion
This review describes and assesses the recent literature about the emerging global issue represented by false-negatives related to pfhrp2, pfhrp3 and pfhrp2/3 deletions in RDT. The study focused on three main concerns. Firstly, the methodology used to study the deletions in pfhrp2 and pfhrp3 genes as the standardization of methodologies could increase the reliability and comparability of results. Secondly, the aggregated prevalence of deletion of pfhrp2, pfhrp3 and pfhrp2/3 genes, with an in-depth analysis for studies conducted in Africa. Thirdly, the implications of pfhrp2, pfhrp3 and pfhrp2/3 genes in false-negative RDT results. As a consequence, this review provides an original global and complete overview of the current situation of these gene deletions. Two previous systematic reviews had partially targeted pfhrp2 and pfhrp3 gene deletions in a specific context (Africa and India) [64, 65], but not worldwide, and a further review was more focused on methodological aspects; however, fewer articles have been included [66].
Methodologies used to study the deletion of pfhrp2, pfhrp3 and pfhrp2/3 genes
Despite the WHO protocol for the surveillance of pfhrp2, pfhrp3 and pfhrp2/3 gene deletion, wide methodological differences were identified between studies [1]. Firstly, studies were conducted in different populations, thus meaning that sociodemographic characteristics might have influenced the prevalence of deletions [66]. For example, it is known that a higher prevalence of suppression has been reported in children [46]. Moreover, although surveillance is recommended only for symptomatic patients, the majority of published studies involved both the asymptomatic and symptomatic population. The WHO recommendation regarding taking samples from febrile patients is based on ensuring good sample quality for the analysis [67]. However, if the deletion involves fitness-cost for the parasites, which has not yet been explored, it is expected that the symptoms will affect the detected deletion prevalence [5].
Another important difference between study populations lies in the malaria transmission season for data collection, which is not provided in all studies. Thus, the prevalence of deletion varies between low and high malaria transmission seasons as malaria is more prevalent during the rainy season [17]. Only one of the articles that reports collection over a whole year stratifies the results by transmission season [25].
Finally, the reliability of results from sample-based studies may also be affected by sample size and geographical location. With regard to sample size, the results varied from too small a sample to provide significant results to a sufficiently large sample to provide significant conclusions, thus limiting the ability to compare statistical significance between studies [64]. Moreover, the varied geographical settings, in terms of location and setting characteristics, make it difficult to assume that studies are representative of the country in which they were conducted, thus also meaning that international comparisons are unsuitable and unreliable.
All studies, except one, included a molecular analysis of deletions (the majority by PCR), with only a few using microsatellites. Although almost all studies used PCR, a wide variety of primers or conditions were used for amplification [64]. Moreover, some articles reported using sequencing, but only for the analysis of sequence variations rather than to confirm gene presence or deletion. This may also have influenced the reliability and comparability of the results.
Another important issue is that some articles only searched for deletions among discordant samples, assuming that most deletions will be found in false-negative RDT results and that it is in this subgroup in which they have an impact. The majority of these studies extrapolated their results to the general sample size [67]. However, this could mean an underestimation of deletion prevalence.
Moreover, for reliable results it is essential to test the quality of DNA. Accordingly, none of the studies that did not test the quality of their samples can assume the reliability of their results [37, 68]. To achieve that, studies should report the amplification of at least two different P. falciparum genes, for example pfmsp1 and pfmsp2 [12, 64, 69]. This is useful to check the quality of the DNA or the concentration to be amplified in order to ensure that, if PCR for pfhrp2 and pfhrp3 does not amplify, there is actually a problem with the DNA used rather than a deletion.
Some studies also analysed parasite density by qPCR, which allows the quality and quantity of the DNA to be tested simultaneously [22, 29]. These studies were able to avoid confusing false-negative RDT results caused by low density with those caused by eliminations [64, 70]. However, as this review shows, the majority of studies (n = 33) did not check the parasite density.
The proven cross-reaction of HRP3, the protein encoded by pfhrp3, with PfHRP2-based RDT means that the deletions in this gene, and the combination of both deletions (pfhrp2/3), must be evaluated to address the performance of RDT [71]. As such, the majority of studies included analysis of the pfhrp3 gene. Moreover, it is relatively common for studies to also include an analysis of flanking genes for pfhrp2 and pfhrp3 [20, 63]. The rapid appearance of deletions in these regions [41], and their possible relation with the multiple origins of the pfhrp2 deletion, has been described [57].
Pooled prevalence of pfhrp2, pfhrp3 and pfhrp2/3 deletions
The WHO recommends not to use HRP2-based RDT as a diagnostic tool when the prevalence of deletions is > 5% given that, in this situation, the efficacy of RDT would be highly compromised [72]. According to the results of the meta-analysis in this study, the prevalence of pfhrp2, pfhrp3 and pfhrp2/3 deletions is only compromised in South and Central America, whereas other WHO Regions report a significantly lower prevalence. Nevertheless, as the map created by WHO showed, the absence of representative studies in all malaria endemic areas makes difficult to assess global conclusions (https://apps.who.int/malaria/maps/threats/). This is important because more than 90% of malaria cases in Africa are caused by P. falciparum, whereas in America the principal aetiological agent is P. vivax [1]. As such, the effects caused by a decrease in the efficacy of HRP2-based RDT would potentially be more critical in Africa.
With regard to the high prevalence of deletions in South America, previous studies have shown that these deletions are not equally distributed, with the Amazonas area concentrating the highest prevalence of deletions [20, 59]. Moreover, various studies have reported an absence of mutations in some regions, thus supporting this unequal distribution [19, 73].
Geographical differences were also found between studies conducted in Africa. This could be explained by the malaria transmission season during sample collection [14], as well as by geographical epidemiological differences in terms of baseline prevalence, resistance, public health strategies [38]. In contrast, according to the results showed in this paper, there were no statistically significant differences in pooled prevalence between Africa’s geographical regions, although this might be related to the lack of more representative studies. Thus, it is essential to correctly characterize the status of deletions in each Africa region due to their different characteristics. To assess the actual national prevalence of deletions, it would be advisable to perform national studies with sufficient geographical representation [46, 74] and during specific seasons [17].
It should also be taken into account that, when the genetic diversity is low (either during the low transmission season or in zones with low prevalence of malaria), deletions will be more likely to be selected, thus making it easier for them to spread [60, 75]. Although malaria is endemic in the majority of countries in Africa, it has different characteristics (transmission, treatment strategy) and there are also countries with strong malaria-control programs in place. This could be a potential scenario for the spread of deletions, which would have severe implications and would stop progress in controlling the disease.
The genetic diversity and geographical distribution give information about the selection process for deletions and their spread. The use of RDT for diagnosis for 10 years could drive up the deletion of pfhrp2 and pfhrp3 genes [27]. In this regard, one study reported that the continued use of HRP2-based RDT alone will quickly result in the selection of deleted parasites [76]. The same article also reported that this pressure decreases when an HRP2 diagnosis is combined with the detection of another protein. In the field, the majority of RDTs currently detect at least two proteins, mainly pLDH and PfHRP2 [77, 78]. This could explain why, although the majority of countries have significantly increased the use of HRP2-based RDT since 2010, there is no agreement about selection pressure. Thus, some articles support the lack of strong evidence about selection pressure for mutants lacking the pfhrp2 and pfhrp3 genes [15, 79], whereas others suggest that the impact of selection pressure favour parasites with deletions [27, 31]. It was also evaluated the possible change in deletions over time but found no relationship between time and deletions, probably because of the lack of a sufficient number of studies to analyse this properly.
The results of the meta-analysis also showed a higher prevalence of deletions among children, as reported previously [17]. This may be because children usually have infections with less genetic diversity, which is especially concerning due to the high mortality in children [80].
Another important issue addressed in this review was the possible differences in deletions between symptomatic and asymptomatic patients. This question was answered by assuming that samples from health centres could be considered to be symptomatic. Previous research suggests the potential fitness costs of the pfhrp2 deletion, thus meaning a greater prevalence of deletion among the asymptomatic population [27]. This was supported by studies reporting a higher prevalence of deletions among asymptomatic patients [28, 46]. In contrast, one study reported no association between pfhrp2/3 mutants and fitness cost [7]. The results of the meta-analysis support the first idea, namely that the asymptomatic population carries this deletion more frequently, as found in studies involving the general population. Either way, there is no sufficiently strong epidemiology or laboratory evidence to support this, probably due to the lack of further studies. To address these concerns, further studies analysing patients according to their clinical characteristics are still required.
Impact of pfhrp2, pfhrp3 and pfhrp2/3 deletions on false-negative RDT results
The major threat of deletions in pfhrp2 and pfhrp3 genes is their effects on the efficacy of RDT. As a result of following the test-based treatment strategy, patients who are mistakenly diagnosed will not receive the correct treatment on time, which is an essential condition for successful recovery and, therefore, for improving malaria control in affected areas [16, 22]. In this regard, it has been suggested that a strong test-based treatment strategy favours the selection of pfhrp2 and pfhrp3 mutants [46]. This hypothesis, which is based on escape from treatment due to a lack of diagnosis, was supported by a previously published simulation model [76].
The majority of studies have reported a relationship between gene deletions and false-negative RDT results by analysing the deletions only among false-negative RDT samples. This approach is useful for that aim but may not be suitable for characterising the deletion status as RDT could be positive, even if pfhrp2 is deleted, due to a cross-reaction with pfhrp3. In addition, a higher prevalence of deletions is expected in these samples [11]. As such, these studies might have underestimated pfhrp2 and pfhrp3 single deletions, the prevalence of which could be important for predicting the future threat to RDT efficacy.
The impact of deletions on HRP2-based RDT results could be related to the lower sensitivity and higher false-negative rate of RDT. The fact that parasites with a pfhrp2/3 deletion are not detected means that the sensitivity of RDT decreases [5]. Although a relationship between RDT and gene deletions has been reported in this review, it have not been possible to determine a directly proportional relationship, probably as a consequence of the high inter-study heterogeneity.
Limitations and strengths of the study
The principal limitation of this review was the high inter-study heterogeneity. Moreover, some articles used a subset of their samples, usually the subgroup of false-negative RDT results, to detect the deletion and calculate the prevalence of deletion in the whole sample using only this value. This could lead to an underestimation of the prevalence. To decrease this heterogeneity, it was performed a subgroup analysis by health facility or general population, which could introduce a bias. Moreover, the wide variety between studies and the way in which their results are reported has made it difficult to interpret and to standardize the results for systematic review and meta-analysis. Finally, although all the obtained results from the publication bias assessment were not significant, some bias related to the absence of complete data or the need to report significant results may nevertheless be present.
In summary, this review represents a complex and global the first approach to characterize the worldwide status of these deletions taking into account different epidemiological variables combined with the main methodological aspects of the different study designs. As such, it could be used as a reference for future studies.
Conclusion
This review highlights the concerning prevalence of pfhrp2, pfhrp3 and pfhrp2/3 gene deletions and their variation worldwide. Although these results are still too limited to reconsider the efficacy of RDT, these deletions represent a serious threat to malaria control as a lack of safe and quality diagnosis means a lack of adequate treatment, thus meaning that uncontrolled infection may increase, along with the morbidity and mortality rates. The impact of these deletions is more severe in regions with a low prevalence of malaria, particularly in those countries close to malaria control or eradication, where regular and systematic surveillance of these deletions as part of national guidelines for malaria control is highly recommended. To combat this threat, this study justifies the need for an RDT that combines two proteins or the combination of HRP2-based RDT with another diagnostic method due to the questionable efficacy of the former. Additionally, it is also highly recommended to combine these strategies with evaluations of diagnostic quality.
This review has also highlighted the need to better characterize the threat posed by these deletions. In this regard, a standardized methodology for all studies could play a key role in increasing the understanding of these deletions, their transmission dynamics and their modifiers and associated effects, thereby allowing a comparison between regions in terms of epidemiological variables. Moreover, more genetic studies could help to better characterize these genes and their dynamics.
It is also important to recommend that the gold standard diagnosis in endemic countries, namely microscopy, should not be neglected. Microscopy has been side-lined in many places due to the introduction of RDTs. However, this diagnostic method is still cheap and gives a malaria diagnosis in less than 30 min. As such, the availability of high quality rapid tests together with quality microscopy would enable diagnostic and reference centres to offer better health care to the population.
Supplementary Information
Additional file 1: Table S1. The PRISMA checklist.
Additional file 2: Table S2. Pooled prevalence of pfhrp2, pfhrp3 and pfhrp2/3 deletions among all P. falciparum cases by WHO region, population symptomatology, age of population and collection season.
Additional file 3: Figure S1. Contour-enhanced funnel plot of studies included in the pfhrp2 deletion meta analysis.
Additional file 4: Figure S2. Contour-enhanced funnel plot of studies included in the pfhrp3 deletion meta analysis.
Additional file 5: Figure S3. Contour-enhanced funnel plot of studies included in the pfhrp2 & pfhrp2 double deletion meta analysis.
Additional file 6: Figure S4. Assessment of inter-study heterogeneity for the pfhrp2 deletion analysis.
Additional file 7: Figure S5. Assessment of inter-study heterogeneity for the pfhrp3 deletion analysis.
Additional file 8: Figure S6. Assessment of inter-study heterogeneity for the pfhrp2 & pfhrp3 double deletion analysis.
Acknowledgements
We would like to thank Prof. Francisco Javier Martínez González (University of Alcalá), university supervisor of the thesis of Irene Molina-de la Fuente, for his advice on the achievement of this study. To the Network Biomedical Research on Tropical Diseases (RICET in Spanish), Madrid, Spain.
Abbreviations
- pfhrp2
Plasmodium falciparum Histidine-rich protein 2
- pfhrp3
Plasmodium falciparum Histidine-rich protein 3
- WHO
World Health Organization
- RDT
Malaria rapid diagnostic test
- HRP2
Histidine-rich protein 2
- HRP3
Histidine-rich protein 3
Authors’ contributions
IMF and PB designed the study and conceive the original idea. IMF and PB review the articles and prepared the data. IMF analysed data. IMF, ZH and AP analysed methodological issues. All authors contributed substantially to the manuscript review. PB and AB are the guarantors. All authors read and approved the final manuscript.
Funding
I.M.F. received a research fellowship from the University of Alcalá that enables her to develop this study.
Availability of data and materials
All data generated during this study are included in this published article and its Additional files. The datasets used and analysed (extraction table) during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. World malaria report 2020. Geneva: World Health Organization; 2020. https://www.who.int/publications/i/item/9789240015791.
- 2.D’Acremont V, Lengeler C, Mshinda H, Mtasiwa D, Tanner M, Genton B. Time to move from presumptive malaria treatment to laboratory-confirmed diagnosis and treatment in African children with fever. PLoS Med. 2009;6:e252. doi: 10.1371/journal.pmed.0050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezennia IJ, Nduka SO, Ekwunife OI. Cost benefit analysis of malaria rapid diagnostic test: the perspective of Nigerian community pharmacists. Malar J. 2017;16:7. doi: 10.1186/s12936-016-1648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuupiel D, Bawontuo V, Drain PK, Gwala N, Mashamba-Thompson TP. Supply chain management and accessibility to point-of-care testing in resource-limited settings: a systematic scoping review. BMC Health Serv Res. 2019;19:519. doi: 10.1186/s12913-019-4351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poti KE, Sullivan DJ, Dondorp AM, Woodrow CJ. HRP2: transforming malaria diagnosis, but with caveats. Trends Parasitol. 2020;36:112–126. doi: 10.1016/j.pt.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Marquart L, Butterworth A, McCarthy JS, Gatton ML. Modelling the dynamics of Plasmodium falciparum histidine-rich protein 2 in human malaria to better understand malaria rapid diagnostic test performance. Malar J. 2012;11:74. doi: 10.1186/1475-2875-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepúlveda N, Phelan J, Diez-benavente E, Clark TG, Hopkins H, Sutherland C, et al. Global analysis of Plasmodium falciparum histidine-rich protein-2 (pfhrp2) and pfhrp3 gene deletions using whole-genome sequencing data and meta-analysis. Infect Genet Evol. 2018;62:211–219. doi: 10.1016/j.meegid.2018.04.039. [DOI] [PubMed] [Google Scholar]
- 8.Kyabayinze DJ, Zongo I, Cunningham J, Gatton M, Angutoko P, Ategeka J, et al. HRP2 and pLDH-based rapid diagnostic tests, expert microscopy, and PCR for detection of malaria infection during pregnancy and at delivery in areas of varied transmission: a prospective cohort study in Burkina Faso and Uganda. PLoS ONE. 2016;11:e0156954. doi: 10.1371/journal.pone.0156954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nderu D, Kimani F, Thiong’o K, Akinyi M, Karanja E, Meyer CG, et al. PfHRP2-PfHRP3 diversity among Kenyan isolates and comparative evaluation of PfHRP2/pLDH malaria RDT with microscopy and nested PCR methodologies. Parasitol Int. 2018;67:793–799. doi: 10.1016/j.parint.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Bwire GM, Ngasala B, Kilonzi M, Mikomangwa WP, Felician FF, Kamuhabwa AAR. Diagnostic performance of CareStartTM malaria HRP2/pLDH test in comparison with standard microscopy for detection of uncomplicated malaria infection among symptomatic patients, Eastern Coast of Tanzania. Malar J. 2019;18:354. doi: 10.1186/s12936-019-2990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozycki CT, Umulisa N, Rulisa S, Mwikarago EI, Musabyimana JP, Habimana JP, et al. False-negative malaria rapid diagnostic tests in Rwanda: impact of Plasmodium falciparum isolates lacking hrp2 and declining malaria transmission. Malar J. 2017;16:123. doi: 10.1186/s12936-017-1768-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson R, Beshir KB, Cunningham J, Baiden F, Bharmal J, Bruxvoort KJ, et al. pfhrp2 and pfhrp3 gene deletions that affect malaria rapid diagnostic tests for Plasmodium falciparum: analysis of archived blood samples from 3 African countries. J Infect Dis. 2019;220:1444–1452. doi: 10.1093/infdis/jiz335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maltha J, Gamboa D, Bendezu J, Sanchez L, Cnops L, Gillet P, et al. Rapid diagnostic tests for malaria diagnosis in the Peruvian Amazon: impact of pfhrp2 gene deletions and cross-reactions. PLoS ONE. 2012;7:e43094. doi: 10.1371/journal.pone.0043094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berhane A, Anderson K, Mihreteab S, Gresty K, Rogier E, Mohamed S, et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis. 2018;24:462–470. doi: 10.3201/eid2403.171723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker J, Ho MF, Pelecanos A, Gatton M, Chen N, Abdullah S, et al. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J. 2010;9:129. doi: 10.1186/1475-2875-9-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wurtz N, Fall B, Bui K, Pascual A, Fall M, Camara C, et al. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar J. 2013;12:34. doi: 10.1186/1475-2875-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson OJ, Verity R, Ghani AC, Garske T, Cunningham J, Tshefu A, et al. Impact of seasonal variations in Plasmodium falciparum malaria transmission on the surveillance of pfhrp2 gene deletions. Elife. 2019;8:e40339. doi: 10.7554/eLife.40339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS ONE. 2010;5:e8091. doi: 10.1371/journal.pone.0008091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdallah JF, Okoth SA, Fontecha GA, Torres RE, Banegas EI, Matute ML, et al. Prevalence of pfhrp2 and pfhrp3 gene deletions in Puerto Lempira, Honduras. Malar J. 2015;14:19. doi: 10.1186/s12936-014-0537-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorado EJ, Okoth SA, Montenegro LM, Diaz G, Barnwell JW, Udhayakumar V, et al. Genetic characterisation of Plasmodium falciparum isolates with deletion of the pfhrp2 and/or pfhrp3 genes in Colombia: the Amazon region, a challenge for malaria diagnosis and control. PLoS ONE. 2016;11:e0163137. doi: 10.1371/journal.pone.0163137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berzosa P, González V, Taravillo L, Mayor A, Romay-Barja M, García L, et al. First evidence of the deletion in the pfhrp2 and pfhrp3 genes in Plasmodium falciparum from Equatorial Guinea. Malar J. 2020;19:99. doi: 10.1186/s12936-020-03178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta H, Matambisso G, Galatas B, Cisteró P, Nhamussua L, Simone W, et al. Molecular surveillance of pfhrp2 and pfhrp3 deletions in Plasmodium falciparum isolates from Mozambique. Malar J. 2017;16:416. doi: 10.1186/s12936-017-2061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menegon M, L’Episcopia M, Nurahmed AM, Talha AA, Nour BYM, Severini C. Identification of Plasmodium falciparum isolates lacking histidine-rich protein 2 and 3 in Eritrea. Infect Genet Evol. 2017;55:131–134. doi: 10.1016/j.meegid.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS ONE. 2016;11:e0157949. doi: 10.1371/journal.pone.0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pati P, Dhangadamajhi G, Bal M, Ranjit M. High proportions of pfhrp2 gene deletion and performance of HRP2-based rapid diagnostic test in Plasmodium falciparum field isolates of Odisha. Malar J. 2018;17:394. doi: 10.1186/s12936-018-2502-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. From malaria control to malaria elimination: a manual for elimination scenario planning. Geneva: World Health Organization; 2014. https://apps.who.int/iris/bitstream/handle/10665/112485/9789241507028_eng.pdf;jsessionid=68D25C39DA9AC95FDBE76572ADD4D28D?sequence=1.
- 27.Watson OJ, Slater HC, Verity R, Parr JB, Mwandagalirwa MK, Tshefu A, et al. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. Elife. 2017;6:e25008. doi: 10.7554/eLife.25008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koita OA, Doumbo OK, Ouattara A, Tall LK, Konaré A, Diakité M, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–198. doi: 10.4269/ajtmh.2012.10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindler T, Deal AC, Fink M, Guirou E, Moser KA, Mwakasungula SM, et al. A multiplex qPCR approach for detection of pfhrp2 and pfhrp3 gene deletions in multiple strain infections of Plasmodium falciparum. Sci Rep. 2019;9:13107. doi: 10.1038/s41598-019-49389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willie N, Zimmerman PA, Mehlotra RK. Plasmodium falciparum histidine-rich protein 2 gene variation in a malaria-endemic area of Papua New Guinea. Am J Trop Med Hyg. 2018;99:697–703. doi: 10.4269/ajtmh.18-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gendrot M, Fawaz R, Dormoi J, Madamet M, Pradines B. Genetic diversity and deletion of Plasmodium falciparum histidine-rich protein 2 and 3: a threat to diagnosis of P. falciparum malaria. Clin Microbiol Infect. 2019;25:580–585. doi: 10.1016/j.cmi.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 32.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45:139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonsohn U, Nelson LD, Simmons JP. P-curve: a key to the file-drawer. J Exp Psychol Gen. 2014;143:534–547. doi: 10.1037/a0033242. [DOI] [PubMed] [Google Scholar]
- 34.Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with {R}: a practical tutorial. Evid Based Ment Health. 2019;22:153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 36.Harrer M, Cuijpers P, Furukawa T, Ebert DD. dmetar: companion R package for the guide ‘doing meta-analysis in R’. R package version 0.0.9000. 2019. http://dmetar.protectlab.org/.
- 37.Ramutton T, Hendriksen IC, Mwanga-Amumpaire J, Mtove G, Olaosebikan R, Tshefu AK, et al. Sequence variation does not confound the measurement of plasma PfHRP2 concentration in African children presenting with severe malaria. Malar J. 2012;11:276. doi: 10.1186/1475-2875-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deme AB, Park DJ, Bei AK, Sarr O, Badiane AS, Gueye PEHO, et al. Analysis of pfhrp2 genetic diversity in Senegal and implications for use of rapid diagnostic tests. Malar J. 2014;13:34. doi: 10.1186/1475-2875-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okoth SA, Abdallah JF, Ceron N, Adhin MR, Chandrabose J, Krishnalall K, et al. Variation in Plasmodium falciparum histidine-rich protein 2 (Pfhrp2) and Plasmodium falciparum histidine-rich protein 3 (Pfhrp3) gene deletions in Guyana and Suriname. PLoS ONE. 2015;10:e0126805. doi: 10.1371/journal.pone.0126805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rachid Viana GM, Okoth SA, Silva-Flannery L, Barbosa DRL, De Oliveira AM, Goldman IF, et al. Histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in Plasmodium falciparum isolates from select sites in Brazil and Bolivia. PLoS ONE. 2017;12:e0171150. doi: 10.1371/journal.pone.0171150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontecha G, Mejía RE, Banegas E, Ade MP, Mendoza L, Ortiz B, et al. Deletions of pfhrp2 and pfhrp3 genes of Plasmodium falciparum from Honduras, Guatemala and Nicaragua. Malar J. 2018;17:320. doi: 10.1186/s12936-018-2470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar N, Singh JPN, Pande V, Mishra N, Srivastava B, Kapoor R, et al. Genetic variation in histidine rich proteins among Indian Plasmodium falciparum population: possible cause of variable sensitivity of malaria rapid diagnostic tests. Malar J. 2012;11:298. doi: 10.1186/1475-2875-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herman C, Huber CS, Jones S, Steinhardt L, Plucinski MM, Lemoine JF, et al. Multiplex malaria antigen detection by bead-based assay and molecular confirmation by PCR shows no evidence of Pfhrp2 and Pfhrp3 deletion in Haiti. Malar J. 2019;18:380. doi: 10.1186/s12936-019-3010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okoth SA, Chenet SM, Arrospide N, Gutierrez S, Cabezas C, Matta JA, et al. Molecular investigation into a malaria outbreak in cusco, Peru: Plasmodium falciparum BV1 lineage is linked to a second outbreak in recent times. Am J Trop Med Hyg. 2016;94:128–131. doi: 10.4269/ajtmh.15-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Xing H, Zhao Z, Yang Z, Cao Y, Yan G, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 in the China-Myanmar border area. Acta Trop. 2015;152:26–31. doi: 10.1016/j.actatropica.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parr JB, Verity R, Doctor SM, Janko M, Carey-Ewend K, Turman BJ, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis. 2017;216:36–44. doi: 10.1093/infdis/jix347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owusu EDA, Djonor SK, Brown CA, Grobusch MP, Mens PF. Plasmodium falciparum diagnostic tools in HIV-positive under-5-year-olds in two ART clinics in Ghana: are there missed infections? Malar J. 2018;17:92. doi: 10.1186/s12936-018-2231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atroosh WM, Al-Mekhlafi HM, Al-Jasari A, Sady H, Al-Delaimy AK, Nasr NA, et al. Genetic variation of pfhrp2 in Plasmodium falciparum isolates from Yemen and the performance of HRP2-based malaria rapid diagnostic test. Parasites Vectors. 2015;8:388. doi: 10.1186/s13071-015-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beshir KB, Sepúlveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep. 2017;7:14718. doi: 10.1038/s41598-017-15031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar N, Pande V, Bhatt RM, Shah NK, Mishra N, Srivastava B, et al. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 2013;125:119–121. doi: 10.1016/j.actatropica.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Plucinski MM, Herman C, Jones S, Dimbu R, Fortes F, Ljolje D, et al. Screening for Pfhrp2/3-deleted Plasmodium falciparum, non-falciparum, and low-density malaria infections by a multiplex antigen assay. J Infect Dis. 2019;219:437–447. doi: 10.1093/infdis/jiy525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi T, Sikalima J, Parr JB, Chaponda M, Stevenson JC, Thuma PE, et al. The search for Plasmodium falciparum histidine-rich protein 2/3 deletions in Zambia and implications for Plasmodium falciparum histidine-rich protein 2-based rapid diagnostic tests. Am J Trop Med Hyg. 2019;100:842–845. doi: 10.4269/ajtmh.18-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranadive N, Kunene S, Darteh S, Ntshalintshali N, Nhlabathi N, Dlamini N, et al. Limitations of rapid diagnostic testing in patients with suspected malaria: a diagnostic accuracy evaluation from Swaziland, a low-endemicity country aiming for malaria elimination. Clin Infect Dis. 2017;64:1221–1228. doi: 10.1093/cid/cix131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laban NM, Kobayashi T, Hamapumbu H, Sullivan D, Mharakurwa S, Thuma PE, et al. Comparison of a PfHRP2-based rapid diagnostic test and PCR for malaria in a low prevalence setting in rural southern Zambia: implications for elimination. Malar J. 2015;14:25. doi: 10.1186/s12936-015-0544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreidenweiss A, Trauner F, Rodi M, Koehne E, Held J, Wyndorps L, et al. Monitoring the threatened utility of malaria rapid diagnostic tests by novel high-throughput detection of Plasmodium falciparum hrp2 and hrp3 deletions: a cross-sectional, diagnostic accuracy study. EBioMedicine. 2019;50:14–22. doi: 10.1016/j.ebiom.2019.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sáenz FE, Morton LC, Okoth SA, Valenzuela G, Vera-Arias CA, Vélez-Álvarez E, et al. Clonal population expansion in an outbreak of Plasmodium falciparum on the northwest coast of Ecuador. Malar J. 2015;14:497. doi: 10.1186/s12936-015-1019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akinyi S, Hayden T, Gamboa D, Torres K, Bendezu J, Abdallah JF, et al. Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru. Sci Rep. 2013;3:2797. doi: 10.1038/srep02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong Y, Liu S, Deng Y, Xu Y, Chen M, Liu Y, et al. Genetic polymorphism of histidine rich protein 2 in Plasmodium falciparum isolates from different infection sources in Yunnan Province, China. Malar J. 2019;18:446. doi: 10.1186/s12936-019-3084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldeviano GC, Okoth SA, Arrospide N, Gonzalez RV, Sánchez JF, Macedo S, et al. Molecular epidemiology of Plasmodium falciparum malaria outbreak, Tumbes, Peru, 2010–2012. Emerg Infect Dis. 2015;21:797–803. doi: 10.3201/eid2105.141427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nderu D, Kimani F, Thiong’o K, Karanja E, Akinyi M, Too E, et al. Plasmodium falciparum histidine-rich protein (PfHRP2 and 3) diversity in Western and Coastal Kenya. Sci Rep. 2019;9:1709. doi: 10.1038/s41598-018-38175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mussa A, Talib M, Mohamed Z, Hajissa K. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. BMC Res Notes. 2019;12:334. doi: 10.1186/s13104-019-4361-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willie N, Mehlotra RK, Howes RE, Rakotomanga TA, Ramboarina S, Ratsimbasoa AC, et al. Insights into the performance of sD bioline malaria ag p.f/Pan rapid diagnostic test and Plasmodium falciparum histidine-rich protein 2 gene variation in Madagascar. Am J Trop Med Hyg. 2018;98:1683–1691. doi: 10.4269/ajtmh.17-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murillo Solano C, Akinyi Okoth S, Abdallah JF, Pava Z, Dorado E, Incardona S, et al. Deletion of Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and histidine-rich protein 3 (pfhrp3) genes in Colombian parasites. PLoS ONE. 2015;10:e0131576. doi: 10.1371/journal.pone.0131576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agaba BB, Yeka A, Nsobya S, Arinaitwe E, Nankabirwa J, Opigo J, et al. Systematic review of the status of pfhrp2 and pfhrp3 gene deletion, approaches and methods used for its estimation and reporting in Plasmodium falciparum populations in Africa: review of published studies 2010–2019. Malar J. 2019;18:335. doi: 10.1186/s12936-019-2987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kojom LP, Singh V. Prevalence of Plasmodium falciparum field isolates with deletions in histidine-rich protein 2 and 3 genes in context with sub-Saharan Africa and India: a systematic review and meta-analysis. Malar J. 2020;19:46. doi: 10.1186/s12936-019-3090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomson R, Parr JB, Cheng Q, Chenet S, Perkins M, Cunningham J. Prevalence of Plasmodium falciparum lacking histidine-rich proteins 2 and 3: a systematic review. Bull World Health Organ. 2020;98:558–568. doi: 10.2471/BLT.20.250621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;13:283. doi: 10.1186/1475-2875-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woodrow CJ, Fanello CI. Pfhrp2 deletions in the Democratic Republic of Congo: evidence of absence, or absence of evidence? J Infect Dis. 2017;216:503–504. doi: 10.1093/infdis/jix345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Funwei R, Nderu D, Nguetse CN, Thomas BN, Falade CO, Velavan TP, et al. Molecular surveillance of pfhrp2 and pfhrp3 genes deletion in Plasmodium falciparum isolates and the implications for rapid diagnostic tests in Nigeria. Acta Trop. 2019;196:121–125. doi: 10.1016/j.actatropica.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 70.Plucinski MM, Dimbu PR, Fortes F, Murphy SC, Smith NT, Cruz KR, et al. Malaria parasite density in individuals with different rapid diagnostic test results and concentrations of HRP2 antigen. Am J Trop Med Hyg. 2019;100:1202–1203. doi: 10.4269/ajtmh.19-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar Bharti P, Singh Chandel H, Krishna S, Nema S, Ahmad A, Udhayakumar V, et al. Sequence variation in Plasmodium falciparum histidine rich proteins 2 and 3 in Indian isolates: implications for malaria rapid diagnostic test performance. Sci Rep. 2017;7:1308. doi: 10.1038/s41598-017-01506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.WHO. False-negative RDT results and P. falciparum histidine-rich protein 2/3 gene deletions. Global Malaria Programme. Geneva: World Health Organization. 2015;2:1–8. https://www.who.int/malaria/publications/atoz/information-note-hrp2-based.
- 73.Trouvay M, Palazon G, Berger F, Volney B, Blanchet D, Faway E, et al. High performance of histidine-rich protein 2 based rapid diagnostic tests in French Guiana are explained by the absence of pfhrp2 gene deletion in P. falciparum. PLoS ONE. 2013;8:e74269. doi: 10.1371/journal.pone.0074269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amoah LE, Abankwa J, Oppong A. Plasmodium falciparum histidine rich protein-2 diversity and the implications for PfHRP 2: based malaria rapid diagnostic tests in Ghana. Malar J. 2016;15:101. doi: 10.1186/s12936-016-1159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maltha J, Guiraud I, Lompo P, Kaboré B, Gillet P, Van Geet C, et al. Accuracy of PfHRP2 versus Pf-pLDH antigen detection by malaria rapid diagnostic tests in hospitalized children in a seasonal hyperendemic malaria transmission area in Burkina Faso. Malar J. 2014;13:20. doi: 10.1186/1475-2875-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gatton ML, Dunn J, Chaudhry A, Ciketic S, Cunningham J, Cheng Q. Implications of parasites lacking Plasmodium falciparum histidine-rich protein 2 on malaria morbidity and control when rapid diagnostic tests are used for diagnosis. J Infect Dis. 2017;215:1156–1166. doi: 10.1093/infdis/jix094. [DOI] [PubMed] [Google Scholar]
- 77.Grigg MJ, William T, Barber BE, Parameswaran U, Bird E, Piera K, et al. Combining parasite lactate dehydrogenase-based and histidine-rich protein 2-based rapid tests to improve specificity for diagnosis of malaria due to Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. J Clin Microbiol. 2014;52:2053–2060. doi: 10.1128/JCM.00181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murungi M, Fulton T, Reyes R, Matte M, Ntaro M, Mulogo E, et al. Improving the specificity of Plasmodium falciparum malaria diagnosis in high-transmission settings with a two-step rapid diagnostic test and microscopy algorithm. J Clin Microbiol. 2017;55:1540–1549. doi: 10.1128/JCM.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fontecha G, Pinto A, Escobar D, Matamoros G, Ortiz B. Genetic variability of Plasmodium falciparum histidine-rich proteins 2 and 3 in Central America. Malar J. 2019;18:31. doi: 10.1186/s12936-019-2668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kiemde F, Bonko MDA, Tahita MC, Lompo P, Rouamba T, Tinto H, et al. Accuracy of a Plasmodium falciparum specific histidine-rich protein 2 rapid diagnostic test in the context of the presence of non-malaria fevers, prior anti-malarial use and seasonal malaria transmission. Malar J. 2017;16:294. doi: 10.1186/s12936-017-1941-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The PRISMA checklist.
Additional file 2: Table S2. Pooled prevalence of pfhrp2, pfhrp3 and pfhrp2/3 deletions among all P. falciparum cases by WHO region, population symptomatology, age of population and collection season.
Additional file 3: Figure S1. Contour-enhanced funnel plot of studies included in the pfhrp2 deletion meta analysis.
Additional file 4: Figure S2. Contour-enhanced funnel plot of studies included in the pfhrp3 deletion meta analysis.
Additional file 5: Figure S3. Contour-enhanced funnel plot of studies included in the pfhrp2 & pfhrp2 double deletion meta analysis.
Additional file 6: Figure S4. Assessment of inter-study heterogeneity for the pfhrp2 deletion analysis.
Additional file 7: Figure S5. Assessment of inter-study heterogeneity for the pfhrp3 deletion analysis.
Additional file 8: Figure S6. Assessment of inter-study heterogeneity for the pfhrp2 & pfhrp3 double deletion analysis.
Data Availability Statement
All data generated during this study are included in this published article and its Additional files. The datasets used and analysed (extraction table) during the current study are available from the corresponding author on reasonable request.